 Open Access
Open Access
ARTICLE
Silvestrol alleviates glioblastoma progression through ERK pathway modulation and MANBA and NRG-1 expression
1 Neurosurgery Department, The First Affiliated Hospital of Chongqing Medical and Pharmaceutical College, Chongqing, China
2 Neurosurgery Department, Changdu People’s Hospital of Tibet, Changdu, China
* Corresponding Authors: BO TIAN. Email: ; FENG YANG. Email:
BIOCELL 2024, 48(7), 1081-1093. https://doi.org/10.32604/biocell.2024.049878
Received 21 January 2024; Accepted 24 April 2024; Issue published 03 July 2024
Abstract
Background: Glioblastoma, a notably malignant tumor within the central nervous system, is distinguished by its aggressive behavior. Silvestrol, a robust inhibitor of the RNA helicase eukaryotic initiation factor 4A (eIF4A), has shown significant potential as an anticancer compound. Yet, the impact of silvestrol on glioblastoma, especially its molecular mechanisms, has not been fully elucidated. Methods: This investigation employed a variety of in vitro assays, such as cell counting kit-8 (CCK-8), clonogenic, 5-ethynyl-2′-deoxyuridine (EDU), wound healing, and flow cytometry, to evaluate cell cycle progression, apoptosis, cell viability, and migration. Western blot analysis was also performed to study the apoptosis and extracellular regulated kinase (ERK) pathways. After the ERK pathway was inhibited, differentially expressed genes (DEGs) in U87 cells were identified, followed by an analysis of target genes using the gene expression profiling interactive analysis (GEPIA) database. Results: Silvestrol significantly suppressed the proliferation, migration, and colony formation of glioma cells. It caused cell cycle arrest and enhanced apoptosis in these cells. Additionally, silvestrol stimulated the ERK pathway, with these effects being reversible by an ERK phosphorylation inhibitor. Transcriptome combined with GEPIA, GSCA, UALCAN, TIMER database screened 4 potential drug targets of silvestrol: chromosome 1 open reading frame 226 (C1ORF226), mannosidase beta A (MANBA), IQ motif and Sec7 domain 2 (IQSEC2), neuregulin 1 (NRG-1). Among them, C1ORF226 was lower risk gene while MANBA, IQSEC2, and NRG-1 were high-risk genes. Furthermore, silvestrol notably reduced MANBA mRNA levels, which could be reversed by inhibiting ERK phosphorylation. Furthermore, silvestrol markedly decreased NRG-1 protein levels, with an additional reduction observed when the ERK pathway was blocked. Conclusion: Silvestrol’s anti-glioma effects are primarily due to the suppression of MANBA expression via the ERK pathway and possibly by hindering the translation of NRG-1 protein, thus reducing its expression. The downregulation of MANBA and NRG-1 proteins may be crucial in hindering glioma development and progression. These results highlight the intricate relationship between the ERK pathway and gene expression regulation in silvestrol’s therapeutic effectiveness against glioma.Keywords
Abbreviations
| eIF4A | Eukaryotic initiation factor 4A |
| EIF4F | Eukaryotic translation initiation factor 4F |
| CCK-8 | Cell counting kit-8 |
| EDU | 5-ethynyl-2′-deoxyuridine |
| ERK | Extracellular regulated kinase |
| C1ORF226 | Chromosome 1 open reading frame 226 |
| MANBA | Mannosidase beta A |
| IQSEC2 | IQ motif and Sec7 domain 2 |
| NRG-1 | Neuregulin 1 |
| TRNP1 | TMF1 regulated nuclear protein 1 |
| TUBA4A | Tubulin alpha 4A |
| GEPIA | Gene expression profiling interactive analysis |
| MEK | Mitogen-activated protein kinase kinase |
| MAPK | Mitogen-activated protein kinases |
Glioblastoma is the most prevalent primary malignant brain tumor, known for its diffuse infiltrative growth [1]. It accounts for approximately 14.5% of all brain tumors, and its incidence stands at approximately 5.6 per 100,000 person-years, predominantly affecting individuals aged between 45 and 75 [2,3]. The standard treatment regimen for glioblastoma in clinical settings includes surgical resection, followed by radiation therapy and chemotherapy [4]. However, the 5-year survival rate for glioblastoma patients remains low at about 6.8%, with median survival spanning between 15 and 18 months [1]. Recent strides have been made in understanding glioblastoma’s molecular mechanisms, tumor microenvironment, and genomics yet progress in treatment and survival outcomes has been modest [4–6].
Silvestrol, a naturally occurring active small molecule, was isolated from the plant Aglaia foveolata in Indonesia [7]. It has been identified as an inhibitor of eukaryotic initiation factor 4A (eIF4A), one of the eukaryotic translation initiation factors, and is noted for its high specificity and tolerability in animal models [8]. The overactivity of translation initiation factors is closely linked to translation dysregulation, a condition that is strongly associated with excessive cell proliferation, angiogenesis, survival, and altered energetics in tumors [9,10]. Consequently, silvestrol has demonstrated the capability to inhibit the growth of numerous cancer cell lines at low nanomolar concentrations [11–14], especially targeting mRNAs with highly structured 5′UTR that are heavily reliant on eIF4A for translation [15]. Notably, oncogenes such as mutant KRAS genes possess long and highly structured 5′UTRs [16]. Inhibitors of eIF4A have displayed significant antitumor activity both in vivo and in vitro in various tumors, including pancreatic ductal adenocarcinoma [17], leukemia [18], hepatocellular carcinoma [19], and have been shown to enhance tumor cell radiosensitivity [20,21]. However, the efficacy of silvestrol against glioblastoma remains uncertain.
Silvestrol is recognized for its role in inhibiting eIF4A, a key component of the eukaryotic translation initiation factor 4F (eIF4F) complex, essential for the initiation of eukaryotic translation. Moreover, research suggests that eIF4F serves as a central node in resistance mechanisms against anti-BRAF and anti-mitogen-activated protein kinase kinase (anti-MEK) cancer therapies [22]. Notably, inhibiting both extracellular regulated kinase (ERK) and eIF4A has been found to exert a synergistic effect [23], with the activation of the mitogen-activated protein kinases (MAPK) pathway also known to boost eIF4A activity [24]. The ERK1/2 intracellular signaling pathway, a classical MAPK transduction pathway, is crucial for mediating extracellular signals in cell proliferation, differentiation, development, stress response, and apoptosis [25]. Recent research has shown that silvestrol inhibits the AKT/mTOR and ERK1/2 signaling pathways, demonstrating anti-tumor effects in Glioblastoma multiforme (GBM) cells [26]. This study focused on the role of hypoxia-inducible factor within these pathways but did not delve deeply into the subsequent gene regulatory networks [26]. External stimuli and receptor engagement activate Ras, leading to the phosphorylation of ERK through the Ras/Raf/MEK/ERK cascade at both threonine and tyrosine sites [27]. Upon activation, ERK rapidly moves from the cytoplasm to the nucleus, phosphorylating various transcription factors and influencing gene expression related to transcription. While the ERK pathway is often overexpressed in tumors and continuously activated to encourage tumor cell proliferation, excessive activation of Ras-ERK signaling has been found to accumulate cyclin-dependent kinase (CDK) inhibitors, including cell cycle arrest [28].
Therefore, this study utilized U87 MG and U251 glioblastoma cell lines to examine silvestrol’s inhibitory effects on glioblastomas in vitro, aiming to determine whether silvestrol’s inhibition relies on the ERK1/2 signaling pathway. Significantly, our research delves into the mechanisms through which silvestrol regulates glioblastoma cell proliferation, employing extensive data mining.
Silvestrol (MCE, cat: HY-13251, NJ, USA) was purchased from MCE, and dissolved in dimethyl sulfoxide (Beyotime, ST038, Shanghai, China). 4% paraformaldehyde (Biosharp, BL539A, GuangDong, China), LY3214996 (MCE, HY-101494, NJ, USA). Antibodies: β-actin, cleaved caspase3, extracellular regulated kinase 1/2 (ERK1/2), p-ERK1/2, mitogen-activated protein kinase kinase (MEK), p-MEK, Goat Anti-Rabbit IgG (H + L) HRP (Affinity, LOT: AF7018, AF7022, AF1015, AF0155, AF8035, AF6385, S0001, WuHan, China), neuregulin 1 (NRG-1) (66492-1-Ig, protein tech, Wuhan, China).
U87 MG and U251 glioblastoma cell lines were obtained from Wuhan Prosser Life Technology Co., Ltd. China. The glioblastoma cell line was cultured in Dulbecco’s modification of Eagle’s medium (BasalMedia, L110KJ, Shanghai, China) medium containing 10% fetal bovine serum (Procell, 164210-500, Wuhan, China) and 1% Penicillin-Streptomycin Solution (Procell, PB180120, Wuhan, China) at 37°C in a 5% CO2 incubator.
The half maximal inhibitory concentration (IC50) of silvestrol on U87 MG and U251 cells for 48 h was determined using the cell counting kit-8 (CCK-8) (DOJINDO, CK04, Kumamoto, Japan). U87 MG and U251 cells were seeded in 96-well plates at 2000 cells per well. After culturing the cells overnight, they were treated with different concentrations of silvestrol for 48 h. 10 μL of CCK-8 solution was then added to each well and incubated for 3 h at 37°C. Subsequently, their optical density values at 450 nm were measured using a microplate reader (BioTek, SYNERGY/H1, Vermont, Germany).
After the IC50 was determined, 0, 2.5, and 10 nM silvestrol were treated with cells for 24, 48, and 72 h, respectively, to assess silvestrol’s impact on cell proliferation, and the rest of the operations were carried out as above.
5-ethynyl-2′-deoxyuridine (EDU) assay
87 MG and U251 cells were plated in 96-well plates at a density of 5 × 104 cells per well. After treatment with drugs for 24 h, 100 μL of EDU (Beyotime, C0088S, Shanghai, China) working solution (20 μM) was added and cells were cultured for an additional 2 h. Following this, cells were fixed with 4% paraformaldehyde (Beyotime, P0099, Shanghai, China) for 15 min at room temperature. Afterward, they were washed with 200 μL of immunostaining strong permeabilizer (Beyotime, P0097, Shanghai, China) and incubated at room temperature for ten minutes. Subsequently, 50 μL of click reaction solution was added, and cells were incubated in the dark at room temperature for 30 min. Finally, cells were counterstained with Hoechst 33342 for 10 min, and blocked, and fluorescence images were captured using a fluorescence microscope (Leica, DMi8, Hessen, Germany).
In 6-well plates, cells were seeded at 1000/well, and after overnight adherence, different concentrations of drugs were added. After 7–14 days, the cell culture medium was removed, washed with PBS, and then washed with 4% paraformaldehyde. The cells were fixed and stained with Giemsa staining solution (Beyotime, C0131, Shanghai, China) for 30 min. Lastly, the cells were washed with PBS three times and photographed.
The apoptosis rate of U87 MG cells was detected using an Annexin V-FITC/PI apoptosis kit (Multi Sciences, C0131, Hangzhou, China). U87 MG cells were harvested following a 24-h drug treatment, washed with pre-cooled PBS at 4°C, and resuspended in a buffer. Subsequently, the Annexin V-FITC/PI mixture was added, gently mixed and incubated for 15 min in the dark. Cell analysis was performed using a flow cytometer (Beckman, CytoFLEX LX, CA, USA).
In 6-well plates, U87 MG and U251 underwent drugs for 24 h and were collected. Cells were washed with PBS, fixed with 75% ethanol, and fixed overnight at 4°C. Before staining, washed with PBS, then added 500 μL PI/RNase Staining Buffer (BD, 550825, NJ, USA) and incubated for 15 min in the dark. Detected using a flow cytometer (Beckman, CytoFLEX LX, CA, USA).
Following a 24-h drug treatment of U87 MG and U251 cells, the cells were collected for Western blot (WB) experiments, with experimental procedures referenced from previous reports [29]. The reagents and materials used in this study are as follows: RIPA lysis buffer (Beyotime, P0013B, Shanghai, China), ultrasensitive multi-function imager (Cytiva, Amersham ImageQuant 800, MA, USA).
In 6-well plates, U251 was cultured until the cell density reached approximately 70%–80%. Slowly create straight scratches in the cell monolayer with a 200 μL pipette tip. After washing away floating cells with PBS, the cells were treated with various drugs in a serum-free medium and then incubated. The microscope was used to capture scratch images of each group at the same location at both 0 and 24 h.
RNA sequencing and data analysis
U87 cell grouping: silvestrol (10 nM) and silvestrol (10 nM) + LY3214996 (ERK inhibitor). Total RNA was extracted using TRIzol reagent (Invitrogen, 15596018CN, CA, USA). Concentrations were quantified using Illumina Novaseq 6000 sequencing platform and cDNA was built libraries by starting material Poly (A)+ RNA, which finally was purified with AMPure XP beads. IlluminaHiSeq platform (BioMarker Technologies, Beijing, China) sequenced the library. Reference gene source: homo_sapiens (http://asia.ensembl.org/Homo_sapiens/Info/Index); Reference genome version: grch38. We normalized fragments per kilobase of transcript per million fragments mapped (FPKM) = cDNA fragments/[mapped fragments (millions) × transcript length (kb)].
Genes upregulated (|FC-1| ≥ 0.5 and p adjust < 0.05) or downregulated (|FC-1| ≥ 0.5 and p adjust < 0.05) by ERK inhibitors (LY3214996) were designated as the RNA-UP and RNA-DOWN gene sets, respectively. Additionally, differentially expressed genes in glioblastoma compared to normal tissues were identified as the GEPIA-UP and GEPIA-DOWN gene sets using the gene expression profiling interactive analysis (GEPIA) database.
The intersection of transcriptomic data and the GEPIA database was obtained to identify uniformly upregulated and downregulated genes for subsequent gene ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment. Furthermore, the association between hub genes and clinical survival rates was analyzed using the tumor immune estimation resource (TIMER) database.
Cell pellets were resuspended in TRNzol Universal Reagent (TianGen, DP424, Beijing, China) and incubated on ice for 30 min. One-fifth of the volume (200 μL) of chloroform was added, followed by gentle inversion for 15 times. The mixture was placed on ice for 10 min to allow phase separation. After centrifugation (4°C, 10000 rpm, 15 min) (Thermo, pico21/21R, CA, USA), the upper aqueous phase was carefully collected and mixed with an equal volume of isopropanol. The mixture was placed on ice for 10 min and then centrifuged (4°C, 10000 rpm, 10 min) to obtain RNA precipitate. The RNA pellet was resuspended in 700 μL of 75% ethanol, followed by centrifugation (4°C, 7500 rpm, 5 min) to wash the RNA. After air-drying on ice, the RNA sample was dissolved in 20 μL of RNase-free water. The PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, RR047A, Kusatsu, Japan) was utilized for the reverse transcription of RNA into cDNA. Real-time fluorescence quantitative PCR was conducted using the GoTaq® qPCR Master Mix (Promega, A6001, WI, USA). The sequence of primers is as follows: β-actin(human)-F: CATGTACGTTGCTATCCAGGC; β-actin(human)-R: CTCCTTAATGTCACGCACGAT; NRG1(human)-F: CGGTGTCCATGCCTTCCAT; NRG1(human)-R: GTGTCACGAGAAGTAGAGGTCT; MANBA(human)-F: TGAGCTGCGTTTCCAGTCAG; MANBA(human)-R: ACATGGCATTCACCCTTCTGC.
After reaching an appropriate cell density in a 96-well plate, U87 cells were fixed with 4% paraformaldehyde for 15 min. Following fixation, cells were washed with PBS and treated with 0.3% Triton X-100 for 10 min. Rinse the wells three times with PBS, then add 50 μL of immunostaining blocking solution (Beyotime, P0102, Shanghai, China), and incubate at room temperature for 60 min. The staining solution containing NRG-1 antibody was added, ensuring thorough wetting of the cells. Incubation was conducted overnight at 4°C, followed by three PBS washes. Subsequently, secondary antibodies (BBI, D110061, and D110090, Shanghai, China) were added and incubated for 1 h, followed by another round of PBS washes. DAPI (Beyotime, C1002, Shanghai, China) was diluted 1:100 in PBS and added (10 μL) for nuclear counterstaining. After 15 min of incubation in the dark, specimens were washed five times with PBST (5 min each) to remove excess DAPI. Finally, the localization and expression of NRG-1 antibody in the cells were observed by a fluorescence microscope (Leica, DMi8, Hessen, Germany).
Comparisons were performed using Prism (Graphpad Software Inc, GraphPad 8.0.1, CA, USA). All data were expressed as the mean ± SD and the one-way ANOVA and Tukey’s multiple comparison test were used to determine the statistical significance of the differences among groups. p < 0.05 was considered to indicate a statistically significant difference.
Silvestrol suppressed the proliferation of brain glioblastoma cells in a dose-dependent manner
Initially, half maximal inhibitory concentration (IC50) of silvestrol was determined in two glioblastoma cell lines over 48 h, revealing IC50 values of 6.379 nM for U251 and 9.154 nM for U87 MG. Compared to the control group, the suppressive effect of silvestrol on brain glioblastoma cells was observed to increase significantly with higher drug concentrations, indicating a clear dose-response relationship (p < 0.05) (Figs. 1A and 1B). Based on these findings, the following experiments were conducted using 10 nM as the high-dose group, 4 nM as the mid-dose group, and 2.5 nM as the low-dose group. Cell proliferation assays demonstrated that silvestrol’s inhibitory effect on both glioblastoma cell types intensified over time and with increasing concentration (Figs. 1C and 1D). Additionally, colony formation assays provided further proof of silvestrol’s suppressive effects, showing a significant decrease in the cloning efficiency of glioblastoma cells at both high and low doses of silvestrol compared to the control group (Figs. 1E and 1F). Silvestrol was also found to hinder the migration of U251 cells, with higher concentrations of silvestrol correlating with a greater percentage of wound closure in a dose-dependent manner (Fig. 1G).
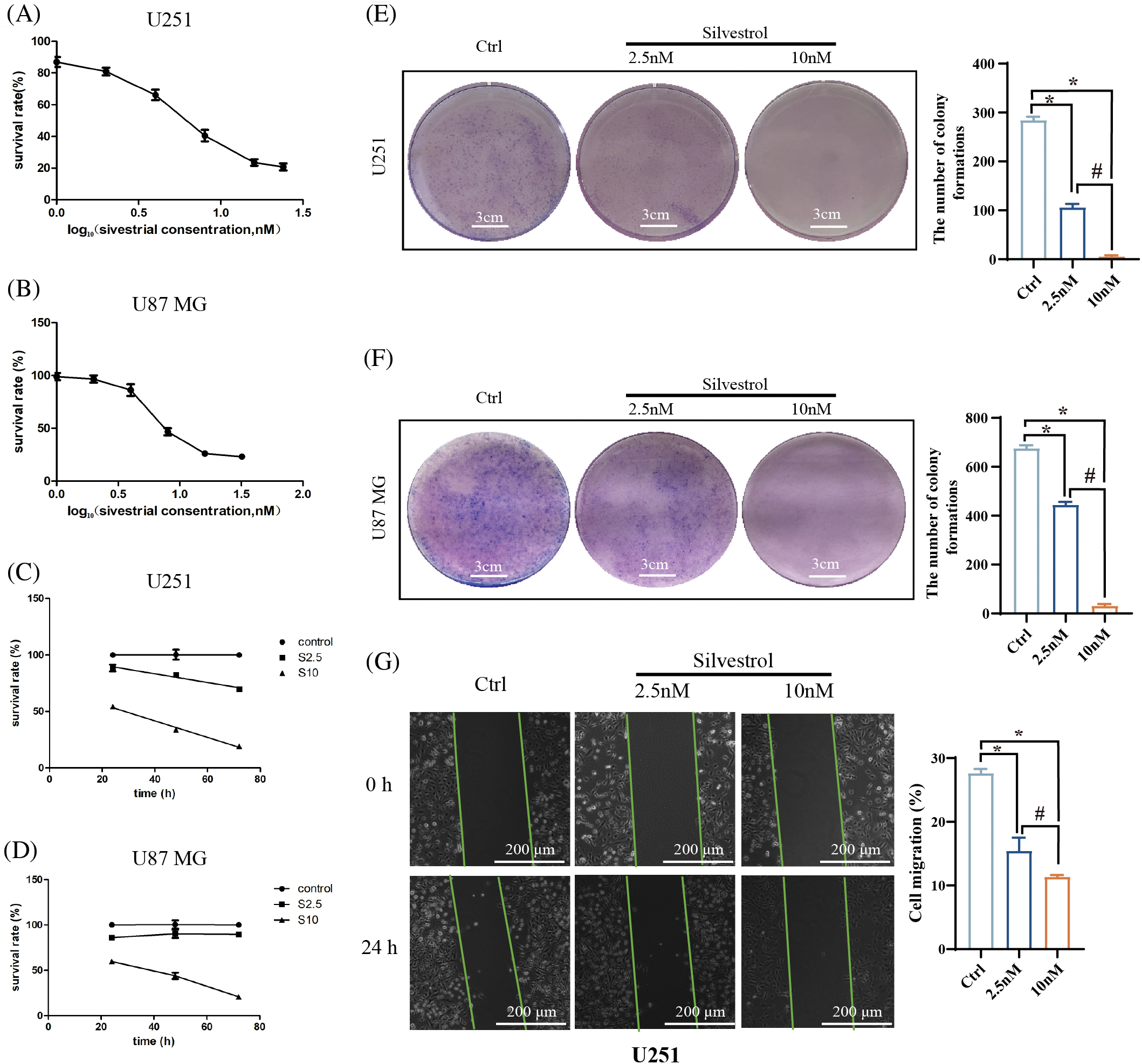
Figure 1: Effects of silvestrol on human GBM cells. Cell viability was assessed using the CCK-8 method. (A) U251 was treated with different concentrations of silvestrol (0, 1, 2, 4, 8, 16, 24 nM) for 48 h. The horizontal axis represents log10(concentrations). (B) U87 MG was treated with different concentrations of silvestrol (0, 1, 2, 4, 8, 16, 32 nM) for 48 h. The horizontal axis represents log10(concentrations). (C) Silvestrol high-dose group (10 nM) and low-dose group (2.5 nM) treated U251 cells for 24, 48, and 72 h. (D) Silvestrol high-dose group and low-dose group treated U87 MG cells for 24, 48, and 72 h. (E and F) Colony formation experiments of silvestrol on U251 (E), U87 MG (F) cells. (G) Scratch experiment of U251 treated with silvestrol. *p < 0.05 vs. ctrl group. #p < 0.05 vs. silvestrol (2.5 nM) group. Scale bar (E and F) = 3 cm. Scale bar (G) = 200 μm.
Silvestrol suppressed proliferation in two brain glioblastoma cell lines by inducing cell cycle arrest and promoting apoptosis in U87 MG cells
EDU proliferation assays revealed that silvestrol decreased the proportion of EDU-positive cells in both U251 and U87 MG cell lines (Figs. 2A and 2B). Flow cytometry analyses showed that silvestrol triggered cell cycle arrest in the G2 phase for U251 cells (Fig. 2C) and in the G1 phase for U87 MG cells (Fig. 2D). This arrest obstructed the seamless transition of cells into subsequent phases, effectively impeding DNA synthesis in both types of glioblastoma cells and disrupting normal cell division. Furthermore, levels of the apoptosis-indicative protein, cleaved caspase3, were significantly increased (Figs. 2E and 2F), with this elevation demonstrating a dose-dependent relationship (p < 0.05). Additional flow cytometry analyses of apoptosis indicated that silvestrol enhanced both early and late apoptosis in U87 MG cells (Fig. 2G). Remarkably, the group treated with a high dose of silvestrol showed a substantially higher total rate of apoptosis compared to both the control and low-dose groups (p < 0.05).
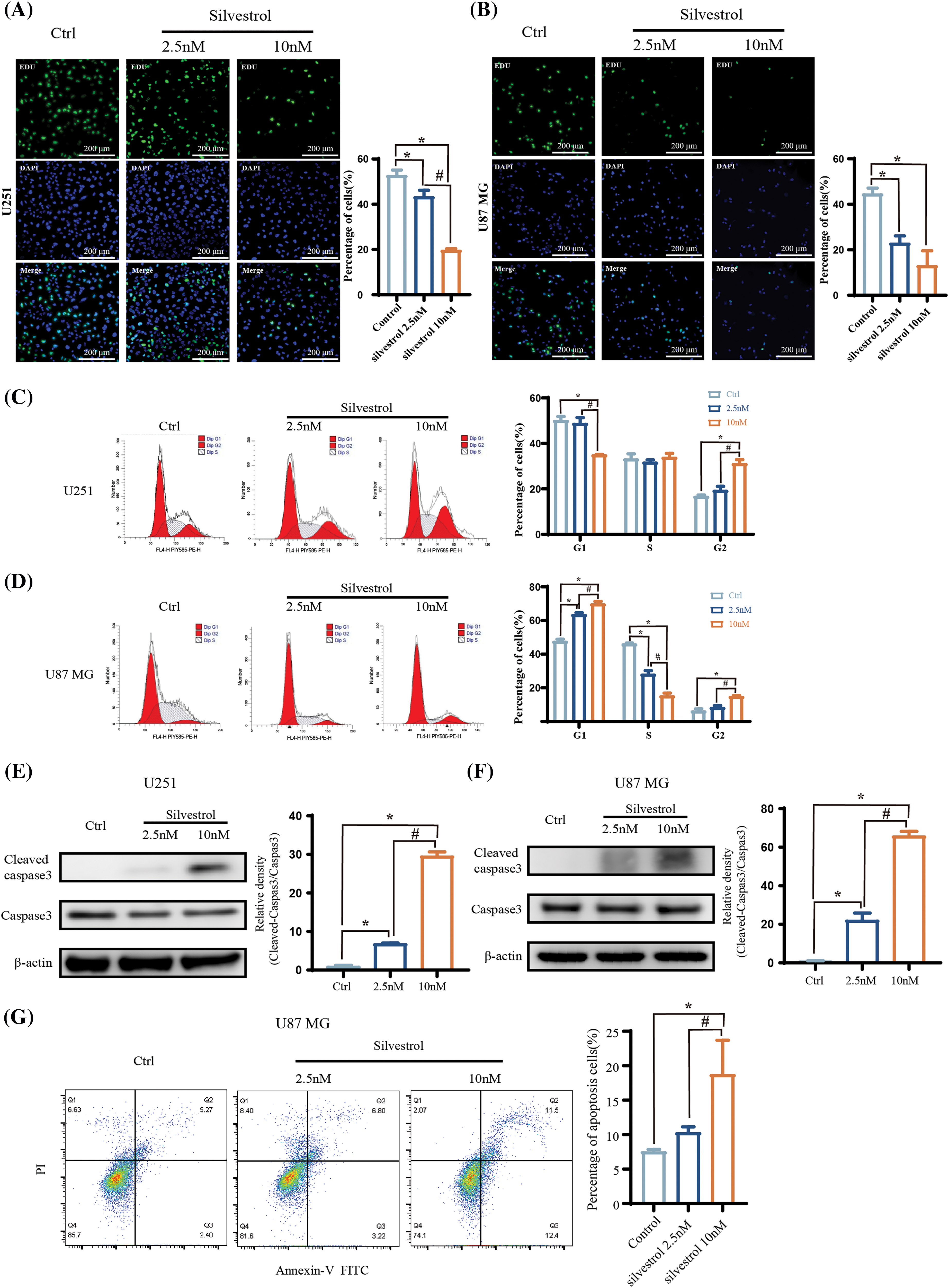
Figure 2: Effects of silvestrol on the proliferation, cycle, and apoptosis of human GBM cells. (A and B) EDU proliferation assay and the percentage of positive proliferation of U251 (A) and U87 MG (B) cells treated with silvestrol high-dose group (10 nM) and low-dose group (2.5 nM) for 24 h. (C and D) Flow cytometry of U251 (C) and U87 MG (D) cells treated with silvestrol high-dose group (10 nM) and low-dose group (2.5 nM) for 24 h. (E and F) Silvestrol high-dose group (10 nM) and low-dose group (2.5 nM) treated U251 (E), U87 MG (F) cells for 24 h, Western blotting to detect the expression levels of cleaved caspase3 and caspase3 proteins. And statistics Ratio of cleaved caspase3/caspase3 protein expression. (G) Silvestrol high-dose group (10 nM) and low-dose group (2.5 nM) were treated with U87 MG cells for 24 h, and cell apoptosis was measured by flow cytometry, and the total apoptosis rate was calculated. The values are performed as mean ± SD (n = 3 in each group). *p < 0.05 vs. ctrl group. #p < 0.05 vs. silvestrol (2.5 nM) group. Bar = 200 μm.
Silvestrol promptly activated the Ras/Raf/MEK/ERK pathway in U87 MG and U251 cell lines
We detected a significant and rapid elevation in extracellular regulated kinase (ERK) phosphorylation levels in both U251 and U87 MG glioblastoma cells within 2 h of treatment with silvestrol at a concentration of 10 nM (p < 0.05). This phosphorylation surge returned to baseline levels after 2 h. Moreover, the phosphorylation of MEK in both U251 and U87 MG cells exhibited a notable increase (p < 0.05) (Figs. 3A and 3B). Additionally, upon administering silvestrol (4 nM) for 1 h, the phosphorylation levels were diminished when an ERK inhibitor, LY3214996, at a concentration of 80 nM was applied (Figs. 3C and 3D). These observations confirm that silvestrol treatment leads to the activation of the Ras/Raf/MEK/ERK signaling pathway and that LY3214996 is effective in inhibiting this activation of the ERK pathway.
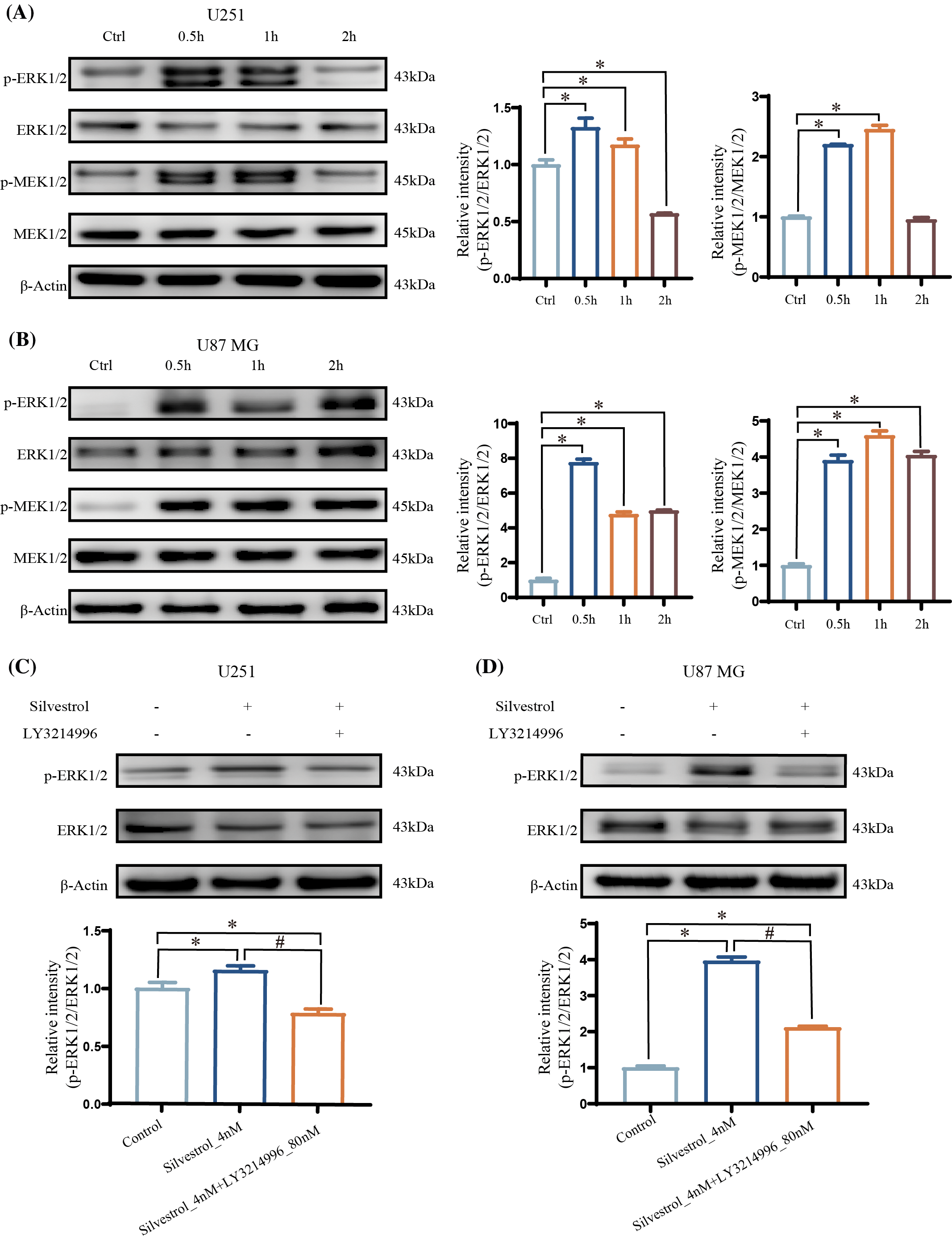
Figure 3: Western blotting to detect the expression levels of ERk, p-ERk, MEk, and p-MEk proteins, and the ratio of p-ERk/ERk and p-MEk/MEk protein expression was calculated. (A and B) The relative expression levels of p-ERk/ERk and p-MEk/MEk in U251 (A) and U87 MG (B) cells treated with silvestrol high-dose group (10 nM) within 2 h (0.5, 1, 2 h). The values are performed as mean ± SD (n = 3 in each group). *p < 0.05 vs. con group. (C and D) Relative expression levels of p-ERk/ERk in silvestrol middle dose group (4 nM) or silvestrol middle dose group (4 nM) + LY3214996 treatment for 1 h. The values are performed as mean ± SD (n = 3 in each group). *p < 0.05 vs. con group. #p < 0.05 vs. silvestrol (4 nM) group.
ERK inhibitors modulate the effects of silvestrol on human GBM cells but do not alter apoptosis induced by silvestrol in U87 MG cells
The application of an ERK inhibitor introduced several significant changes. Firstly, it reduced the antiproliferative effects of silvestrol, as evidenced in Figs. 4A and 4B. It also lessened the cell cycle arrest caused by silvestrol, as illustrated in Figs. 4C and 4D, and improved the colony formation capability of U87 MG cells, as indicated in Fig. 4E. Moreover, the presence of the ERK inhibitor decreased the reduction in U251 cell migration induced by silvestrol, leading to a notably lower percentage of wound healing distance compared to the silvestrol medium-dose group (Fig. 4F). However, it is critical to note that the increase in levels of the apoptosis-related protein, cleaved caspase3, remained largely unaffected by the treatment with silvestrol (4 nM) in conjunction with the ERK inhibitor, as shown in Figs. 4G and 4H. The ERK inhibitor did not significantly influence the early and late apoptosis in U87 MG cells promoted by silvestrol, as depicted in Fig. 4I. Despite this, the overall rate of apoptosis experienced a significant increase (p < 0.05).
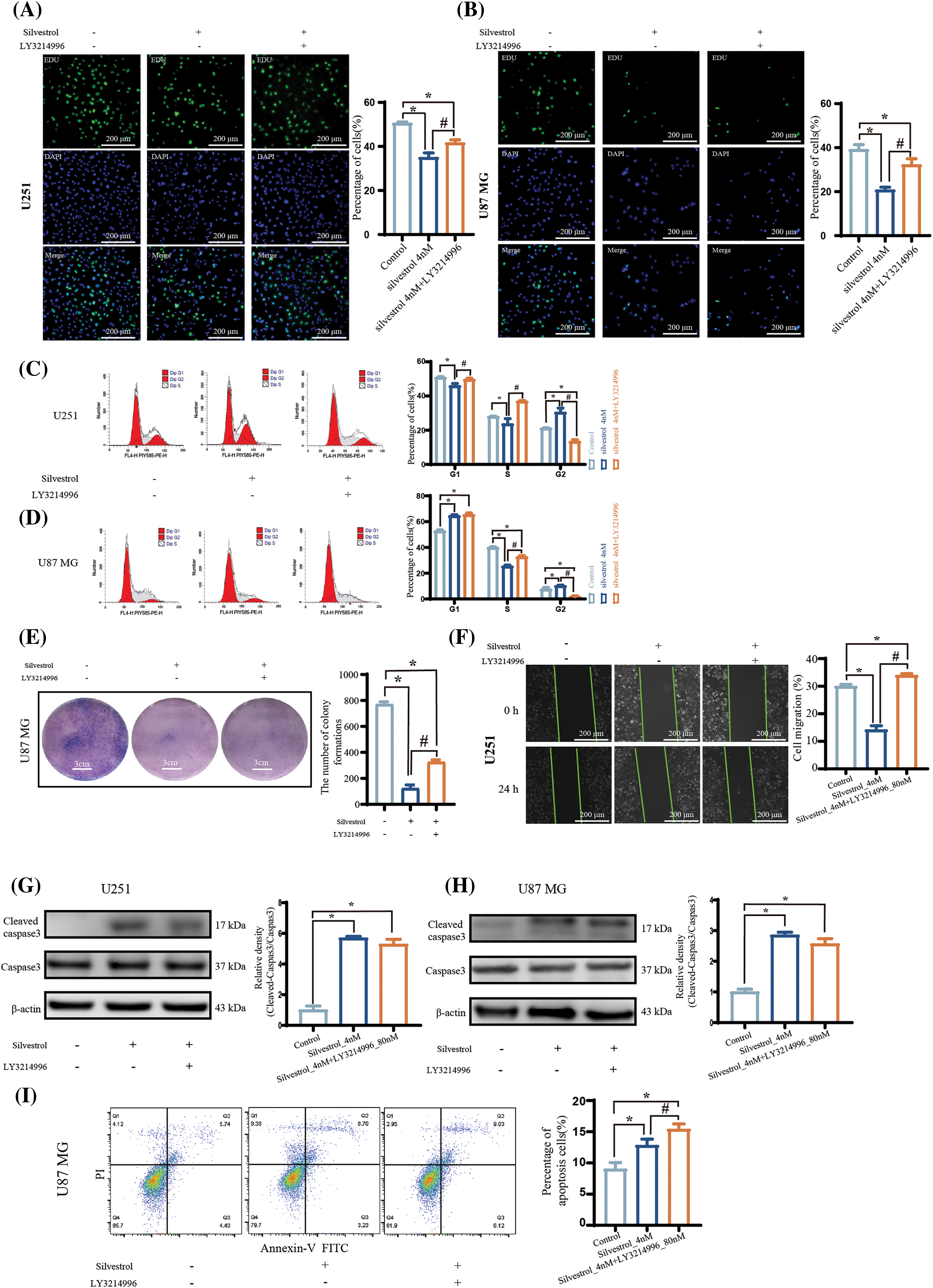
Figure 4: ERK inhibitors affect the regulation of human GBM cells by silvestrol. (A and B) EDU proliferation experiments and percentages of U251 (A) and U87 MG (B) cells treated with silvestrol medium-dose group (4 nM) or silvestrol medium-dose group (4 nM) + ERK inhibitor LY3214996 for 24 h. (C and D) U251 (C) and U87 MG (D) cells treated with silvestrol medium-dose group (4 nM) or silvestrol medium-dose group (4 nM) + LY3214996 for 24 h. and the cell cycle was measured by flow cytometry. (E) Colony formation experiments of U87 MG cells treated with silvestrol medium-dose group (4 nM) or silvestrol medium-dose group (4 nM) + LY3214996. (F) Scratch test of U251 cells treated with silvestrol medium dose group (4 nM) or silvestrol medium dose group (4 nM) + LY3214996. Alteration of the pro-apoptotic effect of silvestrol on human GBM cells after the addition of an ERK inhibitor. (G and H) Silvestrol middle-dose group (4 nM), middle-dose group (4 nM) + LY3214996 treated U251 (A), U87 MG (B) cells for 24 h, Western blotting to detect the expression levels of cleaved caspase3 and caspase3 proteins. And the relative amount of cleaved caspase3/caspase3 was counted. (I) Silvestrol middle-dose group (4 nM) and middle-dose group (4 nM) + LY3214996 were treated with U87 MG cells for 24 h, and the apoptosis was measured by flow cytometry, and the total apoptotic rate was calculated. The values are performed as mean ± SD (n = 3 in each group). *p < 0.05 vs. con group. #p < 0.05 vs. silvestrol (4 nM) group. Scale bar (A, B, and F) = 200 μm. Scale bar (E) = 3 cm.
Silvestrol alters transcriptional profiles in glioblastoma cells by inhibiting the ERK pathway
The overlap between RNA-UP and GEPIA-UP gene sets comprised 37 genes, whereas the intersection of RNA-DOWN and GEPIA-DOWN gene sets included 29 genes (Fig. 5A). Both the upregulated and downregulated hub genes implicated in catalytic activities, binding-related molecular functions, and a range of biological processes (Fig. 5B). In particular, genes that were upregulated were linked to immune diseases, cardiovascular diseases, and genetic information processing. In contrast, downregulated genes were associated with neurodegenerative diseases and resistance to anti-tumor drugs (Fig. 5C). Both hub-up genes and hub-down genes were also verified by UALCAN online tools and GSCA database (Fig. 5D and Table 1).
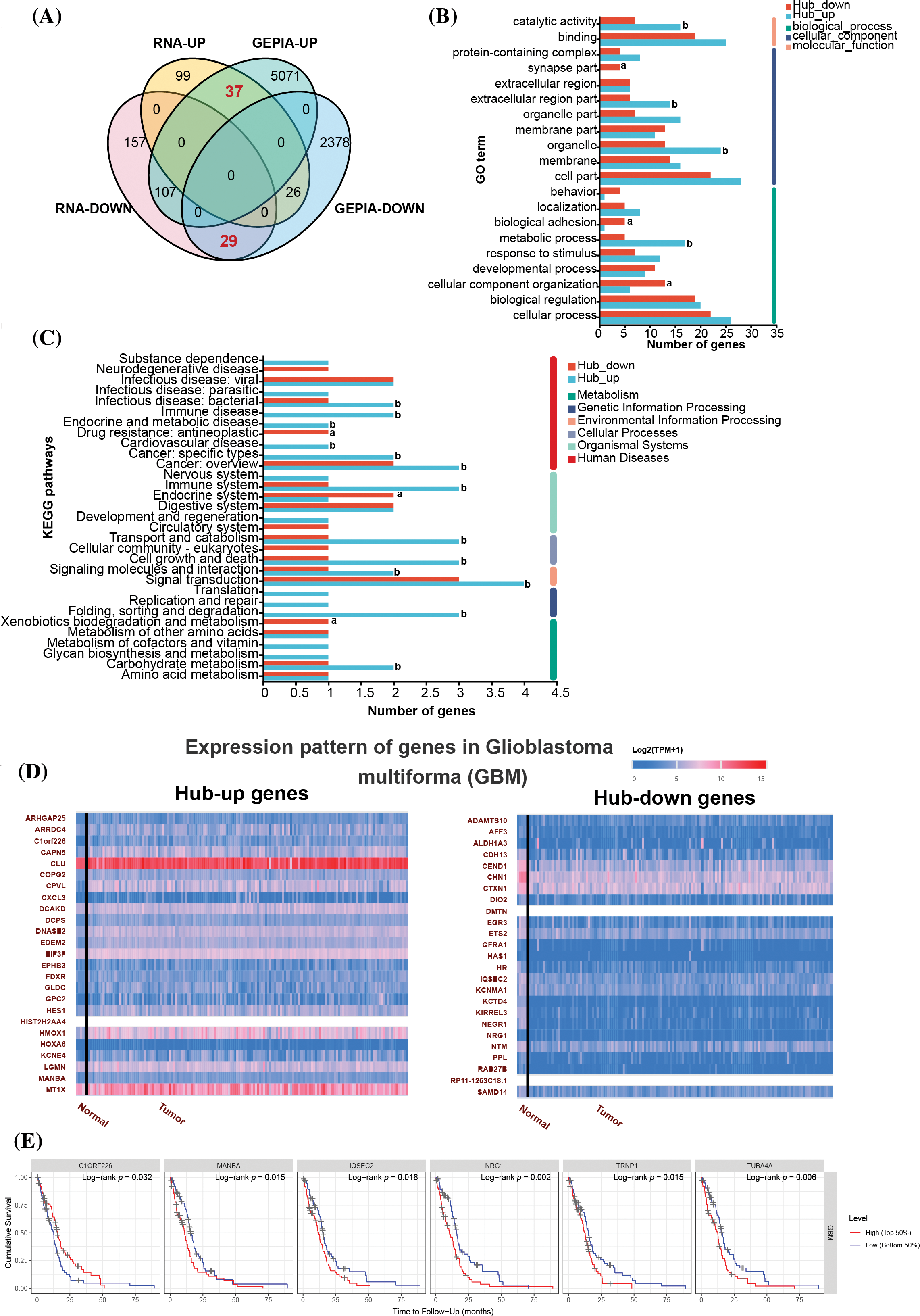
Figure 5: A Comprehensive Analysis of Transcriptomics, GEPIA Database, and Clinical Data Unveiling Hub Genes in Glioblastoma. (A) Venn diagram of RNA-seq and GEPIA databases. The GEPIA-UP and GEPIA-DOWN gene sets represent genes upregulated and downregulated in glioblastoma compared to normal tissues, identified using the GEPIA database. The RNA-UP and RNA-DOWN gene sets consist of genes upregulated and downregulated by ERK inhibitors (LY3214996), reflecting the DEGs between the silvestrol-treated group and the combined silvestrol + LY3214996-treated group. (B) GO results of Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). (C) KEGG pathway enrichment of hub differentially expressed genes (DEGs). (D) Expression pattern of hub genes in glioblastoma multiforma (GBM) using UALCAN online tool (https://ualcan.path.uab.edu/). (E) Cumulative survival is associated with specific gene expression levels in TIMER database.
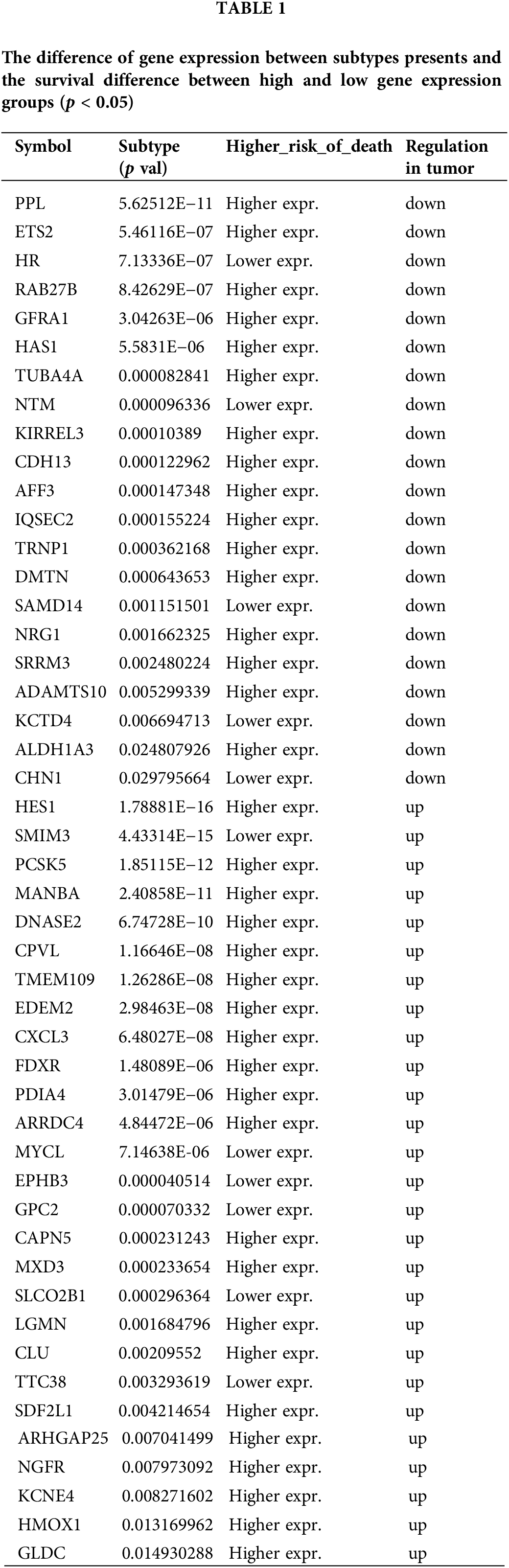
Moreover, TIMER database was used to analyze genes related to cumulative survival rates, and the results revealed that patients with high expression of the chromosome 1 open reading frame 226 (C1ORF226) gene experienced significantly higher survival rates (Log-rank p = 0.032). Conversely, patients with low expression levels of mannosidase beta A (MANBA) (Log-rank p = 0.015), IQ motif and Sec7 domain 2 (IQSEC2) (Log-rank p = 0.018), NRG1 (Log-rank p = 0.002) also showed significantly improved survival rates (Fig. 5E). Notably, C1ORF226 and MANBA were part of the upregulated hub gene set, while IQSEC2 and NRG1 were included in the downregulated hub gene set (Fig. 5D).
Key gene related to the ERK pathway and regulated by silvestrol identified
The expression levels of MANBA mRNA were significantly reduced by silvestrol at a concentration of 4 nM, and this reduction was reversed upon the introduction of LY3214996, as depicted in Fig. 6A. In contrast, silvestrol markedly increased the mRNA levels of NRG-1, and once again, the addition of LY3214996 reversed this effect, as shown in Fig. 6B. Interestingly, Western blot analysis revealed that silvestrol not only significantly decreased the protein levels of NRG-1 but also further reduced these levels upon the addition of LY3214996 (Fig. 6C). Immunofluorescence staining of cells displayed consistent patterns, with similar alterations in the levels of NRG-1 (Fig. 6D).
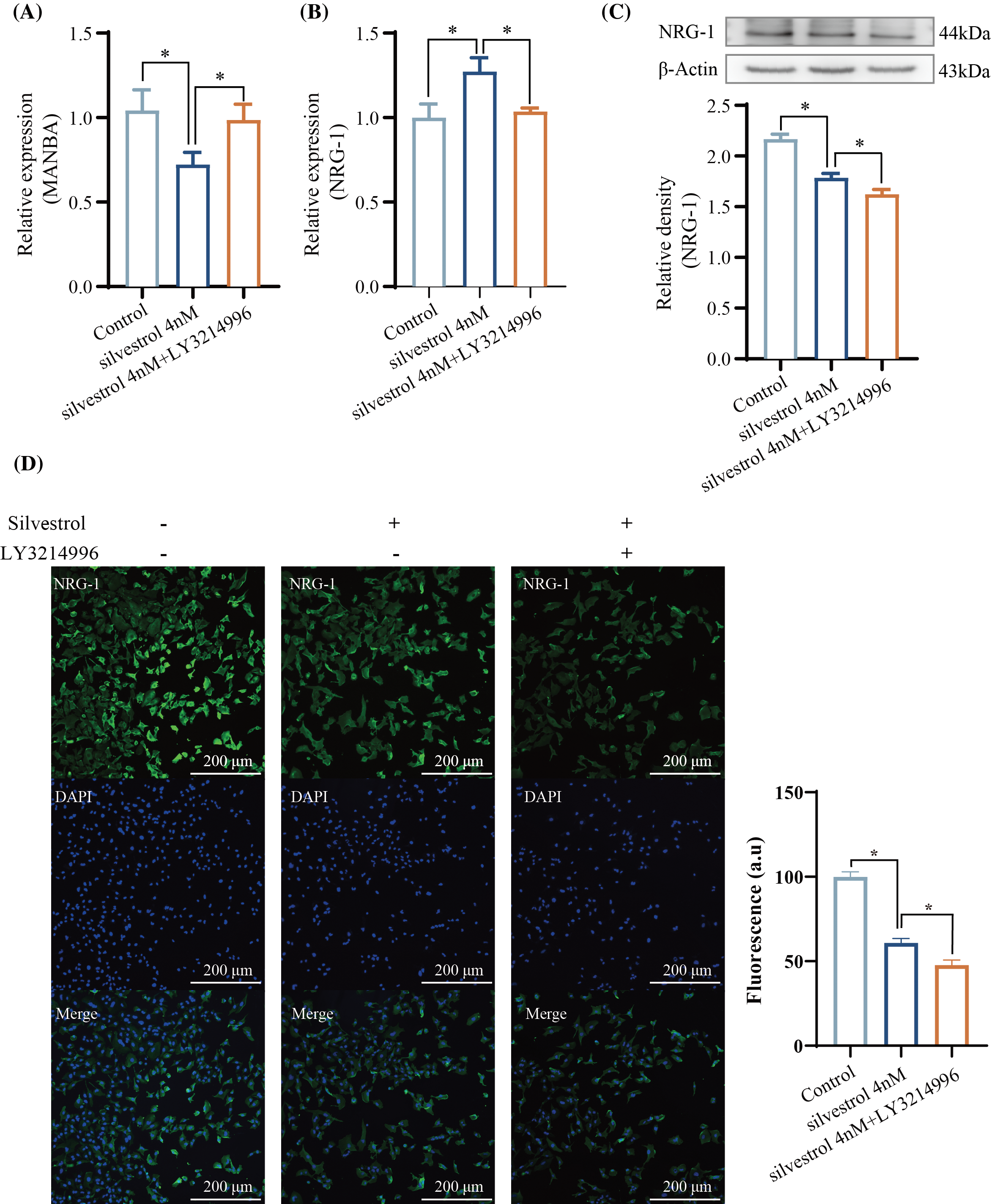
Figure 6: Regulation of MANBA and NRG-1 by Silvestrol. (A) qPCR results of MANBA. (B) qPCR results of NRG-1. (C) Western blotting results of NRG-1. (D) Immunofluorescence staining results of NRG-1. The values are performed as mean ± SD (n = 3 in each group). *p < 0.05 vs. silvestrol_4 nM. Bar = 200 μm.
Silvestrol, a small molecule extracted from natural plants, has demonstrated clear therapeutic effects on tumors, as well as significant inhibitory effects on viruses, including the hepatitis E virus [30], Ebola virus [31], and Chikungunya Virus [32]. As a potent eukaryotic initiation factor 4A (EIF4A) inhibitor, the precise antitumor and antiviral pathways of silvestrol have yet to be fully elucidated. Our findings revealed that silvestrol activates the extracellular regulated kinase (ERK) pathway in a short timeframe.
This study, through a series of experiments, has shown that silvestrol inhibits proliferation (Figs. 1A–1D, 2C and 2D), colony formation (Figs. 1E and 1F), and migration (Fig. 1G) in U87 MG and U251 glioblastoma cell lines, while also promoting apoptosis (Figs. 2E–2G). These results confirm silvestrol’s tumor-suppressive effects on glioblastoma cells. Moreover, we discovered that silvestrol induces phosphorylation of mitogen-activated protein kinase kinase (MEK) and extracellular regulated kinase (ERK) (Fig. 3), shedding light on its mechanism of action. The ERK1/2 pathway, crucial for cell proliferation and entry into the S phase from G1 [28], is primarily activated via the Ras/Raf/MEK/ERK pathway. MEK is the sole specific activator of ERK, with numerous substrates identified for the ERK pathway [33]. To delve deeper into the mechanisms, we utilized the ERK inhibitor LY3214996, which diminishes ERK phosphorylation and blocks ERK pathway activation [34]. The presence of LY3214996 attenuated silvestrol’s inhibitory effects on cell proliferation (Figs. 4A–4D), colony formation (Fig. 4E), and cell migration (Fig. 4F). Additionally, in glioblastoma cells treated with silvestrol, LY3214996 induced transcriptional changes that aligned with the gene expression profile of glioblastoma in the gene expression profiling interactive analysis (GEPIA) database (Fig. 5A), particularly in cell cycle processes (Fig. 5B) and genetic information processing (Fig. 5C). Together, these findings strongly suggest that silvestrol regulates tumor cell proliferation via the ERK pathway.
However, silvestrol’s promotion of apoptosis in glioblastoma cells was not reversed by targeting the ERK pathway (Figs. 4G and 4H). Instead, LY3214996 further intensified apoptosis in U87 MG cells (Fig. 4I), indicating that the ERK pathway might facilitate tumor cell survival, while silvestrol could employ other pathways to induce apoptosis. Further analysis of transcriptomic data and public databases identified two key genes, mannosidase beta A (MANBA) and neuregulin 1 (NRG-1), with notable expression patterns.
The MANBA gene encodes β-mannosidase and has been reported to be overexpressed in liver cancer [35], esophageal squamous cell carcinoma [36], and colorectal cancer in the Swedish population [37], though the specific signaling pathways associated with this overexpression have not been documented. Recent research has found that interfering with the expression of MANBA in glioblastoma cells significantly reduces the clonogenicity, migration, and invasion capabilities of the glioblastoma cells [38]. Public databases indicate that MANBA is overexpressed in glioblastoma patients (Fig. 5D) with high MANBA levels correlated with lower survival rates (p = 0.015) (Fig. 5E). Silvestrol decreased MANBA expression, a process reversed by LY3214996 (Fig. 6A), suggesting silvestrol’s potential to extend glioblastoma patient survival by reducing MANBA levels.
The NRG-1 gene, known to promote proliferation [39], and enhance glioblastoma cell survival [40], showed decreased expression in glioblastoma patients (Fig. 5D), intriguingly correlating lower expression with significantly higher survival rates (p = 0.002) (Fig. 5E). Silvestrol increased NRG1 mRNA levels (Fig. 6B), but decreased NRG-1 protein levels (Fig. 6C), with LY3214996 reversing mRNA changes (Fig. 6B), but further reducing the protein levels (Fig. 6C). This suggests that silvestrol might promote NRG-1 transcription through the ERK pathway but inhibit its translation, explaining the further decrease in protein levels when the ERK pathway is inhibited.
This study has limitations, notably in the speculative nature of the functions of MANBA and NRG-1 proteins, which are based on preliminary research findings and lack further experimental validation. For instance, conducting interference or compensatory experiments targeting these two genes would provide more definitive insights into their roles in silvestrol-mediated inhibition of glioblastoma or the development of glioblastoma.
Silvestrol exerts its anti-glioma effects primarily by inhibiting the expression of MANBA through the ERK pathway and potentially impeding the translation of NRG-1 protein, thereby diminishing its expression. The suppression of MANBA and NRG-1 proteins may play a critical role in inhibiting glioma development and progression. These findings underscore the complex interplay between the ERK pathway and gene expression regulation in the therapeutic efficacy of silvestrol against glioma.
Acknowledgement: None.
Funding Statement: This research was supported by the Chongqing Science and Health Joint Medical Research Project (2020FYYX150).
Author Contributions: Lan Zhou designed the experiments and performed the bioinformatic analysis. Qi Zhang and Lan Zhou performed the experiments. Qi Zhang wrote the paper. Bo Tian and Feng Yang contributed to the final version of the manuscript, as well as read and approved it. All authors read and approved the manuscript.
Availability of Data and Materials: This study did not include data deposited in external repositories. All data presented in this study are included in the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Vargas LA. Glioblastoma in adults: a society for neuro-oncology (SNO) and European society of neuro-oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2021;23(3):502–3. doi:10.1093/neuonc/noaa287. [Google Scholar] [PubMed] [CrossRef]
2. Sun J, Xu L, Zhang Y, Li H, Feng J, Lu X, et al. Long non-coding RNA DPP10-AS1 represses the proliferation and invasiveness of glioblastoma by regulating miR-24-3p/CHD5 signaling pathway. BIOCELL. 2023;47(12):2721–33. doi:10.32604/biocell.2023.043869. [Google Scholar] [CrossRef]
3. Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: a clinical review. JAMA. 2013;310(17):1842–50. doi:10.1001/jama.2013.280319. [Google Scholar] [PubMed] [CrossRef]
4. Schaff LR, Mellinghoff IK. Glioblastoma and other primary brain malignancies in adults: a review. JAMA. 2023;329(7):574–87. doi:10.1001/jama.2023.0023. [Google Scholar] [PubMed] [CrossRef]
5. Smolarska A, Pruszynska I, Wasylko W, Godlewska K, Markowska M, Rybak A, et al. Targeted therapies for glioblastoma treatment. J Physiol Pharmacol. 2023;74(3):251–61. doi:10.26402/jpp.2023.3.01. [Google Scholar] [PubMed] [CrossRef]
6. Ou A, Yung W, Majd N. Molecular mechanisms of treatment resistance in glioblastoma. Int J Mol Sci. 2020;22(1):351. doi:10.3390/ijms22010351. [Google Scholar] [PubMed] [CrossRef]
7. Kinghorn AD, Pan L, Fletcher JN, Chai H. The relevance of higher plants in lead compound discovery programs. J Nat Prod. 2011;74(6):1539–55. doi:10.1021/np200391c. [Google Scholar] [PubMed] [CrossRef]
8. Cencic R, Carrier M, Galicia-Vazquez G, Bordeleau ME, Sukarieh R, Bourdeau A, et al. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4(4):e5223. doi:10.1371/journal.pone.0005223. [Google Scholar] [PubMed] [CrossRef]
9. Song J, Ge Y, Dong M, Guan Q, Ju M, Song X, et al. Molecular interplay between EIF4 family and circular RNAs in cancer: mechanisms and therapeutics. Eur J Pharmacol. 2023;954:175867. doi:10.1016/j.ejphar.2023.175867. [Google Scholar] [PubMed] [CrossRef]
10. Bracic TS, Schatz C, Haybaeck J. Translational regulation in hepatocellular carcinogenesis. Drug Des Devel Ther. 2021;15:4359–69. doi:10.2147/DDDT.S255582. [Google Scholar] [PubMed] [CrossRef]
11. Muller C, Schulte FW, Lange-Grunweller K, Obermann W, Madhugiri R, Pleschka S, et al. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 2018;150(10):123–9. doi:10.1016/j.antiviral.2017.12.010. [Google Scholar] [PubMed] [CrossRef]
12. Schulz G, Victoria C, Kirschning A, Steinmann E. Rocaglamide and silvestrol: a long story from anti-tumor to anti-coronavirus compounds. Nat Prod Rep. 2021;38(1):18–23. doi:10.1039/D0NP00024H. [Google Scholar] [PubMed] [CrossRef]
13. Alachkar H, Santhanam R, Harb JG, Lucas DM, Oaks JJ, Hickey CJ, et al. Silvestrol exhibits significant in vivo and in vitro antileukemic activities and inhibits FLT3 and miR-155 expressions in acute myeloid leukemia. J Hematol Oncol. 2013;6(1):21. doi:10.1186/1756-8722-6-21. [Google Scholar] [PubMed] [CrossRef]
14. Mi Q, Kim S, Hwang BY, Su BN, Chai H, Arbieva ZH, et al. Silvestrol regulates G2/M checkpoint genes independent of p53 activity. Anticancer Res. 2006;26(5A):3349–56. [Google Scholar] [PubMed]
15. Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, et al. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7(3):382–94. doi:10.1017/S135583820100108X. [Google Scholar] [PubMed] [CrossRef]
16. Singh K, Lin J, Lecomte N, Mohan P, Gokce A, Sanghvi VR, et al. Targeting eIF4A-dependent translation of KRAS signaling molecules. Cancer Res. 2021;81(8):2002–14. doi:10.1158/0008-5472.CAN-20-2929. [Google Scholar] [PubMed] [CrossRef]
17. Chan K, Robert F, Oertlin C, Kapeller-Libermann D, Avizonis D, Gutierrez J, et al. eIF4A supports an oncogenic translation program in pancreatic ductal adenocarcinoma. Nat Commun. 2019;10:5151. doi:10.1038/s41467-019-13086-5. [Google Scholar] [PubMed] [CrossRef]
18. Lucas DM, Edwards RB, Lozanski G, West DA, Shin JD, Vargo MA, et al. The novel plant-derived agent silvestrol has B-cell selective activity in chronic lymphocytic leukemia and acute lymphoblastic leukemia in vitro and in vivo. Blood. 2009;113(19):4656–66. doi:10.1182/blood-2008-09-175430. [Google Scholar] [PubMed] [CrossRef]
19. Kogure T, Kinghorn AD, Yan I, Bolon B, Lucas DM, Grever MR, et al. Therapeutic potential of the translation inhibitor silvestrol in hepatocellular cancer. PLoS One. 2013;8(9):e76136. doi:10.1371/journal.pone.0076136. [Google Scholar] [PubMed] [CrossRef]
20. Lehman SL, Wechsler T, Schwartz K, Brown LE, Porco JA, Devine WG, et al. Inhibition of the translation initiation factor eIF4A enhances tumor cell radiosensitivity. Mol Cancer Ther. 2022;21(9):1406–14. doi:10.1158/1535-7163.MCT-22-0037. [Google Scholar] [PubMed] [CrossRef]
21. Webb TE, Davies M, Maher J, Sarker D. The eIF4A inhibitor silvestrol sensitizes T-47D ductal breast carcinoma cells to external-beam radiotherapy. Clin Transl Radiat Oncol. 2020;24(5):123–6. doi:10.1016/j.ctro.2020.07.002. [Google Scholar] [PubMed] [CrossRef]
22. Boussemart L, Malka-Mahieu H, Girault I, Allard D, Hemmingsson O, Tomasic G, et al. eIF4F is a nexus of resistance to anti-BRAF and anti-MEK cancer therapies. Nature. 2014;513(7516):105–9. doi:10.1038/nature13572. [Google Scholar] [PubMed] [CrossRef]
23. Malka-Mahieu H, Girault I, Rubington M, Leriche M, Welsch C, Kamsu-Kom N, et al. Synergistic effects of eIF4A and MEK inhibitors on proliferation of NRAS-mutant melanoma cell lines. Cell Cycle. 2016;15(18):2405–9. doi:10.1080/15384101.2016.1208862. [Google Scholar] [PubMed] [CrossRef]
24. Gerson-Gurwitz A, Young NP, Goel VK, Eam B, Stumpf CR, Chen J, et al. Zotatifin, an eIF4A-selective inhibitor, blocks tumor growth in receptor tyrosine kinase driven tumors. Front Oncol. 2021;11:766298. doi:10.3389/fonc.2021.766298. [Google Scholar] [PubMed] [CrossRef]
25. Martin-Vega A, Cobb MH. Navigating the ERK1/2 MAPK cascade. Biomolecules. 2023;13(10):1555. doi:10.3390/biom13101555. [Google Scholar] [PubMed] [CrossRef]
26. Zhang W, Gong P, Tian Q, Han S, Wang J, He P, et al. The eIF4A inhibitor silvestrol blocks the growth of human glioblastoma cells by inhibiting AKT/mTOR and ERK1/2 signaling pathway. J Oncol. 2022;2022:4396316. doi:10.1155/2022/4396316. [Google Scholar] [PubMed] [CrossRef]
27. McKay MM, Morrison DK. Integrating signals from RTKs to ERK/MAPK. Oncogene. 2007;26(22):3113–21. doi:10.1038/sj.onc.1210394. [Google Scholar] [PubMed] [CrossRef]
28. Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007;26(22):3227–39. doi:10.1038/sj.onc.1210414. [Google Scholar] [PubMed] [CrossRef]
29. Veisaga ML, Ahumada M, Soriano S, Acuna L, Zhang W, Leung I, et al. Anti-proliferative effect of Annona extracts on breast cancer cells. BIOCELL. 2023;47(8):1835–52. doi:10.32604/biocell.2023.029076. [Google Scholar] [CrossRef]
30. Pedroni L, Dellafiora L, Varra MO, Galaverna G, Ghidini S. In silico study on the Hepatitis E virus RNA Helicase and its inhibition by silvestrol, rocaglamide and other flavagline compounds. Sci Rep. 2022;12(1):15512. doi:10.1038/s41598-022-19818-w. [Google Scholar] [PubMed] [CrossRef]
31. Dassanayake MK, Khoo TJ, Chong CH, di Martino P. Molecular docking and in-silico analysis of natural biomolecules against Dengue, Ebola, Zika, SARS-CoV-2 variants of concern and monkeypox virus. Int J Mol Sci. 2022;23(19):11131. doi:10.3390/ijms231911131. [Google Scholar] [PubMed] [CrossRef]
32. Blum L, Geisslinger G, Parnham MJ, Grunweller A, Schiffmann S. Natural antiviral compound silvestrol modulates human monocyte-derived macrophages and dendritic cells. J Cell Mol Med. 2020;24(12):6988–99. doi:10.1111/jcmm.15360. [Google Scholar] [PubMed] [CrossRef]
33. Ullah R, Yin Q, Snell AH, Wan L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 2022;85:123–54. doi:10.1016/j.semcancer.2021.05.010. [Google Scholar] [PubMed] [CrossRef]
34. Kohler J, Zhao Y, Li J, Gokhale PC, Tiv HL, Knott AR, et al. ERK inhibitor LY3214996-based treatment strategies for RAS-driven lung cancer. Mol Cancer Ther. 2021;20(4):641–54. doi:10.1158/1535-7163.MCT-20-0531. [Google Scholar] [PubMed] [CrossRef]
35. Bosmann HB, Spataro AC, Myers MW. Serum and host liver activities of glycosidases and sialyltransferases in animals bearing transplantable tumors. Res Commun Chem Pathol Pharmacol. 1975;12(3):499–512. [Google Scholar] [PubMed]
36. Sud N, Sharma R, Ray R, Chattopadhyay T, Ralhan R. Differential expression of beta mannosidase in human esophageal cancer. Int J Cancer. 2004;112(5):905–7. doi:10.1002/ijc.20469. [Google Scholar] [PubMed] [CrossRef]
37. Gao J, Arbman G, He L, Qiao F, Zhang Z, Zhao Z, et al. MANBA polymorphism was related to increased risk of colorectal cancer in Swedish but not in Chinese populations. Acta Oncol. 2008;47(3):372–8. doi:10.1080/02841860701644052. [Google Scholar] [PubMed] [CrossRef]
38. Xing J, Cai H, Lin Z, Zhao L, Xu H, Song Y, et al. Examining the function of macrophage oxidative stress response and immune system in glioblastoma multiforme through analysis of single-cell transcriptomics. Front Immunol. 2023;14:1288137. doi:10.3389/fimmu.2023.1288137. [Google Scholar] [PubMed] [CrossRef]
39. Sorbini M, Arab S, Soni T, Frisiras A, Mehta S. How can the adult zebrafish and neonatal mice teach us about stimulating cardiac regeneration in the human heart? Regen Med. 2023;18(1):85–99. doi:10.2217/rme-2022-0161. [Google Scholar] [PubMed] [CrossRef]
40. Ritch PS, Carroll SL, Sontheimer H. Neuregulin-1 enhances survival of human astrocytic glioma cells. Glia. 2005;51(3):217–28. doi:10.1002/glia.20197. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools