 Open Access
Open Access
ARTICLE
Bhlhe40 protects cochlear hair cell-like HEI-OC1 cells against HO‑triggered oxidative injury
Department of Otorhinolaryngology, Longgang Otorhinolaryngology Hospital and Shenzhen Key Laboratory of Otorhinolaryngology, Shenzhen Institute of Otorhinolaryngology, Shenzhen, China
* Corresponding Author: XIANHAI ZENG. Email:
BIOCELL 2024, 48(6), 991-999. https://doi.org/10.32604/biocell.2024.050219
Received 31 January 2024; Accepted 21 March 2024; Issue published 10 June 2024
Abstract
Background: Cochlear hair cell injury is a common pathological feature of hearing loss. The basic helix-loop-helix family, member e40 (Bhlhe40), a gene belonging to the basic helix-loop-helix (bHLH) family, exhibits strong transcriptional repression activity. Methods: Oxidative damage, in House Ear Institute-Organ of Corti 1 (HEI‑OC1) cells, was caused using hydrogen peroxide (HO). The Ad-Bhlhe40 particles were constructed to overexpress Bhlhe40 in HEI-OC1 cells. Various assays including cell counting kit-8 (CCK-8), terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling assay (TUNEL), flow cytometry, immunofluorescence, and corresponding commercial kits were employed to investigate the impacts of Bhlhe40 on cell viability, apoptosis, oxidative stress levels, mitochondrial membrane potential and cellular senescence. Additionally, a dual-luciferase reporter assay was performed to confirm the targeting of the histone deacetylases 2 (Hdac2) by Bhlhe40. Results: The results revealed that Bhlhe40 was downregulated in HO‑treated HEI‑OC1 cells, but its overexpression improved cell viability and mitigated HO‑induced oxidative injury in HEI‑OC1 cells with increase of superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx) activities and decrease of reactive oxygen species (ROS) levels. Besides, overexpression of Bhlhe40 suppressed HO‑triggered cell senescence, as evidenced by the fact that the upregulation of P53, P21, and P16 in HEI-OC1 cells treated with HO were all alleviated by Bhlhe40 overexpression. And we further verified that overexpression of Bhlhe40 could inhibit the expression of Hdac2, which may be related to the repression of Hdac2 transcription. Conclusion: This study suggests that Bhlhe40 plays a protective role against senescence and oxidative damage in cochlear hair cells exposed to HO.Graphic Abstract
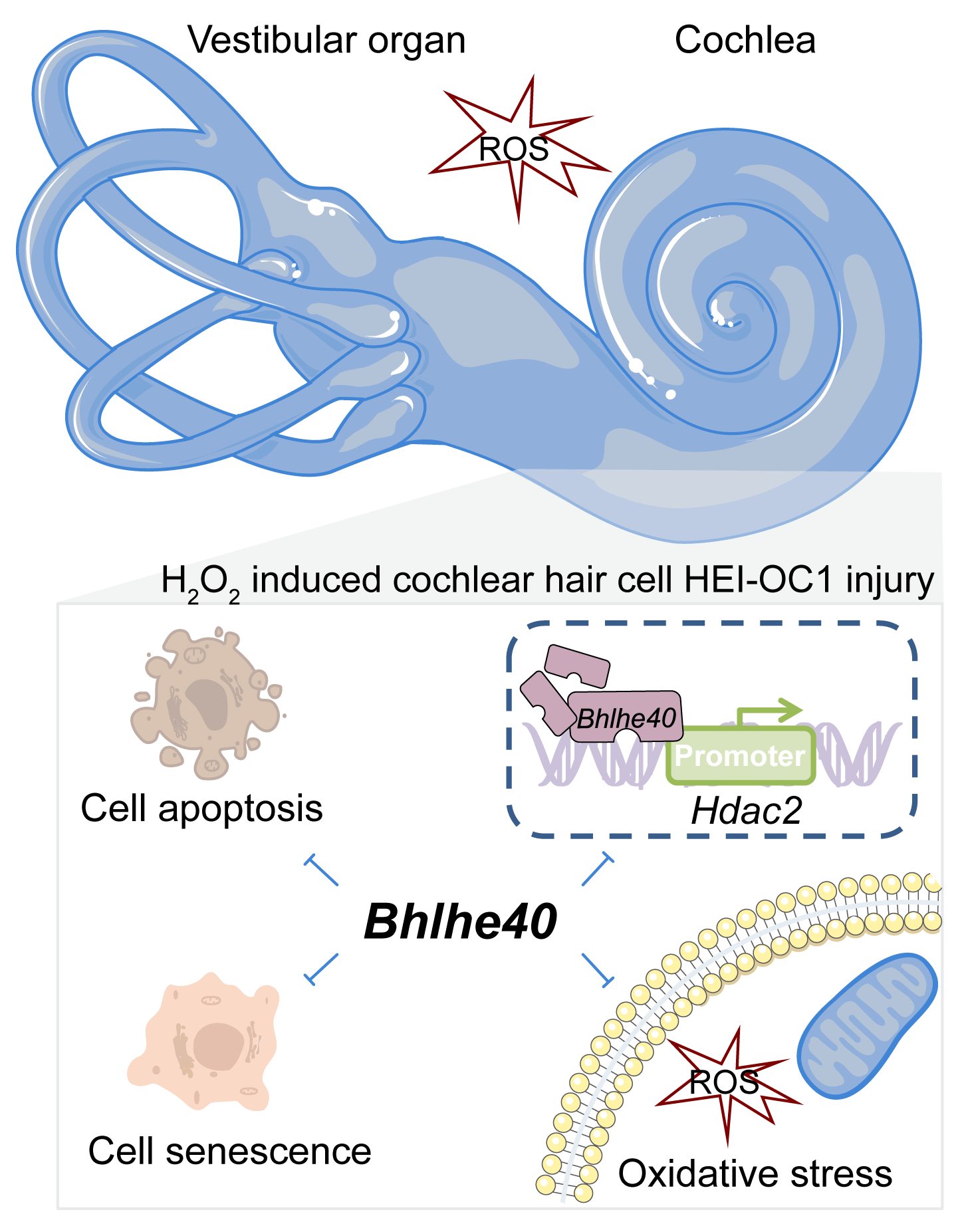
Keywords
Cochlear hair cells are a crucial population of sensory cells in the spiral organ of Corti, responsible for detecting sound waves. And these cells are highly susceptible to the environmental factors like exposure to loud noises or certain medications, as well as genetic factors that may contribute to hearing loss [1,2]. Recent research indicates that cochlear hair cell injury is a significant causative contributing factor to the development of sustained noise-induced hearing loss [3]. Following exposure to traumatic noise, cochlear hair cells experience massive accumulation of free radicals and reactive oxygen species (ROS) [4], leading to oxidative stress and subsequent cellular damage that affects various cellular processes such as cell proliferation, apoptosis and senescence [5]. The cascading effects of oxidative stress induced by ROS have been linked to cochlear damage [6], specifically through the activation of the caspase-mediated apoptosis pathway leading to hair cell loss [7]. Experimental exposure of cells to hydrogen peroxide (H2O2) increases ROS levels [8], and exposure of House Ear Institute-Organ of Corti 1 (HEI-OC1) cells to H2O2 usually serve as a model to study oxidative stress-induced damage [9]. Currently, antioxidant treatment has shown promise in mitigating redox imbalances and preventing sensory hair cell death in the cochlea [10,11].
The basic helix-loop-helix family, member e40 (Bhlhe40) is a basic helix-loop-helix (bHLH) gene known for its transcriptional repression activity through the alpha-helix rich domain in its C terminus [12,13]. Studies have demonstrated that Bhlhe40 is ubiquitously expressed in mammalian tissues [14] and plays a pivotal role in neurogenesis, immune regulation, and apoptosis [15,16]. Notably, Bhlhe40 has been implicated in diseases associated with oxidative stress, with its deficiency leading to signs of oxidative stress and muscle death in mice [17]. Conversely, overexpression of Bhlhe40 enhanced the resistance of podocytes to ROS-induced injury [18]. ROS and its byproducts are well-documented in disrupting the internal connections of molecules, potentially causing damage to cochlear cells or tissues and resulting in hearing loss [3,19]. However, to date, there has been no reported information on whether and how Bhlhe40 contributes to oxidative stress-induced injury in cochlear hair cell.
Additionally, Bhlhe40 has been associated with the regulation of histone deacetylase (Hdac) [20]. Studies have shown decreased levels of histone acetylation and increased levels of histone deacetylases 1, 2, and 3 (Hdac1, Hdac2 and Hdac3) in the cochlear cells of adult mice following exposure to traumatic noise [21]. Given the transcriptional repression function of Bhlhe40, we hypothesize that Bhlhe40 may be involved in the oxidative injury of cochlear hair cells by modulating the transcription of Hdac.
This study aims to explore the impacts of Bhlhe40 on an H2O2‑induced oxidative injury model in cochlear hair cells and elucidate its underlying mechanism.
HEI-OC1 cells (M8-0401, OriCell, Guangzhou, China), a mouse auditory cell line, were utilized in this study as an in vitro system to assess the functional response of Bhlhe40 in oxidative injury to cochlear hair cells. Following a publicly available protocol in Journal of Visualized Experiments (JOVE; http://www.jove.com/) [9,22], HEI-OC1 cells were cultured in the Dulbecco’s Modified Eagle’s Medium (DMEM; G4510, servicebio, Wuhan, China), supplemented with 10% fetal bovine serum (FBS, 11011-8611, Tianhang, Huzhou, China) in the absence of antibiotics, and maintained at 33°C with 10% CO2.
To construct the indicated recombinant adenovirus, the protein-coding DNA sequence (CDS) of Bhlhe40 (NM_011498) was cloned into the shuttle plasmid pDC315 (VT1500, YouBio, Hunan, China) by Auhui General Biology (Chuzhou, China). The recombinant plasmid pDC315, containing Bhlhe40-CDS, was generated by digesting the pDC315 plasmid with Nhel and Sa1I enzymes. HEK-293A cells were then co-transfected with the Bhlhe40-pDC315 plasmid and the adenovirus helper plasmid as per the manufacturer’s instructions to produce purified Bhlhe40-adenovirus (Ad-Bhlhe40) particles. Subsequently, HEI-OC1 cells were infected with Ad-Bhlhe40 particles to achieve Bhlhe40 overexpression.
HEI-OC1 cells were plated at a density of 4 × 103/mL in a 96-well plate (5 × 103 cells/well) and exposed to 2 mM H2O2 for 4, 8 or 12 h in a 33°C incubator with 10% CO2. Following this exposure, the cells were subjected to cell counting kit-8 solution (CCK-8; KGA317, KeyGen Biotech, Nanjing, China) for 2 h at a volume of 10 μL per well. Cell viability was then assessed by measuring the absorbance at 450 nm using a BioTek® 800™ TS Absorbance Reader (BioTek, USA).
RNA extraction and real-time polymerase chain reaction (real-time PCR)
TRIpure reagent (RP1001, BioTeke, Beijing, China) was utilized for total RNA extraction as per the manufacturer’s instructions. The cDNA synthesis was carried out with M-MLV reverse transcriptase (D7160L, Beyotime, Shanghai, China) following the manufacturer’s guidelines. Real-time fluorescence quantitative PCR was conducted on ExicyclerTM 96 (Bioneer, Korea) using SYBR Green (SY1020, Solarbio, Beijing, China) and 2 × Taq PCR MasterMix (PC1150, Solarbio, Beijing, China). β-actin served as a housekeeping gene. The relative expression of Bhlhe40, Hdac1, Hdac2 and Hdac3 were calculated based on 2−△△CT method. The PCR primers are provided as follows:
Mus Bhlhe40 forward primer (FP), AACGGAGCGAAGACAGC; reverse primer (RP), CCAAGTGACCCAAAGTAGTAAG.
Mus Hdac1 FP, GGGCACCAAGAGGAAAG; RP, GCGAATAGAACGCAGGA.
Mus Hdac2 FP, TTTTGGACCAGACTTCA; RP, CTACGACCTCCTTCACC.
Mus Hdac3 FP, ATGTATGAAGTTGGAGCAG; RP, TTTCGGACAGTGTAGCC.
Cells were lysed using RIPA lysis buffer (PR20001, Proteintech Group, Inc., Wuhan China) mixed with 1% protease inhibitor (PR20032, Proteintech Group, Inc., Wuhan, China). The lysate was then centrifuged at 10000 g for 3 min at 4°C to collect resulting supernatant. The concentration of protein was estimated using BCA Protein Concentration Assay Kit (PK10026, Proteintech Group, Inc., Wuhan, China). Samples containing 15–30 μg of proteins were loaded into each well and underwent sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were subsequently transferred to a polyvinylidene fluoride membrane (LC2005, Thermo Scientific, Pittsburgh, PA, USA) and blocked with 5% skimmed milk in Tris-buffer-saline with Tween-20 buffer (TBST; PR20011, Proteintech Group, Inc., Wuhan, China). Protein expression levels were assessed following incubation with primary and secondary antibodies. Specifically, the membranes were probed overnight at 4°C with primary antibodies against BHLHE40 (1: 1000; 17895-1-AP, Proteintech Group, Inc., Wuhan, China), P53 (1: 1000; A0263, ABclonal Technology, Shanghai, China), P21 (1: 1000; A1483, ABclonal Technology, Shanghai, China), P16 (1: 1000; 10883-1-AP, Proteintech Group, Inc., Wuhan, China), Hdac2 (1: 1000; A2084, ABclonal Technology, Shanghai, China) or β-actin (1: 20000; 66009-1-Ig, Proteintech Group, Inc., Wuhan, China). After washing with Western Blot Liquid Detergent (PR20012, Proteintech Group, Inc., Wuhan, China), the membranes were then incubated with the appropriate secondary antibodies HRP-conjugated Affinipure Goat Anti-Rabbit IgG (1: 10000; H + L, SA00001-2, Proteintech Group, Inc., Wuhan, China) or HRP-conjugated Affinipure Goat Anti-Mouse IgG (1: 10000; H + L, SA00001-1, Proteintech Group, Inc., Wuhan, China) for 40 min at 37°C. Following thorough rinsing of the membranes, the immune-reactive bands were visualized utilizing the Hypersensitive ECL Chemiluminescence Detection Kit (PK10003, Proteintech Group, Inc., Rosemont, IL, USA). The density value of bands were analyzed with the Gel-Pro-Analyzer software (Version 4.0, Media Cybernetics, Maryland, USA).
The AnnexinV-FITC Kit (KGA106, Nanjing, China) from KeyGen Biotech was utilized to quantify cell apoptosis. The indicated cells were initially plated in 6-well plates at a density of 5 × 105 cells per well and then exposed to 2 mM H2O2 for 4, 8 or 12 h. Subsequent to centrifugation at 150 g for 5 min, the supernatant was removed, and the cells were washed twice with PBS before being resuspended in 500 μL of binding buffer. Finally, the cells were incubated with Annexin V-FITC/PropidiumIodide (PI) staining reagent for 15 min at room temperature. The apoptotic cell percentage were determined using a flow cytometer (NovoCyte, Agilent, USA).
Terminal deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) assay
Apoptosis in HEI-OC1 cells was evaluate by analyzing DNA fragmentation using an In Situ Cell Death Detection Kit (12156792910, Roche, Basel, Switzerland) following the manufacturer’s guidelines. The cell sections were stained with DAPI solution (D106471, Aladdin, Shanghai, China) for 5 min (protected from light) to visualize the cell nucleus. Following rinsing with PBS, the images were captured using a microscope (BX53, OLYMPUS, Japan).
Apoptosis was assessed by measuring the activity of Caspase-3 in cell culture extracts with the Caspase-3 Activity Detection Kit (C1116, Beyotime, Shanghai, China) as per the manufacturer’s protocol. Based on Caspase-3 catalyze the production of pNA (p-nitroaniline, yellow) from the substrate Ac-DEVD-pNA, Caspase-3 activity was assessed by measuring the absorbance of pNA at 405 nm.
The ROS Assay Kit (KGAF019) from KeyGen Biotech in Nanjing, China utilized the red fluorescent probe dihydroethidium (DHE) to evaluate cytosolic superoxide production. Specifically, after 48 h of adenovirus infection, cells were exposed to 2 mM H2O2 for 8 h. Afterward, the cell culture medium was aspirated, and the cells were incubated with 10 μM of DHE probe at 37°C for 30 min in the absence of light. To ensure the elimination of any residual DHE outside the cells, the incubated cells underwent three washes with serum-free cell culture medium. Finally, the cells were observed under an Olympus DP73 microscopic photography system (OLYMPUS, Tokyo, Japan).
Evaluation the activities of superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPx)
Cell precipitates were resuspended in PBS and lysed through sonication. The protein content determination was carried out using a BCA protein assay kit (P0011, Beyotime, Shanghai, China). SOD (A001), CAT (A007) and GPx (A005) detection were performed with assay kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China) following the manufacturer’s experimental procedure. The optical density (OD) readings were taken at 550 nm for SOD activity, 405 nm for CAT activity, and 412 nm for GPx activity using a UV-Vis spectrophotometer (UV752N, Shanghai Youke Instrument and Meter Co., Ltd., Shanghai, China).
Mitochondrial membrane potential (∆Ψm) detection
JC-1 Mitochondrial Membrane Potential Assay Kit (C2006, Beyotime, Shanghai, China) was employed to determine the ∆Ψm in mitochondria. The specified cells were harvested as per the manufacturer’s experimental procedure, rinsed with PBS, and incubated with 0.5 mL of JC-1 solution for 20 min. After centrifugation at 300 g for 3 min, the cells were washed twice with JC-1 buffer (1×), and then resuspended in 500 μL of JC-1 buffer (1×) for mitochondrial membrane potential analysis using flow cytometry (NovoCyte, Agilent, USA).
Immunofluorescent assessment of BHLHE40 expression in HEI-OC1 cells
Cell climbing sheets were fixed in 4% paraformaldehyde (80096618, Sinoreagen, Shanghai, China) for 15 min and permeabilized with 0.1% tritonX-100 (ST795, Beyotime, Shanghai, China) for 30 min at room temperature. Following three washes with PBS, the specimens were sealed with 1% bovine serum albumin (BSA; A602440-0050, Sangon, Shanghai, China) for 15 min at 25°C. Subsequently, they were incubated with the primary antibody anti-BHLHE40 (1: 100; 17895-1-AP, Proteintech Group, Inc., Wuhan, China) at 4°C overnight. And a secondary Cy3-Goat Anti-Rabbit IgG antibody (1: 200; SA00009-2, Proteintech Group, Inc., Wuhan, China) was then applied under darkness at a concentration of 1:200 for 60 min. DAPI was used for nuclear re-stain. Following a rinsing in PBS, fluorescent visualization of HEI-OC1 cells was performed on a BX53 microscope (Olympus, Tokyo, Japan) with the DP73 photography system.
The activity levels of senescence-associated β-galactosidase (SA-β-gal), a marker of cellular senescence, were confirmed by a β-galactosidase staining kit (G1580, Solarbio, Beijing, China) according to the standard experimental procedure from manufacturer. Briefly, HEI-CO1 cells were fixed with 1 mL β-gal fixative for 15 min at 25°C, followed by incubation with 1 mL staining solution at 37°C overnight. Brightfield images were taken at 100× magnification using an Olympus IX53 microscope (OLYMPUS, Tokyo, Japan).
Dual-luciferase reporter assay
To investigate the potential regulatory of BHLHE40 on Hdac2 transcriptional activity, the luciferase reporter plasmid containing Hdac2 promoter region from −1000 to +100 relative to the transcription start site was constructed by inserting nucleotide fragments into the vector. For the luciferase activity assay, HEK293 cells were co-transfected with this luciferase reporter plasmid, Bhlhe40 overexpressing plasmid, and pRL-TK plasmid (KeyGen Biotech) with renilla luciferase gene. Forty-eight hours later, the luciferase activity was measured using the Luciferase Assay Kit (KGAF040, KeyGen Biotech, Nanjing, China) according to the manufacturer’s protocol.
The experiments were conducted in triplicate, with all values presented as the mean ± standard deviation (SD). Statistical analysis was performed using unpaired student’s t-test for comparisons between two groups and ordinary one-way ANOVA for comparisons among multiple groups. A p-value of less than 0.05 was considered statistically significant.
Bhlhe40 was downregulated in H2O2‑treated HEI‑OC1 cells
The results depicted in Figs. 1A and 1B demonstrated that HEI‑OC1 cells, when exposed to 2 mM H2O2 for 4, 8, and 12 h, exhibited a time-dependent pattern of cell death. Further flow cytometry analysis revealed that the exposure to 2 mM H2O2 led to a significant increase in apoptotic cells, with 31.91% at 8 h and 47.97% at 12 h (Figs. 1C and 1D). Additionally, a decrease in Bhlhe40 expression was observed in a time-dependent manner following H2O2 treatment, particularly evident after 8 h (Figs. 1E and 1F). Immunofluorescent assay results further supported this finding, showing a noticeable reduction in Bhlhe40 levels in HEI-OC1 cells exposed to 2 mM H2O2 for 8 h (Fig. 1G).
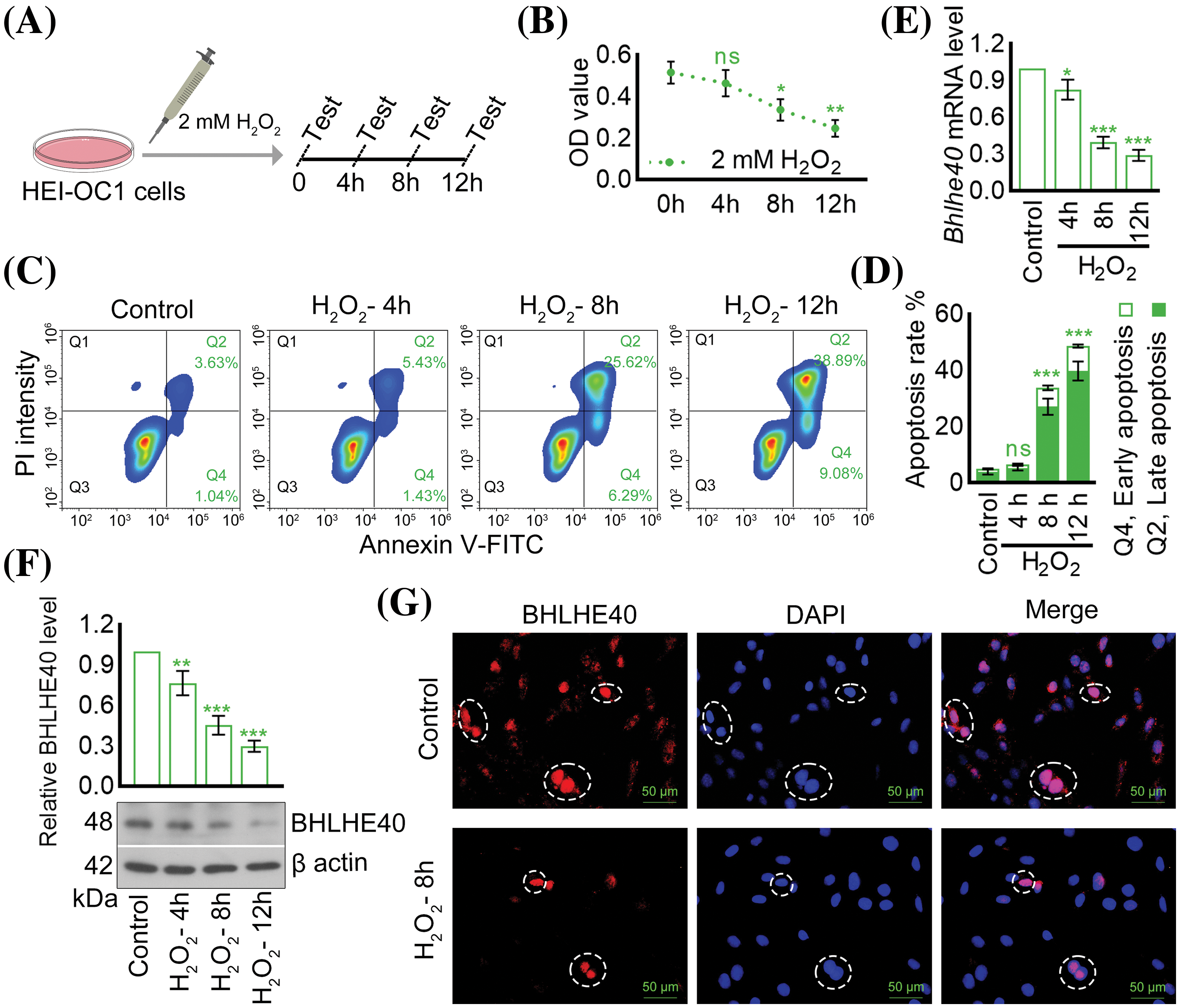
Figure 1: H2O2 treatment resulted in the downregulation of Bhlhe40 in HEI‑OC1 cells. (A) HEI-OC1 cells were exposed to 2 mM H2O2 for specified durations. (B) Cell viability was assessed using the CCK-8 assay. (C and D) Apoptosis was measured though flow cytometry. (E and F) The mRNA and protein levels of Bhlhe40 were determined via real-time PCR and western blotting, respectively, with corresponding gray value analysis of the western blotting results. (G) The BHLHE40 expression in HEI-OC1 cells was examined using immunofluorescent assay, with DAPI-labeled nuclei (blue fluorescent) for visualization. n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001 vs. Control group, while “ns” indicated no statistical significance.
Overexpression of Bhlhe40 inhibited H2O2-triggered HEI‑OC1 cell apoptosis
To investigate the relationship between H2O2-induced apoptosis of HEI-OC1 cells and the expression of Bhlhe40, a Bhlhe40 overexpressing HEI-OC1 cell line was generated using the pDC315 adenoviral vector system (Fig. 2A). The efficiency of Bhlhe40 overexpression was confirmed by real-time PCR and western blotting assay (Figs. 2B and 2C). Subsequent CCK‑8 assay demonstrated a significant reduction in HEI‑OC1 cell viability following H2O2 exposure, which was rescued by Bhlhe40 overexpression (Fig. 3A). Flow cytometry analysis indicated that Bhlhe40 overexpression also hindered H2O2-induced cell apoptosis (Figs. 3B and 3C), a finding further supported by followed TUNEL assays (Fig. 3D). Furthermore, activity detection revealed that H2O2 treatment led to an increase in Caspase-3 activity, which was counteracted by Bhlhe40 overexpression (Fig. 3E).
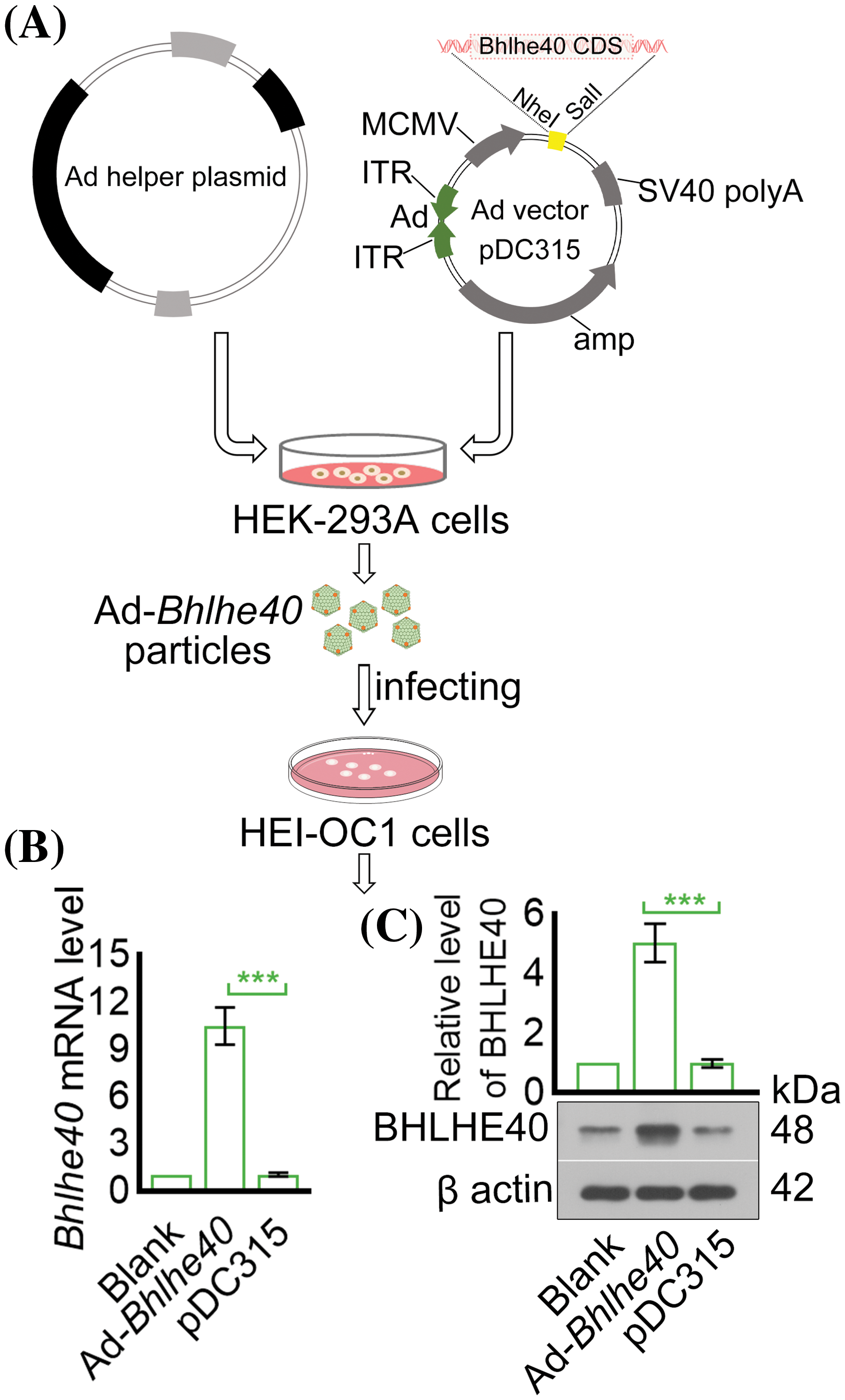
Figure 2: Construction of Bhlhe40 overexpressing HEI-OC1 cells. (A) Construction of recombinant adenovirus. (B and C) The expression of Bhlhe40 in HEI-OC1 cells were detected by real-time PCR and western blotting, with the gray value of BHLHE40 western blotting result measured. n = 3. ***p < 0.001.
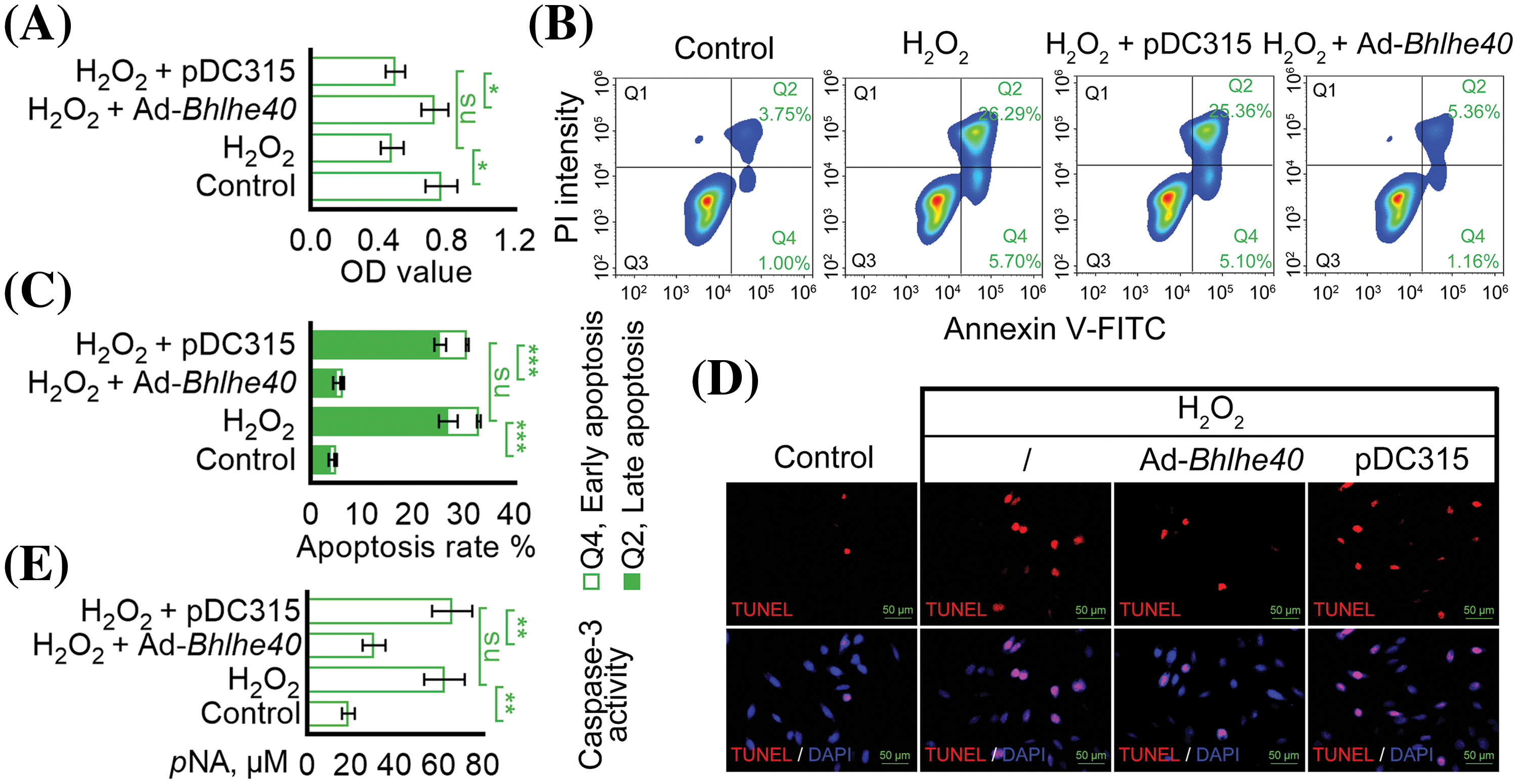
Figure 3: Overexpression of Bhlhe40 inhibited H2O2-induced HEI‑OC1 cell apoptosis. (A) Cell viability was assessed using the CCK-8 assay. (B and C) Apoptosis was detected by flow cytometry. (D) TUNEL apoptotic assay: TUNEL-positive nuclei (red fluorescent) represent apoptotic cells. DAPI-labeled nuclei (blue fluorescent). (E) Caspase-3 activity assay n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001, while “ns” indicated no statistical significance.
Overexpression of Bhlhe40 reduced H2O2‑triggered oxidative stress and mitochondrial injury
Subsequent analysis showed that the oxidative stress markers SOD, CAT and GPx were notably reduced in HEI‑OC1 cells after exposure to H2O2 (Figs. 4A–4C). Conversely, Bhlhe40 overexpression enhanced the activity of SOD, CAT and GPx (Figs. 4A–4C). Additionally, DHE probe detection indicated that Bhlhe40 overexpression alleviated the generation of ROS induced by H2O2 (Fig. 4D). Furthermore, JC-1 analysis revealed that H2O2 treatment led to a decrease in mitochondrial membrane potential, which represented the accelerated apoptosis. Nevertheless, this effect was reversed by Bhlhe40 overexpression (Fig. 4E).
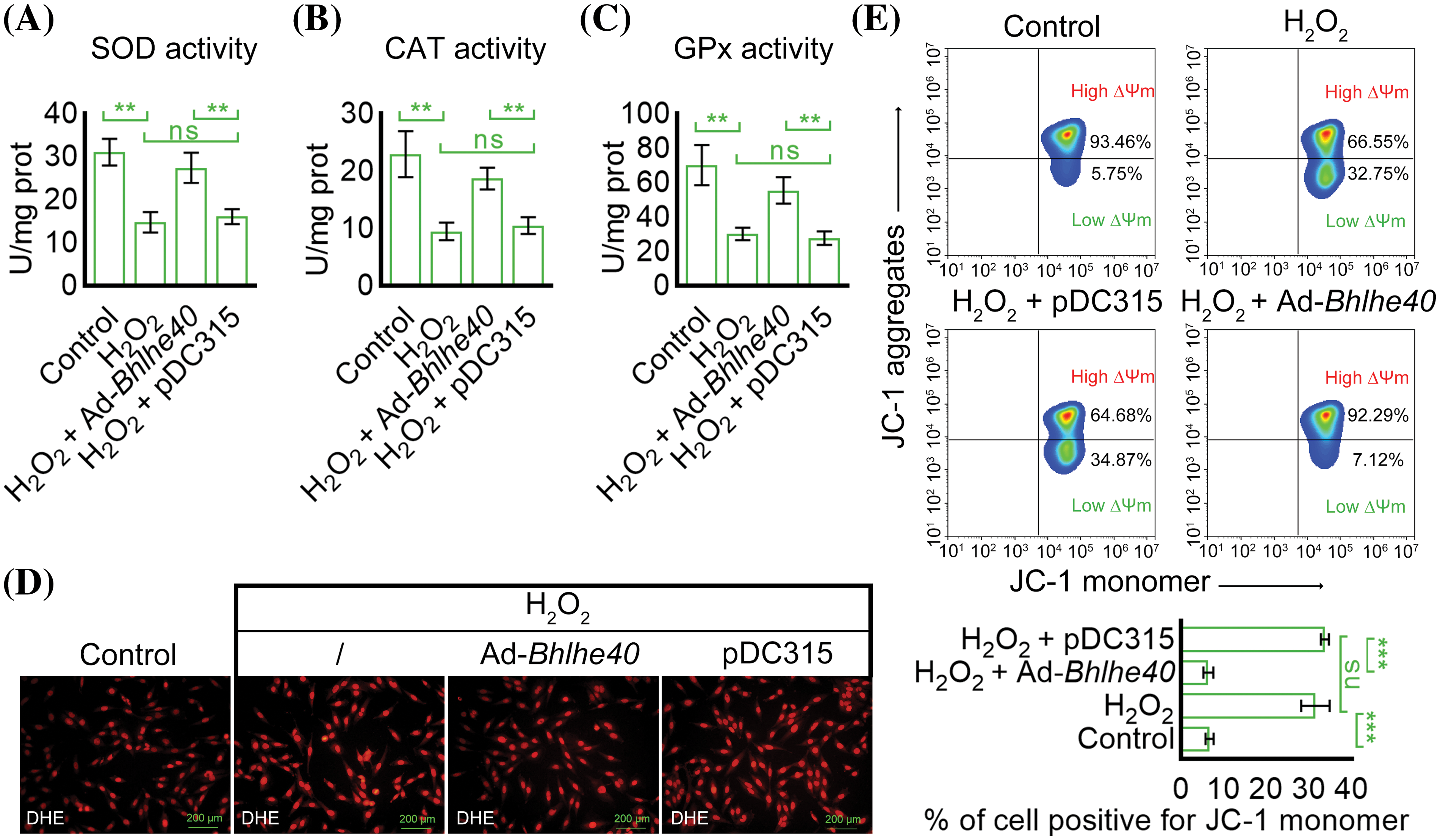
Figure 4: Overexpression of Bhlhe40 reduced H2O2-induced oxidative stress and mitochondrial membrane potential decrease in HEI‑OC1 cells. (A–C) Detection of the activity of oxidative stress markers: SOD, CAT and GPx, respectively. (D) ROS detection with the DHE probe (red fluorescent). (E) The ∆Ψm in mitochondrion was analyzed by the corresponding kit and flow cytometry. n = 3. **p < 0.01 and ***p < 0.001, while “ns” indicated no statistical significance.
Overexpression of Bhlhe40 delayed H2O2‑triggered cell senescence
The SA‑β‑gal staining showed that Bhlhe40 overexpression suppressed H2O2‑induced cell senescence, as evidenced by a decrease in the number of SA-β-gal-positive HEI-OC1 cells (Fig. 5A). Besides, the expression of P53, P21 and P16 was assessed in HEI‑OC1 cells. As depicted in Fig. 5B, the elevated levels of P53, P21 and P16 were observed in cells exposed to H2O2 but decreased after Bhlhe40 overexpression.
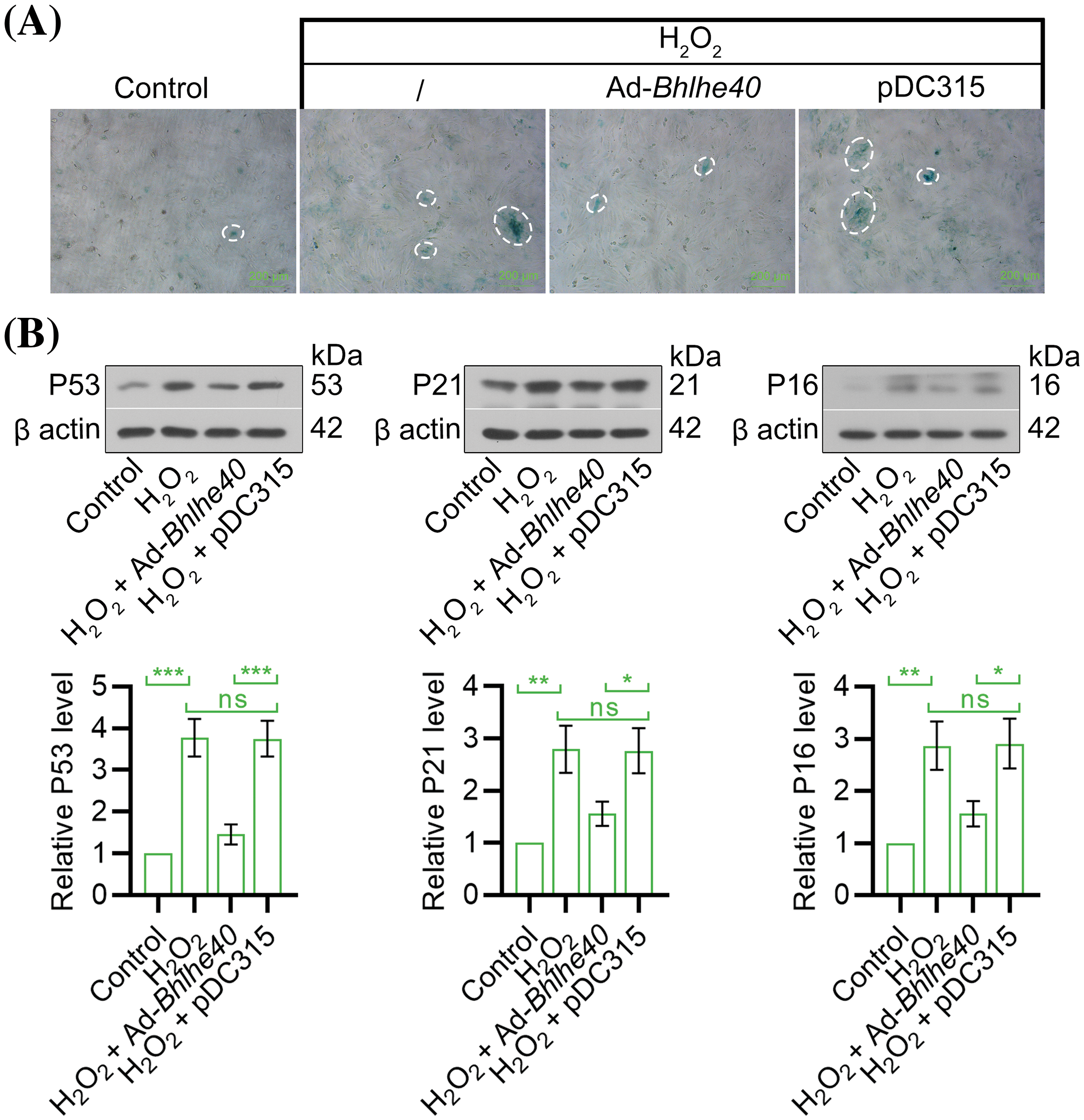
Figure 5: Overexpression of Bhlhe40 alleviated H2O2-induced HEI‑OC1 cell senescence. (A) Cell senescence was evaluated by the SA-β-Gal staining. (B) The expression of senescence‑associated factors (P53, P21 and P16) were detected by western blotting, with corresponding gray value analysis presented n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001, while “ns” indicated no statistical significance.
Overexpression of Bhlhe40 inhibited the transcription of Hdac2
Histone acetylation has been previously identified as playing a role in the pathogenesis of noise-induced cochlear cell death and hearing loss [23]. In this study, we explored the effects of Bhlhe40 on the expression of histone deacetylases 1, 2, and 3 (Hdac1, Hdac2 and Hdac3). Our findings demonstrated that Bhlhe40 overexpression led to the inhibition of H2O2-induced Hdac2 expression, while not significantly affecting the mRNA levels of Hdac1 and Hdac3 (Fig. 6A). Utilizing the JASPAR online database (http://jaspar.genereg.net/), we identified 3 potential BHLHE40-responsive elements in the promoter region of Hdac2 (Fig. 6B). Further western blot analysis confirmed that Bhlhe40 overexpression suppressed the expression of HDAC2 protein (Fig. 6C), possibly due to the repression of Hdac2 transcription (Fig. 6D).
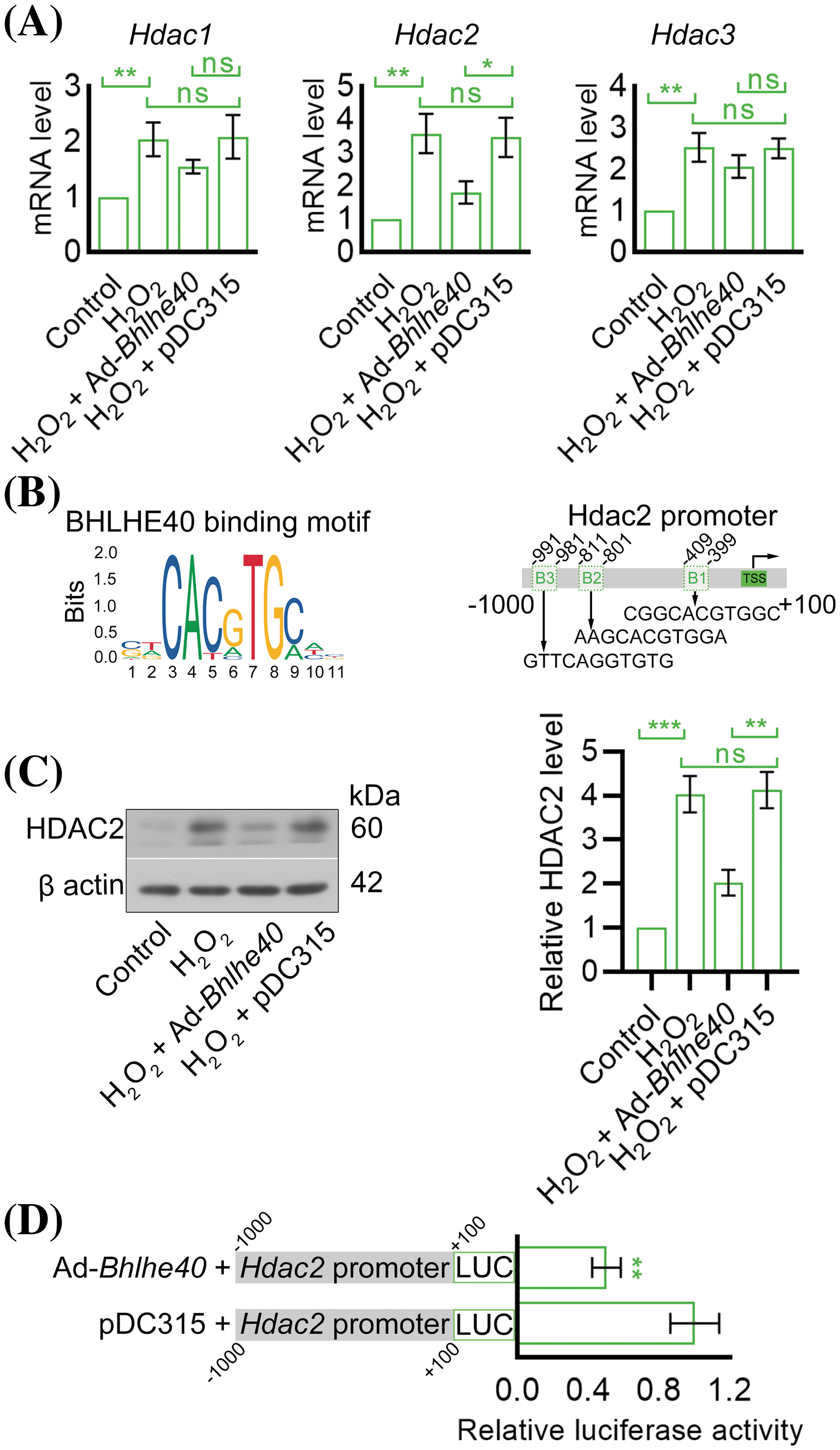
Figure 6: Overexpression of Bhlhe40 curtailed the transcription of Hdac2. (A) The mRNA levels of Hdac1, Hdac2 and Hdac3 in HEI-OC1 cells were detected by real-time PCR. (B) The BHLHE40-binding motif (obtained from JASPAR database) and three potential BHLHE40-responsive elements (B1, B2 and B3) with the JASPAR Score > 8.0 were identified in Hdac2 promoter region. (C) HDAC2 expression in HEI-OC1 cells was detected by western blotting and quantified by grayscale analysis. (D) The Hdac2 promoter activity was determined using a dual-luciferase reporter assay. n = 3. *p < 0.05, **p < 0.01 and ***p < 0.001, while “ns” indicated no statistical significance.
Noise-or drug-induced insults often leads to hearing loss, which is typically associated with damage to cochlear hair cells [3,4]. Given that damaged hair cells in mammals are irreversible [24], understanding the mechanisms implicated in cochlear hair cell injury could be crucial in developing treatments for hearing loss. Oxidative stress has been identified as a key factor in damaging cochlear hair cells, and further contributing to the development of acquired hearing loss [3]. Recently, H2O2 has been commonly used to induce the pathological models of hearing loss caused by various factors, including age‑related hearing loss [25] and noise-induced hearing loss [26]. Therefore, H2O2 was employed in this study to induce injury to cochlear hair cells. The salient results of this research indicate that the downregulation of Bhlhe40 was observed in HEI-OC1 cells treated with H2O2. Furthermore, the overexpression of Bhlhe40 was found to inhibit H2O2-induced cell apoptosis, oxidative stress, and senescence. These effects may be mediated at least in part by the repression of Hdac2 transcription.
Bhlhe40 participates in many crucial processes during differentiation of various cell lineages [27]. Data from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) revealed a significant downregulation of Bhlhe40 in cisplatin-induced mouse cochlear hair cells. Consistent with these findings, our study observed similar results in H2O2-treated HEI-OC1 cells. The regulatory transcription factors of cochlear hair cell regeneration show promise for restoring hearing function. Recent research using single-cell RNA sequencing of adult mouse cochlear hair cells identified multiple key transcriptional regulators, including Bhlhe40, that promoted the transition of hair cells to a more mature outer hair cell-like state [28]. Our investigation demonstrated that Bhlhe40 overexpression effectively inhibited H2O2-triggered HEI-OC1 cell apoptosis. ROS is regarded as harmful byproduct of cellular metabolism [29], with its overproduction and subsequent apoptosis induction has been shown to be an important contributor to some types of sensorineural hearing loss [30,31]. Antioxidant enzymes like SOD, CAT, and GPx are capable of reducing ROS levels to alleviate oxidative stress [32]. Encouragingly, our results showed that Bhlhe40 overexpression restored the decline in SOD, CAT and GPx activities and the increase in ROS levels caused by H2O2, indicating a protective role of Bhlhe40 against H2O2‑induced oxidative stress in HEI‑OC1 cells. Furthermore, we demonstrated that Bhlhe40 overexpression delayed H2O2‑induced cell senescence, as evidenced by the reversal of upregulated P53, P21 and P16 levels in treated HEI‑OC1 cells.
As a transcriptional repressor, Bhlhe40 represses the transcription of specific genes involved in cell growth and terminal differentiation via both Hdac-dependent and Hdac-independent mechanisms [33]. A previous study showed that Hdac1, Hdac2 and Hdac3 were all increased in the cochlear cells of mice exposure to the traumatic noise [21]. Similarly, our study observed upregulation of Hdac1, Hdac2 and Hdac3 in HEI-OC1 cells treated with H2O2. Interestingly, overexpression of Bhlhe40 was able to reverse the elevated levels of Hdac2 level but not Hdac1 or Hdac3. This suggested a potential role of Bhlhe40 in repressing of Hdac2 transcription. The JASPAR online database identified BHLHE40-responsive elements in the promoter region of Hdac2, which was further confirmed by a luciferase reporter assay demonstrating affinity of BHLHE40 for the Hdac2 promoter region. Previous studies have indicated that treatment with Hdac inhibitors like Trichostatin A or Vorinostat can effectively alleviate ototoxic drug-induced damage or noise-induced loss of outer hair cells, thus preventing hearing loss [21,34]. This study examined the negative regulation of Bhlhe40 on Hdac2, however, further investigation is needed to determine if Hdac2 transcriptional repression plays a role in the beneficial impact of Bhlhe40 on cochlear hair cell injury.
Taken together, the current study illustrated that Bhlhe40 has the ability to inhibit H2O2-induced cell apoptosis, oxidative stress, and senescence in HEI‑OC1 cells. This study is the first to uncover the impacts of Bhlhe40, as well as the regulation of Bhlhe40 on Hdac2 in an H2O2‑induced oxidative injury model in cochlear hair cells. These findings lend support to the potential development of a targeted therapy for hearing loss that leverages the molecular mechanism involving Bhlhe40. Moving forward, it is considered that additional investigations be carried out using an in vivo animal model to further explore the role of Bhlhe40 in cochlear hair cell injury and the expression levels of Hdac2.
Acknowledgement: None.
Funding Statement: This research was supported by the Special Fund for Economic and Technological Development of Longgang District, Shenzhen (LGKCYLWS2021000030).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xianhai Zeng; data collection: Liting Wen, Xiaoxia Zeng, Peixiong Chen, Yangyang Li; analysis and interpretation of results: Liting Wen, Xiaoxia Zeng; draft manuscript preparation: Xianhai Zeng, Liting Wen, Dapeng Zhao. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Youm I, Li W. Cochlear hair cell regeneration: an emerging opportunity to cure noise-induced sensorineural hearing loss. Drug Discov Today. 2018;23(8):1564–9. doi:10.1016/j.drudis.2018.05.001 [Google Scholar] [PubMed] [CrossRef]
2. Quan YZ, Wei W, Ergin V, Rameshbabu AP, Huang M, Tian C, et al. Reprogramming by drug-like molecules leads to regeneration of cochlear hair cell-like cells in adult mice. Proc Natl Acad Sci U S A. 2023;120(17):e2215253120. doi:10.1073/pnas.2215253120 [Google Scholar] [PubMed] [CrossRef]
3. Wu F, Xiong H, Sha S. Noise-induced loss of sensory hair cells is mediated by ROS/AMPKα pathway. Redox Biol. 2020;29:101406. doi:10.1016/j.redox.2019.101406 [Google Scholar] [PubMed] [CrossRef]
4. Han WJ, Shi XR, Nuttall A. Noise-induced nitrotyrosine increase and outer hair cell death in guinea pig cochlea. Chin Med J. 2013;126(15):2923–7. doi:10.3760/cma.j.issn.0366-6999.20130838. [Google Scholar] [CrossRef]
5. Zhang J, Zhao Y, Tian Y, Geng M, Liu Y, Zhang W, et al. Genome-wide screening in the haploid system reveals Slc25a43 as a target gene of oxidative toxicity. Cell Death Dis. 2022;13(3):284. doi:10.1038/s41419-022-04738-4 [Google Scholar] [PubMed] [CrossRef]
6. Fetoni AR, Paciello F, Rolesi R, Paludetti G, Troiani D. Targeting dysregulation of redox homeostasis in noise-induced hearing loss: oxidative stress and ROS signaling. Free Radic Biol Med. 2019;135:46–59. doi:10.1016/j.freeradbiomed.2019.02.022 [Google Scholar] [PubMed] [CrossRef]
7. Waqas M, Gao S, us Iram S, Ali MK, Ma Y, Li W. Inner ear hair cell protection in mammals against the noise-induced cochlear damage. Neur Plast. 2018;2018:1–9. doi:10.1155/2018/3170801 [Google Scholar] [PubMed] [CrossRef]
8. Li M, Chen D, Ke J, Zheng R, Su J, Zheng Z, et al. Inhibition of H2O2-induced apoptosis of GC2-spg cells by functionalized selenium nanoparticles with lentinan through ROS-mediated ERK/p53 signaling pathways. BIOCELL. 2023;47(2):401–8. doi:10.32604/biocell.2023.025154. [Google Scholar] [CrossRef]
9. Zhao T, Liu X, Sun Z, Zhang J, Zhang X, Wang C, et al. RNA-seq analysis of potential lncRNAs for age-related hearing loss in a mouse model. Aging. 2020;12(8):7491–510. doi:10.18632/aging.103103 [Google Scholar] [PubMed] [CrossRef]
10. Chen GD, Daszynski DM, Ding D, Jiang H, Woolman T, Blessing K, et al. Novel oral multifunctional antioxidant prevents noise-induced hearing loss and hair cell loss. Hear Res. 2020;388:107880. doi:10.1016/j.heares.2019.107880 [Google Scholar] [PubMed] [CrossRef]
11. Pisani A, Paciello F, Montuoro R, Rolesi R, Galli J, Fetoni AR. Antioxidant therapy as an effective strategy against noise-induced hearing loss: from experimental models to clinic. Life. 2023;13(4):1035. doi:10.3390/life13041035 [Google Scholar] [PubMed] [CrossRef]
12. Tsuyama T, Sato Y, Yoshizawa T, Matsuoka T, Yamagata K. Hypoxia causes pancreatic β-cell dysfunction and impairs insulin secretion by activating the transcriptional repressor BHLHE40. EMBO Rep. 2023;24(8):e56227. doi:10.3390/ijms25084186. [Google Scholar] [CrossRef]
13. Kiss Z, Mudryj M, Ghosh PM. Non-circadian aspects of BHLHE40 cellular function in cancer. Genes Cancer. 2020;11(1–2):1–19. doi:10.3389/fcell.2021.644346. [Google Scholar] [CrossRef]
14. Wesolowski L, Ge J, Castillon L, Sesia D, Dyas A, Hirosue S, et al. The SWI/SNF complex member SMARCB1 supports lineage fidelity in kidney cancer. iScience. 2023;26(8):107360. doi:10.1016/j.isci.2023.107360 [Google Scholar] [PubMed] [CrossRef]
15. Zhao L, Liu D, Ma W, Gu H, Wei X, Luo W, et al. Bhlhe40/Sirt1 axis-regulated mitophagy is implicated in all-trans retinoic acid-induced spina bifida aperta. Front Cell Dev Biol. 2021;9:644346. doi:10.1038/s41419-021-04473-2. [Google Scholar] [CrossRef]
16. Zhong G, Mercado MAB, Li Q, Quick CM, Kim Y, Palmer R, et al. BHLHE40 drives protective polyfunctional CD4 T cell differentiation in the female reproductive tract against Chlamydia. PLoS Pathog. 2024;20(1):e1011983. doi:10.1371/journal.ppat.1011983 [Google Scholar] [PubMed] [CrossRef]
17. Vercherat C, Chung TK, Yalcin S, Gulbagci N, Gopinadhan S, Ghaffari S, et al. Stra13 regulates oxidative stress mediated skeletal muscle degeneration. Hum Mol Genet. 2009;18(22):4304–16. doi:10.1128/MCB.00387-15. [Google Scholar] [CrossRef]
18. Bek MJ, Wahle S, Müller B, Benzing T, Huber TB, Kretzler M, et al. Stra13, a prostaglandin E2-induced gene, regulates the cellular redox state of podocytes. Faseb J. 2003;17(6):682–4. doi:10.1016/j.trsl.2007.01.002. [Google Scholar] [CrossRef]
19. Defourny J, Aghaie A, Perfettini I, Avan P, Delmaghani S, Petit C. Pejvakin-mediated pexophagy protects auditory hair cells against noise-induced damage. Proc Natl Acad Sci U S A. 2019;116(16):8010–7. doi:10.1073/pnas.1821844116 [Google Scholar] [PubMed] [CrossRef]
20. Ivanova AV, Ivanov SV, Danilkovitch-Miagkova A, Lerman MI. Regulation of STRA13 by the von Hippel-Lindau tumor suppressor protein, hypoxia, and the UBC9/ubiquitin proteasome degradation pathway. J Biol Chem. 2001;276(18):15306–15. doi:10.1136/jmg.2004.029835. [Google Scholar] [CrossRef]
21. Chen J, Hill K, Sha SH. Inhibitors of histone deacetylases attenuate noise-induced hearing loss. J Assoc Res Otolaryngol. 2016;17(4):289–302. doi:10.1007/s10162-016-0567-7 [Google Scholar] [PubMed] [CrossRef]
22. Kalinec GM, Park C, Thein P, Kalinec F. Working with auditory HEI-OC1 Cells. J Vis Exp. 2016;(115):54425. doi:10.3791/54425 [Google Scholar] [PubMed] [CrossRef]
23. Xie L, Zhou Q, Chen X, Du X, Liu Z, Fei B, et al. Elucidation of the Hdac2/Sp1/miR-204-5p/Bcl-2 axis as a modulator of cochlear apoptosis via in vivo/in vitro models of acute hearing loss. Mol Ther-Nucleic Acids. 2021;23:1093–109. doi:10.1016/j.omtn.2021.01.017 [Google Scholar] [PubMed] [CrossRef]
24. Shibata SB, West MB, Du X, Iwasa Y, Raphael Y, Kopke RD. Gene therapy for hair cell regeneration: review and new data. Hear Res. 2020;394:107981. doi:10.1016/j.heares.2020.107981 [Google Scholar] [PubMed] [CrossRef]
25. Li L, Xu K, Bai X, Wang Z, Tian X, Chen X. UCHL1 regulated by Sp1 ameliorates cochlear hair cell senescence and oxidative damage. Exp Ther Med. 2023;25(2):94. doi:10.3892/etm.2023.11793 [Google Scholar] [PubMed] [CrossRef]
26. Yang H, Zhu Y, Ye Y, Guan J, Min X, Xiong H. Nitric oxide protects against cochlear hair cell damage and noise-induced hearing loss through glucose metabolic reprogramming. Free Radic Biol Med. 2022;179(2):229–41. doi:10.1016/j.freeradbiomed.2021.11.020 [Google Scholar] [PubMed] [CrossRef]
27. Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11(16):2052–65. doi:10.1101/gad.11.16.2052 [Google Scholar] [PubMed] [CrossRef]
28. Tu S, Zuo J. Systematic single cell RNA sequencing analysis reveals unique transcriptional regulatory networks of Atoh1-mediated hair cell conversion in adult mouse cochleae. PLoS One. 2023;18(12):e0284685. doi:10.1371/journal.pone.0284685 [Google Scholar] [PubMed] [CrossRef]
29. Jomova K, Raptova R, Alomar SY, Alwasel SH, Nepovimova E, Kuca K, et al. Reactive oxygen species, toxicity, oxidative stress, and antioxidants: chronic diseases and aging. Arch Toxicol. 2023;97(10):2499–574. doi:10.1007/s00204-023-03562-9 [Google Scholar] [PubMed] [CrossRef]
30. Rousset F, Nacher-Soler G, Coelho M, Ilmjarv S, Kokje VBC, Marteyn A, et al. Redox activation of excitatory pathways in auditory neurons as mechanism of age-related hearing loss. Redox Biol. 2020;30:101434. doi:10.1016/j.redox.2020.101434 [Google Scholar] [PubMed] [CrossRef]
31. Zhao Z, Han Z, Naveena K, Lei G, Qiu S, Li X, et al. ROS-responsive nanoparticle as a berberine carrier for OHC-targeted therapy of noise-induced hearing loss. ACS Appl Mater Interf. 2021;13(6):7102–14. doi:10.1021/acsami.0c21151 [Google Scholar] [PubMed] [CrossRef]
32. Mosa KA, El-Naggar M, Ramamoorthy K, Alawadhi H, Elnaggar A, Wartanian S, et al. Copper nanoparticles induced genotoxicty, oxidative stress, and changes in superoxide dismutase (SOD) gene expression in cucumber (Cucumis sativus) plants. Front Plant Sci. 2018;9:872. doi:10.3389/fpls.2018.00872 [Google Scholar] [PubMed] [CrossRef]
33. Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci U S A. 2000;97(8):4058–63. doi:10.1073/pnas.070526297 [Google Scholar] [PubMed] [CrossRef]
34. Chen FQ, Schacht J, Sha SH. Aminoglycoside-induced histone deacetylation and hair cell death in the mouse cochlea. J Neurochem. 2009;108(5):1226–36. doi:10.1111/j.1471-4159.2009.05871.x [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools