 Open Access
Open Access
REVIEW
Microphysiological systems for modeling gut-organ interaction
Department of Chemical Engineering, Hongik University, Seoul, Korea
* Corresponding Author: JONG HWAN SUNG. Email:
(This article belongs to the Special Issue: Gut Microbiota in Human Health: Exploring the Complex Interplay)
BIOCELL 2024, 48(8), 1145-1153. https://doi.org/10.32604/biocell.2024.050365
Received 04 February 2024; Accepted 10 May 2024; Issue published 02 August 2024
Abstract
The gut is a digestive organ that absorbs nutrients but also plays a vital role in immune response and defense against external compounds. The complex interaction between the gut microbiota and other organs including the immune system of the host has been known in various contexts, yielding the notion of ‘axes’ between the gut and other organs. While the presence of various gut-organ axes has been reported, the lack of adequate in vitro model systems for studying this interaction has restricted a deeper insight into these phenomena. Recently developed microphysiological systems (MPS), also known as organ-on-a-chip, allow researchers to study complex interactions between diverse organs, and here we provide a review of how recently developed gut-on-a-chip systems are used for building models of various diseases that were difficult to study.Keywords
One of the key steps for orally administered drugs is the process of absorption in the gut and subsequent metabolism, which happens mainly in the liver but also occurs in the intestine, known as first-pass metabolism [1]. In addition, the gut has a close relationship with the whole body, acting as the first layer of defense against external compounds. The human intestine is a highly complex organ with a unique 3D architecture and diverse components comprising the system. It consists of different cell types, such as epithelial (enterocytes), endocrine, immune cells, as well as microbiota, making it a difficult organ to model [2].
Developing models of the human gut that faithfully reproduce the structure and the functions is important in terms of studying drug absorption and metabolism, as well as studying gut-related diseases. Currently, there is a wide range of in vitro models that aim to reproduce the intestinal functions with varying degrees of complexity, for example, a relatively simple, parallel artificial membrane permeability assay (PAMPA) to simulate the gut barrier function on one end [3], and intestinal membrane segments using tissue explants, which conserve the structure and functions of the native tissue [4]. Cell culture models are also widely used, such as Caco-2, which is a colorectal cancer-derived cell line, but have significant limitations in terms of physiological relevance to the native tissues.
Most importantly, these models only aim to reproduce the gut microenvironment and neglect the important aspect of its interaction with other organs in the body. Being the first barrier to orally ingested compounds, an immunological organ, and a host to the gut microbiota implies that the gut has a complex mechanism of action and diverse roles within the body. This has led to the notion of various gut-organ axes. These gut-organ axes play crucial roles in maintaining the body’s homeostasis, with organs such as the brain, liver, and skin being involved [5]. Despite the importance of this concept, it could only be observed in clinical settings or limited animal studies, due to the complex nature of the interaction between different organs in the body. One should also note that the mechanisms of the interaction are diverse, including molecular interaction by systemic circulation as well as neurological pathways between nervous systems [6].
Microphysiological systems (MPS), also known as organ-on-a-chip, are based on microfabrication and microfluidics technology, aimed to reproduce the microenvironment of human tissues to improve the physiological relevance of the models [7]. Initially, organ-on-a-chip technology has been developed to reproduce the microenvironment of specific organs or tissues. For example, a gut-on-a-chip attempts to recapitulate the important aspects of the human intestine, such as the 3D structural architecture [8], mechanical movements of the intestine such as peristalsis [9], and commensal gut microbiome residing in the intestine [10]. Several excellent review papers have been published for readers who are interested in recent developments in organ-on-a-chip technology [11,12].
Particularly, MPS comprising multiple organ components, often known as multi-organ-on-a-chip, can not only reproduce individual organ functions but also recapitulate the interactions between the organs [13]. While most up-to-date research on gut-on-a-chip systems has focused on reproducing the gut microenvironment, a few studies are emerging that capture the crosstalk between organs, although in a primitive manner. In particular, microfluidics-based systems are ideal for capturing the interaction between tissues via a vascular network [14]. Earlier, proof-of-concept studies have shown that reproducing the molecular interaction between organ components is feasible by implementing fluidic connections between them [15], which were followed by more advanced systems enabling more accurate recapitulations of molecular interactions in the body [16]. Although still at a preliminary stage, several important papers have been published showing prominent and interesting findings using such multi-organ-on-a-chip systems. In this review, we report on the recent progress in the development of multi-organ MPS, which we will name as gut-organ-axis-on-a-chip systems, aimed at reproducing the complex interactions of the gut with the whole body. The key element in achieving this goal is a faithful reproduction of the gut physiology and functions.
Orally taken substances are absorbed through the gut epithelium, which is followed by hepatic metabolism in the liver. Known as the first-pass metabolism, this sequential action by the gut and the liver exerts a significant effect on the fate of orally administered drugs [17]. Due to the importance of the first-pass metabolism, several early attempts have been made to reproduce the gut-liver crosstalk, particularly in the context of drugs processed in the body. More recently, the pathological communication between the gut and the liver, for example, non-alcoholic fatty liver disease (NAFLD) or gut inflammation, has been studied, which we will discuss in more detail in the following sections. Here we summarize the recent progress in developing gut-liver axis-on-a-chip systems (Fig. 1).
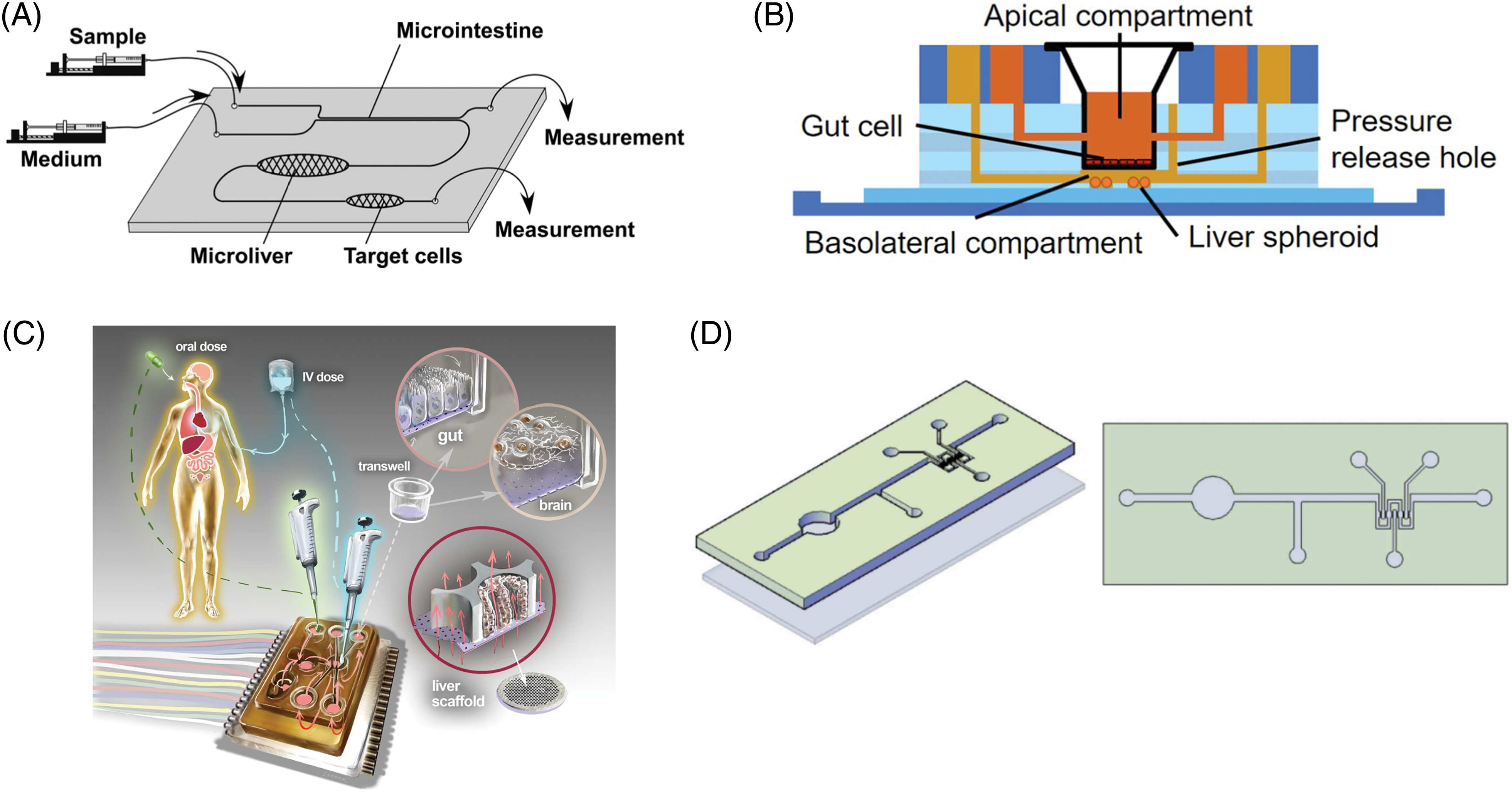
Figure 1: Examples of gut-liver-axis-on-a-chip systems (A) Microfluidic chip connecting the gut, liver, and the target organ, reproduced with permission from [21]. (B) A gut-liver chip for reproducing the PK of drugs, reproduced with permission from [32] (C) An interconnected microphysiological system, containing the gut, liver, and brain, reproduced from [34] with permission under the terms of the Creative Commons CC BY license (D) Intestine-liver axis on-chip for emulating ethanol first-pass metabolism and hepatic damage, reproduced from [47] with permission under the terms of the Creative Commons CC BY license.
The interactions of oral drugs in the gut and the liver have a profound effect on the pharmacokinetic (PK) profiles of the drugs. Therefore, an accurate reproduction of the dynamic interactions between the gut and the liver holds a key to successful drug development, as well as a better understanding of many gut and liver-related diseases. Conventionally, the absorption of drugs in the gut and the metabolism in the liver were assessed in separate processes and then combined to predict the fate of drugs, often with the help of mathematical models such as PK models.
Early primitive experiments to reproduce the first-pass metabolism of drugs by the gut and the liver in a single system were made, although in a very primitive form of co-culturing gut and liver cells in the apical and basal compartments of a Transwell insert, respectively [18]. This early attempt was followed by a more advanced format utilizing a fluidic connection between the gut and liver compartment, where authors observed improved Cytochrome P450 (CYP) metabolic enzyme activity, possibly due to the stimulation by the perfusion of liquids [19]. With the advent of microfluidics technology in the early 2000s, more advanced microfluidic-based gut-liver systems using cell lines have appeared. A micro total bioassay system, developed by Yoshimura et al., was designed to work as a microfluidic system that sequentially assesses intestinal absorption, hepatic metabolism, and bioactivity [20,21]. Cell lines representing the respective organs were used; Caco-2 for the gut, HepG2 for the liver, and MCF-7 for breast carcinoma.
Shuler research group published several pioneering papers on developing multi-organ microfluidic systems [15,22]. Based on these early attempts, a microfluidic chip for studying the effect of orally taken nanoparticles on liver injury was published [23], which was further advanced by utilizing the integration of single-organ chips with modular design [24], and also expanded to accommodate up to 14 compartments [25]. Another study demonstrated a modified chip design mimicking the liver micro-lobe structure while co-culturing primary intestinal and hepatocarcinoma cells for up to 14 days [26].
Bricks et al. published several papers on what they called the Integrated Insert Dynamic Microfluidic Platform (IIDMP), a microfluidic chip for co-culturing gut and liver cells with fluidic connections [27–29]. Liver cells from different origins were compared for their metabolic activity and physiological relevance to humans. In particular, it is notable that a mathematical PK model was employed to calculate key parameters such as intrinsic clearance, which made it easier for authors to compare their results with clinical data. Another research group attempted a four-organ co-culture in a microfluidic setting, including the intestine, liver, skin, and kidney [30]. Although a quantitative study on the pharmacokinetics of drugs is lacking in this study, it is noteworthy that a long-term co-culture was demonstrated successfully. More recently, Choe et al. developed a gut-liver chip with gravity-induced passive flow for reproducing the oral drug’s first-pass metabolism [31]. A simple, but useful two-layer design employed by the authors allowed easy and efficient co-culture of gut and liver cells in a microfluidic device while maintaining proximity between the two cells.
As illustrated by the early example by Prot [29], PK models are useful tools for analyzing the dynamic interaction between different modules in a chipset. The first-pass metabolism of acetaminophen was analyzed using a PK model, with parameters extracted from experimental data using a gut-liver-on-a-chip [32]. In this study, optimal design parameters were suggested that could theoretically enable more accurate reproduction of PK profiles of a model drug. This is an excellent example of utilizing a mathematical and numerical framework for interpreting the experimental data from the chip, as well as exploring various chip design parameters for improved predictions. Since organ-on-a-chip are essentially miniaturized systems of the human body, it inevitably carries the limitation of deviating from the original, due to the disproportionate scaling of different organs. Some researchers suggested theoretical methodologies to approach this issue [33], with some pioneering, proof-of-concept works using model drugs [34–36]. In these papers, mechanistic model-based analysis was used to predict intrinsic PK parameters of diclofenac and hydrocortisone with some success. Although still at a preliminary stage, it has been demonstrated that prediction of key PK parameters using chip systems may be possible [37]. However, more robust and expandable mathematical principles are needed for wider acceptance of such systems.
The gut-liver communication plays important roles in diseased states as well. For example, the gut-liver axis has important implications in terms of NAFLD, with accumulating evidence pointing to a close relationship between the gut microbiome and inflammation [38]. One of the well-known examples is the short-chain fatty acids (SCFAs) produced by the gut commensal microbes suppressing inflammation. The gut-liver axis is also thought to affect the progression of cancer as well as metastasis. Lipopolysaccharides (LPS), which is the component of the outer membrane of Gram-negative bacteria are a ligand of innate immune receptors that can induce inflammatory responses [39]. The difficulty of recapitulating such complex interactions lies not only in the inherent complexity of the interaction, but also in the need to incorporate diverse components, such as the gut epithelium, hepatocytes as well as other supporting cells in the liver, and gut microbiota. Multi-organ-on-a-chip have demonstrated their usefulness for recapitulating the key interactions between different components. Gut and liver cell cultures connected with fluidic circulation recapitulated the migration of colon carcinoma, by reproducing the entry of disseminated cancer cells into circulation leading to the liver [40]. Another critical pathological aspect of interest is an inflammatory response. An integrative gut and liver platform was used to recapitulate the inflammation process [41]. Notably, cells were co-cultured with Kupffer cells and goblet cells improved the physiological relevance. In a subsequent study, the immune component was added to the system with T cells to model ulcerative colitis [42]. This paper is noteworthy in that a detailed study of the mechanism of T-cell-mediated inflammation and the role of SCFAs in this context. Lee et al. developed an in vitro model of gut liver in the context of NAFLD [43,44]. In this work, free fatty acids are absorbed across the epithelium before exerting any effect on the liver cells. Simply by reproducing this sequential action of the free fatty acids, it was possible to demonstrate the antisteatotic activity of different compounds was demonstrated. A subsequent study by the same group further included an immune component by adding macrophages to the system [45]. A gut-liver chip for modeling NAFLD was developed by co-culturing Caco-2 and HepG2 cell lines in a closed-circulation loop fluidic chip [46]. Notably, treating with fatty acids changed the pattern related to endoplasmic reticulum stress, implying that studying the mechanism of NAFLD in a more physiologically relevant context was possible.
The effect of ethanol on hyperpermeability and stromal injury in the intestine was studied using intestine-liver-on-chip, revealing the protective role of the intestine [47]. A gut-liver-on-a-chip was exposed to fine particulate matter (PM2.5) to reproduce the metabolic dysregulation induced by the particles [48]. A significant dysregulation in cholesterol and bile acid metabolism was observed. The gut microbiome is also a critical part of the gut-liver crosstalk. A gut-liver chip with gut epithelial cells, hepatocarcinoma cells in the form of 3D spheroids, was used to examine the effect of microbe-derived metabolites [49]. Interestingly, adding the microbe-derived metabolites seemed to enhance various liver-specific functions, demonstrating their beneficial effect.
Implementation of high-throughput analysis, possibly with robotic liquid handling and chip operation is also an important research area. Satoh et al. published a paper on what authors termed a multi-throughput system, for up to 16 systems in parallel [50]. In a similar study by a different group, the effect of combinations of drugs was evaluated using the gut-liver system [51]. An enterohepatic system was used to examine the fate of triazolam after phase I and II metabolism [52]. Such studies show promise that eventually multi-throughput implementation and automatic analysis of drugs’ action in the body will be possible.
Due to their anatomical proximity, the gut and the liver interact closely during many physiological processes, including digestion, absorption, and metabolism. One of the prominent examples is the role of bile, which is synthesized by the liver, in the digestion and absorption in the intestine. Bile is produced by the liver, stored in the gallbladder, and released through the bile duct to the duodenum. Bile has multiple functions, including the breakdown of fats so that they can be absorbed, the absorption of fat-soluble vitamins, and the removal of metabolic wastes [53]. The majority of bile acids are reabsorbed in the distal ileum and returned to the liver via the portal vein, known as the enterohepatic circulation of bile acids [54]. Several intestinal and hepatic disorders are related to dysregulation in the signaling mediated by the bile, such as the case of bile acid-induced diarrhea caused by a decrease in the bile acid-induced ileal hormone Fibroblast Growth Factor 15/19 (FGF15/19) [55]. Midwood et al. attempted to recapitulate the ileal hormone signaling using a chip platform, by co-culturing the gut and liver tissue slices and exposing the slices to the primary bile acids, which induced the expression of FGF15 in the intestinal slice, resulting in the down-regulation of cytochrome enzyme in the liver [56]. Given the importance of bile metabolism, it is expected that there will be more attempts to develop models of the gut liver involving bile metabolism.
There has been accumulating evidence on the link between the gut and the brain, the gut environment affecting the neurocognitive functions, or vice versa [57]. An interesting feature of the gut-brain axis is that there exist multiple pathways of interaction between the gut and the brain, including biomolecular signaling through blood circulation [58], as well as neurological pathways [59]. It is also noteworthy that extracellular vesicles play important roles in communication between the two organs [60,61] (Fig. 2A). Due to the complex nature of the communication between the gut and the brain, an in vitro system that recapitulates all of the key mechanisms is yet to be developed. However, there are a few examples of preliminary systems that attempted to partially reproduce the gut-brain interaction.
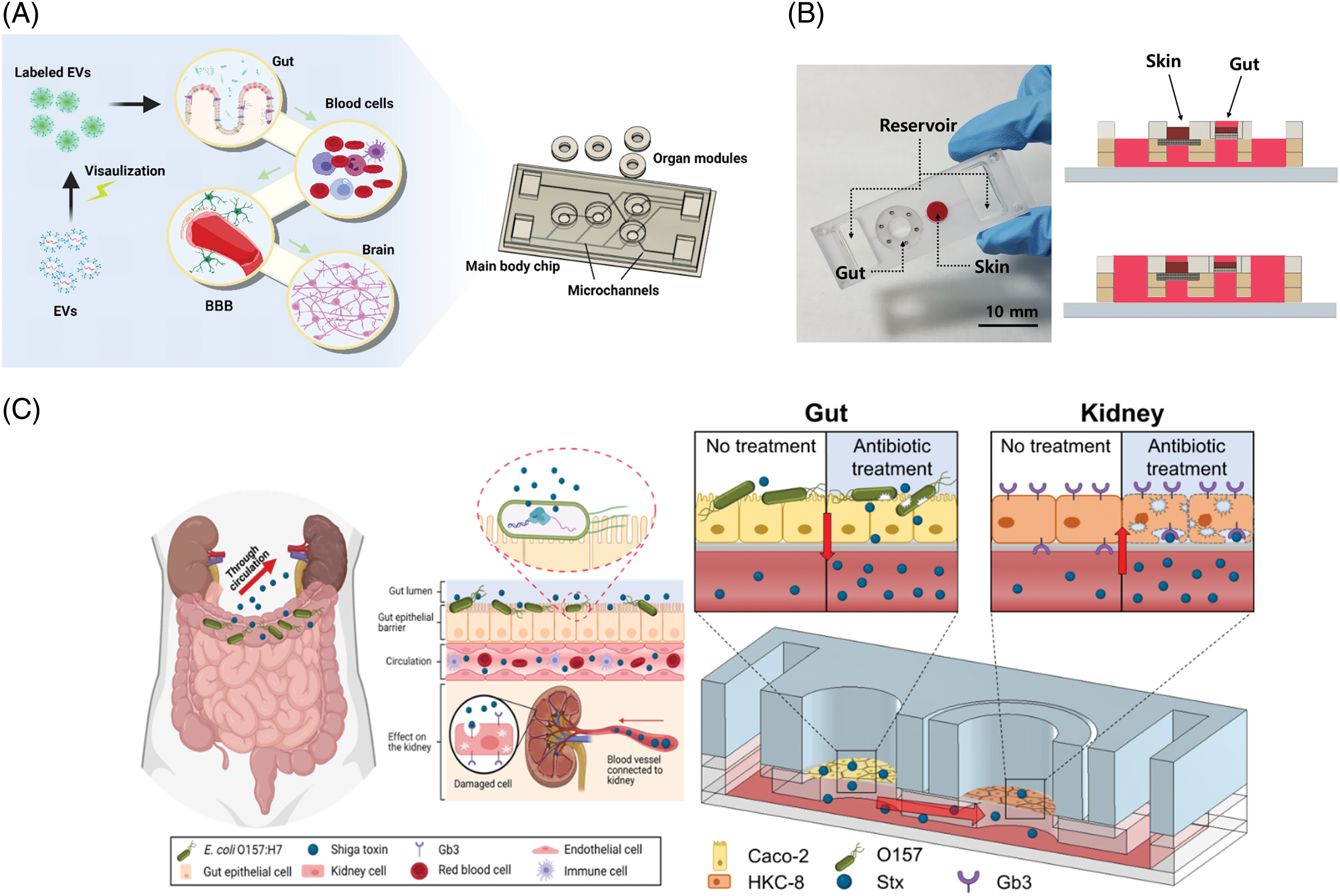
Figure 2: Various gut-organ axis on a chip system (A) Modular multi-organ-on-a-chip system for reproducing the gut-brain axis, reproduced from [60], with permission under the terms of the Creative Commons CC BY license (B) A microfluidic gut-skin-on-a-chip for reproducing the gut-skin interaction, reproduced from [69] with permission (C) Gut-Kidney-on-a-chip platform for studying Hemolytuic Urea Syndrome, reproduced from [76] under an open access Creative Common CC BY license.
One of the key components of the gut-brain axis is the presence of a robust and selective barrier between the systemic circulation and the brain, known as the blood-brain-barrier (BBB), and many chip-based systems have delved into reproducing the structure and the key functions of this barrier [62]. Several prominent examples of miniaturized, microfluidics-based models of BBB have been developed, which are reviewed elsewhere [63]. Based on this recent development, there have been attempts to integrate the ‘gut barrier’ model with the ‘brain barrier’ model. For example, a gut-brain-on-chip was developed with a two-layer microfluidic chip for gut cells and brain microvascular endothelial cells (hBMECs) nearby [64]. The formation of both gut epithelium and BBB inside the chip-enabled the study of the migration of exosomes across the BBB. The model of the gut-brain axis was constructed for Parkinson’s disease, where the gut, liver, and cerebral MPS were connected with containing CD4+ T and T helper cells [65]. One of the superior features of this model was that it contained multiple cell types including the brain, such as neurons, astrocytes, and microglia, containing mutations causing Parkinson’s disease. Short-chain fatty acids (SCFAs) secreted from gut microbes induced several transcriptomic changes, indicating that such a model can be a useful platform for studying the mechanism of gut-brain interaction.
An important player in the context of any organ-organ interaction is the immune component. The gut-brain-immune axis was studied by using a multi-organ chip for co-culturing tissue slices [66]. To represent the immune component, the tissue slices of the Peyer’s patch and mesenteric lymph node were used, and cell migration and cytokine secretion were examined. Another interesting example used cortical neurons and dendritic cells to study neuro-immunological communication [67]. This study is unique in that the authors studied the electrophysiological signal between the two components, linked by synapses. These platforms successfully demonstrate that at least some parts of the features of the gut-brain axis can be recapitulated, which should provide valuable platforms for studying related diseases.
The gut-skin axis is also an active area of research [68]. The gut and the skin have many features in common, including the epithelial barrier, the presence of commensal bacteria, and immune components. Inflammatory responses are deeply involved in pathological conditions in both organs, often observed simultaneously. A preliminary attempt was made to fluidically connect the gut and the skin tissue in a microfluidic chip [69] (Fig. 2B). Although several important components were absent, such as immune cells, challenging the gut compartment with inflammatory signals resulted in skin inflammation, as well as increased expression of a dermal disease marker. Integration of the skin vasculature with this model system is thought to further improve the physiological relevance of the model [70].
It has also been known that inflammatory gastrointestinal diseases have close associations with pulmonary complications [71]. Recent studies also suggest that Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), which causes COVID-19, can also affect the gastrointestinal tract and the gut microbiome [72]. An attempt to develop a model of gut infection by SARS-CoV-2 was made [73]. Although this work did not contain the lung compartment and therefore not exactly a model of the gut-lung axis, it is not difficult to imagine the integration of such a platform with already established lung-on-a-chip platforms [74].
The hemolytic-uremic syndrome (HUS) is an acute disorder characterized by the triad of microangiopathic hemolytic anemia, nephropathy, and thrombocytopenia [75]. One of the major causes of HUS is Shiga-toxin-producing bacteria, such as Escherichia coli (E. coli), often transmitted through ground beef and unpasteurized milk. This pathological interaction between the gut microbe and the kidney failure was reproduced using a microfluidic gut-kidney chip [76] (Fig. 2C). When gut cells were infected by E.coli and treated with antibiotics, Shiga toxins released from the microbes in the gut compartment migrated to the kidney cells via fluidic channels and caused cell death. It is interesting to note that different antibiotics showed different toxic effects on the kidney cells. Ciprofloxacin was more detrimental than Gentamycin, which was consistent with previously reported clinical observations [77].
As a summary of our previous discussion, we included a table of major examples of gut-organ axis models within the past five years (Table 1). Although organ-on-a-chip systems for reproducing various gut-organ axes are still in their preliminary stages, recent studies show great promise in this direction. Because in vitro model systems that can model such interactions between different organs are still lacking, these platforms should find uses in many areas, such as the drug development process and the medical science field by enabling in vitro experiments on complex biological processes in the body. For example, studying the effect of drugs on gut-liver interaction involving bile metabolism would be possible. Biochemical and neurological communication between the gut and the brain consists of complex and multi-faceted mechanisms that are difficult to reproduce using conventional in vitro models, and multi-organ-on-a-chip systems are platforms that can complement traditional systems.

However, there are still many challenges that need to be overcome. In many cases, connecting two organ components alone would not be sufficient to model the whole process. For example, the immune component plays an important role in the progression of many inflammatory diseases involving the gut, skin, liver, etc. Incorporation of immune cells into the organ-on-a-chip systems, or even integration with immunological organs such as the lymph node should yield significantly advanced in vitro models compared to current ones. The presence of proper vascular networks is also important. Co-culturing such multiple components in a single system will inevitably give rise to many problems, such as media compatibility. This will likely require the development of new, universal cell culture media that can support multiple cell types. Novel methodologies for maintaining tissue microenvironment niches for each tissue in a microfluidic system may also be possible, by utilizing the unique advantages of microfluidics [78].
The issue of scalability, mass production, and cost is also important. Since organ-on-a-chip technology has been gaining attention and expanding in the last few years, most of the reported systems are still at the research level and not ready for wider application in the industry. The use of PDMS (polydimethylsiloxane) has contributed significantly to this field by providing a biocompatible, flexible, transparent, and gas-permeable material for fast design and fabrication of microfluidic devices, it is not suitable for mass production, compared to more conventional plastics. More work needs to be done in terms of standardization, automation, and mass production for high-throughput applications in the future.
Another important aspect of organ-on-a-chip technology is how fast it will be accepted by relevant research communities and pharmaceutical companies. This will require support from the regulatory agencies. The recent launching of the Complement-ARIE (Animal Research in Experimentation) program by the NIH (National Institute of Health) council, which aims to catalyze the development, standardization, validation, and use of human-based new approach methodologies is encouraging in this aspect [79]. A wider acceptance in the field and application in the industry will require more thorough validation of the new models, as well as standardization for mass production [11]. Given the recent achievement in this field, it is likely that shortly, we will see more robust and physiologically relevant models being developed and employed in real-world applications.
Acknowledgement: None.
Funding Statement: This work was supported by Korea Technology and Information Promotion Agency (S3316767), National Research Foundation of Korea (2022R1A4A2000748 and 2022M3A9B6018217), Alchemist Project (KEIT 20018560, NTIS 1415184668) by of the Korea Evaluation Institute of Industrial Technology, Korea Institute of Marine Science & Technology Promotion (KIMST, RS-2024-00402200), and Hongik University Research Fund.
Author Contributions: J. H. Sung carried out all of the work to prepare the draft and write this paper.
Availability of Data and Materials: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The author declares that they have no conflicts of interest to report regarding the present study.
References
1. Pang KS, Peng HB, Noh K. The segregated intestinal flow model (SFM) for drug absorption and drug metabolism: implications on intestinal and liver metabolism and drug-drug interactions. Pharmaceutics. 2020;12(4):1–23. [Google Scholar]
2. Yao Y, Shang W, Bao L, Peng Z, Wu C. Epithelial-immune cell crosstalk for intestinal barrier homeostasis. Eur J Immunol. 2024;54:e2350631. [Google Scholar] [PubMed]
3. Bujard A, Sol M, Carrupt PA, Martel S. Predicting both passive intestinal absorption and the dissociation constant toward albumin using the PAMPA technique. Eur J Pharm Sci. 2014;63:36–44. doi:10.1016/j.ejps.2014.06.025. [Google Scholar] [PubMed] [CrossRef]
4. Kaplan SA, Cotler S. Use of cannulated everted intestinal sac for serial sampling as a drug absorbability (permeability) screen. J Pharm Sci. 1972;61(9):1361–5. doi:10.1002/jps.2600610904. [Google Scholar] [PubMed] [CrossRef]
5. Vijay A, Valdes AM. Role of the gut microbiome in chronic diseases: a narrative review. Eur J Clin Nutr. 2022;76(4):489–501. doi:10.1038/s41430-021-00991-6. [Google Scholar] [PubMed] [CrossRef]
6. Rao M, Gershon MD. The bowel and beyond: the enteric nervous system in neurological disorders. Nat Rev Gastroenterol Hepatol. 2016;13(9):517–28. doi:10.1038/nrgastro.2016.107. [Google Scholar] [PubMed] [CrossRef]
7. Zuchowska A, Baranowska P, Flont M, Brzozka Z, Jastrzebska E. Review: 3D cell models for organ-on-a-chip applications. Anal Chim Acta. 2024;1301:342413. doi:10.1016/j.aca.2024.342413. [Google Scholar] [PubMed] [CrossRef]
8. Vera D, Garcia-Diaz M, Torras N, Castillo O, Illa X, Villa R, et al. A 3D bioprinted hydrogel gut-on-chip with integrated electrodes for transepithelial electrical resistance (TEER) measurements. Biofabrication. 2024;16(3):035008. doi:10.1088/1758-5090/ad3aa4. [Google Scholar] [PubMed] [CrossRef]
9. Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A. 2016;113(1):E7–15. [Google Scholar] [PubMed]
10. Lee J, Menon N, Lim CT. Dissecting gut-microbial community interactions using a gut microbiome-on-a-chip. Adv Sci. 2024;7:e2302113. [Google Scholar]
11. Reyes DR, Esch MB, Ewart L, Nasiri R, Herland A, Sung K, et al. From animal testing to in vitro systems: advancing standardization in microphysiological systems. Lab Chip. 2024;24(5):1076–87. doi:10.1039/D3LC00994G. [Google Scholar] [PubMed] [CrossRef]
12. Mansouri M, Lam J, Sung KE. Progress in developing microphysiological systems for biological product assessment. Lab Chip. 2024;24(5):1293–306. doi:10.1039/D3LC00876B. [Google Scholar] [PubMed] [CrossRef]
13. Sung JH. From organ-on-a-chip towards body-on-a-chip. BIOCELL. 2022;46(5):1177–80. doi:10.32604/biocell.2022.019055. [Google Scholar] [CrossRef]
14. Sung JH. Multi-organ-on-a-chip for pharmacokinetics and toxicokinetic study of drugs. Expert Opin Drug Metab Toxicol. 2021;17(8):969–86. doi:10.1080/17425255.2021.1908996. [Google Scholar] [PubMed] [CrossRef]
15. Tatosian DA, Shuler ML. A novel system for evaluation of drug mixtures for potential efficacy in treating multidrug resistant cancers. Biotechnol Bioeng. 2009;103(1):187–98. doi:10.1002/bit.v103:1. [Google Scholar] [CrossRef]
16. Oleaga C, Bernabini C, Smith AS, Srinivasan B, Jackson M, McLamb W, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030. doi:10.1038/srep20030. [Google Scholar] [PubMed] [CrossRef]
17. Alqahtani S, Bukhari I, Albassam A, Alenazi M. An update on the potential role of intestinal first-pass metabolism for the prediction of drug-drug interactions: the role of PBPK modeling. Expert Opin Drug Metab Toxicol. 2018;14(6):625–34. doi:10.1080/17425255.2018.1482277. [Google Scholar] [PubMed] [CrossRef]
18. Lau YY, Chen YH, Liu TT, Li C, Cui X, White RE, et al. Evaluation of a novel in vitro Caco-2 hepatocyte hybrid system for predicting in vivo oral bioavailability. Drug Metab Dispos. 2004;32(9):937–42. [Google Scholar] [PubMed]
19. Choi SH, Fukuda O, Sakoda A, Sakai Y. Enhanced cytochrome P450 capacities of Caco-2 and Hep G2 cells in new coculture system under the static and perfused conditions: evidence for possible organ-to-organ interactions against exogenous stimuli. Mater Sci Eng. 2004;24(3):333–9. doi:10.1016/j.msec.2003.12.002. [Google Scholar] [CrossRef]
20. Imura Y, Yoshimura E, Sato K. Micro total bioassay system for oral drugs: evaluation of gastrointestinal degradation, intestinal absorption, hepatic metabolism, and bioactivity. Anal Sci. 2012;28(3):197–9. doi:10.2116/analsci.28.197. [Google Scholar] [PubMed] [CrossRef]
21. Imura Y, Sato K, Yoshimura E. Micro total bioassay system for ingested substances: assessment of intestinal absorption, hepatic metabolism, and bioactivity. Anal Chem. 2010;82(249983–8. doi:10.1021/ac100806x. [Google Scholar] [PubMed] [CrossRef]
22. Sung JH, Kam C, Shuler ML. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10(4):446–55. doi:10.1039/b917763a. [Google Scholar] [PubMed] [CrossRef]
23. Esch MB, Mahler GJ, Stokol T, Shuler ML. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14(16):3081–92. doi:10.1039/C4LC00371C. [Google Scholar] [PubMed] [CrossRef]
24. Esch MB, Ueno H, Applegate DR, Shuler ML. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip. 2016;16(14):2719–29. doi:10.1039/C6LC00461J. [Google Scholar] [PubMed] [CrossRef]
25. Miller PG, Shuler ML. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol Bioeng. 2016;113(10):2213–27. doi:10.1002/bit.v113.10. [Google Scholar] [CrossRef]
26. Chen HJ, Miller P, Shuler ML. A pumpless body-on-a-chip model using a primary culture of human intestinal cells and a 3D culture of liver cells. Lab Chip. 2018;18(14):2036–46. doi:10.1039/C8LC00111A. [Google Scholar] [PubMed] [CrossRef]
27. Bricks T, Hamon J, Fleury MJ, Jellali R, Merlier F, Herpe YE, et al. Investigation of omeprazole and phenacetin first-pass metabolism in humans using a microscale bioreactor and pharmacokinetic models. Biopharm Drug Dispos. 2015;36(5):275–93. doi:10.1002/bdd.v36.5. [Google Scholar] [CrossRef]
28. Bricks T, Paullier P, Legendre A, Fleury MJ, Zeller P, Merlier F, et al. Development of a new microfluidic platform integrating co-cultures of intestinal and liver cell lines. Toxicol in Vitro. 2014;28(5):885–95. doi:10.1016/j.tiv.2014.02.005. [Google Scholar] [PubMed] [CrossRef]
29. Prot JM, Maciel L, Bricks T, Merlier F, Cotton J, Paullier P, et al. First pass intestinal and liver metabolism of paracetamol in a microfluidic platform coupled with a mathematical modeling as a means of evaluating ADME processes in humans. Biotechnol Bioeng. 2014;111(10):2027–40. doi:10.1002/bit.v111.10. [Google Scholar] [CrossRef]
30. Maschmeyer I, Lorenz AK, Schimek K, Hasenberg T, Ramme AP, Hubner J, et al. A four-organ-chip for interconnected long-term co-culture of human intestine, liver, skin and kidney equivalents. Lab Chip. 2015;15(12):2688–99. doi:10.1039/C5LC00392J. [Google Scholar] [PubMed] [CrossRef]
31. Choe A, Ha SK, Choi I, Choi N, Sung JH. Microfluidic gut-liver chip for reproducing the first pass metabolism. Biomed Microdevices. 2017;19(1):4. doi:10.1007/s10544-016-0143-2. [Google Scholar] [PubMed] [CrossRef]
32. Lee DW, Ha SK, Choi I, Sung JH. 3D gut-liver chip with a PK model for prediction of first-pass metabolism. Biomed Microdev. 2017;19(4):100. doi:10.1007/s10544-017-0242-8. [Google Scholar] [PubMed] [CrossRef]
33. Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol. 2015;7(4):383–91. doi:10.1039/C4IB00292J. [Google Scholar] [PubMed] [CrossRef]
34. Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci Rep. 2018;8(1):4530. doi:10.1038/s41598-018-22749-0. [Google Scholar] [PubMed] [CrossRef]
35. Maass C, Stokes CL, Griffith LG, Cirit M. Multi-functional scaling methodology for translational pharmacokinetic and pharmacodynamic applications using integrated microphysiological systems (MPS). Integr Biol. 2017;9(4):290–302. doi:10.1039/C6IB00243A. [Google Scholar] [PubMed] [CrossRef]
36. Tsamandouras N, Chen WLK, Edington CD, Stokes CL, Griffith LG, Cirit M. Integrated gut and liver microphysiological systems for quantitative in vitro pharmacokinetic studies. AAPS J. 2017;19(5):1499–512. doi:10.1208/s12248-017-0122-4. [Google Scholar] [PubMed] [CrossRef]
37. Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Novak R, et al. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng. 2020;4(4):421–36. doi:10.1038/s41551-019-0498-9. [Google Scholar] [PubMed] [CrossRef]
38. Kirpich IA, Marsano LS, McClain CJ. Gut-liver axis, nutrition, and non-alcoholic fatty liver disease. Clin Biochem. 2015;48(13–14):923–30. [Google Scholar] [PubMed]
39. Ohtani N, Hara E. Gut-liver axis-mediated mechanism of liver cancer: a special focus on the role of gut microbiota. Cancer Sci. 2021;112(11):4433–43. doi:10.1111/cas.v112.11. [Google Scholar] [CrossRef]
40. Skardal A, Devarasetty M, Forsythe S, Atala A, Soker S. A reductionist metastasis-on-a-chip platform for in vitro tumor progression modeling and drug screening. Biotechnol Bioeng. 2016;113(9):2020–32. doi:10.1002/bit.v113.9. [Google Scholar] [CrossRef]
41. Chen WLK, Edington C, Suter E, Yu J, Velazquez JJ, Velazquez JG, et al. Integrated gut/liver microphysiological systems elucidates inflammatory inter-tissue crosstalk. Biotechnol Bioeng. 2017;114(11):2648–59. doi:10.1002/bit.v114.11. [Google Scholar] [CrossRef]
42. Trapecar M, Communal C, Velazquez J, Maass CA, Huang YJ, Schneider K, et al. Gut-liver physiomimetics reveal paradoxical modulation of IBD-related inflammation by short-chain fatty acids. Cell Syst. 2020;10(3):223–39. doi:10.1016/j.cels.2020.02.008. [Google Scholar] [PubMed] [CrossRef]
43. Lee SY, Sung JH. Gut-liver on a chip toward an in vitro model of hepatic steatosis. Biotechnol Bioeng. 2018;115(11):2817–27. doi:10.1002/bit.v115.11. [Google Scholar] [CrossRef]
44. Jeon JW, Lee SH, Kim D, Sung JH. In vitro hepatic steatosis model based on gut-liver-on-a-chip. Biotechnol Prog. 2021;37(3):e3121. doi:10.1002/btpr.3121. [Google Scholar] [PubMed] [CrossRef]
45. Jeon JW, Choi N, Lee SH, Sung JH. Three-tissue microphysiological system for studying inflammatory responses in gut-liver axis. Biomed Microdevices. 2020;22(4):65. doi:10.1007/s10544-020-00519-y. [Google Scholar] [PubMed] [CrossRef]
46. Yang J, Hirai Y, Iida K, Ito S, Trumm M, Terada S, et al. Integrated-gut-liver-on-a-chip platform as an in vitro human model of non-alcoholic fatty liver disease. Commun Biol. 2023;6(1):310. doi:10.1038/s42003-023-04710-8. [Google Scholar] [PubMed] [CrossRef]
47. de Gregorio V, Telesco M, Corrado B, Rosiello V, Urciuolo F, Netti PA, et al. Intestine-liver axis on-chip reveals the intestinal protective role on hepatic damage by emulating ethanol first-pass metabolism. Front Bioeng Biotechnol. 2020;8:163. doi:10.3389/fbioe.2020.00163. [Google Scholar] [PubMed] [CrossRef]
48. Duan X, Zheng L, Zhang X, Wang B, Xiao M, Zhao W, et al. A membrane-free liver-gut-on-chip platform for the assessment on dysregulated mechanisms of cholesterol and bile acid metabolism induced by PM2.5. ACS Sens. 2020;5(11):3483–92. doi:10.1021/acssensors.0c01524. [Google Scholar] [PubMed] [CrossRef]
49. Kang SG, Choi YY, Mo SJ, Kim TH, Ha JH, Hong DK, et al. Effect of gut microbiome-derived metabolites and extracellular vesicles on hepatocyte functions in a gut-liver axis chip. Nano Converg. 2023;10(1):5. doi:10.1186/s40580-022-00350-6. [Google Scholar] [PubMed] [CrossRef]
50. Satoh T, Sugiura S, Shin K, Onuki-Nagasaki R, Ishida S, Kikuchi K, et al. A multi-throughput multi-organ-on-a-chip system on a plate formatted pneumatic pressure-driven medium circulation platform. Lab Chip. 2017;18(1):115–25. [Google Scholar] [PubMed]
51. Jie M, Mao S, Liu H, He Z, Li HF, Lin JM. Evaluation of drug combination for glioblastoma based on an intestine-liver metabolic model on microchip. Analyst. 2017;142(19):3629–38. doi:10.1039/C7AN00453B. [Google Scholar] [PubMed] [CrossRef]
52. Arakawa H, Sugiura S, Kawanishi T, Shin K, Toyoda H, Satoh T, et al. Kinetic analysis of sequential metabolism of triazolam and its extrapolation to humans using an entero-hepatic two-organ microphysiological system. Lab Chip. 2020;20(3):537–47. doi:10.1039/C9LC00884E. [Google Scholar] [PubMed] [CrossRef]
53. Ahmed M. Functional, diagnostic and therapeutic aspects of bile. Clin Exp Gastroenterol. 2022;15:105–20. doi:10.2147/CEG.S360563. [Google Scholar] [PubMed] [CrossRef]
54. Ticho AL, Malhotra P, Dudeja PK, Gill RK, Alrefai WA. Intestinal absorption of bile acids in health and disease. Compr Physiol. 2019;10(1):21–56. [Google Scholar] [PubMed]
55. Lee JM, Ong JR, Vergnes L, de Aguiar Vallim TQ, Nolan J, Cantor RM, et al. Diet1, bile acid diarrhea, and FGF15/19:mouse model and human genetic variants. J Lipid Res. 2018;59(3):429–38. doi:10.1194/jlr.M078279. [Google Scholar] [PubMed] [CrossRef]
56. van Midwoud PM, Merema MT, Verpoorte E, Groothuis GM. A microfluidic approach for in vitro assessment of interorgan interactions in drug metabolism using intestinal and liver slices. Lab Chip. 2010;10(20):2778–86. doi:10.1039/c0lc00043d. [Google Scholar] [PubMed] [CrossRef]
57. Cryan JF, O’Riordan KJ, Cowan CSM, Sandhu KV, Bastiaanssen TFS, Boehme M, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99(4):1877–2013. doi:10.1152/physrev.00018.2018. [Google Scholar] [PubMed] [CrossRef]
58. Banc R, Rusu ME, Filip L, Popa DS. The impact of ellagitannins and their metabolites through gut microbiome on the gut health and brain wellness within the gut-brain axis. Foods. 2023;12(2):1–41. [Google Scholar]
59. Dowling LR, Strazzari MR, Keely S, Kaiko GE. Enteric nervous system and intestinal epithelial regulation of the gut-brain axis. J Allergy Clin Immunol. 2022;150(3):513–22. doi:10.1016/j.jaci.2022.07.015. [Google Scholar] [PubMed] [CrossRef]
60. Kim MH, van Noort D, Sung JH, Park S. Organ-on-a-chip for studying gut-brain interaction mediated by extracellular vesicles in the gut microenvironment. Int J Mol Sci. 2021;22(24):13513. doi:10.3390/ijms222413513. [Google Scholar] [PubMed] [CrossRef]
61. Yadav H, Jaldhi, Bhardwaj R, Anamika, Bakshi A, Gupta S, et al. Unveiling the role of gut-brain axis in regulating neurodegenerative diseases: a comprehensive review. Life Sci. 2023;330:122022. doi:10.1016/j.lfs.2023.122022. [Google Scholar] [PubMed] [CrossRef]
62. Peng B, Hao S, Tong Z, Bai H, Pan S, Lim KL, et al. Blood-brain barrier (BBB)-on-a-chip: a promising breakthrough in brain disease research. Lab Chip. 2022;22(19):3579–602. doi:10.1039/D2LC00305H. [Google Scholar] [PubMed] [CrossRef]
63. Wang X, Hou Y, Ai X, Sun J, Xu B, Meng X, et al. Potential applications of microfluidics based blood brain barrier (BBB)-on-chips for in vitro drug development. Biomed Pharmacother. 2020;132:110822. doi:10.1016/j.biopha.2020.110822. [Google Scholar] [PubMed] [CrossRef]
64. Kim MH, Kim D, Sung JH. A gut-brain axis-on-a-chip for studying transport across epithelial and endothelial barriers. J Ind Eng Chem. 2021;101:126–134. doi:10.1016/j.jiec.2021.06.021. [Google Scholar] [CrossRef]
65. Trapecar M, Wogram E, Svoboda D, Communal C, Omer A, Lungjangwa T, et al. Human physiomimetic model integrating microphysiological systems of the gut, liver, and brain for studies of neurodegenerative diseases. Sci Adv. 2021;7(5):1–22. [Google Scholar]
66. Delong LM, Ross AE. Open multi-organ communication device for easy interrogation of tissue slices. Lab Chip. 2023;23(13):3034–49. doi:10.1039/D3LC00115F. [Google Scholar] [PubMed] [CrossRef]
67. Gabriel-Segard T, Rontard J, Miny L, Dubuisson L, Batut A, Debis D, et al. Proof-of-concept human organ-on-chip study: first step of platform to assess neuro-immunological communication involved in inflammatory bowel diseases. Int J Mol Sci. 2023;24(13):1–13. [Google Scholar]
68. Mahmud MR, Akter S, Tamanna SK, Mazumder L, Esti IZ, Banerjee S, et al. Impact of gut microbiome on skin health: gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes. 2022;14(1):2096995. doi:10.1080/19490976.2022.2096995. [Google Scholar] [PubMed] [CrossRef]
69. Lee HR, Sung JH. Multiorgan-on-a-chip for realization of gut-skin axis. Biotechnol Bioeng. 2022;119(9):2590–601. doi:10.1002/bit.v119.9. [Google Scholar] [CrossRef]
70. Kwak BS, Jin SP, Kim SJ, Kim EJ, Chung JH, Sung JH. Microfluidic skin chip with vasculature for recapitulating the immune response of the skin tissue. Biotechnol Bioeng. 2020;117(6):1853–63. doi:10.1002/bit.v117.6. [Google Scholar] [CrossRef]
71. Saint-Criq V, Lugo-Villarino G, Thomas M. Dysbiosis, malnutrition and enhanced gut-lung axis contribute to age-related respiratory diseases. Ageing Res Rev. 2021;66:101235. doi:10.1016/j.arr.2020.101235. [Google Scholar] [PubMed] [CrossRef]
72. de Oliveira GLV, Oliveira CNS, Pinzan CF, de Salis LVV, Cardoso CRB. Microbiota modulation of the gut-lung axis in COVID-19. Front Immunol. 2021;12:635471. doi:10.3389/fimmu.2021.635471. [Google Scholar] [PubMed] [CrossRef]
73. Guo Y, Luo R, Wang Y, Deng P, Song T, Zhang M, et al. SARS-CoV-2 induced intestinal responses with a biomimetic human gut-on-chip. Sci Bull. 2021;66(8):783–93. doi:10.1016/j.scib.2020.11.015. [Google Scholar] [PubMed] [CrossRef]
74. Francis I, Shrestha J, Paudel KR, Hansbro PM, Warkiani ME, Saha SC. Recent advances in lung-on-a-chip models. Drug Discov Today. 2022;27(9):2593–602. doi:10.1016/j.drudis.2022.06.004. [Google Scholar] [PubMed] [CrossRef]
75. Boyer O, Niaudet P. Hemolytic-uremic syndrome in children. Pediatr Clin North Am. 2022;69(6):1181–97. doi:10.1016/j.pcl.2022.07.006. [Google Scholar] [PubMed] [CrossRef]
76. Lee Y, Kim MH, Alves DR, Kim S, Lee LP, Sung JH, et al. Gut-kidney axis on chip for studying effects of antibiotics on risk of hemolytic uremic syndrome by shiga toxin-producing Escherichia coli. Toxins. 2021;13(11):775. doi:10.3390/toxins13110775. [Google Scholar] [PubMed] [CrossRef]
77. Kakoullis L, Papachristodoulou E, Chra P, Panos G. Shiga toxin-induced haemolytic uraemic syndrome and the role of antibiotics: a global overview. J Infect. 2019;79(2):75–94. doi:10.1016/j.jinf.2019.05.018. [Google Scholar] [PubMed] [CrossRef]
78. Bessy T, Itkin T, Passaro D. Bioengineering the bone marrow vascular niche. Front Cell Dev Biol. 2021;9:645496. doi:10.3389/fcell.2021.645496. [Google Scholar] [PubMed] [CrossRef]
79. Bertagnolli MM. Statement on catalyzing the development of novel alternative methods. Available from: https://www.nih.gov/about-nih/who-we-are/nih-director/statements/statement-catalyzing-development-novel-alternatives-methods. [Accessed 2024]. [Google Scholar]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools