DOI:10.32604/cmc.2021.012632

| Computers, Materials & Continua DOI:10.32604/cmc.2021.012632 |  |
| Article |
A New Multi-Agent Feature Wrapper Machine Learning Approach for Heart Disease Diagnosis
1Department of Computer Science, College of Computer Information Technology, American University in the Emirates, 503000, United Arab Emirates
2College of Computer Science and Information Technology, University of Anbar, Ramadi, 31001, Iraq
3Faculty of Computer Science and Information Technology, Universiti Tun Hussein Onn Malaysia, Johor, 86400, Malaysia
4College of Agriculture, Al-Muthanna University, Samawah, 66001, Iraq
5Department of Software Engineering, College of Computer and Information Sciences, King Saud University, Riyadh, 11451, Saudi Arabia
6eVIDA Lab, University of Deusto, Bilbao, 48007, Spain
7Department of Pure Science, Ministry of Education, General Directorate of Curricula, Baghdad, Iraq
8Department of Medical Laboratory Science, Faculty of Applied Medical Science, King Abdulaziz University, Jeddah, 21589, Saudi Arabia
*Corresponding Author: Mazin Abed Mohammed. Email: mazinalshujeary@uoanbar.edu.iq
Received: 07 July 2020; Accepted: 20 October 2020
Abstract: Heart disease (HD) is a serious widespread life-threatening disease. The heart of patients with HD fails to pump sufficient amounts of blood to the entire body. Diagnosing the occurrence of HD early and efficiently may prevent the manifestation of the debilitating effects of this disease and aid in its effective treatment. Classical methods for diagnosing HD are sometimes unreliable and insufficient in analyzing the related symptoms. As an alternative, noninvasive medical procedures based on machine learning (ML) methods provide reliable HD diagnosis and efficient prediction of HD conditions. However, the existing models of automated ML-based HD diagnostic methods cannot satisfy clinical evaluation criteria because of their inability to recognize anomalies in extracted symptoms represented as classification features from patients with HD. In this study, we propose an automated heart disease diagnosis (AHDD) system that integrates a binary convolutional neural network (CNN) with a new multi-agent feature wrapper (MAFW) model. The MAFW model consists of four software agents that operate a genetic algorithm (GA), a support vector machine (SVM), and Naïve Bayes (NB). The agents instruct the GA to perform a global search on HD features and adjust the weights of SVM and BN during initial classification. A final tuning to CNN is then performed to ensure that the best set of features are included in HD identification. The CNN consists of five layers that categorize patients as healthy or with HD according to the analysis of optimized HD features. We evaluate the classification performance of the proposed AHDD system via 12 common ML techniques and conventional CNN models by using a cross-validation technique and by assessing six evaluation criteria. The AHDD system achieves the highest accuracy of 90.1%, whereas the other ML and conventional CNN models attain only 72.3%–83.8% accuracy on average. Therefore, the AHDD system proposed herein has the highest capability to identify patients with HD. This system can be used by medical practitioners to diagnose HD efficiently.
Keywords: Heart disease; machine learning; multi-agent feature wrapper model; heart disease diagnosis; HD cleveland datasets; convolutional neural network
Heart disease (HD) is a life-threatening disease that can cause heart failure. The heart is responsible for pumping the desired amount of blood to the entire body. The presence of HD may result in insufficient blood supply [1,2]. In many countries, including the United States of America, HD has the highest rate of incidence [3,4]. According to the European Society of Cardiology, over 3.5 million people are diagnosed with HD annually. The total number of patients with HD worldwide is 2.6 million [5], and half of them have lost their lives after the first or second year of diagnosis [6]. The estimated expenditure for HD prevention and treatment is about 3% of the global budget for healthcare [7]. The major symptoms of HD are difficulties in breathing, feelings of fatigue or tiredness, and peripheral edema. These symptoms arise due to abnormalities in cardiac or noncardiac functions [3]. The current methods for diagnosing HD are incapable of identifying HD in its early stages [4]. The severe lack of medical supplies and resources, including specialists and equipment, in developing countries, contribute to inefficient and ineffective HD diagnosis and treatment in these nations [5].
Therefore, appropriate prevention and early diagnostic methods must be developed to minimize the risk of death due to HD [6]. Traditional HD diagnostic methods involve invasive techniques that are time-consuming and tedious. In many cases, the accuracy of these methods is inaccurate. Intelligent noninvasive decision support methods are proposed as an alternative to address the limitations of invasive HD diagnostic methods and reduce the risk of death due to HD. These methods provide an advanced diagnosis by analyzing the medical history of patients with HD patient, examining the physical condition of the patients, and then generating a compressive report on HD cases [8]. To perform data analysis, they utilize data mining and machine learning (ML) techniques, including artificial neural network (ANN), AdaBoost, logistic regression, support vector machine (SVM), Naïve Bayes (NB), fuzzy logic (FL), k-nearest neighbor (K-NN), and decision tree (DT) [9–11]. Various ML methods are used to classify and diagnose HD with reasonable accuracy [12,13]. Numerous researchers have tested their respective proposed ML classification methods by extensively using HD datasets to investigate and predict heart health conditions [14].
Predicting the occurrence of HD in its early stages on the basis of risk factors that could lead to HD, such as diabetes, hypertension, smoking, age and sex [8], provide a potential solution. The common ML methods utilized in analyzing HD risk factors are ANN, DT, NB, and SVM. In a previous study that aimed to predict risks of HD, regression SVM achieved the highest accuracy of 92.1%, whereas DT obtained the lowest accuracy of 89.6% [15]. Previous studies that compared the accuracy and flexibility of various ML techniques in predicting HD reported that associative classification approaches, such as NB, ANN, and DT, are superior to other techniques [16,17]. Another empirical study demonstrated that DT-based methods can produce good accuracy despite its simplicity [18]. Another study tested KNN and NB classifiers on the Cleveland HD database and Stat log Heart HD datasets and found that KNN outperforms NB [19]. A prior study employed ANN with backpropagation training algorithm for HD prediction and achieved convenient accuracy [20]. A previous work proposed an ensemble method to enhance HD diagnosis; results showed that the proposed method achieves a higher accuracy than methods with individual classifiers [21]. Earlier studies also employed ANN to minimize human errors in predicting medical indicators of HD, blood pressure, and blood sugar [22,23].
In predicting HD, most ML classifiers, when combined with other methods, perform better and achieve higher accuracy than when they are used as a standalone method. A previous research presented a hybrid FL-based method that combines genetic algorithm (GA) with ANN [24]. In this hybrid method, FL is used to extract HD features, GA is utilized to optimize feature selection, and ANN is employed to classify HD cases. Experimental results indicated an increase in the overall classification accuracy. Another work described a combination of GA, FL, and ANN for HD diagnosis [25], which performed well in predicting HD. Another study proposed a hybrid approach combining FL and ANN for HD prediction [20]. This approach yields good results with an accuracy of 87.4%. Two previous studies integrated a combination of GA, ANN, and FL into a coactive neuro-fuzzy inference system (CANFIS) [26,27]. In this system, GA is employed to optimize feature selection and automate the tuning of CANFIS parameters. Results demonstrated that CANFIS predicts HD with high accuracy. Prior works combined an SVM model with particle swarm optimization (PSO) to classify heartbeats [28,29]. In this model, PSO is used to optimize and tune SVM parameters. Results demonstrated that the proposed model produces a higher classification accuracy than SVM alone. A related research combined different ML classifiers by adopting an ensemble technique to improve the performance of several classifiers [30]. The authors applied the combined classifiers based on the ensemble technique in healthcare provision and assessed their usefulness in this field. Another study evaluated different HD datasets via several classification models [31], some of which obtained high accuracy. A previous work combined another classification method based on the K-means clustering algorithm with the maximal frequent item set algorithm (MAFIA) to address problems in HD diagnosis [23]. In this classification method, the K-means algorithm is employed for data extraction, whereas MAFIA is utilized for mining frequent patterns. The authors tested the proposed method by using different weights and factors and found that it has a higher accuracy in predicting myocardial infarction than similar basic methods.
Similar to what a previous study conducted [32], the common evolutionary methods used for feature selection are evaluated on the Cleveland HD dataset. Data show that these methods obtain a higher classification accuracy than basic methods. A study integrated a multilayer perceptron (MLP) classifier into SVM methods for HD diagnosis [33]. This classifier achieves a classification accuracy of up to 80.41%. Another study proposed a different classification method on the basis of MLP ANN for HD diagnosis [34]. The classifier is combined with feature selection and backpropagation learning algorithms. A previous work employed DT, NB, and ANN in a medical computer-based tool to help in HD diagnosis [35]. NB produced the best performance with an accuracy of 88.12%, followed by ANN and DT with an accuracy of 86.12% and 80.4%, respectively. Two studies suggested a three-phase ANN-based approach for HD classification [36,37]. Another study utilized a logistic regression classifier in a decision support system for HD case classification [38]. However, the classifier produced a low accuracy of only 77%.
The research of [30] is more similar to the present work than to the aforementioned studies. Both studies comprehensively investigate the performance of various ML methods and propose a hybrid ML model for HD feature selection and classification. In HD feature selection, the features are divided into sets that contain six features, and then the features of the sets are switched. By comparison, in HD classification, classification accuracy is measured to find the best set of features. Nevertheless, the two studies have crucial differences. The work of [30] greatly improves the accuracy of running multiple models. However, the results of this method are applicable to classifying instances with six features only, thereby restricting its generalization. Moreover, its measurement of performance is still limited to a few classification models in which the highest accuracy of 85.48% is achieved by the hybrid model. By contrast, the present study adopts a new feature selection method to determine the best set of features. Furthermore, this study utilizes ML models to improve further the classification accuracy.
In the present study, we propose a new hybrid model for developing an automated heart disease diagnosis (AHDD) system that classifies HD cases on the basis of deep learning and multi-agent paradigms. It includes a multi-agent feature wrapper (MAFW) model for finding a subset of features most relevant to prediction or classification tasks. The MAFW model performs within the framework of the wrapper approach to ensure that the best subset of features is obtained. It is especially useful in performing feature selection for small datasets. The MAFW consists of four types of agents, namely, a data preparation agent ( ), a feature selection agent (
), a feature selection agent ( ), a data classification agent (
), a data classification agent ( ), and a feature evaluation agent (
), and a feature evaluation agent ( ). The model includes two popular classifiers in the data classification agent, namely, SVM and NB. The model also includes a GA for performing primary feature selection. In addition, cross-validation techniques are employed in particular k-fold. Moreover, different evaluation metrics are assessed to evaluate the performance of our proposed method in terms of accuracy, TP, FP, precision, recall, and F1-measure. The HD dataset is also analyzed via data preprocessing methods. Our method is tested on the 2016 Cleveland HD dataset for HD diagnosis. The main contributions of this study are as follows:
). The model includes two popular classifiers in the data classification agent, namely, SVM and NB. The model also includes a GA for performing primary feature selection. In addition, cross-validation techniques are employed in particular k-fold. Moreover, different evaluation metrics are assessed to evaluate the performance of our proposed method in terms of accuracy, TP, FP, precision, recall, and F1-measure. The HD dataset is also analyzed via data preprocessing methods. Our method is tested on the 2016 Cleveland HD dataset for HD diagnosis. The main contributions of this study are as follows:
• A new hybrid model is proposed for developing an AHDD system. The AHDD system integrates a binary CNN model with an MAFW model. The CNN consists of five layers that categorize subjects into healthy individuals or patients with HD by analyzing several HD features. The binary CNN architecture includes an input HD data, a convolution, a pooling activation, and output layers.
• The new MAFW model implements SVM and NB to conduct initial classification to tune the CNN and GA to perform a global search on HD features and adjust the weights of CNN to include the best set of features.
• The performance of all classification ML-based methods is evaluated in terms of prediction accuracy of the overall feature set.
• The performance of all classification ML-based methods is also evaluated in terms of prediction accuracy of the selected features chosen by feature selection via the MAFW model combined with cross-validation (k-fold).
• On the basis of the performance evaluation results, this study recommends the use of a particular classification ML-based method that works well with a certain feature algorithm in designing powerful computer-aided HD prediction systems.
• The performance of various classifications ML-based methods as applied to an HD dataset is compared and analyzed.
The rest of the paper is organized as follows. The materials and methods are described in Section 2. In Section 3, the implementation of the proposed method is presented, and its performance in HD prediction is scrutinized. Lastly, the conclusions and directions for future work are highlighted in Section 4.
Numerous intelligent decision support systems are utilized to aid in various medical and healthcare needs, including diagnosis, patient follow up, disease remediation, and prognosis. To handle data complexity and uncertainty, these intelligent systems combine some of the most successful and widely used intelligent computational algorithms, such as ANN, GA, and K-means clustering [39]. In the context of the learning process, the issue of HD prediction is considered as a clustering classification problem. However, a framework is required to manage different sets of data types. Only one type of class with a restricted HD class set may be classified to address this classification problem. Doing so allows the easy detection of the correct class, resulting in high accuracy.
The Cleveland HD dataset is available from the UCI Machine Learning Repository. The Cleveland HD dataset is extensively used by data miners and researchers on ML for evaluation and analysis purposes. The Cleveland HD dataset is composed of 270 instances and 13 features/attributes, including 6 numeric attributes and 7 categorical attributes. A description of the dataset is shown in Tab. 1.
Table 1: Details and description of Cleveland HD dataset features [40]

The range of age of the patients selected is 29–79 years. A gender value of 1 represents male patients, whereas a gender value of 0 denotes female patients.
Symptoms of HD are associated with four types of chest pain:
1. Heart muscles do not receive the full amount of blood required, resulting in the narrowing of coronary arteries, a condition that causes Angina type 1.
2. Heart muscles do not receive the full amount of blood required, resulting in the narrowing of coronary arteries, a condition that also causes Angina type 2. The main difference is that Angina type 2 is associated with the chest pain felt when experiencing emotional or mental stress.
3. Some chest pains not related to Angina are experienced for various reasons, and this case is not associated with HD.
4. No symptoms reflecting an HD case are noted.
With respect to features, trestbps represents blood pressure reading in the resting position, Chol indicates the level of cholesterol, and Fbs denotes the fasting blood sugar level. If the blood sugar level is less than 120 mg/dl, a value of 1 is assigned; otherwise, this feature is given a value of 0. Furthermore, Restecg represents electrocardiographic results in the resting position, thalach is the maximum value of the heart rate, and exang indicates exercise-induced angina. If pain is felt, then exang is assigned a value of 1; otherwise, it is given a value of 0. Moreover, an old peak represents exercise-induced ST depression. The slope represents the peak slope exercise of ST-segment. In addition, ca is the number of main vessels colored by fluoroscopy, that provides the duration of test exercise in minutes, and num represents the class attribute. With regard to num, a value of 0 is assigned for normal cases; otherwise, a value of 1 is given (for cases with HD abnormality). The desired attribute is classified into four categories: the first three categories reflect HD cases, whereas the fourth category denotes healthy cases. The holdout technique in which the dataset is split into two sets is adopted for training and testing.
The proposed approach aims to properly classify individuals as healthy or with HD. The performance of different ML-based methods is evaluated in terms of accurately diagnosing HD on the basis of complete and selected features. A supervised learning-based method is adopted for classification data availabilities. A diagnostic system for HD is then proposed. The proposed approach includes different ML classifiers to enhance prediction accuracy. Our proposed methodology involves five stages: (1) Preprocessing of HD dataset, (2) A feature selection stage involving the MAFW model, (3) A cross-validation process, (4) Theoretical contexts of 11 ML techniques, and (5) Evaluating ML performance via various techniques. The dataset is split into training and testing sets. The efficiency of the ML classifiers is tested on the dataset described in Fig. 1.

Figure 1: Machine learning-based identification approach for heart disease diagnosis
Data preprocessing is necessary to obtain a suitable data representation for each ML classifier and ensure effective testing and evaluation. Some of these methods are standard scalar method, missing values removal, and MinMax scalar. In the standard scalar method, each feature has a value of 0 for the mean and a value of 1 for the variance, and all features are bridged to a similar factor. The same is true for the MinMax scalar method, in which the data are shifted between 0 and 1 for all features. In the missing values removal method, missing values in each feature row are removed from the entire dataset [41]. The aforementioned data preprocessing methods are adopted in the present study.
2.2.2 Multi-Agent Feature Wrapper
Selection of relevant features is critical for identifying the required classes. It has a positive effect on the efficiency of ML classifiers in terms of prediction accuracy and execution time. By contrast, selecting irrelevant features in the learning process can negatively affect the performance of ML classifiers. In our proposed method, the MAFW model is applied in selecting the important features of targeted classification. HD datasets have over thousands of features but only 13 attributes. Hence, classifying HD is a complex process because of the existence of a wide variety of features and inessential or irrelevant attributes. If the full dataset containing a huge number of features is used, then achieving reliable and accurate results becomes laborious and requires a long computational time. Thus, the size of features must be reduced by selecting proper characteristics as the initial step in the learning process. Doing so helps in understanding the outcomes, thus increasing the classification accuracy while enhancing the performance of classifiers.
The MAFW model is proposed to find a subset of features that are most relevant to prediction or classification tasks. The MAFW model performs within the framework of the wrapper approach to ensure that the best subset of features is obtained. This model is especially useful in performing feature selection for small datasets. The MAFW model consists of four types of agents, namely, a data preparation agent ( ), a feature selection agent (
), a feature selection agent ( ), a data classification agent (
), a data classification agent (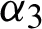 ), and a feature evaluation agent (
), and a feature evaluation agent ( ). The model includes two popular classifiers in the data classification agent, namely, SVM and NB. The model also incorporates a GA for performing primary feature selection. The MAFW model works according to backward elimination mechanism in which it starts with selecting all features and, within its iteration, removes the least important features while maintaining the most relevant ones. The stopping condition is linked to both the number of removed features and the progress of performance improvement of classifiers. Fig. 2 shows the main components of the MAFW model.
). The model includes two popular classifiers in the data classification agent, namely, SVM and NB. The model also incorporates a GA for performing primary feature selection. The MAFW model works according to backward elimination mechanism in which it starts with selecting all features and, within its iteration, removes the least important features while maintaining the most relevant ones. The stopping condition is linked to both the number of removed features and the progress of performance improvement of classifiers. Fig. 2 shows the main components of the MAFW model.

Figure 2: An overview of the MAFW model
In the MAFW model, the agents interact with each other to perform feature selection tasks to reduce the number of features of a given dataset. The agents’ goal is to select features that best improve the prediction performance with a minimum effect on the boundaries of learning generalization of the classifiers. The agents’ roles in the MAFW model are presented below and summarized in Algorithm 1 (Fig. 2):
• 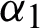 : This agent prepares the feature vector for
: This agent prepares the feature vector for  to perform the feature selection task and prepares the cross-validation data for
to perform the feature selection task and prepares the cross-validation data for  to perform the classification task.
to perform the classification task.
• 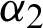 : This agent integrates a GA that applies a binary feature selection operation to produce subsets of features. The GA presents the feature space as a one-dimension binary vector that forms a GA chromosome. This chromosome represents an individual population in which the total population indicates the actual number of features. Each chromosome contains a number of genes that is equal to the actual number of features (e.g., 15 features are represented by 15 chromosomes, and each chromosome is represented by 15 genes). Each gene can hold a binary value (0 or 1) in which assigning a value of 1 to a gene denotes including a feature, whereas assigning a value of 0 to the gene signifies excluding the feature. The first initial population of chromosomes is randomly generated to represent subsets of features.
: This agent integrates a GA that applies a binary feature selection operation to produce subsets of features. The GA presents the feature space as a one-dimension binary vector that forms a GA chromosome. This chromosome represents an individual population in which the total population indicates the actual number of features. Each chromosome contains a number of genes that is equal to the actual number of features (e.g., 15 features are represented by 15 chromosomes, and each chromosome is represented by 15 genes). Each gene can hold a binary value (0 or 1) in which assigning a value of 1 to a gene denotes including a feature, whereas assigning a value of 0 to the gene signifies excluding the feature. The first initial population of chromosomes is randomly generated to represent subsets of features.  passes a copy of the generated subset of features to
passes a copy of the generated subset of features to 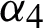 for further evaluation. The GA performs binary crossover to update the genes of two selected chromosomes and binary mutation to shuffle or refine the selection of a particular chromosome. The crossover and mutation decisions are made by
for further evaluation. The GA performs binary crossover to update the genes of two selected chromosomes and binary mutation to shuffle or refine the selection of a particular chromosome. The crossover and mutation decisions are made by  on the basis of feature evaluation results provided by
on the basis of feature evaluation results provided by  (as explained below). It also decides on the stopping condition according to a user-defined setting to the number of features and iteration thresholds.
(as explained below). It also decides on the stopping condition according to a user-defined setting to the number of features and iteration thresholds.
•  : This agent integrates SVM and NB classifiers that perform classification tasks to the provided data according to the subset features evaluated by
: This agent integrates SVM and NB classifiers that perform classification tasks to the provided data according to the subset features evaluated by  .
.  then provides the required classification results to
then provides the required classification results to  for further evaluation of the features.
for further evaluation of the features.
•  : This agent receives the copy of the subset features from
: This agent receives the copy of the subset features from  and the classification results of SVM and NB from
and the classification results of SVM and NB from  .
.  then applies mean decrease in accuracy (MDA) measurements to the classification results for each of SVM and NB to evaluate the subset features. MDA measures the importance of each feature from the variations in accuracy when including or excluding features. On the basis of MDA results,
then applies mean decrease in accuracy (MDA) measurements to the classification results for each of SVM and NB to evaluate the subset features. MDA measures the importance of each feature from the variations in accuracy when including or excluding features. On the basis of MDA results,  updates the independent feature evaluation (IFE) matrix that corresponds to NB results and the dependent feature evaluation (DFE) matrix that corresponds to SVM results.
updates the independent feature evaluation (IFE) matrix that corresponds to NB results and the dependent feature evaluation (DFE) matrix that corresponds to SVM results.  generates an associative feature evaluation (AFE) matrix from mapping between the two IFE and DFE matrixes. On the basis of the AFE matrix, it then checks and compares the importance between the current subset features and other copies of subset features. This operation is performed by checking the maximum weight W of the subset feature F by using the formula MDA
generates an associative feature evaluation (AFE) matrix from mapping between the two IFE and DFE matrixes. On the basis of the AFE matrix, it then checks and compares the importance between the current subset features and other copies of subset features. This operation is performed by checking the maximum weight W of the subset feature F by using the formula MDA  , where n represents the number of features, and wi is the weight of a corresponding feature fi.
, where n represents the number of features, and wi is the weight of a corresponding feature fi.  then passes the best results of the AFE matrix to the other agents for further optimization of the features selected in the following iteration.
then passes the best results of the AFE matrix to the other agents for further optimization of the features selected in the following iteration.

The discussion above describes the main components and provides a complete description of a run cycle of the MAFW model that constitute the main contribution of this paper. The MAFW model differs from existing models by considering the wrapper of an independent feature analysis classifier (i.e., NB) and a dependent feature analysis classifier (i.e., SVM) in selecting the best subset features. They are specifically selected to avoid the overfitting disadvantage of the wrapper feature selection approach. Moreover, the MAFW model applies a multi-agent system that renders the feature selection process more flexible by segregating selection functionalities into four tasks, namely, preparation, selection, classification, and evaluation. These tasks then interact with each other and reason over the input, process, and output of each task and apply the necessary revision to the processes responsible for achieving the tasks during runtime. Given that this model relies on the wrapper feature selection approach, its main limitations are high computational and time complexity [42,43].
2.2.3 Machine Learning and Classification Algorithms
In the context of the learning process, the issue of HD prediction is considered as a clustering classification problem. However, a framework is necessary to manage different sets of available data. Only one type of class with a restricted HD class set may be classified to address this classification problem. Doing so allows the easy detection of the correct class, resulting in high accuracy. In this section, the theoretical contexts of 11 ML classification methods adopted herein are explained. These methods are then compared and analyzed.
• NB is a Bayes theorem classification technique. In general, NB claims that a specific feature present in a specific class is irrelevant to another presented feature [44]. If the fruit is orange, round, and about 10 cm in diameter, then it can be called an orange. If these characteristics are dependent on each other, or they depend on the presence of other characteristics, both features separately lead to an apple fruit probability; hence, this technique is regarded as “Naïve”.
• Stochastic gradient descent (SGD) is a method also employed to find a minima function. SGD is a linear classifier (linear SVM is by default in sklearn) that uses SGD to train (i.e., to scan for loss minima by using SGD). This estimator utilizes SGD with regularized linear model learning: the estimation of each sample at a time by the gradient of the loss and the model is modified along the way with a reduced force schedule (i.e., learning rate) [45].
• The sequential minimal optimization (SMO) algorithm is derived from taking the concept of a decomposition method to its maximum and optimizing at each iteration a minimum subset of just two points. The strength of this technique lies in the fact that an analytical solution is admitted for the optimization problem for two data points, thus eliminating the need to use as part of the algorithm an iterative quadratic programming optimizer [45].
• The voted perceptron method (VPM) is based on the Rosenblatt and Frank perceptron algorithm. This algorithm exploits data with large margins to get the full benefits of linearly separable classes. Compared with Vapnik’s SVM, this approach is easier to apply and also more efficient in terms of computational time. This algorithm can also be implemented with kernel functions in very high dimensional spaces [46].
• KNN or IBK algorithm is a simple supervised learning from the family of ML algorithms. The main idea behind this approach is to find a training sample nearest to the new point at a distance and to estimate the label from those data points [47]. Despite its simplicity, this algorithm suffers from numerous classification and regression problems concerning the nearest neighbors.
• AdaBoostM1 is a shortcut term for adaptive boosting, which is an ML meta-algorithm devised by Yoav Freund and Robert Schapire, who received the 2003 Gödel Prize for this work [48]. Combined with several types of learning algorithms, this meta-algorithm can be utilized to enhance achievement. Other learning algorithm outputs (“weak learners”) are merged with a weighted sum, which indicates the boosted final outperformance of classifiers.
• LogitBoost is a boosting algorithm developed by Jerome Friedman, Robert Tibshirani, and Trevor Hastie on the basis of ML and computational learning theory. Their original work lays a mathematical foundation for the AdaBoost algorithm [49]. If one considers AdaBoost as a generalized additive model, then one can derive the LogitBoost algorithm and then apply the logistic regression cost function.
• MultiClassClassifierUpdateable is a meta classifier for handling multiclass databases with two-class methods. Moreover, this classifier is competent in using error-correcting yield codes for expanded precision. The main method should be an updateable method [50].
• The Hoeffding Tree is an incremental learner of decision tree for big data streams, assuming the distribution of data does not change over time. A decision tree grows incrementally according to the Hoeffding boundary (or Chernoff bound additive) theoretical guarantees. As soon as sufficient statistical evidence is obtained, a node is expanded until an optimal splitting function is achieved, a decision based on the Hoeffding bound, which is independent of distribution [50].
• J48 is an upgrade to ID3. J48 accounts for extra features for the pruning of decision-making trees, continuous attribute value ranges, rule derivation, and missing values. J48 is an execution of open-source Java within the WEKA data mining framework of the C4.5 algorithm. The WEKA tool provides several related choices for tree pruning [50].
• Random Forest (RF) is an ensemble technique typically utilized in the process of classification, whereby the use of different decision trees is employed in data classification [51,52]. Bootstrap templates are built from the main RF numbers, and a raw classification process or regression tree is developed in every bootstrap pattern.
• Hybrid Model for AHDD: A hybrid model for AHDD system is described in this subsection. The AHDD system integrates a binary CNN model with the MAFW model. The CNN consists of five layers that categorize patients into “healthy” or “with HD” on the basis of several HD features. The binary CNN architecture includes input HD data, convolution, pooling activation, and output layers. The MAFW model implements SVM and NB to do initial classification to tune the CNN and GA to perform a global search on the HD features and adjust the weights of the CNN to include the best set of features. Fig. 3 shows the basic model of the AHDD system.

Figure 3: Automated heart disease diagnosis (AHDD) system
The AHDD system process starts with an input layer that receives HD symptoms as inputs in a specific structure, which are then fed to the convolutional layer. Eight convolutional layers (l1–l8) reconstruct the features through a filtering process by using different numbers of kernels (l1:6k, l2:2k, l3:6k, l4:6k, l4:6k, l5:12k, l6:12k, l7:18k, and l8:18k). In conventional CNN, the weights of kernels are randomly initialized. In our hybrid model, the weights are initialized and adjusted on the basis of the MAFW model in which the CNN assigns weights between (0–1) according to the initial classification results of SVM and NB during the data classification phase in the MAFW model. The operation of feature selection is presented in Section 2.2.2. The weights of features are transformed from a 2D matrix into a 1D matrix to be processed by CNN. Subsequently, the pooling layer reduces the dimension of the feature map by calculating the average of kernels in the convolutional layers by using the average pooling function. The activation layer applies the ReLU function to enhance the rate of converging for the learning process. Finally, the output layer classifies the processed cases into “healthy” or “with HD” according to the training process. When the training process converges the best solution results (highest accuracy), the model parameters are set and the model becomes ready for the testing phase. In the testing phase, the weights tuned by CNN are obtained and by which the minimum classification or diagnosis error rate is achieved. Algorithm 2 represents the MAFW optimization to CNN.

2.2.4 Validation of Classifiers
The k-fold cross-validated approach and six metrics are assessed to evaluate the performance of the classifiers. In k-fold cross-validation, the dataset is separated into k of the same size, wherein the classifiers are trained using 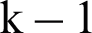 group and the outperformance in each step is checked using the remaining part. The validation cycle is replicated k-times. The performance of 0e classifier is calculated on the basis of k results. Various values of k are chosen for CV.
group and the outperformance in each step is checked using the remaining part. The validation cycle is replicated k-times. The performance of 0e classifier is calculated on the basis of k results. Various values of k are chosen for CV.  is used in our experiment because its performance is good; 90% of the data are utilized for 10-fold CV preparation, whereas 10 percent are employed for research purposes. The procedure is replicated 10 times for every fold of the procedure, and before collecting and testing new sets for the new cycle, both training and evaluation group instances are randomly distributed over the entire data collection [53,54]. Finally, the averages of all output metrics are set at the end of the 10-fold cycle.
is used in our experiment because its performance is good; 90% of the data are utilized for 10-fold CV preparation, whereas 10 percent are employed for research purposes. The procedure is replicated 10 times for every fold of the procedure, and before collecting and testing new sets for the new cycle, both training and evaluation group instances are randomly distributed over the entire data collection [53,54]. Finally, the averages of all output metrics are set at the end of the 10-fold cycle.
2.2.5 Evaluation Metrics of HD Performance
Specific performance evaluation metrics are assessed to evaluate the performance of the classifiers. A confusion matrix, which predicts any observation in exactly one box in the test set, is used (Tab. 2). This matrix is a 2 to 2 matrix because there are two groups of repose. It also provides two forms of proper prediction for classifiers and two types of incorrect prediction classifier.

The following metrics are calculated from the confusion matrix:
• TP: performance is measured as a true positive (TP): the observed subject of HD is classified correctly and the person suffers from HD.
• TN: performance is expected to be a true negative (TN): the observed subject is healthy and classified correctly.
• FP: performance is expected to be a false positive (FP): the observed subject is healthy but wrongly classified as having HD (type 1 error).
• FN: the performance is estimated as a false negative (FN): the observed subject is healthy but wrongly classified as having no HD (type 2 error).
A value of 1 indicates that the positive case is unhealthy, whereas a value of 0 denotes that the negative case is healthy.
The output of each method is evaluated at this phase to determine which method could achieve the best result. The following parameters are evaluated: Accuracy, precision, recall, and F-measure. These parameters are described and calculated as follows:
• Accuracy refers to a measurement’s closeness parameter when reading the data value against the real data values:

• Precision tests the proportion of related subjects. It measures the classifier’s ability to turn down irrelevant subjects:

• Recall measures the proportion of identified related subjects. It tests the classifier’s ability to produce all applicable subject matters:

• F-score can be regarded as an average weight of recall and precision, wherein an F1 score achieves the worst at 0 and the highest value at 1. The precision of relative contribution to the F1 score and recall is equal. The F1 score is calculated as follows:

where the value of precision is obtained on the basis of Eq. (2) and that of recall on the basis of Eq. (3).
2.2.6 Parameter Settings of Machine Learning Techniques
Each classification technique requires one or more parameters that control (effects) the classifier’s predictive outcome. Selecting the best values for those parameters is difficult and requires seeking a trade-off between model complexity and model generalization. In this study, a grid search is used to find the parameter settings. A grid search involves changing a value grid (2D or 3D depending on the number of model parameters) and increasing each parameter by an affixed interval before the values of the optimal parameter are found. The advantage of this approach is that it allows the selection of optimal parameters at specified intervals. However, this approach is considered expensive in terms of computation time. The ML techniques used herein are shown in Tab. 3.
Table 3: Parameter settings of machine-learning techniques

After the step of parameter setting, the dataset is split into testing and training sets according to the cross-validation leave-one-out protocol. The labels are used during the learning process for the supervised approaches. Subsequently, during the test phase, the labels calculated by each classifier are matched with the true labels (reference labels) for calculating the classification performance. Unlike supervised models, unsupervised models are trained using the features extracted only and reference labels are not used. Instead, the labels are only utilized for evaluation purposes of classification. Remember that (1) all the extracted features are used as classifier data, and (2) only the selected features are implemented. Every sub-dataset is considered separately for selecting the most important features.
3 Experimental Results and Discussion
The performance of various ML methods, namely, J48, RF, NB, SGD, SMO algorithm, VPM, IBk, AdaBoostM1, LogitBoost, MultiClassClassifierUpdateable, Hoeffding Tree along with the hybrid model (binary CNN model with the MAFW model), and Cleveland HD dataset, are tested and discussed via different perspectives. The MAFW model and cross-validation k-fold method are adopted for critical feature selection. Several metrics are assessed to evaluate the performance of these methods and test the efficiency of classifiers. All features are standardized and normalized before they are applied to the classifiers. The overall results for the original dataset are obtained and presented in Tab. 4 on the basis of the following 13 features: AGE, SEX, CPT, RBP, SCH, FBS, RES, MHR, EIA, OPK, PES, VCA, and THA. Furthermore, the following parameters are used in the evaluation process: accuracy, TP rate, FP rate, precision, recall, and F-measure.
Table 4: Classification results based on all features

The hybrid model achieves the highest accuracy of 85.3%. Moreover, the hybrid model obtains the highest precision, recall, F-measure values. By contrast, the HoeffdingTree attains the lowest accuracy of 49.1% and the lowest scores for the other parameters. SMO, RF, J48, MultiClassClassifierUpdateable, and SGD have a lower accuracy than the other classifiers. NB, VotedPerceptron, IBk, and LogitBoost achieve an intermediate average accuracy of 80.1%.
Based on the eight features selected by the MAFW model, namely, SEX, CPT, MHR, EIA, OPK, PES, VCA, and THA, the hybrid model has the highest diagnostic accuracy of 90.1% (Tab. 5), followed by SMO and LogitBoost with 83.8%. SMO is also higher than X in terms of precision but not in terms of recall and F-measure. By contrast, RF has the lowest accuracy of 72.3%. Except for J48, the other methods have accuracies above 80%.
Table 5: Classification results based on the features selected by the MAFW model

In general, remarkable improvements are noted in the diagnosis results obtained for the 12 classifiers when the filtered dataset is implemented. The Hoeffding Tree achieves the highest accuracy improvement of 34.4%; thus, it’s performance highly influenced by the proposed feature selection model. The improvement in accuracy is 6.3%, 5.6%, 4.8%, and 3.9% for SGD and MultiClassClassifierUpdateable, SMO, hybrid model, and J48, respectively. By contrast, IBk has the lowest accuracy improvement of 0.3%. The average development in diagnostic accuracy outcomes is up to 6.17%. Furthermore, the TP rate for all classifiers is substantially increased, whereas the FP rate for SMO and LogitBoost is decreased to the minimum level, indicating that the classification results of HD cases are more reliable. In summary, the proposed feature selection model has a considerable effect on accuracy improvement for 11 out of 12 classifiers. This result confirms that the proposed model can work successfully with different types of classification algorithms. The differences in diagnostic accuracy outcomes obtained via the ML models for the two databases is illustrated in Fig. 4.

Figure 4: Differences among the outcomes of all features and selected features
Classification can be precisely conducted by exploring the following aspects: (i) The most excellent technique for diagnosing or predicting a given disease or issue, (ii) The ideal classifier for the assessment and determination of HD features, and (iii) The most excellent parameters for the ML methods based on the HD features selected. Thus, by utilizing various classification methods to HD datasets, the most appropriate and efficient ML method can distinguish healthy individuals from patients with HD. In previous studies, when HD features are decreased, the dataset’s feature vector is evidently improved, the complexity of ML models is reduced, and the precision of diagnosis is enhanced. For example, MultiClass Classifier Updateable, which is a set of methods that can precisely work with complex choice boundaries, frequently exhibit sensitivity to feature determination. Thus, such methods would likely suffer from overfitting issues.
Therefore, the assessment, choice, and ranking of HD features within classifier problems are not guided by settled factors. This procedure is executed on the basis of the properties of HD data learning, type of ML models, and complexity of choice boundaries. The MAFW model applies a multi-agent system that renders the process of feature selection more flexible by segregating selection functionalities into four tasks, namely, preparation, selection, classification, and evaluation. These tasks interact with each other and reason over the input, process, and output of each task and apply the necessary revision to the processes responsible for achieving the tasks during runtime. In this way, this study aims to determine the ideal combination of HD features that can be utilized to obtain adjusted feature selections and to enhance the accuracy of HD identification. The MAFW model selects important HD features to distinguish healthy people from patients with HD. According to the MAFW model, the most important and reasonable features for HD identification are exercise-induced angina, thallium scan, and type of chest pain. Moreover, the model suggests that fasting blood sugar is not appropriate for the identification of patients with HD and healthy individuals. In this study, the classification, feature extraction, dataset preprocessing, validation, and evaluation of classification performance are comprehensively discussed. A complete set of features and a selected set of features are used to evaluate the performance of our system. The complete set of features is reduced to generate the selected set of features. This process greatly affects the accuracy of classification methods and their performance time. The proposed HD diagnosis system can help medical practitioners in identifying patients with HD efficiently.
Benchmarking is the most essential step that needs to be considered in performing research on common medical processes of disease diagnoses. Benchmarking can be used to compare the efficiency and reliability of newly developed approaches and existing ones. Benchmarking is usually conducted either through the use of a standard dataset or different approaches for the same problem domain or application. Moreover, benchmarking is achieved by utilizing the best and modern methods for HD classification based on existing ML approaches and feature selection methods. Tab. 5 summarizes the different benchmarking approaches for several processes.
Table 6: Accuracy of the proposed method versus that of state-of-the-art techniques for HD classification

The limitations of the present work can be summarized as follows:
• The models evaluated herein are 12 different classifiers. Additional classifiers should be tested to provide a more comprehensive evaluation of the results.
• Given that the proposed model relies on the wrapper feature selection approach, its main limitations are high computational cost and time complexity.
• Runtime is not considered as an evaluation criterion.
In this study, an AHDD system for HD diagnosis is proposed. The AHDD system integrates a new MAFW model with a binary CNN model. The MAFW model performs feature selection and optimization tasks, whereas the CNN model conducts classification tasks. The MAFW model consists of four software agents that operate a GA, an SVM, and NB. The agents instruct the GA to perform a global search on HD features and adjust the weights of SVM and BN during the initial classification phase. It chooses imperative features for enhancing the performance of the classifiers. The AHDD system is trained, tested, and validated using the Cleveland HD database. The benchmarking classification models of NB, SGD, SMO, VPM, IBk, AdaBoostM1, LogitBoost, MultiClass Classifier Updateable, Hoeffding Tree, J48, RF, and the hybrid model proposed herein are integrated with the proposed MAFW model for testing and evaluation. The K-fold cross-validation technique is used to evaluate the performance of the ML models and the MAFW model in terms of accuracy, TP rate, FP rate, precision, recall, and F-measure. The MAFW model selects important HD features that increase the accuracy of distinguishing patients with HD from healthy individuals for all the tested classifiers. According to the MAFW model, the strongest features are exercise-induced angina, thallium scan, and type of chest pain, whereas fasting blood sugar is found to be a weak feature. Moreover, the hybrid model achieves the highest accuracy of 90.1%, a high precision of 88.9%, and a high recall of 98.4%. The average accuracy of the benchmarking ML models with the aid of the MAFW model is 75.08%, in which SMO and LogitBoost have the highest accuracy (83.8%) and RF has the lowest accuracy (72.3%). Furthermore, the MAFW model increases the overall accuracy of the benchmarking classifiers by 6.2% and that of the hybrid classifiers by 4.8%. In a follow-up study, we will test the proposed hybrid model and the MAFW model in other multivariate datasets. In addition, we will include training and testing runtime as effective evaluation criteria.
Acknowledgement: The authors would like to acknowledge University of Deusto for providing all facilities and support for this study.
Funding Statement: “This research received funding from Basque Country Government.”
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. A. L. Bui, T. B. Horwich and G. C. Fonarow. (2011). “Epidemiology and risk profile of heart failure,” Nature Reviews Cardiology, vol. 8, no. 1, pp. 30.
2. P. A. Heidenreich, J. G. Trogdon, O. A. Khavjou, J. Butler, K. Dracup et al. (2011). “Forecasting the future of cardiovascular disease in the united states: A policy statement from the American heart association,” Circulation, vol. 123, no. 8, pp. 933–944.
3. M. Durairaj and N. Ramasamy. (2016). “A comparison of the perceptive approaches for preprocessing the data set for predicting fertility success rate,” International Journal of Control Theory and Applications, vol. 9, no. 27, pp. 255–260.
4. J. Mourao-Miranda, A. L. Bokde, C. Born, H. Hampel and M. Stetter. (2005). “Classifying brain states and determining the discriminating activation patterns: Support vector machine on functional MRI data,” NeuroImage, vol. 28, no. 4, pp. 980–995.
5. S. Ghwanmeh, A. Mohammad and A. Al-Ibrahim. (2013). “Innovative artificial neural networks-based decision support system for heart diseases diagnosis,” Journal of Intelligent Learning Systems and Applications, vol. 5, no. 3, pp. 8.
6. I. B. Salah, R. De la Rosa, K. Ouni and R. B. Salah. (2020). “Automatic diagnosis of valvular heart diseases by impedance cardiography signal processing,” Biomedical Signal Processing and Control, vol. 57, pp. 101758.
7. V. L. Roger. (2010). “The heart failure epidemic,” International Journal of Environmental Research and Public Health, vol. 7, no. 4, pp. 1807–1830.
8. K. Vanisree and J. Singaraju. (2011). “Decision support system for congenital heart disease diagnosis based on signs and symptoms using neural networks,” International Journal of Computer Applications, vol. 19, no. 6, pp. 6–12.
9. A. Adeli and M. Neshat. (2010). “A fuzzy expert system for heart disease diagnosis,” in Proc. of Int. Multi Conf. of Engineers and Computer Scientists, Hong Kong, vol. 1, pp. 28–30.
10. M. A. Mohammed, M. K. A. Ghani, N. A. Arunkumar, O. I. Obaid, S. A. Mostafa et al. (2018). “Genetic case-based reasoning for improved mobile phone faults diagnosis,” Computers & Electrical Engineering, vol. 71, pp. 212–222. [Google Scholar]
11. A. Methaila, P. Kansal, H. Arya and P. Kumar. (2014). “Early heart disease prediction using data mining techniques,” Computer Science & Information Technology Journal, vol. 8, no. 2, pp. 53–59. [Google Scholar]
12. O. W. Samuel, G. M. Asogbon, A. K. Sangaiah, P. Fang and G. Li. (2017). “An integrated decision support system based on ANN and fuzzy_AHP for heart failure risk prediction,” Expert Systems with Applications, vol. 68, pp. 163–172. [Google Scholar]
13. P. K. Anooj. (2011). “Clinical decision support system: Risk level prediction of heart disease using weighted fuzzy rules and decision tree rules,” Central European Journal of Computer Science, vol. 1, no. 4, pp. 482–498. [Google Scholar]
14. P. C. Austin, J. V. Tu, J. E. Ho, D. Levy and D. S. Lee. (2013). “Using methods from the data-mining and machine-learning literature for disease classification and prediction: A case study examining classification of heart failure subtypes,” Journal of Clinical Epidemiology, vol. 66, no. 4, pp. 398–407. [Google Scholar]
15. Y. Xing, J. Wang and Z. Zhao. (2007). “Combination data mining methods with new medical data to predicting outcome of coronary heart disease,” in Int. Conf. on Convergence Information Technology, Gyeongju, South Korea, pp. 868–872. [Google Scholar]
16. J. Soni, U. Ansari, D. Sharma and S. Soni. (2011). “Predictive data mining for medical diagnosis: An overview of heart disease prediction,” International Journal of Computer Applications, vol. 17, no. 8, pp. 43–48. [Google Scholar]
17. K. Sudhakar and D. M. Manimekalai. (2014). “Study of heart disease prediction using data mining,” International Journal of Advanced Research in Computer Science and Software Engineering, vol. 4, no. 1, pp. 1157–1160. [Google Scholar]
18. K. Thenmozhi and P. Deepika. (2014). “Heart disease prediction using classification with different decision tree techniques,” International Journal of Engineering Research and General Science, vol. 2, no. 6, pp. 6–11. [Google Scholar]
19. M. S. Maashi. (2020). “Analysis Heart Disease Using Machine Learning,” Multi-Knowledge Electronic Comprehensive Journal for Education and Science Publications (MECSJ), vol. 2, 29. [Google Scholar]
20. N. Guru, A. Dahiya and N. Rajpal. (2007). “Decision support system for heart disease diagnosis using neural network,” Delhi Business Review, vol. 8, no. 1, pp. 99–101. [Google Scholar]
21. B. Fida, M. Nazir, N. Naveed and S. Akram. (2011). “Heart disease classification ensemble optimization using genetic algorithm,” in IEEE 14th Int. Multitopic Conf., Karachi, Pakistan, pp. 19–24. [Google Scholar]
22. J. Nahar, T. Imam, K. S. Tickle and Y. P. P. Chen. (2013). “Computational intelligence for heart disease diagnosis: A medical knowledge driven approach,” Expert Systems with Applications, vol. 40, no. 1, pp. 96–104. [Google Scholar]
23. S. B. Patil and Y. S. Kumaraswamy. (2009). “Extraction of significant patterns from heart disease warehouses for heart attack prediction,” IJCSNS, vol. 9, no. 2, pp. 228–235. [Google Scholar]
24. J. Singh and R. Kaur. (2016). “Cardio vascular disease classification ensemble optimization using genetic algorithm and neural network,” Indian Journal of Scenic and Technology, vol. 9, S1. [Google Scholar]
25. K. Uyar and A. İlhan. (2017). “Diagnosis of heart disease using genetic algorithm based trained recurrent fuzzy neural networks,” Procedia Computer Science, vol. 120, pp. 588–593. [Google Scholar]
26. L. Parthiban and R. Subramanian. (2008). “Intelligent heart disease prediction system using CANFIS and genetic algorithm,” International Journal of Biological, Biomedical and Medical Sciences, vol. 3, no. 3, pp. 157–160. [Google Scholar]
27. S. Chauhan and B. T. Aeri. (2015). “The rising incidence of cardiovascular diseases in India: Assessing its economic impact,” Journal of Preventive Cardiology, vol. 4, no. 4, pp. 735–740. [Google Scholar]
28. A. Khazaee. (2013). “Heart beat classification using particle swarm optimization,” International Journal of Intelligent Systems and Applications, vol. 5, no. 6, pp. 25. [Google Scholar]
29. L. Verma, S. Srivastava and P. C. Negi. (2016). “A hybrid data mining model to predict coronary artery disease cases using non-invasive clinical data,” Journal of Medical Systems, vol. 40, no. 7, pp. 178. [Google Scholar]
30. C. B. C. Latha and S. C. Jeeva. (2019). “Improving the accuracy of prediction of heart disease risk based on ensemble classification techniques,” Informatics in Medicine Unlocked, vol. 16, pp. 100203. [Google Scholar]
31. X. Liu, X. Wang, Q. Su, M. Zhang, Y. Zhu et al. (2017). “A hybrid classification system for heart disease diagnosis based on the RFRS method,” Computational and Mathematical Methods in Medicine, vol. 2017, pp. 11. [Google Scholar]
32. B. Edmonds. (2005). “Using localised ‘Gossip’ to structure distributed learning,” in Proc. of AISB Symposium on Socially Inspired Computing, Hatfield, UK, pp. 1–12. [Google Scholar]
33. M. Gudadhe, K. Wankhade and S. Dongre. (2010). “Decision support system for heart disease based on support vector machine and artificial neural network,” in Int. Conf. on Computer and Communication Technology (ICCCTAllahabad, Uttar Pradesh, India, pp. 741–745. [Google Scholar]
34. M. A. Jabbar, B. L. Deekshatulu and P. Chandra. (2013). “Classification of heart disease using artificial neural network and feature subset selection,” Global Journal of Computer Science and Technology Neural & Artificial Intelligence, vol. 13, no. 3, pp. 4–8. [Google Scholar]
35. S. Palaniappan and R. Awang. (2008). “Intelligent heart disease prediction system using data mining techniques,” in IEEE/ACS Int. Conf. on Computer Systems and Applications, Doha, Qatar, pp. 108–115. [Google Scholar]
36. E. O. Olaniyi, O. K. Oyedotun and K. Adnan. (2015). “Heart diseases diagnosis using neural networks arbitration,” International Journal of Intelligent Systems and Applications, vol. 7, no. 12, pp. 72. [Google Scholar]
37. R. Das, I. Turkoglu and A. Sengur. (2009). “Effective diagnosis of heart disease through neural networks ensembles,” Expert Systems with Applications, vol. 36, no. 4, pp. 7675–7680. [Google Scholar]
38. R. Detrano, A. Janosi, W. Steinbrunn, M. Pfisterer, J. J. Schmid et al. (1989). “International application of a new probability algorithm for the diagnosis of coronary artery disease,” The American Journal of Cardiology, vol. 64, no. 5, pp. 304–310. [Google Scholar]
39. M. K. Abd Ghani, M. A. Mohammed, N. Arunkumar, S. A. Mostafa, D. A. Ibrahim et al. (2020). “Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques,” Neural Computing and Applications, vol. 32, no. 3, pp. 625–638. [Google Scholar]
40. D. Aha and D. Kibler. (1988). “Instance-based prediction of heart-disease presence with the Cleveland database,” University of California, vol. 3, no. 1, pp. 3–2. [Google Scholar]
41. M. A. Mohammed, S. S. Gunasekaran, S. A. Mostafa, A. Mustafa and M. K. Abd Ghani. (2018). “Implementing an agent-based multi-natural language anti-spam model,” in Int. Sym. on Agent, Multi-Agent Systems and Robotics (ISAMSRPutrajaya, Malaysia, pp. 1–5. [Google Scholar]
42. L. Moreno-Alsasua, B. Garcia-Zapirain, J. D. Rodrigo-Carbonero and I. O. Ruiz. (2017). “Primary prevention of asymptomatic cardiovascular disease using physiological sensors connected to an IOS app,” Journal of Medical Systems, vol. 41, no. 12, pp. 191. [Google Scholar]
43. S. A. Mostafa, A. Mustapha, S. H. Khaleefah, M. S. Ahmad and M. A. Mohammed. (2018). “Evaluating the performance of three classification methods in diagnosis of Parkinson’s disease,” in Int. Conf. on Soft Computing and Data Mining, Cham: Springer, vol. 700, pp. 43–52. [Google Scholar]
44. L. Bottou. (2010). “Large-scale machine learning with stochastic gradient descent,” in Proc. of COMPSTAT’2010, Physica-Verlag HD, pp. 177–186. [Google Scholar]
45. Z. Q. Zeng, H. B. Yu, H. R. Xu, Y. Q. Xie and J. Gao. (2008). “Fast training support vector machines using parallel sequential minimal optimization,” in 3rd Int. Conf. on Intelligent System and Knowledge Engineering, Xiamen, China, vol. 1, pp. 997–1001. [Google Scholar]
46. M. A. Mohammed, M. K. Abd Ghani, R. I. Hamed, S. A. Mostafa, D. A. Ibrahim et al. (2017). “Solving vehicle routing problem by using improved K-nearest neighbor algorithm for best solution,” Journal of Computational Science, vol. 21, pp. 232–240. [Google Scholar]
47. E. A. Cortes, M. G. Martinez and N. G. Rubio. (2007). “Multiclass corporate failure prediction by adaboost. M1,” International Advances in Economic Research, vol. 13, no. 3, pp. 301–312. [Google Scholar]
48. J. Otero and L. Sánchez. (2006). “Induction of descriptive fuzzy classifiers with the Logitboost algorithm,” Soft Computing, vol. 10, no. 9, pp. 825–835. [Google Scholar]
49. M. Hall, E. Frank, G. Holmes, B. Pfahringer, P. Reutemann et al. (2009). “The WEKA data mining software: An update,” ACM SIGKDD Explorations Newsletter, vol. 11, no. 1, pp. 10–18. [Google Scholar]
50. V. F. Rodriguez-Galiano, B. Ghimire, J. Rogan, M. Chica-Olmo and J. P. Rigol-Sanchez. (2012). “An assessment of the effectiveness of a random forest classifier for land-cover classification,” ISPRS Journal of Photogrammetry and Remote Sensing, vol. 67, pp. 93–104. [Google Scholar]
51. I. de la Torre Díez, B. Garcia-Zapirain, A. Méndez-Zorrilla and M. (2016). “López-Coronado Monitoring and follow-up of chronic heart failure: A literature review of eHealth applications and systems,” Journal of Medical Systems, vol. 40, no. 7, pp. 179. [Google Scholar]
52. S. A. Mostafa, A. Mustapha, M. A. Mohammed, R. I. Hamed, N. Arunkumar et al. (2019). “Examining multiple feature evaluation and classification methods for improving the diagnosis of Parkinson’s disease,” Cognitive Systems Research, vol. 54, pp. 90–99. [Google Scholar]
53. I. De la Torre-Díez, B. Garcia-Zapirain, M. López-Coronado and J. J. Rodrigues. (2017). “Proposing telecardiology services on cloud for different medical institutions: A model of reference,” Telemedicine and e-Health, vol. 23, no. 8, pp. 654–661. [Google Scholar]
54. M. A. Mohammed, B. Al-Khateeb, A. N. Rashid, D. A. Ibrahim, M. K. Abd Ghani et al. (2018). “Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images,” Computers & Electrical Engineering, vol. 70, pp. 871–882. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |