 Open Access
Open Access
ARTICLE
Revealing the role of honokiol in human glioma cells by RNA-seq analysis
1 The Department of Neurosurgery, First Hospital of Jilin University, Changchun, 130021, China
2 The Department of Anatomy, College of Basic Medical Sciences, Jilin University, Changchun, 130021, China
3 The Third College of Clinical Medicine, Jilin University, Changchun, 130021, China
* Corresponding Authors: YAN LU. Email: ; HAIYAN LIU. Email:
BIOCELL 2024, 48(6), 945-958. https://doi.org/10.32604/biocell.2024.049748
Received 17 January 2024; Accepted 20 March 2024; Issue published 10 June 2024
Abstract
Background: Glioma is a kind of tumor that easily deteriorates and originates from glial cells in nerve tissue. Honokiol is a bisphenol compound that is an essential monomeric compound extracted from the roots and bark of Magnoliaceae plants. It also has anti-infection, antitumor, and immunomodulatory effects. In this study, we found that honokiol induces cell apoptosis in the human glioma cell lines U87-MG and U251-MG. However, the mechanism through which honokiol regulates glioma cell apoptosis is still unknown. Methods: We performed RNA-seq analysis of U251-MG cells treated with honokiol and control cells. Protein-protein interaction (PPI) network analysis was performed, and the 10 top hub unigenes were examined via real-time quantitative PCR. Furthermore, MAPK signaling and ferroptosis were detected via western blotting. Results: 332 differentially expressed genes (DEGs) were found, comprising 163 increased and 169 decreased genes. Analysis of the DEGs revealed that various biological processes were enriched, including ‘response to hypoxia’, ‘cerebellum development cellular response to hypoxia,’ ‘iron ion binding,’ ‘oxygen transporter activity,’ ‘oxygen binding,’ ‘ferric iron binding,’ and ‘structural constituent of cytoskeleton.’ Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the DEGs were enriched in the following pathways: ‘mitogen-activated protein kinases (MAPK)’, ‘Hypoxia-inducible factor 1 (HIF-1)’, ‘ferroptosis,’ ‘Peroxisome proliferator-activated receptor (PPAR),’ ‘Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)- protein kinase B (Akt),’ and ‘phagosome.’ Among these pathways, the MAPK signaling pathway and ferroptosis were verified. Conclusion: This study revealed the potential mechanism by which honokiol induces apoptosis and provided a comprehensive analysis of DEGs in honokiol-treated U251-MG cells and the associated signaling pathways. These data could lead to new ideas for future research and therapy for patients with glioma.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| HK | Honokiol |
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| KEGG | Kyoto encyclopedia of genes and genomes |
| PPI | Protein‒protein interaction |
| MAPK | Mitogen-activated protein kinases |
| HIF-1 | Hypoxia-inducible factor 1 |
| PPARs | Peroxisome proliferator-activated receptors |
| PI3K | Phosphatidylinositol-4,5-bisphosphate 3-kinase |
| Akt | Protein kinase B |
| Bcl-2 | B cell lymphoma 2 |
| BAX | BCL-2 associated X |
| CHOP | C/EBP Homologous Protein |
| p-ERK | p-extracellular signal-regulated kinase |
| BAK | BCL2 antagonist/killer 1 |
| GPX4 | Glutathione peroxidase 4 |
| TF | Transferrin |
| PI | Propidium iodide |
| NFκB | Nuclear factor kappa B |
| H-DMEM | High glucose-Dulbecco’s modified eagle medium |
| PMSF | Phenylmethylsulfonyl fluoride |
| STRING | Search tool for the retrieval of interacting genes/Proteins |
| IC50 | The half maximal inhibitory concentration |
| mTOR | Mechanistic target of rapamycin |
| AMPK | AMP-activated protein kinase |
| ECM | Extracellular matrix |
| Rap1 | Ras-associated protein 1 |
| CT | Computed tomography |
| HMOX1 | Heme-oxygenase 1 |
| VEGFA | Vascular endothelial growth factor A |
| SREBF1 | Sterol regulatory element-binding protein 1 |
| CCL2 | C-C motif chemokine ligand 2 |
| COL1A1 | Collagen type I alpha 1 |
| HMGCS1 | Hydroxy-3-methylglutaryl coenzyme A synthetase 1 |
| FDFT1 | Farnesyl-diphosphate farnesyltransferase |
| SPP1 | Secreted phosphoprotein 1 |
| HMGCR | 3-hydroxy-3-methylglutaryl-CoA reductase |
| LDLR | Low-density lipoprotein receptor |
| SCD | Stearoyl-CoA desaturase |
Glioma, which is strongly invasive and diffuse, causes damage to the surrounding normal brain tissue, thus posing a threat to the health of patients [1,2]. The main treatments for glioma are surgery, chemotherapy and radiotherapy [3]. However, the treatment efficacy is unsatisfactory, and patients easily relapse [4]. Consequently, new methods for improving the prognosis of glioma patients are urgently needed.
Honokiol, an allyl-biphenol-based compound, has a wide spectrum of antibacterial, anti-inflammatory, neuroprotective, and antitumor effects and has high clinical value [5,6]. Honokiol has become a research hotspot in the development and application of active ingredients in traditional Chinese medicine in recent years. In the human lung cancer cell lines A549 and 95-D, honokiol inhibited cell viability and induced cell apoptosis, and the expression of B cell lymphoma 2 (Bcl-2), BCL-2 associated X (BAX), C/EBP Homologous Protein (CHOP), and p-extracellular signal-regulated kinase (p-ERK) [7]. Honokiol also induces caspase-dependent apoptosis, and deregulates many apoptosis related proteins, such as upregulated BAX, and BCL2 antagonist/killer 1 (BAK), as does the downregulation of Bcl-2 in lymphoblastic leukemia, multiple myeloma, and Burkitt lymphoma [8]. Many investigators have reported that honokiol has anticancer effects [9–12]. In summary, studies have demonstrated that honokiol has multiple anticancer effects. For instance, honokiol suppresses cell proliferation, and promotes autophagy and apoptosis.
In addition, honokiol could improve bioavailability in nerve tissue through passing the blood-brain barrier [13]. Several reports have shown that honokiol has neuroprotective effects. For example, Wang et al. demonstrated that honokiol mitigated NaF-induced oxidative stress and mitochondrial dysfunction, which ultimately contributed to neuronal/synaptic injury and cognitive deficits [14]. Consistent with these findings, honokiol could reduce cytokine production and stimulate glial nuclear factor kappa B (NFκB) to eliminate the inflammatory response during cerebral ischemia-reperfusion activity [15]. Moreover, in TgCRND8 mice, Nanohonokiol was more efficacious than free honokiol at improving cognitive impairment [16]. Hydroxyapatite particles also could targeted delivery of honokiol to tumor sites [17]. Thus, honokiol has great application prospects in the treatment of human glioma.
In the current research, we investigated the effect of honokiol on apoptosis in human glioma cells. To further elucidate the potential mechanism through which honokiol affects human glioma cells, RNA sequencing was performed. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses and protein-protein interaction (PPI) network analysis of the DEGs were performed. In addition, several signaling pathways and the top 10 hub unigenes were validated by western blot and qRT-PCR.
U251-MG and U87-MG cells were preserved by the Department of Anatomy, College of Basic Medical Sciences, Jilin University, and cultured in High glucose-Dulbecco’s Modified Eagle Medium (H-DMEM, Cat. SH30249.01, HyClone, Logan, UT, USA) supplemented with 10% Fetal Bovine Serum (FBS, SH30070.03HI, HyClone), and 100 U/mL penicillin-streptomycin (SV30010, HyClone) at 37°C and in the presence of 5% CO2. The following antibodies were used: anti-JNK antibody (Cat. ab179461, Abcam, Cambridge, MA, USA); anti-BAX (Cat. #2774), anti-Bcl-2 (Cat. #2870), anti-p38 (Cat. #8690), anti-p-p38 (Cat. #9216), anti-ERK (Cat. #4696), anti-p-ERK (Cat. #9101), and anti-p-JNK (Cat. #4668) antibodies (Cell Signaling Technology, Beverly, MA, USA); anti-glutathione peroxidase 4 (GPX4, Cat. 67763-1-lg), and anti-transferrin (TF, Cat. 17435-1-AP) antibodies (ProteinTech Group, Chicago, IL, USA). Honokiol purchased from Aladdin (Cat. 1025, Shanghai, China).
Cell counting kit-8 (CCK-8) assay
3000 cells (in 100 mL medium) were planted in a 96-well plate and incubated in an atmosphere (37°C, 5% CO2). In the following 3 days, 100 μL of medium containing one-tenth the volume of CCK-8 (Cat. K1018, ApexBio, Houston, USA) was added to each well, after which the cells were incubated for 1.5 h (37°C, 5% CO2) every day. The samples were tested with a scanning multiwell spectrophotometer (Cat, 30050303, TECAN, CH) at an absorbance of 450 nm (A450).
Cells were collected using trypsin (without ethylene diamine tetraacetic acid, Cat. 15050057, Gibco, Thermo Fisher Scientific, MN, USA). Then, annexin V-FITC and propidium iodide (PI, Cat. APOAF, Sigma) were added (room temperature, 15 min). Cell distribution was detected at an excitation wavelength of 488 nm (annexin V-FITC) and 535 nm (PI) using a flow cytometer (Cat, 651157, BD Biosciences, San Jose, CA, USA).
Cells were washed with precooled 0.01 M PBS 3 times. Then, 300 μL of lysis buffer (Cat. P0013B, Beyotime Institute of Biotechnology, Haimen, China) containing 100 mM phenylmethylsulfonyl fluoride (PMSF, Cat. ZS303, ZOMANBIO, Beijing, China) was added, and the mixture was incubated on ice for 30 min. Then, the cells were scraped off, transferred to a 1.5 mL EP tube and centrifuged. The supernatant was divided into 1.5 mL EP tubes. The protein concentration was detected following the instructions of the BCA Protein Assay Kit (Cat. 23227, Thermo Fisher Scientific, Waltham, MA, USA). Twenty micrograms of protein was added to each lane. The PVDF membrane (Millipore, Billerica, USA) was activated with methanol for 15 s, and the transfer was performed under constant current conditions. The PVDF membrane was then incubated with 5% skim milk powder (in PBST, Cat. SH30256.01, HyClone) for 1 h to block and subsequently incubated in primary antibody diluent (1:1000, PBST) containing 1% skim milk powder at 4°C overnight. On the 2nd day, the PVDF membrane was washed with PBST at room temperature 3 times and then incubated with secondary antibodies (1:3000 dilution, Cat. RGAR001, ProteinTech) for 1 h. The bands were visualized using Immobilon® Western Chemiluminescent HRP Substrate (Cat. WBKLS0500, Millipore).
RNA sequencing was performed to determine the expression profile of the mRNAs (from U251-MG cells treated with DMSO and U251-MG cells treated with honokiol). Total RNA was extracted with TRIzol® Reagent (Cat. 15596026, Invitrogen Life Technologies, NY, USA). Then, the RNA samples were shipped to the Biomarker Technology Company (Beijing, China) for sequencing.
Differential expression analysis
Differential expression analysis of the two groups was performed utilizing the edgeR R package (3.8.6) [12]. The criteria for identifying differentially expressed genes (DEGs) were restricted to those with a p value < 0.05 and a fold change ≥ 1.5.
Functional enrichment analysis
The Wallenius noncentral hypergeometric distribution, which can adjust for gene length bias in DEGs, was utilized for GO enrichment analysis [18]. KEGG [19] is a database resource for understanding high-level functions and utilities of biological systems from molecular-level information (http://www.genome.jp/kegg/). KOBAS [20] software was used to detect the statistical enrichment of DEGs in KEGG pathways.
Protein-protein interactions (PPIs)
The sequences of the DEGs were subjected to BLAST (blastx) against the genome of a related species (Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database: http://string-db.org/) to obtain the predicted PPIs of the DEGs. The PPIs of these DEGs were subsequently visualized via Cytoscape [21].
Real-time fluorescence quantitative polymerase chain reaction (RT-qPCR)
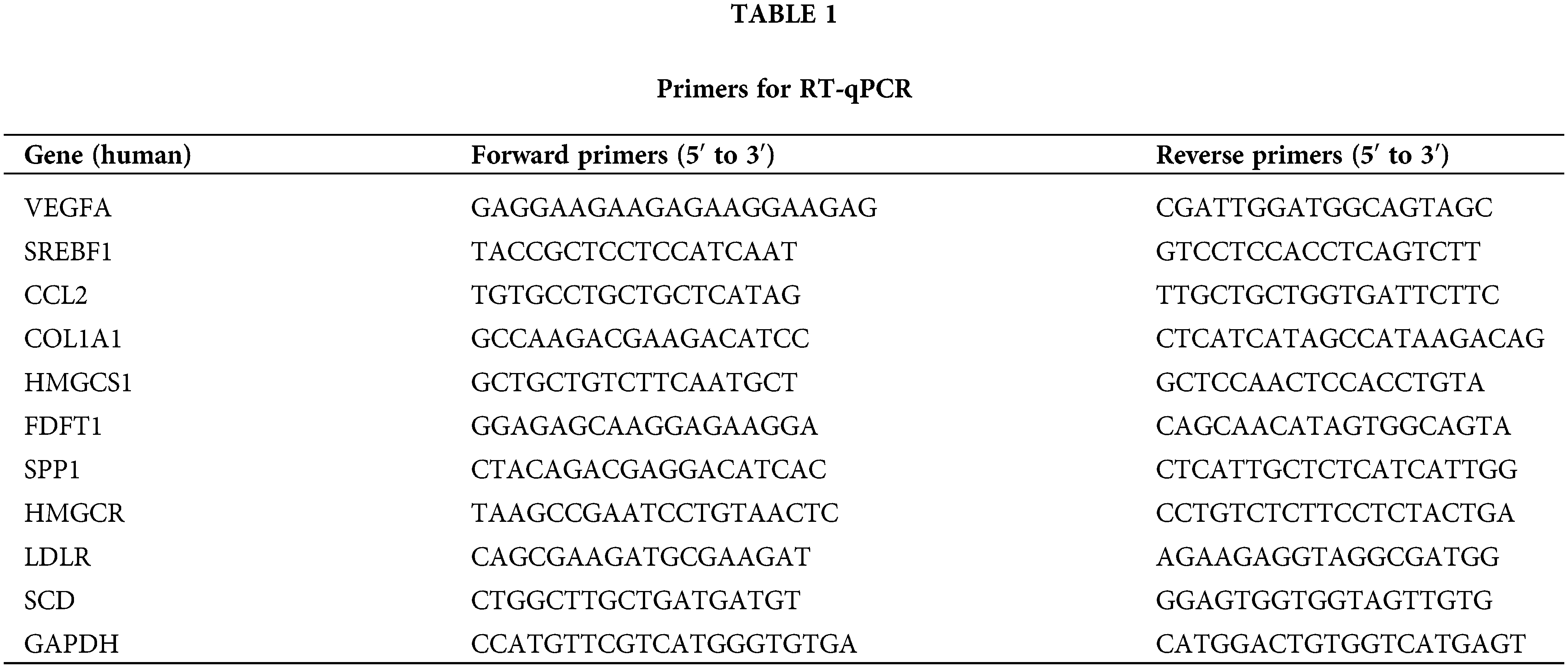
A total of 1 × 106 cells were collected and total RNA was extracted according to the manufacturer’s instructions. The OD values of total RNA at 260 and 280 nm were detected via an ultraviolet spectrophotometer, and the purity and concentration were determined according to the OD260/OD280 values. cDNA was obtained by reverse transcription of RNA according to the instructions of the Thermo Scientific RevertAid First Strand cDNA Synthesis Kit (Cat. K1621). PCR was performed on an ABI7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA) with three replicate wells per sample. GAPDH was used as an endogenous control, and the relative expression was calculated as 2−ΔΔCt using the comparative critical cycle (Ct) method to compare the differences between the two groups. The primers used for RT-qPCR are listed in Table 1.
All the numerical data are presented as the means ± SEMs. The data were analyzed by two-tailed t test with ANOVA (GraphPad Prism 8, San Diego, CA, USA). p < 0.05 was considered to indicate statistical significance.
Honokiol induces apoptosis in human glioma cells
CCK-8 was used to determine the half maximal inhibitory concentration (IC50) of honokiol in U87-MG and U251-MG cells. As a result, the IC50 of honokiol in U87-MG cells was 36.27 μM, and the IC50 of honokiol in U251-MG cells was 59.53 μM (Fig. 1A). As shown in Fig. 1B, honokiol had dose- and time-dependent effects on cell viability. In U87-MG cells, the optimum concentration of honokiol was 20 µM, and the optimum time was 48 h; in U251-MG cells, the optimum concentration of honokiol was 40 µM, and the optimum time was 48 h. In all the following experiments, 20 µM honokiol was applied to U87-MG cells, and 40 µM honokiol was used to U251-MG cells for 48 h.
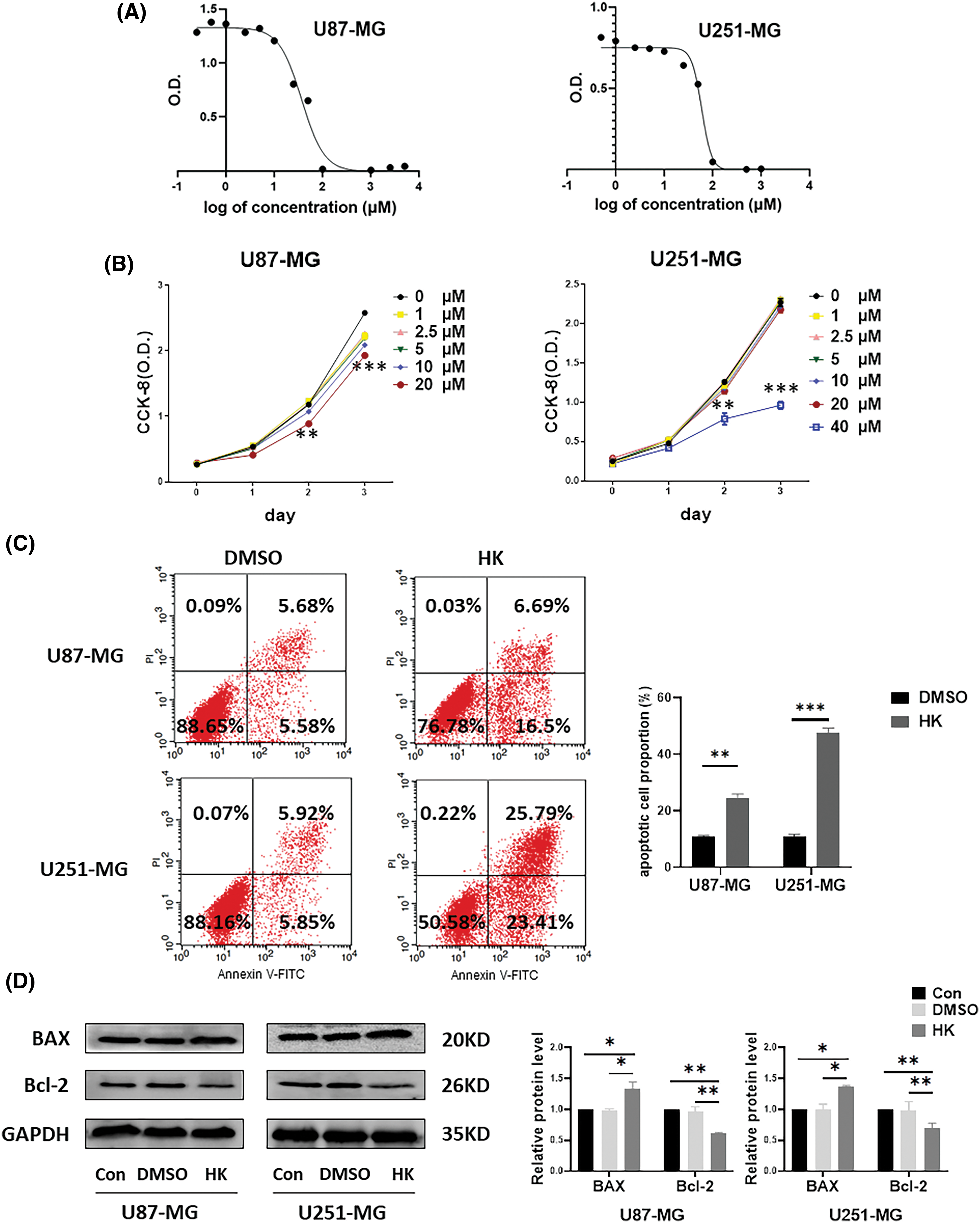
Figure 1: Honokiol induces cell apoptosis in human glioma cells. (A) A CCK-8 assay was performed to determine the IC50 of honokiol on the U87-MG and U251-MG glioma cells. (B) A CCK-8 assay was used to determine the optimal concentration and optimal duration of honokiol treatment in the glioma cell lines U87-MG and U251-MG. (C) An annexin V-FITC/PI assay was utilized to determine the effect of honokiol on human glioma cell apoptosis. (D) Western blot analysis was performed to determine the expression of apoptosis-related proteins (BAX and Bcl-2) in human glioma cells (*p < 0.05; **p < 0.01; ***p < 0.001; Con, U87-MG or U251-MG cells; DMSO, U87-MG or U251-MG cells treated with DMSO; HK, U87-MG or U251-MG cells treated with honokiol. The bars represent the means ± SEMs (n = 3)).
To determine the effect of honokiol on apoptosis, annexin V-FITC/PI staining and western blotting were performed. Annexin V-FITC/PI staining revealed that the percentage of apoptotic cells was greater in both honokiol-treated cell lines than in the control (Fig. 1C). Western blot analysis of the cells revealed that the apoptosis-associated protein BAX was significantly elevated in both honokiol-treated cell lines compared to the control cells, and the apoptosis-associated protein Bcl-2 was suppressed in both honokiol-treated cell lines compared to the control cells (Fig. 1D). These findings demonstrated that honokiol induced apoptosis in the human glioma cell lines U87-MG and U251-MG.
Identification of DEGs in honokiol-treated cells by RNA-seq
To clarify the possible molecular mechanisms by which honokiol induces apoptosis in human glioma cells, RNA-seq was performed. A total of 23,383 genes were detected, and 332 genes, including 163 upregulated and 169 downregulated genes, exhibited differential expression in honokiol-treated U251-MG cells compared with controls (Suppl. Table S1). A difference was considered to be statistically significant only when the fold change was ≥1.5 and p < 0.05. The 100 top DEGs, including 50 upregulated genes and 50 downregulated genes, between honokiol-treated U251-MG cells and the control cells are shown in Fig. 2 and Suppl. Table S2.
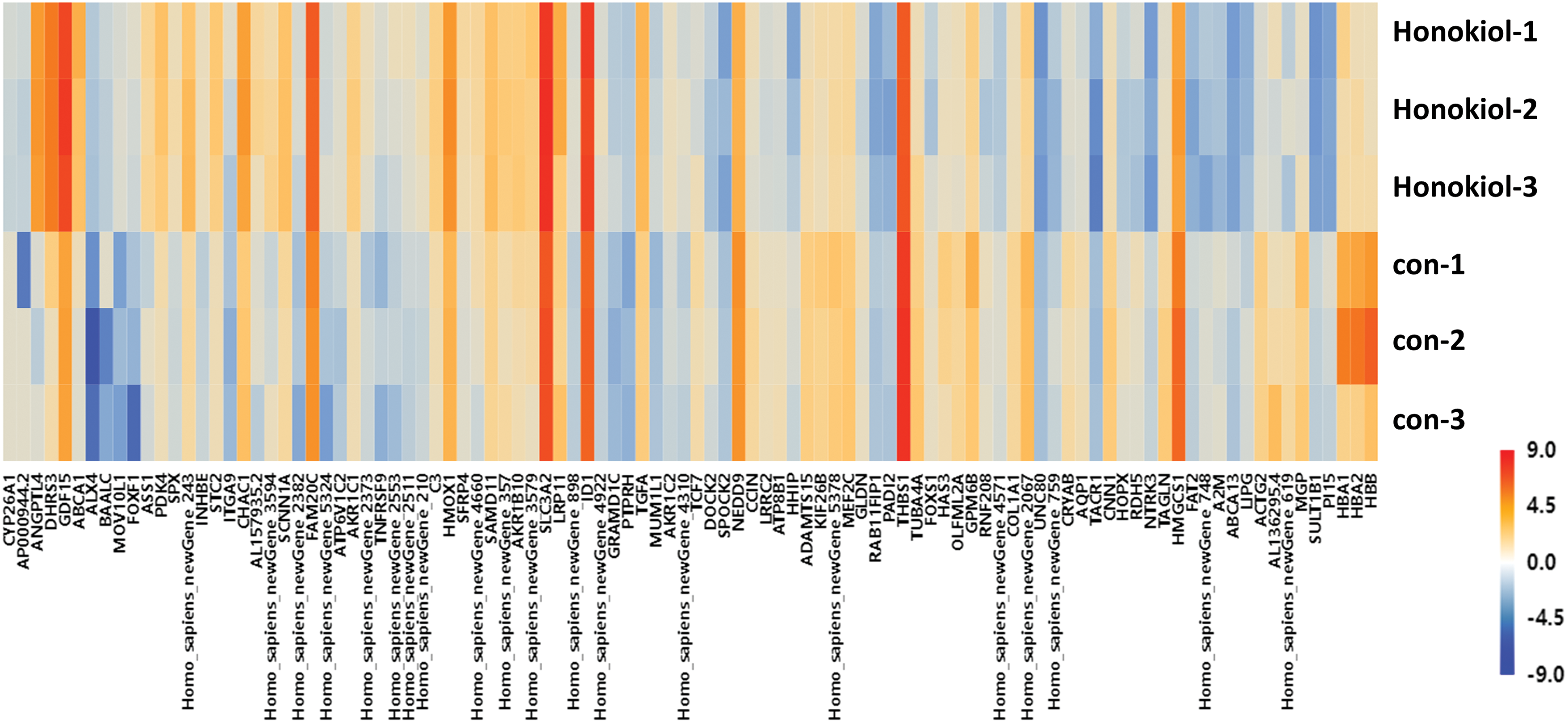
Figure 2: Heatmap of the top 100 DEGs (50 upregulated genes and 50 downregulated genes). Red, upregulated; blue, downregulated. Con, U251-MG cells; Honokiol, U251-MG cells treated with honokiol.
The GO analysis included biological process, cellular component and molecular function. GO analysis of the DEGs associated with biological processes revealed that those related to ‘response to hypoxia’, ‘cholesterol biosynthetic process’, ‘response to nutrient’, ‘cerebellum development cellular response to hypoxia’, ‘intrinsic apoptotic signaling pathway in response to endoplasmic reticulum’, and ‘stress’ were related to ‘response to hypoxia’, ‘cholesterol biosynthetic process via cystathionine’, and ‘response to retinoic acid’, among others (Fig. 3A). Furthermore, the DEGs were enriched mainly in cellular components, for instance, ‘extracellular region extracellular space’, ‘Golgi apparatus’, ‘cell surface’, ‘collagen trimer’, ‘hemoglobin complex’, ‘perikaryon’, and ‘T-tubule’ (Fig. 3B). In addition, GO analysis of molecular function showed that the DEGs were primarily related to ‘oxidoreductase activity growth factor activity’, ‘iron ion binding’, ‘oxygen transporter activity’, ‘oxygen binding’, and ‘ferric iron binding’, with others being enriched in ‘GTP binding’, ‘GTPase activity’, ‘drug binding’, and ‘structural constituent of cytoskeleton’ (Fig. 3C).
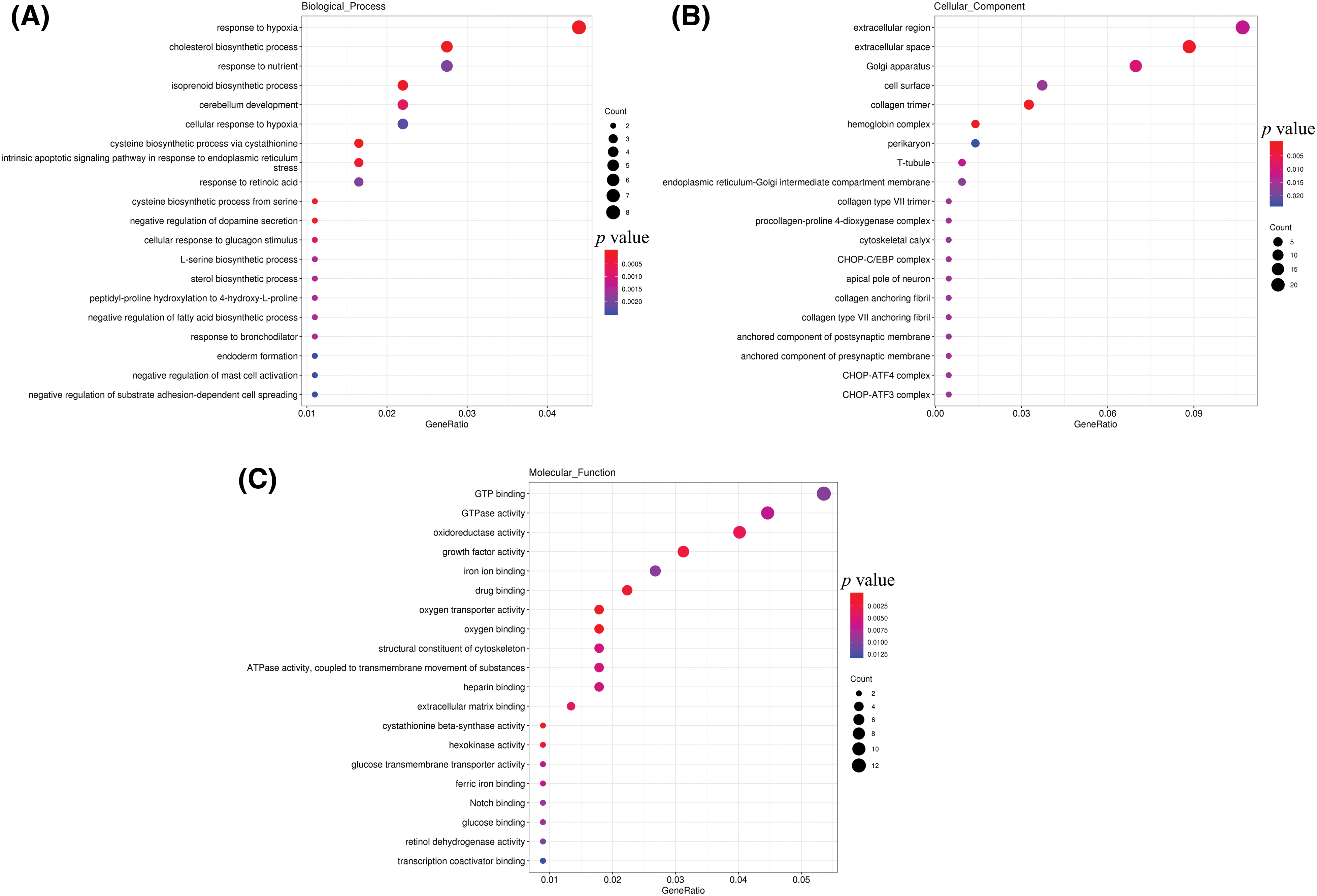
Figure 3: GO analysis of DEGs. (A) Biological process terms; (B) cellular component terms; (C) molecular function terms.
In the KEGG annotation, cellular processes, environmental information processing, human diseases, metabolism, and organismal systems terms were included. KEGG annotation of cellular processes revealed that the DEGs were associated with ‘apoptosis’, ‘phagosome’, ‘focal adhesion’, ‘endocytosis’, and ‘regulation of actin cytoskeleton’. KEGG annotation of the environmental information processing pathway showed that the DEGs were associated with ‘PI3K- Akt signaling pathway’, ‘Ras signaling pathway’, ‘mTOR signaling pathway’, ‘AMPK signaling pathway’, ‘Cytokine-cytokine receptor interaction’, ‘ECM-receptor interaction’, ‘Rap1 signaling pathway’, ‘Neuroactive ligand-receptor interaction’, ‘HIF-1 signaling pathway’, and the ‘MAPK signaling pathway’ (Fig. 4).
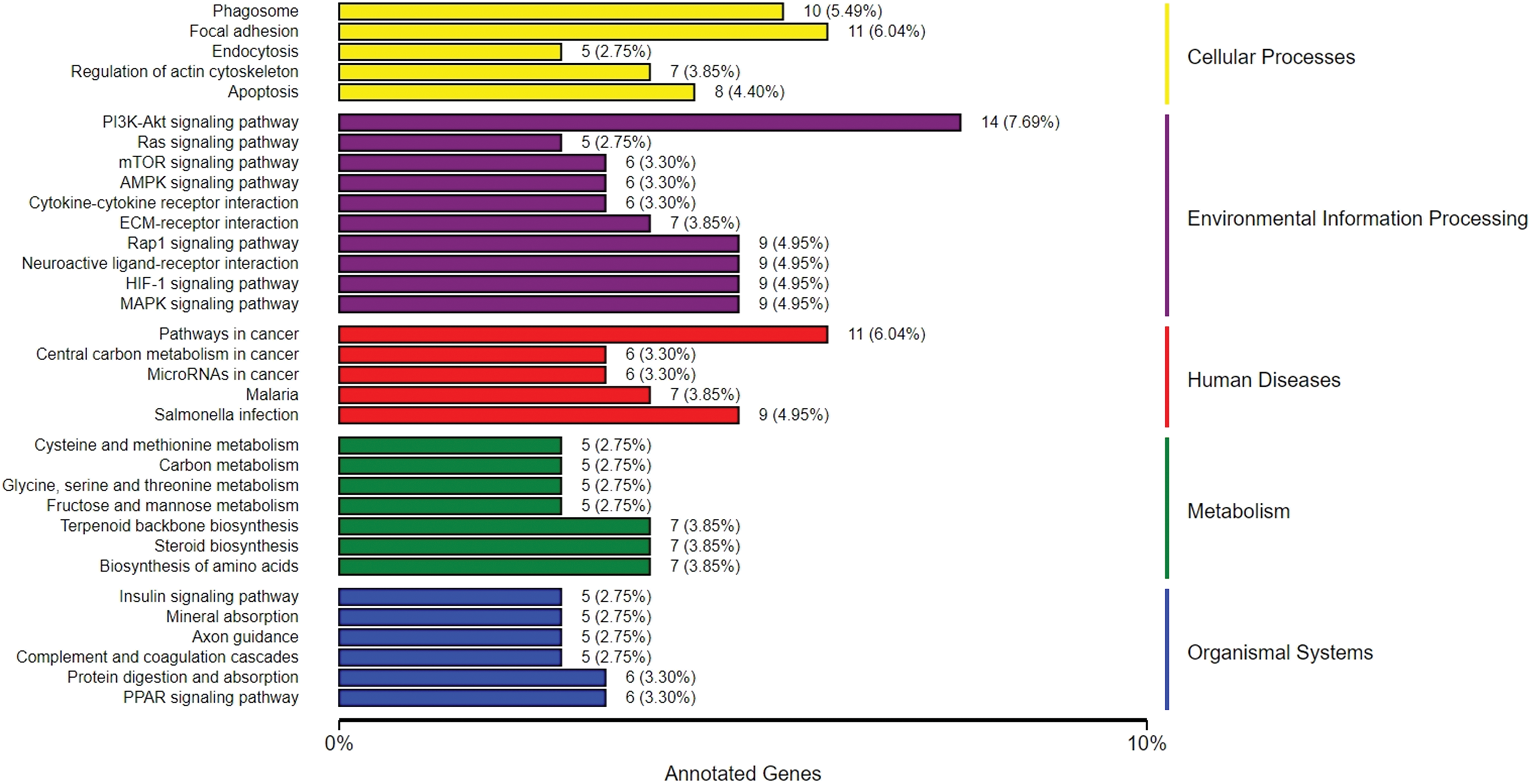
Figure 4: KEGG annotation of DEGs. PI3K, Phosphatidylinositol-4,5-bisphosphate 3-kinase; Akt, protein kinase B; mTOR, mechanistic target of rapamycin; AMPK, AMP-activated protein kinase; ECM, extracellular matrix; Rap1, Ras-associated protein 1; HIF-1, Hypoxia-inducible factor 1; MAPK, mitogen-activated protein kinases.
One of the signaling pathways the ‘MAPK signaling pathway’ was selected for detection, and western blotting was utilized. As a result, there were no differences in the expression of p38, JNK or ERK between honokiol-treated cells and control cells. Moreover, the expression levels of p-p38, p-JNK and p-ERK in honokiol-treated U87-MG and U251-MG cells were significantly cells than those in the control cells (Fig. 5), indicating that honokiol activated cells MAPK signaling pathway in human glioma cells.
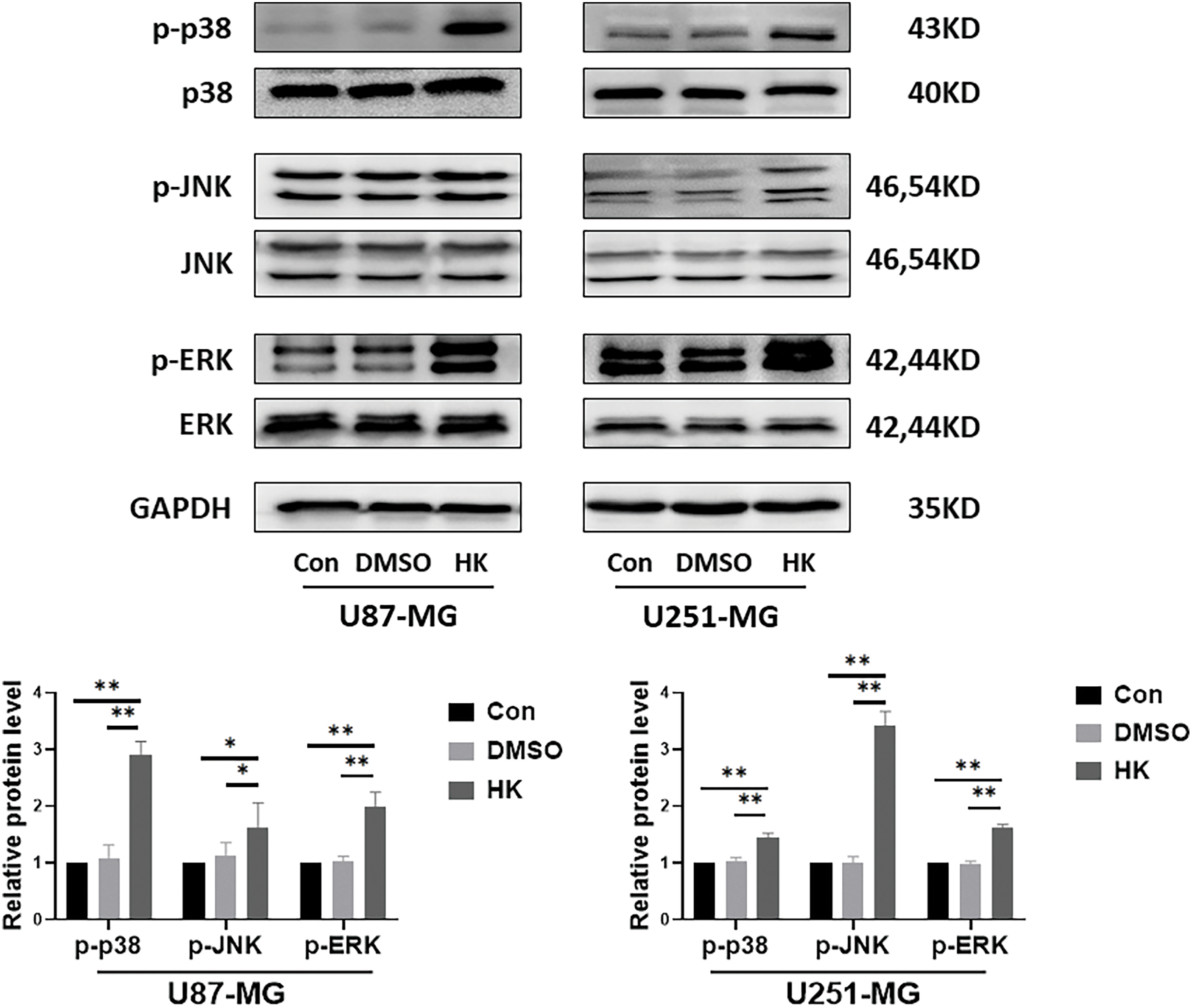
Figure 5: Honokiol activated the MAPK signaling pathway in human glioma cells. Western blot analysis was used to determine the expression of p38, p-p38, JNK, p-JNK, ERK, and p-ERK in human glioma cells. The results of densitometric analysis of relative expression levels after normalization to the loading control GAPDH are presented (*p < 0.05, **p < 0.01; Con, U87-MG or U251-MG cells; DMSO, U87-MG or U251-MG cells treated with DMSO; HK, U87-MG or U251-MG cells treated with honokiol. The bars represent the means ± SEMs (n = 3)).
KEGG pathway analysis was applied to screen the enrichment of signaling pathways. The DEGs were related mainly to the ‘HIF-1 signaling pathway’, ‘ferroptosis’, ‘PPAR signaling pathway’, ‘PI3K- Akt signaling pathway’, and ‘phagosome’ (Fig. 6). Among the signaling pathways, ‘ferroptosis’ was identified by western blot. The results showed that the ferroptosis-associated protein GPX4 was suppressed in honokiol-treated cells compared to control cells. Moreover, the ferroptosis-associated protein TF was upregulated in both honokiol-treated cell lines compared to the control (Fig. 7). The data above illustrated that honokiol induced ferroptosis in human glioma cells.
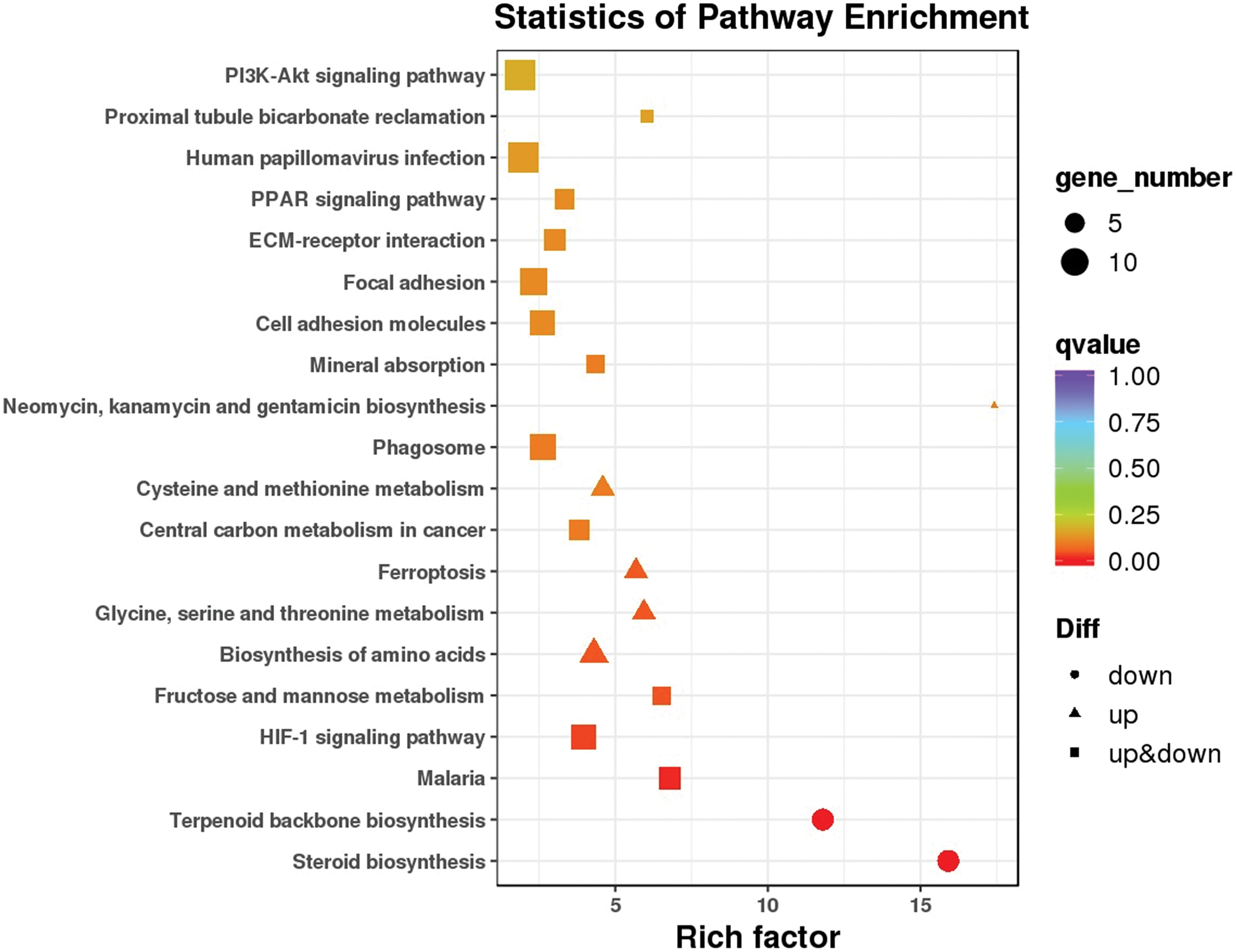
Figure 6: KEGG pathway enrichment analysis. KEGG pathway analysis was applied to screen the enrichment of signaling pathways.
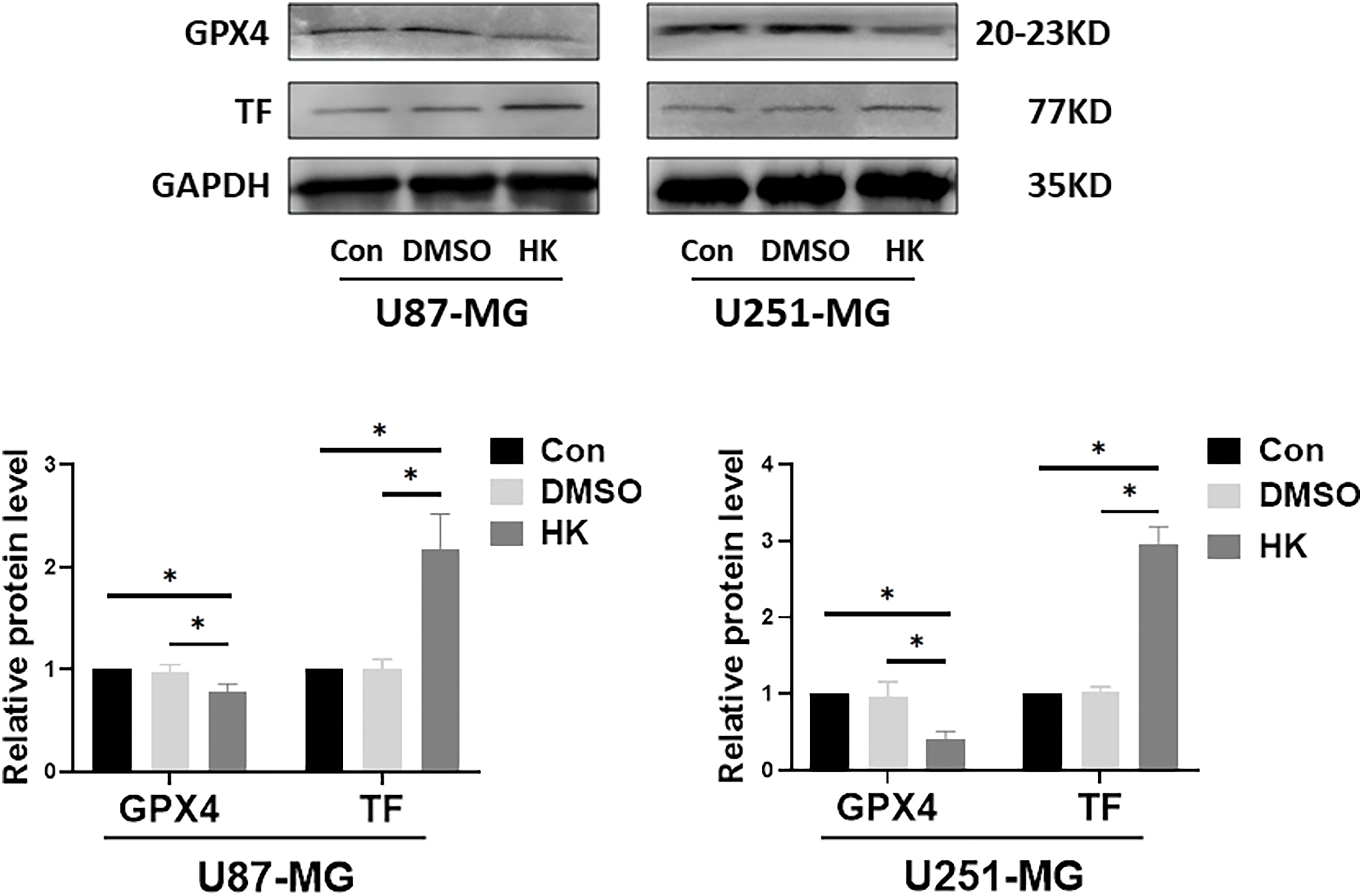
Figure 7: Honokiol activated ferroptosis in human glioma cells. Western blot analysis was performed to detect the expression of ferroptosis-related proteins (GPX4 and TF) in human glioma cells. The results of densitometric analysis of relative expression levels after normalization to the loading control GAPDH are presented (*p < 0.05; Con, U87-MG or U251-MG cells; DMSO, U87-MG or U251-MG cells treated with DMSO; HK, U87-MG or U251-MG cells treated with honokiol. The bars represent the means ± SEMs (n = 3)).
Protein-Protein Interaction (PPI) Network
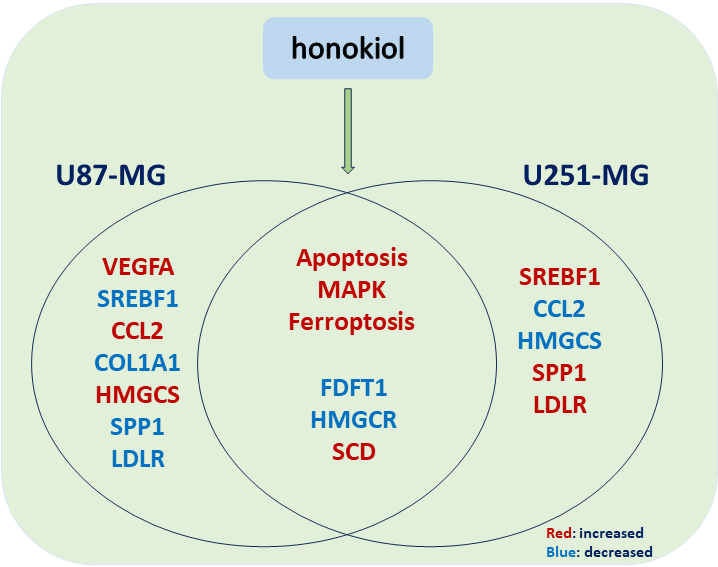
STRING was utilized in this work to predict protein interactions between DEGs. A total of 10 hub genes were obtained based on their association with other proteins, including vascular endothelial growth factor A (VEGFA), sterol regulatory element-binding protein 1 (SREBF1), C-C motif chemokine ligand 2 (CCL2), collagen type I alpha 1 (COL1A1), hydroxy-3-methylglutaryl coenzyme A synthetase 1 (HMGCS1), farnesyl-diphosphate farnesyltransferase (FDFT1), secreted phosphoprotein 1 (SPP1), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR), low density lipoprotein receptor (LDLR), and stearoyl-CoA desaturase (SCD) (Table 2). Among these genes, VEGFA had the highest node degree (48). A PPI was constructed as displayed in Fig. 8. RT-qPCR was subsequently utilized to detect the expression of the 10 hub genes in the honokiol-treated U251-MG cells and in the control cells. Similarly, compared with those in control cells, the expression levels of SREBF1, SPP1, LDLR, and SCD were increased, and the expression levels of CCL2, HMGCS1, FDFT1, and HMGCR were inhibited in honokiol-treated U251-MG cells. Moreover, there was no change in the expression of VEGFA or COL1A1 (Fig. 9). Moreover, the expression of the 10 hub genes in honokiol-treated U87-MG cells was also detected. The results showed that the expression levels of VEGFA, CCL2, HMGCS1, and SCD were increased, while the expression levels of SREBF1, COL1A1, FDFT1, HMGCR, SPP1, and LDLR were suppressed in honokiol-treated U87-MG cells compared with those in control cells. The expression patterns of the 10 hub genes in honokiol-treated U87-MG cells were not consistent with those in honokiol-treated U251-MG cells. Therefore, honokiol might have different effects on U87-MG and U251-MG cells.
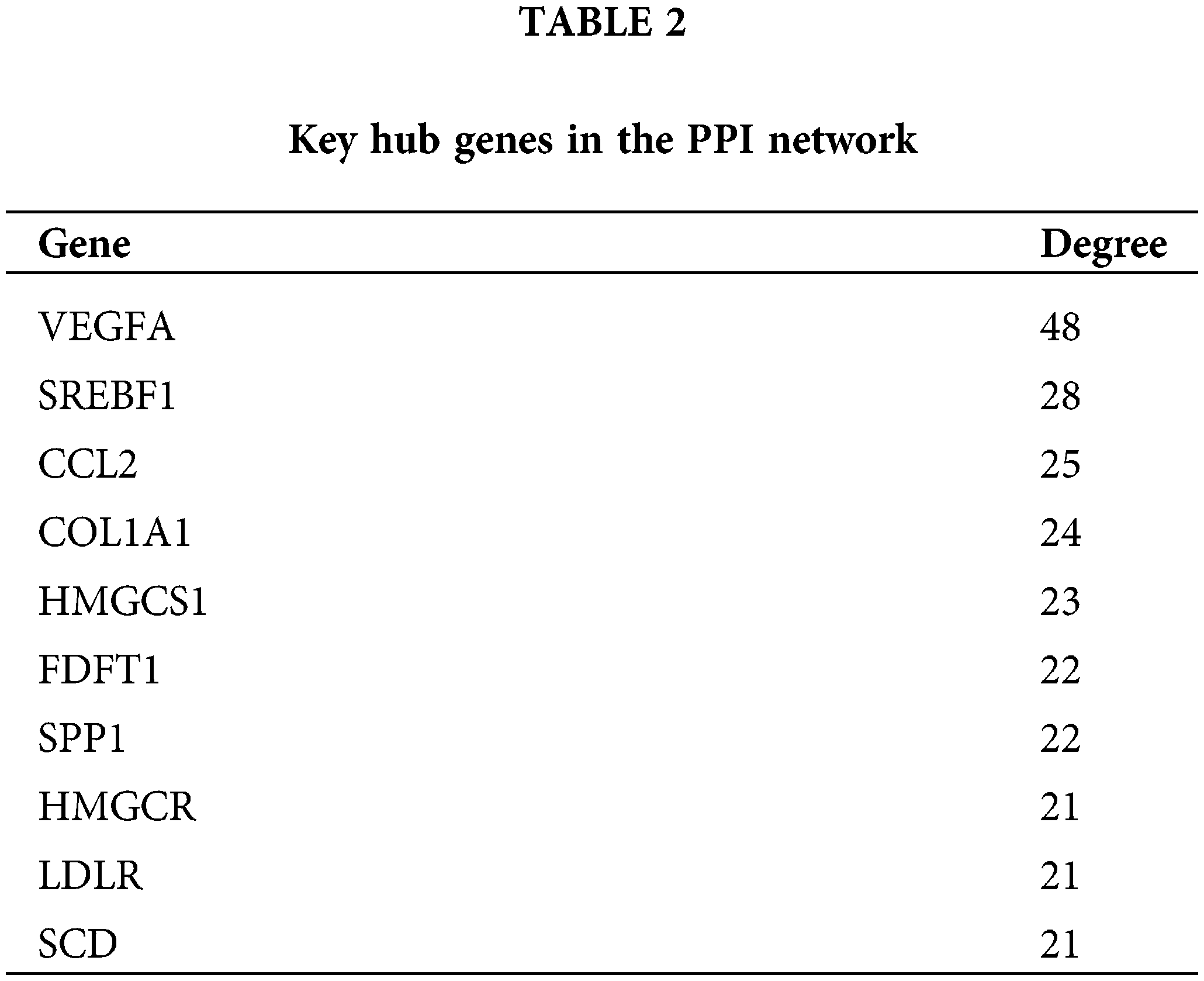
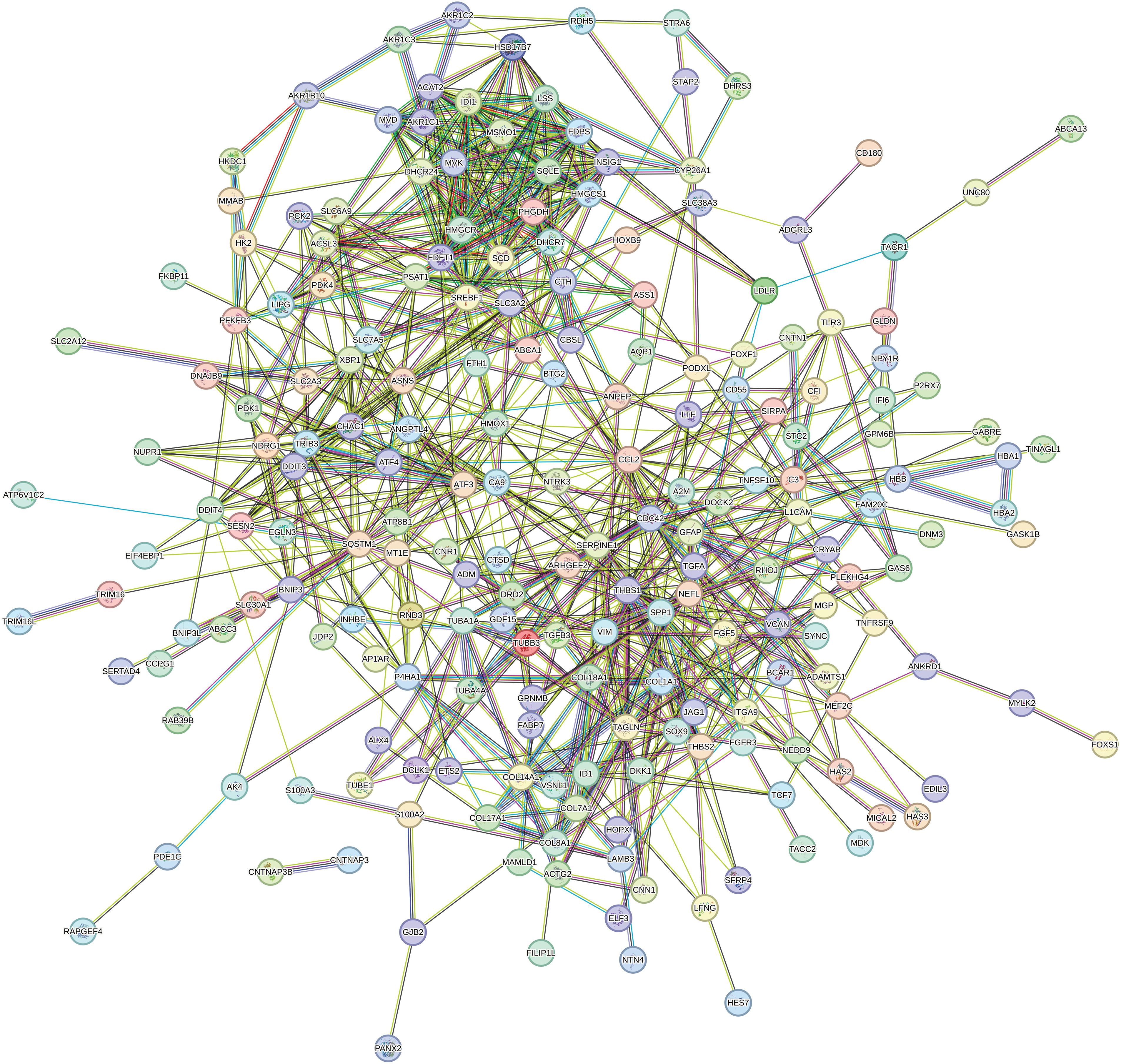
Figure 8: PPI network. PPI network of the DEGs between honokiol-treated U251-MG cells and the control cells. Circles, proteins; lines, strong association between proteins.
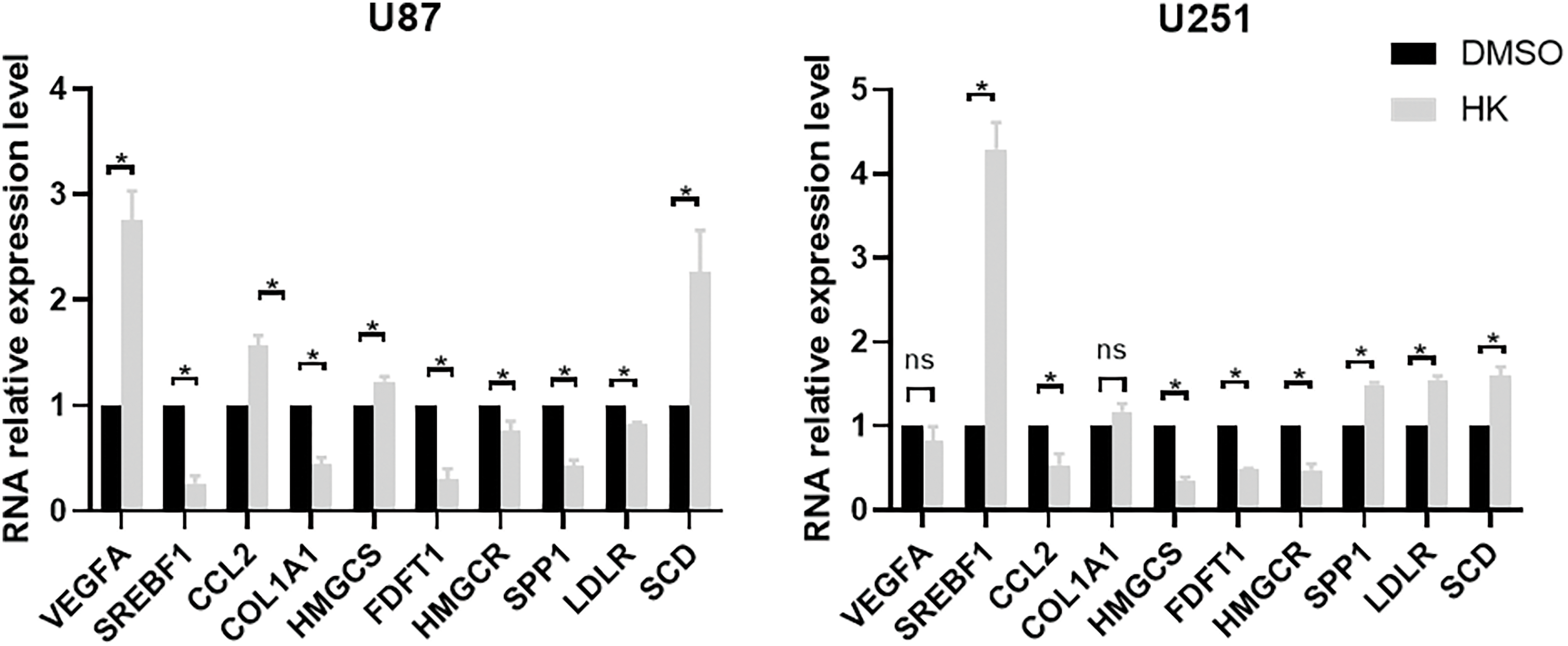
Figure 9: RT-qPCR of the honokiol-treated U251-MG cells and the control cells verified the mRNA expression of the 10 hub genes identified in the PPI network. *p < 0.05, ns, no significance. DMSO, U87-MG or U251-MG cells treated with DMSO; HK, U87-MG or U251-MG cells treated with honokiol. The bars represent the means ± SEMs (n = 3).
Glioma, the most aggressive form of nervous system neoplasm, accounts for approximately 80% of central nervous system malignancies [22]. Moreover, the prognosis of glioma is not ideal. In the United States, the overall ten-year survival rate is 2.9% for gliomas and 44.6% for malignant gliomas [22,23]. Over the years, with the application of computed tomography (CT), occupational exposure and ionizing radiation have led to more brain tumors [24]. Currently, the main treatments for glioma are surgery, chemotherapy, and radiation. However, during recent decades, the prognosis of glioma has not been satisfactory, and new methods are urgently needed to improve patient prognosis [25,26].
Honokiol, a small molecular weight natural product, is isolated and purified from the herb Magnolia officinalis, which is commonly used in Asia. Many investigators have reported that honokiol has anticancer effects, such as suppressing cell growth, inducing cell cycle arrest, and promoting autophagy and apoptosis. Moreover, in recent years, many authorities of food safety have evaluated honokiol and deemed it safe [27]. Hence, honokiol could be a prospective clinical drug for curing glioma.
Annexin V-FITC/PI staining and western blotting were performed to determine the effect of honokiol on apoptosis in human glioma cells. As a result, the proportion of apoptotic cells was greater in the honokiol-treated U87-MG and U251-MG cells than in the control cells. Similarly, compared with that in the control cells, the expression of the apoptosis-associated protein BAX was increased, and the expression of Bcl-2 was suppressed in both honokiol-treated cells, indicating that honokiol induced apoptosis in the human glioma cell lines U87-MG and U251-MG. These data were consistent with previous studies. Fan et al. [28] reported that honokiol inhibited the proliferation, migration and invasion of glioma cells; promoted cell apoptosis with the decreased the expression of Bcl-2 and increased the expression of BAX. Wu et al. [29] showed that honokiol induced the apoptosis of temozolomide (TMZ)-resistant glioma cells by activating caspase 9. Moreover, honokiol promoted TMZ-induced DNA breakage and apoptosis in U87 MG cells, via the promotion of TMZ-induced BAX translocation [30]. In addition, honokiol induces cell apoptosis in many other tumors, such as lung squamous cell carcinoma [31], ovarian carcinoma [32], bladder cancer [33], osteosarcoma [34], and medulloblastoma [35]. However, honokiol could reduce the apoptosis of myocardial cells induced by lipopolysaccharide in septic mice [36] and reduce nerve cell apoptosis in a rat traumatic brain injury model [37].
Due to the extensive use of honokiol, RNA-seq was utilized to demonstrate the possible molecular mechanisms by which honokiol promotes apoptosis in human glioma cells. A total of 332 DEGs were found. GO analysis of the biological processes revealed that DEGs were enriched in ‘response to hypoxia’, ‘cholesterol biosynthetic process’, ‘cerebellum development cellular response to hypoxia’, ‘intrinsic apoptotic signaling pathway in response to endoplasmic reticulum’, and ‘stress’. These biological processes are related to apoptosis. As reported, honokiol inhibited pancreatic β cell apoptosis induced by hypoxia [38]. Inhibition of cholesterol synthesis could induce cell apoptosis in glioblastoma [39]. However, the role of cholesterol in cell apoptosis differs, as cholesterol induces cell apoptosis in many other cells [40,41]. Moreover, researchers have shown that honokiol is an effective radical scavenger that could alleviate oxidative stress on various occasions [38], reduce HIF-1α protein levels and suppress hypoxia-related signaling pathways [42]. In addition, GO analysis of molecular function illustrated that honokiol was related to oxidative stress, ferroptosis, drug binding and the cytoskeleton. Researchers have found that the antioxidant capacity of honokiol is 1000 times greater than that of vitamin E [43]. Similarly, studies have shown that honokiol decreases the apoptotic rate and elevates the viability of podocytes during mouse podocyte apoptosis induced by hydrogen peroxide (H2O2) [44]. Moreover, Hou et al. demonstrated that honokiol prevents H2O2-induced oxidative damage by upregulating the levels of antioxidant enzymes [45]. Furthermore, honokiol was related to ferroptosis. Lai and colleagues [46] reported that honokiol upregulated Heme-oxygenase 1 (HMOX1) to induce ferroptosis in acute myeloid leukemia cells. Nevertheless, Guo C and colleagues [47] demonstrated that honokiol induced ferroptosis by suppressing GPX4 activity in colon cancer cells. In contrast to these findings, Hu et al. [48] reported that honokiol inhibited ferroptosis to attenuate high glucose-induced peripheral neuropathy in Schwann cells.
KEGG annotation of the environmental information processing pathway showed that the DEGs were associated with many signaling pathways. We selected the ‘MAPK signaling pathway’ for verification, as researchers believe that MAPK signaling is related to apoptosis [49,50]. As a result, the MAPK signaling pathway was activated in the honokiol-treated U87-MG and U251-MG cells, which is consistent with the findings of previous studies [35,51,52]. However, honokiol inhibited the MAPK signaling pathway in an immunological model of liver fibrosis [53], in mouse embryonic stem cell-derived endothelial cells [54], and in oral squamous cell carcinoma [55]. The MAPK signaling pathway plays different roles in different cells. KEGG pathway analysis demonstrated that the DEGs were associated mainly with ‘HIF-1 signaling pathway’, ‘ferroptosis’, ‘PPAR signaling pathway’, ‘PI3K-Akt signaling pathway’, and ‘phagosome’. Among the signaling pathways, ‘ferroptosis’ was identified by western blot (downregulation of GPX4 and upregulation of TF), indicating that honokiol induces human glioma cell ferroptosis. Similarly, honokiol induced ferroptosis in acute myeloid leukemia cells [46] and colon cancer cells [47]. However, honokiol inhibited ferroptosis in Schwann cells [48]. Therefore, honokiol could induce or inhibit ferroptosis in different cells. In this study, we also found that the MAPK signaling pathway, an upstream signaling pathway involved in ferroptosis, was activated in honokiol-treated cells. Hence, honokiol might induce ferroptosis in human glioma cells through activating the MAPK signaling pathway.
By constructing a PPI network, we identified a series of hub genes related to honokiol. In this study, the top 10 hub nodes including VEGFA [56–58], SREBF1 [59,60], CCL2 [61,62], COL1A1 [63,64], HMGCS1 [65,66], FDFT1 [67], SPP1 [68], HMGCR [69,70], LDLR [71], and SCD [72], were previously reported to be connected with apoptosis in various cancers. The expression of the top 10 hub genes was verified by RT-qPCR, and the expression of these genes in honokiol-treated U87-MG cells was not consistent with that in honokiol-treated U251-MG cells. For example, SREBF1 was decreased in honokiol-treated U87-MG cells but increased in honokiol-treated U251-MG cells compared to that in the control. It has been reported that ectopic expression of SREBF1could promotes the tumor development [73,74]. However, it has been reported that overexpression of SREBF1 enhances the apoptosis in high glucose-treated podocytes [75]. Therefore, honokiol might induce apoptosis via different pathways in U87-MG and U251-MG cells. The PPI network provides us with ideas for future research on the mechanism of honokiol induced glioma cells cell apoptosis. We would further investigate whether honokiol could regulate apoptosis through the above genes and elucidate the specific underlying molecular mechanisms involved.
In conclusion, this study investigated the role of honokiol in inducing apoptosis in human glioma cells, and provided a comprehensive analysis of DEGs in honokiol-treated U251-MG cells and related signaling pathways, which might be beneficial for the treatment of human glioma. In the current study, we did not deeply explore the mechanism of honokiol inducing apoptosis, but these data provide new ideas for future research and therapy for patients with glioma using honokiol, and we would investigate in the future.
Acknowledgement: We thank all participants involved in this research.
Funding Statement: The study was supported by the Natural Science Foundation of Jilin Province (Grant No. 20200201444JC).
Author Contributions: The study was designed by Haiyan Liu and Yunbao Guo. Yunbao Guo and Xu Liu carried out most of the experiments. Qi Xu performed the statistical analysis. Yan Lu drafted the manuscript. Xiaotong Zhou and Jiawei Liu assisted with the experiments and helped edit the paper. All the authors reviewed the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available in the NCBI repository (https://submit.ncbi.nlm.nih.gov/subs/sra/SUB14098148).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no competing interests regarding the publication of this paper.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/biocell.2024.049748.
References
1. Xu C, Xiao M, Li X, Xin L, Song J, Zhan Q, et al. Origin, activation, and targeted therapy of glioma-associated macrophages. Front Immunol. 2022;13:974996. doi:10.3389/fimmu.2022.974996 [Google Scholar] [PubMed] [CrossRef]
2. Yang XC, Qu JG, Li JJ. Nicotinic acid induces apoptosis of glioma cells via the calcium-dependent endoplasmic reticulum stress pathway. BIOCELL. 2022;46(4):1041–51. doi:10.32604/biocell.2022.017383. [Google Scholar] [CrossRef]
3. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 2021;23(8):1231–51. doi:10.1093/neuonc/noab106 [Google Scholar] [PubMed] [CrossRef]
4. Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–66. doi:10.1016/S1470-2045(09)70025-7 [Google Scholar] [PubMed] [CrossRef]
5. Li H, Li W, Li J, Li S, Kuang L, Pang F, et al. Honokiol microemulsion causes stage-dependent toxicity via dual roles in oxidation-reduction and apoptosis through FoxO signaling pathway. Cells. 2022;11(22):3562. doi:10.3390/cells11223562 [Google Scholar] [PubMed] [CrossRef]
6. Xu T, Tian W, Zhang Q, Liu J, Liu Z, Jin J, et al. Novel 1,3,4-thiadiazole/oxadiazole-linked honokiol derivatives suppress cancer via inducing PI3K/Akt/mTOR-dependent autophagy. Bioorg Chem. 2021;115:105257. doi:10.1016/j.bioorg.2021.105257 [Google Scholar] [PubMed] [CrossRef]
7. Zhu J, Xu S, Gao W, Feng J, Zhao G. Honokiol induces endoplasmic reticulum stress-mediated apoptosis in human lung cancer cells. Life Sci. 2019;221:204–11. doi:10.1016/j.lfs.2019.01.046 [Google Scholar] [PubMed] [CrossRef]
8. Medra A, Witkowska M, Majchrzak A, Cebula-Obrzut B, Bonner MY, Robak T, et al. Pro-apoptotic activity of new honokiol/triphenylmethane analogues in B-cell lymphoid malignancies. Molecules. 2016;21(8):995. doi:10.3390/molecules21080995 [Google Scholar] [PubMed] [CrossRef]
9. Lee YJ, Lee YM, Lee CK, Jung JK, Han SB, Hong JT. Therapeutic applications of compounds in the Magnolia family. Pharmacol Ther. 2011;130(2):157–76. doi:10.1016/j.pharmthera.2011.01.010 [Google Scholar] [PubMed] [CrossRef]
10. Wang J, Liu D, Guan S, Zhu W, Fan L, Zhang Q, et al. Hyaluronic acid-modified liposomal honokiol nanocarrier: enhance anti-metastasis and antitumor efficacy against breast cancer. Carbohydr Polym. 2020;235:115981. doi:10.1016/j.carbpol.2020.115981 [Google Scholar] [PubMed] [CrossRef]
11. Banik K, Ranaware AM, Deshpande V, Nalawade SP, Padmavathi G, Bordoloi D, et al. Honokiol for cancer therapeutics: a traditional medicine that can modulate multiple oncogenic targets. Pharmacol Res. 2019;144:192–209. doi:10.1016/j.phrs.2019.04.004 [Google Scholar] [PubMed] [CrossRef]
12. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinform. 2010;26(1):139–40. doi:10.1093/bioinformatics/btp616 [Google Scholar] [PubMed] [CrossRef]
13. Talarek S, Listos J, Barreca D, Tellone E, Sureda A, Nabavi SF, et al. Neuroprotective effects of honokiol: from chemistry to medicine. Biofactors. 2017;43(6):760–9. doi:10.1002/biof.v43.6. [Google Scholar] [CrossRef]
14. Wang D, Cao L, Zhou X, Wang G, Ma Y, Hao X, et al. Mitigation of honokiol on fluoride-induced mitochondrial oxidative stress, mitochondrial dysfunction, and cognitive deficits through activating AMPK/PGC-1α/Sirt3. J Hazard Mater. 2022;437:129381. doi:10.1016/j.jhazmat.2022.129381 [Google Scholar] [PubMed] [CrossRef]
15. Zhang P, Liu X, Zhu Y, Chen S, Zhou D, Wang Y. Honokiol inhibits the inflammatory reaction during cerebral ischemia reperfusion by suppressing NF-κB activation and cytokine production of glial cells. Neurosci Lett. 2013;534:123–7. doi:10.1016/j.neulet.2012.11.052 [Google Scholar] [PubMed] [CrossRef]
16. Qu C, Li QP, Su ZR, Ip SP, Yuan QJ, Xie YL, et al. Nano-honokiol ameliorates the cognitive deficits in TgCRND8 mice of Alzheimer’s disease via inhibiting neuropathology and modulating gut microbiota. J Adv Res. 2022;35:231–43. doi:10.1016/j.jare.2021.03.012 [Google Scholar] [PubMed] [CrossRef]
17. Lin FH, Hsu YC, Chang KC, Shyong YJ. Porous hydroxyapatite carrier enables localized and sustained delivery of honokiol for glioma treatment. Eur J Pharm Biopharm. 2023;189:224–32. doi:10.1016/j.ejpb.2023.06.016 [Google Scholar] [PubMed] [CrossRef]
18. Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11(2):R14. doi:10.1186/gb-2010-11-2-r14 [Google Scholar] [PubMed] [CrossRef]
19. Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4 [Google Scholar] [PubMed]
20. Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21(19):3787–93. doi:10.1093/bioinformatics/bti430 [Google Scholar] [PubMed] [CrossRef]
21. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi:10.1101/gr.1239303 [Google Scholar] [PubMed] [CrossRef]
22. Ostrom QT, Gittleman H, Liao P, Vecchione-Koval T, Wolinsky Y, Kruchko C, et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro Oncol. 2017;19(suppl_5):v1–88. doi:10.1093/neuonc/nox158 [Google Scholar] [PubMed] [CrossRef]
23. Ostrom QT, Gittleman H, Truitt G, Boscia A, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011–2015. Neuro Oncol. 2018;20(suppl_4):iv1–86. doi:10.1093/neuonc/noy131 [Google Scholar] [PubMed] [CrossRef]
24. Li J, Meng Q, Zhou X, Zhao H, Wang K, Niu H, et al. Gospel of malignant glioma: oncolytic virus therapy. Gene. 2022;818:146217. doi:10.1016/j.gene.2022.146217 [Google Scholar] [PubMed] [CrossRef]
25. Coffin R. Interview with Robert Coffin, inventor of T-VEC: the first oncolytic immunotherapy approved for the treatment of cancer. Immunotherapy. 2016;8(2):103–6. doi:10.2217/imt.15.116 [Google Scholar] [PubMed] [CrossRef]
26. Hamid O, Ismail R, Puzanov I. Intratumoral immunotherapy-update 2019. Oncologist. 2020;25(3):e423–38. doi:10.1634/theoncologist.2019-0438 [Google Scholar] [PubMed] [CrossRef]
27. Sarrica A, Kirika N, Romeo M, Salmona M, Diomede L. Safety and toxicology of magnolol and honokiol. Planta Med. 2018;84(16):1151–64. doi:10.1055/a-0642-1966 [Google Scholar] [PubMed] [CrossRef]
28. Fan Y, Xue W, Schachner M, Zhao W. Honokiol eliminates glioma/glioblastoma stem cell-like cells via JAK-STAT3 signaling and inhibits tumor progression by targeting epidermal growth factor receptor. Cancers. 2018;11(1):22. doi:10.3390/cancers11010022 [Google Scholar] [PubMed] [CrossRef]
29. Wu GJ, Yang ST, Chen RM. Major contribution of caspase-9 to honokiol-induced apoptotic insults to human drug-resistant glioblastoma cells. Molecules. 2020;25(6):1450. doi:10.3390/molecules25061450 [Google Scholar] [PubMed] [CrossRef]
30. Chio CC, Tai YT, Mohanraj M, Liu SH, Yang ST, Chen RM. Honokiol enhances temozolomide-induced apoptotic insults to malignant glioma cells via an intrinsic mitochondrion-dependent pathway. Phytomedicine. 2018;49:41–51. doi:10.1016/j.phymed.2018.06.012 [Google Scholar] [PubMed] [CrossRef]
31. Cen M, Yao Y, Cui L, Yang G, Lu G, Fang L, et al. Honokiol induces apoptosis of lung squamous cell carcinoma by targeting FGF2-FGFR1 autocrine loop. Cancer Med. 2018;7(12):6205–18. doi:10.1002/cam4.2018.7.issue-12. [Google Scholar] [CrossRef]
32. Lee JS, Sul JY, Park JB, Lee MS, Cha EY, Ko YB. Honokiol induces apoptosis and suppresses migration and invasion of ovarian carcinoma cells via AMPK/mTOR signaling pathway. Int J Mol Med. 2019;43(5):1969–78 [Google Scholar] [PubMed]
33. Wang HH, Chen Y, Changchien CY, Chang HH, Lu PJ, Mariadas H, et al. Pharmaceutical evaluation of honokiol and magnolol on apoptosis and migration inhibition in human bladder cancer cells. Front Pharmacol. 2020;11:549338. doi:10.3389/fphar.2020.549338 [Google Scholar] [PubMed] [CrossRef]
34. Huang K, Chen Y, Zhang R, Wu Y, Ma Y, Fang X, et al. Honokiol induces apoptosis and autophagy via the ROS/ERK1/2 signaling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis. 2018;9(2):157. doi:10.1038/s41419-017-0166-5 [Google Scholar] [PubMed] [CrossRef]
35. Li S, Chen J, Fan Y, Wang C, Wang C, Zheng X, et al. Liposomal Honokiol induces ROS-mediated apoptosis via regulation of ERK/p38-MAPK signaling and autophagic inhibition in human medulloblastoma. Signal Transduct Target Ther. 2022;7(1):49. doi:10.1038/s41392-021-00869-w [Google Scholar] [PubMed] [CrossRef]
36. Liu A, Xun S, Zhou G, Zhang Y, Lin L. Honokiol alleviates sepsis-associated cardiac dysfunction via attenuating inflammation, apoptosis and oxidative stress. J Pharm Pharmacol. 2023;75(3):397–406. doi:10.1093/jpp/rgac102 [Google Scholar] [PubMed] [CrossRef]
37. Sun GW, Ding TY, Wang M, Hu CL, Gu JJ, Li J, et al. Honokiol reduces mitochondrial dysfunction and inhibits apoptosis of nerve cells in rats with traumatic brain injury by activating the mitochondrial unfolded protein response. J Mol Neurosci. 2022;72(12):2464–72. doi:10.1007/s12031-022-02089-5 [Google Scholar] [PubMed] [CrossRef]
38. Li CG, Ni CL, Yang M, Tang YZ, Li Z, Zhu YJ, et al. Honokiol protects pancreatic β cell against high glucose and intermittent hypoxia-induced injury by activating Nrf2/ARE pathway in vitro and in vivo. Biomed Pharmacother. 2018;97:1229–37. doi:10.1016/j.biopha.2017.11.063 [Google Scholar] [PubMed] [CrossRef]
39. Yanae M, Tsubaki M, Satou T, Itoh T, Imano M, Yamazoe Y, et al. Statin-induced apoptosis via the suppression of ERK1/2 and Akt activation by inhibition of the geranylgeranyl-pyrophosphate biosynthesis in glioblastoma. J Exp Clin Cancer Res. 2011;30(1):74. doi:10.1186/1756-9966-30-74 [Google Scholar] [PubMed] [CrossRef]
40. Li K, Deng Y, Deng G, Chen P, Wang Y, Wu H, et al. High cholesterol induces apoptosis and autophagy through the ROS-activated AKT/FOXO1 pathway in tendon-derived stem cells. Stem Cell Res Ther. 2020;11(1):131. doi:10.1186/s13287-020-01643-5 [Google Scholar] [PubMed] [CrossRef]
41. Chang H, Wang Y, Wu Y, Ma P, Song Y, Liu C, et al. Cardiac apoptosis caused by elevated cholesterol level in experimental autoimmune myocarditis. Exp Cell Res. 2020;395(1):112169. doi:10.1016/j.yexcr.2020.112169 [Google Scholar] [PubMed] [CrossRef]
42. Lan KL, Lan KH, Sheu ML, Chen MY, Shih YS, Hsu FC, et al. Honokiol inhibits hypoxia-inducible factor-1 pathway. Int J Radiat Biol. 2011;87(6):579–90. doi:10.3109/09553002.2011.568572 [Google Scholar] [PubMed] [CrossRef]
43. Rauf A, Olatunde A, Imran M, Alhumaydhi FA, Aljohani ASM, Khan SA, et al. Honokiol: a review of its pharmacological potential and therapeutic insights. Phytomedicine. 2021;90:153647. doi:10.1016/j.phymed.2021.153647 [Google Scholar] [PubMed] [CrossRef]
44. Wu F, Yao H, Zheng F, Tang S, Lin X, Li L, et al. Protective effects of honokiol against oxidative stress-induced apoptotic signaling in mouse podocytes treated with H2O2. Exp Ther Med. 2018;16(2):1278–84 [Google Scholar] [PubMed]
45. Hou Y, Peng S, Li X, Yao J, Xu J, Fang J. Honokiol alleviates oxidative stress-induced neurotoxicity via activation of Nrf2. ACS Chem Neurosci. 2018;9(12):3108–16. doi:10.1021/acschemneuro.8b00290 [Google Scholar] [PubMed] [CrossRef]
46. Lai X, Sun Y, Zhang X, Wang D, Wang J, Wang H, et al. Honokiol induces ferroptosis by upregulating HMOX1 in acute myeloid leukemia cells. Front Pharmacol. 2022;13:897791. doi:10.3389/fphar.2022.897791 [Google Scholar] [PubMed] [CrossRef]
47. Guo C, Liu P, Deng G, Han Y, Chen Y, Cai C, et al. Honokiol induces ferroptosis in colon cancer cells by regulating GPX4 activity. Am J Cancer Res. 2021;11(6):3039–54 [Google Scholar] [PubMed]
48. Hu M, Jiang W, Ye C, Hu T, Yu Q, Meng M, et al. Honokiol attenuates high glucose-induced peripheral neuropathy via inhibiting ferroptosis and activating AMPK/SIRT1/PGC-1α pathway in Schwann cells. Phytother Res. 2023;37(12):5787–802. doi:10.1002/ptr.v37.12. [Google Scholar] [CrossRef]
49. Guo Y, Liu C, Zhang J, Tian BB, Wang L, Liu GK, et al. A relationship between MAPK/ERK pathway expression and neuronal apoptosis in rats with white matter lesions. Eur Rev Med Pharmacol Sci. 2020;24(8):4412–9 [Google Scholar] [PubMed]
50. Chen NN, Wei F, Wang L, Cui S, Wan Y, Liu S. Tumor necrosis factor alpha induces neural stem cell apoptosis through activating p38 MAPK pathway. Neurochem Res. 2016;41(11):3052–62. doi:10.1007/s11064-016-2024-8 [Google Scholar] [PubMed] [CrossRef]
51. Liu X, Gu Y, Bian Y, Cai D, Li Y, Zhao Y, et al. Honokiol induces paraptosis-like cell death of acute promyelocytic leukemia via mTOR & MAPK signaling pathways activation. Apoptosis. 2021;26(3–4):195–208 [Google Scholar] [PubMed]
52. Zhang Y, Ren X, Shi M, Jiang Z, Wang H, Su Q, et al. Downregulation of STAT3 and activation of MAPK are involved in the induction of apoptosis by HNK in glioblastoma cell line U87. Oncol Rep. 2014;32(5):2038–46. doi:10.3892/or.2014.3434 [Google Scholar] [PubMed] [CrossRef]
53. Elfeky MG, Mantawy EM, Gad AM, Fawzy HM, El-Demerdash E. Mechanistic aspects of antifibrotic effects of honokiol in Con A-induced liver fibrosis in rats: emphasis on TGF-β/SMAD/MAPK signaling pathways. Life Sci. 2020;240:117096. doi:10.1016/j.lfs.2019.117096 [Google Scholar] [PubMed] [CrossRef]
54. Kim GD, Bae SY, Park HJ, Bae K, Lee SK. Honokiol inhibits vascular vessel formation of mouse embryonic stem cell-derived endothelial cells via the suppression of PECAM and MAPK/mTOR signaling pathway. Cell Physiol Biochem. 2012;30(3):758–70. doi:10.1159/000341455 [Google Scholar] [PubMed] [CrossRef]
55. Huang KJ, Kuo CH, Chen SH, Lin CY, Lee YR. Honokiol inhibits in vitro and in vivo growth of oral squamous cell carcinoma through induction of apoptosis, cell cycle arrest and autophagy. J Cell Mol Med. 2018;22(3):1894–908. doi:10.1111/jcmm.2018.22.issue-3. [Google Scholar] [CrossRef]
56. Ma M, Zhang J, Gao X, Yao W, Li Q, Pan Z. miR-361-5p mediates SMAD4 to promote porcine granulosa cell apoptosis through VEGFA. Biomolecules. 2020;10(9):1281. doi:10.3390/biom10091281 [Google Scholar] [PubMed] [CrossRef]
57. Dong B, Zhou B, Sun Z, Huang S, Han L, Nie H, et al. LncRNA-FENDRR mediates VEGFA to promote the apoptosis of brain microvascular endothelial cells via regulating miR-126 in mice with hypertensive intracerebral hemorrhage. Microcirculation. 2018;25(8):e12499. doi:10.1111/micc.2018.25.issue-8. [Google Scholar] [CrossRef]
58. Wei H, Cao C, Wei X, Meng M, Wu B, Meng L, et al. Circular RNA circVEGFC accelerates high glucose-induced vascular endothelial cells apoptosis through miR-338-3p/HIF-1α/VEGFA axis. Aging. 2020;12(14):14365–75. doi:10.18632/aging.v12i14. [Google Scholar] [CrossRef]
59. Sun Y, Guo W, Guo Y, Lin Z, Wang D, Guo Q, et al. Apoptosis induction in human prostate cancer cells related to the fatty acid metabolism by wogonin-mediated regulation of the AKT-SREBP1-FASN signaling network. Food Chem Toxicol. 2022;169:113450. doi:10.1016/j.fct.2022.113450 [Google Scholar] [PubMed] [CrossRef]
60. Jin Y, Chen Z, Dong J, Wang B, Fan S, Yang X, et al. SREBP1/FASN/cholesterol axis facilitates radioresistance in colorectal cancer. FEBS Open Bio. 2021;11(5):1343–52. doi:10.1002/feb4.v11.5. [Google Scholar] [CrossRef]
61. Rong Y, Ji C, Wang Z, Ge X, Wang J, Ye W, et al. Small extracellular vesicles encapsulating CCL2 from activated astrocytes induce microglial activation and neuronal apoptosis after traumatic spinal cord injury. J Neuroinflammation. 2021;18(1):196. doi:10.1186/s12974-021-02268-y [Google Scholar] [PubMed] [CrossRef]
62. Fang S, Tang H, Li MZ, Chu JJ, Yin ZS, Jia QY. Identification of the CCL2 PI3K/Akt axis involved in autophagy and apoptosis after spinal cord injury. Metab Brain Dis. 2023;38(4):1335–49. doi:10.1007/s11011-023-01181-y [Google Scholar] [PubMed] [CrossRef]
63. Liu S, Liao G, Li G. Regulatory effects of COL1A1 on apoptosis induced by radiation in cervical cancer cells. Cancer Cell Int. 2017;17:73. doi:10.1186/s12935-017-0443-5 [Google Scholar] [PubMed] [CrossRef]
64. Fu XH, Chen CZ, Wang Y, Peng YX, Wang WH, Yuan B, et al. COL1A1 affects apoptosis by regulating oxidative stress and autophagy in bovine cumulus cells. Theriogenology. 2019;139:81–9. doi:10.1016/j.theriogenology.2019.07.024 [Google Scholar] [PubMed] [CrossRef]
65. He J, Zhao H, Liu X, Wang D, Wang Y, Ai Y, et al. Sevoflurane suppresses cell viability and invasion and promotes cell apoptosis in colon cancer by modulating exosome‐mediated circ‐HMGCS1 via the miR‐34a‐5p/SGPP1 axis. Oncol Rep. 2020;44(6):2429–42. doi:10.3892/or. [Google Scholar] [CrossRef]
66. Zhou C, Wang Z, Yang S, Li H, Zhao L. Hymeglusin enhances the pro-apoptotic effects of venetoclax in acute myeloid leukemia. Front Oncol. 2022;12:864430. doi:10.3389/fonc.2022.864430 [Google Scholar] [PubMed] [CrossRef]
67. Chattopadhyay T, Mallick B. FDFT1 repression by piR-39980 prevents oncogenesis by regulating proliferation and apoptosis through hypoxia in tongue squamous cell carcinoma. Life Sci. 2023;329:121954. doi:10.1016/j.lfs.2023.121954 [Google Scholar] [PubMed] [CrossRef]
68. Zhang Y, Du W, Chen Z, Xiang C. Upregulation of PD-L1 by SPP1 mediates macrophage polarization and facilitates immune escape in lung adenocarcinoma. Exp Cell Res. 2017;359(2):449–57. doi:10.1016/j.yexcr.2017.08.028 [Google Scholar] [PubMed] [CrossRef]
69. Nie Y, Yun X, Zhang Y, Wang X. Targeting metabolic reprogramming in chronic lymphocytic leukemia. Exp Hematol Oncol. 2022;11(1):39. doi:10.1186/s40164-022-00292-z [Google Scholar] [PubMed] [CrossRef]
70. Li Z, Wang Y, Wu Y, Liu Y, Zhao Y, Chen X, et al. Role of BLACAT1 in IL-1β-induced human articular chondrocyte apoptosis and extracellular matrix degradation via the miR-149-5p/ HMGCR axis. Protein Pept Lett. 2022;29(7):584–94. doi:10.2174/0929866529666220602094940 [Google Scholar] [PubMed] [CrossRef]
71. Yang Q, Wang C, Jin Y, Ma X, Xie T, Wang J, et al. Disocin prevents postmenopausal atherosclerosis in ovariectomized LDLR-/- mice through a PGC-1α/ERα pathway leading to promotion of autophagy and inhibition of oxidative stress, inflammation and apoptosis. Pharmacol Res. 2019;148:104414. doi:10.1016/j.phrs.2019.104414 [Google Scholar] [PubMed] [CrossRef]
72. Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, et al. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79(20):5355–66. doi:10.1158/0008-5472.CAN-19-0369 [Google Scholar] [PubMed] [CrossRef]
73. DeBose-Boyd RA, Ou J, Goldstein JL, Brown MS. Expression of sterol regulatory element-binding protein 1c (SREBP-1c) mRNA in rat hepatoma cells requires endogenous LXR ligands. Proc Natl Acad Sci U S A. 2001;98(4):1477–82. doi:10.1073/pnas.98.4.1477 [Google Scholar] [PubMed] [CrossRef]
74. Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu K, et al. SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int J Mol Sci. 2014;15(5):7124–38. doi:10.3390/ijms15057124 [Google Scholar] [PubMed] [CrossRef]
75. Zhang L, Jia NN, Yang RH, Wang F. Eicosapentaenoic acid reduces inflammation and apoptosis by SREBP1/TLR4/MYD88. Bratisl Lek Listy. 2020;121(11):822–9 [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools