 Open Access
Open Access
ARTICLE
Ultra-conservative noncoding RNA uc.243 confers chemo-resistance by facilitating the efflux of the chemotherapeutic drug in ovarian cancer
1 Department of Gynecology and Obstetrics, The First Affiliated Hospital of Jinan University, Guangzhou, 510180, China
2 Reproductive Medical Center, Boai Hospital of Zhongshan Affiliated to Southern Medical University, Zhongshan, 528400, China
3 Department of Gynecology and Obstetrics, Boai Hospital of Zhongshan Affiliated to Southern Medical University, Zhongshan, 528400, China
4 Department of Gynecology and Obstetrics, Guangzhou First People’s Hospital, Guangzhou Medical University, Guangzhou, 510180, China
* Corresponding Author: JIALI KANG. Email:
(This article belongs to the Special Issue: Subcellular Organelles and Cellular Molecules: Localization, Detection, Prediction, and Diseases)
BIOCELL 2024, 48(8), 1265-1273. https://doi.org/10.32604/biocell.2024.051478
Received 06 March 2024; Accepted 29 April 2024; Issue published 02 August 2024
Abstract
Background: Despite improvements in objective response rates to cisplatin-based combination chemotherapy, the majority of advanced ovarian cancer remains suboptimal, resulting in poor survival. it has been found that non-coding RNAs (ncRNAs) not only participate in the transmission of signals between various cells but also participate in tumor immunity and anti-tumor immune responses, thereby regulating tumor occurrence and development. However, the function and detailed mechanism of ultraconserved RNA (ucRNA) in ovarian cancer chemoresistance is still unclear. Methods: Western blotting assay, Quantitative real-time PCR analysis (qPCR), and Kaplan-Meier Plotter analysis were performed to analyze the expression and prognosis of uc.243 in ovarian carcinoma. Cytotoxicity assay and Annexin V assay were performed to analyze the function of uc.243 in cisplatin resistance in ovarian cancer cells. RNA pull-down and qPCR experiments were performed to explore the molecular mechanism of uc.243 enhancing cisplatin resistance in ovarian cancer cells. Results: Herein, we found that uc.243 was remarkably upregulated and correlated with patient survival in chemoresistance ovarian cancer patients compared with chemo-sensitive ovarian cancer. Functional experiment displayed that uc.243 induced cisplatin resistance on ovarian cancer cells by facilitating the efflux of cisplatin (CDDP); but inhibiting the expression of uc.243 significantly reverses this function. Mechanistically, uc.243 can inhibit the binding of RNA binding protein DGCR8 microprocessor complex subunit to pri-miR-155, thereby inhibiting the cleavage of pri-miR-155 and decrease in mature miR-155, subsequently upregulates the expression of ATP binding cassette subfamily B member (ABCB1, ABCC2). Conclusion: Our research findings indicate that uc.243 can induce chemotherapy resistance in ovarian cancer, suggesting that it may become a new prognostic biomarker for malignant ovarian cancer.Keywords
The incidence rate of ovarian cancer is the third highest among malignant tumors of female reproductive organs, and the mortality rate is the first among malignant tumors of reproductive organs [1,2]. According to the American Cancer Society, there are 25520 new cases and 16090 deaths of ovarian cancer each year. The incidence rate of ovarian cancer in women is 1/70 (1.4%). Usually, most ovarian cancer patients are diagnosed in the middle to late stage [3]. The standard clinical treatment for advanced ovarian cancer patients is tumor cell reduction surgery combined with platinum-based chemotherapy [4]. Although 2/3 of tumor patients exhibit clinical complete remission (CCR) in the combination therapy regimen, up to 75% of patients experience recurrence within 5 years which leads to poor prognosis [5].
Long non-coding RNAs (lncRNA) are a class of non-coding RNAs with a length of more than 200 nucleotides. Due to the lack of an open reading frame, lncRNAs cannot encode proteins. LncRNA was previously thought to be a “noise” generated during the transcription process and did not have biological functions [6]. With the emergence and application of high-throughput sequencing, lncRNA has gradually attracted the attention of researchers. Studies have proved that lncRNA can interact with chromatin, protein, and RNA in the nucleus or cytoplasm, and participate in epigenetic, transcriptional, and post-transcriptional gene expression regulation [7,8]. It plays a role in many biological behaviors of tumors such as proliferation, invasion, migration, and drug resistance [9,10]. A large amount of evidence has shown that LncRNAs play an important role not only in various malignant tumors such as cervical cancer [11], ovarian cancer [12], and hepatocellular carcinoma [13] but also in other human diseases [14,15].
Ultraconserved non-coding RNA (ucRNA) is a new type of long noncoding RNA (lncRNA) discovered in recent years [10,11]. Gill and other researchers used comparative genomics to discover the existence of 481 super (completely) conserved (100%) regions (all >200 bp in length) in the genomes of humans, mice, and rats, named ultraconserved non-coding RNA (ucRNA) [16]. Subsequently, Liz and colleagues demonstrated that ucRNA can regulate the expression of mature miRNAs by binding and inhibiting the cleavage of pri-miRNA, thereby participating in the interaction network ncRNA-ncRNA [11]. UcRNA disorder is associated with various human diseases, such as epithelial differentiation, acute lymphoblastic leukemia, and tumors [17–19]. Therefore, in-depth and systematic research on the biological functions, molecular mechanisms, and clinical significance of ucRNA not only provides a new scientific basis for the occurrence and development of human diseases, but also provides new targets for disease prevention, diagnosis, and treatment.
Chemotherapy, such as cisplatin, is still one of the most effective methods for treating tumors [20]. Tumor patients often exhibit exciting therapeutic effects in the early stages of chemotherapy. However, the subsequent emergence of tumor chemotherapy resistance/drug resistance seriously affects the treatment effect. Clinical manifestations include a high recurrence rate of tumors and a high mortality rate of tumor patients [21–23]. It is reported that approximately 25% of ovarian cancer patients experience recurrence within 6 months; More than 90% of malignant ovarian cancer patients will relapse within 2 years, and the 5-year survival rate is only about 31% [24,25]. Therefore, elucidating the molecular mechanisms of tumor chemotherapy resistance/drug resistance is of great significance for increasing the efficacy of tumors and the survival of tumor patients. Numerous studies have shown that tumors can develop tolerance to cisplatin through various mechanisms and molecules, for example, upregulation of apoptosis-related genes such as BCL2-like 1 (Bcl-xL) [26]. Furthermore, upregulating genes such as ABCB1, ABCC2, and glutamate-cysteine ligase modifier subunit (GCLM) in cancer cells have been reported to promote drug metabolism and pump, and overexpression of RAD50 double-strand break repair protein (RAD50), MRE11A homolog A, double-strand break repair nuclease (MRE11A), and DNA ligase 4 (LIG4) also play important role in enhancing cell viability [27–29].
In our study, we aim to elucidate the function and detailed mechanism of ultraconserved RNA 243 (uc.243) in ovarian cancer chemoresistance, and the identification of effective strategies to overcome drug resistance will have a significant clinical impact on the disease.
Clinical ovarian cancer tissues were collected from 2018 to 2023 and were immediately stored in a liquid nitrogen environment. Moreover, 58 ovarian cancer patients who underwent platinum-based chemotherapy were enrolled in this study, including 27 chemosensitive and 31 chemoresistant patients. All the participants provided informed consent and the study design adhered to the declaration of Helsinki. Furthermore, this study was approved by the Institutional Research Ethics Committee of the Boai Hospital of Zhongshan Affiliated to Southern Medical University (Ethical approval number: KY-2023-011-01).
A2780 (Catalogue No. 93112519) and A2780/cis cell line (Cisplatin-resistant) (Catalogue No. 93112517) were both purchased from the European Collection of Authenticated Cell Cultures (ECACC). A2780/cis was grown in RPMI 1640 + 2 mM Glutamine (56-85-9, Sigma, Saint Louis, MO, USA) + 1 µM cisplatin (CAS 15663-27-1, Sigma, Saint Louis, MO, USA) + 10% Fetal Bovine Serum (FBS, Invitrogen, 10099141, Carlsbad, CA, USA). A2780 was grown in RPMI 1640 supplemented with 10% fetal bovine serum and was in a 5% CO2 atmosphere in a humidified incubator. Short tandem repeat (STR) fingerprinting has been performed.
Plasmid construction and siRNA transfection
The construction vector of ABCB1 and ABCC2 high-expression plasmids is pSIN-EF2-Puro. The PCR primer for plasmid construction were: ABCB1 forward: 5′-atgagtgtcaacttgcaagg-3′; ABCB1 reverse: 5′-tggaacaaagcgccagtga-3′. ABCC2 forward: 5′-atgctggagaagttctgc-3′; ABCC2 reverse 5′- agatcacacaaaccctga-3′. The siRNA sequences for ABCB1 were: cagatgatgtctccaagattaat. for ABCC2 were: atgacatcttcacgtttgtgagt. All plasmids and siRNA transfections in this project were performed using the Lipofectamine 3000 Reagent (L3000015, Invitrogen, CA, USA). We strictly follow the manufacturer’s instructions for the transfection process. Stable cell lines overexpressing uc.243 or miR-155 or repressing uc.243 or miR-155 were selected for 10 days with 0.5 μg/mL puromycin 48 h after infection. The primers for amplifying uc.243 were: forward: 5′-ACAATAAACTTGTACTGGTA-3′;overexpressing uc.243 reverse: 5′-GAAAATAGAATATGAATT-3′; inhibition of uc.243 forward: 5′-atgagtgtcaacttgcaagg-3′; inhibition of uc.243 reverse: 5′-tggaacaaagcgccagtga-3′; The primers for amplifying miR-155 were: up: 5′-TAGGCTTCATCGCCTTGTATCAAGAT-3′; dn: 5′-TACTTAAACCGTCCCATGGTTAAAC-3′.
RNA extraction and quantitative real-time PCR analysis (qPCR)
1 × 106 cells were added with 1 mL Trizol (10296028CN, Invitrogen, USA), shaken at room temperature for 3–5 min, and allowed to stand for 10 min. Then 250 uL of chloroform was added, shaken, and allowed to stand for 15 min. The above solution was centrifuged at 4°C, 12000 rpm for 15 min (Eppendorf, 5427R. USA). The supernatant was reserved and 500 uL of isopropanol was added to the supernatant, shaken, and allowed to stand at room temperature for 15 min. The above solution was centrifuged at 4°C, 12000 rpm for 15 min, the supernatant was removed and 1 mL of 75% ethanol was added to wash the precipitate (RNA). Then the above solution was centrifuged at 44°C, 10000 rpm for 10 min, the supernatant was removed, and the RNA was dissolved with 50 μL DEPC water, used immediately, or stored at −80°C in the refrigerator. Quantitative real-time PCR analysis was performed by using step SYBR® Green RT-qPCR with MMLV & hot-start Taq DNA Polymerase (QR0100, Sigma-Aldrich, Germany). Primers(human) used for PCR are listed following: uc.243-F: 5′-cgcatgtgggcgatttctaga-3′, uc.243-R: 5′ caagcatcaagaccgaagggga 3′; miR-155-F: 5′-gccgcggttaatgctaattgtg-3′, miR-155-R: 5′ cagtgcagggtccgaggt 3′; ABCA9-F: 5′-caagagacgcatgagcgtgggtcag-3′, ABCA9-R: 5′ ttaaactatctacacgtcc′; ABCB1-F: 5′-taggtgctgctttcctgctg-3′, ABCB1-R: 5′ cctcttgatttgcaaatgttca 3′; ABCB11-F: 5′-cagccctctcattgggattggagcag-3′, ABCB11-R: 5′ ttctaattccccaacgctgggc3′; ABCC2-F: 5′-aagccaagatgcccttgtcctgg-3′, ABCC2-R: 5′ tcacttgcaaaggagatcag 3′; ABCD2- F: 5′ - aatctcttacggaggatattat-3′, ABCD2-R: aaaagcctgttctcccaaat′.
RNA pull-down assay was performed by using Pierce™ Magnetic RNA Protein Pull Down Kit (Thermo Fisher, 20164, USA) which was purchased from Thermo Scientific. The specific steps for RNA pull-down are as follows: collect cells (2 × 107), digest them, lyse them with Lysis Buffer, add protease inhibitors and react on ice for 30 min, shake for 30 s every 10 min, centrifuge the supernatant, and measure the protein concentration. After protein collection, immediately store at −40°C. Then, RNA containing desulfurized biotin and Beads containing streptavidin were incubated on a shaking table at 4°C for 3–4 h for binding. Then combine Protein lysates with RNA and incubate overnight at 4°C. Finally, wash and extract the RNA binding protein complex, and collect the supernatant for Western Blot assay.
Western blot assay was performed according to the following steps: total cell protein extraction; BCA method for determining protein content; SDS-PAGE gel electrophoresis; Transfer of the transfer tank to transfer the protein onto the PVDF membrane; Protein immunoassay, using ECL luminescent solution and LAS-4000 mini luminescent imaging analyzer to detect results. In our study, we used anti-cleaved caspase 3antibodies (Abcam, ab32042, 1:500), anti-GAPDH antibodies (Abcam, ab8245, 1:3000), anti-DGCR8 antibodies (Abcam, ab191875, 1:1000), anti-α-tubulin antibody (Abcam, ab7291, Cambridge, MA, USA) for blotting membranes.
Cytotoxicity assay was determined using the (50% inhibitory concentration IC50 assay. Cells were treated with different concentrations of cisplatin (0-100 μM), added 10 μL 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (MTT, 0.15%, 298-93-1, Sigma, Saint Louis, MO, USA) to each hole with a final concentration of 0.5 mg/mL. Incubated the cells at 37°C, 5% CO2 for 4 h. Subsequently all the supernatant from the well to prevent monolayer cell rupture. Added 100 μL formazan solubilization solution to each hole. Placed the board on an oscillator (HZ-9613Y, Changzhou Jintan District Jingda Instrument Manufacturing Co., Ltd., China) and shake for 10 min at 300 rpm/min to dissolve all the impurities. Processed and analyzed the results using). GraphPad Prism (Prism 9, GraphPad Software, USA).
Fluoresceine isothiocyanate (FITC) Annexin V Apoptosis Detection Kit I (Yeasen Biotechnology (Shanghai) Co., Ltd., China; 40306ES50) was used for the evaluation of apoptosis. Prepare cell suspension and count 1 × 106; Resuspension cells with 500 μL binding buffer (as many staining channels as needed, such as in this experiment with two channels, one tube for single staining FITC or Propidium iodide (PI)); Add 5 μL Annexin V-FITC and mix well, then add 5 μL PI and mix well; React at room temperature and avoid light for 15 min, and mix gently during this time; Pass the cells through a 200 mesh filter (or individually packaged 40 μm cell filter), added to a flow tube and placed on ice in a dark place.
High-performance liquid chromatography (HPLC) for the detection of cisplatin
Our project uses high-performance liquid chromatography (LC-20AT, Japan Shimadzu) to detect the concentration of cisplatin in cells. After treatment with cisplatin, tumor cells were washed three times with PBS. The supernatant was removed and 0.5 mL of distilled water was added. The cells were repeatedly frozen and thawed to rupture, mixed well, and then centrifuged for 30 min using a high-speed centrifuge (10000 r/min) (Eppendorf, 5427R. USA). The supernatant was ready for HPLC analysis. The HPLC conditions were carried out using a Zorhax-ODSC-18 column, with a column temperature of 40°C, a mobile phase of methanol and water (1:1, V/V), and a flow rate of 0.7 mL/min detector. SPD-2AS UVdetector. Detection wavelength: 254 nm. Range: 0.005.
Student t test and IBM SPSS 21.0 (IBM Corp., Chicago, IL, USA) statistical software was used in our study for data analysis. *p < 0.05, was supposed to have statistical significance.
uc.243 overexpression correlates with cisplatin resistance and poor prognosis in ovarian cancer
To screen for ucRNAs that play a key role in cisplatin resistance in ovarian cancer, we performed the Real-time PCR analysis and found that the expression level of uc.243 was significantly higher in platinum-resistant ovarian cancer tissue (n = 31) than in platinum-sensitive ovarian cancer tissue (n = 27) (Fig. 1A). Kaplan-Meier Plotter analysis was performed and found that uc.243 overexpression shows shorter overall survival time and progression-free survival for platin-resistant ovarian cancer patients (Fig. 1B).
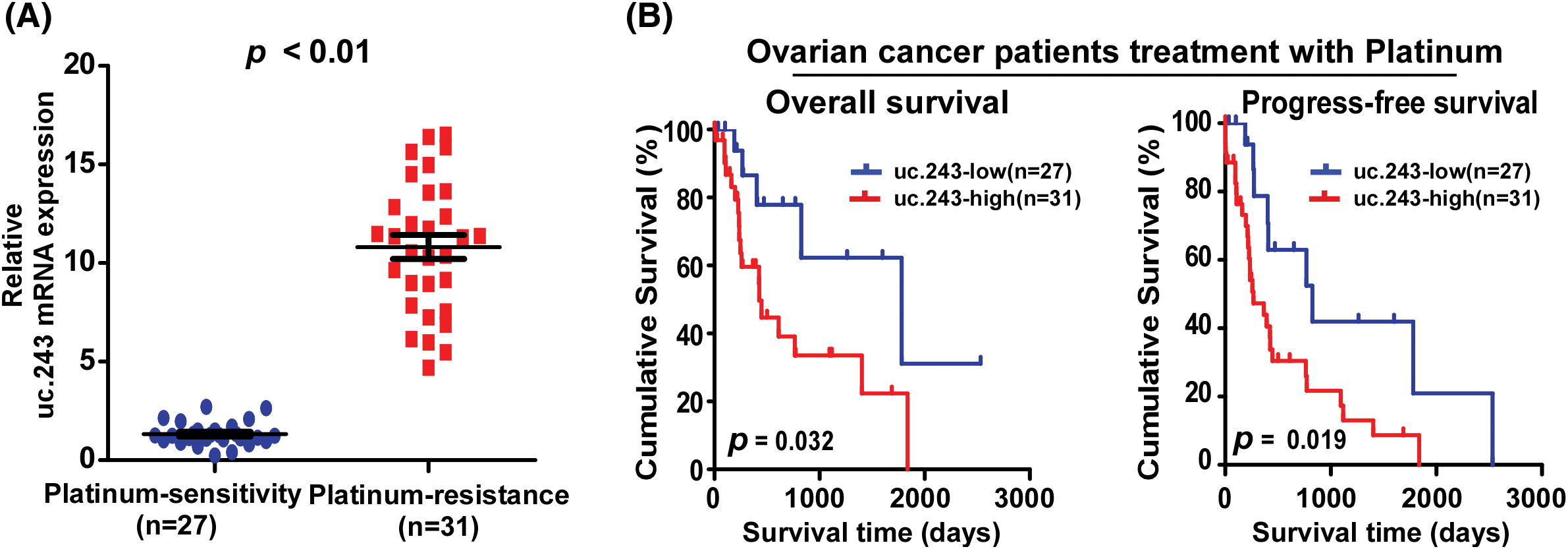
Figure 1: Uc.243 overexpression correlates with cisplatin resistance and poor prognosis in ovarian cancer. (A) mRNA exoression of uc.243 in ovarian cancer tissues. (B) Survival analysis of uc.243 expression in ovarian cancer patients.
Uc.243 amplification correlates with CDDP resistance and poor prognosis in ovarian cancer
Next, we investigate the specific reasons for the increased expression of uc.243 in CDDP-tolerant ovarian cancer. We analyzed ovarian cancer samples from the TCGA database, where the uc.243 locus (chromosome 8 q24.21) showed a highly amplified region (Fig. 2A). Furthermore, the amplification of uc.243 was negatively correlated with overall survival and progression-free survival in ovarian cancer (Fig. 2B). GSEA revealed that uc.243 amplification had a positive correlation with cisplatin resistance, suggesting that amplification of uc.243 may be a potential inducing factors for ovarian cancer CDDP resistance (Fig. 2C).
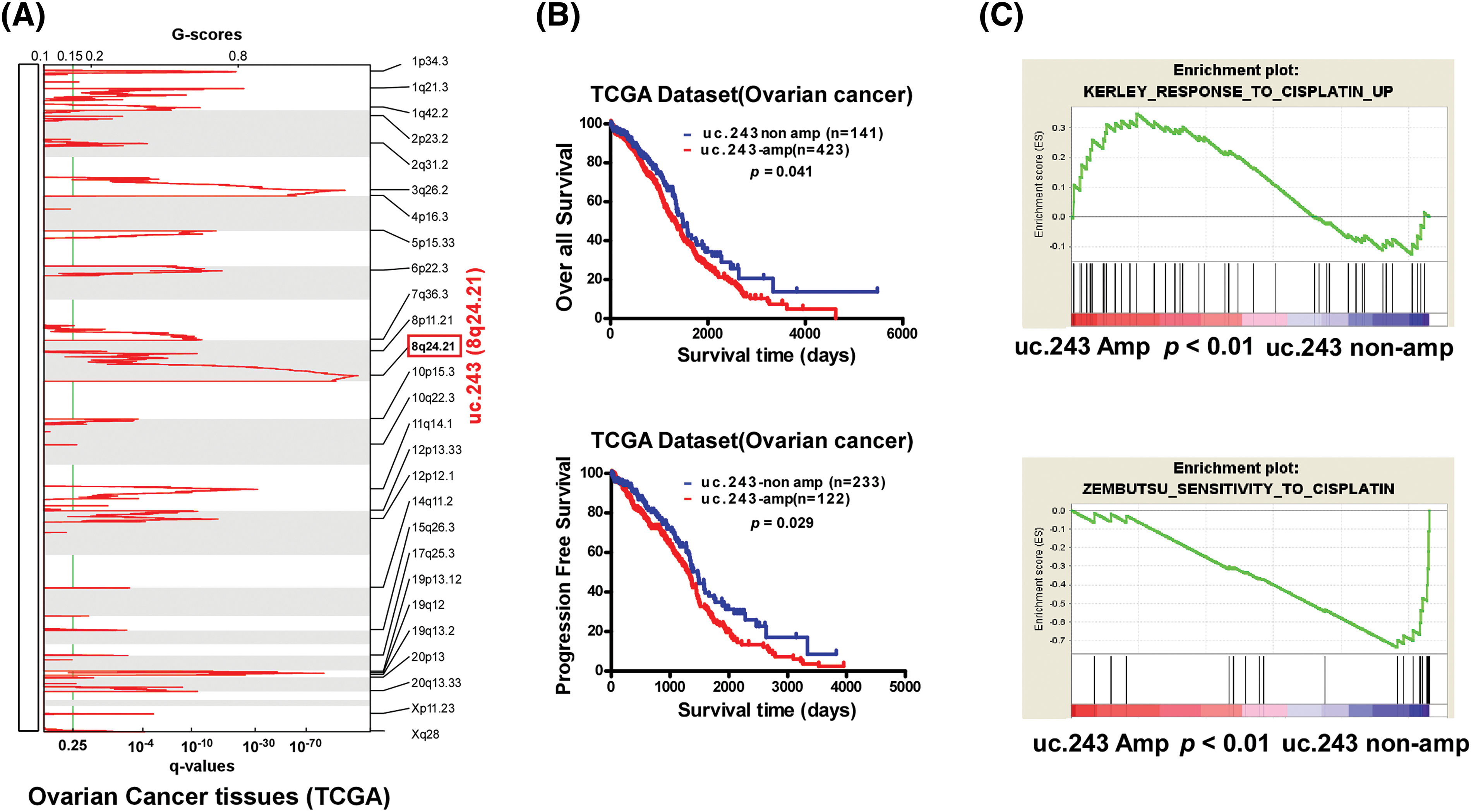
Figure 2: uc.243 amplification correlates with CDDP resistance and poor prognosis in ovarian cancer. (A) Copy number analysis of uc.243 in ovarian cancer in TCGA dataset. (B) Survival analysis of uc.243 in non-amplification and amplification ovarian cancer patients. (C) GSEA analysis of the correlation between uc.243 amplification and cisplatin resistance in the TCGA dataset.
uc.243 promotes cisplatin resistance of ovarian cancer in vitro
Furthermore, A2780 (cisplatin sensitive cell line) stably overexpressed uc.243 and A2780/cis (cisplatin resistance cell line) inhibition of uc.243 were established (Fig. 3A). Firstly, we found that overexpression of uc.243 (IC50: 33.71 μM) significantly improved the IC50 value of cisplatin compared with the vector (IC50: 7.18 μM) in A2780 cells (Fig. 3B). However, inhibition of uc.243 (IC50: 3.35 μM) reduce the IC50 value of cisplatin compared with the control (IC50: 28.78 μM) in A2780/cis cells (Fig. 3B). Secondly, by performing the Annexin V assay we demonstrate that overexpression of uc.243 decreases the ratio of apoptotic cells but inhibiting uc.243 expression increases the ratio of apoptotic cells when treated with CDDP (Fig. 3C). Furthermore, uc.243 overexpression significantly decreases the level of apoptosis-related proteins: cleaved caspase 3 but increases in uc.243 inhibition cells (Fig. 3D).
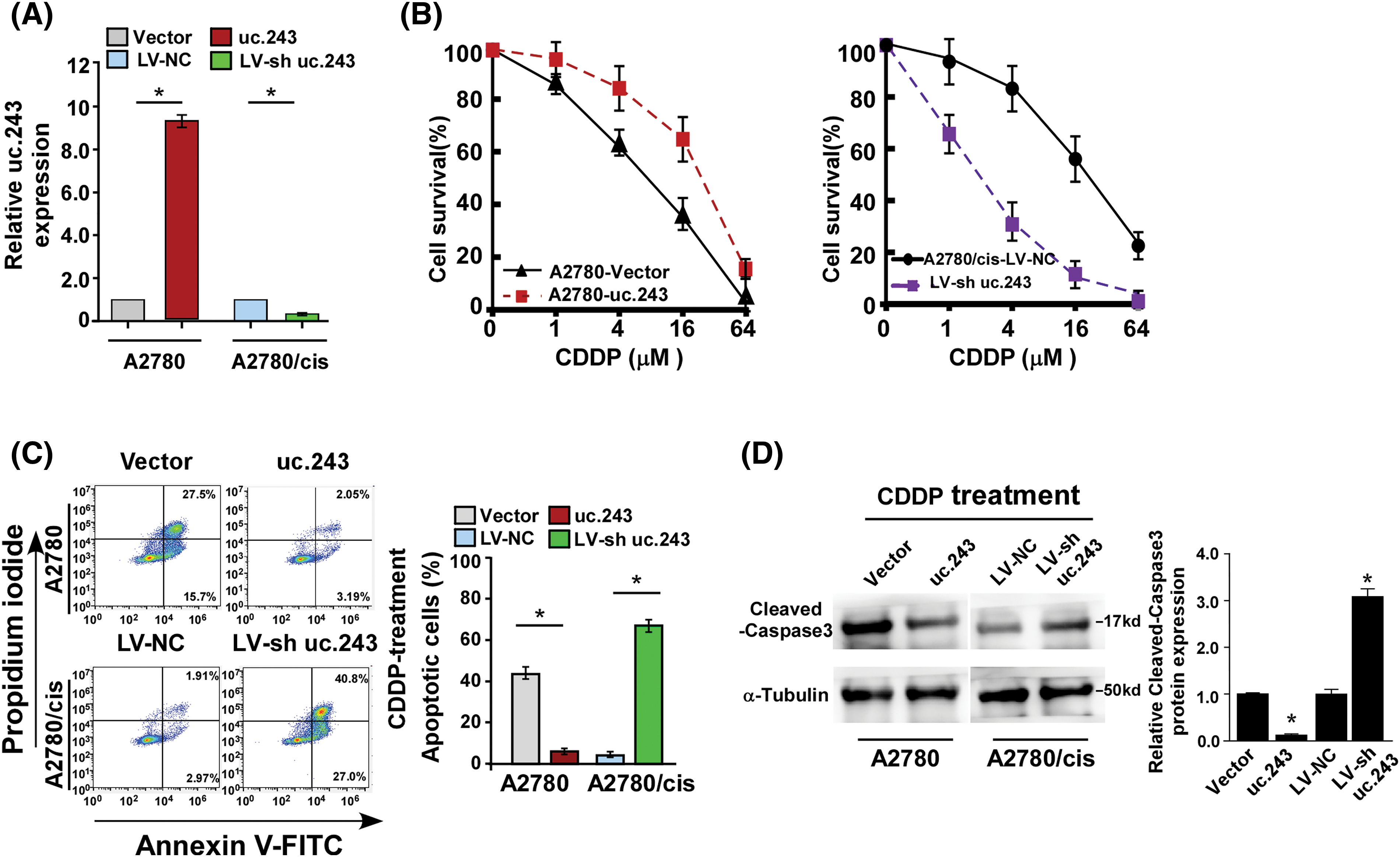
Figure 3: uc.243 promotes cisplatin resistance of ovarian cancer in vitro (A) mRNA expression of uc.243 expression in indicate cells, n = 3 (B) Detection of CDDP IC50 in the indicated cells. *p < 0.05, n = 3 (C) Detection of apoptosis ratio in indicated cells. *p < 0.05, n = 3 (D) Protein expression detection of cleaved caspase3 in the indicated cells.
uc.243 interacts with pri-miR-155 and facilitates efflux of CDDP in ovarian cancer cells
To clarify the molecular mechanism of high expression of uc.243 enhancing cisplatin resistance in ovarian cancer cells, we predict that pri-miR-155 may be associated with uc.243 (Fig. 4A). Furthermore, a combination of uc.243 and pri-miR-155 was demonstrated through RNA pull-down/qPCR experiments (Fig. 4B). We found that overexpression of miR-155 significantly increased the concentration of cisplatin in A2780/uc.243 cells and decreased the rate of cisplatin pumping out by A2780/uc.243 cells (Figs. 4C, 4D). However, inhibition of miR-155 significantly decreased the concentration of cisplatin in A2780/cis-sh-uc.243 cells and increased the rate of cisplatin pumping out by A2780/cis-sh-uc.243 cells (Figs. 4C–4E). Furthermore, through multiple bioinformatics software analyses (RNA22, Target scan, miRanda), we found that miR-155 may act on multiple genes involved in cisplatin metabolism/pumping, such as ABCA9, ABCB1, ABCB11, ABCC2, ABCD2 (Fig. 4F). However, RNA Immunoprecision (RIP) results showed that miR-155 only directly interacts with ABCB1 and ABCC2 in ovarian cancer cells (Fig. 4G). The above results indicate that miR-155 can inhibit the pumping of cisplatin drugs in uc.243 overexpression ovarian cancer cells.
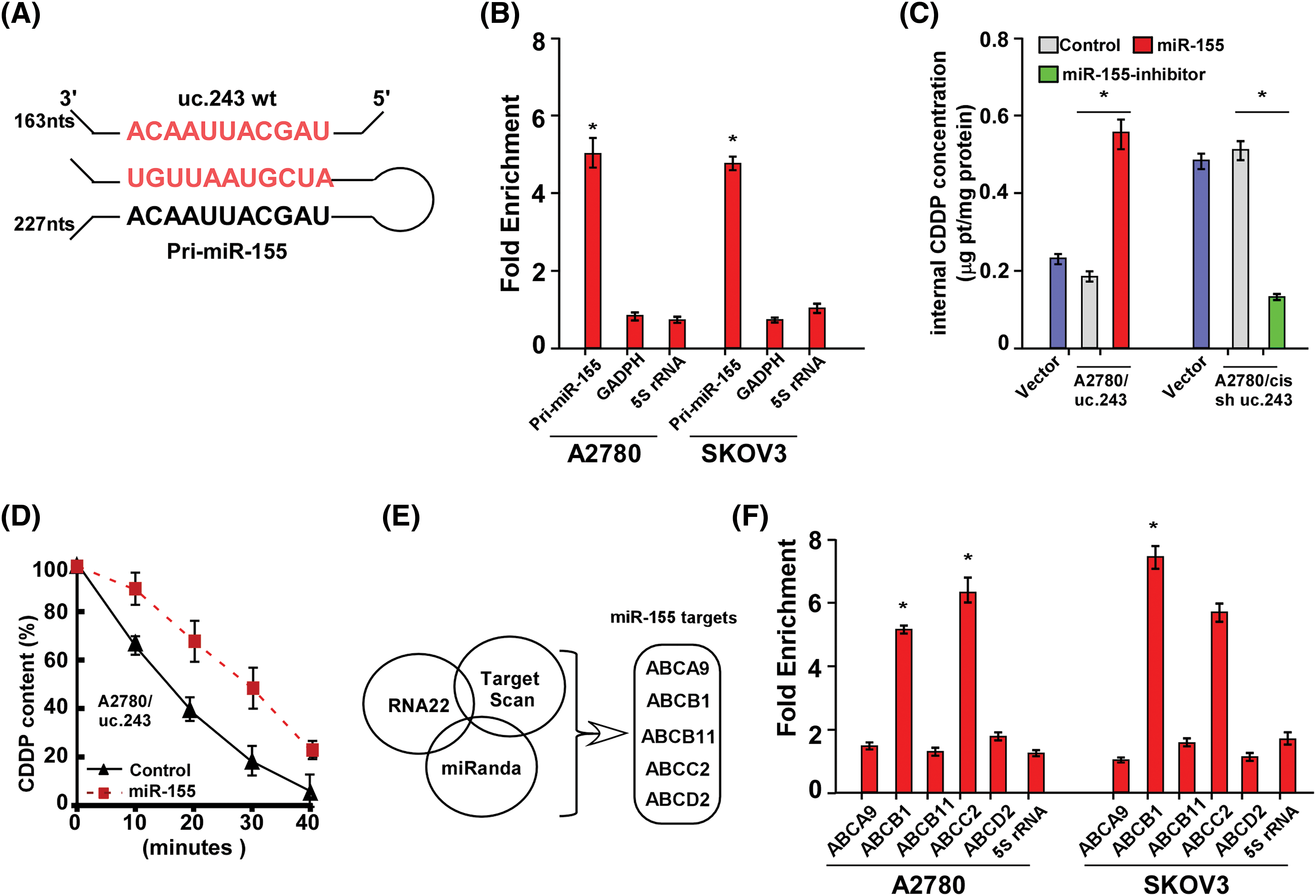
Figure 4: Uc.243 interacts with pri-miR-155 and facilitates efflux of CDDP in ovarian cancer cells. (A) Schematic diagram of pairing between uc.243 and pri-miR-155. (B) RNA pull-down/qPCR analysis of the association between uc.243 and pri-miR-155 in ovarian cancer cells, n = 3.(C) mRNA expression of miR-155. (D) Internal CDDP concentration in indicated cells. (E) CDDP content at different times in indicated cells, n = 3. (F, G) Multiple analysis software predicts the interaction between miR-155 and several ATP-binding cassette subfamily B members. *p < 0.05, n = 3.
ABCB1 and ABCC2 are required for uc.243-induced CDDP resistance on ovarian cancer cells
Next, we investigated whether uc.243 mediated ovarian cancer CDDP resistance through ABCB1 and ABCC2. IC50 assay shows that transfection of ABCB1 or ABCC2 siRNA in uc.243 overexpression-ovarian cancer cells significantly decreases the IC50 value (Fig. 5A). However, overexpression of ABCB1 or ABCC2 open reading frame (ORF) in uc.243-shRNA-ovarian cancer cells significantly increases the IC50 value of ovarian cancer cells (Fig. 5B). Consistently, the Annexin V assay was performed and showed that replenishment of ABCB1 or ABCC2 in uc.243 cells increased the apoptosis ratio on ovarian cancer cell lines (Fig. 5C).
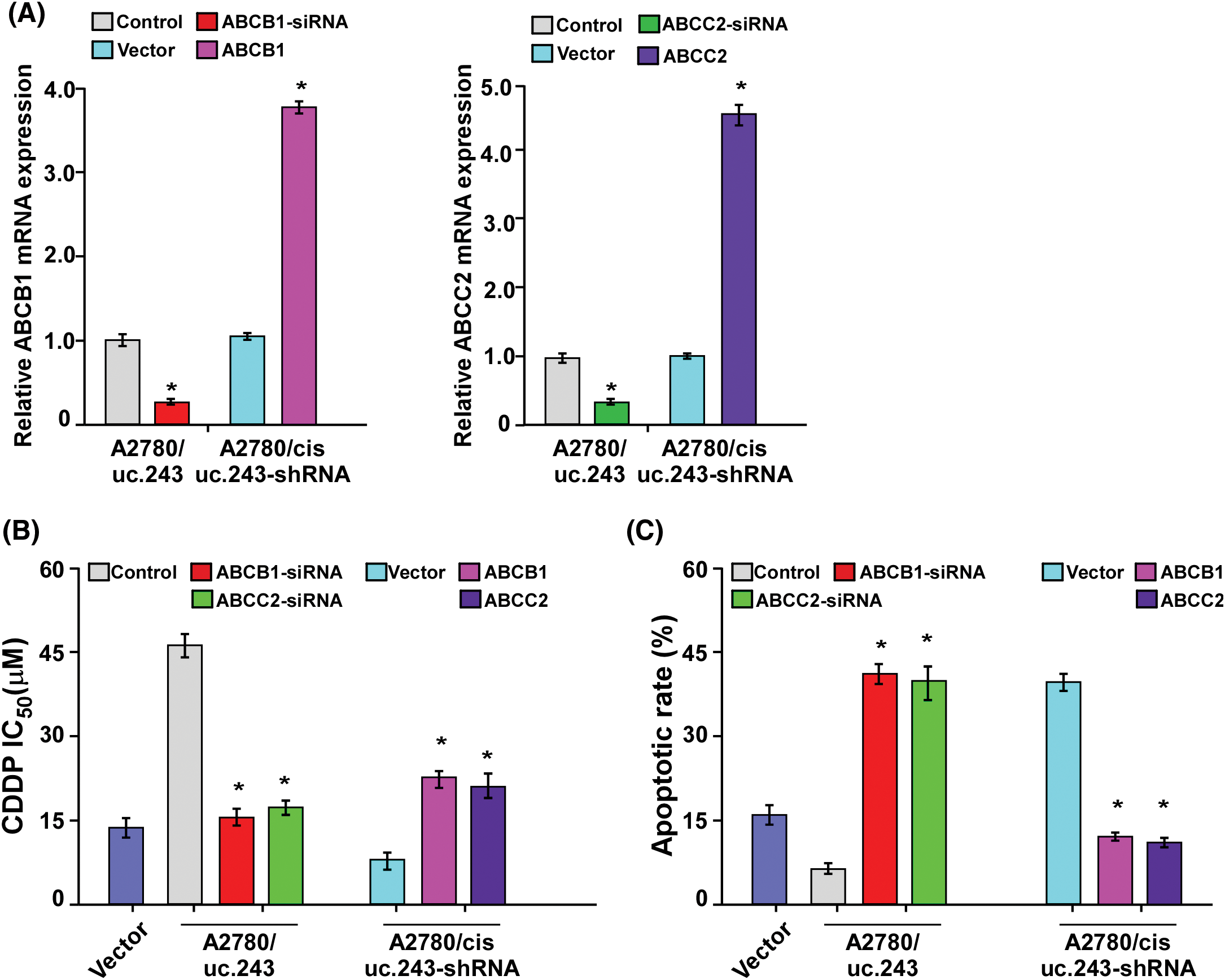
Figure 5: ABCB1 and ABCC2 are required for uc. 243-induced CDDP resistance on ovarian cancer cells. (A) Expression of ABCB1 and ABCC2 in indicated cells, n = 3. (B) Detection of CDDP IC50 in the indicated cells. *p < 0.05. n = 3. (C) Detection of apoptosis ratio in the indicated cells treated with CDDP for 24 h. *p < 0.05, n=3.
uc.243 inhibits the generation of mature miR-155 in ovarian cancer cells
Liz et al. reported that ultra-conservative noncoding RNA can bind to and inhibit pri-microRNA cleavage in the nucleus, subsequently hindering miRNA maturation [11]. Our experiment showed that high expression of uc.243 can inhibit the binding of RNA binding protein DGCR8 to pri-miR-155, thereby inhibiting the cleavage of pri-miR-155 by drosha enzyme and showing a decrease in mature miR-155 (Figs. 6A, 6B). The above results indicate that the super-conserved RNA uc.243 binds and inhibits the generation of mature miR-155 by pri-iR-155.
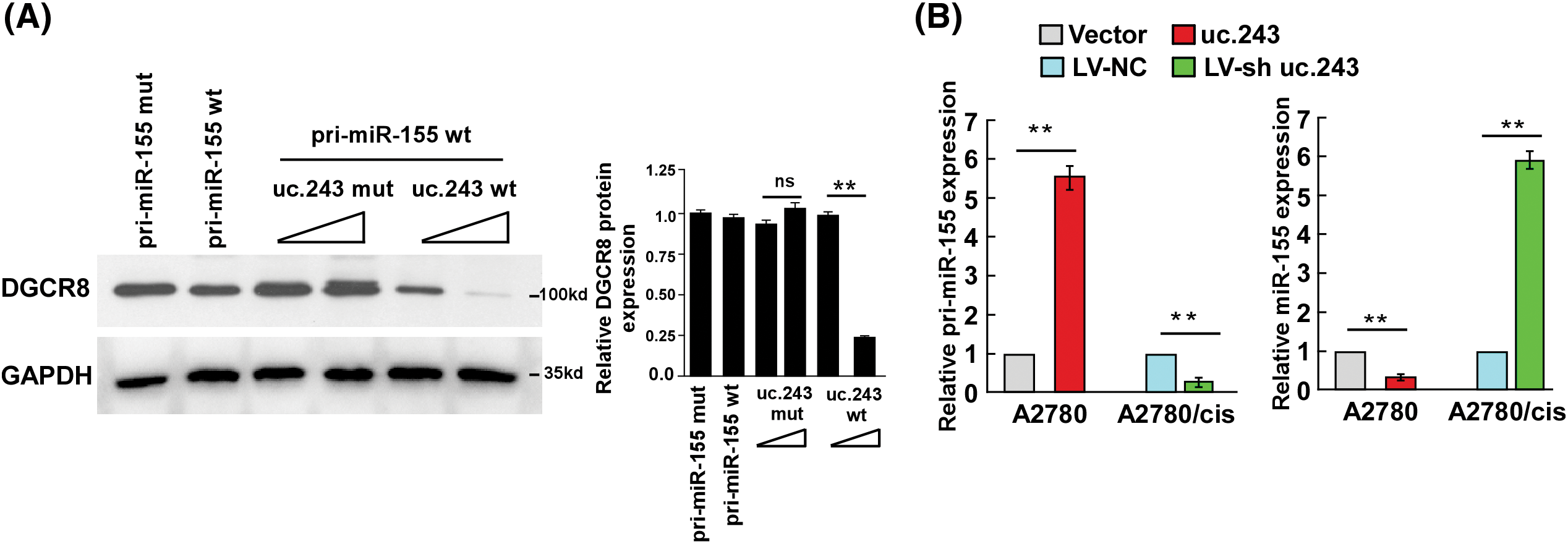
Figure 6: Uc.243 inhibits the generation of mature miR-155 in ovarian cancer cells. (A) Protein expression detection of DGCR8 in the indicated cells. (B) Relative expression of pri-miR-155 and miR-155 in the indicated cells. **p < 0.01, n = 3, ns: no significance.
Herein, we reported that overexpression of uc.243 was found in platinum-resistant ovarian cancer tissue, and uc.243 overexpression was closely negatively correlated with survival prognosis for platin-resistant ovarian cancer patients. Furthermore, overexpression of uc.243 in A2780 cell lines dramatically promoted, but inhibition of uc.243 in A2780/cis cell lines reduced, the chemoresistant capability of ovarian cancer via the promotion of chemodrug efflux in ovarian cancer cells. Mechanistically, we reported that uc.243 can inhibit the binding of RNA binding protein DGCR8 to pri-miR-155, thereby decreasing the levels of mature miR-155, subsequently upregulating the expression of multiple drug transporters (ABCB1, ABCC2). These findings demonstrated the tumor oncogenic role of uc.243 and provided new insights into the mechanism of chemoresistance in ovarian cancer.
Noncoding RNA (ncRNA) is the type of RNA that can be transcribed from the genome but cannot be translated into proteins [30]. Noncoding RNAs play a wide and pivotal role in the malignant development of major human diseases such as malignant tumors, cardiovascular diseases, nervous system diseases, and metabolic endocrine disorders [31–33]. In our study, we found that the overexpression of noncoding RNA uc.243 promotes the efflux of CDDP by inhibiting the binding of RNA binding protein DGCR8 to pri miR-155, thereby inhibiting the cleavage of pri miR-155 and reducing mature miR-155. This upregulates ATP-binding cassette B subfamily members (ABCB1, ABCC2) expression, thereby inducing cisplatin resistance in ovarian cancer cells. The results will provide clinical treatment targets and novel molecular mechanisms for ovarian cancer chemo-resistant.
Although several reports indicate that multiple drug transporter modulators, such as verapamil, VX-710, and XR9576, have been identified and shown to be effective in controlling malignant tumor progression [34–36], however, the clinical trials conducted so far have been disappointing due to the high toxicity of these drugs to the human body, low tolerance doses, and serious side effects [37,38]. Therefore, illuminating the mechanism for the upregulation of drug transporters in tumors would provide new insights for developing therapeutic interventions for cancer. In the current study, we found that uc.243, a novel non-coding RNA was significantly upregulated in chemoresistant ovarian cancer tissues. Enhanced expression of uc.243 significantly enhanced the drug-resistant capability of ovarian cancer by promoting chemodrug efflux. Furthermore, overexpression of uc.243 simultaneous upregulation of multiple drug transporters, such as ABCB1 and ABCC2 by decreasing the levels of mature miR-155 in ovarian cancer Thus, our finding represents a novel mechanism for cancer chemoresistance and suggests that uc.243 levels may be useful determinants for evaluating chemotherapy outcomes for ovarian cancer patients.
Currently, for the clinical treatment of ovarian cancer, failure of conventional chemotherapy is the main reason for significantly reducing the clinical efficacy of chemotherapy drugs in cancer treatment and leading to poor prognosis [39–41]. More importantly, clinical tumors exhibiting resistance to one drug can lead to resistance to many other drugs. The intrinsic or acquired resistance to chemotherapy can be caused by changes in the transport of anticancer drugs, which are mediated by various efflux pumps/transporters, leading to an enhanced ability of cancer cells to promote the efflux of chemotherapy drugs and reduce the intracellular concentration of anticancer drugs [42,43]. Most drug transporters belong to the efflux pump ABC family, which consists of 7 subfamilies and has 49 functional members in humans, ranging from ABCA to ABCG [44,45]. Multiple ABC members, such as ABCB1, ABCC1, and ABCC2, are markedly overexpressed in tumors and involved in the development of cancer cell chemotherapeutics resistance [46–48]. Herein we demonstrated that uc.243 promoted the accumulation of chemodrugs in tumor cells and via simultaneously direct upregulation of multiple drug transporters. Therefore, our results represent a novel mechanism for the overexpression of multiple drug transporters in cancer, and uc.243 may be a novel biomarker for the clinical treatment of ovarian cancer patients.
Limitations: In our study, we validated the characteristic of high expression of uc.243 conferring chemotherapy resistance to ovarian cancer through in vitro experiments. In addition, we elucidated the molecular mechanism by which uc.243 mediates ovarian cancer resistance through biological experiments. However, our study lacks in vivo experiments to further demonstrate the ability of high expression of uc.243 to confer chemotherapy resistance in ovarian cancer. Moreover, if clinical data is used to analyze the correlation between uc.243 and its downstream regulatory genes in clinical ovarian cancer drug-resistant samples, there will be more conclusive evidence to prove that uc.243 may serve as a biomarker for clinical treatment of ovarian cancer drug resistance.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: Jiali Kang and Shan Jiang performed, designed, and analyzed the data, Shan Jiang and Xiufeng Lin performed experiments, analysis, and interpretation of data, Xinning Li collected the specimens, Yanfei Chen and Shan Jiang wrote the manuscript. All authors reviewed the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included either in this article.
Ethics Approval: This study was approved by the Institutional Review Board of the Boai Hospital of Zhongshan Affiliated to Southern Medical University (Ethical approval number: KY-2023-011-01). Informed consent was obtained from all individual participants included in the study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Penny SM. Ovarian cancer: an overview. Radiol Technol. 2020;91(6):561–75. [Google Scholar] [PubMed]
2. Webb PM, Jordan SJ. Global epidemiology of epithelial ovarian cancer. Nat Rev Clin Oncol. 2024;21(5):389–400. doi:10.1038/s41571-024-00881-3. [Google Scholar] [PubMed] [CrossRef]
3. Roett MA, Evans P. Ovarian cancer: an overview. Am Fam Physician. 2009;80(6):609–16. [Google Scholar] [PubMed]
4. Rooth C. Ovarian cancer: risk factors, treatment and management. Br J Nurs. 2013;22(17):S23–30. [Google Scholar] [PubMed]
5. Policarpo R, Sierksma A, de Strooper B, d’Ydewalle C. From junk to function: lncRNAs in CNS health and disease. Front Mol Neurosci. 2021;14:714768. doi:10.3389/fnmol.2021.714768. [Google Scholar] [PubMed] [CrossRef]
6. Herman AB, Tsitsipatis D, Gorospe M. Integrated lncRNA function upon genomic and epigenomic regulation. Mol Cell. 2022;82(12):2252–66. doi:10.1016/j.molcel.2022.05.027. [Google Scholar] [PubMed] [CrossRef]
7. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–5. doi:10.1038/nature20149. [Google Scholar] [PubMed] [CrossRef]
8. Lu L, Huang JJ, Mo JT, Da X, Li QX, Fan M, et al. Exosomal lncRNA TUG1 from cancer-associated fibroblasts promotes liver cancer cell migration, invasion, and glycolysis by regulating the miR-524-5p/SIX1 axis. Cell Mol Biol Lett. 2022;27(1):17. doi:10.1186/s11658-022-00309-9. [Google Scholar] [PubMed] [CrossRef]
9. Singh D, Assaraf YG, Gacche RN. Long non-coding RNA mediated drug resistance in breast cancer. Drug Resist Updat. 2022;63:100851. doi:10.1016/j.drup.2022.100851. [Google Scholar] [PubMed] [CrossRef]
10. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74. doi:10.1038/nrg3074. [Google Scholar] [PubMed] [CrossRef]
11. Liz J, Portela A, Soler M, Gomez A, Ling H, Michlewski G, et al. Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Mol Cell. 2014;55(1):138–47. doi:10.1016/j.molcel.2014.05.005. [Google Scholar] [PubMed] [CrossRef]
12. Zhou F, Xu X, Wei W, Chen X, Sun L. Regulations of exosomal-transmitted AFAP1-AS1 LncRNA on Ovarian cancer cell migration and invasion. Discov Med. 2023;35(178):877–86. doi:10.24976/Discov.Med.202335178.83. [Google Scholar] [PubMed] [CrossRef]
13. Farzaneh M, Ghasemian M, Ghaedrahmati F, Poodineh J, Najafi S, Masoodi T, et al. Functional roles of lncRNA-TUG1 in hepatocellular carcinoma. Life Sci. 2022;308(3):120974. doi:10.1016/j.lfs.2022.120974. [Google Scholar] [PubMed] [CrossRef]
14. Ahmad M, Weiswald LB, Poulain L, Denoyelle C, Meryet-Figuiere M. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: cytoskeleton and ECM crosstalk. J Exp Clin Cancer Res. 2023;42(1):173. doi:10.1186/s13046-023-02741-x. [Google Scholar] [PubMed] [CrossRef]
15. Eptaminitaki GC, Wolff N, Stellas D, Sifakis K, Baritaki S. Long non-coding RNAs (lncRNAs) in response and resistance to cancer immunosurveillance and immunotherapy. Cells. 2021;10(12):3313. doi:10.3390/cells10123313. [Google Scholar] [PubMed] [CrossRef]
16. Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, et al. Ultraconserved elements in the human genome. Science. 2004;304(5675):1321–5. doi:10.1126/science.1098119. [Google Scholar] [PubMed] [CrossRef]
17. das Chagas PF, de Sousa GR, Kodama MH, de Biagi Junior CAO, Yunes JA, Brandalise SR, et al. Ultraconserved long non-coding RNA uc.112 is highly expressed in childhood T versus B-cell acute lymphoblastic leukemia. Hematol Transfus Cell Ther. 2021;43(1):28–34. doi:10.1016/j.htct.2019.12.003. [Google Scholar] [PubMed] [CrossRef]
18. Panatta E, Lena AM, Mancini M, Smirnov A, Marini A, Delli Ponti R, et al. Long non-coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex. EMBO Rep. 2020;21(3):e46734. doi:10.15252/embr.201846734. [Google Scholar] [PubMed] [CrossRef]
19. Wen HJ, Walsh MP, Yan IK, Takahashi K, Fields A, Patel T. Functional modulation of gene expression by ultraconserved long non-coding RNA TUC338 during growth of human hepatocellular carcinoma. iScience. 2018;2:210–20. doi:10.1016/j.isci.2018.03.004. [Google Scholar] [PubMed] [CrossRef]
20. Holohan C, van Schaeybroeck S, Longley DB, Johnston PG. Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 2013;13(10):714–26. doi:10.1038/nrc3599. [Google Scholar] [PubMed] [CrossRef]
21. Gupta S, Nag S, Aggarwal S, Rauthan A, Warrier N. Maintenance therapy for recurrent epithelial ovarian cancer: current therapies and future perspectives—a review. J Ovarian Res. 2019;12(1):103. doi:10.1186/s13048-019-0579-0. [Google Scholar] [PubMed] [CrossRef]
22. Vaughan S, Coward JI, Bast Jr RC, Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11(10):719–25. doi:10.1038/nrc3144. [Google Scholar] [PubMed] [CrossRef]
23. Pinato DJ, Graham J, Gabra H, Sharma R. Evolving concepts in the management of drug resistant ovarian cancer: dose dense chemotherapy and the reversal of clinical platinum resistance. Cancer Treat Rev. 2013;39(2):153–60. doi:10.1016/j.ctrv.2012.04.004. [Google Scholar] [PubMed] [CrossRef]
24. Lheureux S, Gourley C, Vergote I, Oza AM. Epithelial ovarian cancer. Lancet. 2019;393(10177):1240–53. doi:10.1016/S0140-6736(18)32552-2. [Google Scholar] [PubMed] [CrossRef]
25. Wangpaichitr M, Theodoropoulos G, Nguyen DJM, Wu C, Spector SA, Feun LG, et al. Cisplatin resistance and redox-metabolic vulnerability: a second alteration. Int J Mol Sci. 2021;22(14):7379. doi:10.3390/ijms22147379. [Google Scholar] [PubMed] [CrossRef]
26. Alam M, Mishra R. Bcl-xL expression and regulation in the progression, recurrence, and cisplatin resistance of oral cancer. Life Sci. 2021;280:119705. doi:10.1016/j.lfs.2021.119705. [Google Scholar] [PubMed] [CrossRef]
27. Jelen A, Swiechowski R, Zebrowska-Nawrocka M, Salagacka-Kubiak A, Szmajda-Krygier D, Galecki P, et al. Importance of selected ABCB1 SNPs for the level of severity of depressive symptoms and effectiveness of recurrent depressive disorder therapy. Gene. 2023;851:147021. doi:10.1016/j.gene.2022.147021. [Google Scholar] [PubMed] [CrossRef]
28. Oguro S, Imamura H, Yoshimoto J, Ishizaki Y, Kawasaki S. Liver metastases from gastric cancer represent systemic disease in comparison with those from colorectal cancer. J Hepatobiliary Pancreat Sci. 2016;23(6):324–32. doi:10.1002/jhbp.v23.6. [Google Scholar] [CrossRef]
29. Surowiak P, Materna V, Kaplenko I, Spaczynski M, Dolinska-Krajewska B, Gebarowska E, et al. ABCC2 (MRP2, cMOAT) can be localized in the nuclear membrane of ovarian carcinomas and correlates with resistance to cisplatin and clinical outcome. Clin Cancer Res. 2006;12(23):7149–58. doi:10.1158/1078-0432.CCR-06-0564. [Google Scholar] [PubMed] [CrossRef]
30. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006;15(suppl_1):R17–29. doi:10.1093/hmg/ddl046. [Google Scholar] [PubMed] [CrossRef]
31. Hombach S, Kretz M. Non-coding RNAs: classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi:10.1007/978-3-319-42059-2. [Google Scholar] [CrossRef]
32. Ren H, Wang Q. Non-coding RNA and diabetic kidney disease. DNA Cell Biol. 2021;40(4):553–67. doi:10.1089/dna.2020.5973. [Google Scholar] [PubMed] [CrossRef]
33. Yan H, Bu P. Non-coding RNA in cancer. Essays Biochem. 2021;65(4):625–39. doi:10.1042/EBC20200032. [Google Scholar] [PubMed] [CrossRef]
34. Rives ML, Javitch JA, Wickenden AD. Potentiating SLC transporter activity: emerging drug discovery opportunities. Biochem Pharmacol. 2017;135:1–11. doi:10.1016/j.bcp.2017.02.010. [Google Scholar] [PubMed] [CrossRef]
35. Storelli F, Yin M, Kumar AR, Ladumor MK, Evers R, Chothe PP, et al. The next frontier in ADME science: predicting transporter-based drug disposition, tissue concentrations and drug-drug interactions in humans. Pharmacol Ther. 2022;238:108271. doi:10.1016/j.pharmthera.2022.108271. [Google Scholar] [PubMed] [CrossRef]
36. Choi YH, Yu AM. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr Pharm Des. 2014;20(5):793–807. doi:10.2174/138161282005140214165212. [Google Scholar] [PubMed] [CrossRef]
37. Moran MM. TRP channels as potential drug targets. Annu Rev Pharmacol Toxicol. 2018;58(1):309–30. doi:10.1146/annurev-pharmtox-010617-052832. [Google Scholar] [PubMed] [CrossRef]
38. Kaneko Y, Szallasi A. Transient receptor potential (TRP) channels: a clinical perspective. Br J Pharmacol. 2014;171(10):2474–507. doi:10.1111/bph.12414. [Google Scholar] [PubMed] [CrossRef]
39. Xu YZ, Zhu Y. Conventional chemotherapy and emerging targeted therapy for advanced adrenocortical carcinoma. Anticancer Agents Med Chem. 2013;13(2):248–53. doi:10.2174/1871520611313020010. [Google Scholar] [PubMed] [CrossRef]
40. Parmar MK, Ledermann JA, Colombo N, du Bois A, Delaloye JF, Kristensen GB, et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: the ICON4/AGO-OVAR-2.2 trial. Lancet. 2003;361(9375):2099–106. doi:10.1016/S0140-6736(03)13718-X. [Google Scholar] [PubMed] [CrossRef]
41. Fraguas-Sanchez AI, Fernandez-Carballido A, Delie F, Cohen M, Martin-Sabroso C, Mezzanzanica D, et al. Enhancing ovarian cancer conventional chemotherapy through the combination with cannabidiol loaded microparticles. Eur J Pharm Biopharm. 2020;154(234):246–58. doi:10.1016/j.ejpb.2020.07.008. [Google Scholar] [PubMed] [CrossRef]
42. Kopecka J, Trouillas P, Gasparovic AC, Gazzano E, Assaraf YG, Riganti C. Phospholipids and cholesterol: inducers of cancer multidrug resistance and therapeutic targets. Drug Resist Updat. 2020;49:100670. doi:10.1016/j.drup.2019.100670. [Google Scholar] [PubMed] [CrossRef]
43. Rampacci E, Felicetti T, Pietrella D, Sabatini S, Passamonti F. Drug efflux transporters in Staphylococcus pseudintermedius: in silico prediction and characterization of resistance. J Antimicrob Chemother. 2022;77(12):3283–90. doi:10.1186/s13048-019-0579-0. [Google Scholar] [CrossRef]
44. Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, et al. Efflux-mediated antifungal drug resistance. Clin Microbiol Rev. 2009;22(2):291–321. doi:10.1128/CMR.00051-08. [Google Scholar] [PubMed] [CrossRef]
45. Suzuki T, Nishio K, Tanabe S. The MRP family and anticancer drug metabolism. Curr Drug Metab. 2001;2(4):367–77. doi:10.2174/1389200013338289. [Google Scholar] [PubMed] [CrossRef]
46. Mohammad IS, He W, Yin L. Understanding of human ATP binding cassette superfamily and novel multidrug resistance modulators to overcome MDR. Biomed Pharmacother. 2018;100:335–48. doi:10.1016/j.biopha.2018.02.038. [Google Scholar] [PubMed] [CrossRef]
47. Hano M, Tomasova L, Seres M, Pavlikova L, Breier A, Sulova Z. Interplay between P-Glycoprotein expression and resistance to endoplasmic reticulum stressors. Molecules. 2018;23(2):337. doi:10.3390/molecules23020337. [Google Scholar] [PubMed] [CrossRef]
48. Sampath J, Adachi M, Hatse S, Naesens L, Balzarini J, Flatley RM, et al. Role of MRP4 and MRP5 in biology and chemotherapy. AAPS PharmSci. 2002;4(3):14. doi:10.1208/ps040314. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools