DOI:10.32604/cmc.2021.014134

| Computers, Materials & Continua DOI:10.32604/cmc.2021.014134 |  |
| Article |
Identification of Thoracic Diseases by Exploiting Deep Neural Networks
1Department of Information Technology, College of Computer, Qassim University, Buraydah, Saudi Arabia
2Department of Computer Science, Univesity of Gurjat, 52250, Pakistan
3School of Computer Science, Guanzghou University, Guangzhou, 510006, China
4Department of Computer Science & Engineering, Jamia Hamdard, New Delhi, India
5Department of Computer Science, CCSIT, King Faisal University, KSA
*Corresponding Author: Hafiz Tayyab Rauf. Email: hafiztayyabrauf093@gmail.com
Received: 01 September 2020; Accepted: 17 October 2020
Abstract: With the increasing demand for doctors in chest related diseases, there is a 15% performance gap every five years. If this gap is not filled with effective chest disease detection automation, the healthcare industry may face unfavorable consequences. There are only several studies that targeted X-ray images of cardiothoracic diseases. Most of the studies only targeted a single disease, which is inadequate. Although some related studies have provided an identification framework for all classes, the results are not encouraging due to a lack of data and imbalanced data issues. This research provides a significant contribution to Generative Adversarial Network (GAN) based synthetic data and four different types of deep learning-based models that provided comparable results. The models include a ResNet-152 model with image augmentation with an accuracy of 67%, a ResNet-152 model without image augmentation with an accuracy of 62%, transfer learning with Inception-V3 with an accuracy of 68%, and finally ResNet-152 model with image augmentation but targeted only six classes with an accuracy of 83%.
Keywords: GAN; CNN; chest diseases; inception-V3; ResNet152
Cardiothoracic diseases are serious health problems that may lead to disorders Affecting the organs and tissues [1]. These cardiothoracic diseases are Affecting people at an alarming rate due to environmental factors. The air gets more polluted every day, and this pollution is inhaled, causing these diseases to develop [2]. To diagnose these cardiothoracic diseases, a chest X-ray (CXR) is examined by a radiologist [3]. As more people get affected, doctors are becoming scarce, especially in developing countries. However, with the advent of image processing tools, the task of diagnosing these cardiothoracic diseases has seen significant progress [4]. Many researchers have put in work to see how the problems associated with medical images can be mitigated by using neural networks [5,6].
A neural network is a mathematical model of neurons, also defined as a network capable of approximating arbitrary functions mathematically based on the universal approximation theorem [7]. This means that the neural network uses a set of algorithms modeled with the primary purpose of recognizing patterns to classify and cluster a dataset after being trained [8,9]. The training is done by feeding the neural network a large raw dataset, which it uses to recognize numerical patterns in vectors [10].
In this research, we adopted Convolutional Neural Network (CNN) as a class of deep neural networks to propose a generative adversarial network (GAN)-based model to generate synthetic data for training the data as the amount of the data is limited. We will use pre-trained models, which are models that were trained on a large benchmark dataset to solve a problem similar to the one we want to solve. For example, the ResNet-152 model we used was initially trained on the ImageNet dataset.
Other researches in the field of the cardiothoracic disease include: Ganesan et al. [11], who used a Keras framework to classify CXRs to predict lung diseases, reported accuracy of 86.14%, Rajpurkar et al. [12] used pixel-wise annotated DRRs data to learn an unsupervised multi-organ segmentation model on X-ray images, Salehinejad et al. [13] proposed a binary classifier for the detection of pneumonia from frontal-view chest X-rays, Irvin et al. [14] used a DCGAN tailored model designed for training with X-ray images where a generator is trained to generate artificial chest X-rays, Irvin et al. [14] trained several models to detect 14 different cardiothoracic diseases, Chandra et al. [15] used five different models to identify pneumonia and reported 95.631% as the best accuracy. A comprehensive literature review of related works is seen in Tab. 1.
Table 1: A comparison of the chest diseases diagnoses
Previous works used state-of-the-art techniques and got significant results with one or two cardiothoracic diseases but could lead to misclassification. In our work, we adopted GANs to synthesize the chest radiograph (CXR) to augment the training set on multiple cardiothoracic diseases to efficiently diagnose the chest diseases in different classes, as shown in Fig. 1. In this regard, our significant contributions are classifying various cardiothoracic diseases to detect a specific chest disease based on CXR, use the advantage of GANs to overcome the shortages of small training datasets, address the problem of imbalanced data; and implementing optimal deep neural network architecture with different hyper-parameters to improve the model with the best accuracy.
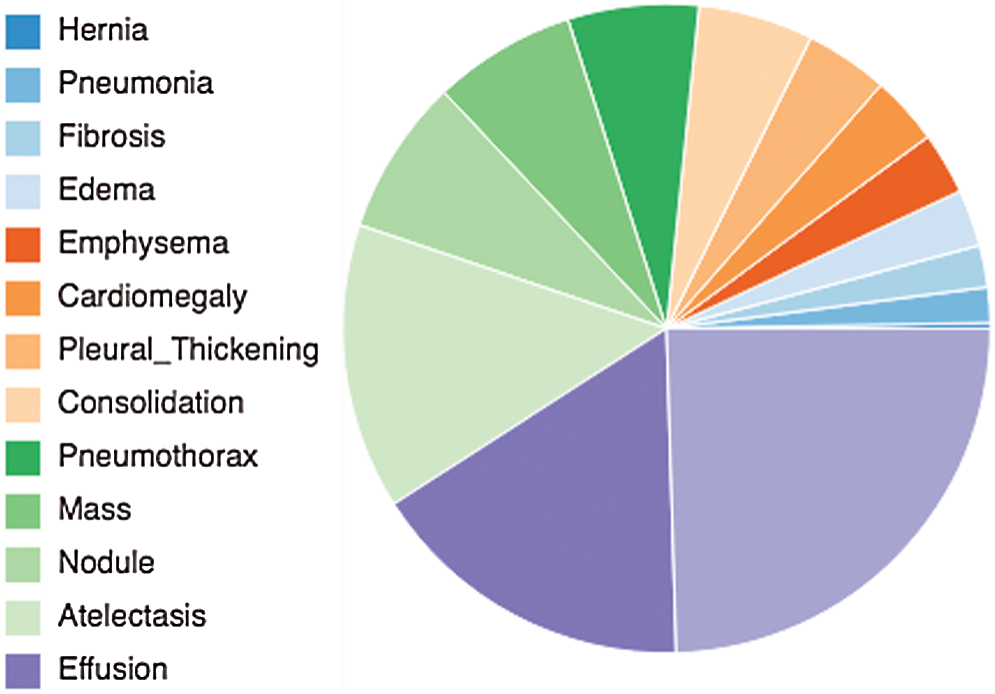
Figure 1: A pie chart showing the 14 underlined diseases related to chest that addressed by this study
The rest of the manuscript is organized as follows: In Section 2, we have reported a dataset and its analytics. Section 3 elaborates on the proposed solution’s overall methodology and shows all deep learning models with different hyper parameters and a comparison of the results. Section 4 discusses the outcomes in the results and analysis section. Finally, the conclusion is drawn in Section 5 with some discussion regarding limitations and future works.
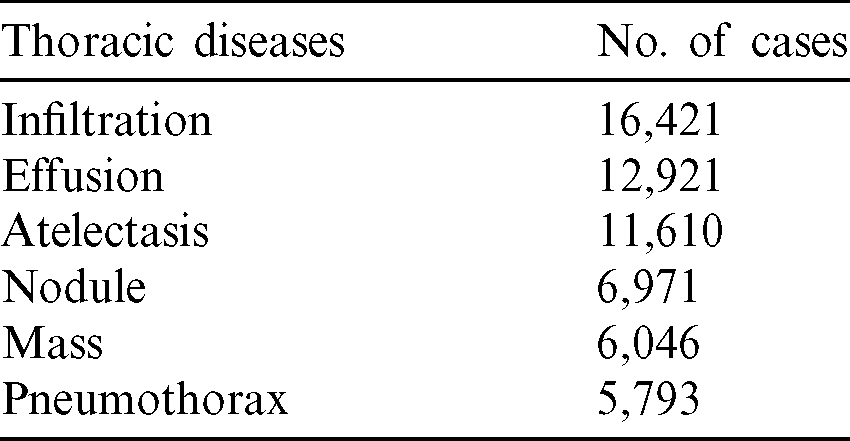
Training of deep learning models requires big data and requires much computational power–-practical training and validation. As more data is gathered for a deep learning model, it becomes more effective and more accurate as the model is prone to over-fitting with insufficient data. This research used a state-of-the-art dataset from [17], and it contains 108,948 frontal-view radiographic images of 32,717 different patients. The dataset is further classified into six major classes of cardiothoracic diseases, as seen in Tab. 2.
Table 2: Six classes of chest related diseases in the dataset used
The model also requires a class for X-rays with no thoracic disease; hence we collected 49,186 images for that. Although the images of remaining classes are not enough for proper training to successfully create it without risk of over-fitting, we were able to resolve the problem by obtaining more images from the dataset of the Kaggle challenge [18] as well as exploiting the synthetic dataset generated by state-of-the-art GAN model. The details can be seen in Section 4. GAN automatically discovers and learns the regularities or patterns in input data in such a way that the model can be used to generate or output new examples that could be drawn from the original dataset, as seen in Fig. 2. This means that GAN will create possible inputs from what it has learned from the inputs from the original dataset making more available for training.
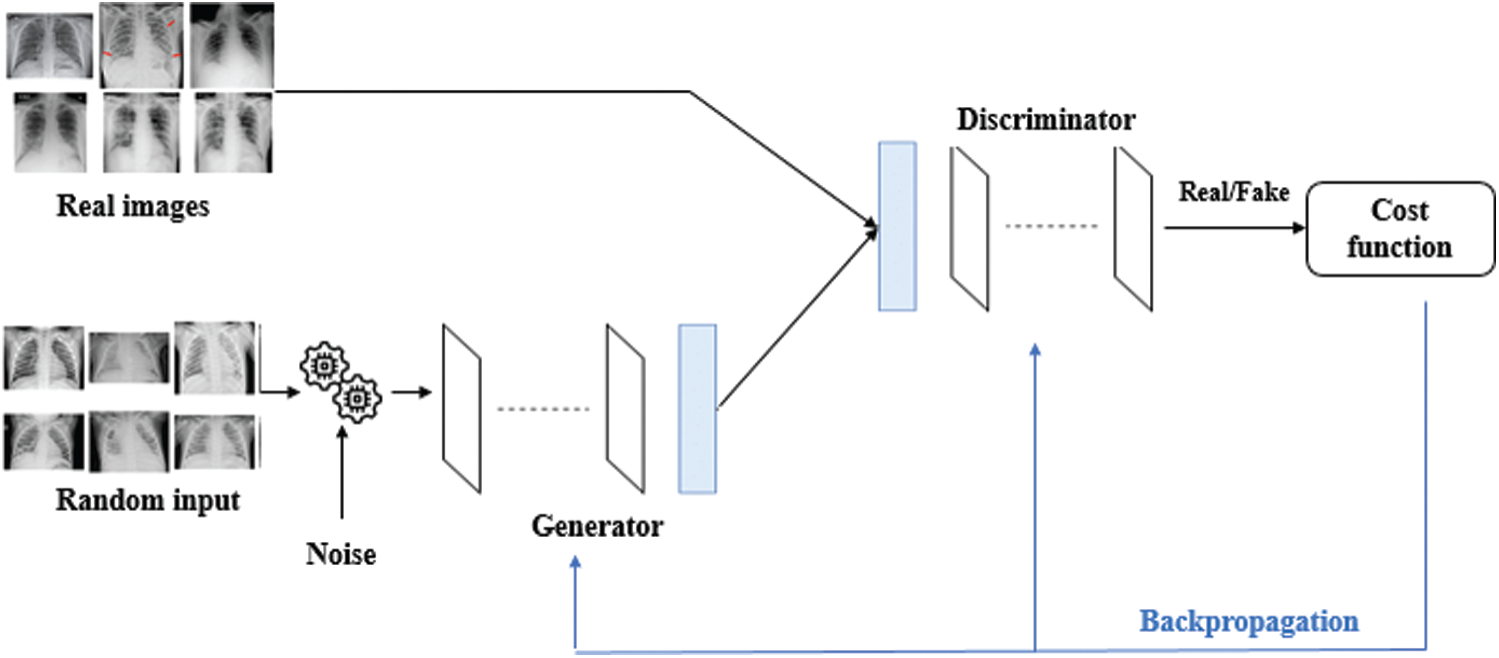
Figure 2: Generative adversarial network (GAN) model
We used python’s matplotlib library to graphically represent the distribution of all the various cardiothoracic diseases and those without the cardiothoracic disease. Furthermore, the familiar X-ray images (without cardiothoracic disease) used in our study have the highest frequency, higher than the combined cardiothoracic diseases. When comparing the cardiothoracic disease frequencies, the infiltration data has the highest frequency, while consolidation has the lowest frequency, as seen in Figs. 3 and 4.
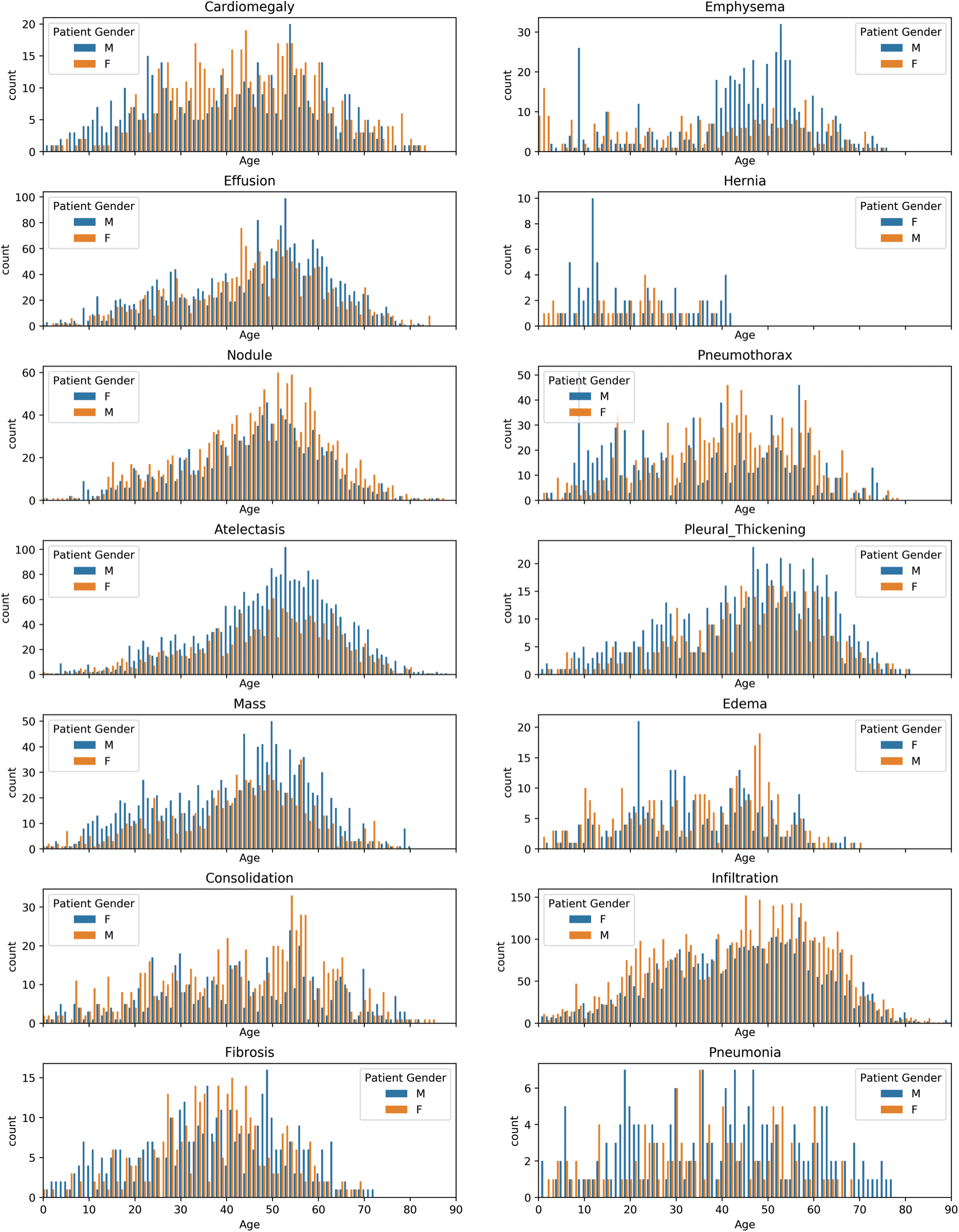
Figure 3: Gender wise distribution of chest related diseases
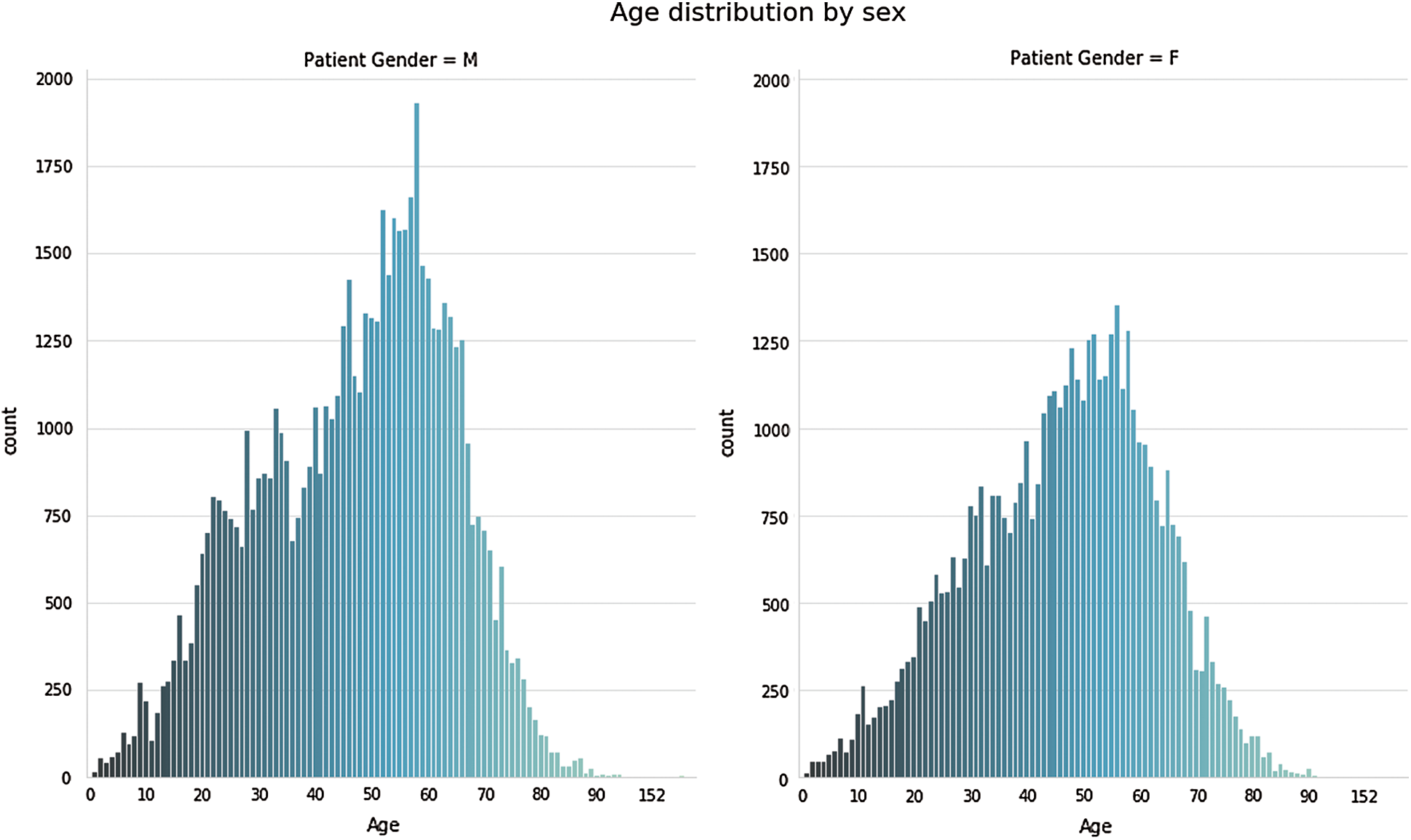
Figure 4: Age wise distribution of chest related diseases
In Fig. 3, we examined each of the pathologies using the pixel distribution. Moreover, the datasets we used are imbalanced, and the following observation can be deduced from Figs. 3 and 4:
• Infiltration, Effusion, Atelectasis, Nodule, Consolidation, Pleural Thickening, Emphysema, and Hernia affected more males compared to females based on the available data used for this analysis.
• Pneumothorax, Cardiomegaly, Edema, and Fibrosis affected more females compared to males.
• Cardiomegaly affects individuals starting around the age of 10 years old but is most common between the ages of 35–60, with a median age of around 50.
• Effusion has a similar distribution with Cardiomegaly but with a median around 55.
• Atelectasis affects individuals mostly between the ages of 25 and upwards. There is one case under the age of 25 which appeared to be an outlier.
• Mass effect mostly affects teenagers and adults with an extreme case occurring in a child.
• Emphysema is mostly seen in adults above 40 years old while those below 40 appeared to an outlier.
The dataset gathered for this research is highly imbalanced as the number of individuals without cardiothoracic disease (49,186) is significantly higher than others. This is an issue as it makes training of the model very difficult to avoid the over-fitting problem. To remedy this problem, a GAN is used as its primary purpose is to learn regularities and make a small dataset useable for training. After the GAN was applied, we went further to seek multiple physicians’ opinions in the cardiothoracic. Field to ensure that there are no errors in the dataset.
Next, we clustered each of our classes of cardiothoracic diseases into separate folders and further divided each folder into training and validation sets. These sets contain the label of each class as it would be used in creating the models. These training and validation sets could not be used the way they are as they have to be reshaped and normalized. Therefore, we reshaped them into (150, 150, 3) and normalized them. The images needed to be rotated in all angles because some images might have been uploaded the wrong way. This was done by augmenting the images. The labels were then encoded using one-hot coding, and the array was shaped into (128, 128, 3). These were saved as pickle files to be used later in the model creation.
As the neural network’s depth increases, the training process becomes more tedious, and the convergence time increases significantly. At the instant, when a deep neural network starts to converge, they are exposed to degradation issues [19]. For example, the network’s accuracy becomes stagnant due to saturation, and eventually, the accuracy starts degrading rapidly.
He et al. [19] proposed a deep neural network to mitigate the issues mentioned above. This study’s basic idea is to incorporate a residual mapping to which the layers of the neural network will fit. The basic building block of ResNet is presented in Fig. 5.
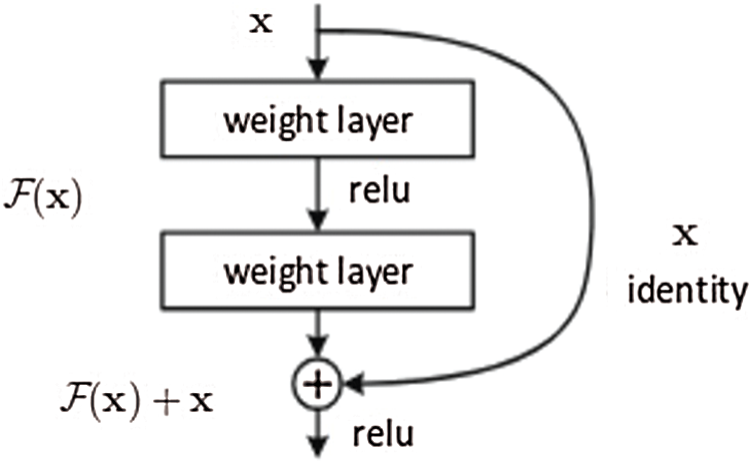
Figure 5: Residual block which is used in the design of ResNet architecture [19]
The residual block serves two purposes. First, when the input and output dimensions are equal, the identity short, i.e., x assists in the computation of the output as presented in (1).

On the contrary, when the dimensions change, the short will perform the identity mapping from the input with zero-padding to increase the dimensions. The dimension shortcut assists in computing the dimension using 1 Ö 1 convolution operation represented as (2).

This architecture makes ResNet very efficient in terms of accuracy, and the depth of the network can be considerably increased. He et al. [19] presented a ResNet with a depth of 152 layers, i.e., approximately eight times the well-known VGG16 [20]. However, the complexity of the former is still less than the later studies [21,22]. Besides, the convergence rate of ResNet is significantly faster, with ResNet-34 achieving a top-five validation error of 5.71% and ResNet-152 achieving a top-five validation error of approximately 4.49%.
3 Experimental Analysis and Results
Different hyper parameters were used, and the various results were compared to improve the proposed model, as shown in Tab. 3. We used ResNet-152 architecture (Fig. 5) in three models that can increase in size without acquiring new images. This is done by duplicating the images with some variation so the model can learn from examples.
Table 3: The proposed models and comparisons of their results
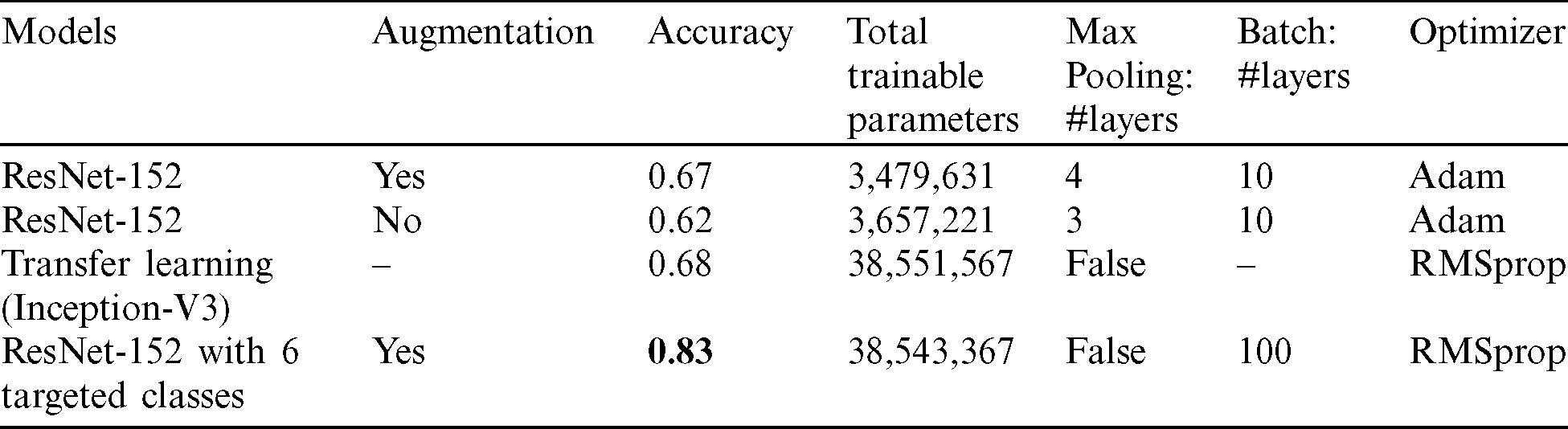
Tab. 3 shows the four proposed models’ comparisons with their experimental analysis and results with 50 epochs in training. Conv2D is implemented in the feature map’s structure in each layer of the proposed CNN to transform the X-ray images into abstract representations. Then, Maxpooling is utilized to decrease the dimensions of the output size. The dropout layer (0.5 or 0.2) is used either before or after the flattened layer to fix the over-fitting problem.
In the first model, we built four layers of convolutions, each followed by a Maxpooling layer. The model has one flattened layer followed by one dropout layer, then finally, two fully connected layers at each end. An Adam optimizer is used. As for the activation function, a leaky ReLU function is used. All these are used to train a multi-class model with 14 classes for our various cardiothoracic diseases with a validation accuracy of 67%. Tab. 3 shows the details of the model that was developed.
The next model uses the same model (ResNet) but without image augmentation. It is clear from Tab. 3 that a deep convolutional model with 512 hidden layers is used with the ReLU activation function. SoftMax is used to activate the output layer with 14 target classes of cardiothoracic disease. Extra batch normalization layers have been included after every Conv2D and dense layers to help distribute normalization in each batch. The validation accuracy is around 62% in predicting cardiothoracic disease in X-rays using a multi-class classification of 14 target labels. The model shows about 5% decline over model 1.
The third model is a transfer learning method utilizing Inception-V3. This transfer learning involves reusing an already developed model for a task as the starting point for a second task model. For this research, we used a pre-trained inception-V3 model, which has 1024 fully connected layers and the ReLU activation function without batch normalization layers as we used more parameters. To remedy the over-fitting issue, a dropout of 0.2 is used, and SoftMax is also used to output 15 classes. The validation accuracy is around 68% in predicting cardiothoracic diseases in X-rays using a multi-class classification of 14 target labels. In general, the validation accuracy of the model is good compared to model 2. However, it is not a significant improvement compared to the original model 1.
The last optimized model targeted six classes of the original 14 classes. A replica of the first model which used image augmentation in the convolutional neural network is used, but the number of target classes is reduced to 6 from the original dataset. This model has all the parameters of the first model for the image augmentation which are as follows: rotation 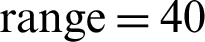 , width shift range 0.2, height shift
, width shift range 0.2, height shift 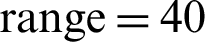 , shear
, shear 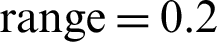 , zoom
, zoom 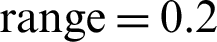 , horizontal
, horizontal 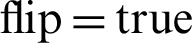 , and fill
, and fill 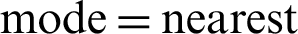 . Notably, this proposed model has an advantage over the previous models, and thus, model 4 of the modified ResNet-152 achieves a much better accuracy at 83%. It is a perfect improvement compared to the previously analysed models. Moreover, the validation accuracy of model 4 increases from around 67% to approximately 83%.
. Notably, this proposed model has an advantage over the previous models, and thus, model 4 of the modified ResNet-152 achieves a much better accuracy at 83%. It is a perfect improvement compared to the previously analysed models. Moreover, the validation accuracy of model 4 increases from around 67% to approximately 83%.
Four different deep learning models were developed for the automatic detection of various cardiothoracic diseases using X-ray images of the chest. After completion of training and validation of the models, we recognize the following:
• Model 4 is a replica of model 1 but with a reduced target class to 6, using all the label samples greater than 100 with a rotation range of 40 and a shift of 0.2. Thus, means model 4 is significantly affected by clustering approach so that the accuracy increased from 0.6721 to 0.83.
• The ResNet-152 without image augmentation has a training accuracy of 99% and a validation accuracy of 62%. This model seems to over fit the training data.
• More training data with balanced classes will significantly increase model accuracy.
• Image augmentation increases the accuracy of the model in predicting cardiothoracic diseases from an X-ray.
• More training data for other rare cases will increase the model performance.
• More training data will increase the model performance in predicting cardiothoracic disease from an X-ray.
• Using a pre-trained model can speed up training and increase model accuracy.
This research employs the advantages of computer vision and medical image analysis to develop an automated model with the clinical potential for early detection of the disease. Using deep learning models, the research aims to evaluate the effectiveness and accuracy of different convolutional neural network models in the automatic diagnosis of cardiothoracic diseases from X-ray images compared to diagnosis by experts in the medical community.
After successfully training and validating the models we developed, ResNet-152 with image augmentation proved to be the best model for the automatic detection of cardiothoracic disease. However, one of the main problems associated with radiographic in-depth learning projects and research is the scarcity and unavailability of enough datasets, a critical component of all deep learning models as they require many data for training. This is why some of our models had image augmentation to increase the number of images without duplication. As more data are collected in chest radiology, the models could be retrained to improve the accuracy of the models as deep learning models improve with more data. The future of Artificial Intelligence in terms of cardiothoracic diseases are unlimited. As more data becomes available, training and testing of different deep learning models will be possible. Multi-classification techniques are required with huge datasets for effective, efficient, and accurate detection of various cardiothoracic diseases. Hence, we are aimed to bring transfer learning in the pre-trained model to use the learning procedures of top deep learning models. We also intended to carry out some optimization algorithms for the automation of hyper parameters in the deep learning models.
Acknowledgement: We would like to thank the Deanship of Scientific Research, Qassim University for funding the publication of this project.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1 M. Bastien, P. Poirier, I. Lemieux and J. P. Despres. (2014). “Overview of epidemiology and contribution of obesity to cardiovascular disease,” Progress in Cardiovascular Diseases, vol. 56, no. 4, pp. 369–381. [Google Scholar]
2 Y. Zhang, S. Miao, T. Mansi and R. Liao. (2020). “Unsupervised X-ray image segmentation with task driven generative adversarial networks,” Medical Image Analysis, vol. 62, 101664. [Google Scholar]
3 K. Ghafoor. (2020). “COVID-19 pneumonia level detection using deep learning algorithm.”. [Google Scholar]
4 L. Huang, R. Han, T. Ai, P. Yu, H. Kang. (2020). et al., “Serial quantitative chest CT assessment of Covid-19: Deep-learning approach,” Radiology: Cardiothoracic Imaging, vol. 2, no. 2, e200075. [Google Scholar]
5 C. Pattichis and A. Constantinides. (1994). “Medical imaging with neural networks,” in Proc. of IEEE Workshop on Neural Networks for Signal Processing, U.K, IEEE. [Google Scholar]
6 N. Suga. (1988). “Neural computation for auditory imaging,” Neural Networks, vol. 1, pp. 276. [Google Scholar]
7 J. Heaton. (2017). “Ian goodfellow, yoshua bengio, and aaron courville: Deep learning,” Genetic Programming and Evolvable Machines, vol. 19, no. 1–2, pp. 305–307. [Google Scholar]
8 G. Currie, B. Iqbal and H. Kiat. (2019). “Intelligent imaging: Radiomics and artificial neural networks in heart failure,” Journal of Medical Imaging and Radiation Sciences, vol. 50, no. 4, pp. 571–574. [Google Scholar]
9 R. Fricks, J. B. Solomon and E. Samei. (2020). “Automatic phantom test pattern,” in Medical Imaging 2020: Physics of Medical Imaging. San Diego, USA: SPIE. [Google Scholar]
10 Knarr. (2019). “The influence of 3D printed prostheses on neural activation patterns,” Case Medical Research, vol. 1. [Google Scholar]
11 S. Ganesan, T. Subashini and K. Jayalakshmi. (2014). “Classification of x-rays using statistical moments and SVM,” in 2014 Int. Conf. on Communication and Signal Processing, Bangkok, IEEE. [Google Scholar]
12 P. Rajpurkar, J. Irvin, R. L. Ball, K. Zhu, B. Yang. (2018). et al., “Deep learning for chest radiograph diagnosis: A retrospective comparison of the CheXNeXt algorithm to practicing radiologists,” PLOS Medicine, vol. 15, no. 11, pp. e1002686. [Google Scholar]
13 H. Salehinejad, S. Valaee, T. Dowdell, E. Colak and J. Barfett. (2018). “Generalization of deep neural networks for chest pathology classification in x-rays using generative adversarial networks,” in 2018 IEEE Int. Conf. on Acoustics, Speech and Signal Processing, Alberta, Canada, IEEE. [Google Scholar]
14 J. Irvin, P. Rajpurkar, M. Ko, Y. Yu, S. Ciurea-Ilcus. (2019). et al., “CheXpert: A large chest radiograph dataset with uncertainty labels and expert comparison,” in Proc. of the AAAI Conf. on Artificial Intelligence, vol. 33, pp. 590–597. [Google Scholar]
15 T. B. Chandra and K. Verma. (2019). “Pneumonia detection on chest X-ray using machine learning paradigm,” in Proc. of 3rd Int. Conf. on Computer Vision and Image Processing, Singapore: Springer, pp. 21–33. [Google Scholar]
16 J. Sivasamy and T. Subashini. (2020). “Classification and predictions of lung diseases from chest x-rays using mobilenet,” International Journal of Analytical and Experimental Modal Analysis, vol. 1.
17 X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri et al. (2019). “ChestX-ray: Hospital-scale chest x-ray database and benchmarks on weakly supervised classification and localization of common thorax diseases,” in Deep Learning and Convolutional Neural Networks for Medical Imaging and Clinical Informatics. Basel, Switzerland: Springer International Publishing, pp. 369–392. [Google Scholar]
18 K. Team. (2015). “Digital pathology classification challenge,” Kaggle, pp. 1, . [online] Available: https://www.kaggle.com/c/digitalpathology. [Google Scholar]
19 K. He, X. Zhang, S. Ren and J. Sun. (2016). “Deep residual learning for image recognition,” in 2016 IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, IEEE. [Google Scholar]
20 K. Simonyan and A. Zisserman. (2014). “Very deep convolutional networks for large-scale image recognition.” arXiv preprint arXiv: 1409.1556. [Google Scholar]
21 M. Arif and G. Wang. (2019). “Fast curvelet transform through genetic algorithm for multimodal medical image fusion,” Soft Computing, vol. 24, no. 3, pp. 1815–1836. [Google Scholar]
22 O. Geman, I. Chiuchisan, I. Ungurean, M. Hagan and M. Arif. (2018). “Ubiquitous healthcare system based on the sensors network and android internet of things gateway,” in 2018 IEEE SmartWorld, Ubiquitous Intelligence & Computing, Advanced & Trusted Computing, Scalable Computing & Communications, Cloud & Big Data Computing, Internet of People and Smart City Innovation (SmartWorld/SCALCOM/UIC/ATC/CBDCom/IOP/SCILeicester, UK. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |