 Open Access
Open Access
REVIEW
HIFs in hypoxic regulation of the extracellular matrix: focus on little-known player HIF-3
Institute of Biomedical Problems of the Russian Academy of Sciences, Khoroshevskoye Shosse 76A, Moscow, 123007, Russia
* Corresponding Author: ELENA ANDREEVA. Email:
(This article belongs to the Special Issue: Cellular Signal Transduction in Biological Activities)
BIOCELL 2024, 48(5), 677-692. https://doi.org/10.32604/biocell.2024.048873
Received 20 December 2023; Accepted 06 March 2024; Issue published 06 May 2024
Abstract
The structural and associated molecules of the extracellular matrix (ECM) complex is an important component of the local milieu of cells, both for maintaining their functions and homeostasis. It is a dynamic structure that is finely tuned to changes in the microenvironment. One of these factors is hypoxia, which can arise in tissues due to physiological or pathological effects. As a result of the hypoxic effect, the properties of the ECM are significantly modified, stiffness increases, the balance between degradation and synthesis of structural proteins shifts, and the deposition of biologically active mediators’ changes. Hypoxia-inducible factors (HIFs) contribute significantly to the modification of the properties of the ECM under hypoxia. Among the three HIF isoforms, HIF-1 is the most studied, with numerous identified target genes encoding proteins that participate in all matrisome compartments. Much less is known about HIF-2 and HIF-3. In the context of the effects of hypoxia on the matrisome, HIF-3 isoform is of particular interest. Unlike the first two isoforms, transiently expressed during the first hours of hypoxia, HIF-3 is activated after around 24 h of exposure to hypoxia and persists for a longer period. In addition, its transcription is more pronounced under hypoxia than are HIFs 1 and 2. HIF-3, rather than the first two isoforms, is possibly responsible for the changes that occur in matrisome during prolonged hypoxic conditions. This review attempts to summarize the available data on the involvement of HIF-3 in the matrisome modification under hypoxia.Keywords
The extracellular matrix (ECM) is a complex macromolecular structure that provides mechanical support to cells and actively regulates tissue physiology. The term “matrisome” was introduced to cover both the ECM structural and ECM-associated molecules [1]. The structural components of the core matrisome present: collagens, glycoproteins, proteoglycans, and glycosaminoglycans. Matrisome-associated molecules comprise regulatory proteins that take part in ECM remodeling, such as enzymes and their inhibitors; deposited biologically active molecules, including cytokines, growth factors, etc., as well as matrisome-associated molecules that are in charge of regulating the tissue microenvironment and modifying the biochemical properties of the matrix [1,2]. The components of the matrisome work cohesively to maintain a three-dimensional extracellular environment and deliver precise mechanical, structural, and biological guidance. Each tissue possesses a matrisome with its distinct composition and topology, resulting from the permanently changing biochemical and biophysical crosstalk between different cellular and extracellular components [3–5].
Changes in matrisome structure and functions are frequently observed in a range of pathological conditions often associated with oxygen deprivation (hypoxia). The role of reduced oxygen levels in ECM metabolism has been studied primarily in the context of vascular disease and tumor progression [6–8]. The adaptation to O2 deprivation results in increased production, alignment, and stiffness of ECM structures. This increase is necessary to activate integrins [9–11], enhance expansion [12], and improve cell survival [13,14]. In particular, collagen I accumulation leads to an increase in tissue fibrosis [15–18], adhesive properties [9,19], and support of cell migration, thus promoting tumor invasion [13,20].
In 2019, the Nobel Prize in physiology or medicine was awarded jointly to W. G. Kaelin Jr, P. J. Ratcliffe, and G. L. Semenza “for their discoveries of how cells sense and adapt to oxygen availability” [21]. The principal signaling pathways of the cellular response to O2 deprivation have been demonstrated to be triggered by a family of the hypoxia-inducible factors (HIFs) represented by three isoforms (HIF-1, -2, -3). The genes encoding matrisome proteins are widely represented among the targets of HIF-1 [22,23]. However, there are blind spots in this story related to the other isoforms, HIF-2 and HIF-3. Very little information has been accumulated on the latter one of these. Several studies suggest the existence of complex mechanisms in the mutual regulation of expression between HIF-1α, HIF-2α, and HIF-3α, including HIF-3α-mediated suppression of the expression of the first two factors [24–27]. This could indicate a potential shift in transcription activity towards HIF-3. This review will focus on the mechanisms of crosstalk between HIF-3 and the other HIF isoforms and the contribution of this to the regulation of matrisome components during cellular adaptation to hypoxia.
Hypoxia-inducible factor-mediated regulation of the cellular response to hypoxia
Hypoxia is one of the most important microenvironmental factors significantly affecting cellular metabolism. The O2 levels in vivo typically range from 0.5% to 14%, depending on the type of tissue [28]. Thus, the O2 concentration in the medullary niche or lymphoid organs is low (ranging from 0.5% to 4.5%) [29], but it reaches high values of up to 14% in kidney tissue [30]. Low tissue O2 levels are physiologically normal for tissues and termed “physioxia”, or “physiological hypoxia”. These concentrations are appropriate for the cells and do not require adaptation. However, for a number of reasons, pathological conditions can occur that are accompanied by oxygen deprivation, such as myocardial infarction, stroke, tumors, сhronic obstructive pulmonary disease, and other ischemic conditions [23,31,32]. Such hypoxic events can lead to adaptive or pathological outcomes, depending on the duration and degree of oxygen deprivation.
HIFs are the master regulators of cellular response to oxygen deprivation, triggering the expression of hundreds of target genes. The discovery of HIF-1 and its control mechanisms marked a significant milestone in studying cell adaptation to hypoxia. Currently, a significant body of knowledge about HIF-1, its regulation and targets has been accumulated, and two other isoforms, HIF-2 and HIF-3, have been discovered. HIFs are heterodimeric proteins consisting of HIF-1α, HIF-2α, or HIF-3α subunit and β-subunit (also known as aryl hydrocarbon receptor nuclear translocator (ARNT)) for each isoform, accordingly: HIF-1β (ARNT) and HIF-2β (ARNT2), while HIF-3β (ARNTL), is classified as a class II basic-helix-loop-helix-Per/Arnt/Sim (bHLH-PAS) factor [33]. The ARNT is expressed constitutively, while the HIF-1α, HIF-2α, and HIF-3α subunits are oxygen-sensitive. Under normoxic conditions, the α-subunit undergoes hydroxylation by the corresponding prolyl hydroxylases (PHD-1, -2, or -3), acquiring a hydroxyl group. This makes possible the binding of von Hippel-Lindau factor (VHL), which is a signal for ubiquitination and subsequent proteasomal degradation. Thus, when oxygen is in sufficient supply, degradation of the α-subunit occurs, and PHD acts as an oxygen sensor [23]. Among most HIF-3α variants, only one proline can be hydroxylated in ODD domains, making them less susceptible to PHD-mediated degradation [34,35]. Later, another oxygen sensor factor inhibiting HIF (FIH) was discovered, which hydroxylates an asparagine residue in the transactivation domain of the HIF-α subunit, thereby blocking the binding of the coactivator proteins p300 and the cAMP response element-binding protein (CREB) [36]. HIF-1α and HIF-2α contain asparagine residues that are hydroxylated by FIH in the C-terminal transactivation domain (C-TAD). As for HIF-3α, it does not include C-TAD [33,34]. Under hypoxia, hydroxylation slows down because PHD and FIH require oxygen for their activity. Therefore, the α-subunit accumulates and translocates to the nucleus. There, it dimerizes with the β-subunit to bind to the coactivator proteins p300 and CBP [23]. Then, binding to hypoxia responsive element (HRE) and activation of transcription of the target genes occurs. Most HRE sites can be recognized by both HIF-1 and HIF-2, indicating that they share common target genes. However, the differences between the N-TAD domains of HIF-1α and HIF-2α support the selective binding of HREs, providing the specificity of target genes for each HIF. Distinct yet very similar HRE sequences were identified for these two HIFs. All these genes showed no difference in their distribution patterns of HIF-1 and HIF-2 HREs. This supports the view that HIF-1/HIF-2 common targets are recognized in a non-competitive and non-compensatory manner in the same regions. A specific motive for HIF-3 has not been proposed to date. Genes with promoter regions enriched in HIF-1 motifs were induced by acute hypoxia and maintained their activity during prolonged hypoxic exposure. Genes with promoter regions enriched in HIF-2 motifs are activated only by prolonged hypoxia. Thus, HIF-1 and HIF-2 do not appear to compete for the same HREs [33,37]. Some of the HIF-targeted genes encoding matrisome-related molecules are presented in Fig. 1.
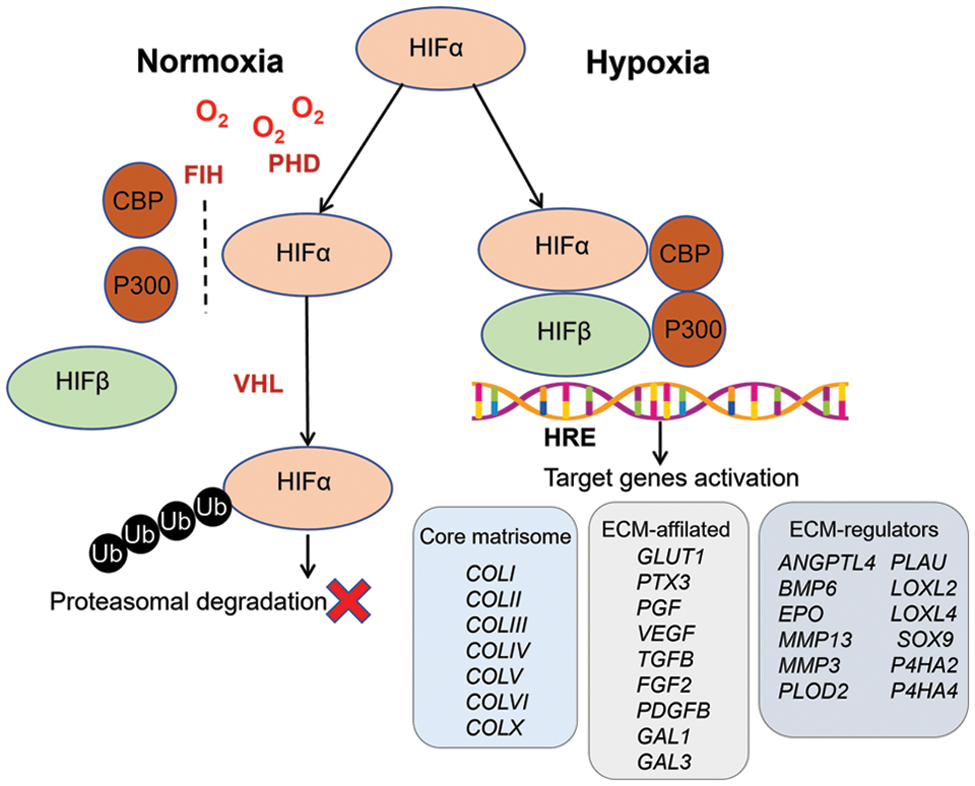
Figure 1: Hypoxia-inducible factors (HIFs) in the regulation of cellular response to hypoxia. HIFs are heterodimeric proteins consisting of oxygen-sensitive HIF-1α, HIF-2α, or HIF-3α subunits and a constitutively expressed HIF-1β subunit. Under normoxic conditions, the α-subunit undergoes hydroxylation by the corresponding prolyl hydroxylases (PHD), binding by von Hippel-Lindau factor (VHL), and subsequent proteasomal degradation. Factor inhibiting HIF (FIH) hydroxylates the C-terminal transactivation domain (C-TAD), blocking the binding of the coactivator proteins p300 and the CREB-binding protein (CBP, where CREB stands for cAMP response element-binding protein). Under hypoxia, hydroxylation slows down because PHD and FIH require oxygen for their activity. Therefore, the α-subunit accumulates and translocates to the nucleus. There it dimerizes with the β-subunit and binds to the coactivator proteins p300 and CBP. Then, this complex binds to hypoxia responsive element (HRE) and consequently, the activation of transcription of the target genes occurs, among other genes encoding matrisome-related molecules.
In addition, the activity of other cue players in the canonical hypoxic pathway may indirectly affect HIFs. For example, prolyl hydroxylase activity may be regulated by several intracellular factors, including reactive oxygen species (ROS) [38], which are in turn negatively modulated by mitochondrial deacetylase sirtuin-3 (SIRT3) [39]. The M2 isoform of pyruvate kinase (PKM2) acts as a coactivator of HIF-1α and HIF-2α. PKM2 expression is known to be induced by HIF-1α. PKM2 directly binds to HIF-1α and HIF-2α, enhances their binding to HREs, and promotes the transactivation of hypoxia-inducible genes by HIF-1 and HIF-2 [40].
Numerous microRNAs participate in HIF regulation and can either repress or activate expression affecting other HIF isoforms or regulatory genes. For instance, hypoxia-induced hsa-miR-147 has been shown to suppress the HIF3A transcript, thus supporting HIF-1 activity in human cervical cancer cells (HeLa) [41]. In turn, direct binding of HIF-3α with miR-630 promoter induces miR-630 expression. A positive feedback loop has been found between HIF-3α-mediated overexpression of miR-630 and elevation of HIF-3α, while the expression of HIF-1α was suppressed by miR-630 [42].
The contribution of hypoxia-inducible factor-1 and -2 in the regulation of physiological processes under hypoxia
Several genes essential for the adaptation and survival of cells at low-oxygen environments are targeted by the HIF factors [23]. More than a thousand HIF-dependent genes have integral roles in a variety of biological processes, including angiogenesis [43–45], cellular metabolism [43,46,47], cell signaling [48,49], cell cycle regulation [50,51], cellular response to damage [44,49,52], stemness [53,54], and tumor progression [52,54,55]. In particular, HIF controls the transcription of vascular endothelial growth factor (VEGF)—the main driver of tissue vascularization. This achieves increased supply as a result of the newly-developed blood vessels [43]. HIF-1 regulates the expression of genes encoding stromal cell-derived factor 1 (SDF-1) [56,57], integrin a4 [10], and matrix metalloproteinases (MMP) [58,59], all of which have had an impact on cell migration. HIF-1α has been shown to be associated with microRNAs that regulate signaling pathways under hypoxia, for instance, with miR-210, which supports the survival and proliferation of stromal progenitors under reduced oxygen levels [60,61]. HIF-1α is also involved in mediating the effects of hypoxia on differentiation [62,63].
HIF-1 and HIF-2 are thought to have overlapping roles, but it cannot be claimed that they are completely redundant [64]. The variations exist in the target genes. The genes encoding heme oxygenase-1, phosphofructokinase, lactate dehydrogenase, and phosphoglycerate kinase were targeted by HIF-1 only [65,66]. Conversely, HIF-2 but not HIF-1, controlled the expression of stemness genes such as those for octamer binding transcription factor (OCT)-)-3/4 and matrix metalloproteinases 2 and 3 [67,68]. Only HIF-2 has been shown to regulate the solute carrier family 11 member 2 gene (SLC11A2), which encodes the iron transporter protein of the divalent metal transporter 1 (DMT1) [69]. The contribution of HIF-1 and HIF-2 in oxygen sensing is uneven: HIF-1 is the more important for survival, as its knockout leads to developmental arrest and lethality [70], whereas HIF-2 knockout mice survived despite the severe defects of vascularization [71]. HIF-1 is a regulator of the response to hypoxia in all eukaryotic cells, whereas HIF-2 is only present in vertebrates. Although HIF-2 is present in many tissues during embryonic development, its expression is less prominent in adult cells, and it has become more specific for well-vascularized tissues, for example, kidney, placenta, and lungs [72,73]. It is considered that erythropoietin (EPO)-a master regulator of erythropoiesis, is controlled by HIFs. Some papers have described HIF-1 involvement in EPO upregulation [23,74]. Meanwhile, there are data indicating that HIF-2 participates in EPO upregulation as well. In the murine liver, EPO has been found to be upregulated by HIF-2, but not by HIF-1 [75]. Studies in mice with postnatal deletion of HIF2A and in Hep3B cells using siRNA have shown that HIF-2, but not HIF-1, plays a key role in the regulation of EPO [76]. Another study reported that it could be regulated by not only HIF-1 and HIF-2, but also by HIF-3 [77].
Other transcription factors may also contribute to HIF-1/2-mediated stimulation of target gene activity under hypoxia. These factors differ depending on the isoform. For instance, signal transducer and activator of transcription 3 (STAT3) binds to promoters of HIF-1 target genes, while upstream transcription factor 2 preferentially binds to promoters of HIF-2 target genes. Transcription of the target gene is then triggered by the formation of complexes with the corresponding alpha subunit [78].
There are some time-course differences in the expression of these factors. For example, the expression of HIF1A is transient and decreases after a few hours of activity. The transcription of HIF2A is more prolonged [79–81].
As mentioned above, of all the factors of the HIF family, the third isoform, HIF-3, is by far the least studied. Many questions still need to be answered about its influence on the regulation of biological processes in the cell, its involvement in pathological processes in the body, and its interaction with the first two isoforms.
The structure of hypoxia-inducible factor-3 and its role in the regulation of the cellular response to hypoxia
The HIF-3α subunit was first described as a novel α-class hypoxia-inducible factor in mice [82]. This isoform was later found in humans [83]. Human HIF-3α is located on chromosome 19q13.13 13.2 (12), which differs from the location of HIF-1α (14q21 24) and HIF-2α (2p16 21) [84]. The structure of HIF-3α differs from the a-subunits of the other two HIFs. For example, HIF-3α contains a leucine zipper (LZIP) that allows for protein-protein interactions. In addition, it has a transactivation domain (TAD) site at its N-terminus only, whereas HIF-1α/2α has these sites on both termini (N and C), and has many alternative splice variants (10 in humans) (Fig. 2). The long splice variants include the entire set of domains, although sometimes with the exception of LZIP. The short splice variants may lack one or more domains.
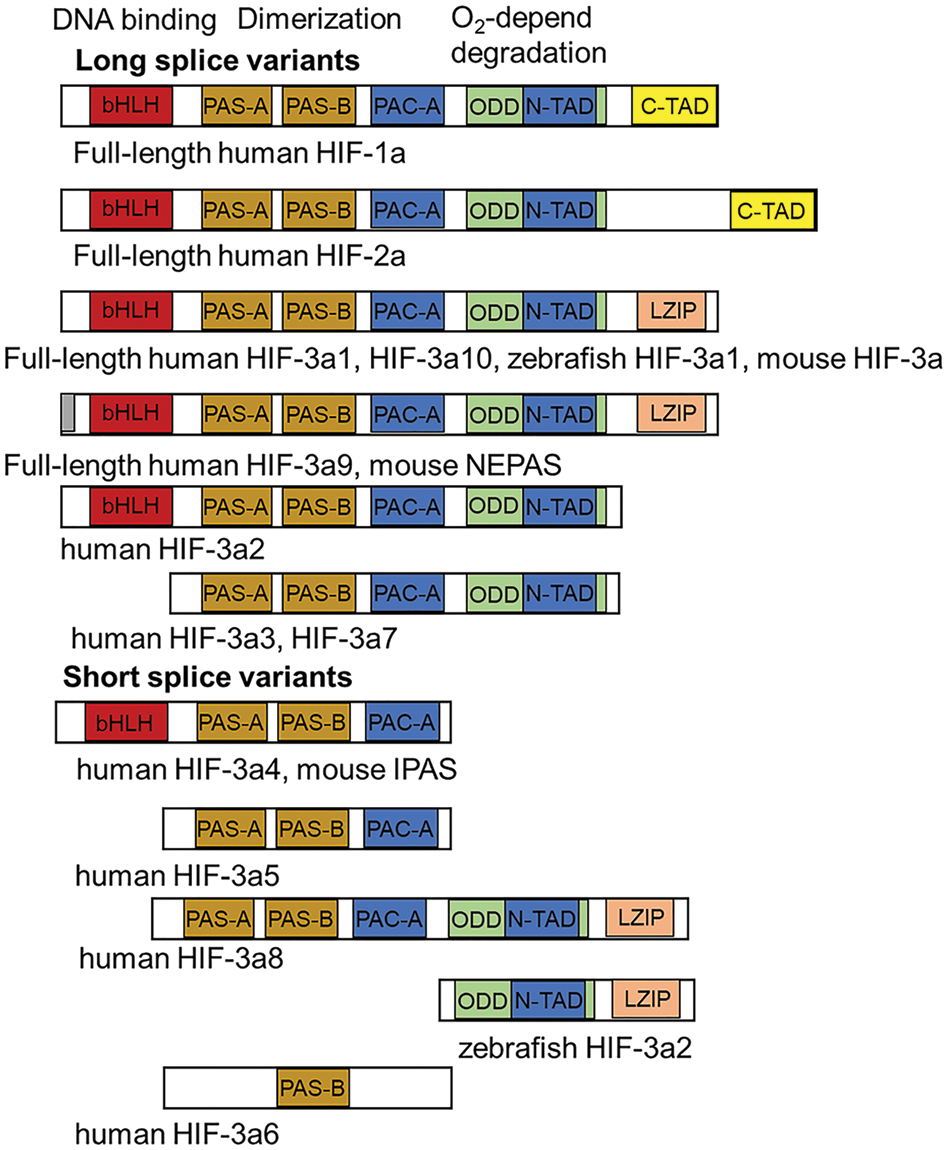
Figure 2: The structure of hypoxia-inducible factors (HIFs). HIF-1/2 isoforms include a basic-helix-loop-helix (bHLH) located at the N-terminus, which is responsible for DNA binding; PAS (Per/Arnt/Sim)-associated COOH-terminal domain (PAC-A). The PAS-A and PAS-B regions dimerize with the bHLH region of the HIF β-subunit (ARNT). Next, the oxygen-dependent degradation domain (ODD) is responsible for oxygen-dependent degradation and transactivation of domain (TAD) at the N-terminus and C-terminus [83]. Long splice variants HIF-3α include a bHLH located at the N-terminus, PAC-A, PAS-A, and PAS-B. Next, the ODD domain and TAD are at the N-terminus [34,83,84]. The C-terminus contains the leucine zipper (LZIP). Both the LZIP and TAD are involved in protein-protein interactions. Among the long splice variants, there are the full-length human HIF-3α1 and the HIF-3α10, the zebrafish HIF-3α1, and the mouse HIF-3α isoforms. Human HIF-3α10 was recently discovered and differs from HIF-3α1 by the presence of a 210-bp intron 1 [26]. Full-length human HIF-3α9 and mouse neonatal embryonic PAS protein (NEPAS) have the same structure as the splice variants described above, except for a difference in some amino acids at the N-terminus. The splice variant human HIF-3α2 lacks the LZIP domain, while human HIF-3α3 and HIF-3α7 also lack the bHLH domain. The short splice variants of human HIF-3α4 and mouse inhibitory PAS domain protein (IPAS) do not include LZIP, ODD and TAD. Human HIF-3α5 lacks bHLH in addition to those mentioned, while human HIF-3α8 lacks only bHLH compared with the full-length variant. There are even shorter splice variants: zebrafish HIF-3α2 carrying only the ODD, TAD, and LZIP domains, and human HIF-3α6 with a PAS-B domain [34].
The presence/absence of certain domains in HIF-3α molecules appears to determine their functions. For example, short splice variants lacking the basic-helix-loop-helix (bHLH), Per/Arnt/Sim (PAS), and PAS-associated COOH-terminal (PAC) domains that are responsible for oxygen sensing and dimerization with the β-subunit, are independent of oxygen levels and therefore perform different functions. Furthermore, various splice variants are often differentially expressed and regulated depending on the tissues concern, the developmental stages, and other factors [26,83]. Certain splice variants have been well studied, while data on some others, mostly the short splice variants, are rather scarce. Moreover, not all studies dealing with the HIF-3α target genes and their biological effects aimed to identify the specific splice variants involved.
Multiple splice variants are not unique to HIF-3α. HIF-1α and HIF-2α also undergo alternative splicing. According to the National Center for Biotechnology Information (NCBI) and Ensemble databases, there may be 12 different variants of the HIF-1α and HIF-2α transcripts, of which the short variants are also poorly studied [35]. Furthermore, the NCBI database indicates there are 19 splice variants predicted for human HIF-3α, 10 of which have been experimentally confirmed.
Pasanen et al. investigated the expression of HIF-3α splice variants in various adult tissues and cell lines. They could not detect HIF-3α3, HIF-3α5, and HIF-3α6. The HIF-3α1, 2, 8, and 9 variants were expressed in all tissues except the placenta. They detected HIF-3α10 in the heart, brain, skeletal muscle, kidney, and pancreas tissues. HIF-3α4 was expressed in the spleen and brain, while HIF-3α7 was detected in the heart, placenta, lung, liver, and skeletal muscles [26]. A similar distribution pattern of the HIF-3α splice variant in tissues has been demonstrated in adult mice [82]. In addition, the expression of inhibitory PAS domain protein (IPAS), which is thought to be a negative regulator of hypoxia-inducible genes, has been detected in the retina and Purkinje cells [85]. Neonatal embryonic PAS protein (NEPAS), associated with embryonic and neonatal development, is highly expressed in embryonic heart, lung, brain, kidney, and liver tissues [24].
In other vertebrates, zebrafish, HIF-3α1 expression has been described in adult ovaries, kidneys, testes, and gills. However, in other tissues, high levels of HIF-3α1 were detected only during embryogenesis [24]. Zebrafish HIF-3α2 is lacking the bHLH, PAS, and PAC domains. It is expressed in both embryonic and adult tissues and could inhibit canonical Wnt through b-catenin binding [86].
HIF-3α is thought to be less involved in carcinogenesis than HIF-1α. Panasen et al. have tested a significant number of tumor cell lines with HIF-1α expression, but only a few have demonstrated the presence of HIF-3α splice variants [26].
Thus, the data from vertebrate studies show a large variation in the distribution of HIF-3α splice variant expression depending on the tissue and developmental stage concerned. HIF-3α differs significantly from the HIF-1α/HIF-2α isoforms in its structure and expression in tissues. The regulatory activity of HIF-3α and its involvement in cell adaptation to the microenvironment have been studied in much less detail, but a number of fundamental differences from the first two HIFs have been revealed.
Similarly to HIF-1 and HIF-2, HIF-3 directly regulates transcription by binding to HREs at promoter sites of target genes. For instance, under 1% hypoxia, chromatin immunoprecipitation analysis has detected HIF-3α2 binding associated with canonical HRE in the promoter regions of EPO and ANGPTL4 in Hep3B hepatoma cells [77] and in the promoter regions of REDD1, MLP3C, and SQRDL in zebrafish embryos provoking the upregulation of above genes (Fig. 3). In addition, HIFs can recruit other factors, including signaling pathway regulators, to improve HIF binding and further transcription. HIF-3α has been found to be a stimulator of osteosarcoma proliferation through lysine demethylase 3A (KDM3A)-dependent demethylation of the SOX9 promoter. Under hypoxic conditions, KDM3A expression was upregulated by HIF-3α, and KDM3A occupied the SRY box transcription factor 9 (SOX9) gene promoter region by H3 lysine 9 demethylation (H3K9me2) [87]. Overexpression of HIF3A in pulmonary artery smooth muscle cells was followed by elevated expression of phosphatidylinositol 3 kinase (PI3K) and protein kinase B (Akt) proteins associated with cell migration, survival, and inactivation of apoptosis [88,89]. In transgenic mice, full-length human HIF-3α bound a conserved HRE site in the Sox2 promoter and weakly transactivated a reporter construct containing the Sox2 promoter region. In this way, HIF-3α significantly contributed to mouse lung development [90]. An increase in the phosphorylation of HIF‑3α-downstream signal transducer Janus kinase and activator of transcription (Jak/STAT) was demonstrated in cytotrophoblast [91]. HIF‑3α regulates cell proliferation and cell apoptosis in extravillous cytotrophoblast, promoting the Fms like tyrosine kinase receptor‑1 expression and phosphorylation levels of Jak2 and STAT3 (Fig. 3).
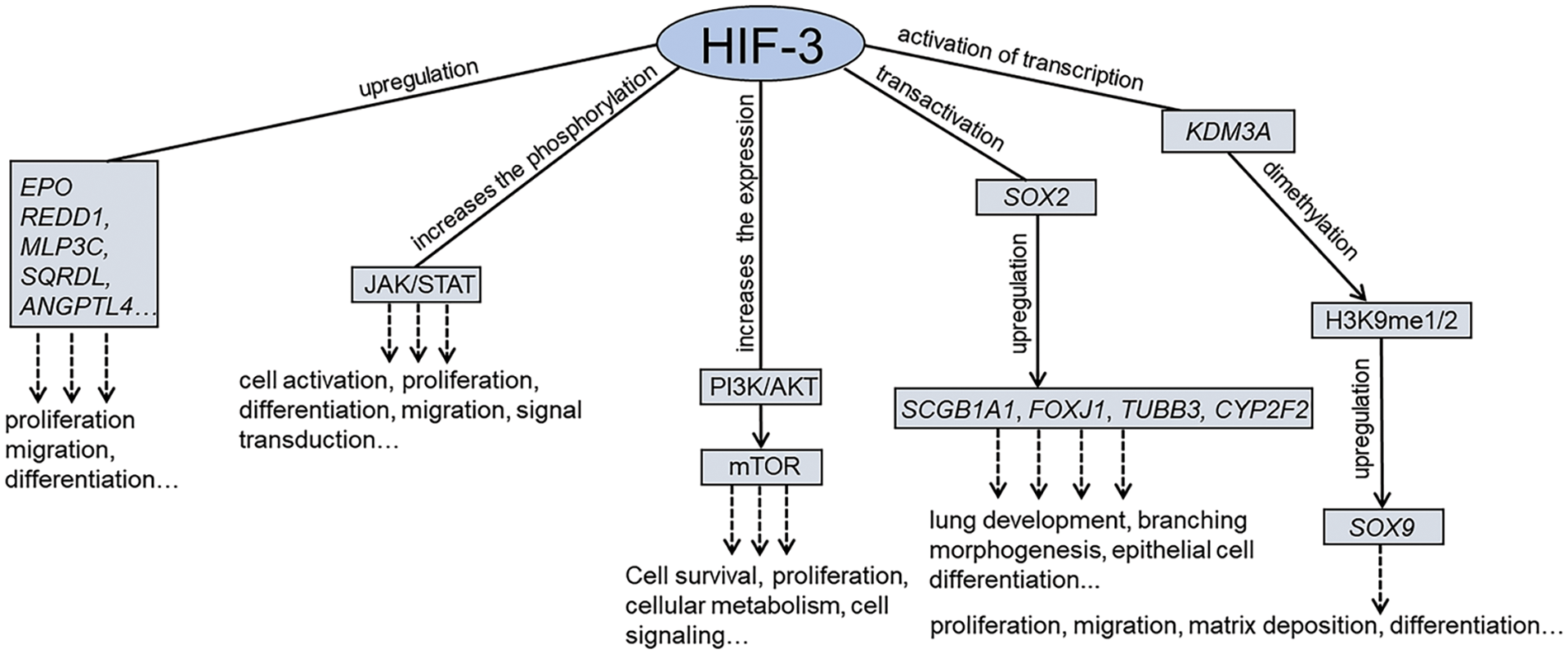
Figure 3: The molecular mechanisms regulated by hypoxia-inducible factors-3 (HIF-3). Similarly to HIF-1 and HIF-2, HIF-3 directly regulates transcription by binding to hypoxia-responsive elements (HREs) at promoter sites of target genes. In addition, HIFs can recruit other factors, including signaling pathway regulators, to improve HIF binding and further transcription.
The target genes of HIF-3 may differ significantly from those of HIF-1 and HIF-2. For example, in rat hepatocytes, HIF3A overexpression caused an increase in the expression of insulin-like growth factor (IGF) binding protein 1 (IGFBP-1), whereas HIF1A upregulation had no such effect [92]. HIF-3α knockdown experiments in zebrafish have shown that upregulation of the insulin-like growth factor binding protein 1 (IGFBP-1B), sulfide quinone reductase-like (SQRDL), microcin LB (MCLB), and zona pellucida glycoprotein 3d tandem duplicate 2 (ZP3V2) genes was induced predominantly by the splice variant HIF-3α1 [93]. Whole genome analysis of zebrafish embryos with stable production of HIF-1α and HIF-3α revealed 690 HIF-1α-dependent upregulated genes. Among these, only 155 genes were upregulated by HIF-3α. In addition, there were only 97 overlapping genes, suggesting the existence of genes regulated by only one of the HIF isoforms. For example, only HIF-3α regulated genes encoding molecules of Janus kinase-STAT and the nucleotide oligomerization domain (NOD)-like receptor signaling pathways, whereas HIF-1α mediated both VEGF and mitogen-activated protein kinase (MAPK) signaling [93].
HIF-1α target genes can be inhibited by short splice variants of HIF-3α. In particular, HIF-3α4 has been shown to suppress the HIF-1α-mediated gene encoding glucose transporter 1 (GLUT1) and VEGF expression in human cancer cells [94]. Similar data were obtained in endothelial and vascular smooth muscle cells when the HIF-3α2 splice variant suppressed the expression of HIF-1-dependent genes encoding VEGF-α and Enolase2 [27]. In addition, the activity of HIF-1α or HIF-2α can be inhibited directly by some splice variants of HIF-3α. For example, in mice, NEPAS binds competitively to the HIF-1β subunit, thereby limiting its availability for HIF-1α and HIF-2α [24]. Direct binding of long human HIF-3α or IPAS to HIF-1α and HIF-2α has been shown to prevent the translocation of the latter into the nucleus [25,85,94]. HIF-3α1 has been shown to downregulate HIF-1 and HIF-2-mediated genes in renal cancer cells. This was due to the competition of HIF-3α with the other isoforms for binding to HIF-1β. As a result, the levels of HIF-1α and HIF-2α decreased, and the transcription of the HIF-1 and HIF-2 target genes was downregulated [83]. Using live-imaging, Kim et al. demonstrated that HIF-3α2/HIF-3α4 splice variants function as negative regulators on the heterodimer formation of HIF-2α with HIF-1β in human prostate cancer cells [95].
The HIF-3α-dependent inhibition of other isoforms can be suggested to not be due to their antagonism but is a step in the adaptation to prolonged hypoxic exposure. Indeed, HIF-α isoforms have been shown to have different time points of activation and time courses of expression. In our laboratory, after exposure of human multipotent mesenchymal stromal cells (MSCs) under 5% O2, HIF1A upregulation was detected for up to 4 h and HIF2A–upregulation – for up to 12 h. HIF3A was upregulated between 12 and 24 h [79]. In lung epithelial cells at 1% O2, HIF1A expression was increased after 30 min but decreased between 8 and 16 h, while HIF3A transcript levels peaked at 8–16 h [96]. In human endothelial cells under 1% O2, HIF1A upregulation was seen up to 4 h during hypoxic exposure, while HIF3A was upregulated after 4 h, with transcription reaching a maximum after 48 h of exposure [97]. In human bladder smooth muscle cells, the upregulation of HIF1A and HIF2A was detected after 2 h of hypoxia but then rapidly declined, while HIF3A was upregulated after 48 h, and remained at high levels for up to 72 h [98].
Both in human cells in vitro and in rodents in vivo, it has been shown that HIF-1α and HIF-2α alone, or together, can stimulate HIF-3α expression [26,27,85,99]. These data suggested a transition of expression activity from transient HIF1A and HIF2A to the prolonged overexpression of HIF3A [26,80]. This mechanism may be critical for cell survival under prolonged hypoxia. Thus, HIF-1 responds first under hypoxia and then “turns on” HIF3A expression in the later steps of adaptation. Bartoszewska et al. have identified that microRNA miR-429 plays a major role in HIF-1a-dependent control of HIF3A expression [80]. In particular, in human endothelial cells, acute hypoxia has been characterized by the accumulation of HIF-1α, while HIF-2α and HIF-3α have been accumulated in the chronic state [100,101].
The oxygen levels required to maintain high expression of O2-inducible HIF-1α and HIF-3α may differ. In lung epithelial cells, HIF3A was maximally upregulated at 3% O2, whereas HIF1A was maximally upregulated at 1% O2 [96]. In addition, HIF-3α responds to hypoxia (or its mimetic CoCl2) and also to inflammatory cytokines and some growth factors. In MSCs, interleukin 6 (IL-6), tumor necrosis factor (TNF-α), monocyte chemoattractant protein-1 (MCP-1), epidermal growth factor (EGF), and VEGF stimulate HIF-3α both under normoxic and hypoxic conditions, while HIF-1α expression was increased only under hypoxia [102].
In conclusion, HIF-3α differs from the other HIF isoforms in its time course and duration of expression, target genes, and tissue specificity. This may be important for adaptation to prolonged hypoxia and the inflammatory microenvironment that often accompanies hypoxia.
The structural and functional compartments of matrisome under hypoxia
Reduced O2 levels impact many processes in cells and, as a consequence, the properties of secreted ECM and ECM-associated mediators. The effects of hypoxia on the matrisome are mainly studied in the context of the tumor microenvironment and ischemic tissues. Changes in matrix stiffness, ECM deposition, and degradation, upon exposure to hypoxia, are largely mediated by HIF factors, as have been detected in different in vitro studies.
It has been clearly demonstrated that resident fibroblasts from different tissues deposit increased levels of collagen I under hypoxic conditions [103–105]. The growth factors, such as the transforming growth factor (TGF-β), actively participate in ECM deposition. In fibroblasts, HIF-1α and HIF-2α have been shown to be involved in TGF-β pathway activation [106,107]. HIF-1α also participates in regulating transcription of the prolyl 4-hydroxylase subunit alpha 1 (P4HA1) and alpha 2 (P4HA2) genes encoding prolyl hydroxylase expression in fibroblasts. When cultured at hypoxia, human corneal fibroblasts have been shown to enhance the deposition of several matrix molecules, including collagens I, V, and VI, plus fibronectin [12]. Pleomorphic adenoma SM-AP1 and SM-AP4 cell lines showed increased production of perlecan and fibronectin under hypoxia. Under normobaric hypoxia in vivo, the deposition of collagens I, III, and IV, plus fibronectin in rat lungs and of fibronectin, fibulins, and aggrecan in mouse vascular endothelial cells have also been demonstrated [108,109].
After 48 h exposure at 1% O2, proteomic profiling of MSCs revealed decreased levels of collagen molecules (COL1a2, COL1a1, and COL3a1) and of ECM organization-related proteins including fibulin 2 (FBLN2), fibulin 1 (FBN1), COL14a2, laminin alpha 5 (LAMA5), ECM protein 1 (ECM1), and of secreted protein acidic and cysteine-rich (SPARC). Reactome annotation of the proteins highlighted pathways related to glycosaminoglycan metabolism, the organization of ECM, including elastin fibers, and the degradation of chondroitin sulfate or dermatan sulfate [110]. Mass spectroscopic analysis of conditioned medium from MSCs after 24 h at 1% O2 showed an increased accumulation of proteins involved in the tensile strength, three-dimensional folding, and mechanical stability of collagen fibrils [111].
After hypoxic exposure, the alteration of post-translational modification of collagens that resulted in enhanced alignment and stiffness of ECM fibers has been described [13,112,113]. Increased expression of prolyl 4-hydroxylase subunit alpha 1 (P4HA1) and procollagen-lysine,2-oxoglutarate 5-dioxygenase l (PLOD1), which are key enzymes for intracellular modification of procollagen molecules, has been reported in various cell types, including tumor and stromal cells [13,114,115]. In addition, hypoxia-dependent extracellular modification of fibrillary collagens through increase the production of lysyl oxidase (LOX) and the lysyl oxidase-like enzymes (LOXL-2 and LOXL-4) ensure the formation of cross-linking between collagenous fibers [13,116]. These thin, aligned fibrils promote cell migration. Furthermore, the upregulation of genes encoding the MMP-2 and MMP-9, as well as of urokinase-type plasminogen activator (PLAU) and urokinase-type plasminogen activator receptor (UPAR), has been described [117–119].
These results demonstrate significant modulation of all matrisome compartments under hypoxic exposure. Our next step will be the analysis of the data with respect to the involvement of hypoxia-inducible factors in these changes.
Hypoxia-inducible factor-1 and -2-mediated regulation of the matrisome
Extracellular matrix deposition and stiffness
Silencing of HIF-1α transcription with siRNA confirmed that core matrix proteins are indeed upregulated by HIF-1α [120]. HIF-1α also participates in regulating P4HA1 and P4HA2, thereby controlling prolyl hydroxylase expression in fibroblasts. The prolyl hydroxylation of collagen leads to its stabilization and prevents its degradation, which in turn promotes its deposition. The lysyl hydroxylases involved in this process are encoded by the PLOD1, PLOD2, and PLOD3 [121,122]; PLOD2 is regulated by HIF-1α. As a result of PLOD2 activation, procollagen α-chains form a triple helix and are secreted into the ECM, due to the hydroxylation of proline and lysine residues [123,124]. LOXs mediate further cross-linking between collagens and elastin, increasing the stiffness of the matrix [125]. As HIF-1α targets, the LOXL2 and LOXL4 genes are upregulated by hypoxia [116,126].
Degradation of extracellular matrix
In addition to increasing stiffness through collagen modifications and cross-linking, hypoxia also affects ECM degradation. This process occurs through the action of proteinases, particularly the MMP family, which makes a significant contribution [127]. MMPs, a family of zinc-dependent enzymes that cleave ECM components, are classified into several types, including gelatinases, stromelysins, collagenases, and cell membrane-associated MMPs, each with specificity for different substrates [7].
Under hypoxia, the upregulation of MMP14, which encodes enzyme cleaves collagen types I, II, and III, has been demonstrated. This gene was shown to be a target for both HIF-1 and HIF-2 [128,129]. MMP2 and MMP9, whose encoding enzymes degrade type IV collagen, were also upregulated under hypoxia. They were targeted by HIF-1 only [117,130]. MMP3 expression was increased in rheumatoid arthritis synoviocytes transfected with a plasmid that expresses HIF-1α. Here, MMP-3 expression was suppressed upon siRNA-mediated silencing the HIF-1α [131]. Hypoxia can upregulate the PLAUR gene in breast cancer cells and the intestinal epithelium. This regulation is mediated by the HIF-1α [118]. The activation of the corresponding receptor produced an effect similar to that of MMP, resulting in the degradation of several ECM components, including the basal membrane [7].
A number of ECM-associated molecules and deposited soluble factors have been found to be targets of HIF-1 or HIF-2. For transforming growth factor (TGF-β), HIF-1α and HIF-2α involvement in TGF-β pathway activation has been demonstrated in fibroblasts and human choriocarcinoma cells cultured under hypoxic conditions [103,132]. HIF-1α-dependent induction of basic fibroblast growth factor (bFGF) under hypoxia has been shown in human endothelial cells [133], as well as induction of bFGF and platelet-derived growth factor-BB (PDGF-BB) in breast cancer cells [134]. VEGF, one of the growth factors that is deposited in the ECM, is a well-known target of HIF-1 and is significantly upregulated under hypoxia [135,136]. Galectins are matrix-associated molecules with immunomodulatory properties. They can also be induced by HIF-1α. Under hypoxia, there are data on the beneficial effects of HIFs on the expression of galectin 1 in human cardiomyocytes and colorectal cancer cells [137,138] and of galectin 3–in human pancreatic cells and canine mammary tumors [139,140].
The cited data indicate that matrisome changes under hypoxia have implications for the physiological state of tissues. Thus, HIF-1-mediated deposition of collagen I and the increased stiffness of the ECM are the main processes leading to fibrosis [7]. Overexpression of HIF-1α induces the progression of fibrotic alterations in primary mouse tubular epithelial cells and in the development of adipose tissue fibrosis in transgenic mice [15,16]. By contrast, HIF-1α-deficient mice showed reduced liver fibrosis [17]. Increased stiffness and concomitant degradation of the ECM contribute significantly to developing osteoarthritis (OA) [141]. Excessive deposition of ECM by tumor-activated fibroblasts and an increase in ECM stiffness, as well as HIF-1α-mediated increase in the cell surface expression of integrins [109,142], facilitate cell migration [7]. In part, this can be related to HIF-1-dependent PLOD2 activity. Blocking PLOD2 decreased the ability of cells to migrate and reduced lung metastasis in mouse models of sarcoma [115]. Thus, numerous data sets have confirmed that both HIF-1 and HIF-2 regulate the expression of genes encoding proteins in all the compartments of the matrisome in a similar manner.
Hypoxia-inducible factor-3-mediated regulation of the matrisome proteins
Based on the available research, the HIF-1- and HIF-2-dependent changes of the matrisome could be reasonably considered as acute response to hypoxic stress. Their expression was upregulated shortly after the O2 deprivation and was sustained for only a relatively short period (a few hours). The involvement of HIF-3 as a long-lasting transcription factor in the prolonged adaptation of cells to moderate hypoxia is of significant interest. However, the data concerning this are scarce and quite contradictory.
As already mentioned, HIF1A and HIF2A were transiently upregulated in cultured human bladder smooth muscle cells after 2 h of exposure to hypoxia, while HIF3A was upregulated after 72 h [98]. The protein levels of HIF-1α and HIF-3α were significantly elevated after 2 and 72 h, respectively. Consistent with these changes, upregulation of genes encoding matrisome components and an increase in the levels of these molecules were observed. The study found that genes encoding collagens I, II, III, and IV plus fibronectin, aggrecan, connective tissue growth factor (CTGF), similar to mothers against decapentaplegic mad (SMAD) 2, and SMAD 3, as well as metallopeptidase inhibitor 1 (TIMP-1) were all upregulated. Additionally, there was a time-dependent increase in total collagenous proteins. The inflammatory cytokines IL-1β, IL-6, TNF-α (mRNA) and TGF-β (protein and mRNA) showed a time-dependent increase in expression, while the anti-inflammatory IL-10 decreased after 72 h. Furthermore, VEGF was upregulated. The signs of smooth muscle cell dedifferentiation, such as an increase in α-smooth muscle actin (αSMA), vimentin, and desmin, could also indicate an increase in the matrix-producing activity of the cells. Therefore, a positive correlation was found in the time-course dynamics of the expression of matrisome proteins, inflammatory cytokines, and HIF-3α expression. The authors hypothesized that HIF-3α may stimulate matrix deposition and subsequent fibrotic changes in the bladder wall [98,143].
However, these studies only show a positive correlation between HIF-3 and the matrisome proteins. They do not provide evidence that HIF-3 regulates the gene expression of these proteins. Li et al. examined the involvement of HIF-3α in ECM production in cultured human chondrocytes from healthy and OA samples by implementing the HIF3A knockdown with miR 210 [141]. The development of OA is directly related to changes in the ECM, these being manifested by increased collagen deposition and cartilage degeneration. In cultured OA chondrocytes, the expression of cartilage-specific collagen type II alpha 1 chain (COL2A1) was downregulated, while the expression of fibrosis-associated collagen type X alpha 1 chain (COL10A1) and MMP13 were upregulated compared to chondrocytes from healthy tissue. These data suggest increased cartilage degradation in OA. MiR-210-mediated knockdown of HIF3A resulted in enhanced proliferation of OA chondrocytes and the upregulation of chondrocyte-specific COL2A1, whereas the expression of COL10A1 and MMP13 mRNA was downregulated. This indicates the involvement of HIF-3 in the pathogenesis of OA through its effects on ECM components [141].
However, the inverse role of HIF-3α in the development of OA has been demonstrated in rat lipopolysaccharide (LPS)-induced OA. In chondrocytes, HIF-3α was shown to be involved in the inhibition of inflammation and matrix degradation via the FOXO1/PRG4/HIF-3α axis. LPS decreased the expression of HIF-3α, the level of forkhead box O1 (FOXO1) and proteoglycan 4 (PRG4), whereas uncu-51 like autophagy activating kinase 1, beclin, and microtubule-associated protein 1 light chain 3 proteins, which are involved in the development of OA, increased. The secretion of inflammatory IL-1β and TNF-α was also stimulated. Upregulation of HIF3A led to degradation of the ECM, increased production of MMP13, of a disintegrin, of metalloproteinase with thrombospondin motifs (ADAMTS5) and collagen type II but decreased the IL-1β and TNF-α levels. Thus, this study concluded that HIF-3α contributes to the degradation of excessively deposited ECM in OA, stimulates chondro-specific collagen II, but reduces the levels of inflammatory cytokines [144]. Comparing their results with those of Li et al. [141], the authors hypothesized that the disparate effects may be due to the use of different experimental models and the fact that in rodents, the mechanisms involving HIF-3α may work differently than in humans.
In the mouse model, upregulation of HIF3A resulted in the downregulation of VEGF, COL101A1 and MMP13 [145]. However, these findings are not consistent with data on collagen I expression in human cells [141] and the upregulation of MMP13 in rats [144]. Therefore, the effects of HIF-3α on the matrisome are not entirely clear, and the explanation for the differences is likely more complex than just species-dependent mechanisms.
The data on the HIF-3α effect on the expression of certain proteins of the matrisome or factors deposited therein are summarized in Table 1.
In Hep3B cells, overexpression of the splice variant HIF-3α2 led to upregulation of genes encoding bone morphogenetic protein 6 (BMP6), thereby enhancing deposition and mineralization of the ECM [150] and overexpression of EPO and placental growth factor (PGF). These growth factors are known to induce the secretion of matrix metalloproteinases [151,152], angiopoietin-like protein (ANGPTL) involved in matrix degradation and inflammation [153], and pentraxin 3 (PTX3), which has anti-inflammatory effects and provides interconnection between matrix structural components [154]. Silencing of HIF-3a with siRNA downregulated the corresponding genes [77]. The relationship between HIF-3α and the expression of αSMA, a marker of myofibroblasts, was investigated by [155]. Myofibroblasts respond to TGF-β with extra collagen production, leading to fibrotic changes. The authors concluded that HIF-3α could potentially be a negative regulator of αSMA expression, thereby attenuating ECM production [155]. Blocking HIF-3α with miR-210 was shown to upregulate collagen type II (COL2) and SRY-related HMG box 9 (SOX9), whose encoded proteins are responsible for maintaining both matrix structure and matrix deposition [146]. Conversely, miR-210 inhibitors reduced ECM deposition. After co-transfection of rat stromal cells with miR-210 inhibitors and HIF-3α siRNA or the transfection of miR-210 inhibitors, there was an increase in the expression of the COLII and SOX9 proteins in the case of co-transfection. Thus, HIF-3α negatively regulates COLII and SOX9 production and decreases ECM deposition, promoting its overall degradation [146].
In human chondrocytes, HIF-3α expression was negatively correlated with the transcription of COL10A1 and MMP13, markers of ECM hypertrophy. These data were confirmed in HIF-3α knockdown experiments, which showed increased COL10A1 and MMP13 expression [148]. In the lungs of mice exposed to cigarette smoke, the deposition of collagen I was HIF-3α-dependent [147]. In a rat model of partial bladder outlet obstruction (pBOO), a concomitant increase in the expression of HIF-3α and collagen types I and III was interpreted as an indicator of fibrosis [156]. In the cartilage of Tnni2K175del mutant mice, HIF-3α was shown to negatively regulate the expression of VEGF, this protein being an ECM-associated growth factor involved in regeneration and vasculogenesis [149,157]. By contrast, HIF-3α4 overexpression in HepG2 and HEK293A cells provoked the downregulation of VEGF and GLUT1 [94,158]. In human umbilical vein endothelial cells (HUVEC), overexpressing HIF-3α2 and HIF-3α3 resulted in significant downregulation of hypoxia-induced VEGFA. When the transcription of HIF-3α was silenced by small hairpin RNA (shRNA), the transcription of VEGFA was restored [27]. In contrast to these data, HIF3A knockdown had no effect on VEGFA transcription in several endothelial lines [100].
In zebrafish embryos, forced expression of HIF-3α by injecting its mRNA accompanied the upregulation of IGFBP1A and IGFBP1B genes for the insulin-like growth factor-binding proteins that activate bioactive matrix molecules, further inducing matrix mineralization via IGF1 [159]. As well as being involved in matrix deposition, these genes are regulated both during development and by the DNA damage response 1 (REDD1) gene, thus potentiating fibronectin production [160]. Dominant-negative inhibition and knockdown experiments resulting in the downregulation of HIF3A, confirmed the precise effect of HIF-3α on the transcription of the above-mentioned genes [93].
To summarize, HIF-3-upregulation and the downregulation of collagen types I and II have been demonstrated under hypoxia. Increased transcription and synthesis of proteins involved in maintaining and remodeling matrix structures, such as those encoded by PTX3, EPO, and PGF have also been described. In addition, the expression of HIF-3α is tissue- and organ-specific, and does not provide a ubiquitous response to hypoxia [34]. Based on the above data, the variation in HIF-3 regulation may be species-dependent. Besides, the functional difference between the splice variants of HIF-3α provides a wide field for studying HIF-3-mediated regulation of the core matrisome and matrisome-associated molecules.
The response to hypoxia is manifested by significant metabolic changes in cells and their matrisome. Hypoxia, as an important microenvironmental cue, influences both the core matrisome and regulatory and matrix-associated molecules. HIFs are pivotal to cellular responses. The HIF-1 and HIF-2 isoforms regulate the matrisome in a similar manner: increasing matrix stiffness, promoting the accumulation of profibrotic collagens, and the deposition of biologically active mediators such as VEGF, TGF-β, bFGF, and PDGF-BB, besides accelerating ECM degradation due to increased metalloproteinase activity.
The available data reflect the ambiguity of the role of HIF-3 in matrix remodeling under hypoxia. Like HIF-1 and HIF-2, HIF-3 can govern profibrotic changes in matrisomes. HIF-3 upregulates the transcription of genes, such as BMP6, COL1A1, and ANGPTL, encoding molecules involved in ECM deposition and mineralization. Suppression of the myofibroblast phenotype and enhanced expression of genes encoding inflammatory molecules have also been demonstrated under HIF-3 control. HIF-3-mediated deposition of profibrotic proteins such as collagens type I or III was demonstrated.
Meanwhile, HIF-3 mediates antifibrotic effects by enhancing tissue-specific collagen II production and preventing OA. Additionally, it upregulates genes encoding molecules that contribute to the maintenance of the tissue-specific structure of the ECM, such as PTX3, EPO, PGF, and COL2A1. These observations are intriguing, and further study could open up wide prospects for treating diseases associated with pathological matrix changes, such as OA and pBOO.
The data presented here support the more intricate mechanism of HIF-3 involvement in matrisome regulation, considering factors such as oxygen levels, splice variant activities, species, and tissue sources. Future studies will focus on the accumulation of data and the identification of the signaling pathways that are involved in this regulation. The involvement of HIF-3 in adapting to prolonged hypoxia and supporting HIF-1 and HIF-2-initiated responses to acute oxygen deprivation broadens our perspective on regulating the matrisome response to hypoxic microenvironments.
Acknowledgement: None.
Funding Statement: This research was funded by the Russian Science Foundation (Grant Number 23-15-00062).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Gornostaeva A. N., Andreeva E. R., Buravkova L. B.; draft manuscript: Gornostaeva A. N., Lobanova M. V.; original draft preparation: Gornostaeva A. N., Andreeva E. R.; writing–review and editing: Gornostaeva A. N., Buravkova L. B., Lobanova M. V., Andreeva E. R. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: The research results highlighted in this review are from publicly available papers.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present review.
References
1. Shao X, Gomez CD, Kapoor N, Considine JM, Grams C, Gao YT, et al. MatrisomeDB 2.0: 2023 updates to the ECM-protein knowledge database. Nucleic Acids Res. 2023;51(D1):D1519–30. doi:10.1093/nar/gkac1009. [Google Scholar] [PubMed] [CrossRef]
2. Buravkova L, Larina I, Andreeva E, Grigoriev A. Microgravity effects on the matrisome. Cells. 2021;10(9):2226. doi:10.3390/cells10092226. [Google Scholar] [PubMed] [CrossRef]
3. Wang T, Nanda SS, Papaefthymiou GC, Yi DK. Mechanophysical cues in extracellular matrix regulation of cell behavior. Chembiochem Eur J Chem Biol. 2020;21(9):1254–64. doi:10.1002/cbic.201900686. [Google Scholar] [PubMed] [CrossRef]
4. Tsiapalis D, de Pieri A, Spanoudes K, Sallent I, Kearns S, Kelly JL, et al. The synergistic effect of low oxygen tension and macromolecular crowding in the development of extracellular matrix-rich tendon equivalents. Biofabrication. 2020;12(2):025018. doi:10.1088/1758-5090/ab6412. [Google Scholar] [PubMed] [CrossRef]
5. Randhawa A, Dutta S, Ganguly K, Patil T, Luthfikasari R, Lim KT. Understanding cell-extracellular matrix interactions for topology-guided tissue regeneration. BIOCELL. 2023;47(4):789–808. doi:10.32604/biocell.2023.026217. [Google Scholar] [CrossRef]
6. Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11(1):5120. doi:10.1038/s41467-020-18794-x. [Google Scholar] [PubMed] [CrossRef]
7. Dekker Y, Le Dévédec SE, Danen EHJ, Liu Q. Crosstalk between hypoxia and extracellular matrix in the tumor microenvironment in breast cancer. Genes. 2022;13(9):1585. doi:10.3390/genes13091585. [Google Scholar] [PubMed] [CrossRef]
8. Nazemi M, Rainero E. Cross-talk between the tumor microenvironment, extracellular matrix, and cell metabolism in cancer. Front Oncol. 2020;10:239. doi:10.3389/fonc.2020.00239. [Google Scholar] [PubMed] [CrossRef]
9. Cowden Dahl KD, Robertson SE, Weaver VM, Simon MC. Hypoxia-inducible factor regulates αvβ3 integrin cell surface expression. Mol Biol Cell. 2005;16(4):1901–12. doi:10.1091/mbc.e04-12-1082. [Google Scholar] [PubMed] [CrossRef]
10. Choi JH, Lee YB, Jung J, Hwang SG, Oh IH, Kim GJ. Hypoxia inducible factor-1α regulates the migration of bone marrow mesenchymal stem cells via integrin α 4. Stem Cells Int. 2016;2016:7932185. doi:10.1155/2016/7932185. [Google Scholar] [PubMed] [CrossRef]
11. Khawaja AA, Chong DLW, Sahota J, Mikolasch TA, Pericleous C, Ripoll VM, et al. Identification of a novel HIF-1α-αMβ2 integrin-NET axis in fibrotic interstitial lung disease. Front Immunol. 2020;11:2190. doi:10.3389/fimmu.2020.02190. [Google Scholar] [PubMed] [CrossRef]
12. Kumar P, Satyam A, Cigognini D, Pandit A, Zeugolis DI. Low oxygen tension and macromolecular crowding accelerate extracellular matrix deposition in human corneal fibroblast culture. J Tissue Eng Regen Med. 2018;12(1):6–18. doi:10.1002/term.2283. [Google Scholar] [PubMed] [CrossRef]
13. Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: drivers of tumour metastasis. Nat Rev Cancer. 2014;14(6):430–9. doi:10.1038/nrc3726. [Google Scholar] [PubMed] [CrossRef]
14. Penolazzi L, Lambertini E, D’Agostino S, Pozzobon M, Notarangelo MP, Greco P, et al. Decellularized extracellular matrix-based scaffold and hypoxic priming: a promising combination to improve the phenotype of degenerate intervertebral disc cells. Life Sci. 2022;301(3):120623. doi:10.1016/j.lfs.2022.120623. [Google Scholar] [PubMed] [CrossRef]
15. Higgins DF, Kimura K, Bernhardt WM, Shrimanker N, Akai Y, Hohenstein B, et al. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J Clin Invest. 2007;117(12):3810–20. doi:10.1172/JCI30487. [Google Scholar] [PubMed] [CrossRef]
16. Halberg N, Khan T, Trujillo ME, Wernstedt-Asterholm I, Attie AD, Sherwani S, et al. Hypoxia-inducible factor 1α induces fibrosis and insulin resistance in white adipose tissue. Mol Cell Biol. 2009;29(16):4467–83. doi:10.1128/MCB.00192-09. [Google Scholar] [PubMed] [CrossRef]
17. Moon JO, Welch TP, Gonzalez FJ, Copple BL. Reduced liver fibrosis in hypoxia-inducible factor-1α-deficient mice. Am J Physiol—Gastrointest Liver Physiol. 2009;296(3):G582–92. doi:10.1152/ajpgi.90368.2008. [Google Scholar] [PubMed] [CrossRef]
18. Valle-Tenney R, Rebolledo DL, Lipson KE, Brandan E. Role of hypoxia in skeletal muscle fibrosis: synergism between hypoxia and TGF-β signaling upregulates CCN2/CTGF expression specifically in muscle fibers. Matrix Biol. 2020;87(6):48–65. doi:10.1016/j.matbio.2019.09.003. [Google Scholar] [PubMed] [CrossRef]
19. Pastwińska J, Walczak-Drzewiecka A, Łukasiak M, Ratajewski M, Dastych J. Hypoxia regulates human mast cell adhesion to fibronectin via the PI3K/AKT signaling pathway. Cell Adhes Migr. 2020;14(1):106–17. doi:10.1080/19336918.2020.1764690. [Google Scholar] [PubMed] [CrossRef]
20. Wang R, Godet I, Yang Y, Salman S, Lu H, Lyu Y, et al. Hypoxia-inducible factor-dependent ADAM12 expression mediates breast cancer invasion and metastasis. Proc Natl Acad Sci USA. 2021;118(19):e2020490118. doi:10.1073/pnas.2020490118. [Google Scholar] [PubMed] [CrossRef]
21. Georgy M, Salhiyyah K, Yacoub MH, Chester AH. Role of hypoxia inducible factor HIF-1α in heart valves. Glob Cardiol Sci Pract. 2023;2023(2):e202309. doi:10.21542/gcsp.2023.9. [Google Scholar] [PubMed] [CrossRef]
22. Wicks EE, Semenza GL. Hypoxia-inducible factors: cancer progression and clinical translation. J Clin Invest. 2023;132(11):e159839. doi:10.1172/JCI159839. [Google Scholar] [PubMed] [CrossRef]
23. Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148(3):399–408. doi:10.1016/j.cell.2012.01.021. [Google Scholar] [PubMed] [CrossRef]
24. Yamashita T, Ohneda O, Nagano M, Iemitsu M, Makino Y, Tanaka H, et al. Abnormal heart development and lung remodeling in mice lacking the hypoxia-inducible factor-related basic helix-loop-helix PAS protein NEPAS. Mol Cell Biol. 2008;28(4):1285–97. doi:10.1128/MCB.01332-07. [Google Scholar] [PubMed] [CrossRef]
25. Heikkilä M, Pasanen A, Kivirikko KI, Myllyharju J. Roles of the human hypoxia-inducible factor (HIF)-3α variants in the hypoxia response. Cell Mol Life Sci CMLS. 2011;68(23):3885–901. doi:10.1007/s00018-011-0679-5. [Google Scholar] [PubMed] [CrossRef]
26. Pasanen A, Heikkilä M, Rautavuoma K, Hirsilä M, Kivirikko KI, Myllyharju J. Hypoxia-inducible factor (HIF)-3α is subject to extensive alternative splicing in human tissues and cancer cells and is regulated by HIF-1 but not HIF-2. Int J Biochem Cell Biol. 2010;42(7):1189–1200. doi:10.1016/j.biocel.2010.04.008. [Google Scholar] [PubMed] [CrossRef]
27. Augstein A, Poitz DM, Braun-Dullaeus RC, Strasser RH, Schmeisser A. Cell-specific and hypoxia-dependent regulation of human HIF-3α: inhibition of the expression of HIF target genes in vascular cells. Cell Mol Life Sci CMLS. 2011;68(15):2627–42. doi:10.1007/s00018-010-0575-4. [Google Scholar] [PubMed] [CrossRef]
28. Sen R, Zhdanov AV, Bastiaanssen TFS, Hirvonen LM, Svihra P, Fitzgerald P, et al. Mapping O2 concentration in ex-vivo tissue samples on a fast PLIM macro-imager. Sci Rep. 2020;10(1):19006. doi:10.1038/s41598-020-75928-3. [Google Scholar] [PubMed] [CrossRef]
29. Narazaki A, Shimizu R, Yoshihara T, Kikuta J, Sakaguchi R, Tobita S, et al. Determination of the physiological range of oxygen tension in bone marrow monocytes using two-photon phosphorescence lifetime imaging microscopy. Sci Rep. 2022;12(1):3497. doi:10.1038/s41598-022-07521-9. [Google Scholar] [PubMed] [CrossRef]
30. Harer MW, Chock VY. Renal tissue oxygenation monitoring-an opportunity to improve kidney outcomes in the vulnerable neonatal population. Front Pediatr. 2020;8:241. doi:10.3389/fped.2020.00241. [Google Scholar] [PubMed] [CrossRef]
31. Zhang G, Wang X, Rothermel BA, Lavandero S, Wang ZV. The integrated stress response in ischemic diseases. Cell Death Differ. 2022;29(4):750–7. doi:10.1038/s41418-021-00889-7. [Google Scholar] [PubMed] [CrossRef]
32. Reyes A, Duarte LF, Farías MA, Tognarelli E, Kalergis AM, Bueno SM, et al. Impact of hypoxia over human viral infections and key cellular processes. Int J Mol Sci. 2021;22(15):7954. doi:10.3390/ijms22157954. [Google Scholar] [PubMed] [CrossRef]
33. Slawski J, Jaśkiewicz M, Barton A, Kozioł S, Collawn JF, Bartoszewski R. Regulation of the HIF switch in human endothelial and cancer cells. Eur J Cell Biol. 2024;103(2):151386. doi:10.1016/j.ejcb.2024.151386. [Google Scholar] [PubMed] [CrossRef]
34. Ravenna L, Salvatori L, Russo MA. HIF3α: the little we know. FEBS J. 2016;283(6):993–1003. doi:10.1111/febs.13572. [Google Scholar] [PubMed] [CrossRef]
35. Duan C. Hypoxia-inducible factor 3 biology: complexities and emerging themes. Am J Physiol Cell Physiol. 2016;310(4):C260–9. doi:10.1152/ajpcell.00315.2015. [Google Scholar] [PubMed] [CrossRef]
36. Yfantis A, Mylonis I, Chachami G, Nikolaidis M, Amoutzias GD, Paraskeva E, et al. Transcriptional response to hypoxia: the role of HIF-1-associated co-regulators. Cells. 2023;12(5):798. doi:10.3390/cells12050798. [Google Scholar] [PubMed] [CrossRef]
37. Orlando IMC, Lafleur VN, Storti F, Spielmann P, Crowther L, Santambrogio S, et al. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica. 2020;105(12):2774–84. doi:10.3324/haematol.2019.236406. [Google Scholar] [PubMed] [CrossRef]
38. Kobayashi Y, Oguro A, Imaoka S. Feedback of hypoxia-inducible factor-1α (HIF-1α) transcriptional activity via redox factor-1 (Ref-1) induction by reactive oxygen species (ROS). Free Radic Res. 2021;55(2):154–64. doi:10.1080/10715762.2020.1870685. [Google Scholar] [PubMed] [CrossRef]
39. Huang B, Ding J, Guo H, Wang H, Xu J, Zheng Q, et al. SIRT3 regulates the ROS-FPR1/HIF-1α axis under hypoxic conditions to influence lung cancer progression. Cell Biochem Biophys. 2023;81(4):813–21. doi:10.1007/s12013-023-01180-x. [Google Scholar] [PubMed] [CrossRef]
40. Sun Q, Chen X, Ma J, Peng H, Wang F, Zha X, et al. Mammalian target of rapamycin up-regulation of pyruvate kinase isoenzyme type M2 is critical for aerobic glycolysis and tumor growth. Proc Nat Acad Sci USA. 2011;108(10):4129–34. doi:10.1073/pnas.1014769108. [Google Scholar] [PubMed] [CrossRef]
41. Wang F, Zhang H, Xu N, Huang N, Tian C, Ye A, et al. A novel hypoxia-induced miR-147a regulates cell proliferation through a positive feedback loop of stabilizing HIF-1α. Cancer Biol Ther. 2016;17(8):790–8. doi:10.1080/15384047.2016.1195040. [Google Scholar] [PubMed] [CrossRef]
42. Gao Q, Ren Z, Jiao S, Guo J, Miao X, Wang J, et al. HIF-3α-induced miR-630 expression promotes cancer hallmarks in cervical cancer cells by forming a positive feedback loop. J Immunol Res. 2022;2022(10167):5262963–19. doi:10.1155/2022/5262963. [Google Scholar] [PubMed] [CrossRef]
43. Chen W, Wu P, Yu F, Luo G, Qing L, Tang J. HIF-1α regulates bone homeostasis and angiogenesis, participating in the occurrence of bone metabolic diseases. Cells. 2022;11(22):3552. doi:10.3390/cells11223552. [Google Scholar] [PubMed] [CrossRef]
44. Sun J, Shen H, Shao L, Teng X, Chen Y, Liu X, et al. HIF-1α overexpression in mesenchymal stem cell-derived exosomes mediates cardioprotection in myocardial infarction by enhanced angiogenesis. Stem Cell Res Ther. 2020;11(1):373. doi:10.1186/s13287-020-01881-7. [Google Scholar] [PubMed] [CrossRef]
45. Cowman SJ, Koh MY. Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer. 2022;8(1):28–42. doi:10.1016/j.trecan.2021.10.004. [Google Scholar] [PubMed] [CrossRef]
46. Kierans SJ, Taylor CT. Regulation of glycolysis by the hypoxia-inducible factor (HIFimplications for cellular physiology. J Physiol. 2021;599(1):23–37. doi:10.1113/JP280572. [Google Scholar] [PubMed] [CrossRef]
47. Taylor CT, Scholz CC. The effect of HIF on metabolism and immunity. Nat Rev Nephrol. 2022;18(9):573–87. doi:10.1038/s41581-022-00587-8. [Google Scholar] [PubMed] [CrossRef]
48. Hu S, He W, Wu G. Hydroxyproline in animal metabolism, nutrition, and cell signaling. Amino Acids. 2022;54(4):513–28. doi:10.1007/s00726-021-03056-x. [Google Scholar] [PubMed] [CrossRef]
49. Sato T, Takeda N. The roles of HIF-1α signaling in cardiovascular diseases. J Cardiol. 2023;81(2):202–8. doi:10.1016/j.jjcc.2022.09.002. [Google Scholar] [PubMed] [CrossRef]
50. Liu J, Huang X, Liu D, Ji K, Tao C, Zhang R, et al. Demethyleneberberine induces cell cycle arrest and cellular senescence of NSCLC cells via c-Myc/HIF-1α pathway. Phytomedicine. 2021;91:153678. doi:10.1016/j.phymed.2021.153678. [Google Scholar] [PubMed] [CrossRef]
51. Chao AC, Chen CH, Wu MH, Hou BY, Yang DI. Roles of Id1/HIF-1 and CDK5/HIF-1 in cell cycle reentry induced by amyloid-beta peptide in post-mitotic cortical neuron. Biochim Biophys Acta BBA. Mol Cell Res. 2020;1867(4):118628. doi:10.1016/j.bbamcr.2019.118628. [Google Scholar] [PubMed] [CrossRef]
52. Wilson JW, Shakir D, Batie M, Frost M, Rocha S. Oxygen-sensing mechanisms in cells. FEBS J. 2020;287(18):3888–906. doi:10.1111/febs.15374. [Google Scholar] [PubMed] [CrossRef]
53. Qiu GZ, Liu Q, Wang XG, Xu GZ, Zhao T, Lou MQ. Hypoxia-induced USP22-BMI1 axis promotes the stemness and malignancy of glioma stem cells via regulation of HIF-1α. Life Sci. 2020;247:117438. doi:10.1016/j.lfs.2020.117438. [Google Scholar] [PubMed] [CrossRef]
54. Hao S, Li F, Jiang P, Gao J. Effect of chronic intermittent hypoxia-induced HIF-1α/ATAD2 expression on lung cancer stemness. Cell Mol Biol Lett. 2022;27(1):44. doi:10.1186/s11658-022-00345-5. [Google Scholar] [PubMed] [CrossRef]
55. Infantino V, Santarsiero A, Convertini P, Todisco S, Iacobazzi V. Cancer cell metabolism in hypoxia: role of HIF-1 as key regulator and therapeutic target. Int J Mol Sci. 2021;22(11):5703. doi:10.3390/ijms22115703. [Google Scholar] [PubMed] [CrossRef]
56. Zhang Y, Zhang YY, Pan ZW, Li QQ, Sun LH, Li X, et al. GDF11 promotes wound healing in diabetic mice via stimulating HIF-1ɑ-VEGF/SDF-1ɑ-mediated endothelial progenitor cell mobilization and neovascularization. Acta Pharmacol Sin. 2023;44(5):999–1013. doi:10.1038/s41401-022-01013-2. [Google Scholar] [PubMed] [CrossRef]
57. Xue Y, Li Z, Wang Y, Zhu X, Hu R, Xu W. Role of the HIF-1α/SDF-1/CXCR4 signaling axis in accelerated fracture healing after craniocerebral injury. Mol Med Rep. 2020;22(4):2767–74. doi:10.3892/mmr.2020.11361. [Google Scholar] [PubMed] [CrossRef]
58. Ito K, Kitajima Y, Kai K, Matsufuji S, Yamada K, Egawa N, et al. Matrix metalloproteinase-1 expression is regulated by HIF-1-dependent and epigenetic mechanisms and serves a tumor-suppressive role in gastric cancer progression. Int J Oncol. 2021;59(6):102. doi:10.3892/ijo.2021.5282. [Google Scholar] [PubMed] [CrossRef]
59. Li H, Huang H, Cui Y, Li W, Zhang S, Chen Y. Study on the mechanism of capillary leakage caused by hypoxia-inducible factor-1α through inducing high expression of matrix metalloproteinase-9. J Oncol. 2021;2021(4):9130650–12. doi:10.1155/2021/9130650. [Google Scholar] [PubMed] [CrossRef]
60. Hui X, Al-Ward H, Shaher F, Liu CY, Liu N. The role of miR-210 in the biological system: a current overview. Hum Hered. 2019;84(6):233–9. doi:10.1159/000509280. [Google Scholar] [PubMed] [CrossRef]
61. Cao G, Fan P, Ma R, Wang Q, He L, Niu H, et al. MiR-210 regulates lung adenocarcinoma by targeting HIF-1α. Heliyon. 2023;9(5):e16079. doi:10.1016/j.heliyon.2023.e16079. [Google Scholar] [PubMed] [CrossRef]
62. Wu J, Yu L, Liu Y, Xiao B, Ye X, Zhao H, et al. Hypoxia regulates adipose mesenchymal stem cells proliferation, migration, and nucleus pulposus-like differentiation by regulating endoplasmic reticulum stress via the HIF-1α pathway. J Orthop Surg. 2023;18(1):339. doi:10.1186/s13018-023-03818-1. [Google Scholar] [PubMed] [CrossRef]
63. Večeřa J, Procházková J, Šumberová V, Pánská V, Paculová H, Lánová MK, et al. Hypoxia/Hif1α prevents premature neuronal differentiation of neural stem cells through the activation of Hes1. Stem Cell Res. 2020;45(4):101770. doi:10.1016/j.scr.2020.101770. [Google Scholar] [PubMed] [CrossRef]
64. Knowles HJ. Distinct roles for the hypoxia-inducible transcription factors HIF-1α and HIF-2α in human osteoclast formation and function. Sci Rep. 2020;10(1):21072. doi:10.1038/s41598-020-78003-z. [Google Scholar] [PubMed] [CrossRef]
65. Raval RR, Lau KW, Tran MGB, Sowter HM, Mandriota SJ, Li JL, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25(13):5675–86. doi:10.1128/MCB.25.13.5675-5686.2005. [Google Scholar] [PubMed] [CrossRef]
66. He Q, Ma Y, Liu J, Zhang D, Ren J, Zhao R, et al. Biological functions and regulatory mechanisms of hypoxia-inducible factor-1α in ischemic stroke. Front Immunol. 2021;12:801985. doi:10.3389/fimmu.2021.801985. [Google Scholar] [PubMed] [CrossRef]
67. Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, et al. HIF-2α regulates Oct-4: effects of hypoxiaon stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20(5):557–70. doi:10.1101/gad.1399906. [Google Scholar] [PubMed] [CrossRef]
68. Keith B, Johnson RS, Simon MC. HIF1α and HIF2α: sibling rivalry in hypoxic tumour growth and progression. Nat Rev Cancer. 2011;12(1):9–22. doi:10.1038/nrc3183. [Google Scholar] [PubMed] [CrossRef]
69. Zhou S, Yan J, Song K, Ge RL. High-Altitude hypoxia induces excessive erythrocytosis in mice via upregulation of the intestinal HIF2a/iron-metabolism pathway. Biomedicine. 2023;11(11):2992. doi:10.3390/biomedicines11112992. [Google Scholar] [PubMed] [CrossRef]
70. Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 1998;12(2):149–62. doi:10.1101/gad.12.2.149. [Google Scholar] [PubMed] [CrossRef]
71. Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1-/- mice. Nat Genet. 2003;35(4):331–40. doi:10.1038/ng1266. [Google Scholar] [PubMed] [CrossRef]
72. Talks KL, Turley H, Gatter KC, Maxwell PH, Pugh CW, Ratcliffe PJ, et al. The expression and distribution of the hypoxia-inducible factors HIF-1α and HIF-2α in normal human tissues, cancers, and tumor-associated macrophages. Am J Pathol. 2000;157(2):411–21. doi:10.1016/S0002-9440(10)64554-3. [Google Scholar] [PubMed] [CrossRef]
73. Prabhakar NR, Semenza GL. Adaptive and maladaptive cardiorespiratory responses to continuous and intermittent hypoxia mediated by hypoxia-inducible factors 1 and 2. Physiol Rev. 2012;92(3):967–1003. doi:10.1152/physrev.00030.2011. [Google Scholar] [PubMed] [CrossRef]
74. Yu Y, Ma L, Zhang H, Sun W, Zheng L, Liu C, et al. EPO could be regulated by HIF-1 and promote osteogenesis and accelerate bone repair. Artif Cells Nanomedicine Biotechnol. 2020;48(1):206–17. doi:10.1080/21691401.2019.1699827. [Google Scholar] [PubMed] [CrossRef]
75. Rankin EB, Biju MP, Liu Q, Unger TL, Rha J, Johnson RS, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117(4):1068–77. doi:10.1172/JCI30117. [Google Scholar] [PubMed] [CrossRef]
76. Gruber M, Hu CJ, Johnson RS, Brown EJ, Keith B, Simon MC. Acute postnatal ablation of Hif-2α results in anemia. Proc Nat Acad Sci USA. 2007;104(7):2301–6. doi:10.1073/pnas.0608382104. [Google Scholar] [PubMed] [CrossRef]
77. Tolonen JP, Heikkilä M, Malinen M, Lee HM, Palvimo JJ, Wei GH, et al. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell Mol Life Sci CMLS. 2020;77(18):3627–42. doi:10.1007/s00018-019-03387-9. [Google Scholar] [PubMed] [CrossRef]
78. Pawlus MR, Wang L, Murakami A, Dai G, Hu CJ. STAT3 or USF2 contributes to HIF target gene specificity. PLoS One. 2013;8(8):e72358. doi:10.1371/journal.pone.0072358. [Google Scholar] [PubMed] [CrossRef]
79. Andreeva ER, Lobanova MV, Udartseva OO, Buravkova LB. Response of adipose tissue-derived stromal cells in tissue-related O2 microenvironment to short-term hypoxic stress. Cells Tissues Organs. 2015;200(5):307–15. doi:10.1159/000438921. [Google Scholar] [PubMed] [CrossRef]
80. Bartoszewska S, Kochan K, Piotrowski A, Kamysz W, Ochocka RJ, Collawn JF, et al. The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1α expression in human endothelial cells through a negative feedback loop. FASEB J Off Publ Fed Am Soc Exp Biol. 2015;29(4):1467–79. doi:10.1096/fj.14-267054. [Google Scholar] [PubMed] [CrossRef]
81. Jaśkiewicz M, Moszyńska A, Króliczewski J, Cabaj A, Bartoszewska S, Charzyńska A, et al. The transition from HIF-1 to HIF-2 during prolonged hypoxia results from reactivation of PHDs and HIF1A mRNA instability. Cell Mol Biol Lett. 2022;27(1):109. doi:10.1186/s11658-022-00408-7. [Google Scholar] [PubMed] [CrossRef]
82. Gu YZ, Moran SM, Hogenesch JB, Wartman L, Bradfield CA. Molecular characterization and chromosomal localization of a third alpha-class hypoxia inducible factor subunit. HIF3α Gene Expr. 1998;7(3):205–13. [Google Scholar] [PubMed]
83. Hara S, Hamada J, Kobayashi C, Kondo Y, Imura N. Expression and characterization of hypoxia-inducible factor (HIF)-3α in human kidney: suppression of HIF-mediated gene expression by HIF-3α. Biochem Biophys Res Commun. 2001;287(4):808–13. doi:10.1006/bbrc.2001.5659. [Google Scholar] [PubMed] [CrossRef]
84. Yang SL, Wu C, Xiong ZF, Fang X. Progress on hypoxia-inducible factor-3: its structure, gene regulation and biological function (Review). Mol Med Rep. 2015;12(2):2411–6. doi:10.3892/mmr.2015.3689. [Google Scholar] [PubMed] [CrossRef]
85. Makino Y, Kanopka A, Wilson WJ, Tanaka H, Poellinger L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J Biol Chem. 2002;277(36):32405–8. doi:10.1074/jbc.C200328200. [Google Scholar] [PubMed] [CrossRef]
86. Zhang P, Bai Y, Lu L, Li Y, Duan C. An oxygen-insensitive Hif-3α isoform inhibits Wnt signaling by destabilizing the nuclear β-catenin complex. eLife. 2016;5:e08996. doi:10.7554/eLife.08996. [Google Scholar] [PubMed] [CrossRef]
87. Li ZF, Meng DD, Liu YY, Bi FG, Tian K, Xu JZ, et al. Hypoxia inducible factor-3α promotes osteosarcoma progression by activating KDM3A-mediated demethylation of SOX9. Chem Biol Interact. 2022;351(Suppl 2):109759. doi:10.1016/j.cbi.2021.109759. [Google Scholar] [PubMed] [CrossRef]
88. Xu Y, Hu T, Ding H, Yuan Y, Chen R. miR-485-5p alleviates obstructive sleep apnea syndrome with hypertension by inhibiting PI3K/AKT signaling pathway via downregulating HIF3A expression. Sleep Breath Schlaf Atm. 2023;27(1):109–19. doi:10.1007/s11325-022-02580-8. [Google Scholar] [PubMed] [CrossRef]
89. Xu J, Hao S, Han K, Yang W, Deng H. How is the AKT/mTOR pathway involved in cell migration and invasion? BIOCELL. 2023;47(4):773–88. doi:10.32604/biocell.2023.026618. [Google Scholar] [CrossRef]
90. Huang Y, Kapere Ochieng J, Kempen MB, Munck AB, Swagemakers S, van Ijcken W, et al. Hypoxia inducible factor 3α plays a critical role in alveolarization and distal epithelial cell differentiation during mouse lung development. PLoS One. 2013;8(2):e57695. doi:10.1371/journal.pone.0057695. [Google Scholar] [PubMed] [CrossRef]
91. Qu H, Yu Q, Jia B, Zhou W, Zhang Y, Mu L. HIF-3α affects preeclampsia development by regulating EVT growth via activation of the Flt-1/JAK/STAT signaling pathway in hypoxia. Mol Med Rep. 2021;23(1):68. doi:10.3892/mmr.2020.11701. [Google Scholar] [PubMed] [CrossRef]
92. Scharf JG, Unterman TG, Kietzmann T. Oxygen-dependent modulation of insulin-like growth factor binding protein biosynthesis in primary cultures of rat hepatocytes. Endocrinology. 2005;146(12):5433–43. doi:10.1210/en.2005-0948. [Google Scholar] [PubMed] [CrossRef]
93. Zhang P, Yao Q, Lu L, Li Y, Chen PJ, Duan C. Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Rep. 2014;6(6):1110–21. doi:10.1016/j.celrep.2014.02.011. [Google Scholar] [PubMed] [CrossRef]
94. Maynard MA, Evans AJ, Hosomi T, Hara S, Jewett MAS, Ohh M. Human HIF-3α4 is a dominant-negative regulator of HIF-1 and is down-regulated in renal cell carcinoma. FASEB J Off Publ Fed Am Soc Exp Biol. 2005;19(11):1396–1406. doi:10.1096/fj.05-3788com. [Google Scholar] [PubMed] [CrossRef]
95. Kim SH, Hwang D, Park H, Yang EG, Chung HS, Kim SY. The action of HIF-3α variants on HIF-2α-HIF-1β heterodimer formation is directly probed in live cells. Exp Cell Res. 2015;336(2):329–37. doi:10.1016/j.yexcr.2015.06.017. [Google Scholar] [PubMed] [CrossRef]
96. Li QF, Wang XR, Yang YW, Lin H. Hypoxia upregulates hypoxia inducible factor (HIF)-3α expression in lung epithelial cells: characterization and comparison with HIF-1alpha. Cell Res. 2006;16(6):548–58. doi:10.1038/sj.cr.7310072. [Google Scholar] [PubMed] [CrossRef]
97. Janaszak-Jasiecka A, Bartoszewska S, Kochan K, Piotrowski A, Kalinowski L, Kamysz W, et al. miR-429 regulates the transition between hypoxia-inducible factor (HIF)1A and HIF3A expression in human endothelial cells. Sci Rep. 2016;6(1):22775. doi:10.1038/srep22775. [Google Scholar] [PubMed] [CrossRef]
98. Wiafe B, Adesida A, Churchill T, Adewuyi EE, Li Z, Metcalfe P. Hypoxia-increased expression of genes involved in inflammation, dedifferentiation, pro-fibrosis, and extracellular matrix remodeling of human bladder smooth muscle cells. In Vitro Cell Dev Biol Anim. 2017;53(1):58–66. doi:10.1007/s11626-016-0085-2. [Google Scholar] [PubMed] [CrossRef]
99. Hatanaka M, Shimba S, Sakaue M, Kondo Y, Kagechika H, Kokame K, et al. Hypoxia-inducible factor-3α functions as an accelerator of 3T3-L1 adipose differentiation. Biol Pharm Bull. 2009;32(7):1166–72. doi:10.1248/bpb.32.1166. [Google Scholar] [PubMed] [CrossRef]
100. Jaskiewicz M, Moszynska A, Serocki M, Króliczewski J, Bartoszewska S, Collawn JF, et al. Hypoxia-inducible factor (HIF)-3α2 serves as an endothelial cell fate executor during chronic hypoxia. EXCLI J. 2022;21:454–69. doi:10.17179/excli2021-4622. [Google Scholar] [PubMed] [CrossRef]
101. Moszyńska A, Jaśkiewicz M, Serocki M, Cabaj A, Crossman DK, Bartoszewska S, et al. The hypoxia-induced changes in miRNA-mRNA in RNA-induced silencing complexes and HIF-2 induced miRNAs in human endothelial cells. FASEB J. 2022;36(7):e22412. doi:10.1096/fj.202101987R. [Google Scholar] [PubMed] [CrossRef]
102. Cuomo F, Coppola A, Botti C, Maione C, Forte A, Scisciola L, et al. Pro-inflammatory cytokines activate hypoxia-inducible factor 3α via epigenetic changes in mesenchymal stromal/stem cells. Sci Rep. 2018;8(1):5842. doi:10.1038/s41598-018-24221-5. [Google Scholar] [PubMed] [CrossRef]
103. Liang M, Si L, Yu Z, Ding H, Wang L, X. C, et al. Intermittent hypoxia induces myofibroblast differentiation and extracellular matrix production of MRC5s via HIF-1α-TGF-β/Smad pathway. Sleep Breath Schlaf Atm. 2023;19:390. doi:10.1007/s11325-023-02889-y. [Google Scholar] [PubMed] [CrossRef]
104. Kang Y, Roh MR, Rajadurai S, Rajadurai A, Kumar R, Njauw CN, et al. Hypoxia and HIF-1α regulate collagen production in keloids. J Invest Dermatol. 2020;140(11):2157–65. doi:10.1016/j.jid.2020.01.036. [Google Scholar] [PubMed] [CrossRef]
105. Hu Y, Zhang T, Chen J, Cheng W, Chen J, Zheng Z, et al. Downregulation of hypoxia-inducible factor-1α by RNA interference alleviates the development of collagen-induced arthritis in rats. Mol Ther Nucleic Acids. 2020;19:1330–42. doi:10.1016/j.omtn.2020.01.014. [Google Scholar] [PubMed] [CrossRef]
106. Mallikarjuna P, Raviprakash TS, Aripaka K, Ljungberg B, Landström M. Interactions between TGF-β type I receptor and hypoxia-inducible factor-α mediates a synergistic crosstalk leading to poor prognosis for patients with clear cell renal cell carcinoma. Cell Cycle Georget Tex. 2019;18(17):2141–56. doi:10.1080/15384101.2019.1642069. [Google Scholar] [PubMed] [CrossRef]
107. Niu G, Zhao Y, Song H, Song Q, Yin X, Zhu Z, et al. Marein ameliorates myocardial fibrosis by inhibiting HIF-1α and TGF-β1/Smad2/3 signaling pathway in isoproterenol-stimulated mice and TGF-β1-stimulated cardiac fibroblasts. Curr Pharm Des. 2023;30(1):71–80. doi:10.2174/0113816128282062231218075341. [Google Scholar] [PubMed] [CrossRef]
108. Berg JT, Breen EC, Fu Z, Mathieu-Costello O, West JB. Alveolar hypoxia increases gene expression of extracellular matrix proteins and platelet-derived growth factor-B in lung parenchyma. Am J Respir Crit Care Med. 1998;158(6):1920–8. doi:10.1164/ajrccm.158.6.9804076. [Google Scholar] [PubMed] [CrossRef]
109. Milner R, Hung S, Erokwu B, Dore-Duffy P, LaManna JC, del Zoppo GJ. Increased expression of fibronectin and the α5β1 integrin in angiogenic cerebral blood vessels of mice subject to hypobaric hypoxia. Mol Cell Neurosci. 2008;38(1):43–52. doi:10.1016/j.mcn.2008.01.013. [Google Scholar] [PubMed] [CrossRef]
110. Braga CL, da Silva LR, Santos RT, de Carvalho LRP, Mandacaru SC, de Oliveira Trugilho MR, et al. Proteomics profile of mesenchymal stromal cells and extracellular vesicles in normoxic and hypoxic conditions. Cytotherapy. 2022;24(12):1211–24. doi:10.1016/j.jcyt.2022.08.009. [Google Scholar] [PubMed] [CrossRef]
111. Riis S, Stensballe A, Emmersen J, Pennisi CP, Birkelund S, Zachar V, et al. Mass spectrometry analysis of adipose-derived stem cells reveals a significant effect of hypoxia on pathways regulating extracellular matrix. Stem Cell Res Ther. 2016;7(1):52. doi:10.1186/s13287-016-0310-7. [Google Scholar] [PubMed] [CrossRef]
112. Manning EP, Ramachandra AB, Schupp JC, Cavinato C, Raredon MSB, Bärnthaler T, et al. Mechanisms of hypoxia-induced pulmonary arterial stiffening in mice revealed by a functional genetics assay of structural, functional, and transcriptomic data. Front Physiol. 2021;12:726253. doi:10.3389/fphys.2021.726253. [Google Scholar] [PubMed] [CrossRef]
113. Matveeva D, Buravkov S, Andreeva E, Buravkova L. Hypoxic extracellular matrix preserves its competence after expansion of human MSCs under physiological hypoxia in vitro. Biomim. 2023;8(6):476. doi:10.3390/biomimetics8060476. [Google Scholar] [PubMed] [CrossRef]
114. Morimoto C, Takedachi M, Kawasaki K, Shimomura J, Murata M, Hirai A, et al. Hypoxia stimulates collagen hydroxylation in gingival fibroblasts and periodontal ligament cells. J Periodontol. 2021;92(11):1635–45. doi:10.1002/JPER.20-0670. [Google Scholar] [PubMed] [CrossRef]
115. Eisinger-Mathason TSK, Zhang M, Qiu Q, Skuli N, Nakazawa MS, Karakasheva T, et al. Hypoxia-dependent modification of collagen networks promotes sarcoma metastasis. Cancer Discov. 2013;3(10):1190–205. doi:10.1158/2159-8290.CD-13-0118. [Google Scholar] [PubMed] [CrossRef]
116. Murdocca M, de Masi C, Pucci S, Mango R, Novelli G, Di Natale C, et al. LOX-1 and cancer: an indissoluble liaison. Cancer Gene Ther. 2021;28(10–11):1088–98. doi:10.1038/s41417-020-00279-0. [Google Scholar] [PubMed] [CrossRef]
117. Choi JY, Jang YS, Min SY, Song JY. Overexpression of MMP-9 and HIF-1α in breast cancer cells under hypoxic conditions. J Breast Cancer. 2011;14(2):88–95. doi:10.4048/jbc.2011.14.2.88. [Google Scholar] [PubMed] [CrossRef]
118. Knyazev E, Maltseva D, Raygorodskaya M, Shkurnikov M. HIF-dependent NFATC1 activation upregulates ITGA5 and PLAUR in intestinal epithelium in inflammatory bowel disease. Front Genet. 2021;12:791640. doi:10.3389/fgene.2021.791640. [Google Scholar] [PubMed] [CrossRef]
119. Kanno Y. The uPA/uPAR System orchestrates the inflammatory response, vascular homeostasis, and immune system in fibrosis progression. Int J Mol Sci. 2023;24(2):1796. doi:10.3390/ijms24021796. [Google Scholar] [PubMed] [CrossRef]
120. Maruyama S, Yamazaki M, Abé T, Cheng J, Saku T, Tanuma J. Hypoxia-induced biosynthesis of the extracellular matrix molecules, perlecan and fibronectin, promotes the growth of pleomorphic adenoma cells in vitro models. Biomedicine. 2023;11(11):2981. doi:10.3390/biomedicines11112981. [Google Scholar] [PubMed] [CrossRef]
121. Zhu X, Liu S, Yang X, Wang W, Shao W, Ji T. P4HA1 as an unfavorable prognostic marker promotes cell migration and invasion of glioblastoma via inducing EMT process under hypoxia microenvironment. Am J Cancer Res. 2021;11(2):590–617. [Google Scholar] [PubMed]
122. Kaluz S, Zhang Q, Kuranaga Y, Yang H, Osuka S, Bhattacharya D, et al. Targeting HIF-activated collagen prolyl 4-hydroxylase expression disrupts collagen deposition and blocks primary and metastatic uveal melanoma growth. Oncogene. 2021;40(33):5182–91. doi:10.1038/s41388-021-01919-x. [Google Scholar] [PubMed] [CrossRef]
123. Du H, Pang M, Hou X, Yuan S, Sun L. PLOD2 in cancer research. Biomed Pharmacother. 2017;90(5):670–6. doi:10.1016/j.biopha.2017.04.023. [Google Scholar] [PubMed] [CrossRef]
124. Liu T, Xiang W, Chen Z, Wang G, Cao R, Zhou F, et al. Hypoxia-induced PLOD2 promotes clear cell renal cell carcinoma progression via modulating EGFR-dependent AKT pathway activation. Cell Death Dis. 2023;14(11):774. doi:10.1038/s41419-023-06298-7. [Google Scholar] [PubMed] [CrossRef]
125. Xing X, Wang Y, Zhang X, Gao X, Li M, Wu S, et al. Matrix stiffness-mediated effects on macrophages polarization and their LOXL2 expression. FEBS J. 2021;288(11):3465–77. doi:10.1111/febs.15566. [Google Scholar] [PubMed] [CrossRef]
126. Li R, Li H, Zhu L, Zhang X, Liu D, Li Q, et al. Reciprocal regulation of LOXL2 and HIF1α drives the Warburg effect to support pancreatic cancer aggressiveness. Cell Death Dis. 2021;12(12):1106. doi:10.1038/s41419-021-04391-3. [Google Scholar] [PubMed] [CrossRef]
127. Wang QM, Lv L, Tang Y, Zhang L, Wang LF. MMP-1 is overexpressed in triple-negative breast cancer tissues and the knockdown of MMP-1 expression inhibits tumor cell malignant behaviors in vitro. Oncol Lett. 2019;17(2):1732–40. doi:10.3892/ol.2018.9779. [Google Scholar] [PubMed] [CrossRef]
128. Sakamoto T, Niiya D, Seiki M. Targeting the Warburg effect that arises in tumor cells expressing membrane type-1 matrix metalloproteinase. J Biol Chem. 2011;286(16):14691–704. doi:10.1074/jbc.M110.188714. [Google Scholar] [PubMed] [CrossRef]
129. Zhang H, Pu J, Qi T, Qi M, Yang C, Li S, et al. MicroRNA-145 inhibits the growth, invasion, metastasis and angiogenesis of neuroblastoma cells through targeting hypoxia-inducible factor 2α. Oncogene. 2014;33(3):387–97. doi:10.1038/onc.2012.574. [Google Scholar] [PubMed] [CrossRef]
130. Yuan J, Zhang Y, Zhang Y, Mo Y, Zhang Q. Effects of metal nanoparticles on tight junction-associated proteins via HIF-1α/miR-29b/MMPs pathway in human epidermal keratinocytes. Part Fibre Toxicol. 2021;18(1):13. doi:10.1186/s12989-021-00405-2. [Google Scholar] [PubMed] [CrossRef]
131. Ahn JK, Koh EM, Cha HS, Lee YS, Kim J, Bae EK, et al. Role of hypoxia-inducible factor-1α in hypoxia-induced expressions of IL-8, MMP-1 and MMP-3 in rheumatoid fibroblast-like synoviocytes. Rheumatol Oxf Engl. 2008;47(6):834–9. doi:10.1093/rheumatology/ken086. [Google Scholar] [PubMed] [CrossRef]
132. Nishi H, Nakada T, Hokamura M, Osakabe Y, Itokazu O, Huang LE, et al. Hypoxia-inducible factor-1 transactivates transforming growth factor-β3 in trophoblast. Endocr. 2004;145(9):4113–8. doi:10.1210/en.2003-1639. [Google Scholar] [PubMed] [CrossRef]
133. Calvani M, Rapisarda A, Uranchimeg B, Shoemaker RH, Melillo G. Hypoxic induction of an HIF-1α–dependent bFGF autocrine loop drives angiogenesis in human endothelial cells. Blood. 2006;107(7):2705–12. doi:10.1182/blood-2005-09-3541. [Google Scholar] [PubMed] [CrossRef]
134. Bos R, van Diest PJ, de Jong JS, van der Groep P, van der Valk P, van der Wall E. Hypoxia-inducible factor-1α is associated with angiogenesis, and expression of bFGF, PDGF-BB, and EGFR in invasive breast cancer. Histopathol. 2005;46(1):31–6. doi:10.1111/j.1365-2559.2005.02045.x. [Google Scholar] [PubMed] [CrossRef]
135. Jensen RL, Ragel BT, Whang K, Gillespie D. Inhibition of hypoxia inducible factor-1α (HIF-1α) decreases vascular endothelial growth factor (VEGF) secretion and tumor growth in malignant gliomas. J Neurooncol. 2006;78(3):233–47. doi:10.1007/s11060-005-9103-z. [Google Scholar] [PubMed] [CrossRef]
136. Ahluwalia AS, Tarnawski A. Critical role of hypoxia sensor- HIF-1α in VEGF gene activation. Implications for angiogenesis and tissue injury healing. Curr Med Chem. 2012;19(1):90–7. doi:10.2174/092986712803413944. [Google Scholar] [PubMed] [CrossRef]
137. Al-Salam S, Hashmi S. Galectin-1 in early acute myocardial infarction. PLoS One. 2014;9(1):e86994. doi:10.1371/journal.pone.0086994. [Google Scholar] [PubMed] [CrossRef]
138. Zhao XY, Chen TT, Xia L, Guo M, Xu Y, Yue F, et al. Hypoxia inducible factor-1 mediates expression of galectin-1: the potential role in migration/invasion of colorectal cancer cells. Carcinog. 2010;31(8):1367–75. doi:10.1093/carcin/bgq116. [Google Scholar] [PubMed] [CrossRef]
139. da Silva Filho AF, de Sousa LM, Consonni SR, da Rocha Pitta MG, Carvalho HF, de Melo Rêgo MJB. Galectin-3 expression in pancreatic cell lines under distinct autophagy-inducing stimulus. Microsc Microanal. 2020;26(6):1187–97. doi:10.1017/S1431927620024526. [Google Scholar] [PubMed] [CrossRef]
140. de Oliveira JT, Ribeiro C, Barros R, Gomes C, de Matos AJ, Reis CA, et al. Hypoxia up-regulates galectin-3 in mammary tumor progression and metastasis. PLoS One. 2015;10(7):e0134458. doi:10.1371/journal.pone.0134458. [Google Scholar] [PubMed] [CrossRef]
141. Li Z, Meng D, Li G, Xu J, Tian K, Li Y. Overexpression of microRNA-210 promotes chondrocyte proliferation and extracellular matrix deposition by targeting HIF-3α in osteoarthritis. Mol Med Rep. 2016;13(3):2769–76. doi:10.3892/mmr.2016.4878. [Google Scholar] [PubMed] [CrossRef]
142. Baril P, Gangeswaran R, Mahon PC, Caulee K, Kocher HM, Harada T, et al. Periostin promotes invasiveness and resistance of pancreatic cancer cells to hypoxia-induced cell death: role of the β4 integrin and the PI3k pathway. Oncogene. 2007;26(14):2082–94. doi:10.1038/sj.onc.1210009. [Google Scholar] [PubMed] [CrossRef]
143. Wiafe B, Adesida AB, Churchill T, Kadam R, Carleton J, Metcalfe PD. Mesenchymal stem cell therapy inhibited inflammatory and profibrotic pathways induced by partial bladder outlet obstruction and prevented high-pressure urine storage. J Pediatr Urol. 2019;15(3):254.E1–254.E10. doi:10.1016/j.jpurol.2019.03.003. [Google Scholar] [PubMed] [CrossRef]
144. Liu F, Yang H, Li D, Wu X, Han Q. Punicalagin attenuates osteoarthritis progression via regulating Foxo1/Prg4/HIF3α axis. Bone. 2021;152(3):116070. doi:10.1016/j.bone.2021.116070. [Google Scholar] [PubMed] [CrossRef]
145. Ruan MZC, Erez A, Guse K, Dawson B, Bertin T, Chen Y, et al. Proteoglycan 4 expression protects against the development of osteoarthritis. Sci Transl Med. 2013;5(176):176ra34. doi:10.1126/scitranslmed.3005409. [Google Scholar] [PubMed] [CrossRef]
146. Yang M, Yan X, Yuan FZ, Ye J, Du MZ, Mao ZM, et al. MicroRNA-210-3p promotes chondrogenic differentiation and inhibits adipogenic differentiation correlated with HIF-3α signalling in bone marrow mesenchymal stem cells. BioMed Res Int. 2021;2021(5):6699910–8. doi:10.1155/2021/6699910. [Google Scholar] [PubMed] [CrossRef]
147. Gu W, Wang L, Deng G, Gu X, Tang Z, Li S, et al. Knockdown of long noncoding RNA MIAT attenuates cigarette smoke-induced airway remodeling by downregulating miR-29c-3p-HIF3A axis. Toxicol Lett. 2022;357:11–9. doi:10.1016/j.toxlet.2021.12.014. [Google Scholar] [PubMed] [CrossRef]
148. Markway BD, Cho H, Zilberman-Rudenko J, Holden P, McAlinden A, Johnstone B. Hypoxia-inducible factor 3-alpha expression is associated with the stable chondrocyte phenotype. J Orthop Res. 2015;33(11):1561–70. doi:10.1002/jor.22930. [Google Scholar] [PubMed] [CrossRef]
149. Zhu X, Wang F, Zhao Y, Yang P, Chen J, Sun H, et al. A gain-of-function mutation in Tnni2 impeded bone development through increasing Hif3a expression in DA2B mice. PLoS Genet. 2014;10(10):e1004589. doi:10.1371/journal.pgen.1004589. [Google Scholar] [PubMed] [CrossRef]
150. Lu CH, Yeh TS, Yeh CL, Fang YHD, Sung LY, Lin SY, et al. Regenerating cartilages by engineered ASCs: prolonged TGF-β3/BMP-6 expression improved articular cartilage formation and restored zonal structure. Mol Ther. 2014;22(1):186–95. doi:10.1038/mt.2013.165. [Google Scholar] [PubMed] [CrossRef]
151. Ricke WA, Smith GW, Smith MF. Matrix metalloproteinase expression and activity following prostaglandin F2α-induced luteolysis. Biol Reprod. 2002;66(3):685–91. doi:10.1095/biolreprod66.3.685. [Google Scholar] [PubMed] [CrossRef]
152. Zhang M, Sui W, Cheng C, Xue F, Tian Z, Cheng J, et al. Erythropoietin promotes abdominal aortic aneurysms in mice through angiogenesis and inflammatory infiltration. Sci Transl Med. 2021;13(603):eaaz4959. doi:10.1126/scitranslmed.aaz4959. [Google Scholar] [PubMed] [CrossRef]
153. Jia C, Li X, Pan J, Ma H, Wu D, Lu H, et al. Silencing of angiopoietin-like protein 4 (Angptl4) decreases inflammation, extracellular matrix degradation, and apoptosis in osteoarthritis via the sirtuin 1/NF-κB pathway. Oxid Med Cell Longev. 2022;2022:1135827–18. doi:10.1155/2022/1135827. [Google Scholar] [PubMed] [CrossRef]
154. Doni A, Garlanda C, Mantovani A. Innate immunity, hemostasis and matrix remodeling: pTX3 as a link. Semin Immunol. 2016;28(6):570–7. doi:10.1016/j.smim.2016.10.012. [Google Scholar] [PubMed] [CrossRef]
155. Aquino-Gálvez A, González-Ávila G, Jiménez-Sánchez LL, Maldonado-Martínez HA, Cisneros J, Toscano-Marquez F, et al. Dysregulated expression of hypoxia-inducible factors augments myofibroblasts differentiation in idiopathic pulmonary fibrosis. Respir Res. 2019;20(1):130. doi:10.1186/s12931-019-1100-4. [Google Scholar] [PubMed] [CrossRef]
156. Wiafe B, Kadam R, Metcalfe PD. Intraperitoneal administration of mesenchymal stem cells is effective at mitigating detrusor deterioration after pBOO. Am J Physiol Renal Physiol. 2020;318(3):F549–F556. doi:10.1152/ajprenal.00486.2019. [Google Scholar] [PubMed] [CrossRef]
157. Park JE, Keller GA, Ferrara N. The vascular endothelial growth factor (VEGF) isoforms: differential deposition into the subepithelial extracellular matrix and bioactivity of extracellular matrix-bound VEGF. Mol Biol Cell. 1993;4(12):1317–26. doi:10.1091/mbc.4.12.1317. [Google Scholar] [PubMed] [CrossRef]
158. Zhang H, Schin M, Saha J, Burke K, Holzman LB, Filipiak W, et al. Podocyte-specific overexpression of GLUT1 surprisingly reduces mesangial matrix expansion in diabetic nephropathy in mice. Am J Physiol-Ren Physiol. 2010;299(1):F91–8. doi:10.1152/ajprenal.00021.2010. [Google Scholar] [PubMed] [CrossRef]
159. Alkharobi H, Alhodhodi A, Hawsawi Y, Alkafaji H, Devine D, El-Gendy R, et al. IGFBP-2 and -3 co-ordinately regulate IGF1 induced matrix mineralisation of differentiating human dental pulp cells. Stem Cell Res. 2016;17(3):517–22. doi:10.1016/j.scr.2016.09.026. [Google Scholar] [PubMed] [CrossRef]
160. Sunilkumar S, Yerlikaya EI, Toro AL, Miller WP, Chen H, Hu K, et al. REDD1 ablation attenuates the development of renal complications in diabetic mice. Diabetes. 2022;71(11):2412–25. doi:10.2337/db22-0402. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF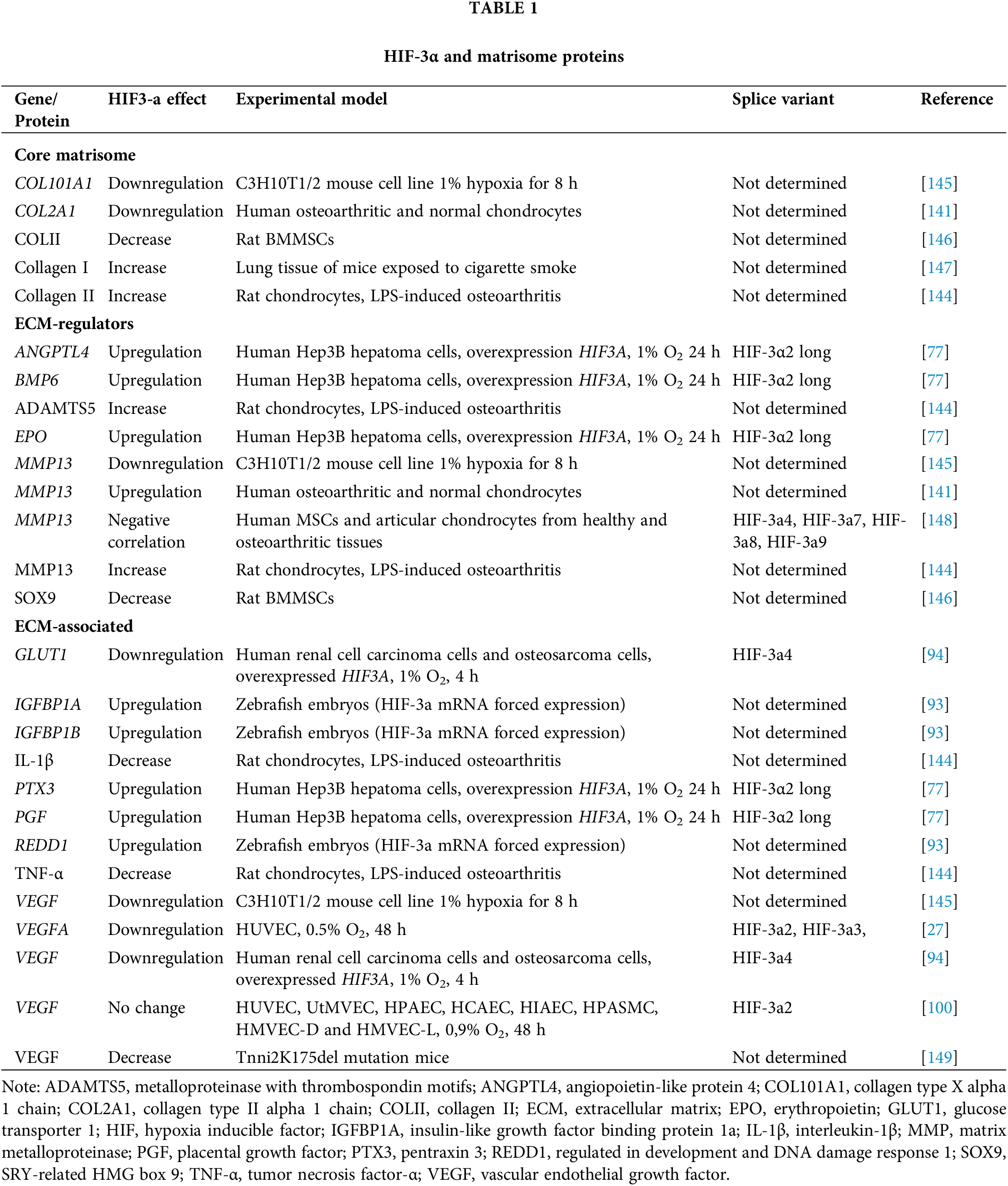
 Downloads
Downloads
 Citation Tools
Citation Tools