 Open Access
Open Access
ARTICLE
DAPK2 promotes autophagy to accelerate the progression of ossification of the posterior longitudinal ligament through the mTORC1 complex
1 Department of Spine Surgery, Shanghai Changzheng Hospital, Shanghai, 200003, China
2 Department of Orthopedics, The 967th Hospital of Chinese People’s Liberation Army, Dalian, 116000, China
* Corresponding Author: JINHAO MIAO. Email:
# These authors contributed equally to this work
BIOCELL 2024, 48(9), 1389-1400. https://doi.org/10.32604/biocell.2024.049562
Received 10 January 2024; Accepted 19 July 2024; Issue published 04 September 2024
Abstract
Background: Ossification of the posterior longitudinal ligament (OPLL) is a prevalent condition in orthopedics. While death-associated protein kinase 2 (DAPK2) is known to play roles in cellular apoptosis and autophagy, its specific contributions to the advancement of OPLL are not well understood. Methods: Ligament fibroblasts were harvested from patients diagnosed with OPLL. Techniques such as real-time reverse transcriptase-polymerase chain reaction (RT-qPCR) and Western blot analysis were employed to assess DAPK2 levels in both ligament tissues and cultured fibroblasts. The extent of osteogenic differentiation in these cells was evaluated using an alizarin red S (ARS) staining. Additionally, the expression of ossification markers and autophagy markers was quantified. The autophagic activity was further analyzed through LC3 immunofluorescence and transmission electron microscopy (TEM). An in vivo heterotopic bone formation assay was conducted in mice to assess the role of DAPK2 in ossification. Results: Elevated DAPK2 expression was confirmed in both OPLL patient tissues and derived fibroblasts, in contrast to non-OPLL controls. Silencing of DAPK2 significantly curtailed osteogenic differentiation and autophagy in these fibroblasts, evidenced by decreased levels of LC3, and Beclin1, and reduced autophagosome formation. Additionally, DAPK2 was found to inhibit the mechanistic target of the rapamycin complex 1 (mTORC1) complex’s activity. In vivo studies demonstrated that DAPK2 facilitates ossification, and this effect could be counteracted by the mTORC1 inhibitor rapamycin. Conclusion: DAPK2 enhances autophagy and osteogenic processes in OPLL through modulation of the mTORC1 pathway.Keywords
Ossification of the posterior longitudinal ligament (OPLL) is a common orthopedic condition characterized by the gradual abnormal calcification of the ligament in the cervical spine [1]. This condition leads to spinal canal narrowing and nerve root compression, causing motor and sensory impairments [2]. OPLL is most commonly observed in the cervical region, followed by the thoracic and lumbar areas, with a male-to-female prevalence ratio of 2:1 [3]. Typically emerging in individuals over 40, OPLL is frequently associated with several coexisting conditions, including diabetes, altered bone mineral density (BMD), and diffuse idiopathic skeletal hyperostosis [4]. Although surgical decompression is routinely employed to relieve symptoms, it carries significant risks and potential for complications [5]. Furthermore, post-surgical recurrence of ossification is a notable challenge [6], underscoring the need for a deeper understanding of OPLL’s pathogenesis and the development of more effective treatments.
Death-associated protein kinase 2 (DAPK2), a member of the death-associated protein kinase (DAPK) family of serine/threonine kinases regulated by Ca2+/calmodulin, influences various cellular functions such as apoptosis, autophagy, and immune response [7]. DAPK2 shares considerable structural similarity in the kinase domain with other DAPK family members like death-associated protein kinase 1 (DAPK1) and death-associated protein kinase 3 (DAPK3) [8]. It is a cytoplasmic protein that promotes the formation of autophagic vesicles when overexpressed [9]. Increasing evidence supports DAPK2’s contribution to the development and progression of many diseases and cancers. For instance, it has been shown to inhibit the progression of epithelial ovarian cancer [10] and regulate lung cancer progression via the nuclear factor-κB (NF-κB) pathway [11]. Furthermore, recent studies [12,13], suggest that DAPK2 may influence the function of the mechanistic target of rapamycin complex 1 (mTORC1) and autophagy, both of which are implicated in OPLL progression.
This study focuses on the role of DAPK2 in OPLL and aims to uncover novel molecular mechanisms and identify possible targets for therapeutic intervention.
Thirty patients who underwent anterior cervical decompression surgery participated in this study. Fifteen patients were identified in this cohort as having OPLL, while the remaining fifteen were diagnosed with cervical disk herniation without presenting with OPLL. Seven patients exhibited the segmental type, six exhibited the local type, and two presented with the mixed type of OPLL. Proximal longitudinal ligament tissue samples were procured and preserved promptly in liquid nitrogen. For each participant, written informed consent was obtained, and the research was approved by the Ethics Committee of Changzheng Hospital in Shanghai, with an ethical approval number of 2021MS13.
Extraction and culture of primary ligament fibroblasts
We isolated primary ligament fibroblasts from collected posterior longitudinal ligament tissues using a protocol adapted from prior research [14]. After removing tissues from non-calcified areas, they were minced, cleansed, and incubated in Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, SH30022.LS, South Logan, UT, USA) enriched with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, A5669801, Waltham, MA, USA) at 37°C. To induce osteogenesis, 10 nM dexamethasone (Sigma-Aldrich, D4902-25MG, St. Louis, MO, USA), 25 μg/mL ascorbic acid (Sigma-Aldrich, A8960-5G, St. Louis, MO, USA), and 10 mM β-glycerophosphate (Sigma-Aldrich, G9422, St. Louis, MO, USA) were added to the culture medium. The cultures were then incubated for 14 days. All utilized cells in this study were free from mycoplasma contamination.
Real-time reverse transcriptase-polymerase chain reaction (RT-qPCR)
RNA was isolated from cells or tissues using TRIzol reagent (Invitrogen, 15596026CN, Carlsbad, CA, USA) 48 h after transfection. The RNA was transcribed into complementary DNA (cDNA) using a Reverse Transcription Kit (Qiagen, RT31-020, Hilden, North Rhine-Westphalia, Germany). The Green Premix Ex Taq II (TaKaRa, RR820Q, Osaka, Japan) was used for quantitative PCR on Step One Plus Real-Time PCR System (Applied Biosystems, 4376600, Foster City, CA, USA). The 2−ΔΔCt method was used to quantify gene expression levels, with normalization to GAPDH. The primer sequences of RNAs were shown as follows: human DAPK2: Forward: 5′-TGCAGCCAAGTTCATCAAGAAGCG-3′, Reverse: 5′-ACACTAGCTCAAGGATGAGCACCA-3′; mice DAPK2: Forward: 5′-TCCTGGATGGGGTGAACTAC-3′, Reverse: 5′-CAGCTTGATGTGTGGAA-3′; human ALP: Forward: 5′-GCCTGGATCTCATCAGTATTTGG-3′, Reverse: 5′-GTTCAGTGCGGTTCCAGACAT-3′; human COL1A1: Forward: 5′-GGGTCTAGACATGTTCAGCTTTGTG-3′, Reverse: 5′-ACCCTTAGGCCATTGTGTATGC-3′; human OSX: Forward: 5′-CTCTCTGCTTGAGGAAGAAG-3′, Reverse: 5′- GTCCATTGGTGCTTGAGAAG-3′; human OCN: Forward: 5′-GGCGCTACCTGTATCAATGG-3′, Reverse: 5′-GTGGTCAGCCAACTCGTCA-3′; human RUNX2: Forward: 5′-CCGGGAATGATGAGAACTA-3′, Reverse: 5′-ACCGTCCACTGTCACTTT-3′; human LC3: Forward: 5′-GAAGTTCAGCCACCTGCCAC-3′, Reverse 5′-TCTGAGGTGGAGGGTCAGTC-3′; human p62: Forward: 5′-GTACCAGGACAGCGAGAGGAA-3′, Reverse: 5′-CCCATGTTGCACGCCAAAC-3′; human Beclin-1: Forward 5′-ATACTGTTCTGGGGGTTTGCG-3′, Reverse 5′-GTCTCTCCTTTTTCCACCTCTTC-3′; human GAPDH: Forward: 5′-AGAAGGTGGTGAAGCAGGCATC-3′, Reverse: 5′-CGAAGGTGGAAGAGTGGGAGTTG-3′.
The cellular components were disrupted using radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, P0013B, Shanghai, China) to obtain the entire protein content 48 h after transfection. Equal quantities of protein were subjected to separation using a 10% SDS-PAGE gel. Afterward, the proteins that had been separated were moved onto polyvinylidene fluoride (PVDF) membranes (Beyotime, P0021S-1L, Shanghai, China). Subsequently, the membranes were blocked and subjected to incubation with primary antibodies including ALP (Abcam, 1/1000, ab307726, Cambridge, MA, USA), COL1A1 (Abcam, 1/1000, ab138492, Cambridge, MA, USA), OSX (Abcam, 1/1000, ab209484, Cambridge, MA, USA), OCN (Abcam, 1/1000, ab133612, Cambridge, MA, USA), RUNX2 (Abcam, 1/1000, ab236639, Cambridge, MA, USA), LC3 (Abcam, 1/2000, ab192890, Cambridge, MA, USA), Beclin 1 (Abcam, 1/2000, ab207612, Cambridge, MA, USA), p62 (Abcam, 1/10000, ab109012, Cambridge, MA, USA), p-Raptor (Cell Signaling Technology, 1/2000, #2083, Massachusetts, USA), Raptor (Abcam, 1/1000, ab40768, Cambridge, MA, USA), p-Thr389 (Abcam, 1/500, ab60948, Cambridge, MA, USA), p70S6K (Abcam, 1/5000, ab32529, Cambridge, MA, USA), p-Thr46 (Abcam, 1/1000, ab278686, Cambridge, MA, USA), 4E-BP1 (Abcam, 1/2000, ab32024, Cambridge, MA, USA), p-mTOR (Abcam, 1/1000, ab109268, Cambridge, USA), mTOR (Abcam, 1/10000, ab134903, Cambridge, MA, USA), ULK1 (Abcam, 1/10000, ab177472, Cambridge, MA, USA), DAPK2 (Invitrogen, 1/1000, MA5-25084, Carlsbad, CA, USA), GAPDH (Abcam, 1/10000, ab181602, Cambridge, MA, USA) overnight at a temperature of 4°C. Afterward, the specimens were subjected to incubation with the secondary antibody (Abcam, 1/2000, ab172730, Cambridge, MA, USA) for a duration of one hour, followed by detection using ECL reagents (Sigma-Aldrich, B8522-1EA, St. Louis, MO, USA). The quantification of band density was performed using ImageJ software (National Institutes of Health, version 23.0, Bethesda, MD, USA).
Short hairpin RNA targeting DAPK2 (sh-DAPK2) and sh-negative control (NC), as well as pcDNA3.1 and pcDNA3.1-DAPK2, were obtained from Gene-Pharma (Shanghai, China). The sequence of sh-DAPK2#1 and sh-DAPK2#2 used in the study were GGAAACGGCUCACAAUCCA and GGAAUUUGUUGCUCCAGAA. Ligament fibroblasts were transfected with sh-DAPK2, sh-NC, pcDNA3.1, or pcDNA3.1-DAPK2 using Lipofectamine 3000 (Invitrogen, L3000150, Carlsbad, CA, USA) as per the guidelines provided by the manufacturer. Following 48 h of transfection, cells were subjected to selection with puromycin to isolate stable transfectants. The selection medium was refreshed every three days, and the selection process was continued for a minimum of three weeks to ensure the elimination of non-transfected cells.
The fibroblasts of the ligament (1 × 105 cells) were cultured on 6-well plates, subsequently treated with 4% paraformaldehyde for fixation, and permeabilized using 0.1% Triton X-100. Then, the cells were blocked in 1% bovine serum albumin (BSA, Sigma-Aldrich, B2064, St. Louis, MO, USA) at room temperature for 30 min. Afterward, the cells were subjected to primary antibody incubation, wherein anti-Vimentin (Abcam, 1/1000, ab16700, Cambridge, MA, USA), anti-DAPK2 (Invitrogen, 1/1000, MA5-25084, Carlsbad, CA, USA), and anti-LC3 (Abcam, 1/1000, ab192890, Cambridge, MA, USA) antibodies were employed and kept at a temperature of 4°C overnight. Subsequently, the cells were subjected to culturing with a secondary antibody Abcam, 1/2000, ab172730, Cambridge, MA, USA) for a duration of one hour at room temperature. The nuclei were stained using DAPI (Sigma-Aldrich, D9542, St. Louis, MO, USA). The observation was conducted using a fluorescence microscope manufactured by Olympus IX70 in Japan.
Ligament fibroblasts (5 × 103 cells) were incubated in a 12-well plate with osteogenic induction medium for two weeks. The cells were treated with 95% ethanol for 30 min to fix them, followed by staining with 0.1% ARS (ScienCell, Catalog #0223, San Diego, CA, USA) for an additional 20 min.
Transmission electron microscopy (TEM)
Ligament fibroblasts were gathered and subjected to centrifugation. Next, they were rinsed with PBS, fixed with 2.5% glutaraldehyde, postfixed with 1% osmic acid, and dehydrated with an acetone gradient. A microtome (Leica Biosystems, HI1220, Nussloch, Germany) was utilized to make the sections. Later, they were double-dyed with uranyl acetate and lead citrate, followed by observation by TEM (Hitachi, HT7800 series, Tokyo, Japan).
Heterotopic bone formation experiments
Sixteen 4-week-old BALB/c homozygous nude mice were acquired from Shanghai Slack Laboratory Animal Co., Ltd. (Shanghai, China) and housed in a specific pathogen-free (SPF) facility with a regulated 12-h light/dark cycle at a constant temperature of 24°C. The animal experiments were conducted by Cyagen (Suzhou, China) Biotechnology Co., Ltd., Suzhou, China. The study protocols were sanctioned by the Institutional Animal Care and Use Committee of Cyagen (Suzhou, China) Biotechnology Co., Ltd., with ethical approval number IACUC-2109025. Fibroblasts, which are prevalent in connective tissues throughout the human body, including skin, have shown capabilities for bone generation both naturally and under pathological conditions, as well as in laboratory-induced osteogenic differentiation [15,16]. Human ligament fibroblasts were genetically modified with sh-DAPK2 and selected using puromycin over a three-week period. The mice then received daily intraperitoneal injections of 4 mg/kg rapamycin (LC Laboratories, R-5000, Woburn, MA, USA) for three weeks, following established guidelines [17]. After a two-week osteogenic induction in culture, the cells were combined with Bio-Oss collagen scaffolds measuring 7 mm × 5 mm × 2 mm (Geistlich, GEWO GmbH, Baden-Baden, Germany) and incubated for two days. The cell-scaffold constructs were then implanted subcutaneously on the dorsal side of the mice. Six weeks post-implantation, the implants were harvested, fixed in 4% paraformaldehyde, and examined using a Quantum FX microCT scanner (PerkinElmer, Shelton, CT, USA). Image analysis and three-dimensional reconstructions were performed using Inveon Research Workplace software (Siemens Healthcare GmbH, Erlangen, Germany), focusing on the designated regions within the scaffolds to assess the bone volume/tissue volume (BV/TV) ratio and bone mineral density (BMD) from the micro-CT data.
Hematoxylin and eosin (HE) staining
The scaffold specimens were decalcified by 10% EDTA for 30 days. After that, specimens were dehydrated, embedded in paraffin, and cut into 5 μm sections. Next, they were dyed with hematoxylin and eosin. A microscope (Olympus IX70, Okaya, Japan) was employed for observation.
The sections were subjected to deparaffinization and rehydration with xylene and graded ethanol. Antigen retrieval was conducted with citrate buffer. Endogenous peroxidase was blocked with 3% H2O2. After blocking nonspecific antigen binding with 5% bovine serum albumin (BSA, Sigma-Aldrich, B2064, St. Louis, MO, USA), sections were cultured with the primary antibody against COL1A1 (Abcam, 1/1500, ab138492, Cambridge, MA, USA) or DAPK2 (Invitrogen, 1/50, MA5-25084, Carlsbad, CA, USA) overnight at 4°C, followed by culturing with secondary antibodies (Abcam, 1/2000, ab172730, Cambridge, MA, USA) for 2 h at room temperature. Subsequently, they underwent the process of dyeing using DAB (Solarbio, DA1010, Beijing, China) and were subsequently counterstained with hematoxylin. Ultimately, an Olympus IX70 microscope (Okaya, Japan) was employed for observation.
Statistical analysis was implemented using GraphPad Prism 8 software (GraphPad, San Diego, CA, USA). The group difference was analyzed with one-way ANOVA and Student’s t test. The results are described as the mean ± SD from three individual repeats. p < 0.05 was considered significant.
DAPK2 expression is upregulated in OPLL
Initially, we assessed the levels of DAPK2 expression in ligament samples obtained from both individuals with OPLL and individuals without OPLL. The RT-qPCR analyses exhibited that the expression of DAPK2 was considerably greater in tissues obtained from patients with OPLL in comparison to the control samples (Fig. 1A). Subsequently, fibroblasts from the primary ligaments were isolated from these tissues. Upon microscopic examination, these cells displayed a morphology resembling fibroblasts, with a spindle-shaped structure (Fig. 1B). Vimentin, commonly used as a fibroblast marker due to its presence in fibroblasts as well as in endothelial and lymphoid cells, was also assessed [18]. Immunofluorescence staining demonstrated both elevated expression and co-localization of Vimentin and DAPK2 in the fibroblasts from the OPLL group (Fig. 1C–E). Further RT-qPCR analysis confirmed the pronounced upregulation of DAPK2 in the ligament fibroblasts from OPLL patients (Fig. 1F).
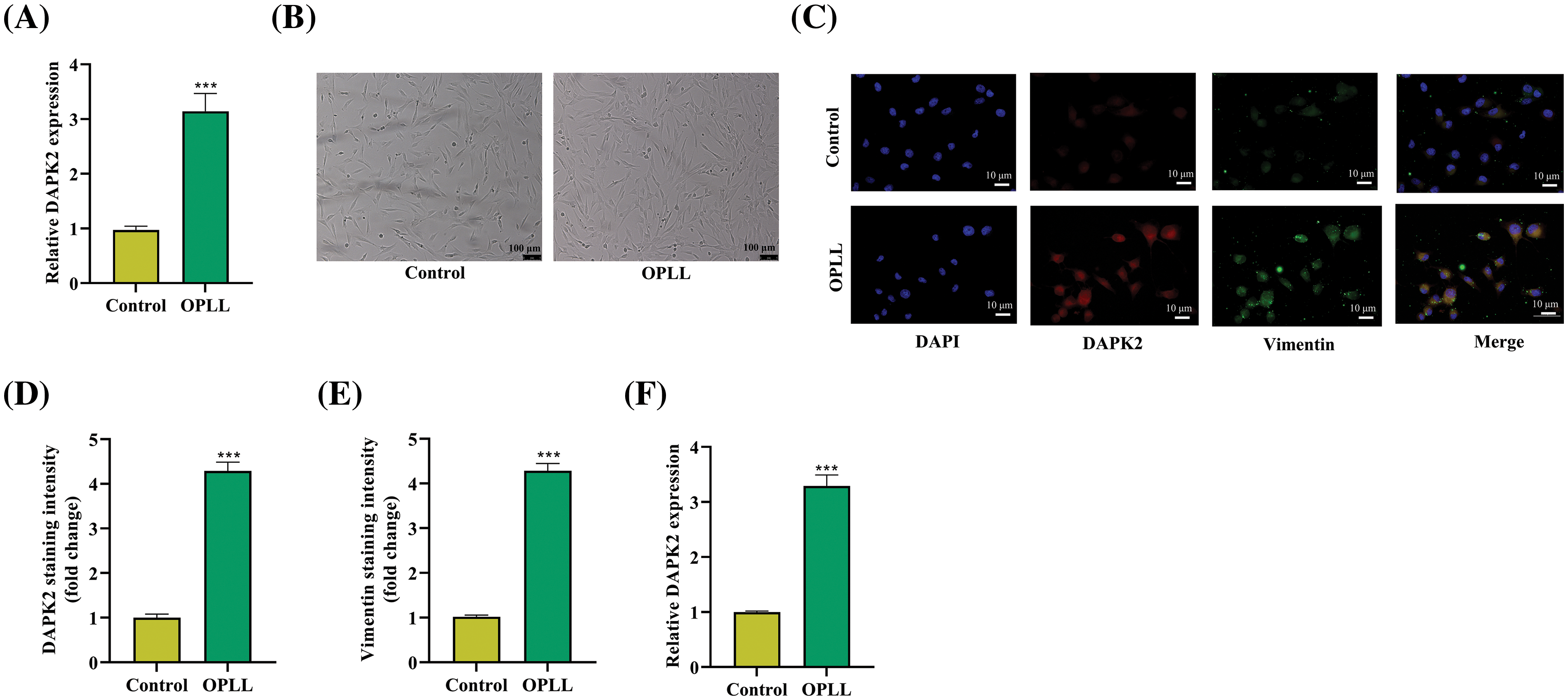
Figure 1: Increased DAPK2 Expression in OPLL. (A) RT-qPCR data showing DAPK2 levels in ligamentous tissues from control and OPLL patients. (B) Microscopic images of isolated fibroblasts from OPLL and control groups. (C–E) Immunofluorescence assays illustrating DAPK2 and Vimentin expression and their co-localization in fibroblasts from both groups. (F) RT-qPCR results demonstrating DAPK2 levels in fibroblasts from control and OPLL patients, indicating significant differences (***p < 0.001). Data presented as mean ± SD (n = 3).
Primary ligament fibroblast ossification is facilitated by DAPK2
We investigated the function of DAPK2 in the ossification process within primary ligament fibroblasts. Initially, DAPK2 expression in fibroblasts from OPLL patients was downregulated using sh-DAPK2 transfection. Both RT-qPCR and Western blot analysis confirmed a substantial decline in DAPK2 mRNA and protein expressions post-transfection (Fig. 2A,B). Subsequently, using Alizarin Red S staining, calcium accumulation was evaluated. This analysis indicated a significant reduction in osteogenic differentiation following DAPK2 suppression (Fig. 2C). Additionally, the levels of ossification markers such as alkaline phosphatase (ALP), collagen type I alpha 1 chain (COL1A1), osterix (OSX), osteocalcin (OCN), and runt-related transcription factor 2 (RUNX2) were evaluated through RT-qPCR and Western blot, showing a lessening in both mRNA and protein levels due to sh-DAPK2 transfection, thereby supporting DAPK2’s role in enhancing ossification (Fig. 2D–E).
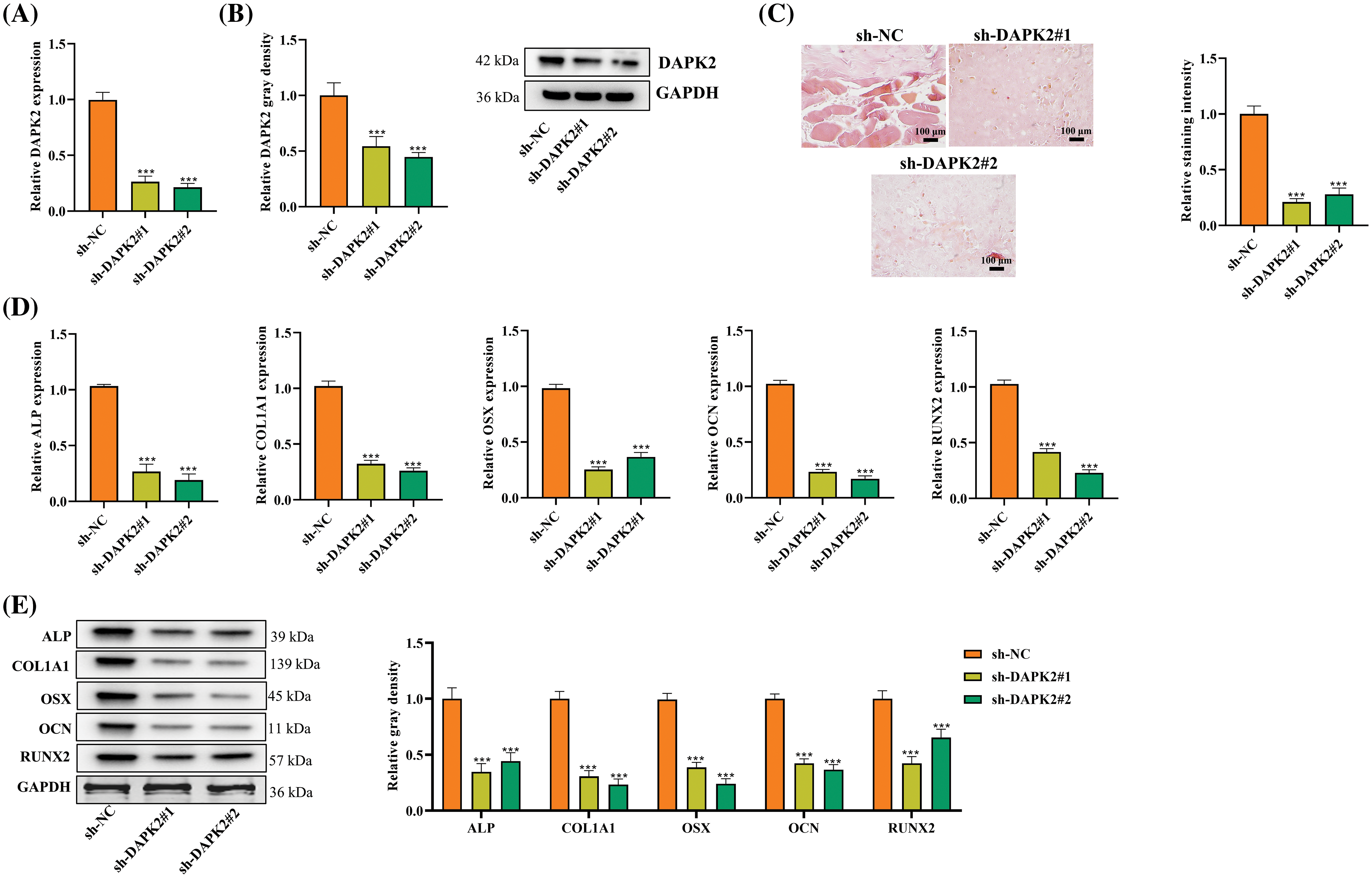
Figure 2: Influence of DAPK2 on Ossification in Ligament Fibroblasts. (A and B) Assessments of DAPK2 silencing efficiency via RT-qPCR and Western blot in OPLL-derived fibroblasts. (C) ARS assay for evaluating osteogenic differentiation after sh-DAPK2 or control transfection. (D and E) Analysis of ossification markers ALP, COL1A1, OSX, OCN, and RUNX2 following DAPK2 suppression, showing significant decreases. Data presented as mean ± SD (n = 3). ***p < 0.001.
Conversely, overexpression of DAPK2 was achieved by transfecting ligament fibroblasts from OPLL patients with pcDNA3.1-DAPK2. Consequently, there was a substantial rise in both mRNA and protein levels of DAPK2, as evidenced by RT-qPCR and Western blot analyses (Fig. A1A,B). Further, ARS staining confirmed that DAPK2 overexpression markedly enhanced osteogenic differentiation (Fig. A1C). Additional tests demonstrated an increase in the levels of ALP, COL1A1, OSX, OCN, and RUNX2, indicating an upregulation of osteogenic markers following DAPK2 overexpression (Fig. A1D,E).
DAPK2 enhances autophagy in primary ligament fibroblasts
Considering the pivotal role of autophagy in OPLL pathogenesis [13], we investigated the influence of DAPK2 on autophagy. LC3, a widely recognized autophagy marker, was analyzed using fluorescence staining. The findings we obtained demonstrated that DAPK2 knockdown resulted in a substantial decrease in LC3 fluorescence intensity, suggesting DAPK2 knockdown decreased autophagy (Fig. 3A). Further analysis of autophagy-related proteins showed that DAPK2 depletion led to a lower LC3-II/I ratio and decreased Beclin1 levels, alongside an increase in p62 accumulation, indicating DAPK2 knockdown blocked autophagy (Fig. 3B). This was validated by RT-qPCR results, which showed that DAPK2 deletion down-regulated LC3 and Beclin1 transcripts and up-regulated p62 mRNA (Fig. 3C). Collectively, these findings implied that DAPK2 positively regulated autophagic flux in primary ligament fibroblasts.
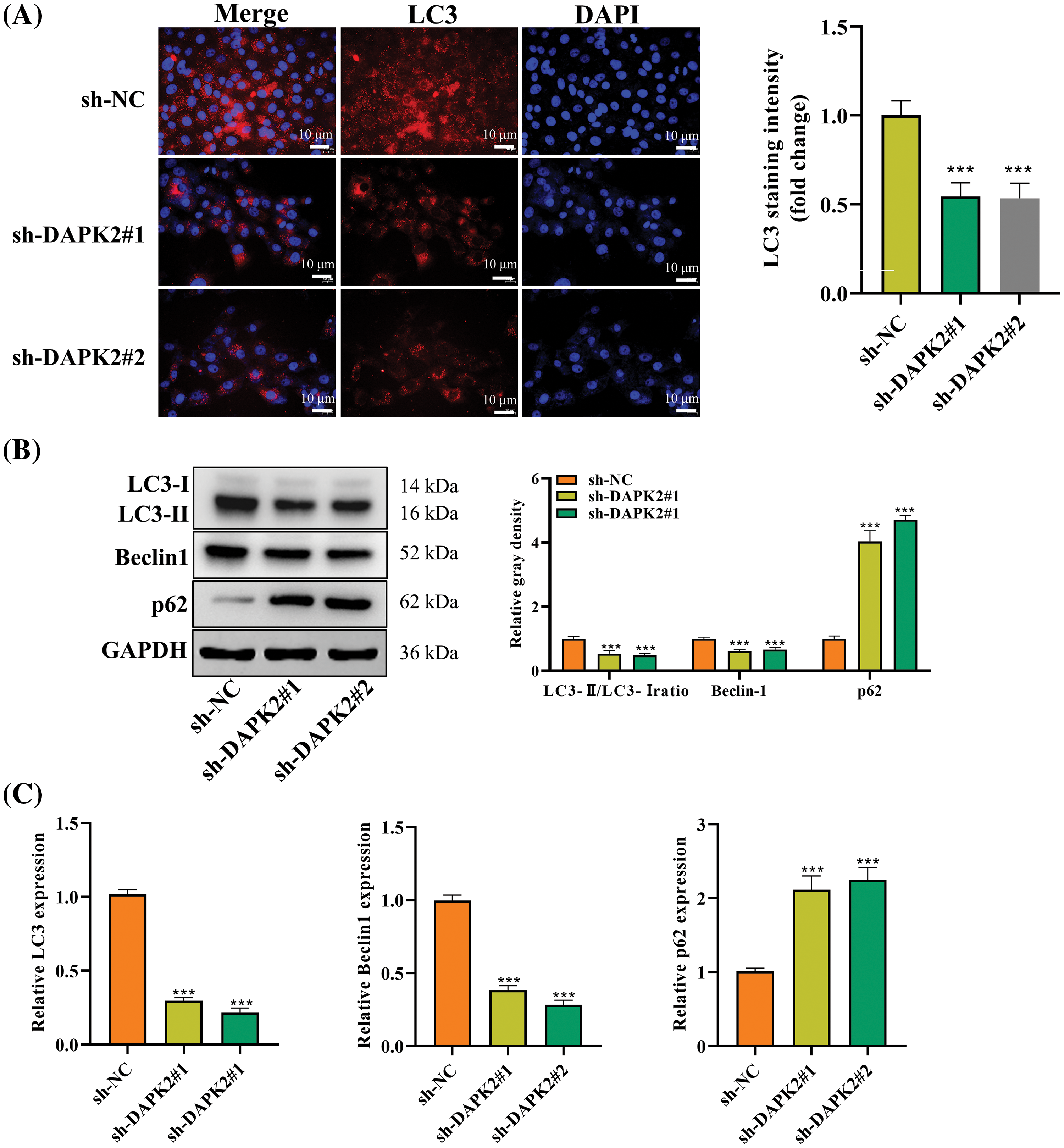
Figure 3: DAPK2 enhances autophagy in primary ligament fibroblasts. (A) An LC3-IF assay was performed to determine the autophagy level in ligament fibroblasts transfected with sh-NC or sh-DAPK2. (B and C) Western blot and RT-qPCR outcomes of LC3, Beclin1, and p62 expression levels in cells. Data presented as mean ± SD (n = 3). ***p < 0.001.
DAPK2 regulates the mTORC1 complex in ligament fibroblasts
The mTORC1 pathway is a key regulator of autophagy and its inhibition is a recognized inducer of autophagy [19]. To determine whether DAPK2 plays a promoting role in autophagy involves the regulation of mTORC1, we assessed the impact of DAPK2 inhibition on the activity of the mTORC1 complex. Western blot analysis revealed that DAPK2 silencing resulted in decreased phosphorylation of Raptor and unc-51 like kinase 1 (ULK1), coupled with enhanced phosphorylation of mTOR and its downstream targets p70S6K, and 4E-binding protein 1 (4E-BP1), which were hallmarks of increased mTORC1 activity (Fig. 4). These changes indicated that inhibition of mTORC1 was alleviated after DAPK2 knockdown, which was consistent with the reduction in autophagy. Hence, we concluded that DAPK2 promoted autophagy in ligament fibroblasts, at least in part, through inhibiting mTORC1 activity.
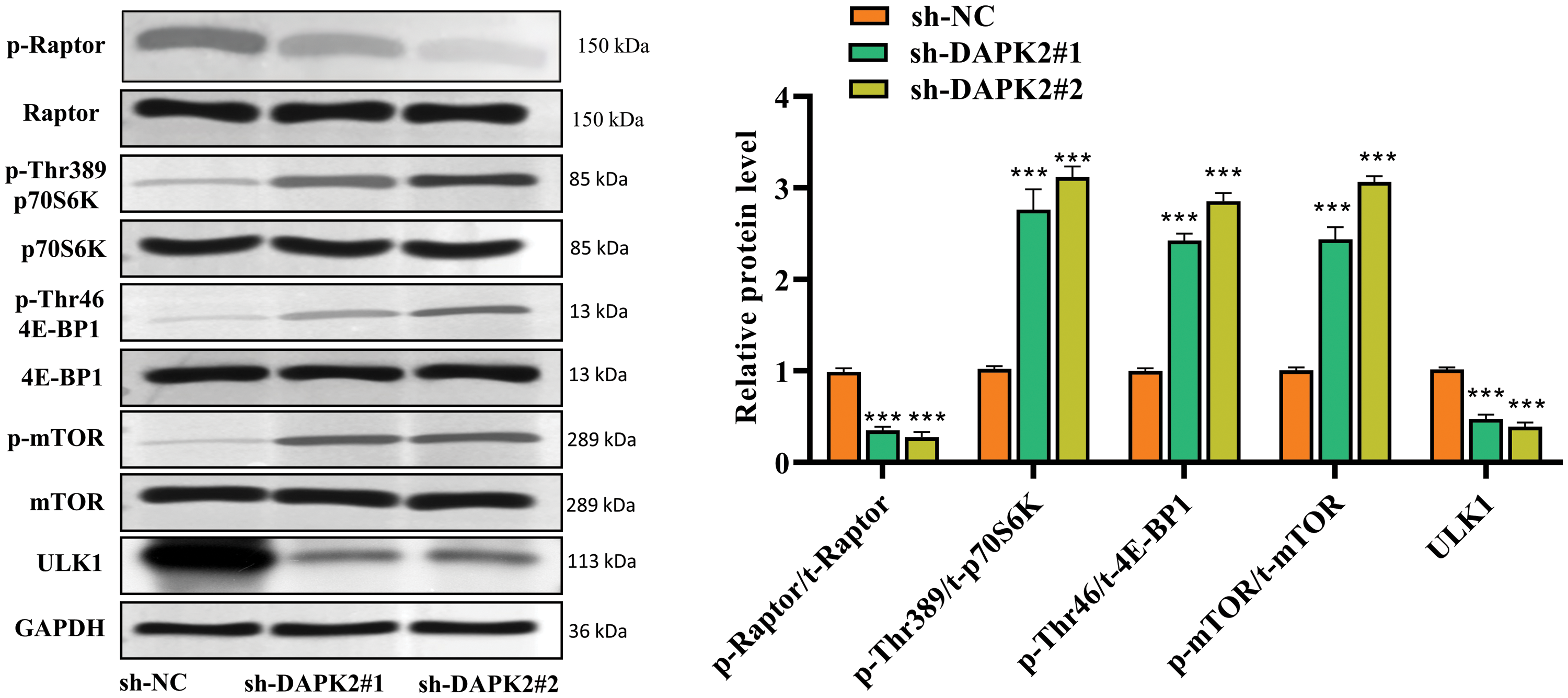
Figure 4: DAPK2 regulates the mTORC1 complex in ligament fibroblasts. Western blot outcomes of p-Raptor, Raptor, p-Thr389-p70S6K, p70S6K, p-Thr46-4E-BP1, 4E-BP1, p-mTOR, mTOR, and ULK1 protein levels in ligament fibroblasts transfected with sh-NC or sh-DAPK2. Data presented as mean ± SD (n = 3). ***p < 0.001.
DAPK2 promotes ossification in vivo
To further analyze the effect of DAPK2 in vivo, we carried out a heterotopic bone formation experiment on mice. Ligament fibroblasts of human stably silenced by DAPK2 were cocultured with Bio-Oss collagen scaffolds for two days. Next, mice were subjected to subcutaneous implantation with the scaffolds on the back for six weeks. Following this, the animals were euthanized eight weeks later, and micro computed tomography (micro-CT) scans were utilized to determine the bone mineral density (BMD) and bone volume/tissue volume (BV/TV). We revealed that the BV/TV rate and BMD were markedly reduced in mice implanted with sh-DAPK2 (Fig. 5A,B). Then, HE staining results illustrated that the formation of lamellar bone tissues was reduced in mice of the DAPK2 knockdown group in comparison to the control group (Fig. 5C). Moreover, IHC assays showed that the numbers of COL1A1-positive cells and DAPK2-positive cells were reduced by DAPK2 silencing (Fig. 5D). Thus, we confirmed that DAPK2 could facilitate ossification of ligament fibroblasts in vivo.
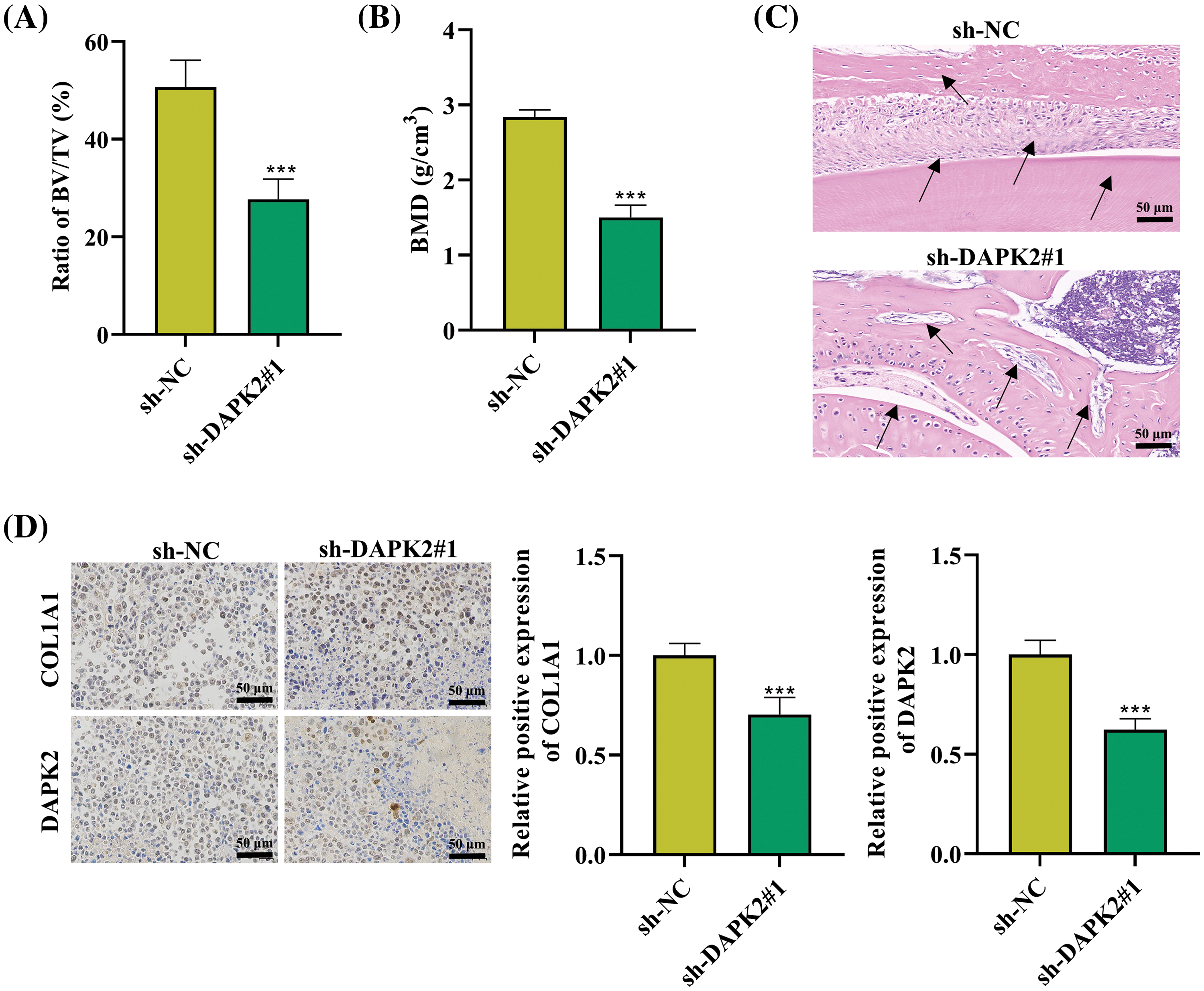
Figure 5: DAPK2 promotes ossification in vivo. (A and B) The BV/TV and BMD ratios were determined. (C) HE staining was conducted to determine the formation of lamellar bone tissues in bone grafts. Black arrows indicate areas of lamellar bone structure. (D) IHC results of COL1A1- and DAPK2-positive cells (6 mice per group). Data presented as mean ± SD (n = 3). ***p < 0.001.
Rapamycin reverse the effect of DAPK2 on ossification in vivo
Moreover, we carried out rescue experiments on mice to verify whether mTORC1 inhibitor promotes bone formation in conjunction with DAPK2 knockdown. It was discovered that the reduced BV/TV rate and BMD in mice implanted with sh-DAPK2#1 were reversed after rapamycin treatment (Fig. A2A,B). Then, HE staining results illustrated that the reduce formation of lamellar bone tissues in mice of the DAPK2 knockdown group was offset after rapamycin treatment (Fig. A2C). Furthermore, IHC assays showed that the reduced numbers of COL1A1-positive cells and DAPK2-positive cells caused by DAPK2 silencing were enhanced after rapamycin treatment (Fig. A2D).
OPLL is a common orthopedic disease that seriously affects the quality of life of patients [2,20]. Currently, OPLL treatment methods are unable to effectively alleviate this disease. The molecular mechanism of OPLL is also unclear. Molecular targeted therapy is a current hot research field [21], and relevant studies have revealed some molecular regulatory mechanisms of OPLL. Chen et al. suggest that connexin 43 expedites OPLL development by activating the extracellular signal-regulated kinases 1 and 2 (ERK1/2) and mitogen-activated protein kinases (MAPK) pathways [22]. The research carried out by Wang et al. confirms the role of long non-coding RNA SNHG1 in promoting the progression of osteogenic differentiation in ligament fibroblasts of OPLL patients [23]. Ectopic bone formation of cervical ligaments is a common characteristic of patients with OPLL [2]. Connective tissue is composed primarily of fibroblasts, which are capable of differentiating into bone cells. The role of osteocytes in the progression of ectopic ossification in a variety of diseases has been established [24–26]. Inhibition of osteogenic differentiation of fibroblasts may be a feasible treatment for OPLL. DAPK2 is categorized as a serine/threonine kinase that has a favorable impact on cell apoptosis and autophagy [27,28]. Accumulating studies have confirmed the regulatory function of DAPK2 in human diseases and cancers [29,30]. Herein, DAPK2 was overexpressed in ligamentous tissues of OPLL patients. Then, we isolated ligament fibroblasts from OPLL ligamentous tissues and demonstrated that DAPK2 was expressed at a high level in cells. Thus, DAPK2 may have been implicated in the pathogenesis of OPLL, according to our initial hypotheses, osteogenic differentiation is often accompanied by upregulation of markers related to bone formation, such as ALP, COL1A1, OSX, OCN, and RUNX2 [31,32]. ALP is an early osteogenic marker that mainly promotes cell maturation and calcification [33]. The higher its activity, the more mature the differentiation of osteoblasts. COL1A1 gene is responsible for producing a subunit of type I collagen, which is the primary organic component of the bone matrix [34]. The transcription factor RUNX2 is essential for controlling the processes of chondrogenesis and osteogenesis [35]. It can stimulate the expression levels of OCN and OSX, which are markers associated with osteogenesis and necessary for the final differentiation of osteoblasts [36]. In this study, we proved that DAPK2 depletion reduced ALP, COL1A1, OSX, OCN, and RUNX2 levels in ligament fibroblasts while DAPK2 overexpression elevated ALP, COL1A1, OSX, OCN, and RUNX2 levels in ligament fibroblasts. DAPK2 depletion also reduced calcium deposition in cells while DAPK2 overexpression enhanced calcium deposition in cells. Therefore, it could be inferred that DAPK2 had the ability to enhance the osteogenic differentiation of ligament fibroblasts in OPLL.
Autophagy is an evolutionarily conserved subcellular degradation pathway. Its function is to degrade defective proteins or organelles in lysosomes and to recover essential components in eukaryotic cells [37]. Increasing research has confirmed the important role of autophagy in the differentiation of bone cells, and its dysfunction is closely related to many orthopedic diseases, such as osteoporosis and osteopenia [38,39]. Zahm et al. prove that autophagic receptors have crucial functions in the activity and differentiation of osteoblasts [40]. LC3 is the most common marker for autophagy and has also been reported to be correlated with osteogenic differentiation [41]. In addition, it has been confirmed that autophagy is enhanced in OPLL ligament fibroblasts, and the autophagy process facilitated by Beclin1 can contribute to the osteogenic differentiation of ligament fibroblasts, thereby promoting the progression of OPLL [13]. The identification of DAPK2 as a novel regulator of mTORC1 activity and autophagy has significant implications in the field [12]. There is evidence that activated AMP-activated protein kinase (AMPK) phosphorylates DAPK2 and increases its catalytic activity [42]. DAPK2 subsequently promotes Beclin-1 phosphorylation, resulting in the induction of autophagy [42]. It has been established that DAPK2 controls the autophagy process in a number of illnesses and malignancies. For instance, it has been documented that DAPK2 controls autophagy to encourage the development of thyroid cancer tumors [9]. MiR-133a-3p suppresses autophagy in cerebral ischemia-reperfusion damage by inhibiting DAPK2 [43]. In this work, we found that in ligament fibroblasts, DAPK2 knockdown significantly decreased LC3 expression and autophagosome formation. Moreover, DAPK2 downregulation increased p62 expression while decreasing the LC3-II/LC3-I ratio and Beclin1 expression. Thus, we were able to verify that DAPK2 promoted autophagy in OPLL ligament fibroblasts.
mTOR, a serine/threonine protein kinase, serves as a crucial inhibitory regulator of autophagy [44]. It has the capability to engage with various proteins, resulting in the formation of two distinct complexes known as mTORC1 and mTORC2 [45]. In addition to the mTOR catalytic subunit, mTORC1 consists of the regulatory-associated protein of mTOR (Raptor). The confirmation of raptor phosphorylation as the mechanism through which upstream kinases regulate mTORC1 activity has been verified in prior research [46,47]. The regulation of mTORC1 on cell growth and metabolism is mainly achieved through phosphorylation of substrates p70S6K and 4E-BP1 [48]. ULK1 is a serine/threonine kinase and crucial for autophagic initiation. Autophagy induction is the result of autophagy stimulation, resulting in inhibition of mTORC1 activity and activation of ULK1 complex [49,50]. Overall, the inactivation of mTORC1 is vital for autophagy, while DAPK2 has been identified as a new modulator of mTORC1 activity [12]. Thus, the regulation of DAPK2 on mTORC1 in OPLL is worth exploring. Evidence has confirmed that DAPK2 can promote autophagy by enhancing the phosphorylation of Raptor, resulting in the inhibition of mTORC1 complex activity [12]. Herein, DAPK2 inhibition notably reduced the levels of Raptor phosphorylation and ULK1, while elevating the phosphorylation levels of mTOR, p70S6K, and 4E-BP1 in ligament fibroblasts, suggesting a decrease in mTORC1 activity by DAPK2. Collectively, we confirmed that DAPK2 could regulate autophagy in ligament fibroblasts through the mTORC1 complex.
In the final animal experiments, we verified the function of DAPK2 in mice with a Bio-Oss collagen scaffold. The experimental results further proved that DAPK2 depletion alleviated ectopic osteogenesis in mice. Moreover, we carried out rescue experiments on mice to verify whether mTORC1 inhibitor promotes bone formation in conjunction with DAPK2 knockdown, and the results indicated that rapamycin reversed the effect of DAPK2 on ossification in vivo.
This study elucidates the role of DAPK2 in the pathogenesis of OPLL. We demonstrated that DAPK2 is overexpressed in OPLL ligamentous tissues and promotes both autophagy and osteogenic differentiation of ligament fibroblasts through regulation of the mTORC1 complex. Our findings answer key research questions regarding the molecular mechanisms underlying OPLL progression and identify DAPK2 as a potential therapeutic target. The significance of this work lies in its contribution to understanding the complex interplay between autophagy and osteogenic differentiation in OPLL, providing new insights into the molecular pathways of this disease and opening avenues for targeted interventions.
However, our study has limitations that should be addressed in future research. The small sample size and lack of clinical validation constrain the generalizability of our findings. Additionally, the mechanism of DAPK2 upregulation in OPLL remains unexplored. Future studies should focus on validating the role of DAPK2 in larger, diverse patient populations, investigating its upstream regulators in OPLL, exploring potential DAPK2-targeted therapies, and examining the broader implications of DAPK2-mediated autophagy in other orthopedic conditions.
In conclusion, our findings underscore the importance of DAPK2 in OPLL pathogenesis and highlight its potential as a therapeutic target. This study contributes to the growing body of knowledge on the molecular mechanisms involved in OPLL and may inform the development of novel treatment strategies for this challenging condition. Further exploration of DAPK2 as a therapeutic target could potentially lead to novel interventions for OPLL and improve the quality of life for affected individuals.
Acknowledgement: None.
Funding Statement: This research received funding from the Natural Science Foundation of Shanghai (Grant No. 20ZR1457600) and the School-Level Basic Medical Project of Naval Medical University (Grant No. 2021MS13).
Author Contributions: Study conception and design: Lei Shi, Jianshi Yin, and Jinhao Miao; data collection: Lei Shi, Jianshi Yin, and Jiangang Shi; analysis and interpretation of results: Lei Shi and Jianshi Yin; draft manuscript preparation: Lei Shi, Jianshi Yin, and Yu Chen. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The corresponding author can provide the data supporting the findings of this study upon a reasonable request.
Ethics Approval: Ethical clearance for the human samples in this study was granted by the Ethics Committee of Shanghai Changzheng Hospital. The ethical approval number was 2021MS13. For each participant, written informed consent was obtained. Ethical clearance for the animal experiments in this study was granted by the Institutional Animal Care and Use Committee of Cyagen (Suzhou) Biotechnology Co., Ltd. The ethical approval number was IACUC-2109025.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Koike Y, Takahata M, Nakajima M, Otomo N, Suetsugu H, Liu X, et al. Genetic insights into ossification of the posterior longitudinal ligament of the spine. eLife. 2023;12:e86514. doi:10.7554/eLife.86514. [Google Scholar] [PubMed] [CrossRef]
2. Le HV, Wick JB, Van BW, Klineberg EO. Ossification of the posterior longitudinal ligament: pathophysiology, diagnosis, and management. J Am Acad Orthop Surg. 2022;30(17):820–30. [Google Scholar] [PubMed]
3. Fujita R, Endo T, Takahata M, Koike Y, Yoneoka D, Suzuki R, et al. High whole-body bone mineral density in ossification of the posterior longitudinal ligament. Spine J. 2023;23(10):1461–70. doi:10.1016/j.spinee.2023.06.400. [Google Scholar] [PubMed] [CrossRef]
4. Doi T, Ohashi S, Ohtomo N, Tozawa K, Nakarai H, Yoshida Y, et al. Evaluation of bone strength using finite-element analysis in patients with ossification of the posterior longitudinal ligament. Spine J. 2022;22(8):1399–407. doi:10.1016/j.spinee.2022.02.018. [Google Scholar] [PubMed] [CrossRef]
5. Li S, Peng J, Xu R, Zheng R, Huang M, Xu Y, et al. Comparison of the surgeries for the ossification of the posterior longitudinal ligament-related cervical spondylosis: a PRISMA-compliant network meta-analysis and literature review. Medicine. 2021;100(9):e24900. doi:10.1097/MD.0000000000024900. [Google Scholar] [PubMed] [CrossRef]
6. Park BJ, Seaman SC, Woodroffe RW, Noeller J, Hitchon PW. Surgical options in treating ossification of the posterior longitudinal ligament: single-center experience. World Neurosurg. 2021;148:617–26. doi:10.1016/j.wneu.2021.01.046. [Google Scholar] [PubMed] [CrossRef]
7. Horvath M, Petrvalska O, Herman P, Obsilova V, Obsil T. 14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites. Commun Biol. 2021;4(1):986. doi:10.1038/s42003-021-02518-y. [Google Scholar] [PubMed] [CrossRef]
8. Takahashi M, Lio CJ, Campeau A, Steger M, Ay F, Mann M, et al. The tumor suppressor kinase DAPK3 drives tumor-intrinsic immunity through the STING-IFN-β pathway. Nat Immunol. 2021;22(4):485–96. doi:10.1038/s41590-021-00896-3. [Google Scholar] [PubMed] [CrossRef]
9. Jiang Y, Liu J, Xu H, Zhou X, He L, Zhu C. DAPK2 activates NF-κB through autophagy-dependent degradation of I-κBα during thyroid cancer development and progression. Ann Transl Med. 2021;9(13):1083. doi:10.21037/atm-21-2062. [Google Scholar] [PubMed] [CrossRef]
10. Zhang J, Liu L, Sun Y, Xiang J, Zhou D, Wang L, et al. MicroRNA-520g promotes epithelial ovarian cancer progression and chemoresistance via DAPK2 repression. Oncotarget. 2016;7(18):26516–34. doi:10.18632/oncotarget.v7i18. [Google Scholar] [CrossRef]
11. Jin M, Li G, Liu W, Wu X, Zhu J, Zhao D, et al. Cigarette smoking induces aberrant N6-methyladenosine of DAPK2 to promote non-small cell lung cancer progression by activating NF-κB pathway. Cancer Lett. 2021;518:214–29. doi:10.1016/j.canlet.2021.07.022. [Google Scholar] [PubMed] [CrossRef]
12. Ber Y, Shiloh R, Gilad Y, Degani N, Bialik S, Kimchi A. DAPK2 is a novel regulator of mTORC1 activity and autophagy. Cell Death Differ. 2015;22(3):465–75. doi:10.1038/cdd.2014.177. [Google Scholar] [PubMed] [CrossRef]
13. Yang Y, Lin Z, Chen J, Ding S, Mao W, Shi S, et al. Autophagy in spinal ligament fibroblasts: evidence and possible implications for ossification of the posterior longitudinal ligament. J Orthop Surg Res. 2020;15(1):490. doi:10.1186/s13018-020-02017-6. [Google Scholar] [PubMed] [CrossRef]
14. Yang HS, Lu XH, Chen DY, Yuan W, Yang LL, Chen Y, et al. Mechanical strain induces Cx43 expression in spinal ligament fibroblasts derived from patients presenting ossification of the posterior longitudinal ligament. Eur Spine J. 2011;20(9):1459–65. doi:10.1007/s00586-011-1767-9. [Google Scholar] [PubMed] [CrossRef]
15. Tillie R, van Kuijk K, Sluimer JC. Fibroblasts in atherosclerosis: heterogeneous and plastic participants. Curr Opin Lipidol. 2020;31(5):273–8. doi:10.1097/MOL.0000000000000700. [Google Scholar] [PubMed] [CrossRef]
16. Claeys L, Bravenboer N, Eekhoff EMW, Micha D. Human fibroblasts as a model for the study of bone disorders. Front Endocrinol. 2020;11:394. doi:10.3389/fendo.2020.00394. [Google Scholar] [PubMed] [CrossRef]
17. Fang Y, Hill CM, Darcy J, Reyes-Ordoñez A, Arauz E, McFadden S, et al. Effects of rapamycin on growth hormone receptor knockout mice. Proc Natl Acad Sci U S A. 2018;115(7):E1495–503. doi:10.1073/pnas.1717065115. [Google Scholar] [PubMed] [CrossRef]
18. Xian H, Wang Y, Bao X, Zhang H, Wei F, Song Y, et al. Hexokinase inhibitor 2-deoxyglucose coordinates citrullination of vimentin and apoptosis of fibroblast-like synoviocytes by inhibiting HK2/mTORC1-induced autophagy. Int Immunopharmacol. 2023;114(5):109556. doi:10.1016/j.intimp.2022.109556. [Google Scholar] [PubMed] [CrossRef]
19. Zhang JL, Zheng HF, Li K, Zhu YP. miR-495-3p depresses cell proliferation and migration by downregulating HMGB1 in colorectal cancer. World J Surg Oncol. 2022;20(1):101. doi:10.1186/s12957-022-02500-w. [Google Scholar] [PubMed] [CrossRef]
20. Sakai K, Yoshii T, Arai Y, Hirai T, Torigoe I, Inose H, et al. Impact of preoperative cervical sagittal alignment for cervical myelopathy caused by ossification of the posterior longitudinal ligament on surgical treatment. J Orthop Sci. 2022;27(6):1208–14. doi:10.1016/j.jos.2021.08.006. [Google Scholar] [PubMed] [CrossRef]
21. Bedard PL, Hyman DM, Davids MS, Siu LL. Small molecules, big impact: 20 years of targeted therapy in oncology. Lancet. 2020;395(10229):1078–88. doi:10.1016/S0140-6736(20)30164-1. [Google Scholar] [PubMed] [CrossRef]
22. Chen D, Liu Y, Yang H, Chen D, Zhang X, Fermandes JC, et al. Connexin 43 promotes ossification of the posterior longitudinal ligament through activation of the ERK1/2 and p38 MAPK pathways. Cell Tissue Res. 2016;363(3):765–73. doi:10.1007/s00441-015-2277-6. [Google Scholar] [PubMed] [CrossRef]
23. Wang J, Zhu P, Toan S, Li R, Ren J, Zhou H. Pum2-Mff axis fine-tunes mitochondrial quality control in acute ischemic kidney injury. Cell Biol Toxicol. 2020;36(4):365–78. doi:10.1007/s10565-020-09513-9. [Google Scholar] [PubMed] [CrossRef]
24. Lendahl U, Muhl L, Betsholtz C. Identification, discrimination and heterogeneity of fibroblasts. Nat Commun. 2022;13(1):3409. doi:10.1038/s41467-022-30633-9. [Google Scholar] [PubMed] [CrossRef]
25. Zhu Z, Ruan S, Jiang Y, Huang F, Xia W, Chen J, et al. Alpha-Klotho released from HK-2 cells inhibits osteogenic differentiation of renal interstitial fibroblasts by inactivating the Wnt-beta-catenin pathway. Cell Mol Life Sci. 2021;78(23):7831–49. doi:10.1007/s00018-021-03972-x. [Google Scholar] [PubMed] [CrossRef]
26. Zeng Y, Wang T, Liu Y, Luo T, Li Q, He Y, et al. Wnt and Smad signaling pathways synergistically regulated the osteogenic differentiation of fibroblasts in ankylosing spondylitis. Tissue Cell. 2022;77:101852. doi:10.1016/j.tice.2022.101852. [Google Scholar] [PubMed] [CrossRef]
27. Villanova L, Barbini C, Piccolo C, Boe A, De Maria R, Fiori ME. miR-1285-3p controls colorectal cancer proliferation and escape from apoptosis through DAPK2. Int J Mol Sci. 2020;21(7):2423. doi:10.3390/ijms21072423. [Google Scholar] [PubMed] [CrossRef]
28. Li C, Yuan Q, Xu G, Yang Q, Hou J, Zheng L, et al. A seven-autophagy-related gene signature for predicting the prognosis of differentiated thyroid carcinoma. World J Surg Oncol. 2022;20(1):129. doi:10.1186/s12957-022-02590-6. [Google Scholar] [PubMed] [CrossRef]
29. Li Y, Xu J, Lu Y, Bian H, Yang L, Wu H, et al. DRAK2 aggravates nonalcoholic fatty liver disease progression through SRSF6-associated RNA alternative splicing. Cell Metab. 2021;33(10):2004–20.e9. doi:10.1016/j.cmet.2021.09.008. [Google Scholar] [PubMed] [CrossRef]
30. Zhang L, Luo B, Lu Y, Chen Y. Targeting death-associated protein kinases for treatment of human diseases: recent advances and future directions. J Med Chem. 2023;66(2):1112–36. doi:10.1021/acs.jmedchem.2c01606. [Google Scholar] [PubMed] [CrossRef]
31. Zhou Y, Xu Z, Wang Y, Song Q, Yin R. LncRNA MALAT1 mediates osteogenic differentiation in osteoporosis by regulating the miR-485-5p/WNT7B axis. Front Endocrinol. 2022;13:922560. doi:10.3389/fendo.2022.922560. [Google Scholar] [PubMed] [CrossRef]
32. Kan T, He Z, Du J, Xu M, Cui J, Han X, et al. Irisin promotes fracture healing by improving osteogenesis and angiogenesis. Journal Orthop Transl. 2022;37:37–45. doi:10.1016/j.jot.2022.07.006. [Google Scholar] [PubMed] [CrossRef]
33. Kanawa M, Igarashi A, Fujimoto K, Saskianti T, Nakashima A, Higashi Y, et al. The identification of marker genes for predicting the osteogenic differentiation potential of mesenchymal stromal cells. Curr Issues Mol Biol. 2021;43(3):2157–66. doi:10.3390/cimb43030150. [Google Scholar] [PubMed] [CrossRef]
34. Junqueira JJM, Lourenco JD, da Silva KR, Cervilha DAB, da Silveira LKR, Correia AT, et al. Decreased bone type I collagen in the early stages of chronic obstructive pulmonary disease (COPD). COPD. 2020;17(5):575–86. doi:10.1080/15412555.2020.1808605. [Google Scholar] [PubMed] [CrossRef]
35. Liu DD, Zhang CY, Liu Y, Li J, Wang YX, Zheng SG. RUNX2 regulates osteoblast differentiation via the BMP4 signaling pathway. J Dent Res. 2022;101(10):1227–37. doi:10.1177/00220345221093518. [Google Scholar] [PubMed] [CrossRef]
36. Xiao Y, Xie X, Chen Z, Yin G, Kong W, Zhou J. Advances in the roles of ATF4 in osteoporosis. Biomed Pharmacother. 2023;169:115864. doi:10.1016/j.biopha.2023.115864. [Google Scholar] [PubMed] [CrossRef]
37. Liu J, Liu Y, Wang Y, Li C, Xie Y, Klionsky DJ, et al. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy. 2023;19(3):945–56. doi:10.1080/15548627.2022.2111635. [Google Scholar] [PubMed] [CrossRef]
38. Bai L, Liu Y, Zhang X, Chen P, Hang R, Xiao Y, et al. Osteoporosis remission via an anti-inflammaging effect by icariin activated autophagy. Biomaterials. 2023;297(4):122125. doi:10.1016/j.biomaterials.2023.122125. [Google Scholar] [PubMed] [CrossRef]
39. Wang J, Zhang Y, Cao J, Wang Y, Anwar N, Zhang Z, et al. The role of autophagy in bone metabolism and clinical significance. Autophagy. 2023;19(9):2409–27. doi:10.1080/15548627.2023.2186112. [Google Scholar] [PubMed] [CrossRef]
40. Zahm AM, Bohensky J, Adams CS, Shapiro IM, Srinivas V. Bone cell autophagy is regulated by environmental factors. Cells Tissues Organs. 2011;194(2–4):274–8. doi:10.1159/000324647. [Google Scholar] [PubMed] [CrossRef]
41. Zhao B, Peng Q, Poon EHL, Chen F, Zhou R, Shang G, et al. Leonurine promotes the osteoblast differentiation of rat BMSCs by activation of autophagy via the PI3K/Akt/mTOR Pathway. Front Bioeng Biotechnol. 2021;9:615191. doi:10.3389/fbioe.2021.615191. [Google Scholar] [PubMed] [CrossRef]
42. Shiloh R, Kimchi A. AMPK activates DAPK2 to promote autophagy. Oncotarget. 2018;9(60):31570–1. doi:10.18632/oncotarget.25842. [Google Scholar] [PubMed] [CrossRef]
43. Yang X, Xu J, Lan S, Tong Z, Chen K, Liu Z, et al. Exosomal miR-133a-3p derived from BMSCs alleviates cerebral ischemia-reperfusion injury via targeting DAPK2. Int J Nanomedicine. 2023;18:65–78. doi:10.2147/IJN.S385395. [Google Scholar] [PubMed] [CrossRef]
44. Pei F, Ma L, Jing J, Feng J, Yuan Y, Guo T, et al. Sensory nerve niche regulates mesenchymal stem cell homeostasis via FGF/mTOR/autophagy axis. Nat Commun. 2023;14(1):344. doi:10.1038/s41467-023-35977-4. [Google Scholar] [PubMed] [CrossRef]
45. Xie Y, Lei X, Zhao G, Guo R, Cui N. mTOR in programmed cell death and its therapeutic implications. Cytokine Growth Factor Rev. 2023;71–72:66–81. doi:10.1016/j.cytogfr.2023.06.002. [Google Scholar] [PubMed] [CrossRef]
46. Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320(5882):1496–501. doi:10.1126/science.1157535. [Google Scholar] [PubMed] [CrossRef]
47. Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J Hematol Oncol. 2019;12(1):71. doi:10.1186/s13045-019-0754-1. [Google Scholar] [PubMed] [CrossRef]
48. Wu ZR, Yan L, Liu YT, Cao L, Guo YH, Zhang Y, et al. Inhibition of mTORC1 by lncRNA H19 via disrupting 4E-BP1/Raptor interaction in pituitary tumours. Nat Commun. 2018;9(1):4624. doi:10.1038/s41467-018-06853-3. [Google Scholar] [PubMed] [CrossRef]
49. Holczer M, Hajdú B, Lőrincz T, Szarka A, Bánhegyi G, Kapuy O. Fine-tuning of AMPK-ULK1-mTORC1 regulatory triangle is crucial for autophagy oscillation. Sci Rep. 2020;10(1):17803. doi:10.1038/s41598-020-75030-8. [Google Scholar] [PubMed] [CrossRef]
50. Zhu X, Sun Y, Yu Q, Wang X, Wang Y, Zhao Y. Exosomal lncRNA GAS5 promotes M1 macrophage polarization in allergic rhinitis via restraining mTORC1/ULK1/ATG13-mediated autophagy and subsequently activating NF-кB signaling. Int Immunopharmacol. 2023;121:110450. doi:10.1016/j.intimp.2023.110450. [Google Scholar] [PubMed] [CrossRef]
Appendix A
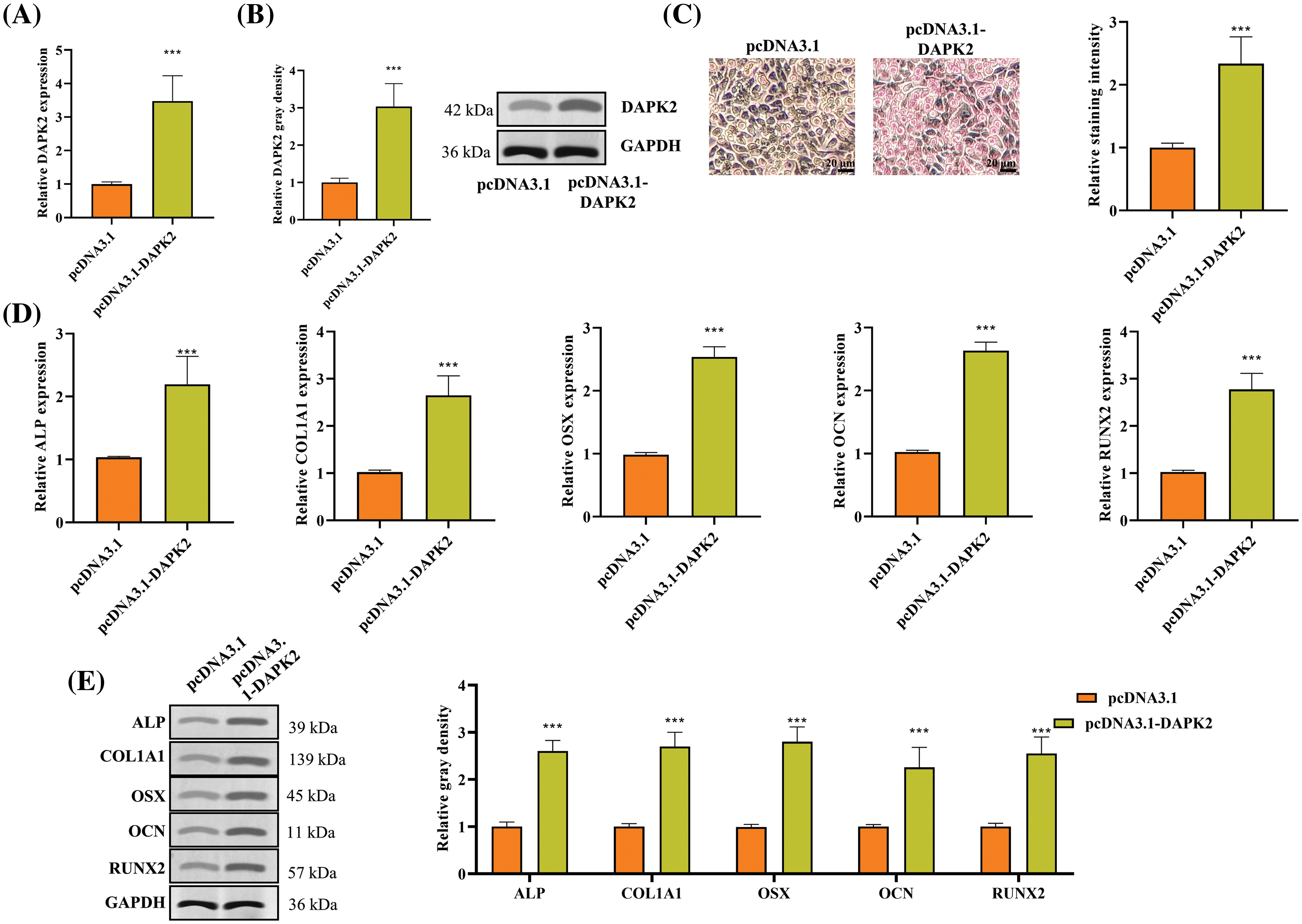
Figure A1: DAPK2 overexpression promotes ossification of ligament fibroblasts. (A and B) RT-qPCR and Western blot outcomes of DAPK2 overexpression efficiency in ligament fibroblasts from OPLL patients. (C) The osteogenic differentiation of ligament fibroblasts transfected with pcDNA3.1 or pcDNA3.1-DAPK2 was assessed using an Alizarin Red S staining assay. (D and E) RT-qPCR and Western blot outcomes of ALP, COL1A1, OSX, OCN, and RUNX2 expression levels in cells. Data presented as mean ± SD (n = 3). ***p < 0.001.
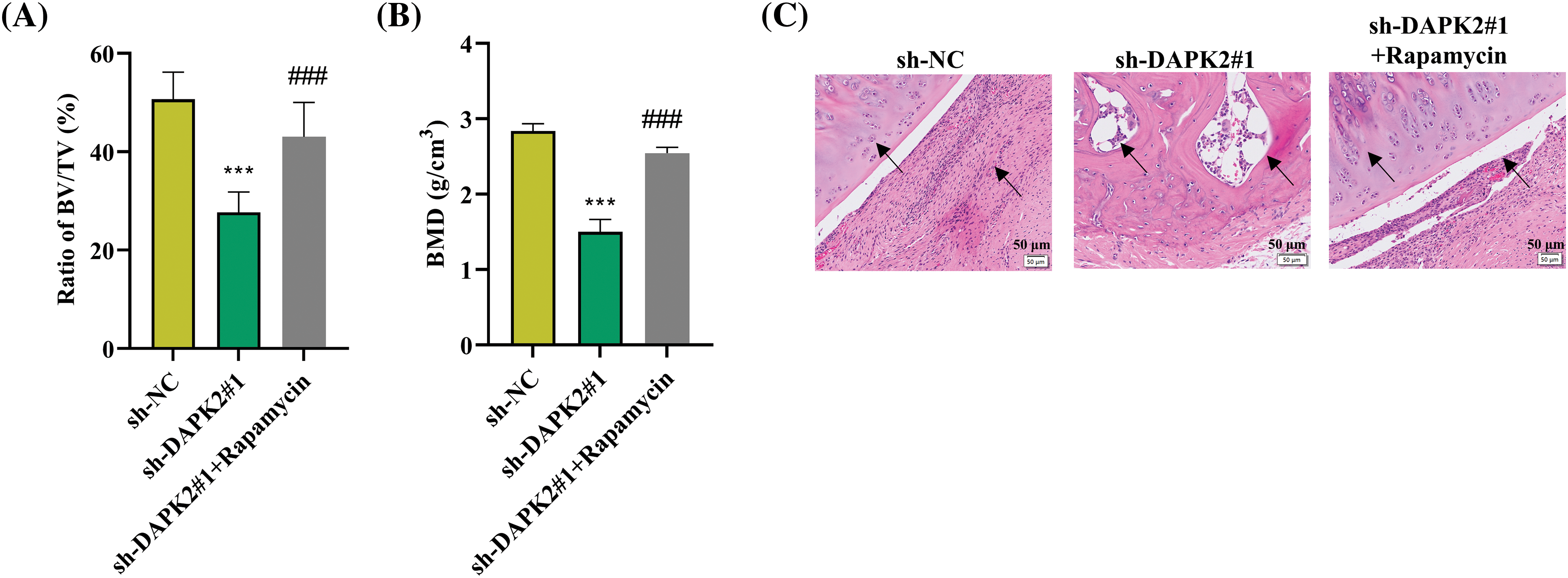
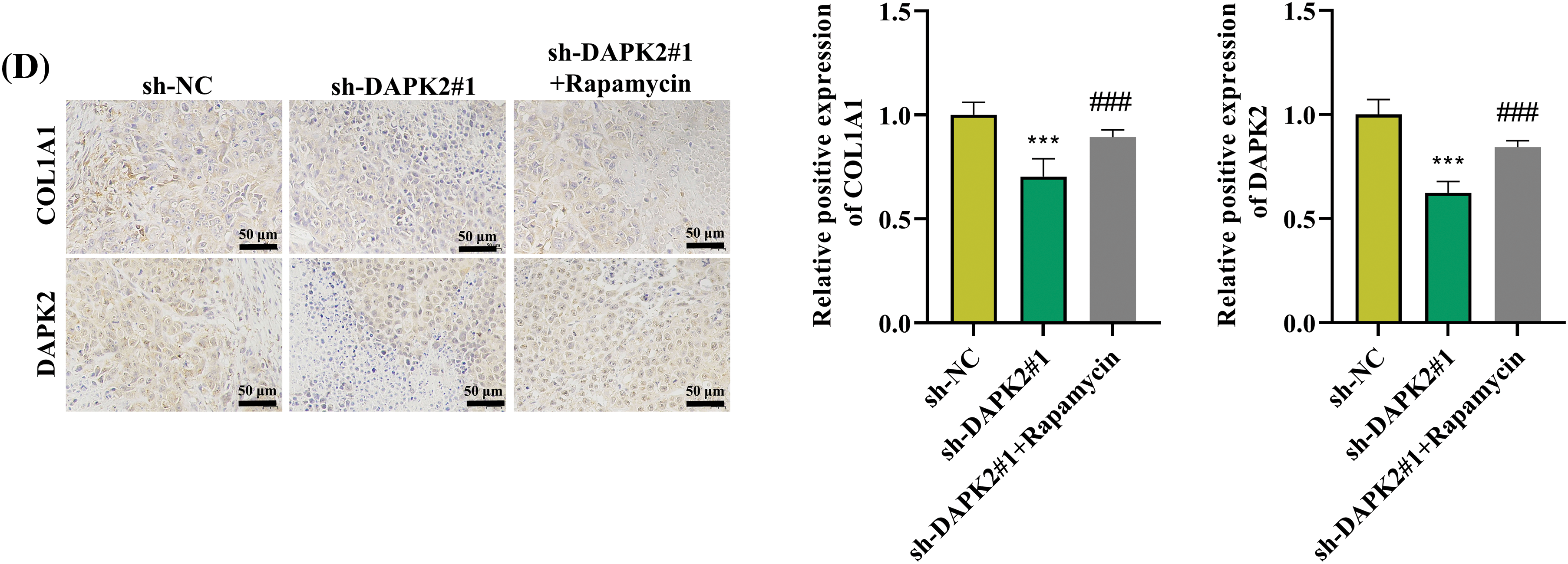
Figure A2: Rapamycin reverses the effect of DAPK2 on ossification in vivo. (A and B) The BV/TV and BMD ratios were determined. (C) HE staining was conducted to determine the formation of lamellar bone tissues in bone grafts. Black arrows indicate areas of lamellar bone structure. (D) IHC results of COL1A1- and DAPK2-positive cells (6 mice per group). Data presented as mean ± SD (n = 3). ***p < 0.001, compared with sh-NC group, ###p < 0.001, compared with sh-DAPK2#1 group.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools