 Open Access
Open Access
ARTICLE
Circular RNA circ_0003609 ameliorates hypertrophied ligamentum flavum by regulating the miR-155/SIRT1 axis
1 Department of Orthopaedics, Baoshan Branch, Ren Ji Hospital, School of Medicine Shanghai Jiao Tong University, Shanghai, 200444, China
2 Department of Orthopaedics, Ren Ji Hospital, School of Medicine Shanghai Jiao Tong University, Shanghai, 200127, China
3 Department of Operation and Anaesthesia, Ren Ji Hospital, School of Medicine Shanghai Jiao Tong University, Shanghai, 200127, China
4 Department of Orthopaedics, Huai Yuan Chang Jiu Hospital, Huaiyuan, 233499, China
* Corresponding Authors: JIANWEI CHEN. Email: ; JUNLING CHEN. Email:
(This article belongs to the Special Issue: MicroRNA as Biomarkers for Disease Diagnosis and Progression)
BIOCELL 2024, 48(6), 1001-1008. https://doi.org/10.32604/biocell.2024.050294
Received 01 February 2024; Accepted 26 March 2024; Issue published 10 June 2024
Abstract
Background: Hypertrophy of the ligamentum flavum (HLF) is a common contributor to spinal stenosis which results in significant neurological impairments. Circular RNA (circRNA) circ_0003609 has been linked to HLF; however, the exact mechanism by which it causes this disease is unclear. Methods: Circ_0003609 expressions were regulated in HLF cells by overexpression vectors and RNA interference. Cell proliferation and fibrosis-related gene expression were checked by the Cell Counting Kit-8 (CCK-8) assay and western blotting. CircBank’s prediction of the association between miR-155 and circ_0003609 was supported by a dual-luciferase reporter experiment. The function of the miR-155/sirtuin 1 (SIRT1) axis in controlling HLF fibrosis was further examined. Results: Overexpression of circ_0003609 suppressed HLF cell propagation and fibrosis compared to its silencing. It was found that circ_0003609 served as the sponge for miR-155 and that the circ_0003609/miR-155 axis controlled the fibrosis of HLF cells. It was found that circ_0003609 acted as a sponge for miR-155, regulating the fibrosis of HLF cells. Further, miR-155 targets SIRT1, and the miR-155/SIRT1 axis promotes HLF cell fibrosis. Conclusion: Circ_0003609 ameliorates hypertrophied ligamentum flavum (LF) by modulating the miR-155/SIRT1 axis, indicating a potential treatment approach for HLF.Keywords
Hypertrophy of ligamentum flavum (HLF) is a prevalent factor in spinal stenosis, resulting in severe neurological impairments [1]. The hypertrophy and fibrosis of the ligamentum flavum (LF) are crucial factors in the reduction of the spinal canal’s, diameter, thus leading to compression [1,2]. Despite advances in surgical interventions and rehabilitation, the treatment option for HLF remains challenging due to its high recurrence rate and postoperative complications. Therefore, it is crucial to comprehend the mechanisms behind the development and emergence of HLF to identify potential treatment targets.
Circular RNAs (circRNAs), which are classified as noncoding RNA, exploit covalent bonds to form closed-loop structures [3,4]. Studies showed that circRNAs significantly impact the progression of fibrotic diseases [5–7]. The study of Zhang et al. demonstrated that circPDK1 promoted the propagation, dissemination, and fibrosis of LF cells [7]. The expression profile of circRNA has revealed a significant downregulation of circ_0003609 in both HLF tissues and cells [8]. However, the precise functions of circ_0003609 in the development of HLF pathology are still not fully understood.
circRNAs function as carriers for microRNAs (miRNAs) to regulate gene expression and have a role in the development of diseases [9,10]. For example, circPDK1 acted as a sponge for miR-4731 and exerted a role in the development of HLF [7]. A previous study demonstrated that the level of miR-155 was significantly elevated in the LF of patients with lumbar spinal stenosis compared to those with lumbar disc herniation [11]. The exact function of miR-155 in HLF and its association with circ_0003609 remain ambiguous.
Small noncoding RNA, such as miRNAs, suppressed the expression of target genes at the translation level by affixing to the 3′ untranslated region (UTR) mRNAs leading to the degradation of the target mRNA [12]. Sirtuin 1 (SIRT1), a deacetylase enzyme that relies on nicotinamide adenine dinucleotide (NAD+), is essential in several cellular mechanisms, like oxidative stress, inflammation, and fibrosis [13–15]. A recent study has shown that miR-34a-5p can induce renal fibrosis by downregulating SIRT1 expression [16]. Moreover, miR-138-5p was found to reduce the migration and growth of human skin fibroblasts by targeting SIRT1 [17]. The study by Li et al. demonstrated a substantial reduction in the levels of mRNA and protein of SIRT1 in humans and mice with HLF [18]. However, research on SIRT1 in HLF is quite limited, and its function in HLF is still incomplete.
The present investigation examines the function of circ_0003609 in HLF and its fundamental mechanism, with particular emphasis on the plausible participation of the miR-155/SIRT1 axis. The level of circ_0003609 was regulated in HLF cells, and subsequent evaluation was performed to determine its impact on cell proliferation and fibrosis. Additionally, we explored the link between circ_0003609 and miR-155, as well as the function of the miR-155/SIRT1 axis in HLF fibrosis.
Collection of ligamentum flavum samples
The present investigation employed LF tissues procured from individuals who had received lumbar spine surgery for lumbar spinal stenosis in the respective hospital. To be eligible for participation in the research, the HLF tissue specimens were required to exhibit LF at the L4/5 level with a thickness greater than 4 mm, as determined by Magnetic Resonance Imaging (MRI). The study excluded individuals with cancer, heart, kidney, rheumatic, and autoimmune diseases. The research was approved by the Ethics Committee of the Baoshan Branch of Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Approval No. 2020-skt-003), and all participating patients gave informed consent.
Isolation of primary HLF cells
The isolation procedure for HLF cells was carried out following a previously described method [19]. Briefly, the HLF tissue was fragmented into small pieces approximately 0.5–1 mm3 and subsequently underwent a digestion process using type I collagenase (250 U/mL; Sigma–Aldrich, SCR103, St Louis, USA) and trypsin (0.25%; Sigma, T4799, USA) in T-25 flasks. Dulbecco’s Modified Eagle Medium (DMEM; 11320033, Gibco, California, USA) with fetal bovine serum (FBS; 10%; Gibco, 10099141C, USA) and 1% antibiotics (penicillin/streptomycin) (Gibco, 15140122, USA) was used to culture cells, and incubation was done in 5% CO2 at 37°C until they achieved confluence. The procedure involved the periodic replacement of the culture medium at three-day intervals, and the detachment of the confluent cells for sub-culturing was accomplished using trypsin. The HLF cells utilized in the experiments were obtained from the third passage.
Short hairpin RNA (shRNA) aimed at circ_0003609 (sh-#1, 5′-GGACCACAUAUUCAAUAAU-3′; sh-#2, 5′-CCGACAAGGAAUUACAAAU-3′; sh-#3, 5′-GAGGGACUCAAAUGGUAUA-3′) and the negative control shRNAs (sh-NC, 5′-CGUACUCAAAGUAGAUUUA-3′) were obtained by GenePharma (Shanghai, China). To upregulate circ_0003609 and SIRT1, the corresponding sequences were integrated into the pcDNA 3.1 expression vector. These plasmids were acquired from Vigenebio (Shandong, China). GenePharma provided the miR-155 inhibitor (5′-ACCCCUAUCACGAUUAGCAUUAA-3′), mimic (5′-UUAAUGCUAAUUGUGAUAGGGGU-3′), and negative controls NC inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′) and NC mimic (5′-UUCUCCGAACGUGUCACGUTT-3′). Transfection protocols were followed by the manufacturer using the Lipofectamine 3000 (Invitrogen, L3000150, Carlsbad, California, USA).
Gene expression analysis via quantitative real time polymerase chain reaction (qRT-PCR)
Trizol reagents (Invitrogen, 15596018CN, USA) were used to obtain the total amount of RNA from the samples and complementary DNA (cDNA) was produced by the cDNA Synthesis Kit (Toyobo, FSQ-301, Osaka, Japan) from the extracted RNA. After that, cDNA was used for the analysis of Polymerase Chain Reaction (PCR) with the SYBR Green qPCR master mix (Qiagen, 330513, Shanghai, China). The PCR reaction system comprised of 50 μL included the following components: 4.0 μL dNTPs (2.5 mM) (TaKaRa, R005A, Beijing, China), 2.0 μL cDNA (20 ng/μL), 5 μL 10 × Pyrobest buffer II (TaKaRa, R005A, Beijing, China), 2.5 μL forward primer (10 pM), 2.5 μL reverse primer (10 pM), 0.5 μL Pyrobest DNA Polymerase (TaKaRa, R005A, Beijing, China) and 33.5 μL sterile water. Primers were procured from Sangon (Shanghai, China) as follows: human circ_0003609: 5′-GCCTGTGCAGTGGAAGGAAAA-3′ (F), 5′-AGGCACTTCATGGGGTATGG-3′ (R); human miR-155: 5′-CCCGTGGAGGCCAAGTGATT-3′ (F), 5′-TTCTTTTGGGAGGCTTCGGT-3′ (R); human SIRT1: 5′-CACCCAGAAAACACAGTGCC-3′ (F), 5′-CTGAAGAATGGACCAGACGGG-3′ (R); human glyceraldehyde-3-phosphate dehydrogenase (GAPDH): 5′-ACCTCAACTACATGGCTGAGAAC-3′ (F), 5′-GTCCACCACTGACACGTTGG-3′ (R); human U6: 5′-GGGCCTATTTCCCATGATTCCT-3′ (F), 5′-GTGTTTCGTCCTTTCCACAAGA-3′ (R). The PCR procedure started with an initial step at 94°C for 4 min. 30 cycles of specific temperature and time intervals (94°C/30 s, 56°C/45 s, 72°C/15 s) were carried out. The procedure was completed with the final step at 72°C for 10 min before terminating the reaction at 4°C. To normalize controls for RNA expression levels both GAPDH and U6 were used, and data were analyzed by the 2−ΔΔCT procedure [20].
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay kit (Beyotime, C0037, Shanghai, China) measures cell proliferation. 5 × 103 HLF cells were allowed to grow in a 96-well plate and transfected with the indicated shRNAs or plasmids. CCK-8 solution (10 μL) was transferred at 24, 48, and 72 h after transfection, and incubation was done for 2 h of at 37°C. The microplate reader (BioTek, EPOCH2, Vermont, USA) detected the absorbance at 450 nm.
Protein extraction was carried out by radioimmunoprecipitation assay (RIPA) buffer (Beyotime, P0013E, Shanghai, China) augmented with phosphatase inhibitors and protease (Beyotime, P1046, Shanghai, China) from HLF cells. The protein concentration was observed through the use of a bicinchoninic acid (BCA) assay (Beyotime, P0012, Shanghai, China). 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), separated the proteins, followed by their deposition onto polyvinylidene fluoride (PVDF) membranes (Millipore, IEVH07850, Billerica, USA). A blocking solution, comprising 5% non-fat milk in Tris-buffered saline and 0.1% Tween-20 (TBST), was used to saturate the PVDF membranes of insignificant protein-binding sites. The membranes were allowed to stay in the blocking solution for 1 h at 25°C. After this, the primary antibodies including collagen I (1:1500, ab279711); collagen III (1:1500, ab6310); transforming growth factor-beta1 (TGF-β1) (1:1500, ab215715); SIRT1(1:2000, ab110304); GAPDH (1:5000, ab8245) and c-myc (1:1000, ab32072; Abcam) were added and incubated for 24 h at 4°C. All these antibodies were purchased from Abcam (Cambridge, UK). Following the TBST washing step, the membranes underwent incubation with Horseradish Peroxidase (HRP) linked secondary antibodies (Proteintech, RGAR001, Wuhan, China) at 25°C for 1 h. The visualization was carried out through the utilization of enhanced chemiluminescence (ECL) reagents (Millipore, WBULS0100, Billerica, USA), and subsequent analysis was carried out via ImageJ software (NIH, Bethesda, MD, USA).
Dual-luciferase reporter assay
Wild-type (WT) and mutant (MUT) plasmids were generated for luciferase detection. To create the recombinant circ_0003609 WT and SIRT1 3′UTR WT plasmids, psiCHECK-2 vectors were used to clone the anticipated miR-155 binding sites from the circ_0003609 or SIRT1 3′UTR sequences (Promega, Madison, WI, USA). Additionally, by incorporating the modified sequence (site mutation in miR-155 binding sites) into the psiCHECK-2 vector, the circ_0003609 MUT and SIRT1 3′UTR MUT plasmids were obtained. Using Lipofectamine 3000 (Invitrogen, L3000150, Carlsbad, California, USA), HLF cells were co-transfected with the miR-155 mimic or NC mimic and the WT or MUT reporter plasmids. We evaluated luciferase activity after 48 h using the Dual-Luciferase Reporter Assay System (Promega, E1910, Madison, WI, USA) as per the instructions. These results were normalized via the activity of Renilla luciferase.
Data were obtained from triplicate experiments and documented in the mean ± standard deviation (SD) format. All statistical tests were conducted via GraphPad Prism 8 (GraphPad Software, La Jolla, CA, USA). Data was statistically analyzed and compared through the student’s t-test or the one-way analysis of variance (ANOVA) followed by Tukey’s post hoc. Statistical significance was attributed to p < 0.05.
Circ_0003609 regulates HLF cell proliferation and fibrosis
To investigate the importance of circ_0003609 in HLF, we modulated its production in HLF cells using shRNAs and overexpression vectors. The efficacy of knockdown (Fig. 1A) and overexpression (Fig. 1B) of circ_0003609 was verified via qRT-PCR. Among the shRNA constructs that were assessed, sh-#1 exhibited the highest level of knockdown efficacy and was consequently employed in subsequent investigations. The silencing of circ_0003609 significantly promoted HLF cell proliferation. In contrast, overexpression of circ_0003609 inhibited cell proliferation, as revealed by the CCK-8 assay (Fig. 1C). Further, overexpression of circ_0003609 decreased the level of the cell proliferation-related protein c-myc in HLF cells (Figs. 1D and 1E). Western blotting analysis displayed that the downregulation of circ_0003609 caused an upsurge in the levels of the TGF-β1, collagen I, and III proteins associated with fibrosis. Conversely, these proteins were downregulated by the overexpression of circ_0003609 (Figs. 1F and 1G).
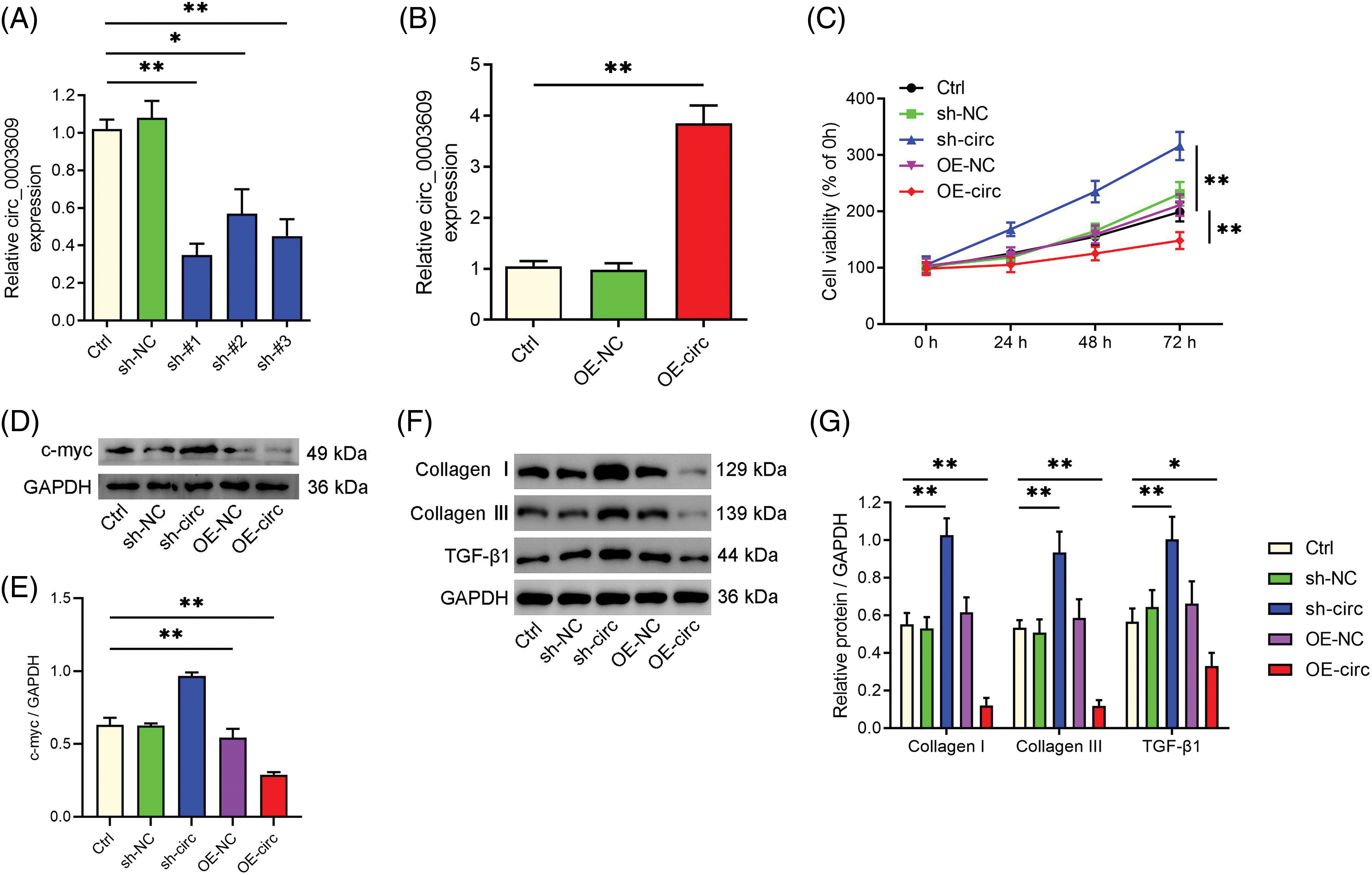
Figure 1: The silencing of circ_0003609 aggravated and overexpression alleviated the proliferation and fibrosis of hypertrophy of ligamentum flavum (HLF) cells in vitro. (A and B) HLF cells transfected with short hairpin RNA (shRNA) against circ_0003609 and circ_0003609 overexpression plasmid, and the expression of circ_0003609 were confirmed by quantitative real-time polymerase chain reaction (qRT-PCR). (C) The growth curves of HLF cells transfected with the specified vector. Cell counting kit-8 (CCK-8) assay was used to analyze cell viability. (D and E) The protein levels of c-myc in HLF cells were measured via western blotting assay. A significant difference was observed in comparison to the control group (Ctrl), with **p < 0.01. (F and G) The protein expression data of fibrosis-related genes (collagen I, collagen III, and TGF-β1) in HLF cells with the specific vectors were confirmed by the western blotting technique. *p < 0.05 and **p < 0.01 compared to the Ctrl group. The results are presented as mean ± standard deviation (SD) (n = 3). Ctrl, control; sh-NC, corresponding negative control of circ_0003609; sh-#1/2/3, the shRNAs against circ_0003609; OE-NC, corresponding negative control of circ_0003609; OE-circ, the overexpression plasmid of circ_0003609; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TGF-β1, transforming growth factor-beta1.
Sponge-like function of circ_0003609 for miR-155
To investigate the mechanism that underlies the impact of circ_0003609 on HLF cell proliferation and fibrosis, we employed the circBank database (http://www.circbank.cn/) to anticipate prospective miRNA binding sites within circ_0003609. Circ_0003609 was observed as the target for miR-155 (Fig. 2A). In HLF cells, transfection results revealed that miR-155 expression was substantially upregulated after transfection with a miR-155 mimic. In contrast, it was considerably downregulated after transfection with a miR-155 inhibitor (Fig. 2B). Assay findings revealed that the luciferase activity of the WT circ_0003609 reporters was reduced after co-transfection with the miR-155 mimic. Still, the treatment unaffected the luciferase activity of the MUT circ_0003609 reporters (Fig. 2C). The data in Fig. 2D illustrates that the expression of miR-155 was found to be increased upon suppression of circ_0003609 and decreased upon overexpression of circ_0003609. The findings indicate that circ_0003609 operates as a miR-155 sponge.
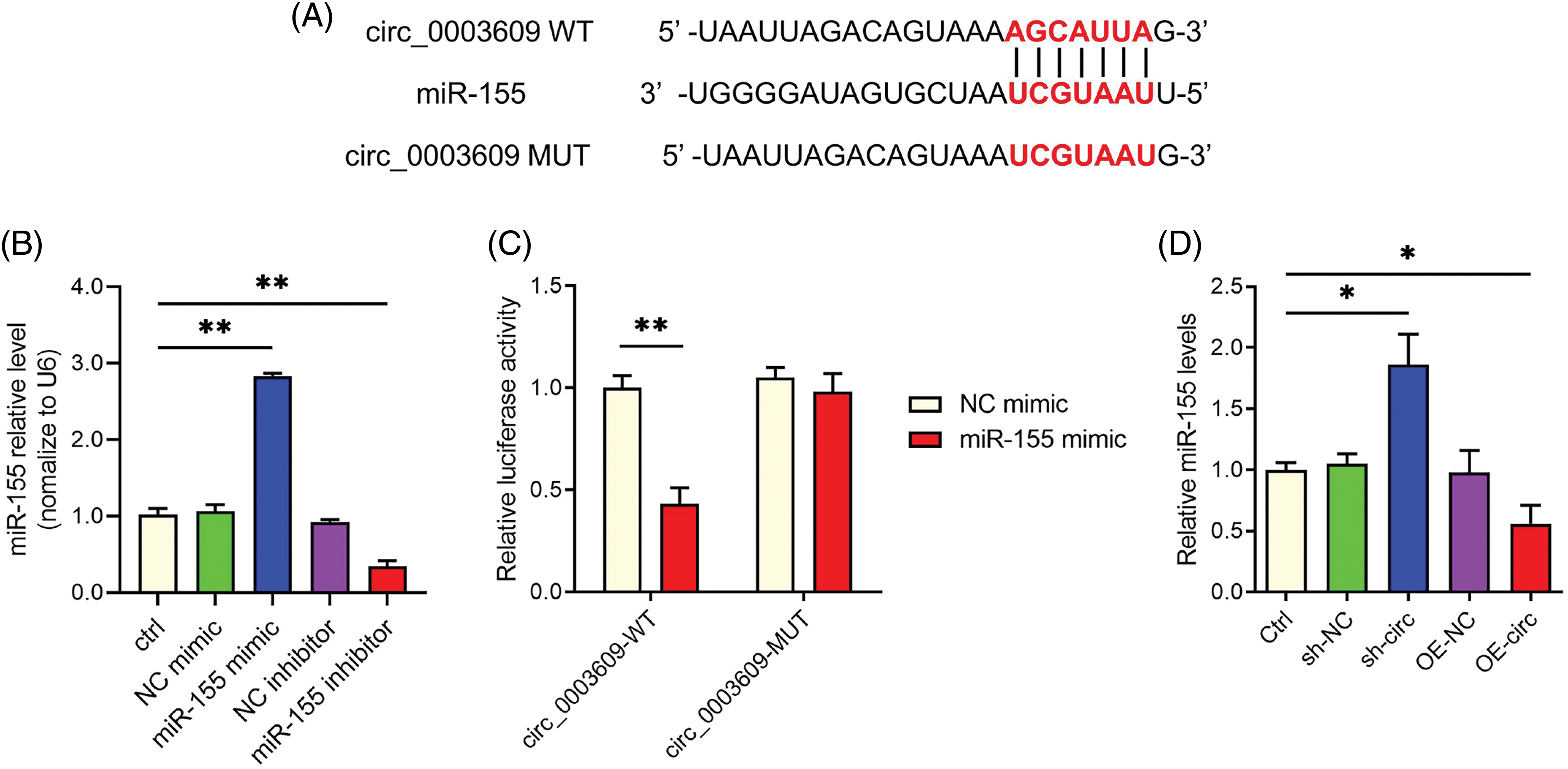
Figure 2: Circ_0003609 functions as a sponge for miR-155. (A) CircBank data show the interaction sites linking circ_0003609 and miR-155. (B) Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the expression of miR-155. **p < 0.01 compared to the Ctrl group. (C) The interaction between circ_0003609 and miR-155 in hypertrophy of ligamentum flavum (HLF) cells was analyzed using a dual-luciferase reporter assay. **p < 0.01 compared to the NC mimic group. (D) The expression level of miR-155 was measured by qRT-PCR after transfection with sh-NC, sh-circ, OE-NC, and OE-circ. *p < 0.05 compared to the Ctrl group. The results are presented as mean ± SD (n = 3). Ctrl, control; NC mimic, corresponding negative control of miR-155 mimic; NC inhibitor, corresponding negative control of miR-155 inhibitor; sh-NC, corresponding negative control of circ_0003609; sh-circ, the shRNA against circ_0003609; OE-NC, corresponding negative control of circ_0003609; OE-circ, the overexpression plasmid of circ_0003609; WT, wild-type; MUT, mutant.
Circ_0003609/miR-155 axis regulated the fibrosis of HLF cells
The experiment involved transfecting HLF cells with an overexpression plasmid of circ_0003609 (OE-circ), OE-circ + miR-155 mimic, and miR-155 mimic, to assess their effects on proteins related to fibrosis, namely TGF-β1, collagen I, and III (Fig. 3). TGF-β1, collagen I, and collagen III was reduced in cells transfected with circ_0003609 OE-circ compared to control cells (Ctrl). However, their expression increased when OE-circ and miR-155 mimics were co-transfected, indicating that the upregulation of miR-155 may offset the fibrotic effects blocked by circ_0003609 overexpression.
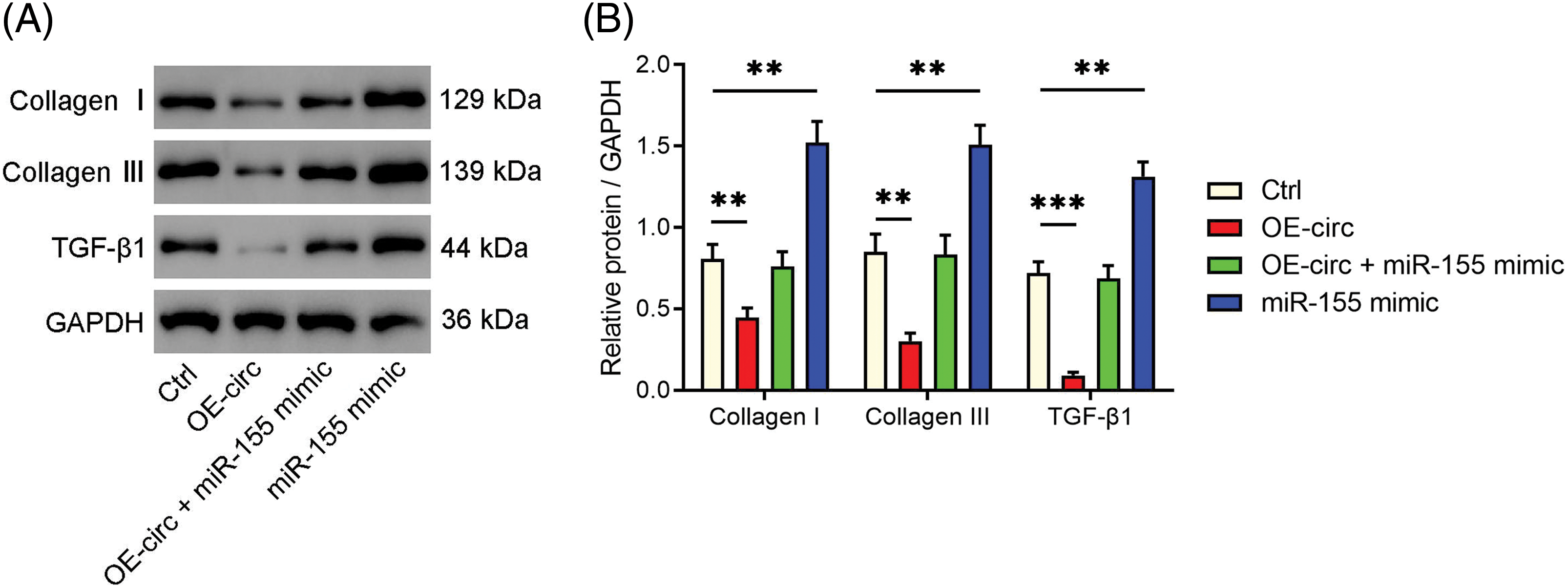
Figure 3: Circ_0003609/miR-155 axis regulated the fibrosis of hypertrophy of ligamentum flavum (HLF) cells. (A and B) HLF cells were transfected with OE-circ, OE-circ + miR-155 mimic, and miR-155 mimic. Western blotting was performed to confirm the protein expression of fibrosis-related genes (collagen I, collagen III, and TGF-β1) in each group. **p < 0.01, and ***p < 0.001 compared to Ctrl group. The results are presented as mean ± SD (n = 3). Ctrl, control; OE-circ, the overexpression plasmid of circ_0003609; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; TGF-β1, transforming growth factor-beta1.
The miR-155/SIRT1 axis regulates HLF fibrosis
Next, we examined miR-155’s target and its connection to HLF fibrosis. SIRT1, a recognized regulator of fibrosis [21], was identified by bioinformatics analysis as a potential miR-155 target (Fig. 4A). Using a dual-luciferase reporter experiment, we verified this interaction (Fig. 4B). Furthermore, we observed that overexpressing miR-155 downregulated SIRT1 levels in HLF cells while inhibiting miR-155 upregulated SIRT1 expression (Figs. 4C and 4D). Importantly, SIRT1 overexpression reversed the pro-fibrotic effect of miR-155 (Figs. 5A–5D), demonstrating that the miR-155/SIRT1 axis controls the development of HLF fibrosis. The impact of circ_0003609 on SIRT1 expression was then validated. Increased circ_0003609 led to higher SIRT1 protein expression, while decreased circ_0003609 resulted in lower SIRT1 protein levels (Figs. 5E and 5F). It was found that circ_0003609 has the potential to reduce hypertrophic LF by modulating the miR-155/SIRT1 axis.
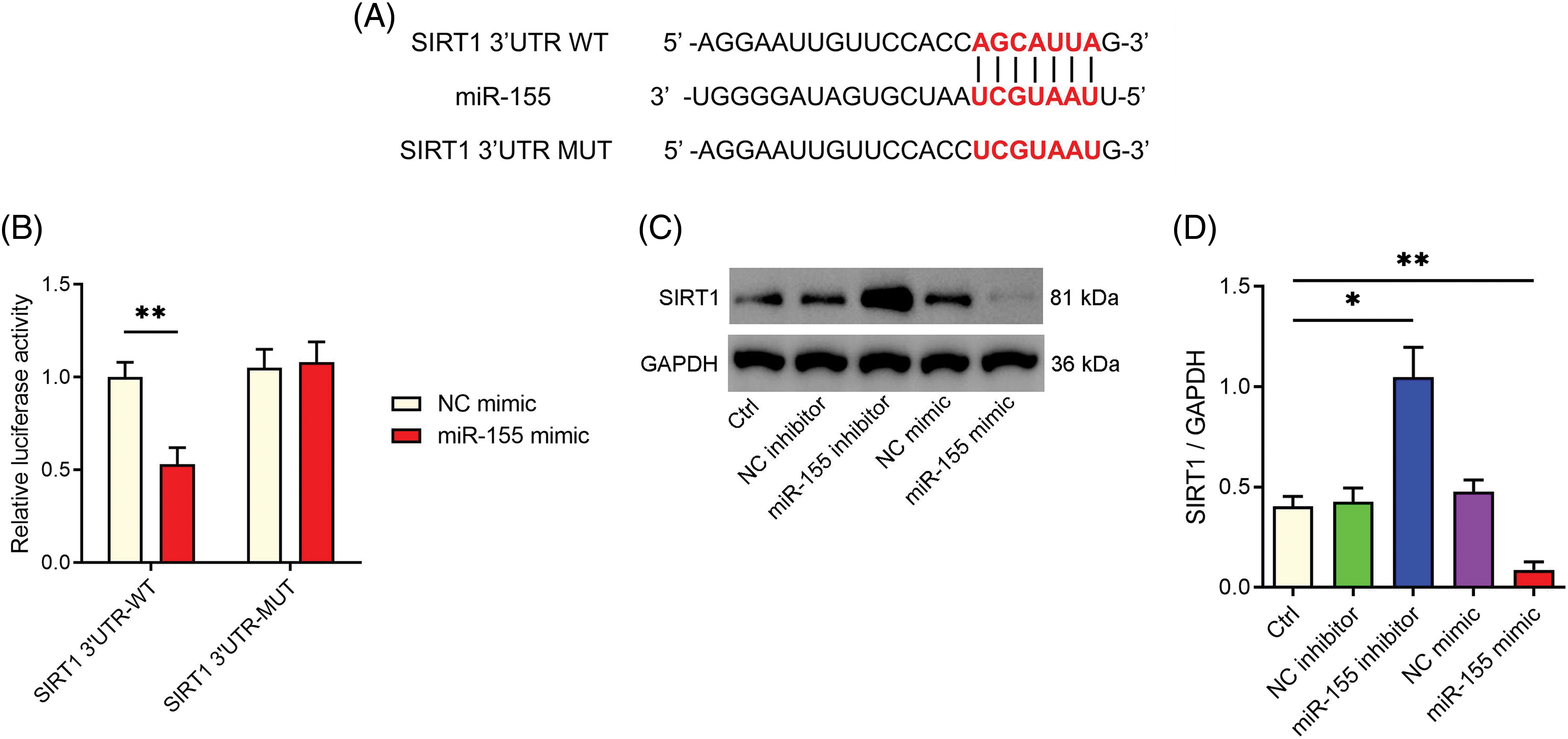
Figure 4: MiR-155 was targeted to SIRT1 in downstream regulation. (A) The starbase (https://rnasysu.com/encori/) predicted the potential binding between SIRT1 and miR-155. (B) The interaction between SIRT1 and miR-155 was confirmed through a dual-luciferase reporter assay. **p < 0.01 compared to the NC mimic group. (C and D) SIRT1 protein levels were determined by western blotting following transfection with miR-155 inhibitor and mimic, as well as their corresponding control groups. *p < 0.05 and **p < 0.01 compared to Ctrl group. The results are presented as mean ± SD (n = 3). Ctrl, control; NC mimic, corresponding negative control of miR-155 mimic; NC inhibitor, corresponding negative control of miR-155 inhibitor; WT, wild-type; MUT, mutant; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SIRT1, sirtuin 1.
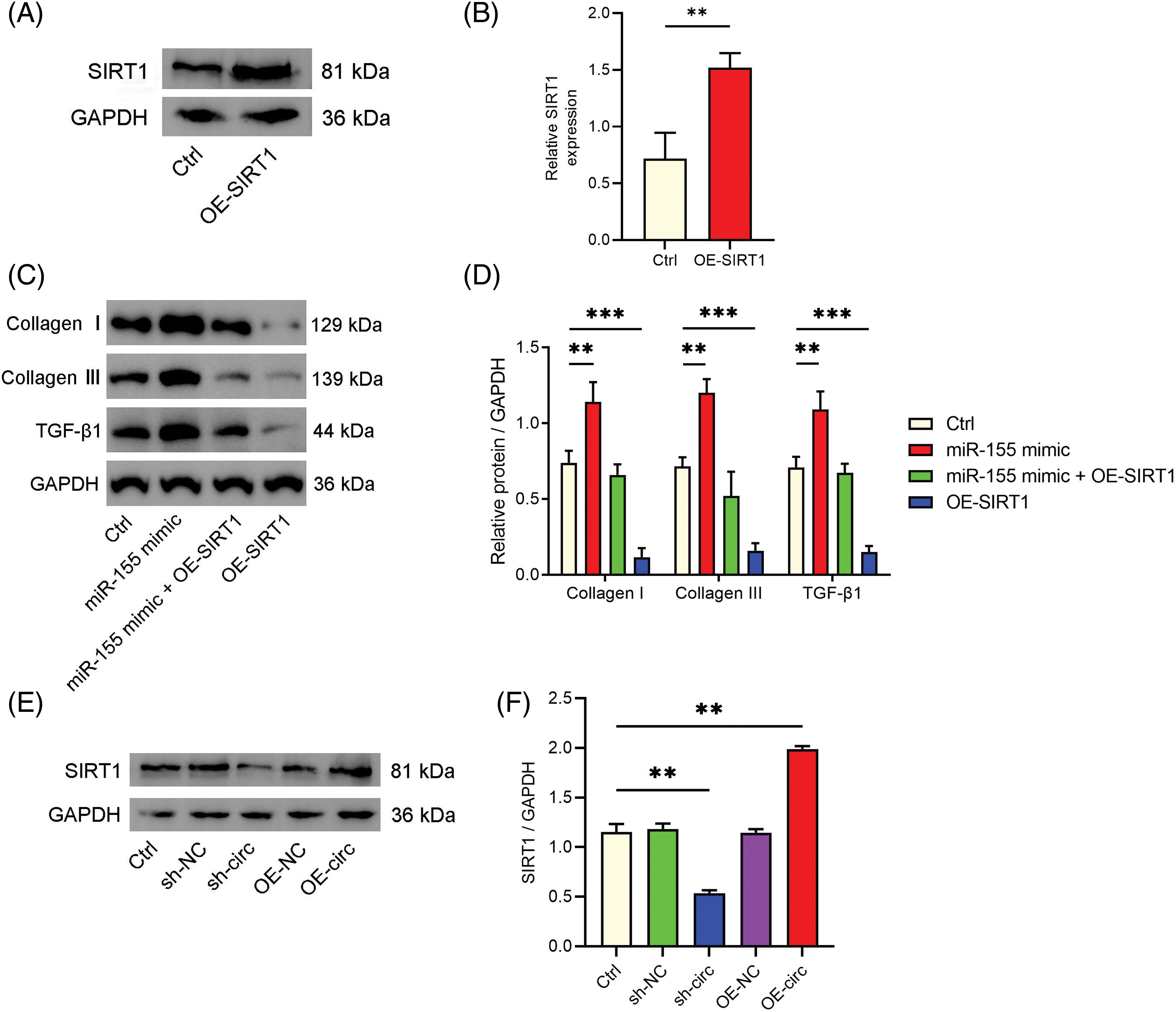
Figure 5: The miR-155/SIRT1 axis was involved in the hypertrophy of ligamentum flavum (HLF) fibrosis. (A and B) Western blotting was used to detect the transfection efficiency of SIRT1 overexpression. (C and D) The protein expression of fibrosis-related genes (collagen I, collagen III, and TGF-β1) in HLF cells transfected with miR-155 mimic, miR-155 mimic + OE-SIRT1, or OE-SIRT1 vectors were confirmed by western blotting. (E and F) The protein expression of SIRT1 was confirmed by western blotting after transfection with sh-NC, sh-circ, OE-NC, and OE-circ. **p < 0.01, and ***p < 0.001 upon comparison to Ctrl group. The results are presented as mean ± SD (n = 3). Ctrl, control; OE-SIRT1, the overexpression plasmid of SIRT1; sh-NC, corresponding negative control of circ_0003609; sh-circ, the shRNA against circ_0003609; OE-NC, corresponding negative control of circ_0003609; OE-circ, the overexpression plasmid of circ_0003609; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; SIRT1, sirtuin 1; TGF-β1, transforming growth factor-beta1.
The present investigation delved into the function of circ_0003609 in HLF and its associated mechanism. Our findings demonstrated that circ_0003609 ameliorates HLF by modulating the miR-155/SIRT1 axis. The aforementioned offer innovative perspectives on the molecular pathways that underlie HLF and propose plausible targets for therapeutic intervention in HLF.
CircRNAs are associated with diverse biological phenomena such as cellular differentiation, proliferation, and programmed cell death [3]. The involvement of circ_0003609 in the pathogenesis of HLF has been previously reported within the relevant literature [8]. However, the precise mechanism by which circ_0003609 contributes to HLF remained unclear. The findings of our investigation indicate that the inhibition of circ_0003609 resulted in an exacerbation of fibrosis and cell proliferation in HLF. Conversely, the upregulation of circ_0003609 had an opposed effect, suggesting a defensive function for circ_0003609 in HLF. Recent studies have established that circRNAs serve a role in modulating cell growth and fibrotic processes, which is consistent with the current findings [22,23]. For example, circ_0000672 was shown to promote cardiac fibrosis by its function as a miR-516a-5p sponge leading to the upregulation of TNF receptor-associated factor 6 (TRAF6), the target gene [24]. The study reported that circPDK1 facilitated the promotion of HLF through sponging miR-4371-5p, resulting in upregulating the target gene, Tenascin XB (TNXB) [7].
CircRNAs are recognized for their role in sequestering miRNAs and regulating the expression of target genes [25]. The present study delved deeper into the molecular mechanism that underlies the impact of circ_0003609 on HLF and subsequently on miR-155 as a plausible target of circ_0003609. MiR-155 has diverse molecular processes, such as immune response, inflammation, and fibrosis [26–28]. Furthermore, prior research has indicated that the suppression of miR-155 can mitigate fibrosis in diverse experimental paradigms [28–30]. The findings indicate circ_0003609 operates as a miR-155 sponge. The co-transfection with miR-155 mimic also mitigated the anti-fibrotic impacts of circ_0003609 overexpression. Based on the findings, the circ_0003609/miR-155 axis has a significant effect on the regulation of HLF fibrosis.
Furthermore, the important part of the miR-155/SIRT1 axis in controlling HLF fibrosis was explored. SIRT1 has been documented to cause modulation of fibrosis across diverse tissues, like the heart [31], liver [32], kidney [16], and lung [33]. We demonstrated that miR-155 is capable of targeting SIRT1 and that the miR-155/SIRT1 axis plays a significant part in the facilitation of fibrosis in HLF cells. The results indicate that the circ_0003609/miR-155/SIRT1 axis could serve as a promising therapeutic option for HLF.
In summary, our research has exhibited that circ_0003609 mitigates hypertrophied LF via the modulation of the miR-155/SIRT1 axis. This study was the first to identify the crucial role of the circ_0003609/miR-155/SIRT1 axis in HLF. This finding offers a novel perspective on the molecular process of HLF and indicates possible targets for medical intervention, paving the way for exploring possibilities in addressing comparable. The previous finding offers a new perspective on the molecular pathways involved in HLF and proposes a potential therapeutic intervention for this ailment. Subsequent research endeavors should probe deeper into the curative capabilities of directing attention toward the circ_0003609/miR-155/SIRT1 axis in HLF.
However, it is important to acknowledge the limitations of this study. Firstly, our research utilized HLF cells exclusively for in vitro experimentation. While cellular studies can offer initial insights into the molecular mechanisms underlying the regulation of HLF by the circ_0003609/miR-155/SIRT1 axis, it is essential to recognize that there may be discrepancies compared to the in vivo setting. Additionally, our investigation focused solely on the interplay between circ_0003609, miR-155, and SIRT1 in HLF regulation, without delving into the downstream signaling pathways. Given these constraints, further refinement of the methodology is imperative for future inquiries.
Acknowledgement: None.
Funding Statement: This research was supported by the Shanghai Natural Science Fund (No. 21ZR1447500) and Shanghai Jiao Tong University School of Medicine Affiliated Renji Hospital Baoshan Branch Medical Key Specialty Construction Project (No. rbzdzk-2023-001).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Jianwei Chen and Junling Chen; data collection: Guibin Zhong, Shurong Wang, Yujin He, Daming Feng, Ke Wei and Yanqiu Yang; analysis and interpretation of results: Guibin Zhong, Shurong Wang, Yujin He, Daming Feng, Ke Wei and Yanqiu Yang; draft manuscript preparation: Guibin Zhong and Shurong Wang. All authors reviewed and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: The research was approved by the Ethics Committee of the Baoshan Branch of Renji Hospital Affiliated to Shanghai Jiao Tong University School of Medicine (Approval No. 2020-skt-003), and all participating patients gave informed consent.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Sun C, Zhang H, Wang X, Liu X. Ligamentum flavum fibrosis and hypertrophy: molecular pathways, cellular mechanisms, and future directions. FASEB J. 2020;34(8):9854–68. doi:10.1096/fj.202000635R [Google Scholar] [PubMed] [CrossRef]
2. Byvaltsev VA, Kalinin AA, Hernandez PA, Shepelev VV, Pestryakov YY, Aliyev MA, et al. Molecular and genetic mechanisms of spinal stenosis formation: systematic review. Int J Mol Sci. 2022;23(21):13479. doi:10.3390/ijms232113479 [Google Scholar] [PubMed] [CrossRef]
3. Jiao L, Liu Y, Yu XY, Pan X, Zhang Y, Tu J, et al. Ribosome biogenesis in disease: new players and therapeutic targets. Signal Transduct Target Ther. 2023;8(1):15. doi:10.1038/s41392-022-01285-4 [Google Scholar] [PubMed] [CrossRef]
4. Li Z, Yu H, Liu Y, Wu W, Zeng H, Li E. Hsa_circ_0036740 in familial adenomatous polyposis: immune regulation and neutrophil effects in CRC based on the high-throughput assay. BIOCELL. 2023;47(11):2409–22. doi:10.32604/biocell.2023.031186 [Google Scholar] [CrossRef]
5. Li T, Xing Y, Zhang G, Wang Y, Wei Y, Cui L, et al. Circular RNA Plasmacytoma Variant Translocation 1 (CircPVT1) knockdown ameliorates hypoxia-induced bladder fibrosis by regulating the miR-203/Suppressor of Cytokine Signaling 3 (SOCS3) signaling axis. Bioengineered. 2022;13(1):1288–303. doi:10.1080/21655979.2021.2001221 [Google Scholar] [PubMed] [CrossRef]
6. Wei L, Liu L, Bai M, Ning X, Sun S. CircRNAs: versatile players and new targets in organ fibrosis. Cell Commun Signal. 2023;21(1):90. doi:10.1186/s12964-023-01051-1 [Google Scholar] [PubMed] [CrossRef]
7. Zhang K, Wang X, Zeng LT, Yang X, Cheng XF, Tian HJ, et al. Circular RNA PDK1 targets miR-4731-5p to enhance TNXB expression in ligamentum flavum hypertrophy. FASEB J. 2023;37(5):e22877. doi:10.1096/fj.202200022RR [Google Scholar] [PubMed] [CrossRef]
8. Chen J, Yu X, Qiu M, Feng F, Liu Z, Zhong G. Circular RNA expression profile in patients with lumbar spinal stenosis associated with hypertrophied ligamentum flavum. Spine. 2021;46(17):E916–25. doi:10.1097/brs.0000000000003975 [Google Scholar] [PubMed] [CrossRef]
9. Grafanaki K, Grammatikakis I, Ghosh A, Gopalan V, Olgun G, Liu H, et al. Noncoding RNA circuitry in melanoma onset, plasticity, and therapeutic response. Pharmacol Ther. 2023;248:108466. doi:10.1016/j.pharmthera.2023.108466 [Google Scholar] [PubMed] [CrossRef]
10. Niu C, Sun R, Li X, Li B, He X. MiR-19a-3p/PTEN axis regulates the anticancer effect of circHIAT1 in breast cancer in vitro. BIOCELL. 2023;47(10):2301–12. doi:10.32604/biocell.2023.029935. [Google Scholar] [CrossRef]
11. Chen J, Liu Z, Zhong G, Qian L, Li Z, Qiao Z, et al. Hypertrophy of ligamentum flavum in lumbar spine stenosis is associated with increased miR-155 level. Dis Markers. 2014;2014:786543. doi:10.1155/2014/786543 [Google Scholar] [PubMed] [CrossRef]
12. Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B, et al. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11(3):345–52. doi:10.7150/ijbs.10921 [Google Scholar] [PubMed] [CrossRef]
13. Seksaria S, Mehan S, Dutta BJ, Gupta GD, Ganti SS, Singh A. Oxymatrine and insulin resistance: focusing on mechanistic intricacies involve in diabetes associated cardiomyopathy via SIRT1/AMPK and TGF-β signaling pathway. J Biochem Mol Toxicol. 2023;37(5):e23330. doi:10.1002/jbt.23330 [Google Scholar] [PubMed] [CrossRef]
14. Han X, Ding C, Sang X, Peng M, Yang Q, Ning Y, et al. Targeting Sirtuin1 to treat aging-related tissue fibrosis: from prevention to therapy. Pharmacol Ther. 2022;229:107983. doi:10.1016/j.pharmthera.2021.107983 [Google Scholar] [PubMed] [CrossRef]
15. Li Y, Zhou J. Roles of silent information regulator 1-serine/arginine-rich splicing factor 10-lipin 1 axis in the pathogenesis of alcohol fatty liver disease. Exp Biol Med. 2017;242(11):1117–25. doi:10.1177/1535370217707729 [Google Scholar] [PubMed] [CrossRef]
16. Huang C, Yu J, Da J, Dong R, Dai L, Yang Y, et al. Dendrobium officinale Kimura & Migo polysaccharide inhibits hyperglycaemia-induced kidney fibrosis via the miRNA-34a-5p/SIRT1 signalling pathway. J Ethnopharmacol. 2023;313:116601. doi:10.1016/j.jep.2023.116601 [Google Scholar] [PubMed] [CrossRef]
17. Zhao W, Zhang R, Zang C, Zhang L, Zhao R, Li Q, et al. Exosome derived from mesenchymal stem cells alleviates pathological scars by inhibiting the proliferation, migration and protein expression of fibroblasts via delivering miR-138-5p to target SIRT1. Int J Nanomedicine. 2022;17:4023–38. doi:10.2147/IJN.S377317 [Google Scholar] [PubMed] [CrossRef]
18. Li P, Fei CS, Chen YL, Chen ZS, Lai ZM, Tan RQ, et al. Revealing the novel autophagy-related genes for ligamentum flavum hypertrophy in patients and mice model. Front Immunol. 2022;13:973799. doi:10.3389/fimmu.2022.973799 [Google Scholar] [PubMed] [CrossRef]
19. Cao Y, Li J, Qiu S, Ni S, Duan Y. LncRNA XIST facilitates hypertrophy of ligamentum flavum by activating VEGFA-mediated autophagy through sponging miR-302b-3p. Biol Direct. 2023;18(1):25. doi:10.1186/s13062-023-00383-9 [Google Scholar] [PubMed] [CrossRef]
20. Larionov A, Krause A, Miller W. A standard curve based method for relative real time PCR data processing. BMC Bioinform. 2005;6(1):62. doi:10.1186/1471-2105-6-62 [Google Scholar] [PubMed] [CrossRef]
21. Lee JA, Shin MR, Choi J, Kim M, Park HJ, Roh SS. Co-treatments of gardeniae fructus and silymarin ameliorates excessive oxidative stress-driven liver fibrosis by regulation of hepatic Sirtuin1 activities using thioacetamide-induced mice model. Antioxid. 2022;12(1):97. doi:10.3390/antiox12010097 [Google Scholar] [PubMed] [CrossRef]
22. Yousefi F, Shabaninejad Z, Vakili S, Derakhshan M, Movahedpour A, Dabiri H, et al. TGF-β and WNT signaling pathways in cardiac fibrosis: non-coding RNAs come into focus. Cell Commun Signal. 2020;18(1):87. doi:10.1186/s12964-020-00555-4 [Google Scholar] [PubMed] [CrossRef]
23. Liu Z, Wang Y, Shu S, Cai J, Tang C, Dong Z. Non-coding RNAs in kidney injury and repair. Am J Physiol Cell Physiol. 2019;317(2):C177–88. doi:10.1152/ajpcell.00048.2019 [Google Scholar] [PubMed] [CrossRef]
24. Liu X, Wu M, He Y, Gui C, Wen W, Jiang Z, et al. Construction and integrated analysis of the ceRNA network hsa_circ_0000672/miR-516a-5p/TRAF6 and its potential function in atrial fibrillation. Sci Rep. 2023;13(1):7701. doi:10.1038/s41598-023-34851-z [Google Scholar] [PubMed] [CrossRef]
25. Zhang B, Chen G, Yang X, Fan T, Chen X, Chen Z. Dysregulation of microRNAs in hypertrophy and ossification of ligamentum flavum: New advances, challenges, and potential directions. Front Genet. 2021;12:641575. doi:10.3389/fgene.2021.641575 [Google Scholar] [PubMed] [CrossRef]
26. Kong G, Chen Y, Liu Z, Wang Y, Li H, Guo C. Adenovirus-IL-10 relieves chronic rejection after mouse heart transplantation by inhibiting miR-155 and activating SOCS5. Int J Med Sci. 2023;20(2):172–85. doi:10.7150/ijms.77093 [Google Scholar] [PubMed] [CrossRef]
27. He L, Chang Q, Zhang Y, Guan X, Ma Z, Chen X, et al. MiR-155-5p aggravated astrocyte activation and glial scarring in a spinal cord injury model by inhibiting Ndfip1 expression and PTEN nuclear translocation. Neurochem Res. 2023;48(6):1912–24. doi:10.1007/s11064-023-03862-7 [Google Scholar] [PubMed] [CrossRef]
28. Wu L, Pu L, Zhuang Z. miR-155-5p/FOXO3a promotes pulmonary fibrosis in rats by mediating NLRP3 inflammasome activation. Immunopharmacol Immunotoxicol. 2023;45(3):257–67. doi:10.1080/08923973.2022.2115923 [Google Scholar] [PubMed] [CrossRef]
29. Dawood AF, Al Humayed S, Momenah MA, El-Sherbiny M, Ashour H, Kamar SS, et al. MiR-155 dysregulation Is associated with the augmentation of ROS/p53 axis of fibrosis in thioacetamide-induced hepatotoxicity and is protected by resveratrol. Diagn. 2022;12(7):1762. doi:10.3390/diagnostics12071762 [Google Scholar] [PubMed] [CrossRef]
30. Zhang C, Hang Y, Tang W, Sil D, Jensen-Smith HC, Bennett RG, et al. Dually active polycation/miRNA nanoparticles for the treatment of fibrosis in alcohol-associated liver disease. Pharm. 2022;14(3):669. doi:10.3390/pharmaceutics14030669 [Google Scholar] [PubMed] [CrossRef]
31. Liu ZH, Zhang Y, Wang X, Fan XF, Zhang Y, Li X, et al. SIRT1 activation attenuates cardiac fibrosis by endothelial-to-mesenchymal transition. Biomed Pharmacother. 2019;118:109227. doi:10.1016/j.biopha.2019.109227 [Google Scholar] [PubMed] [CrossRef]
32. Adjei-Mosi J, Sun Q, Smithson SB, Shealy GL, Amerineni KD, Liang Z, et al. Age-dependent loss of hepatic SIRT1 enhances NLRP3 inflammasome signaling and impairs capacity for liver fibrosis resolution. Aging Cell. 2023;22(5):e13811. doi:10.1111/acel.13811 [Google Scholar] [PubMed] [CrossRef]
33. Liang J, Huang G, Liu X, Taghavifar F, Liu N, Wang Y, et al. The ZIP8/SIRT1 axis regulates alveolar progenitor cell renewal in aging and idiopathic pulmonary fibrosis. J Clin Invest. 2022;132(11):e157338. doi:10.1172/JCI157338 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools