 Open Access
Open Access
ARTICLE
RNF26 up-regulates PD-L1 to regulate the cancer immune response in ccRCC
1 Department of Urology, Hunan Provincial People’s Hospital, Hunan Normal University, Changsha, 410000, China
2 Department of Rehabilitation, The Second Xiangya Hospital, Central South University, Changsha, 410000, China
3 Department of Urology, The Second Xiangya Hospital, Central South University, Changsha, 410000, China
4 Uro-Oncology Institute of Central South University, Changsha, 410000, China
* Corresponding Author: LIANG ZHU. Email:
# These authors contributed equally
BIOCELL 2024, 48(9), 1323-1330. https://doi.org/10.32604/biocell.2024.051747
Received 14 March 2024; Accepted 12 July 2024; Issue published 04 September 2024
Abstract
Background: Clear cell renal cell carcinoma (ccRCC) stands as the most prevalent form of kidney cancer, accounting for a significant proportion of malignancies affecting the kidneys. ccRCC is well known as a type of tumour with immunogenicity. Immune checkpoint inhibitors (ICIs) aim to enhance the anticancer immune response in ccRCC by blocking programmed cell death 1 ligand 1/programmed death 1 (PD-L1/PD-1) pathways. In a previous study, we showed that RING finger protein 26 (RNF26) degrades chromobox 7 (CBX7) to activate the tumor necrosis factor (TNF) in ccRCC. Methods: We analyzed The Cancer Genome Atlas (TCGA) database using the R package ESTIMATE and found that RNF26 was significantly associated with ccRCC immune infiltration. The relationship between RNF26 and the PD1 checkpoint signaling pathway was detected by enrichment analysis. In addition, the molecular mechanism of RNF26 up-regulation of PD-L1 was detected by transcriptome sequencing, RT-qPCR, and Western Blot in ccRCC cell lineages 786-O and A498 cells. The transplantation tumor experiments in C57BL/6 mice were used to test the efficacy of anti-PD1 and knockdown of RNF26 in vivo. Results: We showed that RNF26 suppressed the immune response to ccRCC. Next, we revealed that RNF26 activated the PD-1 checkpoint pathway to suppress the immune response to ccRCC, possibly via the CBX7/PD-L1 axis. Conclusion: The suggestion derived from our results is that targeting RNF26 holds the potential to amplify the efficacy of anticancer immunotherapies in the treatment of ccRCC.Graphic Abstract

Keywords
Supplementary Material
Supplementary Material FileThe most common type of kidney cancer is ccRCC, which makes up a substantial proportion of malignancies affecting the kidneys [1]. The main therapeutic strategy for ccRCC patients is the surgical removal of the tumour [1,2]. Unfortunately, for patients with advanced ccRCC, the prognosis is rarely satisfactory, as surgical treatment is often no longer an option [1]. The prognosis of advanced ccRCC patients has improved due to antiangiogenic therapy [3–5]. New tyrosine kinase inhibitors have been developed based on sunitinib, such as lenvatinib and axitinib, and have already demonstrated efficacy in clinical trials [6]. The development of additional therapeutic modalities is urgently needed, however, because drug resistance often emerges [7,8].
ccRCC is well-known as a type of tumour with immunogenicity [9,10]. Immune checkpoint inhibitors (ICIs) enhance antitumour immunity in the treatment of ccRCC by targeting specific pathways; the effects of ICIs include blocking PD-L1/PD-1, restoring T-cell activation, and reducing T-cell depletion [11]. Several clinical trials have indicated that ICI monotherapies have made some progress in the treatment of patients with ccRCC but do not lead to improved antitumour activity compared to antiangiogenic drugs [12,13]. Thus, a major current goal is improving the efficacy of ICIs in ccRCC.
Combination therapeutic approaches have shown improved efficacy not only in preclinical studies but also in clinical trials; this improvement holds true whether they are immune checkpoint inhibitors or integrative agents such as vascular endothelial growth factor inhibitors or immunomodulators [14]. Therefore, it would be a promising strategy to explore targets to improve therapeutic efficacy to develop new combination therapeutic regimens.
Posttranslational changes are involved in controlling protein activity [15]. Protein ubiquitination, which is mediated by the proteasomal system, accounts for approximately 90% of the regulation of protein stability [16,17]. The ubiquitin-proteasome system contains a major component called the e3 ligase, which specifically recognizes the target protein and facilitates the transport of ubiquitin to the target protein [18,19]. RNF26, an e3 ligase, regulates the immune response after a viral infection [20]. RNF26 can regulate the innate immune system by forming a complex with transmembrane protein 43 (TMEM43), endonuclease domain containing 1 (ENDOD1), transmembrane protein 33 (TMEM33) and transmembrane p24 trafficking protein 1 (TMED1) to act on the cyclic GMP-AMP synthase–stimulator of interferon response cGAMP interactor 1pathway [21]. In addition, we have shown that RNF26 degrades chromobox 7 (CBX7) to activate the TNF signaling pathway in ccRCC [22]. Therefore, RNF26 may modulate the immune response to malignancy. However, the underlying mechanism is still unclear.
In this study, our pursuit was aimed at elucidating the role of RNF26 in modulating the immune susceptibility of urologic tumors. Exploring the possibility that RNF26 is a new focus for improving the effectiveness of ICIs in the treatment of ccRCC.
Proteintech (Wuhan, China) provided all antibodies necessary for Western blot analysis, which included RNF26 (#16802-1-AP), GAPDH (#60004-1-Ig), PD-L1 (#66248-1-Ig), CBX7 (#26278-1-AP), HRP-conjugated AffiniPure Goat Anti-Rabbit IgG (H+L) (#SA00001-2), HRP-conjugated AffiniPure Donkey Anti-Goat IgG (H+L) (#SA00001-3). Lipofectamine 2000 (#11668030), which was acquired from Thermo Fisher Scientific in Shanghai, China, was utilized for transfecting plasmids or shRNAs based on the manufacturer’s instructions. The shControl was specified to use only the lip2000. See supplementary information for sequences of shRNAs (Table S1).
Procell Life Science & Technology (Wuhan, China) provided the ccRCC cell lineages A498 (CL-0254) and Renca (CL-0568). 786-O cells (#TCHu186) were acquired from the Chinese Academy of Sciences Cell Bank Type Culture Conservation Centre located in Shanghai, China. Yuchicell Biology Technology conducted a short tandem repeat (STR) analysis of the abovementioned cell lines. All utilized cells in this study were free from mycoplasma contamination. A498 cells were cultured in Minimum Essential Medium (PM150467) from Procell Life Science & Technology, 786-O cells were grown in Roswell Park Memorial Institute-1640 medium (#12633012) from Gibco in the USA, and Renca cells were cultured in CM-0568 medium provided by Procell Life Science & Technology. The culture medium was supplemented with 10% foetal bovine serum from Shanghai Life-iLab Biotech (AC03L055, China). The cells were all kept in an environment of 37°C and 5% carbon dioxide for incubation.
A protease inhibitor was added to RIPA buffer (Thermo Scientific, China, 89901) for protein extraction from cells. The lysate underwent centrifugation for 15 min at 12,000 × g. We perform protein quantification by BCA Protein Assay Kits (Thermo Scientific, China, 23225). Next, the lysate from the cell was heated in boiling water at 100°C and then electrophoresed on SDS–PAGE gels, which were transferred onto polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific, China, T2234). The PVDF membranes were incubated with the appropriate primary antibodies overnight at 4°C. On the following day, the PVDF membranes were exposed to secondary antibodies (ordinary temperatures, 2 h) and treated with enhanced chemiluminescence reagent (VL002-200) from Sigma‒Aldrich, China.
RT-PCR with real-time quantification
Cells were subjected to RNA extraction utilizing the TRIzol reagent (#AG21102) from Accurate Biotechnology, Hunan, China. Subsequently, RT‒qPCR was performed in accordance with the manufacturer’s instructions, employing reverse transcription kits (#AG11728) and PCR kits (#AG11701), both sourced from Accurate Biotechnology in Hunan, China. The values were standardized to GAPDH, and the fold change was quantified using the 2−ΔΔCt method. The sequences of all primers are listed in Table S2.
The study was carried out according to the principles of the Declaration of Helsinki principles. The Institutional Animal Care and Use Committee (IACUC) of the Second Xiangya Hospital, Central South University, Changsha, China, approved the animal experiments (licence number 20230476). The sex of the mice had no impact on the findings of the research. Six-week-old C57BL/6 mice from Wuhan Yodu Biotechnology Co., Ltd., a subsidiary of Shulaibao Biotech in Wuhan, China, were selected for the study, with an equal number of male and female mice. The animals were maintained in the animal center of the Second Xiangya Hospital by standard conditions, including a humidity level of 60% ± 3% and a controlled environment with a 12-h light/dark cycle at a temperature of 22°C ± 0.5°C. Mice were allowed access to food and water ad libitum. The indicated shRNAs-transfected ccRCC cell lines (5 × 106 cells per mouse) were surgically implanted beneath the skin on the left backs of the mice Once the tumour volume reached 100 mm3, mice with comparable tumours were randomly divided into groups and given either IgG (BioXcell, Clone 2A3) or 200 micrograms of anti-PD-1 antibody (specifically, the BioXcell Clone RMP1-14). The formula (Length × Width2)/2 was used to calculate the tumour volume. A tumour volume of 1500 mm3 was considered the endpoint, and the tumours were obtained and weighed. The conduct of the animal study adhered to the protocols established by the National Institutes of Health for the appropriate care and management of laboratory animals, as outlined in NIH Publications No. 8023 (revised 1978).
Tissue microarrays, specifically identified as (#U081ki01), were acquired from Bioaitech located in Xi’an, China, and then subjected to staining with PD-L1 and RNF26 antibodies. The evaluation of the intensity of staining was carried out following a previously established methodology [23]. The ultimate staining index can be calculated by multiplying the percentage of positively stained cells by the intensity of the staining. CD3 antibodies (#17617-1-AP, Proteintech) were used for immunostaining of the mouse tissue. The method of staining was the same as that utilized for the tissue microarray slides mentioned above.
A498 was transfected with specified shRNF26#1 for 72 h for RNA-seq analysis in GenePlus (Beijing, China). After obtaining count data from the company, we used the DESeq2 R software package (version 1.16.1) for differential expression analysis. In addition, to assess the degree of statistical enrichment of differentially expressed genes in the KEGG (Kyoto Encyclopedia of Genomes), we used the clusterProfiler R package. Three replicates were performed in each group.
The data are reported as the mean accompanied by the standard deviation (SD). A p-value of less than 0.05 is indicative of statistical significance. Statistical significance was assessed using one-way or two-way ANOVA with GRAPHPAD PRISM 5 software from San Diego, California, United States, after detecting the normal distribution of the data.
RNF26 inhibits the immune response to ccRCC
To investigate how RNF26 impacts the immune response to urologic cancers, we analyzed data from three different types of urinary tumours—bladder urothelial carcinoma (BLCA), prostate adenocarcinoma (PRAD), and kidney renal clear cell carcinoma (KIRC)—sourced from TCGA (https://portal.gdc.cancer.gov/, accessed on 1 April 2023). The patients were grouped based on their RNF26 levels (Fig. 1A). The computational analysis revealed that patients with ccRCC displaying higher levels of RNF26 expression exhibited lower immune scores compared to those with lower RNF26 expression levels (Fig. 1A). RNF26 was not correlated with the immune response in prostate cancer or bladder cancer (Fig. 1A).
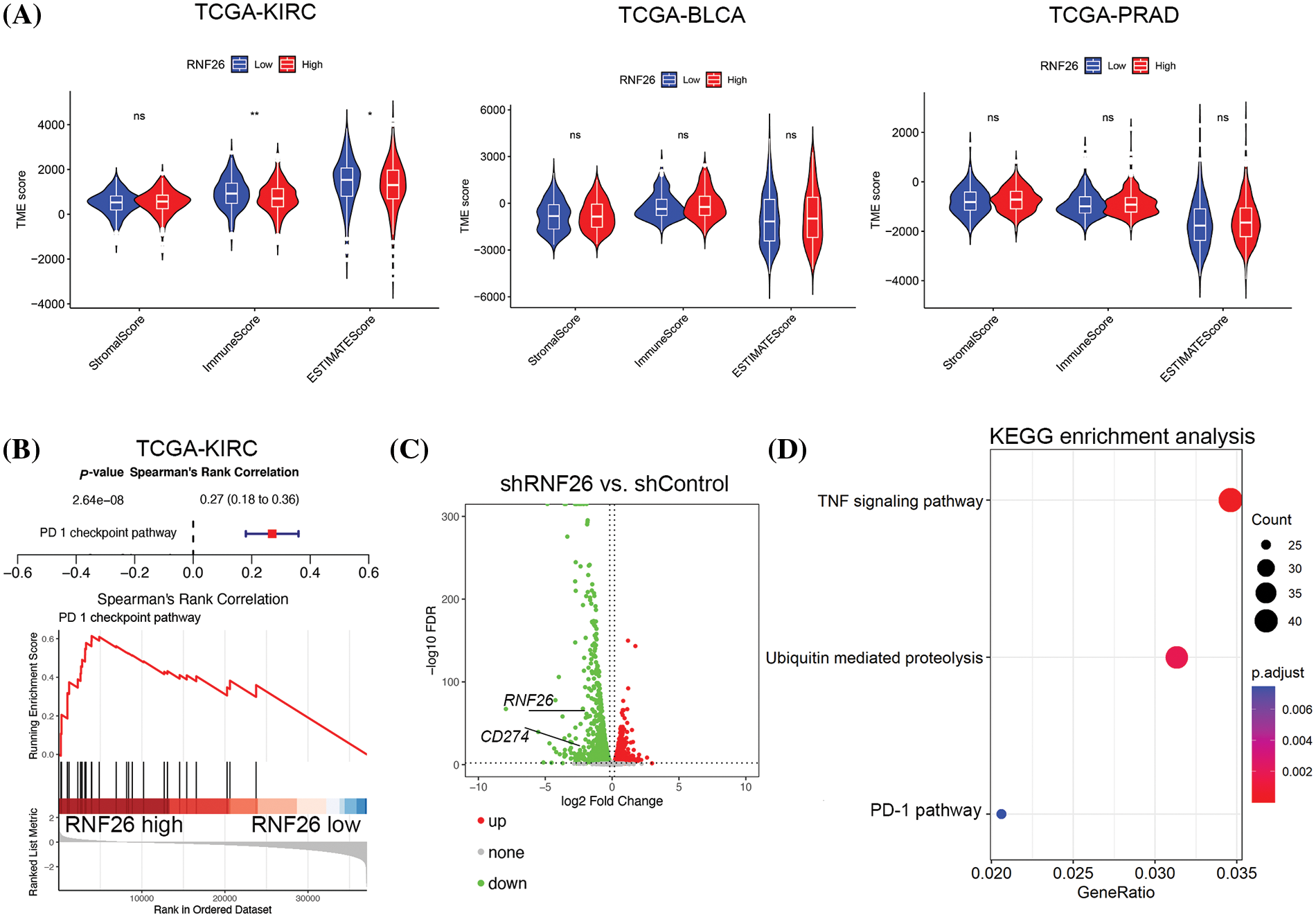
Figure 1: RNF26 inhibits the immune response against ccRCC. (A) We conducted an analysis of the TCGA datasets to explore the association between RNF26 and immune scores. ns, not significant, *p < 0.05; **p < 0.01. (B) GSEA enrichment analysis of the TCGA-KIRC dataset was performed. (C and D) A498 were transfected with shRNF26#1 for 72 h. The transfected cells were then subjected to RNA-seq (C) and KEGG enrichment analysis (D).
As the immune response to ccRCC is well-known that the immune reaction is inhibited by the PD-1 checkpoint pathway [24], we investigated the potential connection between the PD-1 checkpoint pathway and RNF26 expression. KEGG enrichment analysis of TCGA from TCGA KIRC data revealed a notable correlation between elevated RNF26 expression and PD-1 pathway (Fig. 1B). In addition, we performed transcriptome sequencing analysis after depletion of RNF26 in A498 cells (Figs. 1C and S1). Consistent with the patient data, KEGG enrichment analysis revealed that suppression of RNF26 decreased PD-L1 levels in ccRCC (Fig. 1D). In addition, RNF26 knockdown reduced TNF signaling pathway activity and inhibited ubiquitin-mediated proteolysis (Fig. 1D). Our previous findings showed that RNF26 functions as an e3 ligase to degrade CBX7 and trigger TNF signaling [22]. Our findings indicate that RNF26 may inhibit the immune response by controlling the PD-1/PD-L1 checkpoint pathway in ccRCC.
RNF26 increases PD-L1 expression in ccRCC
Validation of the RNA-seq data demonstrated that PD-L1 expression decreased due to RNF26 knockdown (Fig. 1) in both A498 and 786-O cells (Figs. 2A,B and S2A). In contrast, increased RNF26 expression led to increases in PD-L1 protein and mRNA levels in A498 and 786-O cells (Figs. 2C,D and S2B). Additionally, we showed that the abnormal increase in RNF26 could restore the expression of PD-L1 after reducing RNF26 levels in A498 and 786-O cells (Figs. 2E,F and S2C). Meanwhile, we found that shRNF26#1 was better than shRNF26#2, so we used shRNF26 for subsequent experiments. In KIRC, BLCA, and PRAD, mRNA levels of RNF26 and PD-L1 showed a positive association with the TCGA dataset (Fig. 2GI). Furthermore, antibodies against RNF26 and PD-L1 were used to stain a ccRCC tissue microarray (Fig. 2J); The results showed a correlation between high levels of RNF26 protein and PD-L1 protein in ccRCC, as demonstrated in Fig. 2J,K. These data indicate that PD-L1 is elevated by RNF26 administration in patients with ccRCC.
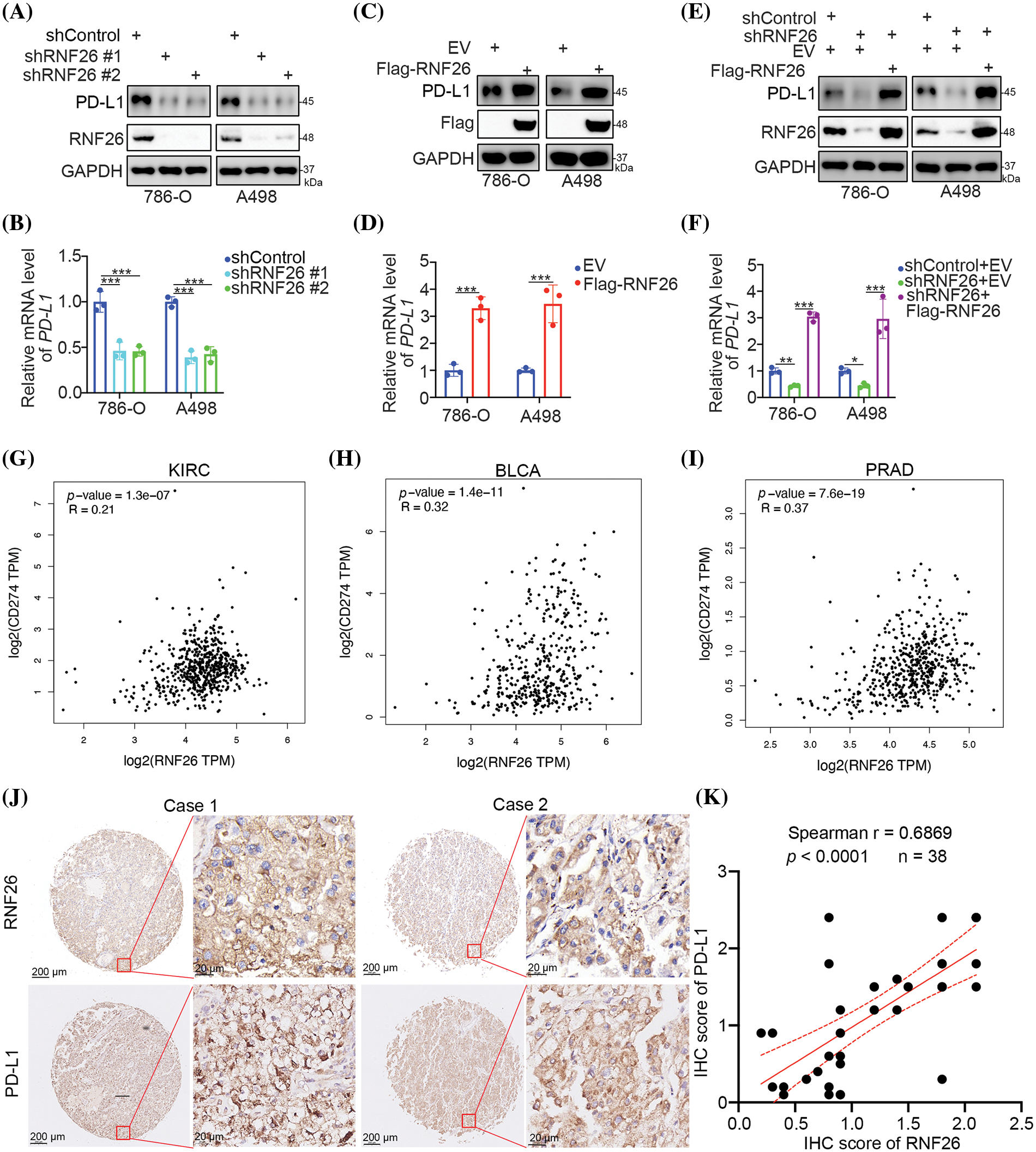
Figure 2: RNF26 increases the expression of PD-L1 in ccRCC cells. (A–F) 786-O and A498 cells were transfected with the indicated shRNAs for 72 or 48 h. Cells were then harvested for Western blot analysis (A, C, E) and RT-qPCR analysis (B, D, F). *, **, and *** corresponding to p < 0.05, p < 0.01, and p < 0.001. n = 3. (G–I) Correlation analysis between RNF26 and PD-L1 (CD274) was performed in KIRC (G) BLCA (H) and PRAD (I) datasets using the GEPIA (http://gepia.cancer-pku.cn, accessed on 1 April 2024). The corresponding p-values and Spearman correlation coefficients (R) are indicated. (J and K) Immunohistochemistry (IHC) staining was performed using antibodies against RNF26 and PD-L1 in tissue microarrays of ccRCC. The associated p-values and Spearman correlation coefficients (r) are provided.
RNF26 modulates the immune response through the PD-1 checkpoint pathway in renal cancer
Given that RNF26 levels have a positive influence on PD-L1 expression, we subsequently aimed to establish if RNF26 modulates the immune responsiveness of ccRCC via PD-L1. Renca cells in which RNF26 was knocked down originated from spontaneous renal adenocarcinoma in mice (Figs. 3A and S3A). Renca cells were subjected to a xenograft assay in immunocompetent C57BL/6 mice with or without PD-1 antibodies (Fig. 3B). RNF26 knockdown inhibited tumor growth and augmented PD-1 blockade’s anticancer activity (Figs. 3C and S3B,C). In addition, RNF26 alone or in combination with PD-1 blockade was sufficient to increase CD3+ T-cell infiltration in mouse tumour tissues (Fig. 3D). RNF26 modulates PD-L1 checkpoint-mediated immune responses to ccRCC, as indicated by these data.
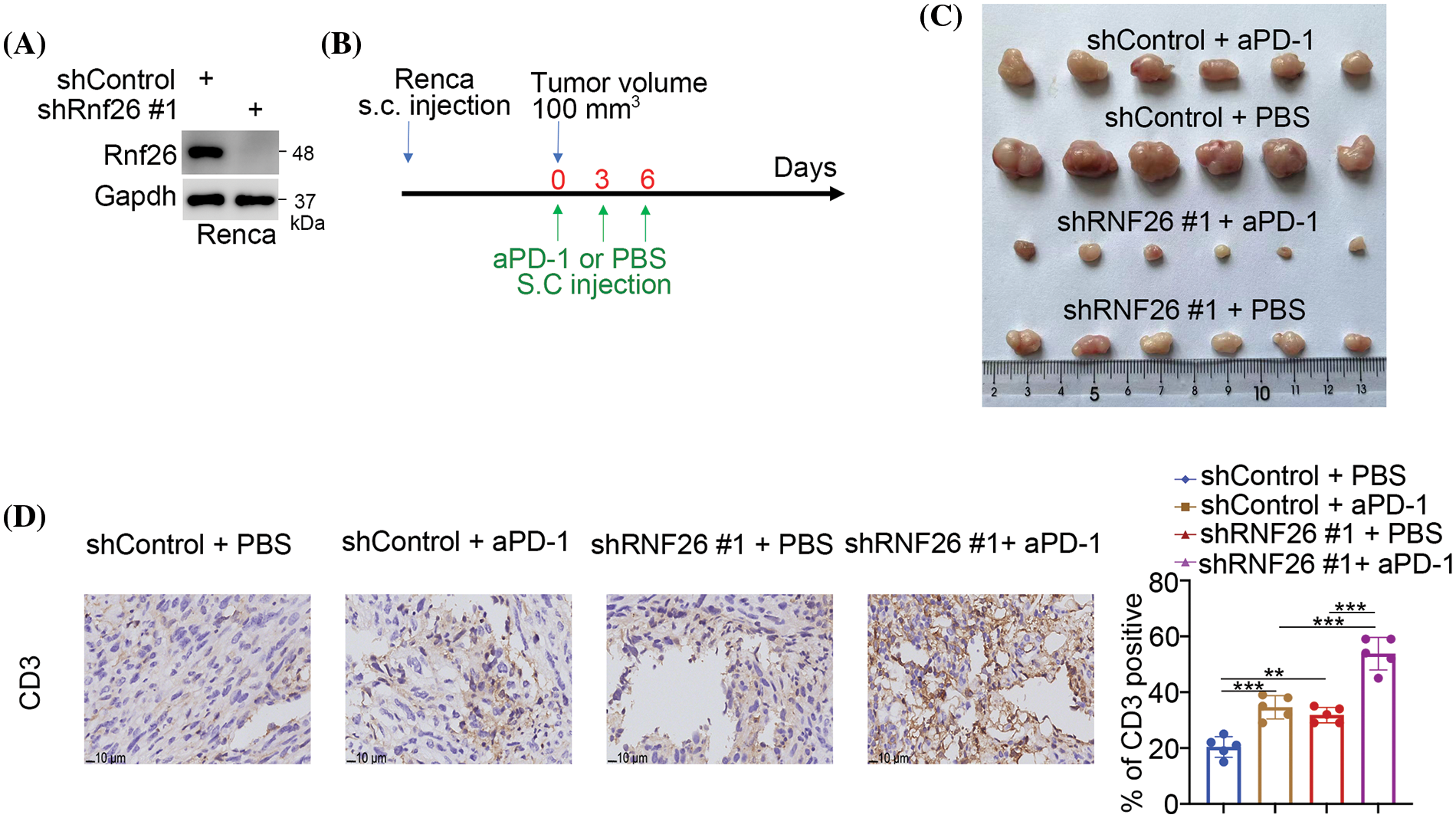
Figure 3: RNF26 modulates the immune response through the PD-1 checkpoint pathway in renal cancer. (A–D) Renca cells were transfected with the shRNF26#1 for 72 h subjected to Western blot (A) and injected subcutaneously into C57BL/6 mice, as outlined in panel (B) The resulting tumors were indicated in panel C (n = 6 per group). The harvested tumors were then stained with a CD3 antibody (n = 5 per group) (D). **p < 0.01; ***p < 0.001.
RNF26 regulates PD-L1 expression in renal cancer cells in part via CBX7
The e3 ligase function of RNF26 directly regulates protein stability at the post-transcription level [25], but RNF26 regulates PD-L1 mRNA levels in kidney carcinoma cells. Notably, in a previous study, CBX7 suppressed the expression of PD-L1 at the mRNA level in bladder cancer through POU class 2 homeobox 2 (POU2F2) [26]. We validated this pathway in ccRCC cells (Fig. S4A). In addition, we reported that RNF26 degrades CBX7 and activates the TNF pathway in renal cancer cells [22]. Thus, we explored whether RNF26 regulated PD-L1 through CBX7 in ccRCC. Initially, blocking CBX7 alone increased the levels of PD-L1 in A498 and 786-O cells (Figs. 4A,B and S4A,B); This result aligns with previous observations made in bladder cancer cells [26]. Subsequently, we discovered that suppressing or increasing RNF26 levels following CBX7 depletion did not lead to additional alterations in PD-L1 levels (Fig. 4A–D). Therefore, these data indicated that the RNF26-induced changes in PD-L1 transcription were mediated by CBX7 in ccRCC.
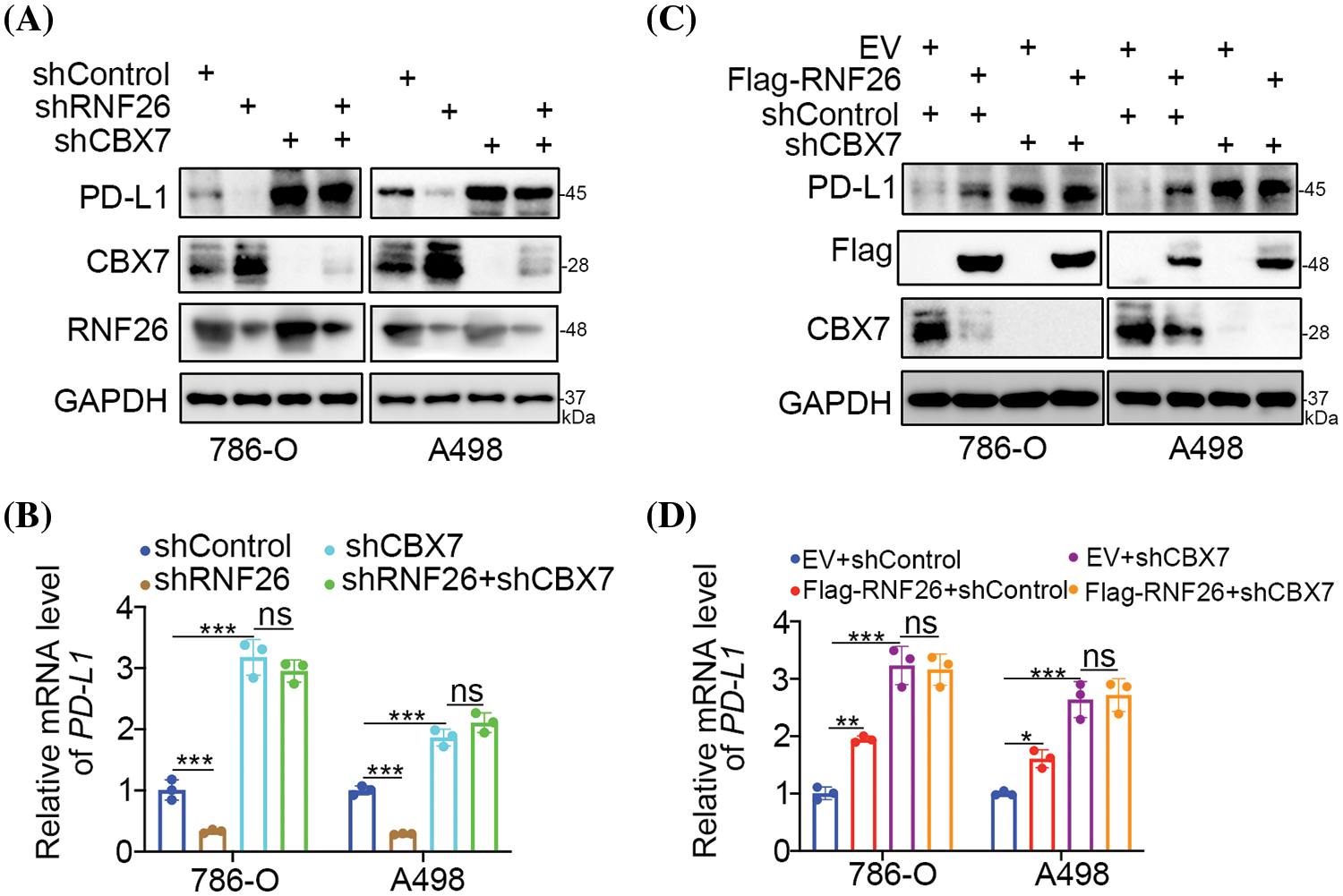
Figure 4: RNF26 regulates PD-L1 expression in part through CBX7 in renal cancer cells. (A–D) 786-O and A498 were transfected with the specified shRNAs for 72 h. Cells were then harvested for Western blot (A, C) and RT-qPCR (B, D). ns, not significant; *p < 0.05; **p < 0.01; ***p < 0.001; n = 3.
The existence of PD-L1 within cancer cells plays a pivotal role in the success of PD-1/PD-L1 inhibition in the treatment of cancers. Elucidating the regulatory pathway governing PD-L1 in ccRCC is helpful for identifying new candidates to improve the antitumour effectiveness of PD-L1 blockade. We previously reported that ribonucleotide reductase regulatory subunit M2 stabilizes annexin A1 to increase PD-L1 and regulate the sensitivity of ccRCC to PD-1 blockade [27]. A recent study has revealed that abnormally elevated levels of nuclear factor of activated T cells-1 (NFAT1) result in augmented PD-L1 expression in ccRCC [23]. RNF26 upregulated PD-L1 to alter immune susceptibility in ccRCC via the PD-1 checkpoint pathway. While based on TCGA database analysis using calculations from the R package ESTIMATE indicated that RNF26 expression was specifically significant in renal cancer, we also discovered a strong association between RNF26 and PD-L1 in prostate and bladder cancers. Therefore, in future work, the relevant findings will be further explored in experiments using two other urological tumours.
Protein homeostasis is regulated by e3 ligases, which are essential for the immune system’s defense against cancer. The quantity of e3 ligases can directly or indirectly modulate PD-L1 expression in carcinoma cells. For instance, the e3 ubiquitin ligase speckle type BTB/POZ protein (binds to PD-L1 and promotes its degradation [28,29]. Shi et al. reported that transmembrane and ubiquitin-like domains containing 1 act as an e3 ligase to destabilize PD-L1 [30]. Moreover, RNF144A can interact with PD-L1 and enhance its degradation in bladder cancer cells [31]. Furthermore, our group reported that expression of e3 ligase F-box and WD repeat domain containing 7 was downregulated in sunitinib-resistant ccRCC cells and that FBW7 is responsible for degrading NFAT1 and indirectly decreasing PD-L1 expression [23]. We similarly demonstrated that RNF26 indirectly increased PD-L1 levels, which is likely to be, at least in part, mediated by CBX7 degradation in ccRCC cells [22]. A further investigation of the related mechanisms in future studies is clearly required to understand the regulation of PD-L1.
Positioned at 11q23 on the human chromosome, the RNF26 gene encodes a 433-residue protein with two distinct domains: a leucine zipper domain at the beginning and a RING finger domain at the end [32]. The RING finger domain confers e3 ligase activity to RNF26 [32]. RNF26 was shown to promote polyubiquitination of lysine 150 at STING by K11 [20], as well as the ubiquitination of sequestosome-1 in cells [25]. Recently, our group identified p57 and CBX7 as novel substrates of RNF26 for e3 ligase-mediated degradation [22,33]. Future studies will likely identify additional substrates of RNF26 and elucidate novel functions of RNF26 in cancer cells.
However, no effective RNF26 inhibitors have been developed. The transcription factor forkhead box M1 (FOXM1) has been documented to enhance the transcriptional activity of RNF26 through its binding to the RNF26 promoter [34]. Previous research has verified that utilizing small molecule inhibitors to target FOXM1 along with anticancer medications could serve as a novel approach for treating cancers that are resistant to chemotherapy [34]. Therefore, indirect regulation of RNF26 by inhibiting FOXM1 might be a good strategy. In addition, siRNA-based gene silencing has now made significant progress in tumor therapy [35]. The research of targeting therapy by delivering siRNF26 through nanocarriers will also be a promising direction.
Overall, we explored the immune-related function of RNF26 in ccRCC. Our data revealed that RNF26 results in immune evasion by ccRCC through the PD-1 checkpoint pathway. Mechanistically, we showed that RNF26 indirectly suppressed PD-L1 expression through CBX7 in ccRCC. Therefore, our results indicate that targeting RNF26 has the potential to augment the efficacy of anticancer immunotherapies in the management of ccRCC.
Acknowledgement: None.
Funding Statement: The study was supported by Natural Science Foundation of Hunan Province of China (Grant No. 2022JJ30870 (Liang Zhu)).
Author Contributions: Study conception and design: Liang Zhu; data collection: Weigang Ren; analysis and interpretation of results: Jing Li; draft manuscript preparation: Ruijiang Zeng. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data sets utilized and/or examined in the current research can be obtained from the corresponding authors upon request.
Ethics Approval: The study was carried out according to the principles of the Declaration of Helsinki principles. The Institutional Animal Care and Use Committee (IACUC) of the Second Xiangya Hospital, Central South University, approved the animal experiments (licence number 20230476).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/biocell.051747.
References
1. Motzer RJ, Jonasch E, Agarwal N, Alva A, Baine M, Beckermann K, et al. Kidney cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(1):71–90. [Google Scholar] [PubMed]
2. Kase AM, George DJ, Ramalingam S. Clear cell renal cell carcinoma: from biology to treatment. Cancers. 2023;15(3):665. [Google Scholar] [PubMed]
3. Schodel J, Grampp S, Maher ER, Moch H, Ratcliffe PJ, Russo P, et al. Hypoxia, hypoxia-inducible transcription factors, and renal cancer. Eur Urol. 2016;69(4):646–57. [Google Scholar] [PubMed]
4. Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature. 2016;539(7627):112–7. [Google Scholar] [PubMed]
5. Wolf MM, Kimryn Rathmell W, Beckermann KE. Modeling clear cell renal cell carcinoma and therapeutic implications. Oncogene. 2020;39(17):3413–26. [Google Scholar] [PubMed]
6. Chen YW, Wang L, Panian J, Dhanji S, Derweesh I, Rose B, et al. Treatment landscape of renal cell carcinoma. Curr Treat Options Oncol. 2023;24(12):1889–916. [Google Scholar] [PubMed]
7. Sekino Y, Teishima J, Liang G, Hinata N. Molecular mechanisms of resistance to tyrosine kinase inhibitor in clear cell renal cell carcinoma. Int J Urol. 2022;29(12):1419–28. [Google Scholar] [PubMed]
8. Makhov P, Joshi S, Ghatalia P, Kutikov A, Uzzo RG, Kolenko VM. Resistance to systemic therapies in clear cell renal cell carcinoma: mechanisms and management strategies. Mol Cancer Ther. 2018;17(7):1355–64. [Google Scholar] [PubMed]
9. Krishna C, DiNatale RG, Kuo F, Srivastava RM, Vuong L, Chowell D, et al. Single-cell sequencing links multiregional immune landscapes and tissue-resident T cells in ccRCC to tumor topology and therapy efficacy. Cancer Cell. 2021;39(5):662–77.e6. [Google Scholar] [PubMed]
10. Braun DA, Street K, Burke KP, Cookmeyer DL, Denize T, Pedersen CB, et al. Progressive immune dysfunction with advancing disease stage in renal cell carcinoma. Cancer Cell. 2021;39(5):632–48.e8. [Google Scholar] [PubMed]
11. Sun C, Mezzadra R, Schumacher TN. Regulation and function of the PD-L1 checkpoint. Immunity. 2018;48(3):434–52. doi:10.1016/j.immuni.2018.03.014. [Google Scholar] [PubMed] [CrossRef]
12. Terry S, Dalban C, Rioux-Leclercq N, Adam J, Meylan M, Buart S, et al. Association of AXL and PD-L1 expression with clinical outcomes in patients with advanced renal cell carcinoma treated with PD-1 blockade. Clin Cancer Res. 2021;27(24):6749–60. doi:10.1158/1078-0432.CCR-21-0972. [Google Scholar] [PubMed] [CrossRef]
13. Msaouel P, Goswami S, Thall PF, Wang X, Yuan Y, Jonasch E, et al. A phase 1-2 trial of sitravatinib and nivolumab in clear cell renal cell carcinoma following progression on antiangiogenic therapy. Sci Transl Med. 2022;14(641):eabm6420. doi:10.1126/scitranslmed.abm6420. [Google Scholar] [PubMed] [CrossRef]
14. Meng L, Collier KA, Wang P, Li Z, Monk P, Mortazavi A, et al. Emerging immunotherapy approaches for advanced clear cell renal cell carcinoma. Cells. 2023;13(1):34. doi:10.3390/cells13010034. [Google Scholar] [PubMed] [CrossRef]
15. Vieira-Coimbra M, Henrique R, Jeronimo C. New insights on chromatin modifiers and histone post-translational modifications in renal cell tumours. Eur J Clin Invest. 2015;45(s1):16–24. doi:10.1111/eci.12360. [Google Scholar] [PubMed] [CrossRef]
16. Dai X, Zhang T, Hua D. Ubiquitination and SUMOylation: protein homeostasis control over cancer. Epigenomics. 2022;14(1):43–58. doi:10.2217/epi-2021-0371. [Google Scholar] [PubMed] [CrossRef]
17. Popovic D, Vucic D, Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20(11):1242–53. doi:10.1038/nm.3739. [Google Scholar] [PubMed] [CrossRef]
18. Berndsen CE, Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21(4):301–7. doi:10.1038/nsmb.2780. [Google Scholar] [PubMed] [CrossRef]
19. Jevtic P, Haakonsen DL, Rape M. An E3 ligase guide to the galaxy of small-molecule-induced protein degradation. Cell Chem Biol. 2021;28(7):1000–13. doi:10.1016/j.chembiol.2021.04.002. [Google Scholar] [PubMed] [CrossRef]
20. Qin Y, Zhou MT, Hu MM, Hu YH, Zhang J, Guo L, et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 2014;10(9):e1004358. doi:10.1371/journal.ppat.1004358. [Google Scholar] [PubMed] [CrossRef]
21. Fenech EJ, Lari F, Charles PD, Fischer R, Laetitia-Thezenas M, Bagola K, et al. Interaction mapping of endoplasmic reticulum ubiquitin ligases identifies modulators of innate immune signalling. Elife. 2020;9. doi:10.7554/eLife.57306.sa2. [Google Scholar] [CrossRef]
22. Liu W, Wang H, Jian C, Li W, Ye K, Ren J, et al. The RNF26/CBX7 axis modulates the TNF pathway to promote cell proliferation and regulate sensitivity to TKIs in ccRCC. Int J Biol Sci. 2022;18(5):2132–45. doi:10.7150/ijbs.69325. [Google Scholar] [PubMed] [CrossRef]
23. Liu W, Ren D, Xiong W, Jin X, Zhu L. A novel FBW7/NFAT1 axis regulates cancer immunity in sunitinib-resistant renal cancer by inducing PD-L1 expression. J Exp Clin Cancer Res. 2022;41(1):38. doi:10.1186/s13046-022-02253-0. [Google Scholar] [PubMed] [CrossRef]
24. Chen YW, Rini BI, Beckermann KE. Emerging targets in clear cell renal cell carcinoma. Cancers. 2022;14(19):4843. doi:10.3390/cancers14194843. [Google Scholar] [PubMed] [CrossRef]
25. Jongsma ML, Berlin I, Wijdeven RH, Janssen L, Janssen GM, Garstka MA, et al. An ER-associated pathway defines endosomal architecture for controlled cargo transport. Cell. 2016;166(1):152–66. doi:10.1016/j.cell.2016.05.078. [Google Scholar] [PubMed] [CrossRef]
26. Ren W, Ren J, Zhang N, Liu X, Deng Y, Jiang Y, et al. CBX7 represses the POU2F2 to inhibit the PD-L1 expression and regulate the immune response in bladder cancer. Biochem Biophys Res Commun. 2022;613:12–8. doi:10.1016/j.bbrc.2022.04.114. [Google Scholar] [PubMed] [CrossRef]
27. Xiong W, Zhang B, Yu H, Zhu L, Yi L, Jin X. RRM2 regulates sensitivity to sunitinib and PD-1 blockade in renal cancer by stabilizing ANXA1 and activating the AKT pathway. Adv Sci. 2021;8(18):e2100881. doi:10.1002/advs.202100881. [Google Scholar] [PubMed] [CrossRef]
28. Zhang J, Bu X, Wang H, Zhu Y, Geng Y, Nihira NT, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature. 2018;553(7686):91–5. doi:10.1038/nature25015. [Google Scholar] [PubMed] [CrossRef]
29. Gao K, Shi Q, Gu Y, Yang W, He Y, Lv Z, et al. SPOP mutations promote tumor immune escape in endometrial cancer via the IRF1-PD-L1 axis. Cell Death Differ. 2023;30(2):475–87. doi:10.1038/s41418-022-01097-7. [Google Scholar] [PubMed] [CrossRef]
30. Shi C, Wang Y, Wu M, Chen Y, Liu F, Shen Z, et al. Promoting anti-tumor immunity by targeting TMUB1 to modulate PD-L1 polyubiquitination and glycosylation. Nat Commun. 2022;13(1):6951. doi:10.1038/s41467-022-34346-x. [Google Scholar] [PubMed] [CrossRef]
31. Ho SR, Lee YC, Ittmann MM, Lin FT, Chan KS, Lin WC. RNF144A deficiency promotes PD-L1 protein stabilization and carcinogen-induced bladder tumorigenesis. Cancer Lett. 2021;520:344–60. [Google Scholar] [PubMed]
32. Katoh M. Molecular cloning and characterization of RNF26 on human chromosome 11q23 region, encoding a novel RING finger protein with leucine zipper. Biochem Biophys Res Commun. 2001;282(4):1038–44. [Google Scholar] [PubMed]
33. Yi L, Wang H, Li W, Ye K, Xiong W, Yu H, et al. The FOXM1/RNF26/p57 axis regulates the cell cycle to promote the aggressiveness of bladder cancer. Cell Death Dis. 2021;12(10):944. [Google Scholar] [PubMed]
34. Raghuwanshi S, Gartel AL. Small-molecule inhibitors targeting FOXM1: current challenges and future perspectives in cancer treatments. Biochim Biophys Acta Rev Cancer. 2023;1878(6):189015. [Google Scholar] [PubMed]
35. Choi Y, Seok SH, Yoon HY, Ryu JH, Kwon IC. Advancing cancer immunotherapy through siRNA-based gene silencing for immune checkpoint blockade. Adv Drug Deliv Rev. 2024;209:115306. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools