 Open Access
Open Access
ARTICLE
Interferon-gamma regulates the progression of neuroblastoma cells through interferon-regulatory factor-1
1 School of Clinical Medicine, Dali University, Dali, 671000, China
2 Department of Radiology, The First Affiliated Hospital of Dali University, Dali University, Dali, 671000, China
3 Department of Pediatric Surgery, The First Affiliated Hospital of Dali University, Dali University, Dali, 671000, China
* Corresponding Author: NING WANG. Email:
# Co-first authors
BIOCELL 2024, 48(9), 1343-1353. https://doi.org/10.32604/biocell.2024.051673
Received 12 March 2024; Accepted 11 June 2024; Issue published 04 September 2024
Abstract
Objective: This study aimed to elucidate the influence of IFN-gamma (IFN-γ) in neuroblastoma (NB) cells and reveal its potential underlying molecular mechanism. Methods: The Cell Counting Kit-8, Transwell apparatus, and flow cytometry were employed to assess cellular viability, migratory capacity, invasive potential, and apoptotic rates, respectively. RNA-seq combined with bioinformatics analysis revealed differentially expressed genes (DEGs) and their possible biological functions. Protein levels were determined by western blot analysis. Results: IFN-γ treatment resulted in diminished cell viability, mitigated migratory and invasive capabilities, and augmented apoptotic activity in the SK-N-BE (2) cell line, whereas it exhibited the opposite effect in SH-SY5Y cells. Furthermore, interferon regulatory factor 1 (IRF-1) was the common DEG in both IFN-γ-treated SK-N-BE (2) and SH-SY5Y cells. Additionally, we found that it was underexpressed in NB tissues. The depletion of IRF-1 promoted the progression of both SK-N-BE (2) and SH-SY5Y cells. Moreover, IRF-1 knockdown effectively counteracted the effects of IFN-γ on SK-N-BE (2) cells, while exacerbating them in SH-SY5Y cells. Conclusion: This study verified that IFN-γ exerted a distinct role in both N-Myc-and non-N-Myc-amplified NB cells, partially by mediating the expression of IRF-1, suggesting that it may serve as a potent agent for treating patients with NB.Keywords
Neuroblastoma (NB) is a common malignancy that frequently occurs in children aged <5 years [1,2]. Among malignant tumors diagnosed in children under 15 years of age, NB accounts for at least 7% [3]. NB typically begins in one of the adrenal glands and metastasizes to multiple parts of the body. Additionally, metastases are discovered in 50%–60% of patients diagnosed with NB [4]. Although various clinical approaches have effectively improved the prognosis of patients with NB, the survival rate of children with advanced NB remains low [5]. Previous studies have demonstrated that NB spontaneously regreses or transforms into a benign form; however, other cases remain unchanged after treatment [6], which poses a substantial challenge for curing NB. Hence, it is vital to explore effective agents for NB to advance pediatric oncology.
Interferon regulatory factor 1 (IRF-1) is a transcription factor that plays a pivotal role in the immune response, particularly in regulating type I interferons and other genes involved in antiviral and antitumor activities. It is constitutively expressed at low levels in many cell types, where it maintains the expression of its target genes, and its expression can be induced by various stimuli, including viral infections and cellular stress [7]. IRF-1 functions downstream of pathogen recognition receptor signaling and modulates the immune response by regulating the activity of immune cells, such as natural killer and T cells. In addition to being recognized as a tumor suppressor gene, IRF-1 exerts antiproliferative and proapoptotic effects on cancer cells, thereby inhibiting tumor growth and progression [8]. The dual role of IRF-1 in innate immunity and cancer suppression highlights the complexity of its function in maintaining cellular homeostasis and defense against pathogens. Recent studies have indicated that IRF-1 upregulates the expression of programmed death ligand 1 (PD-L1), which bears implications for the use of immune checkpoint inhibitors in cancer therapy. These findings contribute to our understanding of the role of IRF-1 in the immune system and its potential therapeutic applications [9].
Diverse transcription factors, such as N-Myc, are implicated in NB development [10]. N-Myc was first discovered in NBs [11] and is mainly expressed in nervous system malignancies, including NB [12]. In NB cells, N-Myc plays a multifaceted role. Additionally, it interacts with aurora kinase A (AURKA), resulting in the stabilization of N-Myc and limiting cell cycle arrest at the G2/M phase. This interaction is crucial for the survival of NB cells because the inhibition of AURKA results in the degradation of N-Myc, which subsequently triggers cell death. Furthermore, N-Myc overexpression has been associated with metabolic reprogramming in NB cells, thereby promoting a glycolytic phenotype, which is a common feature of cancer cells [13]. N-Myc amplification occurs in >40% of patients with NB [14], as well as in patients with NB classified as high-risk [6]. Additionally, N-Myc can accelerate NB cell processes, including proliferation, angiogenesis, and chemotherapy resistance [6,15,16]. Increasing evidence has revealed that N-Myc amplification enhances the ability of cells to resist tumor necrosis factors and inhibits tumor cell apoptosis [17]. Conversely, silencing N-Myc inhibits NB cell proliferation and induces apoptosis [18]. Therefore, reducing N-Myc-amplified cells and increasing non-N-Myc-amplified cells may effectively prevent NB development.
Being the only type II interferon, IFN-gamma (IFN-γ) plays an essential role in the immune response of host cells [19]. IFN-γ activates multiple signaling pathways, thereby contributing to immune stimulation and regulation [20]. Emerging studies have indicated that IFN-γ can inhibit cell proliferation and angiogenesis, facilitate apoptosis, and serve as a tumor suppressor in various cancers [19,21]. Notably, IFN-γ has been demonstrated to suppress N-myc expression in NB cells [22]. Nevertheless, the specific roles and molecular mechanisms of IFN-γ in NB cells require further exploration.
In this study, we investigated the effects of IFN-γ in NB cells and attempted to elucidate the downstream gene involved in the regulation of N-Myc expression by IFN-γ in NB cells. These findings further substantiated the potential of IFN-γ as a clinical agent for treating NB.
Tissue microarray and immunohistochemistry (IHC) assay
The NB and normal adrenal tissue microarray (Zhongke Guanghua (Xi’an) Intelligent Biotechnology Co., Ltd., N264001, Xi’an, Shaanxi, China) included 22 NB tissues and four normal adrenal tissues.
The IHC assays were performed according to the manufacturer’s instructions. The tissue slices (5 μm) were dewaxed, rehydrated, and underwent antigen retrieval by immersion in a container with 0.01 M citrate buffer solution (pH 6.0). After heating in a microwave at medium power for 10 min, the tissue slices were cooled in the buffer for 35 min. They were sequentially rinsed with 1× TBS (ST661-500 ml, Beyotime Biotech. Inc., Shanghai, China) for 1 min per rinse, followed by a 10-min incubation with a 3% H2O2 solution to quench endogenous peroxidase activity at room temperature. Afterward, the tissue slices were rinsed again with 1× TBS, then blocked for 1 h at room temperature in 10% goat serum (C0265, Beyotime Biotech. Inc., Shanghai, China). Subsequently, the tissue slices were incubated with the primary antibody (IRF1 antibody, 11335-1-AP, Proteintech, Chicago, USA) for 1.5 h at room temperature, followed by another series of rinses with 1× TBST (PR20017, Proteintech, Chicago, USA), for 1 min each. Finally, the tissue slices were incubated with a secondary antibody (DD13, Tongling Biological, Xiamen, China) for 30 min at room temperature. And the sections were stained using a DAB Kit (ml095670, Shanghai Enzyme Linked Biotechnology Co., Ltd., Shanghai, China) [3,23].
SK-N-BE (2) (LJS-h380) and SH-SY5Y (LJS-h187) cells were obtained from Wuhan Lingsi Biotechnology Co., Ltd. (Wuhan, China). SK-N-BE (2) cells were maintained in DMEM/F-12 (iCell-0005, iCell Bioscience, Shanghai, China) supplemented with 10% fetal bovine serum (FBS; FB15015; CLARK BIOSCIENCE, Webster, TX, USA) and 1% penicillin-streptomycin (P/S) solution (BL505A, Biosharp, Hefei, Anhui, China). SH-SY5Y cells were incubated in MEM (61100087; Gibco, New York, NY, USA) and Ham’s F-12 (iCell-0006; iCell Bioscience, Shanghai, China) (1:1) supplemented with 10% FBS and 1% P/S solution. All cells were incubated at 37°C in a 5% CO2 atmosphere.
Small interfering RNAs (siRNAs) targeting IRF-1 (siRNA-1, siRNA-2, and siRNA-3) were synthesized by GenePharma (Gene ID: 3659). Following this, 10 μL of siRNA was transfected into cells employing Lipofectamine 2000 (11668-019, Invitrogen, Waltham, MA, USA). After 48 h, the cells were collected, and IRF-1 expression was measured. The siRNA sequences used are listed in Table 1.
Quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA was isolated using an Ultrapure RNA Kit (DNase I) (CW0597S; CWBIO, Taizhou, Jiangsu, China). RNA was reverse transcribed using a kit (AE311, TransGen Biotech, Beijing, China), and RT-qPCR was performed using a kit (AQ131, TransGen Biotech). All protocols were performed according to the manufacturer’s instructions. The 2−ΔΔCT method was employed to calculate the expression of IRF-1. The primers used are listed in Table 2.
Cell counting kit 8 (CCK-8) assay
The cells were seeded in 96-well plates (10,000 cells/well). IFN-γ (0, 500, 750, 1000, or 15000 ng/mL) was subsequently administered to the cells and incubated for 48 h. Next, the 10 μL CCK-8 solution (BS350B, Biosharp, Hefei, Anhui, China) was supplemented into each well for another 1 h. The absorbance at 450 nm was measured using a microplate reader (HBS-1096A, Nanjing De Tie Experimental Equipment Co., Ltd., Nanjing, Jiangsu, China).
A transwell chamber (353097; Falcon, New York, NY, USA) was employed to observe cell migration and invasion. The cells (1,000,000 cells/well) were treated with IFN-γ (750 ng/mL) or/and transfected with si-IRF1. To analyze cell migration, cells (200,000 cells/mL) were inoculated into the upper chamber containing 500 μL of FBS-free medium. Concurrently, 500 μL of the fully supplemented medium was introduced into the lower compartment. Following a 24-h incubation period at 37°C, the cells that had successfully migrated to the lower surface were subjected to staining using a 0.1% solution of crystal violet (A600331-0100, Sangon, Biotech, Shanghai, China). Next, cell images from six random fields were captured using an optical microscope (XD202, Nanjing Jiangnan Novel Optics Co., Ltd., Nanjing, Jiangsu, China). To analyze cell invasion, the chambers were covered with Matrigel (354230, BD Biosciences, Franklin Lakes, NJ, USA), and the other steps were consistent with those performed for cell migration.
An Annexin V-FITC/PI apoptosis detection kit (40302ES20; YEASEN, Shanghai, China) was used to detect apoptotic cells. The cells (1,000,000 cells/well) were treated with IFN-γ (750 ng/mL) or/and transfected with si-IRF1. Subsequently, the cell suspension, within the concentration range of 10,000 to 100,000 cells/mL, was collected into 500 μL of binding buffer and enriched with 5 μL of Annexin V-FITC and 10 μL of propidium iodide (PI). Thereafter, the cells were incubated in darkness for a duration of 5 min, prior to the assessment of apoptosis via flow cytometry (CytoFLEX, Beckman Coulter, Indianapolis, IN, USA).
Differentially expressed genes (DEGs) analysis
In total, 3 μg of total cellular RNA was sent to Wuhan Lingsi Biotechnology Co., Ltd. (Wuhan, China), for RNA sequencing to obtain two sets of transcriptome data. The R software package DESeq was used to analyze DEGs (fold changes >2, q values < 0.01, false discovery rate (FDRs) <0.01) in each transcriptome. A Venn diagram was constructed to summarize the DEGs shared between SK-N-BE (2) and SH-SY5Y cells.
Gene ontology (GO) [24] and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis [25] were performed on DEGs using the ClusterProfiler package. p < 0.05 served as the cutoff standard [26].
Total protein was collected using lysis buffer (P0013B, Beyotime, Shanghai, China) and quantified using a specific kit (PC0020, Solarbio, Beijing, China). Polyvinylidene fluoride (PVDF) membranes (YA1701, Solarbio, Beijing, China) containing proteins were incubated with primary antibody at 4°C overnight. Subsequently, the membranes were treated with secondary antibodies (Table 3) and incubated for 60 min at room temperature. The protein bands were evaluated using a specific reagent (PE0010, Solarbio, Beijing, China) on a gel imaging acquisition and analysis system (4300, Shanghai Qinxiang Scientific Instrument Co., Ltd., Shanghai, China). Finally, the ImageJ software (v1.8.0, NIH, Rockville, MD, USA) was used to quantify the protein bands.
Data from no less than three independent experiments were analyzed using Graphpad Prism 8.0 software (GraphPad Software, San Diego, CA, USA) and are expressed as the mean ± standard deviation. A Student’s t test or analysis of variance was used to analyze differences between two or multiple groups, respectively. *p < 0.05, **p < 0.01, ***p < 0.001, #p < 0.05, ##p < 0.01, ###p < 0.001, &p < 0.05, &&p < 0.01, and &&&p < 0.001 indicated statistical significance.
IFN-γ regulates NB cell progression
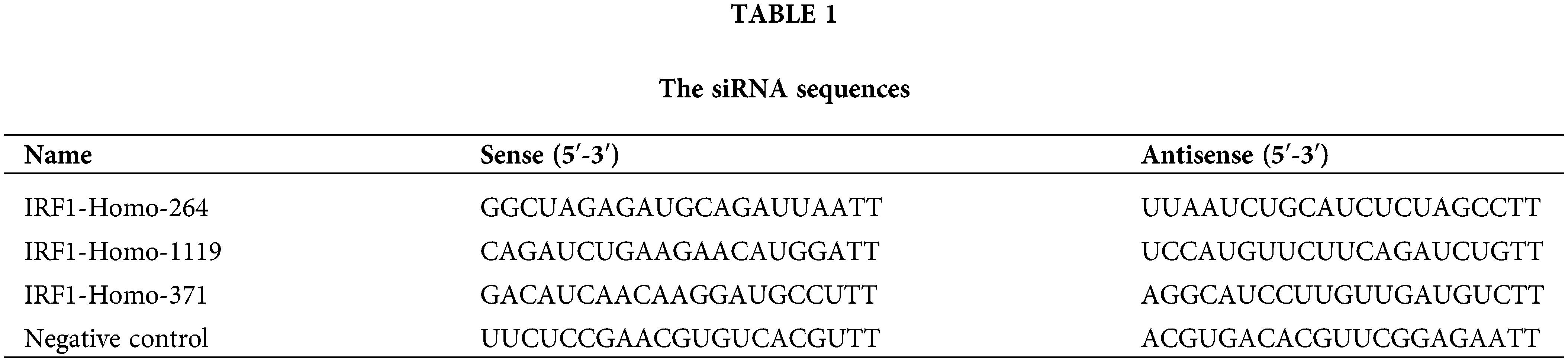
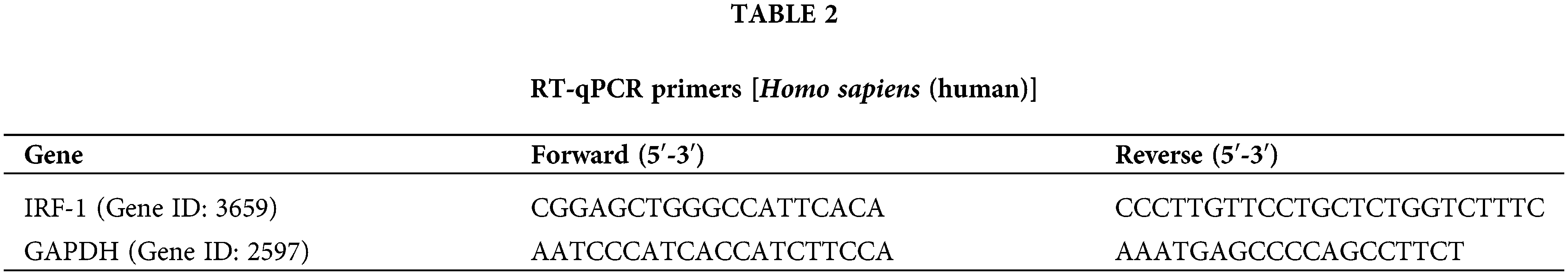
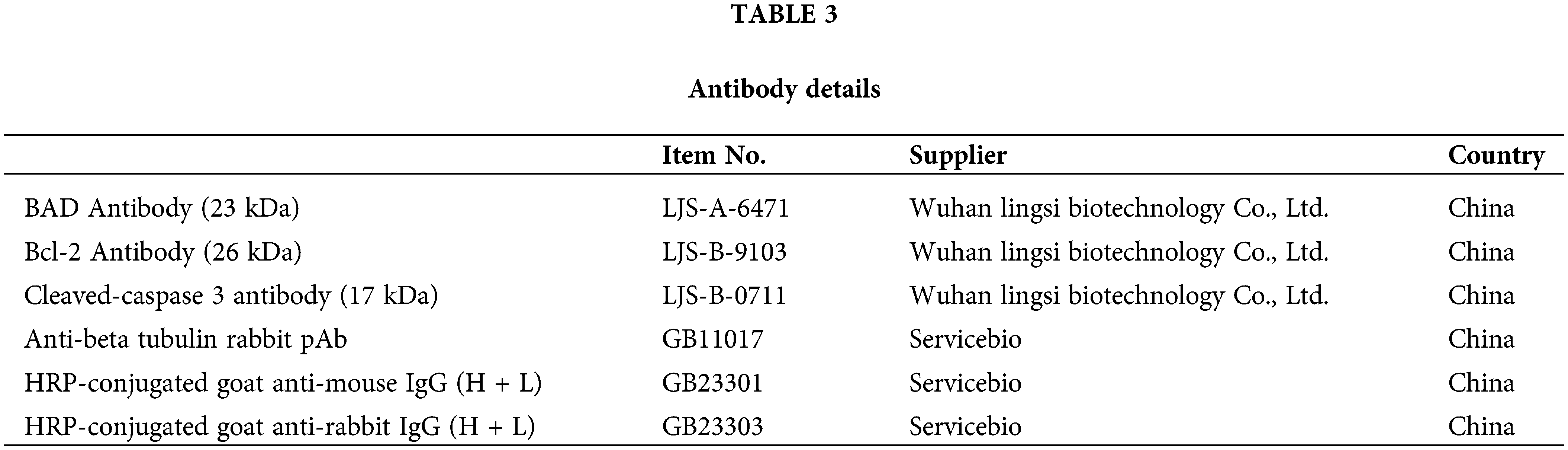
Initially, we treated N-Myc-amplified SK-N-BE (2) and non-N-Myc-amplified SH-SY5Y cells with various doses of IFN-γ (0, 500, 750, 1000, or 15000 ng/mL). The results revealed that IFN-γ administration dramatically decreased the viability of SK-N-BE (2) cells while increasing that of SH-SY5Y cells (Fig. 1A). An intermediate concentration of IFN-γ at 750 ng/mL was chosen for the continuation of the experimental series. The Transwell assay results indicated that IFN-γ effectively curtailed the migratory and invasive activities of SK-N-BE (2) cells, conversely, it stimulated these parameters in SH-SY5Y cells (Fig. 1B,C). Flow cytometry indicated that IFN-γ treatment induced apoptosis in SK-N-BE (2) cells and decreased apoptosis in SH-SY5Y cells (Fig. 1D). These findings demonstrated that IFN-γ exerted a distinct effect on both N-Myc-and non-N-Myc-amplified cells.
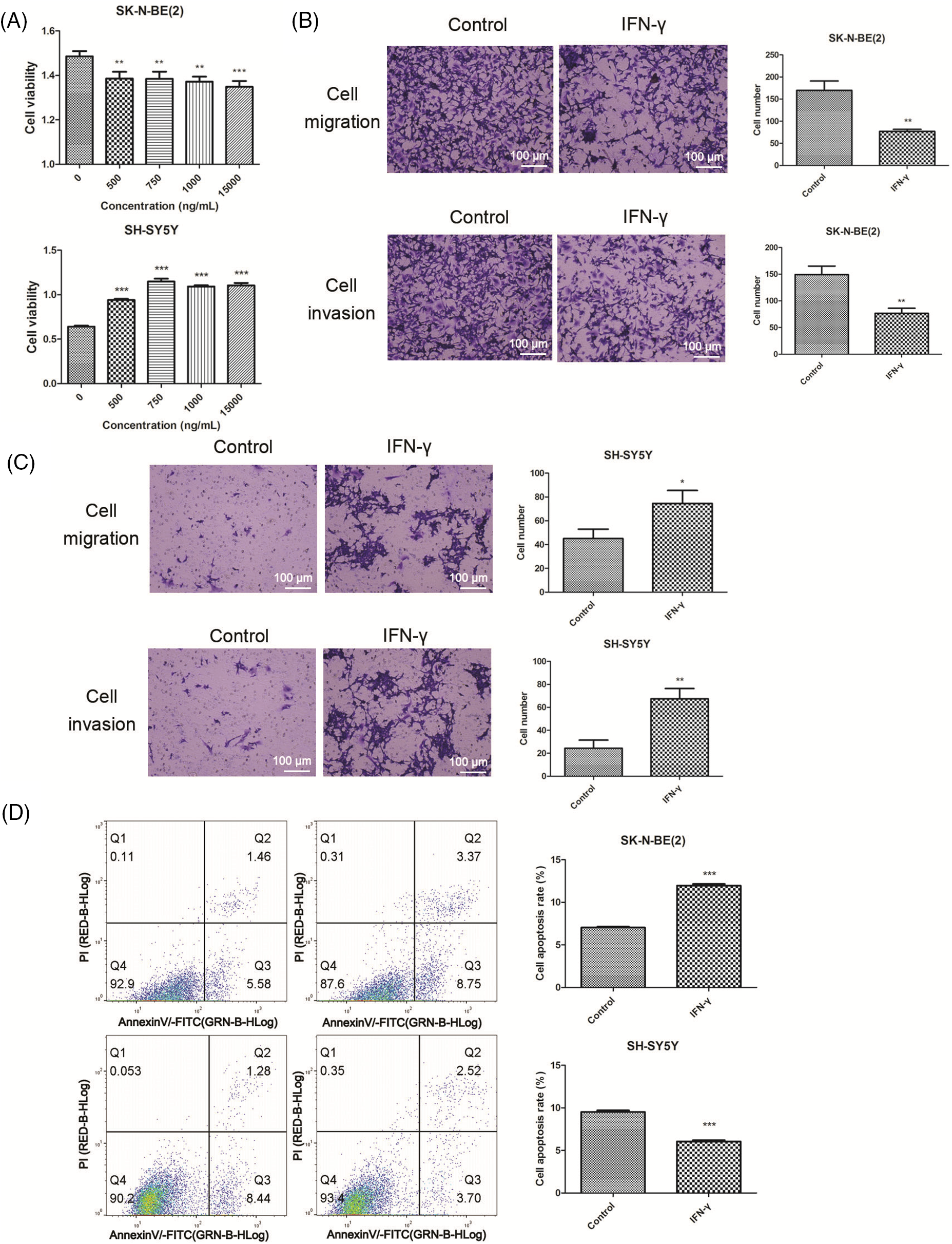
Figure 1: IFN-gamma (IFN-γ) regulates neuroblastoma (NB) cell progression. (A) The viability of SK-N-BE (2) and SH-SY5Y cells. (B and C) Migration and invasion of SK-N-BE (2) (B) and SH-SY5Y cells (C). (D) Apoptosis in SK-N-BE (2) and SH-SY5Y cells. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. 0 or control group.
To investigate the effect of IFN-γ on gene expression in N-Myc-amplified and non-N-Myc-amplified NB cells, we compared mRNA levels in SK-N-BE (2) and SH-SY5Y cells after IFN-γ treatment with those in the respective untreated cells, using R software. As shown in Fig. 2A, 423 upregulated and 11 downregulated genes were identified in SK-N-BE (2) cells (Fig. 2A). In addition, 724 upregulated and 213 downregulated genes were identified in the SH-SY5Y cells (Fig. 2B). Following this, GO and KEGG pathway enrichment analysis of the DEGs was performed. In both SK-N-BE (2) and SH-SY5Y cells, the enriched GO pathways included positive regulation of I-kappaB kinase/NF-kappaB, inflammatory response, and endoplasmic reticulum (ER) to Golgi transport vesicle membrane signaling (Fig. 2C,D). KEGG analysis revealed that Epstein-Barr virus infection, viral myocarditis, and cell adhesion molecule pathways were enriched in both SK-N-BE (2) and SH-SY5Y cells (Fig. 2E,F).
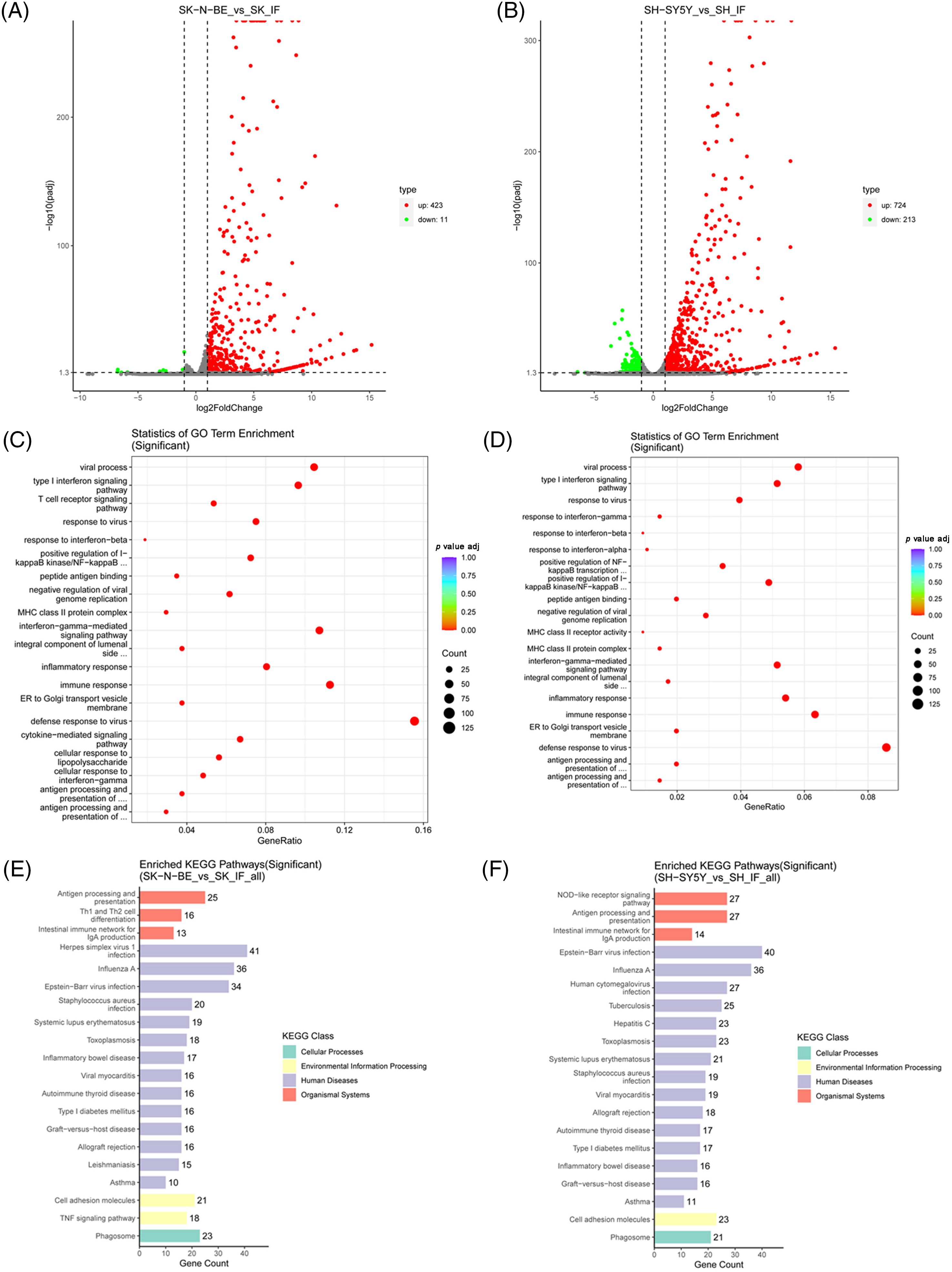
Figure 2: Screening of differentially expressed genes (DEGs). (A and B) Volcano plot of the transcriptome. The up-regulated and down-regulated DEGs were represented using red and green dots, respectively. (C and D) Gene ontology (GO) enrichment analysis for DEGs. (E and F) Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis for DEGs.
IRF-1 is the target of IFN-γ in NB cells
Venn diagram software showed that 311 genes were differentially expressed in SK-N-BE (2) and SH-SY5Y cells after IFN-γ treatment, of which IRF-1 attracted our attention (Fig. 3A). In addition, we performed IHC assays on a tissue microarray (N264001) and verified that IRF-1 expression in normal adrenal tissues (n = 4) was notably higher than that in NB tissues (n = 22) (Fig. 3B), indicating that IRF-1 is likely implicated in the progression of NB. Thus, we silenced IRF-1 in SK-N-BE (2) and SH-SY5Y cells using siRNA targeting IRF-1 and selected siRNA-3 as the most efficient for subsequent experiments (Fig. 3C).
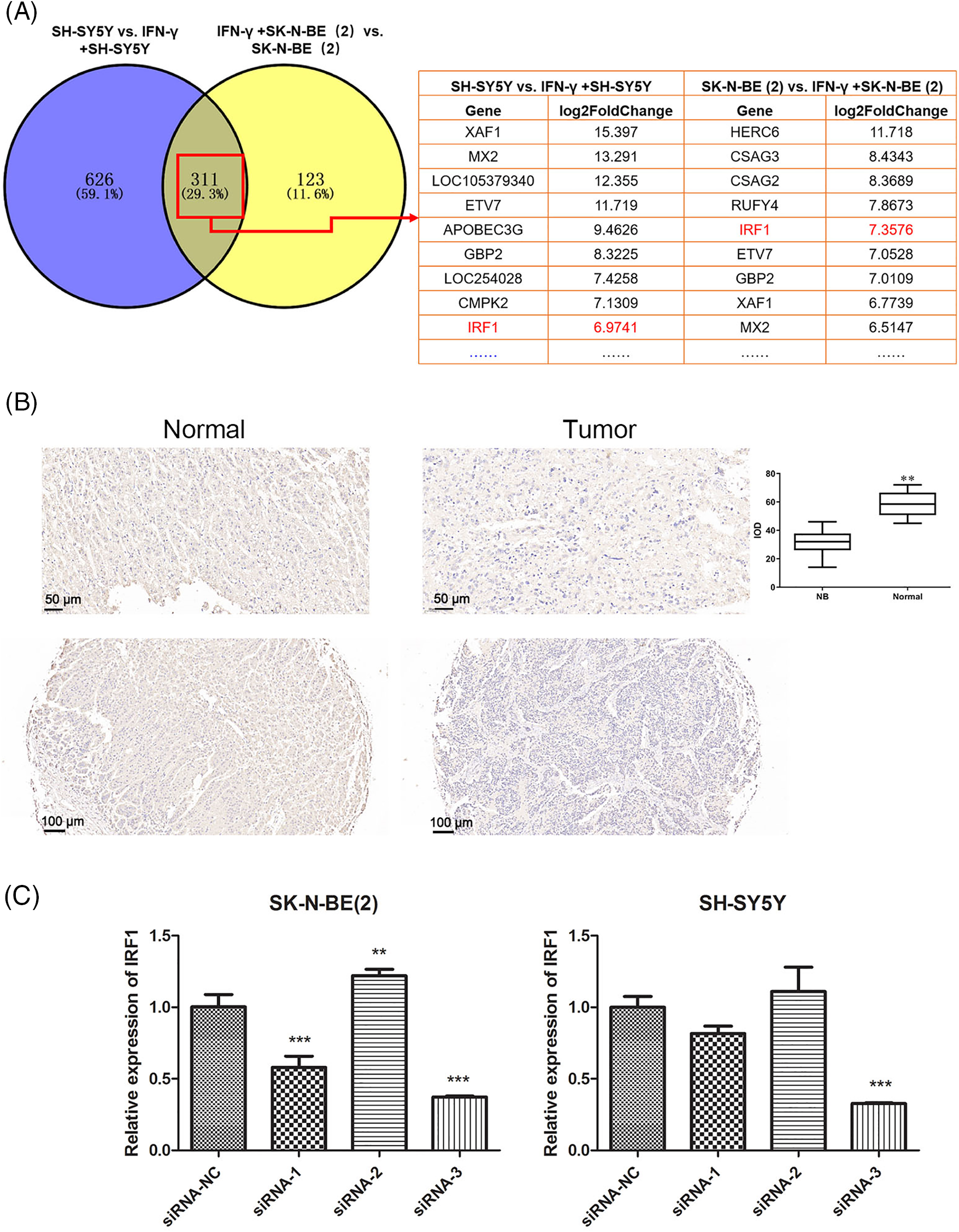
Figure 3: Interferon regulatory factor 1 (IRF-1) is the target of IFN-γ in NB cells. (A) Venn diagram illustrating the overlapping genes in SK-N-BE (2) and SH-SY5Y cells. (B) IRF-1 expression in tissues was measured using an immunohistochemistry (IHC) assay. (C) The IRF-1 expression level was evaluated using a real-time polymerase chain reaction (RT-qPCR) assay. **p < 0.01, ***p < 0.001 vs. the siRNA-NC group.
IRF-1 plays a role in the effects of IFN-γ in NB cells
Further analysis demonstrated that IRF-1 knockdown facilitated the proliferation, migration, and invasion of SK-N-BE (2) and SH-SY5Y cells. Moreover, decreased IRF-1 levels effectively reversed the effects of IFN-γ in SK-N-BE (2) cells, while they enhanced the effects of IFN-γ in SH-SY5Y cells (Fig. 4A–C). Flow cytometry revealed that IFN-γ induced apoptosis in SK-N-BE (2) cells, while conversely inhibiting apoptosis in SH-SY5Y cells. IRF-1 knockdown alone exhibited contrasting effects to IFN-γ in SK-N-BE (2) cells and the same effects in SH-SY5Y cells. Similarly, IRF-1 depletion counteracted the effects of IFN-γ in SK-N-BE (2) cells and further decreased apoptosis in SH-SY5Y cells (Fig. 4D).
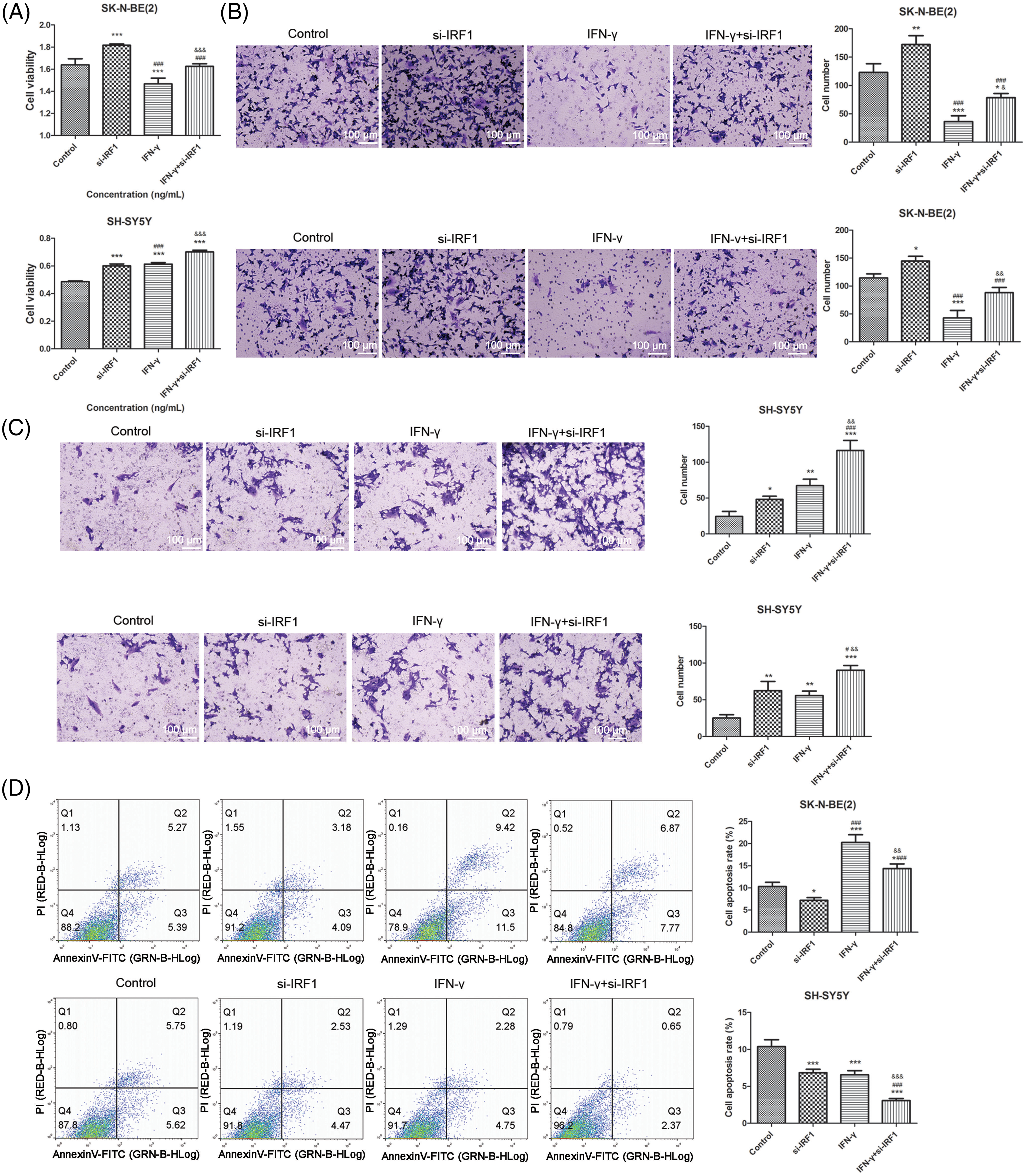
Figure 4: IRF-1 is plays a role in the effects of IFN-γ in NB cells. (A) The viability of SK-N-BE (2) and SH-SY5Y cells. (B) The migration (top) and invasion (bottom) of SK-N-BE (2) cells. (C) The migration (top) and invasion (bottom) of SH-SY5Y cells. (D) The apoptosis of SK-N-BE (2) and SH-SY5Y cells. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. the control group. #p < 0.05 and ###p < 0.001 vs. the si-IRF1 group. &&p < 0.01, &&&p < 0.001 vs. the IFN-γ group.
The caspase and B-cell lymphoma-2 (Bcl-2) family participate in cell apoptosis [27]. As presented in Fig. 5, IFN-γ notably increased the content of Bcl-xL/Bcl-2-associated death promoter (Bad) and caspase-3 while decreasing that of Bcl-2 in SK-N-BE (2) cells. However, IRF-1 knockdown exerted contrasting effects to IFN-γ and partially reversed the effects of IFN-γ in SK-N-BE (2) cells. Furthermore, in SH-SY5Y cells, IFN-γ administration decreased the protein levels of Bad and Caspase-3 and up-regulated Bcl-2 expression. Additionally, IRF-1 depletion further enhanced the effects of IFN-γ on apoptosis-related proteins (Fig. 5). These data highlighted that IRF-1 reversed the effects of IFN-γ in N-Myc-amplified NB cells while enhancing them in non-N-Myc-amplified NB cells.
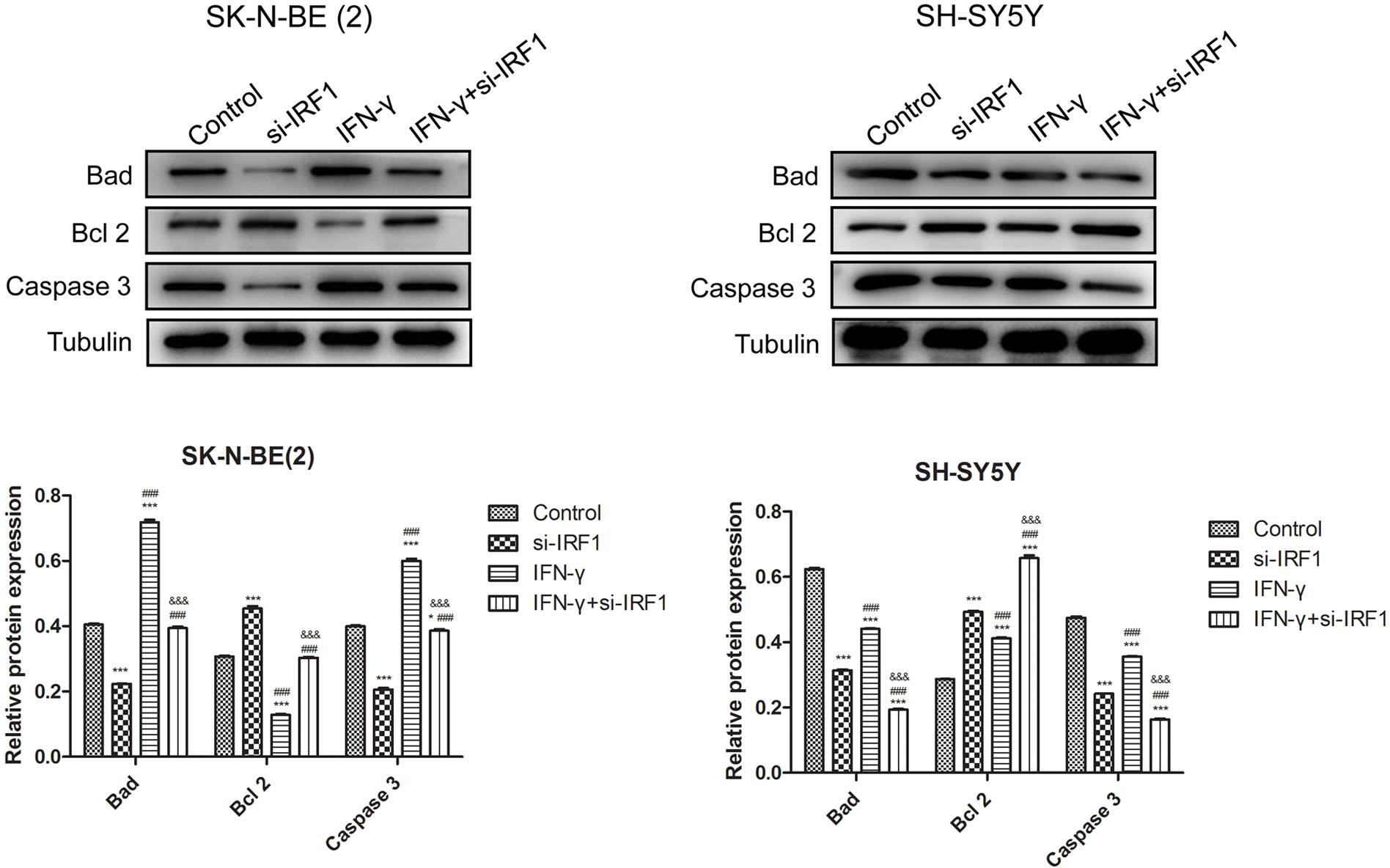
Figure 5: IRF-1 affects apoptosis-related proteins in the presence of IFN-γ. Bad, Bcl-2, caspase-3 content in SK-N-BE (2) SH-SY5Y cells. *p < 0.05 and ***p < 0.001 vs. the control group. ###p < 0.001 vs. the si-IRF1 group. &&&p < 0.001 vs. the IFN-γ group.
This study revealed that IFN-γ treatment reduced cell viability, suppressed migration and invasion, and induced apoptosis in N-Myc-amplified NB cells. Additionally, IFN-γ exhibited a positive role in non-N-Myc-amplified NB cells. Bioinformatics analysis revealed that IRF-1 expression was dramatically down-regulated in both IFN-γ-treated SK-N-BE (2) and SH-SY5Y cells. Furthermore, IRF-1 was insufficiently expressed in NB tissues. Moreover, IRF-1 knockdown promoted cell progression and reduced apoptosis in SK-N-BE (2) and SH-SY5Y cells. Interestingly, in SK-N-BE (2) cells, IRF-1 effectively reversed the effects of IFN-γ, whereas in SH-SY5Y cells, IRF-1 enhanced these effects.
NB is recognized as a serious childhood cancer that poses a threat to the survival of many children [28,29]. Patients with NB are generally divided into four categories: very low-, low-, intermediate-, and high-risk [1,30]. The outcomes for high-risk patients are extremely poor [31,32]. N-Myc amplification is an essential marker of high-risk patients [31,32]. Furthermore, N-Myc expression is strongly associated with poor outcomes in patients with NB [33]. Increasing evidence has shown that N-Myc amplification accelerates the progression of NB by promoting the cell cycle and epithelial-mesenchymal transformation [34]. Consequently, the reduction of N-Myc-amplified cells could constitute a viable therapeutic strategy for neuroblastoma. Nonetheless, this approach is hindered by the absence of a well-defined binding site, no agent has been developed to efficiently target N-Myc-amplified NB cells.
IFN-γ can induce cytotoxic effects and modulate intracellular immune responses [19]. Recent studies have indicated that IFN-γ plays a crucial antitumor role in a variety of cancers [35]. For example, Zhang et al. discovered that IFN-γ suppresses proliferation and migration in pancreatic cancer cells through the Rho GDP dissociation inhibitor 2 (RhoGDI2)/Ras-related C3 botulinum toxin substrate 1 (Rac1)/NF-κB cascade signaling pathway [19]. Cui et al. demonstrated that IFN-γ secreted by NK cells enhances the cytotoxicity of NK-92 cells against colorectal cancer cells [36]. Notably, the latest research demonstrates that IFN-γ exhibits great potential for the treatment of NB [37]. Furthermore, IFN-γ was verified to decrease N-myc levels in NB cells [38]. However, whether IFN-γ affects the ratio of N-Myc-amplified NB cells to non-N-Myc-amplified NB cells remains unclear. In the study, we employed SK-N-BE (2) and SH-SY5Y cells to elucidate the roles of IFN-γ in NB cells. The results demonstrated that IFN-γ inhibited proliferation, suppressed migration and invasion, and induced apoptosis in SK-N-BE (2) cells, while IFN-γ exerted a opposite effect in SH-SY5Y cells.
Additionally, our present study analyzed the transcriptome of SK-N-BE (2) and SH-SY5Y cells after IFN-γ administration, and discovered 434 and 937 DEGs in SK-N-BE (2) and SH-SY5Y cells, respectively. GO and KEGG analyses demonstrated that the DEGs in SK-N-BE (2) and SH-SY5Y cells were enriched in signaling pathways related to viral regulation and immune responses, including the positive regulation of I-kappaB kinase/NF-kappaB, inflammatory response, Epstein-Barr virus infection, and viral myocarditis. These data suggested that IFN-γ was associated with tumor immunity, consistent with previous findings [39,40]. Nevertheless, the molecular mechanisms underlying the regulation of the immune response by IFN-γ in NB cells require further exploration in future studies.
Studies have suggested that the tumor microenvironment plays a significant role in the progression of NB, and IFN-γ, being a potent immunomodulatory cytokine, could potentially influence this environment and the behavior of the tumor. Furthermore, IFN-γ may contribute to the remodeling of the tumor microenvironment. It affects the expression of various molecules involved in cell adhesion, migration, and angiogenesis, all of which are critical processes for tumor growth and metastasis. Theoretically, by altering these processes, IFN-γ could impede the invasive and metastatic potential of NB cells [41].
IRF-1 is a downstream gene of the IFN-γ cascade signal and a classical tumor suppressor gene [9,42]. IRF-1 suppresses this process and induces apoptosis in multiple tumors, including cervical cancer [43] and hepatocellular carcinoma [44]. Recent studies have indicated that IRF-1 plays a positive role in NB inhibition [41]. In this study, we revealed that IRF-1 expression was down-regulated in both IFN-γ-treated SK-N-BE (2) and SH-SY5Y cells. Knockdown of IRF-1 remarkably enhanced proliferation, migration, and invasion, while inhibiting apoptosis in both SK-N-BE (2) and SH-SY5Y cells. In addition, IRF-1 depletion effectively reversed the effects of IFN-γ in SK-N-BE (2) cells; however, it further intensified these effects in SH-SY5Y cells, which might be attributed to the powerful anti-cancer effect of IRF-1 itself.
Given that we only explored the effect of IFN-γ on the development of both N-Myc-amplified and non-N-Myc-amplified NB cells in vitro, the effect of IFN-γ on the progression of these cells in vivo needs further investigation. Additional experiments will be performed to verify the other downstream genes associated with the roles of IFN-γ in NB cells.
This study proved that IFN-γ treatment suppressed proliferation, migration, and invasion and induced apoptosis in N-Myc-amplified NB cells. In contrast, IFN-γ accelerated cell progression and inhibited apoptosis in non-N-Myc-amplified NB cells. Additionally, we demonstrated that IRF-1 exerted a negative effect on the progression of NB cells and played a role in the effect of IFN-γ on NB cells. Consequently, these findings support the application of IFN-γ as a potential therapeutic agent for NB.
Acknowledgement: None.
Funding Statement: This research was funded by the Special Basic Cooperative Research Programs of Yunnan Provincial Undergraduate Universities’ Association (No. 202101BA070001-126).
Author Contributions: Conceptualization and methodology: Jing Wang and Ning Wang; Data curation: Junfeng Xu and Youzheng Qiu; Investigation: Yinran Yang; Formal analysis: Youzheng Qiu and Shanshan Zhang; Writing—original draft preparation: Jing Wang, Junfeng Xu and Yinran Yang; Writing—review and editing: Shanshan Zhang and Ning Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data underlying this article will be shared upon reasonable request from the corresponding authors.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Matthay KK, Maris JM, Schleiermacher G, Nakagawara A, Mackall CL, Diller L, et al. Neuroblastoma. Nat Rev Dis Primers. 2016;2(1):16078. doi:10.1038/nrdp.2016.78. [Google Scholar] [PubMed] [CrossRef]
2. Smith MA, Seibel NL, Altekruse SF, Ries LA, Melbert DL, O’Leary M, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;28(15):2625–34. doi:10.1200/JCO.2009.27.0421. [Google Scholar] [PubMed] [CrossRef]
3. Huang Y, Ma J, Yang C, Wei P, Yang M, Han H, et al. METTL1 promotes neuroblastoma development through m7G tRNA modification and selective oncogenic gene translation. Biomark Res. 2022;10(1):68. doi:10.1186/s40364-022-00414-z. [Google Scholar] [PubMed] [CrossRef]
4. Hsu CL, Yin CF, Chang YW, Fan YC, Lin SH, Wu YC, et al. LncRNA SNHG1 regulates neuroblastoma cell fate via interactions with HDAC1/2. Cell Death Dis. 2022;13(9):809. doi:10.1038/s41419-022-05256-z. [Google Scholar] [PubMed] [CrossRef]
5. He Y, Luo M, Lei S, Zeng Z, Chen T, Wu Y, et al. Luteoloside induces G0/G1 phase arrest of neuroblastoma cells by targeting p38 MAPK. Molecules. 2023;28(4). doi:10.3390/molecules28041748. [Google Scholar] [PubMed] [CrossRef]
6. Zaatiti H, Abdallah J, Nasr Z, Khazen G, Sandler A, Abou-Antoun TJ. Tumorigenic proteins upregulated in the MYCN-amplified IMR-32 human neuroblastoma cells promote proliferation and migration. Int J Oncol. 2018;52(3):787–803. doi:10.3892/ijo.2018.4236. [Google Scholar] [PubMed] [CrossRef]
7. Schmalzl A, Leupold T, Kreiss L, Waldner M, Schürmann S, Neurath MF, et al. Interferon regulatory factor 1 (IRF-1) promotes intestinal group 3 innate lymphoid responses during Citrobacter rodentium infection. Nat Commun. 2022;13(1):5730. doi:10.1038/s41467-022-33326-5. [Google Scholar] [PubMed] [CrossRef]
8. Feng H, Zhang YB, Gui JF, Lemon SM, Yamane D. Interferon regulatory factor 1 (IRF1) and anti-pathogen innate immune responses. PLoS Pathog. 2021;17(1):e1009220. doi:10.1371/journal.ppat.1009220. [Google Scholar] [PubMed] [CrossRef]
9. Yan Y, Zheng L, Du Q, Yazdani H, Dong K, Guo Y, et al. Interferon regulatory factor 1 (IRF-1) activates anti-tumor immunity via CXCL10/CXCR3 axis in hepatocellular carcinoma (HCC). Cancer Lett. 2021;506:95–106. doi:10.1016/j.canlet.2021.03.002. [Google Scholar] [PubMed] [CrossRef]
10. Stafman LL, Beierle EA. Cell proliferation in neuroblastoma. Cancers. 2016;8(1):13. doi:10.3390/cancers8010013. [Google Scholar] [PubMed] [CrossRef]
11. Schwab M, Alitalo K, Klempnauer KH, Varmus HE, Bishop JM, Gilbert F, et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature. 1983;305(5931):245–8. doi:10.1038/305245a0. [Google Scholar] [PubMed] [CrossRef]
12. Hossain MM, Banik NL, Ray SK. N-Myc knockdown and apigenin treatment controlled growth of malignant neuroblastoma cells having N-Myc amplification. Gene. 2013;529(1):27–36. doi:10.1016/j.gene.2013.07.094. [Google Scholar] [PubMed] [CrossRef]
13. Rishfi M, Krols S, Martens F, Bekaert SL, Sanders E, Eggermont A, et al. Targeted AURKA degradation: towards new therapeutic agents for neuroblastoma. Eur J Med Chem. 2023;247:115033. doi:10.1016/j.ejmech.2022.115033. [Google Scholar] [PubMed] [CrossRef]
14. Campbell K, Gastier-Foster JM, Mann M, Naranjo AH, Van Ryn C, Bagatell R. Association of MYCN copy number with clinical features, tumor biology, and outcomes in neuroblastoma: a report from the children’s oncology group. Cancer. 2017;123(21):4224–35. doi:10.1002/cncr.30873. [Google Scholar] [PubMed] [CrossRef]
15. Whittle SB, Smith V, Doherty E, Zhao S, McCarty S, Zage PE. Overview and recent advances in the treatment of neuroblastoma. Expert Rev Anticancer Ther. 2017;17(4):369–86. doi:10.1080/14737140.2017.1285230. [Google Scholar] [PubMed] [CrossRef]
16. Qiu B, Matthay KK. Advancing therapy for neuroblastoma. Nat Rev Clin Oncol. 2022;19(8):515–33. doi:10.1038/s41571-022-00643-z. [Google Scholar] [PubMed] [CrossRef]
17. van Noesel MM, Pieters R, Voûte PA, Versteeg R. The N-myc paradox: n-myc overexpression in neuroblastomas is associated with sensitivity as well as resistance to apoptosis. Cancer Lett. 2003;197(1–2):165–72. doi:10.1016/s0304-3835(03)00101-0. [Google Scholar] [PubMed] [CrossRef]
18. Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351(1):192–7. doi:10.1016/j.bbrc.2006.10.020. [Google Scholar] [PubMed] [CrossRef]
19. Zhang M, Ding G, Zhou L, Shen T, Xu X, Zhao T, et al. Interferon gamma inhibits CXCL8-induced proliferation and migration of pancreatic cancer BxPC-3 cell line via a RhoGDI2/Rac1/NF-κB signaling pathway. J Interferon Cytokine Res. 2018;38(9):413–22. doi:10.1089/jir.2018.0070. [Google Scholar] [PubMed] [CrossRef]
20. Alspach E, Lussier DM, Schreiber RD. Interferon γ and its important roles in promoting and inhibiting spontaneous and therapeutic cancer immunity. Cold Spring Harb Perspect Biol. 2019;11(3). doi:10.1101/cshperspect.a028480. [Google Scholar] [PubMed] [CrossRef]
21. Kammertoens T, Friese C, Arina A, Idel C, Briesemeister D, Rothe M, et al. Tumour ischaemia by interferon-γ resembles physiological blood vessel regression. Nature. 2017;545(7652):98–102. doi:10.1038/nature22311. [Google Scholar] [PubMed] [CrossRef]
22. Cetinkaya C, Hultquist A, Su Y, Wu S, Bahram F, Påhlman S, et al. Combined IFN-γ and retinoic acid treatment targets the N-Myc/Max/Mad1 network resulting in repression of N-Myc target genes in MYCN-amplified neuroblastoma cells. Mol Cancer Ther. 2007;6(10):2634–41. doi:10.1158/1535-7163.MCT-06-0492. [Google Scholar] [PubMed] [CrossRef]
23. Tsapralis D, Panayiotides I, Peros G, Liakakos T, Karamitopoulou E. Human epidermal growth factor receptor-2 gene amplification in gastric cancer using tissue microarray technology. World J Gastroenterol. 2012;18(2):150–5. doi:10.3748/wjg.v18.i2.150. [Google Scholar] [PubMed] [CrossRef]
24. Chen L, Zhang YH, Wang S, Zhang Y, Huang T, Cai YD. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12(9):e0184129. doi:10.1371/journal.pone.0184129. [Google Scholar] [PubMed] [CrossRef]
25. Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi:10.1093/nar/28.1.27. [Google Scholar] [PubMed] [CrossRef]
26. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–7. doi:10.1089/omi.2011.0118. [Google Scholar] [PubMed] [CrossRef]
27. Almasieh M, Wilson AM, Morquette B, Cueva Vargas JL, Di Polo A. The molecular basis of retinal ganglion cell death in glaucoma. Prog Retin Eye Res. 2012;31(2):152–81. doi:10.1016/j.preteyeres.2011.11.002. [Google Scholar] [PubMed] [CrossRef]
28. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654. [Google Scholar] [PubMed] [CrossRef]
29. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7–33. doi:10.3322/caac.21708. [Google Scholar] [PubMed] [CrossRef]
30. Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33(27):3008–17. doi:10.1200/JCO.2014.59.4648. [Google Scholar] [PubMed] [CrossRef]
31. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590. [Google Scholar] [PubMed] [CrossRef]
32. Irwin MS, Naranjo A, Zhang FF, Cohn SL, London WB, Gastier-Foster JM, et al. Revised neuroblastoma risk classification system: a report from the children’s oncology group. J Clin Oncol. 2021;39(29):3229–41. doi:10.1200/jco.21.00278. [Google Scholar] [PubMed] [CrossRef]
33. Otte J, Dyberg C, Pepich A, Johnsen JI. MYCN function in neuroblastoma development. Front Oncol. 2020;10:624079. doi:10.3389/fonc.2020.624079. [Google Scholar] [PubMed] [CrossRef]
34. Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3(10):a014415. doi:10.1101/cshperspect.a014415. [Google Scholar] [PubMed] [CrossRef]
35. Mödl B, Moritsch S, Zwolanek D, Eferl R. Type I and II interferon signaling in colorectal cancer liver metastasis. Cytokine. 2023;161(4):156075. doi:10.1016/j.cyto.2022.156075. [Google Scholar] [PubMed] [CrossRef]
36. Cui F, Qu D, Sun R, Zhang M, Nan K. NK cell-produced IFN-γ regulates cell growth and apoptosis of colorectal cancer by regulating IL-15. Exp Ther Med. 2020;19(2):1400–6. doi:10.3892/etm.2019.8343. [Google Scholar] [PubMed] [CrossRef]
37. Zeki J, Yavuz B, Wood L, Shimada H, Kaplan DL, Chiu B. Concurrent application of interferon-gamma and vincristine inhibits tumor growth in an orthotopic neuroblastoma mouse model. Pediatr Surg Int. 2023;39(1):241. doi:10.1007/s00383-023-05523-w. [Google Scholar] [PubMed] [CrossRef]
38. Wada RK, Pai DS, Huang J, Yamashiro JM, Sidell N. Interferon-γ and retinoic acid down-regulate N-myc in neuroblastoma through complementary mechanisms of action. Cancer Lett. 1997;121(2):181–8. doi:10.1016/S0304-3835(97)00351-0. [Google Scholar] [PubMed] [CrossRef]
39. Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24(8):1143–50. doi:10.1038/s41591-018-0116-5. [Google Scholar] [PubMed] [CrossRef]
40. Ren J, Li N, Pei S, Lian Y, Li L, Peng Y, et al. Histone methyltransferase WHSC1 loss dampens MHC-I antigen presentation pathway to impair IFN-γ-stimulated antitumor immunity. J Clin Invest. 2022;132(8). doi:10.1172/JCI153167. [Google Scholar] [PubMed] [CrossRef]
41. Sha YL, Liu Y, Yang JX, Wang YY, Gong BC, Jin Y, et al. B3GALT4 remodels the tumor microenvironment through GD2-mediated lipid raft formation and the c-met/AKT/mTOR/IRF-1 axis in neuroblastoma. J Exp Clin Cancer Res. 2022;41(1):314. doi:10.1186/s13046-022-02523-x. [Google Scholar] [PubMed] [CrossRef]
42. Meissl K, Macho-Maschler S, Müller M, Strobl B. The good and the bad faces of STAT1 in solid tumours. Cytokine. 2017;89:12–20. doi:10.1016/j.cyto.2015.11.011. [Google Scholar] [PubMed] [CrossRef]
43. Rho SB, Lee SH, Byun HJ, Kim BR, Lee CH. IRF-1 inhibits angiogenic activity of HPV16 E6 oncoprotein in cervical cancer. Int J Mol Sci. 2020;21(20):7622. doi:10.3390/ijms21207622. [Google Scholar] [PubMed] [CrossRef]
44. Yan Y, Zheng L, Du Q, Cui X, Dong K, Guo Y, et al. Interferon regulatory factor 1 (IRF-1) downregulates checkpoint kinase 1 (CHK1) through miR-195 to upregulate apoptosis and PD-L1 expression in Hepatocellular carcinoma (HCC) cells. Br J Cancer. 2021;125(1):101–11. doi:10.1038/s41416-021-01337-6. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools