DOI:10.32604/cmc.2020.013232

| Computers, Materials & Continua DOI:10.32604/cmc.2020.013232 |  |
| Article |
Automatic Detection of COVID-19 Using Chest X-Ray Images and Modified ResNet18-Based Convolution Neural Networks
1University of Babylon, Babylon, 51002, Iraq
2Kufa University, Kufa, 54003, Iraq
*Corresponding Author: Ruaa A. Al-Falluji. Email: fine.ruaa.adeeb@uobabylon.edu.iq
Received: 30 July 2020; Accepted: 12 September 2020
Abstract: The latest studies with radiological imaging techniques indicate that X-ray images provide valuable details on the Coronavirus disease 2019 (COVID-19). The usage of sophisticated artificial intelligence technology (AI) and the radiological images can help in diagnosing the disease reliably and addressing the problem of the shortage of trained doctors in remote villages. In this research, the automated diagnosis of Coronavirus disease was performed using a dataset of X-ray images of patients with severe bacterial pneumonia, reported COVID-19 disease, and normal cases. The goal of the study is to analyze the achievements for medical image recognition of state-of-the-art neural networking architectures. Transfer Learning technique has been implemented in this work. Transfer learning is an ambitious task, but it results in impressive outcomes for identifying distinct patterns in tiny datasets of medical images. The findings indicate that deep learning with X-ray imagery could retrieve important biomarkers relevant for COVID-19 disease detection. Since all diagnostic measures show failure levels that pose questions, the scientific profession should determine the probability of integration of X-rays with the clinical treatment, utilizing the results. The proposed model achieved 96.73% accuracy outperforming the ResNet50 and traditional Resnet18 models. Based on our findings, the proposed system can help the specialist doctors in making verdicts for COVID-19 detection.
Keywords: COVID-19; artificial intelligence; convolutional neural network; chest x-ray images; Resnet18 model
Coronaviruses are a large family of viruses that can infect humans or animals. In humans, numerous coronaviruses are known for causing infections of the respiratory system. The severity of coronavirus infection ranges from cold into much more deadly illnesses like the Middle East Respiratory Syndrome (MERS) and the Severe Acute Respiratory Syndrome (SARS). COVID-19 is an extremely transmissible disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The virus was initiated from Wuhan, China in December 2019 and has subsequently spread all over the world, causing an outbreak in more than 200 countries. The effect is such that the World Health Organization (WHO) has decided to declare the ongoing COVID-19 pandemic as an international public health emergency. Governments are working hard to close borders, track contacts, identify and isolate the affected and suspected cases, isolate the likely cases, but the number of positive COVID-19 cases is growing exponentially in most countries and, unfortunately, is expected to increase until a medicine/vaccine can be developed and made available in the public domain. In the absence of any effective medical treatment of COVID-19, it becomes utmost important to trace and isolate the patients so that virus infection can be restricted. As of May 2020, positive COVID-19 cases are rising rapidly all over the world, as can be observed in Fig. 1. Whereas Fig. 2 lists out the worst ten countries in the world that are affected by COVID-19 [1].
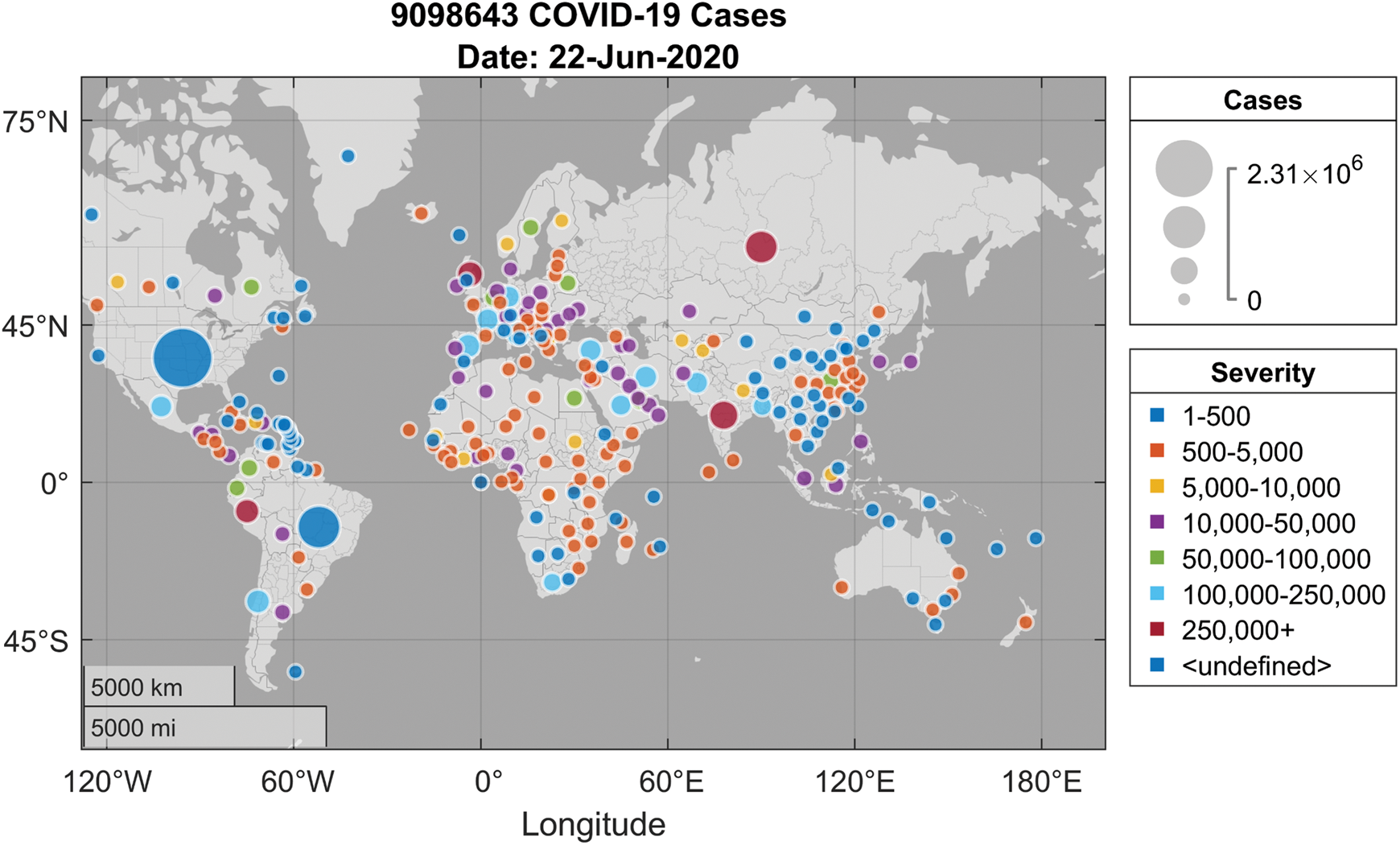
Figure 1: COVID-19 spread around the globe [1]
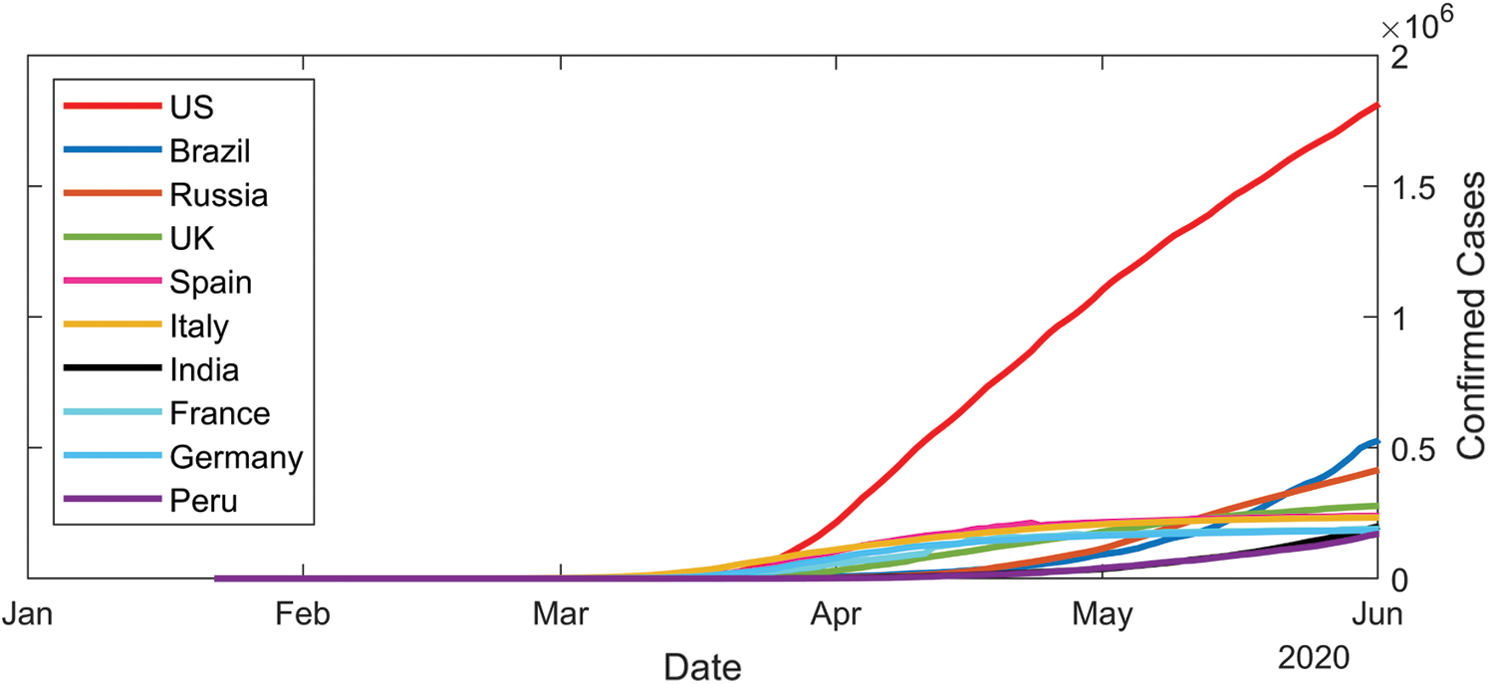
Figure 2: Top ten countries with COVID-19 cases as of June 2020 [1]
Reverse Transcription Polymerase Chain Reaction (RT-PCR) is currently the most used technique for COVID-19 detection in humans. Due to the lack of RT-PCR tests in many countries, the symptoms are found through the examination of radiological images of the patients [2,3].
Computed Tomography (CT) is a sensitive way of detecting pneumonia in COVID-19 that can be viewed as an RT-PCR testing tool [4]. After the symptoms begin, and typically in the first 2 days, patients should report regular CT [5]. CT results can be used to detect if the person suffers from COVID-19 and determine the degree of seriousness at the same time [6]. Chinese health centers initially had inadequate test sets, which also caused a large rate of false-negative tests, such that physicians are advised to diagnosis only based on clinical and thoracic CT findings [5,7]. In places like Turkey, where there are small numbers of test kits at the outset of a pandemic, CT is used extensively for COVID-19 detection. Researchers have stated that CT can help to early detect COVID-19 [7–11]. For COVID-19, radiological images provide valuable diagnostic details. Many important findings about COVID-19 detection using images were obtained by researchers overall the world. Danis et al. [12] observed the existence of unusual opacity in the radiographical image of a COVID-19 patient. The authors in [13] registered a single nodular opacity in the lower left lung area in one of each three patients under review. A further finding is that peripheral focus or multifocal ground-glass opacification (GGOs) can impact both lungs in 50–75% of patients [10]. Similarly, the authors in [3] found that 33% of chest CT scans are circular in the lung. The usage of the machine learning algorithms in automatica detection of diseases has become increasingly popular lately as a supplementary tool for clinicians [14–19].
The artificial intelligence-based research facilitates the development of end-to-end models to get promising results without input being extracted manually [20,21]. In other issues such as the diagnosis of arrhythmias [22,23], skin cancer detection [24,25], breast cancer identification [26,27], brain diseases classification [28], pneumonia diagnosis using chest X-rays images [29], fundus images [30], and pulmonary segmentation [31,32], deep learning strategies have been used effectively. The massive increase of the COVID-19 outbreak encouraged the usage of automatic AI-based detection systems, as they could result in a rapid detection of the infected cases and help in their quick isolation which will in turn reduce the infection spread. Fast, accurate, and rapid AI models will therefore help in overcoming the lack of specialist doctors, and providing the patients with timely support [33,34]. AI approaches can also help in handling some problems, such as inadequate RT-PCR test kits, and the long waiting time for test results. Many types of radiological images were recently used for COVID-19 detection. For the diagnosis of COVID-19 using X-ray images, the authors in [34] suggested a COVIDX-Net model consisting of seven CNN models. Wang and Wong suggested a broad model (COVID-Net) to classify COVID-19 and other pneumonia types. Using 224 COVID-19 verified images, Apostolopoulos et al. [35] established the profound learning model. In both two and three grades, their model reached 98.75% and 93.48% performance respectively. While Narin et al. [36] accomplished 98% COVID-19 identification using chest X-ray imaging in conjunction with the ResNet 50 model.
The features gathered from multiple CNN models utilizing X-ray images with a Support Vector Machine (SVM) classifier were proposed by Sethy et al. [37]. Their analysis indicates the highest results of the ResNet50 model is obtained with the SVM classifier. Finally, a variety of recent research on the COVID-19 prediction utilizing numerous CT images in profound schooling [38–42] has also been published. Different end-to-end designs were proposed without any practical methods of abstraction and including the replication of raw chest X-ray images which have been easily collected. In recovering patients, medical tests performed between 5–13 days have been positive [43]. This indicates that the infection may also transmit to recovering patients. One of the main downsides to the chest X-ray researches is the inability to identify COVID-19 in its early phases since the exposure to GGO detection is not adequate. However, a well-trained network can concentrate and restore the interpretation of points that are non-detectable to humans.
Although many models have been proposed and implemented but more reliable diagnostic approaches are still required. Therefore, the aim of this study is to propose a deep learning framework that automatically detects COVID-19 from X-ray images. The modified CNN network is proposed in the study and it significantly improved the detection of COVID-19. The used data in this study was obtained from various sources and the preprocessing was carried out. The results of this work are summarized below:
a) Develop a deep CNN network to ensure the successful detection of COVID-19 positive cases.
b) One of the primary solutions of this study is to continue validating and investigating the possibility of reducing Resnet18 model complexity, the generalization of the classical machine learning methods, and producing useful features for training instead of the conventional training by using predefined raw data features.
c) The developed system could be used in the hospitals and medical centers to make rapid screening and efficient detection for COVID-19 positive cases.
The main contribution of the modified Resnet18 CNN model is that numerous pre-extraction and features estimation work is reduced to conventional methods, and it is suitable for grayscale images which is fit for X-ray based classification. Our proposed CNN network is more effective than the other reported studies as the results show.
The structure of the paper is as follows. The methodology of the work is illustrated in Section 2. Section 3 defines the used dataset, the CNN architecture, the results of this research as well as the discussions of the obtained results. The conclusion of this analysis is addressed in Section 4.
Through this research, authors have developed a CNN model for the detection of COVID-19 using chest X-ray images, driven by the fact that a deep learning approach could help to properly detect and diagnose COVID-19. In the absence of adequate technology, the radiologists first need to differentiate against COVID-19 X-rays images from normal chest X-ray images, and then, from other viral and bacterial infections. Hence, we select CNN’s architecture to detect one of the following classes:
a) Normal (i.e., no infection),
b) COVID-19,
c) and Pneumonia
The obtained classes from the proposed model can help specialists in making their decisions about the infection (if any) and its type and restricting the RT-PCR test for only questionable cases, which helps in reducing the infection through decreasing contact between the patient and the specialist. The authors have built an end-to-end model by modifying the original residual ResNet18 network structure as it is illustrated in Fig. 3 to classify and predict COVID-19 from the radiographical images. Fig. 4 shows the network pipeline in this paper. The initial ResNet18 model is suitable for the color images, while the modified ResNet18 is more appropriate for the grayscale images which are used to diagnose infections in this study. The average pooling layer of the original ResNet18 is replaced with the Global Average Pooling layer (GAP) and two compression layers are added to support the image rating after a global average pooling layer.
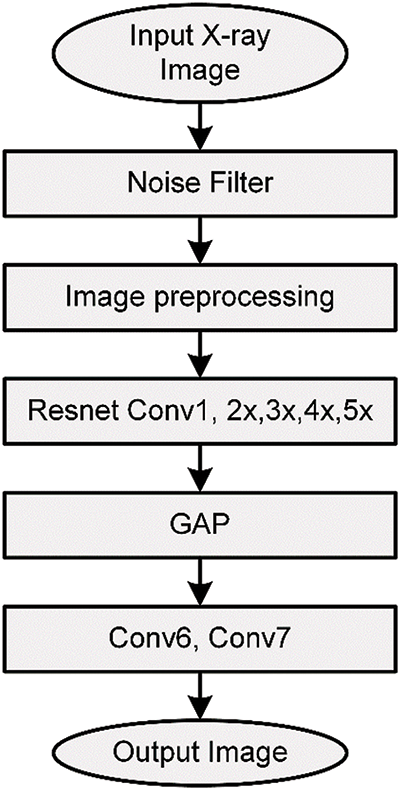
Figure 3: The network operation performed in this work
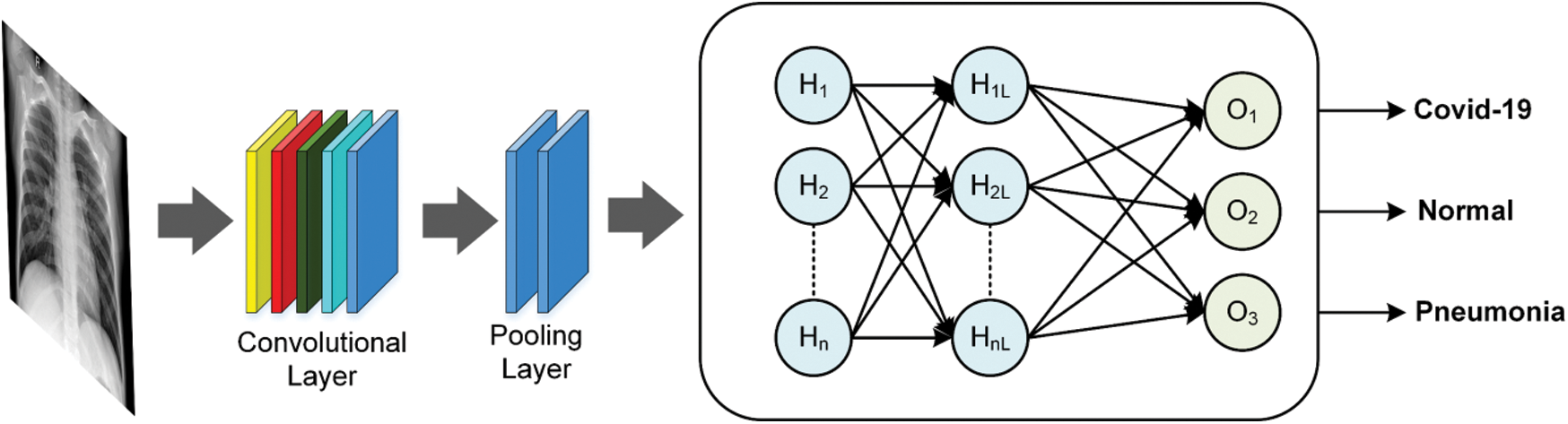
Figure 4: Conventional building blocks of CNN
The model achieves the performance likelihood after the convolutionary layer of each form of label. The cross-entropy loss function is used in this work, to prevent the issue of sluggish learning:
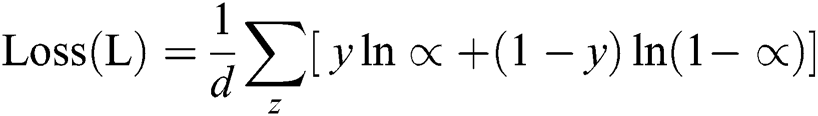
where d is the total number of training data. The summation is performed on all inputs of training, y, z, and their respective target output. The modified network is shown in Fig. 5 and its structure is as follows:
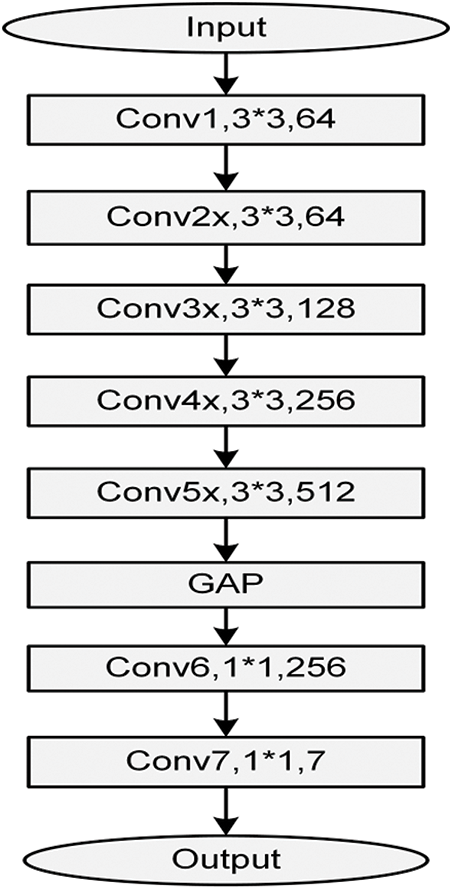
Figure 5: The modified ResNet18 network
It contains image data, and image data are denoted by a 3D matrix. And this needs to be resized into one column. Example: 28 × 28 image is transformed into 784 × 1 before inputs to the system.
The convolutional layer performs an important role in the CNN model. Inside this layer, the features of the image get extracted. Convolution conserves the contiguous correlation in between elements by reading image features getting help from little pores of feeding images. Numbers of filters are used for the convolution process and an activation map is generated to be feed as input to the next layer of CNN.
The pooling layer is used to reduce the size after the convolution of feeding images. It can be used in between two convolutional layers. If applying a fully connected layer after two convolutional layers without applying average or max pooling, then the computations and amount of the parameters will be very high. The pooling layer provides better results against certain transformations.
A fully connected layer contains kernels, weights, and biases. It generally relates a one-layer kernel with a kernel of the next corresponding layer. It is used to categorize images in between certain types via training. A fully connected layer can be referred to as finishing the pooling layer input the features to the ReLU or Softmax activation function (classifier).
It is the final layer of the CNN model that is placed at the last of the fully connected layer.
This is the output layer of CNN and it includes the tag that will be in the shape of single-hard encoded. The used hyper-parameters for training are as follows: learning rates = 0.001, beta = 0.9, and batch size = 16. The network is equipped for 30 epochs.
The authors included two open-source datasets in their study. Chest X-ray images for COVID-19 positive cases were taken from the GitHub repository, prepared by the authors in [44,45]. Samples of the used dataset in this study are shown in Fig. 6.
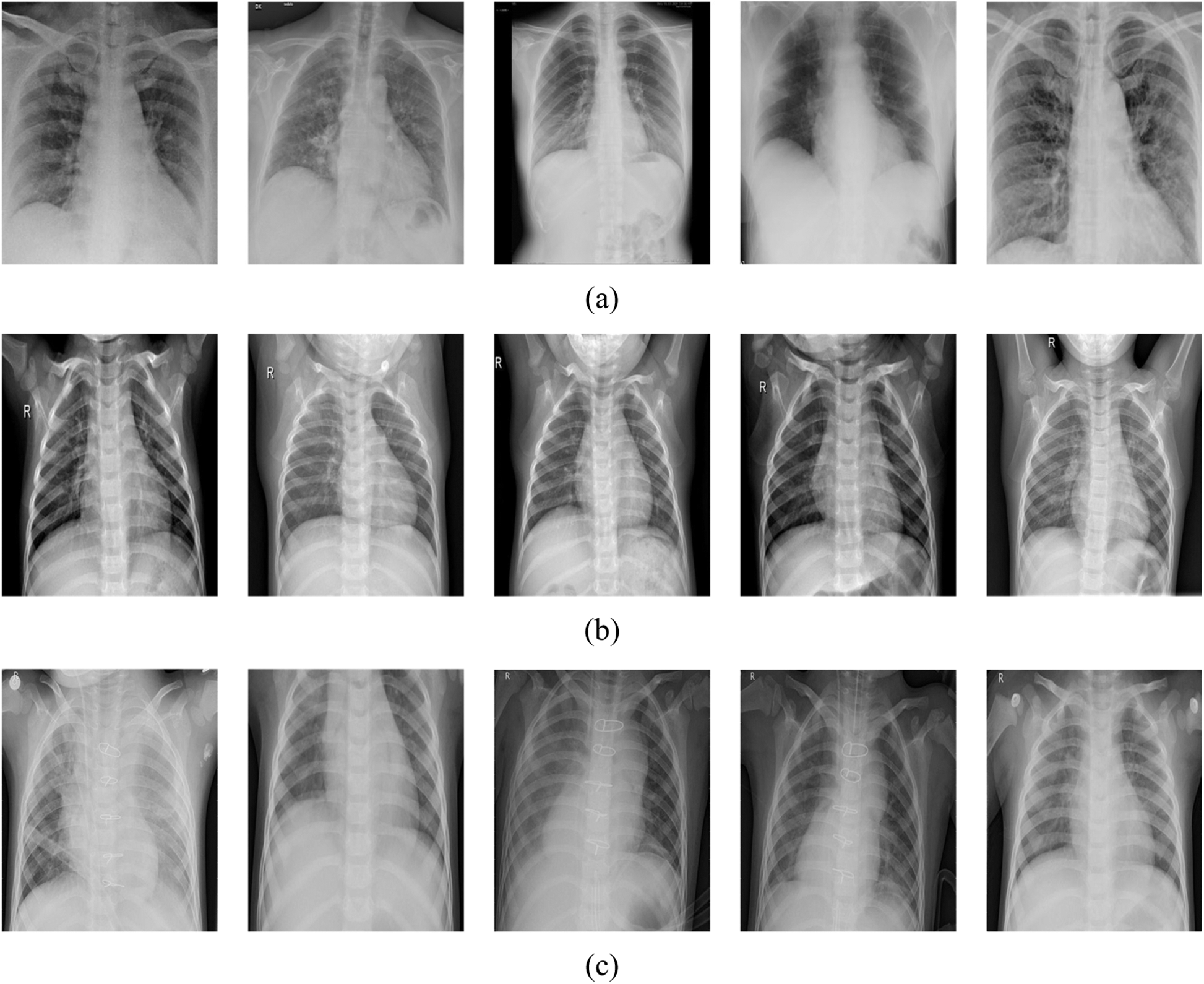
Figure 6: Samples of the used dataset in this project, (a) positive COVID-19, (b) normal, (c) pneumonia
In this research, three classes of images were used. X-ray images of positive COVID-19, ARDS, SARS, Streptococcus, Pneumocystis, and other pneumonia types were employed. The first dataset contains only 180 images for 118 COVID-19 cases and the remaining 42 images for 25 cases of pneumocystis, Streptococcus, and SARS were identified as pneumonia. The second dataset includes 6012 pneumonia cases and 8851 normal cases. As stated, only 180 positive COVID-19 instances are available, which are little data for one class in comparison to other classes. Mixing large numbers of normal and pneumonia images with only few COVID-19 images for training leads to identifying pneumonia and normal cases more effectively by the network than COVID-19 cases due to unbalanced dataset.
Data augmentation is used after handling the size differences of the collected images and converting all the images into the same scale. Data augmentation is a tool used when the dataset is small to artificially produce further examples from the same dataset instead of gathering more data. Since the size of the dataset greatly affects the performance of the machine and deep learning techniques, data augmentation is employed. We implemented some techniques on the original images to get more data such as rotation (left-right, top-down) with a probability of 0.3, resizing with a probability of 0.1, random light with a probability of 0.5, and zooming throughout 0.7.
Although data augmentation is implemented and the whole data are expanded, the problem of unbalanced classes still needs a solution. The average performance cannot be used because the images in normal and pneumonia classes are more than in the COVID-19 class. The easiest approach to address this issue is to align the data collection and supply the network with approximately identical data from each class during the preparation process which enables the network to understand how to classify each class. In this case, since we have no access to other open-source COVID-19 datasets to increase this data, we have chosen the amount of images from normal and pneumonia classes almost equals to the amount of the images of COVID-19 class. In this way, the network receives about the same number of images per batch, thereby helping to boost the identification of COVID-19 with the identification of pneumonia and normal cases as well. There are many benefits to applying this solution. The network integrates more for the other levels of COVID-19 functionality and the identification of normal and pneumonia classes dramatically increases. This approach also makes the network easier to distinguish COVID-19 and not to spot faulty COVID-19 cases.
Popular pre-trained models such as ResNet50, ResNet18, and the proposed modified ResNet18 network have been trained and examined using the chest x-ray images to predict coronavirus disease. The pre-trained model fold-3 confusion matrices are shown in Fig. 7.
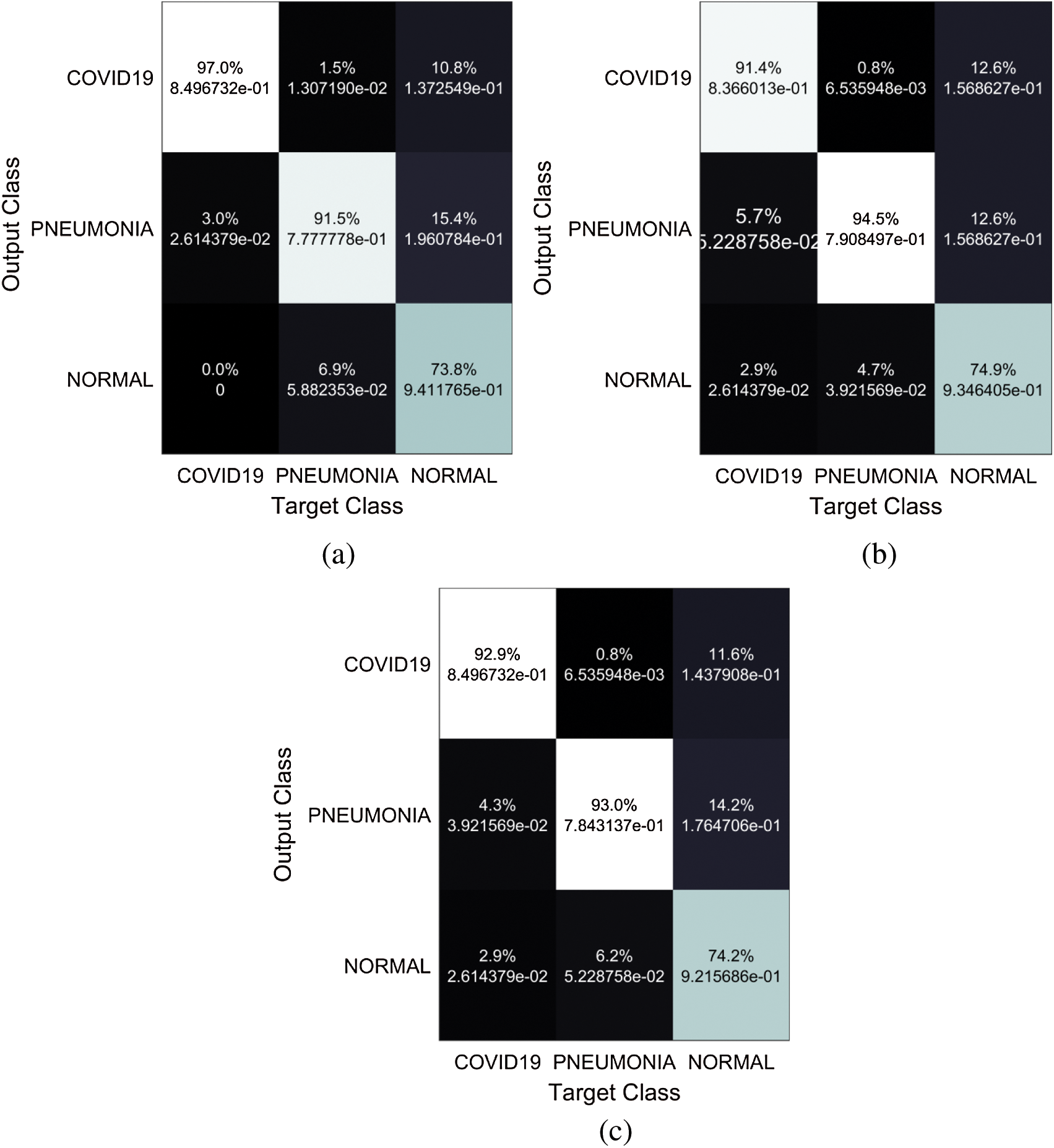
Figure 7: Confusion matrices of the networks (a) proposed network, (b) Resnet 18, (c) Resnet 50
The training stage was implemented for all models until the 30th epoch to prevent overfitting. To evaluate the performance of the proposed model, the accuracy is calculated according to the following equation:
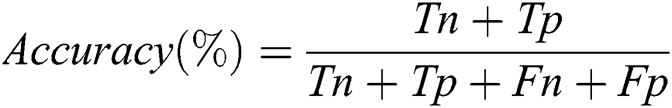
Where Tn is the number of correct predictions that an instance is negative, Fp is the number of incorrect predictions that an instance is positive, Fn is the number of incorrect predictions that an instance is negative, and Tp is the number of correct predictions that an instance is positive [46]. Fig. 8 shows that the highest training accuracy is achieved by the proposed ResNet18 model.
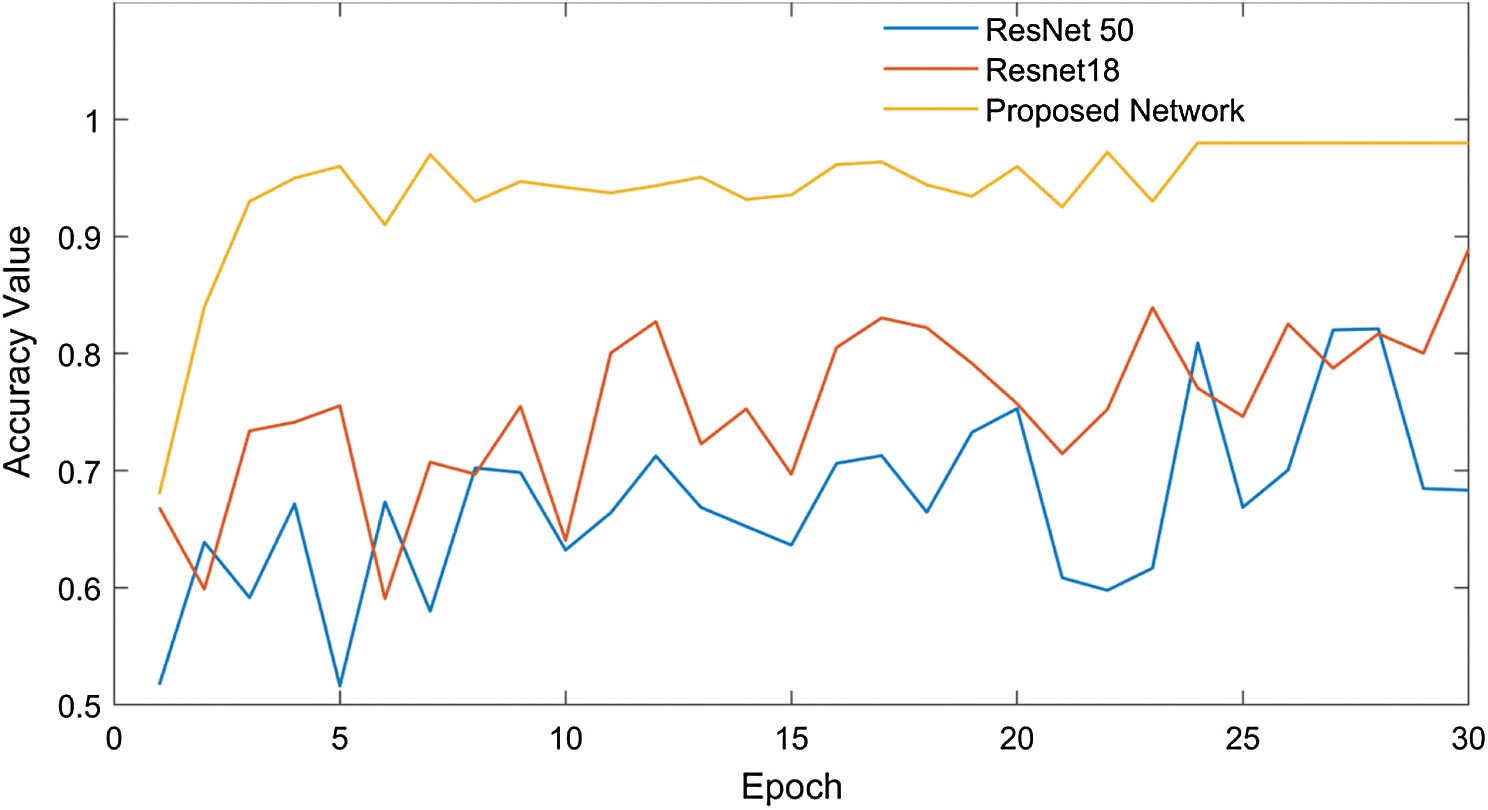
Figure 8: Performance evaluation of the ResNet18 model
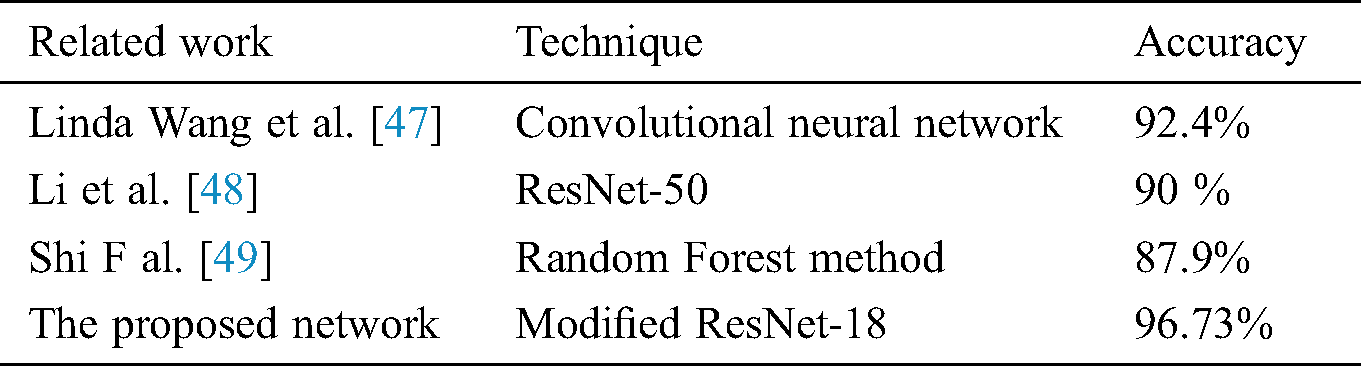
The proposed model has better efficiency and accuracy than the previously reported work as the comparison shows in Tab. 1. The proposed model which is based on a modified ResNet18 method outperforms the previous COVID-19 detection models. It resulted in 96.73% accuracy, 94% recall, and 100% specificity. The lowest performance values were obtained through using the ResNet50 network. The results were 87% for accuracy, 84% for recall, and 90 % for specificity. Therefore, the proposed network performs superiorly in both training and the testing stages when it is compared to the other two models in this study.
Table 1: A comparison of the different CNN network employed to detect COVID-19 with the proposed network
The mentioned matrices indicate that the network concatenated functions are best to identify and perfectly recognize COVID-19 and the other cases. We have evaluated our convolutional neural networks with a large number of images although the problem of unbalanced data, unlike other studies that have identified COVID-19 from few X-ray images (only 25 to 50 images are used in some studies). We presented the results with more practical samples for each class by using data augmentation to overcome the problem of unbalanced samples.
Although the suggested model can diagnose COVID-19 in seconds with high accuracy, but it still has some drawbacks. First of all, the dataset size is not fixed, relatively small, and constantly updated. The generalization of the used method must be checked over a larger dataset which can be implemented if more images are publicly available. Furthermore, the proposed model focuses mainly on the posterior-anterior (PA) view of the chest X-ray images and it cannot be used to distinguish other X-ray views such as the anterior-posterior (AP), the lateral and so on. Also, we did not match the efficiency of our model with the radiologists and specialist doctors.
As the number of COVID-19 cases grows every day, it is a necessity to detect every positive COVID-19 case as soon as possible, although the resources’ deficiency in many countries. The primary and fast diagnosis of COVID-19 infection is very important and plays a vital role in preventing the spread of diseases to other humans. In this article, and as a trail to reduce the direct contact that occurs between the patient and the specialist during the RT-PCR test, we proposed an approach to automatically predict COVID-19 infections using chest X-ray images and deep transfer learning where CNN is used as a classifier. The modifications have been performed on the ResNet18 network to enhance its performance in the classification of chest X-ray images. Performance results show that 96.73% of the accuracy of the three classes (normal, pneumonia, and COVID-19) was obtained using the updated ResNet18 model. Our detailed experimental results show how the proposed architecture outperforms the existing CNN based models for chest X-ray images classification. We expect that our network would be beneficial for medical diagnosis. We also hope that larger COVID-19 datasets will be accessible nearly and the efficiency of our proposed network would be more improved using them.
Funding Statement: The author(s) received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
1. Worldometer coronavirus updates, [Online]. Available: https://www.worldometers.info/coronavirus/ 30 May 2020. [Google Scholar]
2. J. P. Kanne, B. P. Little, J. H. Chung, B. M. Elicker and L. H. Ketai. (2020). “Essentials for radiologists on COVID-19: An update—Radiology scientific expert panel,” Radiology, vol. 296, no. 2, pp. E113–E114. [Google Scholar]
3. X. Xie, Z. Zhong, W. Zhao, C. Zheng. (2020). F. Wanget al., “Chest CT for typical 2019-nCOV pneumonia: Relationship to negative RT-PCR testing,” Radiology, vol. 296, no. 2, pp. E41–E45. [Google Scholar]
4. E. Y. P. Lee, M.Y. Ng and P. L. Khong. (2020). “COVID-19 pneumonia: What has CT taught us?,” Lancet Infectious Diseases, vol. 20, no. 4, pp. 384–385. [Google Scholar]
5. A. Bernheim, X. Mei, M. Hung, Y. Yang. (2020). Z. A. Fayadet al., “Chest CT findings in coronavirus disease-19 (COVID-19Relationship to duration of infection,” Radiology, vol. 295, no. 3, pp. 685–691. [Google Scholar]
6. F. Pan, T. Ye, P. Sun, S. Gui. (2020). B. Lianget al., “Time course of lung changes at chest CT during recovery from coronavirus disease 2019 (COVID-19),” Radiology, vol. 295, no. 3, pp. 715–721. [Google Scholar]
7. M. Abdel-Basset, R. Mohamed, M. Elhoseny, R. K. Chakrabortty and M. Ryan. (2020). “A hybrid COVID-19 detection model using an improved marine predators algorithm and a ranking-based diversity reduction strategy,” IEEE Access, vol. 8, pp. 79521–79540. [Google Scholar]
8. H. Shi, X. Han, N. Jiang, Y. Cao. (2020). O. Alwalidet al., “Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: A descriptive study,” Lancet Infectious Diseases, vol. 20, no. 4, pp. 425–434. [Google Scholar]
9. M. A. Mohammed, K. H. Abdulkareem, A. S. Al-Waisy, S. A. Mostafa. (2020). S. Al-Fahdawiet al., “Benchmarking methodology for selection of optimal COVID-19 diagnostic model based on entropy and TOPSIS methods,” IEEE Access, vol. 8, pp. 99115–99131. [Google Scholar]
10. A. Waheed, M. Goyal, D. Gupta, A. Khanna. (2020). F. Al-Turjman et al., “CovidGAN: Data augmentation using auxiliary classifier GAN for improved COVID-19 detection,” IEEE Access, vol. 8, pp. 91916–91923. [Google Scholar]
11. J. F. W. Chan, S. Yuan, K. H. Kok, K. K. W. To. (2020). H. Chuet al., “A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster,” Lancet, vol. 395, no. 10223, pp. 514–523. [Google Scholar]
12. K. Danis, O. Epaulard, T. Bénet, A. Gaymard. (2020). S. Campoyet al., “Cluster of coronavirus disease 2019 (COVID-19) in the French Alps,” Clinical Infectious Diseases, vol. 71, no. 15, pp. 825–832. [Google Scholar]
13. S. H. Yoon, K. H. Lee, J. Y. Kim, Y. K. Lee, H. Ko. (2020). et al., “Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID-19Analysis of nine patients treated in Korea,” Korean Jounal of Radiology, vol. 21, no. 4, pp. 494–500. [Google Scholar]
14. F. Rustam, A. A. Reshi, A. Mehmood, S. Ullah, W. B. On. (2020). et al., “COVID-19 future forecasting using supervised machine learning models,” IEEE Access, vol. 8, pp. 101489–101499. [Google Scholar]
15. E. Hernández-Orallo, P. Manzoni, C. T. Calafate and J. C. Cano. (2020). “Evaluating how smartphone contact tracing technology can reduce the spread of infectious diseases: The case of COVID-19,” IEEE Access, vol. 8, pp. 99083–99097. [Google Scholar]
16. P. Staszkiewicz, I. Chomiak-Orsa and I. Staszkiewicz. (2020). “Dynamics of the COVID-19 contagion and mortality, country factors, social media, and market response evidence from a global panel analysis,” IEEE Access, vol. 8, pp. 106009–106022. [Google Scholar]
17. J. Fiaidhi. (2020). “Envisioning insight-driven learning based on thick data analytics with focus on healthcare,” IEEE Access, vol. 8, pp. 114998–115004. [Google Scholar]
18. M. K. Abd Ghani, M. A. Mohammed, N. Arunkumr, S. A. Mostafa,D. A. Ibrahim. (2020). et al., “Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques,” Neural Computing and Applications, vol. 32, pp. 625–638. [Google Scholar]
19. Y. LeCun, Y. Bengio and G. Hinton. (2015). “Deep learning,” Nature, vol. 521, pp. 436–444. [Google Scholar]
20. A. Krizhevsky, I. Sutskever and G. E. Hinton. (2017). “ImageNet classification with deep convolutional neural networks,” Communications of the ACM, vol. 60, no. 6, pp. 84–90. [Google Scholar]
21. M. A. Mohammed, K. H. Abdulkareem, S. A. Mostafa, M. K. Abd Ghani,M. S. Maashi. (2020). et al., “Voice pathology detection and classification using convolutional neural network model,” Applied Sciences, vol. 10, no. 11, p. 3723. [Google Scholar]
22. A. Y. Hannun, P. Rajpurkar, M. Haghpanahi, G. H. Tison,C. Bourn. (2019). et al., “Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network,” Nature Medicine, vol. 25, pp. 65–69. [Google Scholar]
23. U. R. Acharya, S. L. Oh, Y. Hagiwara, J. H. Tan,M. Adam. (2017). et al., “A deep convolutional neural network model to classify heartbeats,” Computers in Biology and Medicine, vol. 89, pp. 389–396. [Google Scholar]
24. A. Esteva, B. Kuprel, R. A. Novoa, J. Ko,S. M. Swetter. (2017). et al., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature, vol. 542, pp. 115–118. [Google Scholar]
25. N. C. F. Codella, Q. B. Nguyen, S. Pankanti, D. A. Gutman,B. Helba. (2017). et al., “Deep learning ensembles for melanoma recognition in dermoscopy images,” IBM Journal of Research and Development, vol. 61, no. 4–5, pp. 5:1–5:15. [Google Scholar]
26. Y. Celik, M. Talo, O. Yildirim, M. Karabatak and U. R. Archarya. (2020). “Automated invasive ductal carcinoma detection based using deep transfer learning with whole-slide images,” Pattern Recognition Letters, vol. 133, pp. 232–239. [Google Scholar]
27. A. Cruz-Roa, A. Basavanhally, F. González, H. Gilmore,M. Feldman. (2014). et al., “Automatic detection of invasive ductal carcinoma in whole slide images with convolutional neural networks,” in Proc. SPIE Medical Imaging, San Diego, California, USA. [Google Scholar]
28. M. Talo, O. Yildirim, U. B. Baloglu, G. Aydin,U. R. Archrya. (2019). et al., “Convolutional neural networks for multi-class brain disease detection using MRI images,” Computerized Medical Imaging and Graphics, vol. 78, p. 101673. [Google Scholar]
29. P. Rajpurkar, J. Irvin, K. Zhu, B. Yang,H. Mehtaet al., “CheXNet: Radiologist-level pneumonia detection on chest x-rays with deep learning.” arxiv preprint arxiv: 1711. 05225, 2017. [Google Scholar]
30. J. H. Tan, H. Fujita, S. Sivaprasad, S. V. Bhandary,A. K. Rao. (2017). et al., “Automated segmentation of exudates, haemorrhages, microaneurysms using single convolutional neural network,” Information Sciences, vol. 420, no. c, pp. 66–76. [Google Scholar]
31. G. Gaál, B. Maga and A. Lukács. (2020). “Attention U-Net based adversarial architectures for chest x-ray lung segmentation,” arxiv preprint arxiv:2003.10304. [Google Scholar]
32. J. C. Souza, J. O. B. Diniz, J. L. Ferreira, G. L. F. Silva,A. C. Silva. (2019). et al., “An automatic method for lung segmentation and reconstruction in chest x-ray using deep neural networks,” Computer Methods and Programs in Biomedicine, vol. 177, pp. 285–296. [Google Scholar]
33. F. Caobelli. (2020). “Artificial intelligence in medical imaging: Game over for radiologists?,” European Journal of Radiology, vol. 126, p. 108940. [Google Scholar]
34. E. E. Hemdan, M. A. Shouman and M. E. Karar. (2020). “COVIDX-Net: A framework of deep learning classifiers to diagnose COVID-19 in x-ray images,” arxiv preprint arxiv:2003.11055. [Google Scholar]
35. I. D. Apostolopoulos and T. A. Mpesiana. (2020). “COVID-19: Automatic detection from x-ray images utilizing transfer learning with convolutional neural networks,” Physical and Engineering Sciences in Medicine, vol. 43, no. 2, pp. 635–640. [Google Scholar]
36. A. Narin, C. Kaya and Z. Pamuk. (2020). “Automatic detection of coronavirus disease (COVID-19) using x-ray images and deep convolutional neural networks,” arxiv preprint arxiv:2003.10849. [Google Scholar]
37. P. K. Sethy, S. K. Behera, P. K. Ratha and P. Biswas. (2020). “Detection of coronavirus disease (COVID-19) based on deep features and support vector machine,” International Journal of Mathematical, Engineering and Management Sciences, vol. 5, no. 4, pp. 643–651. [Google Scholar]
38. Y. Song, S. Zheng, L. Li, X. Zhang,X. Zhang. (2020). et al., “Deep learning enables accurate diagnosis of novel coronavirus (COVID-19) with CT images,” Medrxiv. [Google Scholar]
39. S. Wang, B. Kang, J. Ma, X. Zeng,M. Xio. (2020). et al., “A deep learning algorithm using CT images to screen for corona virus disease (COVID-19),” Medrxiv. [Google Scholar]
40. X. Wang, X. Deng, Q. Fu, Q. Zhou, J. Feng, H. Ma, W. Liu and C. Zheng, “A Weakly-Supervised Framework for COVID-19 Classification and Lesion Localization From Chest CT,” IEEE Transactions on Medical Imaging, vol. 39, no. 8, pp. 2615–2625, 2020. [Google Scholar]
41. X. Xu, X. Jiang, C. Ma, P. Du,X. Li. (2020). et al., “Deep learning system to screen Coronavirus disease 2019 pneumonia,” arxiv preprint arxiv:2002.09334. [Google Scholar]
42. M. Barstugan, U. Ozkaya and S. Ozturk. (2020). “Coronavirus (COVID-19) classification using CT images by machine learning methods,” arxiv preprint arxiv:2003.09424. [Google Scholar]
43. L. Lan, D. Xu, G. Ye, C. Xia,S. Wang. (2020). et al., “Positive RT-PCR test results in patients recovered from COVID-19,” Journal of the American Medical Association, vol. 323, no. 15, pp. 1502–1503. [Google Scholar]
44. D. Kermany, K. Zhang and M. Goldbaum. (2018). “Labeled optical coherence tomography (OCT) and chest X-Ray images for classification,” Mendeley data, V2, University of California San Diego, . Available: https://data.mendeley.com/datasets/rscbjbr9sj/2 25 April 2020. [Google Scholar]
45. J. P. Cohen, P. Morrison, L. Dao, K. Roth,T. Q. Duong et al., “COVID-19 image data collection: Prospective predictions are the future,” arxiv preprint arxiv:2006.11988, 2020. Available at: https://github.com/ieee8023/covid-chestxray-dataset 25 April 2020. [Google Scholar]
46. R. Cappelli, D. Maio, D. Maltoni, J. L. Wayman and A. K. Jain. (2006). “Performance evaluation of fingerprint verification systems,” IEEE Transactions on Pattern Analysis and Machine Intelligence, vol. 28, no. 1, pp. 3–18. [Google Scholar]
47. L. Wang and A. Wong. (2020). “COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images,” arXiv preprint arxiv:2003.09871. [Google Scholar]
48. L. Li, L. Qin, Z. Xu, Y. Yin,X. Wang. (2020). et al., “Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT,” Radiology, vol. 296, no. 2, pp. E65–E71. [Google Scholar]
49. F. Shi, L. Xia, F. Shan, D. Wu,Y. Wei. (2020). et al., “Large-scale screening of COVID-19 from community acquired pneumonia using infection size-aware classification,” arXiv preprint arxiv:2003.09860. [Google Scholar]
 | This work is licensed under a Creative Commons Attribution 4.0 International License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. |