 Open Access
Open Access
ARTICLE
UBE2T mediates the stemness properties of breast cancer cells through the mTOR signaling pathway
1 Department of Traumatology, The Second Hospital of Shandong University, Jinan, 250033, China
2 Department of Pathology, Weifang People’s Hospital, Weifang, 261000, China
3 Department of Human Anatomy, Shandong University School of Medicine, Jinan, 250012, China
4 Department of Cell Biology and Key Laboratory of Experimental Teratology, Shandong University School of Medicine, Jinan, 250012, China
* Corresponding Author: MINGXIN WEN. Email:
(This article belongs to the Special Issue: Cell Death and Inflammation in Signaling and Diseases)
BIOCELL 2024, 48(6), 959-970. https://doi.org/10.32604/biocell.2024.049349
Received 04 January 2024; Accepted 03 April 2024; Issue published 10 June 2024
Abstract
Objectives: This study aimed to reveal the role and possible mechanism of the ubiquitin-conjugating enzyme 2T (UBE2T) in the biological activities of breast cancer stem cells (BCSCs). Methods: The specific protein and gene expression were quantified by Western blotting and quantitative real-time polymerase chain reaction, the proportion of BCSCs was examined by flow cytometry, and the self-renewal and proliferation of BCSCs were verified by serial sphere formation and soft agar. Results: Increasing expression of UBE2T was drastically found in breast cancer than that in adjacent tissues. Furthermore, UBE2T overexpression significantly increased the proportion of BCSCs in breast cancer cells and promoted their self-renewal and proliferation. Silent UBE2T exhibited the opposite functions. UBE2T increased the levels of the mammalian target of rapamycin and the phosphorylated mammalian target of rapamycin. Mammalian target of rapamycin (mTOR) inhibitor rapamycin inhibited the function of UBE2T in BCSCs. Conclusion: UBE2T plays a role in BCSCs through mTOR pathway and may suggest a novel therapeutic strategy for breast cancer.Keywords
A higher incidence of breast cancer (BCa) in women has been reported, and BCa is at the top list of cancer-related deaths. It is disputed that cancer stem cells (CSCs) are closely associated with the progression of multiple tumors [1,2]. CSCs are a small number of cells characterized by strong survival, high replication, self-renewal, high differentiation potential, and drug resistance [3]. Abnormal expression of ubiquitin enzymes such as channel intrinsic protein (CHIP) [4], F-box/WD repeat-containing protein 7 (Fbxw7) [5], and tripartite motif-containing protein 16 (TRIM16) [6] plays a significant role in BCSCs. Our study revealed the role and possible mechanism of action of ubiquitin-conjugating enzyme 2T (UBE2T) in breast cancer stem cells (BCSCs), and we suggested a novel target for BCa therapy.
In previous reports, UBE2T was shown to play an essential role in the DNA repair pathway in Fanconi anemia through UBE2T-mediated mono-ubiquitination of Homo sapiens Fanconi anemia, complementation group D2 (FANCD2), resulting in the activation of the Fanconi anemia core pathway [7,8]. It has been reported that elevated expression of UBE2T was found in lung cancer [9,10], bladder cancer [11], BCa [12], and prostate cancer [13]. In addition, UBE2T increased the proliferative rate of BCa cells both in cellular and animal experiments by suppressing the expression of breast cancer susceptibility gene 1 (BRCA1) [14], suggesting poor outcomes in patients with breast cancer [15]. UBE2T also promoted the metastasis of prostate cancer cells through epithelial-mesenchymal transition [13]. Recently, UBE2T was reported to increase the proliferation rate of renal cell carcinoma cells via the phosphoinositide3 (PI3K)-kinase-protein kinase B (AKT) pathway [16] and hepatocarcinoma cells via the p53 pathway [17,18] or the G2/M checkpoint [19]. Although the oncogenic function of UBE2T has been verified in multiple tumors, the specific mechanism remains to be elucidated. In this study, we revealed that UBE2T was effective in maintaining the stemness of BCa cells and suggested a possible pathway through which UBE2T affects the malignancy of BCa.
The mammalian target of rapamycin (mTOR) has been shown to regulate cell growth and many proteins complex as a serine-threonine kinase. Two mTOR complexes 1 and 2 (mTORC1 and mTORC2) have been reported [20]. mTORC1 comprises mTOR, raptor, and mTOR associated protein/LST8 homolog [21] which regulates transcription factors such as ribosomal protein S6 kinase and eIF4E-binding protein [22]. mTORC2 comprises mTOR, DEP domain-containing mTOR-interacting protein, mTOR-associated protein/LST8 homolog, proline-rich protein 5, a rapamycin-insensitive companion of mTOR, and mitogen-activated protein kinase-associated protein 1 [22]. mTOR has been proven to have a positive role in BCSCs [23–25]. However, there is no evidence that UBE2T influences the stemness of BCa cells through the mTOR pathway.
For this work, we attempted to elucidate the role and possible mechanism of UBE2T in BCSCs.
Cell culture and retroviral transduction
We purchased the BCa cell lines from the American Type Culture Collection (Manassas, VA, USA). Cells were cultured according to the manufacturer’s instructions. HCC1428, MDA-MB-231, and BT549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Merck, R6504, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, 12107C, Shanghai, China) and 1% penicillin/streptomycin (Merck, 516106, Shanghai, China) MCF-7 cells were grown in Dulbecco’s modified eagle medium (DMEM)(Merck, D0822, Shanghai, China) supplemented with 10% FBS and 1% penicillin/streptomycin. All the BCa cell lines were incubated at 37°C in 5% CO2.
Cell lines overexpressing or silencing UBE2T were established according to methods described in our previous study [13].
Western blotting (WB) and antibodies
Proteins were extracted using Radioimmunoprecipitation Assay (RIPA) lysis buffer (Merck, 20-188, Shanghai, China). Protein concentration was quantified using a Bicinchoninic Acid (BCA) reagent (Merck, BCA1, Shanghai, Chian). Proteins were separated within a 10% bis-tris precast gel, then were transferred to polyvinylidene fluoride (PVDF) membranes (BESTOPBIO, IPVH00010, Beijing, China), and incubated with primary antibodies overnight at 4°C. The next day, the PVDF membranes were incubated with the secondary antibodies after being washed in TBST (ThermoFisher, 28360, Shanghai, China). Finally, the semiquantitative results of proteins were measured by an enhanced chemiluminescence system.
All antibodies were purchased from Cell Signaling Technology (Danvers, MA, USA) and used at indicated concentrations: anti-actin (1:2000, CST3700), anti-UBE2T (1:1000, CST12992S), anti-SRY-box containing gene 2 (Sox2; 1:500, CST23064), anti-octamer-binding transcription factor (Oct4; 1:500, CST2840), anti-Nanog homeobox (Nanog; 1:1000, CST4893), anti-mTOR (1:500, CST47102), anti-phosphorylated mammalian target of rapamycin (p-mTOR; 1:500, CST5536).
The BCa cells were cultured in 100-mm dishes at 37°C in 5% CO2 and collected at 70–80% density, then incubated with anti-CD44-APC (Abcam, ab81424, Beijing, China) and anti-CD24-PE (Abcam, ab314420, Beijing, China) antibodies or stained with their isotype controls according to the manufacturer’s instructions and examined using a flow cytometry system (BD FACS Melody, ThermoFisher, Shanghai, China). CD24−/CD44+ cells are considered BCSCs.
Quantitative real-time polymerase chain reaction (qRT-PCR)
We used TRIzol reagent (Invitrogen, 15596026, Beijing, China) to extract the total RNA, followed by reverse transcription with First Strand cDNA Synthesis Kit (Invitrogen, K1621, Beijing, China) and real-time quantitative PCR on a PRISM 7900HT system (Applied Biosystems, Shanghai, China). The steps of the qPCR reaction were as follows: denaturation (95°C, 15 min), amplification (40 cycles, 94°C for 40 s), and extension (58°C, 1 min). All oligonucleotide DNA primers utilized in this study were Homo sapiens: UBE2T forward, CAAATATTAGGTGGAGCCAACAC and reverse, TAGATCACCTTGGCAAAGAA.
C; GAPDH forward, TCTTCTTTTGCGTCGCCAG, reverse, TGGAGAAGGCTGGG.
GCT; OCT4A forward, GACAACAATGAGAACCTTCAGGAGA, reverse, CTGGCG.
CCGGTTACAGAACCA; Nanog forward, TTGTGGCCTGAAGAAAACTATCC, reverse, CTGCGTCACACCATTGCTATTCTT; Sox2 forward, CCCCCGGCGGCAAT. AGCA, reverse, TCGGCGCCGGGGAGATACAT.
The RNA-seq samples or cells were stored in liquid nitrogen, and then RNA-seq was performed by PTM Bio (Application registration number: 53112954, Hangzhou, China).
The sphere formation assay was performed as described by Lei Yang et al. with slight modifications [26]. The special culture medium contained mammary epithelial growth medium, B27 (1:50, Gibco, 17504044, Shanghai, China), epidermal growth factor (20 ng/mL, Invitrogen, PHG0311, Beijing, China), basic fibroblast growth factor (20 ng/mL, Invitrogen, 100-18B, Beijing, China), and heparin (10 μg/mL, Sigma, 9041-08-1, Shanghai, China). Single-cell suspensions were plated in ultra-low-attachment 96-well plates (Costar) at 1000 viable cells per well. Cells were grown as spheroids and counted after 7–10 days. The secondary number of spheroids was counted 8–10 days after plating.
The agar (A5431) and agarose (A9045) were purchased from Sigma-Aldrich. The bottom layer was a 0.6% agar mix, and the top layer was 0.35% agarose mixed with 5000 cells that needed to be detected. Then, 2–3 weeks after being incubated at 37°C in 5% CO2, the clones were counted under a microscope (OLYMPUS, BX-36, Shanghai, China).
The 24 pairs of BCa and adjacent tissues were obtained with informed consent. With the approval of the Ethics Committee of Shandong University School of Basic Medical Science (No. ECSBMSSDU2021-2-157). We then determined the expression of UBE2T using WB and qRT-PCR. All tissues were preserved in liquid nitrogen before protein extraction.
All data are expressed as mean ± SD. A p-value of equal to or less than 0.05 based on the Student’s t-test of SPSS (NDTimes, Beijing, China) was considered statistically significant. Each assay was repeated three times.
Elevated expression of UBE2T is found in BCa
To verify whether UBE2T expression levels were elevated in human BCa, RNA-sequence was performed on six paired breast cancer samples. Increased expression of UBE2T was observed in the breast cancer (Figs. 1A and 1B). To confirm this finding, the expression levels of UBE2T in 24 primary BCa and control tissues were detected using WB and qRT-PCR. As shown in the results, both the transcription (Figs. 1C and 1D) and protein (Figs. 1E and 1F) levels of UBE2T in BCa tissues were significantly elevated compared with that in control tissues, suggesting a potentially oncogenic role of UBE2T in BCa. Next, we verified the quantitative levels of UBE2T in several BCa cell lines and human normal mammary epithelial cells (MCF-10A). Similar to the results in BCa tissues and consistent with previous reports [12], the expression levels of UBE2T in BT549, MDA-MB-231, HCC1428, and MCF-7 were higher than those in human normal mammary epithelial cells (MCF-10A) (Figs. 1G and 1H) and were higher in the more invasive BCa cell lines BT549 and MDA-MB231 compared with HCC1428 and MCF7 (Figs. 1G and 1H). These results suggest that UBE2T plays an oncogenic role and contributes to BCa malignancy. Therefore, it would be worthwhile to explore the precise function of UBE2T in BCSCs.
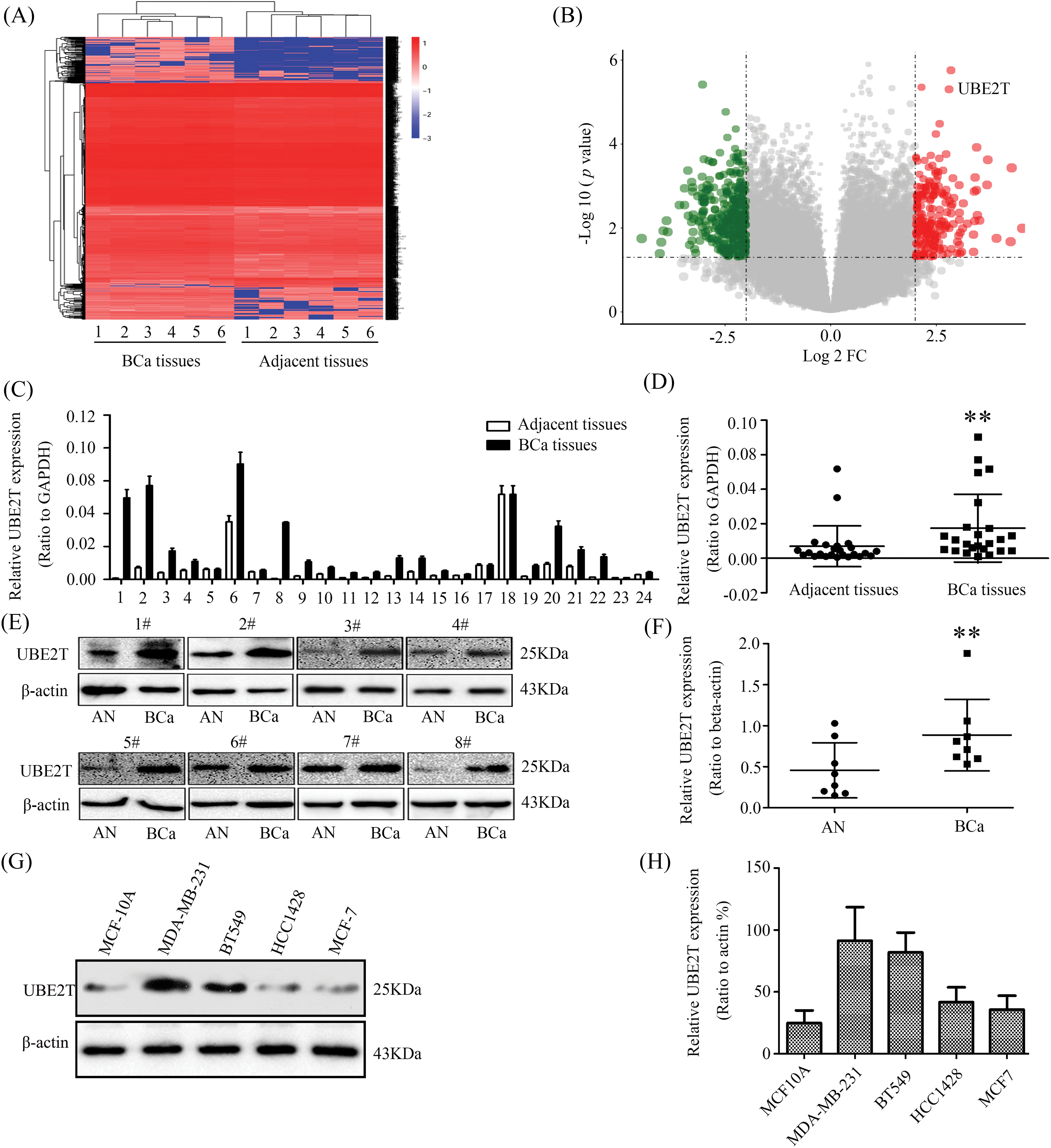
Figure 1: Elevated ubiquitin-conjugating enzyme 2T (UBE2T) is examined in BCa. (A) mRNA-seq of BCa and control. (B) Bioinformatics analysis of the data from (A). (C) qRT-PCR of UBE2T in breast cancer and control. (D) Analysis of UBE2T levels in (C) (**p < 0.01). (E) WB of UBE2T levels in some breast cancer and control. AN: adjacent tissue, BCa: breast cancer. (F) Statistics of UBE2T expressions in BCa and adjacent tissues (**p < 0.01). (G) WB for UBE2T levels in BCa cell lines. (H) Quantification of UBE2T in breast cancer cells. Error bars, SD. Each assay was repeated three times.
Overexpressed UBE2T increases the proportion of BCSCs
Based on the expression levels of UBE2T in BCa tissues and cells and the association of BCSCs with cancer growth, metastasis, and relapse [27,28], we aimed to verify the exact role of UBE2T in BCSCs. First, we retrovirally established breast cancer cells with overexpressed UBE2T or silenced UBE2T which were confirmed by WB (Figs. 2A and 2B). The proportion of BCSCs was analyzed using fluorescence-activated cell sorting. Surprisingly, overexpression of UBE2T significantly increased the proportion of cells marked as CD44+/CD24−/low, which is considered to be the phenotype of BCSCs in MCF-7 and HCC1428 cells, whereas silencing of UBE2T had the opposite effect (Figs. 2C–2F). In summary, UBE2T increased the proportion of BCSCs in BCa cells, suggesting that UBE2T may affect the properties of BCSCs.
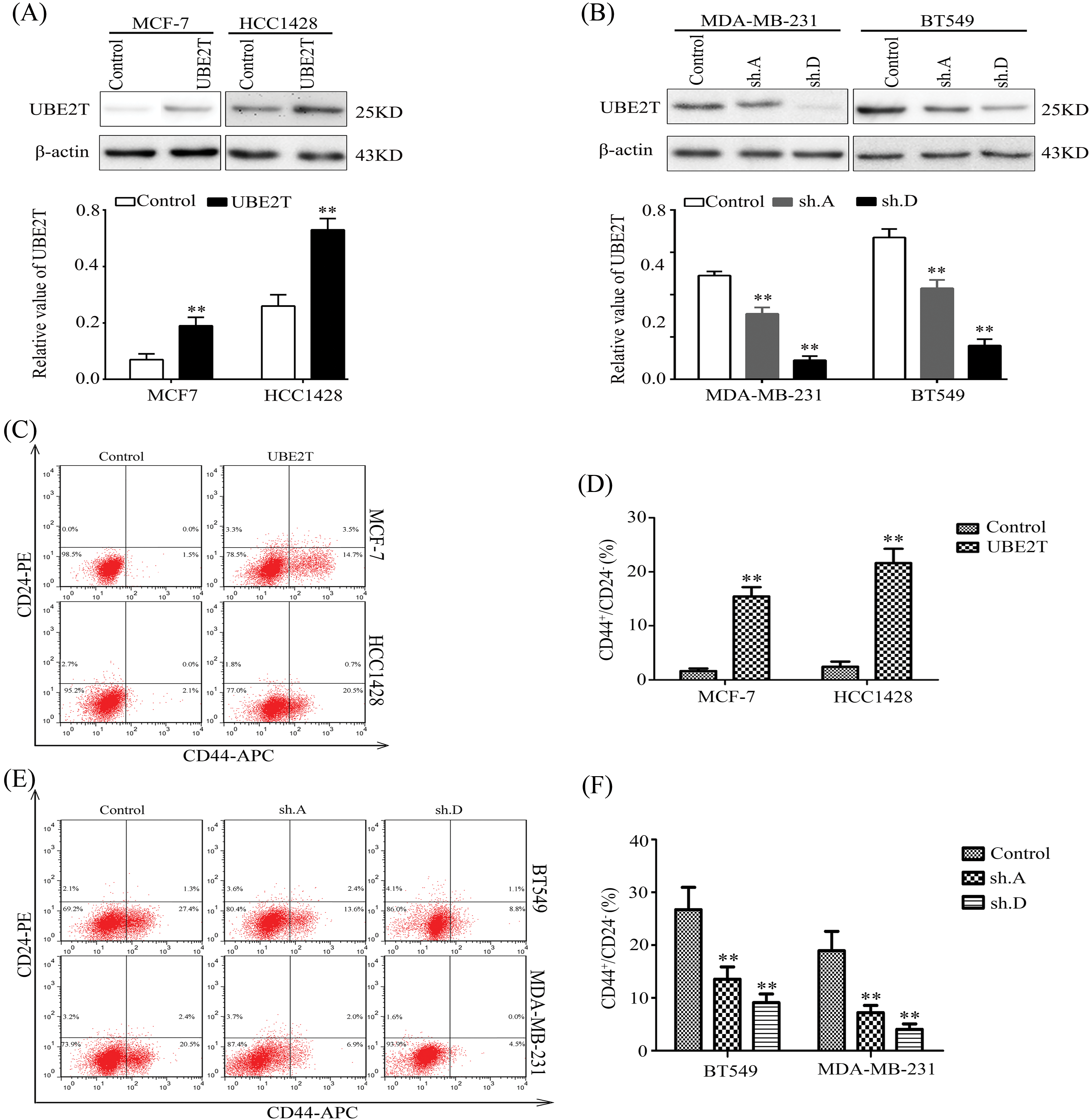
Figure 2: Elevated UBE2T increases the proportion of BCSCs. (A) WB of UBE2T in MCF7 and HCC1428 cells with overexpressed UBE2T (above) and quantitation of UBE2T (below) (**p < 0.01). (B) WB of UBE2T levels in MDA-MB-231 and BT549 cells with silenced UBE2T (above) and quantitation of UBE2T (below) (**p < 0.01). (C and D) Fluorescence-activated cell sorter (FACS) for the proportion of BCSCs in MCF7 and HCC1428 cells with overexpressed UBE2T (**p < 0.01). (E and F) FACS for the proportion of BCSCs in MDA-MB-231 and BT549 cells with silenced UBE2T. In (E and F), we identify 231 CD44-positive cells with higher CD44 expression (**p < 0.01). Error bars, SD. Each assay was repeated three times.
UBE2T regulates the properties of BCSCs
Based on the increased proportion of BCSCs induced by UBE2T overexpression, we verified the influence of UBE2T on the stemness of BCa cells. First, a soft agar assay was used to detect the clonogenic potential of BCSCs. More and larger clones were induced by UBE2T in MCF7 and HCC1428 cells overexpressing UBE2T (Figs. 3A–3D) while silencing of UBE2T showed the opposite effect in BT549 and MDA-MB-231 cells (Figs. 3E–3H). Furthermore, sphere formation for self-renewal of BCSCs verified that overexpression of UBE2T significantly increased the number of primary and secondary spheres formed by MCF7 and HCC1428 cells (Figs. 4A–4D), while silencing of UBE2T showed opposite effect in MDA-MB-231 and BT549 cells (Figs. 4E–4H). Consistent with our hypothesis, these data indicate that UBE2T enhances the properties of BCSCs.
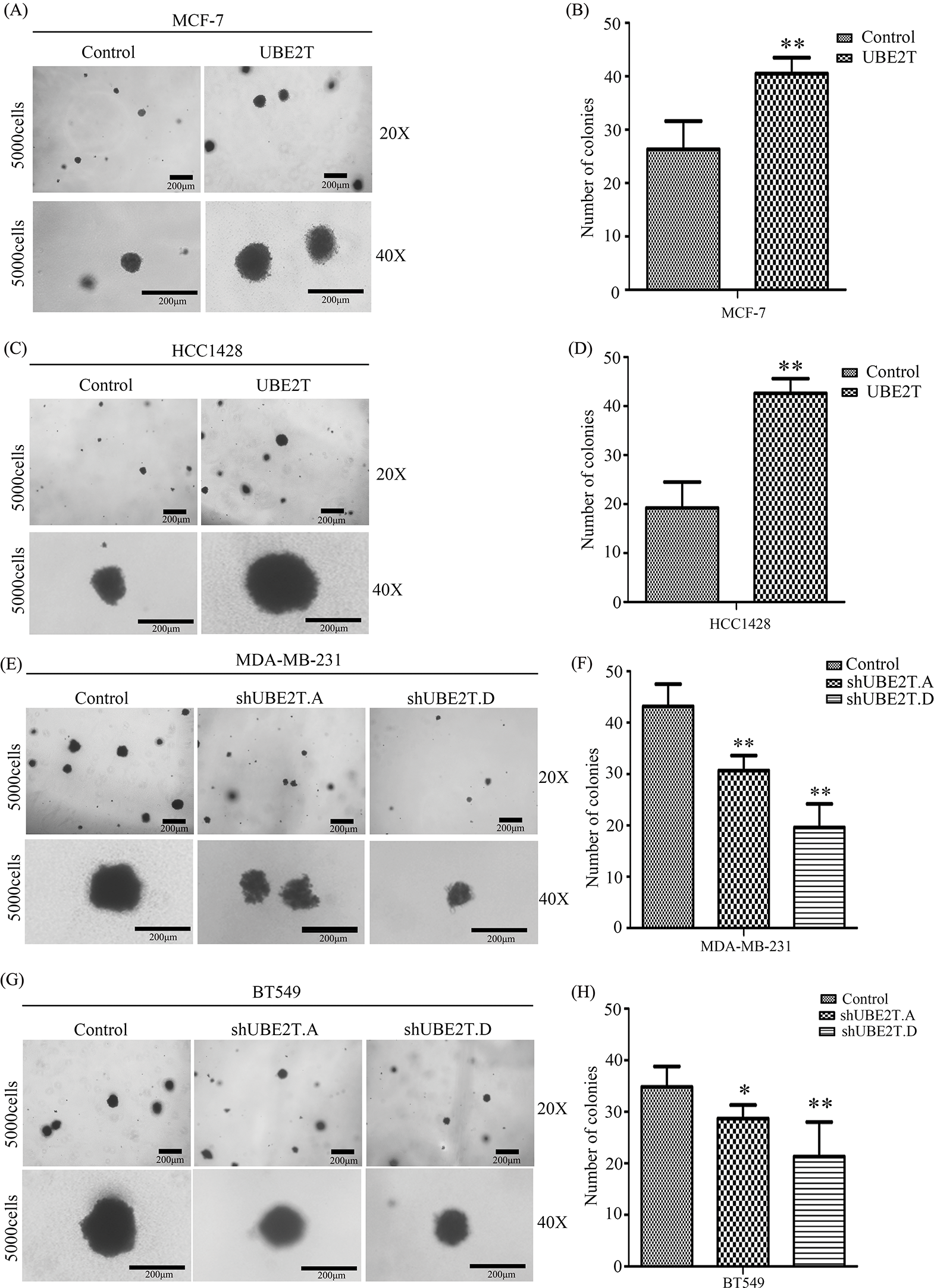
Figure 3: The proliferation of BCSCs with overexpressed UBE2T in soft agar. (A–D) Soft agar assay of MCF7 (A and B) and HCC1428 (C and D) with overexpressed UBE2T (**p < 0.01). (E–H) Soft agar assay of MDA-MB-231 (E and F) and BT549 (G and H) with silenced UBE2T (*p < 0.05, **p < 0.01). Scale bars in A, C, E, G, 200 μm. Error bars, SD. Each assay was repeated three times.
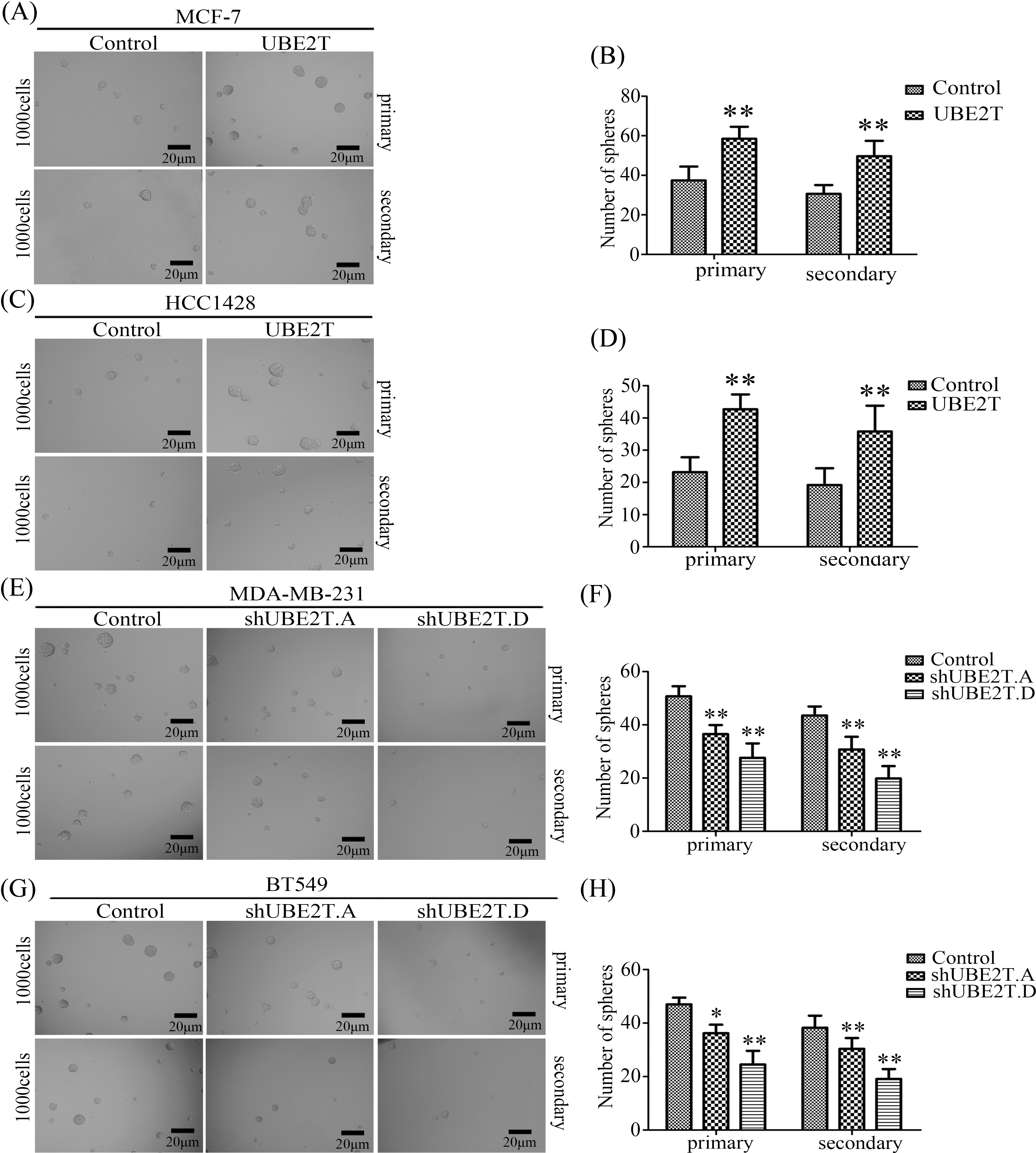
Figure 4: Sphere formation for self-renew of BCSCs. (A–D) Images of spheres formed by MCF7 (A and B) and HCC1428 (C and D) cells with overexpressed UBE2T (**p < 0.01). (E–H) Images of spheres formed by MDA-MB-231 (E and F) and BT549 (G and H) cells with silenced UBE2T expression (*p < 0.05, **p < 0.01). Scale bar, 20 μm. Error bars, SD. Each assay was repeated three times.
Nanog, Oct4, and Sox2 have been reported as pluripotency markers essential for the characteristics of CSCs [29–31]. Therefore, we measured the expression levels of these markers in BCSCs with overexpressed or silenced UBE2T. As shown in Fig. 5, overexpressed UBE2T significantly increased both protein and RNA levels of Sox2, Oct4, and Nanog in MCF7 and HCC1428 cells (Figs. 5A and 5C), whereas silencing UBE2T inhibited the expression levels of these proteins (Figs. 5B and 5D). These data further confirmed that UBE2T regulates the stemness of BCSCs and suggested that UBE2T may regulate the properties of BCSCs by increasing the expression of these CSC-related proteins.

Figure 5: The expression of BCSC markers is affected by UBE2T. (A) WB of BCSC markers in MCF7 and HCC1428 with overexpressed UBE2T (above) and quantitation of these markers (below) (**p < 0.01). (B) WB of BCSC markers in MDA-MB-231 and BT549 with silenced UBE2T (above) and quantitation of these markers (below) (**p < 0.01). (C) qRT-PCR of BCSC markers in MCF7 and HCC1428 with overexpressed UBE2T (*p < 0.05, **p < 0.01). (D) qRT-PCR of BCSC markers in MDA-MB-231 and BT549 with silenced UBE2T (*p < 0.05, **p < 0.01). Error bars, SD. Each assay was repeated three times.
UBE2T contributes to the properties of BCSCs through the mTOR pathway
UBE2T is associated with BCSC properties, but the mechanism involved in this progression remains unknown. Therefore, we attempted to identify the possible mechanism through RNA-seq. As shown in Figs. 6A and 6B, an increased number of genes overlapped with the mTOR pathway, which has been reported to be important in CSCs [32,33]. To verify that whether UBE2T was associated with mTOR pathway, we detected mTOR and p-mTOR expression levels in the indicated cells. As shown in Fig. 6C, elevated levels of p-mTOR were observed in MCF7 and HCC1428 cells overexpressing UBE2T compared with control cells, suggesting that the mTOR pathway was activated. Moreover, UBE2T silencing significantly decreased the p-mTOR levels in BT549 and MDA-MB-213 cells (Fig. 6D). The data above suggested that UBE2T could activate the mTOR pathway. Furthermore, we analyzed the database and found that UBE2T was negatively correlated with phosphatase and tensin homolog (PTEN) (Fig. 6E) and UBE2T inhibited the expression of PTEN (Fig. 6F), which suggests that UBE2T inhibits PTEN and activates mTOR signaling pathway.
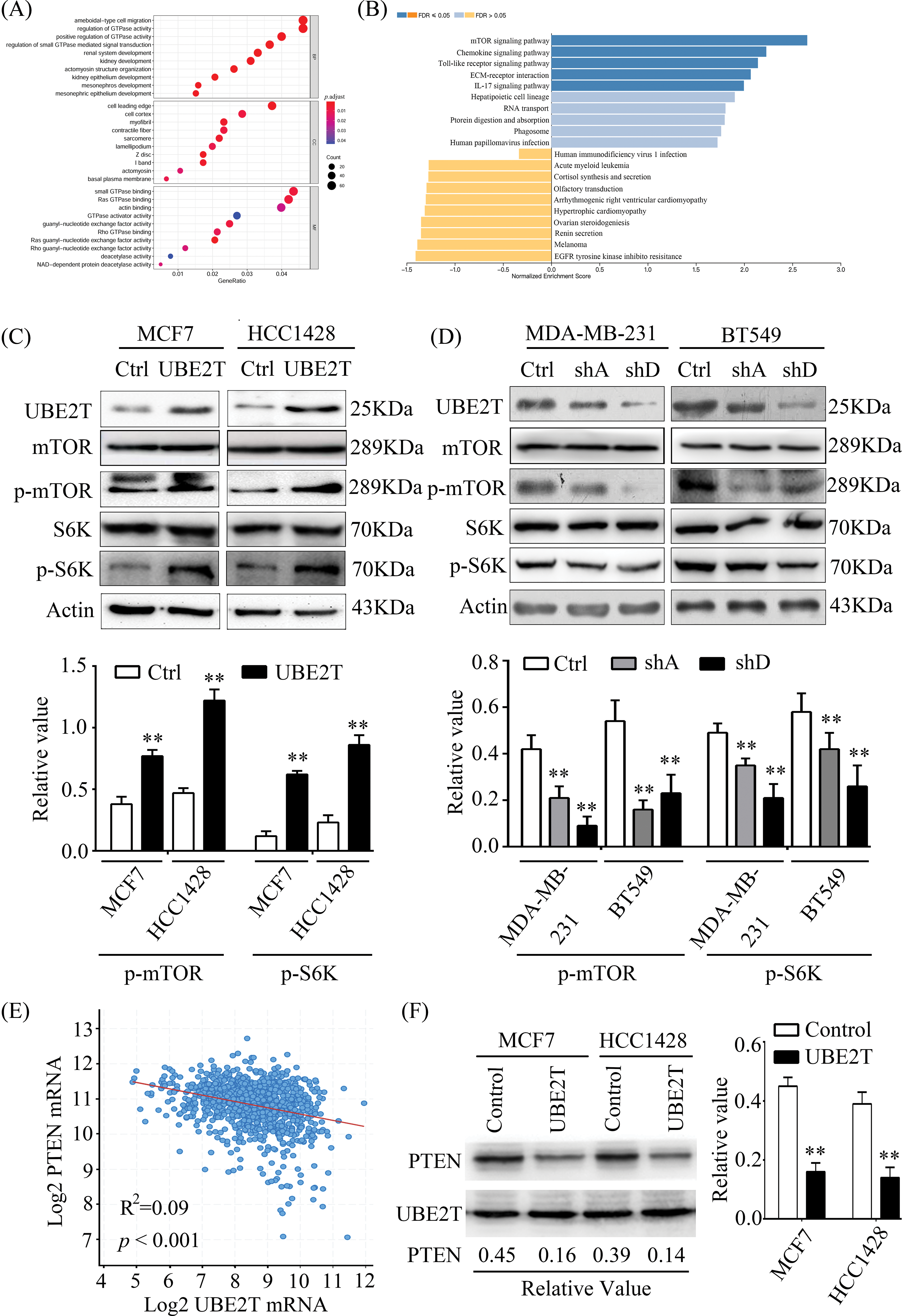
Figure 6: UBE2T promotes the properties of BCSCs through the mTOR pathway. (A) GO analysis of data of mRNA-seq between MCF7 with or without UBE2T overexpression. (B) KEGG analysis of data of mRNA-seq between MCF7 with or without UBE2T overexpression. (C and D) WB for proteins in mTOR signaling pathway in MCF7, HCC1428 with overexpressed UBE2T, and MDA-MB-231 and BT549 with silenced UBE2T (above) and quantification of p-mTOR and p-S6K (below) (**p < 0.01). (E) Linear correlation analysis of UBE2T and PTEN in breast cancer. (F) WB for PTEN in BCa cells with UBE2T overexpression (left) and quantitation of PTEN (right) (**p < 0.01). Each assay was repeated three times.
According to previous reports, mTOR is very important for the biological behavior of CSCs by altering the expression levels of Oct4, Nanog, and Sox2 [34,35]. To confirm whether UBE2T regulated these three proteins through the mTOR pathway in BCSCs, we suppressed the function of mTOR using rapamycin. In test we found that rapamycin significantly inhibited p-mTOR levels in MCF7 and HCC1428 cells overexpressing UBE2T and restrained the upregulated expression of Nanog, Oct4, and Sox2 induced by UBE2T overexpression (Fig. 7A and 7B). Moreover, rapamycin significantly reduced the number of spheres increased by UBE2T overexpression in MCF7 cells (Fig. 7C), and a similar phenomenon was confirmed in HCC1428 cells (Fig. 7D). The preliminary data reveal that UBE2T enhances the stemness of BCa cells through the mTOR pathway.
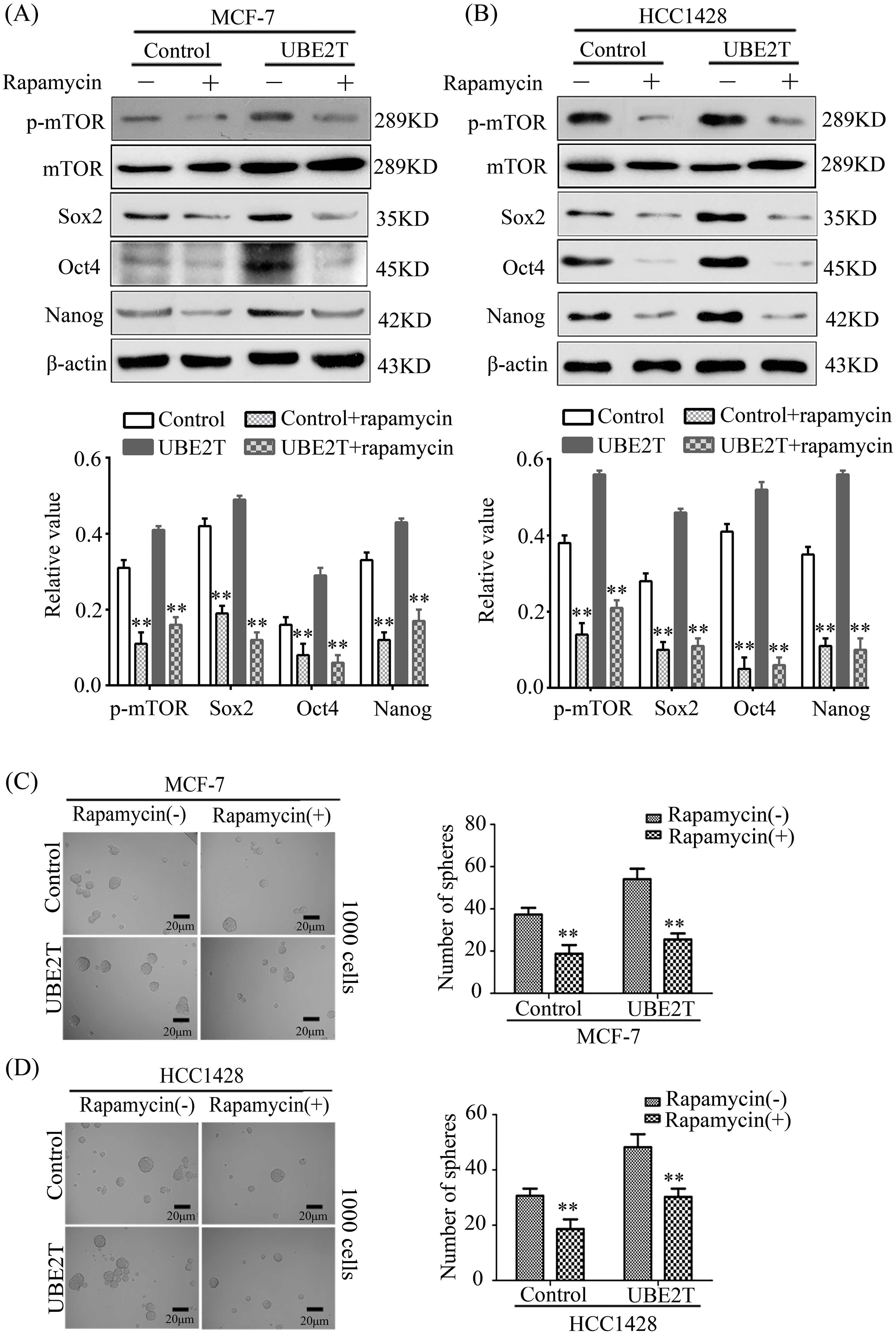
Figure 7: Rapamycin restrains the effect of UBE2T on breast CSCs. (A) WB of BCSC markers in MCF7 and HCC1428 overexpressing UBE2T with mTOR inhibitor rapamycin (above) and quantitation of indicated proteins (below) (**p < 0.01). (B) WB of BCSC markers in MDA-MB-231 and BT549 silencing UBE2T with mTOR inhibitor rapamycin (above) and quantitation of indicated proteins (below) (**p < 0.01). (C and D) Sphere formation assay for the MCF7 and HCC1428 overexpressing UBE2T with mTOR inhibitor rapamycin (**p < 0.01). Scale bar, 20 μm. Error bars, SD. Each assay was repeated three times.
In conclusion, overexpression of UBE2T enhances the function of the mTOR pathway and increases the levels of CSC-related proteins, thus affecting the stemness of BCa cells.
Recently, the incidence of BCa has been increasing in most countries despite efforts to prevent the disease [36,37]. The key role of BCSCs in BCa recurrence has been widely confirmed, and the hypothesis of specifically targeting CSCs sheds new light on the development of BCa treatments and provides the possibility of curing BCa permanently [38]. In this study, we investigated the role of UBE2T in BCSCs. Some assays have revealed that UBE2T significantly increases the proportion of BCSCs and promotes the stemness of BCSCs, including clonogenicity and self-renewal. Mechanistically, UBE2T increased the expression of CSC markers such as Nanog, Oct4, and Sox2 by mediating the mTOR pathway and affecting the stemness of BCa cells. These results suggest a potential therapeutic target for BCa treatment.
The oncogenic role of UBE2T has been reported in the bladder [11], lung [39,40], and prostate cancers [13] and other tumors [41–43]. In this study, UBE2T expression was significantly elevated in human BCa tissues, suggesting that UBE2T may play a role in BCa malignancy. Since BCSCs and stemness have been proven critical for the progression, metastasis, and recurrence of BCa, we examined whether UBE2T affected the stemness of BCa cells. In our results, UBE2T significantly increased the proportion of BCSCs and promoted the self-renewal and clonogenic potential of BCa cells, suggesting that UBE2T regulates the stemness of BCa cells. In other words, BCSCs regulated by UBE2T may be partially responsible for the poor prognosis of BCa.
CSC markers, such as Nanog, Oct4, and Sox2, are responsible for BCSC properties [44–46]. In our study, UBE2T increased the expression of these proteins by upregulating mTOR and p-mTOR expression. Moreover, the mTOR inhibitor, rapamycin, significantly inhibited the function of UBE2T in BCa cells. This was the first time we revealed that UBE2T plays a positive role in BCSCs. Therapies for BCa have been limited, and the prognosis has not been satisfactory because of therapy resistance induced by BCSCs. Herein, we revealed the role and possible mechanism of action of UBE2T in BCSCs and suggested a potential target for treating BCa. However, the mechanism by which UBE2T inhibits PTEN requires further investigation.
UBE2T is associated with BCa malignancy because UBE2T can increase the proportion of BCSCs and promote the clonogenicity and self-renewal properties of BCa cells through the mTOR pathway. These findings suggest a potential target gene for BCa therapy.
Acknowledgement: None.
Funding Statement: This research was partly supported by the Fundamental Research Funds of Shandong University (21510078614097) and the Shandong Natural Science Foundation General Project (ZR2022MC093).
Author Contributions: Mingxin Wen contributed to the conception and design of the study; Jiawei Yin and Yongsheng Wang conducted qRT-PCR, WB, sphere formation, and soft agar assay; Jiawei Yin contributed to cell culture, protein, and RNA extraction; Jiawei Yin and Yongsheng Wang were major contributors to writing the manuscript; Guangwei Wei and Mingxin Wen reviewed the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data and materials supporting the findings of this study are available upon request from the corresponding author.
Ethics Approval: This study was approved by the Ethics Committee of Shandong University School of Basic Medical Science (No. ECSBMSSDU2021-2-157). Twenty-four pairs of samples of patients with breast cancer were obtained with informed consent.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Chen BY, Ahmed SN, Salazar F, Jaiswal S, Farshbaf M, Chean J, et al. Radiopharmaceutical therapy for pancreatic cancer: engineered antiprostate stem cell antigen (PSCA) antibody demonstrates targeted imaging and antitumor effects in a syngeneic mouse model of pancreatic ductal adenocarcinoma (PDAC). Cancer Res. 2024;84(2). doi:10.1158/1538-7445.Panca2023-A088. [Google Scholar] [CrossRef]
2. Sainz BJ, Alcalá S, Villarino L, Ruíz-Cañas L, Couceiro J, Martínez-Calvo M, et al. Targeting cancer stem cell OXPHOS with tailored ruthenium complexes as a new approach to treat pancreatic cancer. Cancer Res. 2024;84(2). doi:10.1158/1538-7445.Panca2023-C079. [Google Scholar] [CrossRef]
3. Li LH, Jensen RA. Understanding and overcoming immunosuppression shaped by cancer stem cells. Cancer Res. 2023;83(13):2096–104. doi:10.1158/0008-5472.Can-23-0230 [Google Scholar] [PubMed] [CrossRef]
4. Tsuchiya M, Nakajima Y, Hirata N, Morishita T, Kishimoto H, Kanda Y, et al. Ubiquitin ligase CHIP suppresses cancer stem cell properties in a population of breast cancer cells. Biochem Biophys Res Commun. 2014;452(4):928–32. doi:10.1016/j.bbrc.2014.09.011 [Google Scholar] [PubMed] [CrossRef]
5. Wang YL, Liu YY, Lu J, Zhang PJ, Wang YS, Xu YY, et al. Rapamycin inhibits FBXW7 loss-induced epithelial-mesenchymal transition and cancer stem cell-like characteristics in colorectal cancer cells. Biochem Bioph Res Commun. 2013;434(2):352–6. doi:10.1016/j.bbrc.2013.03.077 [Google Scholar] [PubMed] [CrossRef]
6. Kim YS, Potashnikova DM, Gisina AM, Kholodenko IV, Kopylov AT, Tikhonova OV, et al. TRIM28 is a novel regulator of CD133 expression associated with cancer stem cell phenotype. Int J Mol Sci. 2022;23(17). doi:10.3390/ijms23179874 [Google Scholar] [PubMed] [CrossRef]
7. Machida YJ, Machida Y, Chen Y, Gurtan AM, Kupfer GM, D'Andrea AD, et al. UBE2T is the E2 in the Fanconi anemia pathway and undergoes negative autoregulation. Mol Cell. 2006;23(4):589–96. doi:10.1016/j.molcel.2006.06.024 [Google Scholar] [PubMed] [CrossRef]
8. Alpi A, Langevin F, Mosedale G, Machida YJ, Dutta A, Patel KJ. UBE2T, the Fanconi anemia core complex, and FANCD2 are recruited independently to chromatin: a basis for the regulation of FANCD2 monoubiquitination. Mol Cell Biol. 2007;27(24):8421–30. doi:10.1128/MCB.00504-07 [Google Scholar] [PubMed] [CrossRef]
9. Perez-Peña J, Corrales-Sánchez V, Amir E, Pandiella A, Ocana A. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7. doi:10.1038/s41598-017-17836-7 [Google Scholar] [PubMed] [CrossRef]
10. Hao J, Xu AJ, Xie XL, Hao JT, Tian T, Gao SG, et al. Elevated expression of UBE2T in lung cancer tumors and cell lines. Tumor Biol. 2008;29(3):195–203. doi:10.1159/000148187 [Google Scholar] [PubMed] [CrossRef]
11. Gong YQ, Peng D, Ning XH, Yang XY, Li XS, Zhou LQ, et al. UBE2T silencing suppresses proliferation and induces cell cycle arrest and apoptosis in bladder cancer cells. Oncol Lett. 2016;12(6):4485–92. doi:10.3892/ol.2016.5237 [Google Scholar] [PubMed] [CrossRef]
12. Ueki T, Park JH, Nishidate T, Kijima K, Hirata K, Nakamura Y, et al. Ubiquitination and downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T overexpression in human breast cancer cells. Cancer Res. 2009;69(22):8752–60. doi:10.1158/0008-5472.CAN-09-1809 [Google Scholar] [PubMed] [CrossRef]
13. Wen M, Kwon Y, Wang Y, Mao JH, Wei G. Elevated expression of UBE2T exhibits oncogenic properties in human prostate cancer. Oncotarget. 2015;6(28):25226–39. doi:10.18632/oncotarget.4712 [Google Scholar] [PubMed] [CrossRef]
14. Cui P, Li H, Wang C, Liu Y, Zhang MJ, Yin Y, et al. UBE2T regulates epithelial-mesenchymal transition through the PI3K-AKT pathway and plays a carcinogenic role in ovarian cancer. J Ovarian Res. 2022;15(1). doi:10.1186/s13048-022-01034-9 [Google Scholar] [PubMed] [CrossRef]
15. Perez-Pena J, Corrales-Sanchez V, Amir E, Pandiella A, Ocana A. Ubiquitin-conjugating enzyme E2T (UBE2T) and denticleless protein homolog (DTL) are linked to poor outcome in breast and lung cancers. Sci Rep. 2017;7(1):17530. doi:10.1038/s41598-017-17836-7 [Google Scholar] [PubMed] [CrossRef]
16. Hao P, Kang B, Li Y, Hao W, Ma F. UBE2T promotes proliferation and regulates PI3K/Akt signaling in renal cell carcinoma. Mol Med Rep. 2019;20(2):1212–20. doi:10.3892/mmr.2019.10322 [Google Scholar] [PubMed] [CrossRef]
17. Liu LP, Yang M, Peng QZ, Li MY, Zhang YS, Guo YH, et al. UBE2T promotes hepatocellular carcinoma cell growth via ubiquitination of p53. Biochem Bioph Res Commun. 2017;493(1):20–7. doi:10.1016/j.bbrc.2017.09.091 [Google Scholar] [PubMed] [CrossRef]
18. Zhu JH, Ao HJ, Liu MD, Cao K, Ma JQ. UBE2T promotes autophagy via the p53/AMPK/mTOR signaling pathway in lung adenocarcinoma. J Transl Med. 2021;19(1). doi:10.1186/s12967-021-03056-1 [Google Scholar] [PubMed] [CrossRef]
19. Liu LL, Zhu JM, Yu XN, Zhu HR, Shi X, Bilegsaikhan E, et al. UBE2T promotes proliferation via G2/M checkpoint in hepatocellular carcinoma. Cancer Manag Res. 2019;11:8359–70. doi:10.2147/CMAR. [Google Scholar] [CrossRef]
20. Nemati S, Rahimi HM, Meyfour A, Pazoki H, Aghdaei HA, Shahrokh S, et al. Evaluation of the mTORC activity in the presence of and azathioprine in human monocyte cell line. BMC Microbiol. 2023;23(1). doi:10.1186/s12866-023-02819-8 [Google Scholar] [PubMed] [CrossRef]
21. Dogan F, Avci CB. Correlation between telomerase and mTOR pathway in cancer stem cells. Gene. 2017. doi:10.1016/j.gene.2017.09.072 [Google Scholar] [PubMed] [CrossRef]
22. Benjamin D, Colombi M, Moroni C, Hall MN. Rapamycin passes the torch: a new generation of mTOR inhibitors. Nat Rev Drug Discov. 2011;10(11):868–80. doi:10.1038/nrd3531 [Google Scholar] [PubMed] [CrossRef]
23. Guerriero C, Manfredelli M, Matera C, Iuzzolino A, Conti L, Dallanoce C, et al. M2 muscarinic receptor stimulation induces autophagy in human glioblastoma cancer stem cells via mTOR complex-1 inhibition. Cancers. 2024;16(1). doi:10.3390/cancers16010025 [Google Scholar] [PubMed] [CrossRef]
24. Luo YS, Zhang KX, Wu JH, Zeng H, Chen JT, Zhang PB, et al. Periosteum-derived mesenchymal stem cell alleviates renal fibrosis through mTOR-mediated Treg differentiation. Ren Fail. 2023;45(1). doi:10.1080/0886022X.2023.2212079 [Google Scholar] [PubMed] [CrossRef]
25. Silva VR, Santos LD, de Castro MVL, Dias RB, Valverde LD, Rocha CAG, et al. A novel ruthenium complex with 5-fluorouracil suppresses colorectal cancer stem cells by inhibiting Akt/mTOR signaling. Cell Death Discov. 2023;9(1). doi:10.1038/s41420-023-01759-6 [Google Scholar] [PubMed] [CrossRef]
26. Yang L, Chen Y, Liu N, Lu YW, Ma WL, Yang ZH, et al. CircMET promotes tumor proliferation by enhancing CDKN2A mRNA decay and upregulating SMAD3. Mol Cancer. 2022;21(1). doi:10.1186/s12943-022-01497-w [Google Scholar] [PubMed] [CrossRef]
27. Korkaya H, Paulson A, Charafe-Jauffret E, Ginestier C, Brown M, Dutcher J, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/β-catenin signaling. PLoS Biol. 2009;7(6):e1000121. doi:10.1371/journal.pbio.1000121 [Google Scholar] [PubMed] [CrossRef]
28. Liu S, Dontu G, Wicha MS. Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res. 2005;7(3):86–95. doi:10.1186/bcr1021 [Google Scholar] [PubMed] [CrossRef]
29. Swain N, Thakur M, Pathak J, Patel S, Hosalkar R, Ghaisas S. Altered immunoexpression of SOX2, OCT4 and Nanog in the normal-appearing oral mucosa of tobacco users. Dent Med Probl. 2022. doi:10.17219/dmp/146485 [Google Scholar] [PubMed] [CrossRef]
30. Verdelli C, Morotti A, Tavanti GS, Silipigni R, Guerneri S, Ferrero S, et al. The core stem genes, and are expressed in human parathyroid tumors and modulated by, and β-catenin pathways activation. Biomedicines. 2021;9(6). doi:10.3390/biomedicines9060637 [Google Scholar] [PubMed] [CrossRef]
31. Rahbaran M, Tayefeh R, Heidari F. The effects of embryo splitting on and gene expression in mouse blastocysts. Iran J Vet Res. 2022;23(4). doi:10.22099/Ijvr.2022.42487.6172 [Google Scholar] [PubMed] [CrossRef]
32. Bouamar H, Broome LE, Lathrop KI, Jatoi I, Brenner AJ, Nazarullah A, et al. mTOR inhibition abrogates human mammary stem cells and early breast cancer progression markers. Breast Cancer Res. 2023;25(1). doi:10.1186/s13058-023-01727-z [Google Scholar] [PubMed] [CrossRef]
33. Michishita M, Ochiai K, Nakahira R, Azakami D, Machida Y, Nagashima T, et al. mTOR pathway as a potential therapeutic target for cancer stem cells in canine mammary carcinoma. Front Oncol. 2023;13. doi:10.3389/fonc.2023.1100602 [Google Scholar] [PubMed] [CrossRef]
34. Yun H, Han GH, Kim J, Chung JY, Kim JH, Cho H. NANOG regulates epithelial-mesenchymal transition via AMPK/mTOR signalling pathway in ovarian cancer SKOV-3 and A2780 cells. J Cell Mol Med. 2022;26(20):5277–91. doi:10.1111/jcmm.17557 [Google Scholar] [PubMed] [CrossRef]
35. Yun H, Han GH, Cho H, Kim S, Kim JH. NANOG regulates tumor metastasis via AMPK/mTOR signaling pathway in patients with epithelial ovarian cancer. Gynecol Oncol. 2022;166:S135–S6. [Google Scholar]
36. Colditz GA, Bohlke K. Priorities for the primary prevention of breast cancer. CA Cancer J Clin. 2014;64(3):186–94. doi:10.3322/caac.21225 [Google Scholar] [PubMed] [CrossRef]
37. Eccles SA, Aboagye EO, Ali S, Anderson AS, Armes J, Berditchevski F, et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res. 2013;15(5):R92. doi:10.1186/bcr3493 [Google Scholar] [PubMed] [CrossRef]
38. Biddle A, Mackenzie IC. Cancer stem cells and EMT in carcinoma. Cancer Metastasis Rev. 2012. doi:10.1007/s10555-012-9345-0 [Google Scholar] [PubMed] [CrossRef]
39. Cao K, Ling XD, Jiang XY, Ma JQ, Zhu JH. Pan-cancer analysis of UBE2T with a focus on prognostic and immunological roles in lung adenocarcinoma. Resp Res. 2022;23(1):714. doi:10.1186/s12931-022-02226-z [Google Scholar] [PubMed] [CrossRef]
40. Yin H, Wang XY, Zhang X, Zeng YY, Xu QY, Wang WB, et al. UBE2T promotes radiation resistance in non-small cell lung cancer via inducing epithelial-mesenchymal transition and the ubiquitination-mediated FOXO1 degradation. Cancer Letters. 2020;494:121. doi:10.1016/j.canlet.2022.215792 [Google Scholar] [PubMed] [CrossRef]
41. Dutta R, Guruvaiah P, Reddi KK, Bugide S, Bandi DSR, Edwards YJK, et al. UBE2T promotes breast cancer tumor growth by suppressing DNA replication stress. Nar Cancer. 2022;4(4). doi:10.1093/narcan/zcac035 [Google Scholar] [PubMed] [CrossRef]
42. Xu N, Cui Y, Shi H, Guo GD, Sun FY, Jian TM, et al. UBE2T/STAT3 signaling promotes the proliferation and tumorigenesis in retinoblastoma. Invest Ophth Vis Sci. 2022;63(9). doi:10.1167/iovs.63.9.20 [Google Scholar] [PubMed] [CrossRef]
43. Zhang JG, Wang JD, Wu JC, Huang JY, Lin ZX, Lin X. UBE2T regulates FANCI monoubiquitination to promote NSCLC progression by activating EMT. Oncol Rep. 2022;48(2). doi:10.3892/or.2022.8350 [Google Scholar] [PubMed] [CrossRef]
44. Takahashi K, Murakami M, Yamanaka S. Role of the phosphoinositide 3-kinase pathway in mouse embryonic stem (ES) cells. Biochem Soc Trans. 2005;33:1522–5. doi:10.1042/BST0331522 [Google Scholar] [PubMed] [CrossRef]
45. Zhou J, Su P, Wang L, Chen J, Zimmermann M, Genbacev O, et al. mTOR supports long-term self-renewal and suppresses mesoderm and endoderm activities of human embryonic stem cells. Proc Natl Acad Sci USA. 2009;106(19):7840–5. doi:10.1073/pnas.0901854106 [Google Scholar] [PubMed] [CrossRef]
46. Corominas-Faja B, Cufi S, Oliveras-Ferraros C, Cuyas E, Lopez-Bonet E, Lupu R, et al. Nuclear reprogramming of luminal-like breast cancer cells generates Sox2-overexpressing cancer stem-like cellular states harboring transcriptional activation of the mTOR pathway. Cell Cycle. 2013;12(18):3109–24. doi:10.4161/cc.26173 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools