 Open Access
Open Access
REVIEW
Modulatory role of plant-derived metabolites on host-microbiota interactions: personalized therapeutics outlook
Department of Biochemistry, Maharshi Dayanand University Rohtak, Rohtak, Haryana, India
* Corresponding Author: NAR SINGH CHAUHAN. Email:
(This article belongs to the Special Issue: Gut Microbiota in Human Health: Exploring the Complex Interplay)
BIOCELL 2024, 48(8), 1127-1143. https://doi.org/10.32604/biocell.2024.051318
Received 02 March 2024; Accepted 10 May 2024; Issue published 02 August 2024
Abstract
A diverse array of microbes in and on the human body constitute the microbiota. These micro-residents continuously interact with the human host through the language of metabolites to dictate the host’s physiology in health and illnesses. Any biotic and abiotic component ensuring a balanced host-microbiota interaction are potential microbiome therapeutic agents to overcome human diseases. Plant metabolites are continually being used to treat various illnesses. These metabolites target the host’s metabolic machinery and host-gut microbiota interactions to overcome human diseases. Despite the paramount therapeutic significance of the factors affecting host-microbiota interactions, a comprehensive overview of the modulatory role of plant-derived metabolites in host-microbiota interactions is lacking. The current review puts an effort into comprehending the role of medicinal plants in gut microbiota modulation to mitigate various human illnesses. It would develop a holistic understanding of host-microbiota interactions and the role of effectors in health and diseases.Graphic Abstract
Keywords
Continuous exposure to microorganisms allowed their settlement in and on the human body, collectively known as human microbiota [1]. The diversity and abundance of these microorganisms vary with human anatomical locations [2]. Among all anatomical locations, the human gut microbiota is highly dense, contributes ~10% of body weight, and extends the protein-coding efficiency of the host by 90% [3]. Despite the abundance, the gut microbiota is represented primarily by Bacteroidetes and Firmicutes, followed by many minor microbial species [4]. Many variables, like host genetics, epigenetics, environmental, dietary preferences, lifestyle, etc., define the host’s gut microbiota composition [5]. Our understanding of this significant ecosystem’s complexity has been entirely transformed by the capacity to characterize the genetic repertoire of bacterial populations without cultivating each bacterium [6,7]. These gut residents catabolize various dietary constituents and release metabolites such as Short-chain fatty acids (SCFAs), polyamines, tryptophan metabolites, etc., in the surrounding environment, which finally reaches the host biological system through absorption [8]. These metabolites alter the host cellular expression profile and influence host physiology. This metabolic crosstalk between microbiota and host regulates various physiological processes to ensure human health [9]. A pathogen invasion or gut microbiota dysbiosis evokes healthy cross-talk. It induces a host inflammatory response [10], leading to the onset of various illnesses and pathologies, including obesity, inflammatory bowel disease (IBD), autoimmune diseases, allergies, respiratory diseases, and neurological conditions [11,12]. Host-microbiota interactions were a prime target for overcoming gut dysbiosis-derived human illnesses [13]. Understanding the importance of host-microbiota crosstalk, any molecule with potential therapeutic properties towards restoring the microbiota equilibrium is critical.
Consumption of plants and plant metabolites generates microbial metabolites such as SCFAs and bio-transformed plant metabolites [14]. These metabolites include carotenoids, alkaloids, polyphenols, organosulfur, and nitrogen-containing compounds [15]. The subsequent absorption of these metabolites into the bloodstream mediates several host-microbe actions and influences host health [16]. Plant and plant-derived compounds have been utilized for a long time for their therapeutic applications to overcome various illnesses [17]. This might be attributed to plant-derived metabolites’ antioxidant, anticancer, antibacterial, and anti-inflammatory properties [17]. Recent studies revealed an association between gut microbiota dysbiosis and the onset of human illness [18,19]. So, if plant and plant-derived compounds effectively treat gut microbiota dysbiosis-induced illnesses, these compounds also regulate microbiota interactions to restore health [17]. Only a few isolated studies indicated their role in host-microbiota interactions and gut epithelial integrity. However, a comprehensive overview of decoding how these microbiota influence host-microbe interactions is still awaited. This review will develop a holistic understanding of host-microbiota interactions in health and diseases and comprehend the mechanistic role of plant metabolites in these molecular bioprocesses.
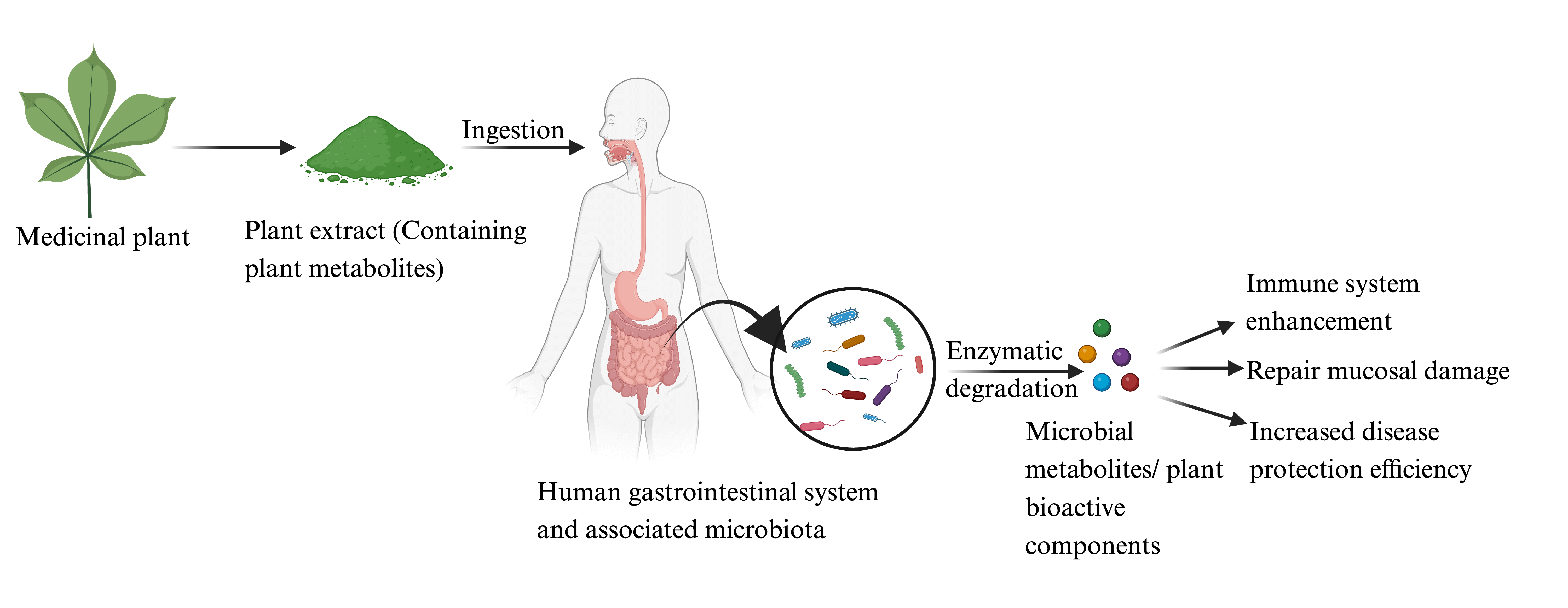
The host-microbe interacts via the gut microbiota-derived metabolites and host immunological response [9]. The human-microbe interaction co-evolves for the benefit of both members, where the human host provides a nutrient-rich safe harbor [20]. At the same time, microbes offer a litany of microbial metabolites, which benefit host physiology and resistance to competitive pathogens [21]. The predominant metabolites are SCFAs, tryptophan metabolites, bile acids, and choline metabolite [22]. These metabolites affect the host physiology by modulating various cellular processes (Fig. 1). SCFAs are derived from bacterial fermentation and play a pivotal role in bodily functioning and the modulation of the immune system [23]. SCFAs, such as acetate, propionate, and butyrate, are synthesized in the gastrointestinal tract and are absorbed by the intestinal epithelial cells (IECs), lining the intestine, to serve as an energy source [24]. They also connect the gut microbiota and the immune system, influencing the development, survival, and function of both IECs and leukocytes [25]. SCFAs exert their effects by activating specific receptors, namely Free fatty acid receptor 2 (FFAR2), FFAR3, G-protein coupled receptor 109a (GPR109a), and Olfactory receptor 78 (Olfr78), and by modulating the activity of enzymes and transcription factors [26]. SCFAs also regulate the functioning of regulatory T cells (Treg) and foster immune tolerance, indicating their potential therapeutic implications for inflammatory disorders [27]. The gut microbiota metabolizes the tryptophan in the intestinal lumen into kynurenine, tryptamine, and indole [28]. Indole and indole-3-propionate (IPA), affect mucosal homeostasis through the pregnane X receptor (PXR) and reduce intestinal permeability. Indole also stimulates enteroendocrine L-cells to release glucagon-like peptide 1 (GLP-1), suppressing appetite, insulin secretion, and gastric emptying [29]. Several tryptophan catabolites modify innate and adaptive immune responses in intestinal immune cells by activating aryl hydrocarbon receptors (AHR), thus maintaining mucosal reactivity [30]. Tryptamine enhances gastrointestinal motility by inducing the enterochromaffin cells to secrete the 5-hydroxy tryptamine (5-HT), acting on enteric nervous system neurons [31]. Gut microbes alter lipid metabolism by producing secondary bile metabolites. Iso-allo Lithocholic acid (LCA), a microbiota-derived metabolite, promotes the Treg population, while cells and 3- oxo Lithocholic acid (3-oxoLCA) inhibit suppression of the T helper-17 (TH17) population [32]. (Fig. 1).
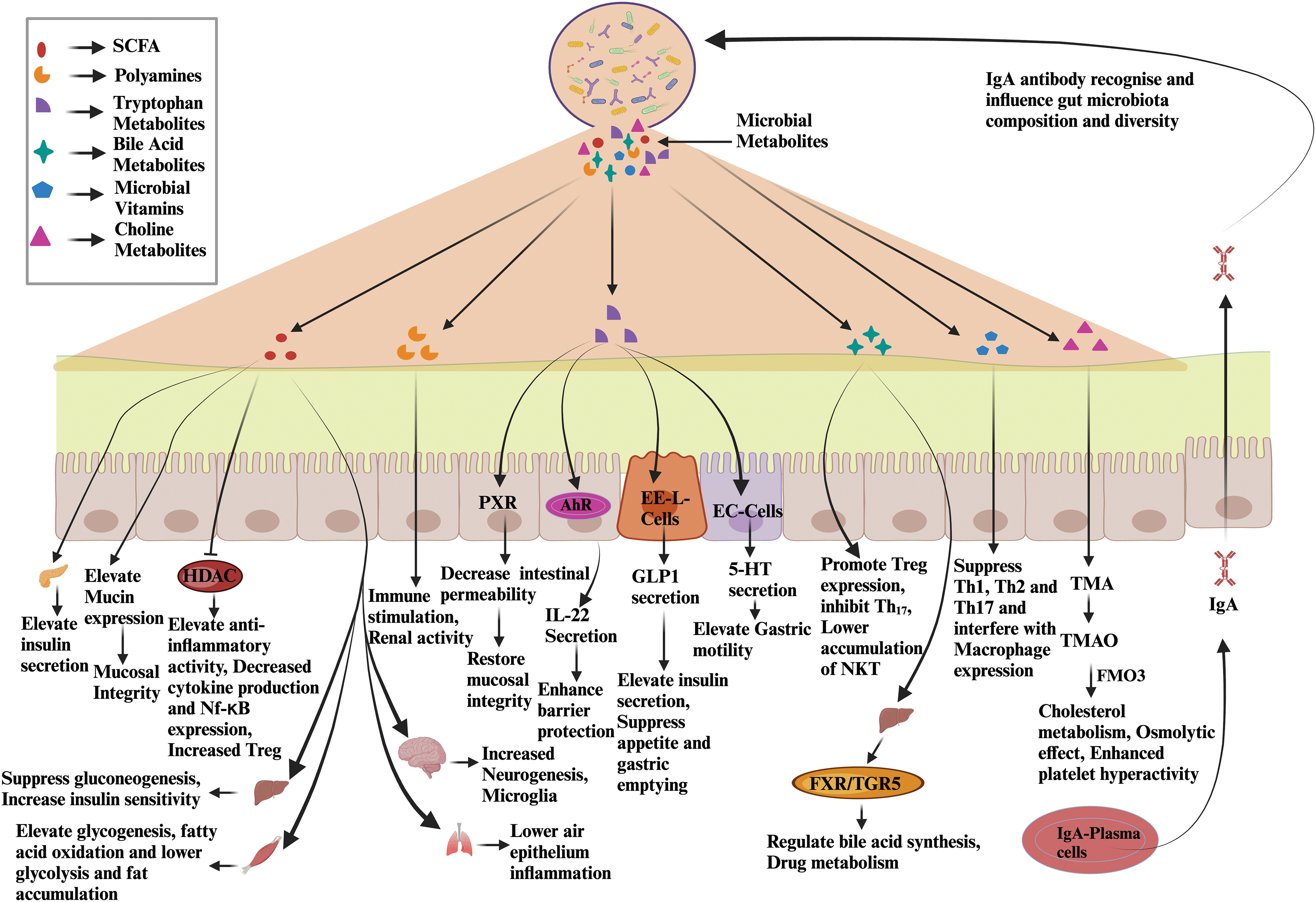
Figure 1: Gut microbiota-derived metabolites influence host physiology. Gut microbiota metabolizes the ingested food particles and produces metabolites like SCFAs, tryptophan metabolites, bile acid metabolites, choline metabolites, and microbial vitamins. These metabolites influence the host physiology by regulating various signaling cascades. Abbreviations: HDAC: Histone deacetylase, Treg: Regulatory T cells, PXR: Pregnane X receptor, AhR: Aryl hydrocarbon receptor, IL-22: Interleukin 22, EE-L-Cells: Enteroendocrine cells, GLP1: Glucagon-like peptide 1, EC-cells: Enterochromaffin-like, 5HT: 5 Hydroxy tryptamine, NKT: Natural killer T cells, FXR: Farnesoid X receptor, TGR5: Takeda G protein-coupled receptor 5, Th1,2: Type 1 T helper cells, Type 2 T helper cells, Th17: T helper 17, TMA: Trimethylamine, TMAO: Trimethylamine N-oxide, FMO3: Flavin-containing dimethylaniline monooxygenase 3, IgA: Immunoglobulin A.
Host immunological response is another facet of the host-microbiota bidirectional cross-talk. The intestinal immune system constantly works to maintain equilibrium between bacterial tolerance and being vigilant against pathogens [33,34]. The immune system and the microbiota communication is mediated by a large set of proteins called pattern recognition receptors (PRRs). These PRRs are synthesized by various hematopoietic cells and IECs [35]. PRRs that identify microbe-associated molecular patterns (MAMPs) include nuclear oligomerization domain-like receptors (NLRs) and toll-like receptors (TLRs) [35,36]. Previous studies have reported that the loss of particular PRRs can result in changes to the composition of microorganisms and intestinal barrier issues, which allows microorganisms to infiltrate systemic organs [37]. TLR deficiency was found to cause longer-term familial changes in microbial composition, although the ileal microbiota composition in TLR2-, TLR4-, TLR5-, and TLR9-deficient mice was demonstrated to stay constant in the short term [38]. It can be deduced that host-microbe interactions are interdependent as NOD-like receptor family pyrin domain containing 6 (NLRP6) deficiency in mice is associated with decreased tissue repair efficiency by interleukin-18 (IL-18)-dependent pathway along with an altered colonic microbiota enriched in the Prevotellaceae and Saccharibacteria TM7 [39]. The adaptive branch of the immune system functions via Immunoglobulin A (IgA) antibodies, which act as a bridge between the innate and immune systems in the intestine [40]. Gene knock-out studies point towards the integrative role of IgA in recognizing the intestinal microbiota, particularly commensals [41]. Bacteria penetrating through the mucus layer leads to class switching of B cells and subsequently produces IgA. These IgA are secreted into the intestinal lumen, which recognizes the microbiota [42]. The host repertoires of IgA are specific to particular bacterial epitopes of commensals and coat most of the intestinal microbiota without causing strong and potentially harmful reactions [43]. The repertoire of secreted IgA is continuously changing to adapt and respond to the fluctuating intestinal microbial environment. This repertoire may be dynamically shaped to mirror the microbiota composition [44]. Other immune cells, i.e., T regulatory type 1 (Tr1) cells, T helper (TH17) cells, and Treg cells, aid the IgA-dependent microbiota interactions [45]. Although the immune system protects the gastric microbiota, the pathogens utilize this for their growth benefit [46]. Immune defense against pathogens leads to intestinal barrier disruption, inflammation, and secretion of inflammatory molecules like tetrathionate and nitrates [47]. These molecules aid the growth of pathogens like Salmonella typhimurium and Escherichia coli [48].
Host-Microbe Interactions in Disease
Host-microbe interactions play a significant role in maintaining gut homeostasis and human health [49]. These interactions alter microbiota dysbiosis, causing chronic inflammation [50]. A leaky gut and inflammation are the primary culprits for the onset of diseases. A leaky gut exposes microbes and microbial antigens to the host’s immune system, disturbing host-microbe interactions [51]. Intestinal epithelial integrity is disturbed by tumor necrosis factor alpha (TNFα)-dependent nuclear factor kappa B (NF-κB) [52]. NF-κB stimulates the myosin light chain kinase (MLCK) promoter, which leads to cytoskeletal contraction and the opening of tight junctions in the intestinal epithelium cells [53]. The compromised barrier integrity allows the exposure of microbial antigens to TLRs to activate the innate immune system [54]. It subsequently creates a pro-inflammatory environment by targeting the immune cells such as macrophages, dendritic cells, and Th17 cells [54]. The TNFα-induced NF-κB pathway significantly impacts inflammatory bowel disease (IBD) pathogenesis [55] (Fig. 2).
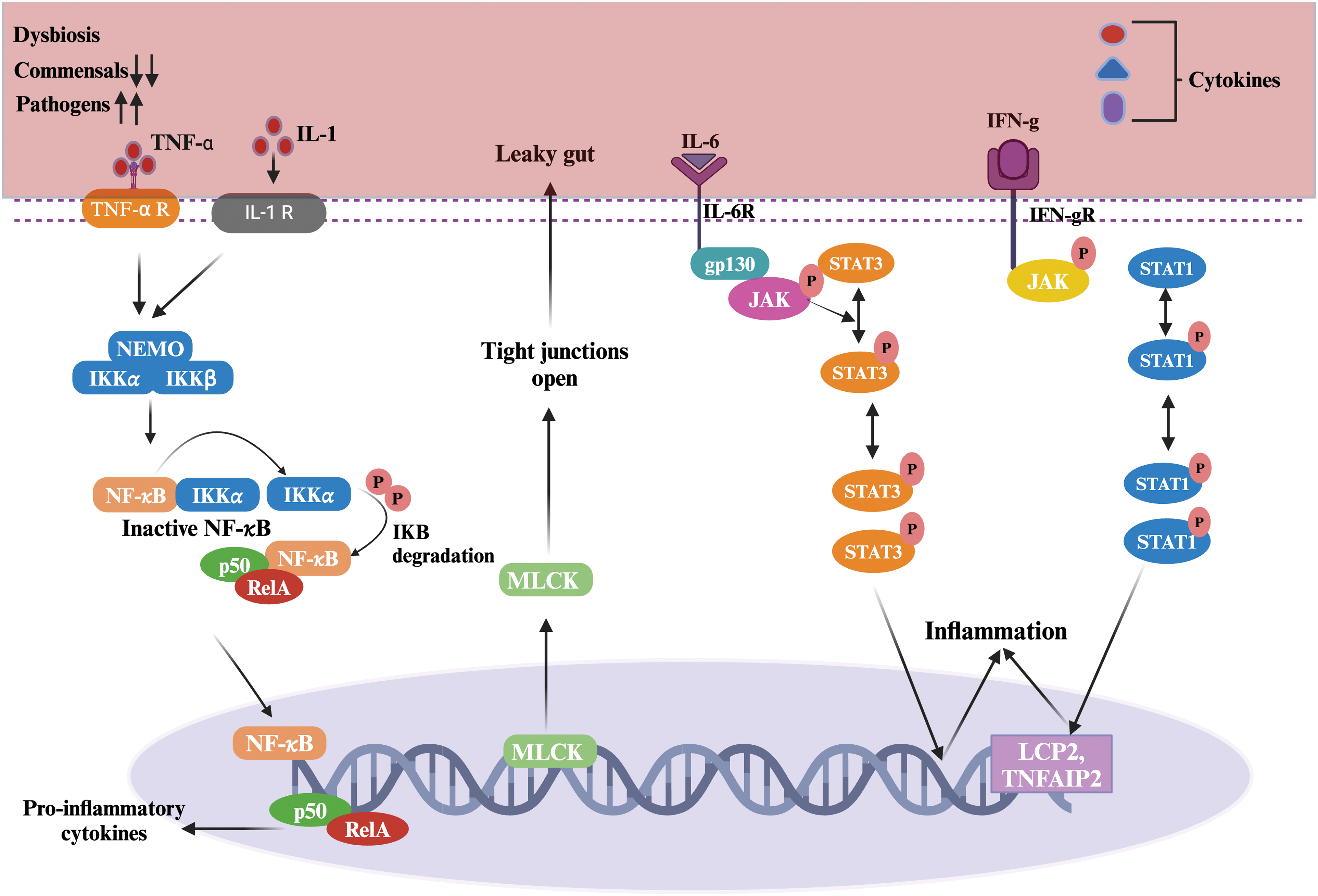
Figure 2: Host-microbe interactions during microbiota dysbiosis associated with human illnesses. The host’s inflammatory pathways are activated in response to antigen stimulation. TNFɑ and IL-1 act binds to TNF-ɑ-Receptor and IL-1 Receptor, respectively. TNFɑ binding to its receptors activates the IKK complex, activating NF-κB. IKK complex phosphorylates IκBα, leading to its degradation. Once IκBα is degraded, NF-κB is translocated to the nucleus. In the nucleus, NF-κB binds to the MLCK gene to regulate target genes, contract tight junction proteins, and promote leaky gut. Similarly, other inflammatory factors, IL-6, and IFN-γ, enhance inflammation by the JAK-STAT pathway. IFN-γ-STAT1 pathway activates LCP2 and TNFAIP2, which induce chronic inflammation. The IL-6 pathway is important for immune response, inflammation, and cell survival in the gut. The binding of IL-6 to its receptor complex activates JAKs associated with gp130. JAKs phosphorylate specific tyrosine residues on gp130, creating docking sites for STAT proteins. Phosphorylated STATs dimerize and translocate to the nucleus. STAT dimers bind to particular DNA sequences and regulate the transcription of target genes. IL-6/STAT signaling promotes the survival and proliferation of intestinal epithelial cells, maintains mucosal barrier integrity, and modulates immune cell responses. Excessive or prolonged IL-6/STAT signaling contributes to chronic inflammation and is involved in the pathology of IBD. IFN-γ binds to its receptor, and downstream activates JAKs, which further phosphorylate and activate STAT-1. STAT-1 dimer translocates to the nucleus and binds to LCP2 which contributes to T-cell activation, and TNFAIP2, which enhances inflammation [56–58]. Abbreviations: IL-1: Interleukin-1, IKK: Inhibitor of nuclear factor kappa B kinase, IFN-γ: Interferon-gamma, JAK-STAT: Janus kinase-Signal transducers and activators of ttranscription, LCP2: Lymphocyte cytosolic protein 2, TNFAIP2: Tumor Necrosis Factor, Alpha-Induced Protein 2, gp130: glycoprotein 130, RelA: v-rel avian reticuloendotheliosis viral oncogene homolog A.
The dysregulation of signaling by NF-κB leads to uncontrolled inflammation and altered immune responses in gastric diseases [52]. Similarly, IL-1β plays a crucial role in intestinal inflammation, particularly in Inflammatory Bowel Disease (IBD), such as Crohn’s disease [59]. In previous studies, IL-1β, produced by NOD-like receptor protein 3 (NLRP3) inflammasome, contributes to the provocation of inflammation in IBD. The NLRP3 inflammasome activates IL-1β, and an exaggerated level of its activity can instigate intestinal inflammation [60]. In the case of active Ulcerative Colitis (UC) and Crohn’s Disease (CD), NLRP3 and IL-1β are upregulated, and the spatial localization of these entities suggests the occurrence of inflammasome-independent processing of IL-1β [60]. IL-1β can suppress the expression of PDZ domain-containing scaffolding protein1 (PDZK1), which regulates intestinal receptors and transporters, by activating NF-κB [61]. Notably, IL-1β also triggers the activation of NF-κB, which subsequently represses gastrin, a pivotal hormonal regulator of acid secretion within the gastrointestinal tract [62]. IL-1β-dependent NF-κB signaling is implicated in intestinal inflammation and regulating diverse gastrointestinal processes, encompassing cytokine production [63]. The IL6-dependent STAT3 activation has also been involved in developing gastric diseases [64]. Moreover, IL-6 has been identified as a necessary target for therapeutic intervention in IBD due to its enhanced expression in the serum and mucosa of patients with active disease [65]. The IL-6/gp130/STAT3 signaling pathway plays a significant role in preserving the integrity of the intestinal barrier to counteract bacterial invasion [66]. However, the dysregulation of IL-6/STAT3 signaling, characterized by an imbalance between STAT3 and suppressor of cytokine signaling 3 (SOCS3), has been observed in IBD-associated dysplasia, indicating a potential disruption in the negative regulation of SOCS3 [66]. The involvement of IFN-γ and STAT1 can be observed in IBD. The association between STAT1, Histone acetyltransferase 300 (EP300), and Histone 3 lysine 27 acetylation (H3K27ac) contributes to the development and progression of Inflammatory Bowel Disease (IBD) [58]. Conclusively, NF-κB-induced inflammasome, oxidative burst, disruption of intestinal epithelial integrity, and intestinal leakage are prominent molecular changes during the onset of various microbiota dysbiosis-associated disorders [67]. These cellular changes have to be reversed to overcome microbiota dysbiosis-associated disorders. Accordingly, multiple therapies are being developed targeting these molecular markers.
Human gut microbiota continuously cross-talk with other organs through the gut- the respective organ axis (Fig. 3) [68]. Hereby, cellular changes affect intestinal physiology and alter the physiology of other organs like the brain, kidney, heart, liver, lungs, and skin [69,70]. The gut-organ axis works as a medium to exchange microbial metabolites between the gut and that organ. Gut microbiota dysbiosis has been reported in various organ-associated diseases [71]. This axis may be the path to dysbiosis-related disease flare-ups [72]. Hereby, any therapy and molecule targeting molecular markers of gut microbiota dysbiosis-associated diseases would help regain healthy physiology in other organs.
Figure 3: Gut microbiota-produced metabolites are exchanged between the gut and organs, which leads to the formation of the Gut-organ Axis.
Plant Metabolites and Host-Microbe Interactions
Plant metabolites play a significant role in the interactions between hosts and microorganisms and in the overall well-being of humans [73]. These metabolites, namely phytochemicals and flavonoids, are inherent compounds in food constituents and possess diverse properties promoting human health [74]. They can be directly absorbed into the bloodstream and modulate the gut microbiota, subsequently impacting the host’s health [75]. The gut microbiota enhances the bioactivity of phytochemicals [76]. Moreover, it generates metabolites from phytochemicals that exhibit therapeutic effects on human ailments [76]. Moreover, bioactive substances sourced from plants can impact the variety of gut bacteria and improve the intestinal barrier integrity, aiding in managing and preventing various illnesses [75].
The significant contribution of plant-derived metabolites to host-microbe interactions is their ability to modulate the immune response and modify the integrity of the gut barrier [77]. These metabolites and their bioactive forms invoke the immune response by stimulating numerous immune cells and regulating cytokines [77]. Similarly, plant metabolites influence the integrity of the gut barrier by modulating the functionality of tight junction proteins, impacting gut microbial diversity, promoting mucin secretion, and alleviating oxidative stress [78].
Plant Metabolites and Gut Microbiota Immune Axis
A balance between pro- and anti-inflammatory mediators in the gut is necessary to preserve the intestinal epithelial barrier [79]. According to a study, immune cells in the bloodstream can penetrate the mucosa and release a range of inflammatory mediators that intensify the gut’s immunological response [80]. Therefore, a successful treatment approach for gastrointestinal disorders is to prevent the invasion of immune cells and inflammatory factors, restoring the integrity of the intestinal partition [81]. Phytochemicals like resveratrol, quercetin, colchicine, genistein, capsaicin, and epigallocatechin-3-gallate can modulate immunity and control critical molecular and cellular interactions [82]. They target several immune response signaling pathways, such as the TNF-α/NF-κB, IL-1/NF-κB, IFN-γ/JAK/STAT, and IL-6/JAK-STAT pathways [83]. Ursolic acid, gingerol, flavopiridol, curcumin, and green tea catechins can inhibit one or more steps of the NF-κB signaling cascade and act as immunomodulatory compounds [84–87]. These phytochemicals help manage cytokine storms since pharmacological profiling reveals strong NF-κB signaling pathway inhibitions [88]. By promoting the immune system’s health and regulating immunological responses, phytochemicals also help establish healthy host-microbiota interactions. They can balance the immune system and control the release of pro-inflammatory cytokines, essential for overcoming gut microbiota-associated disorders [89].
Quercetin inhibits the nuclear translocation of the NF-κB receptors’ p50 and p65 subunits, lowering pro-inflammatory gene production [90]. By preventing Iκ-Bα degradation and p65 nuclear translocation, quercetin also reduced the gene expression and production of IL-1β, IL-6, IL-8, and TNF-α in human mast cells [91]. Various plant compounds inhibit the host NLRP3-mediated inflammation by inhibiting the intermediate effector molecules of the pathway [92]. Apigenin inhibits the production of pro-inflammatory cytokines in lipopolysaccharide (LPS)-induced T cell human leukemia virus positive-1 (THP-1) macrophages, such as TNF-α, IL-1β, and IL-6. When exposed to LPS, apigenin reduces NF-κB activation. Still, it does not affect NLRP3 mRNA or protein levels or apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC) protein levels [93]. Nevertheless, apigenin prevents the development of ASC specks, protein aggregates necessary for the NLRP3 inflammasome to be activated. This implies that apigenin inhibits NLRP3-mediated inflammation by directly targeting ASC [94]. Apigenin can prevent the phosphorylation of two tyrosine kinases that are involved in the phosphorylation of ASC: protein tyrosine kinase 2 (Pyk2) and spleen tyrosine kinase (Syk) [95]. Additionally, apigenin was shown to block IL-1β production mediated by absent in melanoma 2 protein (AIM2), another kind of inflammasome, but not NOD-like receptor family CARD domain containing 4 (NLRC4)-mediated production [96]. This suggests that apigenin inhibits a variety of inflammasome types, indicating that it is a non-selective inflammasome inhibitor. Another phytochemical cardamonin inhibits the NF-κB pathway, a signaling system implicated in inflammation, in RAW 264.7 cells, an immunological cell type [97]. This inhibition occurs by suppressing the inflammatory chemicals prostaglandin-E2 and nitric oxide (NO) [98]. Suppression of these molecules reduces phosphorylation and degradation of Iκ-Bα, a protein that stimulates the NF-κB pathway [99]. The capacity of cardamonin to upregulate the Aryl hydrocarbon receptor (AhR)/Nuclear factor erythroid 2-related factor 2 (Nrf2)/NAD(P)H quinone dehydrogenase (NQO1) signaling pathway, which is known to regulate the NLRP3 inflammasome adversely, is another possible mode of action [94]. Cardamonin has been extensively researched about cancer, but it has also been explored in rheumatoid arthritis (RA), inflammatory bowel disease (IBD), and other disorders [94,100]. Research has shown that the main bioactive component of Polygonimultiflori Radix (PMR) water extract, 2,3,5,4′-tetrahydroxystilbene-2-O-β-D-glucoside (TSG), may increase the level of the anti-inflammatory cytokine IL-10 and reduce the production of pro-inflammatory cytokines, such as TNF-α, IL-1β, and IL-6 [101]. Similarly, as reported, aloe vera administration increased the production of IL-10 while suppressing the expression of pro-inflammatory colonic mediators. This inflammation regulatory role of aloe vera might be attributed to its metabolites, including aloins, emodin, and chrysophanol [102,103]. The traditional Chinese medicine formula Shen-Ling-Bai-Zhu-San (SLBZS), constituted of panaxadiol, disogenin ginsenosides Rg1, neferine, atractylenolide I, II, III, isoliquiritigenin and liquiritigenin, platycodin D. These metabolites may lessen colonic damage, enhance body weight loss brought on by sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), and reduce the disease activity index (DAI) score [104]. Jasoniaglutinosa (L.) DC. (JGD), a traditional herbal remedy referred as rock tea [105], was found to be rich in phytochemicals like pigments (xanthophyll, lutein, β-carotene, chlorophyll b, chlorophyll a, pheophytin a, phenolic substances (hydroxycinnamic acids caffeoylquinic acid, etc.) and flavanols (quercetin-3-O-galactoside) [106]). The investigation revealed that rock tea metabolites had a protective impact on the intestinal barrier and significantly decreased DSS-induced pathological changes.
Pro-inflammatory cytokines such as TNF-α and IL-6 may be produced less often by these components of rock tea [105,106]. Conclusively, plant metabolites hold the potential to control the function of the intestinal barrier and influence the course of the illness by regulating the host inflammatory response essential for equilibrated host-microbiota interactions for good health (Fig. 4).
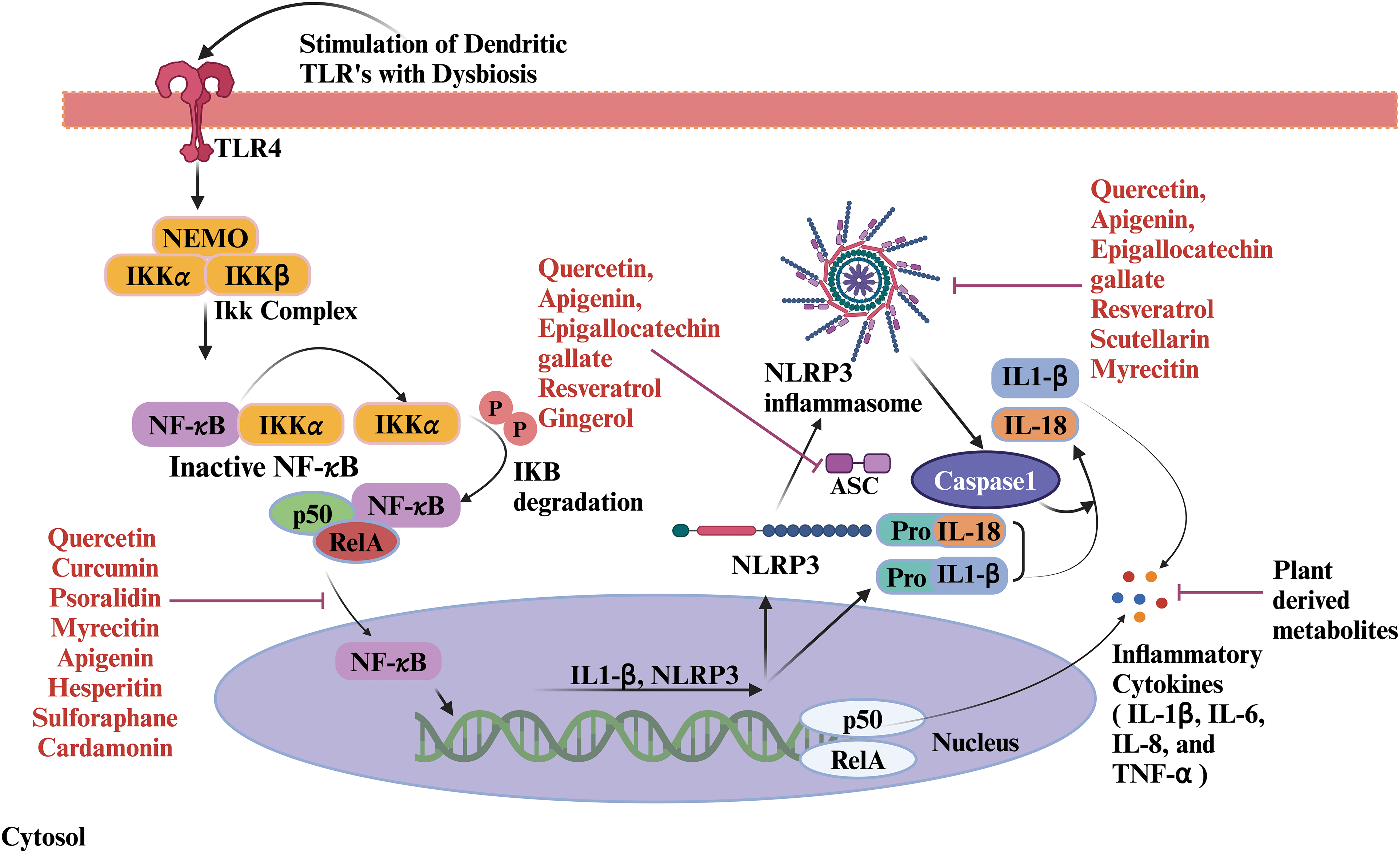
Figure 4: Role of plant metabolites on host immunological response. Gut dysbiosis stimulates TLRs of dendritic cells and inflammatory pathways. TLRs start signaling pathways that activate the NF-κB pathway downstream. TLR signaling induces IκB phosphorylation and degradation, allowing NF-κB to translocate to the nucleus along with p50/RelA. In the nucleus, NF-κB promotes the transcription of inflammatory cytokines. The assembly of ASC specks activates the NLRP3 inflammasome. The assembly of the NLRP3 inflammasome activates the caspase-1. Caspase-1 processes pro-inflammatory cytokines into their active forms. The secretion of IL-1β and IL-18 enhances the inflammatory response. Plant-derived metabolites act as immunomodulators by inhibiting the inflammatory pathways at different stages. Abbreviations: TLR4: Toll-like receptor 4, NEMO: NF-κB essential modulator, IKKα: Inhibitor of nuclear factor kappa B kinase alpha subunit, IKKβ: Inhibitor of nuclear factor kappa B kinase beta subunit, RelA: v-rel avian reticuloendotheliosis viral oncogene homolog A, NLRP 3: NOD-like receptor protein 3, ASC: Apoptosis-associated speck-like protein containing a caspase recruitment domain, IL: Interleukin.
Plant Metabolites and Gut-Barrier Integrity
The intestinal barrier is essential to safeguard stratified gut microbiota in intestinal mucosa and host-microbiota crosstalk [107]. The gut epithelium, the mucus layer, and the microbiota collectively constitute the barrier [108]. These barriers efficiently seal the gaps between neighboring cells [109]. The epithelium’s goblet cells secrete mucin, which forms a mucus coating that prevents potentially pathogenic bacteria from making immediate contact with the epithelium [110]. The commensal microbiota plays a crucial role in host protection by impeding the proliferation of pathogenic organisms through nutritional competition [24]. Furthermore, SCFAs produced by the gut microbiota promote the proliferation of epithelial cells [111]. The biochemical barrier encompasses various bioactive molecules, including gastric acid, regenerating family member 3 gamma (Reg3γ), defensins, secretory immunoglobulin A (sIgA), and antimicrobial peptides generated by intestinal Paneth cells [112]. These bioactive compounds can alter the microbiota’s structure or directly aid in eradicating pathogenic bacteria [113]. The lamina propria of the gut contains a variety of immune cells that make up another part of the biochemical barrier [80]. These cells limit infections by regulating normal immune responses [114]. Disrupted gut barrier integrity has been reported in various gastrointestinal diseases such as IBD, UC, colorectal cancer, etc. [55]. Plant-derived metabolites like berberine, curcumin, lycopene, aloin, polysaccharides, etc., contribute to maintaining gut barrier integrity by modulating tight junction proteins, stimulating mucin production, or altering microbiota diversity [115]. The tight junctions, protein complexes located at the interface connecting cells, serve a vital role in modulating the epithelium’s permeability and structure [116]. Underexpression of tight junction proteins is already observed in UC patients, animals with experimental colitis, and during the onset of IBD [117]. Recent studies have discovered that various traditional medicinal concoctions can improve the integrity of the intestinal barrier function by controlling the expression of tight junction proteins, which can reduce colonic inflammatory situations [107]. In Caco-2 cells, the administration of Dahuang Mudan decoction (DMD), a popular medication for intestinal disorders, markedly elevated the expression of Claudin-1, occluding, and ZO-1 [118]. The primary metabolites of DMD belong to anthraquinones such as emodin, aloe-emodin, rhein, paeoniflorin, and amygdalin [115]. Additionally, it lowers the ratio of natural cytotoxicity receptor (NCR) innate lymphoid cell-(ILC3) and raises the fraction of IL-22 + ILC3 and NCR + ILC3 [118]. Bryophyllum pinnatum (Lamarck) Leaf metabolites kaempferol 3-O-α-L-arabinopyranosyl-(1→2)-α-L-rhamnopyranoside, quercetin 3-O-α-L-arabinopyranosyl-(1→2)-α-L-rhamnopyranoside, and quercetin-3-O-rhamnopyranoside have been shown the anti-UC effects, at least in part due to its regulation of ZO-1 [119]. According to a different study, TSG, an active component of Polygonimultiflori radix (PMR) aqueous extract, may increase the expression of tight junction proteins like occludin and ZO-1, thereby restoring the structure of the intestinal epithelial barrier [101]. In DSS-induced colitis, polysaccharides from Scutellariabaicalensis georgi (SBG) reduced pathological damage to the colon. It improved the integrity of the intestinal barrier by upregulating the expression of tight junction proteins such as claudin-5, ZO-1, and occludin [120]. On the other hand, the precise roles of the chemical components in some conventional decoctions or compounds that occur naturally are still unidentified. Further studies addressing these issues may facilitate the development of new drugs for treating UC by regulating tight junction proteins. Besides tight junction protein, mucin proteins are also affected in intestinal disorders. Mucin deficiency is found to be associated with the onset of infantile IBD [121]. A lack of anterior gradient 2 (AGR2), a disulfide isomerase involved in mucin processing, severely affects the mucus barrier [121,122]. By upregulating phosphorylated Protein kinase C (p-PKC) and phosphorylated extracellular signal-regulated kinase (p-ERK) and downregulating phosphorylated phosphoinositide 3-Kinase (p-PI3K) and phosphorylated- A serine-threonine kinase (p-AKT), aloe vera promoted the production of mucins [102]. Anemone chinensis Bunge (ACB) constituents enhance the production of mucin, specifically mucin-2 and mucin-3A [123]. Furthermore, a correlation between the beneficial effects of Bryophyllum pinnatum (Lamarck) Leaf extract on UC and its ability to stimulate MUC-3 expression has been observed [109]. White Ginseng constituting ginsenosides (Ra1, Rb1, Rb2, Rc, Rh2, Re, Rg1, Rg, vina ginsenoside R2, pseudoginsenoside F11 and majonoside R1), flavonoids, volatile oil has also shown the potential to significantly increase the mRNA expression of Muc2 [124], a prominent mucin found in the intestines of rats. This augmentation can effectively strengthen the intestinal barrier function and improve the gut ecosystem [125].
Plant Metabolites and Microbiota Interactions
Gut microbiota and plant extract, enriched with metabolites or phytochemicals, have cyclic interactions [126]. Exposure of plant phytochemicals to gut microbiota allows their metabolic transformation into different metabolites with improved bioavailability [127]. Simultaneously, plant extracts or phytochemicals modulate the gut microbiota in a variety of ways, including governing the ratio of beneficial to pathogenic bacteria, restoring the diversity of the gut microbiota, enhancing the function of the intestinal barrier, and controlling the metabolites of the gut microbiota (Fig. 5) [128].
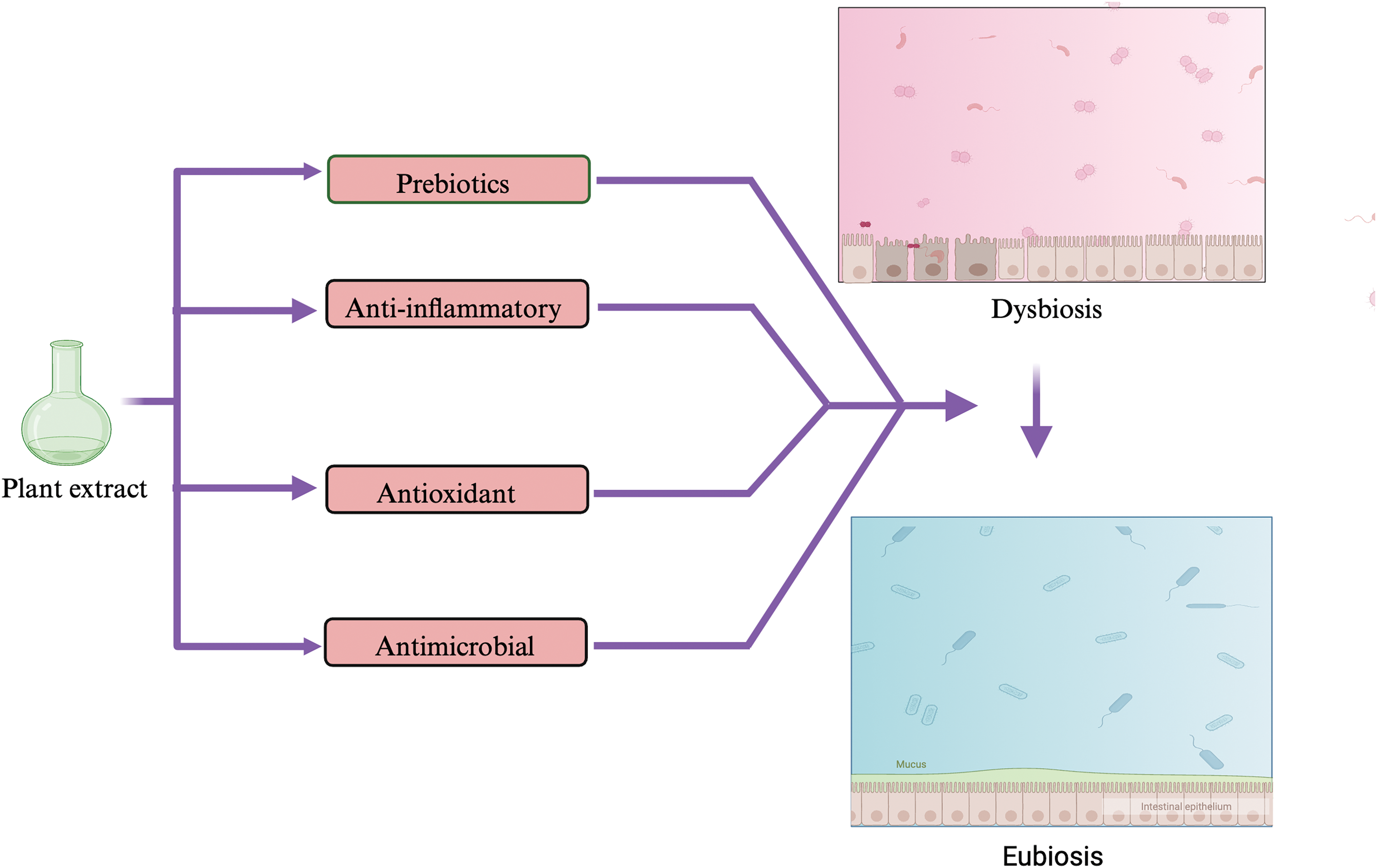
Figure 5: Medicinal plants possess prebiotic, anti-inflammatory, antioxidant, and antimicrobial properties, which help in the restoration of beneficial gut bacteria and restoration of eubiotic conditions in the gut.
Though the mechanism behind plant modulation of gut microbes has not been fully discovered, it can be attributed to the prebiotic, anti-inflammatory, anti-microbial, and antioxidant characteristics of plant constituents, which create a favorable environment for beneficial microbiota flourishing [118,129] (Fig. 5). Medicinal decoctions, loaded with different metabolites from various plants, which work synergistically in a decoction, have been found to alleviate dysbiosis and restore gut microbiota homeostasis [130]. The TSG, a bioactive component of Polygoni multiflori Radix (PMR), increases the abundances of Firmicutes and Bacteroidetes [101]. The bioactive ingredients of Rhubarb, Rhein considerably decreased uric acid which acts as a colonic inflammatory substance [131]. Moreover, Rhein may modify purine metabolism and cause alterations in the gut microbiota’s makeup, both sufficient to reverse the colon damage caused by DSS [131]. The primary metabolites of Jiawei Gegen Qinlian Decoction (JGQD), puerarin, magnolol, baicalein, berberine, and glycyrrhiic acid may cause Escherichia-Shigella to decline and Akkermansia and Romboutsia to grow [132]. In addition to these modifications, the administration of polysaccharide SP2-1 altered the microflora’s composition by considerably increasing the amounts of Firmicutes, Bifidobacterium, Lactobacillus, and Roseburia and reducing the quantities of Bacteroides, Proteobacteria, and Staphylococcus in the gut [133].
Plant-derived metabolites also serve as prebiotics to enhance the diversity and composition of gut microbiota [130]. The polysaccharides of Taraxacum officinale, commonly known as dandelion, have been found to exert the prebiotic effect by enhancing the abundance of Bifidobacterium, Olsenella, and Dialister [134]. Similarly, another medicinal plant, i.e., Garlic, contains allicin, inulin, and fructooligosaccharides, enhances the Bifidobacteria, Lactbacillus acidophillus and suppresses the Clostridia species [135]. The prebiotic properties of plants can be attributed to the high fiber content such as inulin which is digested slowly by gut microbiota and leads to fermentation [136]. Plant metabolites like SCFA also serve as energy sources and are generally used as markers for prebiotic activity [137]. For example, the administration of Huang-Lian Jie-du decoction increased the diversity of gut microbiota, an augmentation in the abundance of Lactobacillus, and a reduction in Firmicutes [138]. Furthermore, the administration of Radix Astragali-Radix Salviae Miltiorrhizae increased the levels of probiotics, such as Lactobacillus and Bifidobacterium [139]. These strains of beneficial bacteria were found to be closely associated with decreased blood pressure and other hypertension-related symptoms [140,141]. The influence of plant extracts and derived metabolites on microbes has also been noted in mental disorders related to the gut-brain axis [142]. Curcumin, a well-known phytochemical, has been discovered to diminish the presence of Prevotella, Coriobacterales, and Ruminococcus, which are significant disease-related microbes [143]. In an animal model of Alzheimer’s disease, the ratio of Firmicutes to Bacteroidetes increased, the abundance of anaerobic and Helicobacter decreased [144], and an enhancement in cognitive function was observed following the administration of curcumin [145]. Humulones and lupulones present in the female flowers of Humulus lupulus L. (commonly known as hop) are utilized as herbal remedies for treating conditions such as anxiety, mood disorders, and sleep disturbances [146].
Inflammation plays a pivotal role in developing dysfunction in the epithelial layer. Disruption of the epithelial barrier allows harmful antigens to pass through, further elevating local inflammation [147]. The inflammatory response recruits immune cells, such as macrophages, to generate reactive oxygen species (ROS) and induce oxidative stress [148]. Consequently, gut inflammation and oxidative stress are interconnected processes [149]. Inflammatory cytokines hinder the proper functioning of the epithelial barrier by weakening the tight junctions between the epithelial cells [150]. These alterations ultimately lead to chronic gut inflammation, as observed in patients suffering from IBD [151]. This chronic inflammation may result in mucosal erosion and ulceration [151]. Hence, the combination of oxidative stress, inflammation, and impaired gut epithelial barrier contributes to dysbiotic conditions [152]. However, introducing antioxidants derived from plants can effectively suppress oxidative stress and inflammation in the mucosal layer of the gut, thereby improving overall gut health [153]. By modulating effector molecules, including superoxide radicals, arachidonate 5-lipoxygenase (5-LOX), inducible nitric oxide synthase (iNOS), nitric oxide (NO), and Cyclooxygenase 2 (COX2), another mechanism implicated in barrier homeostasis maintenance is connected to a decrease in oxidative stress [154]. Various plant-derived metabolites, such as astaxanthin, resveratrol, curcumin, naringenin, quercetin, etc., have been reported to act as antioxidants, thus alleviating oxidative stress [155]. Previous investigations have noted that many plant-derived metabolites like β-carotene enhance the expression of antioxidant enzymes, diminish oxidative byproducts, and enhance the functionality of the gut epithelial barrier (Fig. 6) [156].
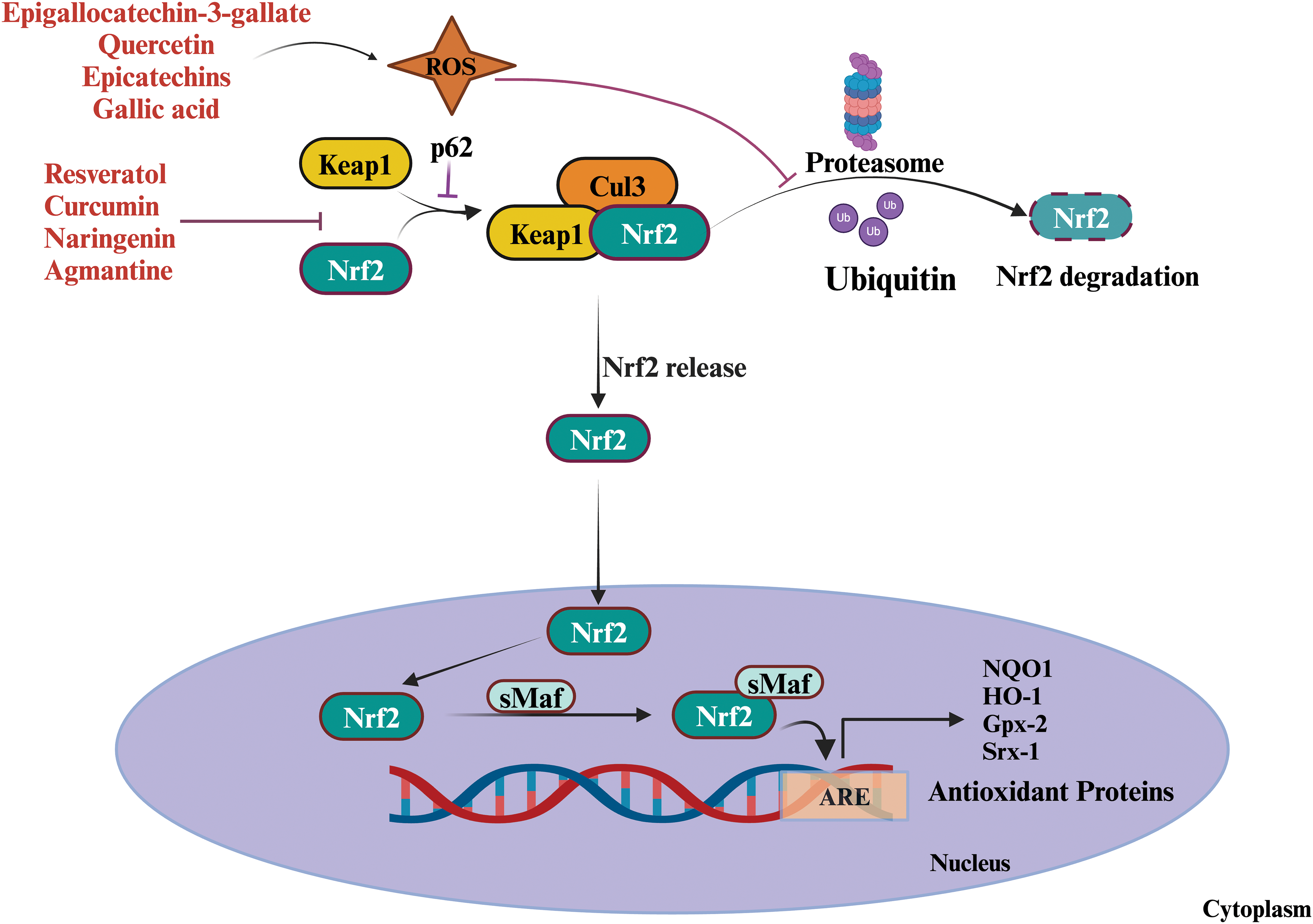
Figure 6: Role of plant-derived metabolites as antioxidants and pro-oxidants. Proxidants increase ROS generation and activate the Nrf2 pathway by inhibiting the proteasomal degradation of Nrf2 which subsequently produces antioxidants. Under normal circumstances, Keap 1, an oxidative stress marker, binds to Nrf2, sequestering it within the cytoplasm and marking it for ubiquitination and subsequent degradation by the proteasome. In response to oxidative stress or electrophilic compounds specific cysteine residues on Keap1 undergo modification disrupting the interaction between Keap1 and Nrf2, thereby preventing Nrf2 degradation. Consequently, Nrf2 becomes stabilized and translocates into the nucleus. Within the nucleus, Nrf2 forms a heterodimer with sMAF proteins. This Nrf2-sMAF complex subsequently binds to the (ARE) located within the promoter regions of target genes, which induces the transcriptional activation of diverse antioxidant and cytoprotective genes. These genes encode proteins such as glutathione S-transferase, heme oxygenase-1, superoxide dismutase, catalase, epigallocatechin-3-gallate (EGCG), quercetin, epicatechins, and gallic acid are involved in removing oxidative stress. While some plant-derived metabolites inhibit the interaction between Keap1 and Nrf2 by modifying the Keap1. Abbreviations : NRF2: Nuclear factor erythroid 2-related factor 2, Keap1: Kelch-like ECH-associated protein, sMAF: small musculoaponeurotic fibrosarcoma, ARE: Antioxidant Response Element, Cul 3: Cullin 3.
Another metabolite, lycopene consumption, shields the intestinal epithelium from deoxynivalenol-induced oxidative stress [157]. The antioxidant enzyme Plasma glutathione peroxidase (GSH-Px) and the antioxidant capacity of plasma experienced a remarkable increase in the colon of colitis-afflicted mice supplemented with the antioxidant-rich Lacto-wolfberry [158]. These advantages primarily arise from the anti-oxidative and anti-inflammatory properties of the ingested compounds [153]. Some phytochemicals have been shown to activate the Nrf2-driven redox pathway [159]. The Nrf2 pathway activates the ARE-dependent antioxidant enzymes [160,161]. The NF-κB activation and cytokines IL-6, IL-8, IL-1β, and TNFα have a pro-inflammatory role in ROS production [160,161]. Curcumin, a metabolite of Turmeric, has been shown to inhibit the NF-κB activation and thus alleviate the oxidative stress [162,163]. Therefore, it can be concluded that antioxidant, prebiotic, and anti-inflammatory are responsible for plant-derived metabolites’ effect.
Plant Metabolites in Personalised Therapy
Personalized therapy (PT), or personalized medicine, involves customizing medical interventions for individual patients by considering their distinctive characteristics like genetics, lifestyle, and diagnostic information [164,165]. The primary goal is to deliver precise treatment to each patient by interindividual heterogeneity, apart from the conventional one-size-fits-all methodology [166]. This strategy leverages advancements in diagnostic imaging, molecular medicine, and artificial intelligence, notably deep learning algorithms such as Convolutional Neural Network (CNN) and recurrent neural network (RNN), to scrutinize patient data and suggest tailored therapies [167]. The need for PT arises due to adverse side effects of conventional medicine, such as multi-drug resistance in gut bacteria. Personalized therapy components are designed to work for a particular genetic repertoire, for example, warfarin and imatinib drugs [168,169]. PT is required to create the drug for a particular mutation, in the case of cystic fibrosis transmembrane conductance regulator (CFTR) gene mutation, the drug Ivacaftor works for a single phenotype [170]. Personalized Therapy can detect the early signs of disease based on personal thresholds of a marker rather than on population thresholds [166]. Similarly, in the case of postoperative colorectal cancer, intake of aspirin leads to increased survival chances in patients who had a somatic mutation in the Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA) gene compared to wild type [171]. The application of personalized treatment shows favorable outcomes in enhancing patient predictions and decreasing healthcare expenses [165].
Any agent that acts as a biomarker for disease and contributes to the development of therapeutics is significant for developing personalized therapy. In this scenario, the human gut microbiota linked to almost every disease can be a substantial agent with microbiome-based therapeutics [172].
The human gut microbiome is a critical factor in personalized therapeutics by impacting drug metabolism, disease development, and treatment methods, offering the potential for personalized therapies [173,174]. Various microbiome-based therapeutic strategies, including prebiotics, probiotics, antibiotics, dietary interventions, fecal microbiota transplantation (FMT), live biotherapeutics, phage therapy, and microbiome mimetics, are being investigated for disease treatment, aided by high-throughput technologies identifying microbial signatures linked to diseases like cancer and inflammatory bowel disease. Many of these techniques have been transitioned from laboratory to clinic, such as ingestible microbiomes (prebiotics, probiotics or postbiotics, and other medicines), which have been approved by the FDA and similar authorities [174,175]. FMT is under preclinical testing from being a strictly regulated biological agent in some countries (USA, Canada, Australia), to a medicinal product or treatment with variable regulation (UK, France, Germany, Switzerland), to no regulation (Austria, Denmark, Sweden, Finland) [176]. Thus, manipulating and understanding the gut microbiome shows potential for advancing personalized therapeutics in medicine.
In this scenario, plant metabolites can effectively influence the microbial population. Plant metabolites play a significant role in microbiome therapeutics by influencing the physiology of endophytic microbes. These metabolites, such as alkaloids, flavonoids, and polyphenols, exhibit antimicrobial properties against various pathogenic bacteria [177]. Additionally, phytochemicals interact with the gut microbiota, modulating its composition and diversity, which in turn impacts human health [79]. Long-term consumption of Ginseng extract rich in ginsenosides enhances the abundance of beneficial bacterial phyla like Bifidobacterium, Allobaculum, Lactobacillus, Clostridium, and Parasutterella [178]. A study reported that a combination of t-resveratrol and ε-viniferin induced changes in microbial functionality and composition, specifically an increase in Enterobacteriales and a decrease in Bacteroidales [179]. Another study investigated the effects of six stilbenoids, including resveratrol, on human fecal microbiota using an in vitro fermentation system. The results showed an increase in the ratio of Bacteroidetes to Firmicutes phyla, a decrease in Clostridium genus and Lachnospiraceae family strains, and an increase in the abundance of Faecalibacterium prausnitzii and Ruminococcus gnavus [180].
Some plant metabolites also function as antibacterial agents against various human pathogens; on the other hand, microbial therapeutics metabolize the plant metabolites, enhancing their bioavailability and efficacy [181]. These metabolites produced by gut microbiota alter the host’s physiology by mimicking the signaling molecules. Bacterial species like Escherichia coli and Blautia produce hydroxylated or glucuronidated metabolites of curcumin, leading to enhanced bioavailability of the compound [182]. Lignan de-glycosylation can be facilitated by Bacteroides and Clostridium, but other important taxa that may be engaged in the subsequent stages include Eubacterium limosum, Blautia producta, Eggerthella lenta, and Acetobacterium dehalogenans [183]. Catechins are broken down by gut bacteria such as Flavonifractor, Eggerthella, and Eubacterium [184] into many small phenolic metabolites are produced, such as γ-valerolactones, hydroxybenzoic acid, hydroxyhippuric acid, pyrogallol, and hydroxyphenyl propionic acid [185]. Another plant metabolite, i.e., ellagitanin is metabolized into urolithins, which regulates the inflammation via suppressing the NF-κB signaling pathway [186]. Overall, plant metabolites play a significant role in shaping microbiome therapeutics through their antimicrobial and health-promoting properties, paving the way for personalized therapy, novel drug discovery, and therapeutic interventions.
Conclusion and Future Perspective
In conclusion, investigating the impact of plant-derived metabolites on the gut microbiota represents a dynamic and evolving field with significant implications for human health. The complex interplay between the bioactive compounds found in plants and the composition of the gut microbiota highlights the potential of plant-based interventions in promoting a well-balanced and resilient microbial community within the digestive system. The observed advantages, from immune system modulation to inflammation mitigation, emphasize plant metabolites’ diverse effects on gut health. As we look toward the future, there is an urgent need for comprehensive mechanistic studies to uncover the specific pathways by which plant compounds influence the gut microbiome. Furthermore, personalized nutrition strategies based on individual gut microbial profiles have the potential to revolutionize our approach to dietary recommendations. Overcoming challenges such as individual variability and standardizing research methodologies will be crucial in translating these findings into practical insights for personalized healthcare. Moreover, it is imperative to integrate sustainability considerations in plant sourcing and cultivation practices into the discussion to ensure the long-term viability of plant-based interventions. The convergence of traditional knowledge and modern scientific approaches is critical to fully realizing the potential of plant formulations in shaping the interactions between the gut microbiota and the host for optimal human health.
Acknowledgement: None.
Funding Statement: The present work was carried out using financial support under Maharshi Dayanand University Rohtak for a Post-Seed Research Grant (DRD/23/75) sanctioned to Dr. NS Chauhan.
Author Contributions: NSC structured the review. NSC and PY wrote the manuscript. All authors edited the manuscript and approved the final draft of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that the review was developed without any commercial or financial association that could be considered a conflict of interest.
References
1. Kumar T, Chauhan NS. Microbiome therapeutics: a path toward sustainable healthcare. In: Glibetic M, editor. Comprehensive Gut Microbiota. Amsterdam, Netherlands: Elsevier; 2022. p. 234–45. [Google Scholar]
2. Yadav M, Chauhan NS, Prasher B, Mukerji M. Dissecting human microbiome for personalized therapy. In: Glibetic M, editor. Comprehensive gut microbiota. Amsterdam, Netherlands: Elsevier; 2022. p. 274–85. [Google Scholar]
3. Wilkins AT, Reimer RA. Obesity, early life gut microbiota, and antibiotics. Microorganisms. 2021;9(2):413. doi:10.3390/microorganisms9020413. [Google Scholar] [PubMed] [CrossRef]
4. Chauhan NS. Microbiome, therapeutics application and epigenetics: conclusion and future prospective. In: Glibetic M, editor. Comprehensive gut microbiota. Amsterdam, Netherlands: Elsevier; 2022. p. 418–27. [Google Scholar]
5. Bubier JA, Chesler EJ, Weinstock GM. Host genetic control of gut microbiome composition. Mamm Genome. 2021;32(4):263–81. doi:10.1007/s00335-021-09884-2. [Google Scholar] [PubMed] [CrossRef]
6. Yen S, Johnson JS. Metagenomics: a path to understanding the gut microbiome. Mamm Genome. 2021;32(4):282–96. doi:10.1007/s00335-021-09889-x. [Google Scholar] [PubMed] [CrossRef]
7. Kumar M, Kumar J, Chauhan NS. Metagenomics: a systemic approach to explore microbial world. In: Kalia VC, editor. Microbial factories. New Delhi: Springer India; 2015. p. 281–98. [Google Scholar]
8. Massey W, Brown JM. The gut microbial endocrine organ in type 2 diabetes. Endocrinology. 2021;162(2):bqaa235. doi:10.1210/endocr/bqaa235. [Google Scholar] [PubMed] [CrossRef]
9. Zhang Y, Chen R, Zhang D, Qi S, Liu Y. Metabolite interactions between host and microbiota during health and disease: which feeds the other? Biomed Pharmacother. 2023;160:114295. doi:10.1016/j.biopha.2023.114295. [Google Scholar] [PubMed] [CrossRef]
10. Li K, Ly K, Mehta S, Braithwaite A. Importance of crosstalk between the microbiota and the neuroimmune system for tissue homeostasis. Clin Transl Immunol. 2022;11(5):e1394. doi:10.1002/cti2.v11.5. [Google Scholar] [CrossRef]
11. Gupta S, Shariff M, Chaturvedi G, Sharma A, Goel N, Yadav M, et al. Comparative analysis of the alveolar microbiome in COPD, ECOPD, sarcoidosis, and ILD patients to identify respiratory illnesses specific microbial signatures. Sci Rep. 2021;11(1):3963. doi:10.1038/s41598-021-83524-2. [Google Scholar] [PubMed] [CrossRef]
12. Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015;26:26191. [Google Scholar] [PubMed]
13. Kandpal M, Indari O, Baral B, Jakhmola S, Tiwari D, Bhandari V, et al. Dysbiosis of gut microbiota from the perspective of the gut-brain axis: role in the provocation of neurological disorders. Metabolites. 2022;12(11):1064. doi:10.3390/metabo12111064. [Google Scholar] [PubMed] [CrossRef]
14. Pferschy-Wenzig EM, Pausan MR, Ardjomand-Woelkart K, Röck S, Ammar RM, Kelber O, et al. Medicinal plants and their impact on the gut microbiome in mental health: a systematic review. Nutrients. 2022;14(10):2111. doi:10.3390/nu14102111. [Google Scholar] [PubMed] [CrossRef]
15. Barbieri R, Coppo E, Marchese A, Daglia M, Sobarzo-Sánchez E, Nabavi SF, et al. Phytochemicals for human disease: an update on plant-derived compounds antibacterial activity. Microbiol Res. 2017;196:44–68. doi:10.1016/j.micres.2016.12.003. [Google Scholar] [PubMed] [CrossRef]
16. Lampe J, Chang J. Interindividual differences in phytochemical metabolism and disposition. Semin Cancer Biol. 2007;17(5):347–53. doi:10.1016/j.semcancer.2007.05.003. [Google Scholar] [PubMed] [CrossRef]
17. Esmeeta A, Adhikary S, Dharshnaa V, Swarnamughi P, Ummul Maqsummiya Z, Banerjee A, et al. Plant-derived bioactive compounds in colon cancer treatment: an updated review. Biomed Pharmacother. 2022;153:113384. doi:10.1016/j.biopha.2022.113384. [Google Scholar] [PubMed] [CrossRef]
18. Waldbaum JDH, Xhumari J, Akinsuyi OS, Arjmandi B, Anton S, Roesch LFW. Association between dysbiosis in the gut microbiota of primary osteoporosis patients and bone loss. Aging Dis. 2023;14(6):2081. doi:10.14336/AD.2023.0425. [Google Scholar] [PubMed] [CrossRef]
19. Kumar J, Kumar M, Pandey R, Chauhan NS. Physiopathology and management of gluten induced celiac disease. J Food Sci. 2017;82(2):270–7. doi:10.1111/jfds.2017.82.issue-2. [Google Scholar] [CrossRef]
20. Yadav M, Kuma T, Maurya R, Pandey R, Chauhan NS. Characterization of Cellulomonas sp. HM71 as potential probiotic strain for human health. Front Cell Infect Microbiol. 2023;12:1082674. doi:10.3389/fcimb.2022.1082674. [Google Scholar] [PubMed] [CrossRef]
21. Feng M, Gao B, Garcia LR, Sun Q. Microbiota-derived metabolites in regulating the development and physiology of Caenorhabditis elegans. Front Microbiol. 2023;14:1035582. doi:10.3389/fmicb.2023.1035582. [Google Scholar] [PubMed] [CrossRef]
22. Ganesan R, Jeong JJ, Kim DJ, Suk KT. Recent trends of microbiota-based microbial metabolites metabolism in liver disease. Front Med. 2022;9:841281. doi:10.3389/fmed.2022.841281. [Google Scholar] [PubMed] [CrossRef]
23. Yadav M, Chauhan NS. Role of gut-microbiota in disease severity and clinical outcomes. Brief Funct Genomics. 2024;23(1):24–37. doi:10.1093/bfgp/elac037. [Google Scholar] [PubMed] [CrossRef]
24. Yadav M, Verma MK, Chauhan NS. A review of metabolic potential of human gut microbiome in human nutrition. Arch Microbiol. 2018;200(2):203–17. doi:10.1007/s00203-017-1459-x. [Google Scholar] [PubMed] [CrossRef]
25. Yoo J, Groer M, Dutra S, Sarkar A, McSkimming D. Gut microbiota and immune system interactions. Microorganisms. 2020;8(10):1587. doi:10.3390/microorganisms8101587. [Google Scholar] [PubMed] [CrossRef]
26. Priyadarshini M, Kotlo KU, Dudeja PK, Layden BT. Role of short chain fatty acid receptors in intestinal physiology and pathophysiology. Compr Physiol. 2018;8(3):1091–115. doi:10.1002/cphy.c170050. [Google Scholar] [PubMed] [CrossRef]
27. Shin Y, Han S, Kwon J, Ju S, Choi T, Kang I, et al. Roles of short-chain fatty acids in inflammatory bowel disease. Nutrients. 2023;15(20):4466. doi:10.3390/nu15204466. [Google Scholar] [PubMed] [CrossRef]
28. Su X, Gao Y, Yang R. Gut microbiota-derived tryptophan metabolites maintain gut and systemic homeostasis. Cells. 2022;11(15):2296. doi:10.3390/cells11152296. [Google Scholar] [PubMed] [CrossRef]
29. Ye X, Li H, Anjum K, Zhong X, Miao S, Zheng G, et al. Dual role of indoles derived from intestinal microbiota on human health. Front Immunol. 2022;13:903526. doi:10.3389/fimmu.2022.903526. [Google Scholar] [PubMed] [CrossRef]
30. Gargaro M, Manni G, Scalisi G, Puccetti P, Fallarino F. Tryptophan metabolites at the crossroad of immune-cell interaction via the aryl hydrocarbon receptor: implications for tumor immunotherapy. Int J Mol Sci. 2021;22(9):4644. doi:10.3390/ijms22094644. [Google Scholar] [PubMed] [CrossRef]
31. Benech N, Rolhion N, Sokol H. Tryptophan metabolites get the gut moving. Cell Host Microbe. 2021;29(2):145–7. doi:10.1016/j.chom.2021.01.009. [Google Scholar] [PubMed] [CrossRef]
32. Yang W, Cong Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell Mol Immunol. 2021;18(4):866–77. doi:10.1038/s41423-021-00661-4. [Google Scholar] [PubMed] [CrossRef]
33. Abraham C, Medzhitov R. Interactions between the host innate immune system and microbes in inflammatory bowel disease. Gastroenterology. 2011;140(6):1729–37. doi:10.1053/j.gastro.2011.02.012. [Google Scholar] [PubMed] [CrossRef]
34. Keogh CE, Rude KM, Gareau MG. Role of pattern recognition receptors and the microbiota in neurological disorders. J Physiol. 2021;599(5):1379–89. doi:10.1113/tjp.v599.5. [Google Scholar] [CrossRef]
35. Wicherska-Pawłowska K, Wróbel T, Rybka J. Toll-like receptors (TLRsNOD-like receptors (NLRsand RIG-I-like receptors (RLRs) in innate immunity. TLRs, NLRs, and RLRs ligands as immunotherapeutic agents for hematopoietic diseases. Int J Mol Sci. 2021;22(24):13397. doi:10.3390/ijms222413397. [Google Scholar] [PubMed] [CrossRef]
36. Silva-Gomes S, Decout A, Nigou J. Pathogen-associated molecular patterns (PAMPs). In: Parnham M, editor. Encyclopedia of inflammatory diseases. Basel: Springer Basel; 2014. p. 1–16. [Google Scholar]
37. Li D, Wu M. Pattern recognition receptors in health and diseases. Sig Transduct Target Ther. 2021;6(1):291. doi:10.1038/s41392-021-00687-0. [Google Scholar] [PubMed] [CrossRef]
38. Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med. 2012;209(8):1445–56. doi:10.1084/jem.20120504. [Google Scholar] [PubMed] [CrossRef]
39. Ghimire L, Paudel S, Jin L, Jeyaseelan S. The NLRP6 inflammasome in health and disease. Mucosal Immunol. 2020;13(3):388–98. doi:10.1038/s41385-020-0256-z. [Google Scholar] [PubMed] [CrossRef]
40. Pabst O, Slack E. IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol. 2020;13(1):12–21. doi:10.1038/s41385-019-0227-4. [Google Scholar] [PubMed] [CrossRef]
41. Takeuchi T, Ohno H. IgA in human health and diseases: potential regulator of commensal microbiota. Front Immunol. 2022;13:1024330. doi:10.3389/fimmu.2022.1024330. [Google Scholar] [PubMed] [CrossRef]
42. Pietrzak B, Tomela K, Olejnik-Schmidt A, Mackiewicz A, Schmidt M. Secretory IgA in intestinal mucosal secretions as an adaptive barrier against microbial cells. Int J Mol Sci. 2020;21(23):9254. doi:10.3390/ijms21239254. [Google Scholar] [PubMed] [CrossRef]
43. Sterlin D, Fadlallah J, Slack E, Gorochov G. The antibody/microbiota interface in health and disease. Mucosal Immunol. 2020;13(1):3–11. doi:10.1038/s41385-019-0192-y. [Google Scholar] [PubMed] [CrossRef]
44. Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4(6):603–11. doi:10.1038/mi.2011.41. [Google Scholar] [PubMed] [CrossRef]
45. León ED, Francino MP. Roles of secretory immunoglobulin a in host-microbiota interactions in the gut ecosystem. Front Microbiol. 2022;13:880484. doi:10.3389/fmicb.2022.880484. [Google Scholar] [PubMed] [CrossRef]
46. Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. doi:10.1016/j.cell.2014.03.011. [Google Scholar] [PubMed] [CrossRef]
47. Khan I, Bai Y, Zha L, Ullah N, Ullah H, Shah SRH, et al. Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front Cell Infect Microbiol. 2021;11:716299. doi:10.3389/fcimb.2021.716299. [Google Scholar] [PubMed] [CrossRef]
48. Patel S, McCormick BA. Mucosal inflammatory response to Salmonella typhimurium infection. Front Immunol. 2014;5:311. [Google Scholar] [PubMed]
49. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020;30(6):492–506. doi:10.1038/s41422-020-0332-7. [Google Scholar] [PubMed] [CrossRef]
50. Christovich A, Luo XM. Gut microbiota, leaky gut, and autoimmune diseases. Front Immunol. 2022;13:946248. doi:10.3389/fimmu.2022.946248. [Google Scholar] [PubMed] [CrossRef]
51. Paray BA, Albeshr MF, Jan AT, Rather IA. Leaky gut and autoimmunity: an intricate balance in individuals health and the diseased state. Int J Mol Sci. 2020;21(24):9770. doi:10.3390/ijms21249770. [Google Scholar] [PubMed] [CrossRef]
52. Ruder B, Atreya R, Becker C. Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int J Mol Sci. 2019;20(8):1887. doi:10.3390/ijms20081887. [Google Scholar] [PubMed] [CrossRef]
53. Cunningham KE, Turner JR. Myosin light chain kinase: pulling the strings of epithelial tight junction function. Ann N Y Acad Sci. 2012;1258(1):34–42. doi:10.1111/j.1749-6632.2012.06526.x. [Google Scholar] [PubMed] [CrossRef]
54. Lavelle EC, Murphy C, O’Neill LAJ, Creagh EM. The role of TLRs, NLRs, and RLRs in mucosal innate immunity and homeostasis. Mucosal Immunol. 2010;3(1):17–28. doi:10.1038/mi.2009.124. [Google Scholar] [PubMed] [CrossRef]
55. Atreya I, Atreya R, Neurath MF. NF-κB in inflammatory bowel disease. J Intern Med. 2008;263(6):591–6. doi:10.1111/jim.2008.263.issue-6. [Google Scholar] [CrossRef]
56. Nighot M, Rawat M, Al-Sadi R, Castillo EF, Nighot P, Ma TY. Lipopolysaccharide-induced increase in intestinal permeability is mediated by TAK-1 activation of IKK and MLCK/MYLK gene. Am J Pathol. 2019;189(4):797–812. doi:10.1016/j.ajpath.2018.12.016. [Google Scholar] [PubMed] [CrossRef]
57. Hu X, Li J, Fu M, Zhao X, Wang W. The JAK/STAT signaling pathway: from bench to clinic. Sig Transduct Target Ther. 2021;6(1):402. doi:10.1038/s41392-021-00791-1. [Google Scholar] [PubMed] [CrossRef]
58. Yu YL, Chen M, Zhu H, Zhuo MX, Chen P, Mao YJ, et al. STAT1 epigenetically regulates LCP2 and TNFAIP2 by recruiting EP300 to contribute to the pathogenesis of inflammatory bowel disease. Clin Epigenet. 2021;13(1):127. doi:10.1186/s13148-021-01101-w. [Google Scholar] [PubMed] [CrossRef]
59. Mao L, Kitani A, Strober W, Fuss IJ. The role of NLRP3 and IL-1β in the pathogenesis of inflammatory bowel disease. Front Immunol. 2018;9:2566. doi:10.3389/fimmu.2018.02566. [Google Scholar] [PubMed] [CrossRef]
60. Ranson N, Veldhuis M, Mitchell B, Fanning S, Cook A, Kunde D, et al. NLRP3-dependent and -independent processing of interleukin (IL)-1β in active ulcerative colitis. Int J Mol Sci. 2018;20(1):57. doi:10.3390/ijms20010057. [Google Scholar] [PubMed] [CrossRef]
61. Luo M, Yeruva S, Liu Y, Chodisetti G, Riederer B, Menon MB, et al. IL-1β-induced downregulation of the multifunctional PDZ adaptor PDZK1 is attenuated by ERK inhibition, RXRα, or PPARα stimulation in enterocytes. Front Physiol. 2017;8:61. [Google Scholar] [PubMed]
62. Datta de D, Datta A, Bhattacharjya S, Roychoudhury S. NF-kappaB mediated transcriptional repression of acid modifying hormone gastrin. PLoS One. 2013;8(8):e73409. doi:10.1371/journal.pone.0073409. [Google Scholar] [PubMed] [CrossRef]
63. Aggeletopoulou I, Kalafateli M, Tsounis EP, Triantos C. Exploring the role of IL-1β in inflammatory bowel disease pathogenesis. Front Med. 2024;11:1307394. doi:10.3389/fmed.2024.1307394. [Google Scholar] [PubMed] [CrossRef]
64. Lin Y, He Z, Ye J, Liu Z, She X, Gao X, et al. Progress in understanding the IL-6/STAT3 pathway in colorectal cancer. OncoTargets Ther. 2020;13:13023–32. doi:10.2147/OTT.S278013. [Google Scholar] [PubMed] [CrossRef]
65. Shahini A, Shahini A. Role of interleukin-6-mediated inflammation in the pathogenesis of inflammatory bowel disease: focus on the available therapeutic approaches and gut microbiome. J Cell Commun Signal. 2023;17(1):55–74. doi:10.1007/s12079-022-00695-x. [Google Scholar] [PubMed] [CrossRef]
66. Gui X, Iacucci M, Ghosh S. Dysregulation of IL6/IL6R-STAT3-SOCS3 signaling pathway in IBD-associated colorectal dysplastic lesions as compared to sporadic colorectal adenomas in non-IBD patients. Pathol Res Pract. 2020;216(11):153211. doi:10.1016/j.prp.2020.153211. [Google Scholar] [PubMed] [CrossRef]
67. Li L, Peng P, Ding N, Jia W, Huang C, Tang Y. Oxidative stress, inflammation, gut dysbiosis: what can polyphenols do in inflammatory bowel disease? Antioxidants. 2023;12(4):967. doi:10.3390/antiox12040967. [Google Scholar] [PubMed] [CrossRef]
68. Guo Y, Chen X, Gong P, Li G, Yao W, Yang W. The gut-organ-axis concept: advances the application of gut-on-chip technology. Int J Mol Sci. 2023;24(4):4089. doi:10.3390/ijms24044089. [Google Scholar] [PubMed] [CrossRef]
69. Chauhan NS. 3.44—Lung microbiome in human health and diseases. In: Glibetic M, editor. Comprehensive gut microbiota. Amsterdam, Netherlands: Elsevier; 2022. p. 497–507. [Google Scholar]
70. Saxami G, Kerezoudi E, Eliopoulos C, Arapoglou D, Kyriacou A. The gut-organ axis within the human body: gut dysbiosis and the role of prebiotics. Life. 2023;13(10):2023. doi:10.3390/life13102023. [Google Scholar] [PubMed] [CrossRef]
71. Morimoto T, Kobayashi T, Kakiuchi T, Esaki M, Tsukamoto M, Yoshihara T, et al. Gut-spine axis: a possible correlation between gut microbiota and spinal degenerative diseases. Front Microbiol. 2023;14:1290858. doi:10.3389/fmicb.2023.1290858. [Google Scholar] [PubMed] [CrossRef]
72. Mukhtar K, Nawaz H, Abid S. Functional gastrointestinal disorders and gut-brain axis: what does the future hold? World J Gastroenterol. 2019;25(5):552–66. doi:10.3748/wjg.v25.i5.552. [Google Scholar] [PubMed] [CrossRef]
73. Roy A, Khan A, Ahmad I, Alghamdi S, Rajab BS, Babalghith AO, et al. Flavonoids a bioactive compound from medicinal plants and its therapeutic applications. Ullah R, editor. BioMed Res Int. 2022;2022:5445291. [Google Scholar] [PubMed]
74. Parker A, Fonseca S, Carding SR. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes. 2020;11(2):135–57. doi:10.1080/19490976.2019.1638722. [Google Scholar] [PubMed] [CrossRef]
75. Kan J, Wu F, Wang F, Zheng J, Cheng J, Li Y, et al. Phytonutrients: sources, bioavailability, interaction with gut microbiota, and their impacts on human health. Front Nutr. 2022;9:960309. doi:10.3389/fnut.2022.960309. [Google Scholar] [PubMed] [CrossRef]
76. Luo B, Wen Y, Ye F, Wu Y, Li N, Farid MS, et al. Bioactive phytochemicals and their potential roles in modulating gut microbiota. J Agric Res. 2023;12:100583. [Google Scholar]
77. Liu J, Tan Y, Cheng H, Zhang D, Feng W, Peng C. Functions of gut microbiota metabolites, current status and future perspectives. Aging Dis. 2022;13(4):1106. doi:10.14336/AD.2022.0104. [Google Scholar] [PubMed] [CrossRef]
78. Ghosh S, Whitley CS, Haribabu B, Jala VR. Regulation of intestinal barrier function by microbial metabolites. Cell Mol Gastroenterol Hepatol. 2021;11(5):1463–82. doi:10.1016/j.jcmgh.2021.02.007. [Google Scholar] [PubMed] [CrossRef]
79. di Vincenzo F, Del Gaudio A, Petito V, Lopetuso LR, Scaldaferri F. Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Intern Emerg Med. 2023;19:275–93. [Google Scholar] [PubMed]
80. Zhang Y, Zhu X, Yu X, Novák P, Gui Q, Yin K. Enhancing intestinal barrier efficiency: a novel metabolic diseases therapy. Front Nutr. 2023;10:1120168. doi:10.3389/fnut.2023.1120168. [Google Scholar] [PubMed] [CrossRef]
81. Bland JS. Application of phytochemicals in immune disorders: their roles beyond antioxidants. Integr Med Encinitas Calif. 2021;20(5):16–21. [Google Scholar]
82. Zebeaman M, Tadesse MG, Bachheti RK, Bachheti A, Gebeyhu R, Chaubey KK. Plants and plant-derived molecules as natural immunomodulators. Li MH, editor. BioMed Research International. 2023;2023:1–14. [Google Scholar]
83. Jit BP, Pradhan B, Dash R, Bhuyan PP, Behera C, Behera RK, et al. Phytochemicals: potential therapeutic modulators of radiation induced signaling pathways. Antioxidants. 2021;11(1):49. doi:10.3390/antiox11010049. [Google Scholar] [PubMed] [CrossRef]
84. Lakshmi SP, Reddy AT, Kodidhela LD, Varadacharyulu NC. The tea catechin epigallocatechin gallate inhibits NF-κB-mediated transcriptional activation by covalent modification. Arch Biochem Biophys. 2020;695:108620. doi:10.1016/j.abb.2020.108620. [Google Scholar] [PubMed] [CrossRef]
85. Fakhri S, Moradi SZ, Yarmohammadi A, Narimani F, Wallace CE, Bishayee A. Modulation of TLR/NF-κB/NLRP signaling by bioactive phytocompounds: a promising strategy to augment cancer chemotherapy and immunotherapy. Front Oncol. 2022;12:834072. doi:10.3389/fonc.2022.834072. [Google Scholar] [PubMed] [CrossRef]
86. Takada Y, Aggarwal BB. Flavopiridol inhibits NF-κB activation induced by various carcinogens and inflammatory agents through inhibition of IκBα kinase and p65 phosphorylation. J Biol Chem. 2004;279(6):4750–9. doi:10.1074/jbc.M304546200. [Google Scholar] [PubMed] [CrossRef]
87. Yücel Ç, Karatoprak GŞ, Açıkara Ö.B, Akkol EK, Barak TH, Sobarzo-Sánchez E, et al. Immunomodulatory and anti-inflammatory therapeutic potential of gingerols and their nanoformulations. Front Pharmacol. 2022;13:902551. doi:10.3389/fphar.2022.902551. [Google Scholar] [PubMed] [CrossRef]
88. Mohammed MA. Fighting cytokine storm and immunomodulatory deficiency: by using natural products therapy up to now. Front Pharmacol. 2023;14:1111329. doi:10.3389/fphar.2023.1111329. [Google Scholar] [PubMed] [CrossRef]
89. Behl T, Kumar K, Brisc C, Rus M, Nistor-Cseppento DC, Bustea C, et al. Exploring the multifocal role of phytochemicals as immunomodulators. Biomed Pharmacother. 2021;133:110959. doi:10.1016/j.biopha.2020.110959. [Google Scholar] [PubMed] [CrossRef]
90. Pei Y, Parks JS, Kang HW. Quercetin alleviates high-fat diet-induced inflammation in brown adipose tissue. J Funct Foods. 2021;85:104614. doi:10.1016/j.jff.2021.104614. [Google Scholar] [CrossRef]
91. Xiong G, Ji W, Wang F, Zhang F, Xue P, Cheng M, et al. Quercetin inhibits inflammatory response induced by LPS from Porphyromonas gingivalis in human gingival fibroblasts via suppressing NF-κB signaling pathway. BioMed Res Int. 2019;2019:1–10. doi:10.1155/2019/6282635. [Google Scholar] [PubMed] [CrossRef]
92. Özenver N, Efferth T. Phytochemical inhibitors of the NLRP3 inflammasome for the treatment of inflammatory diseases. Pharmacol Res. 2021;170:105710. doi:10.1016/j.phrs.2021.105710. [Google Scholar] [PubMed] [CrossRef]
93. Zhang X, Wang G, Gurley EC, Zhou H. Flavonoid apigenin inhibits lipopolysaccharide-induced inflammatory response through multiple mechanisms in macrophages. PLoS One. 2014;9(9):e107072. doi:10.1371/journal.pone.0107072. [Google Scholar] [PubMed] [CrossRef]
94. Blevins HM, Xu Y, Biby S, Zhang S. The NLRP3 inflammasome pathway: a review of mechanisms and inhibitors for the treatment of inflammatory diseases. Front Aging Neurosci. 2022;14:879021. doi:10.3389/fnagi.2022.879021. [Google Scholar] [PubMed] [CrossRef]
95. Lim H, Min DS, Park H, Kim HP. Flavonoids interfere with NLRP3 inflammasome activation. Toxicol Appl Pharmacol. 2018;355:93–102. doi:10.1016/j.taap.2018.06.022. [Google Scholar] [PubMed] [CrossRef]
96. Tanase DM, Valasciuc E, Gosav EM, Ouatu A, Buliga-Finis ON, Floria M, et al. Portrayal of NLRP3 inflammasome in atherosclerosis: current knowledge and therapeutic targets. Int J Mol Sci. 2023;24(9):8162. doi:10.3390/ijms24098162. [Google Scholar] [PubMed] [CrossRef]
97. Ahmad S, Israf DA, Lajis NHJ, Shaari K, Mohamed H, Wahab AA, et al. Cardamonin, inhibits pro-inflammatory mediators in activated RAW 264.7 cells and whole blood. Eur J Pharmacol. 2006;538(1–3):188–94. [Google Scholar] [PubMed]
98. Peng YJ, Lu JW, Lee CH, Lee HS, Chu YH, Ho YJ, et al. Cardamonin attenuates inflammation and oxidative stress in interleukin-1β-stimulated osteoarthritis chondrocyte through the Nrf2 pathway. Antioxidants. 2021;10(6):862. doi:10.3390/antiox10060862. [Google Scholar] [PubMed] [CrossRef]
99. Wang X, Peng H, Huang Y, Kong W, Cui Q, Du J, et al. Post-translational modifications of IκBα: the state of the art. Front Cell Dev Biol. 2020;8:574706. doi:10.3389/fcell.2020.574706. [Google Scholar] [PubMed] [CrossRef]
100. Daimary UD, Parama D, Rana V, Banik K, Kumar A, Harsha C, et al. Emerging roles of cardamonin, a multitargeted nutraceutical in the prevention and treatment of chronic diseases. Curr Res Pharmacol Drug Discov. 2021;2:100008. doi:10.1016/j.crphar.2020.100008. [Google Scholar] [PubMed] [CrossRef]
101. He X, Liu J, Long G, Xia XH. Liu M. 2,3,5,4′-Tetrahydroxystilbene-2-O-β-D-glucoside, a major bioactive component from Polygonimultiflori Radix (Heshouwu) suppresses DSS induced acute colitis in BALb/c mice by modulating gut microbiota. Biomed Pharmacother. 2021;137:111420. doi:10.1016/j.biopha.2021.111420. [Google Scholar] [PubMed] [CrossRef]
102. Shi G, Jiang H, Feng J, Zheng X, Zhang D, Jiang C, et al. Aloe vera mitigates dextran sulfate sodium-induced rat ulcerative colitis by potentiating colon mucus barrier. J Ethnopharmacol. 2021;279:114108. doi:10.1016/j.jep.2021.114108. [Google Scholar] [PubMed] [CrossRef]
103. Lee YS, Ju HK, Kim YJ, Lim TG, Uddin MR, Kim YB, et al. Enhancement of Anti-inflammatory activity of aloe vera adventitious root extracts through the alteration of primary and secondary metabolites via salicylic acid elicitation. PLoS One. 2013;8(12):e82479. doi:10.1371/journal.pone.0082479. [Google Scholar] [PubMed] [CrossRef]
104. Xu X, Wang W, Chen Y, Zhang Q, Li B, Zhong Y, et al. Simultaneous determination of ten Bioactive components from Shenling Baizhu San in rat plasma by UHPLC-MS/MS: applicatiom to a comprehensive pharmacokinetic study in normal and two models of Ulcerative Colitis Rats. Evid-Based Compl Alt. 2021;2021:3518241. [Google Scholar]
105. Valero MS, González M, Ramón-Gimenez M, Andrade PB, Moreo E, Les F, et al. Jasoniaglutinosa (L.) DC., a traditional herbal medicine, reduces inflammation, oxidative stress and protects the intestinal barrier in a murine model of colitis. Inflammopharmacology. 2020;28(6):1717–34. doi:10.1007/s10787-019-00626-0. [Google Scholar] [PubMed] [CrossRef]
106. Les F, Cásedas G, Valero MS, Arbonés-Mainar JM, López V. Rock tea (Jasonia glutinosa (L.) DC.) polyphenolic extract inhibits triglyceride accumulation in 3T3-L1 adipocyte-like cells and obesity related enzymes in vitro. Food Funct. 2020;11(10):8931–8. doi:10.1039/D0FO01497D. [Google Scholar] [PubMed] [CrossRef]
107. Xu Q, Yao Y, Liu Y, Zhang J, Mao L. The mechanism of traditional medicine in alleviating ulcerative colitis: regulating intestinal barrier function. Front Pharmacol. 2023;14:1228969. doi:10.3389/fphar.2023.1228969. [Google Scholar] [PubMed] [CrossRef]
108. Gieryńska M, Szulc-Dąbrowska L, Struzik J, Mielcarska MB, Gregorczyk-Zboroch KP. Integrity of the intestinal barrier: the involvement of epithelial cells and microbiota—A mutual relationship. Animals. 2022;12(2):145. doi:10.3390/ani12020145. [Google Scholar] [PubMed] [CrossRef]
109. Kumar T, Pandey R, Chauhan NS. Hypoxia inducible factor-1α: the curator of gut homeostasis. Front Cell Infect Microbiol. 2020;10:227. doi:10.3389/fcimb.2020.00227. [Google Scholar] [PubMed] [CrossRef]
110. Odenwald MA, Turner JR. The intestinal epithelial barrier: a therapeutic target? Nat Rev Gastroenterol Hepatol. 2017;14(1):9–21. doi:10.1038/nrgastro.2016.169. [Google Scholar] [PubMed] [CrossRef]
111. Nogal A, Valdes AM, Menni C. The role of short-chain fatty acids in the interplay between gut microbiota and diet in cardio-metabolic health. Gut Microbes. 2021;13(1):1897212. doi:10.1080/19490976.2021.1897212. [Google Scholar] [PubMed] [CrossRef]
112. Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020;38(1):23–48. doi:10.1146/annurev-immunol-070119-115104. [Google Scholar] [PubMed] [CrossRef]
113. Sharma BR, Jaiswal S, Ravindra PV. Modulation of gut microbiota by bioactive compounds for prevention and management of type 2 diabetes. Biomed Pharmacother. 2022;152:113148. doi:10.1016/j.biopha.2022.113148. [Google Scholar] [PubMed] [CrossRef]
114. Elkington PTG, O’Kane CM, Friedland JS. The paradox of matrix metalloproteinases in infectious disease. Clin Exp Immunol. 2005;142(1):12–20. doi:10.1111/j.1365-2249.2005.02840.x. [Google Scholar] [PubMed] [CrossRef]
115. Wang D, Wang XH, Yu X, Cao F, Cai X, Chen P, et al. Pharmacokinetics of anthraquinones from medicinal plants. Front Pharmacol. 2021;12:638993. doi:10.3389/fphar.2021.638993. [Google Scholar] [PubMed] [CrossRef]
116. Bhat AA, Uppada S, Achkar IW, Hashem S, Yadav SK, Shanmugakonar M, et al. Tight junction proteins and signaling pathways in cancer and inflammation: a functional crosstalk. Front Physiol. 2019;9:1942. doi:10.3389/fphys.2018.01942. [Google Scholar] [PubMed] [CrossRef]
117. Edelblum KL, Turner JR. The tight junction in inflammatory disease: communication breakdown. Curr Opin Pharmacol. 2009;9(6):715–20. doi:10.1016/j.coph.2009.06.022. [Google Scholar] [PubMed] [CrossRef]
118. Huang S, Wang X, Xie X, Su Y, Pan Z, Li Y, et al. Dahuang Mudan decoction repairs intestinal barrier in chronic colitic mice by regulating the function of ILC3. J Ethnopharmacol. 2022;299:115652. doi:10.1016/j.jep.2022.115652. [Google Scholar] [PubMed] [CrossRef]
119. Andrade AWL, Guerra GCB, De Souza Araújo DF, de Araújo Júnior RF, de Araújo AA, de Carvalho TG, et al. Anti-inflammatory and chemopreventive effects of Bryophyllumpinnatum (Lamarck) leaf extract in experimental colitis models in rodents. Front Pharmacol. 2020;11:998. doi:10.3389/fphar.2020.00998. [Google Scholar] [PubMed] [CrossRef]
120. Cui L, Guan X, Ding W, Luo Y, Wang W, Bu W, et al. Scutellariabaicalensis Georgi polysaccharide ameliorates DSS-induced ulcerative colitis by improving intestinal barrier function and modulating gut microbiota. Int J Biol Macromol. 2021;166:1035–45. doi:10.1016/j.ijbiomac.2020.10.259. [Google Scholar] [PubMed] [CrossRef]
121. Al-Shaibi AA, Abdel-Motal UM, Hubrack SZ, Bullock AN, Al-Marri AA, Agrebi N, et al. Human AGR2 deficiency causes mucus barrier dysfunction and infantile inflammatory bowel disease. Cell Mol Gastroenterol Hepatol. 2021;12(5):1809–30. doi:10.1016/j.jcmgh.2021.07.001. [Google Scholar] [PubMed] [CrossRef]
122. Park MY, Kwon HJ, Sung MK. Dietary aloin, aloesin, or aloe-gel exerts anti-inflammatory activity in a rat colitis model. Life Sci. 2011;88(11–12):486–92. [Google Scholar] [PubMed]
123. Dong L, Fan X, Li Q, Li J, Chen G, Yang D, et al. Anemone chinensis Bunge aqueous enema alleviates dextran sulfate sodium-induced colitis via inhibition of inflammation and regulation of the colonic mucosal microbiota. J Ethnopharmacol. 2022;288:114916. doi:10.1016/j.jep.2021.114916. [Google Scholar] [PubMed] [CrossRef]
124. Han KS, Balan P, Hong HD, Choi WI, Cho CW, Lee YC, et al. Korean ginseng modulates the ileal microbiota and mucin gene expression in the growing rat. Food Funct. 2014;5(7):1506. doi:10.1039/c4fo00087k. [Google Scholar] [PubMed] [CrossRef]
125. Zhao L, Sui M, Zhang T, Zhang K. The interaction between ginseng and gut microbiota. Front Nutr. 2023;10:1301468. doi:10.3389/fnut.2023.1301468. [Google Scholar] [PubMed] [CrossRef]
126. Mirmohammadali SN, Rosenkranz SK. Dietary phytochemicals, gut microbiota composition, and health outcomes in human and animal models. Biosci Microbiota Food Health. 2023;42(3):152–71. doi:10.12938/bmfh.2022-078. [Google Scholar] [PubMed] [CrossRef]
127. de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: mechanistic insights. Gut. 2022;71(5):1020–32. doi:10.1136/gutjnl-2021-326789. [Google Scholar] [PubMed] [CrossRef]
128. Santhiravel S, Bekhit AEDA, Mendis E, Jacobs JL, Dunshea FR, Rajapakse N, et al. The impact of plant phytochemicals on the gut microbiota of humans for a balanced life. Int J Mol Sci. 2022;23(15):8124. doi:10.3390/ijms23158124. [Google Scholar] [PubMed] [CrossRef]
129. Corrêa TAF, Rogero MM, Hassimotto NMA, Lajolo FM. The two-way polyphenols-microbiota interactions and their effects on obesity and related metabolic diseases. Front Nutr. 2019;6:188. doi:10.3389/fnut.2019.00188. [Google Scholar] [PubMed] [CrossRef]
130. Milutinović M, Dimitrijević-Branković S, Rajilić-Stojanović M. Plant extracts rich in polyphenols as potent modulators in the growth of probiotic and pathogenic intestinal microorganisms. Front Nutr. 2021;8:688843. doi:10.3389/fnut.2021.688843. [Google Scholar] [PubMed] [CrossRef]
131. Wu J, Wei Z, Cheng P, Qian C, Xu F, Yang Y, et al. Rhein modulates host purine metabolism in intestine through gut microbiota and ameliorates experimental colitis. Theranostics. 2020;10(23):10665–79. doi:10.7150/thno.43528. [Google Scholar] [PubMed] [CrossRef]
132. Li Q, Cui Y, Xu B, Wang Y, Lv F, Li Z, et al. Main active components of Jiawei Gegen Qinlian decoction protects against ulcerative colitis under different dietary environments in a gut microbiota-dependent manner. Pharmacol Res. 2021;170:105694. doi:10.1016/j.phrs.2021.105694. [Google Scholar] [PubMed] [CrossRef]
133. Xie X, Wu Y, Xie H, Wang H, Zhang X, Yu J, et al. Polysaccharides, next potential agent for the treatment of epilepsy? Front Pharmacol. 2022;13:790136. doi:10.3389/fphar.2022.790136. [Google Scholar] [PubMed] [CrossRef]
134. Cao Z, Ding Y, Liu Z, Liu M, Wu H, Zhao J, et al. Extraction condition optimization and prebiotic potential of dandelion (Taraxacum mongolicum Hand.-Mazz.) polysaccharides. Ind Crops Prod. 2023;194:116318. doi:10.1016/j.indcrop.2023.116318. [Google Scholar] [CrossRef]
135. Qassadi FI, Zhu Z, Monaghan TM. Plant-derived products with therapeutic potential against gastrointestinal bacteria. Pathogens. 2023;12(2):333. doi:10.3390/pathogens12020333. [Google Scholar] [PubMed] [CrossRef]
136. Kaur AP, Bhardwaj S, Dhanjal DS, Nepovimova E, Cruz-Martins N, Kuča K, et al. Plant prebiotics and their role in the amelioration of diseases. Biomolecules. 2021;11(3):440. doi:10.3390/biom11030440. [Google Scholar] [PubMed] [CrossRef]
137. LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact. 2017;16(1):79. doi:10.1186/s12934-017-0691-z. [Google Scholar] [PubMed] [CrossRef]
138. Du H, Zhang G, Yang G, Tian W, Wang W, Wang P, et al. Study on the effect of huang lian ji edu decoction on the composition of gut microflora in SD rats based on 16S rRNA sequencing. Evid-Based Compl Alt. 2020;2020:8872439. doi:10.1155/2020/8872439. [Google Scholar] [PubMed] [CrossRef]
139. Su M, Tang T, Tang W, Long Y, Wang L, Liu M. Astragalus improves intestinal barrier function and immunity by acting on intestibal microbiota to treat T2DM: a research review. Front Immunol. 2023;14:1243834. doi:10.3389/fimmu.2023.1243834. [Google Scholar] [PubMed] [CrossRef]
140. Yan D, Sun Y, Zhou X, Si W, Liu J, Li M, et al. Regulatory effect of gut microbes on blood pressure. Anim Models and Exp Med. 2022;5(6):513–31. doi:10.1002/ame2.v5.6. [Google Scholar] [CrossRef]
141. Li Y, Fu R, Li R, Zeng J, Liu T, Li X, et al. Causality of gut microbiome and hypertension: a bidirectional mendelian randomization study. Front Cardiovasc Med. 2023;10:1167346. doi:10.3389/fcvm.2023.1167346. [Google Scholar] [PubMed] [CrossRef]
142. Kwon C, Ediriweera MK, Kim Cho S. Interplay between phytochemicals and the colonic microbiota. Nutrients. 2023;15(8):1989. doi:10.3390/nu15081989. [Google Scholar] [PubMed] [CrossRef]
143. Shabbir U, Rubab M, Daliri EBM, Chelliah R, Javed A, Curcumin Oh DH, et al. Catechins and metabolic diseases: the role of gut microbiota. Nutrients. 2021;13(1):206. doi:10.3390/nu13010206. [Google Scholar] [CrossRef]
144. Tarawneh R, Penhos E. The gut microbiome and Alzheimer’s disease: complex and bidirectional interactions. Neurosci Biobehav Rev. 2022;141:104814. [Google Scholar] [PubMed]
145. Boronat A, Rodriguez-Morató J, Serreli G, Fitó M, Tyndale RF, Deiana M, et al. Contribution of biotransformations carried out by the microbiota, drug-metabolizing enzymes, and transport proteins to the biological activities of phytochemicals found in the diet. Adv Nutr. 2021;12(6):2172–89. doi:10.1093/advances/nmab085. [Google Scholar] [PubMed] [CrossRef]
146. Korpelainen H, Pietiläinen M. Hop (Humulus lupulus L.traditional and present use, and future potential. Econ Bot. 2021;75(3–4):302–22. [Google Scholar]
147. TCelebi Sozener Z, Ozdel Ozturk B, Cerci P, Turk M, Gorgulu Akin B, Akdis M, et al. Epithelial barrier hypothesis: effect of the external exposome on the microbiome and epithelial barriers in allergic disease. Allergy. 2022;77(5):1418–49. doi:10.1111/all.v77.5. [Google Scholar] [CrossRef]
148. Castaneda OA, Lee SC, Ho CT, Huang TC. Macrophages in oxidative stress and models to evaluate the antioxidant function of dietary natural compounds. J Food Drug Anal. 2017;25(1):111–8. doi:10.1016/j.jfda.2016.11.006. [Google Scholar] [PubMed] [CrossRef]
149. Wang Y, Chen Y, Zhang X, Lu Y, Chen H. New insights in intestinal oxidative stress damage and the health intervention effects of nutrients: a review. J Funct Foods. 2020;75:104248. doi:10.1016/j.jff.2020.104248. [Google Scholar] [CrossRef]
150. Barbara G, Barbaro MR, Fuschi D, Palombo M, Falangone F, Cremon C, et al. Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front Nutr. 2021;8:718356. doi:10.3389/fnut.2021.718356. [Google Scholar] [PubMed] [CrossRef]
151. Khan I, Ullah N, Zha L, Bai Y, Khan A, Zhao T, et al. Alteration of gut microbiota in inflammatory bowel disease (IBDcause or consequence? IBD treatment targeting the gut microbiome. Pathogens. 2019;8(3):126. doi:10.3390/pathogens8030126. [Google Scholar] [PubMed] [CrossRef]
152. Bruewer M, Luegering A, Kucharzik T, Parkos CA, Madara JL, Hopkins AM, et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171(11):6164–72. doi:10.4049/jimmunol.171.11.6164. [Google Scholar] [PubMed] [CrossRef]
153. Riaz Rajoka MS, Thirumdas R, Mehwish HM, Umair M, Khurshid M, Hayat HF, et al. Role of food antioxidants in modulating gut microbial communities: novel understandings in intestinal oxidative stress damage and their impact on host health. Antioxidants. 2021;10(10):1563. doi:10.3390/antiox10101563. [Google Scholar] [PubMed] [CrossRef]
154. Espinoza-Culupú A, Del Santos N, Farfán-López M, Mendes E, da Silva Junior PI, Marques Borges M. In silico and in vitro approach for evaluation of the anti-inflammatory and antioxidant potential of mygalin. IJMS. 2023;24(23):17019. doi:10.3390/ijms242317019. [Google Scholar] [PubMed] [CrossRef]
155. Dewanjee S, Chakraborty P, Mukherjee B, de Feo V. Plant-based antidiabetic nanoformulations: the emerging paradigm for effective therapy. Int J Mol Sci. 2020;21(6):2217. doi:10.3390/ijms21062217. [Google Scholar] [PubMed] [CrossRef]
156. Eroglu A, Al’Abri IS, Kopec RE, Crook N, Bohn T. Carotenoids and their health benefits as derived via their interactions with gut microbiota. Adv Nutr. 2023;14(2):238–55. doi:10.1016/j.advnut.2022.10.007. [Google Scholar] [PubMed] [CrossRef]
157. Rajput SA, Liang SJ, Wang XQ, Yan HC. Lycopene protects intestinal epithelium from deoxynivalenol-induced oxidative damage via regulating Keap1/Nrf2 signaling. Antioxidants. 2021;10(9):1493. doi:10.3390/antiox10091493. [Google Scholar] [PubMed] [CrossRef]
158. Philippe D. Anti-inflammatory effects of lacto-wolfberry in a mouse model of experimental colitis. World J Gastroenterol. 2012;18(38):5351. doi:10.3748/wjg.v18.i38.5351. [Google Scholar] [PubMed] [CrossRef]
159. Saleh HA, Yousef MH, Abdelnaser A. The anti-inflammatory properties of phytochemicals and their effects on epigenetic mechanisms involved in TLR4/NF-κB-mediated inflammation. Front Immunol. 2021;12:606069. doi:10.3389/fimmu.2021.606069. [Google Scholar] [PubMed] [CrossRef]
160. Zhang M, An C, Gao Y, Leak RK, Chen J, Zhang F. Emerging roles of Nrf2 and phase II antioxidant enzymes in neuroprotection. Prog Neurobiol. 2013;100:30–47. doi:10.1016/j.pneurobio.2012.09.003. [Google Scholar] [PubMed] [CrossRef]
161. Liu T, Zhang L, Joo D, Sun SC. NF-κB signaling in inflammation. Sig Transduct Target Ther. 2017;2(1):17023. doi:10.1038/sigtrans.2017.23. [Google Scholar] [PubMed] [CrossRef]
162. Sharifi-Rad J, Rayess YE, Rizk AA, Sadaka C, Zgheib R, Zam W, et al. Turmeric and its major compound curcumin on health: bioactive effects and safety profiles for food. Pharmaceutical Biotechnol Med Appl Front Pharmacol. 2020;11:01021. [Google Scholar]
163. Mittal A, Kumar N, Chauhan NS. Curcumin encapsulated PEGylated nanoliposomes: a potential anti-infective therapeutic agent. Indian J Microbiol. 2019;59(3):336–43. doi:10.1007/s12088-019-00811-3. [Google Scholar] [PubMed] [CrossRef]
164. Chauhan NS, Kumar S. Microbiome therapeutics: personalized therapy beyond conventional approaches. Amsterdam, Netherlands: Elsevier; 2023. [Google Scholar]
165. Goetz LH, Schork NJ. Personalized medicine: motivation, challenges, and progress. Fertil Steril. 2018;109(6):952–63. doi:10.1016/j.fertnstert.2018.05.006. [Google Scholar] [PubMed] [CrossRef]
166. Wang RC, Wang Z. Precision medicine: disease subtyping and tailored treatment. Cancers. 2023;15(15):3837. doi:10.3390/cancers15153837. [Google Scholar] [PubMed] [CrossRef]
167. Najjar R. Redefining radiology: a review of artificial intelligence integration in medical imaging. Diagnostics. 2023;13(17):2760. doi:10.3390/diagnostics13172760. [Google Scholar] [PubMed] [CrossRef]
168. Wigle T, Jansen L, Teft W, Kim R. Pharmacogenomics guided-personalization of warfarin and tamoxifen. J Pers Med. 2017;7(4):20. doi:10.3390/jpm7040020. [Google Scholar] [PubMed] [CrossRef]
169. Druker BJ. Imatinib as a paradigm of targeted therapies. Adv Cancer Res. 2004;91:1–30. doi:10.1016/S0065-230X(04)91001-9. [Google Scholar] [PubMed] [CrossRef]
170. Sermet-Gaudelus I. Ivacaftor treatment in patients with cystic fibrosis and the G551D-CFTR mutation. Eur Respir Rev. 2013;22(127):66–71. doi:10.1183/09059180.00008512. [Google Scholar] [PubMed] [CrossRef]
171. Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367(17):1596–606. doi:10.1056/NEJMoa1207756. [Google Scholar] [PubMed] [CrossRef]
172. Chauhan NS. Personalized nutrition, personalized medicine, and microbiome therapeutics. In: Chauhan NS, Kumar S, editors. Microbiome therapeutics: personalized therapy beyond conventional approaches. Cambridge, England: Academic Press; 2023. p. 387–99. [Google Scholar]
173. Yadav M, Chauhan NS. Microbiome therapeutics: exploring the present scenario and challenges. Gastroenterol Rep. 2022;10:goab046. doi:10.1093/gastro/goab046. [Google Scholar] [PubMed] [CrossRef]
174. Chauhan NS, Kumar S. Conclusion and future perspective. In: Chauhan NS, Kumar S, editor. Microbiome therapeutics: personalized therapy beyond conventional approaches. Cambridge, England: Academic Press; 2023. p. 501–5. [Google Scholar]
175. Singh A, Midha V, Chauhan NS, Sood A. Current perspectives on fecal microbiota transplantation in inflammatory bowel disease. Indian J Gastroenterol. 2024;43(1):129–44. doi:10.1007/s12664-023-01516-8. [Google Scholar] [PubMed] [CrossRef]
176. Gulliver EL, Young RB, Chonwerawong M, D’Adamo GL, Thomason T, Widdop JT, et al. Review article: the future of microbiome-based therapeutics. Aliment Pharmacol Ther. 2022l;56(2):192–208. doi:10.1111/apt.17049. [Google Scholar] [PubMed] [CrossRef]
177. Singh M, Kumar A, Singh R, Pandey KD. Endophytic bacteria: a new source of bioactive compounds. 3 Biotech. 2017;7(5):315. doi:10.1007/s13205-017-0942-z. [Google Scholar] [PubMed] [CrossRef]
178. St-Laurent T, Hammami R. The untapped potential of ginsenosides and American ginseng berry in promoting mental health via the gut-brain axis. Nutrients. 2022;14(12):2523. doi:10.3390/nu14122523. [Google Scholar] [PubMed] [CrossRef]
179. Kim TT, Parajuli N, Sung MM, Bairwa SC, Levasseur J, Soltys CLM, et al. Fecal transplant from resveratrol-fed donors improves glycaemia and cardiovascular features of the metabolic syndrome in mice. Am J Physiol-Endocrinol and Metab. 2018;315(4):511–9. doi:10.1152/ajpendo.00471.2017. [Google Scholar] [PubMed] [CrossRef]
180. Jaimes J, Jarosova V, Vesely O, Mekadim C, Mrazek J, Marsik P, et al. Effect of selected stilbenoids on human fecal microbiota. Molecules. 2019;24(4):744184. [Google Scholar]
181. Yadav M, Chauhan NS. Overview of the rules of the microbial engagement in gut microbiome: a step towards microbiome therapeutics. J Appl Microbiol. 2021;130(6):1425–41. [Google Scholar] [PubMed]
182. Dei Cas M, Ghidoni R. Dietary curcumin: correlation between bioavailability and health potential. Nutrients. 2019;11(9):2147. doi:10.3390/nu11092147. [Google Scholar] [PubMed] [CrossRef]
183. Clavel T, Doré J, Blaut M. Bioavailability of lignans in human subjects. Nutr Res Rev. 2006;19(2):187–96. doi:10.1017/S0954422407249704. [Google Scholar] [PubMed] [CrossRef]
184. Takagaki A, Nanjo F. Catabolism of (+)-catechin and (−)-epicatechin by rat intestinal microbiota. J Agric Food Chem. 2013;61(20):4927–35. doi:10.1021/jf304431v. [Google Scholar] [PubMed] [CrossRef]
185. van Duynhoven J, Vaughan EE, van Dorsten F, Gomez-Roldan V, de Vos R, Vervoort J, et al. Interactions of black tea polyphenols with human gut microbiota: implications for gut and cardiovascular health. Am J Clin Nutr. 2013;98(6):1631S–41S. doi:10.3945/ajcn.113.058263. [Google Scholar] [PubMed] [CrossRef]
186. Djedjibegovic J, Marjanovic A, Panieri E, Saso L. Ellagic acid-derived urolithins as modulators of oxidative stress. Oxid Med Cell Longev. 2020;2020:1–15. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools