 Open Access
Open Access
REVIEW
The Potential of Circular RNAs as Biomarkers in Pulmonary Arterial Hypertension Related to Congenital Heart Disease
1 Doctorate Program of Medical and Health Science, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, 55281, Indonesia
2 Department of Biochemistry, Faculty of Medicine, Universitas Riau, Pekanbaru, 28133, Indonesia
3 Department of Histology and Cell Biology, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada, Yogyakarta, 55281, Indonesia
4 Department of Cardiology and Vascular Medicine, Faculty of Medicine, Public Health and Nursing, Universitas Gadjah Mada Dr. Sardjito Hospital, Yogyakarta, 55281, Indonesia
* Corresponding Author: Anggoro Budi Hartopo. Email:
Congenital Heart Disease 2024, 19(4), 375-388. https://doi.org/10.32604/chd.2024.054742
Received 06 June 2024; Accepted 29 September 2024; Issue published 31 October 2024
Abstract
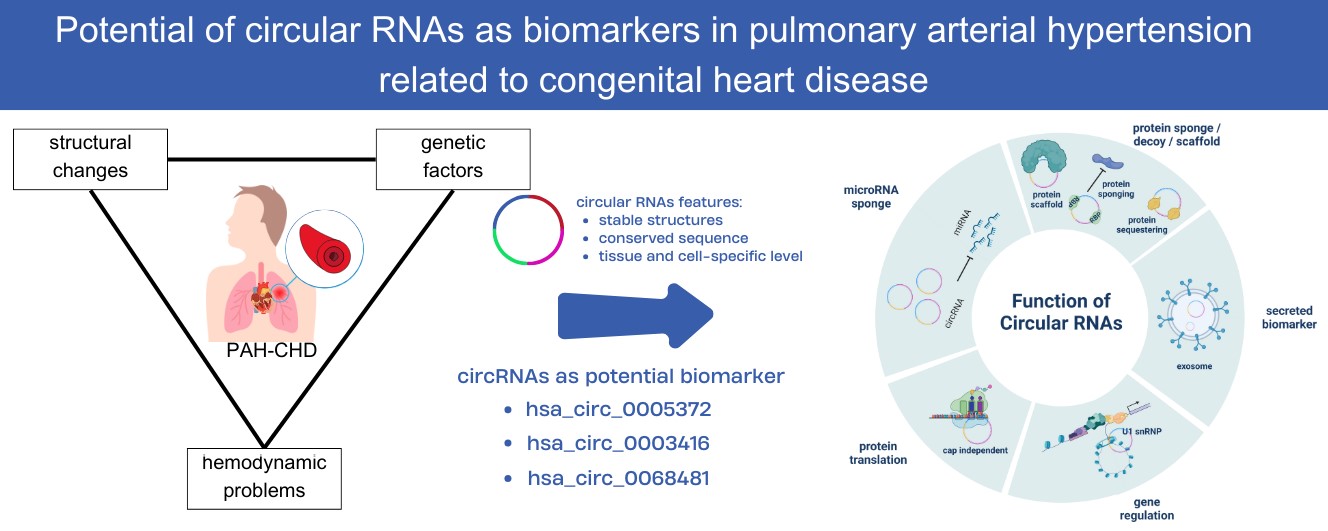
A particular type of endogenous noncoding RNAs known as circular RNAs (circRNAs) has now become possible biomarkers for several diseases because of their stability and tissue-specific expression patterns. CircRNAs might play a role in various of biological processes. The identification of particular circRNAs dysregulated in pulmonary arterial hypertension (PAH) raises the possibility of these molecules serving as biomarkers for the disease’s early diagnosis and treatment. This review mainly summarizes the role and potential of circRNA as a future biomarker in PAH related to congenital heart disease. This study presented several potential circRNA targets as diagnostic biomarkers for PAH, discussed their biological functions, and addressed the challenges that need to be considered for their application in clinical settings.Graphic Abstract
Keywords
Nomenclature
| ASD | Atrial septal defect |
| AUC | Area under the curve |
| BMPR2 | Bone morphogenetic protein receptor-2 |
| BSJs | Back-splice junctions |
| CCND1 | Cyclin D1 |
| CeRNAs | Competing endogenous RNAs |
| CHD | Congenital heart disease |
| CI | Cardiac output index |
| CircRNAs | Circular RNAs |
| COPD | Chronic obstructive pulmonary disease |
| COX-2 | Cyclooxygenase-2 |
| CTEPH | Chronic thromboembolic pulmonary hypertension |
| ddRT-HRCA | Reverse-transcription-hyperbranched rolling circle amplification |
| EYA3 | Eyes absent transcriptional coactivator and phosphatase 3 |
| KCNK3 | Potassium channel subfamily K member 3 |
| lncRNAs | Long noncoding RNAs |
| miRNAs | MicroRNAs |
| mPAP | Mean pulmonary artery pressure |
| mRNAs | Messenger RNAs |
| NT-proBNP | N-terminal prohormone of brain natriuretic peptide |
| PAH | Pulmonary arterial hypertension |
| PASMCs | Pulmonary artery smooth muscle cells |
| PVR | Pulmonary vascular resistance |
| RNA | Ribonucleic acid |
| ROC | Receiver operating characteristic |
| RT-qPCR | Reverse-transcription quantitative polymerase chain reaction |
| RVH | Right ventricular hypertrophy |
| SOX17 | SRY-related HMG-box transcription factor 17 |
| TBX4 | T-box transcription factor 4 |
| VSD | Ventricular septal defect |
Pulmonary arterial hypertension (PAH) is a relatively rare disease, with an estimated global incidence of 15–50 instances per million individuals [1]. However, it is more frequent in some groups, such as those with congenital heart disease (CHD). The increased blood flow and systemic pressure in the pulmonary circulation of patients with CHD put this group at higher risk of PAH [2]. According to the Indonesian registry, PAH develops in 66.9% of patients with CHD, and the most prevalent type of CHD is secundum atrial septal defect (ASD) [3]. Although it may occur in people of any age, PAH is typically identified in patients aged 30–60 years, and women are more likely than men to be diagnosed [1].
Patients and healthcare providers both suffer several difficulties because of the complicated nature of PAH. Through developments in medical care, the prognosis for PAH has improved recently; however, survival and quality of life are still significantly affected [4]. The 5-year survival rate of approximately 70% underscores the need for early and accurate detection and the development of effective biomarkers for monitoring and prognosis [5,6].
A particular type of endogenous noncoding RNAs known as circular RNAs (circRNAs) has now become possible biomarkers for several diseases owing to their stability and tissue-specific expression patterns [7]. CircRNAs have a closed-loop shape, giving them more strength and resistance to degradation than linear RNAs [8]. The identification of particular circRNAs dysregulated in PAH raises the potential of these molecules as biomarkers for early diagnosis and treatment [9–11].
CircRNAs might play a role in a wide range of biological processes [12]. CircRNAs serve an intriguing role in signaling networks, microRNA sponging, gene expression modulation, and cellular function regulation [13]. In PAH, circRNAs have been implicated in the pathogenesis of pulmonary vascular remodeling, a key feature of the disease. They regulate gene expression and protein translation through mechanisms such as microRNA sponging and binding with RNA-binding proteins [14]. For example, in chronic obstructive pulmonary disease-associated pulmonary hypertension (COPD-PH), specific circRNAs such as hsa_circ_0007608 and hsa_circ_0064656 are involved in cellular proliferation, migration, and endothelial-to-mesenchymal transition, which are critical processes in disease progression [15]. Gaining a thorough knowledge of the particular functions of circRNAs in the pathophysiology of PAH related to CHD can shed important light on the processes underlying the disease [16]. Furthermore, new diagnostic and prognostic approaches that can improve patient treatment, and outcomes might develop from investigating and analyzing circRNAs in patients with PAH related to CHD.
Despite these findings, the role of circRNAs in PAH, particularly in CHD, is still not comprehensively elucidated. Most studies have focused on idiopathic or other types of PAH, and PAH related to CHD received limited attention [17–19]. This review will fill this gap by explicitly focusing on the role and potential of circRNAs as biomarkers in PAH related to CHD. By synthesizing current research and highlighting novel circRNA targets, this review will provide a detailed overview of the current state of knowledge and identify future research directions. In summary, this review presents several potential circRNA targets as diagnostic biomarkers for PAH, discusses their biological functions, and addresses the challenges that need to be considered for their application in clinical settings. This review aimed to advance knowledge in this critical area and promote the development of effective diagnostic and prognostic strategies for PAH related to CHD in the context of circular RNA biomarkers.
2 Pathogenesis of Pulmonary Arterial Hypertension in Congenital Heart Disease
A confluence of structural, hemodynamic, and genetic problems causes PAH related to CHD, a complex condition [2]. Often, this condition is caused by abnormalities in the left-to-right shunt that are not corrected. Poor functioning of the cells lining the blood vessels, anomalies in the usual signaling pathways inside the cells, and altered blood vessel structure are only a few of the complex interplay of factors that cause PAH related to CHD [20].
Genetic predisposition plays a significant role in PAH development in patients with CHD. Mutations in genes related to endothelium function, proliferation of smooth muscle cells, and fibroblast activity in the pulmonary vasculature have been linked to disease onset. Studies have identified numerous genetic variants linked to PAH susceptibility and development [21]. By modifying cyclooxygenase-2 (COX-2) expression levels, polymorphisms in COX-2 and associated miRNAs, such as rs20417, rs5275, miR-26b-5p, and miR-146a-5p, have been demonstrated to affect PAH development [22]. Furthermore, genetic studies have shown significant genetic heterogeneity in PAH, with disease development being linked to genes such as BMPR2, KCNK3, TBX4, and SOX17 [23,24]. These genetic mutations disrupt normal signaling pathways, leading to vascular remodeling and increased pulmonary vascular resistance (PVR), which are hallmarks of PAH.
Through several pathways, structural defects in CHD result in PAH. In CHD with left-to-right shunting, increased pulmonary blood flow results in endothelial dysfunction and vascular remodeling, which increases PVR and sets off a series of proliferative and inflammatory reactions in the pulmonary vasculature [2,25]. Large systemic-to-pulmonary shunts, such as those seen in untreated ventricular septal defects (VSDs) and ASDs aggravate PAH by contributing to the steady increase in pulmonary arterial pressure (PAP) caused by this aberrant vascular homeostasis [25]. Vascular resistance and PAP increase with age in specific CHD lesions, including VSDs, atrioventricular septal defects, and transposition of the great arteries. If these defects are not repaired early, the persistent high flow and pressure can lead to irreversible pulmonary vascular disease [2]. Unlike the tetralogy of Fallot with significant aortopulmonary collateral arteries, postoperative PAH related to CHD can be caused by thrombosis, hypoplasia, or pulmonary agenesis, underscoring the multiplicity of anatomical and hemodynamic characteristics that contribute to PAH [26].
The complex interaction of genetic predisposition changes in hemodynamics and vascular remodeling highlights the complicated pathophysiology of PAH in CHD [25,27]. Hemodynamic indices, including mean PAP (mPAP), PVR, right atrial pressure, and cardiac output index, are significantly associated with clinical outcomes in patients with PAH. Changes in these indices, particularly mPAP and PVR, have high explanatory power for predicting adverse clinical events and mortality, underscoring their importance in risk stratification and treatment monitoring, to consider more contributing elements. These can influence PAH onset and course and include oxidative stress, metabolic abnormalities, and epigenetic changes [21]. In addition, noncoding RNAs, histone acetylation, and DNA methylation are some epigenetic changes that can affect gene expression patterns and exacerbate pathways linked to PVR and vascular remodeling, indicating the involvement of both genetic and epigenetic mechanisms [28].
3 Current PAH Related to CHD Biomarkers in Clinical Research
Currently, numerous biomarkers that have shown promise in diagnosing and monitoring PAH related to CHD have been identified [29]. These biomarkers include endothelin-1 [30], cardiac troponins [31], natriuretic peptides [32], and microRNAs [33]. These biomarkers can give important information for risk stratification, prognosis, and therapy response in patients with PAH related to CHD and are linked to the disease severity [34].
The N-terminal pro-B-type natriuretic peptide (NT-proBNP) has attracted interest as one of the essential indicators in this area [32]. This biomarker can give important information regarding the prognosis of patients with CHD and has shown the potential to assist in evaluating PAH severity. Jin et al identified Serum S-adenosyl methionine, guanine, and NT-proBNP as among the panel of serum biomarkers promising to differentiate children with PAH related to CHD from those with CHD alone. The combined use of these indicators showed good predictive accuracy, making them potentially helpful instruments for screening and diagnosis of patients with PAH related to CHD [35].
Another possible biomarker for PAH related to CHD is endothelin-1. Endothelin-1 levels in patients with PAH are high and are linked to both disease severity and higher PVR [30,36]. Other promising biomarkers in PAH related to CHD research are microRNAs, small noncoding RNAs involved in the control of gene expression, and cardiac troponins, markers of myocardial damage [33]. Studies have demonstrated that the development and progression of PAH are caused by the deregulation of specific pathways, notably the BMPR2 signaling pathway [37]. Biomarkers linked to these pathways might shed important light on the etiology of diseases and possible targets for treatments.
Biomarkers are still being used in clinical practice for PAH related to CHD, and more studies are required to confirm their efficacy and create common standards for their application. In this context, early diagnosis, prognosis prediction, and construction of customized treatment plans depend on discovering new biomarkers [38].
4 Roles of Circular RNAs in PAH Related to CHD
CircRNAs are an unusual type of endogenous noncoding RNAs. These molecules are formed without 3′ poly-(A) tails or 5′ cap structures by covalently closed-loop-like structures produced by back-splicing during pre-mRNA processing [13]. After their synthesis in the nucleus via back splicing, circRNAs are carried across the nuclear membrane. RNAs can circularize in both the nucleus and cytoplasm, with minor spliceosome perhaps mediating cytoplasmic biogenesis [39].
CircRNAs are involved in many biological processes, and diseases, including PAH related to CHD, have been linked to their aberrant control. These noncoding RNAs have been connected to controlling gene expression, serving as microRNA sponges, modifying protein functions, and affecting several cellular functions [40] (Fig. 1).
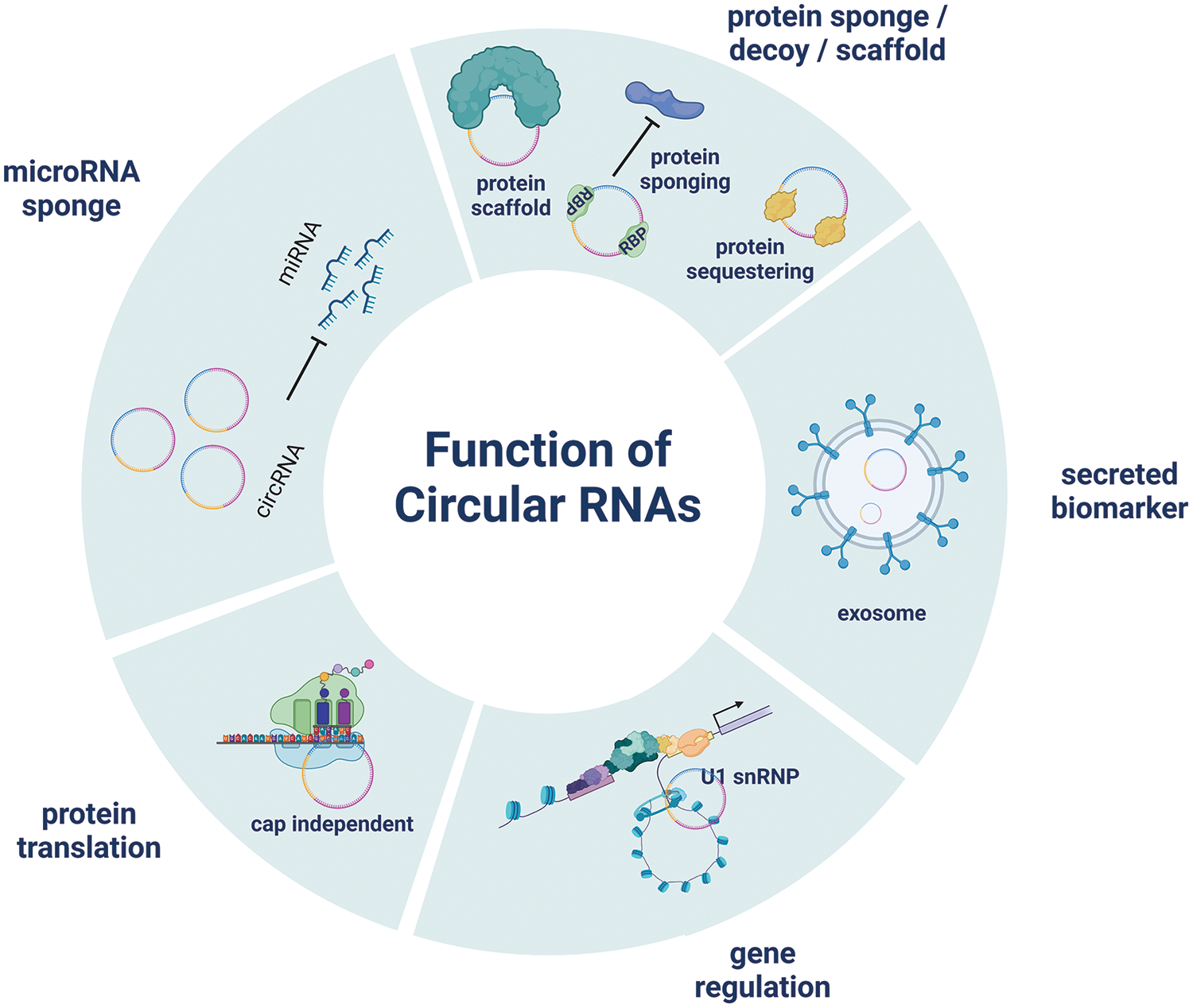
Figure 1: CircRNA plays a regulating role in gene expression, and an essential role in the process of biological development
Among the most engaging roles of circRNAs is that of miRNA sponges [41]. MicroRNAs, small noncoding RNA molecules are essential to the post-transcriptional control of gene expression. MicroRNAs can be competitively bound by circRNAs, blocking their interactions with target mRNAs [42]. CircRNAs control the expression of miRNA target genes, affecting several biological pathways by sequestering miRNAs [43].
CircRNAs have also been reported to control transcription, affect alternative splicing, and interact with RNA-binding proteins to change cellular processes. CircRNAs have numerous modes of action, so they are significant participants in the complex networks of regulation seen in cells [39,44]. Because of their special stability and abundance, circRNAs in body fluids are compelling candidates for noninvasive diagnostic and prognostic tools [45,46]. Significantly, circRNAs demonstrate higher tissue specificity than mRNAs of their host genes. This indicates that circRNAs could provide superior analytical validity, encompassing better analytical specificity, robustness, reproducibility, and repeatability when utilized as biomarker molecules [47,48]. A recent study examining circRNA expression in 20 clinically relevant tissue samples highlighted the potential of circRNAs as disease biomarkers, reinforcing the findings that circRNA expression is highly tissue-specific [49]. These findings underscore the significant potential of circRNAs as biomarkers in various diseases, including cancers, neurodegenerative disorders, and cardiovascular diseases, highlighting their importance in advancing diagnostic and therapeutic strategies in clinical research.
Knowledge of the precise functions of circRNAs in endothelial dysfunction, vascular remodeling, and aberrant cellular signaling pathways will provide crucial new information on the pathophysiology of PAH related to CHD [45]. In recent research, numerous potential circRNAs demonstrated different expression in the PAH group than in the control group (Table 1). These circRNAs, such as has_circ_0003416 and has_circ_0068481, may be candidate biomarkers for early diagnosis and prognostic evaluation of PAH in CHD patients.
Has_circ_005372 is identified as significantly upregulated and potentially involved in regulating gene expression in PAH. It binds to various miRNAs associated with cell proliferation, migration, and differentiation, such as miR-3125, miR-216a-5p, and miR-365a-5p. In addition, has_circ_005372 is linked to terms such as cellular response to retinoic acid and platelet alpha granule, indicating its potential role in biological processes and cellular components. However, this study only examined the expression of has_circ_005372 in a relatively small sample, comparing the CHD group (32 patients) and the CHD-related PAH group (16 patients), without conducting a diagnostic analysis [10].
Has_circ_0003416 has been identified as significantly downregulated in pediatric PAH related to CHD, as reported by Huang et al. [9] and Su et al. [10]. The receiver operating characteristic (ROC) analysis was conducted to assess the diagnostic potential of plasma has_circ_0003416 levels for PAH related to CHD. In their study, the area under the curve (AUC) for plasma has_circ_0003416 was 0.721 (95% confidence interval = 0.585–0.857, p = 0.004), with a cutoff value of 0.99. Furthermore, the sensitivity and specificity were 0.66 and 0.70, respectively [9].
Has_circ_0003416 also showed a negative association with the B-type natriuretic peptide. The mechanism underlying the aberrant expression of has_circ_0003416 in PAH has not yet been elucidated; however, in silico analysis demonstrated full-length has_circ_0003416 to be 124-bp long, encoded by TMSB4X. TMSB4X is an important regulator of angiogenesis. miR-199a/b-3p, miR-3194-5p, and miR-486-3p are targets of circRNA has_circ_003416 in its downregulated form [9]. Therefore, further studies are needed for its validation. The expression profile and diagnostic value of has_circ_0003416 reflect its importance as a potential biomarker for identifying and monitoring PAH related to CHD [9,10].
High serum has_circ_0068481 levels show promise as a diagnostic and prognostic indicator of idiopathic PAH. A study has found that has_circ_0068481 expression was significantly higher in patients with PAH, correlating with disease severity, right heart failure (RHF), and poorer clinical outcomes. The diagnostic capability of serum circ_0068481 levels for identifying PAH and RHF in PAH was evaluated through ROC curve analysis. At an optimal expression cutoff value of 0.995 for identifying PAH, the sensitivity, specificity, and AUC were 74.39%, 98.78%, and 0.895, respectively. For identifying PAH with RHF, at an optimal expression cutoff value of 2.97, the sensitivity, specificity, and AUC were 94.59%, 92.45%, and 0.978, respectively [11]. The findings by Guo et al. further reinforce these results in a larger sample, demonstrating that has_circ_0068481 could be a biomarker for right ventricular hypertrophy (RVH) in patients with PAH, with an AUC of 0.974 [51].
In addition, has_circ_0068481 levels are associated with heart function, 6-min walk distance, and biochemical markers such as NT-proBNP, indicating its potential as a diagnostic tool [11]. Furthermore, has_circRNA_0068481 upregulates EYA3 expression while downregulating the expression levels of miR-646, miR-570, and miR-885, thereby promoting RVH development. This interaction among has_circ_0068481, miRNAs, and EYA3 highlights the mechanistic role of has_circ_0068481 in the diagnosis and progression of RVH in patients with PAH [51]. These findings collectively highlight the utility of serum has_circ_0068481 levels in diagnosing and predicting outcomes in patients with PAH [11,51].
Has_circ_0016070 significantly contributes to the pathogenesis of PAH by affecting vascular remodeling and promoting the proliferation of pulmonary artery smooth muscle cells (PASMCs). Has_circ_0016070, identified through circRNA microarray analysis, is linked to an increased PAH risk in patients with COPD [18]. One of the key mechanisms involves has_circ_0016070 as a competing endogenous RNA (ceRNA) for miR-340-5p, thereby promoting the TCF4/β-catenin/TWIST1 complex, which aggravates PAH progression [52]. It also operates by modulating the miR-942/CCND1 axis, wherein has_circ_0016070 downregulates miR-942 expression, leading to the upregulation of CCND1, a gene crucial for cell cycle regulation and proliferation [18]. However, observations in the present study are constrained by its single-center, cross-sectional design, small sample size, and absence of evidence linking has_circ_0016070 serum levels with clinical outcomes [18].
Miao et al. [19] reported that has_circ_0046159 may play a significant role in chronic thromboembolic pulmonary hypertension (CTEPH) by interacting with miR-1226-3p and potentially regulating ATP2A2. The expression of has_circ_0046159 is notably increased in patients with CTEPH compared with that in normal individuals. This circRNA is involved in a ceRNA network and may contribute to CTEPH development. However, this study was validated in a relatively small sample size (4 cases vs. 4 controls), and the specific roles of has_circ_0046159 in PAH were not directly investigated [19].
Has_circNFXL1_009 is significantly reduced in patients with PAH associated with COPD, highlighting its crucial role in the disease process [17]. Microarray and reverse-transcription quantitative polymerase chain reaction (RT-qPCR) analyses revealed that has_circNFXL1_009 is notably downregulated in patients with PAH related COPD, identifying 158 dysregulated circRNAs. Among these, has_circNFXL1_009 demonstrated the highest area under the ROC with a value of 0.941, as well as high sensitivity (90.48%) and specificity (95.24%). Functionally, has_circNFXL1_009 regulates abnormal proliferation, apoptosis, and migration of human PASMCs under hypoxic conditions. It binds to has-miR-29b-2-5p, which helps regulate the expression of the potassium channel gene KCNB1, essential for maintaining normal cell function. Therefore, has_circNFXL1_009 could be a valuable biomarker for diagnosing patients with PAH and a potential therapeutic target to inhibit PAH progression [17].
5 Challenges and Opportunities in circRNA Detection
CircRNAs are detected and quantified using various methods, each with strengths and limitations. RNA sequencing (RNA-seq) is a fundamental technique for detecting circRNAs, employing computational tools to identify their distinctive back-splice junctions (BSJs). However, RNA-seq may yield false positives because of sequencing artifacts, requiring meticulous algorithmic management to reduce these errors [53]. In contrast, microarray technology improves circRNA detection and profiling by specifically targeting their unique BSJs. A major advantage of microarray technology is its ability to simultaneously examine the expression profiles of thousands of circRNAs, offering a comprehensive view of their differential expression across various conditions. A significant drawback is its reliance on predesigned probes targeting known back-splice sites, which restricts its ability to discover novel circRNAs. This limitation contrasts with RNA-seq, which can identify new circRNAs even without prior knowledge of their sequences [54].
A benchmarking study of 16 circRNA detection tools revealed high precision but variable sensitivities, highlighting the importance of tool selection and the potential benefit of combining multiple tools to enhance detection sensitivity while minimizing false discoveries [55]. For validation, RT-qPCR is commonly used; however, linear RNA contaminants can confound it. To address this, Rnase R treatment is employed to degrade linear RNAs; however, the protocol must be optimized to avoid partial degradation of circRNAs [56].
An advanced method, RT-hyperbranched rolling circle amplification (RT-HRCA), and its droplet digital variant (ddRT-HRCA), offer sensitive and quantitative detection of circRNAs, achieving low detection limits and high selectivity without the need for Rnase R enrichment [57]. Despite these advancements, traditional RT-qPCR remains limited by potential strand substitution during RT, which can hinder accurate quantification. In addition, the tissue-specific expression and evolutionary conservation of circRNAs necessitate precise and sensitive detection platforms to fully understand their biological roles and potential as disease biomarkers [58]. Overall, while RNA-seq and RT-qPCR are foundational techniques in circRNA research, emerging methods such as ddRT-HRCA provide promising alternatives for more accurate and sensitive circRNA quantification, addressing some of the limitations inherent in conventional approaches [53,56–58].
Furthermore, circRNAs are known for their stability owing to their closed-loop structures, which makes them resistant to exonuclease-mediated degradation. This stability makes circRNAs promising candidates for clinical biomarkers [46]. However, sample collection and processing can still affect such stability. For example, circRNAs remain stable during sample processing delays, showing minimal expression changes up to 48 h at 4°C, whereas linear mRNAs and lncRNAs degrade significantly after 12 h [59]. The half-life of circRNAs in blood samples is approximately 24.56 ± 5.2 h, longer than mRNAs (16.4 h) and other types of noncoding RNAs such as miRNAs (16.42 h) and lncRNAs (17.46 h) [60]. Although the half-life of circRNAs is better than that of linear RNAs, appropriate and professional methods must be employed for preserving blood samples to ensure the accuracy and reliability of test results. This is important in clinical trials where obtaining high-quality experimental samples is essential. Therefore, the effects of specimen analysis and processing must be carefully considered [61].
The feasibility of translating circRNA findings into clinical practice is supported by advanced techniques such as qRT-PCR and bioinformatics, which allow for precise detection and functional analysis of circRNAs. Moreover, the circRNA-miRNA-mRNA regulatory networks provide a comprehensive understanding of disease mechanisms, offering new avenues for targeted therapies [62]. Overall, the use of circRNAs as biomarkers in PAH related to CHD presents several advantages over existing biomarkers, including higher stability, specificity, and ability to reflect complex regulatory networks, making them highly feasible for clinical application and potentially improving current diagnostic and therapeutic strategies.
6 Future Directions for Research
Prior research has demonstrated that circRNAs undergo substantial dysregulation and may serve as potential biomarkers in CHD [63,64], various types of PAH [11,18,19], and PAH related to CHD [9,10]. However, the identification of circRNAs in PAH related to CHD remains limited, highlighting a significant gap in the current understanding and diagnostic capabilities for this condition. Thus, a multifaceted approach is necessary to identify and validate a broader spectrum of circRNAs comprehensively. This could include high-throughput technologies, such as RNA-seq [65] or microarray [54], followed by rigorous validations using RT-qPCR and functional assays [66]. Integrating these findings with other omics data, such as transcriptomics, proteomics, and metabolomics, could provide a more holistic view of the molecular landscape of PAH related to CHD. In addition, leveraging advanced bioinformatics tools to analyze circRNA-miRNA-mRNA interaction networks could elucidate the regulatory mechanisms underlying PAH related to CHD, offering new therapeutic targets [39,53,67]. Encouraging collaborative research efforts and longitudinal studies is crucial in establishing the diagnostic accuracy and clinical relevance of these biomarkers.
In addition, future studies should not only look at the molecular underpinnings of circRNAs but also at developing noninvasive techniques for identifying and tracking circRNA biomarkers in PAH related to CHD. This might include investigating circRNA signatures in biofluids such as serum or blood, providing a less intrusive method of determining the disease state and course [68].
Reproducibility factors for each circRNA target must be further validated to ensure their reliability across different studies and clinical settings. To the best of our knowledge, in PAH, only has_circ_0003416 and has_circ_0068481 have been reported based on findings from two studies, although conducted from the same population. Moreover, validating these biomarkers in large, well-characterized cohorts of patients with CHD, both with and without PAH, would enhance their clinical relevance and utility [69]. A multi-omics approach could uncover complex regulatory networks and potential crosstalk across various molecular pathways, identifying new targets for therapeutic intervention and personalized treatment plans [70].
To overcome sample size constraints, establishing a sample storage system in conjunction with registries such as COHARD PH [3] is essential for advancing future research and understanding this rare, life-limiting disease. Consequently, each center must develop such registries and sample banks to manage and utilize valuable data and samples effectively. In addition, confounding factors that may influence circRNA expression must be considered. Although previous studies [9,11] have not found significant differences in sex, age, and body mass index between cases and controls, information regarding other comorbidities, such as diabetes or other diseases, was not provided. This gap underscores the need to apply strict criteria to minimize the effects of confounding factors.
The multifaceted roles of circRNAs in biological processes underscore their significance in molecular biology and their potential for therapeutic applications. The potential of circRNAs as diagnostic and therapeutic targets in the PAH related to CHD is a promising area for future exploration. It may pave the way for advancements in managing this complex disease. Although the AUC values for circRNAs as a diagnostic biomarker are high, confirmation and validation through longitudinal multicenter studies involving large, well-characterized patient cohorts and unbiased molecular phenotyping are still required.
Acknowledgement: We thank the PAH team and Klinik Bahasa from FKKMK UGM for their valuable support of this study.
Funding Statement: This work was supported by “Hibah Penelitian Dana Masyarakat FKKMK UGM” grant No. 1528/UN1/FKKMK/PPKE/PT/2024 and “Riset dan Inovasi Untuk Indonesia Maju Gelombang 4” grant No. 172/IV/KS/11/2023-6815/UN1/DITLIT/Dit-Lit/KP.01.03/2023 received by Anggoro Budi Hartopo.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Fajri Marindra Siregar, Sofia Mubarika Haryana and Anggoro Budi Hartopo; draft manuscript preparation: Fajri Marindra Siregar and Anggoro Budi Hartopo; supervision: Sofia Mubarika Haryana, Dyah Wulan Anggrahini, Lucia Kris Dinarti and Anggoro Budi Hartopo. All authors reviewed and approved the final version of the manuscript.
Availability of Data and Materials: The data set used for this review will be shared upon request from the study authors.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Levine DJ. Pulmonary arterial hypertension: updates in epidemiology and evaluation of patients. Am J Manag Care. 2021;27(3 Suppl):S35–41. doi:10.37765/ajmc. [Google Scholar] [PubMed] [CrossRef]
2. D’Alto M, Mahadevan VS. Pulmonary arterial hypertension associated with congenital heart disease. Eur Respir Rev. 2012;21(126):328–37. doi:10.1183/09059180.00004712. [Google Scholar] [CrossRef]
3. Dinarti LK, Hartopo AB, Kusuma AD, Satwiko MG, Hadwiono MR, Pradana AD, et al. The Congenital heart Disease in adult and Pulmonary Hypertension (COHARD-PH) registry: a descriptive study from single-center hospital registry of adult congenital heart disease and pulmonary hypertension in Indonesia. BMC Cardiovasc Disord. 2020;20(1):163. doi:10.1186/s12872-020-01434-z. [Google Scholar] [PubMed] [CrossRef]
4. Aguirre-Camacho A. Understanding the perspective of patients with pulmonary arterial hypertension: looking beyond health-related quality of life. Ther Adv Rare Dis. 2021;2. doi:10.1177/2633004020986166. [Google Scholar] [PubMed] [CrossRef]
5. Marques-Alves P, Baptista R, Marinho da Silva A, Pêgo M, Castro G. Real-world, long-term survival of incident patients with pulmonary arterial hypertension. Rev Port Pneumol. 2017;23(3):124–31. doi:10.1016/j.rppnen.2017.01.006. [Google Scholar] [PubMed] [CrossRef]
6. Hojda SE, Chis IC, Clichici S. Biomarkers in pulmonary arterial hypertension. Diagnostics. 2022;12(12):1–23. doi:10.3390/diagnostics12123033. [Google Scholar] [PubMed] [CrossRef]
7. Jiao S, Wu S, Huang S, Liu M, Gao B. Advances in the identification of circular RNAs and research into circRNAs in human diseases. Front Genet. 2021;12:665233. doi:10.3389/fgene.2021.665233. [Google Scholar] [PubMed] [CrossRef]
8. Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–61. doi:10.1038/nbt.2890. [Google Scholar] [PubMed] [CrossRef]
9. Huang Y, Su D, Ye B, Huang Y, Qin S, Chen C, et al. Expression and clinical significance of circular RNA hsa_circ_0003416 in pediatric pulmonary arterial hypertension associated with congenital heart disease. J Clin Lab Anal. 2022;36(4):e24273. doi:10.1002/jcla.24273. [Google Scholar] [PubMed] [CrossRef]
10. Su D, Huang Y, Liu D, Huang Y, Ye B, Qin S, et al. Bioinformatic analysis of dysregulated circular RNAs in pediatric pulmonary hypertension linked congenital heart disease. Transl Pediatr. 2022;11(5):715–27. doi:10.21037/tp-22-117. [Google Scholar] [PubMed] [CrossRef]
11. Zhang Y, Chen Y, Yao H, Lie Z, Chen G, Tan H, et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ. 2019;9(4). doi:10.1177/2045894019888416. [Google Scholar] [PubMed] [CrossRef]
12. Chen L-L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17(4):205–11. doi:10.1038/nrm.2015.32. [Google Scholar] [PubMed] [CrossRef]
13. Wilusz JE. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA. 2018;9(4):e1478. doi:10.1002/wrna.1478. [Google Scholar] [PubMed] [CrossRef]
14. Ali MK, Schimmel K, Zhao L, Chen C-K, Dua K, Nicolls MR, et al. The role of circular RNAs in pulmonary hypertension. Eur Respir J. 2022;60(6):1–14. [Google Scholar]
15. Yu J, Huang S, Shen W, Zhang Z, Ye S, Chen Y, et al. Expression profiles of circRNAs and identification of hsa_circ_0007608 and hsa_circ_0064656 as potential biomarkers for COPD-PH patients. Int J Chron Obstruct Pulmon Dis. 2023;18:2457–71. doi:10.2147/COPD.S424712. [Google Scholar] [PubMed] [CrossRef]
16. Wang Q, Sun Y, Zhao Q, Wu W, Wang L, Miao Y, et al. Circular RNAs in pulmonary hypertension: emerging biological concepts and potential mechanism. Anim Models Exp Med. 2022;5(1):38–47. doi:10.1002/ame2.v5.1. [Google Scholar] [CrossRef]
17. Jin X, Xu Y, Guo M, Sun Y, Ding J, Li L, et al. hsa_circNFXL1_009 modulates apoptosis, proliferation, migration, and potassium channel activation in pulmonary hypertension. Mol Ther Nucleic Acids. 2021;23:1007–19. doi:10.1016/j.omtn.2020.09.029. [Google Scholar] [PubMed] [CrossRef]
18. Zhou S, Jiang H, Li M, Wu P, Sun L, Liu Y, et al. Circular RNA hsa_circ_0016070 is associated with pulmonary arterial hypertension by promoting PASMC proliferation. Mol Ther Nucleic Acids. 2019;18:275–84. doi:10.1016/j.omtn.2019.08.026. [Google Scholar] [PubMed] [CrossRef]
19. Miao R, Gong J, Zhang C, Wang Y, Guo X, Li J, et al. Hsa_circ_0046159 is involved in the development of chronic thromboembolic pulmonary hypertension. J Thromb Thrombolysis. 2020;49(3):386–94. doi:10.1007/s11239-019-01998-4. [Google Scholar] [PubMed] [CrossRef]
20. Handoyo, Gunawijaya E, Yantie NPVK. Factors associated with the uncorrectable congenital heart disease in children with pulmonary arterial hypertension. Med J Indones. 2023;32(1):38–44. doi:10.13181/mji.oa.236582. [Google Scholar] [CrossRef]
21. Bousseau S, Sobrano Fais R, Gu S, Frump A, Lahm T. Pathophysiology and new advances in pulmonary hypertension. BMJ Med. 2023;2(1):e000137. doi:10.1136/bmjmed-2022-000137. [Google Scholar] [PubMed] [CrossRef]
22. Fredenburgh LE, Ma J, Perrella MA. Cyclooxygenase-2 inhibition and hypoxia-induced pulmonary hypertension: effects on pulmonary vascular remodeling and contractility. Trends Cardiovasc Med. 2009;19(2):31–7. doi:10.1016/j.tcm.2009.04.003. [Google Scholar] [PubMed] [CrossRef]
23. Navas P, Tenorio J, Quezada CA, Barrios E, Gordo G, Arias P, et al. Molecular analysis of BMPR2, TBX4, and KCNK3 and genotype-phenotype correlations in spanish patients and families with idiopathic and hereditary pulmonary arterial hypertension. Rev Esp Cardiol. 2016;69(11):1011–9. doi:10.1016/j.recesp.2016.03.031. [Google Scholar] [CrossRef]
24. Hartopo AB, Anggrahini DW, Emoto N, Dinarti LK. The BMPR2, ALK1 and ENG genes mutation in congenital heart disease associated pulmonary artery hypertension. Acta Cardiol Indones. 2019;5(2):145–9. [Google Scholar]
25. Wacker J, Joye R, Genecand L, Lador F, Beghetti M. Pulmonary vascular disease as a complication of pediatric congenital heart diseases. Transl Pediatr. 2023;12(5):1041–52. doi:10.21037/tp-23-64. [Google Scholar] [PubMed] [CrossRef]
26. Yasuhara J, Yamagishi H. Pulmonary arterial hypertension associated with tetralogy of Fallot. Int Heart J. 2015;56:S17–21. [Google Scholar] [PubMed]
27. Jone P-N, Ivy DD, Hauck A, Karamlou T, Truong U, Coleman RD, et al. Pulmonary hypertension in congenital heart disease: a scientific statement from the American Heart Association. Circ Heart Fail. 2023;16(7):e00080. [Google Scholar] [PubMed]
28. Dave J, Jagana V, Janostiak R, Bisserier M. Unraveling the epigenetic landscape of pulmonary arterial hypertension: implications for personalized medicine development. J Transl Med. 2023;21(1):477. doi:10.1186/s12967-023-04339-5. [Google Scholar] [PubMed] [CrossRef]
29. Correale M, Tricarico L, Bevere EML, Chirivì F, Croella F, Severino P, et al. Circulating biomarkers in pulmonary arterial hypertension: an update. Biomolecules. 2024;14(5):1–19. [Google Scholar]
30. Chester AH, Yacoub MH. The role of endothelin-1 in pulmonary arterial hypertension. Glob Cardiol Sci Pract. 2014;2014(2):62–78. [Google Scholar] [PubMed]
31. Heresi GA, Tang WHW, Aytekin M, Hammel J, Hazen SL, Dweik RA. Sensitive cardiac troponin I predicts poor outcomes in pulmonary arterial hypertension. Eur Respir J. 2012;39(4):939–44. doi:10.1183/09031936.00067011. [Google Scholar] [PubMed] [CrossRef]
32. Lewis RA, Durrington C, Condliffe R, Kiely DG. BNP/NT-proBNP in pulmonary arterial hypertension: time for point-of-care testing? Eur Respir Rev. 2020;29(156):1–14. doi:10.1183/16000617.0009-2020. [Google Scholar] [PubMed] [CrossRef]
33. Siregar FM, Hartopo AB, Mubarika S. Recent progress in the roles of microRNAs in pulmonary arterial hypertension associated with congenital heart disease. Narra J. 2024;4(1):e579. doi:10.52225/narra.v4i1.579. [Google Scholar] [PubMed] [CrossRef]
34. Smits AJ, Botros L, Mol MAE, Ziesemer KA, Wilkins MR, Vonk Noordegraaf A, et al. A systematic review with meta-analysis of biomarkers for detection of pulmonary arterial hypertension. ERJ Open Res. 2022;8(2):1–14. doi:10.1183/23120541.00009-2022. [Google Scholar] [PubMed] [CrossRef]
35. Jin N, Yu M, Du X, Wu Z, Zhai C, Pan H, et al. Identification of potential serum biomarkers for congenital heart disease children with pulmonary arterial hypertension by metabonomics. BMC Cardiovasc Disord. 2023;23(1):167. doi:10.1186/s12872-023-03171-5. [Google Scholar] [PubMed] [CrossRef]
36. Dinarti LK, Hartopo AB, Anggrahini DW, Sadewa AH, Setianto BY, Wahab AS. Profile of endothelin-1, nitric oxide, and prostacyclin levels in pulmonary arterial hypertension related to uncorrected atrial septal defect: results from a single center study in Indonesia. Cardiol Res Pract. 2020;2020:7526508. [Google Scholar] [PubMed]
37. Zhang Y, Peng B, Han Y. MiR-23a regulates the proliferation and migration of human pulmonary artery smooth muscle cells (HPASMCs) through targeting BMPR2/Smad1 signaling. Biomed Pharmacother. 2018;103:1279–86. doi:10.1016/j.biopha.2018.04.172. [Google Scholar] [PubMed] [CrossRef]
38. Hemnes A, Rothman AMK, Swift AJ, Zisman LS. Role of biomarkers in evaluation, treatment and clinical studies of pulmonary arterial hypertension. Pulm Circ. 2020;10(4). doi:10.1177/2045894020957234. [Google Scholar] [PubMed] [CrossRef]
39. Li X, Yang L, Chen L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–42. doi:10.1016/j.molcel.2018.06.034. [Google Scholar] [PubMed] [CrossRef]
40. Zhou W-Y, Cai Z-R, Liu J, Wang D-S, Ju H-Q, Xu R-H. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. doi:10.1186/s12943-020-01286-3. [Google Scholar] [PubMed] [CrossRef]
41. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8. doi:10.1038/nature11993. [Google Scholar] [PubMed] [CrossRef]
42. Ranganathan K, Sivasankar V. MicroRNAs-Biology and clinical applications. J Oral Maxillofac Pathol. 2014;18(2):229–34. doi:10.4103/0973-029X.140762. [Google Scholar] [PubMed] [CrossRef]
43. Min X, Liu D-L, Xiong X-D. Circular rnas as competing endogenous rnas in cardiovascular and cerebrovascular diseases: molecular mechanisms and clinical implications. Front Cardiovasc Med. 2021;8:682357. doi:10.3389/fcvm.2021.682357. [Google Scholar] [PubMed] [CrossRef]
44. Sharma AR, Bhattacharya M, Bhakta S, Saha A, Lee S-S, Chakraborty C. Recent research progress on circular RNAs: biogenesis, properties, functions, and therapeutic potential. Mol Ther Nucleic Acids. 2021;25:355–71. doi:10.1016/j.omtn.2021.05.022. [Google Scholar] [PubMed] [CrossRef]
45. Zhang Z, Guo R, Wang Y, Huang H, Liu J, Wang C, et al. Diagnostic value of circRNAs as effective biomarkers in human cardiovascular disease: an updated meta-analysis. Int J Med Sci. 2022;19(3):446–59. doi:10.7150/ijms.67094. [Google Scholar] [PubMed] [CrossRef]
46. Wen G, Zhou T, Gu W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell. 2021;12(12):911–46. doi:10.1007/s13238-020-00799-3. [Google Scholar] [PubMed] [CrossRef]
47. Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17(5):257–71. doi:10.1038/nrg.2016.10. [Google Scholar] [PubMed] [CrossRef]
48. Xu T, Wu J, Han P, Zhao Z, Song X. Circular RNA expression profiles and features in human tissues: a study using RNA-seq data. BMC Genomics. 2017;18(Suppl 6):680. doi:10.1186/s12864-017-4029-3. [Google Scholar] [PubMed] [CrossRef]
49. Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med. 2017;95(11):1179–89. doi:10.1007/s00109-017-1582-9. [Google Scholar] [PubMed] [CrossRef]
50. Wang F, Li W, Li B, Xie L, Tong Y, Xu X. cRNAsp12 web server for the prediction of circular RNA secondary structures and stabilities. Int J Mol Sci. 2023;24(4):1–12. doi:10.3390/ijms24043822. [Google Scholar] [PubMed] [CrossRef]
51. Guo H-M, Liu Z-P. Up-regulation of circRNA_0068481 promotes right ventricular hypertrophy in PAH patients via regulating miR-646/miR-570/miR-885. J Cell Mol Med. 2021;25(8):3735–43. doi:10.1111/jcmm.16164. [Google Scholar] [PubMed] [CrossRef]
52. Huang C-X, Jiang Z-X, Du D-Y, Zhang Z-M, Liu Y, Li Y-T. Hsa_circ_0016070/micro-340-5p axis accelerates pulmonary arterial hypertension progression by upregulating TWIST1 transcription via TCF4/β-Catenin complex. J Am Heart Assoc. 2022;11(14):e024147. doi:10.1161/JAHA.121.024147. [Google Scholar] [PubMed] [CrossRef]
53. Digby B, Finn S, Ó Broin P. Computational approaches and challenges in the analysis of circRNA data. BMC Genomics. 2024;25(1):527. doi:10.1186/s12864-024-10420-0. [Google Scholar] [PubMed] [CrossRef]
54. Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinf. 2019;20(4):1420–33. doi:10.1093/bib/bby006. [Google Scholar] [PubMed] [CrossRef]
55. Vromman M, Anckaert J, Bortoluzzi S, Buratin A, Chen C-Y, Chu Q, et al. Large-scale benchmarking of circRNA detection tools reveals large differences in sensitivity but not in precision. Nat Methods. 2023;20(8):1159–69. doi:10.1038/s41592-023-01944-6. [Google Scholar] [PubMed] [CrossRef]
56. Vromman M, Yigit N, Verniers K, Lefever S, Vandesompele J, Volders P-J. Validation of circular RNAs using RT-qPCR after effective removal of linear RNAs by ribonuclease R. Curr Protoc. 2021;1(7):e181. doi:10.1002/cpz1.181. [Google Scholar] [PubMed] [CrossRef]
57. Wang K, Bai X, Xue Y, Luo X, Dong J, Yang G, et al. Absolute quantification of circRNA using digital reverse transcription-hyperbranched rolling circle amplification. Sensors and Act B: Chem. 2023;375:132893. doi:10.1016/j.snb.2022.132893. [Google Scholar] [CrossRef]
58. Dong J, Zeng Z, Huang Y, Chen C, Cheng Z, Zhu Q. Challenges and opportunities for circRNA identification and delivery. Crit Rev Biochem Mol Biol. 2023;58(1):19–35. doi:10.1080/10409238.2023.2185764. [Google Scholar] [PubMed] [CrossRef]
59. Wen G, Gu W. Circular RNAs in peripheral blood mononuclear cells are more stable than linear RNAs upon sample processing delay. J Cell Mol Med. 2022;26(19):5021–32. doi:10.1111/jcmm.17525. [Google Scholar] [PubMed] [CrossRef]
60. Wang C, Liu H. Factors influencing degradation kinetics of mRNAs and half-lives of microRNAs, circRNAs, lncRNAs in blood in vitro using quantitative PCR. Sci Rep. 2022;12(1):7259. doi:10.1038/s41598-022-11339-w. [Google Scholar] [PubMed] [CrossRef]
61. Seidl C, Huber HM, Müller-Kuller T, Sireis W, Aquilina A, Barotine-Toth K, et al. Blood collection and processing. Quality guidelines and standards reflecting common best practice standards referring to the EuBIS manual and guide. ISBT Sci Ser. 2012;7(1):16–23. doi:10.1111/j.1751-2824.2012.01612.x. [Google Scholar] [CrossRef]
62. Zhang Z, Qian H, Wang L, Tao Z, Cheng K, Wang K, et al. Construction of a circRNA-miRNA-mRNA regulatory network for coronary artery disease by bioinformatics analysis. Cardiol Res Pract. 2022;2022(11):4017082. doi:10.1155/2022/4017082. [Google Scholar] [PubMed] [CrossRef]
63. Wu J, Li J, Liu H, Yin J, Zhang M, Yu Z, et al. Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal. 2019;33(9):e22998. doi:10.1002/jcla.22998. [Google Scholar] [PubMed] [CrossRef]
64. Yu H, Wang X, Cao H. Construction and investigation of a circRNA-associated ceRNA regulatory network in Tetralogy of Fallot. BMC Cardiovasc Disord. 2021;21(1):437. doi:10.1186/s12872-021-02217-w. [Google Scholar] [PubMed] [CrossRef]
65. López-Jiménez E, Rojas AM, Andrés-León E. RNA sequencing and prediction tools for circular RNAs analysis. Adv Exp Med Biol. 2018;1087:17–33. doi:10.1007/978-981-13-1426-1. [Google Scholar] [CrossRef]
66. Panda AC, Gorospe M. Detection and analysis of circular RNAs by RT-PCR. Bio Protoc. 2018;8(6):1–9. [Google Scholar]
67. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–92. doi:10.1038/nrg.2016.114. [Google Scholar] [PubMed] [CrossRef]
68. Shi H, Zhou Y, Jia E, Liu Z, Pan M, Bai Y, et al. Comparative analysis of circular RNA enrichment methods. RNA Biol. 2022;19(1):55–67. doi:10.1080/15476286.2021.2012632. [Google Scholar] [PubMed] [CrossRef]
69. Tang Y, Bao J, Hu J, Liu L, Xu D-Y. Circular RNA in cardiovascular disease: expression, mechanisms and clinical prospects. J Cell Mol Med. 2021;25(4):1817–24. doi:10.1111/jcmm.v25.4. [Google Scholar] [CrossRef]
70. Fazal S, Bisserier M, Hadri L. Molecular and genetic profiling for precision medicines in pulmonary arterial hypertension. Cells. 2021;10(3):1–32. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools