 Open Access
Open Access
ARTICLE
NUDT21 Functions as a Pro-Tumorigenic Gene in Colorectal Cancer by Upregulating the TAZ Protein Expression
1 Department of Colorectal and Anal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, 200092, China
2 Department of Thoracic Surgery, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200011, China
3 Department of Gastrointestinal Surgery, Tiantai People’s Hospital, Taizhou, 317200, China
* Corresponding Authors: Yun Liu. Email: ; Zhongchuan Wang. Email:
# These authors have contributed equally to this work
(This article belongs to the Special Issue: Genetic Biomarkers of Cancer: Insights into Molecular and Cellular Mechanisms)
BIOCELL 2025, 49(3), 503-518. https://doi.org/10.32604/biocell.2025.059286
Received 03 October 2024; Accepted 20 February 2025; Issue published 31 March 2025
Abstract
Background: Nudix Hydrolase 21 (NUDT21) is crucial for the regulation of alternative polyadenylation, with its reduced expression frequently resulting in a shortened mRNA 3′ untranslated region (UTR), thereby enhancing the protein levels of downstream genes. Although NUDT21 is widely recognized for its tumor-suppressive function in various cancers, its involvement in colorectal cancer (CRC) remains poorly understood. Methods: The expression of NUDT21 in CRC and adjacent normal tissues was analyzed through qPCR, Western blot, and immunohistochemistry (IHC). Additionally, we investigated the correlation between NUDT21 expression and patient prognosis. With Cell Counting Kit-8 assay and Transwell assay, we detected the exact role of NUDT21 in the development and progress of CRC. We used RNA-Seq analysis to explore the downstream target gene of NUDT21. Meanwhile, DaPars analysis revealed the Hippo pathway as a critical pathway in CRC progress. Western blotting and luciferase assays were used to investigate the specific mechanisms by which NUDT21 regulates the Hippo pathway. In addition, the interaction between NUDT21 and TAZ was confirmed by co-immunoprecipitation and the correlation between the two expressions was verified by IHC. Results: In CRC, elevated NUDT21 expression has been identified as a predictor of poor prognosis, as well as a promoter of CRC cell proliferation and migration. Combined with RNA-seq and DaPars analyses, we identified the Hippo pathway as an important downstream target of NUDT21 in CRC. NUDT21 knockdown significantly downregulated the YAP, TAZ, and TEAD1 protein levels. Mechanistically, shortening of the 3′UTR of TAZ by NUDT21 knockdown suppressed TAZ protein expression. A positive correlation was observed between NUDT21 and TAZ proteins in CRC. Conclusion: NUDT21 plays a pro-tumorigenic role in CRC by upregulating TAZ protein expression, revealing a new regulatory mechanism of the Hippo pathway and providing insight into the biological effect of mRNA 3′UTR shortening.Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| NUDT21 | Nudix Hydrolase 21 |
| CRC | Colorectal cancer |
| APA | Alternative polyadenylation |
| PAS | Polyadenylation sites |
| NF-κB | Nuclear factor kappa B |
| HCC | Hepatocellular carcinoma |
| IHC | Immunohistochemistry |
| CCK8 | Cell Counting Kit-8 |
| Co-IP | Co-immunoprecipitation |
| PDUI | Distal polyA site usage index |
| PABP | Poly (A)-binding protein. |
Globally, cancer is the second most common cause of death, and among its various forms, colorectal cancer (CRC) stands out as one of the most widespread [1]. In 2020, CRC emerged as the third most common cancer, accounting for 1.93 million new cases, or approximately 10% of all cancers. Furthermore, it ranked second in cancer-related mortality, with 935,000 deaths, representing 9.4% of total cancer fatalities [2]. This data was provided by the latest report from the International Agency for Research on Cancer (IARC), an organization under the World Health Organization, which also highlighted a global total of 19.29 million new cancer diagnoses and 9.96 million deaths attributed to cancer. Approximately 25% of CRC patients have metastasis at the time of the initial diagnosis, and once metastasis occurs, the prognosis of patients is often poor [3–5]. Early diagnosis and timely surgical treatment are key to the prognosis of CRC; therefore, identifying new biomarkers and exploring new mechanisms of CRC development and progression is essential for early screening and diagnosis and prevention or intervention of CRC metastasis.
Alternative polyadenylation (APA) is an essential mechanism in RNA processing, influencing critical gene regulation events, including mRNA maturation, stabilization, and the production of proteins [6–8]. APA uses different polyadenylation sites (PAS) in the 3′UTR region of pre-mRNA to produce mRNA heterodimers containing distinct 3′UTR lengths [9]. Dysregulation of APA is frequently noted in cancer and leads to altered downstream gene expression and activation or inactivation of signaling pathways, suggesting that APA regulation is a potential target for cancer therapy [10–12]. NUDT21, a conserved factor involved in cleavage and polyadenylation, is instrumental in the regulation of APA [13]. Knockdown of NUDT21 triggers the selection of the proximal poly(A) site, which reduces the length of the 3′UTR [14]. NUDT21 is recognized for its potent tumor-suppressive properties across various malignancies. In bladder cancer, it modulates key genes such as ANXA2 and LIMK2 by regulating the Wnt/β-catenin and NF-κB signaling networks, thereby exerting a decisive inhibitory effect on tumor progression [15]. Reduced NUDT21 expression is observed in glioblastoma and is associated with poor survival [16]. NUDT21 is downregulated in hepatocellular carcinoma (HCC) and mediates tumor suppression. The loss of NUDT21 hampers circRNA cyclization, thereby disrupting the competitive endogenous RNA (ceRNA) network and the intricate circRNA–miRNA–mRNA interactions. This disruption enhances the miRNA-mediated repression of tumor suppressor genes, facilitating tumor progression [17]. In cervical cancer, NUDT21 serves a tumor-suppressive function by regulating fatty acid metabolism and influencing both the Wnt and NF-κB signaling pathways, with these effects mediated through the mechanism of APA [18]. A recent study highlighted the tumor-promoting function of NUDT21 in driving the growth of gastric cancer [19]. However, there is a paucity of research on the specific functions and molecular mechanisms of NUDT21 in CRC.
Using RNA-seq and DaPars analyses, we observed that the reduction of NUDT21 inhibited the transcription of downstream target genes of the Hippo pathway and shortened the 3′UTR of key Hippo pathway genes, including Yes-related protein (YAP), WW domain-containing transcriptional regulator 1 (WWTR1, also known as TAZ), and TEA domain transcription factor 1 (TEAD1). The Hippo signaling pathway has garnered significant attention in oncology research in recent years, given its crucial role in tumorigenesis, metastasis, and immune regulation [20–23]. YAP and TAZ serve as pivotal mediators of the Hippo pathway, modulating gene expression through their interactions with transcription factors from the TEAD family [24]. Dysregulated control of the Hippo signaling cascade and the persistent activation of the YAP/TAZ-TEAD transcriptional complex are closely associated with the onset and progression of various cancers. Elevated levels of YAP, TAZ, and TEAD promote the transcription of numerous oncogenes, thereby driving tumor cell proliferation, migration, and invasiveness [25–27].
The current study exhibits that NUDT21 is significantly overexpressed in CRC tumor tissues and is linked to an unfavorable prognosis. In contrast to the widely reported tumor suppressor function in various cancers, NUDT21 facilitates the initiation and advancement of CRC. Through RNA-seq and DaPars analyses, we identified a regulatory link between NUDT21 and the Hippo pathway. Specifically, the knockdown of NUDT21 caused a marked reduction in the protein levels of YAP, TAZ, and TEAD1, underscoring its influence on this essential signaling network. Thus, we hypothesized that NUDT21 upregulates TAZ expression to promote CRC development and progression. Moreover, we found that NUDT21 interacts with TAZ; however, this interaction did not affect the degradation of TAZ. Finally, we validated the strong correlation between NUDT21 and TAZ expression in clinical CRC samples. In conclusion, we identified the pro-tumorigenic role of NUDT21 in CRC and the specific mechanism by which NUDT21 regulates the Hippo pathway to promote CRC development through its APA regulatory function.
2.1 Cell Culture and Transfection
Human Embryonic Kidney (HEK) 293T Cells, Human CRC cell lines HCT116, SW620, DLD1, HT29, LoVo, SW480, and RKO, along with the epithelial cell line NCM460 and CCD841, which were derived from normal human colon were all purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). LoVo was cultured in Kaighn’s Modification of Ham’s F-12 Medium (F12-K) (Gibco, C11765500BT, Suzhou, China), and the other cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (Hyclone, SH30022, Logan, UT, USA) all supplemented with 10% fetal bovine serum (Gibco, Fetal Bovine Serum, Premium Plus A5669701) and 1% penicillin/streptomycin 37°C with 5% CO2. Mycoplasma testing is routinely performed monthly in our laboratory to avoid mycoplasma contamination. Stable NUDT21 knockdown cell lines, shNUDT21-1 and shNUDT21-2, were engineered through the deployment of the pLKO.1 vector system. Human NUDT21 cDNA was cloned into a pQCXIH vector for NUDT21 overexpression, which was purchased from the Bio-research Innovation Center Suzhou (Suzhou, China). The following virus packaging and transfection were performed as previously described [18]. The NUDT21 shRNA sequences are listed in Table S1. Similarly, HEK 293T cells were transfected with specific siRNA targeting NUDT21 mRNA to reduce NUDT21 expression. The sequences of siRNA are listed in Table S1.
2.2 Patient Samples’ Tissue Array and Immunohistochemistry (IHC)
Human CRC samples for tissue array were collected from June 2008 to December 2018 at Department of Colorectal and Anal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine. Informed consent was obtained from all participants or their legal guardians and this project was approved by Xinhua Hospital Ethics Committee, Affiliated with Shanghai Jiaotong University School of Medicine (XHEC-NSFC-2021-326). All methods were performed in accordance with the Declarations of Helsinki. IHC staining was performed on the tissue array following established protocols to assess NUDT21 and TAZ expression. Semi-quantitative analysis was conducted based on previously described criteria, evaluating staining intensity and percentage [28]. The analysis was conducted by experienced pathologists who were unaware of the clinical information, to avoid any potential bias. Staining scores below 6 were classified into the low expression group, while scores of 6 or higher were designated as the high expression group. A total of 16 pairs of fresh normal colon and colorectal cancer (CRC) samples were collected to assess the mRNA levels of NUDT21, with 12 of these pairs also being used to evaluate protein expression.
2.3 Western Blotting, Co-Immunoprecipitation, and Cycloheximide (CHX) Assay
Western blotting and co-immunoprecipitation were conducted following the methods outlined in previous studies [29]. The primary antibody included NUDT21 (Proteintech, 10322-1-AP, 1:1000, Wuhan, China), Flag (CST, D6W5B 14793T, 1:2000, Danvers, MA, USA), HA (CST C29F4 3724T, 1:1000), YAP (sc-101199, 1:1000, Santa Cruz, CA, USA) and TAZ (Abcam, ab242313, 1:1000, Cambridge, MA, USA). Briefly, exogenous plasmids were transfected in HEK-293T for 48 h, and then cells were harvested using FLAG-magnetic beads (Selleck, B26101, Houston, TX, USA) or HA-magnetic beads (Selleck B26201) for the subsequently western blot detection. Assay for CHX Chase, Cells were treated with 10 mM CHX (MCE, HY-12320, Monmouth Junction, NJ, USA) for the indicated time points and harvested for the following analysis by Western blotting. Proteasome inhibitor MG132 (MCE, HY-13259) is used to block the proteasome-dependent degradation pathway.
2.4 RNA-Seq Data Analysis and qPCR
Total RNA was extracted utilizing TRIZOL reagent (Invitrogen, 15596026CN), following the protocol outlined by the manufacturer. For RNA-seq, total RNA was extracted from HCT116 cells transfected with pLKO.1 and sh_NUDT21, respectively, and subsequently sequenced using the HiSeq 2500 platform. Differentially expressed genes were analyzed by Deseq2 Rpackage (https://github.com/thelovelab/DESeq2) (accessed on 19 February 2025). The data were deposited in Gene Expression Omnibus (GSE223415). To pinpoint genes that undergo alternative polyadenylation (APA) upon NUDT21 depletion, we employed the widely recognized DaPars algorithm (https://github.com/ZhengXia/DaPars (accessed on 19 February 2025); Xia et al. [7]), which predicts proximal APA sites and quantifies the relative abundance of both long and short 3′UTR isoforms. Next, the distal polyadenylation site usage index (PDUI) was computed for each transcript, representing the percentage of distal site utilization [14]. For qPCR, 500 ng of total RNA was reverse-transcribed into cDNA using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme R323-01, Nanjing, China). cDNA was quantified by RT-PCR and the data were acquired with ChamQ Universal SYBR qPCR Master Mix (Vazyme, Q711) using Applied Biosystems 7500 instrument (Thermo Fisher Scientific Inc, Waltham, MA, USA). β-actin was used as an internal control, and the ΔΔCt method was applied to calculate relative gene expression. The primers are listed in Table S1.
2.5 Cell Proliferation, Colony Formation, and Transwell Migration Assays
The methodology employed for colony formation and transwell assays was the previously described approach outlined in reference [27]. Cell proliferation was evaluated using the Cell Counting Kit-8 (CCK-8) assay, with stable cells seeded in 96-well plates at a density of 1000 cells per well to assess their growth over time. Cell viability was assessed over a five-day period using the Cell Counting Kit-8 (Vazyme A311-01).
The TAZ long 3′UTR (chr3:149517235-149520804) and short 3′UTR (chr3:149520788-149520804) were cloned into the pmirGLO Vector. Cells were plated in 24-well plates and incubated overnight to achieve around 70% confluence. They were then co-transfected with a luciferase reporter plasmid, which carried either the long or short 3′UTR of TAZ. A Renilla luciferase plasmid served as the control for normalization. The above plasmids were purchased from the Bio-research Innovation Center Suzhou. Following a 24 to 36-h incubation, luciferase activity was assessed through a dual-luciferase reporter assay (Promega E1980, Madison, WI, USA), with values expressed as a ratio compared to Renilla luciferase activity.
Statistical analyses were performed using GraphPad Prism (version 8.0, La Jolla, CA, USA) and SPSS (version 22.0, IBM, Armonk, NY, USA). Pearson’s correlation coefficient was calculated to assess the relationships between variables. Data are presented as mean ± standard deviation (SD), and p-values below 0.05 were deemed statistically significant. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). Each experiment was repeated three times independently.
3.1 Overexpression of NUDT21 in CRC Indicates a Poor Prognosis
Based on the Cancer Genome Atlas (TCGA) dataset, we found that NUDT21 mRNA expression was upregulated in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) tissues compared with adjacent normal tissues (Fig. 1a). To validate this result, we compared the protein and mRNA levels of NUDT21 in CRC tumor tissues with those in their paired adjacent normal tissues. In agreement with the TCGA results, we observed overexpression of NUDT21 mRNA and protein in CRC tumor tissues (Fig. 1b,c).
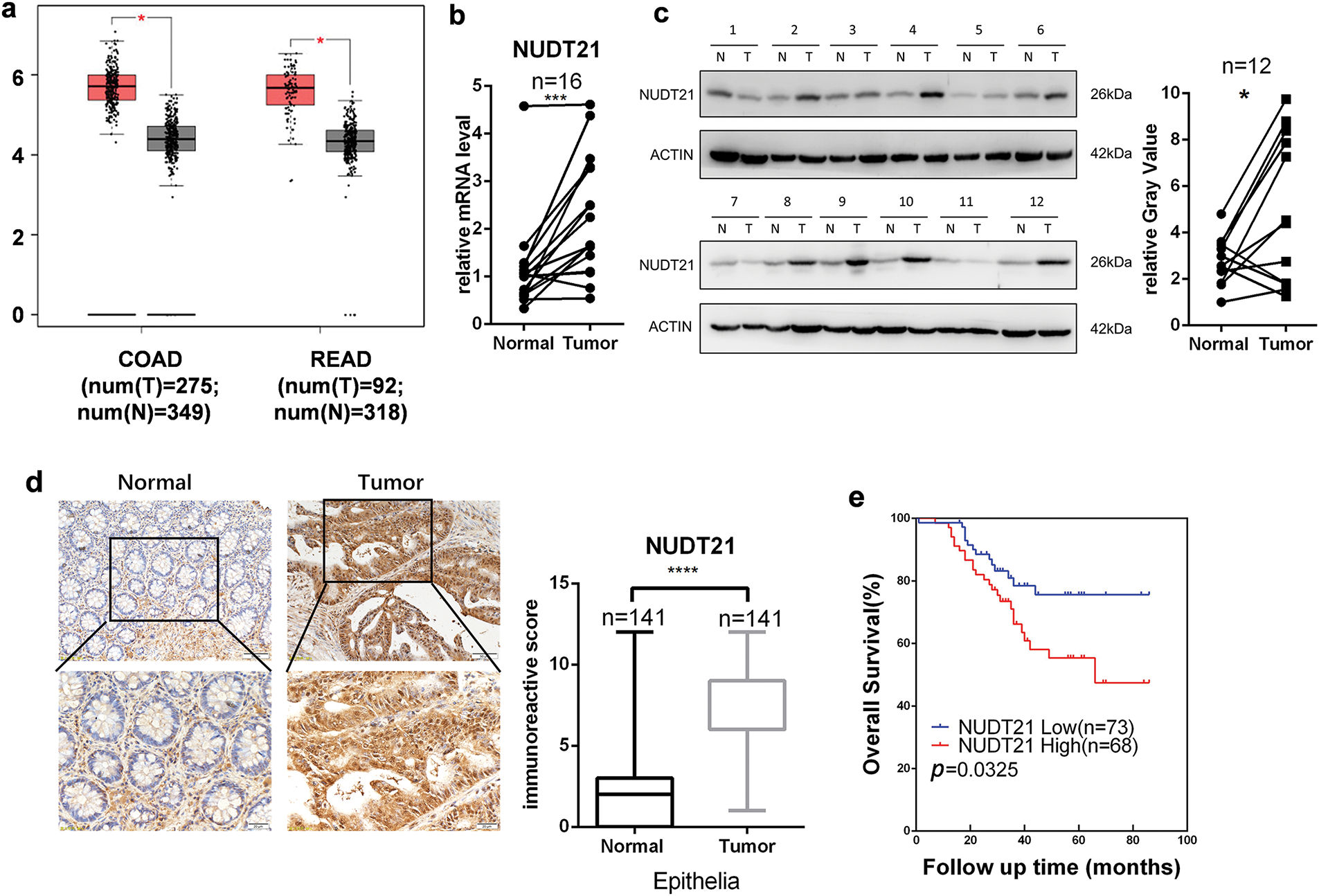
Figure 1: Overexpression of NUDT21 in colorectal cancer indicates poor prognosis. a. The mRNA expression levels of NUDT21 in colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) tissues compared with them in paired normal tissues from the TCGA dataset. The red box is tumor tissue; the gray box is paired with normal tissue. b. qRT-PCR accessing mRNA levels of NUDT21 in CRC (n = 16) and adjacent normal tissues (n = 16). c. Western blot accessing protein levels of NUDT21 in CRC (n = 12) and adjacent normal tissues (n = 12). N, normal tissue; T, tumor tissue. d. NUDT21 expression in both CRC tissues and adjacent non-tumorous tissues was analyzed using immunohistochemistry (IHC) (n = 141). The bar chart presents the statistical analysis of IHC staining intensity in the training cohort. e. Kaplan-Meier survival analysis was performed for CRC patients, stratified based on their NUDT21 IHC staining scores, to assess overall survival. *p < 0.05, ***p < 0.001, ****p < 0.0001
To further explore the expression pattern of NUDT21 in CRC and its correlation with prognosis, we performed immunohistochemical staining of tissue microarrays containing 141 pairs of CRC intestinal cancer and adjacent normal tissues. The results showed that NUDT21 was barely expressed in normal mucosa, while NUDT21 was significantly expressed in tumor tissues (Fig. 1d). Based on the NUDT21 histochemical score of tumor tissues, we divided 141 patients into a NUDT21 high group (n = 68) and a NUDT21 low group (n = 73). Our analysis revealed that the overall survival was notably reduced in the NUDT21 high-expression group compared to the low-expression group (Fig. 1e). In conclusion, we established that NUDT21 expression is significantly elevated in CRC tissues relative to normal tissues. Furthermore, the overexpression of NUDT21 was strongly linked to poor prognosis in CRC, implying a potential pro-tumorigenic function of NUDT21.
3.2 NUDT21 Promotes the Proliferation and Migration of CRC Cells
We then detected NUDT21 protein expression in several CRC and normal colon cell lines. Compared to normal colon cell lines (NCM460 and CCD841), the protein levels of NUDT21 increased in CRC cell lines (HCT116, SW620, DLD1, HT29, SW480, and RKO) (Fig. 2a). We explored the function of NUDT21 in CRC cells by generating stable NUDT21 overexpression lines in HCT116 and LoVo cells, along with stable knockdown lines in HCT116 and SW620 cells. Using shRNA specifically targeting NUDT21, we found that NUDT21 protein was downregulated by more than 70% in HCT116 and SW620 cells (Fig. 2b). Meanwhile, overexpression of exogenous NUDT21 protein containing the Flag tag increased NUDT21 protein 30-fold in HCT116 cells and 2-fold in LoVo cells (Fig. 2c). Upon NUDT21 depletion, we observed a significant inhibition in the proliferation of SW620 cells with CCK8 assay (Fig. 2d). Using clone formation methods, it was found that NUDT21 knockdown could lead to diminished proliferative capacity of SW620 cells, but overexpression of NUDT21 could promote the proliferation of HCT116 cells (Fig. 2e) and a notable decrease in the migration ability of HCT116 cells (Fig. 2f). Conversely, ectopic expression of NUDT21 in LoVo greatly enhanced cell migration (Fig. 2f).
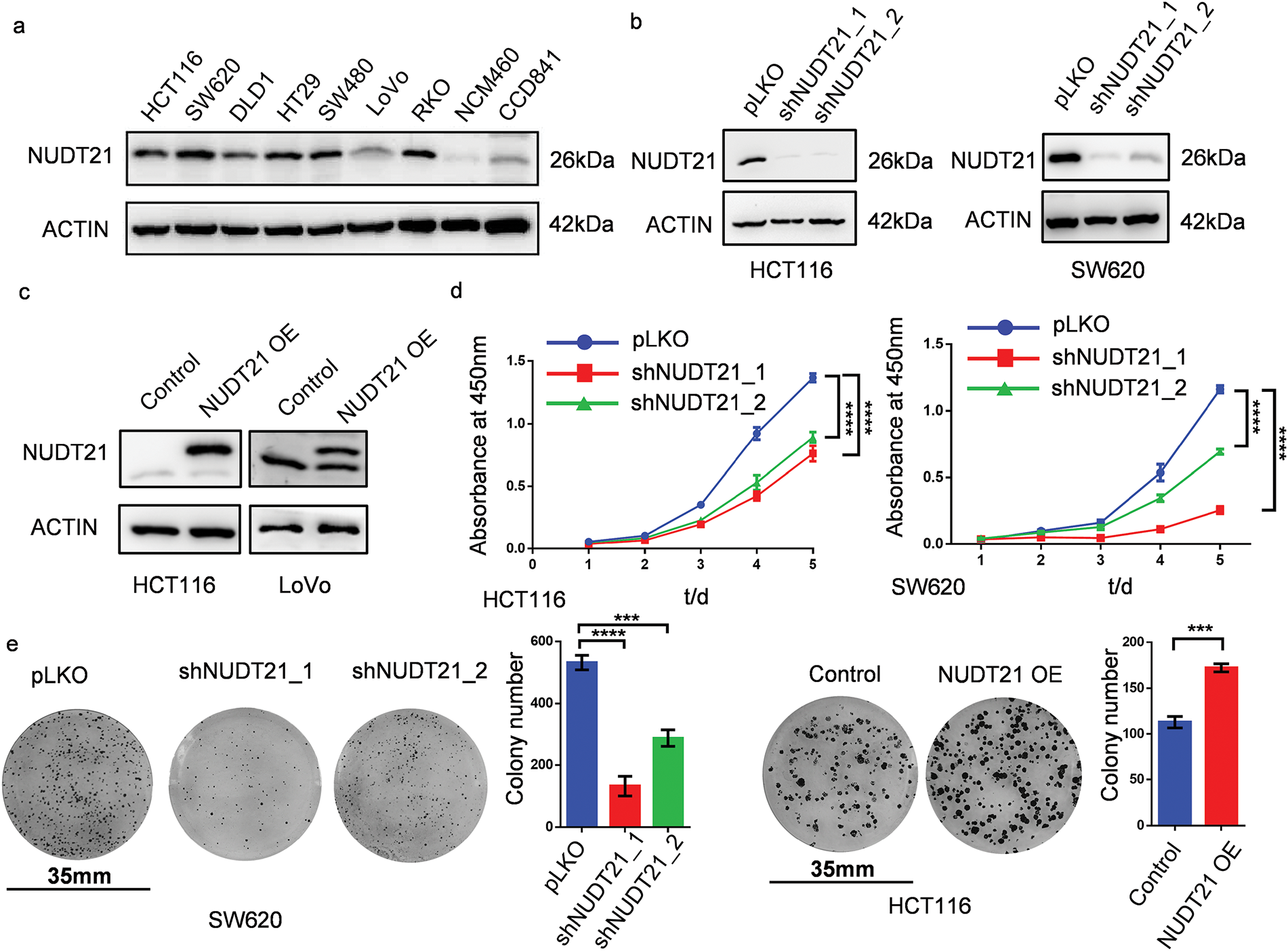
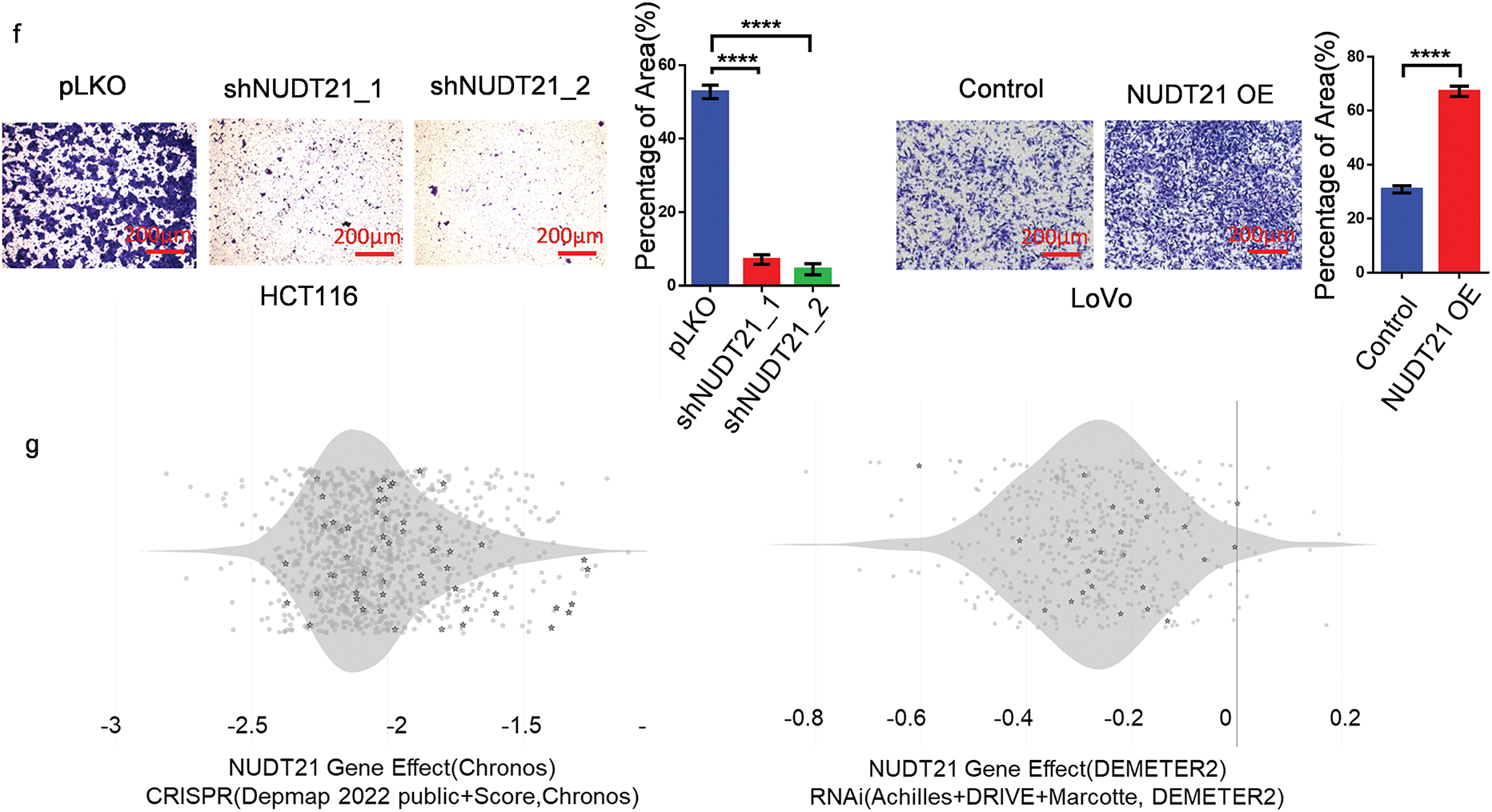
Figure 2: NUDT21 promotes the proliferation and migration of CRC cells. a. Assessment of NUDT21 expression in CRC and normal colon cell lines using Western blotting. b. Western blotting analysis of NUDT21 expression in HCT116 and SW620 cells stably expressing NUDT21 shRNA. c. Evaluation of NUDT21 expression in HCT116 and LoVo cells stably transfected with ectopic NUDT21 cDNA using Western blotting. d. CCK8 assay showing of the proliferation of HCT116 and SW620 cells with NUDT21 knockdown. ****p < 0.0001. e. Colony formation assay and statistical analysis of the proliferation of SW620 cells with NUDT21 knockdown and HCT116 cells with NUDT21 overexpression. Scale bars, 35 mm. ***p < 0.001, ****p < 0.0001. f. Evaluation of cell migration in HCT116 cells with NUDT21 knockdown and LoVo cells with NUDT21 overexpression was performed using a Transwell assay, followed by statistical analysis. ****p < 0.0001. g. DEPMAP database showing the function of NUDT21 knockout or knockdown to inhibit the proliferation of CRC cell lines. Each dot represents a different cell line, whereas each pentagram represents a CRC cell line
Additionally, we leveraged the DEPMAP database (www.depmap.org/) (accessed on 19 February 2025) to explore the biological roles of NUDT21 across various cancer cell lines. Briefly, the DEPMAP database contains information on the proliferative capacity of tumor cells after CRISPR/RNAi knockdown in different cell lines, where a negative GENE effect value indicates that tumor cell proliferation is inhibited by the knockdown of the gene, and a positive GENE effect value indicates that proliferation is enhanced, thus reflecting the effect of the gene on tumor cells. The results of CRISPR data showed that gene effect values of NUDT21 in various CRC cell lines are less than −1, indicating that NUDT21 knockout inhibits cell proliferation. The RNAi results also showed that NUDT21 knockdown had a suppressive effect on the growth of CRC cells (Fig. 2g). Collectively, our findings underscore the pivotal role of NUDT21 in driving both the proliferation and migration of CRC cells.
3.3 NUDT21 Knockdown Promotes 3′UTR Shortening and Downregulates YAP/TAZ Target Genes’ Expression in CRC
Given the critical oncogenic effect of NUDT21 in CRC, we studied its exact mechanism and downstream target genes. RNA-seq was conducted on NUDT21 knockdown and control HCT116 cells, revealing that 699 genes were downregulated and 640 genes were upregulated (FC > 2, p < 0.05) in the NUDT21 knockdown cells compared to the control group (Table S2). Notably, we observed the downregulation of a series of YAP/TAZ target genes, including ANKRD1, CYR61, and CTGF (Fig. 3a). To validate this result, we detected the mRNA levels of the Hippo target gene in NUDT21 knockdown stable cell lines using qPCR. NUDT21 knockdown significantly suppressed the transcription of YAP/TAZ target genes (Fig. 3b). Similar results were obtained for SW620 NUDT21 KD cells (Fig. 3c).
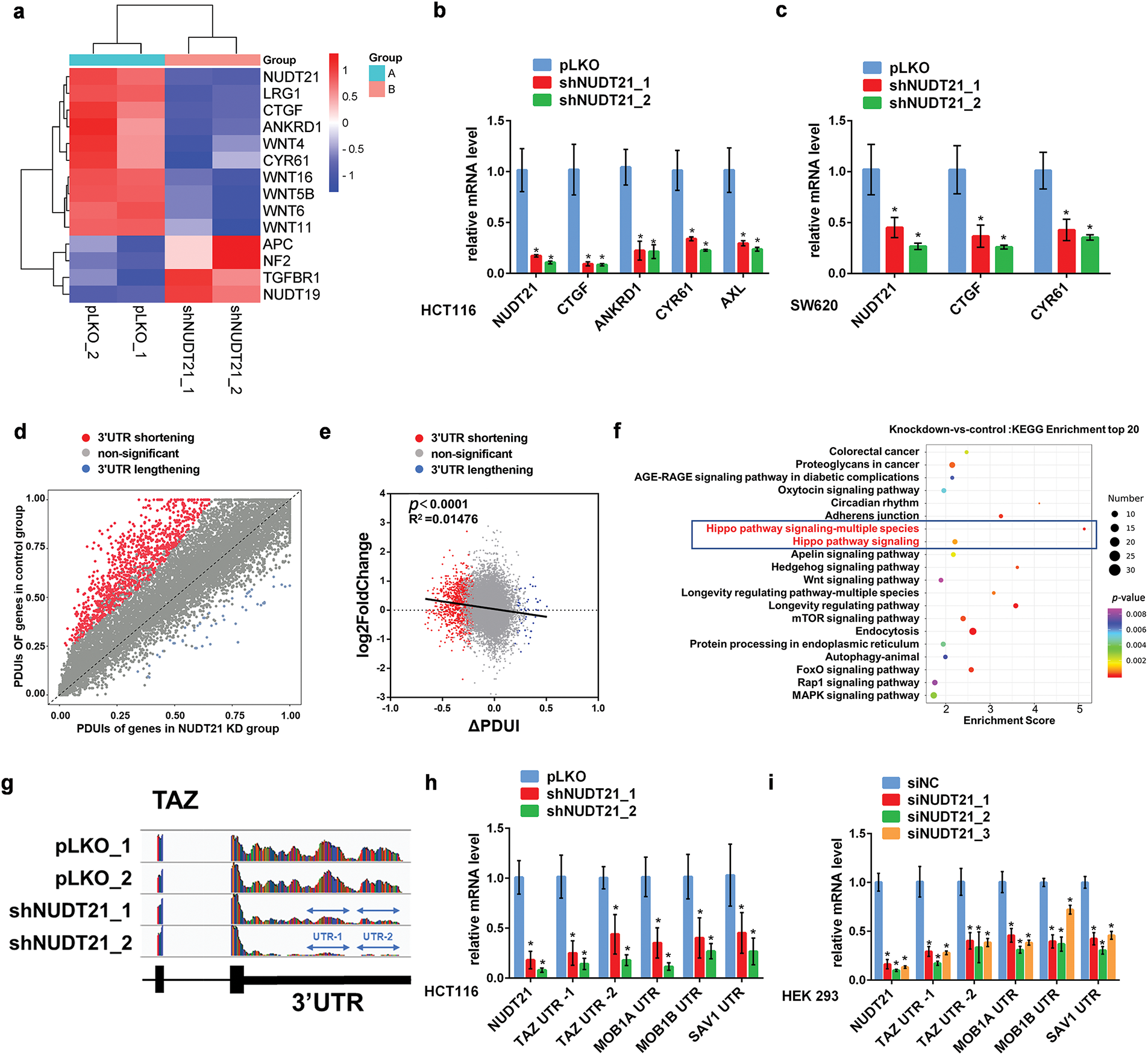
Figure 3: NUDT21 knockdown promotes 3′UTR shortening and downregulates YAP/TAZ target genes’ expression in CRC. a. Heat map showing significant downregulation of mRNA levels of YAP/TAZ target genes in NUDT21 knockdown HCT116 cells, as detected by RNA-seq. Z-cores were calculated for each sample and displayed as a heat map. b. qRT-PCR verifying YAP/TAZ target genes’ mRNA expression in NUDT21 knockdown HCT116 cells. *p < 0.05. c. qRT-PCR verifying YAP/TAZ target genes’ mRNA expression in NUDT21 knockdown SW620 cells. *p < 0.05. d. Scatterplot of PDUIs in control and NUDT21KD group. Red dots, mRNA shortening (n = 1326); blue dots, mRNA lengthening (n = 55). e. The relationship between dPAS site utilization and gene expression levels in control vs. NUDT21 knockdown cells was examined. f. KEGG enrichment analysis showing significant alterations in the 3′UTR length of Hippo pathway-related genes’ transcript. g. Representative RNA-seq density plots for TAZ. h. qRT-PCR verified the significant downregulation of TAZ, MOB1A, MOB1B, and SAV1 3′UTR in HCT116 NUDT21 knockdown cells. Primers were designed for specific sequences of the 3′UTR of the above genes and are provided in Table S1. *p < 0.05. i. qRT-PCR verifying a significant downregulation of TAZ, MOB1A, MOB1B, SAV1 3′UTR in HEK293 cells transfected with siRNA specific for NUDT21. *p < 0.05
Considering the important role of NUDT21 in 3′UTR shortening, we performed DaPars analysis of the RNA-seq data to compare the length of the transcript 3′UTR in NUDT21 knockdown cells with that in control cells. Consistent with previous studies in Hela cells, a human cell line derived from a patient with cervical cancer, the 3′UTR of 1326 transcripts in the NUDT21 knockdown group underwent shortening, while the 3′UTR of 55 transcripts underwent lengthening (p ≤ 0.05, |ΔPDUI| ≥ 0.2 and at least two-fold-change of distal polyA site usage index (PDUIs) between NUDT21 KD and control) (Table S3), which further supports the hypothesis that knockdown of NUDT21 results in 3′UTR shortening (Fig. 3d) [6]. Furthermore, we found a weak correlation between different levels of gene expression (log2FC) and change in 3′UTR length (ΔPDUI) (R2 = 0.01476, p < 0.0001) (Fig. 3e). Interestingly, KEGG enrichment analysis revealed that genes with a significant change in 3′UTR transcript lengths upon NUDT21 knockdown, including YAP, TAZ, MOB1, SAV1, and TEAD1, were enriched in the Hippo pathway (Fig. 3f). Among the studied transcripts, the TAZ transcript had the most pronounced 3′UTR shortening (Fig. 3g). Then, we designed quantitative PCR primers for specific sequences in the 3′UTR and verified using qPCR that the 3′UTR of TAZ, MOB1A, MOB1B, and SAV1 were shortened in the NUDT21 knockdown group compared to the control group, indicating that NUDT21 knockdown promoted the usage of proximal PAS (Fig. 3h). Shortening of the 3′UTR of TAZ, MOB1A, MOB1B, and SAV1 was also observed in HEK293 cells with NUDT21 knockdown (Fig. 3i). Collectively, these findings suggest that NUDT21 could promote the transcription of YAP/TAZ target genes by modulating the shortening of their 3′UTR.
3.4 NUDT21 Knockdown Downregulates the Protein Expression of YAP, TAZ, and TEAD1
YAP, TAZ, and TEAD1 can form a transcriptional complex, which is a downstream effector of the Hippo pathway. Next, we explored the effects of NUDT21 knockdown on the protein levels of YAP, TAZ, and TEAD1. Consistent with the decreased mRNA levels of YAP/TAZ target genes, NUDT21 knockdown significantly downregulated the protein expression of YAP, TAZ, and TEAD1 in HCT116 cells (Fig. 4a). Increased TAZ expression was also observed after exogenous NUDT21 overexpression in HCT116 cells (Fig. 4b). Similarly, NUDT21 knockdown downregulated TAZ protein levels, whereas NUDT21 overexpression upregulated TAZ protein levels in HEK293 cells (Fig. 4c and d).
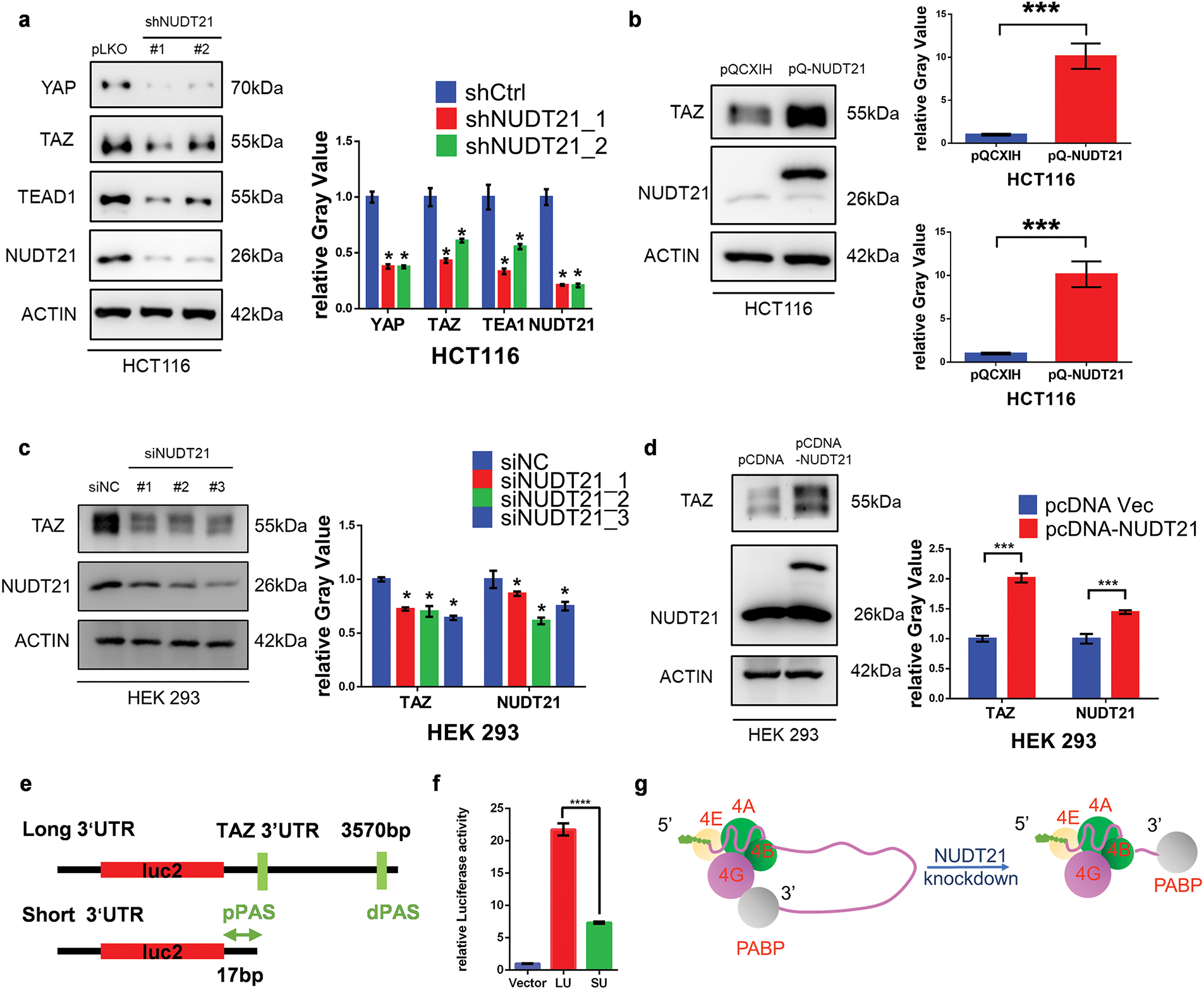
Figure 4: NUDT21 knockdown downregulates the protein expression of YAP, TAZ, and TEAD1. a. Western blotting analysis of YAP, TAZ, and TEAD1 expression in HCT116 cells stably expressing NUDT21 shRNA. *p < 0.05. b. Western blot analysis was conducted to assess TAZ expression in HCT116 cells stably transfected with ectopic NUDT21 cDNA. ***p < 0.001. c. Western blotting analysis of TAZ expression in HEK 293 cells transfected with NUDT21 siRNA. *p < 0.05. d. Western blotting analysis of TAZ expression in HEK 293 cells transfected with pCDNA-NUDT21. ***p < 0.001. e. Schematic diagram of a luciferase plasmid containing a longer TAZ 3′UTR or a shortened TAZ 3′UTR. The original TAZ 3′UTR was cloned from the human genome and is approximately 3570 bp. The short 3′UTR is 17 bp within the stop codon and proximal APA site. The proximal and distal poly(A) sites are indicated by pPAS and dPAS. f. Luciferase assay showing the shortened TAZ 3′UTR reporter exhibited lower luciferase activity than the longer TAZ 3′UTR reporter. ****p < 0.0001. g. The excessively short 3′UTR due to NUDT21 knockdown may prevent the eIF4 complex from forming the loop required for translation with PABP
Normally, a shortened 3′UTR length leads to increased stability and translation efficiency of mRNA [30,31]. However, we observed that NUDT21 knockdown significantly downregulated the protein expression of YAP, TAZ, and TEAD1 with a shortened 3′UTR (Fig. 4a). To explore this contradictory observation, we investigated the 3′UTR sequence using DaPars analysis. We determined the location of the proximal PAS as well as the distal PAS in the TAZ 3′UTR region. DaPars analysis showed that the original YAP 3′UTR was approximately 3 400 bp in length which was shortened to 70 bp after the NUDT21 knockdown. Similarly, the original TAZ 3′UTR was approximately 3570 bp long and was shortened to only 17 bp after the NUDT21 knockdown. The very short 3′UTR of TAZ attracted our attention. Protein translation involves multiple initiation factors. The 5′ cap of the mRNA binds to the initiation factor eIF4, while the 3′ polyA tail binds to poly (A)-binding protein (PABP). In addition, eIF4 and PABP bind via eIF4G, allowing the 5′ cap and 3′ polyA tail of the mRNA to join and form a loop structure. This structure can stimulate translation and promote the recruitment of the 43S pre-initiation complex to mRNA, thereby improving the efficiency of translation initiation [32]. We speculated that the excessively short 3′UTR might suppress protein translation. To test this hypothesis, we constructed luciferase plasmids containing either the longer TAZ 3′UTR or the shortened TAZ 3′UTR (Fig. 4e). The luciferase assay showed that the shortened TAZ 3′UTR reporter exhibited lower luciferase activity than the longer TAZ 3′UTR reporter (Fig. 4f). Thus, the very short 3′UTR of TAZ might lead to decreased protein translation and expression; however, further experimental verification is required (Fig. 4g). Overall, the TAZ 3vUTR shortening due to NUDT21 knockdown may decrease TAZ protein expression.
3.5 NUDT21 Interacts with TAZ and Does Not Affect TAZ Protein Stability
Proteins exhibit a dynamic balance between degradation and synthesis within the cells. We further explored the effect of NUDT21 on TAZ degradation. First, we noticed that NUDT21 possesses a PPXY motif (154PPQY157), which mediates its binding to the WW domain [33]. Co-immunoprecipitation (Co-IP) assay revealed a strong interaction between NUDT21 and TAZ (Fig. 5a). Previous studies have demonstrated that TAZ undergoes proteasomal degradation through β-transducin repeat-containing protein (β-TrCP)-mediated ubiquitination [34]. Therefore, we further investigated whether the interaction between NUDT21 and TAZ protects TAZ from degradation. The Co-IP assay showed that NUDT21 did not affect the binding between TAZ and β-TrCP (Fig. 5b). Meanwhile, overexpression of NUDT21 did not affect the level of TAZ ubiquitination (Fig. 5c). We also examined the effect of NUDT21 knockdown on intracellular β-TrCP protein expression and found no significant differences between the control and NDUT21 knockdown groups (Fig. 5d). When co-incubated with the proteasome inhibitor MG132, TAZ protein degradation was inhibited in both NUDT21 knockdown and control cells. However, even after MG132 treatment, the TAZ protein levels in the NUDT21 knockdown group remained reduced compared to the control group, indicating that NUDT21 does not safeguard TAZ from proteasomal degradation (Fig. 5e).
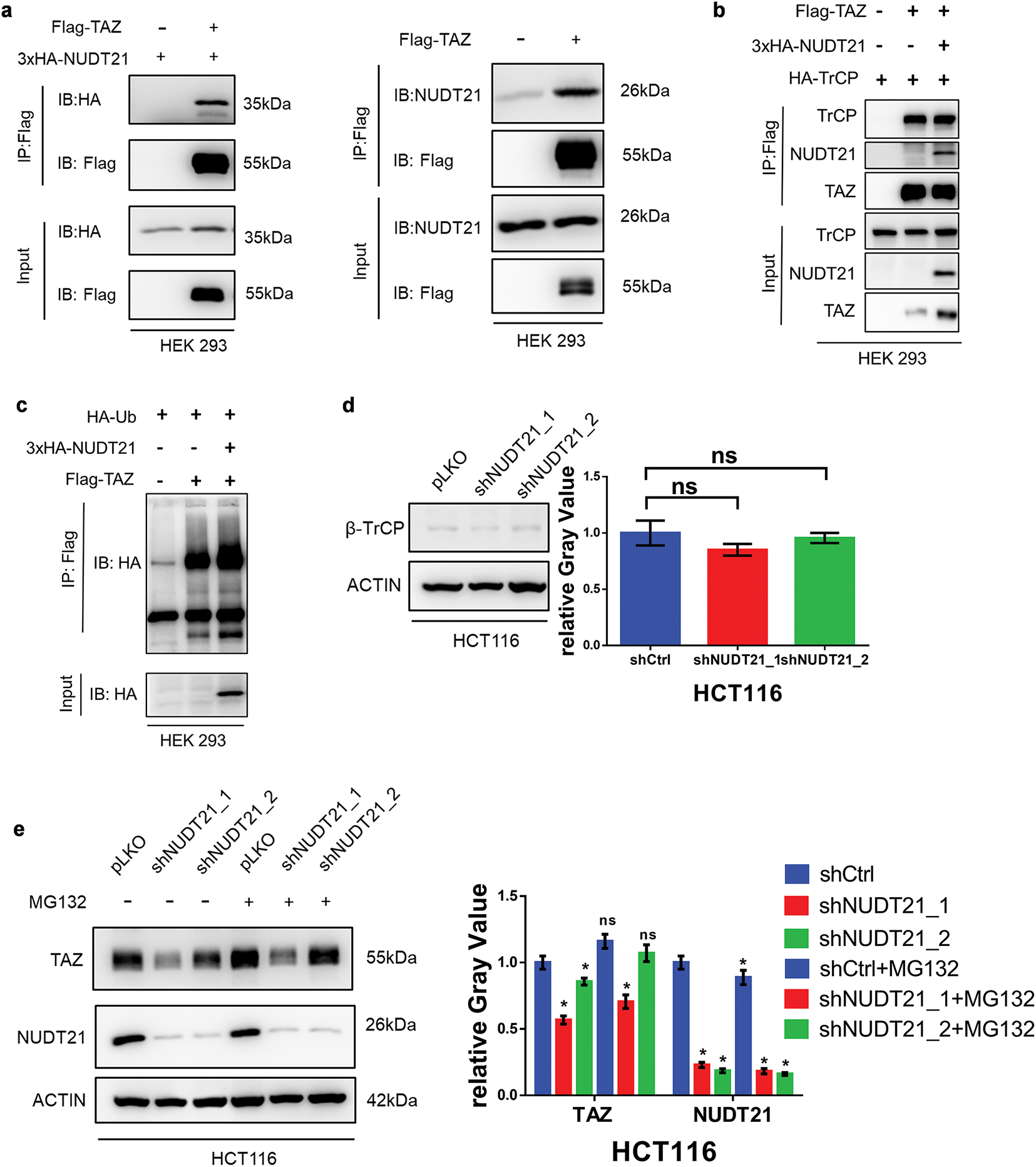
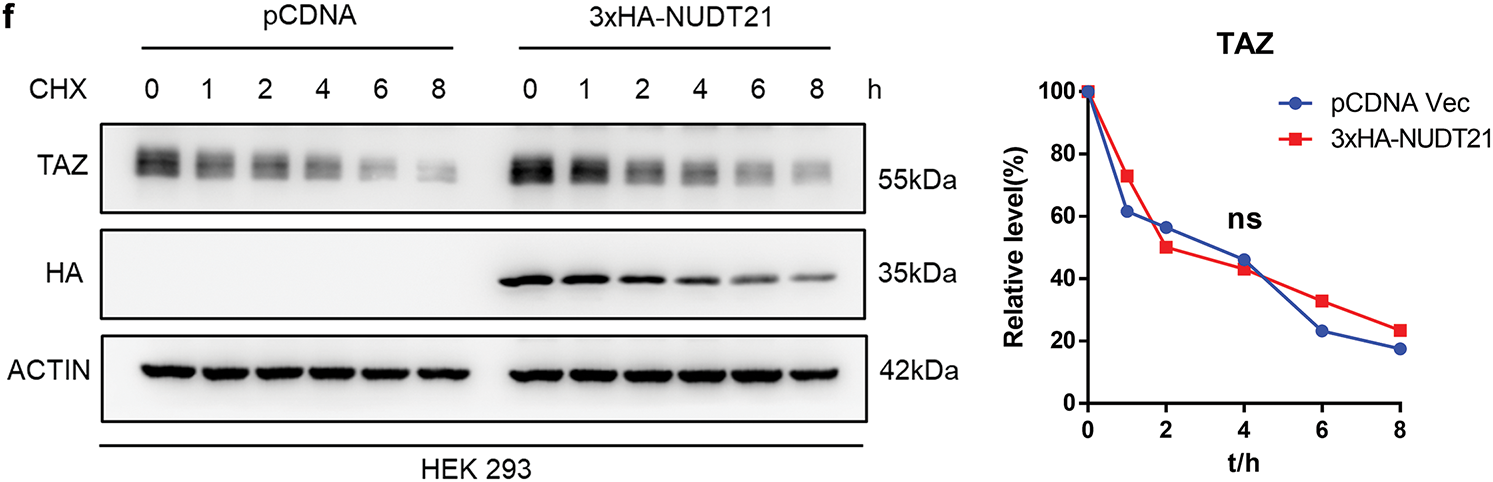
Figure 5: NUDT21 interacts with TAZ and does not affect TAZ protein stability. a. Co-IP of exogenous Flag-TAZ and HA-NUDT21 in HEK 293 cells. Semi-endogenous co-IP of exogenous Flag-TAZ and endogenous NUDT21 in HEK 293 cells. b. NUDT21 did not affect the interaction between TAZ and β-TrCP. Co-IP of exogenous HA-TrCP and Flag-TAZ in HEK-293T cells with or without overexpression of HA-NUDT21. c. NUDT21 did not affect the level of TAZ ubiquitination. Western blot analysis was conducted to evaluate the ubiquitination levels of TAZ with or without HA-NUDT21 overexpression. d. Western blot analysis was performed to assess β-TrCP expression in HCT116 cells stably expressing NUDT21 shRNA. e. Western blot analysis was conducted to examine TAZ expression in HCT116 cells stably expressing NUDT21 shRNA, with or without treatment with the proteasome inhibitor MG132. *p < 0.05. f. TAZ expression in HEK293 cells was assessed by Western blotting, comparing cells with and without HA-NUDT21 overexpression. Cells were treated with 10 mM cycloheximide at the indicated time and then analyzed by Western blotting analysis. The intensity of TAZ protein was quantified using a densitometer. ns: p > 0.05
We further compared the effects of NUDT21 on the rate of TAZ protein degradation following cycloheximide (CHX) treatment. CHX is a commonly used inhibitor of protein synthesis. It selectively blocks the translocation step of eukaryotic ribosomes, thereby stopping protein synthesis. Protein synthesis was inhibited after CHX treatment, and the protein expression levels after different treatment times reflected the total protein degradation rate. The results showed that NUDT21 overexpression did not affect the TAZ degradation rate (Fig. 5f). Based on the above results, NUDT21 upregulates TAZ protein expression, but not by reducing TAZ degradation, suggesting that the binding of NUDT21 and TAZ may have other roles.
3.6 Clinical Relevance of NUDT21-TAZ Axis in CRC
Finally, we investigated the clinical implications of the NUDT21-TAZ axis in CRC samples. Similarly, we performed immunohistochemical staining of previous tissue microarrays containing paired CRC tissues and adjacent normal tissues using YAP- and TAZ-specific antibodies. We found a significant positive correlation between NUDT21 and TAZ expression (R2 = 0.1390, p < 0.0001) (Fig. 6). Taken together, our results indicate that NUDT21 overexpression enhances TAZ expression, which consequently contributes to CRC development and progression.
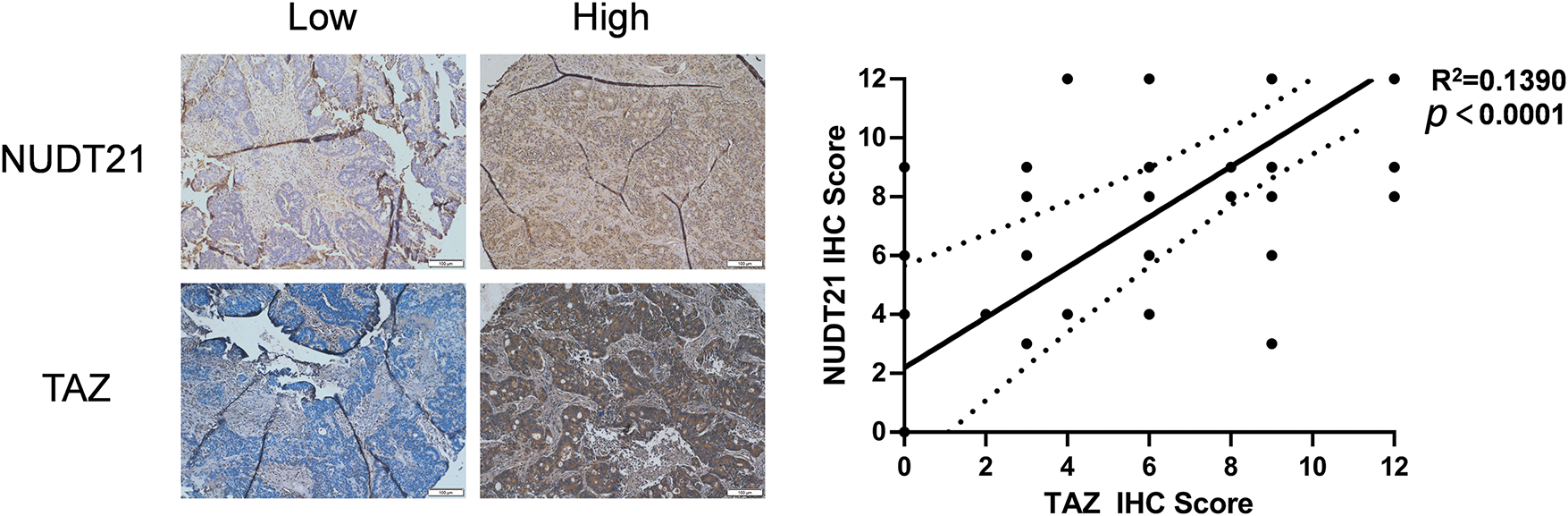
Figure 6: Clinical relevance of NUDT21-TAZ axis in CRC. Correlation between NUDT21 protein expression and TAZ protein expression was detected in the 141 CRC samples. Spearman’s rank correlation coefficient was employed to assess the strength and direction of the relationship between variables
In our study, we found that NUDT21 is overexpressed in CRC tumor tissues, and its high expression indicates a poor prognosis. We investigated its function in CRC cells. Consistent with its expression pattern, NUDT21 knockdown markedly inhibited the proliferation and migration of CRC cells, suggesting that NUDT21 plays an oncogene role in CRC.
NUDT21 is an important APA effector molecule and can inhibit shearing of the proximal PAS. Many experiments have been conducted to specifically investigate the function of NUDT21 in different tumor types. Chu et al. demonstrated that NUDT21 is reduced in glioblastoma, and NUDT21 knockdown can promote glioblastoma progression by promoting the expression of the downstream oncogene PAK1 through 3′UTR APA [16]. In a comparable study, Tan et al. revealed that depletion of NUDT21 elevates the expression of PSMB2 and CXXC5 in hepatocellular carcinoma, a process driven by enhanced utilization of the proximal polyadenylation site within the 3′UTR [35]. Some studies have also reported that NUDT21 exerts tumor-suppressive effects by regulating APA in bladder and cervical cancers [15,18]. These studies suggest that a reduction in NUDT21 leads to the shortening of the downstream oncogene 3′UTR, thereby allowing the oncogene to escape mi-RNA-mediated translational repression.
Our study demonstrated that the truncation of the 3′UTR in target genes caused by NUDT21 knockdown did not consistently lead to an upregulation of protein levels in the corresponding genes. We observed that the 3′UTRs of the key effector molecules of the Hippo pathway, YAP, TAZ, and TEAD1, were shortened by NUDT21 knockdown and their protein expression was downregulated. Therefore, we linked NUDT21 regulation of CRC progression to the Hippo pathway. This indicates that APA regulates protein output via other mechanisms. Based on the DaPars analysis, the 3′UTR of TAZ was extremely shortened from 3,570 bp to 17 bp. We suggest that the 3′UTR of TAZ may be too short to form a translational loop after the NUDT21 knockdown, providing new insights into APA-regulated protein translation. However, there is a lack of specific experiments in our study to further confirm the hypothesis.
In conclusion, NUDT21 is overexpressed in CRC tumor tissues and is positively associated with a worse prognosis. NUDT21 can upregulate the TAZ protein levels, a key effector molecule of the Hippo pathway, thereby promoting the transcription of TAZ downstream target genes and eventually contributing to the development of CRC. In addition, we observed a protein interaction between NUDT21 and TAZ, providing new insights into the development of CRC.
Acknowledgement: We acknowledge the study participants, collaborators, and team members.
Funding Statement: This work was funded by grants from the National Natural Science Foundation of China (82073056) and the Shanghai Pujiang Program (19PJ1407600).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Xiaojian Chen and Zhongchuan Wang; data collection: Xiaojian Chen, Zhujiang Dai, and Qiang Wang; analysis and interpretation of results: Xiaojian Chen, Zhujiang Dai, Wei Chen, and Yun Liu; draft manuscript preparation: Xiaojian Chen and Zhujiang Dai. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GSE223415).
Ethics Approval: Human CRC samples for tissue array were collected from June 2008 to December 2018 at Department of Colorectal and Anal Surgery, Xinhua Hospital, Shanghai Jiaotong University School of Medicine. Informed consent was obtained from all participants or their legal guardians and this project was approved by Xinhua Hospital Ethics Committee, Affiliated with Shanghai Jiaotong University School of Medicine (XHEC-NSFC-2021-326). All methods were performed in accordance with the Declarations of Helsinki.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/biocell.2025.059286.
References
1. Ioffe D, Dotan E. Evidence-based care of older adults with metastatic colorectal cancer: insights from landmark clinical trials. J Clin Oncol. 2023;41(34):5228–36. doi:10.1200/JCO.23.01337. [Google Scholar] [CrossRef]
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi:10.3322/caac.21660. [Google Scholar] [PubMed] [CrossRef]
3. Li R, Liu X, Huang X, Zhang D, Chen Z, Zhang J, et al. Single-cell transcriptomic analysis deciphers heterogenous cancer stem-like cells in colorectal cancer and their organ-specific metastasis. Gut. 2024;73:470–84. doi:10.1136/gutjnl-2023-330243. [Google Scholar] [PubMed] [CrossRef]
4. Zhou H, Liu Z, Wang Y, Wen X, Amador EH, Yuan L, et al. Colorectal liver metastasis: molecular mechanism and interventional therapy. Signal Transduct Target Ther. 2022;7(1):70. doi:10.1038/s41392-022-00922-2. [Google Scholar] [PubMed] [CrossRef]
5. Cañellas-Socias A, Sancho E, Batlle E. Mechanisms of metastatic colorectal cancer. Nat Rev Gastroenterol Hepatol. 2024;21:609–25. [Google Scholar]
6. Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14(7):496–506. doi:10.1038/nrg3482. [Google Scholar] [PubMed] [CrossRef]
7. Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3′-UTR landscape across seven tumour types. Nat Commun. 2014;5(1):5274. doi:10.1038/ncomms6274. [Google Scholar] [PubMed] [CrossRef]
8. Mitschka S, Mayr C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat Rev Mol Cell Biol. 2022;23(12):779–96. doi:10.1038/s41580-022-00507-5. [Google Scholar] [PubMed] [CrossRef]
9. Tang P, Yang Y, Li G, Huang L, Wen M, Ruan W, et al. Alternative polyadenylation by sequential activation of distal and proximal PolyA sites. Nat Struct Mol Biol. 2022;29(1):21–31. doi:10.1038/s41594-021-00709-z. [Google Scholar] [PubMed] [CrossRef]
10. Zhang Y, Liu L, Qiu Q, Zhou Q, Ding J, Lu Y, et al. Alternative polyadenylation: methods, mechanism, function, and role in cancer. J Exp Clin Cancer Res. 2021;40(1):51. doi:10.1186/s13046-021-01852-7. [Google Scholar] [PubMed] [CrossRef]
11. Gruber AJ, Zavolan M. Alternative cleavage and polyadenylation in health and disease. Nat Rev Genet. 2019;20(10):599–614. doi:10.1038/s41576-019-0145-z. [Google Scholar] [PubMed] [CrossRef]
12. Li B, Cai Y, Chen C, Li G, Zhang M, Lu Z, et al. Genetic variants that impact alternative polyadenylation in cancer represent candidate causal risk loci. Cancer Res. 2023;83(21):3650–66. doi:10.1158/0008-5472.CAN-23-0251. [Google Scholar] [PubMed] [CrossRef]
13. Jafari Najaf Abadi MH, Shafabakhsh R, Asemi Z, Mirzaei HR, Sahebnasagh R, Mirzaei H, et al. CFIm25 and alternative polyadenylation: conflicting roles in cancer. Cancer Lett. 2019 Sep;459:112–21. doi:10.1016/j.canlet.2019.114430. [Google Scholar] [PubMed] [CrossRef]
14. Masamha CP, Xia Z, Yang J, Albrecht TR, Li M, Shyu AB, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510(7505):412–6. doi:10.1038/nature13261. [Google Scholar] [PubMed] [CrossRef]
15. Xiong M, Chen L, Zhou L, Ding Y, Kazobinka G, Chen Z, et al. NUDT21 inhibits bladder cancer progression through ANXA2 and LIMK2 by alternative polyadenylation. Theranostics. 2019;9(24):7156–67. doi:10.7150/thno.36030. [Google Scholar] [PubMed] [CrossRef]
16. Chu Y, Elrod ND, Wang C, Li L, Chen T, Routh A, et al. Nudt21 regulates the alternative polyadenylation of Pak1 and is predictive in the prognosis of glioblastoma patients. Oncogene. 2019;38(21):4154–68. doi:10.1038/s41388-019-0714-9. [Google Scholar] [PubMed] [CrossRef]
17. Sun M, Ding J, Li D, Yang G, Cheng Z, Zhu Q. NUDT21 regulates 3′-UTR length and microRNA-mediated gene silencing in hepatocellular carcinoma. Cancer Lett. 2017;410(0):158–68. doi:10.1016/j.canlet.2017.09.026. [Google Scholar] [PubMed] [CrossRef]
18. Xing Y, Chen L, Gu H, Yang C, Zhao J, Chen Z, et al. Downregulation of NUDT21 contributes to cervical cancer progression through alternative polyadenylation. Oncogene. 2021;40(11):2051–64. doi:10.1038/s41388-021-01693-w. [Google Scholar] [PubMed] [CrossRef]
19. Zhu Y, Zhang R, Zhang Y, Cheng X, Li L, Wu Z, et al. NUDT21 promotes tumor growth and metastasis through modulating SGPP2 in human gastric cancer. Front Oncol. 2021;11:670353. doi:10.3389/fonc.2021.670353. [Google Scholar] [PubMed] [CrossRef]
20. Fu M, Hu Y, Lan T, Guan KL, Luo T, Luo M. The hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther. 2022;7(1):1–20. doi:10.1038/s41392-022-01191-9. [Google Scholar] [PubMed] [CrossRef]
21. Zhong Z, Jiao Z, Yu FX. The Hippo signaling pathway in development and regeneration. Cell Rep. 2024;43(3):113926–6. doi:10.1016/j.celrep.2024.113926. [Google Scholar] [PubMed] [CrossRef]
22. Wang S, Zhou L, Ling L, Meng X, Chu F, Zhang S, et al. The crosstalk between hippo-YAP pathway and innate immunity. Front Immunol. 2020 Feb 27;11:323. doi:10.3389/fimmu.2020.00323. [Google Scholar] [PubMed] [CrossRef]
23. Liu X, Chen X, Liu L, Xia J, Zhang H. AKAP12 inhibits the proliferation of ovarian cancer by activating the Hippo pathway. Oncologie. 2023;26(1):105–16. doi:10.1515/oncologie-2023-0242. [Google Scholar] [CrossRef]
24. Zanconato F, Cordenonsi M, Piccolo S. YAP/TAZ at the roots of cancer. Cancer Cell. 2016;29(6):783–803. doi:10.1016/j.ccell.2016.05.005. [Google Scholar] [PubMed] [CrossRef]
25. Liu Y, Wang G, Yang Y, Zhong M, Liang Z, Cui A, et al. Increased TEAD4 expression and nuclear localization in colorectal cancer promote epithelial-mesenchymal transition and metastasis in a YAP-independent manner. Oncogene. 2015;35(21):2789–800. doi:10.1038/onc.2015.342. [Google Scholar] [CrossRef]
26. Piccolo S, Panciera T, Contessotto P, Cordenonsi M. YAP/TAZ as master regulators in cancer: modulation, function and therapeutic approaches. Nat Cancer. 2023;4(1):9–26. [Google Scholar]
27. Luo J, Zou H, Guo Y, Tong T, Chen Y, Xiao Y, et al. The oncogenic roles and clinical implications of YAP/TAZ in breast cancer. Br J Cancer. 2023;128:1611–24. [Google Scholar] [PubMed]
28. Gu C, Huang Z, Chen X, Liu C, Rocco G, Zhao S, et al. TEAD4 promotes tumor development in patients with lung adenocarcinoma via ERK signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2020;1866(12):165921. doi:10.1016/j.bbadis.2020.165921. [Google Scholar] [PubMed] [CrossRef]
29. Guo Y, Zhu Z, Huang Z, Cui L, Yu W, Hong W, et al. CK2-induced cooperation of HHEX with the YAP-TEAD4 complex promotes colorectal tumorigenesis. Nat Commun. 2022;13(1):4995. doi:10.1038/s41467-022-32674-6. [Google Scholar] [PubMed] [CrossRef]
30. Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138(4):673–84. doi:10.1016/j.cell.2009.06.016. [Google Scholar] [PubMed] [CrossRef]
31. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–79. doi:10.1146/annurev-biochem-060308-103103. [Google Scholar] [PubMed] [CrossRef]
32. Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11(2):113–27. doi:10.1038/nrm2838. [Google Scholar] [CrossRef]
33. Salah Z, Aqeilan RI. WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis. 2011;2(6):e172–2. doi:10.1038/cddis.2011.53. [Google Scholar] [PubMed] [CrossRef]
34. Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF-TRCP. Genes Dev. 2010;24(1):72–85. doi:10.1101/gad.1843810. [Google Scholar] [PubMed] [CrossRef]
35. Tan S, Li H, Zhang W, Shao Y, Liu Y, Guan H, et al. NUDT21 negatively regulates PSMB2 and CXXC5 by alternative polyadenylation and contributes to hepatocellular carcinoma suppression. Oncogene. 2018;37(35):4887–900. doi:10.1038/s41388-018-0280-6. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools