 Open Access
Open Access
REVIEW
Non-coding RNA as future target for diagnose and treatment of perineural invasion in cancers
1 Department of Pathophysiology, School of Basic Medical Sciences, Zhengzhou University, Zhengzhou, China
2 Collaborative Innovation Center of Henan Province for Cancer Chemoprevention, Zhengzhou, China
3 State Key Laboratory of Esophageal Cancer Prevention and Treatment, Zhengzhou, China
4 Department of Research and Development, Henan Tianxing Education and Media Company Limited, Zhengzhou, China
* Corresponding Author: SAIJUN MO. Email:
(This article belongs to the Special Issue: Advances in Biomarker Research: Unveiling the Pathways to Precision Medicine)
BIOCELL 2024, 48(6), 923-934. https://doi.org/10.32604/biocell.2024.049160
Received 29 December 2023; Accepted 21 February 2024; Issue published 10 June 2024
Abstract
Perineural invasion (PNI), a particularly insidious form of tumor metastasis distinct from hematogenous or lymphatic spread, has the capacity to extend well beyond the primary tumor site, infiltrating distant regions devoid of lymphatic or vascular structures. PNI often heralds a decrease in patient survival rates and is recognized as an indicator of an unfavorable prognosis across a variety of cancers. Despite its clinical significance, the underlying molecular mechanisms of PNI remain elusive, complicating the development of specific and efficacious diagnostic and therapeutic strategies. In the realm of cancer research, non-coding RNAs (ncRNAs) have attracted considerable attention due to their multifaceted roles and cancer-specific expression profiles, positioning them as promising candidates for applications in cancer diagnostics, prognostics, and treatment. Among the various types of ncRNAs, microRNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs) have emerged as influential players in PNI. Their involvement is increasingly recognized as a contributing factor to tumor progression and therapeutic resistance. Our study synthesizes and explores the diverse functions and mechanisms of ncRNAs in relation to PNI in cancer. This comprehensive review aims to shed light on cutting-edge perspectives that could pave the way for innovative diagnostic and therapeutic approaches to address the challenges posed by PNI in oncology.Keywords
In the world, cancer is a major public health problem and is the first or second leading cause of death before the age of 70 years in most of countries [1,2]. The continuously growing of cancer incidence and mortality are leading to an ever-increasing economic burden on society and families. A variety of factors can increase its risk, including aging, lifestyle habits (like smoking and alcohol), exposure to certain chemicals and substances, certain behaviors (such as diet and physical activity), exposure to some types of radiation, certain infections, and a family history of cancer [3,4]. Despite advancements in cancer diagnosis and treatments, the overall treatment effect is not satisfying.
The poor prognosis of cancer patients is predominantly due to the advanced stage of the disease coupled with metastasis, which contributes to approximately 90% of cancer-associated fatalities [5]. Perineural involvement, which includes perineural spread (PNS) and perineural invasion (PNI), is often a contributing component in cancer metastasis. PNS is a clinical and radiographic phenotype characterized by extensive cancerous growth within and alongside nerves that it becomes identifiable through radiographic imaging [6,7]. In contrast, PNI, the histopathological correlate of PNS, is broadly delineated as the infiltration of cancerous cells into the perineural and intraneural spaces [6–8]. Microscopic examination is required to identify PNI, signifying the infiltration of cancer within neural structures. Efforts to improve the identification of the more elusive manifestations of PNI involve the deployment of comprehensive immunostaining techniques, which include the employment of S100 protein markers. The widely recognized framework for PNI categorization hinges on the spatial relation of cancer cells to the nerves, bifurcating into two subtypes: extratumoral and intratumoral PNI [7]. Intratumoral PNI is typified by the direct encroachment of malignant cells into the nerve fibers themselves, leading to their compromisation. In contrast, extratumoral PNI is characterized by the presence of cancer cells in the regions surrounding the nerve, without invading the nerve fibers. Moreover, PNI may be further subclassified as either focal or extensive, denoting the restriction of cancer cells to a discrete, localized region, or indicating a widespread or multifocal pattern of cancer involvement within the nerve structures [7].
PNI occurs frequently across a wide spectrum of cancers and serves as a significant prognostic marker, often heralding aggressive malignancy and correlating with diminished survival rates in a variety of tumors, including those of the head and neck [8], colon [9], prostate [10,11], and pancreas [12,13], etc. When cancer cells exploit the perineural space for growth, following the path of least resistance along nerve fibers, cancer pain was happened [12,14]. Beyond its role in pain, PNI is the subject of extensive research, with emerging studies shedding light on the intricate and evolving nature of its progression. For example, the relationship between PNI and inflammation has been found to be nuanced and reciprocal, with inflammation fostering conditions that favor PNI, while PNI, in turn, can exacerbate inflammatory responses [15,16]. Additionally, PNI is associated with metabolic alterations in both neoplastic and neuronal cells, with these changes potentially influencing, or being influenced by, the occurrence of PNI [17]. Consequently, the identification of PNI can prompt a reassessment of clinical management, potentially leading to broader surgical resections, the strategic application of radiotherapy, or the initiation of systemic chemotherapy to combat the insidious spread of microscopic disease.
There are several factors, such as cancer type, cancer microenvironment, and genetic and epigenetic regulatory, are known to impact PNI. Comprehending these factors is crucial for developing strategies to predict, prevent, and treat PNI. Notably, recent studies have shed light on the significant role of epigenetic factors, particularly Non-coding RNAs (ncRNAs), in the context of cancer-related PNI [9,11,13]. NcRNAs represent an extensive category of RNA molecules pivotal in modulating gene expression at both transcriptional and post-transcriptional levels. This group encompasses microRNA (miRNA), long ncRNA (lncRNA), circular RNA (circRNA), endogenous small interfering RNA, Piwi-associated small RNA [18]. These ncRNAs are integral to a vast array of biological functions, ensuring cellular integrity and developmental processes. The dysregulation of ncRNAs has been markedly linked to a spectrum of pathologies, including neurological [19], cardiovascular [20], metabolic disorders [21], and cancer [22–24]. Within the cancer landscape, ncRNAs can function as either oncogenes or tumor suppressors, influencing the disease course by modulating cell proliferation, apoptosis, metastasis, and angiogenesis. Alterations in the expression patterns or mutations of ncRNAs have been associated with a variety of cancers, positioning them as promising biomarkers and targets for diagnostics, prognostics, and therapeutics [25,26]. Among the ncRNAs, miRNA, lncRNA and circRNA have garnered significant interest due to their established connection with PNI [9,15,27]. This review aims to elucidate the roles and mechanisms of ncRNAs in PNI, an endeavor that holds promise for refining cancer staging approaches, introducing novel therapeutic techniques, and potentially catalyzing a shift in the clinical management of patients.
PNI is a Distinctive Pathway of Cancer Metastasis
PNI is a unique metastasis pathway associated with nerves, capable of invading and spreading to regions distant from the original tumor. The accurate detection and verification of PNI are paramount for precise cancer staging, prognostic assessment, and devising a tailored therapeutic approach. PNI is formally characterized by the proximity of tumor cells to neural elements, specifically when a tumor encases at least one-third of the nerve’s circumference, or when cancer cells are identified within any of the three layers of the nerve sheath: the epineurium, perineurium, or endoneurium [28].
The methods to detect PNI in cancer
The primary screening method for PNI involves the use of hematoxylin and eosin (HE) staining, with immunohistochemistry (IHC) employing S100 markers enhancing the detection accuracy. Research indicates that shortcomings in identifying PNI via HE can be addressed through S100 immunostaining, thereby quantification of PNI by S100 is a potential method for prognosis prediction [29,30]. Advances have also been made in molecular methodologies that leverage markers associated with PNI. For instance, Lu et al. [31] developed GAP-43Ra-ICG-PEG, an active-targeting near-infrared fluorescent (NIRF) probe, which binds selectively to GAP-43-expressing neural cells in PNI models. This probe has demonstrated efficacy in illuminating PNI in preclinical pancreatic cancer models, thereby enhancing the prospects for NIRF-assisted pancreatic surgeries, especially in patients with PNI. Additionally, nerve conduction studies and electromyography (EMG) may offer indirect indications of PNI by detecting nerve function anomalies [32]. Accurate detection of PNI is paramount, underscoring the necessity for the advancement and refinement of diagnostic methods and technologies.
PNI holds considerable significance in the diagnosis, treatment, and prognosis of cancer
PNI is an established feature in a multitude of cancers, including pancreatic cancer (PCa) with an occurrence rate of 80%–100% [33], salivary adenoid cystic carcinoma (SACC) ranging from 26%–96% [34], head and neck cancers affecting 25%–80% of cases [35], prostate cancer seen in 7%–90% [36], gastric cancer (GC) in 7%–76% [37], esophageal cancer in 5%–66% [38], and colorectal cancer (CRC) in 9%–39% [9]. Given the high incidence and pivotal role of PNI across these tumor types, it significantly impacts the degree of malignancy and the overall prognosis of the affected patients. Consequently, PNI holds considerable weight as a crucial factor for diagnosis and prognostication. For instance, in oral squamous cell carcinoma (OSCC), PNI is strongly linked with tumors originating in the tongue and the emergence of lymph node metastases, although it shows minimal correlation with the number of foci, scale, and precise location of PNI involvement. The presence of larger affected nerves may indicate a more advanced tumor stage [39]. Moreover, OSCC patients burdened with PNI typically exhibit compromised survival outcomes, including overall survival (OS), disease-free survival (DFS), disease-specific survival (DSS), and failure-free survival (FFS), and an increased likelihood of recurrence. These findings underscore PNI as a harbinger of poor prognosis in OSCC [40].
The presence of PNI also effect the cancer therapies, including surgical approaches, adjuvant therapy decisions, and the overall treatment strategy [16,41]. Therefore, improved treatment methods and plans are highly likely to enhance efficacy. Recent studies suggest that integrating adjuvant chemotherapy with radiotherapeutic strategies, such as postoperative radiotherapy (PORT), may enhance DFS in OSCC patients with PNI. This points to the potential benefit of adjuvant chemoradiation in improving patient outcomes in the context of PNI, a premise that warrants further corroboration through prospective clinical trials [42].
Furthermore, PNI is frequently associated with the occurrence of pain in affected individuals, profoundly detracting from their overall well-being. Pain stemming from cancer is a prevalent affliction among patients with malignancies, with a majority reporting intense pain at some juncture of their condition. PNI not only exacerbates this cancer-related pain but also emerges as a predictive pathological indicator of early metastatic activity, providing vital insights into the prognosis of the disease [12,14,43,44]. Notably, researchers have pinpointed pain as a key variable intricately tied to PNI across various tumor localizations in patients with head and neck cancer (HNC), regardless of whether the tumors originate from the oropharynx or other non-oropharyngeal regions [45].
The regulatory mechanism of PNI in cancer
PNI is a complex process influenced by a multitude of factors that can affect its incidence and progression in cancer. The tumor microenvironment, stands as a pivotal determinant in cancer progression, is governed by a complex interplay among various cellular constituents. Among these, cancer cells, nerve cells (with a predominance of Schwann cells), stromal cells, and fibroblasts have been most rigorously investigated. These cellular entities are central themes in a vast expanse of academic literature, reflecting their significant roles in tumorigenesis and progression [8,12,13]. Contemporary research efforts have pivoted towards elucidating the impact of immune cell infiltration on PNI. A study on ovarian cancer tissue’s transcriptome unearthed an inverse correlation between PNI presence and the penetration of cytotoxic immune cells, encompassing macrophages, central memory CD4+ T-cells, natural killer cells, and monocytes [15]. Similarly, in pancreatic ductal adenocarcinoma (PDAC), a distinct infiltration pattern of immune cells is observed to influence PNI development. Specifically, samples exhibiting a higher degree of PNI were characterized by a heightened presence of mast cells, myeloid-derived suppressor cells (MDSCs), and regulatory T cells, in contrast to those with a less pronounced degree of PNI [15]. These findings underscore the critical role of immune dynamics in the progression of PNI and spotlight potential avenues for targeted cancer immunotherapy.
Molecular determinants constitute the second principal modulator of PNI. The molecular underpinnings of PNI are primarily interwoven with oncogenes, tumor suppressor genes, neurotrophins, and cell adhesion molecules, each playing a pivotal role in its regulation and manifestation [11,12,40]. However, intriguingly, the outcomes vary between different studies, highlighting the complexity of the mechanisms involved. For instance, mutations in the oncogene Kirsten rats arcomaviral oncogene homolog (K-ras) are strongly linked with PNI in colon cancer and intrahepatic cholangiocarcinoma [46,47]. In contrast, in PDAC, high-degree PNI samples have a significantly lower total mutation burden compared to low-degree PNI samples, yet there is no appreciable difference in K-ras mutation frequency. This finding also extends to other genes such as Tumor Protein 53 (TP53), cyclin-dependent kinase inhibitor 2A (CDKN2A), Small mothers against decapentaplegic homolog 4 (SMAD4), guanine nucleotide-binding protein α-stimulating activity polypeptide(GNAS), ring finger protein 43 (RNF43), transforming growth factor beta receptor 2 (TGFBR2), protein polybromo-1 (PBRM1), ras responsive element binding protein 1 (RREB1), and AT rich interaction domain 1A (ARID1A), as well as the levels of DNA hypermethylation between the two PNI groups in PDAC [15]. Additionally, stem cell markers appear to play a pivotal role in PNI. Octamer binding transcription factor 4 (Oct4) and Nanog, both vital for the pluripotency of embryonic stem cells, have been significantly associated with PNI and desmoplasia in human salivary gland mucoepidermoid carcinoma [48]. And with a positive correlation to cancer stem cell-like properties, elevated levels of integrin beta1 (ITGβ1) have been implicated as key contributors to PNI and the sustenance of radioresistance in OSCC cells [49]. These findings suggest genetic factors in PNI have great potential implications for developing biomarkers and therapeutic targets specific to the PNI of cancer.
Despite a wealth of emerging discoveries, a holistic grasp of the molecular mechanisms driving PNI remains elusive. In recent times, the spotlight has shifted toward epigenetic regulation, with a special focus on ncRNAs. These ncRNAs are gaining recognition for their superior potential over genetic regulation in the contexts of tumor diagnosis, treatment, and prognosis. Their regulatory roles and relatively stable nature present promising avenues for clinical applications, potentially leading to more refined strategies in managing and combating cancer.
The role and mechanism of ncRNA on PNI in cancers
In the realm of oncology, specific ncRNAs, notably miRNAs, lncRNAs, and circRNAs, have been identified as integral contributors to both the initiation and advancement of cancer, including phenomena like PNI. They play a dual role in oncogenesis and cancer suppression by modulating gene expression, inducing epigenetic and genetic modifications, influencing the tumor microenvironment, and driving the metabolic reprogramming of cancer cells. Their multifaceted involvement in cancer biology underscores their potential as therapeutic targets and biomarkers for diagnosis and prognosis.
The advanced methods for detecting ncRNA
NcRNAs are a broad class of RNA molecules that have crucial roles in the regulation of gene expression and the preservation of cellular integrity. This diverse group includes several prominent subclasses such as miRNAs, lncRNAs, circRNAs, small interfering RNAs (siRNAs), piwi-Interacting RNAs, small nucleolar RNAs (snoRNAs). As for the detection method of ncRNA, it is completely consistent with the research method of nucleic acid. Bioinformatic analyses leveraging datasets from resources such as the Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) are integral to the prediction and screening of ncRNAs for scientific inquiry [50–53]. The methodologies for ncRNA detection are in line with conventional nucleic acid research approaches. Quantitative reverse transcription PCR (qRT-PCR) is widely utilized to measure and quantify ncRNA expression levels. Additionally, methods like in situ hybridization (ISH) and fluorescence in situ hybridization (FISH) not only enable the detection of ncRNA expression but also elucidate the precise cellular and tissue localization of these molecules [54–57]. For a more extensive evaluation of ncRNA expression alterations across tissues and cells, whole transcriptome sequencing and ncRNA-specific microarrays provide a comprehensive survey of the ncRNA expression landscape [58–61].
MiRNAs involving in PNI in cancers
The role of miRNAs in the processes of tumorigenesis and cancer progression has become increasingly evident over recent years, highlighting their value as diagnostic biomarkers and prospective therapeutic agents. Functioning as either oncogenes or tumor suppressors, miRNAs are pivotal in regulating PNI, affecting diverse molecular pathways leveraged by cancer cells to permeate peripheral nerves in a variety of cancers, including PDAC, PCa, SACC, head and neck squamous cell carcinoma (HNSCC), OSCC, and CRC, as delineated in Table 1. For example, in PCa, the levels of miR-17-5p, miR-210, miR-1, miR-10, miR-30c, miR-100, miR-125b, and miR-224 were elevated in cancerous tissues compared to adjacent normal tissues [54,55,62].
Interestingly, these miRNAs also demonstrated heightened expression in cases of PCa with PNI(+), in stark contrast to PNI(−) cases, whereas miR-130 exhibited inverse trends [63]. In OSCC, miR-7, miR-370, miR-190, miR-204, and miR-377 showed reduced expression in cancer tissues and were negatively associated with PNI, while miR-196a, miR-196b, and miR-424 played opposing roles [52,69,70]. Beyond these observations, these miRNAs also exhibited correlations with other clinicopathological characteristics. Apart from PNI, the upregulation of miR-21, miR-23a, and miR-27a in PDAC correlated positively with microscopic tumor infiltration at resection margins, poorer OS post-surgery, and enhanced cancer cell proliferation, as well as increased xenograft tumor growth [50]. Moreover, a significant decline in miR-370 expression in OSCC has been associated with lymphovascular invasion, and its downregulation also appears to facilitate cell migration and anchorage-independent growth [70]. Notably, although intricately linked to PNI, the majority of miRNAs tend to exhibit a consistent relationship with both lymphovascular invasion and lymph node metastasis across various cancer types. MiRNAs such as miR-210 [62], miR-130 [63], miR-205 [51], miR-7 [52], miR-370 [70], miR-9-3p [72], miR-21 [73], miR-128-3p [61], miR-200a [66], and miR-373 [66], including miR-30c [66], have been documented to possess such parallel associations, underscoring their multifaceted roles within the metastatic cascade.
MiRNAs consistently play a pivotal biological role in PNI by modulating the expression of their target genes and related signaling pathways. For instance, miR-187, which is downregulated in the progression of SACC and shows a decreasing trend at the frontiers of nerve invasion, is suppressed by C-X-C chemokine receptor type 5 (CXCR5). CXCR5 is found in increased levels in PNI positive samples compared to PNI negative, and its inhibition has been demonstrated to curb PNI in a SACC xenograft model. CXCR5 can trigger the differentiation of SACC cells into schwann-like cells by inhibiting miR-187, which disinhibited S100 calcium binding protein A4 (S100A4), a schwann cell marker, thus promotes SACC PNI [57]. Another illustrative example is miR-491-5p, which is downregulated in GC and plays a role in restraining cell proliferation, invasion, and migration. The link between miR-491-5p and PNI may depend on the Foxi1 (forkhead box I1)/miR-491-5p/Wnt3a axis. Foxi1 is instrumental in the transcriptional control of miR-491-5p, and miR-491-5p serves as a tumor suppressor by targeting the Wnt3a, strongly correlating with an increased incidence of PNI in GC patients [75].
Additionally, miRNA regulatory mechanisms are likely to be implicated in cancer metastasis with particular reference to miRNA function in PNI. For instance, miR-128-3p, stimulated by IL-6 (interleukin 6), plays a crucial role in epithelial-mesenchymal transition (EMT) in CRC by targeting FOXO4 (forkhead box O4) through the TGF-β/SMAD (transforming growth factor-β/small mothers against decapentaplegic) and JAK/STAT3 (Januskinase/signal transducers and activators of transcription 3) signaling pathways [61]. Notably, high levels of miR-128-3p are detected in exosomes from EMT-induced HCT-116 cells following IL-6 stimulation. Overexpression of miR-128-3p results in the downregulation of FOXO4 and the activation of TGF-β/SMAD and JAK/STAT3 signaling pathways in CRC cells and xenografted tumors, leading to EMT. Meanwhile, miR-370, which is downregulated in OSCC, targets the insulin receptor substrate-1 (IRS-1), which in turn regulates OSCC cell migration and anchorage-independent growth (AIG), both of which are intricately associated with PNI [70].
LncRNAs involving in PNI in cancer
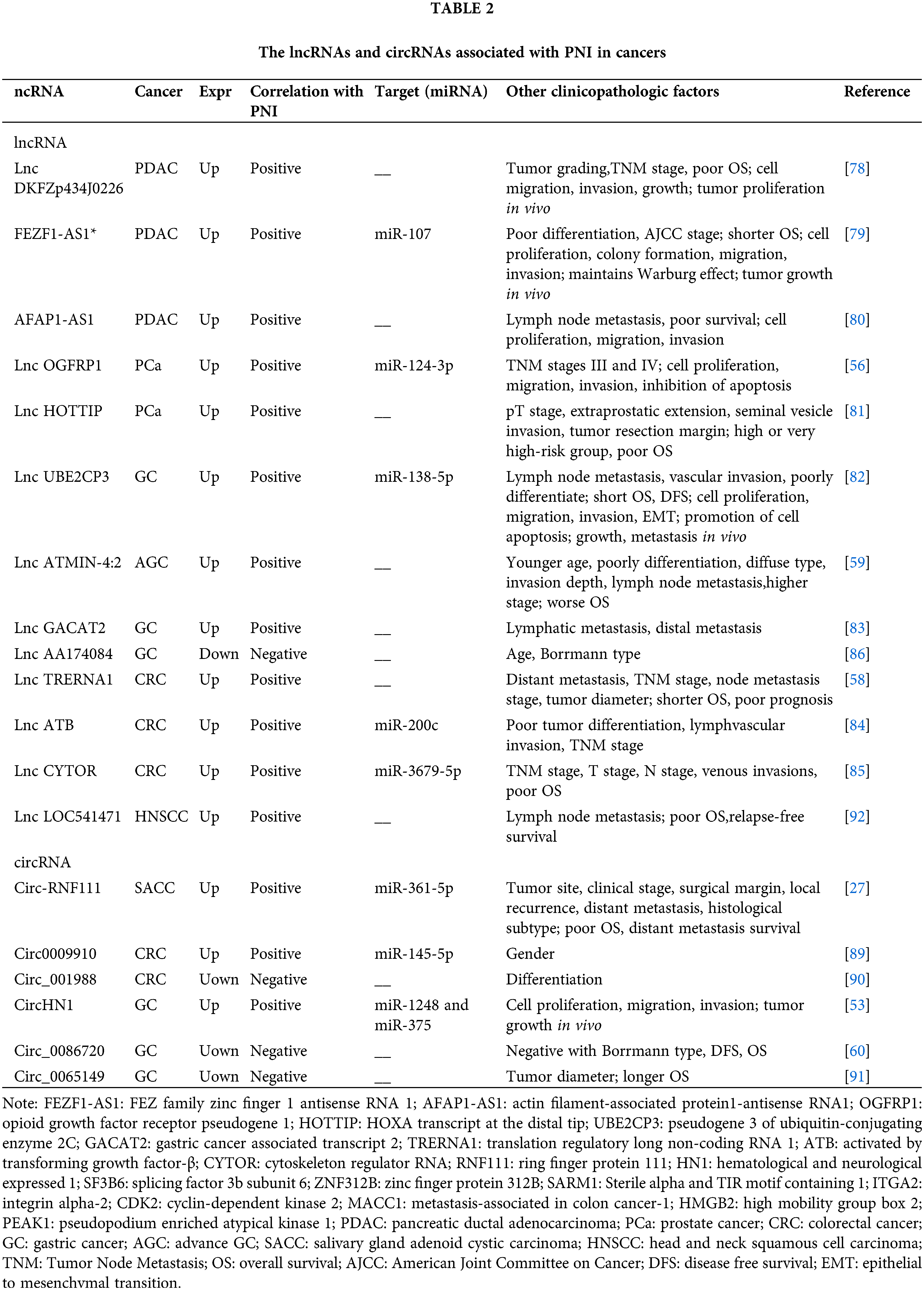
LncRNAs, characterized by their length of over 200 nucleotides, are pivotal in modulating gene expression throughout development and have been implicated in a multitude of diseases, including various forms of cancer [26,77]. Within the cancer paradigm, lncRNAs may act as either oncogenes or tumor suppressors, thus serving as critical regulators of a myriad of biological processes such as cell proliferation, apoptosis, metastasis, and angiogenesis. Numerous lncRNAs have been identified with elevated expression levels in cancerous tissues and are significantly associated with PNI (Table 2). For instance, lncRNAs such as DKFZp434J0226, FEZ family zinc finger 1 antisense RNA 1 (FEZF1-AS1), and actin filament-associated protein1-antisense RNA1 (AFAP1-AS1) in PDAC [78–80]; opioid growth factor receptor pseudogene 1 (OGFRP1) and HOXA transcript at the distal tip (HOTTIP) in Pca [56,81]; pseudogene 3 of ubiquitin-conjugating enzyme 2C (UBE2CP3), ATMIN-4:2, and gastric cancer associated transcript 2 (GACAT2) in GC [59,82,83]; and translation regulatory long non-coding RNA 1 (TRERNA1), activated by transforming growth factor-β (ATB), and cytoskeleton regulator RNA (CYTOR) in CRC [58,84,85] not only demonstrate high expression in tumor tissues but also exhibit a strong correlation with negative clinical prognostic indicators, such as PNI, OS, lymphovascular invasion, and metastasis. Conversely, the lncRNA AA174084 represents an exception to this trend. It was found to be downregulated in GC and exhibits a negative correlation with PNI, as well as patient age and Borrmann type, suggesting a potential protective role against cancer progression [86].
LncRNAs implicated in PNI can function as competitive endogenous RNAs (ceRNAs) or “molecular sponges,” effectively sequestering miRNAs to prevent them from binding to their target mRNAs. As a ceRNA, for example, the overexpression of lncOGFRP1 in PCa cells acts as a decoy for miR-124-3p, a miRNA known to be downregulated in PCa and capable of suppressing cancer cell proliferation, invasion, and migration. MiR-124-3p itself negatively regulates the expression of SARM1, which exerts effects contrary to those of miR-124-3p in PCa. It has been found that miR-124-3p contributes to the malignant progression of cancer, including behaviors such as migration and invasion, and is also involved in the recovery of neural functions, which is closely associated with the development of PNI [87]. Moreover, the lncRNA ATB is known to sponges miR-200c, and a higher expression of ATB combined with a lower expression of miR-200c has been observed more frequently in CRC populations characterized by PNI [84]. The lncRNA ATB/miR-200c/CDK2 axis has been identified as a regulatory pathway for CRC cell proliferation and apoptosis, while its connection to cell invasion and metastasis has yet to be reported. Furthermore, a dysregulated ceRNA network, UBE2CP3/miR-138-5p/integrin alpha-2 (ITGA2), has been unearthed in GC: the lncRNA UBE2CP3 upregulates ITGA2 expression by sponging miR-138, and the profound inhibitory effect of UBE2CP3 depletion on GC cell migration and invasion can be counteracted by miR-138 inhibitors and synthetic siRNAs targeting ITGA2. Additionally, UBE2CP3 expression is negatively controlled by the transcription factor Early Flowering 3 (ELF3) and positively stabilized through interactions with ELF3 protein by insulin-like growth factor binding protein 7 (IGFBP7) mRNA [82]. This highlights the complexity of the upstream regulatory mechanisms governing ncRNAs.
CircRNAs involving in PNI in cancers
CircRNAs are unique in that they form covalently closed loop structures without the 5’ or 3’ ends typical of linear RNAs [26,88]. This circular configuration endows circRNAs with remarkable stability, as it renders them resistant to degradation by exonucleases. With advancements in high-throughput RNA sequencing and bioinformatics, circRNAs have gained prominence for their roles in various biological processes, including those related to cancer development and progression. To date, researchers have identified six circRNAs with significant correlations to PNI. These include circ-ring finger protein 111 (RNF111) in SACC, circ0009910 in CRC, and circ hematological and neurological expressed 1 (HN1) in GC, all of which exhibit overexpression in cancer tissues and are positively associated with PNI [27,89]. In contrast, lower expressions of circ_001988 in CRC, along with circ_0086720 and circ_0065149 in GC, have been found to correlate negatively with PNI [53,60,90,91]. However, the impact of these circRNAs extends beyond their association with PNI; they also influence other clinicopathological factors. For instance, high levels of circ-RNF111 are indicative of an association with multiple clinical parameters in SACC, including tumor site, clinical stage, surgical margins, local recurrence, distant metastasis, histological subtype, and poorer OS [27]. Similarly, in GC, elevated circ_0086720 levels are negatively associated with the Borrmann type, DFS, and OS, highlighting the potential of circRNAs as multifaceted biomarkers in cancer prognosis and as possible targets for therapeutic intervention [60].
Within the context of PNI, circRNAs also are known to play critical roles as miRNA sponges, effectively binding to and sequestering miRNAs, which subsequently modulates gene expression. In SACC, the role of circ-RNF111 in facilitating PNI has been elucidated across various experimental platforms, including analyses of tumor tissues, cell line studies, and xenograft models. This particular circRNA promotes PNI by engaging the miR-361-5p/high mobility group box 2 (HMGB2) regulatory axis [27]. In the case of SACC-LM cells, which are characterized by a high metastatic potential, suppression of circ-RNF111 has been shown to inhibit cell proliferation and reduce the cells’ invasive capabilities towards neuronal BV2 cells, a hallmark characteristic of PNI. Moreover, the biological functions associated with PNI can be modulated by targeting the miR-361-5p sponge effect of circ-RNF111 as well as its downstream target, HMGB2, suggesting a potential therapeutic strategy to counteract PNI in cancer.
Conclusion and Future Perspectives
PNI is broadly recognized as an indicator of advanced tumor malignancy, associated with increased pain, higher recurrence rates, and poorer prognostic outcomes [6,7,12]. Despite this recognition, the complexity of PNI poses persistent challenges in its detection, understanding, and therapeutic intervention [7,15,93]. Within the intricate landscape of PNI, ncRNAs, particularly miRNAs, lncRNAs, and circRNAs, were identified to play a crucial role in the molecular crosstalk that permits the incursion of cancer cells into nerve [9–11,13]. Those ncRNAs’ influence is exerted through either upregulation or downregulation in the cancerous tissues or bodily fluids of patients. Consequently, ncRNAs have the potential to significantly enhance clinical diagnostics, treatment and prognostic assessments for PNI.
The predominant hurdles in utilizing ncRNAs for diagnostic and prognostic purposes are the lack of widely endorsed protocols for their isolation and quantification, as well as a lack of consensus on methods for data interpretation [19,21,26]. Recent advancements in technologies such as RNA sequencing, quantitative reverse transcription PCR (qRT-PCR), alongside improvements in imaging and biopsy techniques, have significantly boosted our ability to detect and measure ncRNAs with remarkable sensitivity and precision [29,31,32,94]. However, in spite of these innovations, the direct application of ncRNAs as clinical tools for cancer diagnostics and prognostication, particularly with regard to PNI, is still in its developmental stages.
Likewise, there are substantial challenges in the therapeutics application of ncRNA for PNI, as well as cancers and other diseases. To this day, only two siRNA-based drugs, patisiran and givosiran, have received approval from the Food and Drug Administration (FDA). And out of a limited 10 miRNAs have made it to clinical trial stages, yet none have progressed to phase III trials as per the clinicaltrials.gov database [23,95]. Moreover, the development of targeted drugs for lncRNAs and circRNAs remains in its infancy, with no significant updates on their advancement. The main reason why the application of ncRNA in clinical treatment is difficult to advance may be related to the instability of ncRNA drugs, their lack of target specificity, the challenges in achieving efficient delivery to tumor cells, and the potential for unwanted off-target effects [22]. Recent discoveries have unveiled that ncRNAs are both regulators and targets of immune molecules, which highlights their significant role in the onset and advancement of PNI. These insights have opened up fresh avenues targeted therapeutic strategies for PNI, such as combining ncRNA-based approaches with immunotherapy [15,16,57,94]. Additionally, traditional lncRNAs have been distinguished by their apparent lack of protein-coding capacity. However, recent research has challenged this view, revealing that some lncRNAs and circRNAs might have the potential to code for small, functional proteins or peptides. Although the full spectrum of activities of these peptides is still under investigation, emerging evidence points to their involvement in diverse cellular functions including gene expression regulation, signaling pathways, and cellular metabolism [96–98]. The identification of these bioactive peptides complicates the classic distinction between coding and non-coding RNAs and heralds new horizons for the exploration of gene regulatory complexity and proteomics.
In summary, owing to their distinctive characteristics such as accessibility, disease-specificity, and sensitivity, ncRNAs possess substantial clinical value in the context of PNI, presenting avenues for enhanced diagnostic and prognostic accuracy, as well as the creation of targeted therapies. This review contributes to the deepening of our comprehension of PNI, with the potential to improve patient outcomes. Beyond tackling the inherent challenges of ncRNAs as drug targets, it is noteworthy that the newly discovered functions and effects of ncRNAs might broaden the spectrum of function in cancer development and inhibition. This emerging field ncRNA research is poised to uncover new aspects of tumor biology, potentially leading to innovative strategies for cancer management.
Acknowledgement: The authors would like to thank Dr. Yang Hai (Zhongyuan University of Technology, Zhengzhou, Henan) for carefully checking and improving the English writing.
Funding Statement: Foundation of Henan Educational Committee, Grant Number 22A310024; Natural Science Foundation for Young Teachers’ Basic Research of Zhengzhou University, Grant Number JC202035025.
Author Contributions: Study conception and design: Saijun Mo, Wenbo Cao; Draft manuscript preparation: Bingjie Li, Wenbo Cao; Data collection: Bingjie Li, Jingjin Xiao, Yixiao Chen, Qiying Wei; Review and editing: Mingjin Yue, Saijun Mo. All authors reviewed and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Ju W, Zheng R, Zhang S, Zeng H, Sun K, Wang S, et al. Cancer statistics in Chinese older people, 2022: current burden, time trends, and comparisons with the US, Japan, and the Republic of Korea. Sci China Life Sci. 2023;66(5):1079–91. doi:10.1007/s11427-022-2218-x [Google Scholar] [PubMed] [CrossRef]
2. Xiang D, Hu S, Mai T, Zhang X, Zhang L, Wang S, et al. Worldwide cancer statistics of adults over 75 years old in 2019: a systematic analysis of the global burden of disease study 2019. BMC Public Health. 2022;22(1):1979. doi:10.1186/s12889-022-14412-1 [Google Scholar] [PubMed] [CrossRef]
3. Zheng RS, Zhang SW, Sun KX, Chen R, Wang SM, Li L, et al. Cancer statistics in China, 2016. Zhonghua Zhong Liu Za Zhi. 2023;45(3):212–20 (In Chinese) [Google Scholar] [PubMed]
4. Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. doi:10.3322/caac.v73.1. [Google Scholar] [CrossRef]
5. Dillekas H, Rogers MS, Straume O. Are 90% of deaths from cancer caused by metastases? Cancer Med. 2019;8(12):5574–6 [Google Scholar] [PubMed]
6. Tu W, Gottumukkala RV, Schieda N, Lavallee L, Adam BA, Silverman SG. Perineural invasion and spread in common abdominopelvic diseases: imaging diagnosis and clinical significance. Radiographics. 2023;43(7):e220148. doi:10.1148/rg.220148 [Google Scholar] [PubMed] [CrossRef]
7. Tong JY, Huilgol SC, James C, Rajak S, Selva D. Perineural invasion and perineural spread in periocular squamous cell carcinoma. Eye. 2023;37(5):875–84. doi:10.1038/s41433-022-02306-w [Google Scholar] [PubMed] [CrossRef]
8. Goswami PR, Singh G. Perineural invasion (PNI) definition, histopathological parameters of PNI in oral squamous cell carcinoma with molecular insight and prognostic significance. Cureus. 2023;15(6):e40165 [Google Scholar] [PubMed]
9. Wang H, Huo R, He K, Cheng L, Zhang S, Yu M, et al. Perineural invasion in colorectal cancer: mechanisms of action and clinical relevance. Cell Oncol. 2024;47(1):1–17. [Google Scholar]
10. Niu Y, Forster S, Muders M. The role of perineural invasion in prostate cancer and its prognostic significance. Cancers. 2022;14(17):4065. doi:10.3390/cancers14174065 [Google Scholar] [PubMed] [CrossRef]
11. Liu Q, Ma Z, Cao Q, Zhao H, Guo Y, Liu T, et al. Perineural invasion-associated biomarkers for tumor development. Biomed Pharmacother. 2022;155:113691. doi:10.1016/j.biopha.2022.113691 [Google Scholar] [PubMed] [CrossRef]
12. Shi RJ, Ke BW, Tang YL, Liang XH. Perineural invasion: a potential driver of cancer-induced pain. Biochem Pharmacol. 2023;215:115692. doi:10.1016/j.bcp.2023.115692 [Google Scholar] [PubMed] [CrossRef]
13. Selvaggi F, Melchiorre E, Casari I, Cinalli S, Cinalli M, Aceto GM, et al. Perineural invasion in pancreatic ductal adenocarcinoma: from molecules towards drugs of clinical relevance. Cancers. 2022;14(23):5793. doi:10.3390/cancers14235793 [Google Scholar] [PubMed] [CrossRef]
14. Wang J, Chen Y, Li X, Zou X. Perineural invasion and associated pain transmission in pancreatic cancer. Cancers. 2021;13(18):4594. doi:10.3390/cancers13184594 [Google Scholar] [PubMed] [CrossRef]
15. Jiang SH, Li RK, Liu DJ, Xue JL, Yu MH, Zhang S, et al. The genomic, transcriptomic, and immunological profiles of perineural invasion in pancreatic ductal adenocarcinoma. Sci China Life Sci. 2023;66(1):183–6. doi:10.1007/s11427-022-2146-5 [Google Scholar] [PubMed] [CrossRef]
16. Li X, Wang Y, Zhai Z, Mao Q, Chen D, Xiao L, et al. Predicting response to immunotherapy in gastric cancer via assessing perineural invasion-mediated inflammation in tumor microenvironment. J Exp Clin Cancer Res. 2023;42(1):206. doi:10.1186/s13046-023-02730-0 [Google Scholar] [PubMed] [CrossRef]
17. Zhang L, Zhang W, Zhang X, Min Y, Zhao Y, Wang B, et al. High‐glucose microenvironment promotes perineural invasion of pancreatic cancer via activation of hypoxia inducible factor 1alpha. Oncol Rep. 2022;47(4):64. doi:10.3892/or. [Google Scholar] [CrossRef]
18. Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–65. doi:10.1038/nrd4140 [Google Scholar] [PubMed] [CrossRef]
19. Zhu K, Wang T, Li S, Liu Z, Zhan Y, Zhang Q. NcRNA: key and potential in hearing loss. Front Neurosci. 2023;17:1333131 [Google Scholar] [PubMed]
20. Blokhin I, Khorkova O, Hsiao J, Wahlestedt C. Developments in lncRNA drug discovery: where are we heading? Expert Opin Drug Discov. 2018;13(9):837–49. doi:10.1080/17460441.2018.1501024 [Google Scholar] [PubMed] [CrossRef]
21. Gao D, Ren L, Hao YD, Schaduangrat N, Liu XW, Yuan SS, et al. The role of ncRNA regulatory mechanisms in diseases-case on gestational diabetes. Brief Bioinform. 2023;25(1):bbad489. doi:10.1093/bib/bbad489 [Google Scholar] [PubMed] [CrossRef]
22. Cannataro R, Cione E. miRNA as drug: antagomir and beyond. Curr Pharm Des. 2023;29(6):462–5. doi:10.2174/1381612829666230220123150 [Google Scholar] [PubMed] [CrossRef]
23. Zhang S, Cheng Z, Wang Y, Han T. The risks of miRNA therapeutics: in a drug target perspective. Drug Des Devel Ther. 2021;15:721–33. doi:10.2147/DDDT.S288859 [Google Scholar] [PubMed] [CrossRef]
24. Du L, Dou K, Liang N, Sun J, Bai RU. MiR-194-5p suppresses the warburg effect in ovarian cancer cells through the IGF1R/PI3K/AKT axis. BIOCELL. 2023;47(4):547. doi:10.32604/biocell.2023.025048. [Google Scholar] [CrossRef]
25. Yang B, Wang YW, Zhang K. Interactions between circRNA and protein in breast cancer. Gene. 2023;895:148019 [Google Scholar] [PubMed]
26. Ma B, Wang S, Wu W, Shan P, Chen Y, Meng J, et al. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed Pharmacother. 2023;162:114672. doi:10.1016/j.biopha.2023.114672 [Google Scholar] [PubMed] [CrossRef]
27. Su R, Zhong S, Wang P, Lin Z. Induction of perineural invasion in salivary adenoid cystic carcinoma by circular RNA RNF111. Clin Transl Oncol. 2023;25(11):3152–64. doi:10.1007/s12094-023-03182-w [Google Scholar] [PubMed] [CrossRef]
28. Liebig C, Ayala G, Wilks JA, Berger DH, Albo D. Perineural invasion in cancer: a review of the literature. Cancer. 2009;115(15):3379–91. doi:10.1002/cncr.v115:15. [Google Scholar] [CrossRef]
29. Fan KH, Kang CJ, Lin CY, Ng SH, Wang HM, Hsieh CH, et al. Quantitative measurement of perineural invasion for prognosis analysis of oral cavity cancer treated by radical surgery with or without adjuvant therapy. Technol Cancer Res Treat. 2023;22:15330338231176366 [Google Scholar] [PubMed]
30. Fukuda Y, Tanaka Y, Eto K, Ukai N, Sonobe S, Takahashi H, et al. S100-stained perineural invasion is associated with worse prognosis in stage I/II colorectal cancer: its possible association with immunosuppression in the tumor. Pathol Int. 2022;72(2):117–27. doi:10.1111/pin.v72.2. [Google Scholar] [CrossRef]
31. Lu WL, Kuang H, Gu J, Hu X, Chen B, Fan Y. GAP-43 targeted indocyanine green-loaded near-infrared fluorescent probe for real-time mapping of perineural invasion lesions in pancreatic cancer in vivo. Nanomed. 2023;50(7):102671. [Google Scholar]
32. Ceyhun S, Hulya T, Erdem T, Meryem GG, Fugen VA. Determination of paraneoplastic neuropathy in newly diagnosed breast tumor patients. eNeurologicalSci. 2020;21(6):100265 [Google Scholar] [PubMed]
33. Ferdoushi A, Griffin N, Marsland M, Xu X, Faulkner S, Gao F, et al. Tumor innervation and clinical outcome in pancreatic cancer. Sci Rep. 2021;11(1):7390. doi:10.1038/s41598-021-86831-w [Google Scholar] [PubMed] [CrossRef]
34. Barrett AW, Speight PM. Perineural invasion in adenoid cystic carcinoma of the salivary glands: a valid prognostic indicator? Oral Oncol. 2009;45(11):936–40. doi:10.1016/j.oraloncology.2009.07.001 [Google Scholar] [PubMed] [CrossRef]
35. Bakst RL, Glastonbury CM, Parvathaneni U, Katabi N, Hu KS, Yom SS. Perineural invasion and perineural tumor spread in head and neck cancer. Int J Radiat Oncol Biol Phys. 2019;103(5):1109–24. doi:10.1016/j.ijrobp.2018.12.009 [Google Scholar] [PubMed] [CrossRef]
36. Zhao J, Chen J, Zhang M, Tang X, Sun G, Zhu S, et al. The clinical significance of perineural invasion in patients with de novo metastatic prostate cancer. Andrology. 2019;7(2):184–92. doi:10.1111/andr.2019.7.issue-2. [Google Scholar] [CrossRef]
37. Zhao B, Lv W, Mei D, Luo R, Bao S, Huang B, et al. Perineural invasion as a predictive factor for survival outcome in gastric cancer patients: a systematic review and meta-analysis. J Clin Pathol. 2020;73(9):544–51. doi:10.1136/jclinpath-2019-206372 [Google Scholar] [PubMed] [CrossRef]
38. Bai L, Yan L, Guo Y, He L, Sun Z, Cao W, et al. Perineural invasion is a significant indicator of high malignant degree and poor prognosis in esophageal cancer: a systematic review and meta-analysis. Front Oncol. 2022;12:816270. doi:10.3389/fonc.2022.816270 [Google Scholar] [PubMed] [CrossRef]
39. Arun I, Roy P, Jain PV, Hameed S, Manikantan K, Arun P. Subcategorization of perineural invasion and its impact on survival in patients with oral squamous cell carcinoma. Head Neck Pathol. 2023;17(2):383–92 [Google Scholar] [PubMed]
40. Binmadi N, Alsharif M, Almazrooa S, Aljohani S, Akeel S, Osailan S, et al. Perineural invasion is a significant prognostic factor in oral squamous cell carcinoma: a systematic review and meta-analysis. Diagnostics. 2023;13(21):3339. doi:10.3390/diagnostics13213339 [Google Scholar] [PubMed] [CrossRef]
41. Xu W, Liu J, Zhang J, Lu J, Guo J. Tumor microenvironment crosstalk between tumors and the nervous system in pancreatic cancer: molecular mechanisms and clinical perspectives. Biochim Biophys Acta Rev Cancer. 2024;1879(1):189032. doi:10.1016/j.bbcan.2023.189032 [Google Scholar] [PubMed] [CrossRef]
42. Festa BM, Costantino A, Pace GM, Petruzzi G, Campo F, Pellini R, et al. Impact of adjuvant radiotherapy in squamous cell carcinoma of the oral cavity with perineural invasion. Laryngoscope. 2023;17. doi:10.1002/lary.31148 [Google Scholar] [PubMed] [CrossRef]
43. Fahmy MD, Clegg D, Belcastro A, Smith BD, Eric Heidel R, Carlson ER, et al. Are throat pain and otalgia predictive of perineural invasion in squamous cell carcinoma of the oropharynx? J Oral Maxillofac Surg. 2022;80(2):363–71. doi:10.1016/j.joms.2021.08.265 [Google Scholar] [PubMed] [CrossRef]
44. Pyne JH, Myint E, Clark SP, Clifopoulos C, Fishburn P, Gorji M, et al. Squamous cell carcinoma: pain as a clue to increased tumour diameter, increased invasion depth, the grade of differentiation, acantholysis and perineural invasion. Clin Exp Dermatol. 2020;45(2):180–6. doi:10.1111/ced.v45.2. [Google Scholar] [CrossRef]
45. Scheff NN, Harris AL, Li J, Horan NL, Kubik MW, Kim SW, et al. Pretreatment pain predicts perineural invasion in patients with head and neck squamous cell carcinoma. Support Care Cancer. 2023;31(7):405. doi:10.1007/s00520-023-07872-7 [Google Scholar] [PubMed] [CrossRef]
46. Chen TC, Jan YY, Yeh TS. K-ras mutation is strongly associated with perineural invasion and represents an independent prognostic factor of intrahepatic cholangiocarcinoma after hepatectomy. Ann Surg Oncol. 2012;19(Suppl 3):S675–81 [Google Scholar] [PubMed]
47. Li Y, Eresen A, Shangguan J, Yang J, Benson3rd AB, Yaghmai V, et al. Preoperative prediction of perineural invasion and KRAS mutation in colon cancer using machine learning. J Cancer Res Clin Oncol. 2020;146(12):3165–74. doi:10.1007/s00432-020-03354-z [Google Scholar] [PubMed] [CrossRef]
48. Destro Rodrigues MF, Sedassari BT, Esteves CM, de Andrade NP, Altemani A, de Sousa SC, et al. Embryonic stem cells markers Oct4 and Nanog correlate with perineural invasion in human salivary gland mucoepidermoid carcinoma. J Oral Pathol Med. 2017;46(2):112–20. doi:10.1111/jop.2017.46.issue-2. [Google Scholar] [CrossRef]
49. Park SJ, Min HJ, Yoon C, Kim SH, Kim JH, Lee SY. Integrin beta1 regulates the perineural invasion and radioresistance of oral squamous carcinoma cells by modulating cancer cell stemness. Cell Signal. 2023;110:110808. doi:10.1016/j.cellsig.2023.110808 [Google Scholar] [PubMed] [CrossRef]
50. Frampton AE, Castellano L, Colombo T, Giovannetti E, Krell J, Jacob J, et al. Integrated molecular analysis to investigate the role of microRNAs in pancreatic tumour growth and progression. Lancet. 2015;385(Suppl 1):S37 [Google Scholar] [PubMed]
51. Kolenda T, Guglas K, Teresiak A, Blizniak R, Lamperska K. Low let-7d and high miR-205 expression levels positively influence HNSCC patient outcome. J Biomed Sci. 2019;26(1):17. doi:10.1186/s12929-019-0511-3 [Google Scholar] [PubMed] [CrossRef]
52. Dou Z, Gao L, Ren W, Zhang H, Wang X, Li S, et al. CiRS-7 functions as a ceRNA of RAF-1/PIK3CD to promote metastatic progression of oral squamous cell carcinoma via MAPK/AKT signaling pathways. Exp Cell Res. 2020;396(2):112290. doi:10.1016/j.yexcr.2020.112290 [Google Scholar] [PubMed] [CrossRef]
53. Zhang Y, Wang M, Zang X, Mao Z, Chen Y, Mao F, et al. CircHN1 affects cell proliferation and migration in gastric cancer. J Clin Lab Anal. 2020;34(10):e23433. doi:10.1002/jcla.v34.10. [Google Scholar] [CrossRef]
54. Stoen MJ, Andersen S, Rakaee M, Pedersen MI, Ingebriktsen LM, Bremnes RM, et al. High expression of miR-17-5p in tumor epithelium is a predictor for poor prognosis for prostate cancer patients. Sci Rep. 2021;11(1):13864. doi:10.1038/s41598-021-93208-6 [Google Scholar] [PubMed] [CrossRef]
55. Prueitt RL, Yi M, Hudson RS, Wallace TA, Howe TM, Yfantis HG, et al. Expression of microRNAs and protein-coding genes associated with perineural invasion in prostate cancer. Prostate. 2008;68(11):1152–64. doi:10.1002/pros.v68:11. [Google Scholar] [CrossRef]
56. Yan K, Hou L, Liu T, Jiao W, Ma Q, Fang Z, et al. lncRNA OGFRP1 functions as a ceRNA to promote the progression of prostate cancer by regulating SARM1 level via miR-124-3p. Aging. 2020;12(10):8880–92. doi:10.18632/aging.v12i10. [Google Scholar] [CrossRef]
57. Zhang M, Wu JS, Xian HC, Chen BJ, Wang HF, Yu XH, et al. CXCR5 induces perineural invasion of salivary adenoid cystic carcinoma by inhibiting microRNA-187. Aging. 2021;13(11):15384–99. doi:10.18632/aging.v13i11. [Google Scholar] [CrossRef]
58. Wang W, Tang X, Qu H, He Q. Translation regulatory long non-coding RNA 1 represents a potential prognostic biomarker for colorectal cancer. Oncol Lett. 2020;19(6):4077–87 [Google Scholar] [PubMed]
59. Kim E, Kim H, Yeo MK, Kim CH, Kim JY, Park S, et al. Identification of a novel long non-coding RNA, lnc-ATMIN-4: 2, and its clinicopathological and prognostic significance in advanced gastric cancer. Cancer Genom Proteom. 2022;19(6):761–72. doi:10.21873/cgp.20358 [Google Scholar] [PubMed] [CrossRef]
60. Shao Y, Qi C, Yan J, Lu R, Ye G, Guo J. Biological and clinical implications of hsa_circ_0086720 in gastric cancer and its clinical application. J Clin Lab Anal. 2022;36(5):e24369. doi:10.1002/jcla.v36.5. [Google Scholar] [CrossRef]
61. Bai J, Zhang X, Shi D, Xiang Z, Wang S, Yang C, et al. Exosomal miR-128-3p promotes epithelial-to-mesenchymal transition in colorectal cancer cells by targeting FOXO4 via TGF-β/SMAD and JAK/STAT3 signaling. Front Cell Dev Biol. 2021;9:568738. doi:10.3389/fcell.2021.568738 [Google Scholar] [PubMed] [CrossRef]
62. Andersen S, Richardsen E, Moi L, Donnem T, Nordby Y, Ness N, et al. Fibroblast miR-210 overexpression is independently associated with clinical failure in prostate cancer—a multicenter (in situ hybridization) study. Sci Rep. 2016;6(1):36573. doi:10.1038/srep36573 [Google Scholar] [PubMed] [CrossRef]
63. Bozgeyik E, Ceylan O. Distinct expression signatures of miR-130a, miR-301a, miR-454 in formalin fixed paraffin embedded tissue samples of prostate cancer patients. Pathol Res Pract. 2022;234:153897. doi:10.1016/j.prp.2022.153897 [Google Scholar] [PubMed] [CrossRef]
64. Zanon MF, Scapulatempo-Neto C, Gama RR, Marques MMC, Reis RM, Evangelista AF. Identification of microRNA expression profiles related to the aggressiveness of salivary gland adenoid cystic carcinomas. Genes. 2023;14(6):1220. doi:10.3390/genes14061220 [Google Scholar] [PubMed] [CrossRef]
65. Feng X, Matsuo K, Zhang T, Hu Y, Mays AC, Browne JD, et al. MicroRNA profiling and target genes related to metastasis of salivary adenoid cystic carcinoma. Anticancer Res. 2017;37(7):3473–81 [Google Scholar] [PubMed]
66. Koparal M, Bozgeyik E, Ceylan O, Ege B, Kurt MY, Yumrutas O, et al. Salivary gland tumors exhibit distinct miRNA signatures involved in Wnt/β-Catenin signaling in formalin fixed paraffin embedded tissue samples. Pathol Res Pract. 2022;238:154119. doi:10.1016/j.prp.2022.154119 [Google Scholar] [PubMed] [CrossRef]
67. Čelešnik H, Büdefeld T, Čizmarević B, Švagan M, Potočnik U. MIR137/MIR2682 locus is associated with perineural invasiveness in head and neck cancer. J Oral Pathol Med. 2021;50(9):874–81. doi:10.1111/jop.v50.9. [Google Scholar] [CrossRef]
68. Zhang Q, Chen Y, Hu SQ, Pu YM, Zhang K, Wang YX. A HPV16-related prognostic indicator for head and neck squamous cell carcinoma. Ann Transl Med. 2020;8(22):1492. doi:10.21037/atm. [Google Scholar] [CrossRef]
69. Rajan C, Roshan VGD, Khan I, Manasa VG, Himal I, Kattoor J, et al. MiRNA expression profiling and emergence of new prognostic signature for oral squamous cell carcinoma. Sci Rep. 2021;11(1):7298. doi:10.1038/s41598-021-86316-w [Google Scholar] [PubMed] [CrossRef]
70. Chang KW, Chu TH, Gong NR, Chiang WF, Yang CC, Liu CJ, et al. miR-370 modulates insulin receptor substrate-1 expression and inhibits the tumor phenotypes of oral carcinoma. Oral Dis. 2013;19(6):611–9. doi:10.1111/odi.2013.19.issue-6. [Google Scholar] [CrossRef]
71. Brito BL, Lourenco SV, Damascena AS, Kowalski LP, Soares FA, Coutinho-Camillo CM. Expression of stem cell-regulating miRNAs in oral cavity and oropharynx squamous cell carcinoma. J Oral Pathol Med. 2016;45(9):647–54. doi:10.1111/jop.2016.45.issue-9. [Google Scholar] [CrossRef]
72. Kovarikova J, Baranova I, Laco J, Rozkosova K, Vosmikova H, Vosmik M, et al. Deregulation of selected micrornas in sinonasal squamous cell carcinoma: searching for potential prognostic biomarkers. Folia Biol. 2019;65(3):142–51. doi:10.14712/fb2019065030142. [Google Scholar] [CrossRef]
73. Fukada M, Matsuhashi N, Takahashi T, Sugito N, Heishima K, Akao Y, et al. Tumor tissue MIR92a and plasma MIRs21 and 29a as predictive biomarkers associated with clinicopathological features and surgical resection in a prospective study on colorectal cancer patients. J Clin Med. 2020;9(8):2509. doi:10.3390/jcm9082509 [Google Scholar] [PubMed] [CrossRef]
74. Niculae AM, Dobre M, Herlea V, Manuc TE, Trandafir B, Milanesi E, et al. Let-7 microRNAs are possibly associated with perineural invasion in colorectal cancer by targeting IGF axis. Life. 2022;12(10):1638. doi:10.3390/life12101638 [Google Scholar] [PubMed] [CrossRef]
75. Sun R, Liu Z, Tong D, Yang Y, Guo B, Wang X, et al. miR-491-5p, mediated by Foxi1, functions as a tumor suppressor by targeting Wnt3a/β-catenin signaling in the development of gastric cancer. Cell Death Dis. 2017;8(3):e2714. doi:10.1038/cddis.2017.134 [Google Scholar] [PubMed] [CrossRef]
76. Cañueto J, Cardeñoso-Álvarez E, García-Hernández JL, Galindo-Villardón P, Vicente-Galindo P, Vicente-Villardón JL, et al. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br J Dermatol. 2017;177(1):168–78. doi:10.1111/bjd.2017.177.issue-1. [Google Scholar] [CrossRef]
77. Li J, Kang R, Tang D. Cellular and molecular mechanisms of perineural invasion of pancreatic ductal adenocarcinoma. Cancer Commun. 2021;41(8):642–60. doi:10.1002/cac2.v41.8. [Google Scholar] [CrossRef]
78. Li J, Tong H, Li D, Jiang Q, Zhang Y, Tang W, et al. The long non-coding RNA DKFZp434J0226 regulates the alternative splicing process through phosphorylation of SF3B6 in PDAC. Mol Med. 2021;27(1):95. doi:10.1186/s10020-021-00347-7 [Google Scholar] [PubMed] [CrossRef]
79. Ye H, Zhou Q, Zheng S, Li G, Lin Q, Ye L, et al. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(2):34. doi:10.1038/s41419-017-0052-1 [Google Scholar] [PubMed] [CrossRef]
80. Ye Y, Chen J, Zhou Y, Fu Z, Zhou Q, Wang Y, et al. High expression of AFAP1-AS1 is associated with poor survival and short-term recurrence in pancreatic ductal adenocarcinoma. J Transl Med. 2015;13(1):137. doi:10.1186/s12967-015-0490-4 [Google Scholar] [PubMed] [CrossRef]
81. Lee YJ, Oh H, Kim E, Ahn B, Lee JH, Lee Y, et al. Long noncoding RNA HOTTIP overexpression: a potential prognostic biomarker in prostate cancer. Pathol Res Pract. 2019;215(11):152649. doi:10.1016/j.prp.2019.152649 [Google Scholar] [PubMed] [CrossRef]
82. Li D, She J, Hu X, Zhang M, Sun R, Qin S. The ELF3-regulated lncRNA UBE2CP3 is over-stabilized by RNA-RNA interactions and drives gastric cancer metastasis via miR-138-5p/ITGA2 axis. Oncogene. 2021;40(35):5403–15. doi:10.1038/s41388-021-01948-6 [Google Scholar] [PubMed] [CrossRef]
83. Tan L, Yang Y, Shao Y, Zhang H, Guo J. Plasma lncRNA-GACAT2 is a valuable marker for the screening of gastric cancer. Oncol Lett. 2016;12(6):4845–9. doi:10.3892/ol.2016.5297 [Google Scholar] [PubMed] [CrossRef]
84. Gao Z, Zhou H, Wang Y, Chen J, Ou Y. Regulatory effects of lncRNA ATB targeting miR-200c on proliferation and apoptosis of colorectal cancer cells. J Cell Biochem. 2020;121(1):332–43. doi:10.1002/jcb.v121.1. [Google Scholar] [CrossRef]
85. Li M, Wang Q, Xue F, Wu Y. lncRNA-CYTOR works as an oncogene through the CYTOR/miR-3679-5p/MACC1 axis in colorectal cancer. DNA Cell Biol. 2019;38(6):572–82. doi:10.1089/dna.2018.4548 [Google Scholar] [PubMed] [CrossRef]
86. Shao Y, Ye M, Jiang X, Sun W, Ding X, Liu Z, et al. Gastric juice long noncoding RNA used as a tumor marker for screening gastric cancer. Cancer. 2014;120(21):3320–8. doi:10.1002/cncr.v120.21. [Google Scholar] [CrossRef]
87. Li Q, Liu S, Yan J, Sun MZ, Greenaway FT. The potential role of miR-124-3p in tumorigenesis and other related diseases. Mol Biol Rep. 2021;48(4):3579–91. doi:10.1007/s11033-021-06347-4 [Google Scholar] [PubMed] [CrossRef]
88. Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11(1):98. doi:10.1186/s13045-018-0643-z [Google Scholar] [PubMed] [CrossRef]
89. Kadkhoda S, Taslimi R, Noorbakhsh F, Darbeheshti F, Bazzaz JT, Ghafouri-Fard S, et al. Importance of Circ0009910 in colorectal cancer pathogenesis as a possible regulator of miR-145 and PEAK1. World J Surg Oncol. 2021;19(1):265. doi:10.1186/s12957-021-02378-0 [Google Scholar] [PubMed] [CrossRef]
90. Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–5 [Google Scholar] [PubMed]
91. Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, et al. Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res. 2020;26(3):1475–82. doi:10.1007/s12253-019-00716-y [Google Scholar] [PubMed] [CrossRef]
92. Wu H, Yu DH, Wu MH, Huang T. Long non-coding RNA LOC541471: a novel prognostic biomarker for head and neck squamous cell carcinoma. Oncol Lett. 2019;17(2):2457–64 [Google Scholar] [PubMed]
93. D’Silva NJ, Perez-Pacheco C, Schmitd LB. The 3D’s of neural phenotypes in oral cancer: distance, diameter, and density. Adv Biol. 2023;7(2):e2200188. doi:10.1002/adbi.v7.2. [Google Scholar] [CrossRef]
94. Zheng Z, Li X, Chen G, Chen J, Zhu X, Teng Y. Transcriptome analyses reveal new insights on key determinants of perineural invasion in high-grade serous ovarian cancer. Front Cell Dev Biol. 2023;11:1109710. doi:10.3389/fcell.2023.1109710 [Google Scholar] [PubMed] [CrossRef]
95. Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: a promising therapeutic target in cancer. Int J Mol Sci. 2022;23(19):11502. doi:10.3390/ijms231911502 [Google Scholar] [PubMed] [CrossRef]
96. Cai T, Zhang Q, Wu B, Wang J, Li N, Zhang T, et al. LncRNA-encoded microproteins: a new form of cargo in cell culture-derived and circulating extracellular vesicles. J Extracell Vesicles. 2021;10(9):e12123. doi:10.1002/jev2.v10.9. [Google Scholar] [CrossRef]
97. Chen J, Brunner AD, Cogan JZ, Nunez JK, Fields AP, Adamson B, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020;367(6482):1140–6. doi:10.1126/science.aay0262 [Google Scholar] [PubMed] [CrossRef]
98. Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang H, et al. circ-EIF6 encodes EIF6-224aa to promote TNBC progression via stabilizing MYH9 and activating the Wnt/β-catenin pathway. Mol Ther. 2022;30(1):415–30. doi:10.1016/j.ymthe.2021.08.026 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF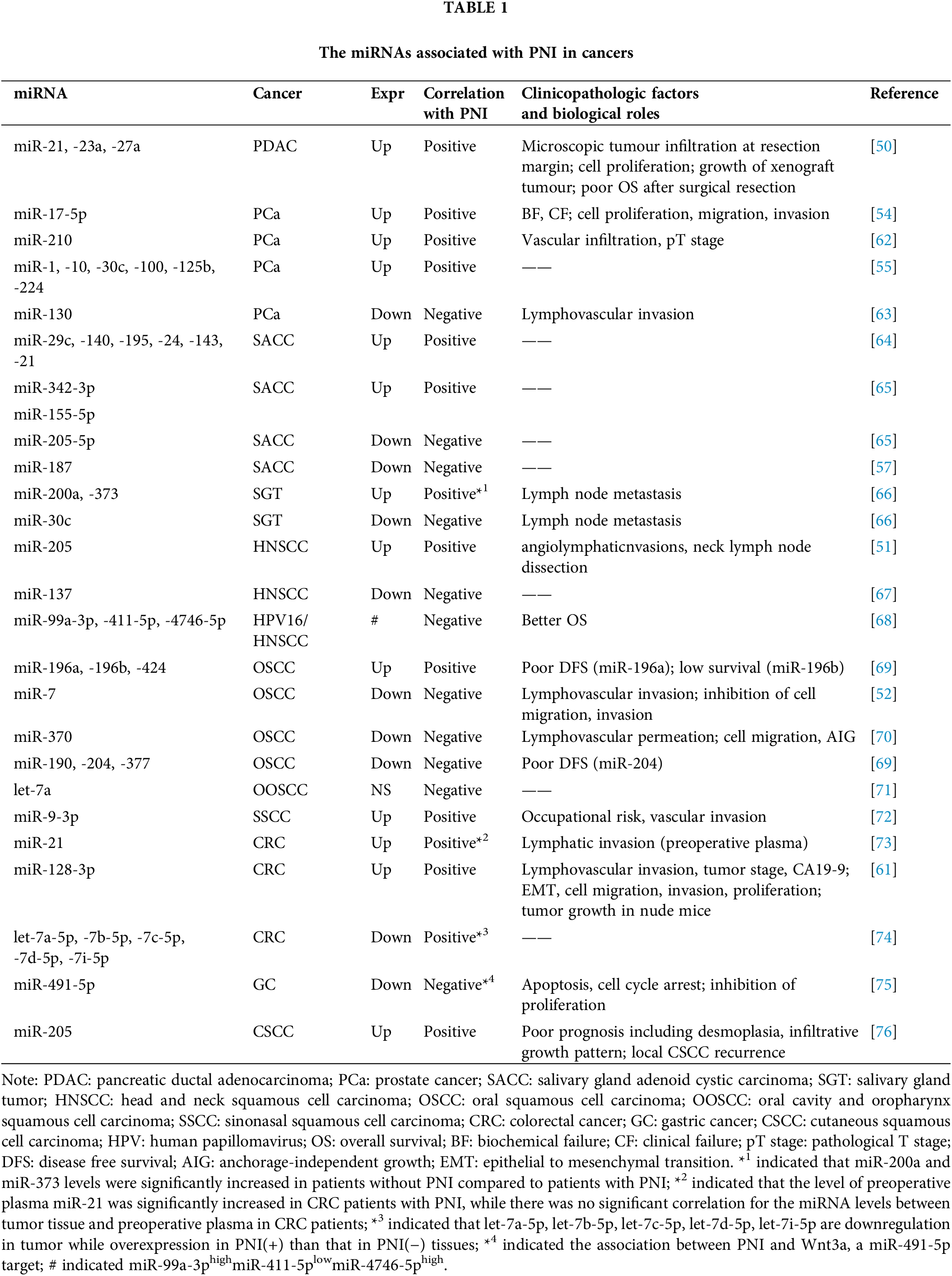
 Downloads
Downloads
 Citation Tools
Citation Tools