 Open Access
Open Access
REVIEW
Metabolic engineering and genome editing strategies for enhanced lipid production in microalgae
1 Department of Biotechnology, Vignan’s Foundation Science, Technology and Research, Vadlamudi, Andhra Pradesh, 522213, India
2 ICAR-Directorate of Floricultural Research, Pune, 412307, India
* Corresponding Author: NUNE SATYA SAMPATH KUMAR. Email:
BIOCELL 2024, 48(8), 1181-1195. https://doi.org/10.32604/biocell.2024.050540
Received 09 February 2024; Accepted 14 June 2024; Issue published 02 August 2024
Abstract
Depleting global petroleum reserves and skyrocketing prices coupled with succinct supply have been a grave concern, which needs alternative sources to conventional fuels. Oleaginous microalgae have been explored for enhanced lipid production, leading towards biodiesel production. These microalgae have short life cycles, require less labor, and space, and are easy to scale up. Triacylglycerol, the primary source of lipids needed to produce biodiesel, is accumulated by most microalgae. The article focuses on different types of oleaginous microalgae, which can be used as a feedstock to produce biodiesel. Lipid biosynthesis in microalgae occurs through fatty acid synthesis and TAG synthesis approaches. In-depth discussions are held regarding other efficient methods for enhancing fatty acid and TAG synthesis, regulating TAG biosynthesis bypass methods, blocking competing pathways, multigene approach, and genome editing. The most potential targets for gene transformation are hypothesized to be a malic enzyme and diacylglycerol acyltransferase while lowering phosphoenolpyruvate carboxylase activity is reported to be advantageous for lipid synthesis.Keywords
Abbreviations
| ACC | Acetyl-CoA carboxylase |
| Acyl-ACP thioesterases | Acyl–acyl carrier protein thioesterases |
| ADP-glucose pyrophosphorylase | Adenosine diphosphoglucose pyrophosphorylase |
| AGPase | ADP-glucose pyrophosphorylase |
| BE | Branching enzymes |
| cphA | Cyanophycin synthetase |
| CrDGAT2–4 gene | Diacylglycerol acyltransferase 2–4 gene |
| CRISPRs | Clustered regularly interspaced short palindromic repeats |
| DGAT | Diacylglycerol acyltransferase |
| FAME | Fatty acid methyl ester |
| FAS | Fatty acid synthetase |
| FAT | Acyl-ACP-thioesterase |
| G-1-P | Glucose 1-phosphate |
| G-6-P | Glucose 6-phosphate |
| G6PD | Glucose-6-phosphate dehydrogenase |
| GAP | Glyceraldehyde-3-phosphate |
| GPAT | Glycerol-3-phosphate acetyltransferases gene |
| LPAAT | Lysophosphatidic acetyltransferase gene |
| MAT | Malonyl CoA: ACP transacylase |
| ME | Malic enzyme |
| MLDP | Major lipid droplet protein |
| PAM | Protospacer adjacent motif |
| PEPC | Phosphoenolpyruvate carboxylase |
| PPi | Inorganic pyrophosphate |
| PK | Pyruvate kinase |
| PTA | Phosphotransacetylase |
| SS | Starch synthase |
| TAG | Triacylglycerol |
| TALENs | Transcription activator-like effector nuclease |
| tracrRNA | Trans-activating CRISPR RNA |
| ZFN | Zinc finger nucleases |
The cumulative effect of dwindling fossil fuels, the impact on the environment due to their excess consumption, the exponential rise in prices, infrastructure, storage, and supply issues in conventional fossil fuel-driven energy economy, etc., have triggered the exploration for clean, renewable, and bio-based fuels. The potential of biodiesel as an alternative fuel has attracted much attention among the extensively researched biofuels. Biodiesel is a mixture of fatty acid methyl esters and is typically made from vegetable oils or animal fats by transesterification of triacylglycerols (TAGs). Biodiesel has no/negligible net sulphur or carbon dioxide contribution to the atmosphere and a meagre gaseous pollutants emission compared to diesel. However, the production of biodiesel from plant oil is an expensive and tedious process of which 70%–85% of the total expense is due to the prices of the source which require enormous energy and land to produce adequate oilseed crops [1].
Conversely, animal fats are extremely viscous and generally in solid form at ambient temperature because of their high saturated fatty acids. Much emphasis has been laid on the production of microbial oils to reduce the cost of oil raw materials. It is reported that a variety of microorganisms, including microalgae, yeast, bacteria, and fungi, can accumulate oils under ideal cultivation conditions. The advantages of microbes include a rapid growth rate, cheap and abundant raw materials, less labour requirement, minimal susceptibility to variations in habitat/culture conditions, and easy scalability in the production process [2]. Among the aforementioned microorganisms, microalgae are considered candidates of great potential for biodiesel production owing to their negligible or no known adverse impact on the environment and food security.
The major obstacle to the commercialization of microalgal biofuel is its cost compared to other conventional sources; thus, various strategies to enhance biomass and lipid content are being implemented. Selection of suitable strains, improvement of these strains through mutagenesis and genetic engineering approaches, manipulation of nutritional and environmental factors, use of phytohormones, co-cultivation of microalgae with bacteria or yeasts, and genome editing are various strategies that are being implemented to enhance lipid content. This review includes detailed information about strategies reported for lipid production, improvement of fatty acid synthesis, and prospects of genome editing in oleaginous microalgae.
Oleaginous Microorganisms for Biodiesel Production
Despite many microbes being potent enough to deposit lipids in their membranous structure, emphasis has shifted towards oleaginous microbes, as they can accumulate lipids up to 20% of their biomass weight. These microbes can even accumulate lipids upto 70%–80% of their biomass under stress conditions [3]. The oils produced by oleaginous microbes such as fungi, yeast, microalgae, and bacteria are referred to as microbial oils [4]. Eukaryotic yeast, microalgae, and molds produce triacylglycerols (TAGs) that have compositional similarities with vegetable oils while bacteria produce specific lipids. During transesterification, these TAGs combine with alcohols in the presence of an acid or alkali catalyst to form fatty acid methyl ester (FAME, biodiesel), with glycerol as a by-product [5]. Microbial oils are gaining more importance than plant and animal oils because of their fast growth, non-reliance on seasonal and climatic changes, minimal labour, relatively low spatial footprint, and ease of scale-up [6]. Because of these features, microbial oils are emerging as a potential feedstock for biodiesel production.
Microalgae are one of the efficient feedstocks for biodiesel production, as they can accumulate lipids up to 60%–77% of their dry weight under specific conditions [7]. Apart from biodiesel production, microalgae are also utilized in the generation of various valuable products. The residual biomass after lipid extraction can be further utilized as animal feed or as a source for the synthesis of biomethane [8]. In view of these advantages, microalgae are considered to be a sustainable feedstock in biodiesel production.
Galán et al. [9] reported 80% lipid content in Schizochytrium sp. which is 770 times higher than oleaginous plants such as sunflower, colza, etc. Microalgae belonging to Bacillariophyceae and Chlorophyta produce abundant amounts of oil rich in C16 and C18 fatty acids [10]. These oils are generally composed of polyunsaturated fatty acids with more than four double bonds, distinguishing them from vegetable oil. Chlorella is currently drawing the attention of international energy research institutions due to its suitability for large-scale breeding for biodiesel generation [11]. Furthermore, substantial research is underway addressing microalgae such as Chlorella kessleri, Galdieria partita, Chlorococcum littorale, Nannochloropsis oculata, Synechococcus PCC7942 and Chlorella sorokiniana BTA 9031 that can fix CO2 from the gases and produce biodiesel [12].
Temperature, intensity of light, pH & salinity of the medium, availability of nitrogen, minerals, etc., are some of the parameters that affect the generation of oil in microalgae [13]. Besides, nutrient limitation, stress-induced conditions, and co-cultivation with other microorganisms also improve the lipid content in microalgae. Al-Lwayzy et al. [14] blended the biodiesel produced from Chlorella protothecoides with petroleum biodiesel and the efficiency of the blend was determined from the emission of a 25.8 kW agricultural tractor engine. Mostafa et al. [15] studied the physicochemical properties of blends (B2, B5, B10, and B20) prepared from S. platensis biodiesel and petro-diesel and found that the characteristics corroborated with petro-diesel that is being marketed in Egypt. An insight into various strains of oleaginous microalgae, their lipid content along with composition of saturated, mono, and polyunsaturated fatty acids under specific culture conditions have been shown in Table 1.
Autotrophic microalgae can be converted into heterotrophic organisms by changing growing conditions and employing genetic engineering approaches. The use of ‘‘engineered microalgae’’ with economic and ecological value/importance can improve oil accumulation.
Strategies for lipid production
Lipid enhancement in microalgae can be broadly classified into five approaches such as:
1) Enhancement of FAS approach
2) Enhancement of TAG synthesis approach
3) Blockage of competitive pathways
4) Regulation of bypass approaches of TAG synthesis pathways
5) Multi-gene approach
Enhancement of fatty acid synthesis approach
Triacylglycerols (TAGs) are the lipids that are vital constituents in biodiesel production. Synthesis of TAG in microalgae can be accomplished by various approaches such as:
a) Overexpression of enzymes in fatty acid and TAG synthesis pathways
b) Multi-gene approaches
Fatty acid synthesis (FAS) approach
Microalgae use both organic (glucose, acetate) and inorganic (CO2) carbon sources to produce lipids, which are further converted to acetyl-CoA that can produce other organic molecules [24]. In prokaryotes, TAG synthesis takes place in the cytosol, whereas in eukaryotes TAG synthesis occurs in endoplasmic reticulum and mitochondria. The predominant pathway for TAG biosynthesis is known as the Kennedy Pathway or Glycerol 3-phosphate pathway. Since FAS is one of the prominent pathways for lipid synthesis, exploring the pathway and its enzymes can certainly enhance lipid synthesis. Several research reports have illustrated that enzymes like acetyl-CoA carboxylase (ACC), fatty acid synthetase (FAS), and acyl-ACP-thioesterase (FAT) are vital for enhanced lipid synthesis in microalgae.
Among three enzymes, research data profoundly advocates the utilization of ACC for enhanced lipid synthesis. ACC facilitates the conversion of acetyl-CoA into malonyl-CoA, whereas malonyl-CoA to malonyl-ACP is catalyzed by malonyl CoA: ACP transacylase (MAT). In microalgae such as Haematococcus pulvialis, the overexpression of genes like ACP (acyl-carrier-protein), KAS (Beta-Ketoacyl-Acyl-carrier-protein Synthase), and FAT correlate with the synthesis of mono-and polyunsaturated fatty acids. Modification of these genes may lead to improved production of microalgal biofuels [25].
Acetyl-CoA carboxylase (ACC) approach
A study on the expression of E. coli ACC alone showed no significant improvement in fatty acid production. This implies that the later steps involved in the pathway could limit the flux, resulting in an insignificant increase in fatty acids [26]. However, co-expression of ACC and thioesterase I (tesA gene) resulted in enhanced fatty acid synthesis by six-fold. This indicates that the ACC catalyzing the committed step, is the rate-limiting step for fatty acid biosynthesis in E. coli [27].
Acyl-ACP feedback inhibition by the ACC enzyme may lead to enhanced fatty acid synthesis. However, the tesA gene may be critical in regulating this inhibition by generating free fatty acids, which enhances lipid production [28]. Another potential strategy for reducing feedback inhibition is the expression of a non-native gene (ACC), which is not recognized by the acyl-ACP of E. coli. Lazaro et al. [29] reported a three-fold increase in lipid content levels by heterologous expression of ACC in E. coli from Acinetobacter calcoaceticus.
In eukaryotes, total lipid accumulation has been hindered because of regulatory and metabolic control executed by fatty acids on the ACC gene. The role of the ACC1 enzyme is to provide malonyl-CoA, which is used in the fatty acid synthesis of cellular cytoplasm. Attempts have been made to enhance the lipid content by heterologous expression of the ACC1 gene derived from the oleaginous fungus Mucor rouxii into Hansenula polymorpha a non-oleaginous yeast, resulting in a 40% increase in fatty acid content [30]. Furthermore, overexpression of the ACC gene in Aspergillus oryza showed no significant increase in fatty acids or TAGs productivities compared to the parental strain [31]. An exception was found in Yarrowia lipolytica, where overexpression of the endogenous ACC1 gene resulted in a two-fold rise in the overall lipid content [32].
Acyl-ACP-thioesterase is a group of enzymes that hydrolyze acyl-ACPs into ACP and free fatty acids, synthesized as a result of the FAT gene insertion. FAT breaks the cycle of fatty acid elongation and releases the free fatty acids. Azimov et al. [33] reported that the introduction of Umbellularia californica FAT enzyme into E. coli deficient in fatty acid degradation resulted in increased hydrolytic activity, leading to the accumulation of medium-chain fatty acids. This highlights the feedback inhibition of the acyl-ACP intermediate for fatty acid synthesis. Insertion of FAT genes obtained from various plants into E. coli strain ML103 resulted in fatty acid production that varied in both quantity and composition. The strains carrying the FAT genes from Ricinus communis and Jatropha produced C14, C16:1, and C16 fatty acids at levels of about 40%, 35%, and 20%, respectively [34]. The results infer that the FAT has a profound influence on the accumulation of free fatty acid.
Enhancement of TAG synthesis approach
Acyl-CoA: glycerol-sn-3-phosphate acyl-transferase (GPAT)
The first reaction of TAG synthesis where lysophosphatidate (LPA) is formed via the Kennedy pathway catalyzed by GPAT. Additionally, LPA is also formed by the acylation of dihydroxyacetone phosphate (DHAP) by acyl-CoA: DHAP acyltransferase (DHAPAT) followed by reduction. Major GPAT activities in S. cerevisiae are coded by GAT1 and GAT2 genes. Among these, the GAT1 gene product uses both glycerol-3-phosphate (G3P) and DHAP with similar efficiencies, whereas the GAT2 gene product prefers G3P to DHAP with a higher preference for C16 fatty acyl chains. Metabolic studies revealed a 50% decrease in TAG synthesis in GAT2 yeast, demonstrating that GAT2 initiates the crucial route for TAG [35]. It was also reported that the expression of the GPAT gene of plastidial safflower and E. coli raised oil content from 8% to 29% in Arabidopsis seeds [36]. The GPD1 gene of yeast, which codes for cytosolic glycerol-3-phosphate dehydrogenase (Gly3PDH) has been expressed in oilseed rape under the influence of a napin promoter. The resulting transgenic plants showed two-fold improvement in Gly3PDH activity, 3–4 fold enhanced Gly3P levels, along with a 40% increase in total seed oil content [36].
Lysophosphatidate acyl-transferase (LPAT)
LPAT acylates lysophosphatidate (LPA) to phosphatidate (PA). The incorporation of the yeast SLC1-1 gene, which encodes a variant LPAT, resulted in a remarkable shift in the volume and composition of the fatty acids. Improved expression of glycerolipid acyltransferases in seeds leads to a greater flux of intermediates and better accumulation of TAG, as evidenced by the reported increase in oil content from 8% to 48% in rapeseed and Arabidopsis [36].
Acyl-CoA: diacylglycerol acyl-transferase (DGAT)
The last step of TAG synthesis is catalyzed by the enzyme DGAT. In yeasts and plants, DGAT1/DGAT2 and phospholipid: diacylglycerol acyltransferase (PDAT) are responsible for TAG formation. LRO1 encodes PDAT in yeast, rendering it incapable of synthesizing sterols. Deletion of LRO1 in Y. lipolytica resulted in a 40% loss of TAG [37]. Gao et al. [38] reported that upon incorporation of Arabidopsis DGAT, the yeast showed a 3- to 9-fold enhanced TAG accumulation. Upon overexpression of DGAT, it was observed that rather than phospholipid formation, TAG synthesis increased significantly. Guo et al. [39] inferred that DGAT might be instrumental in the synthesis of TAG, as a strong relation between DGAR activity and TAG content is prominent.
Glycerol 3-phosphate dehydrogenase (GPDH)
GPD1 is an isoform of GPDH that reduces DHAP to G3P, facilitating glycerol for TAG. Upon expressing the gene responsible for GPD1 into Brassica napus L., with the specific promoter, a 40% enhancement in lipid content has been observed [36]. The GUT2 gene codes for GPDH isomer that catalyses the formation of DHAP from G3P, was deleted in Y. lipolytica. This deletion boosted the availability of G3P and increased lipid accumulation 3-folds [40].
Regulation of bypass approaches of TAG synthesis pathways
Besides the mentioned enzymes, there are a few more enzymes that are not directly involved in lipid metabolism but enhance the essential metabolites for lipid biosynthesis. The enzymes that fall under this category are acetyl-CoA synthase (ACS), malic enzyme (ME), and ATP: citrate lyase (ACL). Increased ACS levels enhance activation of acetyl-CoA, which further increases the rate of fatty acid synthesis.
ME is involved in the irreversible decarboxylation reaction where malate is converted back to pyruvate. Nicotinamide adenine dinucleotide phosphate hydrogen (NADPH), which is required for the biosynthesis of fatty acids, is synthesized during this reaction. Enhanced NADPH synthesis from overexpression of ME is utilized by TAG synthesis-related enzymes ACC, FAS, and ATP: citrate lyase (ACL) to produce more lipids [41]. Overexpression of genes encoding for ME and its isoforms in Mucor circinelloides resulted in increased lipid accumulation by 2.5 folds and biomass from 12% to 30% when compared with leucine auxotrophic strain [41].
ACL is involved in the conversion of citrate to acetyl-CoA and oxaloacetate. It is regarded as one of the key enzymes for lipid production in mammals, fungi, and oleaginous yeasts. This enzyme catalyzes a rate-limiting reaction for lipid biosynthesis. Enhanced expression of ACL in Aspergillus oryzae resulted in a 1.7-fold increase in fatty acid productivity and a 1.9-fold improvement in TAG compared to the parental strain [42].
Overexpression of enzymes of lipid biosynthesis
Fatty acid and TAG biosynthesis involve different steps catalyzed by various enzymes, among which acetyl-CoA carboxylase (ACCase) plays a key role. ACCase catalyzes the carboxylation of acetyl-CoA to malonyl-CoA thereby directing carbon flux towards fatty acid synthesis. A number of studies have emphasized that overexpression of ACCase increases the availability of malonyl CoA, resulting in an increase in fatty acid biosynthesis in chloroplasts [43,44]. Its role is significantly explored in lipid enhancement studies of both plants and microalgae.
Rawat et al. [45] reported that by integrating and expressing PtACC2 in the chloroplast of Phaeodactylum tricornutum strain ACCase increased 3.3-fold, significantly enhancing lipid content up to 1.77-fold, reaching 40.8% of dry weight. Similarly, Chen et al. [46] cloned two strains of Chlamydomonas reinhardtii for overexpression of ACCase, resulting in 55.45% and 56.15% unsaturated fatty acids content, which is 1.16 fold higher than the wild strain. However, in a few species like Navicula saprophila and Cyclotella cryptic, expression of the ACCase gene only increased the activity of ACCase, not the synthesis of fatty acids [47]. These reports confirmed that random integration of target genes or non-specific expression of ACCase will not enhance fatty acid biosynthesis, even though strains are genetically modified. Therefore, identifying and employing a potential genetic engineering tool will improve the overproduction of fatty acids in oleaginous microalgae [47].
The additional enzymes targeted for increasing fatty acid and TAG synthesis in microalgae are fatty acid synthatase (FAS), malic enzyme (ME), and acetyl-CoA synthase (ACS). Out of these enzymes, Xue et al. [48] reported a 2.5-fold enhancement of lipid content in Phaeodactylum tricornutum by over-expressing ME without tampering with the cell growth. Besides, a couple of proteins also play a key role in enhancing fatty acid and TAG synthesis. For instance, in Cyanidioschyzon merolae, even under normal growth conditions, the overexpression of CmGPAT1 led to a noticeable accumulation of TAG, with a maximum TAG productivity that was 56.1 times higher than that of the control strain [49].
Lipid synthesis can be enhanced using metabolic engineering approaches by blocking competitive pathways. These approaches are mainly aimed at channeling the metabolic flux towards TAG biosynthesis, thereby enhancing lipid production. The main competitive pathways to be repressed are the β-oxidation pathway, phospholipid biosynthesis, phosphoenolpyruvate carboxylase (PEPC), and starch biosynthesis. Among these pathways, repression of PEPC and starch biosynthesis are widely explored, through which enhanced lipid content has been reported.
Lipid accumulation can be enhanced by reducing the lipid breakdown and downregulating the genes responsible for the synthesis of fatty acids and TAG, as well as genes involved in β-oxidation. Dysfunction of acyl-CoA oxidase (AOX) in S. cerevisiae leads to inactivation of the β-oxidation process, resulting in the accumulation of cellular fatty acids. Feedback regulation of the oxidation process affects hydrophobic substrate transporters and controls the diffusion process. Hence, non-oleaginous yeast such as S. cerevisiae cannot accumulate lipids on par with the oleaginous microbes [50]. On the other hand, Yarrowia lipolytica, an oleaginous yeast, accumulates lipids up to 50% of its dry weight [51]. This organism contains several AOX proteins encoded by (plant homeobox) POX1–POX6 genes, which catalyze the limiting step of peroxisomal β-oxidation. Alterations in the POX genotype prevent lipid degradation and enhance lipid accumulation. Additionally, phospholipid biosynthesis is an alternative competitive pathway for TAG formation as it competes against TAG biosynthesis for a common substrate, phosphatidate (PA) [52]. PA enters the pathway leading to the synthesis of phospholipids if it is transformed into CDP-diacylglycerol. The production of long fatty acids is caused by inhibition of phospholipid synthesis.
Phosphoenolpyruvate carboxylase repression
The substrate used in the production of proteins and lipids is phosphoenolpyruvate (PEP). Animals and fungi do not synthesize PEPC, an enzyme that changes pyruvate into oxaloacetate. It is primarily produced by plants and photosynthetic microalgae such as cyanobacteria. In TAG biosynthesis, PEP is produced through a series of reactions that result in pyruvate, acetyl-CoA, malonyl CoA, and fatty acids [53]. PEPC is a strategic enzyme that readily converts the substrate phosphoenol pyruvate into oxaloacetate and directly plays a role in protein anabolism (Fig. 1). In contrast, in the lipid synthesis process, PEP is converted into pyruvate by pyruvate kinase (PK). This pyruvate is converted into acetyl-CoA using the pyruvate dehydrogenase enzyme, ultimately leading to the formation of fatty acid. To enhance lipid content, inhibition of PEPC activity is desired. In Chlamydomonas reinhardtii, knocking down PEPC and overexpression of GroELS chaperon resulted in enhanced biomass and lipid content of 2.56 g/L and 23.5 mg/L, respectively [54].
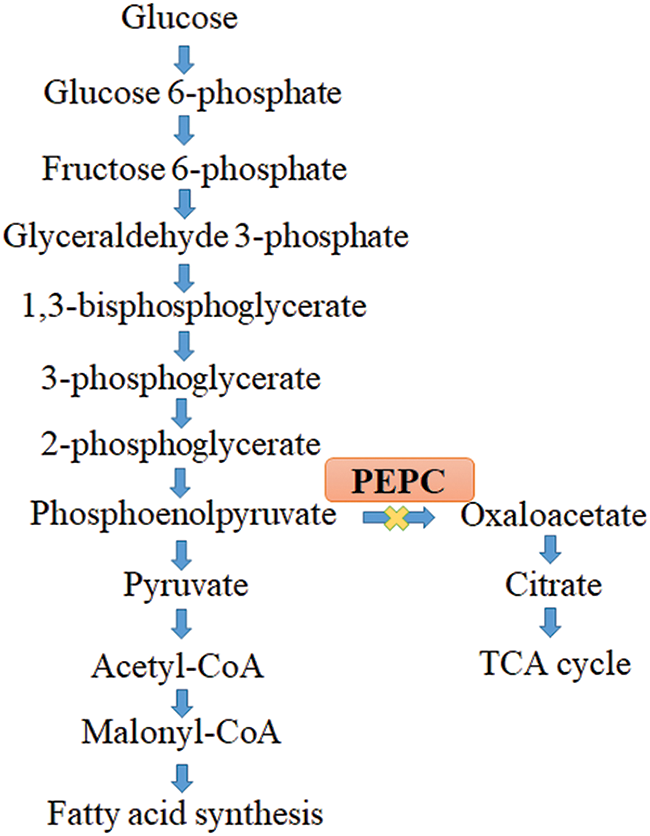
Figure 1: Role of strategic enzyme phosphoenol pyruvate carboxylase (PEPC) in the fatty acid biosynthesis pathway.
Research studies on antisense PEPC expressed in B. napus resulted in a 6.4%–18% increase in oil content [53]. Moreover, significant increase in lipid contents have been reported in the transgenic soybean lines containing the construct of an anti-PEPC gene [55]. These studies substantiate that lipid enhancement can be achieved by repressing the PEPC gene in plants.
However, in microalgae, thorough investigations must be conducted to validate the significance of PEPC in fatty acid synthesis regulation. A recent study in Anabaena sp. has reported that an increase in lipid content (46.9%) was observed by repression of PEPC activity, which implies the engineering prospects for enhanced lipid content [56].
Starch biosynthesis repression
A homopolymer of α-D glucose units, starch acts as a storehouse for carbon and energy, especially during times of stress. As carbon dioxide is assimilated during starch biosynthesis, glyceraldehyde-3-phosphate, glucose-6-phosphate, and glucose-1-phosphate are produced. Conversion of glucose-1-phosphate to ADP-glucose and Pi catalyzed by ADP-glucose pyrophosphorylase (AGPase) is the rate-limiting step of starch synthesis in plastids (Fig. 2). Consequently, ADP-glucose is utilized by starch synthases (SS) and branching enzymes (BE) to elongate glucan chains of starch polymer. To increase the lipid content, blocking starch synthesis is one of the strategic approaches in the green microalga Chlamydomonas reinhardtii rather than manipulation of the fatty acid pathway.
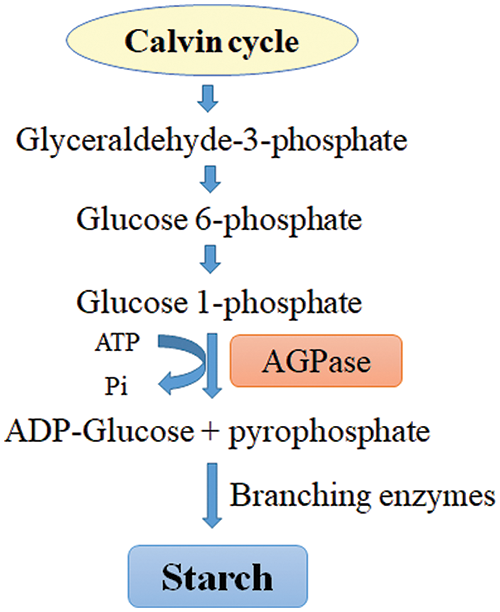
Figure 2: Starch biosynthesis pathway in the chloroplast of microalgae. ADP-glucose pyrophosphorylase (AGPase) catalyzes the rate-limiting step of the pathway.
Li et al. [57] reported a 10-fold increase in TAG synthesis by the inactivation of AGPase in a starchless mutant (Chlamydomonas reinhardtii), illustrating the streamlining of photosynthetic carbon from starch synthesis to TAG formation [57]. In a separate study, the lipid content of one of the C. reinhardtii mutants (BAFJ5), defective in the small subunit of AGPase, was 46.4% higher than that of the wild type.
The essential genetic alterations of microalgae have recently been possible due to advancements in the field of genetic engineering and improved understanding of biochemical processes [58]. Numerous investigations have revealed that TAG productivity could be increased by genetic engineering by expressing key GAP pathway enzymes [59,60] (Table 2).
TAG synthesis starts with the acylation of glycerol-3-phosphate (G3P) and undergoes a five step process as shown in the Fig. 3 which is coordinated via four enzymes viz., glycerol 3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAT), phosphatidic acid phosphatase (PAP) and diacylglycerol acyltransferase (DGAT) [70]. Over-expression of genes encoding these enzymes individually or collectively in microalgae has been experimentally proven to increase in lipid content by various researchers. For instance, over-expression of GPAT in Phaeodactylum tricornutum showed 2-fold rise in fatty acid content compared to wild strain which accounted for 42.6% lipid content per dry weight [71]. TAG levels were increased by 13% and 20% in P. tricornutum and Chlamydomonas reinhardtii by over-expression of LPAT and DGAT, respectively [72].
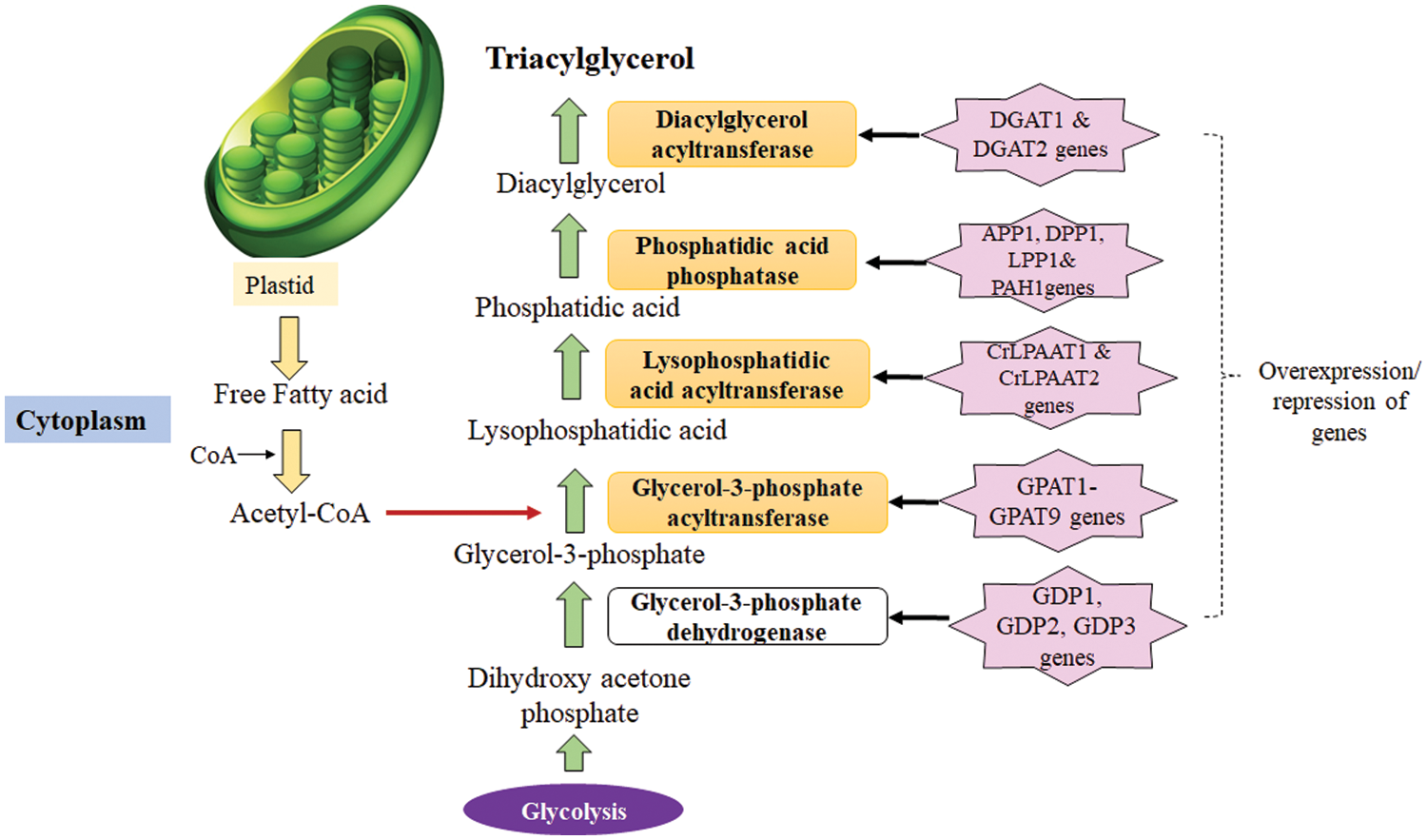
Figure 3: Triacylglycerol biosynthesis in microalgae (Key enzymes in the TAG synthesis have been highlighted).
Muñoz et al. [72] found that overexpression of single and multiple encoding genes for LPAT, GPAT, and DGAT in N. oleoabundans produced 45% and 39% of triacylglycerols which is almost 1.3 folds higher than that of wild strain. Above all over-expressing multiple genes-LPAT, GPAT, and DGAT improved lipid production even during undernourishment with no significant effect on growth [72]. Similarly, main regulatory genes for fatty acid biosynthesis pathway in H. pluvialis was linked to the enzymes: acyl carrier protein, 3-ketoacyl-ACP-synthase, and acyl-ACP thioesterase because higher expression resulted in increased synthesis of mono unsaturated and polyunsaturated fatty acids [73]. Because acyl-CoA intermediates function as both product and feedback inhibitors in the fatty acid (upstream) pathway and as important precursors in the TAG (downstream) pathway, many gene approaches may be promising options for improving quality and increasing fatty acid production. So, by depleting acyl-CoA intermediates and increasing the rate of TAG accumulation in lipid bodies, up-regulation of the downstream pathway produces a driving force by properly regulating the necessary genes simultaneously.
Altering fatty acid chain length for improved lipid quality
Composition of the microalgal lipids determines the properties of biodiesel. Various strategies must be followed to increase the quality of the lipids so as to meet the specific standards of the biodiesel [74]. Saturated and monounsaturated fatty acids with carbon length of 12:0, 14:0, 16:0, 16:1, 18:0 and 18:1 are suitable for biodiesel production. Acyl-ACP thioesterases control the release of fatty acid chains of various lengths from fatty acid synthase [75]. As per the metabolic needs, various organisms synthesize thioesterases that are specific for particular fatty acids chain. Hence, transformation of thioesterases genes into microalgae could enrich the fatty acid composition desirable for standard fuel properties. Upon transforming the fatty acid acyl-ACP thioesterases from Cinnamomum camphora and Umbellularia californic into Phaeodactylum tricornutum, the diatom produced lipids with increased percentage of lauric and myristic acids [58]. From previously reported studies, it is evident that alteration in the length of fatty acid chain has influence on the composition of the lipids.
Effect of Environmental Factors on Metabolic Engineering of Microalgae
Environmental factors play a significant role in metabolic engineering strategies aimed at enhancing lipid production in microalgae. These factors can influence various aspects of microalgal physiology and metabolism, ultimately affecting lipid biosynthesis. Among various environmental factors-light, temperature, nutrients, CO2, stress conditions, pH and salinity are recognized as crucial parameters to impact during metabolic engineering in microalgae [13].
• Light intensity and quality: Light is a crucial environmental factor that directly affects photosynthesis, biomass growth, and lipid accumulation in microalgae. Optimizing light intensity, photoperiod, and spectral quality can enhance lipid productivity [17,76]. Low light intensity stimulates the formation of TAGs, which make up the majority of non-polar lipids found in biodiesel. Conversely, low light levels cause polar lipid concentration to decrease in order to safeguard the photosystem. A higher concentration of light stimulates the production of oxidizing agents that break down polyunsaturated fatty acids, lowering the amount of polar lipids and raising the amount of non-polar lipids. Most of the microalgae species show optimal growth at a light intensity of 200–400 μmol photons m−2s−1 [77]. According to Nzayisenga et al. [78], when Scenedesmus obliquus was exposed to 300 μmol photons m−2s−1 of light, its fatty acid content doubled from 5.8% to 11.6%. Research conducted on Chlorella sp. and Monoraphidium sp. revealed that their neutral lipid content rose when they were grown in light intensities lower than 400 μmol photons m−2s−1 [79].
• Temperature: Temperature influences the metabolic activity and growth rate of microalgae, thereby impacting lipid production. Lower temperatures often promote lipid accumulation, while higher temperatures can enhance biomass productivity. According to Wu et al. [80], Monoraphidium sp. thrived well at temperatures between 25°C and 35°C, reaching its maximum lipid output of 29 mg L−1d−1 at 30°C. At 40°C, the cells began to degrade, producing very little biomass and no lipids as compared to cells that were under 25°C–35°C. An additional investigation into Chaetoceros sp. revealed that it grows best at 25°C and has a lipid content of roughly 20.42% [81]. Lipid buildup in Chlorella vulgaris and Nannochloropsis oculata rose from 7.90% to 15.31% and 5.90%–16.41% respectively at 25°C–30°C [82].
• Nutrient availability: Nutrients such as nitrogen, phosphorus, and micronutrients are essential for microalgal growth and lipid biosynthesis. Nutrient limitation, particularly nitrogen deprivation, induces lipid accumulation in many microalgae. A two-fold increase in TAG yield (from 23.47 to 46.19 mg L−1d−1) has been observed in Neochloris oleoabundans when cultivated under nitrogen-deprived conditions [83]. Similarly, Yang et al. [84] reported that Chlamydomonas reinhardtii cultivated in a medium without nitrogen and with various concentrations of phosphorous showed the highest total fatty acid (TFA) concentration of 105 μg/mg (i.e., 104.7% higher than the control) in the media deprived of both nitrogen and phosphorous.
• CO2 concentration: Elevated CO2 levels can stimulate photosynthesis and redirect carbon flux towards enhanced lipid biosynthesis pathways. Furthermore, coupling microalgal cultivation with CO2 capture technologies can mitigate greenhouse gas emissions while promoting lipid production. Carbon content influences fatty acid composition and saturation [85]. Certain strains produce elevated levels of long-chain fatty acids (C18:0, C18:1, and C20:0) when exposed to a 15% increase in CO2 concentration [86]. For instance, Chorella vulgaris and Gloeothece membranacea produced lipid contents of 25.6% and 36%, respectively, when the CO2 concentration was raised to 15% in a photobioreactor under blue light [86]. Different CO2 concentrations and nitrogen: phosphorus (N:P) ratios were used in another study to grow Scenedesmus dimorphus, where a lipid content of 31.6% dry weight was found at 6% CO2 [87].
• pH and salinity: pH and salinity affect cellular physiology and metabolic processes in microalgae. Optimal pH and salinity conditions vary among microalgal species and can influence lipid accumulation. The most suitable pH range for microalgal growth is 6 to 8.76. Chlorella sp., Nannochloropsis salina, and Pavlova lutheri exhibited optimal growth at pH 8, with lipid content of 23%, 24.75%, and 35%, respectively [88,89]. Under optimum conditions, lipid content increased from 150 to 190 g lipid kg−1 biomass in Amphora subtropica, and from 190 to 280 g lipid kg−1 biomass in Dunaliella sp. [90]. However, the ideal pH and salinity ranges vary depending on the microalgae strain and their individual requirements for lipid productivity.
• Stress conditions: Environmental stress conditions viz., drought, high light intensity, and oxidative stress, can induce lipid accumulation as a protective mechanism in microalgae. Under salinity stress condition, the lipid content of Chlorella vulgaris increased from 60% to 70% by increasing the NaCl concentration from 0.5 to 1.0 M [91]. This is due to an increase in ACCase and a decrease in AGPase [92].
The cost-intensive steps in microalgal biodiesel production are harvesting biomass and extracting lipids from the biomass. Genetic modification is an economic strategy that enables the secretion of lipids from microalgae into the medium. Singh et al. [93] introduced acyl-acyl carrier thioeseterase gene into wild type Synechocystis sp., enabling the secretion of fatty acids up to 1.8 ± 0.06 mg/L. The transformed cyanobacteria secreted 197 ± 14 mg/L of fatty acids, which is 109 times higher than that secreted by the wild strain. Furthermore, by deleting the competitive genes responsible for fatty acid synthesis and removing the genes specific for surface proteins, fatty acid synthesis was enhanced in microalgae. These genetic modification strategies are not only energy-efficient but also non-toxic, as they do not involve the use of toxic chemicals for lipid extraction. Despite these advantages, the drawbacks of this technique includes cell fragility and reduced CO2 aeration, which affect cell photosynthesis. Therefore, this strategy required substantial modification for improved lipid yield at industrial scale.
Prospects of transcription factors and genome editing for enhanced lipid production
Metabolic engineering has showed potential for enhanced lipid content, but several drawbacks still hinder maximizing the technique’s potential. For the past two decades, zinc finger nucleases (ZFNs) and transcription activator-like effector nuclease (TALENs) have been used as promising genome editing tools [94]. The zinc-finger domain comprises of 3–6 nucleotide triplets and DNA cleavage domain (nucleases) that function as dimers. Some successful studies on genome editing using ZFN in Chlamydomonas have been reported [95]. Although ZFNs offer few off-target effects, identifying 3 and 6 triple base targets for cleavage may not be possible in some instances, making the method laborious and challenging to apply for each editing study [95]. Unlike ZFNs, TALENs recognize single nucleotide and can amend any sequence [96]. However, the TALENs process is susceptible to cytosine methylation, requires a separate design for each nuclease, and has limited for off-target effects [97].
Transcription factor-mediated strategies, such as using GmDof4 from soybean, LEAFY COTYLEDON2 (LEC2) and WRINKLED1 (WRI1) from Arabidopsis, have been observed to enhance lipid content in several microalgae. For instance, in C. reinhardtii, over-expression of stress-induced transcription factors was observed to produce a 50% increase in lipid yield [98]. Over-expression of LEC2 transcription factor in tobacco under the control of acetaldehyde inducible promoter (Alc) led to an increase in lipid content from 2.9% to 6.8% [99].
In comparison to ZFNs and TALENs, recent investigations have shown that clustered regularly interspaced short palindromic repeats (CRISPRs) have greater potential for making desirable changes in the DNA for enhanced lipid content. CRISPRs are loci of DNA with tandem repeats of base sequences and are mostly associated with Cas genes, which code for CRISPRs related proteins. This system is a prokaryotic immune system that provide resistance to phages and plasmids [100].
CRISPR functions by identifying the spacer sequences recognizable and excising these exogenous genetic elements, similar to the RNAi mechanism [99]. Among eleven types of CRISPRs, type II CRISPR/Cas systems are widely adapted as a genome editing (GE) tools and differ from others by the presence of protospacer adjacent motif (PAM) sequence and a second RNA called trans-acting CRISPR RNA (tracrRNA), which together act in the maturation and redirecting of the Cas9 nuclease to DNA [101]. Furthermore, crRNA and tracrRNA are fused to form a ‘guide’ RNA (sgRNA or gRNA), which has huge potential in genome editing [102]. In genome editing process, the CRISPR/Cas9 system use gRNA to target specific nucleic acid for degradation through nuclease. Unlike ZFN and TALEN, gRNA synthesis and designare easier and more cost-effective than the other protein domains of CRISPR [103]. Besides, the CRISPR/Cas system has shown improved efficiency over ZFNs and TALENs with desirable mutations and can also be employed to introduce multiple genes by using multiple gRNAs (multiplexed mutations). Although the CRISPR/Cas system has great advantages over ZFNs and TALENs, it has the drawback of causing mutations at non-specific loci (off-target) [103].
Cas9 expression in cyanobacteria S. elongatus UTEX2973 and PCC7942 and in the eukaryotic microalgae C. reinhardtii showed toxicity due to consistent off-target expression. Off-site targeting often results in cell toxicity and hampers the technology’s potential [104]. A recent study comparing chromosomal and episomal expression of CRISPR/Cas9 in P. trocornutum showed that in both methods, mutation ratio is similar; however, episomal mutants required more growth time, while chromosomal expression of Cas9 showed re-editing genes with small indels [105]. This implies that the design for efficient Cas protein coupled with sgRNA expression is important to consider. To overcome this problem, fusing codon optimized Cas gene with nuclear localization signals could be a viable strategy for efficient nuclear DNA targeting [106]. Additionally, to increase the specificity, sgRNA can be constructed with flanking self-cleaving ribozyme ends or engineered hairpin structures of sgRNA spacer, or highly efficient sgRNA sequences can be selected using web tools. Besides designing constructs, delivery method and regulatory sequences could significantly enhance genome editing efficiency [107].
In a recent study, a genome editing efficiency of 30% was achieved for the adenine phosphoribosyl transferase (Apt9) gene in C. reinhardtii through cell agitation with glass bead before particle bombardment [108]. Furthermore, Tanwar et al. [109] reported 60% genome editing efficiency in P. tricornutum using sgRNA, Cas9, antibiotic-marker elements expressed within a plasmid containing endogenous promoters, terminators and introns. In addition to CRISPR/Cas9, other strategies such as Cas12a variant and Cas9 ribonucleoprotein complexes (RNP) could be deployed for higher genome editing efficiency [110].
Integrating Omics with Gene Editing Tools
Integrating data generated through various omics tools during genome-scale metabolic modelling has significant implications for enhancing lipid production in microalgae. Rapid advances in high throughput analytical techniques for sequencing genome of organisms and measuring gene expression, protein, lipid and other metabolites have fundamentally altered metabolic modelling paradigms. Above all, the cost of data creation and analysis has significantly decreased, enabling exponential growth in omics data generation for an organism within a very short period [111]. The effect of omics data in genome-scale metabolic modelling of microalgae can be useful in the following contexts.
• Identification of Lipid Biosynthesis Pathways: Transcriptomics and proteomics data are particularly useful in identifying genes and enzymes involved in biosynthesis pathways. This approach has already been successful in fungal strains [112] and similar approach can be extended to microalgae. Analyzing gene expression and protein abundance data helps researchers pinpoint the key enzymes responsible for lipid synthesis during fatty acid biosynthesis and triacylglycerol assembly.
• Quantitative Assessment of Metabolic Fluxes: Omics data provide quantitative information about the expression levels of genes, proteins, and metabolites involved in lipid metabolism. Integrating this data into metabolic models allows for the estimation of metabolic fluxes through lipid biosynthesis pathways under different growth conditions. This information is crucial for predicting lipid production rates and optimizing metabolic engineering strategies [113].
• Optimization of Growth Conditions: Omics data provide researchers with necessary understanding of the impact of various culture conditions (e.g., nutrient availability, light intensity, temperature) on lipid metabolism in microalgae. Such information helps optimize growth conditions to enhance lipid production [114].
• Dynamic Modelling of Lipid Production: Omics data enable the development of dynamic models that can simulate lipid production over time. As reported by Wang et al. [106], integrating time-resolved omics data into dynamic modeling approaches helps predict lipid metabolism based on dynamic changes in environmental conditions or genetic perturbations for sustained lipid production in microalgae cultures.
• Flux Balance Analysis (FBA) is a computational method used to analyse the flow of metabolites and predict the behaviour of biochemical pathways in various organisms, including microalgae. TAG biosynthesis is a vital pathway for lipid production, which has applications in the biofuel and nutraceutical industries. To perform FBA for TAG synthesis, a metabolic network model comprising all relevant biochemical reactions (such as glycolysis, carbon fixation, pentose phosphate pathway, fatty acid synthesis and TAG assembly) need to be constructed [115]. This should be followed by defining certain constraints (like nutrient and light availability, rate of CO2 uptake) and objectives (maximizing TAG and biomass production) for the FBA simulation [116]. Through mathematical optimization techniques, FBA calculates the fluxes/rates of all biochemical reactions that satisfies the defined constraints and optimizes the objective function. The output provides insights into the functioning of metabolic network under optimized conditions and identify strategies for enhancing TAG production. Several software tools like COBRA Toolbox in MATLAB, COBRApy in Python are available to perform flux balance analysis.
Challenges and Future Perspective
Metabolic engineering of microalgae for lipid production presents several challenges, but with ongoing research and technological advancements, promising future perspectives are emerging. Many microalgae species have inherently low lipid productivity, making it challenging to achieve economically viable lipid yields for industrial applications. Additionally, microalgal metabolism is highly complex, with interconnected metabolic pathways. Identifying and engineering key metabolic pathways for increased lipid production while minimizing undesirable side effects is another significant challenge.
Lipid production in microalgae mostly depends upon the availability of nutrients and light intensity. Deviation from the required concentration may lead to a drastic reduction in the lipid production, so optimizing nutrient utilization and developing strategies for nutrient recycling are important for sustainable lipid production. Furthermore, many microalgal species lack robust genetic tools and efficient transformation methods, hindering the ability to engineer them for improved lipid production.
Developing genetic engineering tools, optimizing transformation protocols, utilizing bioinformatics tools and machine learning algorithms for data analysis and model prediction for a wide range of microalgae species are essential. Additionally, screening natural biodiversity and selecting promising strains for further engineering can accelerate the development of high-yielding lipid-producing microalgae strains. Overall, overcoming the challenges associated with metabolic engineering of microalgae for lipid production requires interdisciplinary collaboration and continued innovation across multiple fields.
The exploitation of microalgae as a source for quality lipids is highly desirable due to advantages like shorter incubation time, higher lipid content. Traditionally biochemical approaches have resulted in meager lipid content. To improve lipid content, metabolic engineering approaches coupled with genome editing technique could be useful. Targeting and overexpression of genes responsible for FAS and TAG biosynthesis could result in higher lipid content, particularly through the co-expression of ACC and tesA genes. Additionally, repression of competitive pathways, such as AGP-ase and PEPC genes, leads to enhanced TAG formation. This process is important for streamlining photosynthetic carbon towards lipid synthesis rather than direct manipulation of lipid synthesis.
Moreover, a multigene approach using CRISPR-Cas9 technique could be employed for an efficient genome editing process. Genome editing and metabolic engineering techniques are further envisioned to improve the designed strains. Embracing advanced techniques coupled with fermentation strategies could increase the lipid content in microalgae. Unprecedentedly, the energy demand could be satiated with microalgae to bolster sustainable energy. Furthermore, biorefinery development could alleviate the cost incurred in biodiesel production, thus creating a circular economy, which is inevitable for the commercial viability of the technology.
Acknowledgement: The authors acknowledge DST, SERB Vignan’s Foundation for Science, Technology and Research, Vadlamudi, Guntur, and the Director, ICAR-Directorate of Floricultural Research, Pune for facilitating research conducive environment.
Funding Statement: This work is partially supported by Department of Science and Technology, Science and Engineering Research Board under Teachers Associateship for Research Excellence (TARE) Scheme (File Number TAR/2023/000036).
Author Contributions: Manuscript drafting/preparation: Anjani Devi Chintagunta; study conception and design: Nune Satya Sampath Kumar; reviewed literature and meticulously corrected the draft: Samudrala Prashant Jeevan Kumar. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Singh D, Sharma D, Soni SL, Sharma S, Sharma PK, Jhalani A. A review on feedstocks, production processes, and yield for different generations of biodiesel. Fuel. 2020;262:116553. doi:10.1016/j.fuel.2019.116553. [Google Scholar] [CrossRef]
2. Sarwar W, Ali Q, Ahmed S. Microscopic visualization of the antibiofilm potential of essential oils against Staphylococcus aureus and Klebsiella pneumoniae. Microsc Res Tech. 2022;85(12):3921–31. doi:10.1002/jemt.24243. [Google Scholar] [PubMed] [CrossRef]
3. Christophe G, Kumar V, Nouaille R, Gaudet G, Fontanille P, Pandey A, et al. Recent developments in microbial oils production: a possible alternative to vegetable oils for biodiesel without competition with human food? Braz Arch Biol Technol. 2012;55(1):29–46. doi:10.1590/S1516-89132012000100004. [Google Scholar] [CrossRef]
4. Ochsenreither K, Glück C, Stressler T, Fischer L, Syldatk C. Production strategies and applications of microbial single cell oils. Front Microbiol. 2016;7:209534. doi:10.3389/fmicb.2016.01539. [Google Scholar] [PubMed] [CrossRef]
5. Chintagunta AD, Zuccaro G, Kumar M, Kumar SJ, Garlapati VK, Postemsky PD, et al. Biodiesel production from lignocellulosic biomass using oleaginous microbes: prospects for integrated biofuel production. Front Microbiol. 2021;12:658284. doi:10.3389/fmicb.2021.658284. [Google Scholar] [PubMed] [CrossRef]
6. Mhlongo SI, Ezeokoli OT, Roopnarain A, Ndaba B, Sekoai PT, Habimana O, et al. The potential of single-cell oils derived from filamentous fungi as alternative feedstock sources for biodiesel production. Front Microbiol. 2021;12:637381. doi:10.3389/fmicb.2021.637381. [Google Scholar] [PubMed] [CrossRef]
7. Chhandama MV, Ruatpuia JV, Ao S, Chetia AC, Satyan KB, Rokhum SL. Microalgae as a sustainable feedstock for biodiesel and other production industries: prospects and challenges. Energy Nexus. 2023:100255. doi:10.1016/j.nexus.2023.100255. [Google Scholar] [CrossRef]
8. Jacob S, Chintagunta AD, Banerjee R. Selective digestion of industrial potato wastes for efficient biomethanation: a sustainable solution for safe environmental disposal. Int J Environ Sci Te. 2016;13(10):2363–74. doi:10.1007/s13762-016-1051-y. [Google Scholar] [CrossRef]
9. Galán B, Santos-Merino M, Nogales J, de la Cruz F, García JL. Microbial oils as nutraceuticals and animal feeds. In: Health consequences of microbial interactions with hydrocarbons, oils, and lipids. 2020;192(2):401–45. doi:10.1007/978-3-030-15147-8_34. [Google Scholar] [CrossRef]
10. Maltsev Y, Maltseva K. Fatty acids of microalgae: diversity and applications. Rev Environ Sci Bio. 2021:515–47. doi:10.1007/s11157-021-09571-3. [Google Scholar] [CrossRef]
11. Patel A, Gami B, Patel P, Patel B. Biodiesel production from microalgae Dunaliella tertiolecta: a study on economic feasibility on large-scale cultivation systems. Biomass Convers Bior. 2023;13(2):1071–85. doi:10.1007/s13399-020-01191-1. [Google Scholar] [CrossRef]
12. Mondal M, Ghosh A, Sharma AS, Tiwari ON, Gayen K, Mandal MK, et al. Mixotrophic cultivation of Chlorella sp. BTA, 9031 and Chlamydomonas sp. BTA, 9032 isolated from coal field using various carbon sources for biodiesel production. Energy Convers Manag. 2016;124(1):297–304. doi:10.1016/j.enconman.2016.07.033. [Google Scholar] [CrossRef]
13. Chowdury KH, Nahar N, Deb UK. The growth factors involved in microalgae cultivation for biofuel production: a review. Comput Water Energy Environ Eng. 2020;9(4):185–215. doi:10.4236/cweee.2020.94012. [Google Scholar] [CrossRef]
14. Al-Lwayzy SH, Yusaf T. Chlorella protothecoides microalgae as an alternative fuel for tractor diesel engines. Energies. 2013;6(2):766–83. doi:10.3390/en6020766. [Google Scholar] [CrossRef]
15. Mostafa SS, El-Gendy NS. Evaluation of fuel properties for microalgae Spirulina platensis bio-diesel and its blends with Egyptian petro-diesel. Arab J Chem. 2017;10(4):S2040–50. doi:10.1016/j.arabjc.2013.07.034. [Google Scholar] [CrossRef]
16. Feng Y, Li C, Zhang D. Lipid production of Chlorella vulgaris cultured in artificial wastewater medium. Bioresour Technol. 2011;102(1):101–5. doi:10.1016/j.biortech.2010.06.016. [Google Scholar] [PubMed] [CrossRef]
17. Rodolfi L, Chini Zittelli G, Bassi N, Padovani G, Biondi N, Bonini G, et al. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng. 2009;102(1):100–12. doi:10.1002/bit.22033. [Google Scholar] [PubMed] [CrossRef]
18. Lee CH, Chae HS, Lee SH, Kim HS. Growth characteristics and lipid content of three Korean isolates of Botryococcus braunii (Trebouxiophyceae). J Ecol Env. 2015;38(1):67–74. doi:10.5141/ecoenv.2015.007. [Google Scholar] [CrossRef]
19. Morales-Sánchez D, Tinoco-Valencia R, Kyndt J, Martinez A. Heterotrophic growth of Neochloris oleoabundans using glucose as a carbon source. Biotechnol Biofuels. 2013;6(1):1–3. doi:10.1186/1754-6834-6-100. [Google Scholar] [PubMed] [CrossRef]
20. Liang MH, Jiang JG. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog Lipid Res. 2013;52(4):395–408. doi:10.1016/j.plipres.2013.05.002. [Google Scholar] [PubMed] [CrossRef]
21. Mondal M, Goswami S, Ghosh A, Oinam G, Tiwari ON, Das P, et al. Production of biodiesel from microalgae through biological carbon capture: a review. 3 Biotech. 2017;7(2):1–21. doi:10.1007/s13205-017-0727-4. [Google Scholar] [PubMed] [CrossRef]
22. Araujo GS, Matos LJ, Gonçalves LR, Fernandes FA, Farias WR. Bioprospecting for oil producing microalgal strains: evaluation of oil and biomass production for ten microalgal strains. Bioresour Technol. 2011;102(8):5248–50. doi:10.1016/j.biortech.2011.01.089. [Google Scholar] [PubMed] [CrossRef]
23. Gao C, Zhai Y, Ding Y, Wu Q. Application of sweet sorghum for biodiesel production by heterotrophic microalga Chlorella protothecoides. Appl Energy. 2010;87(3):756–61. doi:10.1016/j.apenergy.2009.09.006. [Google Scholar] [CrossRef]
24. Wu W, Tan L, Chang H, Zhang C, Tan X, Liao Q, et al. Advancements on process regulation for microalgae-based carbon neutrality and biodiesel production. Renew Sustain Energy Rev. 2023;171:112969. doi:10.1016/j.rser.2022.112969. [Google Scholar] [CrossRef]
25. Santin A, Russo MT, Ferrante MI, Balzano S, Orefice I, Sardo A. Highly valuable polyunsaturated fatty acids from microalgae: strategies to improve their yields and their potential exploitation in aquaculture. Molecules. 2021;26(24):7697. doi:10.3390/molecules26247697. [Google Scholar] [PubMed] [CrossRef]
26. Satoh S, Ozaki M, Matsumoto S, Nabatame T, Kaku M, Shudo T, et al. Enhancement of fatty acid biosynthesis by exogenous acetyl-CoA carboxylase and pantothenate kinase in Escherichia coli. Biotechnol Lett. 2020;42(12):2595–605. doi:10.1007/s10529-020-02996-w. [Google Scholar] [PubMed] [CrossRef]
27. Davis MS, Solbiati J, Cronan JE. Overproduction of acetyl-CoA carboxylase activity increases the rate of fatty acid biosynthesis in Escherichia coli. J Biol Chem. 2000;275(37):28593–8. doi:10.1074/jbc.M004756200. [Google Scholar] [PubMed] [CrossRef]
28. Davis MS, Cronan JEJr. Inhibition of Escherichia coli acetyl coenzyme A carboxylase by acyl-acyl carrier protein. J Bacteriol. 2001;183(4):1499–503. doi:10.1128/jb.183.4.1499-1503.2001. [Google Scholar] [PubMed] [CrossRef]
29. Lazaro B, Villa JA, Santin O, Cabezas M, Milagre CD, de la Cruz F, et al. Heterologous expression of a thermophilic diacylglycerol acyltransferase triggers triglyceride accumulation in Escherichia coli. PLoS One. 2017;12(4):e0176520. doi:10.1371/journal.pone.0176520. [Google Scholar] [PubMed] [CrossRef]
30. Szczepańska P, Hapeta P, Lazar Z. Advances in production of high-value lipids by oleaginous yeasts. Crit Rev Biotechnol. 2022;2(1):1–22. doi:10.1080/07388551.2021.1922353. [Google Scholar] [PubMed] [CrossRef]
31. Wong PS, Tamano K, Aburatani S. Improvement of free fatty acid secretory productivity in Aspergillus oryzae by comprehensive analysis on time-series gene expression. Front Microbiol. 2021;12:605095. doi:10.3389/fmicb.2021.605095. [Google Scholar] [PubMed] [CrossRef]
32. Tai M, Stephanopoulos G. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab Eng. 2013;15:1–9. doi:10.1016/j.ymben.2012.08.007. [Google Scholar] [PubMed] [CrossRef]
33. Azimov U, Okoro V, Hernandez HH. Recent progress and trends in the development of microbial biofuels from solid waste—a review. Energies. 2021;14(19):6011. doi:10.3390/en14196011. [Google Scholar] [CrossRef]
34. Zhang X, Li M, Agrawal A, San KY. Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab Eng. 2011;13(6):713–22. doi:10.1016/j.ymben.2011.09.007. [Google Scholar] [PubMed] [CrossRef]
35. Zaremberg V, McMaster CR. Differential partitioning of lipids metabolized by separate yeast glycerol-3-phosphate acyltransferases reveals that phospholipase D generation of phosphatidic acid mediates sensitivity to choline-containing lysolipids and drugs. J Biol Chem. 2002;277(41):39035–44. doi:10.1074/jbc.M207753200. [Google Scholar] [PubMed] [CrossRef]
36. Vigeolas H, Waldeck P, Zank T, Geigenberger P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J. 2007;5(3):431–41. doi:10.1111/j.1467-7652.2007.00252.x. [Google Scholar] [PubMed] [CrossRef]
37. Urbanikova V, Park YK, Krajciova D, Tachekort M, Certik M, Grigoras I, et al. Yarrowia lipolytica as a platform for punicic acid production. Int J Mol Sci. 2023;24(10):8823. doi:10.3390/ijms24108823. [Google Scholar] [PubMed] [CrossRef]
38. Gao Y, Sun Y, Gao H, Chen Y, Wang X, Xue J, et al. Ectopic overexpression of a type-II DGAT (CeDGAT2-2) derived from oil-rich tuber of Cyperus esculentus enhances accumulation of oil and oleic acid in tobacco leaves. Biotechnol Biofuels. 2021;14(1):1–6. doi:10.1186/s13068-021-01928-8. [Google Scholar] [PubMed] [CrossRef]
39. Guo P, Yao X, Jin X, Xv Y, Zhang J, Li Q, et al. Interference with DGAT gene inhibited TAG accumulation and lipid droplet synthesis in bovine preadipocytes. Animals. 2023;13(13):2223. doi:10.3390/ani13132223. [Google Scholar] [PubMed] [CrossRef]
40. Beopoulos A, Mrozova Z, Thevenieau F, Le Dall MT, Hapala I, Papanikolaou S, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74(24):7779–89. doi:10.1128/AEM.01412-08. [Google Scholar] [PubMed] [CrossRef]
41. Fazili AB, Shah AM, Zan X, Naz T, Nosheen S, Nazir Y, et al. Mucor circinelloides: a model organism for oleaginous fungi and its potential applications in bioactive lipid production. Microb Cell Fact. 2022;21(1):29. doi:10.1186/s12934-022-01758-9. [Google Scholar] [PubMed] [CrossRef]
42. Chang L, Lu H, Chen H, Tang X, Zhao J, Zhang H, et al. Lipid metabolism research in oleaginous fungus Mortierella alpina: current progress and future prospects. Biotechnol Adv. 2022;54(3):107794. doi:10.1016/j.biotechadv.2021.107794. [Google Scholar] [PubMed] [CrossRef]
43. Ye Y, Nikovics K, To A, Lepiniec L, Fedosejevs ET, van Doren SR, et al. Docking of acetyl-CoA carboxylase to the plastid envelope membrane attenuates fatty acid production in plants. Nat Commun. 2020;11(1):6191. doi:10.1038/s41467-020-20014-5. [Google Scholar] [PubMed] [CrossRef]
44. Wang M, Garneau MG, Poudel AN, Lamm D, Koo AJ, Bates PD, et al. Overexpression of pea α-carboxyltransferase in Arabidopsis and camelina increases fatty acid synthesis leading to improved seed oil content. Plant J. 2022;110(4):1035–46. [Google Scholar] [PubMed]
45. Rawat J, Gupta PK, Pandit S, Priya K, Agarwal D, Pant M, et al. Latest expansions in lipid enhancement of microalgae for biodiesel production: an update. Energies. 2022;15(4):1550. doi:10.3390/en15041550. [Google Scholar] [CrossRef]
46. Chen D, Yuan X, Liang L, Liu K, Ye H, Liu Z, et al. Overexpression of acetyl-CoA carboxylase increases fatty acid production in the green alga Chlamydomonas reinhardtii. Biotechnol Lett. 2019;41(10):1133–45. doi:10.1007/s10529-019-02715-0. [Google Scholar] [PubMed] [CrossRef]
47. Dunahay TG, Jarvis EE, Dais SS, Roessler PG. Manipulation of microalgal lipid production using genetic engineering. Appl Biochem Biotechnol. 1996;57(1):223–31. doi:10.1007/BF02941703. [Google Scholar] [CrossRef]
48. Xue J, Niu YF, Huang T, Yang WD, Liu JS, Li HY. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab Eng. 2015;27:1–9. doi:10.1016/j.ymben.2014.10.002. [Google Scholar] [PubMed] [CrossRef]
49. Fukuda S, Hirasawa E, Takemura T, Takahashi S, Chokshi K, Pancha I, et al. Accelerated triacylglycerol production without growth inhibition by overexpression of a glycerol-3-phosphate acyltransferase in the unicellular red alga Cyanidioschyzon merolae. Sci Rep. 2018;8(1):12410. doi:10.1038/s41598-018-30809-8. [Google Scholar] [PubMed] [CrossRef]
50. Cho HU, Park JM. Biodiesel production by various oleaginous microorganisms from organic wastes. Bioresour Technol. 2018;256:502–8. doi:10.1016/j.biortech.2018.02.010. [Google Scholar] [PubMed] [CrossRef]
51. Naveira-Pazos C, Veiga MC, Kennes C. Accumulation of lipids by the oleaginous yeast Yarrowia lipolytica grown on carboxylic acids simulating syngas and carbon dioxide fermentation. Bioresour Technol. 2022;360(3):127649. doi:10.1016/j.biortech.2022.127649. [Google Scholar] [PubMed] [CrossRef]
52. Li J, Mu X, Dong W, Chen Y, Kang Q, Zhao G, et al. A non-carboxylative route for the efficient synthesis of central metabolite malonyl-CoA and its derived products. Nat Catal. 2024:1–4. doi:10.1038/s41929-023-01103-2. [Google Scholar] [CrossRef]
53. Chen JQ, Lang CX, Hu ZH, Liu ZH, Huang RZ. Antisense PEP gene regulates to ratio of protein and lipid content in Brassica napus seeds. J Agric Biotechnol. 1999;7(4):316–20 (In Chinese). [Google Scholar]
54. Lin JY, Ng IS. Enhanced carbon capture, lipid and lutein production in Chlamydomonas reinhardtii under meso-thermophilic conditions using chaperone and CRISPRi system. Bioresour Technol. 2023;384(1):129340. doi:10.1016/j.biortech.2023.129340. [Google Scholar] [PubMed] [CrossRef]
55. Sayanova O, Napier AJ. Metabolic engineering of microalgae for sustainable production of omega-3 long chain polyunsaturated fatty acids. Curr Biotechnol. 2016;5(3):198–212. [Google Scholar]
56. Kuang T, Lu C, Zhang L, Hou L, Jia X, Shi D, et al. Expression of PEPC gene, lipid content and photosynthesis in Anabaena 7120. In: Photosynthesis research for food, fuel and the future, Berlin Heidelberg: Springer; 2013. p. 335–8. [Google Scholar]
57. Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, et al. Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng. 2010;12(4):387–91. doi:10.1016/j.ymben.2010.02.002. [Google Scholar] [PubMed] [CrossRef]
58. Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng. 2011;13(1):89–95. doi:10.1016/j.ymben.2010.10.003. [Google Scholar] [PubMed] [CrossRef]
59. Fayyaz M, Chew KW, Show PL, Ling TC, Ng IS, Chang JS. Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol Adv. 2020;43:107554. doi:10.1016/j.biotechadv.2020.107554. [Google Scholar] [PubMed] [CrossRef]
60. Fakas S. Lipid biosynthesis in yeasts: a comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica. Eng Life Sci. 2017;17(3):292–302. doi:10.1002/elsc.201600040. [Google Scholar] [PubMed] [CrossRef]
61. Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell. 2010;9(1):97–106. doi:10.1128/ec.00203-09. [Google Scholar] [PubMed] [CrossRef]
62. Sun H, Wu T, Chen SH, Ren Y, Yang S, Huang J, et al. Powerful tools for productivity improvements in microalgal production. Renew Sustain Energy Rev. 2021;152(2):111609. doi:10.1016/j.rser.2021.111609. [Google Scholar] [CrossRef]
63. Deng XD, Gu B, Li YJ, Hu XW, Guo JC, Fei XW. The roles of acyl-CoA: diacylglycerol acyltransferase 2 genes in the biosynthesis of triacylglycerols by the green algae Chlamydomonas reinhardtii. Molecular Plant. 2012;5(4):945–7. doi:10.1093/mp/sss040. [Google Scholar] [PubMed] [CrossRef]
64. Hsieh HJ, Su CH, Chien LJ. Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J Microbiol. 2012;50(3):526–34. doi:10.1007/s12275-012-2041-5. [Google Scholar] [PubMed] [CrossRef]
65. Hao X, Luo L, Jouhet J, Rébeillé F, Maréchal E, Hu H, et al. Enhanced triacylglycerol production in the diatom Phaeodactylum tricornutum by inactivation of a Hotdog-fold thioesterase gene using TALEN-based targeted mutagenesis. Biotechnol Biofuels. 2018;11(1):1–8. doi:10.1186/s13068-018-1309-3. [Google Scholar] [PubMed] [CrossRef]
66. Yoo D, Hong SJ, Yun S, Kang MJ, Cho BK, Lee H, et al. Metabolic engineering for redirecting carbon to enhance the fatty acid content of Synechocystis sp. PCC6803. Biotechnol Biofuels. 2023;28(2):274–80. doi:10.1007/s12257-020-0386-x. [Google Scholar] [CrossRef]
67. Carpine R, Du W, Olivieri G, Pollio A, Hellingwerf KJ, Marzocchella A, et al. Genetic engineering of Synechocystis sp. PCC6803 for poly-β-hydroxybutyrate overproduction. Algal Res. 2017;25:117–27. doi:10.1016/j.algal.2017.05.013. [Google Scholar] [CrossRef]
68. Kao PH, Ng IS. CRISPRi mediated phosphoenolpyruvate carboxylase regulation to enhance the production of lipid in Chlamydomonas reinhardtii. Bioresour Technol. 2017;245(4):1527–37. doi:10.1016/j.biortech.2017.04.111. [Google Scholar] [PubMed] [CrossRef]
69. Villanova V, Singh D, Pagliardini J, Fell D, Le Monnier A, Finazzi G, et al. Boosting biomass quantity and quality by improved mixotrophic culture of the diatom Phaeodactylum tricornutum. Front Plant Sci. 2021;12:642199. doi:10.3389/fpls.2021.642199. [Google Scholar] [PubMed] [CrossRef]
70. Zhou Y, Huang X, Hu T, Chen S, Wang Y, Shi X, et al. Genome-wide analysis of glycerol-3-phosphate acyltransferase (GPAT) family in perilla frutescens and functional characterization of PfGPAT9 crucial for biosynthesis of storage oils rich in high-value lipids. Int J Mol Sci. 2023;24(20):15106. doi:10.3390/ijms242015106. [Google Scholar] [PubMed] [CrossRef]
71. Niu YF, Wang X, Hu DX, Balamurugan S, Li DW, Yang WD, et al. Molecular characterization of a glycerol-3-phosphate acyltransferase reveals key features essential for triacylglycerol production in Phaeodactylum tricornutum. Biotechnol Biofuels. 2016;9(1):60. doi:10.1186/s13068-016-0478-1. [Google Scholar] [PubMed] [CrossRef]
72. Muñoz CF, Weusthuis RA, D’Adamo S, Wijffels RH. Effect of single and combined expression of lysophosphatidic acid acyltransferase, glycerol-3-phosphate acyltransferase, and diacylglycerol acyltransferase on lipid accumulation and composition in Neochloris oleoabundans. Front Plant Sci. 2019;10:1573. doi:10.3389/fpls.2019.01573. [Google Scholar] [PubMed] [CrossRef]
73. Lei A, Chen H, Shen G, Hu Z, Chen L, Wang J. Expression of fatty acid synthesis genes and fatty acid accumulation in Haematococcus pluvialis under different stressors. Biotechnol Biofuels. 2012;5(1):18. doi:10.1186/1754-6834-5-18. [Google Scholar] [PubMed] [CrossRef]
74. Mathew GM, Raina D, Narisetty V, Kumar V, Saran S, Pugazhendi A, et al. Recent advances in biodiesel production: challenges and solutions. Sci Total Environ. 2021;794(1):148751. doi:10.1016/j.scitotenv.2021.148751. [Google Scholar] [PubMed] [CrossRef]
75. Kazaz S, Miray R, Lepiniec L, Baud S. Plant monounsaturated fatty acids: diversity, biosynthesis, functions and uses. Prog Lipid Res. 2022;85(21):101138. doi:10.1016/j.plipres.2021.101138. [Google Scholar] [PubMed] [CrossRef]
76. Zheng S, Sun S, Zou S, Song J, Hua L, Chen H, et al. Effects of culture temperature and light regimes on the biomass and lipid accumulation of Chlamydomonas reinhardtii under carbon-rich and nitrogen-limited conditions. Bioresour Technol. 2024;399(1):130613. doi:10.1016/j.biortech.2024.130613. [Google Scholar] [PubMed] [CrossRef]
77. Shekh A, Sharma A, Schenk PM, Kumar G, Mudliar S. Microalgae cultivation: photobioreactors, CO2 utilization, and value-added products of industrial importance. J Chem Technol Biotechnol. 2022;97(5):1064–85. doi:10.1002/jctb.6902. [Google Scholar] [CrossRef]
78. Nzayisenga JC, Farge X, Groll SL, Sellstedt A. Effects of light intensity on growth and lipid production in microalgae grown in wastewater. Biotechnol Biofuels. 2020;13(1):1–8. doi:10.1186/s13068-019-1646-x. [Google Scholar] [PubMed] [CrossRef]
79. He Q, Yang H, Wu L, Hu C. Effect of light intensity on physiological changes, carbon allocation and neutral lipid accumulation in oleaginous microalgae. Bioresour Technol. 2015;191(7):219–28. doi:10.1016/j.biortech.2015.05.021. [Google Scholar] [PubMed] [CrossRef]
80. Wu LF, Chen PC, Lee CM. The effects of nitrogen sources and temperature on cell growth and lipid accumulation of microalgae. Int Biodeterior Biodegrad. 2013;85:506–10. doi:10.1016/j.ibiod.2013.05.016. [Google Scholar] [CrossRef]
81. Chaisutyakorn P, Praiboon J, Kaewsuralikhit C. The effect of temperature on growth and lipid and fatty acid composition on marine microalgae used for biodiesel production. J Appl Phycol. 2018;30(1):37–45. doi:10.1007/s10811-017-1186-3. [Google Scholar] [CrossRef]
82. Converti A, Casazza AA, Ortiz EY, Perego P, Del Borghi M. Effect of temperature and nitrogen concentration on the growth and lipid content of Nannochloropsis oculata and Chlorella vulgaris for biodiesel production. Chem Eng Process. 2009;48(6):1146–51. doi:10.1016/j.cep.2009.03.006. [Google Scholar] [CrossRef]
83. Sun X, Cao Y, Xu H, Liu Y, Sun J, Qiao D, et al. Effect of nitrogen-starvation, light intensity and iron on triacylglyceride/carbohydrate production and fatty acid profile of Neochloris oleoabundans HK-129 by a two-stage process. Bioresour Technol. 2014;155(3):204–12. doi:10.1016/j.biortech.2013.12.109. [Google Scholar] [PubMed] [CrossRef]
84. Yang L, Chen J, Qin S, Zeng M, Jiang Y, Hu L, et al. Growth and lipid accumulation by different nutrients in the microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2018;11(1):40. doi:10.1186/s13068-018-1041-z. [Google Scholar] [PubMed] [CrossRef]
85. Sajjadi B, Chen WY, Raman AA, Ibrahim S. Microalgae lipid and biomass for biofuel production: a comprehensive review on lipid enhancement strategies and their effects on fatty acid composition. Renew Sustain Energy Rev. 2018;97:200–32. doi:10.1016/j.rser.2018.07.050. [Google Scholar] [CrossRef]
86. Mohsenpour SF, Willoughby N. Effect of CO2 aeration on cultivation of microalgae in luminescent photobioreactors. Biomass Bioenergy. 2016;85(1–3):168–77. doi:10.1016/j.biombioe.2015.12.002. [Google Scholar] [CrossRef]
87. Omar Faruque M, Ilyas M, Mozahar Hossain M, Abdur Razzak S. Influence of nitrogen to phosphorus ratio and CO2 concentration on lipids accumulation of Scenedesmus dimorphus for bioenergy production and CO2 biofixation. Chem Asian J. 2020;15(24):4307–20. doi:10.1002/asia.202001063. [Google Scholar] [PubMed] [CrossRef]
88. Bartley ML, Boeing WJ, Dungan BN, Holguin FO, Schaub T. pH effects on growth and lipid accumulation of the biofuel microalgae Nannochloropsis salina and invading organisms. J Appl Phycol. 2014;26(3):1431–7. doi:10.1007/s10811-013-0177-2. [Google Scholar] [CrossRef]
89. Shah SMU, Che Radziah C, Ibrahim S, Latiff F, Othman MF, Abdullah MA. Effects of photoperiod, salinity and pH on cell growth and lipid content of Pavlova lutheri. Ann Microbiol. 2014;64(1):157–64. doi:10.1007/s13213-013-0645-6. [Google Scholar] [CrossRef]
90. BenMoussa-Dahmen I, Chtourou H, Rezgui F, Sayadi S, Dhouib A. Salinity stress increases lipid, secondary metabolites and enzyme activity in Amphora subtropica and Dunaliella sp. for biodiesel production. Bioresour Technol. 2016;218:816–25. doi:10.1016/j.biortech.2016.07.022. [Google Scholar] [PubMed] [CrossRef]
91. Li X, Song M, Yu Z, Wang C, Sun J, Su K, et al. Comparison of heterotrophic and mixotrophic Chlorella pyrenoidosa cultivation for the growth and lipid accumulation through acetic acid as a carbon source. J Environ Chem Eng. 2022;10(1):107054. doi:10.1016/j.jece.2021.107054. [Google Scholar] [CrossRef]
92. Shi TQ, Wang LR, Zhang ZX, Sun XM, Huang H. Stresses as first-line tools for enhancing lipid and carotenoid production in microalgae. Front Bioeng Biotechnol. 2020;8:610. doi:10.3389/fbioe.2020.00610. [Google Scholar] [PubMed] [CrossRef]
93. Singh P, Kumari S, Guldhe A, Misra R, Rawat I, Bux F. Trends and novel strategies for enhancing lipid accumulation and quality in microalgae. Renew Sustain Energy Rev. 2016;55:1–6. doi:10.1016/j.rser.2015.11.001. [Google Scholar] [CrossRef]
94. Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther. 2020;5(1):1. doi:10.1038/s41392-019-0089-y. [Google Scholar] [PubMed] [CrossRef]
95. Ghribi M, Nouemssi SB, Meddeb-Mouelhi F, Desgagné-Penix I. Genome editing by CRISPR-Cas: a game change in the genetic manipulation of Chlamydomonas. Life. 2020;10(11):295. doi:10.3390/life10110295. [Google Scholar] [PubMed] [CrossRef]
96. Fine EJ, Cradick TJ, Bao G. Strategies to determine off-target effects of engineered nucleases. In: Cathomen T, Hirsch M, Porteus M, editors. Genome Editing. Advances in experimental medicine and biology. New York: Springer; 2016. p. 187–222. doi:10.1007/978-1-4939-3509-3_11. [Google Scholar] [CrossRef]
97. Shamshirgaran Y, Liu J, Sumer H, Verma PJ, Taheri-Ghahfarokhi A. Tools for efficient genome editing; ZFN, TALEN, and CRISPR. In: Verma PJ, Sumer H, Liu J, editors. Applications of genome modulation and editing. Methods in molecular biology, vol. 2495. New York: Humana; 2022. p. 29–46. doi:10.1007/978-1-0716-2301-5_2. [Google Scholar] [PubMed] [CrossRef]
98. Munawar N, Ahmad A. CRISPR/Cas system: an introduction. CRISPR crops: the future of food security. Springer Nature. 2021;1–35. doi:10.1007/978-981-15-7142-8_1. [Google Scholar] [CrossRef]
99. Andrianov V, Borisjuk N, Pogrebnyak N, Brinker A, Dixon J, Spitsin S, et al. Tobacco as a production platform for biofuel: overexpression of Arabidopsis DGAT and LEC2 genes increases accumulation and shifts the composition of lipids in green biomass. Plant Biotech J. 2010;8:277–87. [Google Scholar]
100. Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Annu Rev Microbiol. 2010;64(1):475–93. doi:10.1146/annurev.micro.112408.134123. [Google Scholar] [PubMed] [CrossRef]
101. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi:10.1038/s41580-019-0131-5. [Google Scholar] [PubMed] [CrossRef]
102. Asmamaw M, Zawdie B. Mechanism and applications of CRISPR/Cas-9-mediated genome editing. Biologics. 2021;15:353–61. doi:10.2147/BTT.S326422. [Google Scholar] [PubMed] [CrossRef]
103. Gaj T, Gersbach CA, Barbas C.F. ZFN. TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi:10.1016/j.tibtech.2013.04.004. [Google Scholar] [PubMed] [CrossRef]
104. Wendt KE, Ungerer J, Cobb RE, Zhao H, Pakrasi HB. CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX 2973. Microb Cell Fact. 2016;15(1):115. doi:10.1186/s12934-016-0514-7. [Google Scholar] [PubMed] [CrossRef]
105. Stukenberg D, Zauner S, Dell’Aquila G, Maier UG. Optimizing CRISPR/Cas9 for the Diatom Phaeodactylum tricornutum. Front Plant Sci. 2018;9:740. doi:10.3389/fpls.2018.00740. [Google Scholar] [PubMed] [CrossRef]
106. Wang G, Li M, Zhang C, Cheng H, Gao Y, Deng W, et al. Transcriptome and proteome analyses reveal the regulatory networks and metabolite biosynthesis pathways during the development of Tolypocladium guangdongense. Comput Struct Biotechnol J. 2020;18:2081–94. [Google Scholar] [PubMed]
107. Guha TK, Wai A, Hausner G. Programmable genome editing tools and their regulation for efficient genome engineering. Comput Struct Biotechnol J. 2017;15(6):146–60. doi:10.1016/j.csbj.2016.12.006. [Google Scholar] [PubMed] [CrossRef]
108. Guzmán-Zapata D, Sandoval-Vargas JM, Macedo-Osorio KS, Salgado-Manjarrez E, Castrejón-Flores JL, Oliver-Salvador MDC, et al. Efficient editing of the nuclear APT reporter gene in Chlamydomonas reinhardtii via expression of a CRISPR-Cas9 module. Int J Mol Sci. 2019 Mar 12;20(5):1247. doi:10.3390/ijms20051247. [Google Scholar] [PubMed] [CrossRef]
109. Tanwar A, Sharma S, Kumar S. Targeted genome editing in algae using CRISPR/Cas9. Indian J Plant Physiol. 2018;23(4):653–69. doi:10.1007/s40502-018-0423-3. [Google Scholar] [CrossRef]
110. Ma E, Chen K, Shi H, Stahl EC, Adler B, Trinidad M, et al. Improved genome editing by an engineered CRISPR-Cas12a. Nucleic Acids Res. 2022;50(22):12689–701. doi:10.1093/nar/gkac1192. [Google Scholar] [PubMed] [CrossRef]
111. Rai A, Saito K. Omics data input for metabolic modeling. Curr Opin Biotechnol. 2016;37:127–34. doi:10.1016/j.copbio.2015.10.010. [Google Scholar] [PubMed] [CrossRef]
112. Tan KC, Ipcho SV, Trengove RD, Oliver RP, Solomon PS. Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol Plant Pathol. 2009;10(5):703–15. doi:10.1111/j.1364-3703.2009.00565.x. [Google Scholar] [PubMed] [CrossRef]
113. Sen P, Orešič M. Integrating omics data in genome-scale metabolic modeling: a methodological perspective for precision medicine. Metabolites. 2023;13(7):855. doi:10.3390/metabo13070855. [Google Scholar] [PubMed] [CrossRef]
114. Arora N, Pienkos PT, Pruthi V, Poluri KM, Guarnieri MT. Leveraging algal omics to reveal potential targets for augmenting TAG accumulation. Biotechnol Adv. 2018;36(4):1274–92. doi:10.1016/j.biotechadv.2018.04.005. [Google Scholar] [PubMed] [CrossRef]
115. Kloska SM, Pałczyński K, Marciniak T, Talaśka T, Wysocki BJ, Davis P, et al. Integrating glycolysis, citric acid cycle, pentose phosphate pathway, and fatty acid beta-oxidation into a single computational model. Sci Rep. 2023;13(1):14484. doi:10.1038/s41598-023-41765-3. [Google Scholar] [PubMed] [CrossRef]
116. de Bhowmick G, Koduru L, Sen R. Metabolic pathway engineering towards enhancing microalgal lipid biosynthesis for biofuel application–a review. Renew Sustain Energy Rev. 2015;50:1239–53. doi:10.1016/j.rser.2015.04.131. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF

 Downloads
Downloads
 Citation Tools
Citation Tools