 Open Access
Open Access
ARTICLE
miR-557 suppresses hepatocellular carcinoma cell proliferation and migration via downregulating CBX4
1 Department of Orthopedics, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200080, China
2 Department of General Surgery, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200080, China
3 Department of Thoracic Surgery, Shanghai Chest Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, 200030, China
4 Department of Radiology, Zhongda Hospital, Southeast University School of Medicine, Nanjing, 210009, China
* Corresponding Author: ZHIAN FANG. Email:
(This article belongs to the Special Issue: New Perspectives on Inflammatory Cancer Transformation)
BIOCELL 2024, 48(7), 1071-1079. https://doi.org/10.32604/biocell.2024.050519
Received 08 February 2024; Accepted 15 March 2024; Issue published 03 July 2024
Abstract
Introduction: Hepatocellular carcinoma (HCC), a prevalent malignancy, poses significant challenges with high tumor heterogeneity and poor prognosis. MicroRNAs (miRNAs) play a pivotal role in hepatocarcinogenesis. Although abnormalities in microRNA-557 (miR-557) expression have been implicated in various cancer types, its role in HCC remains unclear. Therefore, there is a need to explore the function of microRNA-557 in HCC. Methods: Candidate miRNAs were identified through screening in GSE108724 and GSE20077. Real-time PCR was employed to analyze the expression level of miR-557 in hepatoma cell lines and tissues. Cell viability and migration assays were applied to assess the impact of miR-557 on HCC cell lines. Furthermore, the miR-557 target was predicted through three algorithms (Targetscan, miRWalk, and miRanda), and this was confirmed through luciferase assay and Western blotting. Results: In this study, miR-557 was identified in two datasets and expressed at a low level in both hepatoma cell lines and tissues. Notably, high expression of miR-557 in HCC cells inhibited oncogenesis. Conversely, low expression of miR-557 enhanced tumor proliferation and migration. Polycomb chromobox 4 (CBX4) was identified as a direct target of miR-557. Silencing CBX4 influenced the functional impact of miR-557 on HCC cell migration. Conclusion: Taken together, our study contributed to elucidating the hepatoma molecular heterogeneity and provided novel insights into miR-557 role and its target CBX4 in HCC, suggesting its potential as a future effectively druggable target for HCC intervention.Graphic Abstract
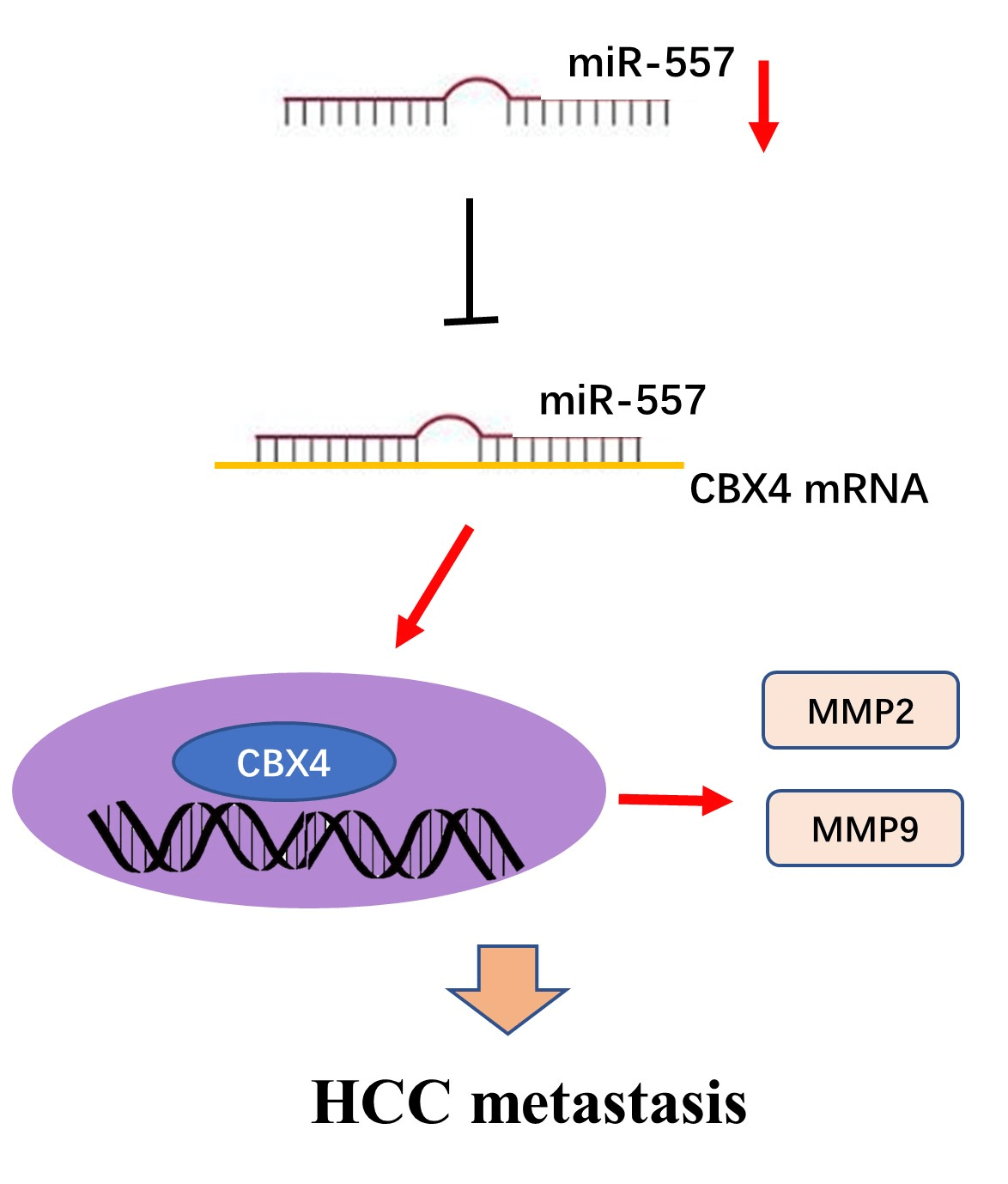
Keywords
Hepatocellular carcinoma (HCC), the most frequent primary malignancy of liver cancer, which affects over 700,000 individuals worldwide each year, has an overall 5-year overall survival (OS) rate of less than 20% [1,2]. Currently, hepatectomy and liver transplantation are the best curative options for HCC. However, unpredictable recurrences or metastases still occur frequently, resulting in poor prognosis [3]. Even in early-stage liver cancer patients, the postoperative recurrence rate remains high, and the majority of patients lose the opportunity for surgery by the time of diagnosis, leaving them with only options like radiotherapy, chemotherapy, and targeted therapy [4,5]. Furthermore, liver transplantation is one of the most efficacious strategies for liver cancer, but the 5-year tumor recurrence rate after liver cancer transplantation ranges from 20% to 57.8% [6]. Hence, exploring the molecular mechanisms involved in hepatocarcinogenesis is essential to develop more effective therapeutic interventions for HCC.
MicroRNAs (miRNAs), an important and evolutionarily conserved class of small noncoding RNAs, primarily act as pivotal regulator of gene expression through mRNA degradation or translational repression [7–9]. They are involved in regulating fundamental cellular processes and play a role in various hepatic disease (e.g., viral hepatitis, liver cirrhosis, and hepatoma) [10]. Accumulating evidence indicated that miRNAs are particularly important in driving hepatocarcinogenesis. For instance, miR-223 could modulate HCC progression by enhancing apoptosis via Ras-related protein Rab-1 (Rab1)/mammalian target of rapamycin (mTOR) pathway, whereas miR-21 may contribute to cell invasion and proliferation in HCC [11,12]. Moreover, miRNAs could promote or suppress antitumor immunity, implicating their involvement in the regulation of the tumor immune microenvironment (TIME) [13–15]. In HCC, miR-148b and miR-155 closely correlate with tumor-associated macrophages (TAMs) infiltration as well as miR-125a [16]. Of note, aberration in miRNAs expression is closely correlated with different HCC prognoses, although the mechanisms underlying this process are unclear. Clinically, miRNAs represent potential targets for HCC management, including diagnosis and therapy, such as miR-34, miR-193, and miR-199 [17–19]. Recently, abnormal miR-557 expression has been implicated in various malignancies (e.g., gastric cancer, breast cancer and lung cancer) [20–22]. However, there was limited research on the functional role of miR-557 in HCC.
Emerging evidence has suggested that polycomb chromobox 4 (CBX4) is involved in regulating TME in various tumor, which significantly affects patients’ prognosis in various tumors, such as colorectal cancer (CRC) and HCC [23,24]. Functionally, CBX4 facilitates HCC progression by sumoylating hypoxia inducible factor-1 alpha (HIF-1α). This leads to the upregulation of vascular endothelial growth factor (VEGF) [25]. Additionally, CRC metastasis was obviously suppressed by CBX4 via the recruitment of histone deacetylase 3 (HDAC3) to the Runt-related transcription factor 2 (Runx2) [26]. Nevertheless, the involvement of CBX4 in HCC pathogenesis remains ambiguous.
Here, the pattern of miR-557 expression was determined in hepatoma tissues. Furthermore, the impact of miR-557 on HCC was characterized in vitro, particularly focusing on its effects on proliferation and invasiveness. This study aims to provide functional insights into the role of miR-557 in HCC progression. These insights can contribute to identifying the molecular basis of liver carcinogenesis, illustrating the high heterogeneity of HCC, and identifying novel therapeutic targets for HCC intervention strategies.
Cell culture and tissue specimens
THLE3, normal liver epithelial cells, and HCC cell lines, including BEL-7402, MHCC97L, SMMC-7721, HepG2, Hep3B, and Huh7, were obtained from iCell Bioscience (Shanghai, China) and included our study. Dulbecco’s modified Eagle’s medium (DMEM, cat# C11965500BT, Gibco, Waltham, MA, USA), supplemented with 10% fetal bovine serum (FBS, cat# 10099158, Gibco, Grand Island, NY, USA), and 1% penicillin-streptomycin (cat# 15140122, Gibco, Grand Island, NY, USA), was used for culturing above cells at 37°C with 5% CO2. HCC tissues from patients undergoing hepatectomy were collected from Zhongda Hospital (Southeast University, Jiangsu, China), along with adjacent non-cancerous tissues (hepatic tissues). Informed consent was obtained from all patients with HCC. All tissues were utilized to investigate patterns of gene expression in HCC. The study received approval from the Ethics Committee of Zhongda Hospital, Southeast University (Approval no. 2022-10).
Two miRNA microarray datasets (GSE108724 and GSE20077), which included 15 HCC tissue samples and their corresponding adjacent hepatic tissues, were acquired from the GEO repository. GEO2R was used to identify differentially expressed miRNAs (threshold: p value less than 0.05 and |logFC| greater than 1) between these two groups, and the results were visualized using Venn diagrams.
Construction of stable cell lines
The genomic pre-miR-557 gene was cloned into the the plasmid pMSCV-pur from GeneChem (Shanghai, China) to generate the newly created pMSCV-miR-557 used as the miR-557 expression plasmid. Subsequently, such indicated plasmid was co-transfected into 293T cells with the pIK plasmid following a previously established protocol [27]. After 36 h of co-transfection, supernatants were harvested and subjected to a 24-h incubation with HCC cells to promote infection. Stably transduced cells were then selected over a period of 10 days using puromycin obtained from Sigma (cat# 15140122, USA).
The miR-557 mimic (5′-GUUUGCACGGGUGGGCCUUGUCU-3′) and miR-557 inhibitor (miR-557-in, 5′-AGACAAGGCCCACCCGUGCAAAC-3′) were obtained as well as a negative control (NC, 5′-UCACAACCUCCUAGAAAGAGUAGA-3′) from GeneChem in Shanghai, China. Additionally, to deplete CBX4, a siRNA (human) with the sequence AGATGAAGATAGTCAAGAA was synthesized and purified. The delivery of above siRNAs was achieved through transfection using Lipofectamine 2000 (cat# 11668030, Invitrogen, USA) as well as the oligonucleotides. Hepatoma cells were transfected following the previously established experimental protocol.
The amplification of 3′UTR segments of CBX4 were performed to generate luciferase reporter vectors and the details of the primers were provided below: CBX4-3′UTR forward, 5′-CTCGAGAACTGCCTCACCGTTACTT-3′, and reverse, 5′-GCGGCCGCAATATTTACATTCAAGCAGG-3′. Mutant inserts were generated by PCR and the details of the primers were provided below: CBX4-3′UTR-mut forward, 5′-GCGGCCGCAACTGCCTCACCGTTACTT-3′, and reverse, 5′-CTCGAGAATATTTACATTCAAGCAGG-3′. These above specific products were then inserted into the modified pGL3 control vector, positioned immediately downstream of the luciferase gene’s stop codon. Sequencing was conducted to verify both wild-type (WT) or mutated (mut). HCC cells were seeded in a 24-well plate. Following that, pGL3-CBX4-3′UTR-luciferase plasmid or mut plasmid was introduced into HCC cells via transfection. Luciferase activity was measured 48 h post-transfection utilizing the Dual Luciferase Reporter Assay Kit (cat# E1910, Promega, USA).
Total RNA was isolated from HCC cells, employing Trizol reagent (cat# 15596026, Invitrogen, Carlsbad, CA, USA), followed by conversion into cDNA utilizing the Reverse Transcription System (cat# 05091284001, Roche, Germany). Quantitative PCR was conducted as described for the previous experiment, including denaturation step (95°C with 30 s), cycle reaction (95°C with 10 s and 60°C with 30 s; 40 cycles), melting curve (95°C with 15 s, 60°C with 30 s, and 95°C with 15 s). The expressed levels were determined according to the ΔΔCT method. The GAPDH served as the internal control for mRNA, while small nuclear RNA U6 acted as the internal control for microRNA. The details of the primers (human) were provided below: MMP2 forward: 5′-AGGCCAAGTGGTCCGTGT
GA-3′; MMP2 reverse: 5′-TAGGTGGTGGAGCACCAGAG-3′; MMP9 forward: 5′-ATCCGGCACCTCTATGGTCCTC-3′; MMP9 reverse:
5′-GCACAGTAGTGGCCGTAGAAGG-3′; CBX4 forward: 5′-GCAGA GTGGAGTATCTGGTGA-3′; CBX4 reverse: 5′-AGCTTGGCACGGTT GTCAG-3′; GAPDH forward: 5′-ACAGTCAGCCGCATCTTCTT-3′;
GAPDH reverse: 5′-GACAAGCTTCCCGTTCTCAG-3′; miR-557 forward: 5′-GTTTGCACGGGTGGGC-3′; miR-557 reverse: 5′-GAAC ATGTCTGCGTATCTC-3′; U6 forward: 5′-GCTTCGGCAGCACATAT
ACTAAAAT-3′; U6 reverse: 5′-CGCTTCACGAATTTGCGT-3′.
Cell viability and migration assays
For cell viability study, HCC cells were plated in 96-well plates and incubated for 24 h. Following 2 h incubation with Cell Counting Kit-8 (CCK-8) reagent (cat# JE603, Dojindo, Kumamoto, Japan) under the same cell culture conditions as described above, the absorbance at 450 nm was measured to obtian optical density (OD) values. In the wound-healing assay, HCC cells were plated and maintained in 6-well plate for 48 h. A wound track was then generated with a sterile 10 μL tip, followed by wash (2 times) with phosphate-buffered saline (PBS), and fresh DMEM was substituted for the existing DMEM. Gap length of the wound, including the initial (0 h) and the residual (24 h) gap length, were measured using National Institutes of Health (NIH) ImageJ software (USA). For transwell assay, HCC cells were cultured onto 24-well inserts in serum-free medium, with 20% FBS, a high serum-medium, added to the lower chamber. After allowing for 24 h migration, cells that did not migrate were eliminated by scraping the upper surface of the membrane. HCC cells migrated were fixed with paraformaldehyde (cat# P0099, Beyotime, Shanghai, China), colored with crystal violet (cat# C0121, Beyotime, Shanghai, China), and counted with high-power microscopy (AE2000, Motic Electric Group, Xiamen, China).
Western blotting (WB) was developed to validate the target protein expression level, as previously described [28]. Briefly, total proteins were extracted from HCC cells and loaded onto gels, followed by transfer onto a polyvinyl difluoride (PVDF) membrane (cat# FFP39, Millipore, Darmstadt, Germany), which was then blocked with 5% skimmed milk (1 h). This membrane was subsequently probed with specific antibodies (4°C with overnight). The primary antibodies used were the following: anti-CBX4 (rabbit, cat# ab242149) and anti-GAPDH (mouse, cat# ab8245) from Abcam, USA. For secondary antibody, the membrane was incubated with anti-rabbit (cat# BA1055) and anti-mouse IgG (cat# BA1051) from Boster, China, at room temperature (2 h). The immunoblots were visualized with LI-COR reader (USA).
Experimental data were expressed as the mean ± standard deviation and analyzed with SPSS software, V.24.0 (Chicago, USA). All the experiments were replicated 3 times. Significance analysis was assessed using Student’s t-test or one-way analysis of variance (ANOVA) as appropriate, with significance set at p < 0.05.
Markedly downregulated miR-557 in HCC
To identify miRNA species potentially involved in HCC progression, we initially selected two miRNA microarray datasets (GSE108724 and GSE20077) to identify differentially expressed miRNAs (miR-532-5p, miR-324-5p, miR-146b-5p, miR-383-3p, miR-175, and miR-557), as shown in Fig. 1A. Among these, the role of miR-557 in hepatocarcinogenesis remains largely unknown. We analyzed the pattern of this candidate expression in hepatoma. Fig. 1B illustrated a significantly lower expression level of miR-557 in HCC cell lines, including BEL-7402, MHCC97L, SMMC-7721, HepG2, Hep3B, and Huh7, when compared to THLE3. Furthermore, the expression of miR-557 consistently showed downregulation in HCC specimens compared to the control group (Fig. 1C). In summary, miR-557 was found to be less expressed in HCC, suggesting its potential contribution to HCC initiation and development.
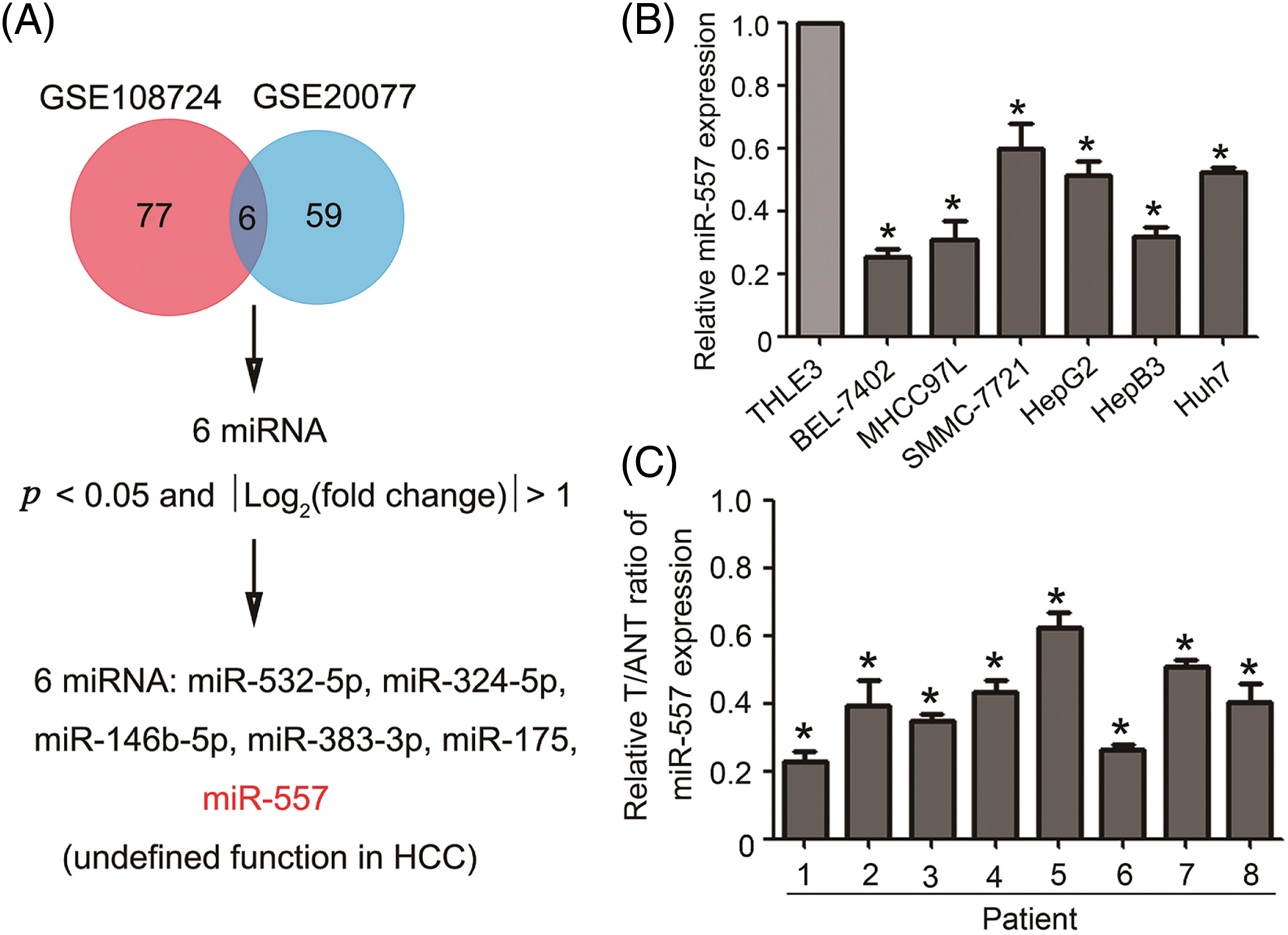
Figure 1: Downregulated expressed miR-557 in both HCC cell lines and tissues. (A) Screening strategy for differentially expressed miRNAs in HCC patients from GSE108724 and GSE20077. Real-time PCR analysis of miR-557 expression in (B) HCC cell lines and (C) hepatoma tissues and matching hepatic tissues (n = 3). T, tumor tissues; ANT, adjacent non-tumor tissues; *p < 0.05.
Ectopic miR-557 expression restrains migration and invasion capacity
For an in-depth study of the pathogenic role of miR-557 in HCC, we established stable miR-557-expressing SMMC-7721 and HepG2 cell lines (Fig. 2A). Compared to control cells, ectopically expressed miR-557 obviously suppressed cell viability in HCC (Fig. 2B). Additionally, migration assays, including wound-healing and transwell assays, revealed a substantial reduction in the migratory capacity of HCC cells upon ectopic expression of miR-557 (Figs. 2C, 2D). Given the pivotal roles of MMP2 and MMP9 in tumor invasion, which could degrade extracellular matrix (ECM), we assessed the expressed level of MMP2 and MMP9 mRNA. The findings indicated that ectopic miR-557 could downregulate MMP2 and MMP9 expression levels (Fig. 2E). The above results collectively suggested that high expression of miR-557 could remarkably inhibit the migration and invasion of HCC cells.
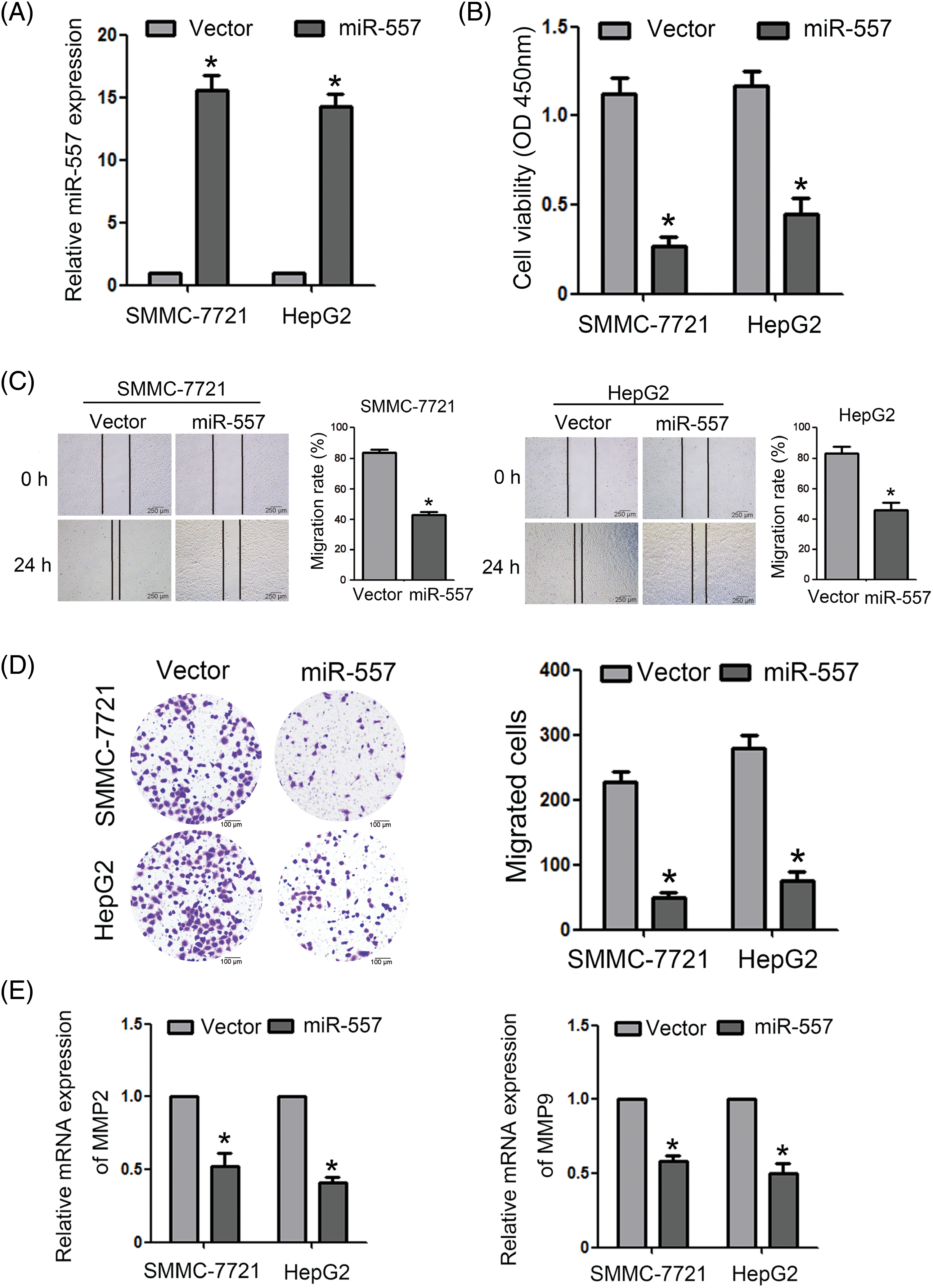
Figure 2: Ectopic miR-55 expression restrains migration and invasion capacity. (A) The expressed miR-557 level in indicated cells (n = 3). (B) Cell viability was assessed using the CCK-8 assay (n = 3). Cell migration was evaluated through both (C) wound-healingand (D) transwell assays (n = 3). (E) The expressed MMP2 and MMP9 levels in indicated cells (n = 3). *p < 0.05.
MiR-557 inhibition enhanced cell migration and invasion
To investigate the involvement of endogenous miR-557 in impeding the migration and invasion of HCC cells, loss-of-function study were carried out with a miR-557-in (Fig. 3A). The impact of miR-557 on HCC cell proliferation was evaluated via CCK-8 assays, which demonstrated that the suppression of miR-557 considerably heightened the proliferative capacity of both SMMC-7721 and HepG2 cells (Fig. 3B). Moreover, both wound-healing and transwell assays (Figs. 3C, 3D) disclosed that the inhibition of miR-557 expedited HCC migration and invasion properties of HCC cells. Consistently, the expressed levels of MMP2 and MMP9 were also upregulated by the miR-557-in (Fig. 3E).
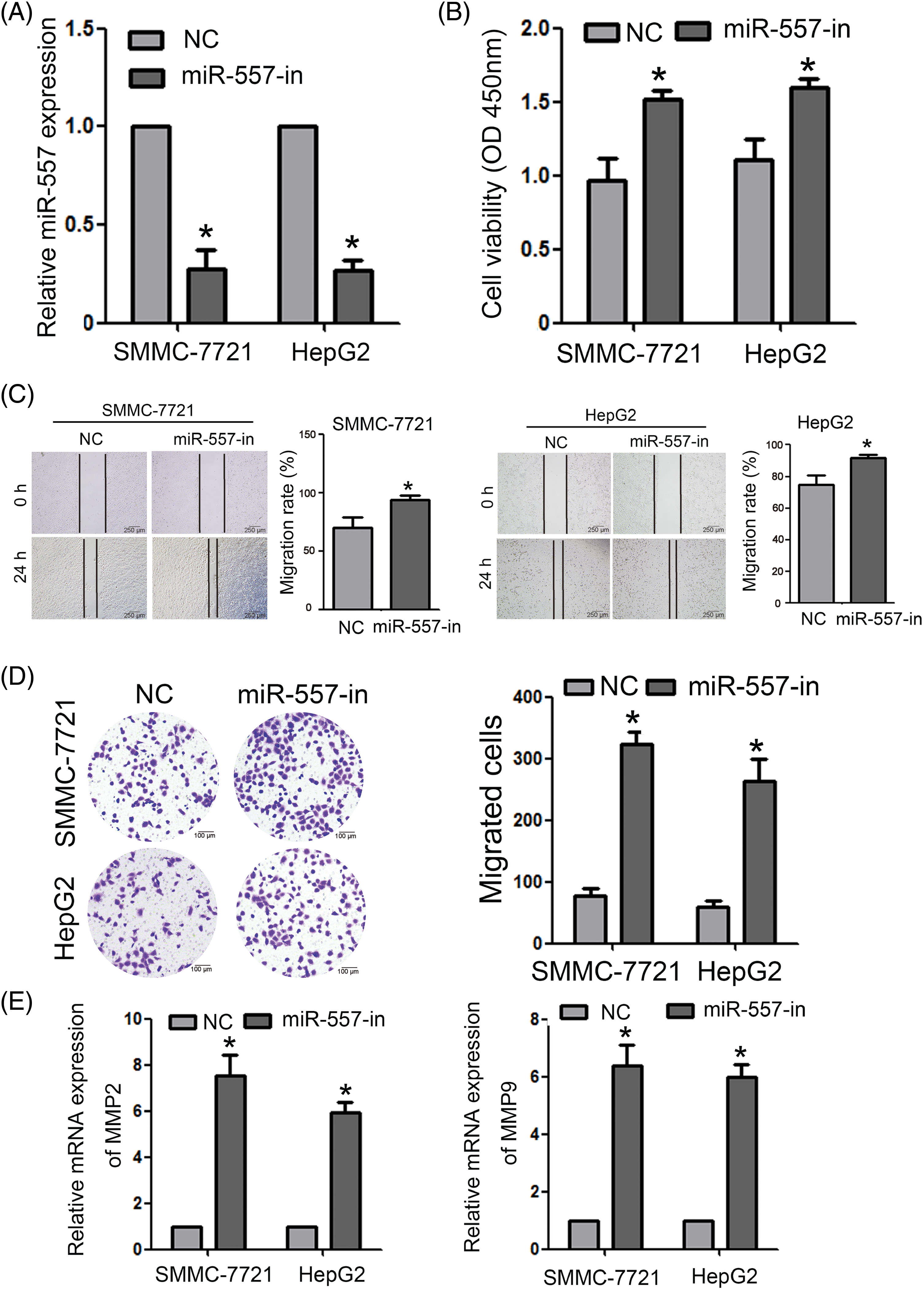
Figure 3: MiR-557 inhibition enhanced HCC cell invasion and migration. (A) The expressed miR-557 level in inhibiting miR-557 SMMC-7721 and HepG2 cells (n = 3). (B) Cell viability was assessed using the CCK-8 assay (n = 3). Cell migration was evaluated through both (C) wound-healing and (D) transwell assays (n = 3). (E) The expressed MMP2 and MMP9 levels in indicated cells (n = 3). miR-557 inhibitor, miR-557-in; negative control, NC; *p < 0.05.
MiR-557 directly targeted CBX4
To delve deeper into the molecular impact of miR-557 on HCC cells, we employed target prediction software, including Targetscan, miRWalk, and miRanda [29]. These tools collectively identified CBX4 as a predicted target for miR-557. The sequences for luciferase reporter vectors, encompassing either the WT or mut 3′UTRs of CBX4, were depicted in Fig. 4A. WB analysis demonstrated a decrease in CBX4 expression in SMMC-7721 and HepG2 cells overexpressing miR-557, while its expression showed an increase in miR-557-in transfected cells (Fig. 4B). To validate that CBX4 could be targeted directly by miR-557, we constructed the pGL3-CBX4-3′UTR-luciferase reporter, which harbored two potential miR-557 binding sites. The aforementioned finding indicated that ectopic miR-557 expression led to a decrease in luciferase activity, whereas inhibition of miR-557 resulted in an increase in the activity of the pGL3-CBX4-3′UTR-luciferase reporter. Conversely, a miR-557 mut did not to show any inhibitory effect on luciferase activity (Fig. 4C). In summary, our findings strongly suggest that miR-557 directly targets and downregulates CBX4.
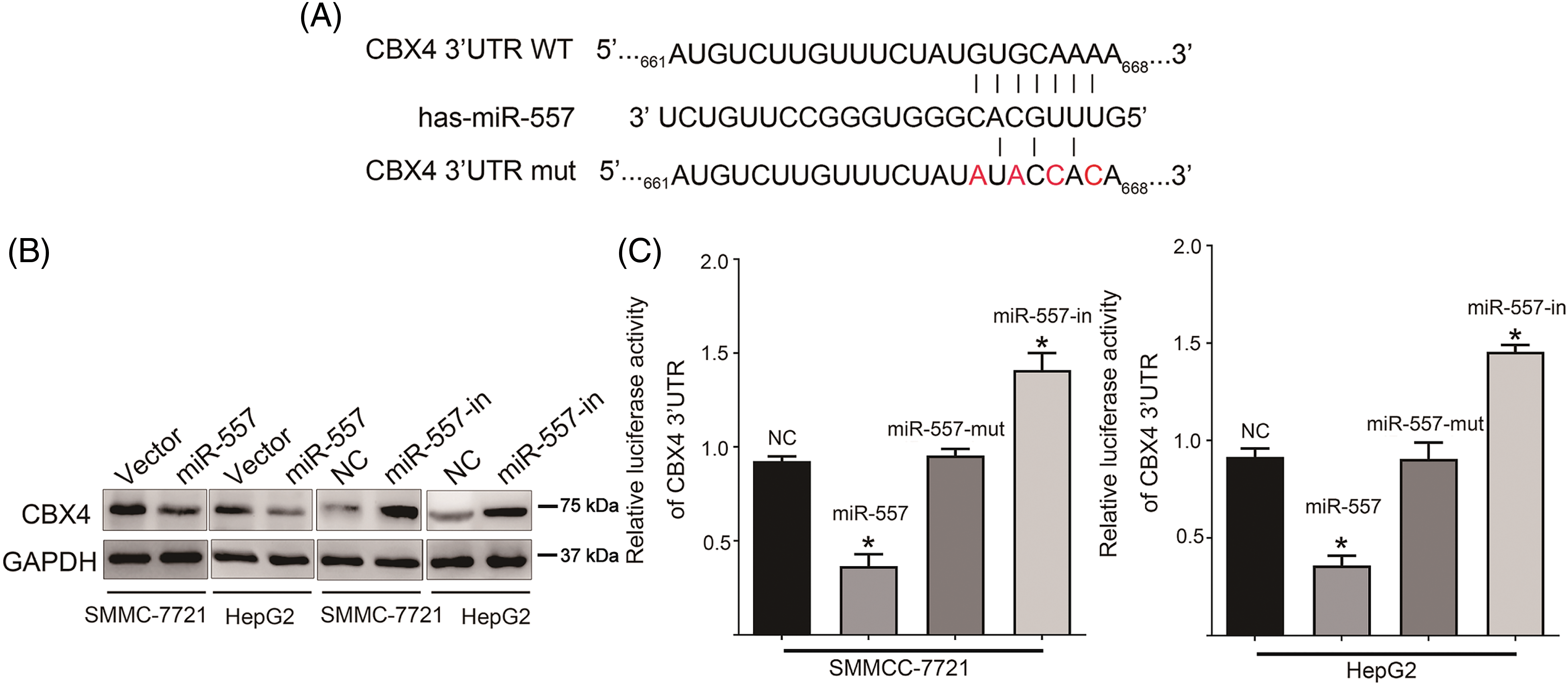
Figure 4: CBX4 was a target of miR-557. (A) Sequences of CBX4 3′UTR containing WT or mut miR-557 binding sites. (B) CBX4 protein level in HCC cells overexpressing or inhibiting miR-557 (n = 3). (C) Luciferase assay of pGL3-CBX4-3′UTR reporter cotransfected the miR-557 mimic or miR-557-mut in indicated cells, or miR-557 inhibitor (n = 3). Wild-type, WT; mutated, mut; *p < 0.05.
The crucial role of CBX4 in miR-557-inhibited HCC cell
To assess the impact of CBX4 suppression on miR-557-inhibiting migration and invasion in HCC, we utilized a CBX4-specific siRNA to suppress endogenous CBX4 expression. WB confirmed the downregulation of CBX4 in HCC cells following CBX4 silencing (Fig. 5A). Furthermore, silencing CBX4 in cells transfected with miR-557-in or NC led to decreased HCC proliferation, as described in Fig. 5B. Moreover, inhibition of CBX4 markedly attenuated miR-557-inhibited HCC cells’ migration capabilities (Fig. 5C). In line with above findings, the expressed MMP2 and MMP9 level exhibited similar patterns (Fig. 5D), indicating that CBX4 silencing reversed the promoting effect of miR-557-in on ECM degradation. Combined, these results further underscored the essential role of CBX4 suppression in mediating miR-557-inhibited cell migration and invasion in hepatoma.
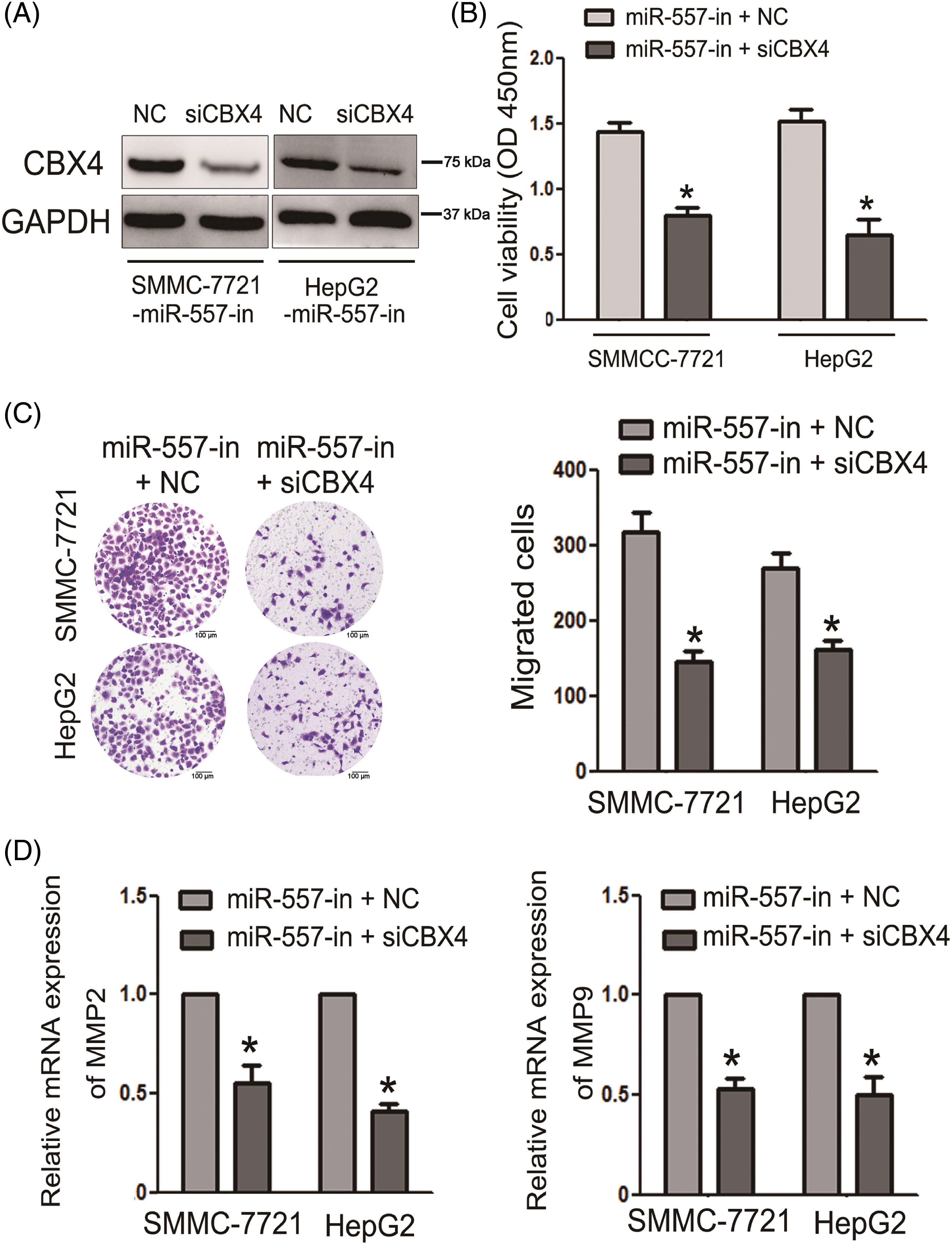
Figure 5: CBX4 suppression was essential for miR-557-inhibited HCC invasion and migration. (A) CBX4 protein level in miR-557-inhibitor transfected HCC cells that were transfected with siCBX4 (n = 3). (B) Cell viability was assessed using the CCK-8 assay (n = 3). (C) Cell migration was evaluated through transwell assays (n = 3). (D) The expressed MMP2 and MMP9 levels in indicated cells (n = 3). *p < 0.05.
HCC features an inflammatory background and multi-step carcinogenesis process characterized by the accumulation of changes in gene expression profiles and disruptions in intracellular signal pathways, representing the third leading cause of cancer-related mortality [2,5]. For most HCC patients, there is no chance of surgery at the time of diagnosis due to an advance stage. Furthermore, the notable characteristics of HCC, including tumor heterogeneity and drug resistance, contribute to a particularly poor prognosis [4,30]. Even the most advanced immunotherapy strategies also have limited efficacy owing to its complex pathogenesis [31]. Therefore, elucidating the underlying molecular mechanism of HCC is urgent and needs to be addressed.
Recently, miRNAs have been shown to influence hepatocarcinogenesis and have great potential in the diagnosis and therapy of tumors [7,32]. In the clinic setting, a panel of plasma microRNA (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) has been used to diagnose in HCC [33]. In HCC patients, evaluated miR-22 exhibits a longer OS (p = 0.0002), increased activity in metabolic signaling, and decreased signaling in interleukin 17 (IL17) pro-inflammatory pathways. Surprisingly, in comparison to lenvatinib, HCC treated with miR-22 had no apparent drug-related toxicities [9,34]. Prior research has established the pivotal role of miR-557 in the progression of diverse types of tumors, with the exception of HCC. Here, we focused our investigation on the role of miR-577 in HCC pathogenesis.
Changes in expression patterns of the key gene could lead to HCC progression [28,35]. In hepatoma cell lines and tissues, miR-557 was expressed at low levels, indicating that miR-557 might involve in HCC deterioration and a worsening prognosis. Further exploration of its function revealed that ectopically expressed miR-557 suppressed the HCC cells’ proliferation and invasion abilities, while the inhibition of miR-557 enhanced HCC cells’ invasive characteristic. Mechanistically, miR-557 could promote the malignant phenotype of HCC cells. Moreover, ectopic miR-557 expression resulted in the downregulation of MMP2 and MMP9, which have been extensively implicated in tumor metastasis through ECM degradation [36]. Conversely, the miR-557-in led to the up-expressed levels of MMP2 and MMP9. These observed findings suggested that miR-557 functioned as a tumor suppressor and merged as a promising therapeutic target for HCC, potentially inhibiting cancer cell invasion by modulating the expressed levels of MMP2 and MMP9.
To further dissect the tumor-suppressive mechanism of miR-557 in HCC, we employed Targetscan, miRWalk, and miRanda to identify potential miR-557 targets for hepatoma, revealing binding sequences between the 3′UTR of CBX4 and miR-557. CBX4, a member of the polycomb group proteins, is implicated in tumorigenesis and cell cycle. Previous reports have indicated that CBX4 expression is closely associated HCC prognosis and sorafenib resistance [24,37]. Herein, the above results unveiled CBX4 as a direct miR-557 target, with miR-557 exerting transcriptional inhibition on expressed CBX4 level by binding to the 3′UTR of its mRNA. Notably, the suppression of CBX4 expression emerged as a crucial factor in mediating the tumor-suppressive effects of miR-557. In future research, we would explore the miR-557 targeting of CBX4 downstream signaling pathways. Nevertheless, our study provided novel insights into the molecular mechanisms of miR-557 and its relation to CBX4.
There were certain limitations to this present study. Firstly, due to the complexities of TME, further research is warranted to explore the network mechanisms of miR-577 impact in HCC. Moreover, currently, clinical trials are primarily focused on exploring the diagnostic and predictive value of miRNAs in tumors, such as ChiCTR2100052195, ChiCTR2100043969, ChiCTR2000036653, and ChiCTR2100052722. Future efforts should focus on developing specific clinical applications for the novel target miR-577 in HCC. Lastly, although the results of our study were supported by functional experiments with cell-line models, additional evidence from well-designed experiments, including animal models (nude mice), single-cell sequencing and organoid experiments, could further reinforce our conclusions.
In summary, our study shed light on the first comprehensive understanding of miR-557 role in hepatocarcinogenesis. Furthermore, the exact mechanism involved with miR-557 in HCC development remained to be elucidated thus far. Our study also established that miR-557 negatively regulated the oncogenic CBX4. These findings indicated that miR-557 could potentially serve as a prognostic biomarker in HCC progression and a promising molecular for therapeutic interventions.
Acknowledgement: None.
Funding Statement: The authors did not receive specific funding for this study.
Author Contributions: Study conception and design: Zhian Fang; data collection: Xulong Sun, Wentao Ding, and Chao Jiang; analysis and interpretation of results: Xulong Sun and Chao Jiang; draft manuscript preparation: Xulong Sun and Wentao Ding. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The corresponding author can be contacted to request access to the datasets generated and/or analyzed during the current study.
Ethics Approval: This study was approved by the Ethics Committee of Zhongda Hospital, Southeast University (Approval no. 2022-10) and informed consent was obtained from all patients.
Conflicts of Interest: There are no conflicts of interest to disclose.
References
1. Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20(4):203–22. doi:10.1038/s41575-022-00704-9. [Google Scholar] [PubMed] [CrossRef]
2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–14. doi:10.1016/S0140-6736(18)30010-2. [Google Scholar] [PubMed] [CrossRef]
3. Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. 2023;42(3):629–52. doi:10.1007/s10555-023-10084-4. [Google Scholar] [PubMed] [CrossRef]
4. Sun Y, Wu P, Zhang Z, Wang Z, Zhou K, Song M, et al. Integrated multi-omics profiling to dissect the spatiotemporal evolution of metastatic hepatocellular carcinoma. Cancer Cell. 2024;42(1):135–56. doi:10.1016/j.ccell.2023.11.010. [Google Scholar] [PubMed] [CrossRef]
5. Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni A, Kamel IR, Cloyd JM, et al. Management of hepatocellular carcinoma: a review. JAMA Surg. 2023;158(4):410–20. doi:10.1001/jamasurg.2022.7989. [Google Scholar] [PubMed] [CrossRef]
6. Tang SC, Zhang KL, Lin KY, Tang YD, Fu J, Zhou WP, et al. A multicenter propensity score analysis of significance of hepatic resection type for early-stage hepatocellular carcinoma. Hepatol Int. 2023;18:623–35. [Google Scholar] [PubMed]
7. Sell MC, Ramlogan-Steel CA, Steel JC, Dhungel BP. MicroRNAs in cancer metastasis: biological and therapeutic implications. Expert Rev Mol Med. 2023;25:e14. doi:10.1017/erm.2023.7. [Google Scholar] [PubMed] [CrossRef]
8. Panicker S, Chengizkhan G, Gor R, Ramachandran I, Ramalingam S. Exploring the relationship between fusion genes and microRNAs in cancer. Cells. 2023;12(20):2467. doi:10.3390/cells12202467. [Google Scholar] [PubMed] [CrossRef]
9. Hu Y, Setayesh T, Vaziri F, Wu X, Hwang ST, Chen X, et al. miR-22 gene therapy treats HCC by promoting anti-tumor immunity and enhancing metabolism. Mol Ther. 2023;31(6):1829–45. doi:10.1016/j.ymthe.2023.04.019. [Google Scholar] [PubMed] [CrossRef]
10. Mohr R, Özdirik B, Lambrecht J, Demir M, Eschrich J, Geisler L, et al. From liver cirrhosis to cancer: the role of micro-RNAs in hepatocarcinogenesis. Int J Mol Sci. 2021;22(3):1492. doi:10.3390/ijms22031492. [Google Scholar] [PubMed] [CrossRef]
11. Correia de Sousa M, Calo N, Sobolewski C, Gjorgjieva M, Clément S, Maeder C, et al. Mir-21 Suppression promotes mouse hepatocarcinogenesis. Cancers. 2021;13(19):4983. doi:10.3390/cancers13194983. [Google Scholar] [PubMed] [CrossRef]
12. Dong Z, Qi R, Guo X, Zhao X, Li Y, Zeng Z, et al. MiR-223 modulates hepatocellular carcinoma cell proliferation through promoting apoptosis via the Rab1-mediated mTOR activation. Biochem Biophys Res Commun. 2017;483(1):630–7. doi:10.1016/j.bbrc.2016.12.091. [Google Scholar] [PubMed] [CrossRef]
13. Zhang X, Yu C, Zhao S, Wang M, Shang L, Zhou J, et al. The role of tumor-associated macrophages in hepatocellular carcinoma progression: a narrative review. Cancer Med. 2023;12(24):22109–29. doi:10.1002/cam4.v12.24. [Google Scholar] [CrossRef]
14. Xu SJ, Chen JH, Chang S, Li HL. The role of miRNAs in T helper cell development, activation, fate decisions and tumor immunity. Front Immunol. 2023;14:1320305. [Google Scholar] [PubMed]
15. Cao Y, Hu L, Tang Y. Hepatitis B virus X protein-mediated upregulation of miR-221 activates the CXCL12-CXCR4 axis to promote NKT cells in HBV-related hepatocellular carcinoma. Biocell. 2023;47(7):1537–48. doi:10.32604/biocell.2023.027205. [Google Scholar] [CrossRef]
16. Pan Z, Tian Y, Niu G, Cao C. Role of microRNAs in remodeling the tumor microenvironment (Review). Int J Oncol. 2020;56(2):407–16. [Google Scholar] [PubMed]
17. Lou G, Chen L, Xia C, Wang W, Qi J, Li A, et al. MiR-199a-modified exosomes from adipose tissue-derived mesenchymal stem cells improve hepatocellular carcinoma chemosensitivity through mTOR pathway. J Exp Clin Cancer Res. 2020;39(1):4. doi:10.1186/s13046-019-1512-5. [Google Scholar] [PubMed] [CrossRef]
18. Roy S, Hooiveld GJ, Seehawer M, Caruso S, Heinzmann F, Schneider AT, et al. microRNA 193a-5p regulates levels of nucleolar-and spindle-associated protein 1 to suppress hepatocarcinogenesis. Gastroenterol. 2018;155(6):1951–66.e26. [Google Scholar]
19. Chen Q, Li L, Tu Y, Zheng LL, Liu W, Zuo XY, et al. MiR-34a regulates apoptosis in liver cells by targeting the KLF4 gene. Cell Mol Biol Lett. 2014;19(1):52–64. [Google Scholar] [PubMed]
20. Zhu K, Yi C, Tong C. circ_0058063 promotes breast cancer progression by upregulating DLGAP5 via sponging miR-557. Cancer Biomark. 2024;39(1):1–13. doi:10.3233/CBM-220410. [Google Scholar] [PubMed] [CrossRef]
21. Qiu J, Hao Y, Huang S, Ma Y, Li X, Li D, et al. MiR-557 works as a tumor suppressor in human lung cancers by negatively regulating LEF1 expression. Tumour Biol. 2017;39(6):1010428317709467. [Google Scholar] [PubMed]
22. Jiang HB, Yang TJ, Lu P, Ma YJ. Gene expression profiling of gastric cancer. Eur Rev Med Pharmacol Sci. 2014;18(15):2109–15. [Google Scholar] [PubMed]
23. Ren L, Li Z, Zhou Y, Zhang J, Zhao Z, Wu Z, et al. CBX4 promotes antitumor immunity by suppressing Pdcd1 expression in T cells. Mol Oncol. 2023;17(12):2694–708. doi:10.1002/mol2.v17.12. [Google Scholar] [CrossRef]
24. Zhao W, Ma B, Tian Z, Han H, Tang J, Dong B, et al. Inhibiting CBX4 efficiently protects hepatocellular carcinoma cells against sorafenib resistance. Br J Cancer. 2021;124(7):1237–48. doi:10.1038/s41416-020-01240-6. [Google Scholar] [PubMed] [CrossRef]
25. Li J, Xu Y, Long XD, Wang W, Jiao HK, Mei Z, et al. Cbx4 governs HIF-1α to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 2014;25(1):118–31. doi:10.1016/j.ccr.2013.12.008. [Google Scholar] [PubMed] [CrossRef]
26. Wang X, Li L, Wu Y, Zhang R, Zhang M, Liao D, et al. CBX4 suppresses metastasis via recruitment of HDAC3 to the Runx2 promoter in colorectal carcinoma. Cancer Res. 2016;76(24):7277–89. doi:10.1158/0008-5472.CAN-16-2100. [Google Scholar] [PubMed] [CrossRef]
27. Shen L, Gu P, Qiu C, Ding WT, Zhang L, Cao WY, et al. Lysophosphatidylcholine acyltransferase 1 promotes epithelial-mesenchymal transition of hepatocellular carcinoma via the Wnt/β-catenin signaling pathway. Ann Hepatol. 2022;27(3):100680. doi:10.1016/j.aohep.2022.100680. [Google Scholar] [PubMed] [CrossRef]
28. Chen D, Lou Y, Lu J, Fan X, Zhu Q, Sun H. Characterization of the clinical significance and immunological landscapes of a novel TMEMs signature in hepatocellular carcinoma and the contribution of TMEM201 to hepatocarcinogenesis. Int J Mol Sci. 2023;24(12):10285. doi:10.3390/ijms241210285. [Google Scholar] [PubMed] [CrossRef]
29. Ma L, Ma Y, Lian A. Involvement of miR-769-5p/Retinoic acid receptor responder 1 Axis in the progression of osteosarcoma: characterization of potential therapeutic targets. Pharmacol. 2022;107(3–4):179–87. [Google Scholar]
30. Chen L, Fan Z, Zhao Y, Yang H, Lv G. Genetic factors in the clinical predictive model for hepatocellular carcinoma: evidence from genetic association analyses. J Hepatol. 2023;79(1):E33–5. doi:10.1016/j.jhep.2022.12.024. [Google Scholar] [PubMed] [CrossRef]
31. Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022;19(3):151–72. doi:10.1038/s41571-021-00573-2. [Google Scholar] [PubMed] [CrossRef]
32. Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Shen TW, Zhao Y, et al. Serum microRNA profiles and pathways in hepatitis B-Associated hepatocellular carcinoma: a South African study. Int J Mol Sci. 2024;25(2):975. doi:10.3390/ijms25020975. [Google Scholar] [PubMed] [CrossRef]
33. Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, et al. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29(36):4781–8. doi:10.1200/JCO.2011.38.2697. [Google Scholar] [PubMed] [CrossRef]
34. Zhang L, Yang P, Wang J, Liu Q, Wang T, Wang Y, et al. MiR-22 regulated T cell differentiation and hepatocellular carcinoma growth by directly targeting Jarid2. Am J Cancer Res. 2021;11(5):2159–73. [Google Scholar] [PubMed]
35. Chen D, Zhu Q, Li T, Fan X, Lou Y, Zhang Y, et al. KLF4 loss in hepatocellular carcinoma: improving prognostic prediction and correlating immune infiltrates. Front Genet. 2023;14:1106952. doi:10.3389/fgene.2023.1106952. [Google Scholar] [PubMed] [CrossRef]
36. Chen R, Cui J, Xu C, Xue T, Guo K, Gao D, et al. The significance of MMP-9 over MMP-2 in HCC invasiveness and recurrence of hepatocellular carcinoma after curative resection. Ann Surg Oncol. 2012;19:375–84. doi:10.1245/s10434-011-1836-7. [Google Scholar] [PubMed] [CrossRef]
37. Jiao HK, Xu Y, Li J, Wang W, Mei Z, Long XD, et al. Prognostic significance of Cbx4 expression and its beneficial effect for transarterial chemoembolization in hepatocellular carcinoma. Cell Death Dis. 2015;6(3):e1689. doi:10.1038/cddis.2015.57. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools