 Open Access
Open Access
ARTICLE
LA-D-B1, a novel Abemaciclib derivative, exerts anti-breast cancer effects through CDK4/6
1 Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening, College of Pharmacy, Jiangsu Ocean University, Lianyungang, 222000, China
2 School of Medicine, Xiamen University, Xiamen, 361102, China
* Corresponding Authors: XIUJUN WANG. Email: ; JING JI. Email:
# Ling Ma and Zirui Jiang are co-first authors and contributed equally to this paper
(This article belongs to the Special Issue: Navigating the Interplay of Cancer, Autophagy, ER Stress, Cell Cycle and Apoptosis: Mechanisms, Therapies, and Future Directions)
BIOCELL 2024, 48(5), 847-860. https://doi.org/10.32604/biocell.2024.050868
Received 20 February 2024; Accepted 11 March 2024; Issue published 06 May 2024
Abstract
Background: Regulatory proteins involved in human cellular division and proliferation, cyclin-dependent kinases 4 and 6 (CDK4/6) are overexpressed in numerous cancers, including triple-negative breast cancer (TNBC). TNBC is a common pathological subtype of breast cancer that is prone to recurrence and metastasis, and has a single treatment method. As one of the CDK4/6 inhibitors, abemaciclib can effectively inhibit the growth of breast tumors. In this study, we synthesized LA-D-B1, a derivative of Abemaciclib, and investigated its anti-tumor effects in breast cancer. Methods: Cellular viability was assessed using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Cell cloning and migration abilities were determined by colony formation assay and wound healing assay. Cell invasion abilities and adhesion were determined by cell invasion assay and cell adhesion assay. The impact of compound LA-D-B1 on cell proliferation and the cell cycle was analyzed through Western blotting, which quantified the levels of proteins associated with the cyclin-dependent kinase (CDK) 4/6-cyclin D-Rb-E2F pathway. The in vivo anti-tumor activity of compound LA-D-B1 was investigated using a chick chorioallantoic membrane (CAM) model. Results: The study demonstrated that LA-D-B1 effectively suppressed breast cancer cell proliferation, induced apoptosis, and caused cell cycle arrest. Furthermore, LA-D-B1 reduced the expression of key proteins in the CDK4/6-cyclin D-Rb-E2F pathway, including CDK4, CDK6 and E2F1. The results also indicated significant antitumor activity of LA-D-B1 in a transplanted tumor model. Conclusion: In this study, LA-D-B1 demonstrated a potent anti-tumor effect by effectively suppressing cell proliferation and inhibiting cell cycle progression in breast cancer. These findings highlight the potential of LA-D-B1 as a valuable compound for enhancing therapeutic outcomes and controlling the progression of breast cancer.Keywords
Breast cancer (BC) is a leading cause of cancer-related deaths among women worldwide [1]. Recently reported, breast cancer has replaced lung cancer as the most common cancer worldwide [2]. It is caused by the uncontrolled proliferation of breast epithelial cells due to various carcinogenic factors. Globally, breast cancer represents approximately 25% of new cancer cases in women, with one in eight women being diagnosed with breast cancer during their lifetime [3]. There is increasing evidence that the prognosis of breast cancer is poor and that the majority of breast cancer patients die from breast cancer progression [4]. Among the different molecular subtypes of breast cancer, triple-negative breast cancer (TNBC) is particularly prevalent [5]. TNBC is known to have a higher incidence in younger females and is associated with a significantly elevated mortality rate of up to 40%, as well as recurrence rates of up to 25% within 5 years of diagnosis [6,7]. Remedy choices for TNBC are restrained, and the mainstay of treatment remains chemotherapy [8]. Dysregulation of the cell cycle machinery is not an unusual phenomenon discovered in lots of cancers, leading to out-of-control cell proliferation, a typical hallmark of cancer [9]. This dysregulation is often attributed to abnormal activation of cyclin-dependent kinase (CDK) [10]. CDK is a kinase system composed of about 20 serine/threonine kinases that participate in cell cycle regulation by initiating G1 (gap1), S (synthesis phase), G2 (gap2), and M (mitosis) phases of the cellular cycle [11].
Under permissive conditions of mitosis, hormones, and growth factors, cyclin D binds to cyclin-dependent kinase CDK4/6, leading to the phosphorylation of retinoblastoma protein (Rb) and the formation of a phospho-Rb complex. This complex influences the dissociation of the transcription factor E2F. Once free, E2F binds to DNA, driving DNA replication, regulating gene expression and cell division, and facilitating the transition from the G1 to the S phase [12]. Cell-cycle dysregulation in cancer is often caused by overexpression of CDKs and cyclins, gain-of-function mutations, and reduced expression and efficacy of endogenous inhibitors [13]. The dysregulation of the CDK4/6-cyclin D-Rb-E2F pathway, caused by the overexpression of CDK4/6, is implicated as a key factor in the pathogenesis of breast cancer [14]. Consequently, the use of CDK4/6 inhibitors has become a potent anti-tumor strategy for breast cancer treatment [15]. CDK4/6 inhibitors effectively hinder tumor cell proliferation [16]. It inhibits cellular cycle development by inhibiting the phosphorylation of RB1, thereby inducing cell cycle arrest [17]. The Food and Drug Administration (FDA) has granted approval for the use of four CDK4/6 inhibitors, which include Palbociclib, Abemaciclib, Ribociclib, and Trilaciclib, in the treatment of breast cancer [18]. The search for more selective CDK4/6 inhibitors remains a challenge for the development of novel anti-cancer cures.
In our current study, we synthesized a novel CDK4/6 inhibitor, LA-D-B1, and further investigated it in breast cancer. Using molecular docking, we found that LA-D-B1 has a sturdy binding potential to CDK4/6, and we speculate that LA-D-B1 has a similar inhibitory effect on CDKs as Abemaciclib. This study explored the viable anti-tumor impact of compound LA-D-B1 on TNBC and its mechanism, providing a promising drug target for the remedy of TNBC [19].
We utilized a WRS-1B virtual melting point meter (temperature uncorrected) from Shanghai Precision Clinical Gadgets Co. Ltd. (Shanghai, China) to measure the melting points of the compounds. NMR spectra were obtained using an Avance III 500 MHz liquid NMR spectrometry (tetramethylsilane as inner trendy, Bruker, Germany). Silica gel column chromatography (200–300 mesh, 50–71 μm), silica gel thin layer chromatography plate GF254 used products from Qingdao Marine Chemical Company (Qingdao, Shandong, China). DMSO-d6 was used as the solvent, and the method used to be determined through Thermo Fisher Scientific (Shanghai, China) Q Exactive HF LC/MS. The other reagents used were of analytical grade, which may be used once after purchase.
For all molecular docking tests, we used the Viglen Genie Intel(R) Core(TM) i5-7300HQ CPU running Windows 10 1909 at 2.50 GHz (Lenovo (Shanghai) Electronic Technology Co., Ltd., China). Molecular modeling was performed using Pymol 1.7.0.0.win32-py2.7 (Delano Scientific LLC, San Francisco, CA, USA) and AutoDockTools 1.5.6 (Scripps Research Institute, La Jolla, CA, USA). The PDB database (http://www.rcsb.org/; PDB code 5L2S) provides the X-ray eutectic structure of the cyclin-dependent kinase CDK4/CDK6. Prior to docking, Pymol was used for protein pre-processing, including the removal of metallic ions and water molecules (HOH). Chem3D was used to construct the ligand structure, and a genetic algorithm was used for the molecular docking study. The molecular docking results were sorted and analyzed.
Purchased from the School of Basic Medical Sciences, Cell Resource Center, Peking Union Medical College, and Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China), breast cancer cells, Hela cells, and A498 cells were cultured in minimum essential medium (MEM) or dulbecco’s modified eagle medium (DMEM). Breast cancer cells and Hela cells were cultured in DMEM (E600003-0500, Sanon BioEngineering (Shanghai) Co., Ltd., China) polysaccharide medium supplemented with 1% penicillin/streptomycin (E607011-0100, Sanon BioEngineering (Shanghai) Co., Ltd., China) and 10% fetal bovine serum (FBS-CE500, Nanjing Wobo Biotechnology Co., Ltd., China), or A498 cells were grown in MEM (KGM41500-500, Jiangsu Keji Biotechnology Co., Ltd., China) incomplete medium supplemented with 1% penicillin/streptomycin A498 and 10% fetal bovine serum (FBS). Cells were cultured at 37°C in an incubator (Thermo Fisher Scientific, BB150) with 5% carbon dioxide. When the cell fusion rate was 70% to 80%, 0.25% trypsin (VC2005, Vicmed, Xuzhou, China) was added for digestion, passage, and culture. Cells with a better growth status were decided on for further exploration.
In subculture plates with 96 wells, 1 × 104 cells were cultured in each well. After cellular attachment, the culture medium in the 96-subculture plates (NEST, Nantong, China) was aspirated, and then cell culture medium containing diverse doses of the compound LA-D-B1 and the positive drug was delivered to each culture plate. Cells were treated for 24 h. After adding 10 µL of methyl thiazolyl tetrazolium (MTT) solution, it was incubated for an additional 4 h (this operation was done in a dark room because the MTT solution was light-sensitive). Following the removal of the supernatant (with caution to avoid touching the cell layer), dimethyl sulfoxide (DMSO) was added in accordance with the drug concentration, totaling 100 μL, to facilitate color development. The microplate detector (BioTek, Synergy Neo2, USA) was used to measure the optical density (OD) of each group at 490 nm. The concentration inducing 50% cell growth inhibition used to be decided graphically by nonlinear regression with the use of the curve fitting algorithm of GraphPad Prism 9 (GraphPad Software, La Jolla, CA, USA).
Cell viability assays for siRNA cell lines
Lentiviral human siRNA was purchased from Sigma. The target sequence of the plasmid providing the siRNA was as follows: CDK4-siRNA: 5′-CCCCGCCCAGTGCAGTCGGTGGTACCTGAGATGGAGGAGTCGGGAGCACAGCTGCTGCTGGAAATGCTGACTTTT-3′; CDK6-siRNA: 5′-CCGCAGAACATTCTGGTGACCAGCAGTGGACAGATAAAGCTGGCTGACTTTGGCCTTGCCCGCATCTATAGTTTT-3′. In accord with the certain transfection the method, 3 μg of siRNA was used to transfect cells. Different concentrations of drug LA-D-B1 were set according to IC50, then the corresponding cells were added and transferred to 96-well plates for 24 h. The medium was eliminated, and subsequent treatment of the cells involved exposure to MTT (6 mg/mL) for a duration of 3 h. Lysis of the cells was achieved by the addition of 100 μL of DMSO. The absorbance of each group was obtained at 490 nm using a microplate detector (BioTek, Synergy Neo2, USA), and using GraphPad Prism 9, data were analyzed and cell viability was counted.
Morphological observation was used to assess the impact of compound LA-D-B1 on the morphology of 293T and MDA-MB-231 cells. The cells were seeded in 96-well plates at a density of 1 × 104 cells/well for 293T cells and in 12-well plates at a density of 1 × 105 cells/well for MDA-MB-231 cells. The plates were then incubated overnight at 37°C with 5% CO2. Subsequently, the various concentrations of LA-D-B1 (0.05, 0.1, 0.2, and 0.4 µM) and Abemacicilb (0.4 µM) were added to the cells. Images documenting cellular morphology were captured at both the 0 and 24-h time points. Images were taken using a biomicroscope (Nanjing Jiangnan Yongxin Optics Co., Ltd., China).
A total of 1000 cells in the exponential growth phase were added to each well after MDA-MB-231 cells in good growth state were seeded into 6-well culture plates, while the 12-well culture plates were populated with MCF-7 cells. In an incubator with 5% CO2 and 37°C, the cell culture plates were evenly positioned and cultivated. After the cells were attached, medium containing compound LA-D-B1 at each concentration or effective control drug was added to each well. The cell culture medium was changed to nomal medium after the 12 h culture period. The medium was refreshed at 3-day intervals throughout the following 2-week incubation. After clone formation, the cells were decontaminated by rinsing with sterile PBS. Following this, the cells were fixed by treating them with 4% paraformaldehyde. The culture plates underwent a dropwise application of crystal violet, followed by air drying, and then visualized for the formation of colonies. In the end, the culture plates were photographed upside down and then counted using ImageJ software.
MDA-MB-231 and MCF-7 cells were seeded in 12-well subculture plates and cultured at 5% CO2 and 37°C for 24 h, respectively. After removing the supernatant, medium containing various concentrations of compound LA-B-D1 and the effective drug Abemaciclib was added. The mixture was then incubated for an additional 24 h. The cells were then collected using a high-speed refrigerated centrifuge (Centrifuge 5424 R, Eppendorf, Germany) and centrifuged at 1200 rpm for 3 min. Cells were seeded at a density of 10,000 cells per 96-well plate precoated with matrix gel (Corning BioCoatings, 354248). After an hour, the supernatant was discarded, and 10 µL of MTT solution was added and incubated for an additional 4 h. Following aspiration and removal of the supernatant, the addition of 100 μL of DMSO to develop color. The adhesion rate was assessed by examining the optical density (OD) values of each group at 490 nm using a microwell detector with multiple functions (BioTek, Synergy Neo2, USA).
Thawed matrix gels (Corning BioCoatings, 354248) were diluted to a concentration of 220 μg/mL using serum-free DMEM in accordance with the instructions provided by the manufacturer. A 100 µL dilution was applied to the bottom surface of the Transwell upper chamber and air-dried for 4 h in a 5% CO2 incubator at 37°C. Various concentrations of the compound LA-D-B1 and Abemaciclib were added to MCF-7 and MDA-MB-231 cells. After 24 h, the lower chamber was filled with 650 µL of media containing 10% FBS as a chemotactic invasion inducer. Simultaneously, a 100 µL single-cell suspension was prepared by mixing cells with serum-free medium supplemented with 0.2% BSA. This suspension was then added to the top chamber of the Transwell. After incubating for 24 to 48 h, the cells that had invaded through the matrix gel and reached the lower chamber of the Transwell were subjected to fixation using paraformaldehyde for a duration of 15 min. The upper outer side of the Transwell was gently swabbed to remove any remaining cells. The plates were washed with sterile PBS and then left to color for 30 min with a drop addition of crystal violet (VS1003, Vicmed, Xuzhou, China). They were then washed with deionized water and dried. The filming area was selected appropriately, and the plates were photographed using a biological inverted microscope (Jiangnan XD-202). The invasive cell density was examined with the ImageJ software program.
Wound healing scratch analysis
To achieve satisfactory growth conditions, 1.5 × 105 cells/well of breast cancer cells were incubated overnight on 12-well cell culture plates. To make a scratch, the center of each well’s cell layer was scribed using a sterile pipette tip. Subsequently, cell debris was removed with the use of sterilization-treated PBS. Various concentrations of LA-D-B1, or the effective drug Abemaciclib, were brought to each well of cells in whole medium for co-incubation, and photographs were taken after 0, 24, and 48 h. ImageJ software was used to calculate the scratch healing rate at 24 and 48 h after the scratch.
Breast cancer cells were cultured in 12-well plates, followed by the addition of varying concentrations of the compound LA-D-B1, or the effective drug to the cell wells, after cell adhesion. After 24 h of incubation in a cell incubator, the cells were harvested. The cells were fully lysed by blowing with an appropriate amount of sterile PBS, centrifuged (12000 rpm, 1 min) in a high-speed refrigerated centrifuge (Centrifuge 5424 R, Eppendorf, Germany), the supernatant used to be aspirated, and the protein molecular density used to be quantified using the BCA kit (P0011, Shanghai Biyuntian Biotechnology Co., Ltd., China). A suitable quantity of quantitative protein was used for sample preparation, and 10% or 12% preformed glue was used for spot sampling. Electrophoresis was performed in 1 × SDS-PAGE buffer (VIC364, Vicmed, Xuzhou, China), and proteins were then transferred to methanol-treated PVDF membranes via the “wet turn” approach. After blocking the membranes with 5% skim milk for 45 min, an appropriate amount of Tween 20 (A600560-0500, Sangon Biotech, Shanghai, China) was added to PBS solution (1 × PBST), the membrane was washed with 1 × PBST for 3 min, and then the following primary antibody diluent (1:1000 volume dilution) was added: CDK4 (A0366, ABclonal), CDK6 (A0106, ABclonal), Cyclin E1 (11554-1-AP; Proteintech), E2F1 (AF6756, Beyotime), Rb (AF1564, Beyotime), p-Rb (AF1135, Beyotime), Cyclin D3 (26755-1-AP, Proteintech), Bcl-2 (A0208, ABclonal), BAX (50599-2-Ig, Proteintech), Cleaved-Caspase-3 (AF7022, Affinity), Caspase-3 (AC030, Beyotime), β-actin (AF0003, Beyotime), and the incubation box was placed on a shaker at 4°C overnight. After 12 h, the antibody diluent was recovered, and the membranes were repeatedly rinsed 4 times with 1 × PBST for 5 min each. Followed by the addition of the accordingly secondary antibody diluent (1:1000 volume dilution), and the incubation was kept at room temperature for an additional 50 min. 1 × PBST was used to properly rinse the membranes once more. The developing droplets were added to the membrane and visualized using HRP-ECL luminescence (180-501, Shanghai Tianneng Life Science Co., Ltd., China). The molecular weight and density values of the target protein bands were analyzed using ImageJ software.
Chicken embryo chorioallantoic membrane model
Breeding eggs were bought from Hanshan Ecological Farm, Shuyang, Jiangsu, China. Eggs exhibiting high hatching potential were chosen. These selected eggs were then placed in a controlled and sterile environment with a temperature maintained at 37°C to facilitate the hatching process. After 7 days, the hatching status of the eggs was observed, and then a window was opened on the egg surface to transfer the air chamber to a position approximately 1 cm from the vessel. To maintain the sterility of the environment, the windows were sealed using sterile plastic film. Cells were resuspended with the use of a blended suspension of Matrigel (40183ES08, Yeasen Biotechnology (Shanghai) Co., Ltd., China) and DMEM (1:1) and altered to 1.1–1.6 × 106 cells/mL. The cell mixture was injected into the vessel using the same-sized spear tip. Four days after injection, different concentrations of LA-D-B1 and a positive drug were added to the chick embryo allantoic membrane. They were then incubated for 3 days, and then the tumors in each group were removed, photographed, and weighed for analysis. The animal study was reviewed and approved by the Experiment Ethics Review Committee of Jiangsu Ocean University on November 04, 2022 (No. Jou2022110402).
Data analysis was performed using GraphPad Prism 9 software. The statistical comparison between the two groups was conducted using a t-test. The three independent tests’ mean ± standard deviation is shown as the result. Statistical significance was determined by a p-value of less than 0.05.
Synthetic route of compound LA-D-B1
6-((2-chloro-4-((4-(4-fluoro-1-isopropyl-2-methyl-1H-benzo[d]imidazol-6-yl)pyrimidin-2-yl)amino)phenoxy)methyl)picolinonitrile.
The experiment commenced by adding 6-((4-amino-2-chlorophenoxy)methyl)picolinonitrile (0.570 mmol, 116 mg) and 6-(2-chloropyrimidin-4-yl)-4-fluoro-1-isopropyl-2-methyl-1H-benzo[d]imidazole (0.475 mmol, 150 mg) to a 250-ml three-necked flask. Subsequently, Pd(OAc)2 (0.123 mmol, 28 mg), Cs2CO3 (1.476 mmol, 481 mg), XantPhos (0.246 mmol, 153 mg), and 1,4-dioxane (25 ml) were introduced into the flask. The mixture was purged with nitrogen three times to create an inert atmosphere. The reactants were then refluxed at 100°C and mixed for 5 h. The progress of the response used to be monitored via thin layer chromatography (TLC), which indicated the complete consumption of the starting material (eluent: DCM:MeOH in a ratio of 20:1). The reaction mixture was then subjected to rotary evaporation followed by column chromatography (eluent: DCM:MeOH in a ratio of 80:1). This resulted in the isolation of a light yellow solid weighing 110 mg, rate of production was 43.86% (Fig. 1). Hydrogen spectrum of compound LA-D-B1 (Suppl. Fig. S1): 1H NMR (500 MHz, DMSO) δ 9.72 (s, 1H), 8.56 (d, J = 5.3 Hz, 1H), 8.30 (d, J = 1.2 Hz, 1H), 8.17 (t, J = 7.9 Hz, 1H), 8.13 (d, J = 2.6 Hz, 1H), 8.04 (d, J = 7.6 Hz, 1H), 7.92 (d, J = 7.8 Hz, 1H), 7.83 (d, J = 12.2 Hz, 1H), 7.68 (dd, J = 9.0, 2.6 Hz, 1H), 7.59 (d, J = 5.3 Hz, 1H), 7.21 (d, J = 9.1 Hz, 1H), 5.33 (s, 2H), 4.91 – 4.81 (m, 1H), 2.65 (s, 3H), 1.64 (d, J = 6.9 Hz, 6H); Carbon spectrum of compound LA-D-B1 (Suppl. Fig. S2): 13C NMR (126 MHz, DMSO) δ 160.28, 159.32, 154.56, 148.18, 139.56, 135.80, 132.46, 128.69, 125.88, 121.74, 120.87, 119.18, 117.79, 115.24, 108.73, 71.02, 48.56, 40.50, 40.43, 40.34, 40.26, 40.17, 40.00, 39.83, 39.67, 39.50, 21.52, 15.21.
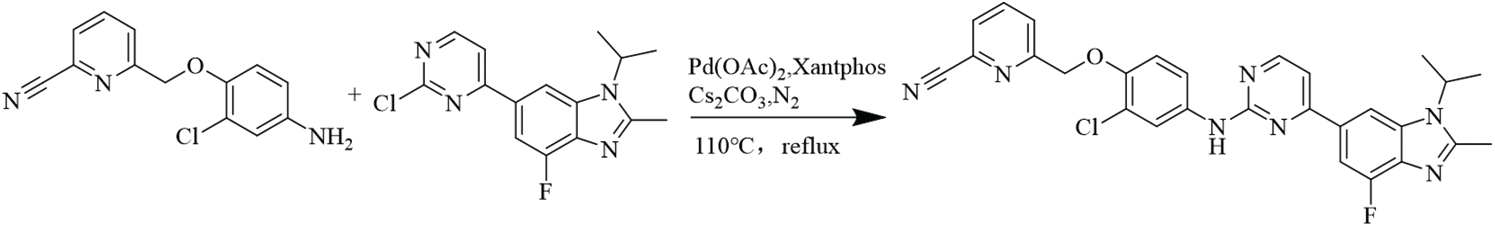
Figure 1: Equation for the synthesis of compound LA-D-B1.
Molecular docking of compound LA-D-B1
Molecular docking studies were conducted using AutoDockTools 1.5.6 to investigate the interaction between CDK4 and CDK6 with compound LA-D-B1. The docking site results of compound LA-D-B1 and CDK4 protein kinase showed (Fig. 2A) that the compound LA-D-B1 was tightly bound to CDK4 to a certain extent by forming three hydrogen bond forces. These three hydrogen bond forces are -NH and -N- with VAL-96, -N- with LYS-35. The binding site of compound LA-D-B1 to the CDK6 protein kinase (Fig. 2B) revealed that the pyridine cease of compound LA-D-B1 prolonged into a groove at the lateral side of the pocket. Meanwhile, compound LA-D-B1 was tightly bound to CDK6 by three hydrogen bonding forces, including -NH with ASP-104, GLU-21 with -N- and -O-. The results confirmed that compound LA-D-B1 could strongly bind to CDK4 and CDK6 to a certain extent.
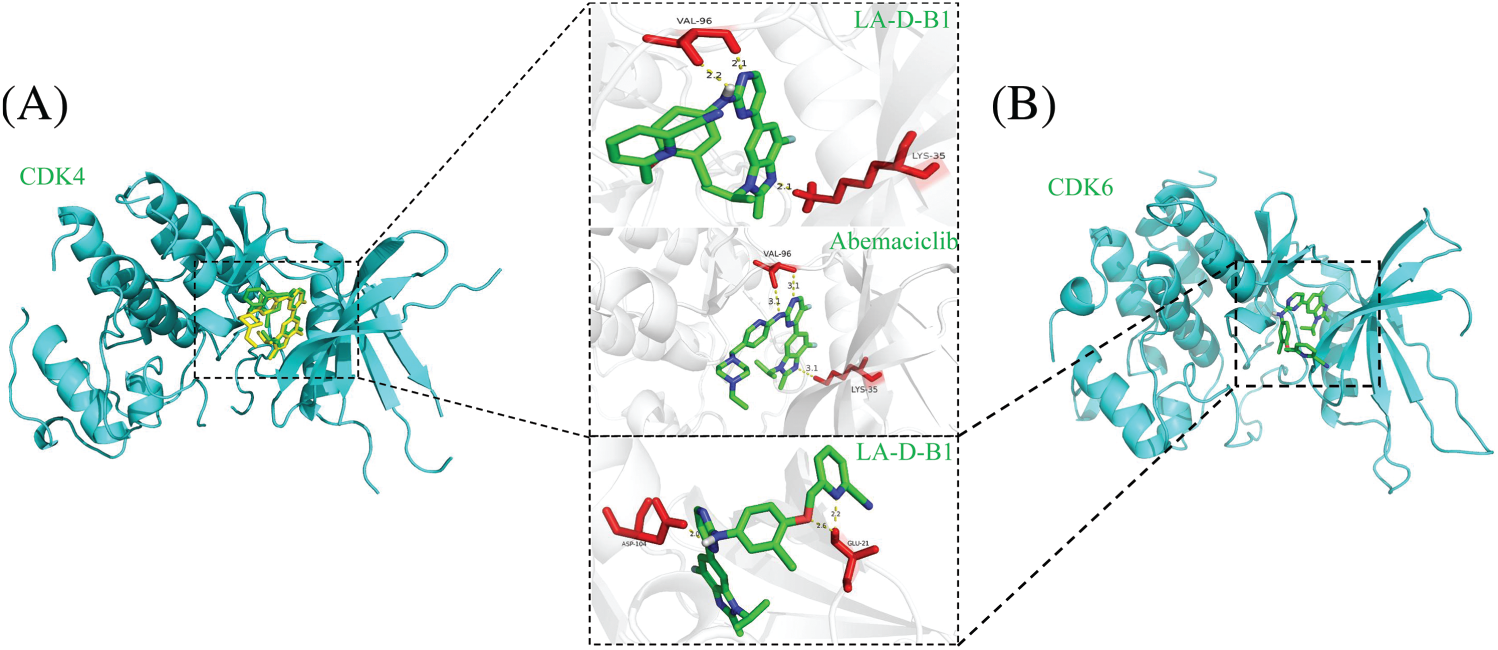
Figure 2: Molecular docking of compound LA-D-B1 with CDK4 and CDK6. (A) Interaction diagram between CDK4 and compound LA-D-B1. (B) Diagram of the interaction of CDK6 with compound LA-D-B1.
Additionally, we superimposed the binding conformations of LA-D-B1 and Abemaciclib in the CDK4 binding pocket for comparison. The consequences indicated that both Abemaciclib and LA-D-B1 maintained a similar conformation in the identical binding pocket.
Cytotoxicity of the compound LA-D-B1
MTT assays were performed to evaluate the antitumor activity of compound LA-D-B1 in vitro. Several cell lines, including MDA-MB-231, Hela, A498, and MCF-7 cells, were selected to investigate the cytotoxicity of the compound LA-D-B1. The cytotoxic effects of LA-D-B1 and the drug Abemaciclib were assessed on various cell strains, including MDA-MB-231, Hela, A498, and MCF-7. The results of the cytotoxicity assays are shown in Table 1 below. It was shown that in MDA-MB-231 cells, compound LA-D-B1 had a lower IC50 (1.64 ± 2.66 μM) than Abemaciclib (2.27 ± 1.59 μM). Meantime, morphological observation was used to assess the impact of compound LA-D-B1 on the morphology of normal 293T cells and breast cancer cells. After being seeded into culture plates, 293T and MDA-MB-231 cells were kept for an overnight period at 37°C and 5% CO2. Subsequently, cells were exposed to varying doses of LA-D-B1 (0.05, 0.1, 0.2, and 0.4 µM) and Abemacicilb (0.4 µM), and pictures of cell morphology were taken using an inverted biomicroscope after 0 and 24 h. After 24 h, the morphology of 293T cells was observed. Compared with the effective drug Abemaciclib at the identical concentration, compound LA-D-B1 caused less damage to the morphology of 293T cells. The results showed that the compound LA-D-B1 was less toxic to normal somatic cells than Abemaciclib at the identical concentration (Fig. 3A). Remedy of MDA-MB-231 cells with diverse concentrations of LA-D-B1 produced morphological changes, with a decrease in cell density after 24 h. The compound LA-D-B1 caused more significant morphological damage to breast cancer cells than Abemaciclib at the identical concentration (Fig. 3B). In conclusion, compound LA-D-B1 was less toxic to normal 293T cells but more sensitive and more toxic to breast cancer cells than Abemaciclib at the identical concentration. These results indicate that the compound LA-D-B1 is highly selective and can significantly inhibit the growth activity of breast cancer cells.
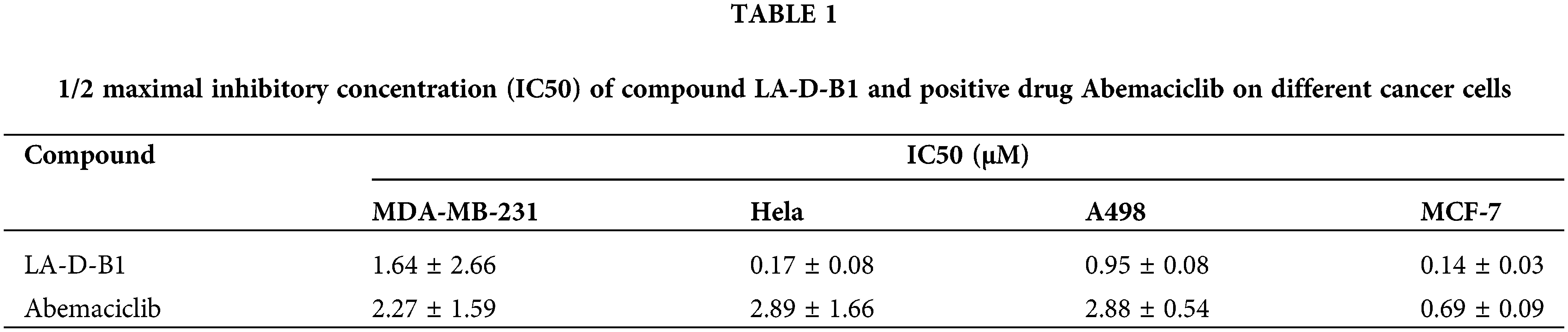
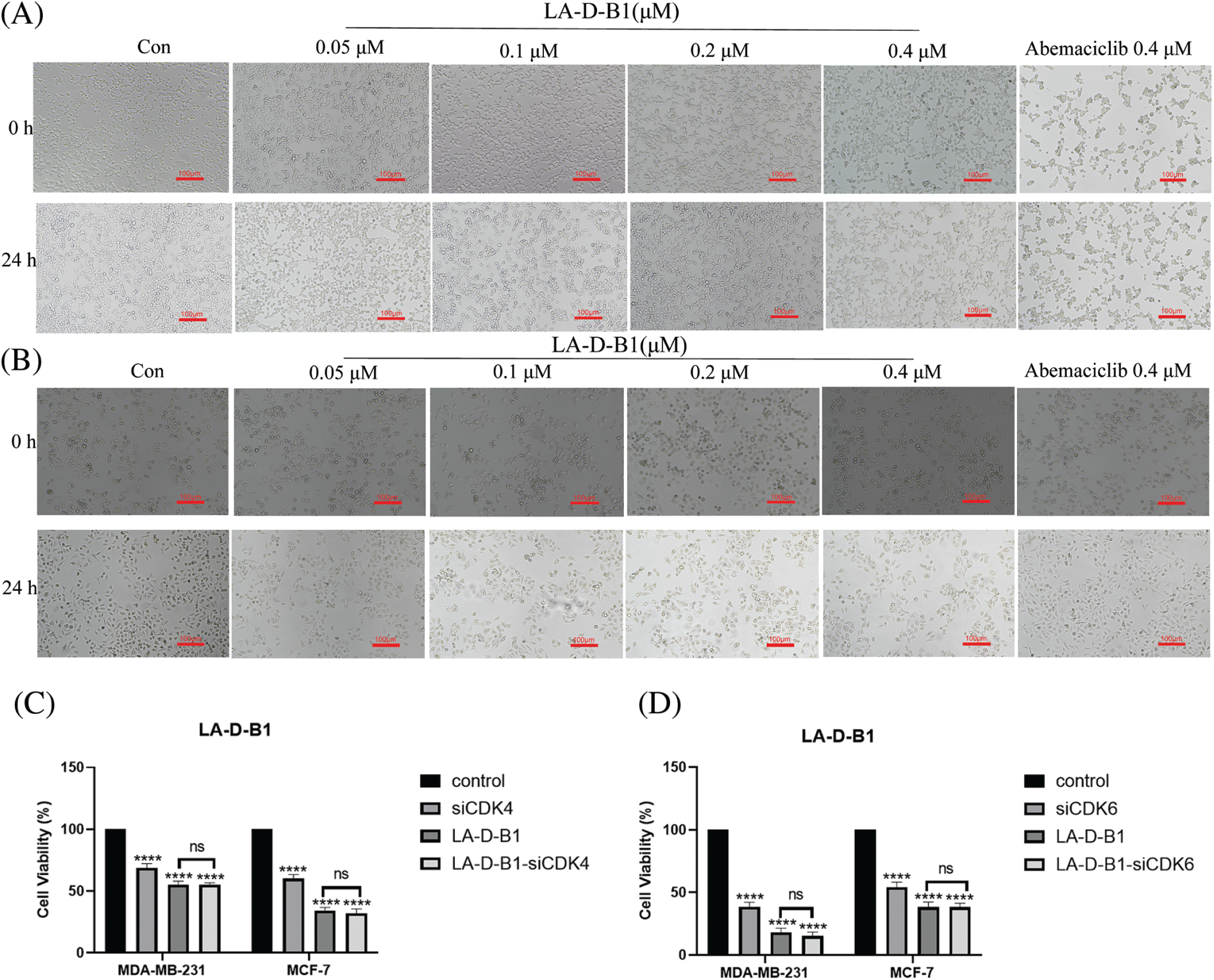
Figure 3: Cytotoxicity and cellular morphological changes of compound LA-D-B1. (A) Morphological effect of LA-D-B1 on 293T cells (n = 3). (B) Morphological effect of LA-D-B1 on MDA-MB-231 cells (n = 3). (C and D) The effect of LA-D-B1 on breast cancer cell viability after RNA silencing was detected (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 vs. control. ns: no significance.
In order to delve deeper into the toxic effects of LA-D-B1 on breast tumor cells, silencing RNA cell lines were constructed and transfected with MDA-MB-231 and MCF-7 cell lines according to the specific transfection protocol. After cell culture for 24 h, different groups were set up. According to the IC50 values, the compound LA-D-B1 of 0.4 and 0.1 μM was added to MDA-MB-231 and MCF-7 cells. Cell viability was measured by MTT assay and analyzed by GraphPad Prism 9 software. The results showed (Figs. 3C and 3D) that the cell growth activity was decreased in both the siRNA group and the LA-D-B1 group as compared to the control group. After the use of CDK4/6-siRNA in the LA-D-B1 group, there was no significant difference in cell growth activity between the LA-D-B1-siRNA group and the LA-D-B1 group, suggesting that CDK4/6 is involved in the antitumor effect of compound LA-D-B1.
Data are as the mean ± SE of dose-response curves from as a minimum three impartial experiments, each with three determinations
Compound LA-D-B1 inhibited the proliferation potential of MCF-7 and MDA-MB-231 cells
We next performed colony-forming assays to explore the impact of compound LA-D-B1 on the proliferative ability of breast cancer cells. Breast cancer cells were incubated with different concentrations of LA-D-B1 and Abemaciclib to allow for the formation of colonies. The results demonstrated that LA-D-B1 significantly reduced the proliferation capacity of MCF-7 and MDA-MB-231 cells compared to Abemaciclib (Figs. 4A–4D). The percentages of cell proliferation at different concentrations of LA-D-B1 were (75.27 ± 1.98%, 77.04 ± 18.21%), (47.29 ± 1.66%, 49.14 ± 14.25%), (27.16 ± 5.32%, 17.52 ± 9.89%), and (6.72 ± 3.68%, 1.99 ± 1.57%). Additionally, LA-D-B1 exhibited a more pronounced inhibitory effect on cell cloning compared to Abemaciclib (82.63 ± 7.73%, 80.89 ± 11.69%). These results indicate that compound LA-D-B1 significantly decreases the proliferative capacity of MCF-7 and MDA-MB-231 cells.
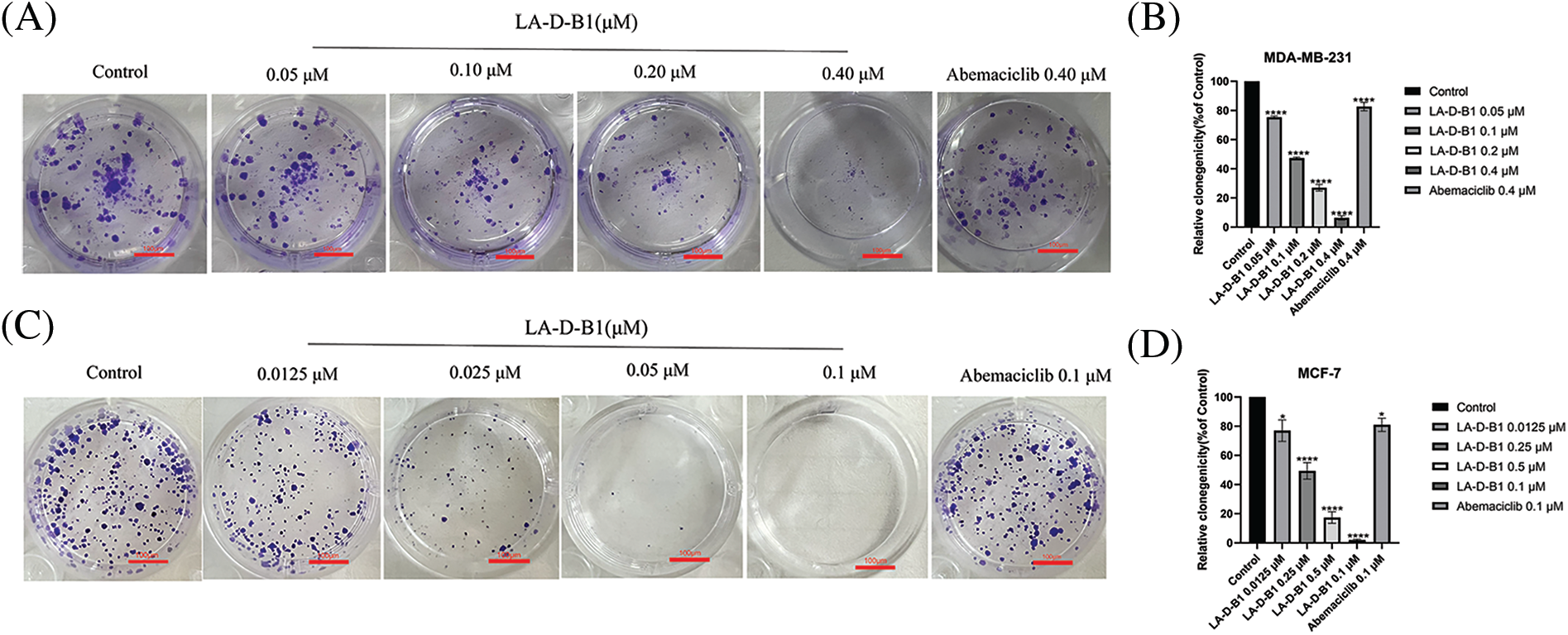
Figure 4: The compound LA-D-B1 inhibited MDA-MB-231 and MCF-7 cell clones. (A and B) Effect of compounds LA-D-B1 and Abemaciclib on colony formation of MDA-MB-231 cells (n = 3). (C and D) Result of compounds LA-D-B1 and Abemaciclib on colony formation of MCF-7 cells (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 vs. control.
Compound LA-D-B1 inhibited the migration, invasion, or adhesion of MCF-7 and MDA-MB-231 cells
Within the present examination, we further investigated the results of compound LA-D-B1 at the migration, adhesion, and invasion of breast cancer cells, which were treated with compound LA-D-B1 and positive drug at different concentrations, and found that, compared with the same concentration of positive drug (92.04 ± 5.80%, 81.77 ± 15.65%), compound LA-D-B1 (34.68 ± 5.74%, 12.57 ± 0.64%) showed the most significant inhibitory effect on adhesion of breast cancer cells (Figs. 5A–5D). Wound-healing assays were conducted to assess the ability of LA-D-B1 to inhibit the migration of cancer cells. The results demonstrated that LA-D-B1 at different concentrations significantly inhibited the wound-healing capability of cancer cells compared with the control. Furthermore, the inhibitory effect of LA-D-B1 was superior to that of Abemaciclib at the same concentration (Figs. 5E–5H). Invasion assays were performed to assess whether the compound LA-D-B1 could lessen the potential of tumor cells to invade the surrounding tissues. The consequences reflect that different concentrations of the compound LA-D-B1 could inhibit the invasion of tumor cells. The inhibitory capacity was (35.39 ± 12.89%, 77.52 ± 9.84%), (12.46 ± 4.55%, 56.96 ± 21.27%), (7.45 ± 4.00%, 22.54 ± 6.16%), and (1.84 ± 1.33%, 5.46 ± 1.90%), respectively. The inhibitory effect used to be better than the effective drug Abemaciclib at the identical concentration (52.14 ± 15.10%, 70.81 ± 15.74%) (Figs. 5I–5L). Overall, these findings suggest that compound LA-D-B1 is effective in inhibiting breast cancer cell adhesion, migration, and invasion.
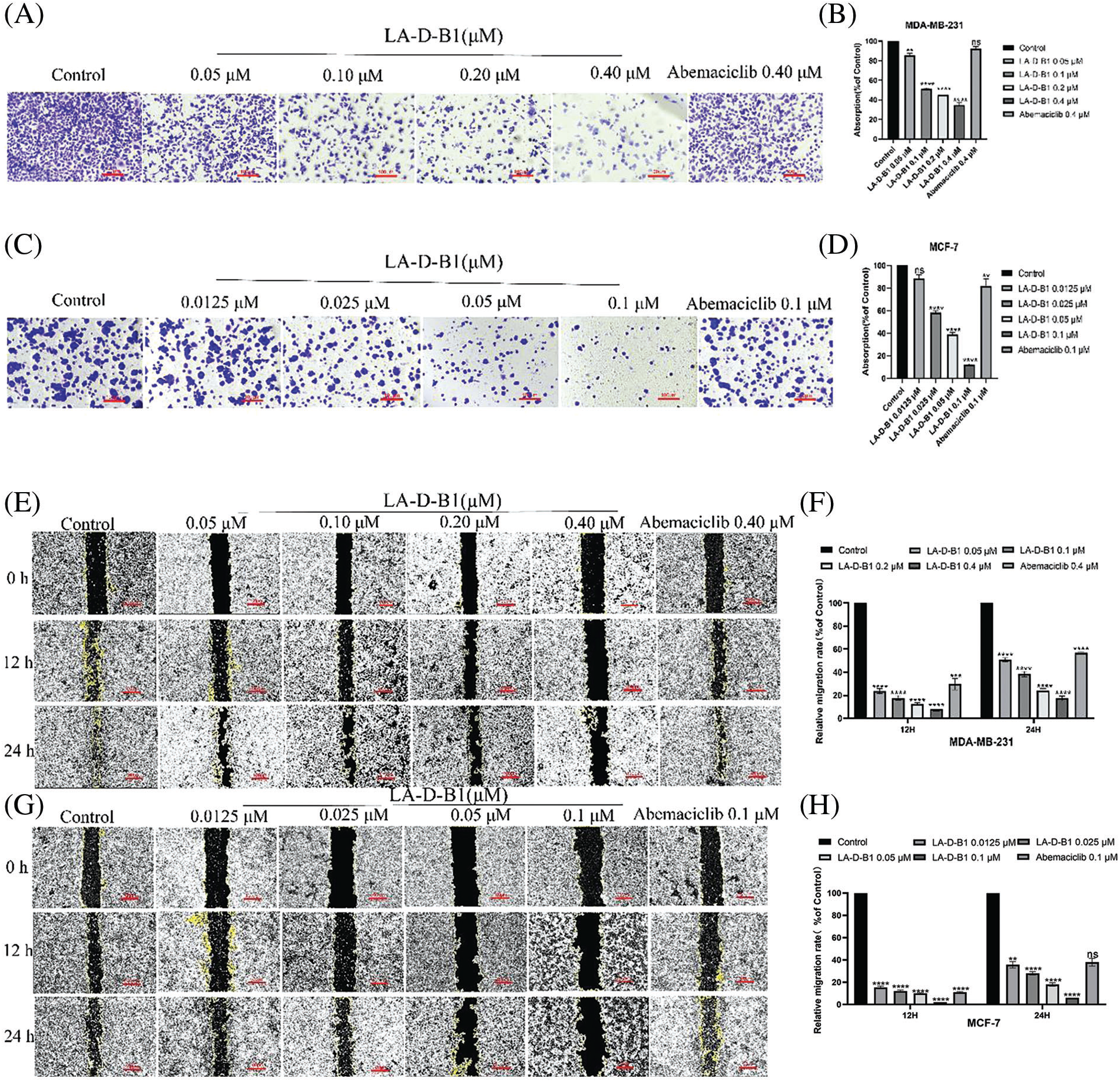
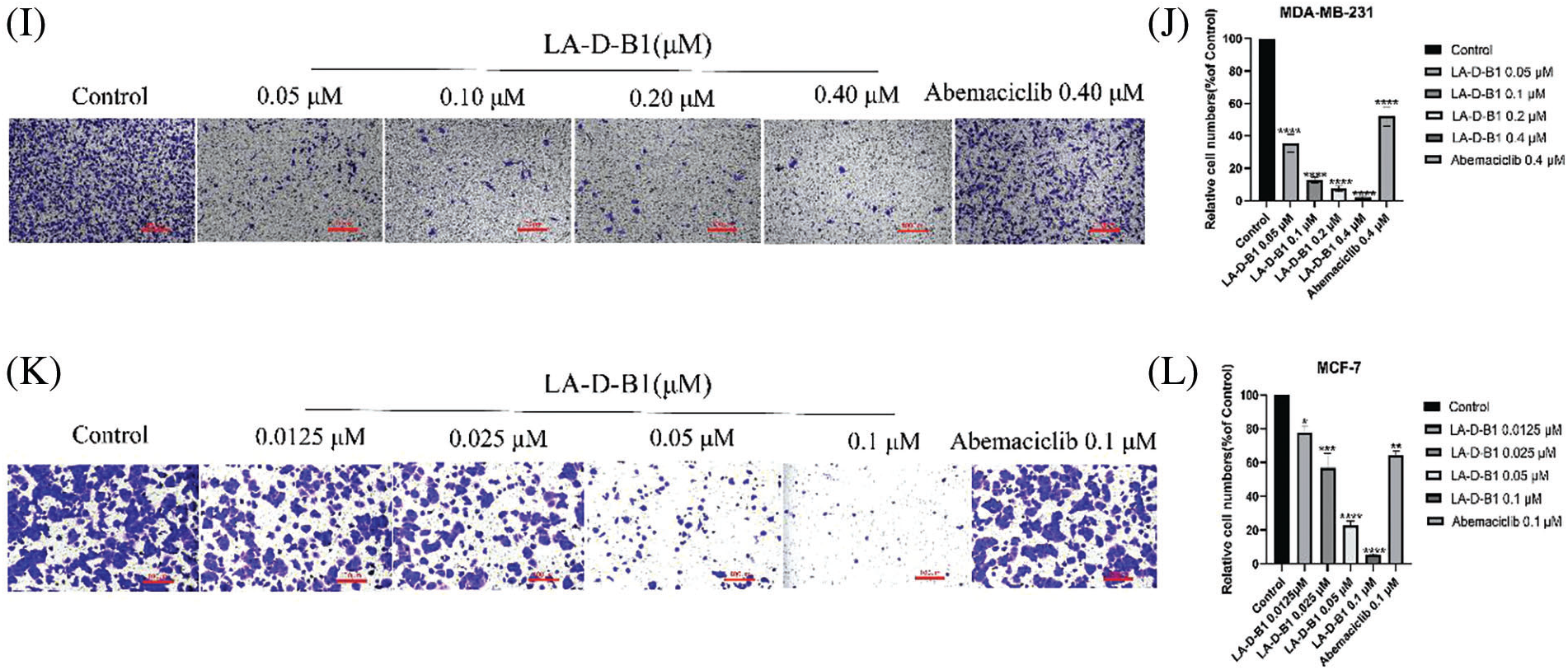
Figure 5: Compound LA-D-B1 hindered the inhibited migration, invasion, or adhesion of MCF-7 and MDA-MB-231 cells. (A–D) Effect of compounds LA-D-B1 and Abemaciclib on the adhesion of MCF-7 and MDA-MB-231 cells (n = 3). (E–H) Effect of the compounds LA-D-B1 and Abemaciclib on the migration of MCF-7 and MDA-MB-231 cells (n = 3). (I–L) Effect of the compounds LA-D-B1 and Abemaciclib on the invasive capability of MCF-7 and MDA-MB-231 cells (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 vs. control.
The compound LA-D-B1 regulates the expression of related cyclins in MCF-7 and MDA-MB-231 cells
To investigate the mechanism underlying the anti-tumor effect of compound LA-D-B1, Western blotting was conducted to analyze changes in cell cycle proteins in MDA-MB-231 and MCF-7 cells after treatment with LA-D-B1. Cells were treated with different concentrations of LA-D-B1 and the positive drug Abemaciclib, and then subjected to Western blot analysis after 24 h. The results showed that the levels of CDK4, CDK6, CyclinE1, Cyclin D3, E2F1, and p-Rb/Rb were significantly suppressed after treatment with LA-D-B1 in a dose-dependent manner (Figs. 6A and 6B). Meanwhile, the consequences reflected that compound LA-D-B1 increased the expression levels of BAX, Caspase-3, and cleaved Caspase-3 and inhibited the expression of the anti-apoptotic protein Bcl-2 (Figs. 6A–6D). Among them, BAX is a pro-apoptotic protein, and Caspase-3 and cleaved Caspase-3 are the main detection signals of apoptosis. The above results indicate that the compound LA-D-B1 can induce apoptosis and cycle arrest in cancer cells. Therefore, our results confirmed that LA-D-B1 prevented Rb phosphorylation and E2F1 release by inhibiting the expression levels of cell cyclin-dependent kinases CDK4 and CDK6, and the excessive proliferation and abnormal replication of tumor cells were prevented.
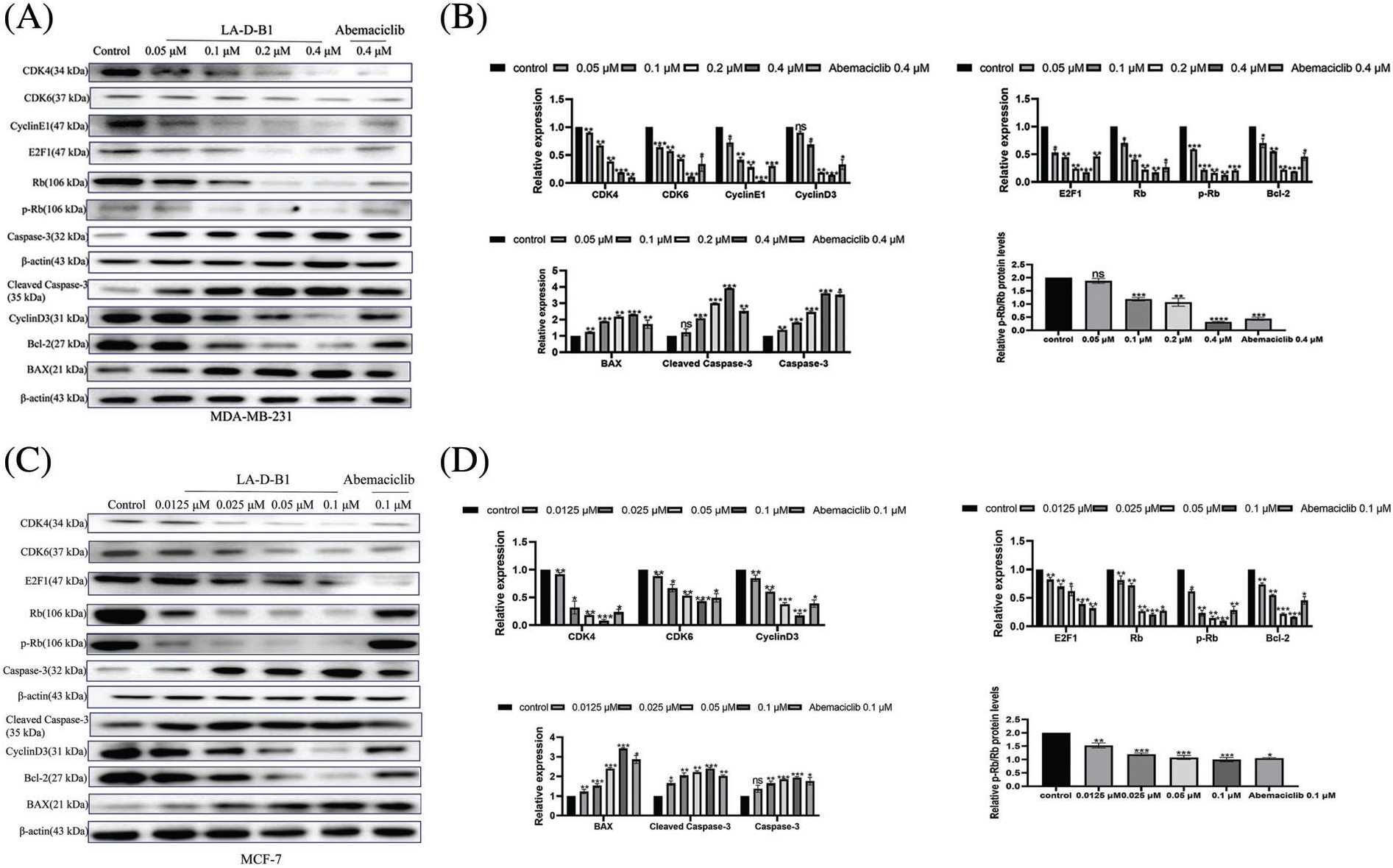
Figure 6: Compound LA-D-B1 causes cellular cycle arrest and induces apoptosis by affecting related proteins in breast cancer cells. (A–D) Western blot analysis of breast cancer cells treated with diverse concentrations of the compound LA-B-D1 (n = 3). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 vs. control.
Antitumor effect of compound LA-D-B1 in vivo
The anti-tumor activity of compound LA-D-B1 was further investigated in vivo using a chick embryo xenograft tumor model established with breast cancer cells. After treatment with different concentrations of LA-D-B1, the number of angiogenesis in each group was analyzed using ImageJ software. The angiogenesis rates were (90.75 ± 5.90%, 90.79 ± 7.23%), (78.94 ± 5.45%, 69.40 ± 5.48%), and (54.13 ± 6.92%, 50.65 ± 0.86%). The results showed that as the concentration of LA-D-B1 increased, tumor angiogenesis decreased (Figs. 7A–7D). This indicates that LA-D-B1 possesses a strong ability to inhibit tumor angiogenesis, which is more potent than the effective drug Abemaciclib at the identical concentration.
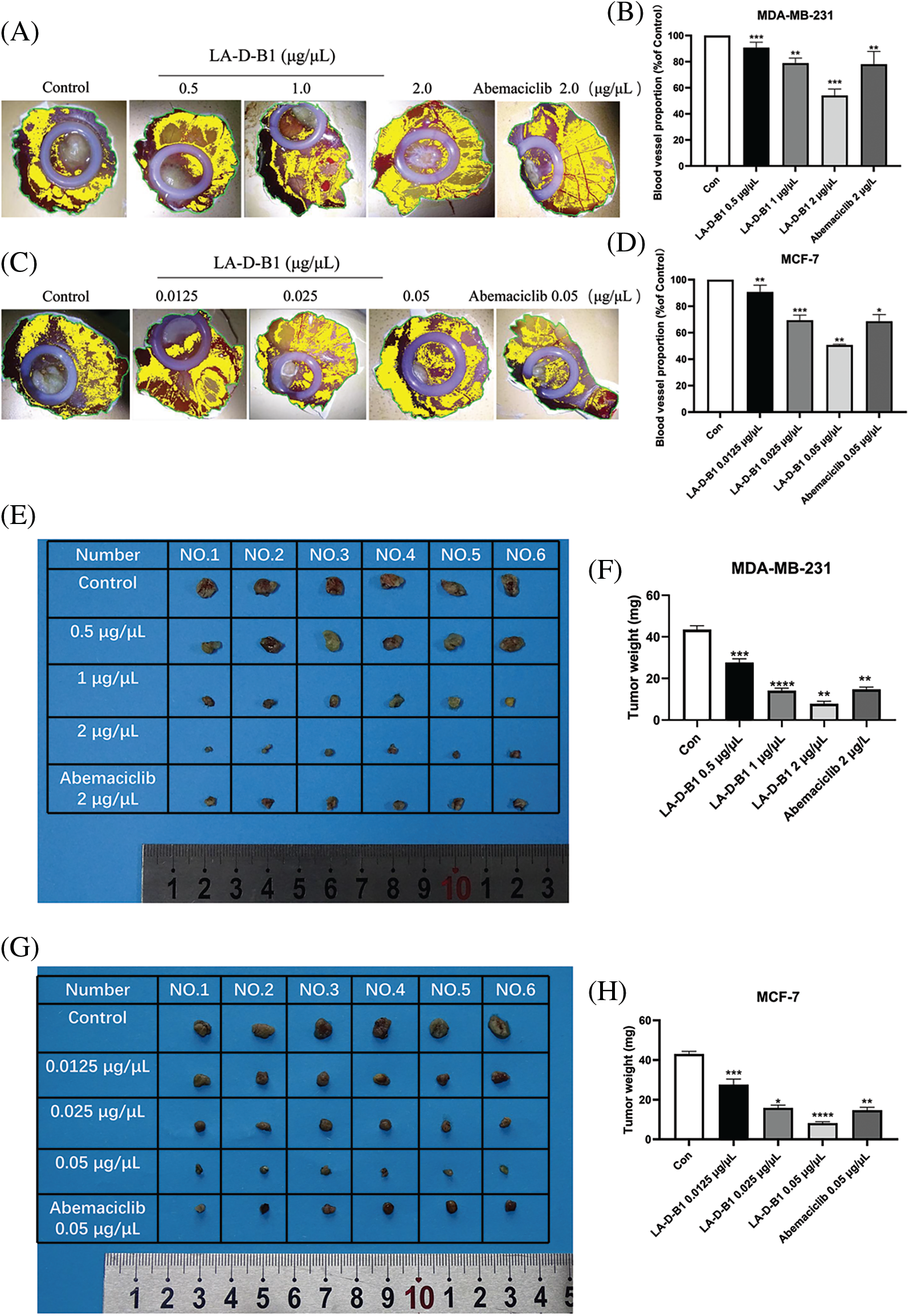
Figure 7: Compound LA-D-B1 inhibits tumor proliferation in vivo. (A–D) Statistical diagram of blood vessel distribution and number (n = 3). (E–H) Plot of tumor anatomy and number statistics (n = 6). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 vs. control
Furthermore, the results demonstrated that compared to the control group (43.50 ± 4.18 mg, 43.00 ± 3.16 mg), the weight of the chick embryo tumors decreased to (27.67 ± 3.92, 27.67 ± 6.11 mg), (14.17 ± 2.61, 15.83 ± 3.29 mg), and (7.83 ± 2.61, 8.17 ± 1.68 mg) after treatment with different concentrations of LA-D-B1. Additionally, the tumor size became smaller as the concentration of LA-D-B1 increased (Figs. 7E–7H).
These findings suggest that tumor growth was arrested with increasing concentrations of LA-D-B1 treatment. In conclusion, the compound LA-D-B1 demonstrated a significant inhibition of breast tumor growth in the chick embryo xenograft tumor model, highlighting its efficacy in impeding in vivo tumor cell proliferation.
The cell cycle is a fundamental process in cellular life activities, and its proper progression requires the balance of regulatory factors [20]. CDKs play a critical role in regulating the cell cycle [21]. CDK4/6 binds to CyclinD to form a CDK4/6-CyclinD complex, leading to the phosphorylation of pRB. This phosphorylation inhibits the binding of p-RB to E2F transcription factors to activate and transcribe genes required for S-phase entry [22,23]. As key regulatory enzymes, CDKs are concerned with the regulation of cellular proliferation [24]. Dysregulation of CDKs leads to uncontrolled cellular proliferation, which promotes the development of malignancies, including breast cancer [25]. Therefore, it is considered a potential goal in most cancer remedies. Progress in the remedy of TNBC, which has a poor prognosis and limited remedy options compared with other BC subtypes, remains an important challenge [26]. The FDA has granted approval for three CDK4/6 inhibitors, namely Abemaciclib, Palbociclib, and Ribociclib, in the context of breast cancer treatment [27]. These inhibitors effectively block the cellular cycle from G1 to S section via blocking off the phosphorylation of Rb protein, thus inhibiting the proliferation of tumor cells [28–30]. Although these drugs have achieved remarkable effects in the remedy of breast cancer, sufferers will develop drug resistance after a lengthy period of use [31]. In this examination, we designed and synthesized LA-D-B1, a singular Abemaciclib by-product, and further investigated its antitumor impact on breast cancer. Studies have shown that LA-D-B1 has an extensively higher toxic effect on breast cancer cells than the effective drug Abemaciclib and can reduce resistance to single-endocrine therapy.
In breast cancer, the CyclinD-CDK4/6-RB pathway is dysregulated to promote tumor progression [32]. Previous studies have shown that CDK inhibitors have made great progress in clinical applications [33]. The development of a novel CDK4/6 inhibitor with low toxicity and side effects could provide insights into the development of cancer treatment strategies. The present study showed that compound LA-D-B1 inhibited proliferation, migration, invasion, and colony formation of breast cancer cells. Western blot analysis was conducted to evaluate the effects of different concentrations of LA-D-B1 and the positive drug Abemaciclib on the expression of cell cycle-related proteins in breast cancer cells. The results showed that compound LA-D-B1 blocked the expression of CDK4, CDK6, CyclinE1, CyclinD3, E2F1, Rb, p-Rb, Bcl-2 and enhanced the expression of BAX, Cleaved Caspase-3 and Caspase-3. Additionally, the ratio of p-Rb/Rb also decreased with the increase of LA-D-B1 concentration. Compound LA-D-B1 significantly inhibited Rb expression, thereby reducing Rb phosphorylation, and the inhibitory effect was better than Abemaciclib at the identical concentration. Furthermore, LA-D-B1 demonstrated significant inhibition of tumor growth in the chick embryo chorioallantois model, further highlighting its potent anti-tumor proliferation effect. Our findings suggest that compound LA-D-B1, similar to abemaciclib, exerts anti-breast cancer effects by targeting the CDK4/6-cyclinD-Rb-E2F pathway. Interestingly, LA-D-B1 demonstrated superior anti-tumor efficacy compared to abemaciclib at the same dose. This unexpected result led us to hypothesize that LA-D-B1 may exert its anti-tumor effects through additional target pathways in breast cancer. Further exploration of these potential pathways is warranted. Therefore, LA-D-B1 holds significant potential as a novel CDK4/6 inhibitor for targeted therapy of triple-negative breast cancer (TNBC). Nevertheless, it is crucial to acknowledge the constraints of our present study concerning the evaluation of the anti-breast cancer efficacy of LA-D-B1. These limitations encompass the utilization of rudimentary research methodologies and the exclusive reliance on a single breed of experimental animals. Further investigation into the clinical pharmacological effects of LA-D-B1 is crucial.
This study shows that LA-D-B1 has anti-tumor effects on breast cancer cells both in vitro and in vivo, suggesting that LA-D-B1 is a potential targeted drug for the treatment of triple-negative breast cancer (TNBC), and its mechanism of action needs to be further studied.
Acknowledgement: This project is gratuitated to the Jiangsu Province Key Laboratory for Screening of Marine Pharmaceutical Active Molecules for support of this study.
Funding Statement: This study was supported by the National Natural Science Foundation of China (82273167, 82104174, 81602626, 12271068, 82172558 and 82373112), Jiangsu Province Basic Research Program Natural Science Foundation (Outstanding Youth Fund Project, BK20220063), the Key Program of Basic Science (Natural Science) of Jiangsu Province (22KJA350001), “Huaguo Mountain Talent Plan” of Lianyungang City (Innovative Talents Liu Bin), Qing Lan Project of Jiangsu Universities (Outstanding Young Backbone Teachers, Ji Jing), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (No. 20KJB350008), Priority Academic Program Development of Jiangsu Higher Education Institutions, College Students’ Innovative Entrepreneurial Training Plan Program (Project Nos. SY202211641640011, SY202311641640002 and SZ202311641640002), Chen Xiao-Ping Foundation for the Development of Science and Technology of Hubei Province (Project No. CXPJJH123003-027), the Distinguished Young Scholars of Nanjing (JQX20008) and Scientific Research Foundation for Returned Scholars of Tongji Hospital (Project 2022hgry021).
Author Contributions: Study conception and design: Jing Ji, Xiujun Wang, Ling Ma, Yuting Xu, Ziyun Chen, Shaojie Ma, Geng Zhang; data collection: Jing Ji, Ling Ma, Zirui Jiang, Xiao Hou, Siyi Zhang, Hanxue Li; analysis and interpretation of results: Jing Ji, Ling Ma, Zirui Jiang, Xiao Hou, Siyi Zhang, Hanxue Li; draft manuscript preparation: Ling Ma. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: The animal study was reviewed and approved by Experiment Ethics Review Committee of Jiangsu Ocean University on November 04, 2022 (No. Jou2022110402).
Conflicts of Interest: All authors have no conflict of interests to declare.
References
1. Luo J, Zou H, Guo Y, Tong T, Ye L, Zhu C, et al. SRC kinase-mediated signaling pathways and targeted therapies in breast cancer. Breast Cancer Res. 2022;24(1):99. [Google Scholar] [PubMed]
2. Yang XI, Wu H, Xiong H, Zhao C, Liu BO, et al. Comprehensive bioinformatics analysis of CYB561 expression in breast cancer: link between prognosis and immune infiltration. BIOCELL. 2023;47:1021–37. doi:10.32604/biocell.2023.027103. [Google Scholar] [CrossRef]
3. Houghton SC, Hankinson SE. Cancer progress and priorities: breast cancer. Cancer Epidemiol Biomarkers Prev. 2021;30(5):822–44. [Google Scholar] [PubMed]
4. Fan K, Weng J. The progress of combination therapy with immune checkpoint inhibitors in breast cancer. Biocell. 2023;47(6):1199–211. [Google Scholar]
5. Sukumar J, Gast K, Quiroga D, Lustberg M, Williams N. Triple-negative breast cancer: promising prognostic biomarkers currently in development. Expert Rev Anticancer Ther. 2021;21(2):135–48. [Google Scholar] [PubMed]
6. Lee J. Current treatment landscape for early triple-negative breast cancer (TNBC). J Clin Med. 2023;12(4):1524. [Google Scholar] [PubMed]
7. Lehrberg A, Davis MB, Baidoun F, Petersen L, Susick L, Jenkins B, et al. Outcome of African-American compared to white-American patients with early-stage breast cancer, stratified by phenotype. Breast J. 2021;27(7):573–80. [Google Scholar] [PubMed]
8. Bou Zerdan M, Ghorayeb T, Saliba F, Allam S, Bou Zerdan M, Yaghi M, et al. Triple negative breast cancer: updates on classification and treatment in 2021. Cancers. 2022;14(5):1253. [Google Scholar] [PubMed]
9. Chong QY, Kok ZH, Bui NL, Xiang X, Wong AL, Yong WP, et al. A unique CDK4/6 inhibitor: current and future therapeutic strategies of abemaciclib. Pharmacol Res. 2020;156:104686. [Google Scholar] [PubMed]
10. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Dong P, Gassler N, Taheri M, et al. A review on the role of cyclin dependent kinases in cancers. Cancer Cell Int. 2022;22(1):325. [Google Scholar] [PubMed]
11. Chou J, Quigley DA, Robinson TM, Feng FY, Ashworth A. Transcription-associated cyclin-dependent kinases as targets and biomarkers for cancer therapy. Cancer Discov. 2020;10(3):351–70. [Google Scholar] [PubMed]
12. Cornwell JA, Crncec A, Afifi MM, Tang K, Amin R, Cappell SD. Loss of CDK4/6 activity in S/G2 phase leads to cell cycle reversal. Nat. 2023;619(7969):363–70. [Google Scholar]
13. Ammazzalorso A, Agamennone M, de Filippis B, Fantacuzzi M. Development of CDK4/6 inhibitors: a five years update. Mol. 2021;26(5):1488. [Google Scholar]
14. Goel S, Bergholz JS, Zhao JJ. Targeting CDK4 and CDK6 in cancer. Nat Rev Cancer. 2022;22(6):356–72. [Google Scholar] [PubMed]
15. Pavlovic D, Niciforovic D, Papic D, Milojevic K, Markovic M. CDK4/6 inhibitors: basics, pros, and major cons in breast cancer treatment with specific regard to cardiotoxicity–a narrative review. Ther Adv Med Oncol. 2023;15:17588359231205848. [Google Scholar] [PubMed]
16. Braal CL, Jongbloed EM, Wilting SM, Mathijssen RHJ, Koolen SLW, Jager A. Inhibiting CDK4/6 in breast cancer with Palbociclib, Ribociclib, and Abemaciclib: similarities and differences. Drugs. 2021;81(3):317–31. [Google Scholar] [PubMed]
17. Papadimitriou MC, Pazaiti A, Iliakopoulos K, Markouli M, Michalaki V, Papadimitriou CA. Resistance to CDK4/6 inhibition: mechanisms and strategies to overcome a therapeutic problem in the treatment of hormone receptor-positive metastatic breast cancer. Biochim Biophys Acta Mol Cell Res. 2022;1869(12):119346. [Google Scholar] [PubMed]
18. Mughal MJ, Bhadresha K, Kwok HF. CDK inhibitors from past to present: a new wave of cancer therapy. Semin Cancer Biol. 2023;88:106–22. [Google Scholar] [PubMed]
19. Ji J, Zhang Z, He X, Pan G, Li G, Lv J, et al. A novel ribociclib derivative WXJ-103 exerts anti-breast cancer effect through CDK4/6. Anticancer Drugs. 2023;34(7):803–15. [Google Scholar] [PubMed]
20. Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29(5):946–60. [Google Scholar] [PubMed]
21. Kashyap D, Garg VK, Sandberg EN, Goel N, Bishayee A. Oncogenic and tumor suppressive components of the cell cycle in breast cancer progression and prognosis. Pharma. 2021;13(4):569. [Google Scholar]
22. Koirala N, Dey N, Aske J, de P. Targeting cell cycle progression in HER2+ breast cancer: an emerging treatment opportunity. Int J Mol Sci. 2022;23(12):6547. [Google Scholar] [PubMed]
23. Yousuf M, Alam M, Shamsi A, Khan P, Hasan GM, Rizwanul Haque QM, et al. Structure-guided design and development of cyclin-dependent kinase 4/6 inhibitors: a review on therapeutic implications. Int J Biol Macromol. 2022;218:394–408. [Google Scholar] [PubMed]
24. Sofi S, Mehraj U, Qayoom H, Aisha S, Asdaq SMB, Almilaibary A, et al. Cyclin-dependent kinases in breast cancer: expression pattern and therapeutic implications. Med Oncol. 2022;39(6):106. [Google Scholar] [PubMed]
25. Ding L, Cao J, Lin W, Chen H, Xiong X, Ao H, et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020;21(6):1960. [Google Scholar] [PubMed]
26. Vagia E, Mahalingam D, Cristofanilli M. The landscape of targeted therapies in TNBC. Cancers. 2020;12(4):916. [Google Scholar] [PubMed]
27. Hassan MA, Ates-Alagoz Z. Cyclin-dependent kinase 4/6 inhibitors against breast cancer. Mini Rev Med Chem. 2023;23(4):412–28. [Google Scholar] [PubMed]
28. Gomes I, Abreu C, Costa L, Casimiro S. The evolving pathways of the efficacy of and resistance to CDK4/6 inhibitors in breast cancer. Cancers. 2023;15(19):4835. doi:10.3390/cancers15194835. [Google Scholar] [PubMed] [CrossRef]
29. Hermansyah D, Firsty NN, Alhudawy MN, Nasution RA. The combination of CDK 4/6 inhibitors plus endocrine treatment versus endocrine treatment alone in hormone-receptor (HR)-positive breast cancer: a systematic review and meta-analysis. Med Arch. 2022;76(5):333–42. [Google Scholar] [PubMed]
30. Jerzak KJ, Bouganim N, Brezden-Masley C, Edwards S, Gelmon K, Henning JW, et al. HR+/HER2– advanced breast cancer treatment in the first-line setting. Expert Review. Curr Oncol. 2023;30(6):5425–47. [Google Scholar] [PubMed]
31. Huang J, Zheng L, Sun Z, Li J. CDK4/6 inhibitor resistance mechanisms and treatment strategies. Int J Mol Med. 2022;50(4):128. [Google Scholar] [PubMed]
32. Krasniqi E, Goeman F, Pulito C, Palcau AC, Ciuffreda L, Di Lisa FS, et al. Biomarkers of response and resistance to CDK4/6 inhibitors in breast cancer: hints from liquid biopsy and microRNA exploration. Int J Mol Sci. 2022;23(23):14534. [Google Scholar] [PubMed]
33. Zhang M, Zhang L, Hei R, Li X, Cai H, Wu X, et al. CDK inhibitors in cancer therapy, an overview of recent development. Am J Cancer Res. 2021;11(5):1913–35. [Google Scholar] [PubMed]
Supplementary Material
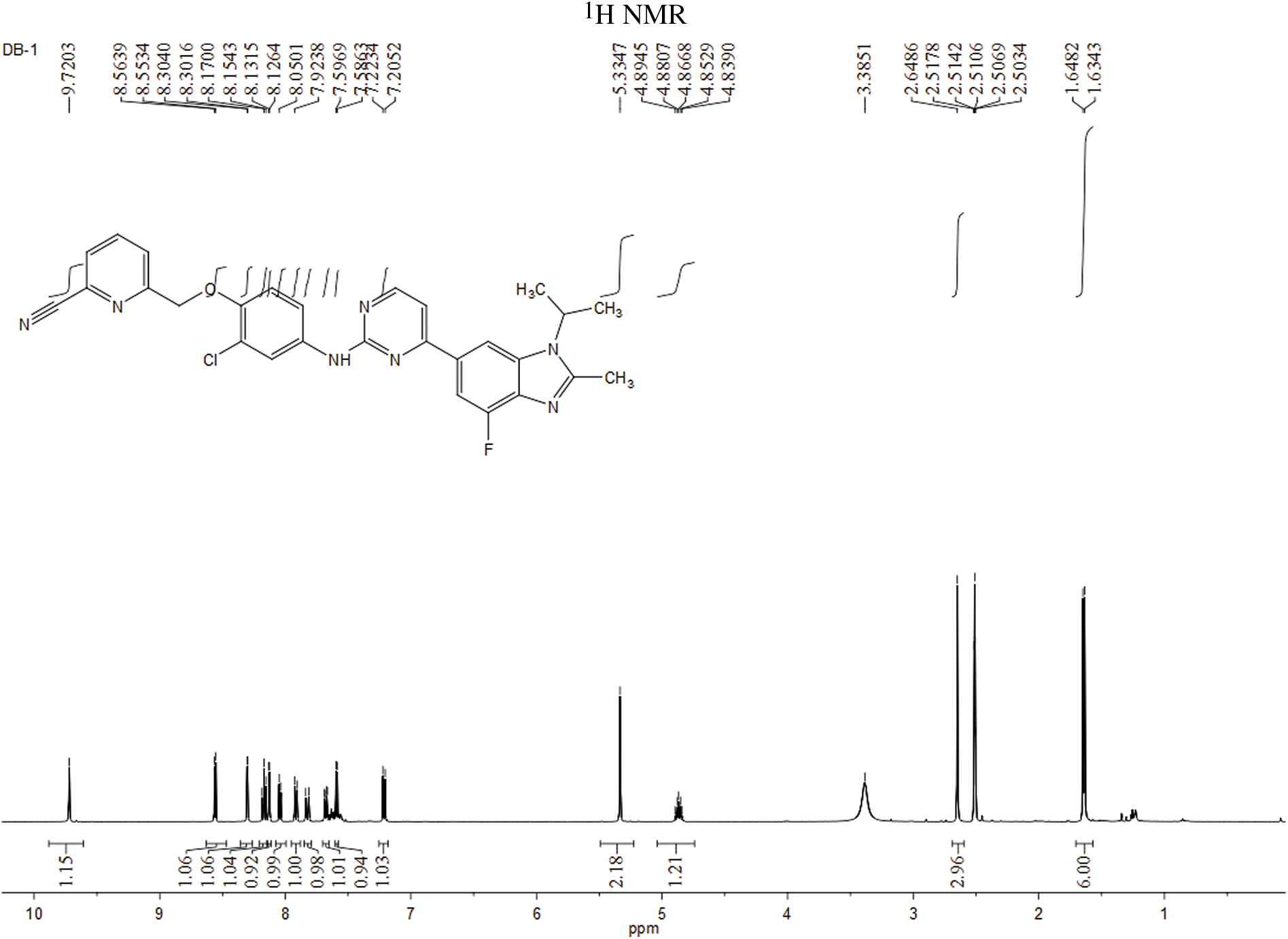
Figure S1: Hydrogen spectrum of compound LA-D-B1.
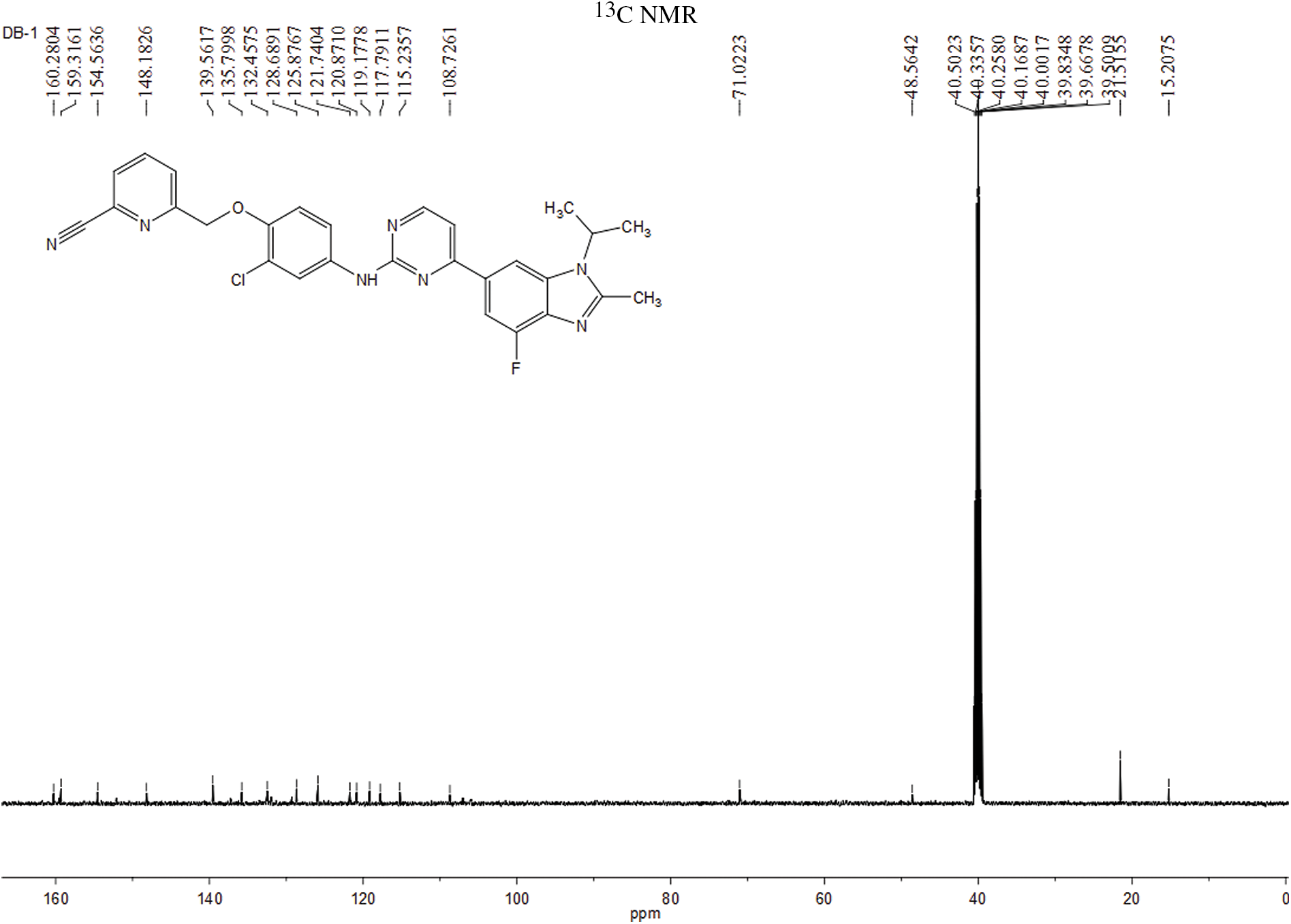
Figure S2: Carbon spectrum of the compound LA-D-B1.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools