DOI:10.32604/cmc.2021.012874

| Computers, Materials & Continua DOI:10.32604/cmc.2021.012874 |  |
| Article |
A Comprehensive Investigation of Machine Learning Feature Extraction and Classification Methods for Automated Diagnosis of COVID-19 Based on X-ray Images
1College of Computer Science and Information Technology, University of Anbar, Ramadi, 31001, Iraq
2College of Agriculture, Al-Muthanna University, Samawah, 66001, Iraq
3eVIDA Lab, University of Deusto, Bilbao, 48007, Spain
4Faculty of Computer Science and Information Technology, Universiti Tun Hussein Onn Malaysia, Johor, 86400, Malaysia
5Software Engineering Department, College of Computer and Information Sciences, King Saud University, Riyadh, 11451, Saudi Arabia
6Faculty of Engineering and Built Environment, Universiti Kebangsaan Malaysia, Bangi, 43600, Malaysia
7Pure Science Department, Ministry of Education, General Directorate of Curricula, Baghdad, 10, Iraq
8Institute of Research and Development, Duy Tan University, Da Nang, 550000, Vietnam
9Faculty of Information Technology, Duy Tan University, Da Nang, 550000, Vietnam
*Corresponding Author: Dac-Nhuong Le. Email: ledacnhuong@duytan.edu.vn
Received: 15 July 2020; Accepted: 12 August 2020
Abstract: The quick spread of the Coronavirus Disease (COVID-19) infection around the world considered a real danger for global health. The biological structure and symptoms of COVID-19 are similar to other viral chest maladies, which makes it challenging and a big issue to improve approaches for efficient identification of COVID-19 disease. In this study, an automatic prediction of COVID-19 identification is proposed to automatically discriminate between healthy and COVID-19 infected subjects in X-ray images using two successful moderns are traditional machine learning methods (e.g., artificial neural network (ANN), support vector machine (SVM), linear kernel and radial basis function (RBF), k-nearest neighbor (k-NN), Decision Tree (DT), and CN 2 rule inducer techniques) and deep learning models (e.g., MobileNets V2, ResNet50, GoogleNet, DarkNet and Xception). A large X-ray dataset has been created and developed, namely the COVID-19 vs. Normal (400 healthy cases, and 400 COVID cases). To the best of our knowledge, it is currently the largest publicly accessible COVID-19 dataset with the largest number of X-ray images of confirmed COVID-19 infection cases. Based on the results obtained from the experiments, it can be concluded that all the models performed well, deep learning models had achieved the optimum accuracy of 98.8% in ResNet50 model. In comparison, in traditional machine learning techniques, the SVM demonstrated the best result for an accuracy of 95% and RBF accuracy 94% for the prediction of coronavirus disease 2019.
Keywords: Coronavirus disease; COVID-19 diagnosis; machine learning; convolutional neural networks; resnet50; artificial neural network; support vector machine; X-ray images; feature transfer learning
The most recent and popular disease is that caused by the coronavirus which belongs to the family of viruses causing a wide range of illnesses, including the common cold and other chronic diseases like Severe Acute Respiratory Syndrome (SARS-CoV), and Middle East Respiratory Syndrome (MERS-CoV). The novel coronavirus (COVID-19) is a new strain which has never been detected in humans. They have previously been known to infect only animals with diseases. The newly identified coronavirus has caused cold-like symptoms in most of the affected patients, with two other kinds of coronavirus [Middle East respiratory syndrome (Mers) and severe acute respiratory syndrome (Sars)] causing more chronic diseases in humans. These two other coronaviruses have killed over 1,500 people since the year 2002 [1]. The dangerous nature of the novel virus referred to as COVID-19 can be seen in its rapid spread and the number of people affected. So far, about 20% of confirmed cases have been classed as severe or critical, and the current death rate stands at about 2%, which is far lesser than the rates of Sars (10%) and Mers (30%). Despite, the low rate, it is still regarded as a significant threat that must be handled with care. It is believed that the coronavirus originated from a “wet market” in Wuhan, where both live and dead animals are sold, including birds and fish. In this kind of market, the risk of the virus being transferred from animals to humans is high since it is difficult to maintain hygiene standards in an environment where live animals are kept and also slaughtered at the same time [2]. However, the animal source of the latest global outbreak is yet to be identified but is suspected that the original host of the virus is bats. Even though there were no bats sold at the Wuhan market, but they may have infected live chickens and other animals sold there. There is a wide range of zoonotic viruses hosted by bats, and some of them include rabies, Ebola and HIV. WHO and NHS have spelt the major symptoms of the novel coronavirus, and they include high temperature, cough and shortness of breath? These health organizations have noted that there are not many disparities between these symptoms and other respiratory diseases like common cold and flu [3].
Machine Learning techniques are a section of the Artificial Intelligence field that are firmly related to statistics domain. It focuses on developing techniques and algorithms that allow computers to utilize experience to learn and obtain intelligence. In contrast, Convolutional Neural Network (CNN) is among the most established methods in deep learning. A CNN is composed of two core structures, a convolution layer, and a pooling layer. The data in the convolutional layers are organized as feature maps, where each of which connected to the previous layer through a set of weights. The outcomes of the sum of these weights are then passed to a non-linear unit, for example, rectified linear units or (ReLU). In contrast, the role of the pooling layers is concerned with the merging of the features with semantic similarity [4,5]. Here, learning means the system can recognize and comprehend input cases as (data) that can be used for decision-making and predictions. The first step of the learning process involves data gathering from different sources using different means. The next step involves the preparation of data by pre-processing it to make it free of error, reduce the dimensionality of the space through the elimination of choosing the data of interest or irrelevant data. Because a large amount of data is used for learning, decision-making becomes a challenging task for the system. For this reason, the use of statistics, logic, probability, control theory, etc. are employed in the design of algorithms to facilitate the analysis of data and retrieval of information from the previous experiences. In the next process, the model is tested to determine the system’s performance, as well as accuracy. Lastly, the system is optimized by using the new dataset or rules to improvise the model. Machine learning techniques are used to predict, classify and recognize patterns. There are several areas in which the use of machine learning can be employed, and some of the areas include email filtering, search engine, face tagging and recognition, web page ranking, gaming, character recognition, robotics, prediction of disease, and management of traffic [5].
Recently, machine learning is used to analyse biomedical data that is highly dimensional automatically. Some examples of the biomedical applications of machine learning include risk assessment of cardiovascular diseases, diagnosis of skin lesions, diagnosis of nasopharyngeal carcinoma, brain cancer segmentation, and evaluation and classification methods for enhancing the diagnosis of Parkinson’s disease [6]. In the study carried out by [7], the ANN approach was successfully implemented for the diagnosis of breast cancer disease. The use of classification models was employed in work done by Mohammed et al. [8] who aimed at diagnosing nasopharyngeal carcinoma based on Decision-level fusion scheme using machine learning methods. Some of the classification models include Genetic algorithm, SVM, and ANN [9,10].
Nevertheless, to the best of our knowledge, these AI systems are not publicly available, and hence this limits the exposure for adequate research and investigation. Many researchers have recently enriched the open sources of COVID-19 diagnosis solutions for AI and ML that have used X-ray screening. The COVID-19 Chest X-ray data collection is now available and open to the public. In order to ensure that the principal reliability and complexity criteria are met, the standard of AI and ML classifiers used to diagnose COVID-19 should be evaluated and benchmarked. According to [9], radiographically driven AI and ML models could deliver a precise, effective COVID-19 solution that helps to make early diagnoses. The implementation of the new model that is proposed in this study is done utilizing machine learning methods in estimation and prediction modelling of coronavirus disease dataset. In this study, two machine learning approaches are implemented, traditional machine learning and deep learning. In the first approach, the performances of seven models have been examined based on ANN, SVM, RBF, (k-NN), DT, and CN 2 rule inducer techniques-based prediction approach for increasing the accuracy of automated coronavirus disease diagnosis. While in the second approach, five pre-trained models have been adapted, MobileNets V2, ResNet50, GoogleNet, DarkNet and Xception. These models have been retrained with the proposed COVID-19 dataset, where both the feature extraction process and the classification tasks are performed automatically. Three main challenges are by this study as follows: (i) Limited publicly available image dataset for COVID-19 case; (ii) Different models of transfer learning are available in the literature, and (iii) Different diagnosing models are proposed for tackling the prediction of COVID-19 cases. The main contributions and development of this study can be summarized as follows:
• • An automatic prediction of COVID-19 identification is proposed to automatically discriminate between healthy and COVID-19 infected subjects in X-ray images using two successful moderns are traditional machine learning methods (e.g., ANN, SVM, RBF, k-NN, DT, and CN 2 rule inducer technique) and deep learning models (e.g., MobileNets V2, ResNet50, GoogleNet, DarkNet and Xception). To the authors’ best knowledge, this is the first effort that examines the possibility of utilizing traditional and deep learning models in an integrated approach to detect the COVID-19 infection by learning high discriminative features representations from X-ray images.
• In comparison to most current studies, which use only one trained model to produce a final prediction approach. The proposed COVID identification method is developed by fusing the results from two separate traditional machine learning methods and deep-learning models which were trained from scratch with large datasets. Here, a parallel architecture is considered, which enables radiologists to differentiate between a healthy and COVID-19 diagnosed subjects with a high degree of confidence.
• A large X-ray dataset has been created and developed, namely the COVID-19 vs. Normal. To the best of our knowledge, it is currently the largest publicly accessible COVID 19 dataset with the largest number of X-ray images of confirmed COVID-19 infection cases.
• One of the main findings of this work is to continue validating and exploring the possibility of reducing algorithm complexity, generalization of traditional machine learning and deep learning approaches by applying transfer learning and producing valuable features for training instead of the conventional training by using predefined raw data features.
• The aim of this study is also to increase the ability to generalize the proposed approach of COVID recognition to avoid the question of overfitting with distinctive training methodologies backed by a variety of training policies such as AdaGrad algorithm, and data enhancement.
• In this study, all the methods used have an end-to-end scheme that provides the models to acquire a good accuracy using transfer learning method without manual feature extraction and selection techniques.
• The experimental results demonstrated that ResNet50 model is an efficient and useful pre-trained model between other five pre-trained models and seven traditional machine learning methods.
• The pre-trained models and traditional machine learning methods are presented provide very high outcomes and accuracy in large COVID dataset (400 healthy cases, and 400 COVID cases).
• The effectiveness and helpfulness of the COVID-19 identification procedure are identified, along with the possibility that the COVID-19 virus will be used in the hospitals and medical centres with less than 3 s per case for early diagnostics in order to achieve the necessary outcomes.
The rest organization of our research are including of six sections: Section 2 contains the related studies that show the COVID-19 diagnostic with its models used. Section 3 contains the materials and methods that are used in this study such as COVID dataset, five CNN pre-trained models and seven traditional machine learning methods. The proposed methodology with two main stages for transfer learning selection stage and diagnosis model selection stage are presented in Section 4. Section 5 demonstrates the outcomes of five CNN pre-trained models and seven traditional machine learning methods. Finally, in Section 6, the conclusion has been presented.
In December 2019, a new Coronavirus, so called (COVID-19) is appeared in China, particularly in Wuhan. The COVID-19 is spread around the world caused disastrous effects and might leads to death. COVID-19 pandemic gets great attention from the global and health institutions as it has no-cure is available yet [1]. COVID-19 biologically consists of RNA-type with positive-oriented single-stranded which makes finding the treatment is challenging because of having a mutating characteristic. There is a hard effort done by scientists and researchers around the world in attempting to discover an effective treatment for COVID-19. According to the global statistical data, China, USA, Brazil and Italy, Spain, Iran, the UK, and many other countries have lost over thousands of persons due to the COVID-19 disease. The Coronavirus family have different types and they can be infected the animals. This is included the COVID-19 which can be seen in bat, poultry, rodent, cat, dog, pig and human as well. The common COVID-19 symptoms are fever, dry cough, headache, sore throat, runny nose and muscles pains. Infected people with COVID-19 who have weak immune systems are potential to death [1,11]. Healthy People can be infected by COVID-19 thought physical contact such as breath, mucous and hand contacts with infected people [11]. Fig. 1 shows the spread of COVID-19 around the world [12].
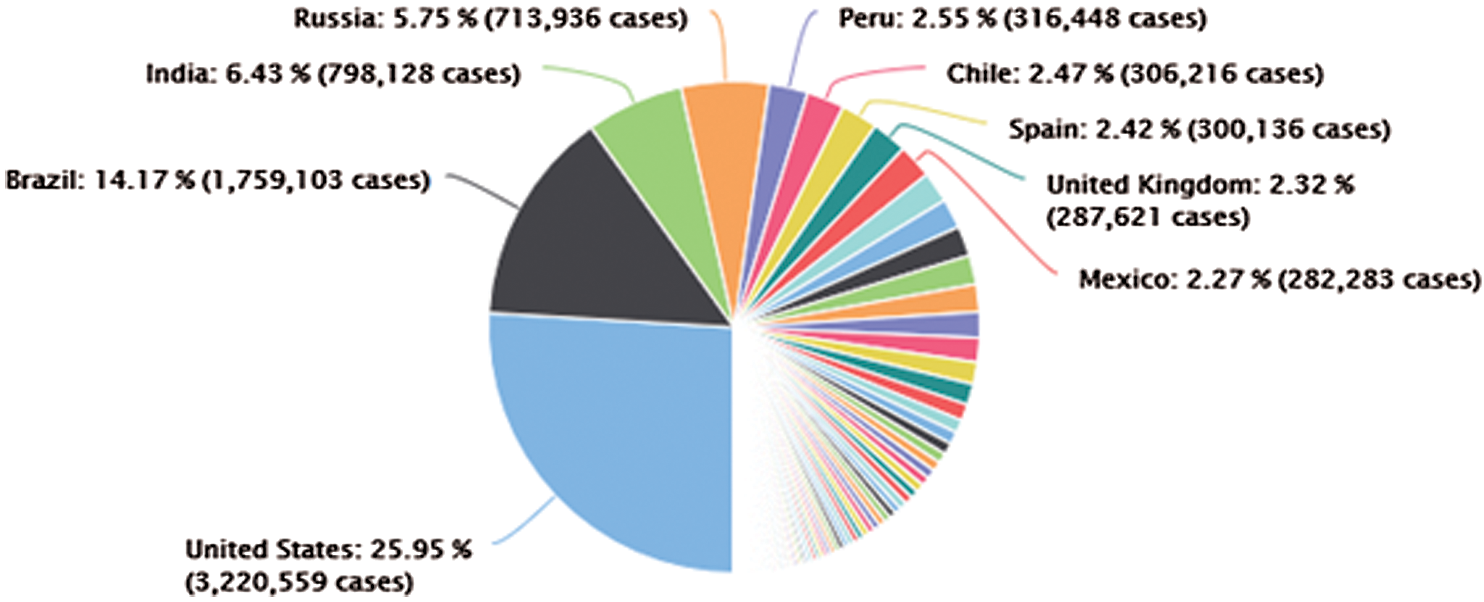
Figure 1: The distribution of the confirmed COVID-19 cases around the world (04 November 2020)
The major COVID-19 examination is reverse transcription-polymerase chain reaction (RT-PCR) is commonly used to diagnosing the disease [1]. For the regions that early attacked by the COVID-19 pandemic, RT-PCR can be an inappropriate examination. As reported in [13], some lab COVID-19 examinations are deficient in sensitivity with 71%. A reason for that is based on many factors including the quality control and sample preparation [14]. There are massive studies are presented recently that proposed detection and prediction methods for COVID-19 diagnosis [1]. The chest X-ray images can be considered less touchy than CT scan imaging, in spite of being the ordinary first-line imaging methodology utilized for patients beneath examination of COVID-19. The latest studies detailed that X-ray appears typical in early or gentle infection [15]. In specific, abnormal chest objects were detected in 69% of the patients at the first time of the investigation, and in 80% of the patients at some point after amid hospitalization [15].
The identification of COVID-19 from other symptoms, healthy subjects and pneumonia have been investigated. [16] proposed a Bayesian CNN to evaluation the finding uncertainty in COVID-19 classification. They used 70 X-ray images for COVID-19 public dataset of patients infected by this virus [17], and healthy cases were acquired from Kaggle’s Chest X-ray Images. The outcomes illustration that Bayesian model enhances the recognition precision of the typical VGG16 method from 85.7% to 92.9%. The authors further generate saliency maps to illustrate the locations focused by the deep network, to improve the understanding of deep learning results and facilitate a more informed decision-making process. In their study, they produce saliency maps to demonstrate the regions centred by CNN model, to enhance the understanding of CNN model outcomes and encourage a more accurate decision-support. Another study carried out by [18] proposed three diverse CNN models, such as Inception-ResNetV2, InceptionV3, and ResNet50, to identify COVID-19 infection in X-ray cases. They used small COVID-19 dataset obtained from Kaggle’s Chest X-ray Images and [17]. This dataset contains 50 images classified as 25 normal cases and 25 COVID cases. The assessment outcomes demonstrated that the ResNet50 method succeeds the best detection and identification evaluation by 98.0% accuracy metric, compared to 97.0% accuracy by InceptionV3 and 87% accuracy by Inception-ResNetV2. According to [19], the authors proposed a ResNet model to identify COVID-19 with X-ray images. This study with this method has two processes, firstly identification between healthy cases and COVID-19, secondly, anomaly identify. The anomaly identification process provides an anomaly score to enhance the COVID-19 scores utilized for the identification task. Two datasets have been used in this study that includes 1008 normal pneumonia patients and 70 X-ray images for COVID cases. The evaluation of the experimental results shows the specificity, sensitivity, and AUC metrics achieve are 70.7%, 96.0%, and 0.952, respectively.
Generally, the most recent researches utilize X-ray images to detecting and classifying among healthy cases and COVID-19 and other pneumonia cases. The main sources of these images from two online public databases that include only 50 COVID-19 cases. However, the limited cases of COVID samples, it’s deficiently to assess the strength of the models additionally postures questions to the generalizability concerning uses in other hospitals and medical departments. Therefore, one of the contributions of this study is a large X-ray dataset has been created and developed, namely the COVID-19 vs. Normal. To the best of our knowledge, it is currently the largest publicly accessible COVID 19 dataset with the largest number of X-ray images of confirmed COVID-19 infection cases. In addition, the pre-trained models and traditional machine learning methods are presented provide very high outcomes and accuracy in large COVID dataset (400 healthy cases, and 400 COVID cases).
In this research, many X-ray images from various sources were carefully chosen to establish the relatively large COVID-19 X-rays of confirmed cases, namely COVID-19 vs. Normal-datasets, which are reported as infected cases. It is then mixed with some normal X-ray images for a more confident diagnosis of COVID-19. The COVID-19 vs. Normal-dataset was collected from the following sources:
• The first collection includes 200 X-ray images with confirmed COVID-19 infection cases that collect from Cohen’s GitHub repository [20].
• The second collection includes 200 COVID-19 X-ray images with confirmed COVID-19 infection cases collected from three various sources are RSNA, SIRM, and Radiopaedia dataset.
• The third collection includes 400 chest X-ray images of normal cases is gathered from Kaggle’s chest X-ray images (Pneumonia) dataset.
Fig. 2 shows COVID-19 samples and normal large-scale COVID-19 X-ray cases. The number of cases of COVID-19 is regularly updated with new X-ray imaging with a reported COVID-19 infection available, and the full dataset is publicly available at https:/gitub.com/AlaaSulaiman/COVID19-vs-normal-dataset for academic purposes. To prevent overfitting problems and to increase the potential for generalization of the last trained model, a data augmentation technique is used. Next, the image’s size is rescaled to (224 to 224) pixels, and then five random image areas of ( ) pixels were extracted from each image. A horizontal image flip and rotation of 5-degrees (e.g., clockwise and counterclockwise) are then applied to each image within the dataset. Consequently, in both classes (for example, COVID-19 and normal pictures), a total of 24,000 X-ray images of the size (128 to 128) pixels were extracted. To prevent biased predictive results, the COVID-19 vs. Normal was divided into three mutually exclusive sets (e.g., t, validation and test set) followed by the data augmentation procedure.
) pixels were extracted from each image. A horizontal image flip and rotation of 5-degrees (e.g., clockwise and counterclockwise) are then applied to each image within the dataset. Consequently, in both classes (for example, COVID-19 and normal pictures), a total of 24,000 X-ray images of the size (128 to 128) pixels were extracted. To prevent biased predictive results, the COVID-19 vs. Normal was divided into three mutually exclusive sets (e.g., t, validation and test set) followed by the data augmentation procedure.
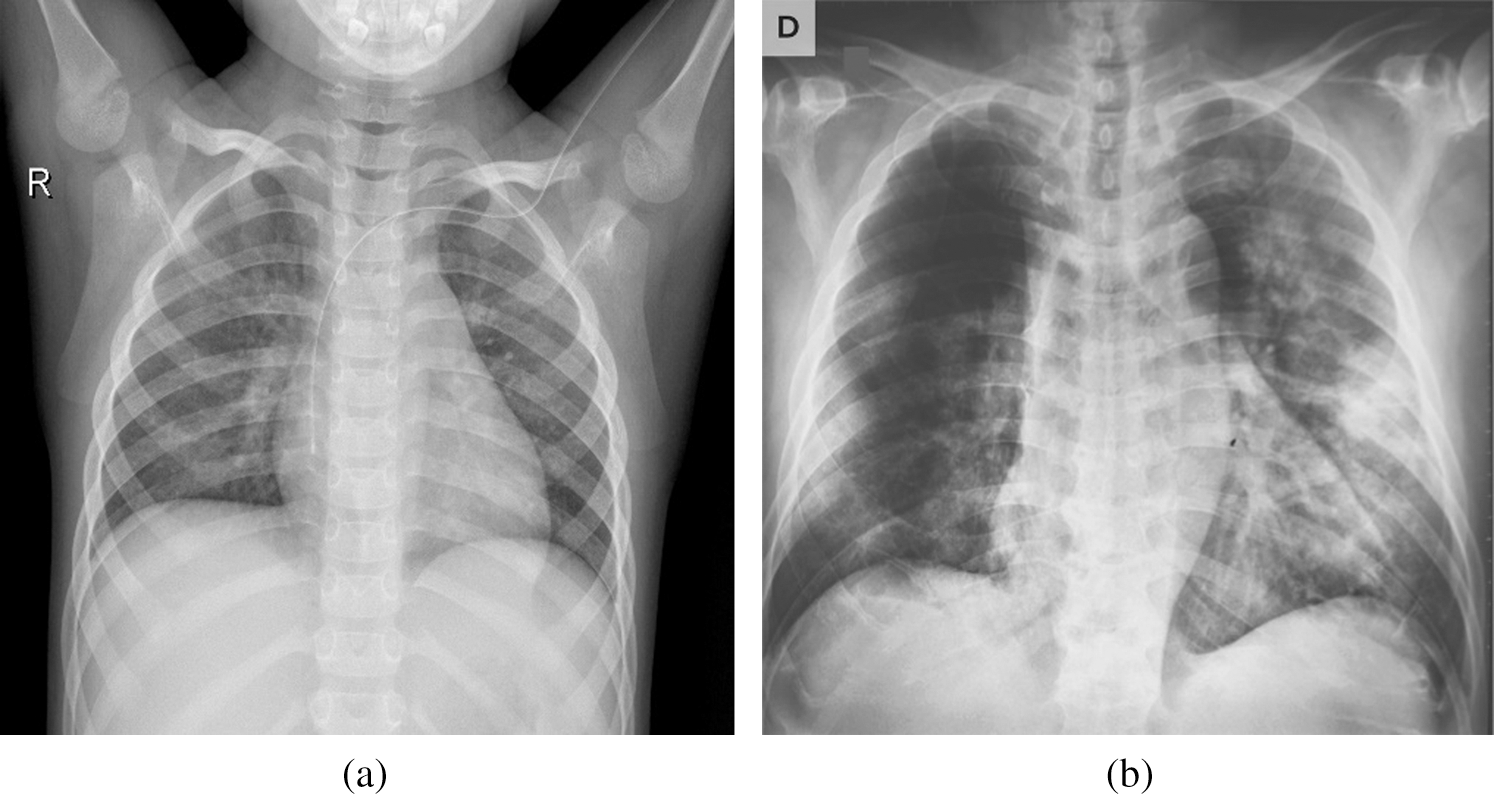
Figure 2: Samples of X-ray dataset (a) normal case (b) positive COVID-19 case
The relevance of transfer learning to several applications is awakened by the advancement of deep learning. This is especially true for the field of medical imaging, where the standard practice requires combining an extant architecture that is specially tailored for natural image datasets like ImagNet, alongside corresponding pre-trained weights (e.g., ResNet [21], Inception [22]), and then fine-tune the model on the medical imaging data [23]. This adoption of this standard formula can be seen in different parts of the globe, especially in different medical areas of specialization [24–33]. More so, the methodology has also been applied in the early detection of Alzheimer’s Disease [34], skin cancer identification based on dermatologist level photographs [35], and even determining the quality of human embryo for IVF procedures [36].
In such cases, the learning performance can be improved through knowledge transfer, whereby time-consuming data labelling efforts are not required. This problem can be solved through the use of transfer learning as novel learning frameworks [37–40]. An example of transfer learning applied to medical images can be seen in Fig. 3 from [40]. In this work, the utilize of original InceptionV3 [41] method have been utilized in several medical images uses is employed as a feature extraction process [28,29]. Particularly, this model has been used for the detection of Pneumonia through Chest X-ray [40]. To provide decent results and efficiency google improved the architecture of Inceptionv3 with weights that have been pre-trained on ImageNet, being an updated sort of InceptionV1. Formerly, the training of the model was performed on more than one million images from 1,000 classes using some very powerful machines. Repeating blocks which are referred to as Inception Block in deep networks are contained in the InceptionV3, where hyper-tuning of the parameter can be done. After each iteration, the results are produced by multiple convolutional filters ranging from  to
to  , thereby preventing localized information from being lost. This way, there will be a decrease in the number of parameters that require training as compared to other extant algorithms [42]. The SqueezeNet was founded by DeepScale, UC Berkeley and Stanford University for mage Classification process used in this study. The SqueezeNet Architectural Design methods are to Replace
, thereby preventing localized information from being lost. This way, there will be a decrease in the number of parameters that require training as compared to other extant algorithms [42]. The SqueezeNet was founded by DeepScale, UC Berkeley and Stanford University for mage Classification process used in this study. The SqueezeNet Architectural Design methods are to Replace  filters with
filters with  filter, reduce the number of input channels to
filter, reduce the number of input channels to  filters to provide good accuracy and reduce the time-consuming in convolution layers [43]. The VGG16 is a CNN model founded by K. Simonyan and A. Zisserman from the University of Oxford used in image classification field used in this study. This model accomplishes 92.7% top-5 testing exactness in ImageNet, which is a dataset of more than 14 million cases having a place with 1000 classes. It was one of the renowned models used to ILSVRC-2014. It makes the development over AlexNet by supplanting enormous piece measured channels (11 and 5 in the first and second convolutional layer, separately) with different
filters to provide good accuracy and reduce the time-consuming in convolution layers [43]. The VGG16 is a CNN model founded by K. Simonyan and A. Zisserman from the University of Oxford used in image classification field used in this study. This model accomplishes 92.7% top-5 testing exactness in ImageNet, which is a dataset of more than 14 million cases having a place with 1000 classes. It was one of the renowned models used to ILSVRC-2014. It makes the development over AlexNet by supplanting enormous piece measured channels (11 and 5 in the first and second convolutional layer, separately) with different  part estimated channels in a steady progression [44].
part estimated channels in a steady progression [44].
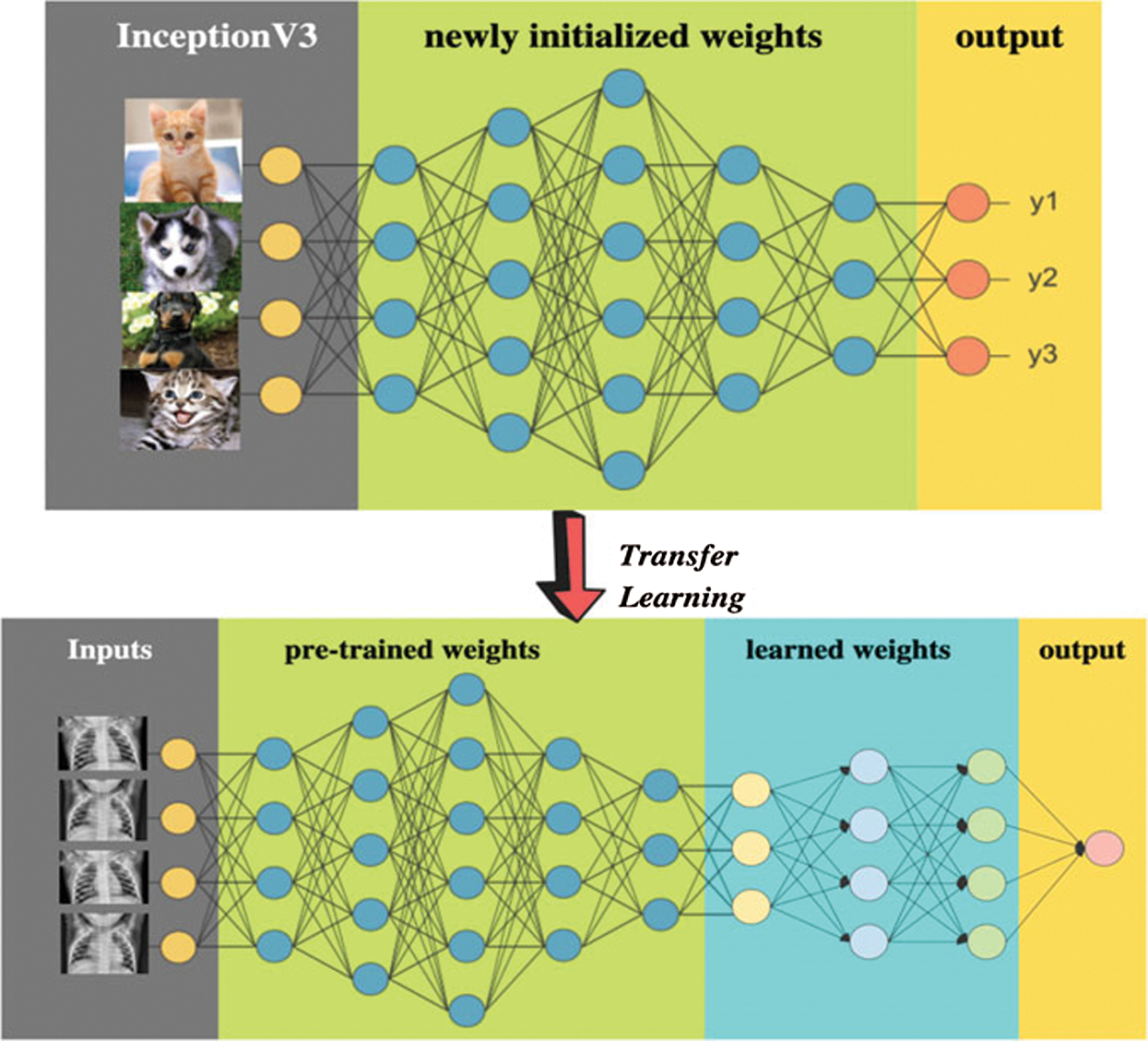
Figure 3: Example of transfer learning in medical images
3.3 Machine Learning Techniques
To perform binary classification of the Novel Corona Virus 2019 Dataset alongside its attributes, a supervised machine learning method was utilized. Seven different algorithms including, including CN2 rule inducer ANN, RBF, DT, k-NN, and SVM were used to determine if a patient has coronavirus or not. The seven algorithms are explained in the subsequent subsections.
• Artificial Neural Network (ANN): is a family of models that can effectively solve problems associated with function approximation, classification, pattern recognition and clustering. Biological neural networks of the brain-inspired artificial neural networks. Attempts have been made to mimic the biological neural networks of the brain in a manner that is simplified [45].
• The Support Vector Machine (SVM): is a machine learning algorithm that performs the analysis of data for classification, and regression analysis. SVM is a supervised learning method that looks at data and sorts it into one of two categories. After sorting and categorizing the data, the SVM produces a map of the sorted data with the margins between the two categories as far apart as possible [1,46].
• Radial Basis Function (RBF) kernel SVM: The efficiency of the SVM has been proven on both linear data and non-linear data. Non-linear data classification has been achieved through the implementation of the radial base function with this algorithm [1]. The role played by the Kernel function is important, as it puts data into feature space.
• K-Nearest Neighbour’s Algorithm (KNN): In the area of patterns classification or discrimination, k-NN is a technique that enables the classification of the object in terms of the nearest training cases within the feature space. KNN is a kind of instance-based learning or lazy learning that involves the local approximation of the capacity, and all calculations are conceded until the point of recognition task [47].
• Random Forests (RF): which is a widely used ensemble method, is extensively adopted in the processes of classification, in which a wide range of decision trees is used for the classification of data. In this method, the building of bootstrap is done from the random forest original data, and a regression tree or raw classification is grown for each bootstrap template [48].
• The Decision Tree (DT): is described as a simple, easy-to-interpret and effective supervised method of classification [49]. A DT founds a non-linear correlation between a system’s inputs and outputs.
• CN2 Rule Inducer Algorithm: An example of a learning algorithm, if the CN2 induction algorithm, which is used for rule induction, and it is specially tailored to work even during imperfect training data. The idea of the algorithm is inspired by the concepts from the ID3 algorithm and AQ algorithm [50].
Convolutional Neural Network (CNN) is among the most established methods in deep learning. CNNs are composed of two core structures, a convolution layer, and a pooling layer. The data in the convolutional layers are organized as feature maps, where each of which connected to the previous layer through a set of weights. The outcomes of the sum of these weights are then passed to a non-linear unit, for example, rectified linear units or (ReLU). At the same time, the role of the pooling layers is concerned with the merging of the features with semantic similarity. The convolutional layer shares many weights, and the pooling layer samples the overall layer output and decreases the data rate from the next layer [51,52]. Five pre-trained models are chosen for their performance and adaptability, namely MobileNets V2, ResNet50, GoogleNet, DarkNet, and Xception.
• MobileNetsV2: Compared to the previous MobileNetV1 version introduced by Google, MobileNetV2 is suitable for mobile devices or any device with low computational power and a Depth-wise Separable Convolution which dramatically reduces the complexity and model size of networks. A better module with an inverted residual structure is introduced in MobileNetV2. [53].
• ResNet: is a traditional neural network used as backbones for many computer visions tasks, the term ResNet is short for Residual Networks. This model won the ImageNet competition in 2015. ResNet’s breakthrough enabled us to train extremely deep neural networks successfully with more than 150 layers. The problem of the vanishing gradients was very difficult before the ResNet training of very deep neural networks [51].
• GoogleNet: was the first version of inception models developed by Google as the names suggest. The GoogleNet Architecture is 22 layers deep, with 27 pooling layers included. There are 9 inception modules stacked linearly in total. The ends of the inception modules are connected to the global average pooling layer. Below is a zoomed-out image of the full GoogleNet architecture.
• Darknet: is a framework written in C and CUDA for open source neural networks. The CPU and GPU calculation are quick and easy to install. It features a state-of-the-art, real-time object detection system called You Only Look Once (YOLO). Using a Titan X, images processor at 40-90 FPS with a 78,6% mAP on VOC 2007 and 44,0% mAP COCO test-dev datasets. Upon the 1000-class ImageNet challenge, users may use Darknet to classify images [54].
• Xception: is a deep neural network architecture involving Depthwise Separable Convolutions. It was developed by scientists from Google. Xception has a 71-convolution layer deep neural network. A pre-trained version of the network can be loaded from the ImageNet database, which was trained with over a million images [54].
In this study, we aimed at obtaining better outcomes by using highly correlated variables. Here, input data means testing the coronavirus disease data utilized for the prediction and confusion matrix of the machine learning-based prediction approach. The methodology of this study is based on two main stages, as shown in Fig. 4. The two stages are implemented to solve the current challenges relevant to the process of employing the most optimal diagnosis model.
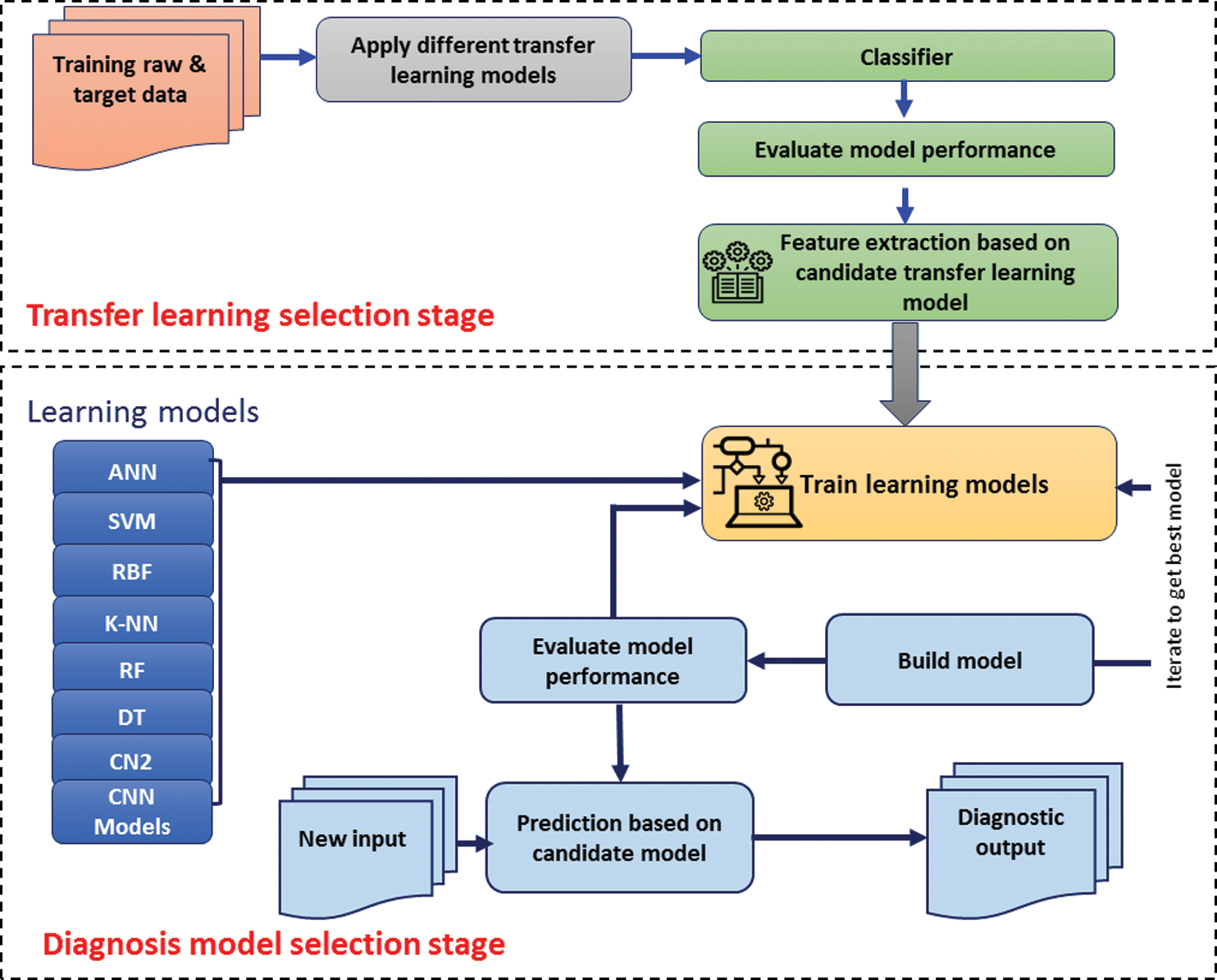
Figure 4: The framework of the COVID-19 prediction models
4.1 Transfer Learning Selection Stage
According to the variation between transfer learning models in terms of feature extraction number, it is quite challenging to determine which transfer learning model is the optimal one. Therefore, a solution is needed to tackle this issue. Based on the dataset mentioned in Section 2.1, the development of three transfer learning models will be conduct. Each of inceptionV3, VGG16, and SqueeZnet models are selected for comparison purpose. Due to the popularity of the SVM model, so it is selected as a classifier that will evaluate the performance transfer, learning models. Each one of three models will be used as a feature extractor model for SVM classifier. After evaluation for SVM based on features extracted from each transfer-learning model, the highest accuracy ratio will identify the candidate transfer-learning model for feature extraction purpose.
4.2 Diagnosis Model Selection Stage
Based on the candidate transfer learning model identified in Section 4.1, the stage of selecting the optimal diagnosis COVID-19 model is conducted. First, the dataset has been identified in Section 3.1 also have been used in this stage. Second, several models for the detection of coronavirus disease 2019 (COVID-19) have been developed using supervised algorithms. For this purpose, ANN, SVM, RBF, k-NN, Random Forest, Decision Tree, and CN2 rule inducer are used. Tab. 1 presents the various machine learning techniques that have been trained on coronavirus disease 2019 (COVID-19) dataset with improved ML techniques parameters. The experiments for all the classification methods were performed on one platform. The data set was divided into two parts, which are training and testing. The models are trained using random sampling, with 70% of the dataset, while the remaining 30% was used to test the model. Furthermore, four cross-validation schemes are included in the training and testing phases, namely, 2-folds, 3-folds, 5-folds, 10-folds. Third, each of training, prediction, and evaluation processes are conducted iteratively to define all related performance measurements for all classifiers. The arithmetic mean will be calculated for all accuracy values for a specific classifier based on the random sampling and cross-validation schemes. Then, based descending order for the accuracy values for each classifier, the rank for classifiers will be identified. Finally, based on the rank for all classifiers, the candidate classifier for diagnosing COVID-19 cases is determined. Then the candidate model will be recommended for a specific COVID-19 diagnosing task.
Table 1: The specification of the COVID-19 prediction models

It is very important to estimate the transmission risk of COVID-19 as well as its epidemic trend because these can trigger the vigilance of health providers, policymakers and the society at large. This way, adequate resources can be channelled towards efficient and rapid control and prevention. The quality of patients’ life can be improved, and their life expectancy can be enhanced through the early detection of coronavirus disease 2019 (COVID-19).
5.1 Traditional Machine Learning Models Results
In the first stage of selection, three transfer learning models have been applied as feature extraction shown in Tab. 2. Each one of these models has different properties, such as the number of layers, the number of extracted features.
Table 2: The results of the transfer learning models and classification
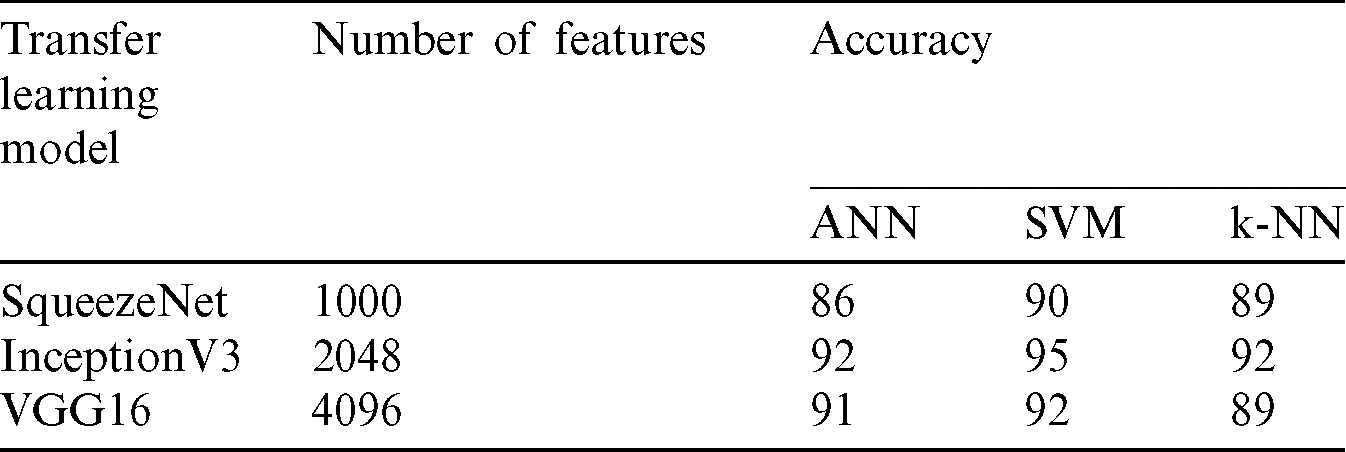
Based on the above results of Tab. 2, the highest accuracy is obtained by the three classifiers ANN, SVM and k-NN when we used the InceptionV3 for the feature extraction purpose, which produces 2048 features. On the other hand, the highest number of features are extracted by the VGG16 model, but surpassingly the accuracy score is lower than the InceptionV3. The SqueezeNet produces the lower number of features of 1000 features and scores considerably the lowest accuracy results. However, the SqueezeNet is might be more suitable thanVGG16 since it can achieve good accuracy with the minimum number of features which less the computational load for the classifier, but in terms of accuracy this model still quite far from the score of the InceptionV3. Hence, in this work, the InceptionV3 is considered the most suitable transfer learning model among three and selected in the seven COVID-19 prediction models.
Based on Tab. 3, which shows all the parameters used for the evaluation of all the models, it can be observed that the ANN model demonstrated an accuracy of 0.92. The accuracy of SVM 0.95. For RBF model accuracy is found to be 0.94, while for KNN it is 0.92. Moreover, the random forest-based model demonstrated an accuracy of 0.88. The accuracy for CN2 rule inducer model was found to be 0.73, while for that of Decision Tree was found to be 0.84. As for the Recall, which is indicative of the properly-recognized percentage of actual positives coronavirus cases, the ANN approach demonstrated a Recall value of 0.91, and for SVM and RBF methods, it is 0.92 and 0.93, respectively. For k-NN, RF, DT and CN2 rule inducer-based methods, the recall ratios are initiated to be 0.90, 0.85, 0.81 and 0.70 individually. The precision of ANN, SVM, RBF, k-NN, RF, DT and CN2 rule inducer-based methods are presented as 0.90, 0.93, 0.92, 0.89, 0.86, 0.80, and 0.71 respectively. The execution of the ML techniques has been measured by calculating the area under the curve (AUC). The AUC of ANN approach is achieved 0.91, whereas that of SVM and RBF methods are achieved 0.94 and 0.92, while k-NN, RF, DT and CN2 rule inducer-based methods are achieved 0.90, 0.90, 0.83, and 0.81 correspondingly. F1 score of ANN, SVM, RBF, k-NN, RF, DT and CN2 rule inducer-based methods are achieved 0.92, 0.93, 0.92, 0.90, 0.87, 0.83, and 0.70 respectively.
Table 3: Evaluation metrics of the COVID-19 prediction models

We used four schemes of 2, 3, 5, and 10 folds cross-validation to test and evaluate the machine learning models. This technique validates the effectiveness of the models in which it ensures that every observation from the original COVID-19 dataset appears in the training and testing sets. It is observed from the results in Tab. 3 that the stability of accuracy for a certain model in all schemes of four cross-validations. Each method affected the ratio from one-fold to another such as random forest algorithm started in 2-folds with 0.82 but in 3-folds increased to 0.84, while in 5-fold decreased to 0.85, in 10-fold repeated to increased again to 0.88. The stability of accuracy for a certain model in all schemes of four cross-validations it has affected based on the number of the folds. Tab. 4 shows cross-validation accuracy results of the COVID-19 prediction models.
Table 4: The accuracy results of cross-validation
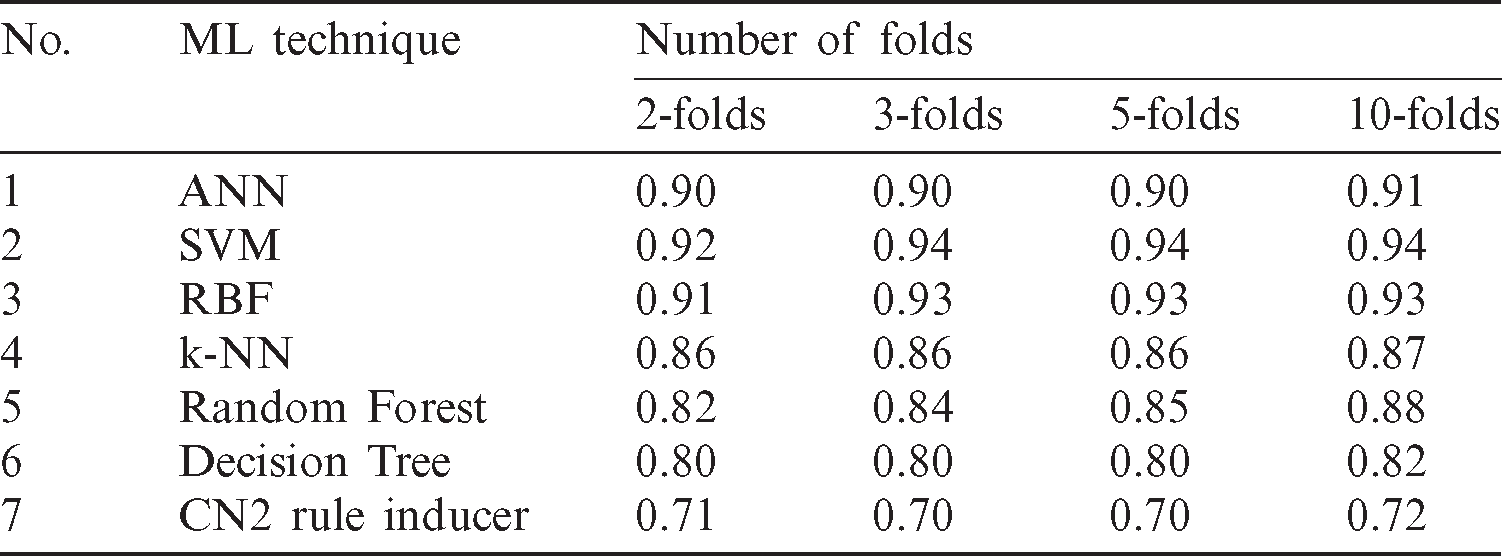
Tab. 5 confirmed the result that obtained in Tabs. 2 and 3 which indicates that each of SVM and ANN has scored the highest rank value while each of Tree and CN2 rule induces classifiers have obtained the worst rank values. So, based on the all mentioned results in this section, the most recommended classifiers for diagnosing of COVID-19 cases based on X-ray dataset are SVM and ANN.
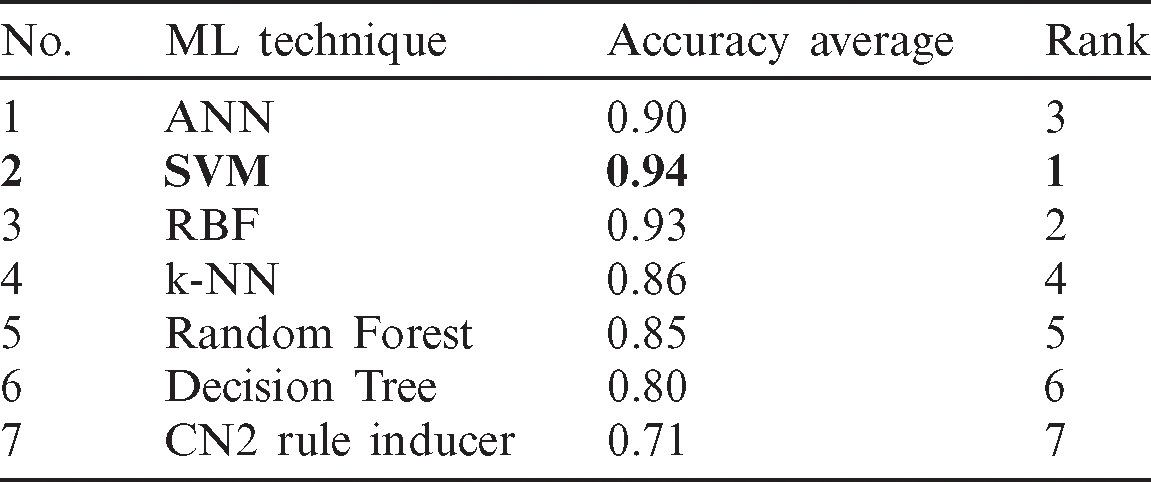
Fig. 5 shows the Receiver Operating Characteristics (ROC) curves for each of the tested seven models. It serves as a mean of comparing the prediction ability of the classification models. In the ROC measures, the x-axis represents the False Positive (FP) rate or 1-specificity (i.e., the probability of predicting true result when the actual result is false). At the same time, the y-axis represents the True Positive (TP) rate or sensitivity (i.e., the probability of predicting true result and the actual result is true too). Based on the results of the ROC analysis, the ANN, SVM and RBF can draw the closer curves of the ROC space and hence represent the most accurate classifiers. Moreover, it plots that the SVM represents the optimal classifier for the COVID-19 cases.
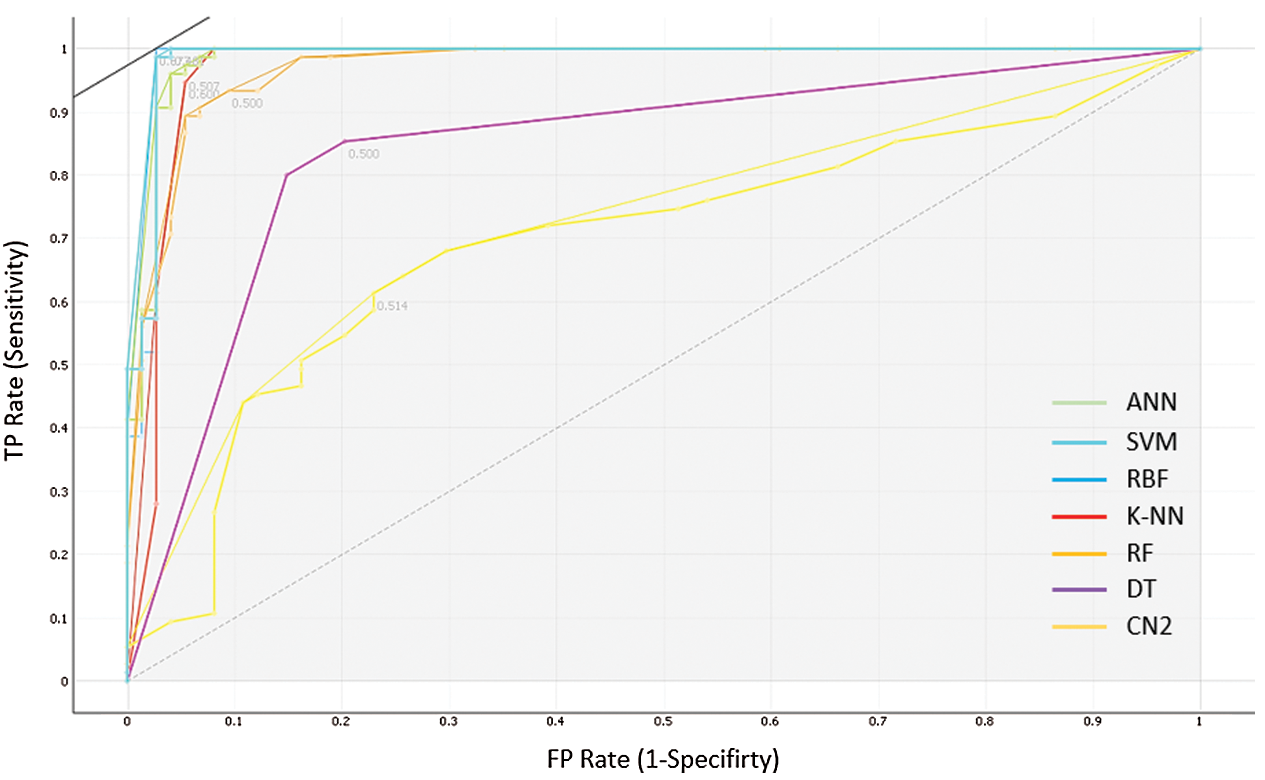
Figure 5: The ROC analysis for the performance of the seven prediction models
Ultimately, the results of this study reveal that in terms of all the parameters used in measuring the performance of the models, the ANN and SVM are the two best models that can be used in determining if a patient is affected or unaffected by a coronavirus. Besides, it can be observed that in terms of precision and accuracy, the ANN outperforms the SVM model. Nevertheless, the ANN and SVM models demonstrated the same AUC. In the case of a balanced class, the good performance of a binary classifier may not necessarily be indicated by accuracy alone. F1 score is a better indicator of a classifier’s performance in the case of balanced class distribution. Thus, great attention must be paid to the F1 score. Also, it can be observed that the ANN and SVM demonstrated high AUC values, which shows that the SVM approach and RBF method are the best classifiers for the COVID-19 dataset.
5.2 Convolutional Neural Networks Models Results
In deep learning architectures, every CNN model is composed of two tasks to be performed, feature extraction and classification. Feature extraction includes several convolution layers followed by max-pooling and an activation function. In contrast, the classification is carried out by applying fully connected layers. In this research, five pre-trained models were selected to evaluate the performance of the proposed dataset, namely, Mobilenets V2, GoogleNet, DarkNet, Xception, and ResNet50. These pre-trained models contain trained weights for the network. Hence if a network pre-trained for some classification task is used, the number of steps for the output to converge reduces. It is because generally, for the classification task, the features extracted will be similar. Instead of initializing the model with random weights, initializing it with the pre-trained weights reduces the training time and hence is more efficient. Tab. 6 shows the characteristics and performance analysis of each model. The training evaluation is shown in Figs. 6 and 7, where the training accuracies and loss for each of the five trained models.
Table 6: CNN models performance analysis
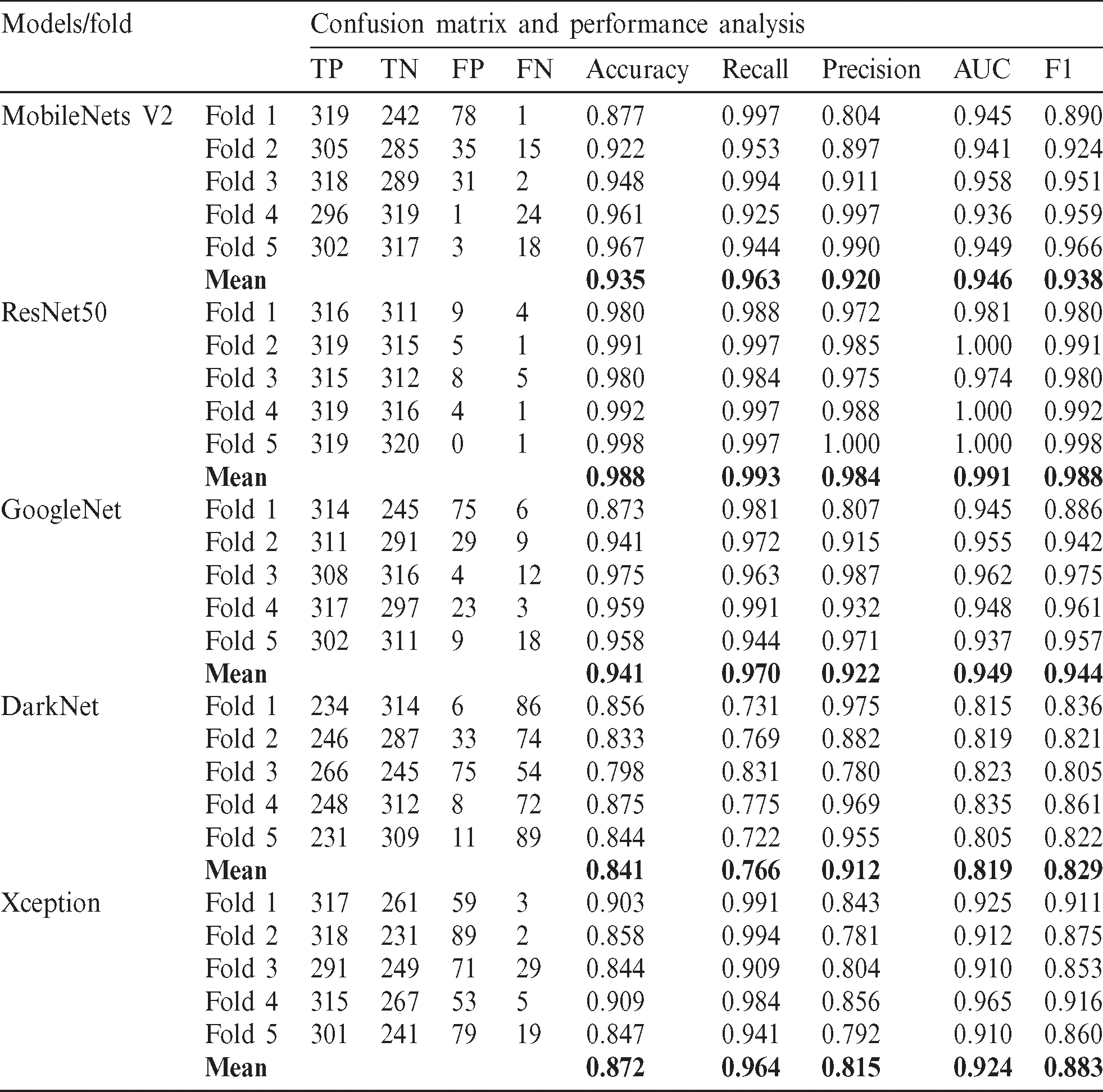

Figure 6: Training Accuracies of the deep learning trained models (a) MobieNets V2 training accuracy (b) ResNet50 training accuracy (c) GoogleNet training accuracy. (d) Darknet training accuracy (e) Xception training accuracy
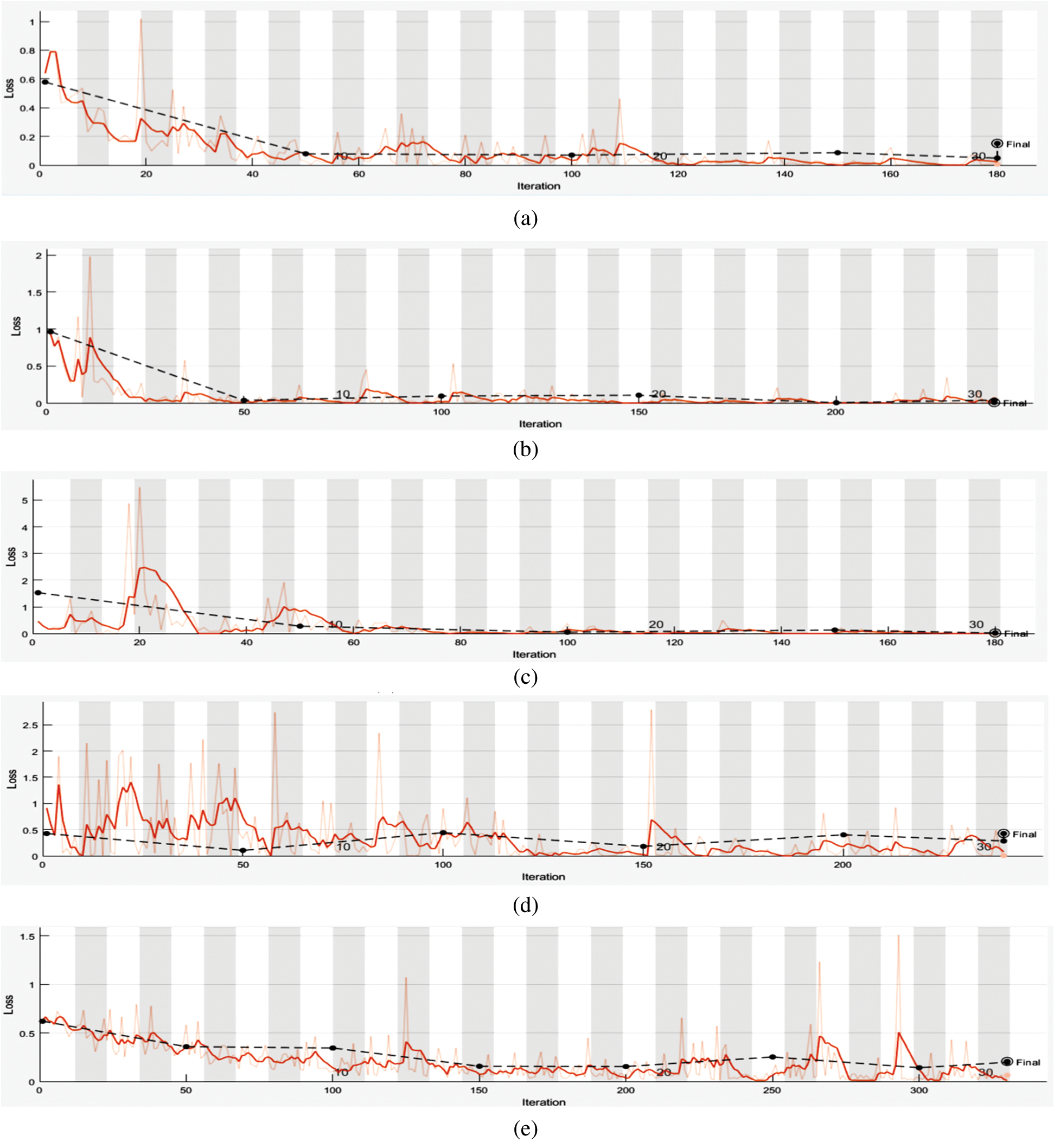
Figure 7: Training loss of the deep learning trained models (a) training loss for MobileNetsV2 (b) training loss for ResNet50 (c) training loss for GoogleNet (d) training loss for DarkNet (e) training loss for Xception
All models were evaluated using 5-validation method with 320 images used for training for each class (640 images) while the remaining 80 images per class are used for testing. The overall evaluation is considered by taking the mean of the evaluation metrics, as seen in Tab. 6.
From Tab. 6, it can be seen that ResNet50 has the best performance among the other four models with accuracy and F1 score of 98.8% respectively. This is due to the model architecture and its adaptability to relatively small datasets. For better performance analysis, the ROC curve is shown in Fig. 8.

Figure 8: The ROC analysis for the performance of the deep learning models
The current Limitations of our study can be summaries as follows; firstly, the limited dataset that publicly available to do more comprehensive evaluation experiments. More investigation is needed for deep learning models that applied for diagnosing COVID-19 cases. Also, more criteria are needed to consider in terms of selection for the most suitable diagnosing model for COVID-19 cases. The results of the study did not reflect the performance of transfer learning models and classifiers in terms of time complexity. Finally, the overall results are restricted only to the x-ray covid19 images dataset.
Several challenges are addressed and examined based on the experiment and deep analysis. First, limited publically available image dataset for COIVD-19 as a solution the only dataset provides is the X-ray image with small samples. However, this issue draws another challenge which is a small sample for feature extraction and training purposes. Secondly, transfer learning models appear as a solution for the previous challenge, but the different number of models made a new challenge to select the most suitable one. Thirdly, a comparison among three transfer models have been made based on the experiments, and the result revealed that the InceptionV3 is the most suitable model. Fourthly, in terms of a different number of classifiers, this issue has been solved based selection process in stage two. In this study, traditional machine learning and deep convolutional neural networks have been followed. In traditional machine learning, seven models have been developed to aid the detection of coronavirus (ANN, SVM with linear RBF, k-NN, DT, and CN 2 rule inducer). In such cases, the learning performance can be improved through knowledge transfer, whereby time-consuming data labelling efforts are not required. This problem can be solved through the use of transfer learning as novel learning frameworks. Here, the extraction of knowledge is done from different source tasks, and the application of the knowledge is made to a different target task. While in deep convolutional neural networks, five pre-trained models were selected, MobileNets V2, ResNet50, GoogleNet, DarkNet, and Xception. These pre-trained models have been nominated for their performance in classification and automatic feature extraction. The evaluations of all the models are done in terms of different parameters such as accuracy, precision, recall, AUC and F1 score. Based on the results obtained from the experiments, it can be concluded that all the models performed well, deep learning models had achieved the optimum accuracy of 98.8% in ResNet50 model. In comparison, in traditional machine learning techniques, the SVM demonstrated the best result for an accuracy of 95 % and RBF accuracy 94% for the prediction of coronavirus disease 2019. Also, it can be seen that ResNet50 has the best performance among the other four models with accuracy and F1 score of 98.8% respectively. Future work will include a hybrid multimodal deep learning system for improving COVID-19 pneumonia detection to try to obtain better results.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1 M. A. Mohammed, K. H. Abdulkareem, A. S. Al-Waisy, S. A. Mostafa, S. Al-Fahdawi. (2020). et al., “Benchmarking methodology for selection of optimal COVID-19 diagnostic model based on entropy and topsis methods,” IEEE Access, vol. 8, pp. 99115–99131. [Google Scholar]
2 World Health Organization. (2020). “Coronavirus disease 2019 (COVID-19Situation Report,” . [Online]. Available: https://apps.who.int/iris/handle/10665/331475./ [Google Scholar]
3 H. Chen, J. Guo, C. Wang, F. Luo, X. Yu. (2020). et al., “Clinical characteristics and intrauterine vertical transmission potential of Covid-19 infection in nine pregnant women: A retrospective review of medical records,” Lancet, vol. 395, no. 10226, pp. 809–815. [Google Scholar]
4 M. K. Abd Ghani, M. A. Mohammed, N. Arunkumar, S. A. Mostafa, D. A. Ibrahim. (2020). et al., “Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques,” Neural Computing and Applications, vol. 32, no. 3, pp. 625–638. [Google Scholar]
5 K. H. Abdulkareem, M. A. Mohammed, S. S. Gunasekaran, M. N. Al-Mhiqani, A. A. Mutlag. (2019). et al., “A review of fog computing and machine learning: Concepts, applications, challenges, and open issues,” IEEE Access, vol. 7, pp. 153123–153140. [Google Scholar]
6 S. A. Mostafa, A. Mustapha, M. A. Mohammed, R. I. Hamed, N. Arunkumar. (2019). et al., “Examining multiple feature evaluation and classification methods for improving the diagnosis of Parkinson’s disease,” Cognitive Systems Research, vol. 54, pp. 90–99. [Google Scholar]
7 M. A. Mohammed, B. Al-Khateeb, A. N. Rashid, D. A. Ibrahim, M. K. Abd Ghani. (2018). et al., “Neural network and multi-fractal dimension features for breast cancer classification from ultrasound images,” Computers & Electrical Engineering, vol. 70, pp. 871–882. [Google Scholar]
8 M. A. Mohammed, M. K. Abd Ghani, N. Arunkumar, R. I. Hamed, M. K. Abdullah. (2018). et al., “A real time computer aided object detection of nasopharyngeal carcinoma using genetic algorithm and artificial neural network based on Haar feature fear,” Future Generation Computer Systems, vol. 89, pp. 539–547. [Google Scholar]
9 N. Arunkumar, M. A. Mohammed, S. A. Mostafa, D. A. Ibrahim, J. J. Rodrigues. (2020). et al., “Fully automatic model-based segmentation and classification approach for mri brain tumor using artificial neural networks,” Concurrency and Computation: Practice and Experience, vol. 32, no. 1, e4962. [Google Scholar]
10 O. I. Obaid, M. A. Mohammed, M. K. Abd Ghani, S. A. Mostafa and F. Taha. (2018). “Evaluating the performance of machine learning techniques in the classification of Wisconsin Breast Cancer,” International Journal of Engineering & Technology, vol. 7, no. 4.36, pp. 160–166. [Google Scholar]
11 F. Shi, J. Wang, J. Shi, Z. Wu, Q. Wang. (2020). et al., “Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID-19,” IEEE Reviews in Biomedical Engineering, pp. 1–13. [Google Scholar]
12 D. Dansana, R. Kumar, J. Das Adhikari, M. Mohapatra, R. Sharma. (2020). et al., “Global forecasting confirmed and fatal cases of COVID-19 outbreak using autoregressive integrated moving average model,” Frontiers in Public Health, vol. 8, 580327. [Google Scholar]
13 Y. Fang, H. Zhang, J. Xie, M. Lin, L. Ying. (2020). et al., “Sensitivity of chest ct for COVID-19: Comparison to RT-PCR,” Radiology, vol. 296, no. 2, pp. 115–117. [Google Scholar]
14 T. Liang. (2020). Handbook of COVID-19 Prevention and Treatment. The First Affiliated Hospital, Zhejiang University School of Medicine, Zhejiang University, China. [Google Scholar]
15 H. Y. F. Wong, H. Y. S. Lam, A. H. T. Fong, S. T. Leung, T. W. Y. Chin. (2020). et al., “Frequency and distribution of chest radiographic findings in covid-19 positive patients,” Radiology, vol. 296, no. 2, 201160. [Google Scholar]
16 L. Brunese, F. Mercaldo, A. Reginelli and A. Santone. (2020). “Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays,” Computer Methods and Programs in Biomedicine, vol. 196, 105608. [Google Scholar]
17 J. P. Cohen, P. Morrison, L. Dao, K. Roth, T. Q. Duong. (2020). et al., “Covid-19 image data collection: Prospective predictions are the future,” arXiv preprint arXiv: 2006.11988, (Preprint). [Google Scholar]
18 C. L. Van, V. Puri, N. T. Thao and D. Le. (2021). “Detecting lumbar implant and diagnosing scoliosis from vietnamese X-ray imaging using the pre-trained api models and transfer learning,” Computers, Materials & Continua, vol. 66, no. 1, pp. 17–33. [Google Scholar]
19 S. H. Yoo, H. Geng, T. L. Chiu, S. K. Yu, D. C. Cho. (2020). et al., “Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging,” Frontiers in Medicine, vol. 7, 427. [Google Scholar]
20 J. P. Cohen, P. Morrison and L. Dao. (2020). “COVID-19 image data collection,” . [Online]. Available: https://github.com/ieee8023/covid-chestxray-dataset. [Google Scholar]
21 K. He, X. Zhang, S. Ren and J. Sun. (2016). “Deep residual learning for image recognition,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, pp. 770–778. [Google Scholar]
22 C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed. (2015). et al., “Going deeper with convolutions,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Boston, MA, USA, pp. 1–9. [Google Scholar]
23 M. Raghu, C. Zhang, J. Kleinberg and S. Bengio. (2019). “Transfusion: Understanding transfer learning for medical imaging,” in Advances in Neural Information Processing Systems, Vancouver, Canada, pp. 3342–3352. [Google Scholar]
24 P. Rajpurkar, J. Irvin, K. Zhu, B. Yang, H. Mehta. (2017). et al., “Chexnet: Radiologist-level pneumonia detection on chest X-rays with deep learning,” arXiv preprint arXiv: 1711.05225. [Google Scholar]
25 X. Wang, Y. Peng, L. Lu, Z. Lu, M. Bagheri. (2017). et al., “Chest X-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Honolulu, HI, USA, pp. 2097–2106.
26 S. S. Yadav and S. M. Jadhav. (2019). “Deep convolutional neural network based medical image classification for disease diagnosis,” Journal of Big Data, vol. 6, no. 1, pp. 113.
27 R. Vaishya, M. Javaid, I. H. Khan and A. Haleem. (2020). “Artificial intelligence (AI) applications for COVID-19 pandemic,” Diabetes & Metabolic Syndrome: Clinical Research & Reviews, vol. 14, no. 4, pp. 337–339.
28 M. D. Abràmoff, Y. Lou, A. Erginay, W. Clarida, R. Amelon. (2016). et al., “Improved automated detection of diabetic retinopathy on a publicly available dataset through integration of deep learning,” Investigative Ophthalmology & Visual Science, vol. 57, no. 13, pp. 5200–5206. [Google Scholar]
29 V. Gulshan, L. Peng, M. Coram, M. C. Stumpe, D. Wu. (2016). et al., “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” JAMA, vol. 316, no. 22, pp. 2402–2410. [Google Scholar]
30 J. De Fauw, J. R. Ledsam, B. Romera-Paredes, S. Nikolov, N. Tomasev. (2018). et al., “Clinically applicable deep learning for diagnosis and referral in retinal disease,” Nature Medicine, vol. 24, no. 9, pp. 1342–1350.
31 M. Raghu, K. Blumer, R. Sayres, Z. Obermeyer, R. Kleinberg. (2019). et al., “Direct uncertainty prediction for medical second opinions,” in Int. Conf. on Machine Learning, California, USA, pp. 5281–5290.
32 E. J. Topol. (2019). “High-performance medicine: The convergence of human and artificial intelligence,” Nature Medicine, vol. 25, no. 1, pp. 44–56.
33 A. A. Van Der Heijden, M. D. Abramoff, F. Verbraak, M. V. Van Hecke, A. Liem. (2018). et al., “Validation of automated screening for referable diabetic retinopathy with the idx-dr device in the hoorn diabetes care system,” Acta Ophthalmologica, vol. 96, no. 1, pp. 63–68. [Google Scholar]
34 Y. Ding, J. H. Sohn, M. G. Kawczynski, H. Trivedi, R. Harnish. (2019). et al., “A deep learning model to predict a diagnosis of Alzheimer disease by using 18f-fdg pet of the brain,” Radiology, vol. 290, no. 2, pp. 456–464. [Google Scholar]
35 A. Esteva, B. Kuprel, R. A. Novoa, J. Ko, S. M. Swetter. (2017). et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, no. 7639, pp. 115–118. [Google Scholar]
36 P. Khosravi, E. Kazemi, Q. Zhan, J. E. Malmsten, M. Toschi. (2019). et al., “Deep learning enables robust assessment and selection of human blastocysts after in vitro fertilization,” NPJ Digital Medicine, vol. 2, no. 1, pp. 1–9. [Google Scholar]
37 S. J. Pan and Q. Yang. (2009). “A survey on transfer learning,” IEEE Transactions on Knowledge and Data Engineering, vol. 22, no. 10, pp. 1345–1359. [Google Scholar]
38 L. Rampasek and A. Goldenberg. (2018). “Learning from everyday images enables expert-like diagnosis of retinal diseases,” Cell, vol. 172, no. 5, pp. 893–895.
39 A. Samanta, A. Saha, S. C. Satapathy, S. L. Fernandes and Y. D. Zhang. (2020). “Automated detection of diabetic retinopathy using convolutional neural networks on a small dataset,” Pattern Recognition Letters, vol. 135, pp. 293–298.
40 P. Chhikara, P. Singh, P. Gupta and T. Bhatia. (2020). “Deep convolutional neural network with transfer learning for detecting pneumonia on chest x-rays,” in Advances in Bioinformatics, Multimedia, and Electronics Circuits and Signals. Singapore: Springer, pp. 155–168. [Google Scholar]
41 C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna. (2016). “Rethinking the inception architecture for computer vision,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, pp. 2818–2826. [Google Scholar]
42 C. Szegedy, S. Ioffe, V. Vanhoucke and A. A. Alemi. (2017). “Inception-v4, inception-resnet and the impact of residual connections on learning,” in Proc. of the Thirty-First AAAI Conf. on Artificial Intelligence, California, USA, pp. 4278–4284. [Google Scholar]
43 F. Ucar and D. Korkmaz. (2020). “COVIDiagnosis-Net: Deep Bayes-SqueezeNet based diagnostic of the coronavirus disease 2019 (COVID-19) from X-ray images,” Medical Hypotheses, vol. 140, p. 109761. [Google Scholar]
44 T. Le Jordan. (2019). “The improvement of machine learning accuracies through transfer learning,” Bachelor dissertation, University Honors. [Google Scholar]
45 N. Arunkumar, M. A. Mohammed, M. K. Abd Ghani, D. A. Ibrahim, E. Abdulhay. (2019). et al., “K-means clustering and neural network for object detecting and identifying abnormality of brain tumor,” Soft Computing, vol. 23, no. 19, pp. 9083–9096. [Google Scholar]
46 M. A. Mohammed, M. K. Abd Ghani, N. Arunkumar, S. A. Mostafa, M. K. Abdullah. (2018). et al., “Trainable model for segmenting and identifying Nasopharyngeal carcinoma,” Computers & Electrical Engineering, vol. 71, pp. 372–387. [Google Scholar]
47 M. A. Mohammed, M. K. Abd Ghani, R. I. Hamed, S. A. Mostafa, D. A. Ibrahim. (2017). et al., “Solving vehicle routing problem by using improved K-nearest neighbor algorithm for best solution,” Journal of Computational Science, vol. 21, pp. 232–240. [Google Scholar]
48 A. Cutler, D. R. Cutler and J. R. Stevens. (2012). “Random forests,” in Ensemble Machine Learning. Boston, USA: Springer, pp. 157–175. [Google Scholar]
49 M. A. Friedl and C. E. Brodley. (1997). “Decision tree classification of land cover from remotely sensed data,” Remote Sensing of Environment, vol. 61, no. 3, pp. 399–409. [Google Scholar]
50 A. Wilcox and G. Hripcsak. (1999). “Classification algorithms applied to narrative reports,” in Proc. of the Amia Sym., Washington DC, USA: American Medical Informatics Association, pp. 455–459. [Google Scholar]
51 M. A. Mohammed, K. H. Abdulkareem, S. A. Mostafa, M. K. Abd Abd Ghani, M. S. Maashi. (2020). et al., “Voice pathology detection and classification using convolutional neural network model,” Applied Sciences, vol. 10, no. 11, pp. 3723. [Google Scholar]
52 S. Zahia, D. Sierra-Sosa, B. Garcia-Zapirain and A. Elmaghraby. (2019). “Tissue classification and segmentation of pressure injuries using convolutional neural networks,” Computer Methods and Programs in Biomedicine, vol. 159, pp. 51–58. [Google Scholar]
53 M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. C. Chen. (2018). “Mobilenetv2: Inverted residuals and linear bottlenecks,” in Proc. of the IEEE Conf. on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 4510–4520. [Google Scholar]
54 H. Takahashi, H. Tampo, Y. Arai, Y. Inoue and H. Kawashima. (2017). “Applying artificial intelligence to disease staging: Deep learning for improved staging of diabetic retinopathy,” PLoS One, vol. 12, no. 6, e0179790. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |