DOI:10.32604/cmc.2021.014347

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014347 |  |
| Article |
Automatic Segmentation of Liver from Abdominal Computed Tomography Images Using Energy Feature
1University Department of Computer Science Engineering, Anna University, Chennai, 600025, India
2University Department of Information Technology, MIT Campus, Chennai, 600044, India
*Corresponding Author: Shiloah Elizabeth Darmanayagam. Email: shiloah@annauniv.edu
Received: 15 September 2020; Accepted: 28 October 2020
Abstract: Liver Segmentation is one of the challenging tasks in detecting and classifying liver tumors from Computed Tomography (CT) images. The segmentation of hepatic organ is more intricate task, owing to the fact that it possesses a sizeable quantum of vascularization. This paper proposes an algorithm for automatic seed point selection using energy feature for use in level set algorithm for segmentation of liver region in CT scans. The effectiveness of the method can be determined when used in a model to classify the liver CT images as tumorous or not. This involves segmentation of the region of interest (ROI) from the segmented liver, extraction of the shape and texture features from the segmented ROI and classification of the ROIs as tumorous or not by using a classifier based on the extracted features. In this work, the proposed seed point selection technique has been used in level set algorithm for segmentation of liver region in CT scans and the ROIs have been extracted using Fuzzy C Means clustering (FCM) which is one of the algorithms to segment the images. The dataset used in this method has been collected from various repositories and scan centers. The outcome of this proposed segmentation model has reduced the area overlap error that could offer the intended accuracy and consistency. It gives better results when compared with other existing algorithms. Fast execution in short span of time is another advantage of this method which in turns helps the radiologist to ascertain the abnormalities instantly.
Keywords: Liver segmentation; automatic seed point; tumor segmentation; classification; fuzzy C means clustering
Liver is an auburn gland that carries out more than 500 essential functions of human life. Evolution of Chronic Liver Disease (CLD) is witnessed by many varying stages having distinct pathological characteristics. Steatosis otherwise known as Fatty liver infiltration is the preliminary stage of the liver anomaly that crop up due to surge of fat deposits in hepatocytes. Hepatic cirrhosis is an acute advanced malady of the hepatic organ liver that is characterized by metastasized damage to liver with scaring of the tissue and aggregation of cells. The extremity of cirrhosis is the Hepatocellular Carcinoma (HCC) or hepatoma is an initial malignancy of liver. Hepatitis B and Hepatitis C appears to be the most significant etiology of hepatocellular carcinoma. The high resolution techniques that are intended in the therapeutic field is decisively considered to be beneficial one. It offers the physician a better diagnosis and follows up in the patient’s ailments.
Computer Aided Diagnosis (CAD) is a technical approach which assists the radiologists to elucidate the images more precisely and to discern the potential findings so as to exclude the fallacious interpretation. The CT images possess higher signal to noise ratio, so the detections are more exact. CT images are immensely preferred for hepatic anomaly diagnosis because they offer gross sectional image. Therefore, CAD procedures give the doctors a second opinion in the diagnosis of liver diseases.
Manual segmentation of liver has various problems. Because of the intricate nature of anatomy of human body, the ambit of liver cannot be ascertained to the exact. Often, different interventional radiologists identify it differently. These pitfalls lead to under segmentation or over segmentation [1]. The goal of medical image segmentation is to delineate the image areas depicting the different anatomies. This prevails as an arduous task because of the intersection and overlap of delicate tissues with meaty intra organ variation and parallel voxel intensities of the adjacent organs. Liver is an organ with high degree of vascularization, which implies that the liver encompasses many blood vessels within the tissue [2]. Owing to this fact, the conventional perceiving of the hepatic tumours from the similar gray intensity is inordinately cumbersome task. Liver segmentation is usually considered the toughest because of the similar intensities of neighboring organs.
The following are the four steps involved in CAD System.
Pre-processing–-It is done to eliminate the unwanted noise level and to ameliorate consistency of the image.
Segmentation–-It is the technique of extricating liver images from the ventral section of computed tomography images. Then the tumor section is disengaged from the segmented liver called region of interest (ROI).
Feature Extraction–-From the segmented tumor region, texture and shape features are extracted for the purpose of classification.
Classification–-Where classifiers are adopted to ascertain the hepatic ailments based on the extracted elements.
The remainder of the paper is organized as follows: Section 2 discusses the related work, Section 3 explains the architecture of the proposed work, Section 4 presents the experimental result and analysis, and Section 5 presents the conclusion and future work.
The CT image dataset used in this work have been collected from scan centers and repositories. MATLAB 2018 has been used to implement the proposed work.
This work focuses on automatic seed point selection using energy feature for segmentation of liver.
Cheema et al. [3] have suggested a method that effects the segmentation of liver with greater particularity by debarring the noise from CT images. The noiseless images are extracted by modulating the residual convolutional neural network (LER-CN) [3] that flecks and bring outs a liver model with ultimate noiseless which could enable better interpretation. This convoluted noise removal component (NRC) discharge the extraction by transmitting the Low Dose CT (LDCT) images which is noise removed to structural preservation component (SPC) in order to revamp and collate the acquired image by sustaining its entire modules. LER-CN differentiated the fringes of liver followed by the discrimination of the organ’s texture to the required level. The working model of the NRC and SPC is kept synchronized so that the input and output images do well with the mapped estimation. This LER-CN inculcates fast output by endorsing last in first out (LIFO) in which the last NRC layer is linked to the initial layer of SPC and the first NRC layers are connected with the last SPC layers. By adopting LIFO, the LER-CN is ensured with a fine flow of data. This LDCT images are extracted with the usage successive de-convolutional layer, so that the output images are tuned with input images. The specified dimensions of overlapped patches are inculcated in order to augment the training samples.
Song et al. [4] have proposed the improved confidence connected livers segmentation method based on the imaging characteristics in three views [4] that are the reducing noise, automatic seed point selection and finally combining the liver contours that are extracted from coronal, sagittal and cross sectional images. This method reduces the frequency of the wrong classification that is caused due to the closeness of the neighboring organs. Further this method enhances the accuracy and automatic segmentation without user intervention. Also it efficiently excludes the complexity of classifying the edges of the liver.
Elaziz et al. [5] have proposed an advanced algorithm for automatic segmentation of liver and hepatic tumor from abdominal CT images. Their proposed algorithm [5] abates the computation time by excluding the sections of the other structures as most strategy in present day are extremely time exhausting. Region growing method is adopted from the segmented liver that initiated from a seed point which is selected automatically.
Peng et al. [6] have put forth the level set method for segmentation of the input image based on the local section gradient. In some of the existing method, because of similar intensity and weak edges, the proper results can’t achieve. So they have considered local gradient information to overcome this issue. At first, the correspondence between the segmented object and image gradient to local minima and maxima all over the picture element are presented based on two assumptions, from which new pixel classification method is introduced based on weight of Euclidean distance [6]. Secondly, to improve the anti-noise capacity of the proposed gradient information based model, a combination of variational level set and image spatial adjacent information is implemented. Thirdly a new promulgation procedure with the edge indicator function is encompassed into the level set function to classify the picture element in similar regions of the segmentation object and also to make the proposed method more insensitive to origin contours and static numerical implementation.
Li et al. [7] have proposed a neoteric embark on automatic liver segmentation, which potentially coalesce the shape based initialization and deformable graph cut methodology along with shape limitation that intercepts the arising snag which soared on by the specific oddity of the liver’s anatomical pattern and caliber of the picture element [7]. As on exemplification it’s higher rendition, 50 CT scan images have been put up in a communal accessible domain. The tentative outcome of their method was efficacious and impeccable for progressive perception of the liver surface. They asserted that their proposed method can identify the hepatic surface with lower error and can successfully cope with under-segmentation and over segmentation.
Singh et al. [8] have proposed segmentation of liver using Hybrid K-means clustering and Level set. Their method [8] uses crossbreed approach that is the hybrid clustering algorithm to sniff out the auburn gland. The hybrid clustering is accomplished by determining the specified number of clusters in the image using K-means clustering technique and following to that Ant Colony Optimization (ACO) is exerted to treat the fallacious graded k-clusters. Thus applying level set after clustering increases accuracy of segmentation.
Altarawneh et al. [9] have proposed a revamped model that is the modified distance regularized level set that adds on innovative balloon force to the existing Distance regularized level set estimation (DRLSE) model to superintend the orientation of the growing silhouette’s by the various preferred overtures. The newly included balloon force [9] discourages the evolving contour from exceeding the liver boundary or leaking at a region that is associated with a weak edge, or does not have an edge. This model deals with over-segmentation more effectively compared to DRLSE model.
Heckel et al. [10] have proposed a swift approach for partial volume correction for solid lesions in CT scans. Their algorithm [10] is an inductive reasoning of SPVA (Segmentation-based Partial Volume Analysis) proposed by Kuhnigk. Also it incorporates the augmented Kuhnigk’s approach, that it can be administered to any designated segmentation outcomes after the manual evolution of any rigid firm target that comprises non-analogous abscess and lesions with the non-identical physical encompassing. The accuracy of the volumetric measurement is highly improved the veracity of the volumetric quantification, obviously that has been shown on various phantom data. The algorithm has apparently pronounced to be more authentic method for volume estimation when relate with the voxel counting for the most normal abscess or tumors which are assessed during all kinds of therapeutic treatment including chemotherapy. It is compatible to other solid abscess, since all vital information’s are extricated from the given CT image, based on a given segmentation mask.
Li et al. [11] have put forward a supervised variational level set to mobilize the statistical energy function with a weighted probability approximation that is applied in liver segmentation. For better estimation of absolute potential distribution [11] and discriminate the statistical intensity differences in all dimensional aspects. The prevailing impreciseness in the potential distribution is resolved by the numerical function for all its rear end and the user interface.
Liu et al. [12] have put forth a technique in which pixel altogether with the region level conditions are frequently clustered by the suggested region level Hidden Markov Random Field-Fuzzy C Means (HMRF-FCM) algorithm, in which region-level contextual information is structured by Region level Markov Random Field (RMRF), and is further amplified by the average template of memberships. The HMRF-FCM algorithm [12] has been enhanced in accordance with abstracting potential outcomes into a level-one-region where voluminous parameters are modeled by the MRF and are enhanced by the region-level mean template of fuzzy memberships.
Liu et al. [13] have proposed an advanced unsupervised FCM-based image segmentation modus by holding scrupulous study in the selection of local information. For better monitoring over the strength and latitude of the responding pixel, they have integrated the region level local information into the fuzzy clustering technique. First, a novel variant task is initiated by merging region based and pixel based distance functions together. This is done in order to enhance the relationship between pixels which have similar local characteristics. Secondly, a probability function is propounded by assimilating the variances between neighboring region into the mean model of the fuzzy membership function which is accustomed to select the local spatial limitations [13] by a tradeoff weight in accordance with whether the pixel belongs to homogeneous region or not. The proposed modus intensified the prevalence between the pixel within the parallel region and averts the segment fringes becoming smooth by integrating the section based information into the spatially extend limitations.
From the related work, it is found that fixing the seed point automatically is a challenging task because many neighboring organs are connected. So, in this paper an automatic seed point selection strategy using energy feature has been proposed. This segmentation approach has resulted in a reduction in area overlapping error and execution time as compared to the existing work.
The proposed model for automatic seed point selection can be used in level set segmentation technique which will automatically detect liver from CT scans and a classifier can be used to classify whether tumor is present or not. The system requires CT images as input. A guided filter is used for pre-processing of the CT images. With the help of energy feature which is one of the texture features, the seed point is selected automatically and the value of the x and y axis of the seed point is given as the initial starting point of the level set algorithm. This method helps to segment the liver and then the segmented liver portion is stored separately. A fuzzy c-means algorithm is now applied on the stored liver part to segment the tumors. Additional features are extracted for the tumor. The extracted features are labelled with class labels to form the training set. Finally the SVM classifier is made ready for classification. Fig. 1 shows an architecture of automatic liver tumor sementation.

Figure 1: Architecture of the Automatic Liver Tumor Segmentation Model
The preprocessing is the procedure done before taking an image into the system. The original CT images of various sessions possess different contrast. So an adjustment of the contrast is done using the histogram of liver intensity. A guided filter is used as an edge preserving smoothing operator like the desired bilateral filter. It has a better behavior around edges of an object.
Segmentation of an image is the method of fixing a label to each and every pixel in an intended object. The segmentation of liver is basically the toughest part, since the intensity levels of the neighboring organs and liver are quite similar in a CT scan [14]. Here, automatically the seed point is selected with the help of energy feature and a modified level set algorithm is used for liver segmentation. This is the most important process as any under segmentation or over segmentation will result in missing a part or whole of the tumors.
3.2.1 Energy Based Seed Point Selection
The first step in liver segmentation is to choose the position of the seed point from where the contour grows. This is done using the energy feature.

Numerous region based level set methods have been suggested for segmenting a particular image. The image driven forces advances to all its desired boundary of the target. The user specifies an initial guess for the contour, which is then moved by image driven forces to the boundaries of the desired objects. So the seed point generated by energy based seed point selection algorithm is considered as initial point. In this method, two different kinds of forces are taken into account, an internal force which ascertain within the curves are made to retain the model in leveled up manner during the deformation process. While the external force that are determined from the pooling image data are ascertained to advance the model towards the target or specified features within the image. A proposed modified distance regularized level-set algorithm, shows the existence of balloon force which controls the growth of the internal forces within the curve along rough edges.
3.3 Tumor Candidate Segmentation
Fuzzy C-Means Clustering (FCM) is administered on the segmented liver image to extract tumor portions from it. FCM is commonly used for segmentation. It assigns the membership to image pixels related to every cluster midpoint on the grounding of the extension between the cluster point and image pixels. Then, the segmented tumor portions are saved.
Working of the FCM:
This algorithm works by grouping or clustering data points related to the center of clusters. Here, an aggregation of every data point membership it must be equal to one. Then after each epoch membership and center of clusters should be re-corrected by using the given formula:


where,
‘d’–-The number of data points.
‘vj’–-jth cluster center
‘f’–-The index of fuzziness m with the limit of [1, 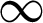 ].
].
‘n’–-The total number of cluster center
‘Mij’–-The membership of ith data to jth cluster center.
‘aij’–-The Euclidean distance between ith data and jth cluster center.
The ultimate aim of FCM algorithm is to minimize:

where, ‘||xi – vj||’–-The Euclidean distance between jth cluster center and ith data.
1. The cluster centers ‘n’ are selected randomly.
2. Determine the fuzzy membership ‘Mij’ by the given formula given in Eq. (1).
3. Calculate the updated cluster centers ‘ ’ using the formula given in Eq. (2).
’ using the formula given in Eq. (2).
4. Continue the previous two steps until the minimal ‘J’ value is attained
‘J’–-objective function.
Segmentation of ROI
Input: Segmented liver image (.jpeg),
Output: Tumor Candidate (if any),
1. Choose the number of clusters c between 2 to n, and the exponential weight Mu (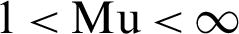 ), and termination criteria and partition matrix, U.
), and termination criteria and partition matrix, U.
2. Determine the cluster center of fuzzy 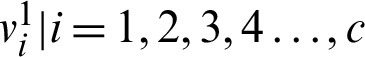 by using U1.
by using U1.
3. Estimate the new partition matrix Ul+1,
4. Find the updated partition matrix 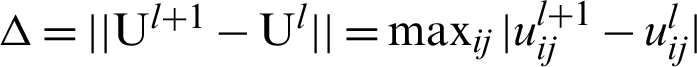 .
.
4 Experimental Results and Analysis
MATLAB has been used for the implementation of the proposed work. The abdominal CT image is fed as input. The CT image is used to generate seed point using the proposed automatic seed point selection algorithm.
A sample input CT image is shown in Fig. 2. It is processed using guided filter to smoothen the boundaries to facilitate more accurate level set segmentation. The processed CT image shown in Fig. 4 is now used for level set segmentation where segmented liver region is obtained.

Figure 2: Abdominal CT input image
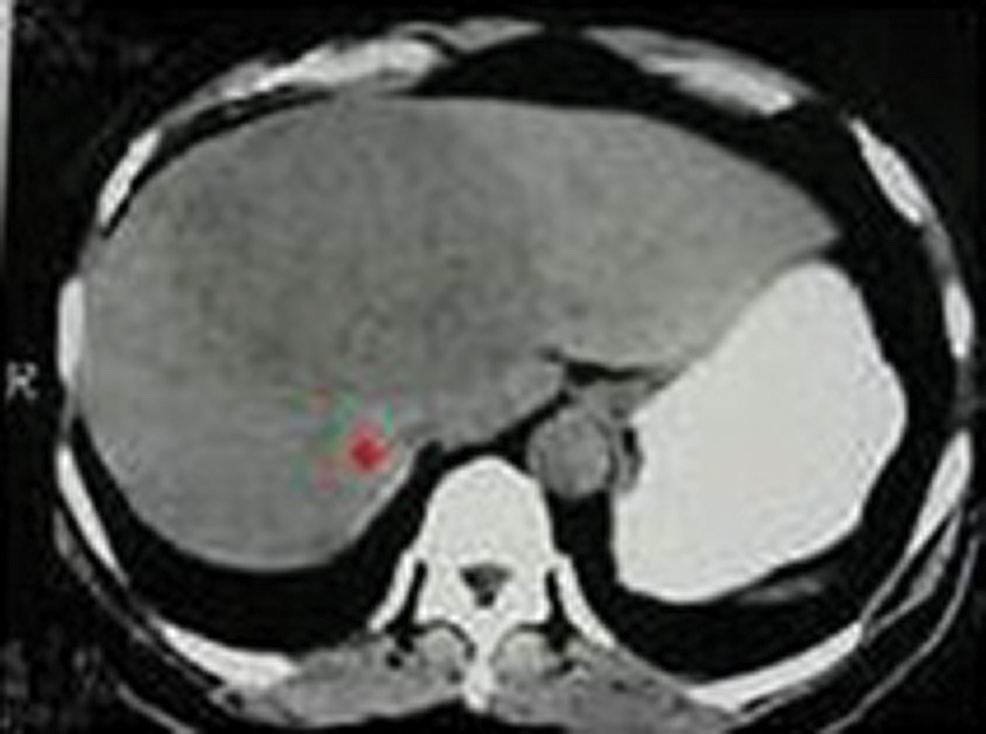
Figure 3: Seed point selection using the proposed algorithm

Figure 4: Processing of the input CT method being applied image using guided filter
The above Fig. 3 shows the image with seed point selection marked in it and Fig. 5 shows the growing contour which covers the liver image.

Figure 5: Level set method being applied to the processed liver image
After isolating the liver region from the abdominal CT image which is shown in Fig. 6. FCM algorithm is applied on it and the mask of the tumor region is obtained. Then the masked region is cut from the liver image to get the segmented liver tumor region shown in Fig. 7.
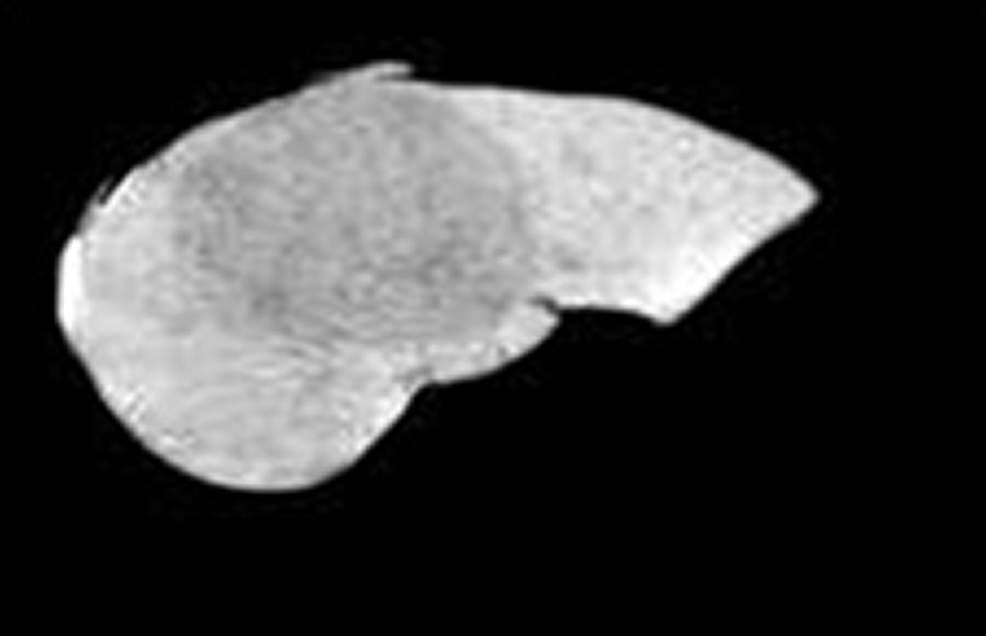
Figure 6: Segmented liver region

Figure 7: Segmented tumor region
Performance Analysis
Different parameters have been used to analyze the performance of the segmentation of liver, segmentation of tumor and classification of healthy and tumorous image.
The calculated results are mentioned in Tabs. 1–3, which gives the comparison between the existing and proposed techniques on the basis of Area Overlap Error and execution time. The results were taken for 50 CT scan images. The images were processed using MATLAB.
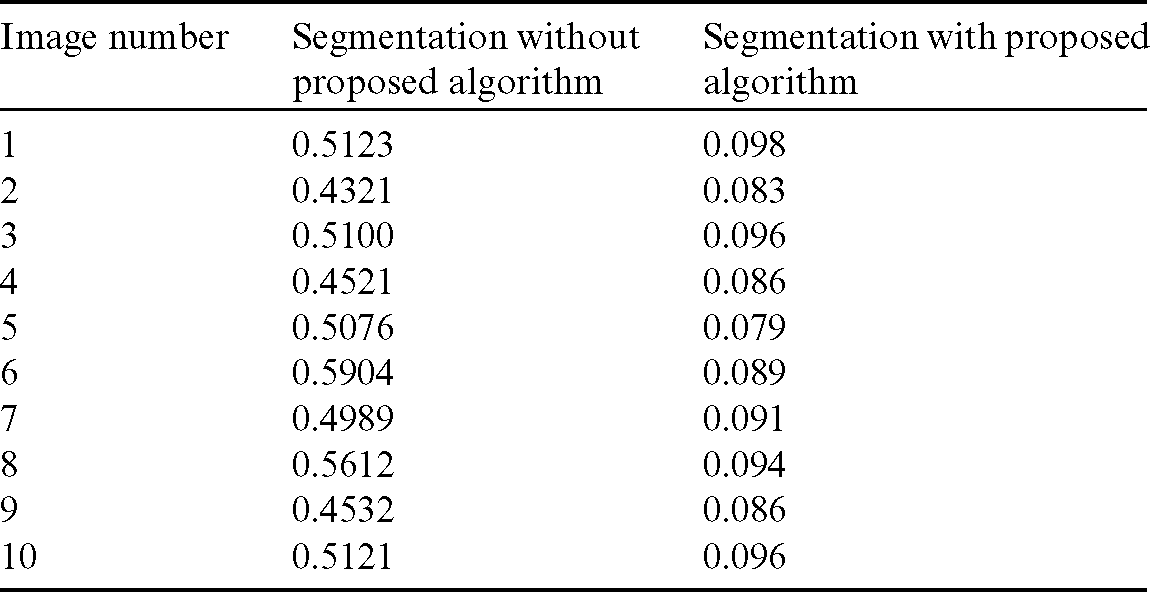
Table 1: Comparison of area overlap error of segmented liver image obtained from level set with the proposed automatic seed point selection and those obtained using level set without automatic seed point selection
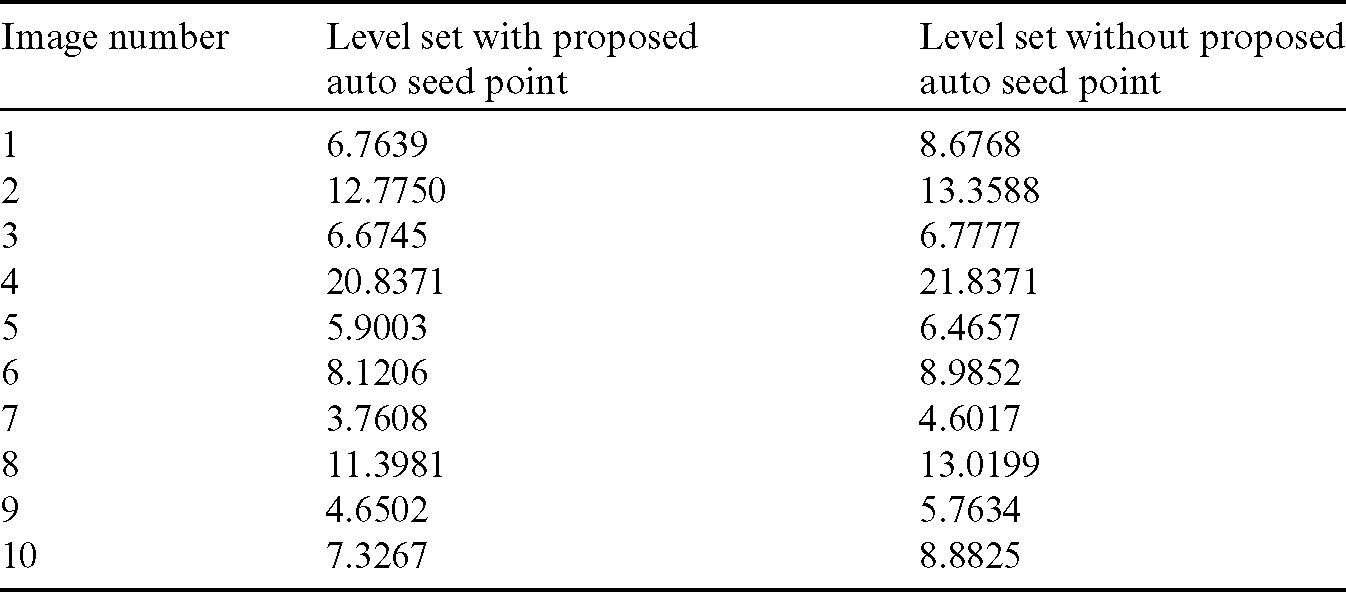
Table 2: Comparison of area overlap error of segmented liver image obtained from level set with the proposed automatic seed point selection and existing algorithms
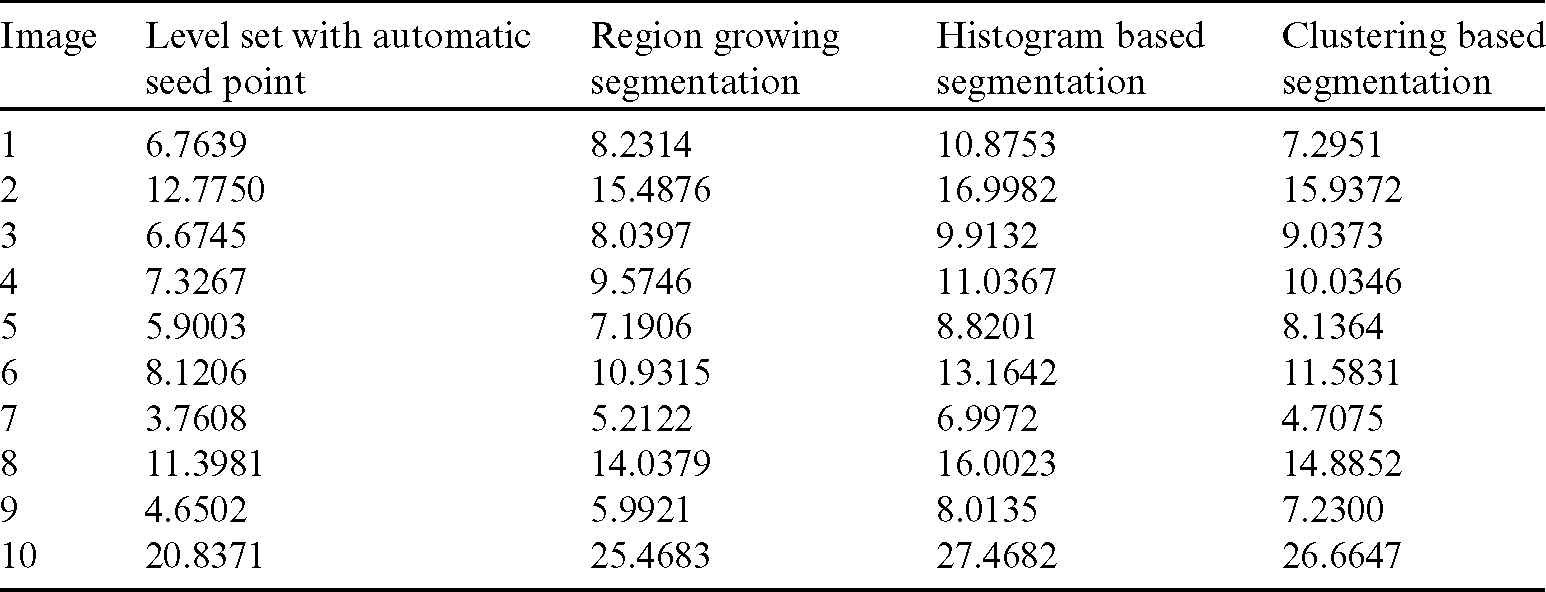
Table 3: Comparison of the existing and the proposed technique on basis of the execution time (ms)
Tab. 3 represents execution time of the entire process with and without the proposed algorithm. Though an automatic seed point selection algorithm is appended, the execution time has reduced, because it segments the liver region properly. Execution time will increase if under or over segmentation happens. But here this problem has been rectified and this has reduced the execution time.
Area Overlap Error
Area Overlap error is used to compare the accuracy of the image segmented using the proposed method and the ground truth. Area Overlap error can be computed using the formula.

where A and B are the areas of the liver image obtained from the proposed segmentation method and ground truth respectively.
The comparison of area overlap error in level set algorithm with and without automatic seed point selection is mentioned in Tab. 1. Similarly, Tab. 2 represents the area overlap error of proposed method with other existing techniques. An execution time of overall system mentioned in Tab. 3.
The above Fig. 8 shows comparison graph of Area overlap error of segmented liver image obtained from level set with the proposed automatic seed point selection and those obtained using level set without automatic seed point selection. Similarly, Fig. 9 represents, comparison graph of the proposed and existing algorithms with respect to area overlap error. And Fig. 10 represents the comparison graph of overall system execution time.
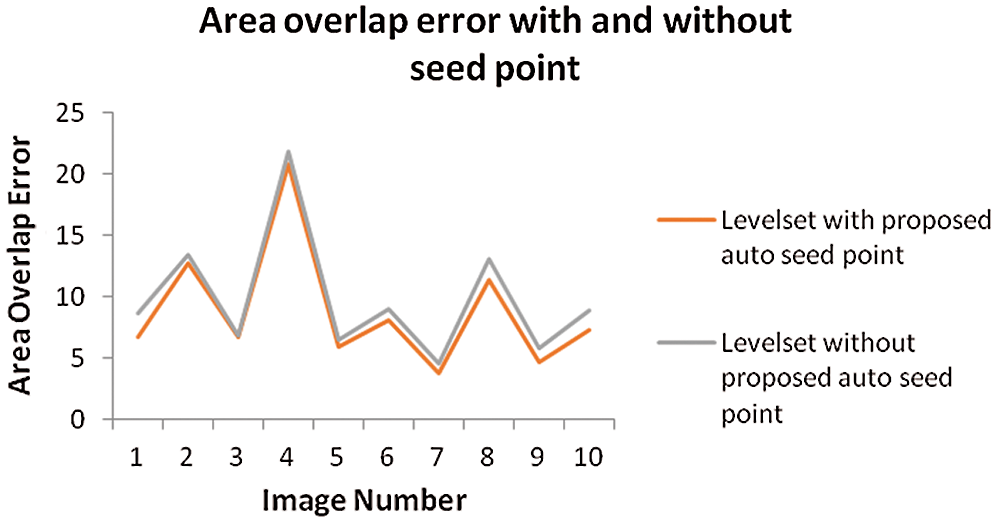
Figure 8: Comparison graph of area overlap error of segmented liver image obtained from level set with the proposed automatic seed point selection and those obtained using level set without automatic seed point selection

Figure 9: Comparison graph of area overlap error of segmented liver image obtained from level set with the proposed automatic seed point selection and existing algorithms

Figure 10: Comparison graph between with and without proposed algorithm of execution time
The automatic detection of liver tumors from CT scans has been proposed keeping in mind the challenges prevailing in the field of medical imaging in detecting the cancer at an early stage. Early detection of cancer is important in the field of cancer diagnosis and treatment. The early the cancer is detected, higher the chances of cure. This paper has been visualized in a holistic approach considering the critical issues that are daunting in the domain. The proposed framework for automatic detection of liver tumors has been developed in such a way that it is as easy as possible to implement. The results are fairly consistent when tested with different datasets. Experimental results show that the proposed work is as good as existing systems with lower area overlap error and lesser time for operation. In the future work, a better system may be developed that reduces the error rates and further strengthens the chances of detection and classification. Also, this proposed system can be extended to work for 3D images and also images from different medical imaging modalities such as MRI, Ultrasound. Moreover, the shape and texture features can be extracted from the segmented ROI and those features can be used to classify the healthy and tumorous images using Support Vector Machine (SVM).
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Y. Song, W. Cai, H. Huang, X. Wang, Y. Zhou et al. (2014). , “Lesion detection and characterization with context driven approximation in thoracic FDG PET-CT images of NSCLC studies,” IEEE Transactions on Medical Imaging, vol. 33, no. 2, pp. 408–421.
2. W. Huang, Y. Yang, Z. Lin, G. B. Huang, J. Zhou et al. (2014). , “Random feature subspace ensemble based extreme learning machine for liver tumor detection and segmentation,” IEEE Transactions on Medical Imaging, vol. 4, no. 1, pp. 4675–4678.
3. M. N. Cheema, A. Nazrir, B. Sheng, P. Li, J. Qin et al. (2019). , “Liver extraction using residual convolution neural networks from low dose CT images,” IEEE Transactions on Biomedical Engineering, vol. 66, no. 9, pp. 2641–2650.
4. X. Song, G. Deng, Y. Zhuang and N. Zeng. (2019). “An improved confidence connected liver segmentation method based on three views of CT images,” IEEE Access, vol. 7, pp. 58429–58434.
5. O. F. Elaziz, M. S. Sayed and M. I. Abdullah. (2014). “Liver tumors segmentation from abdominal CT images using region growing and morphological processing,” IEEE Transactions on Image Processing, vol. 2, no. 1, pp. 1–6.
6. Y. Peng and Y. Ma. (2018). “A level set method to image segmentation based on local direction gradient,” KSII Transactions on Internet and Information Systems, vol. 12, no. 4, pp. 1760–1778.
7. G. Li, X. Chen, F. Shi, W. Zhu, J. Tian et al. (2015). , “Automatic liver segmentation based on shape constraints and deformable graph cut in CT images,” IEEE Transactions on Image Processing, vol. 24, no. 8, pp. 5315–5329.
8. I. Singh and N. Gupta. (2015). “Segmentation of liver using hybrid K-means clustering and level set,” IEEE Transactions on Medical Imaging, vol. 5, no. 8, pp. 742–746.
9. N. M. Altarawneh, S. Luo, B. Regan and C. Sun. (2015). “A modified distance regularized level set model for liver segmentation from CT images,” IEEE Transactions on Biomedical Imaging, vol. 6, no. 1, pp. 1–11.
10. F. Heckel, H. Meine, H. Moltz, J. Kuhnigk, J. T. Heverhagen et al. (2014). , “Segmentation based partial volume correction for volume estimation of solid lesions in CT,” IEEE Transactions on Medical Imaging, vol. 33, no. 2, pp. 462–479. [Google Scholar]
11. C. Li, X. Wang, S. Eberl, M. Fulham, Y. Yin et al. (2015). , “Supervised variational model with statistical inference and its application in medical image segmentation,” IEEE Transactions on Biomedical Engineering, vol. 62, no. 1, pp. 196–207. [Google Scholar]
12. G. Liu, Z. Zhao and Y. Zhang. (2015). “Image fuzzy clustering based on the region level markov random field model,” IEEE Geoscience and Remote Sensing Letters, vol. 12, no. 8, pp. 1770–1774. [Google Scholar]
13. G. Liu, Y. Zhang and A. Wang. (2015). “Incorporating adaptive local information into fuzzy clustering for image segmentation,” IEEE Transactions on Image Processing, vol. 24, no. 11, pp. 3990–4000. [Google Scholar]
14. E. Rikxoort, Y. Arzhaeva and B. Ginneken. (2007). “Automatic segmentation of the liver in computed tomography scans with voxel classification and atlas matching,” in Proc. MICCAI Workshop 3-D Segmentation, Clinic: A Grand Challenge, Brisbane, Australia, pp. 101–108. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |