 Open Access
Open Access
ARTICLE
Knockdown of RCN1 contributes to the apoptosis of colorectal cancer via regulating IP3R1
1 Department of Gastroenterology, The First Affiliated Hospital of Dalian Medical University, Dalian, China
2 Department of Gastroenterology, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, China
3 The First Clinical College, Dalian Medical University, Dalian, China
* Corresponding Author: AIXIA GONG. Email:
(This article belongs to the Special Issue: Navigating the Interplay of Cancer, Autophagy, ER Stress, Cell Cycle and Apoptosis: Mechanisms, Therapies, and Future Directions)
BIOCELL 2024, 48(5), 835-845. https://doi.org/10.32604/biocell.2024.048076
Received 27 November 2023; Accepted 06 February 2024; Issue published 06 May 2024
Abstract
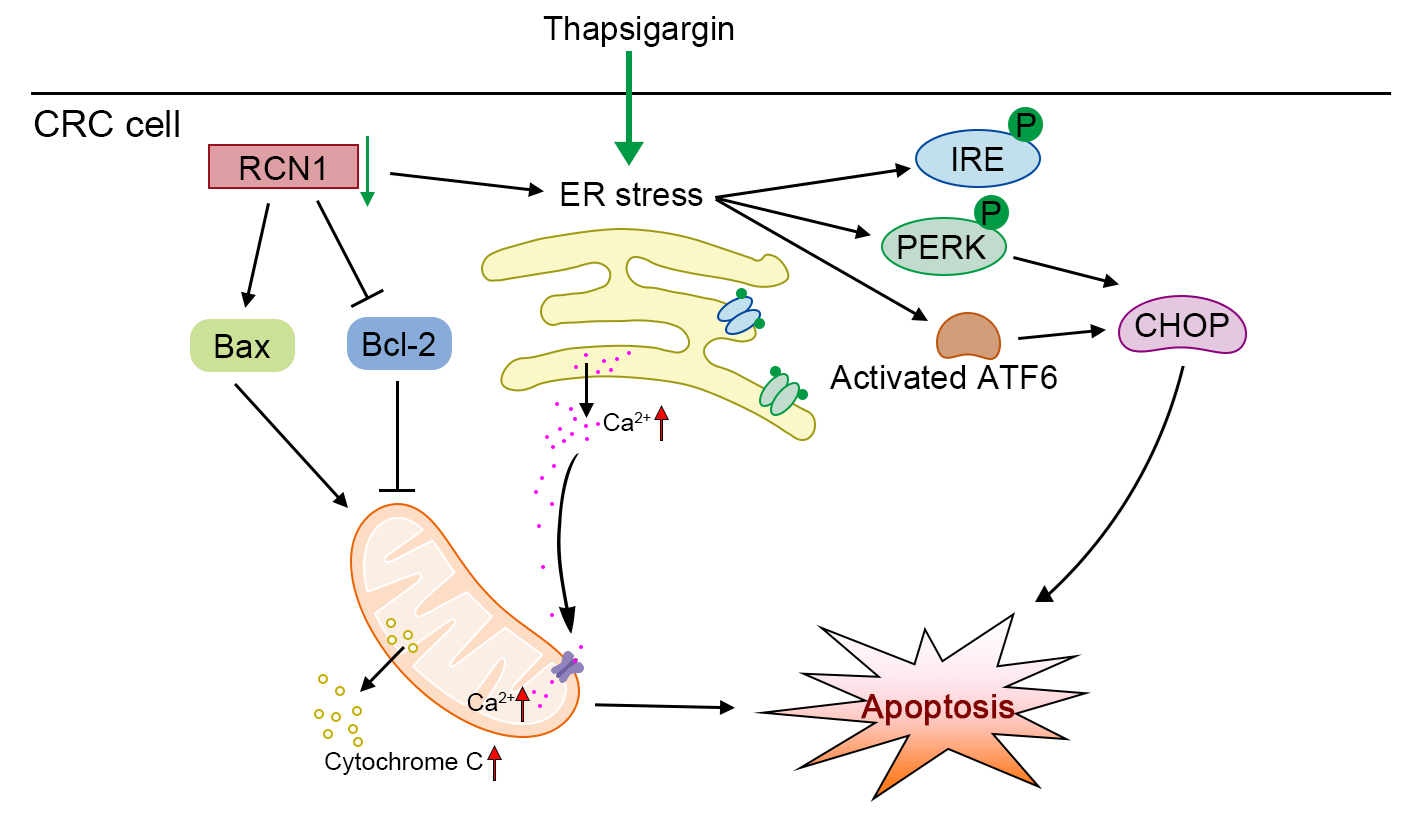
Background: The incidence of colorectal cancer (CRC) has been increasing in recent years. Thus, the discovery of factors that can assist in alleviating CRC is urgently warranted. Methods: To identify a potential factor involved in the development of CRC, we screened the upregulated genes in tumor tissues through four datasets from an online database. The expression of reticulocalbin 1 (RCN1), a Ca-binding protein, was upregulated in the four datasets. Based on loss-of-function experiments, the effect of RCN1 on cell viability was assessed by Cell Counting Kit-8 (CCK-8) assay. The regulatory effect of RCN1 on apoptosis was evaluated through Annexin V-fluorescein 5-isothiocyanate (FITC)/propidium iodide (PI) staining assay and terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay in RKO and SW480 cells. Activation of endoplasmic reticulum (ER) stress signaling pathways was confirmed by estimating the phosphorylation and expression of PRKR-like ER kinase (PERK), inositol-requiring kinase-1 (IRE1), transcription factor 6 (ACT6), and CCAAT/enhancer-binding protein-homologous protein (CHOP). The intracellular Ca homeostasis regulated by RCN1 was determined through the detection of Ca concentration and mitochondrial membrane potential (MMP) measurement. Moreover, whether inositol 1,4,5-trisphosphate receptor type 1 (IP3R1) was involved in the regulation of RCN1 in CRC was verified through the depletion of IP3R1 in RKO cells. Results: Knockdown of RCN1 reduced cell viability and facilitated apoptosis in RKO and SW480 cells. Phosphorylation of PERK and IRE1, activation of ATF6, and upregulation of CHOP were induced by the absence of RCN1, suggesting that the unfolded protein response (UPR) was activated in CRC cells. The concentration of Ca in mitochondria was increased after RCN1 depletion, followed by reduction in the MMP and release of cytochrome c from mitochondria to the cytoplasm in RKO and SW480 cells. Moreover, it was demonstrated that IP3R1 mediates the effect of RCN1 on apoptosis induced by ER stress in CRC cells. The downregulation of IP3R1 restored the RCN1 loss-induced apoptosis and the increased Ca concentration. Conclusion: Taken together, our results confirmed that silencing of RCN1 disrupted intracellular Ca homeostasis and promoted cell apoptosis caused by TG-induced ER stress by regulating IP3R1 and activating the UPR signaling pathways.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileAbbreviations
| ATF6 | Transcription factor 6 |
| Bax | Bcl-2-associated X |
| Bcl-2 | B-cell lymphoma-2 |
| CALU | Calumenin |
| CCK-8 | Cell Counting Kit-8 |
| CHOP | CCAAT/enhancer-binding protein-homologous protein |
| COAD | Colon adenocarcinoma |
| CRC | Colorectal cancer |
| DEGs | Differentially expressed genes |
| ER | Endoplasmic reticulum |
| FITC | Fluorescein 5-isothiocyanate |
| GEO | Gene expression omnibus |
| GO | Gene ontology |
| IP3R1 | Inositol 1,4,5-trisphosphate receptor type 1 |
| IRE1 | Inositol-requiring kinase-1 |
| MMP | Mitochondrial membrane potential |
| PERK | PRKR-like ER kinase |
| PI | Propidium iodide |
| RCN | Reticulocalbin |
| READ | Rectum adenocarcinoma |
| SDF4 | Stromal cell derived factor 4 |
| shRNA | Short hairpin RNA |
| TCGA | The cancer genome atlas |
| TG | Thapsigargin |
| TUNEL | Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling |
| UPR | Unfolded protein response |
Colorectal cancer (CRC) mostly originates from adenomas in the colon mucosa, driving high cancer-related recurrence and mortality rates worldwide [1]. The development of the malignant phenotype of CRC is controlled by multiple genetic alterations [2]. A large number of patients are already in the middle and late stages when diagnosed, thus missing the optimal window of opportunity for surgical treatment [3]. The other major treatments, including radiotherapy and chemotherapy, have limited impact on the curative effect and prognosis in patients with CRC [4]. Therefore, this study focuses on a factor that can regulate the development of CRC, providing a potential approach for the precise diagnosis and good prognosis of CRC.
Apoptosis is a programmed cell death process regulated by specific factors, which is used to describe the spontaneous death of cells after stimulation [5], including endoplasmic reticulum (ER) stress. The activation of apoptosis is one of the necessary approaches to suppressing the development of various cancers [6]. ER stress-induced unfolded protein response (UPR) to establish ER homeostasis and cytoprotective mechanisms [7,8]. However, when sustained and irreversible ER stress occurs, UPR was activated to induce further ER stress and cell apoptosis through several signaling pathways [9], including the PRKR-like ER kinase (PERK), inositol-requiring kinase-1 (IRE1) and activating transcription factor 6 (ATF6) signaling pathways. The disruption of ER homeostasis and promotes cancer cell apoptosis were also caused by persistent ER stress [10]. Blocking the UPR induced by ER stress may effectively promote apoptosis and inhibit the development of cancer. Previous reports revealed that ER stress-induced apoptosis may inhibit tumor growth in vivo in CRC model mice [11]. Thapsigargin (TG), an inhibitor that induces a sustained increase in cytosolic Ca2+ concentration by inhibiting sarcoplasmic/ER Ca2+-ATPase pumps, disrupts Ca2+ homeostasis in the ER [12]. It has been widely used to induce ER stress and Ca2+ homeostasis disturbances in a variety of cells. Previous reports have demonstrated that TG induced apoptosis through activating ER stress and impairing mitochondrial pathway [13,14]. Therefore, this study focused on exploring the molecules that promote cell apoptosis by regulating the ER stress.
Reticulocalbin 1 (RCN1) localized in the ER is a member of the CREC family, including Ca2+-binding protein, reticulocalbin (RCNs: RCN1, RCN2, RCN3), stromal cell derived factor 4 (SDF4), ER calcium-binding protein, and calumenin (CALU) [15]. Almost all these proteins contain multiple EF-hand motifs, which present a high affinity to Ca2+. RCN1 has been reported to regulate Ca2+ activity in the ER lumen and cytoplasm [16,17]. Multiple researches have confirmed that differentially expressed RCN1 is related to the progression of a variety of cancer types. It was found that RCN1 was upregulated in hepatocellular carcinoma [18], lung cancer [19], and kidney cancer [20]. RCN1 promotes drug resistance and a malignant phenotype by activating the c-MYC signaling pathway in hepatocellular carcinoma [21]. Notably, RCN1 has been identified to inhibit ER stress-induced apoptosis in hepatocellular carcinoma cells by regulating Ca2+ homeostasis and the PRKR-like ER kinase (PERK)-CCAAT/enhancer-binding protein-homologous protein (CHOP) pathway [22]. Similar effects of RCN1 on apoptosis have been reported in glioblastoma [23], nasopharyngeal carcinoma [24], and prostate cancer [17] cells. Moreover, it has been shown that RCN1 expression is upregulated in CRC tissues [25]. However, the effects of RCN1 on the apoptosis of CRC cells have not been fully elucidated. Therefore, the purpose of this study was to investigate the regulatory role of RCN1 in ER stress-induced apoptosis of CRC cells.
Through the loss-of-function experiments, knockdown of RCN1 was identified to suppress cell viability and facilitate TG-induced apoptosis in RKO and SW480. The activation of PERK, IRE1 and ATF6 signaling pathways proved that the deletion of RCN1 promoted apoptosis through inducing ER stress and activating UPR. Additionally, intracellular Ca2+ homeostasis was disrupted by silencing of RCN1. Inositol 1,4,5-trisphosphate receptor type 1 (IP3R1) meditated the effect of RCN1 on apoptosis of CRC cells.
The CRC datasets GSE32323 [26] and GSE26571 [27] in Gene Expression Omnibus (GEO) database, and colon adenocarcinoma (COAD) and rectum adenocarcinoma (READ) datasets in The Cancer Genome Atlas (TCGA) database were downloaded to analyze the expression levels of differentially expressed genes (DEG) were in normal tissues and tumor tissues. Based on the condition of log2fold-change > 1 and p < 0.05, the upregulated DEG were obtained and shown as a Venn diagram.
The Database for Annotation, Visualization and Integrated Discovery online tool was used for gene ontology (GO) analysis. The top 10 terms in biological process, cellular compartment, and molecular function with minimal significance were plotted.
The CRC samples and adjacent non-tumor tissues (n = 40) were collected from CRC patients, and stored at −80°C after freezing in liquid nitrogen for the subsequent experiments. This study has been approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University (approval number: KYLL202375). All patients provided informed consents for taking part in this study.
Cell lines and culture conditions
HCT-116, RKO, SW480, DLD-1 and NCM460 cells were purchased from iCell Bioscience Inc. in Shanghai, China. HCT-116 cells were cultured in McCOY’s 5A medium (Catalog Number [No.] PM150710) supplied by Procell Life Science & Technology Co., Ltd. (Procell) in Wuhan, China. RKO cells were cultured in minimum essential medium (No. 41500, Solarbio, Beijing, China). The RPMI-1640 medium (No. 31800, Solarbio) was used for culturing DLD-1 and NCM460. The above cells were lived in the medium with 1% penicillin (No. C8251, Solarbio), 1% streptomycin (No. S8290, Solarbio) and 10% fetal bovine serum (No. 11011-8611, Tianhang Biotechnology, Huzhou, China) in an incubator with 5% CO2 at 37°C. L15 medium (No. PM151010, Procell) containing 100 kU/L penicillin, 100 mg/L streptomycin was used for SW480 cells under 100% air conditions. The sequences of short hairpin RNAs (shRNAs) targeting RCN1 were as follows: shRCN1-1: 5′-GGATGAGAAGCTAACTAAA-3′, shRCN1-2: 5′-GGACGGGAAGTTAGACAAA-3′. The sequence targeting shRNA targeting IP3R1 (shIP3R1) was 5′-GATAGAGATTGTCAGATTA-3′. RKO and SW480 were transfected with shRNAs using Lipofectamine 3000 (No. L3000015) purchased from Invitrogen (Carlsbad, CA, USA). At 24 h after transfection, apoptosis of CRC cells was induced by 1 µM TG (No. T863962, Macklin, Shanghai, China).
TRIpure lysis (No. RP1001, BioTeke, Beijing, China) was used to isolate RNA from cells and tissues. These RNA samples were reversely transcribed to cDNA using BeyoRT II M-MLV reverse transcriptase (No. D7160L) purchased from Beyotime (Shanghai, China). Real-time PCR was performed using 2 × Taq PCR MasterMix (No. PC1150, Solarbio) and SYBR Green (No. SY1020, Solarbio) according to the manufacturer’s instructions. Sequences of primers used for relative RCN1 mRNA expression detection were listed in Suppl. Table S1. The thermocycling protocol was shown in Suppl. Table S2.
The harvested cells were mixed with the RIPA lysis (No. R0010, Solarbio) and 1 mM PMSF (No. P0100, Solarbio) for extracting proteins. Protein samples were separated through SDS-PAGE and transferred onto a polyvinylidene fluoride (No. IPVH00010, Millipore, Bedford, MA, USA) membrane. The membrane was incubated at 4°C overnight with primary antibodies after blocking for 1 h. Subsequently, the membrane was incubated with secondary antibodies at 37°C for 1 h. The details of antibodies were listed in Suppl. Table S3. Thereafter, the protein bands on the membrane were visualized by employing enhanced chemiluminescence (No. PE0010, Solarbio).
Cells were collected and seeded in a 96-well plate (5 × 103 cells per well). After treatment with TG for 24 h, cells in each well were incubated with CCK-8 solution (10 μL) (No. KGA317, KeyGEN BioTECH, Nanjing, China) at 37°C for 2 h. The optical density value of cells was assessed at 450 nm using a microplate reader (BioTek, Winooski, VT, USA).
Annexin V-fluorescein 5-isothiocyanate (FITC)/propidium iodide (PI) staining assay
Cell apoptosis was determined using an Annexin V-FITC/PI Apoptosis Detection Kit (No. KGA1102, KeyGEN BioTECH). After centrifugation at 150×g for 5 min, cells from each group were harvested and mixed in binding buffer (500 µL). Cells were then stained with Annexin V-FITC (5 μL) followed by mixing with PI (5 μL) for 15 min in the dark. NovoCyte flow cytometer (Agilent, Santa Clara, CA, USA) was used for measurement of apoptosis rate.
Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay
Apoptotic cells were stained by the mixture of enzyme solution and label solution in In Situ Cell Death Detection Kit (Red) (No. 12156792910, Roche, Basel, Switzerland) at 37°C for 1 h in the dark. Following the rinsing by phosphate buffer saline, cell sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (No. D106471-5mg, Aladdin, Shanghai, China) for 5 min. Next, the cells were observed and captured using a microscope (Olympus, Tokyo, Japan).
Detection of Ca2+ concentration
The intracellular Ca2+ concentration was detected by a Fluo-4 AM fluorescent probe (No. MX4504, Shanghai Maokang Biotechnology Co., Ltd. (Maokang), Shanghai, China). Cells in each group were centrifugated at 150×g for 5 min. Then, they were incubated in serum-free medium containing Fluo-4 AM (4 μM) at 37°C for 30 min. Subsequently, cells were photographed using a BX53 fluorescence microscope (Olympus). For the quantification of Ca2+ concentration, harvested cells was evaluated by flow cytometry.
RhoD-2AM (Red) and Mito-Tracker Green fluorescent probes (No. MX4507, Maokang) were used to label mitochondrial Ca2+ in CRC cells. Cells were incubated in serum-free medium with Rhod-2 AM (4 μM) and Mito-Tracker Green (100 nM, No. MX4309, Maokang) at 37°C for 30 min. Thereafter, cells were observed and captured using the aforementioned fluorescence microscope.
The mitochondrial membrane potential (MMP) in RKO and SW480 cells was detected using the JC-1 staining kit (No. C2006, Beyotime) according to the instructions provided by the manufacturers. Briefly, cells in each group were stained by JC-1 solution (0.5 mL) for 20 min. Cells were collected and resuspended by JC-1 staining buffer (1×) (200 μL). Finally, the MMP was examined by flow cytometry.
Cells were fixed with 4% polyformaldehyde for 15 min. After rinsing by phosphate buffer saline for three times, cells were covered by 0.1% tritonX-100 (No. ST795, Beyotime) for 30 min and then incubated by 1% bull serum albumin (No. A602440-0050, Sangon Biotech, Shanghai, China) for 15 min. Next, cells were incubated with primary antibodies at 4˚C overnight, including rabbit anti-RCN1 antibody (dilution 1:500, No. ab210404, Abcam) and mouse anti-IP3R1 antibody (dilution 1:50, No. sc-271197, Santa Cruz). Cells were then incubated with the same dilution ratio (1:1000) of Cy3-conjugated goat anti-rabbit IgG antibody (No. ab6939, Abcam) and FITC-conjugated goat anti-mouse IgG antibody (No. ab6785, Abcam). Next, nuclei of RKO and SW480 cells were counterstained with DAPI (No. D106471-5mg, Aladdin). After treatment with mounting medium (No. S2100, Solarbio), fluorescence in cells was recorded using the aforementioned fluorescence microscope.
Statistical analysis was performed using GraphPad Prism software 8.0 (GraphPad, San Diego, CA, USA). Student’s t-test was used for analyzing differences between two groups. One-way analysis of variances was performed to evaluate differences among three or more groups. The p-values < 0.05 indicated statistically significant differences. All data were displayed as the mean ± SD.
RCN1 was highly expressed in CRC tissues
To identify genes that potentially regulate the phonotypes of CRC tissues, we downloaded the GSE32323, GSE26571, COAD and READ datasets from the GEO and TCGA databases. The data from these datasets showed that 35 genes were upregulated in CRC tissues (Fig. 1A). The top 10 GO pathways in biological process, cellular compartment, and molecular function significantly enriched for these genes are presented in Fig. 1B. Previous studies have reported that the CREC protein family participates in the process of multiple diseases, and is associated with the activity and transport of Ca2+ in the cytosol [16]. The expression levels of RCN1, RCN2, RCN3, SDF4 and CALU in GSE32323 are presented as a heatmap. RCN1 showed high expression in CRC tissues (Fig. 1C), while its effect on the apoptosis of CRC cells remains unclear. The location of the CREC family genes on the chromosome is displayed in Fig. 1D. Additionally, the RCN1 expression in tumor tissue was significantly upregulated in GSE32323 and GSE26571 datasets (Fig. 1E). Relative RCN1 mRNA level was assessed in 40 pairs of tumors and non-tumor tissues. RCN1 expression was significantly increased in CRC tumor tissues (Figs. 1F and 1G). Therefore, we confirmed that RCN1 was highly expressed in RCR tissues.
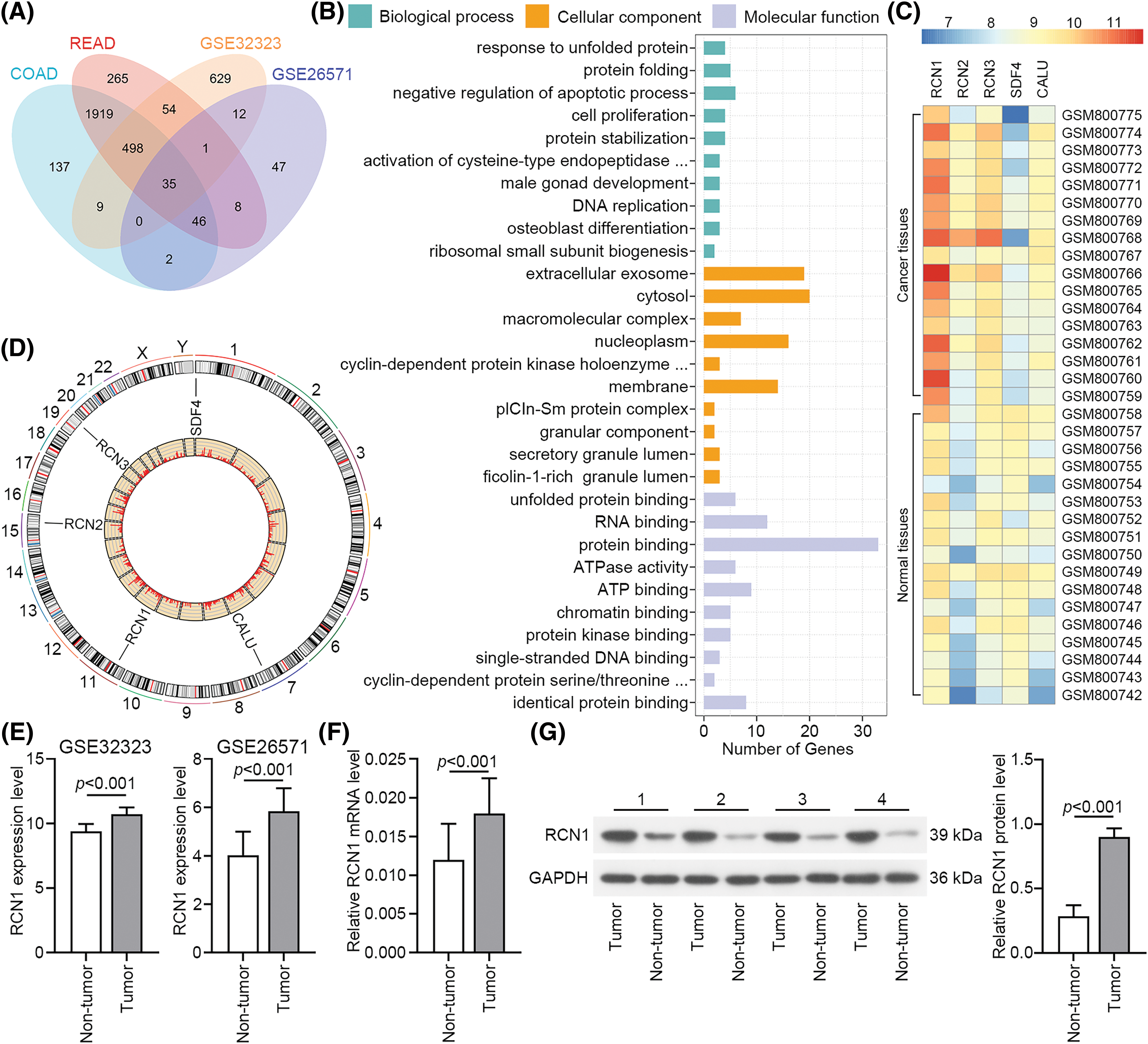
Figure 1: RCN1 shows high expression in colorectal cancer (CRC) tissues. (A) The overlap of upregulated differentially expressed genes (DEGs) in CRC tissues, whose data came from COAD, READ, GSE32323 and GSE26571 datasets. (B) Gene ontology (GO) enrichment terms of upregulated DEGs. (C) Heatmap showed the expression of CREC family genes in GSE32323. (D) The location of CREC family genes in genome. (E) RCN1 expression in GSE32323 and GSE26571 datasets. (F and G) RCN1 expressions were estimated in tumor and adjacent non-tumor samples.
Knockdown of RCN1 facilitated the ER stress‑induced apoptosis of CRC cells
Compared with NCM460 cells, RCN1 expression was obviously increased in CRC cell lines at mRNA and protein levels (Fig. 2A). RKO and SW480 cells were transfected with two pieces of shRCN1-1 and shRCN1-2. In the two types of CRC cells, relative mRNA and protein levels of RCN1 were reduced significantly (Figs. 2B and 2C). Additionally, previous reports demonstrated that the apoptosis of RKO and SW480 was promoted by TG-induced ER stress [28,29]. The viability of RKO and SW480 cells was suppressed by the absence of RCN1 (Fig. 2D).
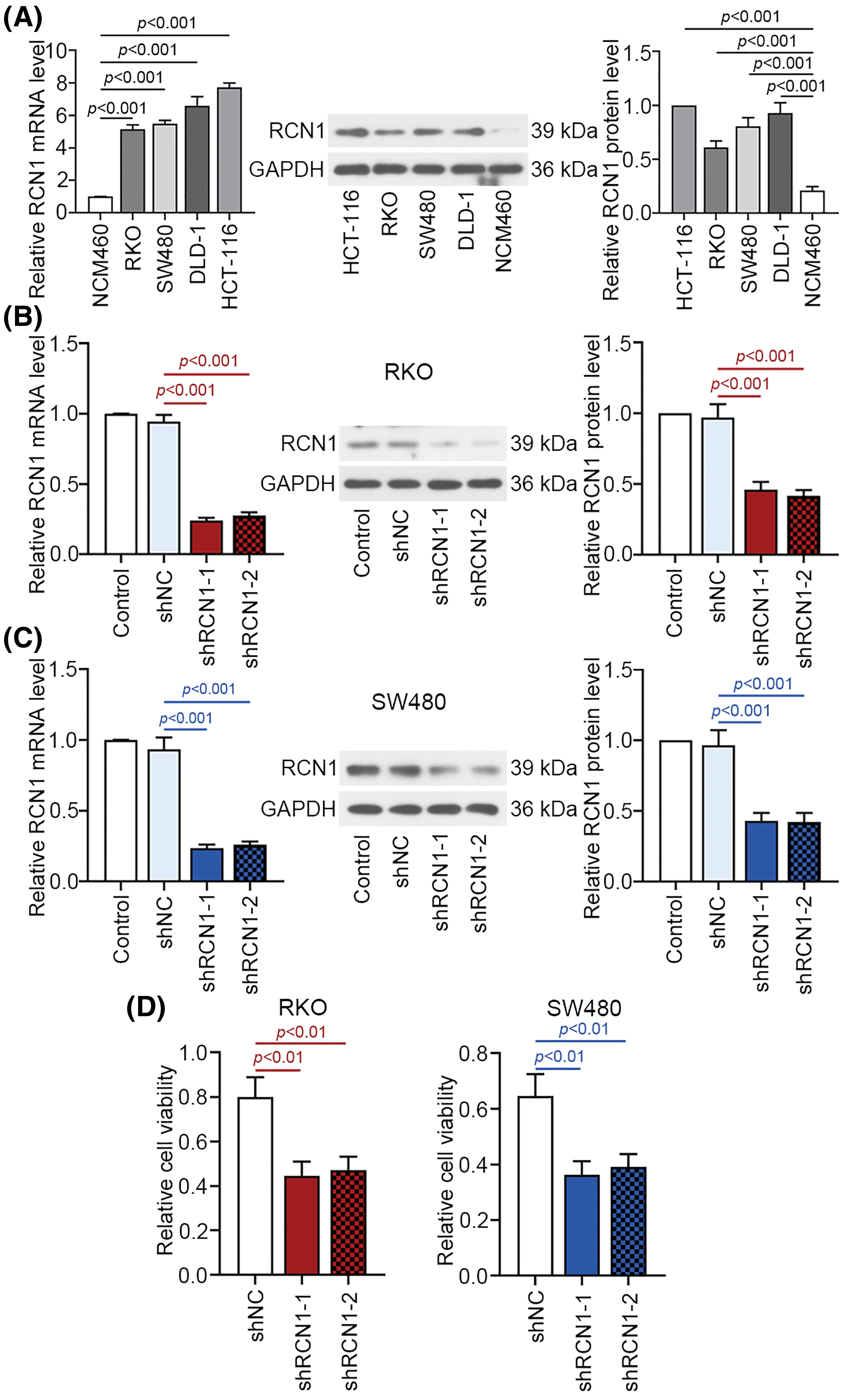
Figure 2: Knockdown of RCN1 suppresses cell viability of CRC cells. (A) RCN1 expression was estimated in NCM460, RKO, SW480, DLD-1 and HCT-116 cell lines using real time PCR. (B and C) The inhibition efficiency of shRCN1-1 and shRCN1-2 was examined in RKO and SW480 cells. (D) Viability of RKO and SW480 cells was assessed using CCK-8 assay.
Moreover, as displayed in Fig. 3A, knockdown of RCN1 obviously promoted the apoptosis of RKO and SW480 cell lines. B-cell lymphoma-2 (Bcl-2) protein expression was markedly suppressed by silencing RCN1. In contrast, absence of RCN1 increased the protein expression of Bcl-2-associated X (Bax) (Fig. 3B). Furthermore, after transfected with shRCN1s, TUNEL-positive cell rate upregulated remarkably (Fig. 3C). Collectively, these findings revealed that loss of RCN1 function suppressed cell viability and further promoted ER stress‑induced apoptosis of CRC cells.
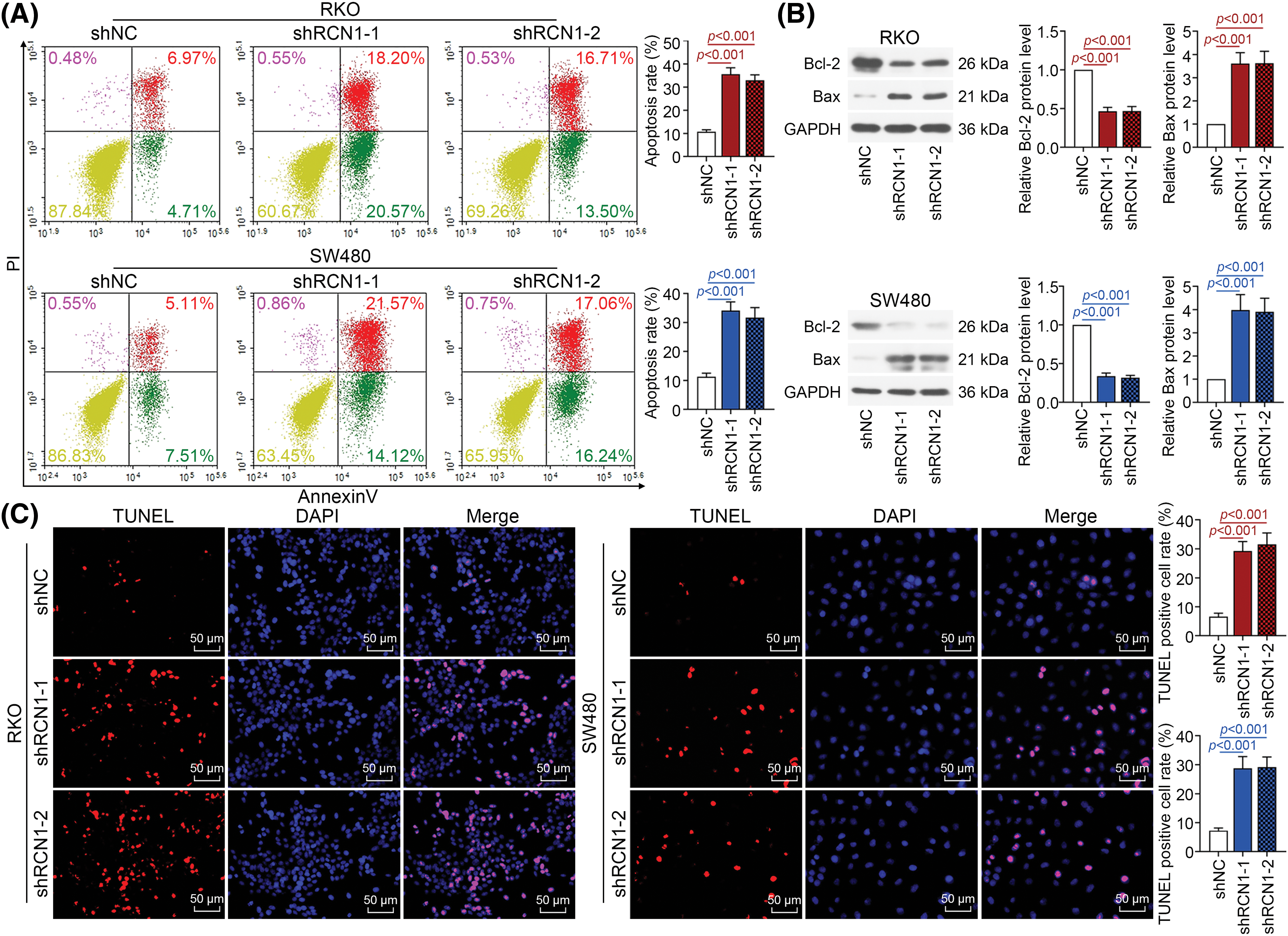
Figure 3: Downregulation of RCN1 promotes apoptosis induced by endoplasmic reticulum (ER) stress in CRC cells. (A) The apoptotic CRC cells stained by Annexin V-FITC/PI was detected by flow cytometry. (B) The regulatory effect of RCN1 on Bcl-2 and Bax was assessed by western blot. (C) TUNEL assay examined the regulation of RCN1 downregulation in cell apoptosis. Scale bar = 50 μm.
Depletion of RCN1 promoted ER stress-induced UPR
When defective proteins accumulate abnormally, the PERK, IRE1 and ATF6 signaling pathways are activated to induce further ER stress and cell apoptosis. PERK, IRE1 and ATF6 are crucial factors in the three pathways. These ER stress pathways are activated to mediate the UPR and subsequent cellular stress [30]. Depletion of RCN1 markedly induced the phosphorylation of PERK and IRE1 (Figs. 4A and 4B). Moreover, ATF6 was activated in RKO and SW480 cells. Knockdown of RCN1 also increased the levels of CHOP (Figs. 4C and 4D). The above findings suggested that absence of RCN1 activated the PERK, IRE1 and ATF6 pathways and facilitated UPR in CRC cells.
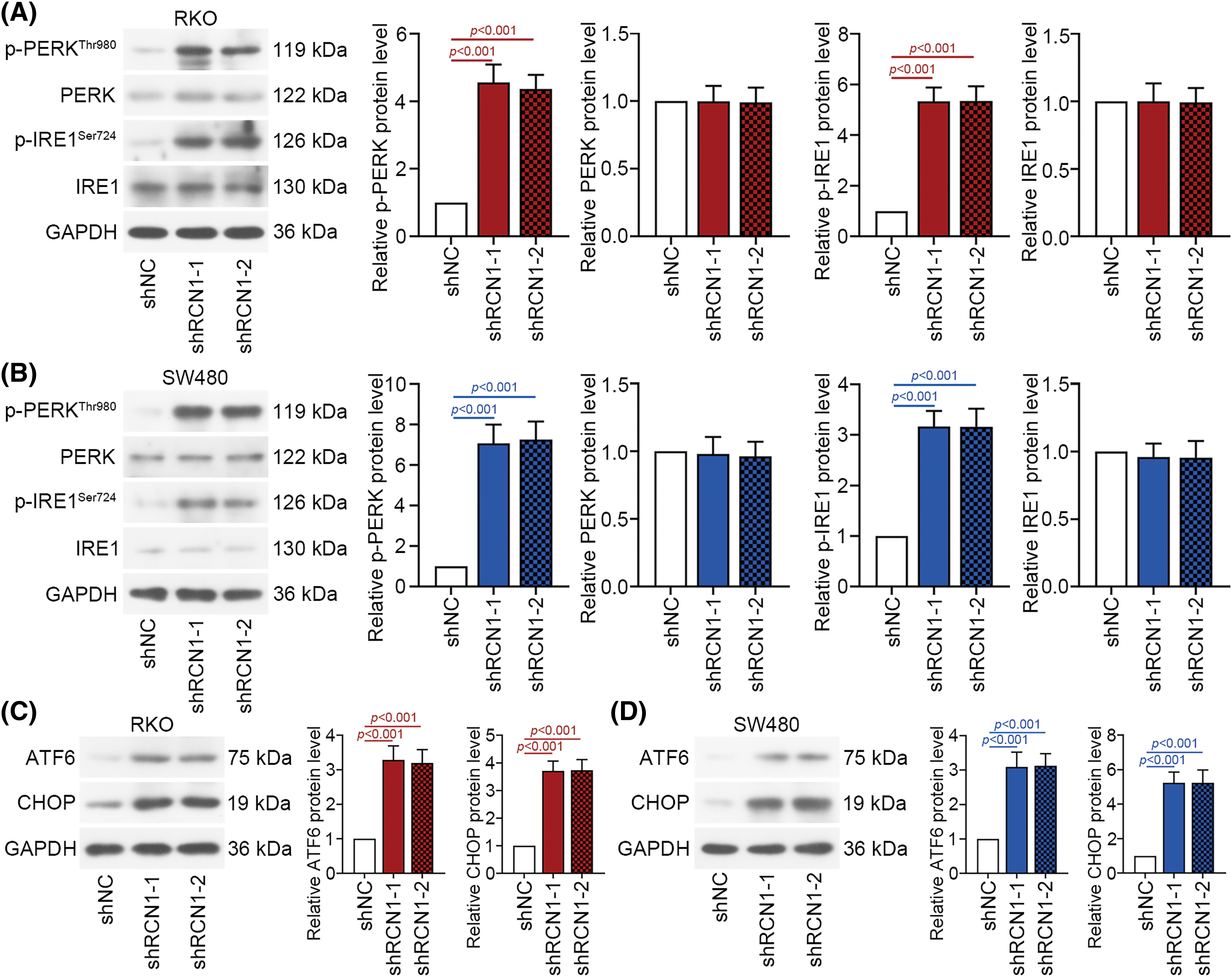
Figure 4: Depletion of RCN1 facilitates ER stress-induced UPR. (A and B) Activation of the PERK and IRE1 signaling pathways regulated by knocking down RCN1 was estimated by their phosphorylation levels. (C and D) Activation of the ATF6 signaling pathway was determined by the protein expression of ATF6 and CHOP.
Silencing of RCN1 disrupted intracellular Ca2+ homeostasis
Concentration of Ca2+ has been considered an important factor in cell apoptosis [31]. Intracellular Ca2+ homeostasis is the basis for maintaining normal cellular structure and function [32]. As depicted in Figs. 5A and 5B, intracellular Ca2+ was labeled with Fluo-4 AM fluorescent probes in RKO and SW480 cells. The intracellular Ca2+ concentration was significantly upregulated by the absence of RCN1, suggesting that a decrease in RCN1 expression disrupted the intracellular Ca2+ homeostasis in CRC cells. Additionally, the mitochondrial Ca2+ was stained with Rhod-2AM, and the staining results demonstrated that mitochondrial Ca2+ was obviously increased in RCN1-silenced RKO and SW480 cells (Figs. 5C and 5D). Moreover, a reduction in MMP was observed after knockdown of RCN1 in CRC cells. Notably, downregulation of shRCN1s significantly increased the MMP in cells with low MMP (Fig. 5E). The expression of mitochondrial cytochrome c was reduced in RKO and SW480 cells by knocking down RCN1. Meanwhile, cytochrome c in cytoplasm was significantly upregulated by depletion of RCN1 (Fig. 5F). Therefore, the intracellular Ca2+ homeostasis was disrupted by the downregulation of RCN1 in CRC cells.

Figure 5: Knockdown of RCN1 increases Ca2+ concentration and reduces mitochondrial membrane potential (MMP). (A and B) The intracellular Ca2+ was stained with Fluo-4 AM fluorescent probe in RCN1 silenced-RKO and SW480 cells. Flow cytometry detected the fluorescence intensity. Scale bar = 200 μm. (C and D) The RKO and SW480 cells with shRNAs were stained with Rhod-2AM and Mito-Tracker Green fluorescent probes, which marked intracellular Ca2+ and mitochondrial Ca2+. Scale bar = 50 μm. (E) JC-1 assay measured the MMP levels of CRC cells, and cells with low MMP were quantified. (F) The activation of cytochrome c was assessed by detecting its protein expression in mitochondria and cytoplasm.
IP3R1 mediated the regulatory effect of RCN1 on ER stress-induced apoptosis
Previous research has demonstrated that IP3R1 is involved in the release of ER Ca2+ through interaction with RCN1 [22]. Therefore, we investigated whether the RCN1 regulates ER stress-induced apoptosis by affecting IP3R1. The results of fluorescence co-localization analysis revealed that both RCN1 and IP3R1 were abundant in the cytoplasm of untreated RKO and SW480 cells (Fig. 6A). As displayed in Fig. 6B, RCN1 and IP3R1 were significantly downregulated by knocking down RCN1, and IP3R1 expression was obviously reduced by silencing IP3R1 in RKO cells. The depletion of IP3R1 further downregulated the protein expression of RCN1 and IP3R1 in the RCN1-reduced RKO cells. Cell apoptosis induced by knocking down RCN1 was suppressed in CRC cells after reduction of IP3R1 (Fig. 6C). RCN1 silencing-induced the damage to intracellular Ca2+ homeostasis induced by shRCN1 was reversed by the silencing of IP3R1 (Fig. 6D). In conclusion, the findings demonstrated that IP3R1 mediates ER stress-induced apoptosis promoted by RCN1 reduction.
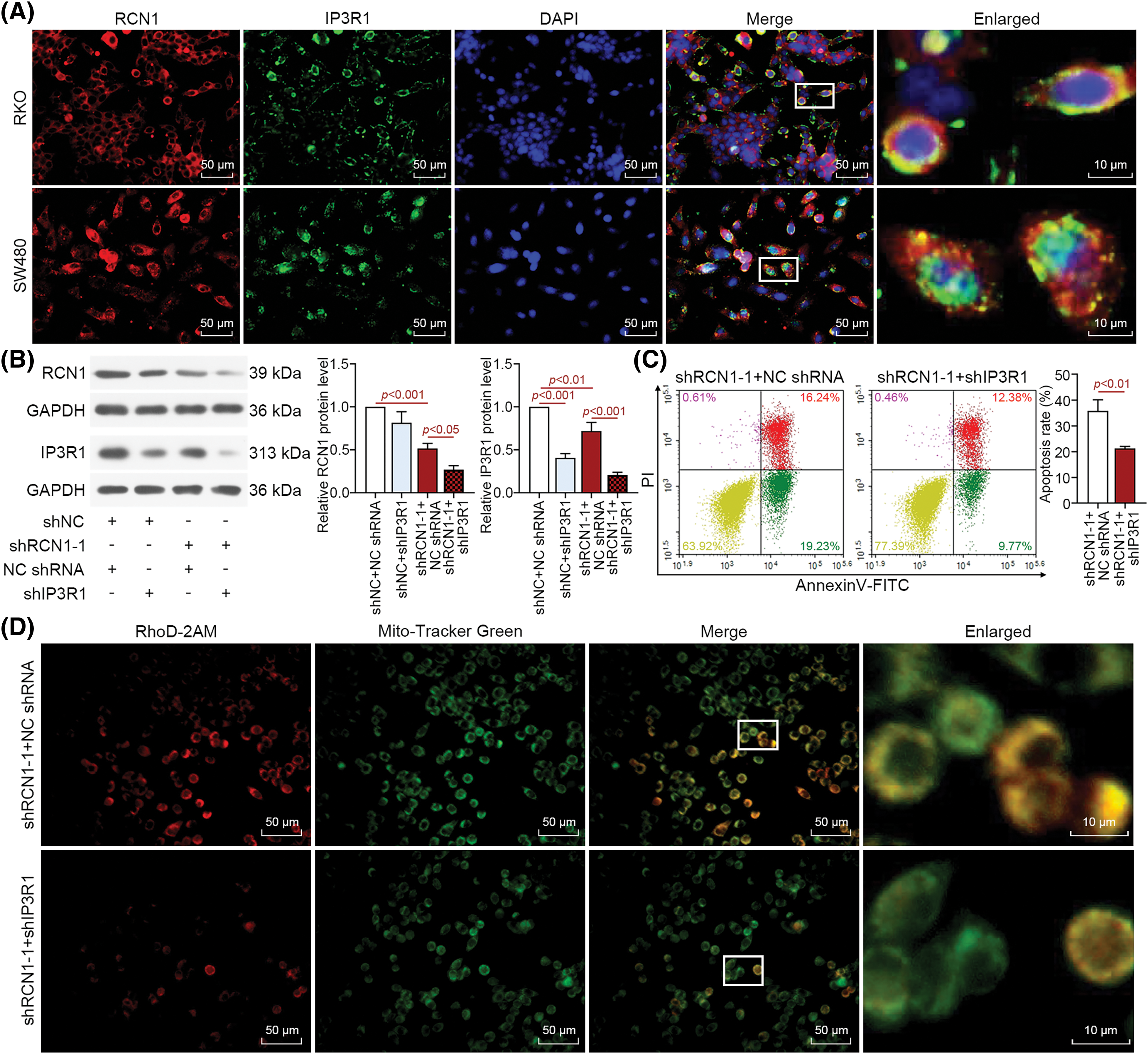
Figure 6: RCN1 regulates ER stress-induced apoptosis through IP3R1. (A) The co-localization of RCN1 and IP3R1 was assessed by immunofluorescence double staining in untreated RKO and SW480 cells. Scale bar = 50 μm. (B) RCN1 and IP3R1 expression levels were detected in RKO cell line under the absence of RCN1 or IP3R1 condition. (C) The effect of knockdown RCN1 or IP3R1 on cell apoptosis was estimated by flow cytometry. (D) The mitochondrial Ca2+ was labeled and evaluated by Rhod-2AM and Mito-Tracker Green fluorescent probe. Scale bar = 50 μm.
Findings in this report revealed that RCN1 was upregulated in CRC tissues. Downregulation of RCN1 facilitated the ER stress-induced apoptosis of RKO and SW480 cells. Depletion of RCN1 in CRC cells, and resulted in the disturbance of intracellular Ca2+ homeostasis and MMP. IP3R1 mediates the effect of RCN1 on cell apoptosis. The present findings may provide an available therapeutic approach to intercept the development of CRC.
RCN1 functions as a Ca2+-binding protein that is stored in ER [33]. It has been reported that a decrease in RCN1 expression obstructs the progress of lung cancer cells [19]. Results from a proteomic analysis confirms that RCN1 was upregulated in renal cell carcinoma tissues and clarifies that RCN1 serves as a potential molecule to regulate the progress of renal cell carcinoma [20]. Moreover, loss of RCN1 function has been reported to induce the apoptosis of prostate cancer cells [17]. Downregulation of RCN1 facilitates apoptosis induced by ER stress in nasopharyngeal carcinoma [24]. RCN1 has been identified as a novel candidate for CRC therapy [34]. However, the function of RCN1 in human CRC warrants further investigation. Through loss-of-function experiments, RCN1 was downregulated in CRC cells. At present, there is no reliable evidence regarding the use of this method in clinical practice. The sequences of shRCN1-1 and shRCN1-2 differed, and both significantly suppressed RCN1 expression in CRC cells. Absence of RCN1 suppressed cell viability and promoted apoptosis induced by ER stress. The flow cytometry results demonstrated that the rate of late apoptosis of RCN1-silenced cells was obviously increased. Therefore, downregulation of RCN1 may serve as an effective approach to inhibiting the development of CRC. These findings indicated that RCN1 was implicated in the malignant phonotypes of CRC.
Whether ER stress-induced apoptosis is regulated by RCN1 in RKO and SW480 cells is still unknown. Therefore, efficient induction of ER stress is necessary in this study. Previous literatures suggests that TG treatment promotes the expression of 78 kDa glucose-regulated protein and cleaved caspase-3 in SW480 cells, and disrupts ER homeostasis, as well as facilitating cell apoptosis [28]. Additionally, the viability of RKO cells treated with TG is inhibited compared with that of control cells. Treatment with TG remarkably induces apoptosis of RKO cells [29]. According to these reports, TG as an effective inducer for ER stress was used in RKO and SW480 cells. Knockdown of RCN1 was involved in ER stress-induced apoptosis of TG-treated CRC cells.
ER is a main storage site for intracellular Ca2+. ER stress caused by excessive accumulation of defective proteins activates the UPR to induce apoptosis [35]. During the UPR process, the PERK, IRE1, and ATF6 was activated, accompanied by promoting the expression of pro-apoptotic factor CHOP [36]. Our results demonstrated that knockdown of RCN1 in RKO and SW480 cells led to phosphorylation of PERK and IRE1, as well as activation of the ATF6 (Fig. 4). Upregulation of CHOP induced by the depletion of RCN1 activated Bax and suppressed Bcl-2 in CRC cells. Our analysis verified that downregulation of RCN1 facilitated apoptosis induced by ER stress via activating the PERK, IRE1, and ATF6 pathways. Bax is an essential factor facilitating mitochondrial membrane permeabilization [30]. Low RCN1 expression induced a reduction of MMP in CRC cells.
IP3R1 is one of the ER-resident Ca2+ release channels [37]. The alteration of Ca2+ concentration in ER has been identified as an important cause of ER stress [38]. RCN1 interacts with IP3R1 at the ER lumen via two EF-hand Ca2+-binding motifs to suppress the release of ER Ca2+ and cell apoptosis. RCN1 depletion activated UPR through the regulation of IP3R1 and further affected the expression of CHOP [22]. Results in this report confirmed that IP3R1 downregulation suppressed the accumulation of Ca2+ in the mitochondria of RCN1-silenced CRC cells. Moreover, it reversed the promotive effect of RCN1 loss on ER stress-induced apoptosis. It has been reported that IP3R1 promoted the accumulation of mitochondrial Ca2+ to activate the release of cytochrome c and induced cell apoptosis [39]. Our results suggested that downregulation of RCN1 promoted the activation of cytochrome c in RKO and SW480 cells, indicating that RCN1 participated in the apoptosis of CRC cells through affecting IP3R1.
Based on the present findings, RCN1 was identified as a crucial factor for regulating cell death in CRC. Silencing RCN1 may be an effective therapeutic approach to suppressing the development of CRC. Further investigation on RCN1 is warranted to determine its importance as a molecular target and promote its use in clinical practice. Moreover, it is necessary to elucidate the mechanism underlying of RCN1 and its effects on the progression of CRC. This is the direction of our research in the future.
Collectively, the findings displayed in this study clearly demonstrated that depletion of RCN1 inhibited cell viability and promoted ER stress-induced apoptosis via regulating IP3R1 and activating the PERK, IRE1, and ATF6 signaling pathways.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Xuan Shi, Aixia Gong; data collection: Xuan Shi, Yufen Wang; analysis and interpretation of results: Chenyu Li, Wangshu Fu, Xinyue Zhang; draft manuscript preparation: Xuan Shi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: The study has been approved by the Ethics Committee of the First Affiliated Hospital of Jinzhou Medical University (approval number: KYLL202375). All patients provided informed consents for taking part in this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary materials are available online at https://doi.org/10.32604/biocell.2024.048076.
References
1. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17. [Google Scholar] [PubMed]
2. Uzozie AC, Selevsek N, Wahlander A, Nanni P, Grossmann J, Weber A, et al. Targeted proteomics for multiplexed verification of markers of colorectal tumorigenesis. Mol Cell Proteomics. 2017;16(3):407–27. [Google Scholar] [PubMed]
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [Google Scholar] [PubMed]
4. Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. 2020;70(3):145–64. [Google Scholar] [PubMed]
5. Kerr JF, Harmon BV. Definition and incidence of apoptosis: an historical perspective. In: Tomei LD, Cope FO, editors. Apoptosis: the molecular basis of cell death. vol. 3. New York: Cold Spring Harbor Laboratory Press; 1991. p. 5–29. [Google Scholar]
6. Kashyap D, Garg VK, Goel N. Intrinsic and extrinsic pathways of apoptosis: role in cancer development and prognosis. Adv Protein Chem Struct Biol. 2021;125:73–120. [Google Scholar] [PubMed]
7. Liu C, Xu J, Guo C, Chen X, Qian C, Zhang X, et al. Gambogenic acid induces endoplasmic reticulum stress in colorectal cancer via the aurora a pathway. Front Cell Dev Biol. 2021;9:736350. [Google Scholar] [PubMed]
8. Bonsignore G, Martinotti S, Ranzato E. Endoplasmic reticulum stress and cancer: could unfolded protein response be a druggable target for cancer therapy? Int J Mol Sci. 2023;24(2):1566. [Google Scholar] [PubMed]
9. Ren J, Bi Y, Sowers JR, Hetz C, Zhang Y. Endoplasmic reticulum stress and unfolded protein response in cardiovascular diseases. Nat Rev Cardiol. 2021;18(7):499–521. [Google Scholar] [PubMed]
10. Zhang R, Bian C, Gao J, Ren H. Endoplasmic reticulum stress in diabetic kidney disease: adaptation and apoptosis after three UPR pathways. Apoptosis. 2023;28(7–8):977–96. [Google Scholar] [PubMed]
11. Choi SS, Lee SK, Kim JK, Park HK, Lee E, Jang J, et al. Flightless-1 inhibits ER stress-induced apoptosis in colorectal cancer cells by regulating Ca2+ homeostasis. Exp Mol Med. 2020;52(6):940–50. [Google Scholar] [PubMed]
12. Oslowski CM, Urano F. Measuring ER stress and the unfolded protein response using mammalian tissue culture system. Methods Enzymol. 2011;490(2):71–92. [Google Scholar] [PubMed]
13. Wang C, Li T, Tang S, Zhao D, Zhang C, Zhang S, et al. Thapsigargin induces apoptosis when autophagy is inhibited in HepG2 cells and both processes are regulated by ROS-dependent pathway. Environ Toxicol Pharmacol. 2016;41:167–79. [Google Scholar] [PubMed]
14. Wang F, Liu DZ, Xu H, Li Y, Wang W, Liu BL, et al. Thapsigargin induces apoptosis by impairing cytoskeleton dynamics in human lung adenocarcinoma cells. Sci World J. 2014;2014:619050. [Google Scholar]
15. Wang Y, Zhou X, Wang H, Sun L, Wang B, Jiang Y, et al. The role of Eimeria tenella EtCab protein in the attachment and invasion of host cells. Vet Parasitol. 2021;292:109415. [Google Scholar] [PubMed]
16. Honoré B. The rapidly expanding CREC protein family: members, localization, function, and role in disease. Bioessays. 2009;31(3):262–77. [Google Scholar]
17. Liu X, Zhang N, Wang D, Zhu D, Yuan Q, Zhang X, et al. Downregulation of reticulocalbin-1 differentially facilitates apoptosis and necroptosis in human prostate cancer cells. Cancer Sci. 2018;109(4):1147–57. [Google Scholar] [PubMed]
18. Yu LR, Zeng R, Shao XX, Wang N, Xu YH, Xia QC. Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophor. 2000;21(14):3058–68. [Google Scholar]
19. Chen X, Shao W, Huang H, Feng X, Yao S, Ke H. Overexpression of RCN1 correlates with poor prognosis and progression in non-small cell lung cancer. Hum Pathol. 2019;83:140–8. [Google Scholar] [PubMed]
20. Giribaldi G, Barbero G, Mandili G, Daniele L, Khadjavi A, Notarpietro A, et al. Proteomic identification of Reticulocalbin 1 as potential tumor marker in renal cell carcinoma. J Proteomics. 2013;91:385–92. [Google Scholar] [PubMed]
21. Wang JW, Ma L, Liang Y, Yang XJ, Wei S, Peng H, et al. RCN1 induces sorafenib resistance and malignancy in hepatocellular carcinoma by activating c-MYC signaling via the IRE1α-XBP1s pathway. Cell Death Discov. 2021;7(1):298. [Google Scholar] [PubMed]
22. Xu S, Xu Y, Chen L, Fang Q, Song S, Chen J, et al. RCN1 suppresses ER stress-induced apoptosis via calcium homeostasis and PERK-CHOP signaling. Oncogenesis. 2017;6(3):e304. [Google Scholar] [PubMed]
23. Gomez J, Areeb Z, Stuart SF, Nguyen HPT, Paradiso L, Zulkifli A, et al. EGFRvIII promotes cell survival during endoplasmic reticulum stress through a reticulocalbin 1-dependent mechanism. Cancers. 2021;13(6). [Google Scholar]
24. Huang ZH, Qiao J, Feng YY, Qiu MT, Cheng T, Wang J, et al. Reticulocalbin-1 knockdown increases the sensitivity of cells to Adriamycin in nasopharyngeal carcinoma and promotes endoplasmic reticulum stress-induced cell apoptosis. Cell Cycle. 2020;19(13):1576–89. [Google Scholar] [PubMed]
25. Ning J, Liu M, Shen J, Wang D, Gao L, Li H, et al. Expression signature and prognostic value of CREC gene family in human colorectal cancer. BMC Cancer. 2023;23(1):878. [Google Scholar] [PubMed]
26. Khamas A, Ishikawa T, Shimokawa K, Mogushi K, Iida S, Ishiguro M, et al. Screening for epigenetically masked genes in colorectal cancer using 5-Aza-2'-deoxycytidine, microarray and gene expression profile. Cancer Genomics Proteomics. 2012;9(2):67–75. [Google Scholar] [PubMed]
27. Leydold SM, Seewald M, Stratowa C, Kaserer K, Sommergruber W, Kraut N, et al. Peroxireduxin-4 is over-expressed in colon cancer and its down-regulation leads to apoptosis. Cancer Growth and Metastasis. 2011;4:CGM.S6584. [Google Scholar]
28. Liang G, Fang X, Yang Y, Song Y. Knockdown of CEMIP suppresses proliferation and induces apoptosis in colorectal cancer cells: downregulation of GRP78 and attenuation of unfolded protein response. Biochem Cell Biol. 2018;96(3):332–41. [Google Scholar] [PubMed]
29. Wang G, Han J, Wang G, Wu X, Huang Y, Wu M, et al. ERO1α mediates endoplasmic reticulum stress-induced apoptosis via microRNA-101/EZH2 axis in colon cancer RKO and HT-29 cells. Hum Cell. 2021;34(3):932–44. [Google Scholar] [PubMed]
30. Keramidas P, Papachristou E, Papi RM, Mantsou A, Choli-Papadopoulou T. Inhibition of PERK kinase, an orchestrator of the unfolded protein response (UPRsignificantly reduces apoptosis and inflammation of lung epithelial cells triggered by SARS-CoV-2 ORF3a protein. Biomedicines. 2023;11(6):1585. [Google Scholar] [PubMed]
31. Bahar E, Kim H, Yoon H. ER stress-mediated signaling: action potential and Ca2+ as key players. Int J Mol Sci. 2016;17(9):1558. [Google Scholar] [PubMed]
32. Luan S, Wang C. Calcium signaling mechanisms across kingdoms. Annu Rev Cell Dev Biol. 2021;37:311–40. [Google Scholar] [PubMed]
33. Rybarski M, Mrohs D, Osenberg K, Hemmersbach M, Pfeffel K, Steinkamp J, et al. Loss of parkin causes endoplasmic reticulum calcium dyshomeostasis by upregulation of reticulocalbin 1. Eur J Neurosci. 2023;57(5):739–61. [Google Scholar] [PubMed]
34. Watanabe M, Takemasa I, Kawaguchi N, Miyake M, Nishimura N, Matsubara T, et al. An application of the 2-nitrobenzenesulfenyl method to proteomic profiling of human colorectal carcinoma: a novel approach for biomarker discovery. Proteomics Clin Appl. 2008;2(6):925–35. [Google Scholar] [PubMed]
35. Marciniak SJ, Chambers JE, Ron D. Pharmacological targeting of endoplasmic reticulum stress in disease. Nat Rev Drug Discov. 2022;21(2):115–40. [Google Scholar] [PubMed]
36. Wek RC. Role of eIF2α kinases in translational control and adaptation to cellular stress. Cold Spring Harb Perspect Biol. 2018;10(7):a03287. [Google Scholar]
37. Rieusset J, Fauconnier J, Paillard M, Belaidi E, Tubbs E, Chauvin MA, et al. Disruption of calcium transfer from ER to mitochondria links alterations of mitochondria-associated ER membrane integrity to hepatic insulin resistance. Diabetologia. 2016;59(3):614–23. [Google Scholar] [PubMed]
38. Zhou F, Gao H, Shang L, Li J, Zhang M, Wang S, et al. Oridonin promotes endoplasmic reticulum stress via TP53-repressed TCF4 transactivation in colorectal cancer. J Exp Clin Cancer Res. 2023;42(1):150. [Google Scholar] [PubMed]
39. Yuan M, Gong M, He J, Xie B, Zhang Z, Meng L, et al. IP3R1/GRP75/VDAC1 complex mediates endoplasmic reticulum stress-mitochondrial oxidative stress in diabetic atrial remodeling. Redox Biol. 2022;52:102289. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools