 Open Access
Open Access
ARTICLE
Single-Nucleus RNA Sequencing Reveals Cardiac Macrophage Landscape in Hypoplastic Left Heart Syndrome
1 First Clinical Medical College, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, 210008, China
2 Department of Geriatrics, Affiliated Hospital of Nanjing University of Chinese Medicine, Nanjing, 210029, China
* Corresponding Author: Xu Han. Email:
Congenital Heart Disease 2024, 19(2), 233-246. https://doi.org/10.32604/chd.2024.050231
Received 31 January 2024; Accepted 16 April 2024; Issue published 16 May 2024
Abstract
Background: Hypoplastic left heart syndrome (HLHS) is one of the most challenging congenital heart diseases in clinical treatment. In cardiac tissues, resident macrophages fulfill critical functions in maintaining a stable cardiac state and have strong regenerative capacity and organ specificity. However, the molecular mechanisms of macrophages in HLHS remained unclear. Methods: Single-nucleus RNA sequencing (snRNA-seq) data of HLHS and healthy control (donors) samples obtained from the Gene Expression Omnibus (GEO) database were normalized and clustered using the Seurat package. The “FindMarkers” function was used to screen differentially expressed genes (DEGs) between the HLHS and donor groups and to analyze the functional enrichment of the set of genes of interest. Finally, cell-cell communication, pseudotime, and single-cell regulatory network inference and clustering (SCENIC) analyses were used to study the mechanisms of macrophages in HLHS. Results: Based on the snRNA-seq data of HLHS and donors, we identified a total of 9 cell clusters, among which the proportion of macrophages was significantly less in the HLHS group than in the control group. Subdivision of macrophage subpopulations (Macrophages 1, 2, and 3) showed that Macrophages 1 was mainly involved in nervous system development, angiogenesis, and apoptotic processes. In addition, analysis of communication between Macrophages 1 and cardiomyocytes revealed that ligand-acceptor pairs such as GAS6/AXL, IL6, IGF1, THY1, and L1CAM were present only in the donor group. Finally, pesudotime and SCENIC analyses demonstrated that FOXO3 and ELF2 played a critical role for Macrophages 1 to maintain cardiac function in patients with HLHS. Conclusion: Our study improved the current understanding of the molecular mechanisms of macrophage development in HLHS, showing that manipulating the regulatory role of macrophages in the heart can be a novel treatment for HLHS.Graphic Abstract
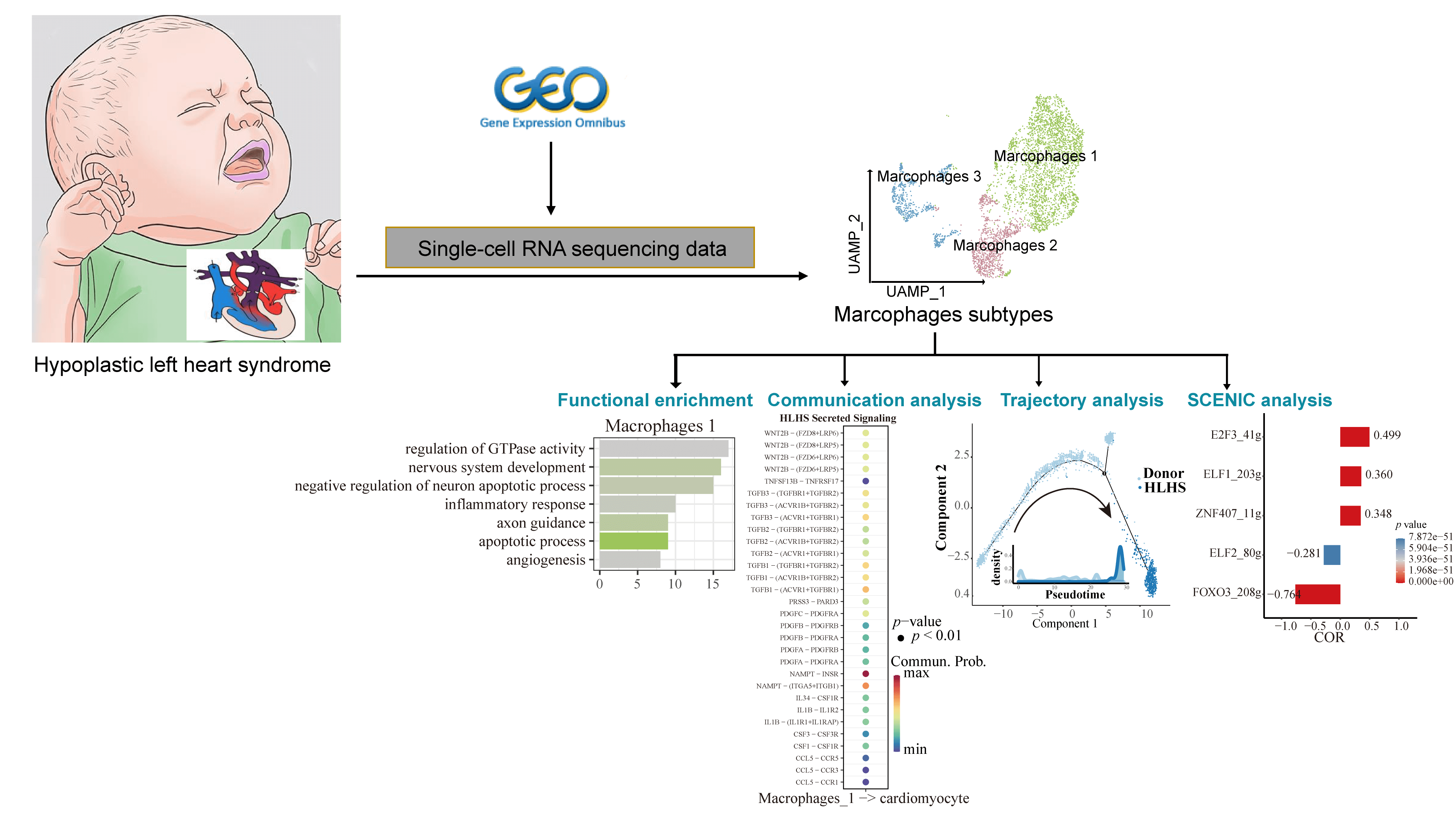
Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| HLHS | Hypoplastic left heart syndrome |
| GEO | Gene expression omnibus |
| snRNA-seq | Single-nucleus RNA sequencing |
| DEGs | Differentially expressed genes |
| SCENIC | Single-cell regulatory network inference and clustering |
| PCA | Principal component analysis |
| UMAP | Uniform manifold approximation, and projection |
| GO | Gene ontology |
| BP | Biological processes |
| GnRH | Gonadotropin-releasing hormone |
| TFs | Transcription factors |
| IGF1 | Insulin-like growth factor 1 |
| GAS6 | Growth arrest-specific protein 6 |
| FOXO3 | Forkhead box O3 |
| ELF2 | E74-like factor 2 |
Hypoplastic left heart syndrome (HLHS) is a congenital heart disease with a high mortality, especially during the early neonatal period and surgical treatment [1]. HLHS is mainly characterized by severe hypoplasia of the mitral valve, aortic valve, left ventricle, and ascending aorta. Children born with HLHS require staged surgery, which, however, only works to reconfigure the structure and blood flow of the heart. Even after surgical treatment, children with HLHS often have poor exercise capacity and might develop right ventricular failure [2]. Currently, a broad spectrum of variations and molecular pathways correlated with the etiology of HLHS have been discovered [3], but the intricate cellular interactions that result in HLHS remain to be comprehensively studied.
Macrophages as an important component of the innate and adaptive immune responses play crucial roles in maintaining the normal functions of the cardiovascular system. Normally, macrophages eliminate apoptotic cells to protect the organism from pathogens, participating in electrical conduction in arterial tone regulation, and heart, and vessel patrolling [4,5]. The predominant cell population in a healthy heart consists of resident macrophages with only a limited proportion of macrophages derived from monocytes [6]. However, when the heart is injured, cardiomyocytes experience necrosis to attract circulating monocytes expressing the CCR2 receptor, and these monocytes, along with the monocyte-derived macrophages, play a crucial role in eliminating dead cells and debris from the injured site. Moreover, they release pro-inflammatory cytokines to promote the degradation of extracellular matrix during the initial phases of cardiac injury, and then a specific subgroup of resident macrophages will secrete substantial quantities of anti-inflammatory cytokines to enhance tissue repair at the inflammation subsides [7–9]. The highly plastic nature of macrophages often blurs the boundaries of different phenotypes, which is largely correlated with the tissue microenvironment and macrophage-specific functions [5]. At present, the molecular mechanisms through which macrophages influence the progression of HLHS remain unclear, which requires a comprehensive analysis of macrophage functions in HLHS to help discover effective therapeutic targets.
Advances in single-cell and single-nucleus RNA sequencing (snRNA-seq) technologies allow for a better understanding of the underlying transcriptional profiles of healthy and diseased human hearts [10]. In this study, snRNA-seq data were employed to analyze the macrophage landscape in HLHS and to explore its molecular mechanisms. The workflow diagram is shown in Fig. S1. The snRNA-seq data of HLHS were collected from public databases to define macrophage subtypes by performing clustering analysis. Trajectory analysis of macrophage subpopulations and transcription factor regulatory networks were used to characterize the dynamic changes of macrophage from healthy state to HLHS. In conclusion, our study confirmed that macrophages played a key role in the development of HLHS, providing a new direction for the treatment of HLHS patients.
2.1 Downloading Single-Nucleus RNA Sequencing (snRNA-seq) Data
The snRNA-seq data for 4 HLHS and 12 normal healthy control samples (donors) were obtained from GSE203274 (https://www.ncbi.nlm.nih.gov/geo/) in the Gene Expression Omnibus (GEO) database. Clinical information of the samples is shown in Table S1.
2.2 Pre-Processing of the snRNA-seq Data
Firstly, we used the “Read10X” function in the Seurat package of R software to read the snRNA-seq data of each sample [11]. Cells with nuclear genes in the range of 700–10,000 and mitochondria no more than 20% were retained. Next, data were normalized using the “NormalizeData” function, and the batch effect was removed by the harmony package after principal component analysis (PCA) downscaling. The uniform manifold approximation and projection (UMAP) analysis was performed using the “RunUMAP” function to acquire the top 20 principal components. Cell clustering was performed by applying “FindNeighbors” and “FindClusters” functions. All HLHS cells and macrophages were initially clustered under the parameters of dims = 1:20 at resolution = 0.1. Finally, marker genes were collected from the CellMarker 2.0 database (http://bio-bigdata.hrbmu.edu.cn/CellMarker/) to annotate HLHS cell clusters.
2.3 Differential Expression Analysis between Different HLHS Cell Clusters
The heterogeneity of gene expression patterns among different cell subpopulations was analyzed. Specifically, differentially expressed genes (DEGs) in each cell subpopulation were calculated using the “FindAllMarkers” function. The DEGs between different HLHS cell clusters were identified under the parameters of logfc.threshold = 0.25, min.pct = 0.25, and only.pos = TRUE, and for identifying those between different macrophage clusters, the parameters were set at |avg_log2FC| > 0.25 and p_val_adj < 0.01.
2.4 Functional Enrichment Analysis
To investigate the biological processes (BP) associated with the specific cell subtypes, the Gene Ontology (GO) enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/). The p-value < 0.05 was defined as the threshold value.
2.5 Ligand–Receptor Expression and Cell Interactions
Cell-to-cell communication analysis was conducted in the CellChat package [12] to explore whether macrophage subtypes were involved in regulating cardiomyocytes and the differences between the HLHS group and the donor group. The possibility of secreted signaling and cell-cell contact in different cell subtypes was quantitatively inferred based on the CellChatDB database [13], and overexpressed ligands or receptors in these subtypes were identified. The netVisual_bubble package to visualize the bubble diagram for receptor-ligand interactions between macrophages and cardiomyocytes.
2.6 Pseudotime Trajectory Analysis
After quality control, developmental trajectories of differential genes in macrophage subtypes between the HLHS and donor groups were reconstructed using the Monocle2 package [14]. Subsequently, these data were downscaled by applying the “DDRTree” method with default parameters, and the cells were sorted according to pseudotime.
2.7 Single-Cell Regulatory Network Inference and Clustering (SCENIC) Analysis
Following the official tutorial available on the SCENIC website (https://github.com/aertslab/SCENIC), the GENIE3 method was used to select potential target genes associated with each transcription factor (TFs) [15]. Subsequently, we adopted the top10perTarget approach to establish a transcription factor regulatory network. Afterward, pairs of TF-gene relationships with a strong level of certainty were selected and the AUCell tool was employed to evaluate regulon activity within each cell. The regulon signified a TF and its specific collection of genes responsible for directly controlling target genes. The SCENIC package was used to measure the activity of each regulon within individual cells, with a higher score indicating a stronger activation for the respective gene set.
In this study, all biological analyses and computational processes were performed using R software. The degree of correlation between the activity of TFs and pseudotime was calculated by the Pearson method under the thresholds of |R| > 0.25 and p < 0.05. Specifically, p < 0.01 was considered as statistically significant.
3.1 Single-Cell Profiling Analysis of HLHS Patients
After normalizing, downscaling, and clustering the snRNA-seq data from the GSE203274 dataset, we obtained a total of 75,136 cells (Fig. S2). Based on the expression levels of all genes, these cells were categorized and annotated into 9 cell clusters, including cardiac fibroblast, cardiomyocyte, endocardial cell, endothelial cell, macrophage, neuron, pericyte, T cell, and vascular smooth muscle cells (Fig. 1A). The expression of specific marker genes for the nine cell types is shown in Fig. 1B. DEGs between different cell clusters are shown in Table S2. We observed that the cellular proportion of macrophages, neurons, pericytes, T cells, and vascular smooth muscle cells in the HLHS group was reduced when compared to donors (Fig. 1C). As shown in Fig. 1D, the percentage of cardiomyocytes reached 53.95% in the whole sample, followed by endothelial cells (16.57%) and cardiac fibroblast (11.78%). Macrophages have immune-independent tissue-specific functions that could produce a critical impact on the heart [16]. For this reason, the role of macrophages in HLHS was further explored.
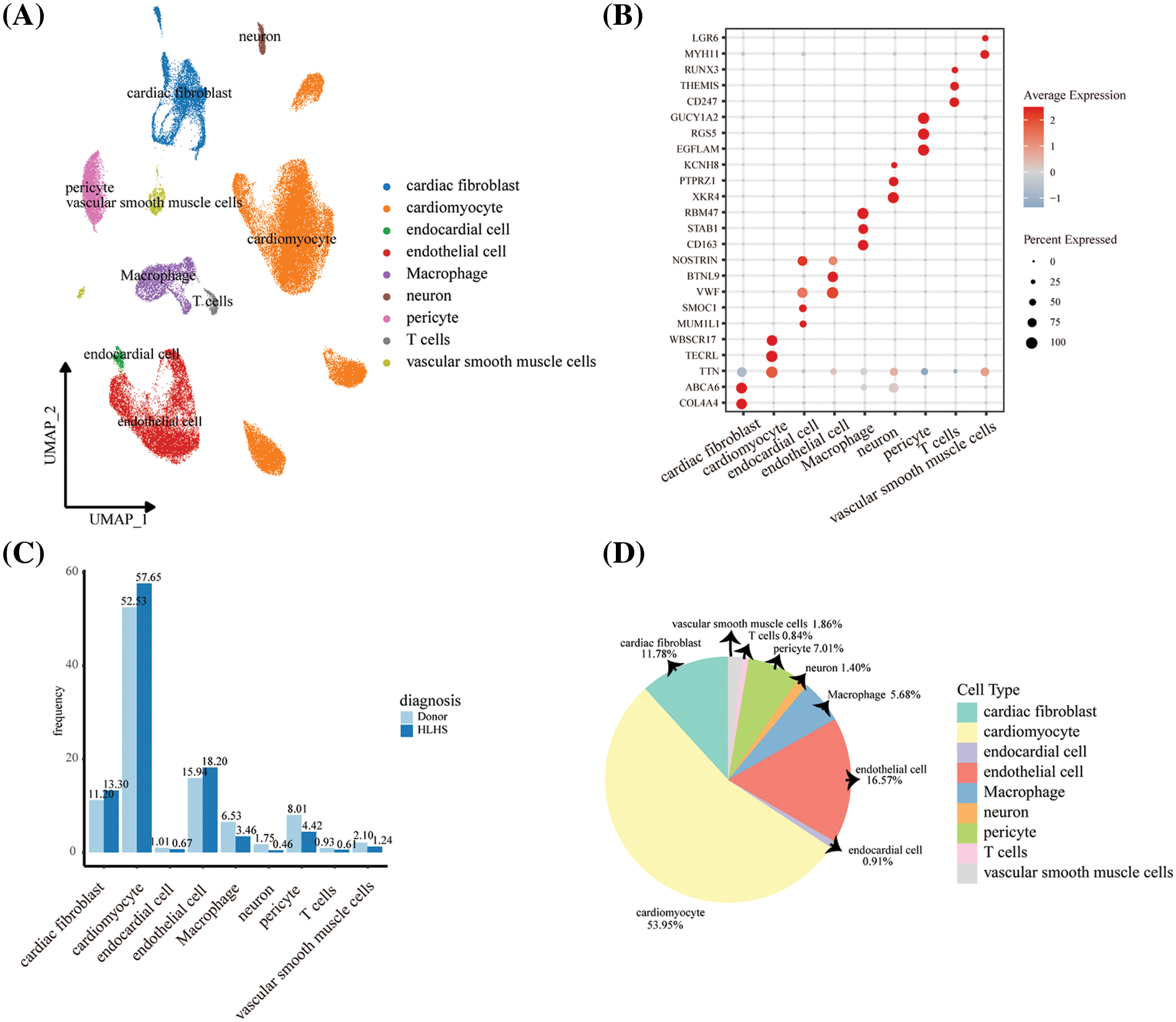
Figure 1: Single-cell profiles of patients with HLHS. (A) UMAP plot showing nuclei in HLHS and healthy control tissue samples annotated and clustered with 9 cell clusters. (B) Bubble plot showing expression levels of marker genes in 9 cell clusters. (C) Differences in the proportion of different cell clusters between HLHS and healthy donors. (D) The proportion of 9 cell clusters in all HLHS samples
3.2 Identification of Macrophage Subpopulations in Cardiac Tissues
To further explore the heterogeneity of macrophages in HLHS patients and healthy donors, we screened macrophages in HLHS and normal controls after quality control and obtained a total of 4,237 macrophages. Using these macrophages, 3 macrophage subpopulations in HLHS were defined, including Macrophages 1, Macrophages 2, and Macrophages 3 (Fig. 2A). Marker genes for the 3 macrophage subpopulations are shown in Fig. 2B. Specifically, ENPP2, FGF13, NPAS3, AVIL, RYR1, and DAB2 were high-expressed in Macrophages 1; HLA-DQA1, ITGAX, FLT3, IL18R1, IL1R2, and FCN1 were high-expressed in Macrophages 2; CDK1, AURKB, ESCO2, SPC25, BIRC5, and CDCA3 were high-expressed in Macrophages 3. Analysis of the biological processes of the 3 macrophage subpopulations was performed. Macrophages 1 was mainly associated with pathways such as nervous system development, angiogenesis, and apoptotic processes; Macrophages 2 was closely associated with pathways such as the T cell receptor signaling pathway and the immune response; Macrophages 3 was mainly enriched in pathways related to cell proliferation and the cell cycle (Fig. 2C). Notably, we found that Macrophages 1 and Macrophages 2 had proximity in the UMAP map, suggesting that the biological roles of these two macrophage subpopulations were similar. In the functional enrichment analysis, we also found that both the two macrophage subpopulations were enriched in immune-related pathways. These results demonstrated that Macrophages 1 may have functions similar to cardiac tissue macrophages [16,17].
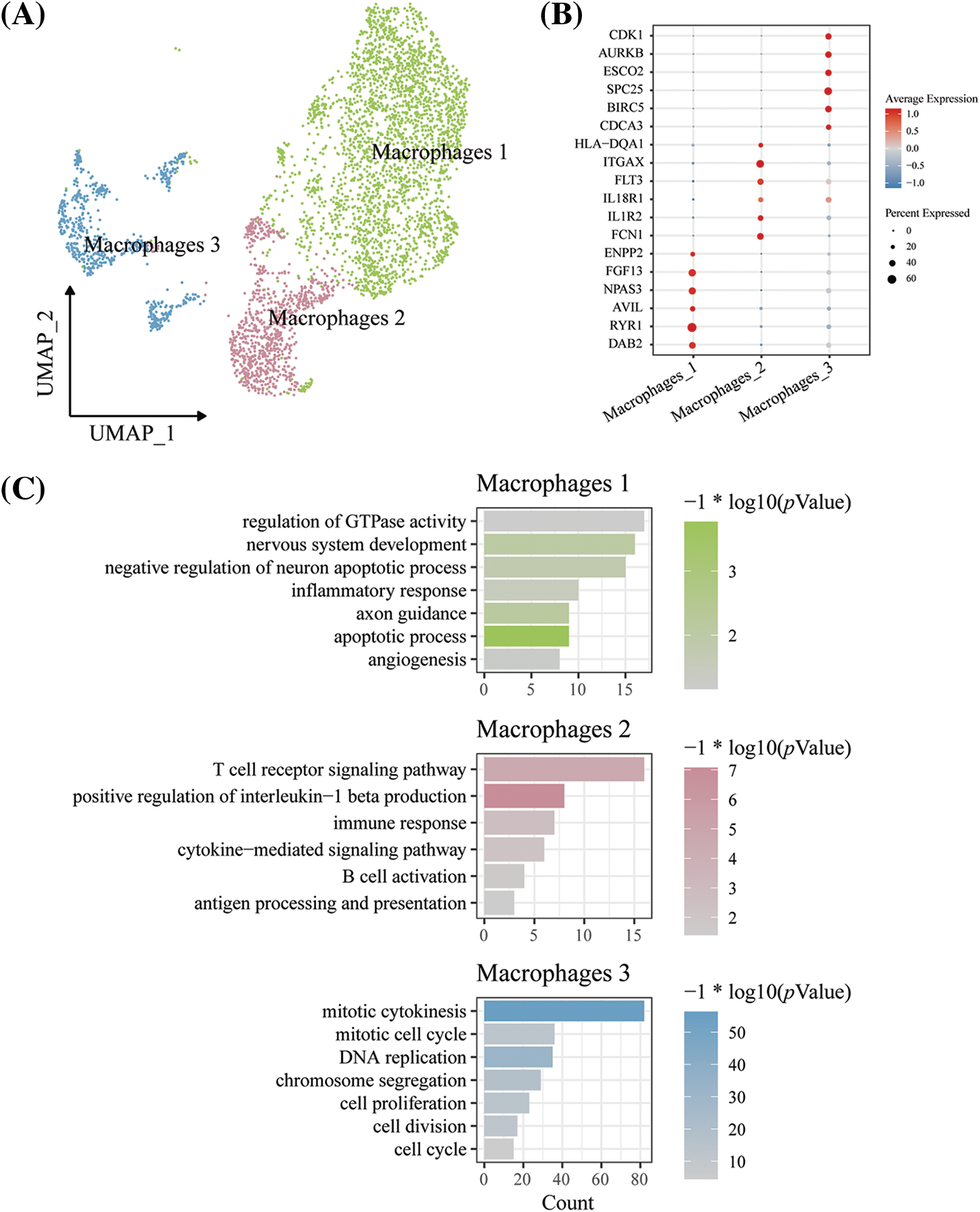
Figure 2: Analysis of macrophage heterogeneity in HLHS and its control samples. (A) UMAP diagram for the macrophage subpopulations in HLHS. (B) Differences in expression levels of marker genes in the 3 macrophage subpopulations. (C) Enrichment analysis of biological processes in DEGs between the macrophage subpopulations
3.3 Analysis of Cell-Cell Communication between Macrophages 1 and Cardiomyocytes
Communication between Macrophages 1 and cardiomyocytes in HLHS patients and healthy donors was further explored to analyze the effects of macrophages on the heart in HLHS. As shown in Figs. 3A and 3B, the ligand-receptor pairs were compared according to the secreted signals in the HLHS group and the donor group, and we observed that GAS6-ASL, IL6, and IGF1 were only present in the healthy donor group rather than in the HLHS group. The results of ligand-receptor pair analysis in cell-cell contact showed that compared to HLHS patients, THY1 and L1CAM were only high-expressed in the healthy donor group. (Figs. 3C and 3D). These results indicated that Macrophages 1 fulfilled the role of cardiomyocytes to maintain their homeostasis and function during the development of HLHS. However, the intricate cellular interactions that resulted in the abnormalities of HLHS remained unclear.
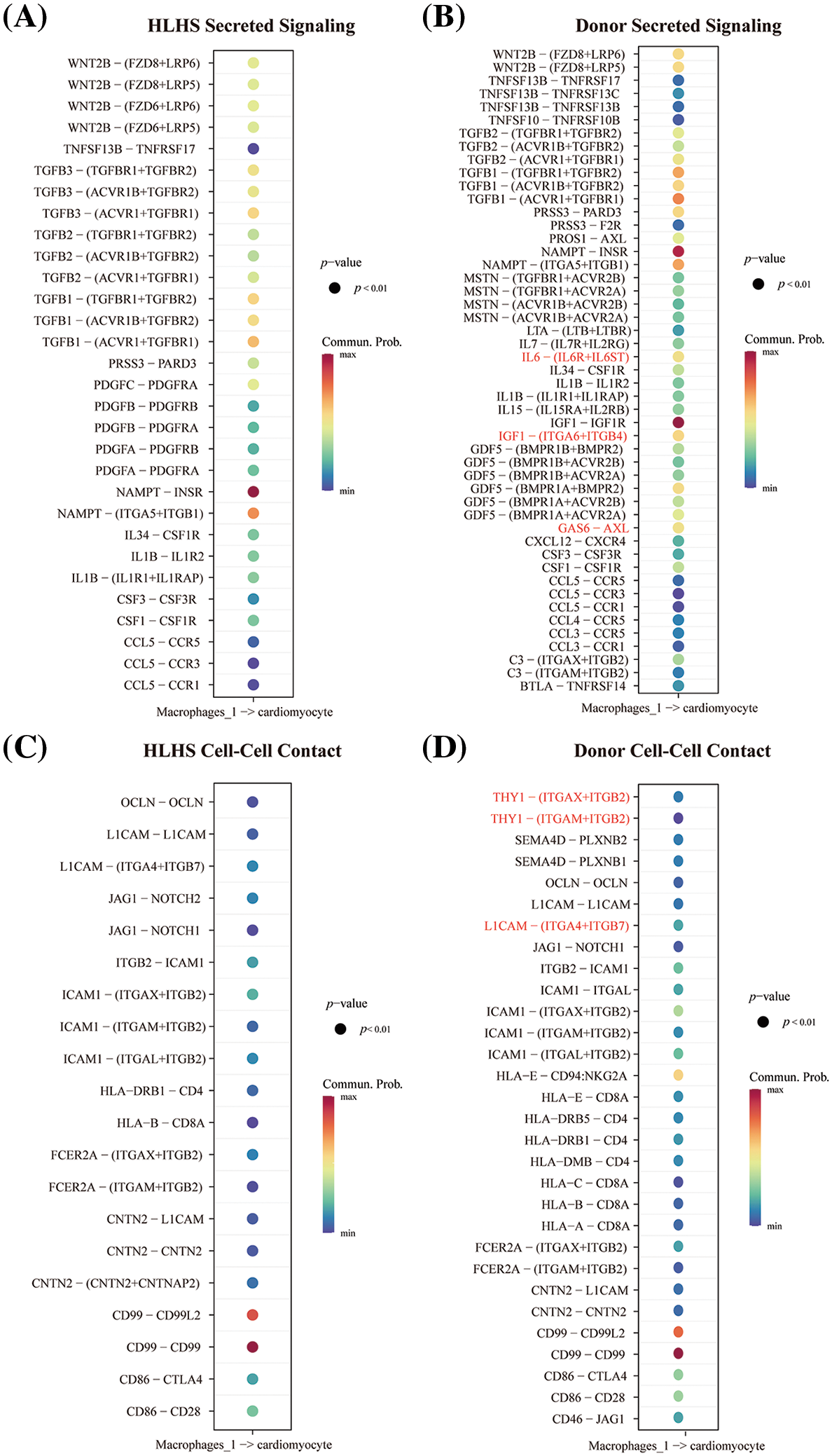
Figure 3: Analysis of cell-cell communication between macrophages and cardiomyocytes in HLHS. (A and B) Analysis of secretory signal communication between macrophages and cardiomyocytes in HLHS (A) and donor (B) groups. (C and D) Cell-cell chat analysis between macrophages and cardiomyocytes in the HLHS group (C) and the donor group (D)
3.4 Cell Trajectory Analysis of Macrophages 1
The evolution of the Macrophages 1 subpopulation from donor to HLHS was reconstructed by mapping its differentiation trajectory with the use of the monocle package to observe the dynamics of the gene expression patterns (Fig. 4A). Subsequently, the variation of DEGs with pseudotime between the two sets of samples from HLHS and donor were analyzed and two cell clusters (cluster1 and cluster2) were classified accordingly. As shown in Fig. 4B, cluster1 was mainly enriched in pathways related to nervous system development, heart development, and multicellular organism growth, while cluster2 was mainly enriched in negative regulation concerning cell proliferation, cell growth and angiogenesis, and apoptosis-related pathways. Next, pseudotime trajectory changes in genes related to cardiac development and nervous system development were analyzed. Macrophages 1 subpopulation was significantly reduced in HLHS tissues, and its biological function changed along the disease progression. As shown in Figs. 4C and 4D, we found that the expression levels of genes related to cardiac development and neurodevelopment were downregulated in Macrophages 1, suggesting that macrophages were no longer effective in supporting normal cardiac development and functional maintenance in the HLHS heart.
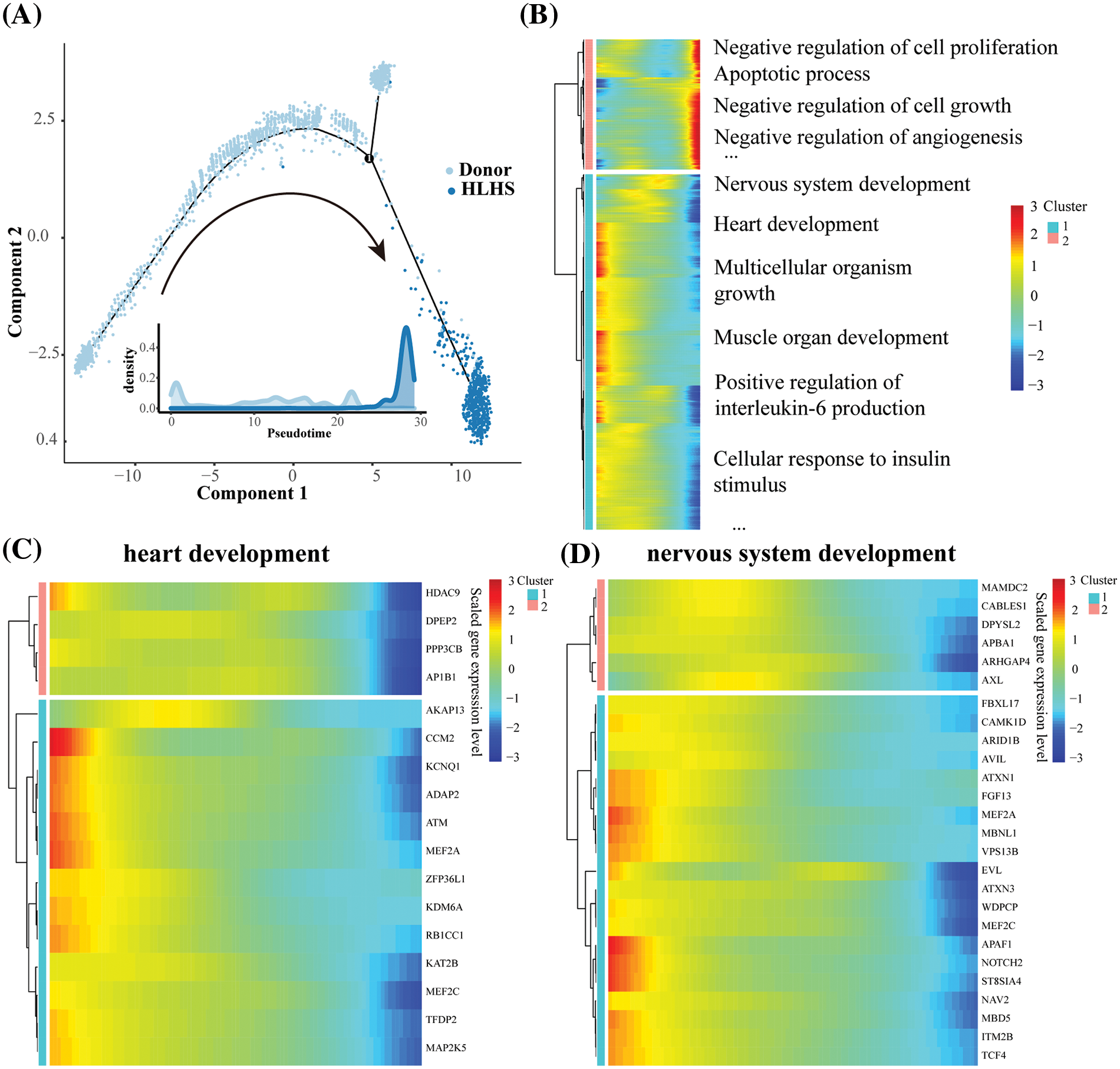
Figure 4: Cellular trajectory analysis of Macrophages 1 in HLHS. (A) Pseudotime trajectory analysis for Macrophages 1. (B) Expression profiles of pseudotime genes of DEGs in Macrophages 1 cells. (C) Pseudotime gene expression profiles of cardiac development-related genes in Macrophages 1 cells. (D) Pseudotime gene expression profiles of nervous system development-related genes in Macrophages 1 cells
3.5 Identification of Markers with Potential Influence on Macrophages in HLHS
To further identify key TFs with regulatory roles in the Macrophages 1 subpopulation, we used the SCENIC package to construct a network of TFs that regulated target genes and identified regulons in this subpopulation. We calculated the AUCell score for each regulon, with a higher score indicating a higher activity of the TF. As shown in Fig. 5A, analysis of the relationship between each TF and pesudotime demonstrated that E2F3, ELF1, and ZNF407 were significantly positively correlated with pesudotime, while ELF2 and FOXO3 were significantly negatively correlated with pesudotime. Notably, FOXO3 was the most negatively correlated with pesudotime (R = −0.764, p < 2.2e-16) (Fig. 5B). To further determine the roles of FOXO3 and ELF2, we screened target genes for pathways related to cardiac development, WNT signaling pathway, cell cycle, and signal transduction, respectively, and found that the expression levels of these genes were lower in the HLHS group than in the healthy controls (Fig. 5C and 5D, Table S3). These results demonstrated that the TFs such as FOXO3 and ELF2 played a crucial role in Macrophages 1 subpopulations to promote cardiac functional homeostasis in HLHS.
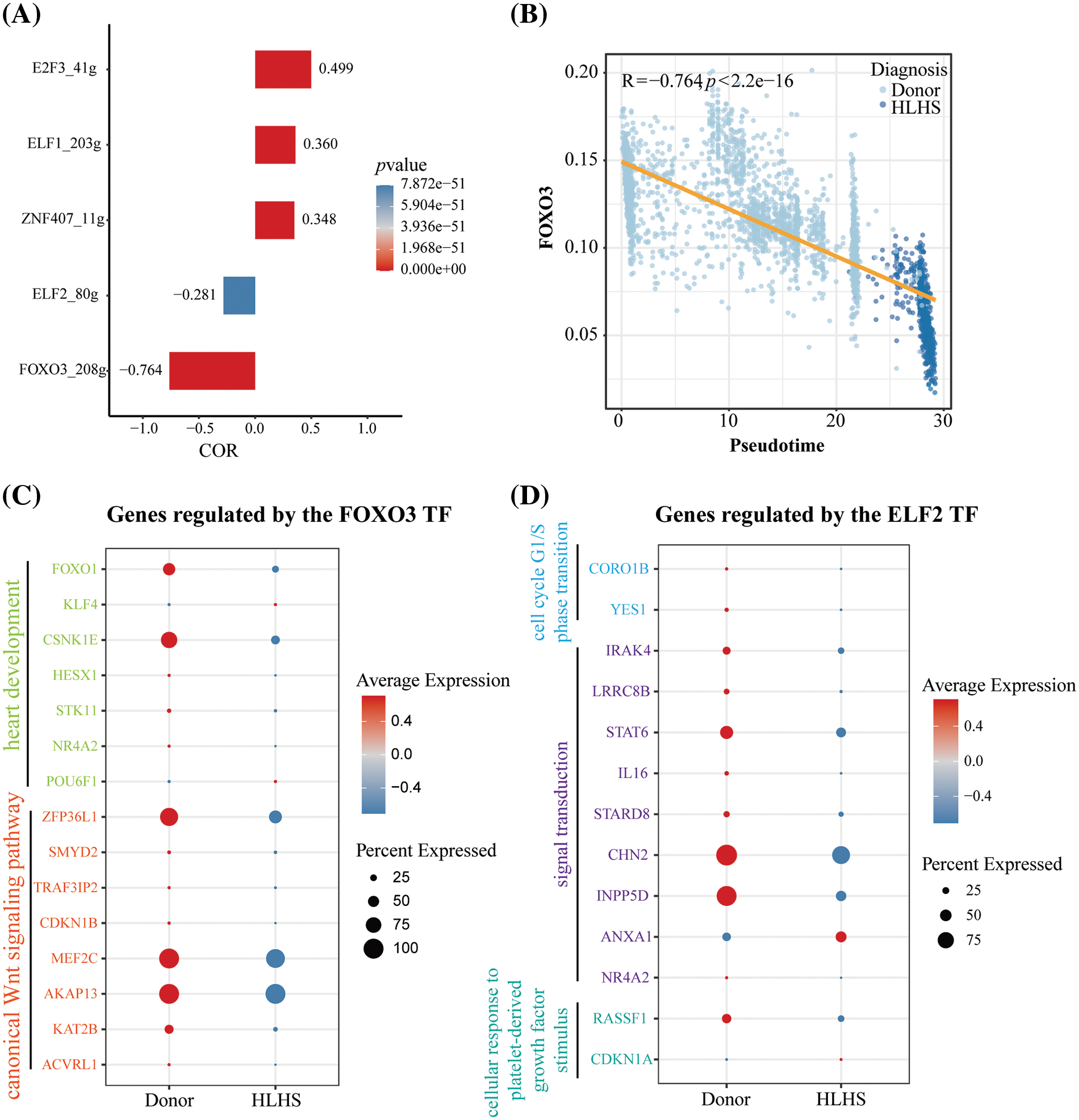
Figure 5: Transcription factor regulatory network of Macrophages 1 cells in HLHS. (A) Correlation of AUCell scores and proposed times for regulon within Macrophages 1. (B) Scatterplot of FOXO3 and pesudotime correlation in Macrophages 1. (C) Expression level analysis of target genes associated with FOXO3 and cardiac development and WNT signaling pathway. (D) Expression level analysis of target genes associated with ELF2 and cell cycle, signal transduction, and cellular response to platelet-derived growth factor stimulus pathway
HLHS refers to an anatomical and functional inadequacy of the left side of the heart and an inability of the left ventricle to perform systemic perfusion [18]. Macrophages have prominent immune-independent tissue-specific functions in the heart. In particular, for example, tissue-resident macrophages could prevent fibrosis, increase electrical conduction in the heart, and facilitate wound healing [4,16]. However, the extreme plasticity of macrophages often blurs the boundaries between their different phenotypes. Macrophage expression in different tissues can be distinguished by different gene expression profiles and protein levels [5,19–21]. Therefore, we used snRNA-seq data to further analyze the molecular mechanisms of macrophages in HLHS.
It was found that the proportion of macrophage clusters was reduced in HLHS patients compared to donors. To further explore the heterogeneity of macrophages in HLHS, macrophages were divided into 3 cellular subpopulations (Macrophages 1, 2, and 3). Macrophages 1 were largely involved in nervous system developmental, angiogenic, and apoptotic pathways. The heart is highly innervated by autonomic nerve cells, and dynamic autoregulation of the heart and blood vessels is essential for maintaining normal life activities in mammals [22]. Fujiu et al. showed that in mice with cardiac pressure overload, sympathetic activation activates cells within the kidney in which cytokine CSF2 causes secretion between tissue macrophages and endothelial cells and then stimulates cardiac-resident macrophages. This dynamic interaction also highlights the homeostatic function of tissue macrophages and the sympathetic nervous system in the context of cardiovascular disease [23]. HLHS exhibits pathological changes such as empty areas within the heart muscle, formation of vascular channels, and infiltration of mononuclear cells along the septum of the ventricle [24]. Analysis of the communication between Macrophages 1 and cardiomyocytes showed that some ligand-acceptors were absent in the HLHS group but not in donors, which suggested that these ligands and receptors functioned critically in the development of HLHS. For example, AXL/GAS6 is essential for the secretion of gonadotropin-releasing hormone (GnRH) to promote survival and migration of hypothalamic neurons. AXL enhances this process by facilitating Rac1-mediated chemotactic migration of the developing GnRH neurons [25,26]. IL6, a pleiotropic cytokine, plays a key role in tissue regeneration and cell differentiation [27–30]. Hunter and his colleagues found that the functions of IL6 and its receptors vary when IL6 is expressed in different cells or activated at different times or modes [31]. Although IGF1 is structurally and functionally related to insulin, it has a high growth-promoting activity. By stimulating glucose transport in bone-derived osteoblasts, IGF1 acts at a much lower concentration than insulin, which contributes to synapse maturation [32,33]. In addition, in cell-cell contact, THY1 also appeared only in the donor group and was absent in the HLHS group. During synaptogenesis and other events in the brain, THY1 also plays an important role in cell-ligand or cell-cell interactions [34]. These results tentatively confirmed that the Macrophages 1 subpopulation identified in this study can promote nervous system development, tissue regeneration, and angiogenesis, which were similar to the functions of cardiac tissue-resident macrophages [17,35]. Hence, we speculated that Macrophages 1 may be a cardiac tissue-resident macrophages.
Our pseudotime-based analyses demonstrated that Macrophages 1 had the same potential to promote the nervous system and cardiac development and the growth of multiple tissues. In addition, cardiac and neurodevelopment-related genes progressively reduced in Macrophages 1 along the pseudotime trajectories. In particular, we observed that two TFs (FOXO3 and ELF2) were significantly negatively correlated with sham time, and that the target genes associated with pathways such as cardiac development, WNT, cell cycle, and signaling were all significantly reduced in the HLHS group. FOXO3 could promote gene-gene interactions, changes in chromatin conformation, and interactions of topologically related structural domains, which are involved in the regulation of several genes in cellular resilience processes such as stress response, autophagy, cell proliferation, and apoptosis [36,37]. Hill et al. observed a state of congenital heart disease-specific cells in cardiomyocytes with increased signaling of FOXO [38]. FOXO3 has been shown to have a decisive effect on cardiomyocytes, and some researchers indicated that microRNAs (miR-122) can facilitate cardiomyocyte hypertrophy progression partially by directly regulating the FoxO3-calcineurin pathway [39]. In addition, Willcox et al. showed that the main reason for the protective allele of FOXO3 to extend human lifespan is its protection against coronary death [40]. Similarly, Chen et al. demonstrated that the FOXO3-associated longevity genotype could also extend the lifespan of high-risk patients with cardiometabolic diseases by protecting the heart under metabolic stress [41]. As a member of the Ets family of transcription factors, ELF2 participates in the regulatory regions of lymphocyte development and functional genes [42]. Guan et al. identified ELF2 isoforms as important regulators of cell proliferation, cell cycle, and apoptosis [43]. Consistently, we found that the expression levels of the relevant cell cycle target genes of ELF2 in Macrophages 1 were downregulated in patients with HLHS. These results reconfirmed that the critical role of the Macrophages 1 subpopulation in promoting homeostasis of cardiac function may be achieved by targeting FOXO3 and ELF2.
However, there were some limitations in our study. Firstly, the data in this paper were obtained mainly from public databases, and due to the rarity of cardiac explants in children, we were unable to obtain snRNA-seq derived from fresh tissues of healthy controls. Hence, our future study will increase the diversity of datasets in order to provide a more comprehensive analysis of biosignatures and disease mechanisms. In addition, an animal model of HLHS and fresh tissue samples will be used to verify the reliability and accuracy of the current results.
In summary, we analyzed snRNA-seq data to classify macrophage subtypes that maintained the cardiac functions in HLHS and revealed the differentiation status of Macrophages 1 and the activity of TFs. Our study confirmed that the role of Macrophages 1 in maintaining functional homeostasis of cardiomyocytes was possibly realized through regulating FOXO3 and ELF2 or targeting their genes. The present findings provided novel insights into the heterogeneity of macrophages in HLHS and the molecular mechanisms of macrophages in the development of HLHS.
Acknowledgement: None.
Funding Statement: This work was supported by Jiangsu Province Postgraduate Practice Innovation Program (SJCX22_0766), Natural Science Foundation of Jiangsu Province (BK20231378), Leader of Geriatric Clinical Technology Application Research Project of Jiangsu Provincial Health Commission (LR2022002).
Author Contributions: The authors confirm contributions to the paper as follows: study conception and design: Xiaozhuo Xu, Yilin Huang, Xu Han; data collection: Yilin Huang; analysis and interpretation of results: Xu Han, Yilin Huang. Xiaozhuo Xu; draft manuscript preparation: Yilin Huang, Xiaozhuo Xu, Huiqing Yu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available in the [GSE203274] repository (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203274).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/chd.2024.050231.
References
1. Giugliano P, Ciamarra P, de Simone M, Feola A, Zangani P, Campobasso CP, et al. Hypoplastic left heart syndrome: about a postnatal death. Diagnostics. 2023;13(5):821. doi:10.3390/diagnostics13050821. [Google Scholar] [PubMed] [CrossRef]
2. Bejjani AT, Wary N, Gu M. Hypoplastic left heart syndrome (HLHSmolecular pathogenesis and emerging drug targets for cardiac repair and regeneration. Expert Opin Ther Targets. 2021;25(8):621–32. doi:10.1080/14728222.2021.1978069. [Google Scholar] [PubMed] [CrossRef]
3. Datta S, Cao W, Skillman M, Wu M. Hypoplastic left heart syndrome: signaling & molecular perspectives, and the road ahead. Int J Mol Sci. 2023;24(20):15249. doi:10.3390/ijms242015249. [Google Scholar] [PubMed] [CrossRef]
4. Hulsmans M, Clauss S, Xiao L, Aguirre AD, King KR, Hanley A, et al. Macrophages facilitate electrical conduction in the heart. Cell. 2017;169(3):510–22.e20. doi:10.1016/j.cell.2017.03.050. [Google Scholar] [PubMed] [CrossRef]
5. Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11(11):723–37. doi:10.1038/nri3073. [Google Scholar] [PubMed] [CrossRef]
6. Molawi K, Wolf Y, Kandalla PK, Favret J, Hagemeyer N, Frenzel K, et al. Progressive replacement of embryo-derived cardiac macrophages with age. J Exp Med. 2014;211(11):2151–8. doi:10.1084/jem.20140639. [Google Scholar] [PubMed] [CrossRef]
7. O’Rourke SA, Dunne A, Monaghan MG. The role of macrophages in the infarcted myocardium: orchestrators of ECM remodeling. Front Cardiovasc Med. 2019;6:101. doi:10.3389/fcvm.2019.00101. [Google Scholar] [PubMed] [CrossRef]
8. Epelman S, Lavine KJ, Beaudin AE, Sojka DK, Carrero JA, Calderon B, et al. Embryonic and adult-derived resident cardiac macrophages are maintained through distinct mechanisms at steady state and during inflammation. Immunity. 2014;40(1):91–104. doi:10.1016/j.immuni.2013.11.019. [Google Scholar] [PubMed] [CrossRef]
9. Heidt T, Courties G, Dutta P, Sager HB, Sebas M, Iwamoto Y, et al. Differential contribution of monocytes to heart macrophages in steady-state and after myocardial infarction. Circ Res. 2014;115(2):284–95. doi:10.1161/CIRCRESAHA.115.303567. [Google Scholar] [PubMed] [CrossRef]
10. Simonson B, Chaffin M, Hill MC, Atwa O, Guedira Y, Bhasin H, et al. Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure. Cell Rep. 2023;42(2):112086. doi:10.1016/j.celrep.2023.112086. [Google Scholar] [PubMed] [CrossRef]
11. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck 3rd WM, et al. Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–902.e21. doi:10.1016/j.cell.2019.05.031. [Google Scholar] [PubMed] [CrossRef]
12. Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun. 2021;12(1):1088. doi:10.1038/s41467-021-21246-9. [Google Scholar] [PubMed] [CrossRef]
13. Lin J, Cai Y, Wang Z, Ma Y, Pan J, Liu Y, et al. Novel biomarkers predict prognosis and drug-induced neuroendocrine differentiation in patients with prostate cancer. Front Endocrinol. 2022;13:1005916. doi:10.3389/fendo.2022.1005916. [Google Scholar] [PubMed] [CrossRef]
14. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat Methods. 2017;14(3):309–15. doi:10.1038/nmeth.4150. [Google Scholar] [PubMed] [CrossRef]
15. Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–6. doi:10.1038/nmeth.4463. [Google Scholar] [PubMed] [CrossRef]
16. Nicolás-Ávila JA, Lechuga-Vieco AV, Esteban-Martínez L, Sánchez-Díaz M, Díaz-García E, Santiago DJ, et al. A network of macrophages supports mitochondrial homeostasis in the heart. Cell. 2020;183(1):94–109.e23. doi:10.1016/j.cell.2020.08.031. [Google Scholar] [PubMed] [CrossRef]
17. Moskalik A, Niderla-Bielińska J, Ratajska A. Multiple roles of cardiac macrophages in heart homeostasis and failure. Heart Fail Rev. 2022;27(4):1413–30. doi:10.1007/s10741-021-10156-z. [Google Scholar] [PubMed] [CrossRef]
18. Metcalf MK, Rychik J. Outcomes in hypoplastic left heart syndrome. Pediatr Clin North Am. 2020;67(5):945–62. doi:10.1016/j.pcl.2020.06.008. [Google Scholar] [PubMed] [CrossRef]
19. Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159(6):1312–26. doi:10.1016/j.cell.2014.11.018. [Google Scholar] [PubMed] [CrossRef]
20. Artyomov MN, Sergushichev A, Schilling JD. Integrating immunometabolism and macrophage diversity. Semin Immunol. 2016;28(5):417–24. doi:10.1016/j.smim.2016.10.004. [Google Scholar] [PubMed] [CrossRef]
21. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13(11):1118–28. doi:10.1038/ni.2419. [Google Scholar] [PubMed] [CrossRef]
22. Fujiu K, Manabe I. Nerve-macrophage interactions in cardiovascular disease. Int Immunol. 2022;34(2):81–95. doi:10.1093/intimm/dxab036. [Google Scholar] [PubMed] [CrossRef]
23. Fujiu K, Shibata M, Nakayama Y, Ogata F, Matsumoto S, Noshita K, et al. A heart-brain-kidney network controls adaptation to cardiac stress through tissue macrophage activation. Nat Med. 2017;23(5):611–22. doi:10.1038/nm.4326. [Google Scholar] [PubMed] [CrossRef]
24. Norwood WI, Lang P, Casteneda AR, Campbell DN. Experience with operations for hypoplastic left heart syndrome. J Thorac Cardiovasc Surg. 1981;82(4):511–9. [Google Scholar] [PubMed]
25. Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338(6211):161–4. doi:10.1038/338161a0. [Google Scholar] [PubMed] [CrossRef]
26. Allen MP, Linseman DA, Udo H, Xu M, Schaack JB, Varnum B, et al. Novel mechanism for gonadotropin-releasing hormone neuronal migration involving Gas6/Ark signaling to p38 mitogen-activated protein kinase. Mol Cell Biol. 2002;22(2):599–613. doi:10.1128/MCB.22.2.599-613.2002. [Google Scholar] [PubMed] [CrossRef]
27. Ng J, Hynes K, White G, Sivanathan KN, Vandyke K, Bartold PM, et al. Immunomodulatory properties of induced pluripotent stem cell-derived mesenchymal cells. J Cell Biochem. 2016;117(12):2844–53. doi:10.1002/jcb.25596. [Google Scholar] [PubMed] [CrossRef]
28. Xie Z, Tang S, Ye G, Wang P, Li J, Liu W, et al. Interleukin-6/interleukin-6 receptor complex promotes osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Stem Cell Res Ther. 2018;9(1):13. doi:10.1186/s13287-017-0766-0. [Google Scholar] [PubMed] [CrossRef]
29. Kondo M, Yamaoka K, Sakata K, Sonomoto K, Lin L, Nakano K, et al. Contribution of the interleukin-6/STAT-3 signaling pathway to chondrogenic differentiation of human mesenchymal stem cells. Arthritis Rheumatol. 2015;67(5):1250–60. doi:10.1002/art.39036. [Google Scholar] [PubMed] [CrossRef]
30. Deng W, Chen H, Su H, Wu X, Xie Z, Wu Y, et al. IL6 receptor facilitates adipogenesis differentiation of human mesenchymal stem cells through activating P38 pathway. Int J Stem Cells. 2020;13(1):142–50. doi:10.15283/ijsc19073. [Google Scholar] [PubMed] [CrossRef]
31. Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16(5):448–57. doi:10.1038/ni.3153. [Google Scholar] [PubMed] [CrossRef]
32. Zoidis E, Ghirlanda-Keller C, Schmid C. Stimulation of glucose transport in osteoblastic cells by parathyroid hormone and insulin-like growth factor I. Mol Cell Biochem. 2011;348(1–2):33–42. doi:10.1007/s11010-010-0634-z. [Google Scholar] [PubMed] [CrossRef]
33. Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503(7475):267–71. doi:10.1038/nature12618. [Google Scholar] [PubMed] [CrossRef]
34. Sedov E, Koren E, Chopra S, Ankawa R, Yosefzon Y, Yusupova M, et al. THY1-mediated mechanisms converge to drive YAP activation in skin homeostasis and repair. Nat Cell Biol. 2022;24(7):1049–63. doi:10.1038/s41556-022-00944-6. [Google Scholar] [PubMed] [CrossRef]
35. Yang P, Chen Z, Huang W, Zhang J, Zou L, Wang H. Communications between macrophages and cardiomyocytes. Cell Commun Signal. 2023;21:206. [Google Scholar] [PubMed]
36. Donlon TA, Morris BJ, Chen R, Masaki KH, Allsopp RC, Willcox DC, et al. FOXO3 longevity interactome on chromosome 6. Aging Cell. 2017;16(5):1016–25. doi:10.1111/acel.12625. [Google Scholar] [PubMed] [CrossRef]
37. Donlon TA, Willcox BJ, Morris BJ. FOXO3 cell resilience gene neighborhood. Aging. 2017;9(12):2467–8. doi:10.18632/aging.101349. [Google Scholar] [PubMed] [CrossRef]
38. Hill MC, Kadow ZA, Long H, Morikawa Y, Martin TJ, Birks EJ, et al. Integrated multi-omic characterization of congenital heart disease. Nature. 2022;608(7921):181–91. doi:10.1038/s41586-022-04989-3. [Google Scholar] [PubMed] [CrossRef]
39. Song G, Zhu L, Ruan Z, Wang R, Shen Y. MicroRNA-122 promotes cardiomyocyte hypertrophy via targeting FoxO3. Biochem Biophys Res Commun. 2019;519(4):682–8. doi:10.1016/j.bbrc.2019.09.035. [Google Scholar] [PubMed] [CrossRef]
40. Willcox BJ, Tranah GJ, Chen R, Morris BJ, Masaki KH, He Q, et al. The FoxO3 gene and cause-specific mortality. Aging Cell. 2016;15(4):617–24. doi:10.1111/acel.12452. [Google Scholar] [PubMed] [CrossRef]
41. Chen R, Morris BJ, Donlon TA, Masaki KH, Willcox DC, Davy PMC, et al. FOXO3 longevity genotype mitigates the increased mortality risk in men with a cardiometabolic disease. Aging. 2020;12(23):23509–24. doi:10.18632/aging.202175. [Google Scholar] [PubMed] [CrossRef]
42. Cho JY, Akbarali Y, Zerbini LF, Gu X, Boltax J, Wang Y, et al. Isoforms of the Ets transcription factor NERF/ELF-2 physically interact with AML1 and mediate opposing effects on AML1-mediated transcription of the B cell-specific blk gene. J Biol Chem. 2004;279(19):19512–22. doi:10.1074/jbc.M309074200. [Google Scholar] [PubMed] [CrossRef]
43. Guan FH, Bailey CG, Metierre C, O’Young P, Gao D, Khoo TL, et al. The antiproliferative ELF2 isoform, ELF2B, induces apoptosis in vitro and perturbs early lymphocytic development in vivo. J Hematol Oncol. 2017;10(1):75. doi:10.1186/s13045-017-0446-7. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools