DOI:10.32604/cmc.2021.014387

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014387 |  |
| Article |
Technology Landscape for Epidemiological Prediction and Diagnosis of COVID-19
1Department of Instrumentation and Control, Netaji Subhas University of Technology, New Delhi, 110078, India
2Department of Computer Science & Engineering, Maharaja Surajmal Institute of Technology, New Delhi, 110058, India
3Department of Information Technology, Netaji Subhas University of Technology, New Delhi, 110078, India
4Research Center for AI and IoT, Near East University, Nicosia, Mersin, 10, Turkey
5Computer Science Department, Camerino University, Camerino, 62032, Italy
*Corresponding Author: Deepak Kumar Sharma. Email: dk.sharma1982@yahoo.com
Received: 17 September 2020; Accepted: 23 October 2020
Abstract: The COVID-19 outbreak initiated from the Chinese city of Wuhan and eventually affected almost every nation around the globe. From China, the disease started spreading to the rest of the world. After China, Italy became the next epicentre of the virus and witnessed a very high death toll. Soon nations like the USA became severely hit by SARS-CoV-2 virus. The World Health Organisation, on 11th March 2020, declared COVID-19 a pandemic. To combat the epidemic, the nations from every corner of the world has instituted various policies like physical distancing, isolation of infected population and researching on the potential vaccine of SARS-CoV-2. To identify the impact of various policies implemented by the affected countries on the pandemic spread, a myriad of AI-based models have been presented to analyse and predict the epidemiological trends of COVID-19. In this work, the authors present a detailed study of different artificial intelligence frameworks applied for predictive analysis of COVID-19 patient record. The forecasting models acquire information from records to detect the pandemic spreading and thus enabling an opportunity to take immediate actions to reduce the spread of the virus. This paper addresses the research issues and corresponding solutions associated with the prediction and detection of infectious diseases like COVID-19. It further focuses on the study of vaccinations to cope with the pandemic. Finally, the research challenges in terms of data availability, reliability, the accuracy of the existing prediction models and other open issues are discussed to outline the future course of this study.
Keywords: COVID-19; diagnosis; deep learning; forecasting models; machine learning; metaheuristics; prediction; big data; pandemic
The COVID-19 disease, caused by SARS-CoV-2, was initially reported on 31st December 2019, in Wuhan from the Hubei province in China. Since then, the disease started spreading rapidly to all other continents. The SARS-CoV-2 disease is highly infectious and exhibited fast-spreading capacity, endangering the life of a vast number of people across the world. As on 27 July 2020, the number of total COVID-19 infected patients are 1,64,80,485 and number of deceased are 6,54,036 [1]. This requires rapid action in order the attenuate the infectious spread of the disease. The incubation period of COVID-19 where an individual is exposed to the disease but not infected can vary from 14 to 21 days [2]. Even during this period, that particular individual is capable of spreading the disease to others. The uncertainty in the disease transmission scenario and chances of recovery makes it difficult for the nations to prepare for appropriate responses in terms of medical, social and economic facilities to be provided to the affected population. To effectively prepare the countries for the pandemic, it is essential to estimate the disease spread curve. Due to the unknown nature of the new virus, providing an accurate estimate, in the beginning, is a challenging task. To accomplish this objective, scientists have focussed on the time series models. The time series models [3] focus on the past trends followed by infectious disease to predict the future course of an epidemic outbreak. A time-series comprises of discrete data collected at certain interval of time. The prediction of future outcomes of a time series can be made through the use of specific statistical and machine learning models. To choose the appropriate model it is essential to identify the certain traits of the time series. The foremost criteria are to examine whether the time series is univariate or multivariate [3]. In addition, it is essential to determine whether the time series exhibits linearity and is stationary [4]. The prediction models on time series data can be used to analyze present and historical records to make future forecasts [5] of the outbreak pattern of nCoV2019. The process flow of prediction (shown in Fig. 1) is a chain of methods that transform one or more inputs (present and past records) into one or more outputs (future outcome(s)).

Figure 1: Prediction process chart
Quantitative and mathematical analyses of time series data of COVID-19 patients are required for estimating the number of the affected individual in this case. The spread of infectious diseases among the population is a complex transmission process. Predictive models can be built to examine the diffusion of this extremely infectious disease. Prediction of time series data of COVID-19 pandemic outbreak may be achieved through a variety of models like machine learning, statistical, and epidemiological models. Prediction estimates the future trend of the epidemic, however, to curb the menace of the virus infection tracing and identifying vaccination strategies using AI framework, are equally important. RT-PCR is usually used for detection of COVID-19. However, non-abundance of test kits and false-negative results requires a supplementary method for detection of SARS-CoV-2 infection. Chest X-ray and CT scan are being used along with medical conditions of an individual to detect COVID-19 infection, where deep learning methods are deployed for image analysis and correlating with clinical symptoms. The human trial for COVID-19 vaccine has started.
In this paper, the authors review existing work pertaining to artificial intelligence-based predictive analysis of COVID-19, disease prediction and diagnosis. Further, the authors assess the prediction models used in each study and prepare a comparative analysis of the same with other predictive models. The paper is ordered as follows. Background research on different predictive models available for infectious diseases is presented in Section 2. Section 3 summarizes a detailed review of the COVID-19 prediction models. Section 4 presents a detailed analysis of the review. The paper is concluded by describing the impending issues and prospects of this research on the research trajectory.
For successful management of an epidemic, precise estimation and prediction of a disease outbreak are important. Many researchers have deployed different time series models for forecasting of infectious disease using machine learning algorithms, mathematical models and disease-based models. Study of these models is significant for perceiving the review work presented in this literature.
2.1 Machine Learning-Based Infection Tracing and Predictive Modelling
Machine learning a part of Artificial Intelligence which can be deployed to learn important information from a huge data set. The applications of the field of machine learning span a vast range of fields including engineering and technology, criminal forensics, medicine, statistics, engineering technology [6]. The ML algorithms work better than simple statistical models when the data set exhibits complex characteristics including nonlinearity, missing values, large dimension. Similarly, in case of diseases like nCoV-2019 which is a zoonotic disease, it is necessary to understand the pattern of the outbreak from the transmission history of the disease. Machine Learning Algorithms are categorized broadly into supervised, unsupervised and reinforcement learning. The supervised learning technique is most suitable for dealing with COVID-19 outbreak in terms of infection detection, forecasting and identification of potential vaccine components. For infection detection, CT images of chests of patients are being used. For this supervised deep learning, a part of AI-based ML methods is being deployed. Deep Learning architectures are widely used for medical image analysis. “Deep” in deep learning denotes the use of several heterogeneous layers in the network. Deep learning varies from primitive ML techniques in various ways like feature extraction is an integral component of deep learning and it is an end to end process which is implemented optimally. Even outbreak prediction can be performed by deep learning models like multilayer perceptron, recurrent neural network and long short-term memory (LSTM) models which draw information from the past trends and identify the possible nonlinear relation between the input (historical data) and the output (future trend). Several other supervised learning techniques have also been deployed for COVID-19 prediction like support vector machine, polynomial linear regression. The LSTM models (a type of RNN) are also being used for drug and vaccine component identification for infectious diseases.
2.2 Statistical Predictive Models
Numerous statistical techniques are available for time series prediction. Statistical models have been built for identifying the dependencies between the input variables and the outputs using a variety of stochastic models to forecast the possible transmission pattern of this epidemic. The statistical models need prior information about the distribution of data in the given time series. The popular statistical time series prediction models, used for forecasting infectious diseases like COVID-19 using one-dimensional time series dataset are Exponential Smoothing (ES) [7] and Autoregressive Integrated Moving Average model (ARIMA) [8].
Epidemiological models [9] are a type of mathematics (difference) based infectious disease forecasting models. In the SEIR disease model, the population is categorized as S (susceptible), E (exposed), I (infected) or R (recovered/dead). Individuals may move from one category to another depending on the stage of infection. Initially in a pandemic, the entire population is susceptible and one infected individual is present, who spreads the infection to the population. In diseases with no incubation state, the E category is not used. Once the individual is detected with the virus, the person is in an infectious state and may either recover or decease. According to medical research, a person who recovered from COVID-19 infection is no longer susceptible to the disease. To underscore the effectiveness of studies in this domain it’s pivotal we understand the dynamic SEIR model. Fig. 2 illustrates the four tiers of a conventional SEIR model that is used for epidemiological analysis. The three parameters of the models are “ ” which determines the speed of spreading of the virus, The reproduction rate has greatly varied in studies so far and usually is an estimate based on previous values or a predicted value based on other models, “
” which determines the speed of spreading of the virus, The reproduction rate has greatly varied in studies so far and usually is an estimate based on previous values or a predicted value based on other models, “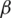 ” denotes the incubation rate, this is a measure of the latent person becoming infected. Essentially,
” denotes the incubation rate, this is a measure of the latent person becoming infected. Essentially,  /p, where p is the incubation period which medians between 5–6 days however can go as long as 14 days for COVID-19, “
/p, where p is the incubation period which medians between 5–6 days however can go as long as 14 days for COVID-19, “ ” denotes the recovery rate, this is governed by the amount of time taken by an infected individual to person recover from the virus.
” denotes the recovery rate, this is governed by the amount of time taken by an infected individual to person recover from the virus.  , where g denotes the duration of recovery post which the individual is pushed to removed phase.
, where g denotes the duration of recovery post which the individual is pushed to removed phase.

Figure 2: Traditional SEIR model for epidemiological analysis
3 Systematic Reviews of Literature and Analysis
A myriad of statistics based schemes has been traditionally employed in epidemiological surveys and predictive assessment for epidemics and spread of viral infraction. With the growing number of COVID-19 cases, experts and public health officials have been scrambling to scout for a model that enables them to make informed decisions and implement appropriate measures to address the pandemic. However, these schemes are deprived of pivotal data, general robustness and exhibit a lower accuracy with respect to long term predictions. Machine Learning (ML) based approach establishes itself as a promising avenue to address the impending crisis and benchmarks its potential as a tool to model and make a predictive assessment in the COVID crisis on numerous fronts. Efficacy of traditional statistics epidemiological surveys and models rely on various factors which are often not favourable in a novel situation such as this where we are still learning and discovering the behaviour of the virus and its spread.
Ardabili et al. [10] in their paper, noted that the SEIR model showed relative inaccuracy for countries where individuals practised preventive measures such as social distancing and voluntary quarantine such as Spain, China, Italy, Hungary and France and the accuracy was higher for the United States where there was a delay in containment measures. The SEIR model presumes that the “p” is a random variable and there exists a disease-free equilibrium. These models do not work very well where networks of contact and social minimizing is non-stationary with respect to time, something that’s prevalent with COVID-19. The two parameters contribute to determining the reproduction rate (R0) value.
Owing to the complexities of disease models, ML-based models have been proposed for the outbreak prediction and assessment of epidemiological surveys. The subsequent paragraphs expand on various ML-based approaches that model the pandemic data and make suitable considerations for providing relevant insights. Further Tab. 1 gives the comparative assessment for some of the important techniques. Iwendi et al. [11], modelled patient data using a boosted rendition of the Random Forest Algorithm (RFA) to make health predictions for COVID-19 patients. The proposed model employed the Adaboost algorithm to ameliorate the conventional random forest classifier algorithm and factors in demographic, travel, health and geographic data to estimate the severity of the case and probable outcome based on that. The model exhibited an accuracy of 94% with an F score of 0.86. The authors used Novel Coronavirus 2019 Dataset which is compiled from the data by JHU and WHO. The authors noted that their boosted RFA was accurate even on imbalanced datasets, additionally, the rate of deaths was higher in native people of Wuhan in contrast to non-natives. Benvenuto et al. [12], in a bid to map epidemiological trends for the novel coronavirus, employed an econometric model for estimating probable evolution. The authors employed an ARIMA model for their work on the data set collected from John Hopkins University’s (JHU) repository from 20th January to 10th February 2020 using 22 number determinations. The results echoed an epidemic plateau nature and the increase in the number of cases was a variable non-constant increase.
Table 1: Comparative assessment of prediction and diagnostics based studies on COVID-19

Nature Inspired Algorithms (NIA) and metaheuristic approaches have been used to solve numerous optimization and research problem statements from a myriad of domains. Martínez-Álvarez et al. [13], proposed a nature-inspired metaheuristic approach to map the spread of coronavirus in their algorithm titled as Coronavirus Optimization Algorithm abbreviated as CVOA. The scheme also combines the metaheuristics with deep learning paradigm to set the optimal hyperparameters of the LSTM. The CVOA makes use of relevant data regarding the virus such as the known rates of infection of patients from patient zero. It carries conventional three-tier segregation for the population-based SIR model, where an individual is either susceptible, infected or recovered from the activity of the virus. Using a bio-inspired approach has several benefits as it addresses large search spaces and finds sub-optimal solutions in reasonable computation time. This is particularly beneficial since the case data in here is continuous and ever-increasing. Although re-transmission is considered in the work but owing to lack of definite data a certain degree of ambiguity is attributed because of that.
CoronaTracker, a data analytics and prediction model, was proposed by the CoronaTracker Community Research Group composed of professionals and academicians from a plethora of backgrounds and institutions [14]. Through predictive modelling the research group aimed to forecast SEIR model—susceptibility, infected, exposed and recovered/removed due to the novel coronavirus. Additionally, the work employed sentimental modelling to map patterns of spread of public health information and socio-economic impact pertaining to the virus. The python based microservices and scrapper developed by the authors fetches data from JHU, WHO and other trusted news agencies.
Alzab et al. proposed a deep learning-based approach and uses VGG16 vision model architecture to diagnose COVID-19 and make a predictive assessment of its spread. The authors employ an artificial intelligence paradigm based on Convolution Neural Network that is implemented on chest X-rays to identify COVID-19 patients [15]. The model leveraged hidden patterns in chest X-ray images to perform COVID-19 diagnosis, the dataset used is a Kaggle database on COVID-19 from Jordan and Australia respectively. The proposed model shows promising results with high accuracy with F measure ranging between 95% and 99%. Further, to map the incidence of COVID-19 three forecasting schemes are used, the schemes are Prophet Algorithm (PA), ARIMA and LSTM. The models showed promising results for Jordan and Australia with accuracy soaring at 88.43% and 94.80%. The authors observed improved performance when employing augmentation. Among several early pieces of research, Al-qaness et al. [16] proposed an optimization scheme for COVID-19 forecasting cases that are confirmed for China and the United States. The study was among the early computational intelligence studies done in this domain. The proposed scheme forecasted the COVID cases for a period of 10 days based on previous data using Adaptive Neuro-Fuzzy Inference System (ANFIS) in conjunction with a nature-inspired algorithm based on pollination of a flower which is augmented using the Salp Swarm Algorithm (SSA). The simulation results show a high correlation between and proposed method and COVID 19 with the value of R2 soaring at 0.97.
Car et al. [17] proposed a model of the spread of COVID-19 infection employing a Multi-Layer Perceptron(MLP)—a type of feedforward Artificial Neural Network (ANN). The authors of this paper made use of a time-series data set which was manipulated into a regression model that is employed to train the MLP ANN. The dataset from the repository is maintained by the JHU Center for Systems Science and Engineering and Applied Physics Team. Using a grid search algorithm, the hyperparameters are varied and the combination amounts to 5376. The authors noted a 4 neuron in 4 HL configuration which employed a ReLU function along with limited memory-BFGS solver where the determination coefficient amounted to 0.9859, 099943 and 0.97941 for each of the three groups. The model displayed high fidelity for the two of three groups with the execution being the recovered patients. Li et al. [18] proposed a study on the propagation and prediction of COVID-19, the scope of the study was narrowed to the Hubei province of China. The study made consideration of transmission of the epidemic as the various stages and associated characteristics using the Gaussian Distribution Theory to build an experimental contemporary transmission model for the coronavirus.
The COVID-19 outbreak emanating from the Hubei province also clashed with mass migration associated with the annual spring festival Chunyun in the region, followed by unprecedented measures by Chinese authorities to curb the spread and break the transmission chain. Yang et al. [19], in their paper, aimed to do an impact assessment of these policies on the epidemic and show how these policies contained and controlled the same. The data pertaining to population migration prior to 23rd January was integrated with the latest data regarding COVID epidemic and plugged into the SEIR model to ascertain a new curve for the epidemic. Further a recurrent neural network, LSTM, was used to make further consideration; the model was trained on the SARS epidemic from 2003. The modified SEIR model predicted that the epidemic will peak by the end of February and gradually decay towards the end of April. Piccolomiini et al. [20] developed a modified SEIRD model to predict the transmission of COVID-19 in Italy. This study joins the rank of several regional studies conducted with a regional perspective at initial hotspots of the then epidemic which enabled the policymakers to make tangible steps to curb the transmission. The authors use the data from Italian Protezione Civile for 24th February and using their model forecast the spread of COVID in regions of Italy, specifically Lombardia and Emilia–Romagna, through their differential SEIRD model. The authors have emulated the infection rate as a function of time, susceptible to the policies of Italian authorities to curb the spread. The results encompass the impact of successive lockdown restrictions by the government and the output is a very good fit to data with marginal errors.
In a bid to harness computational intelligence in clinical decision-making Jiang et al. [21], proposed an AI-based data-driven system that determines the medical severity of a COVID case. The authors proposed a Predictive Analysis (PA) based scheme to provide rapid and critical decision making for COVID cases [21]. The scheme uses a filter method that makes entropic consideration for attributes which encompasses their variance and its impact on ascertaining the final label. The model considers rise in alanine aminotransferase (ALT) level, rise in haemoglobin level l and myalgias level as clinical indicators for the model. The proposed model showed 70%–80% accuracy in ascertaining the severe cases. Yudistira proposed a multivariable LSTM paradigm to predict correlation of COVID cases’ growth globally. In order to deal with the non-linear and complex nature of the COVID time-series curve an LSTM based RNN is used with sigmoid activation and dropout regularization [22]. The use of multivariable dataset is beneficial as the pandemic growth is affected by a litany of factors. A MinMaxScaler is used to normalize the data owing to the high sensitivity of LSTM to data pre-processing and normalization. Chimmula et al. [23] also worked with RNN based models to propose their rendition which aims to use the time series forecasting for the transmission of COVID-19 through LSTM networks for Canadian regions. Their deep learning-based model uses data from the JHU repository and Canadian Health Authority (CHA). Their model predicted that the spread of infection will die out by June 2020. Tomar et al. [24], in their paper, proposed a data-driven model using LSTM and curve fitting for the prediction of COVID cases in India for a period of 30 days to monitor the effect of preventive policies including lockdowns and physical distancing. Their model employs MATLAB environment and data from official data from COVID-19 in to train and validate its model.
Using CT scan of patients, Chen et al. [25] proposed a deep learning-based method to diagnose COVID-19. The proposed scheme makes use of chest CT scan images and identifies segmented lesions through a nested U-Net architecture, abbreviated as UNet++. The proposed model reduces the diagnosis time by nearly 0.65 times. In contrast to using the image information that is segmented, Zheng et al. [26] make use of a deep learning scheme for the diagnosis of COVID-19. The authors perform lung segmentation using a U-Net based paradigm whose results are fed as input to a 3D Convolution Neural Network (CNN) so as to ascertain COVID-19 in a probabilistic form. A sensitivity of 90% is achieved by the proposed scheme. Li et al. [27] designed a 3D deep learning model to detect the presence of SARS-CoV-2 in an individual from CT scan images of the chest. Heatmaps of the images are used by the model in determining infected individuals. An AI framework is presented by Harmon et al. [28] to categorize CT scan image of patients as positive or negative for COVID-19 induced Pneumonia. The primary training of the model was done on COVID positive patients in RT-PCR kit. A framework for identifying acoustic biomarkers including articulation, phonation, and respirationin COVID-19 patients (including asymptomatic) is proposed in [29]. Laguarta et al. [30] propose a CNN based framework to determine the voice biomarkers for COVID-19 from pre-recorded coughing sounds. An analysis of COVID-19 patients in terms of travel history, virus spread, gender and age are presented in [31]. Ahuja et al. [32] discuss the use of an AI framework to handle the COVID-19 pandemic in diverse ways. The authors mention the use of AI-based search algorithms which are capable of finding compound which can be a potential adjuvant for COVID-19 vaccine. In addition, the importance of deploying chat systems for public interaction and identification of integrative medicine for COVId-19 using AI framework, for managing this pandemic is also specified. A dataset referred as “CoronaDB-AI” comprising of compounds, has been prepared by Arshadi et al. [33]. This dataset can be used to train AI models for extracting drug components that can be potentially used for treating COVID-19.
4 Discussion and Performance Assessment
4.1 Applicability of Computation Intelligence in COVID Research
The above research survey has highlighted a wide array of work in various domains of combating COVID. With vaccine trials still underway, the fact of the matter remains that modelling of this viral disease remains of critical importance in understanding the impact. Analysis based on pure statistics and stochastic approaches fails to consider the intricacies within the data. Computational Intelligence (CI) based schemes such as AI and ML in addition to “learning” from the general trend also factor in the entanglement and deliver higher quality models. Fig. 3 illustrates the technology landscape in COVID-19 sector; the illustration represents the key avenues of research and associated technologies used during the first half of this year in COVID research, primarily in forecasting and clinical decision making.

Figure 3: Technology landscape and primary avenues for COVID-19 research
4.2 Challenges Associated with Computational Intelligence-Based Techniques in COVID-19 Research
The above-surveyed articles attest to the accelerating nature of CI-based applications in various domains of medicine. But their clinical applicability as effective techniques is limited. Even COVID-19 research has gained significant traction with the implementation of computational intelligence based paradigms.
4.2.1 Contention between Retrospectivity and Prospectivity in Research
As seen above, most of the research conducted above utilized a great number of data of COVID patients with relevant benchmarking via expert performance. Through them, we have seen a great majority of these studies that use a retrospective approach though employing historical data to train and validate the various AI and ML-based models. It has been argued that only through a prospective approach the actual ability of these models and systems can be understood, these studies often experienced a deteriorated performance when dealing with real-world data which is dissimilar to the one used in algorithm training. With COVID-19 pandemic, the regional and global nature has been extremely transient, and the data from a few months ago is extremely dissimilar from a trend perspective to the current data. This can likely be attributed to the fact we are still learning about this novel infection and different steps and therapeutics are being taken on an ad-hoc basis that is impacting the same.
4.2.2 Absence of Peer-Review Studies and Randomised Controlled Trials
A great majority of AI and CI-based studies, including the ones on COVID surveyed above are primarily published on preprint avenues as opposed to being submitted in peer-reviewed journals. A peer review study holds more importance and trust which consequently leads to a smoother adoption within the medical diaspora. Further, even fewer studies considered Randomised Controlled Trials (RCT), considered as a “gold standard” in the field of medicine, of CI-based models to date. This is even more obscure for CI models on COVID-19.
4.2.3 Bridging the Gap between Parameters and Medical Application
Clinical applicability has plagued the community and been an impending endeavour for the computational intelligence researchers from an applicability perspective. This “chasm” of AI becomes evident when we weigh in efficacy from a clinical point and compare it with accuracy. The area under operating characteristics of a receiver is not best reflective of clinical applicability despite the prevalence of machine learning-based research and also perplexes clinical experts.
4.2.4 Impediments to Comparing Dissimilar Algorithms
Research studies often encounter several impediments in objectively comparing algorithms and schemes, especially for COVID-19 as these models employ different methods, dissimilar population sets and varying distribution of a sample. In order for the comparison to be fair these studies needed to be subjected to similar data and similar metrics for performance, in absence of this the medical experts were not be able to identify the best-suited model or scheme for their patients. Hence, independent testing is required to detail a comparative assessment that is in congruence to clinical standards.
4.2.5 Factoring Shift in the Dataset and Quality Control
Algorithms based on electronic health records often neglect the fact the input data is produced from an environment that is non-stationary in nature where there is a significant shift in the population of the patients. This, in particular, is prevalent with COVID-19 where the practice on operation and clinical is dynamically evolving with time. In addition, a precursor to deployment is a robust regulatory framework that is necessary for the safety and effectiveness of CI-based systems. This is particularly challenging amidst the “novel” coronavirus pandemic. While learning of CI-based models is perpetual but system updates that are periodic are preferred for any clinical application, hence this dynamism needs to be factored in regulatory approvals in light of continual calibrations.
4.2.6 Impediments to Generalizations to Novel Settings and Populations and Algorithm Bias
Generalizability is often one front the AI-based models are unable to achieve which is a precursor to clinical applicability. There exist several blind spots in a model that if not identified through rigorous testing impedes in translation into actual use and contributes to incorrect decisions. Based on the above-conducted survey, these differences can be attributed to dissimilarity in e-health records, sensing equipment and associated technical and administrative incongruity. Algorithm bias present in the models can have three components; model bias, model variance and outcome notes.
4.2.7 Logistical Impediments and Susceptibility to Security Issues
Susceptibility of adversarial attacks is somewhat theoretical within the scope of this paper but it’s more amenable to real-world scenarios. CI-based systems have a myriad of underlying security issues that require conflict resolution before they can be safely deployed. This entails system related, privacy-related and authentication-related issues. This underscores the importance of uniform format of data collection and a system-wide coherence with regards to COVID-19 patient records.
4.2.8 Simulation and Performance Analysis
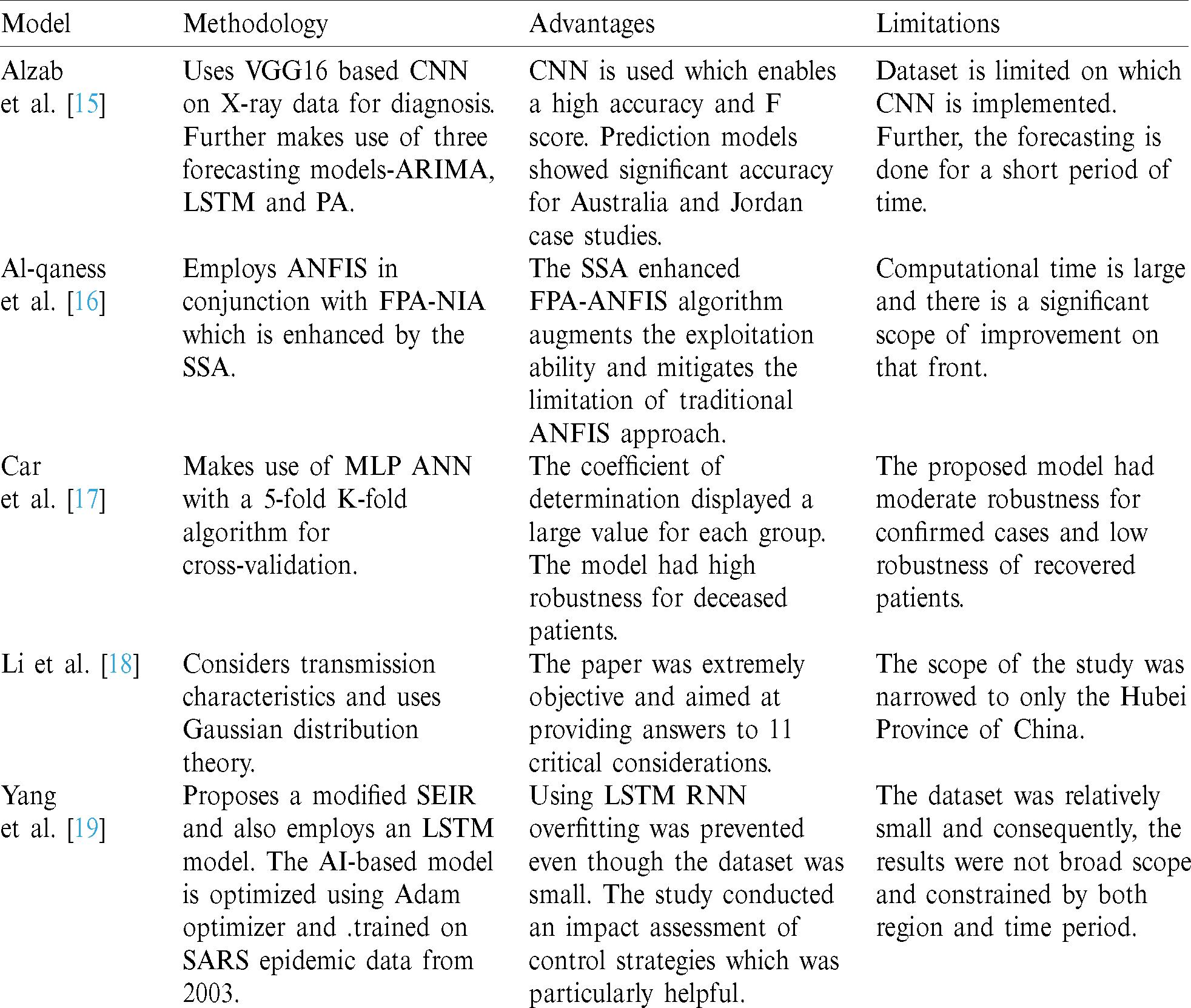
There has been a myriad of metrics, clinical and non-clinical, used in the above described techniques to enable forecasting of COVID cases. The metrics can be clustered as per Tab. 2. These metrics are some of the commonly available parameters from the prevalent databases around the world.
Table 2: Primary of the metrics employed in research scheme
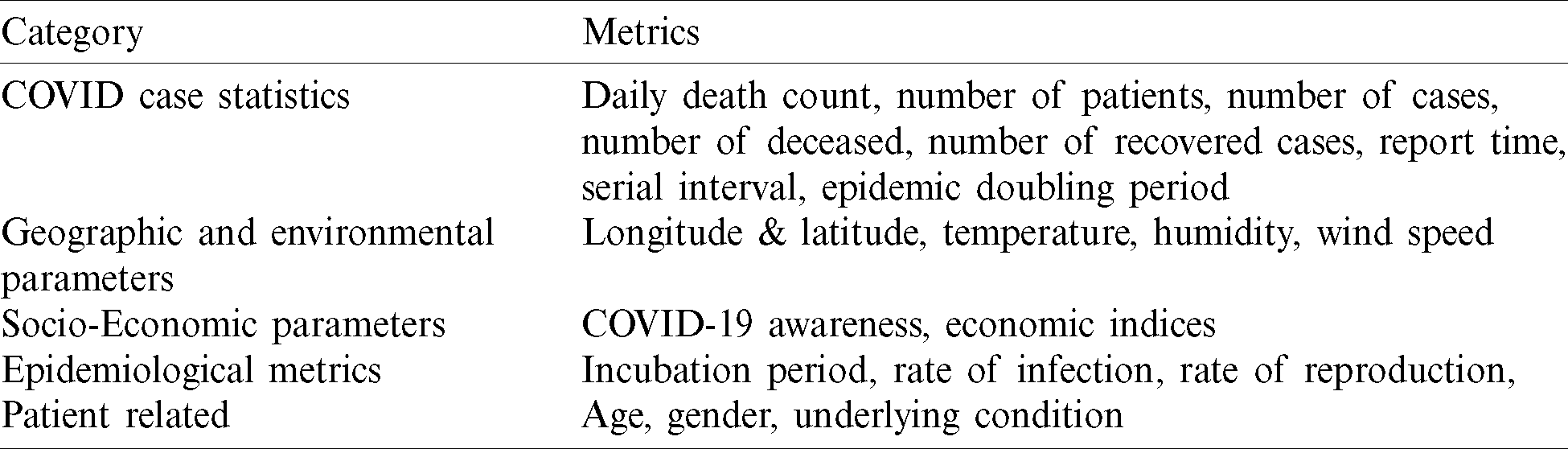
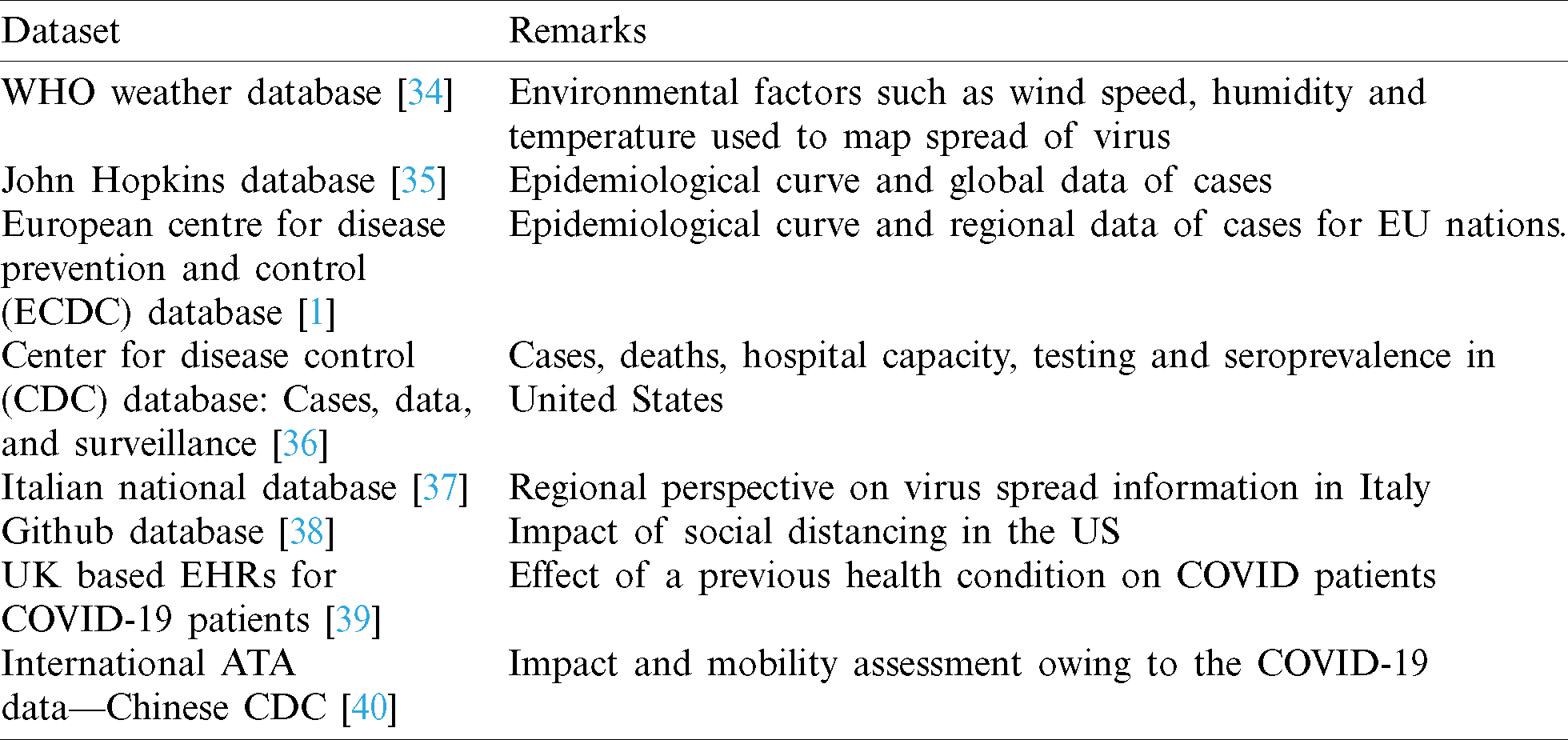
Tab. 3 lists out a few databases that are used in prediction and diagnosis techniques, as authors the purpose to list out them is such that we provide an appropriate starting point for our work.
Table 3: Uses of various datasets for COVID research
To under the scope of various model and techniques, a comparative assessment of various predictive techniques is performed. A times series database of worldwide COVID-19 cases starting from 22nd January, 2020 to 27 July, 2020 has been acquired from the ECDC database [39] in “.csv” format. We evaluate the performance of LSTM, linear regression; support vector regression (SVR), ARIMA and SEIR for prediction of COVID, the comparative assessment of the simulated models has been illustrated in Fig. 4 on Python 3.7 platform. The LSTM model is built with 2 LSTM layers and “RELU” activation with Adam optimizer is used with the dropout value of 0.2. For statistical analysis of the time series data ARIMA (0, 2, 4) is used. The degree of the SVR is set at 4 and a polynomial kernel is used. For linear regression, the basic model with a polynomial degree of 3 is used. For the SEIR model, the initial value of ‘N’ is set at 7 billion (approximate world population count) and the susceptible population is (N–I) and ‘I’ is 555 (as on 22.1.2020). The initial value of the basic reproduction rate of the virus is set to 3.6.

Figure 4: Comparative assessment for case prediction for COVID-19 graphs
The models are trained and tested with the above-mentioned dataset using holdout and cross-validation techniques. The RMSE (Root Mean Square Error), MAPE (Mean Absolute Percentage Error) and RMSLE (Root Mean Square Logarithmic Error) values obtained are shown in Tab. 4.
Table 4: Performance evaluation of models
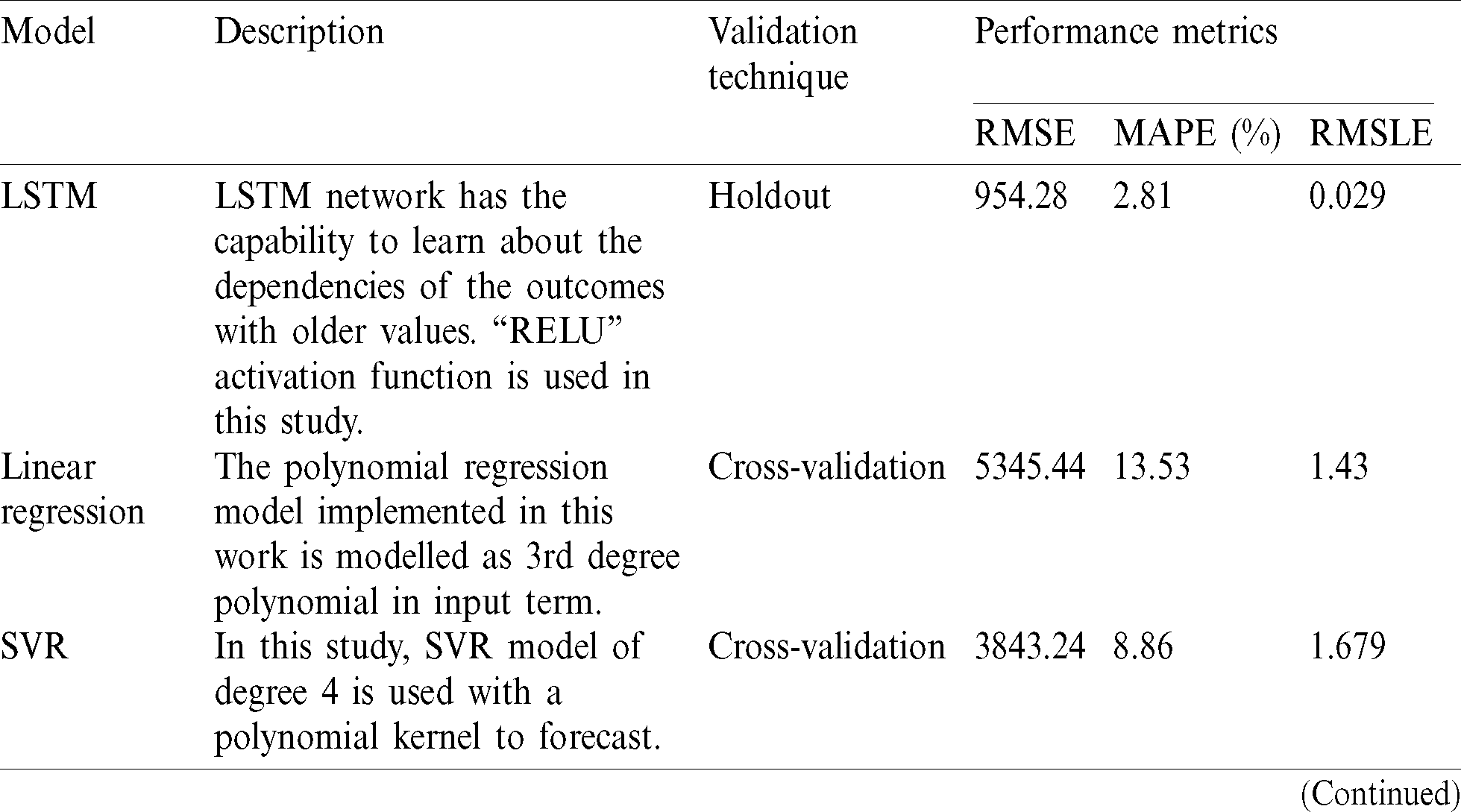
RMSE exhibits distributed property and serves as a better indicator of fit. The MAPE provides the extent of the difference between predicted and actual outcome. The RMSLE counts the fraction between the actual and predicted outcome. The true performance can be evaluated by using RMSE. Tab. 4 clearly shows that deep learning models like LSTM outperform the other models. Moreover, it is seen that ML-based models show better learning ability and have a superior prediction capacity as compared to the statistical and epidemiological model. The simple SEIR model can only capture the rising and flattening trend of the pandemic. LSTM and RNNs have better handling capacity for non-linear datasets in comparison to ARIMA models and given number of COVID-19 cases follow a non-linear form thus explain the lower RMSE value signifying better performance.
This technical composition provides a comprehensive review of the research trajectory in light of COVID-19 pandemic with special focus on computational intelligence-based paradigms in forecasting and diagnosis. This paper serves as a reference point for prospective researchers to orient future research work in this domain. AI and ML have proven to be a promising research avenue with wide-ranging applicability in various fields of medicine and beyond. From dealing with large search space, ascertaining optimal solution and providing data-driven intelligent techniques, CI has proven and continues to assert itself as a prospective area in medicine that provides state of art solutions to the improving the diagnostics and medical domain. The work surveyed above in light of the coronavirus pandemic attests to the same. The authors here chart out various computational intelligence-based methodologies used for forecasting and diagnosis of COVID-19, in particular, the learning and data-driven techniques with a critical focus on machine learning and AI.
Based on the analysis, this paper also identifies key impediments from a practical standpoint that impedes the translation of this research into application. This confluence of computational intelligence and medicine requires a deliberative and holistic approach so the insights from the models can be employed to combat this and future pandemics. Under performance review, this paper assesses the accuracy of various models, namely LSTM, Linear Prediction, SVR, ARIMA and SEIR in a comparative assessment across RMSE. The RSME value of LSTM based model showed the lowest RMSE value at 954.28 thus making it stand out in the comparative assessment whereas traditional SIER model showed the least accuracy where the RMSE stood at 12833.48 thereby attesting to the need to use ML-AI driven approaches for the forecasting of cases for COVID-19. Further LSTM is the only technique used in detection, prediction and therapeutic research of COVID-19.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. H. C. Johnson, C. M. Gossner, E. Colzani, J. Kinsman, L. Alexaks et al. (2020). , “Potential scenarios for the progression of a COVID-19 epidemic in the European Union and the European Economic Area,” Eurosurveillance, vol. 25, no. 9, pp. 1–5. [Google Scholar]
2. F. A. L. Marson and M. M. Ortega. (2020). “COVID-19 in Brazil,” Pulmonology, vol. 26, no. 4, pp. 241–256. [Google Scholar]
3. J. Geweke and S. Porter-Hudak. (1983). “The estimation and application of long memory time series models,” Journal of Time Series Analysis, vol. 4, pp. 221–238. [Google Scholar]
4. D. Kwiatkowski, P. C. Phillips, P. Schmidt and Y. Shin. (1992). “Testing the null hypothesis of stationarity against the alternative of a unit root,” Journal of Econometrics, vol. 54, no. 1–3, pp. 159–178. [Google Scholar]
5. S. Finlay. (2014). Predictive Analytics, Data Mining and Big Data: Myths, Misconceptions and Methods, 1 ed. UK: Palgrave Macmillan, pp. 21–34. [Google Scholar]
6. N. K. Ahmed, A. F. Atiya, N. E. Gayar and H. El-Shishiny. (2010). “An empirical comparison of machine learning models for time series forecasting,” Econometric Reviews, vol. 29, no. 5–6, pp. 594–621. [Google Scholar]
7. E. S. Gardner Jr. (1985). “Exponential smoothing: The state of the art,” Journal of Forecasting, vol. 4, no. 1, pp. 1–28. [Google Scholar]
8. G. E. Box, G. M. Jenkins, G. C. Reinsel and G. M. Ljung. (2015). Time Series Analysis: Forecasting and Control, 5 ed. Hoboken, New Jersey: John Wiley & Sons, pp. 305–340. [Google Scholar]
9. W. O. Kermack and A. G. McKendrick. (1991). “Contributions to the mathematical theory of epidemics—I,” Bulletin of Mathematical Biology, vol. 53, no. 1–2, pp. 33–55. [Google Scholar]
10. S. F. Ardabili, A. Mosavi, P. Ghamisi, F. Ferdinand, A. R. Varkonyi-Koczy et al. (2020). , “Covid-19 outbreak prediction with machine learning,” Algorithms, vol. 13, no. 10, pp. 1–38. [Google Scholar]
11. C. Iwendi, A. K. Bashir, A. Peshkar, R. Sujatha, J. M. Chatterjee et al. (2020). , “COVID-19 patient health prediction using boosted random forest algorithm,” Frontiers in Public Health, vol. 8, pp. 357. [Google Scholar]
12. D. Benvenuto, M. Giovanetti, L. Vassallo, S. Angeletti and M. Ciccozzi. (2020). “Application of the ARIMA model on the COVID-2019 epidemic dataset,” Data in Brief, vol. 29, pp. 1–4. [Google Scholar]
13. F. Martínez-Álvarez, G. Asencio-Cortés, J. F. Torres, D. Gutiérrez-Avilés, L. Melgar-García et al. (2020). , “Coronavirus optimization algorithm: A bioinspired metaheuristic based on the COVID-19 propagation model,” Big Data, vol. 8, no. 4, pp. 1–8. [Google Scholar]
14. F. B. Hamzah, C. Lau, H. Nazri, D. V. Ligot, G. Lee et al. (2020). , “CoronaTracker: Worldwide COVID-19 outbreak data analysis and prediction,” Bulletin of the World Health Organization, pp. 1–33. [Google Scholar]
15. M. Alazab, A. Awajan, A. Mesleh, A. Abraham, V. Jatana et al. (2020). , “COVID-19 prediction and detection using deep learning,” International Journal of Computer Information Systems and Industrial Management Applications, vol. 12, pp. 168–181. [Google Scholar]
16. M. A. Al-Qaness, A. A. Ewees, H. Fan and M. Abd El Aziz. (2020). “Optimization method for forecasting confirmed cases of COVID-19 in China,” Journal of Clinical Medicine, vol. 9, no. 3, pp. 1–15. [Google Scholar]
17. Z. Car, S. Baressi Šegota, N. Andelić, I. Lorencin and V. Mrzljak. (2020). “Modeling the spread of COVID-19 infection using a multilayer perceptron,” Computational and Mathematical Methods in Medicine, vol. 2020, pp. 1–10. [Google Scholar]
18. L. Li, Z. Yang, Z. Dang, C. Meng, J. Huang et al. (2020). , “Propagation analysis and prediction of the COVID-19,” Infectious Disease Modelling, vol. 5, pp. 282–292. [Google Scholar]
19. Z. Yang, Z. Zeng, K. Wang, S. Wong, W. Liang et al. (2020). , “Modified SEIR and AI prediction of the epidemics trend of COVID-19 in China under public health interventions,” Journal of Thoracic Disease, vol. 12, no. 3, pp. 165–174. [Google Scholar]
20. E. L. Piccolomiini and F. Zama. (2020). “Monitoring Italian COVID-19 spread by an adaptive SEIRD model,” PLoS One, vol. 15, no. 8, pp. 1–15. [Google Scholar]
21. X. Jiang, M. Coffee, A. Bari, J. Wang, X. Jiang et al. (2020). , “Towards an artificial intelligence framework for data-driven prediction of coronavirus clinical severity,” Computers, Materials & Continua, vol. 62, no. 3, pp. 537–551. [Google Scholar]
22. N. Yudistira. (2020). “COVID-19 growth prediction using multivariate long short term memory,” IAENG International Journal of Computer Science, vol. 47, no. 4, pp. 829–837. [Google Scholar]
23. V. K. R. Chimmula and L. Zhang. (2020). “Time series forecasting of COVID-19 transmission in Canada using LSTM networks,” Chaos, Solitons & Fractals, vol. 135, 109864, pp. 1–6. [Google Scholar]
24. A. Tomar and N. Gupta. (2020). “Prediction for the spread of COVID-19 in India and effectiveness of preventive measures,” Science of the Total Environment, vol. 728, no. 138762, pp. 1–6. [Google Scholar]
25. J. Chen, L. Wu, J. Zhang, L. Zhang, D. Gong et al. (2020). , “Deep learning-based model for detecting 2019 novel coronavirus pneumonia on high-resolution computed tomography: A prospective study,” medRxiv, pp. 1–27. [Google Scholar]
26. C. Zheng, X. Deng, Q. Fu, Q. Zhou, J. Feng et al. (2020). , “Deep learning-based detection for COVID-19 from chest CT using weak label,” medRxiv, pp. 1–13. [Google Scholar]
27. L. Li, L. Qin and Z. Xu. (2020). “Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: Evaluation of the diagnostic accuracy,” Radiology, Medline, vol. 296, no. 2, pp. 65–71. [Google Scholar]
28. S. A. Harmon, T. H. Sanford and S. Xu. (2020). “Artificial intelligence for the detection of COVID-19 pneumonia on chest CT using multinational datasets,” Nature Communications, vol. 11, no. 4080, pp. 1–7. [Google Scholar]
29. T. F. Quatieri, T. Talkar and J. S. Palmer. (2020). “A framework for biomarkers of COVID-19 based on coordination of speech-production subsystems,” IEEE Open Journal of Engineering in Medicine and Biology, vol. 1, pp. 203–206. [Google Scholar]
30. J. Laguarta, F. Hueto and B. Subirana. (2020). “COVID-19 artificial intelligence diagnosis using only cough recordings,” IEEE Open Journal of Engineering in Medicine and Biology, vol. 1, pp. 1. [Google Scholar]
31. V. Bhatnagar, R. C. Poonia, P. Nagar, S. Kumar, V. Singh et al. (2020). , “Descriptive analysis of COVID-19 patients in the context of India,” Journal of Interdisciplinary Mathematics, pp. 1–16. [Google Scholar]
32. A. S. Ahuja, V. P. Reddy and O. Marques. (2020). “Artificial intelligence and COVID-19: A multidisciplinary approach,” Integrative medicine research, Integrative Medicine Research, vol. 9, no. 3, pp. 100434–110043. [Google Scholar]
33. A. K. Arshadi, J. Webb, M. Salem, E. Cruz, S. Calad-Thomson et al. (2020). , “Artificial intelligence for COVID-19 drug discovery and vaccine development,” Frontiers Artificial. Intelligence, vol. 3, no. 65, pp. 1–13. [Google Scholar]
34. S. Bhattacharjee. (2020). “Statistical investigation of relationship between spread of coronavirus disease (COVID-19) and environmental factors based on study of four mostly affected places of China and five mostly affected places of Italy,” arXiv-CS Cornell University. [Google Scholar]
35. E. Dong, H. Du and L. Gardner. (2020). “An interactive web-based dashboard to track COVID-19 in real time,” The Lancet Infectious Diseases, vol. 20, no. 5, pp. 533–534. [Google Scholar]
36. T. M. McMichael, D. W. Currie, S. Clark, S. Pogosjans, M. Kay et al. (2020). , “Epidemiology of Covid-19 in a long-term care facility in King County, Washington,” New England Journal of Medicine, vol. 382, no. 21, pp. 2005–2011. [Google Scholar]
37. G. Giordano, F. Blanchini, R. Bruno, P. Colaneri, A. Di Filippo et al. (2020). , “Modelling the COVID-19 epidemic and implementation of population-wide interventions in Italy,” Nature Medicine, vol. 26, no. 1, pp. 855–860. [Google Scholar]
38. J. Bayham and E. P. Fenichel. (2020). “The impact of school closure for COVID- 19 on the US healthcare workforce and the net mortality effects,” The Lancet-Public Health, vol. 5, no. 5, pp. 271–278. [Google Scholar]
39. A. Banerjee, L. Pasea, S. Harris, A. Gonzalez-Izquierdo, A. Torralbo et al. (2020). , “Estimating excess 1-year mortality from COVID-19 according to underlying conditions and age in England: A rapid analysis using NHS health records in 3.8 million adults,” Lancet, vol. 395, no. 10238, pp. 1715–1725. [Google Scholar]
40. M. Hossain, A. Junus, X. Zhu, P. Jia, T. H. Wen et al. (2020). , “The effects of border control and quarantine measures on global spread of COVID-19,” Epidemics, vol. 32, no. 1, pp. 1755–4365. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |