 Open Access
Open Access
ARTICLE
Revealing the role of honokiol in human glioma cells by RNA-seq analysis
1 The Department of Neurosurgery, First Hospital of Jilin University, Changchun, 130021, China
2 The Department of Anatomy, College of Basic Medical Sciences, Jilin University, Changchun, 130021, China
3 The Third College of Clinical Medicine, Jilin University, Changchun, 130021, China
* Corresponding Authors: YAN LU. Email: ; HAIYAN LIU. Email:
BIOCELL 2024, 48(6), 945-958. https://doi.org/10.32604/biocell.2024.049748
Received 17 January 2024; Accepted 20 March 2024; Issue published 10 June 2024
Abstract
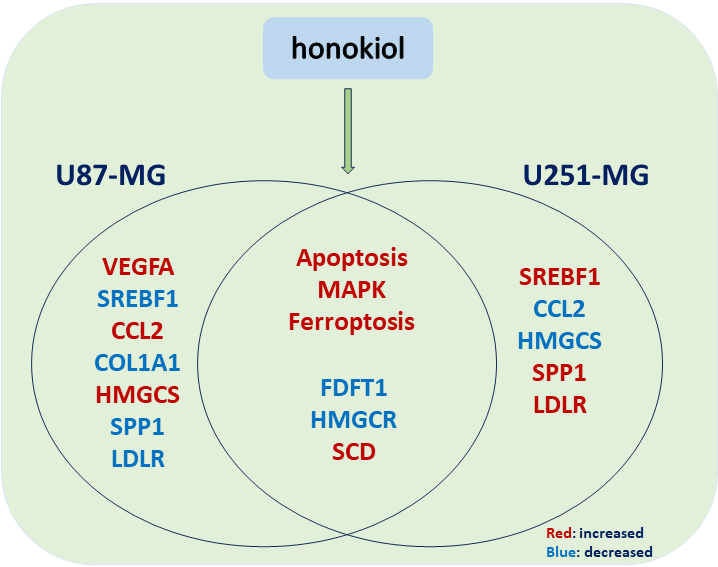
Background: Glioma is a kind of tumor that easily deteriorates and originates from glial cells in nerve tissue. Honokiol is a bisphenol compound that is an essential monomeric compound extracted from the roots and bark of Magnoliaceae plants. It also has anti-infection, antitumor, and immunomodulatory effects. In this study, we found that honokiol induces cell apoptosis in the human glioma cell lines U87-MG and U251-MG. However, the mechanism through which honokiol regulates glioma cell apoptosis is still unknown. Methods: We performed RNA-seq analysis of U251-MG cells treated with honokiol and control cells. Protein-protein interaction (PPI) network analysis was performed, and the 10 top hub unigenes were examined via real-time quantitative PCR. Furthermore, MAPK signaling and ferroptosis were detected via western blotting. Results: 332 differentially expressed genes (DEGs) were found, comprising 163 increased and 169 decreased genes. Analysis of the DEGs revealed that various biological processes were enriched, including ‘response to hypoxia’, ‘cerebellum development cellular response to hypoxia,’ ‘iron ion binding,’ ‘oxygen transporter activity,’ ‘oxygen binding,’ ‘ferric iron binding,’ and ‘structural constituent of cytoskeleton.’ Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that the DEGs were enriched in the following pathways: ‘mitogen-activated protein kinases (MAPK)’, ‘Hypoxia-inducible factor 1 (HIF-1)’, ‘ferroptosis,’ ‘Peroxisome proliferator-activated receptor (PPAR),’ ‘Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K)- protein kinase B (Akt),’ and ‘phagosome.’ Among these pathways, the MAPK signaling pathway and ferroptosis were verified. Conclusion: This study revealed the potential mechanism by which honokiol induces apoptosis and provided a comprehensive analysis of DEGs in honokiol-treated U251-MG cells and the associated signaling pathways. These data could lead to new ideas for future research and therapy for patients with glioma.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileCite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools