 Open Access
Open Access
ARTICLE
PKHD1L1 blocks the malignant behavior of lung adenocarcinoma cells and restricts tumor growth by regulating CBX7
1 Department of Pulmonary and Critical Care Medicine, The Affiliated Changzhou Second People’s Hospital of Nanjing Medical University, Changzhou, 213004, China
2 Department of Pulmonary and Critical Care Medicine, Shanghai Tenth Clinical Medical College of Nanjing Medical University, Shanghai, 200072, China
* Corresponding Author: CHANGHUI WANG. Email:
BIOCELL 2024, 48(8), 1209-1221. https://doi.org/10.32604/biocell.2024.049626
Received 12 January 2024; Accepted 15 May 2024; Issue published 02 August 2024
Abstract
Objective: To explore the role of polycystic kidney and hepatic disease 1-like 1 (PKHD1L1) in lung adenocarcinoma (LUAD). Methods: Bioinformatics tools were utilized to examine the clinical profile of PKHD1L1 and chromobox protein homolog 7 (CBX7) in LUAD. The Cell Counting Kit-8, colony formation, terminal deoxynucleotidyl transferase dUTP nick end labeling, Transwell, and wound-healing assays were carried out to assess the proliferative, apoptotic, invasive, and migrative capacities of the cells. Furthermore, the interrelation between PKHD1L1 and CBX7 was validated using a co-immunoprecipitation assay. A LUAD mice model was constructed by subcutaneous injection of A549 cells. Finally, immunohistochemical staining was performed to evaluate CBX7 and Ki67 expression. Results: PKHD1L1 was downregulated in LUAD and predicted dismal outcomes in patients with LUAD. PKHD1L1 upregulation repressed the proliferative, invasive, and migrative capabilities of A549 cells and exacerbated the apoptotic rate. Additionally, PKHD1L1 may bind to CBX7 and positively modulate CBX7 expression. CBX7 deletion partly abrogated the effects of PKHD1L1 upregulation on the cellular biological activities in A549 cells. Furthermore, the PKHD1L1/CBX7 axis regulates the Hippo signaling pathway in A549 cells. PKHD1L1 restricted tumor growth in LUAD xenograft mice; this was partly abolished by CBX7 knockdown. Conclusion: PKHD1L1 can hinder LUAD progression by regulating CBX7-mediated Hippo signaling.Keywords
Lung cancer is a prevailing reason for cancer-related mortality globally [1]. Non-small cell lung carcinoma (NSCLC) is the major histological form of lung cancer, comprising 85% of cases. As the main histological variant of NSCLC, lung adenocarcinoma (LUAD) accounts for 50% of NSCLC cases [2]. Despite significant advances in surgery, radiotherapy, chemical therapy, targeted therapy, and immunotherapy, the clinical results for patients with LUAD remain unsatisfactory, and the average 5-year survival rate is still below 20% [3]. Therefore, exploring the molecular etiology underlying LUAD progression is urgently warranted to develop novel biomarkers and drug targets for treating patients with LUAD.
Polycystic kidney and hepatic disease 1-like 1 (PKHD1L1) is a macromolecular extracellular protein comprising 4249 amino acids; it comprises a transmembrane domain and an intracellular C-terminal structure. PKHD1L1 is a newly identified coat protein of hair cell stereocilia, which is essential for hearing in mice [4,5]. Meanwhile, PKHD1L1 encodes fibrocystin-L and a receptor with inducible T lymphocyte expression, playing a vital role in cellular immunity [6,7]. In addition, high glucose levels can regulate PKHD1L1, playing an important role in diabetic retinopathy [8]. Although the role of PKHD1L1 has long been discussed, its exact role in human cancer remains poorly understood. To date, a few relevant studies have revealed the importance of PKHD1L1 in esophageal squamous cell carcinoma, breast cancer, and thyroid cancer. The downregulation of PKHD1L1 can aggravate cell proliferation and invasion in thyroid cancer, highlighting the potential of PKHD1L1 in suppressing thyroid cancer [9–11]. Furthermore, in another study involving patients with LUAD, researchers conducted single nucleotide polymorphism gene enrichment and protein-protein interaction network analyses and identified a mutation in PKHD1L1. PKHD1L1 rs768349010 was discovered to be associated with LUAD prognosis. This suggests the involvement of PKHD1L1 in LUAD development [12]. Nevertheless, the precise role and potential regulation of PKHD1L1 in LUAD should be further explored.
Coincidentally, by searching LinkedOmics (https://kb.linkedomics.org/, accessed on 12/03/2024) and Genemania (http://genemania.org/, accessed on 12/03/2024) websites, we observed a potential protein-protein interaction between PKHD1L1 and chromobox protein homolog 7 (CBX7), with PKHD1L1 exhibiting a positive correlation with CBX7 in LUAD. CBX7, a member of the chromobox protein family, is implicated in generating polycomb repressive complex 1, a modulator in epigenetics and targeted genes via chromatin modification [13]. The aberrant expression of CBX7 results in gene expression imbalance, thereby affecting cancer occurrence and development. Previous reports have manifested that CBX7 exerts an antioncogenic function in urinary bladder and cervical cancers [14,15]; however, in gastric and prostate cancers, it serves as an oncogenic gene [16,17]. Thus, CBX7 plays a dual role in tumorigenesis. Importantly, CBX7 is downregulated in LUAD, and CBX7 overexpression abolishes the promoting effect of miR-19 on cancer development; this suggests that CBX7 elicits tumor-suppressive properties in LUAD [18,19]. Further evidence reveals that CBX7 is a repressor of the transcription coactivator Yes-Associated Protein (YAP), an inhibitory target of Hippo signaling and that CBX7 can exert antitumor activity in glioblastoma by activating Hippo signaling [20]. The highly conserved Hippo signaling was first identified in Drosophila melanogaster in 1995. This pathway is a major signaling pathway in mediating organ size, balancing tissue homeostasis, and repressing tumor growth. Accumulating evidence suggests the participation of the dysregulated Hippo pathway in lung cancer [21]. Considering the potential correlation between PKHD1L1 and CBX7, studies should be undertaken to determine whether PKHD1L1 interacts with CBX7 to participate in LUAD progression and whether it is associated with the Hippo pathway.
Therefore, in this study, we explored the role of PKHD1L1 and CBX7 and the potential regulatory mechanism in LUAD progression. Our study will shed novel insights into the advancement of LUAD treatment.
Starbase (https://rnasysu.com/encori/, accessed on 12/03/2024) [22] was utilized to observe the expression patterns of PKHD1L1 and CBX7 in cancer (n = 526) and normal (n = 59) samples from patients with LUAD and analyze the overall survival for PKHD1L1 and CBX7 in patients with LUAD. Based on gene functions, the Genemania database (http://genemania.org/) [23] was utilized to predict the meshwork of the interrelated proteins of PKHD1L1. The LinkedOmics database (https://kb.linkedomics.org/) [24] was used to determine the interrelation between CBX7 and PKHD1L1 in LUAD.
Human bronchoepithelioid cells 16HBE (FH1013) and the human LUAD cell line A549 (FH0045) were supplied by Shanghai Fuheng Biotechnology Co., Ltd. (Shanghai, China). 16HBE cells were cultured in keratinocyte medium (Sciencell, #2101, Carlsbad, CA, USA), whereas A549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, C11875500BT, Grand Island, NY, USA) in the presence of 10% fetal bovine serum (FBS; Beyotime Biotechnology, C0251, Shanghai, China) and 1% streptomycin-penicillin (KeyGEN BioTECH, KGY0023, Nanjing, China) under a 5% CO2 atmosphere at 37°C.
The pcDNA3.1 PKHD1L1 overexpression plasmid (Oe-PKHD1L1), negative control plasmid (Oe-NC), shRNAs specific to CBX7 (sh-CBX7-1, 5′-ATAGGAAGAGAGGTCCGAAAC-3′ and sh-CBX7-2, 5′-CGGAAGGGTAAAGTCGAGTAT-3′), and scrambled shRNA (sh-NC) were supplied by GenePharma (Shanghai, China). Lipofectamine 2000 reagent (Thermo Fisher Scientific, 11668019, Waltham, MA, USA) was utilized to transfect A549 cells at 60%–70% confluence with the abovementioned plasmids or shRNAs. After incubation at 37°C for 48 h, the transfected cells were used for the following research.
Quantitative real-time polymerase chain reaction (qRT-PCR)
TRIzol reagent (Cowin Bio, CW0580S, Taizhou, China) was utilized for the extraction of total RNA. The HiFiScript cDNA Synthesis Kit (Cowin Bio, CW2569M, Taizhou, China) was used for the synthesis of complementary DNA (cDNA). PCR was performed by adopting the UltraSYBR Mixture (Cowin Bio, CW2601S, Taizhou, China). The following primer sequences for Homo sapiens genes were utilized: PKHD1L1, forward, 5′-GTTAGTGTTGTGGTGGGAAGTA-3′, reverse, 5′-GCTTCCAGAAGTTGGTGATAGA-3′; CBX7, forward, 5′-GAGTATCTGGTGAAGTGGAAAGG-3′, reverse, 5′-TTCGGACCTCTCTTCCTATACC-3′; and GAPDH, forward, 5′-CGGGAAATCGTGCGTGAC-3′, reverse, 5′-CAGGAAGGAAGGCTGGAAG-3′. The ΔΔCt method was used, and the data were calibrated using GAPDH as the constitutive marker.
RIPA lysis reagent (Beyotime Biotechnology, P1003E, Shanghai, China) was utilized for the extraction of total proteins, whereas Nuclear and Cytoplasmic Protein Extraction Kit (Beyotime Biotechnology, P0027, Shanghai, China) was utilized for the extraction of nuclear proteins. Protein quantification was achieved by adopting the BCA method (Beyotime Biotechnology, P0012, Shanghai, China). The same amount of protein was separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The separated proteins underwent transfer onto polyvinylidene fluoride (PVDF) membranes (Millipore, IPVH00010, Burlington, MA, USA). The membrane was probed overnight with primary antibodies against PKHD1L1 (Abnova, H00093035-A01, Taiwan), CBX7 (26278-1-AP, Proteintech, Wuhan, China), Ki67 (Abcam, ab16667, Cambridge, UK), PCNA (Abcam, ab18197, Cambridge, UK), Bcl-2 (Abcam, ab32124, Cambridge, UK), Bax (Abcam, ab32503, Cambridge, UK), cleaved-caspase3 (Cell Signaling Technology, #9664, Boston, MA, USA), cleaved-caspase9 (Proteintech, 10380-1-AP, Wuhan, China), phosphorylated (p)-LATS1 (Cell Signaling Technology, #9157, Boston, MA, USA), LATS1 (Cell Signaling Technology, #9153, Boston, MA, USA), p-YAP (Cell Signaling Technology, #75784, Boston, MA, USA), YAP (Cell Signaling Technology, #4912, Boston, MA, USA), lamin A (Abcam, ab26300, Cambridge, UK), and GAPDH (Proteintech, 10494-1-AP, Wuhan, China) at 4°C. Thereafter, the membrane was incubated, at room temperature for 2 h, with a goat anti-rabbit (Abbkine, A21020, Wuhan, China) or goat anti-mouse (Abbkine, A21010, Wuhan, China) HRP-linked IgG secondary antibodies. Eventually, an enhanced chemiluminescence kit (180501, Tanon, Suzhou, China) was adopted for developing the blots.
Cell counting kit-8 (CCK-8) assay
A549 cells were inoculated into 96-well plates at a density of 5 × 103 cells/well. To every well, 10 μL of CCK-8 reagent (Vazyme Biotech Co., Ltd., A311-02, Nanjing, China) was added at varying time points. The cells were then cultured for another 2 h. Quantification of the colored solution was obtained utilizing a microplate reader (Bio-Rad, Hercules, CA, USA) at 450 nm.
A549 cells (1.5 × 103 cells/well) were inoculated into 6-well plates and incubated under 5% CO2 atmosphere for 10 days. Then, the cells were immobilized with 4% paraformaldehyde (Biosharp, BL539A, Hefei, China) for 20 min, followed by staining with 0.01% crystal violet (Beyotime Biotechnology, C0121, Shanghai, China) for 20 min. Finally, the colonies formed were counted.
5 × 106 A549 cells were inoculated into 6-well plates. Upon reaching 100% confluency, the scratched cell-free areas were established using an aseptic pipette tip. After removing the cell debris, the adherent cells were washed and cultured in a fresh medium without serum. The wounds were imaged utilizing a light microscope (CX41, Olympus, Tokyo, Japan) at 0, 24 h after wounding.
100 μL of cell suspension in serum-deprived medium was added to each apical chamber of a Matrigel (Beyotime Biotechnology, C0372, Shanghai, China)-precoated 24-well Transwell (Millipore, PI8P01250, Burlington, MA, USA). The basolateral chamber comprised 500 μL of complete medium in the presence of 10% FBS. Following 24 h of cultivation, the non-invaded cells were eliminated. The adhering cells underwent staining with crystal violet solution for 5 min. Images were acquired using a light microscope (CX41, Olympus, Tokyo, Japan).
Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining
The TUNEL Apoptosis Detection Kit (Yeasen, 40306ES60, Shanghai, China) was utilized to assess the cellular apoptotic rate. 1 × 105 A549 cells on chamber slides were immobilized with 4% paraformaldehyde and then received permeabilization with 0.2% Triton X-100. Next, cells were labeled with the TUNEL reaction buffer for 60 min at 37°C away from the light, followed by incubation with DAPI (Beyotime Biotechnology, C1006, Shanghai, China). The fluorescent signals were captured under a fluorescence microscope (IX53, Olympus, Tokyo, Japan).
HDOCK server (http://hdock.phys.hust.edu.cn/, accessed on 12/03/2024) [25], a web server for protein−DNA/RNA and protein−protein docking, was adopted to predict the interrelation between PKHD1L1 and CBX7, which was further validated by conducting co-immunoprecipitation (Co-IP) assay using an immunoprecipitation kit (Absin, abs955, Shanghai, China). In brief, cell protein lysates were subjected to overnight incubation with anti-PKHD1L1/anti-CBX7 or anti-IgG at 4°C. Untreated proteins were used as an input control. After supplementing protein A/G to the lysates, the immunoprecipitated protein complex was formed, which was then subjected to western blotting using anti-CBX7/anti-PKHD1L1 antibodies for IP detection.
Twenty BALB/c nude mice (female, 18−22 g) were supplied by Cavens Biogle Model Animal Research Co., Ltd. (Suzhou, China). All mice were kept in a specific sterile environment with ad libitum access to food and water. After adaption for 1 week, BALB/c mice (n = 5 each group) received subcutaneous injection, at the right flanks, with 1 × 107 A549 cells transfected with lentivirus harboring Oe-PKHD1L1 with or without sh-NC/sh-CBX7. Mouse weight and tumor volume were measured every 3 days. Finally, all mice were sacrificed using 1% sodium pentobarbital (50 mg/kg) via intraperitoneal injection. The tumors were collected after excision. All procedures involving animals were conducted according to the ARRIVE guidelines, with ethical acceptance by The Affiliated Changzhou Second People’s Hospital of Nanjing Medical University (Approval No. IACUC-20210915-01). A part of the tumor was fixed in 10% formalin, and the remaining part was stored in a −80°C refrigerator for western blotting.
Tumor tissues were immobilized with 10% formalin, dehydrated in alcohol, transparent with xylene II, and then subjected to embedment in paraffin. Then, slices with 4-μm thickness were obtained. After deparaffinization using xylene and rehydration in alcohol, the slices were processed for antigen retrieval and blockage of endogenous peroxidase activity in 3% H2O2. After incubation with 10% goat serum (Absin, abs933, Shanghai, China) for 30 min, the slices were successively probed overnight at 4°C with anti-ki67 (Abcam, ab16667, Cambridge, UK) and anti-CBX7 (Proteintech, 26278-1-AP, Wuhan, China) antibodies, and subsequent secondary antibody (Abbkine A21020, Wuhan, China) at room temperature for 2 h. The sections were stained with diaminobenzidine and counterstained with hematoxylin. The final observation was conducted under a light microscope (CX41, Olympus, Tokyo, Japan).
The analysis of experimental data was conducted depending on GraphPad Prism 8.0 (GraphPad Software, USA). Data were displayed as mean ± standard deviation of three independent measurements. Significant differences were evaluated with the application of Student’s t-test or one-way analysis of variance (ANOVA) along with Tukey’s post hoc test. A p-value of < 0.05 was considered as statistical significance.
First, PKHD1L1 expression in LUAD was investigated using the Starbase database. PKHD1L1 expression was downregulated in LUAD tumor samples (p < 0.001; Fig. 1A). Furthermore, downregulated PKHD1L1 expression was positively correlated with the dismal outcomes of patients with LUAD (p = 0.00021; Fig. 1B). These findings suggest the correlation between PKHD1L1 and LUAD development. In addition, PKHD1L1 expression was examined in 16HBE and A549 cells. Compared with 16HBE cells, PKHD1L1 expression at the mRNA expression (p = 0.0282; Fig. 1C) and protein level (p = 0.0014; Fig. 1D) were decreased in A549 cells. Collectively, PKHD1L1 is downregulated in LUAD.
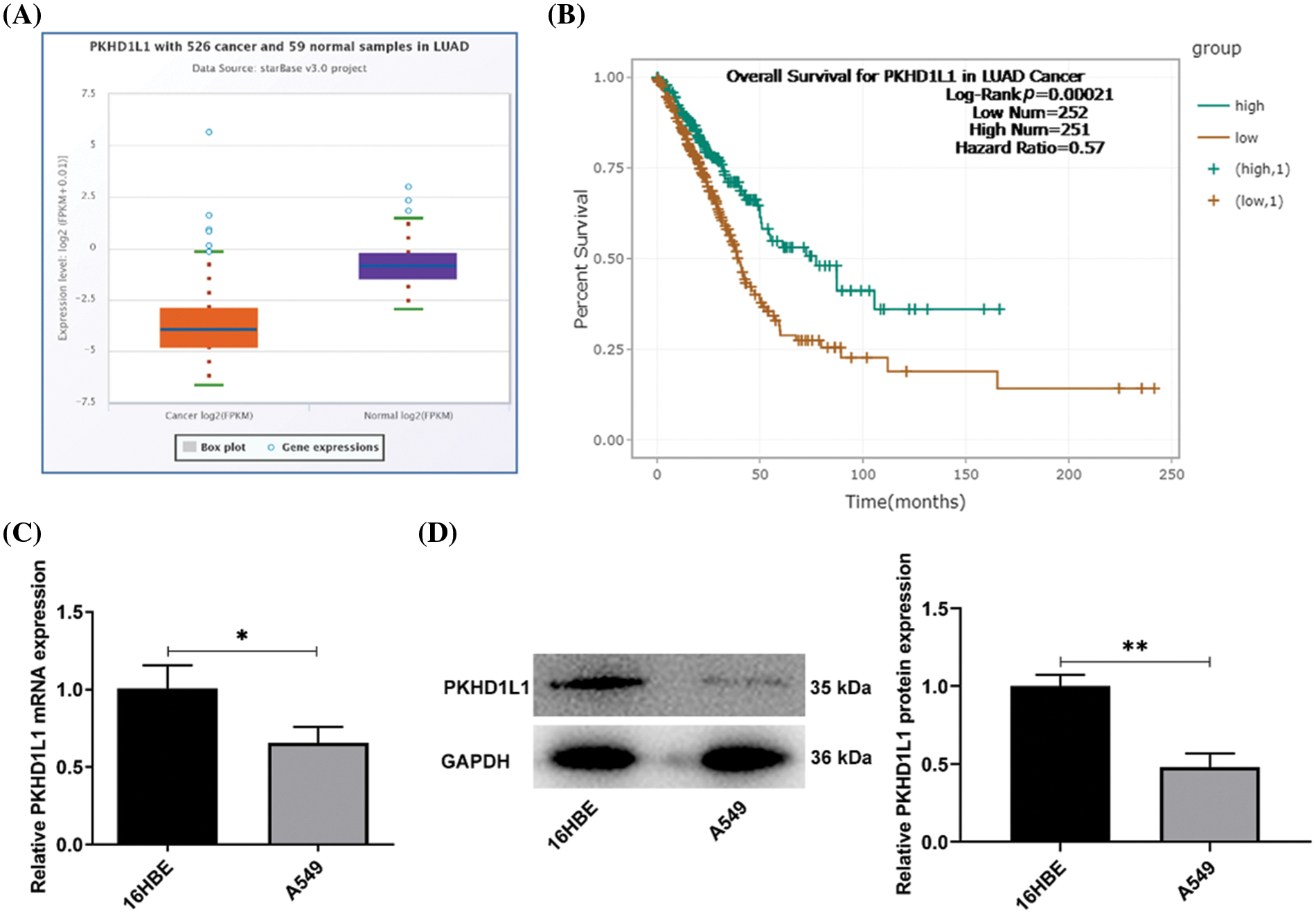
Figure 1: PKHD1L1 is depleted in LUAD. (A) The Starbase database was used to investigate PKHD1L1 expression in LUAD samples. (B) The Starbase database was utilized to predict the overall survival of patients with LUAD and low/high PKHD1L1 expression. (C) qRT-PCR was carried out to examine the mRNA expression of PKHD1L1 in 16HBE and A549 cells. (D) Western blotting was carried out to measure PKHD1L1 protein levels in 16HBE and A549 cells. Results are displayed as mean ± standard deviation. Student’s t-test was applied to assess significant differences. n = 3. *p < 0.05, **p < 0.01.
PKHD1L1 suppresses the proliferative, migrative, and invasive capacities of A549 cells
To delineate the specific effects of PKHD1L1 on LUAD, Oe-NC, and Oe-PKHD1L1 were transfected into A549 cells. PKHD1L1 expression was substantially increased in the Oe-PKHD1L1 group in comparison to the Oe-NC group (Fig. 2A,B). Subsequently, as observed from colony formation and CCK-8 assays, PKHD1L1 overexpression remarkably reduced cell activity at varying time points and hindered colony formation (Fig. 2C,D). This suggests that PKHD1L1 overexpression can lower the proliferation capacity of A549 cells, which was further verified by the decrease of Ki67 and proliferating cell nuclear antigen (PCNA) protein levels in the Oe-PKHD1L1 group (Fig. 2E). Thereafter, PKHD1L1 upregulation decreased the number of migrated and invaded cells in the Oe-PKHD1L1 group in comparison to the Oe-NC group (Fig. 2F,G), suggesting that PKHD1L1 overexpression can hinder the migrative and invasive capacities of A549 cells. Taken together, PKHD1L1 overexpression abates the aggressive phenotypes of A549 cells.
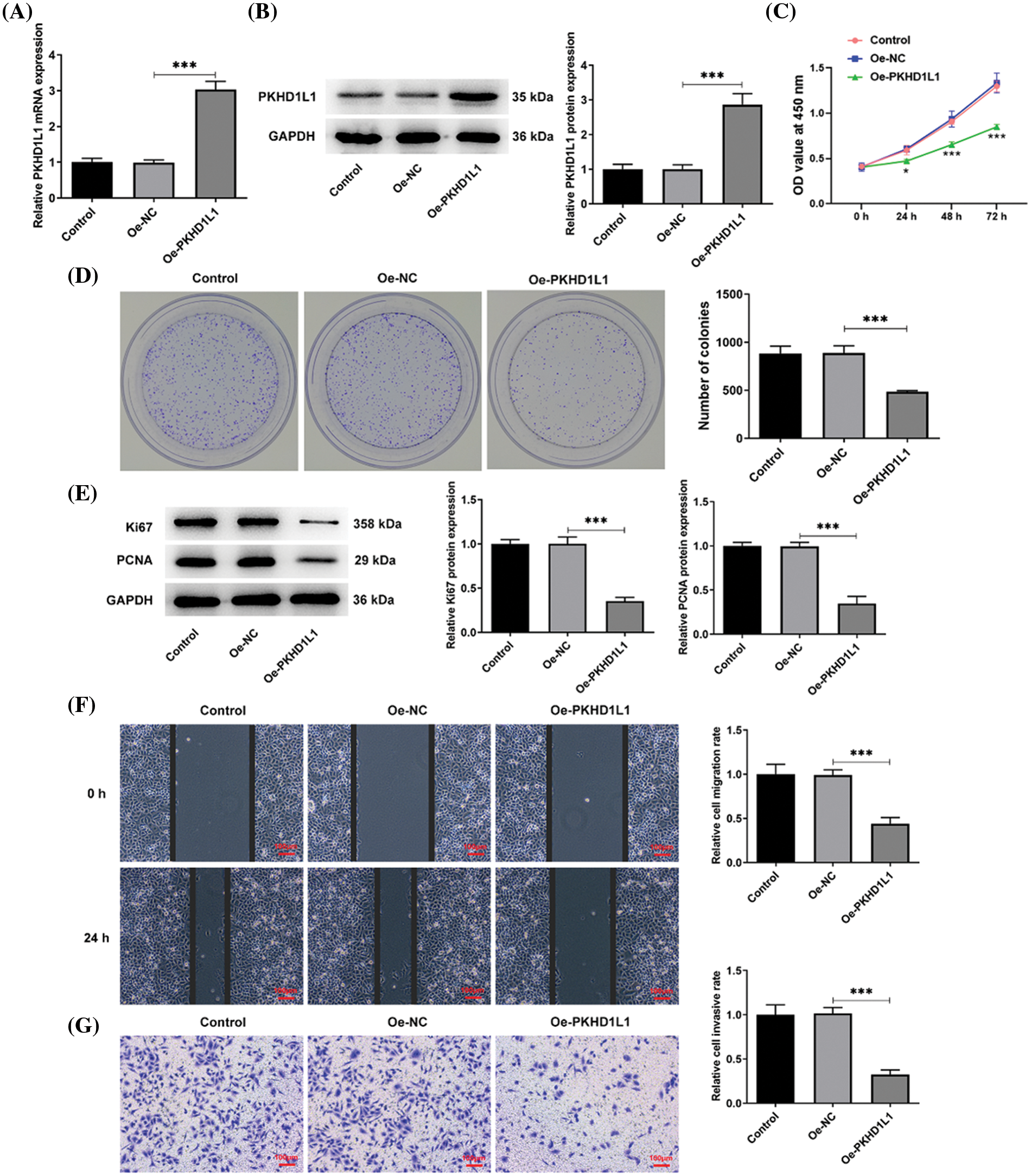
Figure 2: PKHD1L1 suppresses the proliferative, migrative, and invasive capacities of A549 cells. (A) After transfection with Oe-NC and Oe-PKHD1L1, qRT-PCR was performed to examine the mRNA expression of PKHD1L1. (B) Western blotting was carried out to measure PKHD1L1 protein levels. ***p < 0.001. (C) CCK-8 assay to examine cell activity at varying time points. *p < 0.05, ***p < 0.001 vs. Oe-NC. (D) Cell proliferation was evaluated using a colony formation assay. (E) Western blotting to measure Ki67 and PCNA protein levels. (F) Wound-healing assay to assess the migrative capacity. (G) Transwell assay to assess the invasive capacity. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. ***p < 0.001.
PKHD1L1 facilitates the apoptosis rate of LUAD cells
Next, we evaluated the effects of PKHD1L1 on the apoptotic conditions in LUAD. Fig. 3A illustrates higher TUNEL-positive cells in the Oe-PKHD1L1 group than in the Oe-NC group. This indicates that PKHD1L1 overexpression promotes the apoptotic rate of A549 cells. The protein levels of Bcl-2, an antiapoptotic protein, were considerably reduced; however, the protein levels of cleaved caspase-3, cleaved caspase-9, and Bax were greatly elevated following PKHD1L1 overexpression (Fig. 3B). This further demonstrates the proapoptotic property of PKHD1L1 in A549 cells.
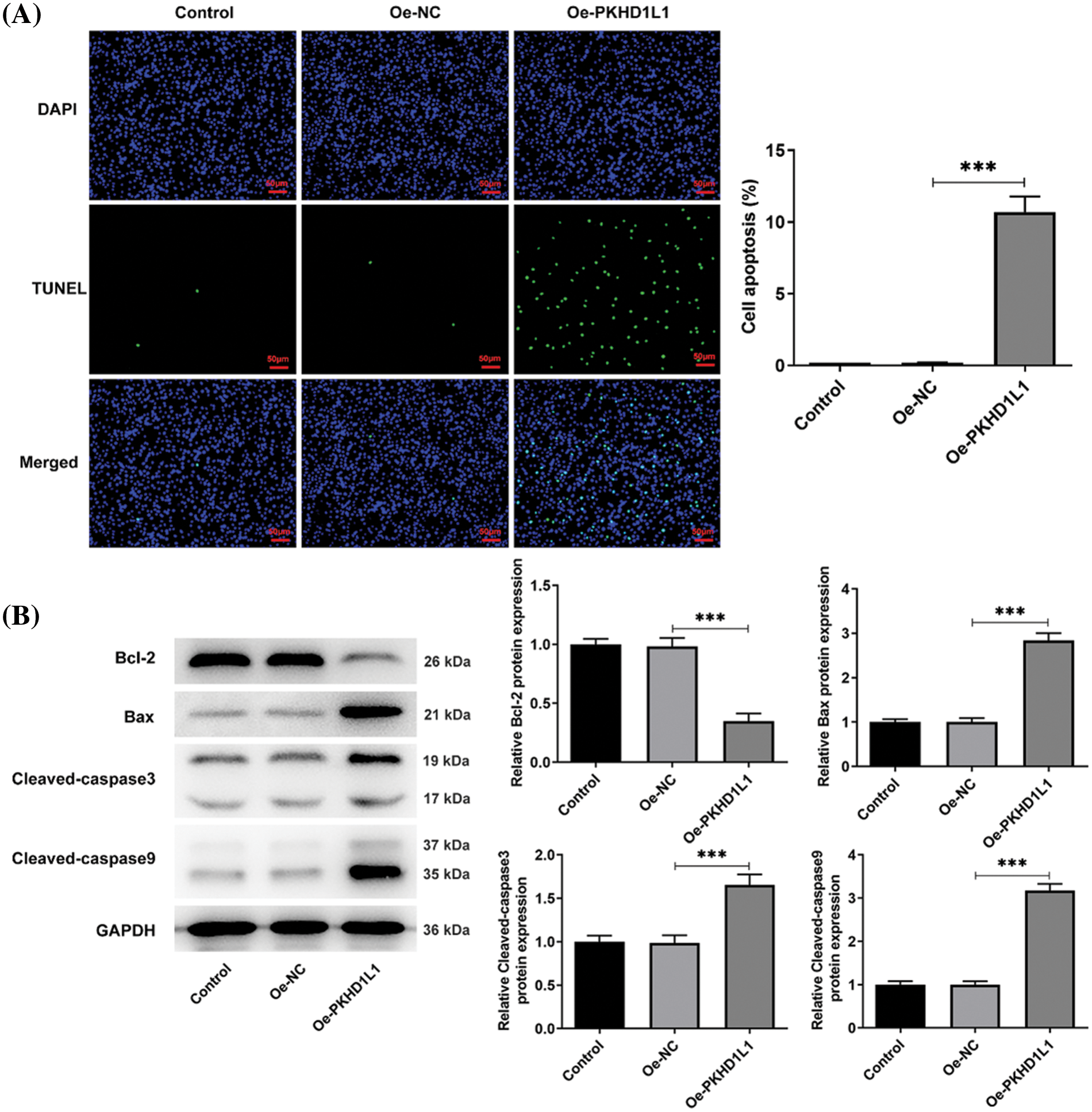
Figure 3: PKHD1L1 promotes the apoptosis rate of LUAD cells. (A) After transfection, a TUNEL assay was performed to determine the cellular apoptosis rate. (B) Western blotting was performed to measure apoptosis-related proteins. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. ***p < 0.001.
CBX7 is depleted in LUAD and is positively correlated with PKHD1L1
Next, based on Starbase analysis, we observed the downregulation of CBX7 in LUAD tumor samples (p < 0.001; Fig. 4A), with the declined CBX7 expression correlating with the dismal prognosis of patients with LUAD (p = 0.0028; Fig. 4B). Genemania database analysis revealed a potential protein-protein interaction between CBX7 and PKHD1L1 (Fig. 4C). Then, bioinformatics analysis using the LinkedOmics database displayed a positive interrelation between CBX7 and PKHD1L1 expression in LUAD (r = 0.4267, p < 0.001; Fig. 4D). To verify the abovementioned prediction and analysis, CBX7 expression was measured in A549 and 16HBE cells. In comparison to 16HBE cells, the mRNA expression and protein level of CBX7 were markedly lowered in A549 cells (p = 0.0031 in Fig. 4E; p = 0.0009 in Fig. 4F). Using the HDOCK server, we observed the binding sites between CBX7 and PKHD1L1, with an HDOCK score of −279.22 (Fig. 4G). This confirms that PKHD1L1 can directly bind to CBX7. Moreover, the Co-IP assay revealed that CBX7 and PKHD1L1 can interact with each other in A549 cells (Fig. 4H). Further, to expound the detailed interrelation between CBX7 and PKHD1L1 in LUAD, CBX7 expression was measured in A549 cells following Oe-PKHD1L1 transfection. As demonstrated in Fig. 4I,J, PKHD1L1 overexpression significantly elevated CBX7 expression in A549 cells. This confirms that PKHD1L1 can interact with CBX7 and positively regulate CBX7 expression.
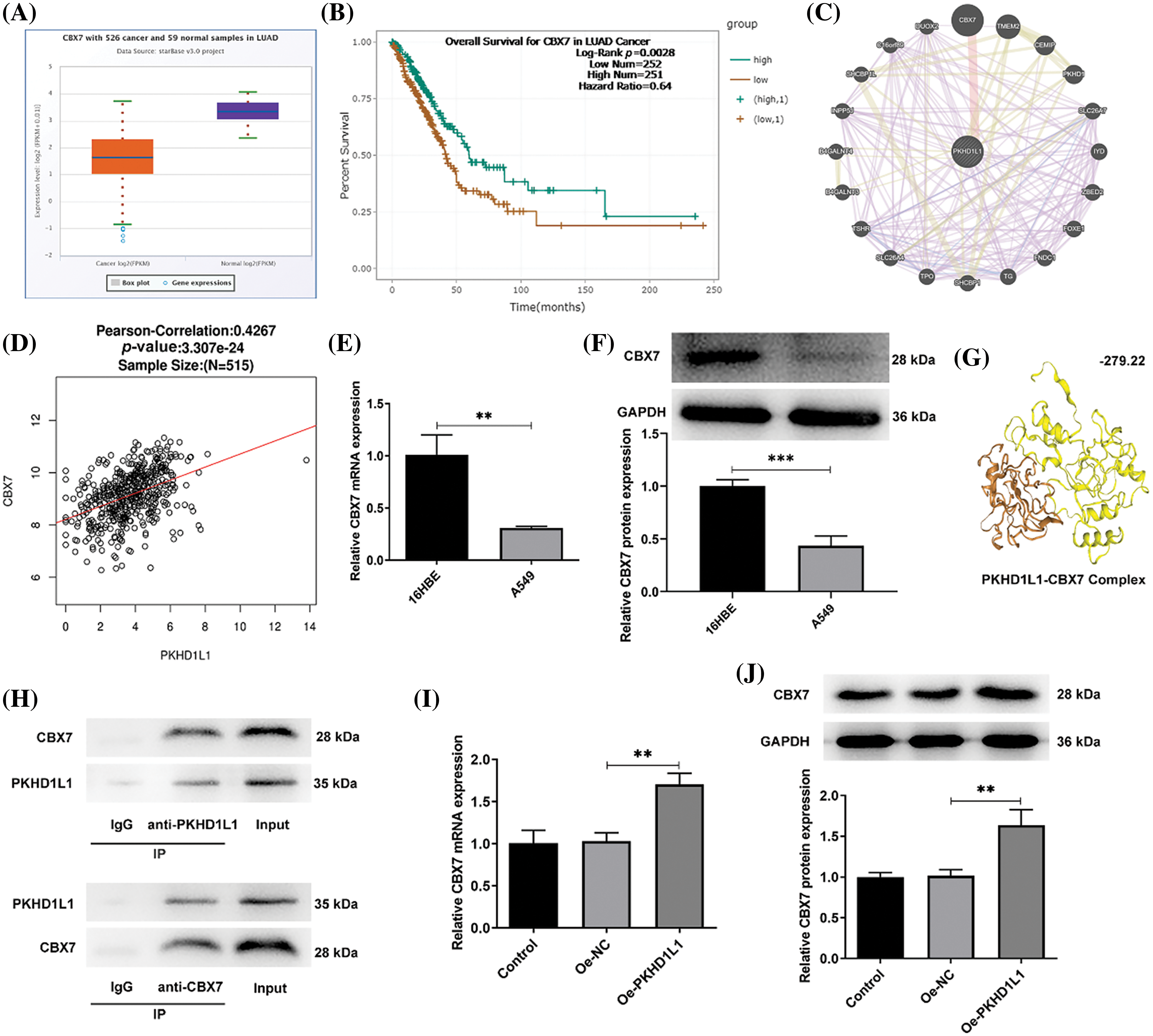
Figure 4: CBX7 is depleted in LUAD and is positively correlated with PKHD1L1. (A) The Starbase database was used to predict CBX7 expression in LUAD samples. (B) The Starbase database was utilized to predict the overall survival of patients with LUAD and low/high CBX7 expression. (C) The Genemania database (http://genemania.org/) was used to predict the possible interrelation between CBX7 and PKHD1L1. (D) The correlation between CBX7 and PKHD1L1 expression in LUAD was analyzed using the LinkedOmics (https://kb.linkedomics.org/) database. (E) qRT-PCR to examine CBX7 expression in A549 and 16HBE cells. (F) Western blotting to measure CBX7 levels in 16HBE and A549 cells. Results are displayed as mean ± standard deviation. Student’s t-test was applied to assess significant differences. (G) Graphical representation of the molecular binding between CBX7 and PKHD1L1 using the HDOCK Server. (H) Co-IP assay for validation of the interrelation between CBX7 and PKHD1L1. (I) After transfection, qRT-PCR was performed to measure CBX7 mRNA expression. (J) Western blotting to measure CBX7 protein levels. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. **p < 0.01, ***p < 0.001.
CBX7 knockdown weakens the repressive effects of PKHD1L1 on the aggressive behaviors of LUAD cells
To investigate the mechanism by which PKHD1L1 plays an anticancer role by regulating CBX7, functional assays were performed. At first, the mRNA expression and protein level of CBX7 declined in the sh-CBX7-1 and sh-CBX7-2 groups in comparison to the sh-NC group (Fig. 5A,B). Owing to a better transfection efficacy, sh-CBX7-2 was adopted in the follow-up assays. Next, only Oe-PKHD1L1 or Oe-PKHD1L1 and sh-NC/sh-CBX7-2 were transfected into A549 cells. We observed that PKHD1L1 overexpression significantly increased CBX7 expression; however, simultaneous CBX7 knockdown partly abolished this phenomenon (Fig. 5C,D). Subsequently, the colony formation and CCK-8 assays revealed that CBX7 deficiency attenuated the inhibitory effects of PKHD1L1 overexpression on the colony-forming ability and cell viability of A549 cells (Fig. 5E,F). Meanwhile, compared with the Oe-PKHD1L + sh-NC group, Ki67 and PCNA expression were significantly elevated in the Oe-PKHD1L1 + sh-CBX7 group (Fig. 5G).
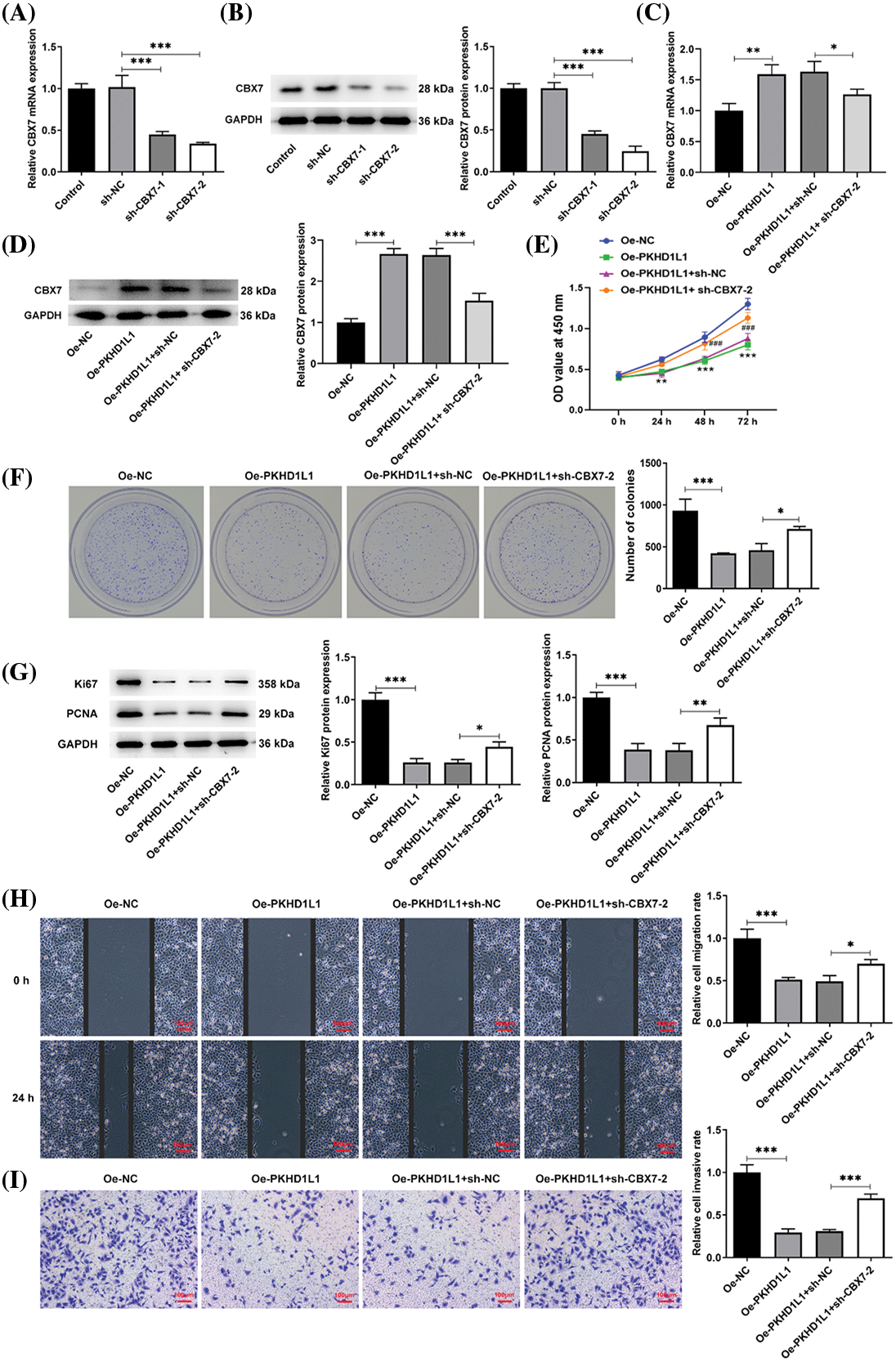
Figure 5: CBX7 knockdown weakens the repressive effects of PKHD1L1 on the aggressive behavior of LUAD cells. (A) After transfection with sh-NC or sh-CBX7-1/2, qRT-PCR was carried out to measure CBX7 mRNA expression. (B) Western blotting was performed to measure CBX7 protein levels. (C) Only Oe-PKHD1L1 or Oe-PKHD1L1 and sh-NC/sh-CBX7-2 were transfected into A549 cells, and qRT-PCR was carried out to measure CBX7 mRNA expression. (D) Western blotting to measure CBX7 protein levels. *p < 0.05, **p < 0.01, and ***p < 0.001. (E) The CCK-8 assay measures cell activity at varying time points. **p < 0.01, ***p < 0.001 vs. Oe-NC; ###p < 0.001 vs. Oe-PKHD1L1 + sh-NC. (F) Colony formation assay to estimate cell proliferation. (G) Western blotting to measure Ki67 and PCNA protein levels. (H) Wound-healing assay to assess the migrative capacity. (I) Transwell assay to assess the invasive capacity. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. *p < 0.05, **p < 0.01, and ***p < 0.001.
Moreover, the wound-healing and Transwell assays illustrated that PKHD1L1 overexpression suppressed the migrative and invasive capacities of A549 cells; this was partly abolished by CBX7 knockdown (Fig. 5H,I). Furthermore, the TUNEL assay revealed that the elevated TUNEL-positive cells caused by PKHD1L1 overexpression were partially restored by CBX7 knockdown (Fig. 6A). This suggests that the proapoptotic role of PKHD1L1 in A549 cells is hindered by CBX7 knockdown. This finding was further verified by elevated Bcl-2 levels and reduced cleaved caspase-3, cleaved caspase-9, and Bax levels in the Oe-PKHD1L1 + sh-CBX7 group in comparison to the Oe-PKHD1L1 + sh-NC group (Fig. 6B). Taken together, the abovementioned findings suggest that CBX7 knockdown partially hinders the repressive effects of PKHD1L1 overexpression on cell proliferative, migrative, and invasive capacities and the promoting effect of PKHD1L1 overexpression on cell apoptosis.
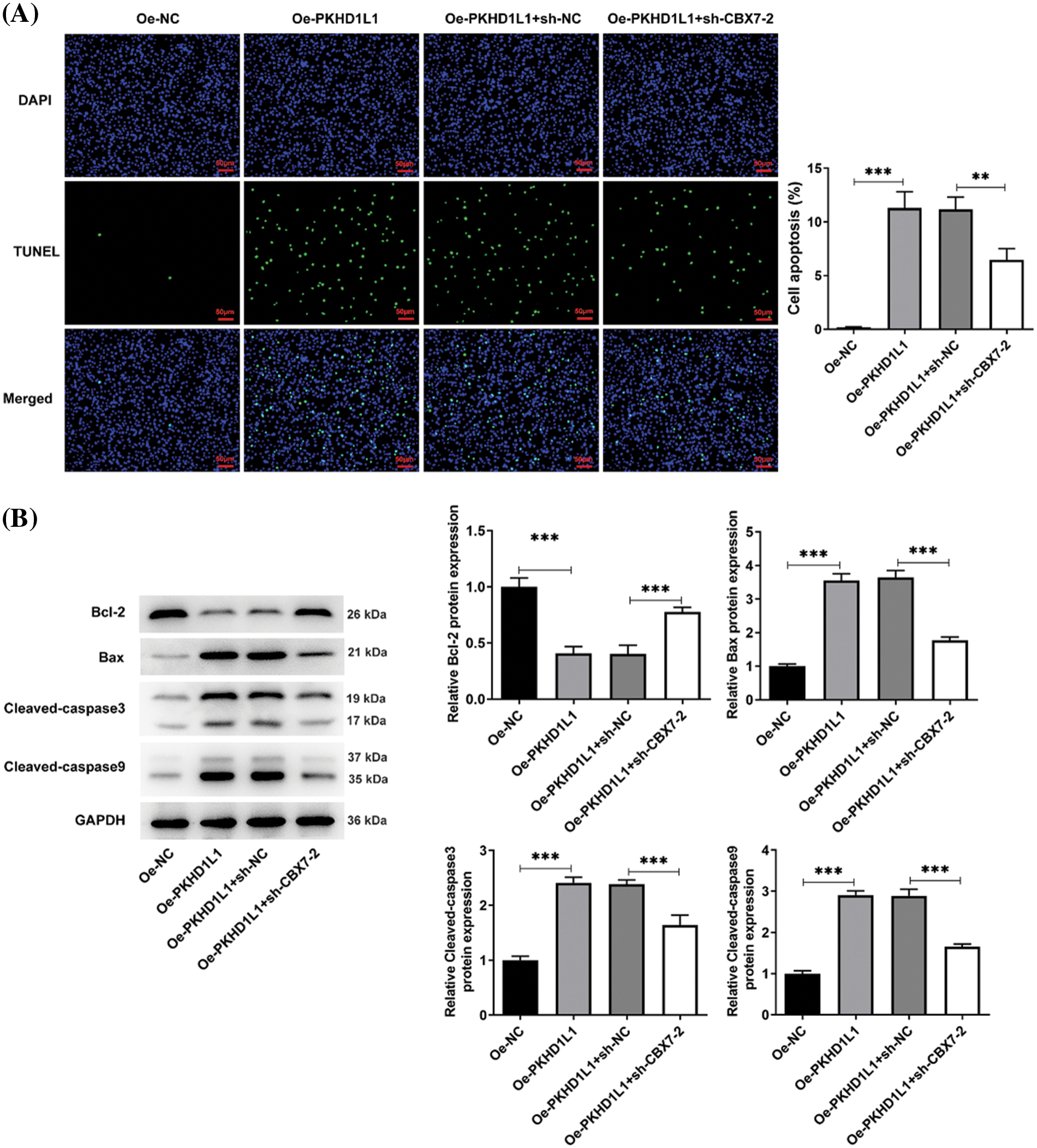
Figure 6: CBX7 knockdown weakens the stimulatory effects of PKHD1L1 on the apoptotic rate of LUAD cells. (A) Only Oe-PKHD1L1 or Oe-PKHD1L1 and sh-NC/sh-CBX7-2 were transfected into A549 cells. TUNEL assay was performed to determine the cellular apoptosis rate. (B) Western blotting to measure apoptosis-related proteins. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. **p < 0.01, ***p < 0.001.
Hippo signaling is regulated by the PKHD1L1/CBX7 axis in A549 cells
Further research was conducted to elucidate how the PKHD1L1/CBX7 axis regulates LUAD development. Considering that dysregulated Hippo signaling contributes to lung carcinogenesis and CBX7 can exert antitumor effects by activating Hippo signaling [20,21], we speculated whether PKHD1L1/CBX7 regulation in A549 cells was associated with the Hippo pathway. Therefore, we investigated the changes in the Hippo signaling pathway during the regulation of the PKHD1L1/CBX7 axis in A549 cells. As demonstrated in Fig. 7, PKHD1L1 overexpression distinctly promoted p-YAP and p-LATS1 expression but declined nuclear YAP expression. This indicates that PKHD1L1 can inhibit the YAP translocation into the nucleus and thereby activate the Hippo signaling pathway. In contrast, CBX7 interference partly abolished these changes, hinting that the Hippo signaling pathway is mediated by the PKHD1L1/CBX7 axis in A549 cells.
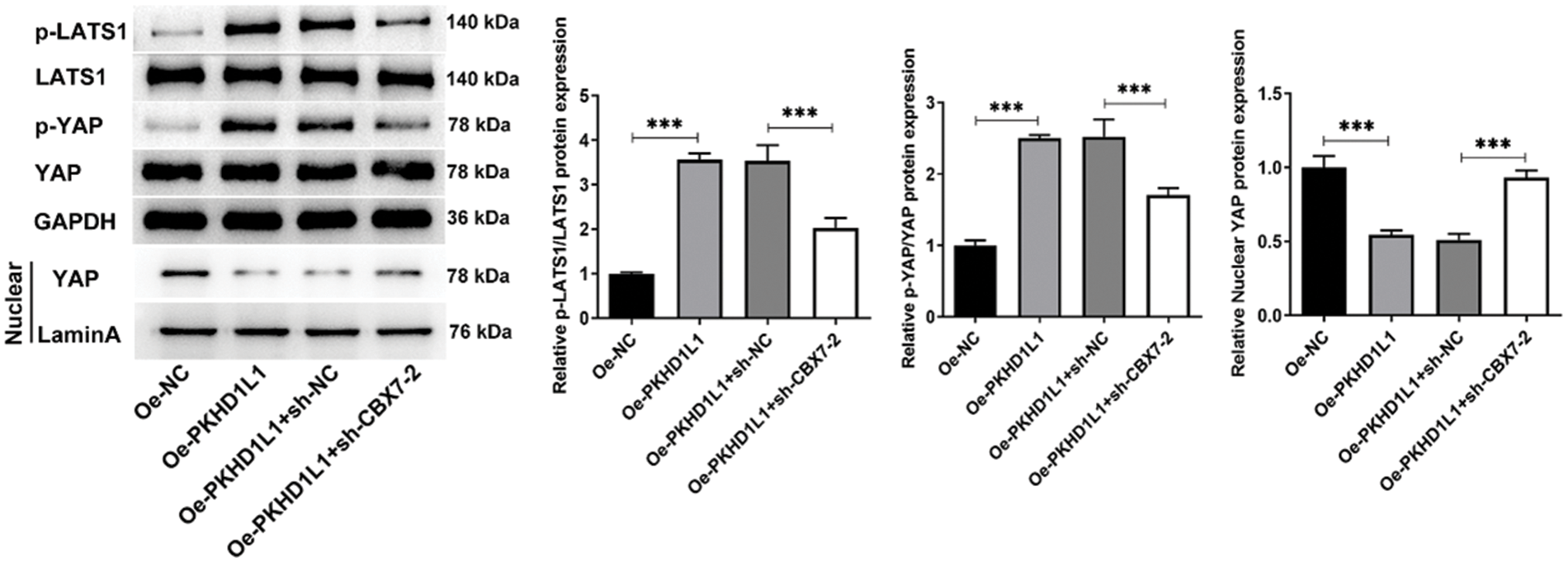
Figure 7: Hippo signaling is regulated by the PKHD1L1/CBX7 axis in A549 cells. After transfection with only Oe-PKHD1L1 or Oe-PKHD1L1 and sh-NC/sh-CBX7-2, western blotting was carried out to measure Hippo signaling-related proteins. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 3. ***p < 0.001.
CBX7 knockdown restricts the suppressive influence of PKHD1L1 on tumor growth in mice
Finally, we verified our findings concerning the regulatory role of PKHD1L1/CBX7 in LUAD in vivo. As demonstrated in Fig. 8A,B, PKHD1L1 overexpression significantly lowered tumor volume and hindered tumor growth; this was partially abolished by CBX7 knockdown. Furthermore, the weight of the harvested tumor was significantly reduced following PKHD1L1 overexpression; this was also partly reversed by additional CBX7 knockdown (Fig. 8C). Meanwhile, IHC staining revealed that PKHD1L1 overexpression significantly promoted CBX7 expression in tumor samples; this was partly hindered by CBX7 knockdown (Fig. 8D). Moreover, IHC staining images revealed that PKHD1L1 overexpression remarkably reduced Ki67 expression in tumor tissues in comparison to the Oe-NC group; however, this reduction was partly hindered upon simultaneous PKHD1L1 overexpression and CBX7 knockdown (Fig. 8E). Moreover, western blotting revealed that CBX7 knockdown weakened the influence of PKHD1L1 overexpression on downregulating Bcl-2 expression and upregulating Bax expression, confirming that the proapoptotic activity of PKHD1L1 in tumor tissues is restricted by CBX7 knockdown (Fig. 8F). Taken together, the inhibitory role of PKHD1L1 against tumor growth in LUAD-bearing mice may be partially restricted by CBX7 knockdown.
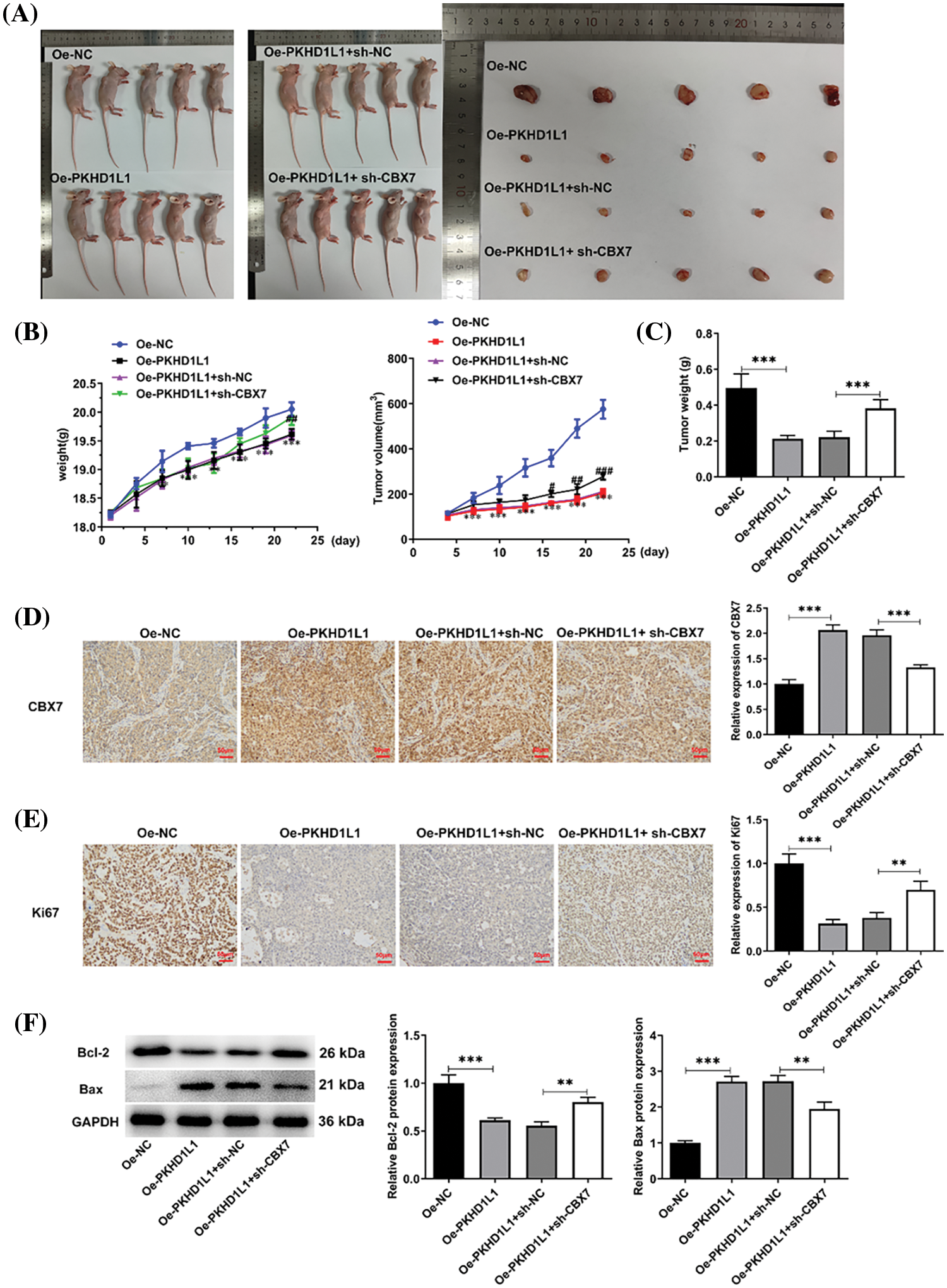
Figure 8: CBX7 knockdown restricts the suppressive effect of PKHD1L1 on tumor growth. (A) A549 cells were subcutaneously injected into BALB/c mice to construct an in vivo LUAD model. After sacrificing the animals, the tumors were excised and photographed. (B) Mouse weight and tumor volume were measured every 3 days. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 5 mice per group. ***p < 0.001 vs. Oe-NC; #p < 0.05, ##p < 0.01, and ###p < 0.001 vs. Oe-PKHD1L1 + sh-NC. (C) Tumor weight measurements. (D) IHC staining to measure CBX7 expression in tumor samples. (E) IHC staining to measure Ki67 expression in tumor samples. (F) Western blotting to measure Bcl-2 and Bax levels in tumor samples. Results are displayed as mean ± standard deviation. ANOVA along with Tukey’s post hoc test was applied to assess significant differences. n = 5 mice per group. **p < 0.01, ***p < 0.001.
Lung cancer remains the prevailing reason for cancer-related mortalities, with NSCLC accounting for 85% of mortalities. LUAD is a common histological form of NSCLC, accounting for approximately 40% of all lung cancers, with a proportion as high as 11.6% among all cancers [26,27]. To date, although tremendous efforts have been undertaken to identify tumor suppressors or oncogenes that may elicit critical activities in the course of LUAD, the precise molecular mechanism underlying multiple clinical characteristics remains incompletely elucidated. In this study, we disclosed the downregulation of PKHD1L1 and CBX7 in LUAD and elucidated that PKHD1L1 may suppress the proliferative, migrative, and invasive capabilities of LUAD cells while facilitating the cellular apoptotic cascade partly by upregulating CBX7, accompanied with the involvement of Hippo signaling.
Uncontrolled proliferation and invasion and abnormal apoptosis are important features and contributing factors to metastatic malignant tumors, with metastasis being the prevailing reason for cancer-related mortalities [28]. Dachshund homolog 1 (DACH1) functions as a tumor suppressor because its silencing enhances the proliferative and invasive capabilities of A549 cells while weakening their spontaneous apoptosis [29]; β, β-Dimethyl-acryl-alkannin (ALCAP2) can inhibit invasiveness and metastasis of LUAD cells, thereby serving as a tumor suppressor in LUAD [30]. Therefore, the modulators of cell proliferation, invasion, and apoptosis play important roles in tumorigenesis and cancer progression. In the current study, we observed that PKHD1L1 markedly weakened the proliferative, migrative, and invasive abilities and aggravated cell apoptosis rate in A549 cells, proving that PKHD1L1 can play an antioncogenic role in LUAD, consistent with its tumor-suppressing role in thyroid cancer [11].
Growing evidence suggests the importance of protein-protein interactions in numerous biological processes in living cells because proteins prefer to function as team players in a dynamic network [31]. For instance, employing the Co-IP assay, Lan et al. demonstrated the interrelation between P68/DEAD box protein 5 (DDX5) and KRAB domain-associated protein 1 (KAP-1), and disclosed that KAP-1 silencing repressed cell proliferative, migrative, and invasive capabilities by degrading DDX5, thereby hindering thyroid cancer development [32]. Furthermore, Tian et al. proved an interaction between Sirtuin 6 and phosphatase and tensin homolog (PTEN), and Sirtuin 6 inhibited colon cancer progression by modulating PTEN/protein kinase B (Akt) signaling [33]. Consistently, we also confirmed a remarkable interaction between PKHD1L1 and CBX7 by employing the Co-IP assay. Meanwhile, we proved that CBX7 expression positively regulates PKHD1L1. Additional functional assays revealed that the tumor-suppressive activity of PKHD1L1 was partially weakened owing to CBX7 knockdown. This suggests that PKHD1L1 represses LUAD development by partially upregulating CBX7.
The Hippo signaling pathway in mammals comprises a kinase cascade: upstream regulatory elements (MST1/2, LATS1/2) and a downstream effector (YAP). When Hippo signaling is activated, MST kinases phosphorylate and activate LATS1/2, followed by the subsequent YAP phosphorylation. Phosphorylated YAP remains in the cytoplasm for ubiquitination and degradation. Once Hippo signaling is inhibited, which leaves YAP unphosphorylated, YAP can enter the nucleus to induce transcription factor activity, thereby activating multiple downstream genes that are associated with cell proliferation, survival, mobility, stemness, and differentiation, ultimately mediating tumor development [34,35]. In recent years, accumulating evidence confirms that the Hippo signaling pathway is vital for lung cancer development. For instance, WW domain binding protein-2 (WBP2) represses Hippo signaling by reducing LATS1 phosphorylation and facilitating YAP nuclear translocation, eventually contributing to aggressive lung cancer progression [36]. Additionally, FERM-domain-containing protein-1 (FRMPD1) has been discovered to activate the Hippo pathway, accompanied by elevated p-LATS1 levels, which can inhibit the invasive and proliferative capacities of lung cancer cells [37]. Thus, controlling the Hippo pathway may be an alternative method to discover targeted strategies for lung cancer treatment. Consistently, in this study, we observed that p-LATS1 and p-YAP were upregulated but nuclear YAP was significantly downregulated following PKHD1L1 overexpression, revealing that PKHD1L1 may hinder the nuclear translocation of YAP by promoting LATS1 phosphorylation, ultimately activating the Hippo signaling pathway. Therefore, PKHD1L1 may suppress the malignant behaviors of LUAD cells by activating the Hippo pathway. Furthermore, existing evidence suggests that CBX7 is a repressor of the transcription coactivator YAP/TAZ. Exogenous CBX7 expression inhibited YAP/TAZ-dependent transcription, thereby inhibiting glioma cell migration. This indicates that CBX7 exerts a regulatory function in cancer by regulating the Hippo signaling pathway [20]. In this study, the nuclear translocation of YAP was inhibited by PKHD1L1 overexpression but was partly restored by CBX7 silencing. This suggests that PKHD1L1 modulates Hippo signaling partly depending on its interaction with CBX7 in LUAD.
The present study has some limitations that should be considered. First, the expression patterns and vital role of PKHD1L1 and CBX7 in LUAD should be validated in clinical trials. Second, in addition to CBX7, diverse CBX family members play important roles in LUAD [38]. Therefore, whether PKHD1L1 can also regulate other CBX family members remains unclear, deserving investigation in our future work. Third, the exact mechanism of the involvement of Hippo signaling via the PKHD1L1/CBX7 axis during LUAD progression should be further studied. Finally, to improve the clinical relevance of this study, a CBX7 inhibitor (such as UNC3866 [39]) should be examined in an in vivo assay in our future work.
To summarize, this is the first study in which the antitumor role of PKHD1L1 against LUAD has been confirmed and its molecular mechanism has been elucidated. In detail, PKHD1L1 can suppress the malignant behavior of A549 cells and restrict tumor growth in vivo. This may be partly achieved by regulating the CBX7-mediated Hippo signaling pathway. Our study findings offer a new perspective for understanding and treating LUAD.
Acknowledgement: None.
Funding Statement: This study was supported by Major Clinical Research Projects of Shanghai Tenth Clinical Medical College of Nanjing Medical University (SHDC2020CR3068B).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Changhui Wang; data collection: Kewei Cheng, Lei Shi, Caiwen Shi, Shuanshuan Xie; analysis and interpretation of results: Kewei Cheng, Lei Shi, Caiwen Shi; draft manuscript preparation: Kewei Cheng, Changhui Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All datasets/data generated or analyzed during this study are available from the corresponding author upon reasonable request.
Ethics Approval: All animal experiments were conducted according to the ARRIVE guidelines and were approved by the Ethics Committee of the Affiliated Changzhou Second People’s Hospital of Nanjing Medical University (Approval No. IACUC-20210915-01).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi:10.3322/caac.21763. [Google Scholar] [PubMed] [CrossRef]
2. Zhang Z, Ma Y, Guo X, Du Y, Zhu Q, Wang X, et al. FDX1 can impact the prognosis and mediate the metabolism of lung adenocarcinoma. Front Pharmacol. 2021;12:749134. doi:10.3389/fphar.2021.749134. [Google Scholar] [PubMed] [CrossRef]
3. Lin SH, Lu JW, Hsieh WT, Chou YE, Su TC, Tsai TJ, et al. Evaluation of the clinical significance of long non-coding RNA MALAT1 genetic variants in human lung adenocarcinoma. Aging. 2024;16(6):5740–50. doi:10.18632/aging.205675. [Google Scholar] [PubMed] [CrossRef]
4. Wu X, Ivanchenko MV, Al Jandal H, Cicconet M, Indzhykulian AA, Corey DP. PKHD1L1 is a coat protein of hair-cell stereocilia and is required for normal hearing. Nat Commun. 2019;10(1):3801. doi:10.1038/s41467-019-11712-w. [Google Scholar] [PubMed] [CrossRef]
5. Ivanchenko MV, Cicconet M, Jandal HA, Wu X, Corey DP, Indzhykulian AA. Serial scanning electron microscopy of anti-PKHD1L1 immuno-gold labeled mouse hair cell stereocilia bundles. Sci Data. 2020;7(1):182. doi:10.1038/s41597-020-0509-4. [Google Scholar] [PubMed] [CrossRef]
6. Hogan MC, Griffin MD, Rossetti S, Torres VE, Ward CJ, Harris PC. PKHDL1, a homolog of the autosomal recessive polycystic kidney disease gene, encodes a receptor with inducible T lymphocyte expression. Hum Mol Genet. 2003;12(6):685–98. doi:10.1093/hmg/ddg068. [Google Scholar] [CrossRef]
7. Lian PW, Fu YL, Li A, Dai BZ, Ding ZW, Li L, et al. Preparation and characterization of a polyclonal antibody against human Fibrocystin-L. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2011;27(1):78–81. [Google Scholar] [PubMed]
8. Ung C, Sanchez AV, Shen L, Davoudi S, Ahmadi T, Navarro-Gomez D, et al. Whole exome sequencing identification of novel candidate genes in patients with proliferative diabetic retinopathy. Vision Res. 2017;139(12):168–76. doi:10.1016/j.visres.2017.03.007. [Google Scholar] [PubMed] [CrossRef]
9. Yuan P, Rao W, Lin Z, Liu S, Lin X, Wu C, et al. Genomic analyses reveal SCN7A is associated with the prognosis of esophageal squamous cell carcinoma. Esophagus. 2022;19(2):303–15. doi:10.1007/s10388-021-00898-y. [Google Scholar] [PubMed] [CrossRef]
10. Saravia CH, Flores C, Schwarz LJ, Bravo L, Zavaleta J, Araujo J, et al. Patterns of mutation enrichment in metastatic triple-negative breast cancer. Clin Med Insights Oncol. 2019;13:1179554919868482. doi:10.1177/1179554919868482. [Google Scholar] [PubMed] [CrossRef]
11. Zheng C, Quan R, Xia EJ, Bhandari A, Zhang X. Original tumour suppressor gene polycystic kidney and hepatic disease 1-like 1 is associated with thyroid cancer cell progression. Oncol Lett. 2019;18(3):3227–35. doi:10.3892/ol.2019.10632. [Google Scholar] [PubMed] [CrossRef]
12. Wang Y, Zhang L, Chen Y, Li M, Ha M, Li S. Screening and identification of biomarkers associated with the diagnosis and prognosis of lung adenocarcinoma. J Clin Lab Anal. 2020;34(10):e23450. doi:10.1002/jcla.23450. [Google Scholar] [PubMed] [CrossRef]
13. Ma RG, Zhang Y, Sun TT, Cheng B. Epigenetic regulation by polycomb group complexes: focus on roles of CBX proteins. J Zhejiang Univ Sci B. 2014;15(5):412–28. doi:10.1631/jzus.B1400077. [Google Scholar] [PubMed] [CrossRef]
14. Tian P, Deng J, Ma C, Miershali A, Maimaitirexiati G, Yan Q, et al. CBX7 is involved in the progression of cervical cancer through the ITGβ3/TGFβ1/AKT pathway. Oncol Lett. 2024;27(1):14. doi:10.3892/ol.2023.14147. [Google Scholar] [PubMed] [CrossRef]
15. Huang Z, Yan Y, Zhu Z, Liu J, He X, Dalangood S, et al. CBX7 suppresses urinary bladder cancer progression via modulating AKR1B10-ERK signaling. Cell Death Dis. 2021;12(6):537. doi:10.1038/s41419-021-03819-0. [Google Scholar] [PubMed] [CrossRef]
16. Gong L, Tang Y, Jiang L, Tang W, Luo S. Regulation of circGOLPH3 and its binding protein CBX7 on the proliferation and apoptosis of prostate cancer cells. Biosci Rep. 2020;40(12):792. doi:10.1042/BSR20200936. [Google Scholar] [PubMed] [CrossRef]
17. Ni SJ, Zhao LQ, Wang XF, Wu ZH, Hua RX, Wan CH, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018;11(1):17. doi:10.1186/s13045-018-0562-z. [Google Scholar] [PubMed] [CrossRef]
18. Yang Y, Hu Z, Sun H, Yu Q, Yang L, Yin F, et al. CBX7, a potential prognostic biomarker in lung adenocarcinoma. Onco Targets Ther. 2021;14:5477–92. doi:10.2147/OTT.S325203. [Google Scholar] [PubMed] [CrossRef]
19. Peng X, Guan L, Gao B. miRNA-19 promotes non-small-cell lung cancer cell proliferation via inhibiting CBX7 expression. Onco Targets Ther. 2018;11:8865–74. doi:10.2147/OTT. [Google Scholar] [CrossRef]
20. Nawaz Z, Patil V, Arora A, Hegde AS, Arivazhagan A, Santosh V, et al. Cbx7 is epigenetically silenced in glioblastoma and inhibits cell migration by targeting YAP/TAZ-dependent transcription. Sci Rep. 2016;6(1):27753. doi:10.1038/srep27753. [Google Scholar] [PubMed] [CrossRef]
21. Ghafouri-Fard S, Poornajaf Y, Hussen BM, Tavakkoli Avval S, Taheri M, Mokhtari M. Deciphering the role of Hippo pathway in lung cancer. Pathol Res Pract. 2023;243(1):154339. doi:10.1016/j.prp.2023.154339. [Google Scholar] [PubMed] [CrossRef]
22. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–7. doi:10.1093/nar/gkt1248. [Google Scholar] [PubMed] [CrossRef]
23. Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60–4. doi:10.1093/nar/gky311. [Google Scholar] [PubMed] [CrossRef]
24. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–63. doi:10.1093/nar/gkx1090. [Google Scholar] [PubMed] [CrossRef]
25. Yan Y, Zhang D, Zhou P, Li B, Huang SY. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–73. doi:10.1093/nar/gkx407. [Google Scholar] [PubMed] [CrossRef]
26. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1):505. doi:10.5334/aogh.2419. [Google Scholar] [PubMed] [CrossRef]
27. Caso R, Sanchez-Vega F, Tan KS, Mastrogiacomo B, Zhou J, Jones GD, et al. The underlying tumor genomics of predominant histologic subtypes in lung adenocarcinoma. J Thorac Oncol. 2020;15(12):1844–56. doi:10.1016/j.jtho.2020.08.005. [Google Scholar] [PubMed] [CrossRef]
28. Zeeshan R, Mutahir Z. Cancer metastasis-tricks of the trade. Bosn J Basic Med Sci. 2017;17(3):172–82. doi:10.17305/bjbms.2017.1908. [Google Scholar] [PubMed] [CrossRef]
29. Yu J, Jiang P, Zhao K, Chen Z, Zuo T, Chen B. Role of DACH1 on proliferation, invasion, and apoptosis in human lung adenocarcinoma cells. Curr Mol Med. 2021;21(9):806–11. doi:10.2174/1566524021666210119094633. [Google Scholar] [PubMed] [CrossRef]
30. Zhang W, Zhang R, Zeng Y, Li Y, Chen Y, Zhou J, et al. ALCAP2 inhibits lung adenocarcinoma cell proliferation, migration and invasion via the ubiquitination of β-catenin by upregulating the E3 ligase NEDD4L. Cell Death Dis. 2021;12(8):755. doi:10.1038/s41419-021-04043-6. [Google Scholar] [PubMed] [CrossRef]
31. Lin JS, Lai EM. Protein-protein interactions: co-immunoprecipitation. Methods Mol Biol. 2017;1615:211–9. [Google Scholar] [PubMed]
32. Lan H, Lin C, Yuan H. Knockdown of KRAB domain-associated protein 1 suppresses the proliferation, migration and invasion of thyroid cancer cells by regulating P68/DEAD box protein 5. Bioengineered. 2022;13(5):11945–57. [Google Scholar] [PubMed]
33. Tian J, Yuan L. Sirtuin 6 inhibits colon cancer progression by modulating PTEN/AKT signaling. Biomed Pharmacother. 2018;106:109–16. [Google Scholar] [PubMed]
34. Fu M, Hu Y, Lan T, Guan KL, Luo T, Luo M. The Hippo signalling pathway and its implications in human health and diseases. Signal Transduct Target Ther. 2022;7(1):376. [Google Scholar] [PubMed]
35. Lv L, Zhou X. Targeting Hippo signaling in cancer: novel perspectives and therapeutic potential. MedComm. 2023;4(5):e375. [Google Scholar] [PubMed]
36. Han Q, Rong X, Lin X, Zhang X, Fan C, Zhao H, et al. WBP2 negatively regulates the Hippo pathway by competitively binding to WWC3 with LATS1 to promote non-small cell lung cancer progression. Cell Death Dis. 2021;12(4):384. doi:10.1038/s41419-021-03600-3. [Google Scholar] [PubMed] [CrossRef]
37. Rong X, Han Q, Lin X, Kremerskothen J, Wang E. FRMPD1 activates the Hippo pathway via interaction with WWC3 to suppress the proliferation and invasiveness of lung cancer cells. Cancer Manag Res. 2019;11:3395–410. doi:10.2147/CMAR. [Google Scholar] [CrossRef]
38. Xie X, Ning Y, Long J, Wang H, Chen X. Diverse CBX family members as potential prognostic biomarkers in non-small-cell lung cancer. FEBS Open Bio. 2020;10(10):2206–15. doi:10.1002/2211-5463.12971. [Google Scholar] [PubMed] [CrossRef]
39. Liu H, Li Z, Li L. The molecular selectivity of UNC3866 inhibitor for Polycomb CBX7 protein from molecular dynamics simulation. Comput Biol Chem. 2018;74(Suppl. 3):339–46. doi:10.1016/j.compbiolchem.2018.04.005. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools