 Open Access
Open Access
ARTICLE
Single-Cell RNA Sequencing Reveals Potential for Endothelial-to-Mesenchymal Transition in Tetralogy of Fallot
1 State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia, Urumqi, 830002, China
2 Department 5 of Cardiology, Affiliated Hospital of Traditional Chinese Medicine of Xinjiang Medical University, Urumqi, 830002, China
3 Department of Cardiology, The First Affiliated Hospital of Jinan University, Guangzhou, 510630, China
* Corresponding Author: Baojian Zhang. Email:
# Equal contribution
Congenital Heart Disease 2023, 18(6), 611-625. https://doi.org/10.32604/chd.2023.047689
Received 14 November 2023; Accepted 26 December 2023; Issue published 19 January 2024
Abstract
Background: Tetralogy of Fallot (TOF) is a very common cyanotic congenital heart disease. Endothelial-to-mesenchymal transition (EndoMT) is recognized as a physiological mechanism involved in embryonic heart development and endothelial formation. However, there is still a gap in the reports related to the mechanism of EndoMT development in TOF. Methods: First, transcriptomic data of single cell nuclei of TOF and Donor were obtained based on the Gene Expression Omnibus (GEO) database, and the data were normalized and clustered by dimensionality reduction using the Seurat package. Subsequently, differentially expressed genes (DEGs) between TOF and Donor were screened using the “FindMarkers” function, and the gene sets of interest were enriched. Finally, to characterize the dynamics of EndoMT occurrence in TOF, we performed pseudotime cell trajectory inference as well as utilized SCENIC analysis to probe the gene regulatory networks (GRNs) dominated by transcription factors (TFs) in endothelial cells. Results: We identified a total of six cell clusters based on single-cell nuclear transcriptome data from TOF and Donor. We found that 611 genes with up-regulated expression within TOF showed conversion to mesenchyme. By subdividing endothelial cell subtypes, endothelial cells 2 were shown to be involved in cell adhesion, migration and extracellular matrix processes. Pseudo-time and SCENIC analyses showed that endothelial cell 2 has EndoMT potential. In addition, ERG and TEAD1 are TFs that play key regulatory roles in this subtype, and both of their target genes are also highly expressed in TOF. This demonstrates that ERG and TEAD1 effectively promote the EndoMT process. Conclusion: Our study reveals the molecular mechanisms underlying the development of EndoMT in TOF, which demonstrates that manipulating the endothelial-to-mesenchymal transition may offer unprecedented therapeutic potential for the treatment of TOF.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| TOF | Tetralogy of fallot |
| CHD | Congenital heart disease |
| EndoMT | Endothelial-to-mesenchymal transition |
| DEGs | Differentially expressed genes |
| GRNs | Gene regulatory networks |
| TFs | Transcription factors |
| PCA | Principal component analysis |
| GEO | Gene Expression Omnibus |
| FC | Fold change |
| UMAP | Uniform manifold approximation and projection |
| ERG | ETS-related gene |
| TGF-β | Transforming growth factor-beta |
| TEAD1 | Transcriptional enhancer activator domain 1 |
| YAP | Yes-associated protein |
Congenital heart disease (CHD) is a common congenital disability and a leading cause of infant mortality, with a global prevalence of 0.6%–9.4% [1]. Tetralogy of Fallot (TOF) is the most common cyanotic CHD and is clinically characterized by overriding aorta, right ventricular hypertrophy, ventricular septal defect, and pulmonary stenosis with a morbidity of 3.9/10,000 births [2,3]. With current surgical techniques for CHD are improving the mortality has decreased from early 25% to less than 2% [4]. However for patients with complex CHD, especially TOF infants, continue to experience high postoperative morbidity and mortality [5–7].
Endothelial cells are the most numerous types of mesenchymal cells in the heart [8]. Although endothelial cells in the heart come from different sources, they share many similarities in biological functions. For example, there are abundant cell connections that can effectively block the entry of macromolecules and particles into cells, serving as a good barrier between circulating blood and myocardial cells [9]. Secondly, compared to endothelial cells in other organs such as the lungs, kidneys, and liver, endothelial cells in the heart have higher angiogenic potential and metabolic efficiency, providing sufficient oxygen and energy for normal growth and functional maintenance of the heart [10–12]. Recently, analysis of single-cell sequencing technology has revealed that endothelial cells in the heart play an important role in cardiac remodeling and heart failure [13,14]. Compared to endothelial cells in organs such as the lungs, kidneys, and liver, endothelial cells in the heart have a higher angiogenic potential and metabolic efficiency, which provides sufficient oxygen and energy for normal growth and maintenance of heart function [10]. Endothelial cells also undergo phenotypic changes after stimulation with various growth factors, such as the adoption of more mesenchymal-like features [15]. In particular, endothelial-mesenchymal transition (EndoMT) serves as a unique form of epithelial-mesenchymal transition from endothelial cells to mesenchymal cells. In addition, EndoMT is also considered to be an intermediate state of vascular fibers, and endothelial cells primarily pass through EndoMT in order to regulate myocardial fibrosis, which is a type of ischemic cardiomyopathy [16,17]. Zeisberg et al. found that during myocardial fibrosis induced by a transverse aortic constriction, a large number of cardiomyocytes came to be overactivated by EndoMT of endothelial cells [18]. EndoMT hyperactivation of endothelial cells was present in angiotensin II as well as in isoprenaline-induced myocardial fibrosis [19,20]. Besides, EndoMT in tumors is also thought to contribute to the development of myocardial fibrosis and atherosclerosis following cardiac stress or injury [21,22].
Given the complexity of cardiac development, the development of TOF during cardiogenesis is affected in multiple ways. However, the mechanism regarding the occurrence of EndoMT has not been reported in TOF. Here, we demonstrated the ability of endothelial cells to transform to mesenchyme in TOF by single-cell analysis technique. We obtained TOF single-cell RNA sequencing (scRNA-seq) data based on public databases and identified the endothelial cell subpopulations most capable of transformation to mesenchyme by clustering. Subsequently, the trajectory analysis and transcription factor regulatory network of endothelial cell subpopulations were constructed in order to identify key markers that promote endothelial-to-mesenchymal transition. Our study provides new insights into the treatment of TOF.
2.1 Acquisition of Single-Cell Transcriptome Data
The scRNA-seq data for TOF and donors were obtained from the Gene Expression Omnibus (GEO) database from GSE203274 (https://www.ncbi.nlm.nih.gov/geo/) (GSE203274_AlNuclei_snRNA_ metadata.csv.gz). Subsequently, 10× Genomics to be used to construct sequencing libraries for TOF.
2.2 Processing of Single-Cell Transcriptome Data
We read the data using the “Read10X” function in the Seurat package [23] and retained cells with mitochondrial gene ratios of less than 0.2% and gene expression numbers between 200 and 10,000. Next, the data are normalized using the “SCTransform” function. Removing batch effects between different samples using harmony package [24] after the principal component analysis (PCA) dimensionality reduction. In addition, the top 30 principal components (PCs) were analyzed by uniform manifold approximation and projection (UMAP) analysis for dimension reduction. “FindNeighbors” and “FindClusters” functions for clustering different cell clusters. For the initial clustering of all TOF cells, we set the resolution to 0.15. And for the clustering of endothelial cells, the resolution was set to 0.1. Finally, we annotated the cell type of each cell according to the marker genes provided by Cell Taxonomy (https://ngdc.cncb.ac.cn/celltaxonomy/), PanglaoDB (https://panglaodb.se/index.html), and CellMarker 2.0 (http://bio-bigdata.hrbmu.edu.cn/CellMarker/) databases.
2.3 Differential Expression Analysis in Endothelial Cells
In order to further analyze the differences between the endothelial cell subtypes of TOF, we first extracted all endothelial cells using the “subset” function, and screened the differentially expressed genes (DEGs) between TOF and its donors using the “FindMarkers” function. The filtering criteria were |avg logFC| > 0.25 and p val adj < 0.05.
We uploaded the gene sets of interest into the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) and analyzed them based on the Gene Ontology (GO) enrichment analysis to explore the biological process (BP) of the relevant cell subtypes. Additionally, gene sets were also uploaded into Kyoto Encyclopedia of Genes and Genomes (KEGG) database for pathway enrichment analysis. And p-value < 0.05 was set as the threshold values.
2.5 Trajectory Analysis of Cell Subtypes
To explore the fate of cell differentiation in TOF, we performed pseudotime analysis of cell types of interest using Monocle 2. We first use Monocle 2 to read the count data of the expression matrix and merge the phenotypic information of the cells, and use the “newCellDataSet” function to construct the Monocle object. Subsequently, we filtered out cells with less than 10 expressed genes and utilized the “FindAllmarkers” function to calculate the DEGs between TOF and donors. Next, we use the “reduceDimension” function to reduce the dimensionality of the above data (max_components = 2, method = “DDRTree”), and use the “orderCells” function to order the cells to complete the construction of the trajectories. We set more branches of cells within the donor as the starting point of the trajectory. The “differentialGeneTset” function (fullModelFormulaStr = “~sm.ns(Pseudotime)”) was used to calculate the pseudotime-related genes (q val < 0.01), and the “plot_pseudotime_heatmap” function was used to plot the heatmap of these genes with respect to pseudotime expression. Finally, we utilize the “plot_genes_in_pseudotime” function to plot the scatterplot of the gene of interest with respect to pseudotime expression.
2.6 Prediction of Core Transcription Factors (TFs)
To infer the core TFs for each branch based on Monocle identification, we used the SCENIC algorithm to infer the regulatory activity in each cell state [25,26]. We used the GENIE3 method to calculate the potential target genes for each TFs according to the official tutorial provided on the SCENIC website (https://github.com/aertslab/SCENIC), and used the top10perTarget approach to construct the transcription factor regulatory network. Subsequently, we identified TF-target gene relationship pairs with high confidence and utilized the AUCell package to calculate the degree of regulon activity within each cell. Each regulon is a TF and its set of genes that directly regulate target genes, and the SCENIC package scores the activity of each regulon in individual cells. A higher score represents a higher level of activation for that gene set. Finally, we used the cytoscape package [27] to demonstrate the target gene network regulated by TFs.
2.7 Potential Drugs Targeting Core TFs
Comparative Toxicogenomics Database (CTD) was used for searching and download drugs which targeting core TFs.
In this study, we compared the differences between the two groups of continuous variables by using the Wilcoxon rank sum test. The Pearson algorithm was used to evaluate the correlation between the AUCell score and pseudotime for each cellular transcription factor. All computational procedures and plots were done using the R software (version 4.3.1). p-values < 0.05 was considered as indicating significant.
3.1 TOF scRNA-seq Data Revealed 6 Cell Clusters
In this study, we obtained a total of 62,214 cells for analysis after quality control and normalization of scRNA-seq data (Fig. S1A). After gene expression had been normalized and PCA performed, we co-clustered and identified TOF cell types. These cell clusters are divided into six known cell types by being labeled with genes (Fig. 1A): cardiomyocytes (69.5%, marked with FABP3 and TNNT2), endothelial cells (10.33%, marked with VWF and CDH5), fibroblasts (12.81%, marked with DCN and COL1A1), smooth muscle cells (3.97%, marked with ACTA2 and MYLK), macrophages (2.45%, marked with CTSC and C1QA), and neuronal cells (0.95%, marked with NRXN1 and RELN) (Figs. 1B–1D, Fig. S1B). The detailed cell information could be seen in (Figs. S1C–S1E). In addition, compared with the donor group, the percentage of cardiomyocytes and fibroblasts was significantly higher in TOF (p < 0.05), whereas the percentage of endothelial cells, smooth muscle cells, macrophages, and neuronal cells was lower (p < 0.05, Fig. 1D).
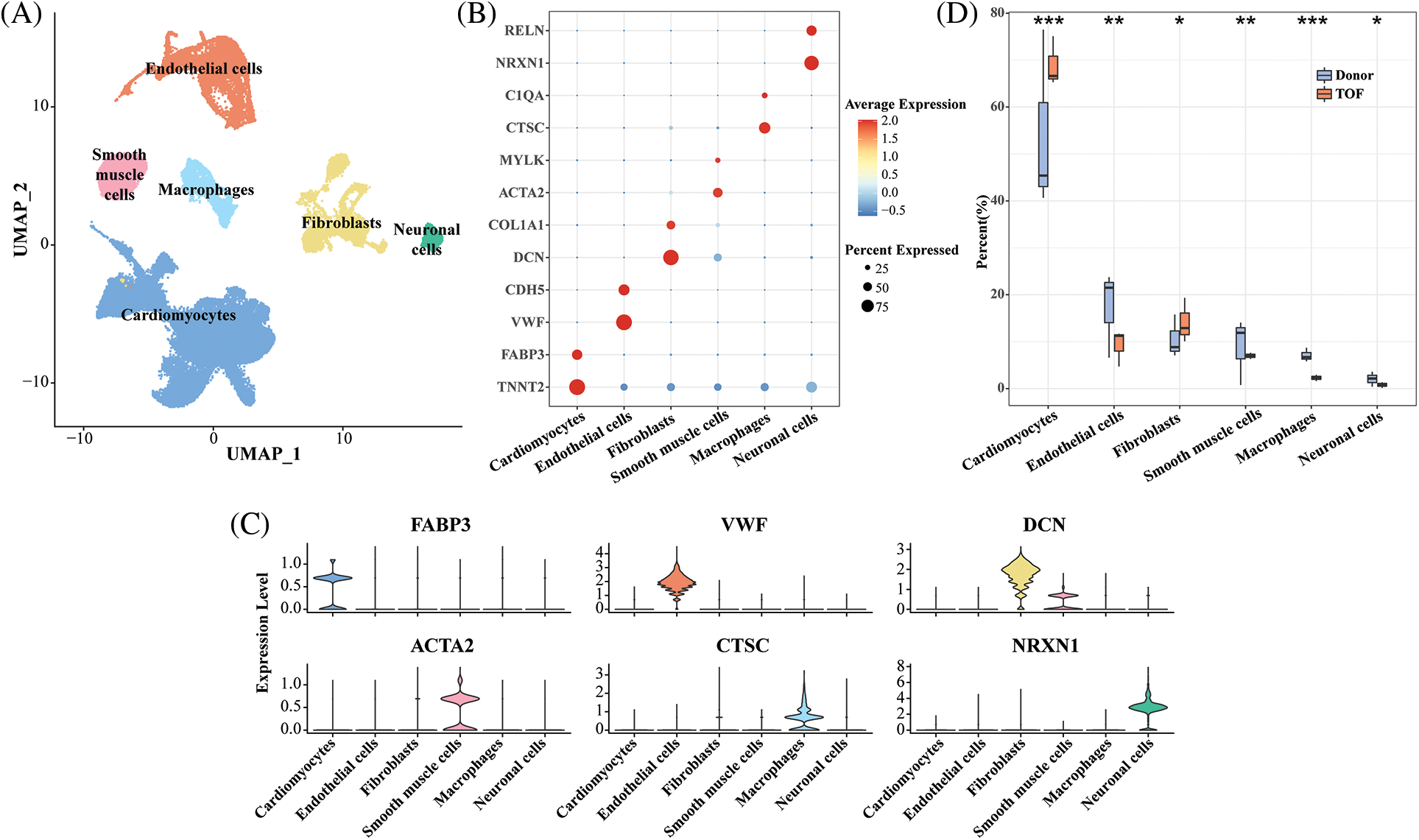
Figure 1: Single-cell landscape and molecular characterization of TOF samples. (A) UMAP plot showing the distribution of six annotated cell types in 62,214 TOF cells based on CellMarker database; (B) Bubble plots showing highly expressed marker gene expression levels for each cell type; (C) Violin plot showing specific marker gene expression for six annotated cell types; (D) Cell proportion difference of each cell cluster in each sample within TOF and Donor groups. *p < 0.05, **p < 0.01, ***p < 0.001
3.2 TOF Endothelial Cells Have the Potential to Transform to Mesenchyme
Endothelial cells in the heart have an important role in the remodeling of the heart. To further investigate the changes in endothelial gene expression after TOF, we calculated 611 up-regulated genes and 25 down-regulated genes based on the “FindMarkers” function (Fig. 2A; Table S1). Subsequently, results based on GO enrichment analysis showed that DEGs with up-regulated expression were mainly enriched in biological processes associated with cell migration, cell division, angiogenesis, Wnt signaling pathway, and epithelial-to-mesenchymal transition (EMT) (Fig. 2B). As shown in Fig. 2C, DEGs with down-regulated expression in TOF were mainly enriched in angiogenesis, stem cell and endothelial cell proliferation, and receptor internalization, among other pathways. Notably, since the entry for GO enrichment analysis was absent regarding the EndoMT pathway, in this study we demonstrated that endothelial cells have the potential to transform to mesenchyme by verifying that endothelial cells are enriched in the EMT pathway. For this purpose, we extracted genes with up-regulated expression in TOF endothelial cells for conversion to mesenchyme. As shown in Fig. 2D, a total of 12 genes were involved in the EndoMT process of TOF, including ITGAV, RAB1A, NCKAP1, LAMB1, WWC2, FLT1, ENG, FGFR1, NOTCH4, ROCK1, DDX5, and DDX17.
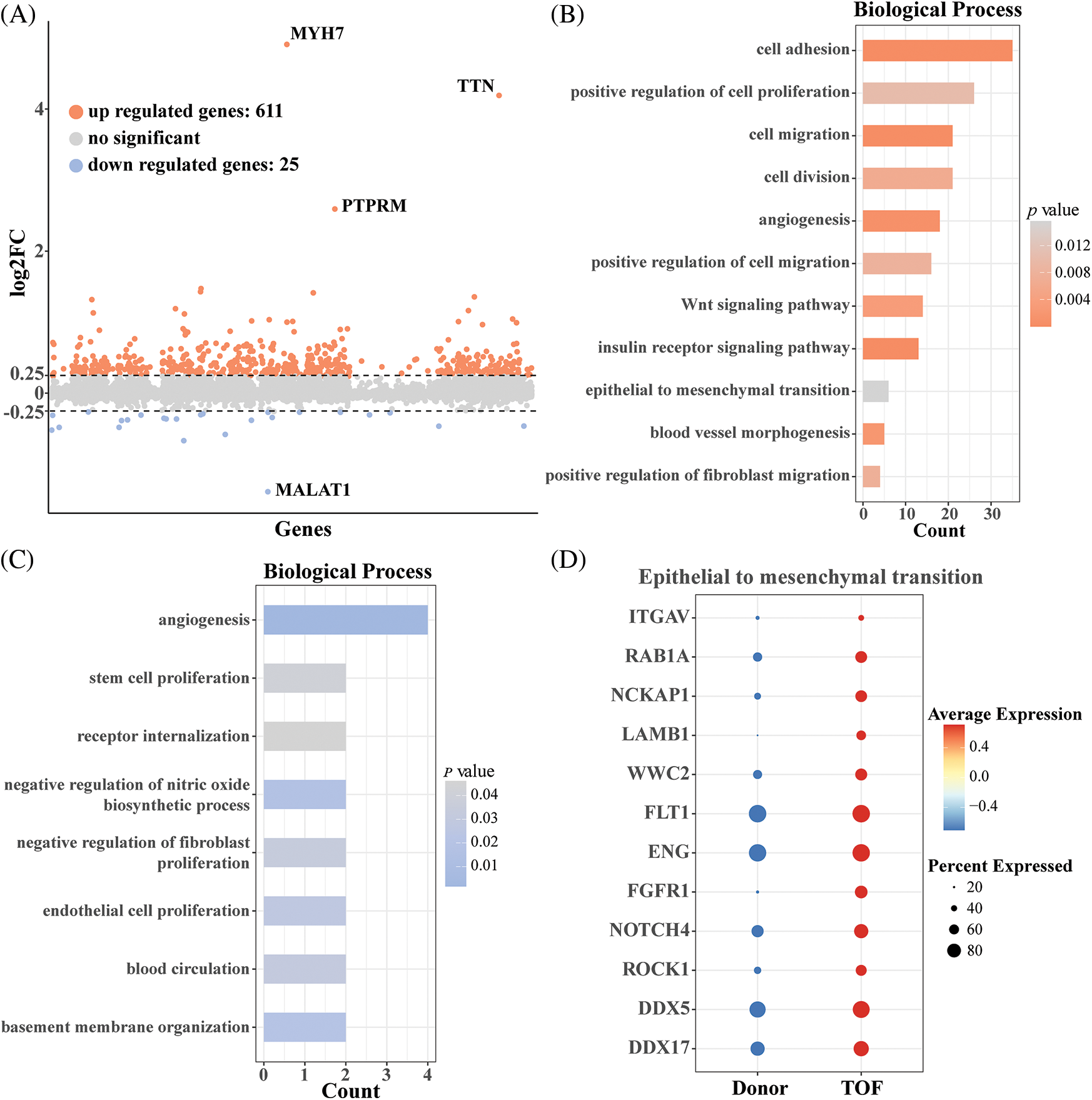
Figure 2: Differential expression analysis of endothelial cells between TOF and Donor groups. (A) Volcano plot of differentially expressed genes (DEGs) in annotated endothelial cells between TOF and Donor groups; (B) biological processes enriched for up-regulated DEGs between two groups; (C) biological processes enriched for down-regulated DEGs between two groups; (D) expression levels of genes involving in transformation to mesenchyme in TOF and Donor epithelial cells
3.3 Identification of Endothelial Cell Subtypes in TOF
To further explore the heterogeneity of endothelial cells at the gene expression level between TOF and Donor, we normalized and down-clustered the scRNA-seq of endothelial cells and obtained five endothelial cell subtypes (Fig. 3A). Subsequently, compared to Donor, we found that endothelial cell 2 (32.73%) and endothelial cell 4 (14.17%) accounted for a higher proportion of the TOF (Fig. 3B). This implies that endothelial cells 2 and 4 are closely related to the development of TOF. For this purpose, we identified genes that were highly expressed in endothelial cells 2 and 4, respectively. THSD4, ELN, PTPRJ, GCNT2, NTRK2, NOS1AP, and GJA5 were significantly expressed in endothelial cells 2. ACTC1, CRYAB, TNNT2, LAMB1, DES, LPL and MYH6 are highly expressed in endothelial cells 4 (Fig. 3C). DEGs between different endothelial cell subpopulations are shown in Table S2. Enrichment analysis showed that endothelial cells 4 was mainly enriched in pathways such as cardiac muscle contraction and cardiac myofibril assembly, suggesting that its biological function is similar to that of cardiomyocytes. Similar results could also be seen in Table S3. Importantly, we found that endothelial cells 2 were mainly enriched in pathways such as cell adhesion, cell migration and extracellular matrix organization (Fig. 3D). This implies that endothelial cell 2 may have the ability to transform to mesenchyme and play a key role in TOF progression.
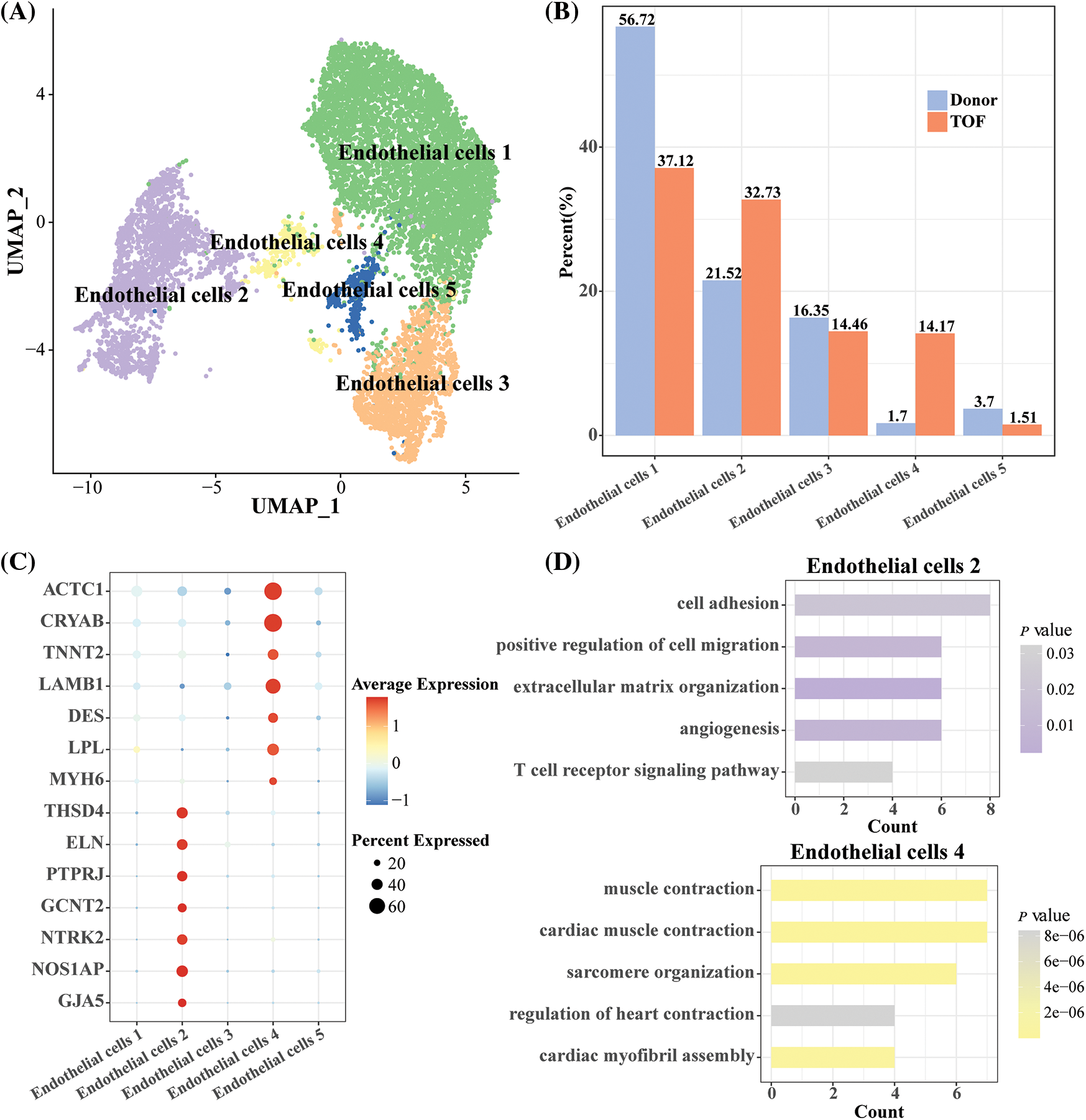
Figure 3: Heterogeneity analysis of endothelial cell subgroups in TOF and Donor groups. (A) UMAP plot displaying the classification of endothelial cell subpopulations by specific highly expressed genes; (B) cell proportions of each endothelial cell subpopulation between TOF and Donor groups; (C) bubble diagram demonstrating specific highly expressed genes in endothelial cell subpopulations 2 and 4; (D) GO enrichment analysis for biological processes of specific highly expressed genes in endothelial cell 2 and 4
3.4 Trajectory of Endothelial Cell Differentiation in TOF
To gain insight into the evolution of the endothelial cell 2 subtype from donor to TOF, we mapped the differentiation trajectory of this subtype using the monocle package to observe the dynamic changes in gene expression patterns therein (Fig. 4A). Subsequently, we found that with the prolongation of pesudotime, the expression level of genes related to angiogenesis, endothelial cell migration, Wnt and TGF-β receptor signaling pathway, and positive regulation of cell proliferation were gradually increased (Fig. 4B). This suggests that in TOF, the ability of endothelial cells to transform into mesenchyme increases progressively as the disease progresses. Subsequently, we found that genes within both the TGF-β receptor signaling pathway and the WNT signaling pathway exhibited a conversion process from donor to TOF, which again endothelial cells have the ability to convert to mesenchyme and are closely related to the TGF-β receptor signaling pathway and the WNT signaling pathway (Figs. 4C and 4D).
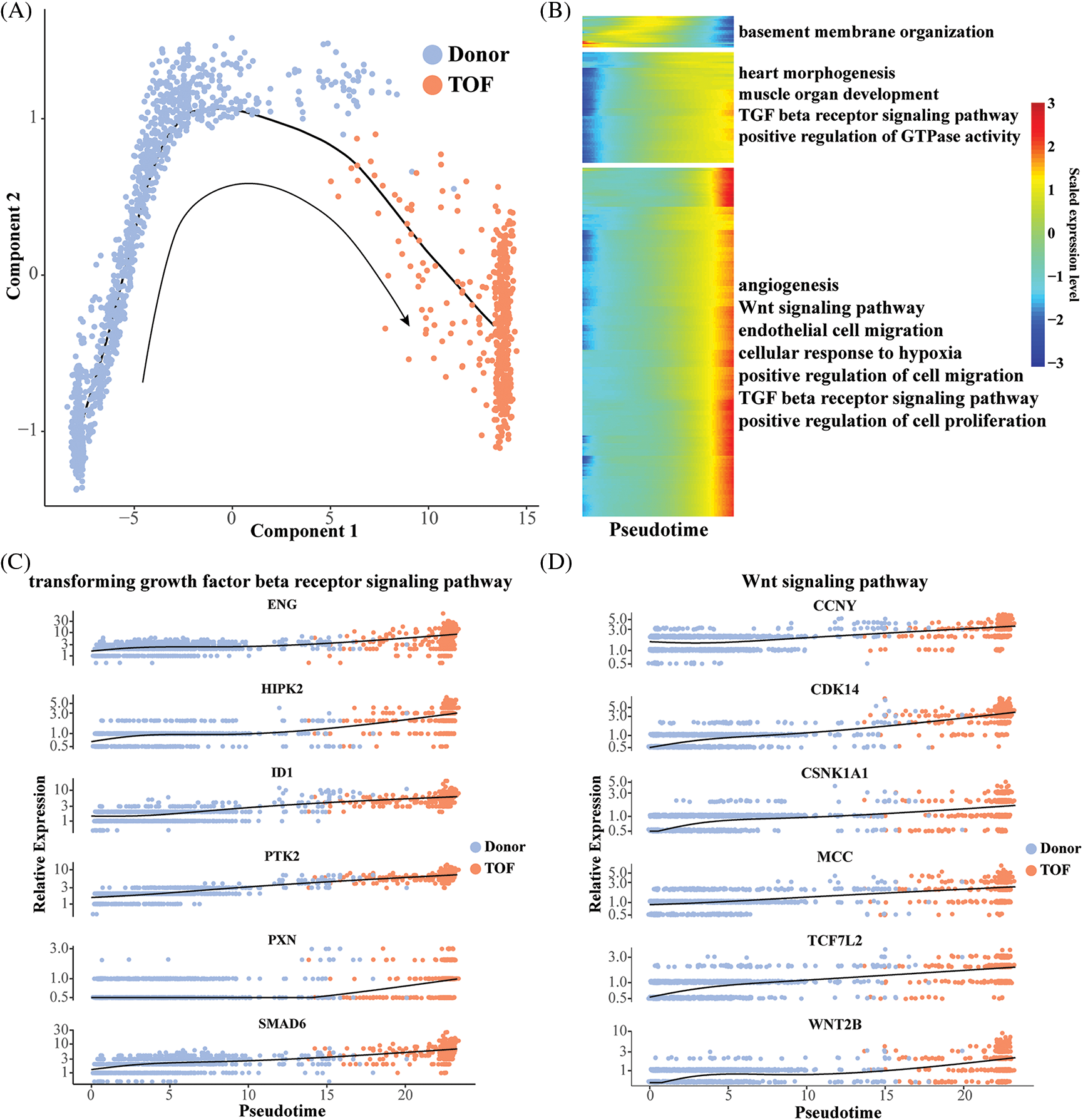
Figure 4: Pseudo-time analysis of endothelial cell 2. (A) Differentiation trajectory describing the trajectory of endothelial cell 2 conversion from Donor to TOF status; (B) heatmap of pseudotime-related genes with pseudotime changes and involved pathways; (C, D) scatterplot displaying the expression levels changes of genes participating in the TGF-β receptor (C) and Wnt (D) signaling pathway during pseudotime analysis
3.5 Identification of Markers with the Potential to Promote EndoMT
To further identify TFs that play a regulatory role during EndoMT, we first constructed gene regulatory networks (GRNs) of TFs within endothelial cell 2 subtypes using the SCENIC package. Subsequently, the activity scores of each TFs were calculated by the AUCell algorithm, where a higher score represents a stronger transcriptional regulatory role played by this TF within that cell subpopulation. Notably, we found the highest correlation coefficients of AUCell scores with pseudotime for REG (R = 0.6391) and TEAD1 (R = 0.4539) (Figs. 5A and 5B). In addition, ERG target genes including TEAD4, ACTB, SS18, DOCK4, DOCK9, SH2D3C, BCL2L1, GIT1, and PIK3R4, which are mainly involved in the positive regulation of stem cell population maintenance, regulation of cytokinesis, and GTPase mediated signal transduction (Fig. 5C). As shown in Fig. 5D, TEAD1 and target genes including USP6NL, GNAS, EV15, SMAD3, LTBP4, LATS2, RRAS2, ITGA5, PIAS1, and KHDRBS1, which are primarily involved in the G1/S transition of mitotic cell cycle, positive regulation of GTPase activity, positive regulation of cell migration, and regulation of TGF-β receptor signaling pathway. Among them, the pairs of regulatory relationships between TFs and target genes are shown in Table S4. Finally, in comparison to Donor, we found that ERG, TEAD, and related target genes were all significantly overexpressed in TOF patients (Figs. 5E and 5F). These results confirm the potential of REG and TEAD1 to effectively promote the transformation of endothelial cell subpopulation 2 to mesenchyme in TOF. For a supplement, we also predicted the potential drugs that could reduce the expressions for REG and TEAD1 (Table S5), hoping to provide a direction in drug targets research for TOF disease.
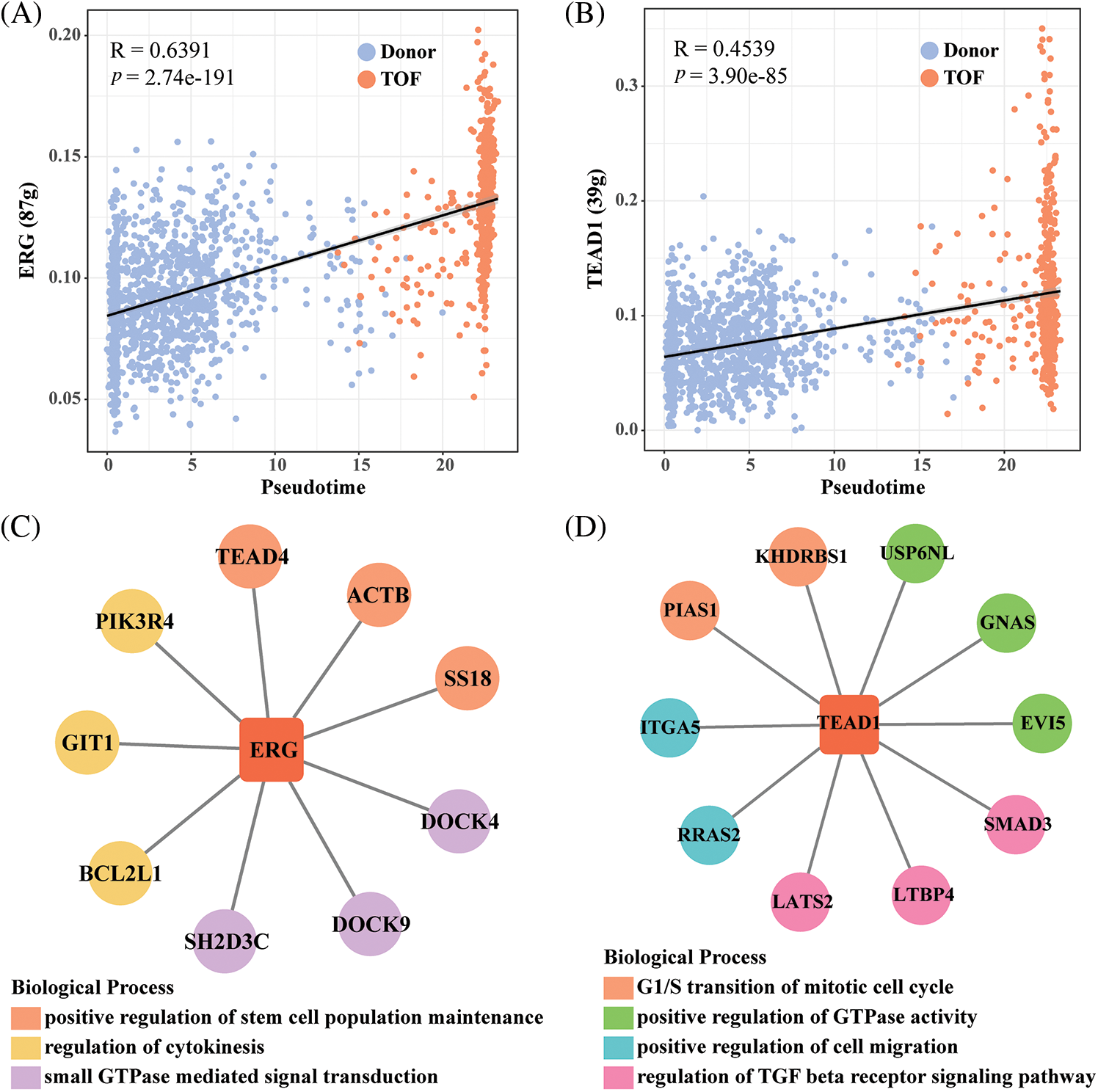
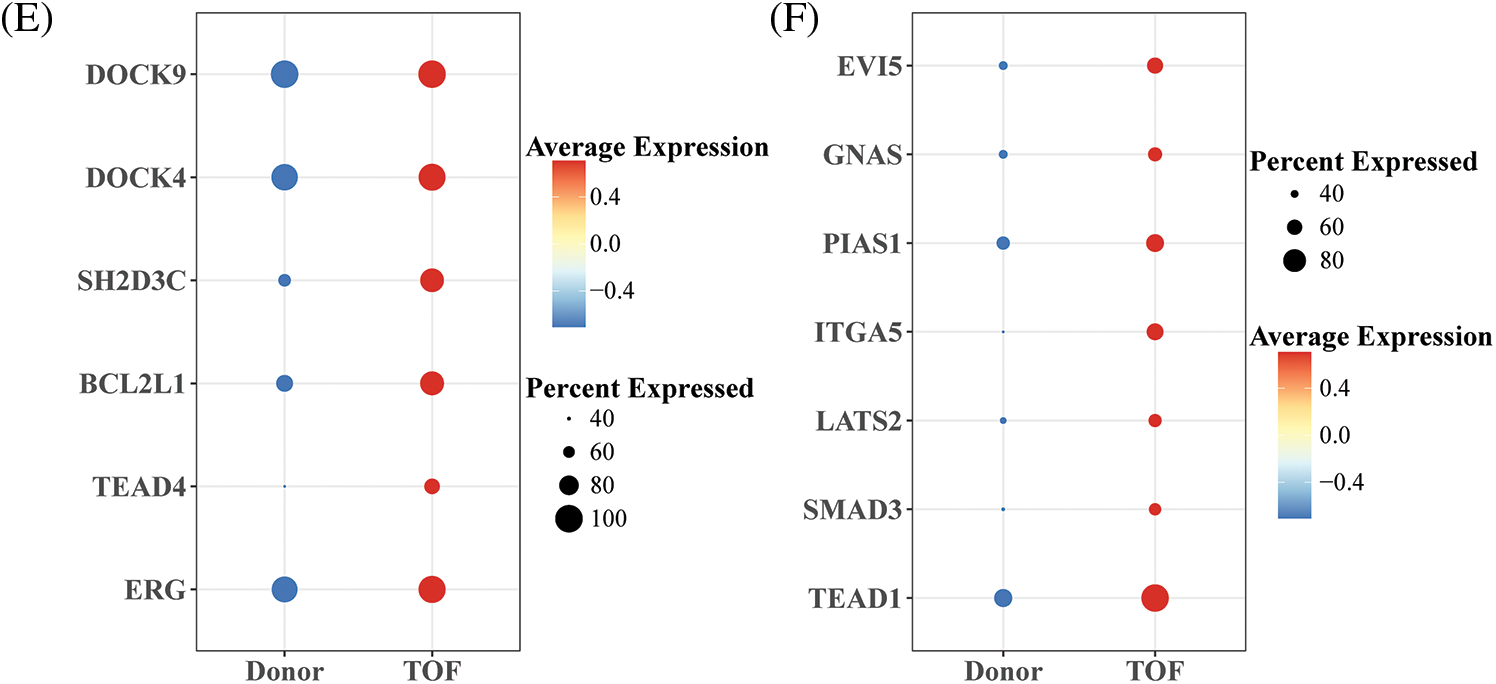
Figure 5: Identification of TFs of endothelial cell 2 subpopulations in TOF. (A, B) Scatter plots of correlation between AUCell score and pseudotime for ERG (A) and TEAD1(B); (C, D) GRNs of ERG (C) and TEAD1 (D) and their target genes (each color means a kind of biological functions); (E, F) differences in expression levels of ERG and TEAD1 and their target genes between the TOF and Donor groups
TOF is a typical CHD which accounts for 7%–10% of all CHDs [28]. Endothelial cells are essential for maintaining normal heart function, and damage to them can potentially lead to myocardial infarction and subsequent heart failure [29]. EndoMT is a process that endothelial cells are required to undergo, a process that is accompanied by a series of molecular and cellular changes, which in turn ultimately lead to alterations in the phenotype of mesenchymal cells [30]. Therefore, we based the single-cell nuclear data on TOF in order to explore the mechanism of EndoMT development.
In this study, we found that the percentage of endothelial cell clusters was reduced in TOF patients compared to Donor. To further explore the heterogeneity of endothelial cells in TOF, we categorized endothelial cells into five cellular subpopulations. Endothelial cells 2 seemed to involve in cell adhesion, cell migration, and extracellular matrix processes. EndoMT has been shown to alter the levels of mucins attached to cells and contribute to the remodeling of the extracellular matrix [28,31]. EndoMT is complex in the cardiovascular system, where endothelial cells in the cardiovascular lose intercellular adhesion throughout EndoMT and also acquire a strong ability to migrate and invade [32]. In addition, we found that PTPRJ, GCNT2, and GJA5 were highly expressed in endothelial cell 2 compared to other endothelial cell subpopulations. Among them, negative regulation of PTPRJ expression in endothelial cells can be accomplished by controlling the COX-2/PGE2 signaling pathway [33]. Zhang et al. found in breast cancer that GCNT2 was also able to promote endothelial cell adhesion and cell migration and invasion in vitro, it was also able to participate in the signaling process of EMT and TGF-β, which is a comprehensive mechanism in cancer cell metastasis [34]. Lu and his colleagues found that decreased GJA5 expression in arterial endothelial cells could damage arterial generation in acute ischemic cardiovascular disease [35]. Collectively, these results suggest that reduced endothelial cells in TOF may account for their abilities to transform to mesenchyme by high expression of relevant genes (PTPRJ, GCNT2, and GJA5), decreasing intercellular adhesion as well as promoting cell migration, which in turn promotes the development of TOF.
EndoMT is thought to be important for valve formation and cardiac separation during cardiac development, and its activation requires the involvement of many signaling pathways, including the TGF-β and Wnt signaling pathways [22,32]. The TGF-β signaling pathway is the most common pathway studied for induction of EndoMT. Fan et al. found that it is through activation of the TGF-β/SMAD2 pathway that lactic acid aims to disrupt the function of endothelial cells [36]. When the receptor for TGF-β is activated, TGF-β signaling is transmitted from the cell membrane to the nucleus via transduction of SMAD proteins [37]. Importantly, an important inhibitory protein in SAMD proteins, SMAD6, was identified in our study, which is progressively expressed at elevated levels with prolonged pesudotime in the development of TOF. SMAD6 expression levels are tightly correlated with the development of CHD [38]. Ruter et al. showed that SMAD6 is also a necessary element for endothelial cell flow-mediated responses [39]. Kulikauskas et al., on the other hand, showed that SMAD6 is able to maintain normal vascular development by regulating the balance of ALK1 signaling [40]. Notably, both TGF and Wnt signaling converge in the nucleus, where they will work together to coordinate the transcriptional control of shared target genes [41]. Wnt signaling, on the other hand, is extremely critical for EndoMT in endocardial cushions in the developing heart [42]. Zhong et al. showed that Wnt/β-catenin signaling in EndoMT has the ability to promote myofibrillar production in the valve [43]. MiR-222 was also able to inhibit cardiac fibrosis in diabetic mice by regulating Wnt/β-catenin-mediated EndoMT [44]. These results all further confirm that development in TOF may be accomplished through the EndoMT process mediated by TGF-β and Wnt signaling.
Based on SCIENIC analysis, we identified ERG and TEAD1 as key TFs that promote the transformation of endothelial cell 2 subtypes to mesenchymal in TOF. The ETS family is one of the members of TFs in which ETS-related genes (ERGs) have been shown to be specifically highly expressed in endothelial cells [45]. ERG is one of the factors important for the maintenance of endothelial homeostasis [46,47]. ERG has been shown to protect endothelial cells from EndoMT by regulating the TGF-β/SMAD signaling pathway [48]. Nagai et al. found that ERG expression is down-regulated in tumor endothelial cells and suggested that ERG acts as an activator of key transcription of specific genes on endothelial cells [49]. In addition, as a core component on the Hippo pathway, which is important for cardiac development and regeneration, yes-associated protein 1 (YAP1) requires binding to TFs to function because it lacks a DNA-binding structural domain [50–52]. To date, the importance of TEAD1 in cardiomyocytes, which contributes to cardiomyocyte proliferation, has been established [53]. Liu et al. showed that TEAD1 deficiency leads to acute dilated cardiomyopathy in adults. This is mainly due to the fact that TEAD1 autoregulates cardiomyocyte mitochondrial OXPHOS to result in diminished aerobic respiration and maximal mitochondrial oxygen consumption [54]. Yamada et al. indicated that dysregulation of TEAD1 target genes was observed in cardiac tissues of patients with dilated cardiomyopathy with LMNA gene mutations [55]. Furthermore, in human endothelial cells, TEAD1 can also interact with vestigial-like family member 4 (VGLL4) to promote the production of key TFs by endothelial cells and contribute to the enhancement of endothelial cell lineage specification [56]. These results confirm that the two key TFs we identified, including ERG and TEAD1 are important in CHD and even in TOF. However, in the present study, we found that ERG and TEAD1 and related target genes were progressively up-regulated during the development of TOF and had a contributory role in the EndoMT process. Therefore, in future studies, in-depth exploration of the molecular mechanisms of EndoMT of ERG and TEAD1 in TOF still needs to be further verified by in vivo and in vitro experiments. In addition, our study was based only on scRNA-seq data from the GEO database, which still requires a larger sample size and TOF of clinical samples to validate our results.
Limitations in this study should be noted. All the analysis were conducted based on online database and bioinformatics tools. In the future, web experiment for the role of selected genes on EndoMT process as well as clinical detections on the expression of interested genes are needed to validate our finding.
In summary, we analyzed single-cell transcriptome data to identify endothelial cell subtypes in TOF that exhibit a marked ability to transform to the mesenchyme. Based on the analysis of scRNA-seq data, we identified six cell clusters, screened five endothelial cell subtypes associated with EndoMT in TOF, and revealed the differentiation status of endothelial cells 2 and the activity of TFs. Furthermore, enrichment analysis revealed that endothelial cell 2 may mediate the development of EndoMT in TOF by decreasing intercellular adhesion capacity and promoting cell migration, as well as through TGF-β and Wnt signaling pathways. Importantly, we identified two key TFs (ERG and TEAD1) with the potential to promote EndoMT. This not only emphasizes the heterogeneity of endothelial cells present in TOF, but also reveals the molecular mechanism of EndoMT in TOF.
Acknowledgement: None.
Funding Statement: This study was supported by The National Natural Science Foundation of China (No. 82160050) and State Key Laboratory of Pathogenesis, Prevention and Treatment of High Incidence Diseases in Central Asia (No. SKL-HIDCA-2021-24).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Aisa Zulibiya, Jing Wen, Huiqing Yu; data collection: Xiaoming Chen; analysis and interpretation of results: Lei Xu, Xiao Ma, Aisa Zulibiya; draft manuscript preparation: Baojian Zhang, Aisa Zulibiya, Jing Wen, Huiqing Yu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available in the [GSE203274] repository, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE203274.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/chd.2023.047689.
References
1. Hoffman, J. I., Kaplan, S. (2002). The incidence of congenital heart disease. Journal of The American College of Cardiology, 39(12), 1890–1900. [Google Scholar] [PubMed]
2. Apitz, C., Webb, G. D., Redington, A. N. (2009). Tetralogy of fallot. Lancet, 374(9699), 1462–1471. [Google Scholar] [PubMed]
3. Smith, C. A., McCracken, C., Thomas, A. S., Spector, L. G., St Louis, J. D. et al. (2019). Long-term outcomes of tetralogy of fallot: A study from the pediatric cardiac care consortium. Journal of the American Medical Association Cardiol, 4(1), 34–41. [Google Scholar]
4. Forman, J., Beech, R., Slugantz, L., Donnellan, A. (2019). A review of tetralogy of fallot and postoperative management. Critical Care Nursing Clinics of North America, 31(3), 315–328. [Google Scholar] [PubMed]
5. Guo, Z., Li, B., Tian, P., Li, D., Zhang, Y. et al. (2019). DGCR8 expression is altered in children with congenital heart defects. Clinica Chimica Acta, 495, 25–28. [Google Scholar]
6. Müller, N., Rothkegel, S. T., Boerter, N., Breuer, J., Freudenthal, N. J. (2020). Urinary values of NT-proBNP in children with congenital heart disease–Is it feasible? Clinica Chimica Acta, 509, 224–227. [Google Scholar]
7. Müller, N., Rothkegel, S. T., Boerter, N., Sumaria, K., Breuer, J. et al. (2021). Perioperative urinary NT-ProBNP values and their usefulness as diagnostic and prognostic markers in children with congenital heart disease. Clinica Chimica Acta, 518, 28–32. [Google Scholar]
8. Pinto, A. R., Ilinykh, A., Ivey, M. J., Kuwabara, J. T., D’Antoni, M. L. et al. (2016). Revisiting cardiac cellular composition. Circulation Research, 118(3), 400–409. [Google Scholar] [PubMed]
9. Marcu, R., Choi, Y. J., Xue, J., Fortin, C. L., Wang, Y. et al. (2018). Human organ-specific endothelial cell heterogeneity. iScience, 4, 20–35. [Google Scholar] [PubMed]
10. Rafii, S., Butler, J. M., Ding, B. S. (2016). Angiocrine functions of organ-specific endothelial cells. Nature, 529(7586), 316–325. [Google Scholar] [PubMed]
11. Shi, J., Chen, Y., Peng, C., Kuang, L., Zhang, Z. et al. (2022). Advances in targeted therapy against driver mutations and epigenetic alterations in non-small cell lung cancer. Oncologie, 24(4), 613–648. [Google Scholar]
12. Yan, D., Li, C., Zhou, Y., Yan, X., Zhi, W. et al. (2022). Exploration of combinational therapeutic strategies for HCC based on TCGA HCC database. Oncologie, 24(1), 101–111. [Google Scholar]
13. Ren, Z., Yu, P., Li, D., Li, Z., Liao, Y. et al. (2020). Single-cell reconstruction of progression trajectory reveals intervention principles in pathological cardiac hypertrophy. Circulation, 141(21), 1704–1719. [Google Scholar] [PubMed]
14. Wang, L., Yu, P., Zhou, B., Song, J., Li, Z. et al. (2020). Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nature Cell Biology, 22(1), 108–119. [Google Scholar] [PubMed]
15. Wardman, R., Keles, M., Pachkiv, I., Hemanna, S., Grein, S. et al. (2023). RNA-Binding proteins regulate post-transcriptional responses to TGF-β to coordinate function and mesenchymal activation of murine endothelial cells. Arteriosclerosis, Thrombosis, and Vascular Biology, 43(10), 1967–1989. [Google Scholar] [PubMed]
16. Wesseling, M., Sakkers, T. R., de Jager, S. C. A., Pasterkamp, G., Goumans, M. J. (2018). The morphological and molecular mechanisms of epithelial/endothelial-to-mesenchymal transition and its involvement in atherosclerosis. Vascular Pharmacology, 106, 1–8. [Google Scholar] [PubMed]
17. Cui, X., Lu, Y. W., Lee, V., Kim, D., Dorsey, T. et al. (2015). Venous endothelial marker COUP-TFII regulates the distinct pathologic potentials of adult arteries and veins. Scientific Reports, 5, 16193. [Google Scholar] [PubMed]
18. Zeisberg, E. M., Tarnavski, O., Zeisberg, M., Dorfman, A. L., McMullen, J. R. et al. (2007). Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nature Medicine, 13(8), 952–961. [Google Scholar] [PubMed]
19. Liu, Z. H., Zhang, Y., Wang, X., Fan, X. F., Zhang, Y. et al. (2019). SIRT1 activation attenuates cardiac fibrosis by endothelial-to-mesenchymal transition. Biomedicine & Pharmacotherapy, 118, 109227. [Google Scholar]
20. Xu, L., Fu, M., Chen, D., Han, W., Ostrowski, M. C. et al. (2019). Endothelial-specific deletion of Ets-1 attenuates angiotensin II-induced cardiac fibrosis via suppression of endothelial-to-mesenchymal transition. BMB Reports, 52(10), 595–600. [Google Scholar] [PubMed]
21. Chen, P. Y., Qin, L., Baeyens, N., Li, G., Afolabi, T. et al. (2015). Endothelial-to-mesenchymal transition drives atherosclerosis progression. Journal of Clinical Investigation, 125(12), 4514–4528. [Google Scholar] [PubMed]
22. Piera-Velazquez, S., Jimenez, S. A. (2019). Endothelial to mesenchymal transition: Role in physiology and in the pathogenesis of human diseases. Physiological Reviews, 99(2), 1281–1324. [Google Scholar] [PubMed]
23. Stuart, T., Butler, A., Hoffman, P., Hafemeister, C., Papalexi, E. et al. (2019). Comprehensive integration of single-cell data. Cell, 177(7), 1888–1902.E21. [Google Scholar] [PubMed]
24. Korsunsky, I., Millard, N., Fan, J., Slowikowski, K., Zhang, F. et al. (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods, 16(12), 1289–1296. [Google Scholar] [PubMed]
25. Ge, W., Tan, S. J., Wang, S. H., Li, L., Sun, X. F. et al. (2020). Single-cell transcriptome profiling reveals dermal and epithelial cell fate decisions during embryonic hair follicle development. Theranostics, 10(17), 7581–7598. [Google Scholar] [PubMed]
26. Aibar, S., González-Blas, C. B., Moerman, T., Huynh-Thu, V. A., Imrichova, H. et al. (2017). SCENIC: Single-cell regulatory network inference and clustering. Nature Methods, 14(11), 1083–1086. [Google Scholar] [PubMed]
27. Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T. et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. [Google Scholar] [PubMed]
28. Wiktorska, M., Sacewicz-Hofman, I., Niewiarowska, J. (2023). The endothelial-to-mesenchymal transition changes the focal adhesion site proteins levels and the SLRP-lumican level in HMEC-1 cell line. Experimental Cell Research, 430(1), 113692. [Google Scholar] [PubMed]
29. Davidson, S. M. (2010). Endothelial mitochondria and heart disease. Cardiovascular Research, 88(1), 58–66. [Google Scholar] [PubMed]
30. Kovacic, J. C., Dimmeler, S., Harvey, R. P., Finkel, T., Aikawa, E. et al. (2019). Endothelial to mesenchymal transition in cardiovascular disease: JACC State-of-the-art review. Journal of the American College of Cardiology, 73(2), 190–209. [Google Scholar] [PubMed]
31. Auersperg, N., Pan, J., Grove, B. D., Peterson, T., Fisher, J. et al. (1999). E-cadherin induces mesenchymal-to-epithelial transition in human ovarian surface epithelium. Proceedings of the National Academy of Sciences of the United Atates of America, 96(11), 6249–6254. [Google Scholar] [PubMed]
32. Islam, S., Boström, K. I., di Carlo, D., Simmons, C. A., Tintut, Y. et al. (2021). The mechanobiology of endothelial-to-mesenchymal transition in cardiovascular disease. Frontiers in Physiology, 12, 734215. [Google Scholar] [PubMed]
33. Xu, X., Lan, W., Jin, X., Wang, B., Yan, H. et al. (2014). Regulated expression of PTPRJ by COX-2/PGE2 axis in endothelial cells. PLoS One, 9(12), e114996. [Google Scholar] [PubMed]
34. Zhang, H., Meng, F., Wu, S., Kreike, B., Sethi, S. et al. (2011). Engagement of I-branching β-1, 6-N-acetylglucosaminyltransferase 2 in breast cancer metastasis and TGF-β signaling. Cancer Research, 71(14), 4846–4856. [Google Scholar] [PubMed]
35. Lu, X. J., Wang, H. T. (2017). Reduced Gja5 expression in arterial endothelial cells impairs arteriogenesis during acute ischemic cardiovascular disease. Experimental And Therapeutic Medicine, 14(5), 4339–4343. [Google Scholar] [PubMed]
36. Fan, M., Yang, K., Wang, X., Chen, L., Gill, P. S. et al. (2023). Lactate promotes endothelial-to-mesenchymal transition via Snail1 lactylation after myocardial infarction. Science Advances, 9(5), eadc9465. [Google Scholar] [PubMed]
37. Xie, F., Zhang, Z., van Dam, H., Zhang, L., Zhou, F. (2014). Regulation of TGF-β superfamily signaling by SMAD Mono-Ubiquitination. Cells, 3(4), 981–993. [Google Scholar] [PubMed]
38. Luyckx, I., Verstraeten, A., Goumans, M. J., Loeys, B. (2022). SMAD6-deficiency in human genetic disorders. npj Genomic Medicine, 7(1), 68. [Google Scholar] [PubMed]
39. Ruter, D. L., Liu, Z., Ngo, K. M., S., X., Marvin, A. et al. (2021). SMAD6 transduces endothelial cell flow responses required for blood vessel homeostasis. Angiogenesis, 24(2), 387–398. [Google Scholar] [PubMed]
40. Kulikauskas, M. R., Oatley, M., Yu, T., Liu, Z., Matsumura, L. et al. (2023). Endothelial cell SMAD6 balances Alk1 function to regulate adherens junctions and hepatic vascular development. Development, 150(21), dev201811. [Google Scholar] [PubMed]
41. Cadigan, K. M., Waterman, M. L. (2012). TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harbor Perspectives in Biology, 4(11), a007906. [Google Scholar] [PubMed]
42. Liebner, S., Cattelino, A., Gallini, R., Rudini, N., Iurlaro, M. et al. (2004). β-catenin is required for endothelial-mesenchymal transformation during heart cushion development in the mouse. The Journal of Cell Biology, 166(3), 359–367. [Google Scholar] [PubMed]
43. Zhong, A., Mirzaei, Z., Simmons, C. A. (2018). The roles of matrix stiffness and ß-catenin signaling in endothelial-to-mesenchymal transition of aortic valve endothelial cells. Cardiovascular Engineering and Technology, 9(2), 158–167. [Google Scholar] [PubMed]
44. Wang, Z., Wang, Z., Gao, L., Xiao, L., Yao, R. et al. (2020). miR-222 inhibits cardiac fibrosis in diabetic mice heart via regulating Wnt/β-catenin-mediated endothelium to mesenchymal transition. Journal of Cellular Physiology, 235(3), 2149–2160. [Google Scholar] [PubMed]
45. Hollenhorst, P. C., Jones, D. A., Graves, B. J. (2004). Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucleic Acids Research, 32(18), 5693–5702. [Google Scholar] [PubMed]
46. Shah, A. V., Birdsey, G. M., Randi, A. M. (2016). Regulation of endothelial homeostasis, vascular development and angiogenesis by the transcription factor ERG. Vascular Pharmacology, 86, 3–13. [Google Scholar] [PubMed]
47. Kalna, V., Yang, Y., Peghaire, C. R., Frudd, K., Hannah, R. et al. (2019). The transcription factor ERG regulates super-enhancers associated with an endothelial-specific gene expression program. Circulation Research, 124(9), 1337–1349. [Google Scholar] [PubMed]
48. Dufton, N. P., Peghaire, C. R., Osuna-Almagro, L., Raimondi, C., Kalna, V. et al. (2017). Dynamic regulation of canonical TGFβ signalling by endothelial transcription factor ERG protects from liver fibrogenesis. Nature Communications, 8(1), 895. [Google Scholar] [PubMed]
49. Nagai, N., Ohguchi, H., Nakaki, R., Matsumura, Y., Kanki, Y. et al. (2018). Downregulation of ERG and FLI1 expression in endothelial cells triggers endothelial-to-mesenchymal transition. PLoS Genetics, 14(11), e1007826. [Google Scholar] [PubMed]
50. Wang, J., Liu, S., Heallen, T., Martin, J. F. (2018). The Hippo pathway in the heart: Pivotal roles in development, disease, and regeneration. Nature Reviews Cardiology, 15(11), 672–684. [Google Scholar] [PubMed]
51. Wackerhage, H., Del Re, D. P., Judson, R. N., Sudol, M., Sadoshima, J. (2014). The Hippo signal transduction network in skeletal and cardiac muscle. Science Signaling, 7(337), re4. [Google Scholar] [PubMed]
52. Lin, K. C., Park, H. W., Guan, K. L. (2017). Regulation of the hippo pathway transcription factor TEAD. Trends in Biochemical Sciences, 42(11), 862–872. [Google Scholar] [PubMed]
53. Liu, R., Jagannathan, R., Li, F., Lee, J., Balasubramanyam, N. et al. (2019). Tead1 is required for perinatal cardiomyocyte proliferation. PLoS One, 14(2), e0212017. [Google Scholar] [PubMed]
54. Liu, R., Jagannathan, R., Sun, L., Li, F., Yang, P. et al. (2020). Tead1 is essential for mitochondrial function in cardiomyocytes. American Journal of Physiology-Heart and Circulatory Physiology, 319(1), H89–H99. [Google Scholar] [PubMed]
55. Yamada, S., Ko, T., Ito, M., Sassa, T., Nomura, S. et al. (2023). TEAD1 trapping by the Q353R-Lamin A/C causes dilated cardiomyopathy. Science Advances, 9(15), eade7047. [Google Scholar] [PubMed]
56. Quan, Y., Hu, M., Jiang, J., Jin, P., Fan, J. et al. (2023). VGLL4 promotes vascular endothelium specification via TEAD1 in the vascular organoids and human pluripotent stem cells-derived endothelium model. Cellular and Molecular Life Sciences, 80(8), 215. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools