 Open Access
Open Access
ARTICLE
miR-30a-5p/PHTF2 axis regulates the tumorigenesis and metastasis of lung adenocarcinoma
1 Department of Rehabilitation and Palliative Medicine, The Third Affiliated Hospital of Kunming Medical University, Yunnan Cancer Hospital, Kunming, China
2 Department of Pathology, The Third Affiliated Hospital of Kunming Medical University, Yunnan Cancer Hospital, Kunming, China
3 Emergency Department (Outpatient Chemotherapy Center), The Third Affiliated Hospital of Kunming Medical University, Yunnan Cancer Hospital, Kunming, China
4 Department of Cancer BioTreatment Center, The Third Affiliated Hospital of Kunming Medical University, Yunnan Cancer Hospital, Kunming, China
5 Department of Cancer Institute, The Third Affiliated Hospital of Kunming Medical University, Yunnan Cancer Hospital, Kunming, China
* Corresponding Author: XICAI WANG. Email:
# Contributed equally
(This article belongs to the Special Issue: MicroRNA as Biomarkers for Disease Diagnosis and Progression)
BIOCELL 2024, 48(4), 581-590. https://doi.org/10.32604/biocell.2024.047260
Received 31 October 2023; Accepted 23 January 2024; Issue published 09 April 2024
Abstract
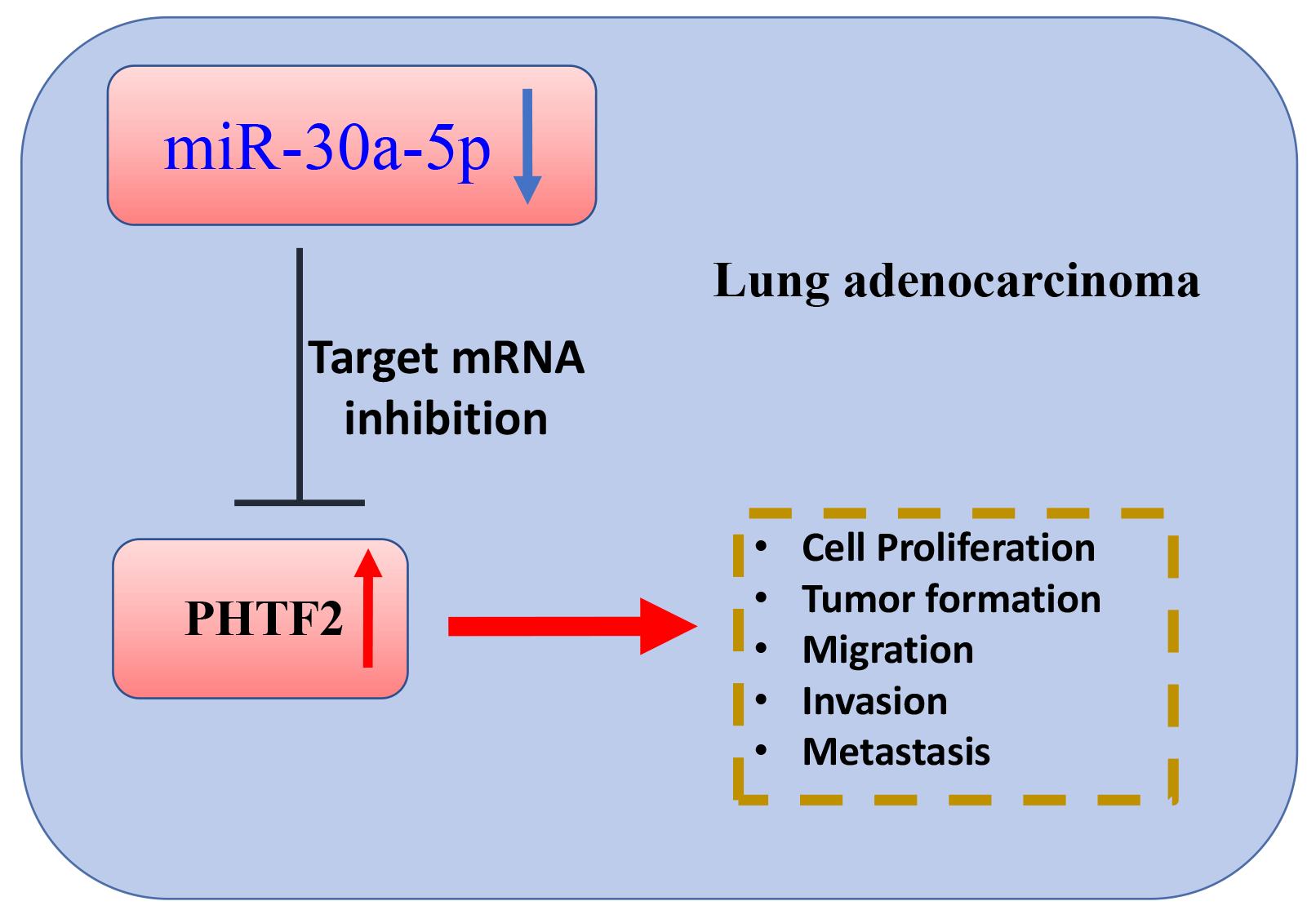
Background: Lung adenocarcinoma is a very pervasive histological form of lung cancers, and inhibiting metastasis is crucial for effective treatment. In this investigation, we explored the functional interaction of miR-30a-5p and the putative transcription factor 2 of the homeodomain (PHTF2) in dictating the aggressiveness and metastasis of lung adenocarcinoma. Method: We collected clinical samples to evaluate the expression patterns of miR-30a-5p and PHTF2 in lung adenocarcinoma along with normal tissues. Cellular experiments including cell count kit (CCK)-8 growth assay, apoptosis analysis, migration and invasion examinations were performed to assess the aggressiveness of lung adenocarcinoma cells. Furthermore, we examined tumorigenesis and metastasis in a nude mouse model. Results: MiR-30a-5p exhibited downregulation pattern in lung adenocarcinoma samples. Transfection of miR-30a-5p mimic in lung adenocarcinoma cells resulted in the suppression of malignant characteristics. Notably, the administration of miR-30a-5p mimic also curbed tumorigenesis and metastasis of lung adenocarcinoma cells in animal model. Moreover, PHTF2 was found to be a molecular target of miR-30a-5p. Upregulating PHTF2 counteracted the tumor-suppressive effect of the miR-30a-5p mimic. Conclusion: miR-30a-5p functions as a tumor-suppressive molecule while PHTF2 acts as an oncogenic factor in the development and metastasis of lung adenocarcinoma. Therefore, targeting miR-30a-5p and PHTF2 could be developed into a promising therapeutic approach for inhibiting metastasis in lung adenocarcinoma.Graphic Abstract
Keywords
Abbreviations
| ALL | acute lymphoblastic leukemia |
| CCK | cell count kit |
| E2F7 | E2F Transcription Factor |
| EMT | epithelial mesenchymal transition |
| LDH | Lactate dehydrogenase |
| LUAD | Lung adenocarcinoma |
| miRNA | micro RNA |
| miR-NC | miRNA control |
| MUT | mutated |
| PHTF2 | The putative transcription factor 2 of the homeodomain |
| TCGA | The Cancer Genome Atlas |
| UTR | untranslated region |
| WT | wild-type |
Lung cancer has become a primary cause of cancer-associated death globally, and its overall survival rate over a 5-year period is estimated to be 15% [1]. Among the different forms of pulmonary tumors, lung adenocarcinoma is the most common histological subtype [2,3]. This particular form of cancer primarily originates from the mucosal glands in the lung tissues and takes up about 40% of all lung cancer diagnoses [4]. Typically, lung adenocarcinoma initiates in the peripheral regions of the lung, although it can also be found in areas with chronic inflammation or scars [5]. Smoking is considered as a major identifiable risk for lung adenocarcinoma development, although other factors such as a family history of lung cancer and occupational exposure to carcinogens have been implicated in its occurrence [6]. Despite the implementation of various treatment approaches, including chemotherapy, radiotherapy, targeted therapy, and immunotherapy, managing lung adenocarcinoma is challenging due to its inherent heterogeneity at the histological, cellular, and molecular levels [7–10]. The development of drug resistance and frequent metastasis during tumor progression are the major obstacles in achieving effective therapeutic outcomes [11,12]. Consequently, investigating the molecular changes that occur during tumor progression is crucial for developing improved therapeutic strategies.
MicroRNAs (miRNAs) are extensively studied non-coding RNAs which are engaged in controlling various cellular events, encompassing cell growth, differentiation, mobility, and the aggressiveness of cancer [13,14]. miRNAs interact with specific regions in the target mRNA molecules, and this interaction leads to the formation of double-stranded RNA structures that can be recognized and degraded by a complex involved in miRNA-mediated silencing [15]. Interestingly, more than 50% of miRNA coding sequences are located within genomic domains associated with cancer, implying that the dysregulation of miRNAs may be implicated in the pathogenic development of cancerous diseases [16]. Of note, miRNAs can either promote oncogenic activities or act as tumor suppressors, depending on the specific downstream targets and the context of tumors [17,18].
In lung adenocarcinoma, the regulation of the epithelial mesenchymal transition (EMT) by miRNAs varies depending on the targets they affect. This process ultimately facilitates the invasion and metastasis of cancer cells [19]. For instance, miRNA-520a has been shown to inhibit the pathogenic advancement of lung cancer [20]. On the other hand, miRNA-155-5p boosts cell growth, motility, and tumorigenesis of lung squamous cell carcinoma [21]. Additionally, miR-30a-5p is an emerging tumor suppressor in cancer biology that was reported to repress cell growth and metastasis in several tumors, including renal cell carcinoma, breast tumors, esophageal cancer, gallbladder cancer, and gastric cancer [22–26]. However, it remains to be explored in-depth how miR-30a-5p regulates lung adenocarcinoma progression.
The putative transcription factor 2 of the homeodomain (PHTF2) is a recently discovered transcription factor with limited understanding of its biological role. In gastric cancer, PHTF2 was identified as the central gene in the protein-protein interaction network of genes with differential expressions [27]. PHTF2 exhibited increased expression in various subtypes of gastric tumor tissues according to the analyses of publicly available datasets and clinical samples. Furthermore, elevated levels of PHTF2 were shown to be linked with the dismal prognosis within cancer patients [27]. Despite these findings, the biological function of PHTF2 in other cancer types, including lung adenocarcinoma, is poorly understood. In this investigation, we studied the engagement of miR-30a-5p and PHTF2 in lung adenocarcinoma, revealing their functional interplay and the joint contribution to cell aggressiveness and metastasis in the cell and animal models.
Surgery was performed to obtain 60 pairs of lung adenocarcinoma specimens and normal tissues adjacent to the cancerous area at Yunnan Provincial Cancer Hospital. Two separate pathologists conducted evaluations on all of the collected samples. The utilization of human samples had been granted the approval by the Medical Research Ethics Committee of Yunnan Provincial Cancer Hospital (Approval number: KYLX2021-124). The informed consent was signed by every participating patient. This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Kunming Medical University (No. Kmmu20220583).
Cell culture and stable PHTF2 cell line generation
Human lung adenocarcinoma cell lines (XWLC-05 cells, Kanglang Biotech, Shanghai, China; and NCI-H1650 cells, Cobioer Bioscience, Nanjing, China), human pulmonary epithelial cell line (BEAS-2B cells, Xingyu Biotech, Shanghai, China), and human embryonic kidney cell line (HEK293 cells, Procell, Wuhan, China) were cultivated using Dulbecco’s Modification of Eagle’s Medium (PM150210, Procell, Wuhan, China) supplemented with 10% fetal bovine serum (SH3007003, GE™ Hyclone, San Diego, CA, USA), 1% penicillin/streptomycin antibiotic solution (PB180120, Procell, Wuhan, China) at 37°C and 5% CO2. The empty vector (pLVTHM empty vector) and PHTF2 expression plasmid (pLVTHM-PHTF2) were produced by GenePhama (Shanghai, China). To generate the control and PHTF2 expressing lentivirus, pSPAX2 and pMD2.g packaging plasmids were co-delivered into HEK293 cells using Lipofectamine 3000 (L3000001, Invirtogen, Shanghai, China). 48 h post-transfection, the lentiviral particle-containing supernatant was collected and used to transduce XWLC-05 and NCI-H1650 cells in the presence of polybrene (HY-112735, MedChemExpress, Shanghai, China). Following transduction, cell culture medium was prepared with 200 μg/ml puromycin (HY-15680, MedChemExpress, Shanghai, China) for 14 days. This selection process resulted in the generation of stable OE-NC cells (cells carrying the empty vector) and PHTF2-OE cells (cells stably expressing PHTF2).
MiR-NC oligos, miRNA mimic, inhibitor NC, and miRNA inhibitor were produced by MedChemExpress (Shanghai, China). Lipofectamine 3000 transfection reagent was utilized according to the supplier’s protocols for transfecting the molecules into XWLC-05 and NCI-H1650 cells in a 6-well plate. After 48 h of transfection, transfected cells were collected for further functional assays.
Cell count kit (CCK)-8 cell growth assay
To examine cell proliferation in XWLC-05 and NCI-H1650 cells, the CCK-8 kit (CA1210, Solarbio, Beijing, China) was employed. 2,000 cells/per well were inoculated in the 96-well plates. Following the designated culturing period, 15 μL/well of CCK8 reagent was introduced into every sample, followed by a 3-h reaction at 37°C. Light absorbance (OD value) under each condition was measured at 450 nm.
Lactate dehydrogenase (LDH) activity measurement
An LDH activity analysis kit (C0016, Beyotime Biotechnology, Beijing, China) was utilized in cytotoxicity assessment by detecting the release of LDH into the culture medium. In brief, a volume of 50 µL LDH assay buffer was added into each well in the assay plate, and subsequently 50 µL sample was loaded into each well. Next 50 µL of substrate solution was introduced into each well for an incubation at 37°C for 45 min. Following a gentle mixing, the signal was promptly assessed at OD 490 nm at 37°C.
XWLC-05 and NCI-H1650 cells were seeded into a 6-well plate and cultivated until around 90% coverage. A scratch was made in the middle of the cell layer and floating cells were discarded by replacing the old medium with fresh medium. The attached cells were then incubated at a temperature of 37°C for a period of 24–48 h. The Leica AM6000 microscope (Leica, Wetzlar, Germany) was used to capture images of the cells.
The ability of cell invasion was assessed using a transwell cassette of 12 mm size and 8.0 µm pore membrane. The upper cassette was spread with Matrigel (356234, BD Biosciences, Franklin Lakes, NJ, USA) at 37°C for 30 min prior to cell seeding. XWLC-05 and NCI-H1650 cells with designated treatment were inoculated at a concentration of 2 × 105 cells in the upper compartment using the medium without FBS. The lower cassette was loaded with complete medium. Following a 24-h incubation period, the invading cells on the membrane insert were fixed using ice-cold ethanol and labeled by 0.1% crystal violet (C0121, Beyotime, Beijing, China) for 15 min. The images of invading cells were then captured using a Leica AM6000 microscope (Leica, Wetzlar, Germany).
Molecular interaction analysis by luciferase assay
The fragment of wild-type (WT) or mutated (MUT) interaction sequences was respectively inserted into Luciferase-pcDNA3 (Plasmid #18964) obtained from Addgene (Watertown, MA, USA). Following the supplier’s protocol, the reporter plasmid was co-delivered into the cells together with either miR-30a-5p mimic or miR-NC (negative control for miR-30a-5p) using Lipofectamine 3000 transfection kit. The relative activity of WT and MUT reporter was determined using the Fire-Lumi Luciferase Assay Kit purchased from GenScript (L00877C, Nanjing, China) based on the provided protocols.
The purification of RNA sample was conducted by a Trizol-based extraction kit (ZY-6901, Zeye Biotech, Shanghai, China). Subsequently, RNA samples were transcribed in reverse to produce cDNA using either the BeyoRT II reverse transcription Kit (D7168S, Beyotime Biotechnology, Beijing, China) or the EZB-miRT2 microRNA RT Kit (EZB-miRT2-S, Ezbioscience, Roseville MN, USA). For real-time qPCR analysis of cDNA samples, the TB Green® Fast qPCR Mix (RR430B, Takara, Dalian, China) was utilized under the thermal conditions: 98°C for 5 min, 40 cycles of 98°C for 15 s, 60°C for 15 s, and 72°C for 45 s. Target gene expression was analyzed through the 2–∆∆Ct approach, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 snRNA as internal references. Primer sequences used: miR-30a-5p: F-CGCGATGTTGAAACATCCTCGAC, R-ATCCAGTGCAGGGTCCGAGG. PHTF2: F-AATGGTCCTAGCAAAGATACCCA, R-CCTGTCCACATGACGACGTAA. GAPDH: F-GGAGCGAGATCCCTCCAAAAT, R-GGCTGTTGTCATACTTCTCATGG. U6: F-CTCGCTTCGGCAGCACAT, R-TTTGCGTGTCATCCTTGCG.
To collect protein samples from XWLC-05/NCI-H1650 cells or tissues, we utilized RIPA cell lysis buffer (P0013c, Beyotime Biotechnology, Beijing China). Sample concentration analysis was performed by the BCA Protein Assay Kit (ZY-6125, Shanghai, China) from Zeye Biotech. Electrophoresis was conducted to separate the denatured protein samples. Following membrane transfer and blocking, detection of target proteins occurred at 4°C for 18 h using the PHTF2 (1:1500, ab1867474, Abcam, Cambridge, UK) or actin antibody (1:2000, ab8226, Abcam). The membrane was subsequently labeled using a secondary detection antibody (1:3500, ab97051, Abcam) at 25°C for 1 h. Finally, the signals of protein bands were visualized using the BeyoECL plus kit (P0018S) from Beyotime Biotechnology.
To perform the tumorigenesis assay, XWLC-05 cells (either OE-NC or PHTF2-OE cells, 2.5 × 106 cells per animal) in 250 μL PBS were subcutaneously injected into nude mice (8-week-old; male; procured from Weitong Lihua Experimental Animal Technology Co., Ltd., Beijing, China) (n = 6 per group). Further, either miRNA control (miR-NC) or miR-30a-5p mimic was administrated at a dosage of 1 mg/kg per week using EntransterTM-in vivo-RNA transfection reagent (18668-11-1, Engreen Biosystem Co., Ltd., Beijing, China) in accordance with the supplier’s protocols. Tumor formation in each experimental group was followed for 4 weeks, and on day 28 the xenograft tumor samples were collected from euthanized mice for weight determination and subsequent analyses. For the metastasis analysis, we injected XWLC-05 cells carrying the luciferase (OE-NC or PHTF2-OE cells, 2.5 × 106 cells per animal) into the nude mice via the tail vein. 4 weeks after cell inoculation, 100 µL of D-luciferin solution (HY-12591B, MedChemExpress, Shanghai, China) was intraperitoneally injected at 150 mg/kg into each mouse for bioluminescence imaging. An IVIS Imaging System 100 (Xenogen/Caliper Life Sciences, Alameda, CA, USA) was adopted to capture luminescent images. The images were recorded 5 min after D-luciferin administration, with a sequential image recording at a rate of one image per minute until 40 min post-injection.
In order to examine in situ Ki-67 expression by immuno-staining, 5-μm xenograft tumor tissues sections were produced for the initial de-paraffinization and re-hydration processes, followed by antigen retrieval at 98°C for 10 min in the citrate antigen recovery solution (P0081, Beyotime Biotechnology, Beijing, China). The sections were permeabilized using 0.05% TritonX100 (P0096, Beyotime Biotechnology, Beijing, China) for 15 min and then blocked using 5% goat serum (C0265, Beyotime Biotechnology, Beijing, China) for 1 h at ambient temperature. To block peroxidase activity, a 3% hydrogen peroxide solution was applied to the sections for 10 min. After rinsing with Tris-buffered saline with 0.1% Tween® 20 buffer, a primary antibody targeting Ki-67 (ab16667, 1:600, Abcam), and subsequently labeled with the SignalStain® Boost Detection Reagent (Rabbit #8114, Cell Signaling Technologies, Boston, MA, USA) for 5 to 10 min at 25°C. Finally, the stained samples were imaged under a Leica AM6000 microscope (Leica, Wetzlar, Germany).
The experimental results were summarized and processed using SPSS software version 19.0 (IBM, New York, NY, USA). The data were displayed as the mean with standard deviation. Either the Wilcoxon test or Student’s t-test was adopted to compare quantitative results between two conditions. For the analyses of multiple conditions, one-way ANOVA was employed with p < 0.05 as the cutoff for the determination of statistical significance.
Reduced expression of miR-30a-5p in lung adenocarcinoma
To investigate the expression pattern of miR-30a-5p in lung adenocarcinoma, we obtained the dataset from The Cancer Genome Atlas (TCGA)-lung adenocarcinoma (LUAD) project miRNA-seq. The analysis revealed a notable decrease of miR-30a-5p levels within the tumor samples when compared to the normal tissues (Fig. 1A). Additionally, we procured 60 sets of lung adenocarcinoma specimens together with the corresponding para-cancerous normal tissues, and RT-qPCR analysis further showed the decreased expression of miR-30a-5p in lung adenocarcinoma (Fig. 1B). Further, miR-30a-5p also displayed downregulation in lung adenocarcinoma cells (XWLC-05 and NCI-H1650) as compared to a normal lung epithelial cell (BEAS-2B) (Fig. 1C).
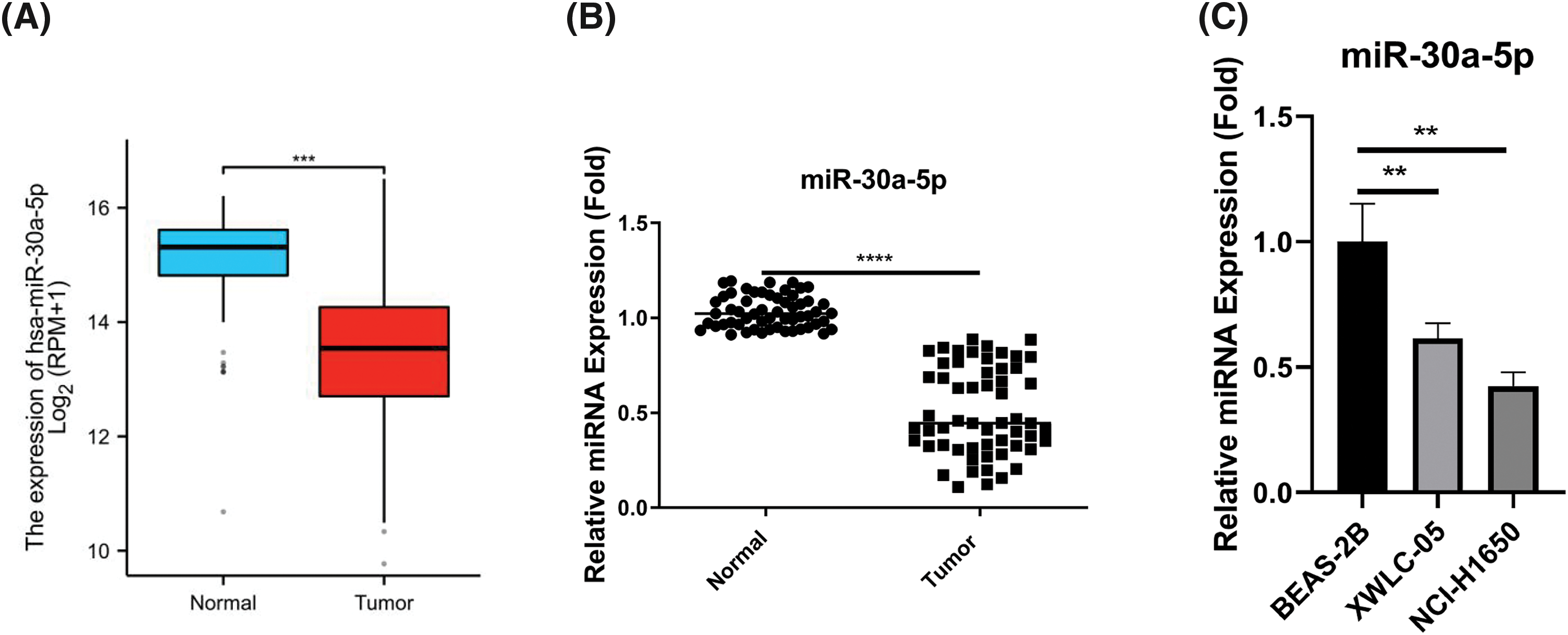
Figure 1: The reduced expression of miR-30a-5p in lung adenocarcinoma samples and cell lines. (A) The Cancer Genome Atlas (TCGA)-lung adenocarcinoma (LUAD) project miRNA-seq data was analyzed to show the relative levels of miR-30a-5p in both lung adenocarcinoma and normal tissues. (B) RT-qPCR analysis was conducted on 60 pairs of lung adenocarcinoma specimens and their respective para-cancerous specimens to examine the relative miR-30a-5p levels. (C) RT-qPCR detection of miR-30a-5p levels in lung adenocarcinoma cells (XWLC-05 and NCI-H1650) and normal lung cells (BEAS-2B), n = 3. **p < 0.01; ***p < 0.001; ****p < 0.0001.
The transfection of miR-30a-5p mimic suppresses aggressive phenotype in lung adenocarcinoma cells
To further decipher the functional roles of miR-30a-5p in cells of lung adenocarcinoma, we conducted further analyses using XWLC-05 and NCI-H1650 cells. These cells were transfected with either miR-NC or miR-30a-5p mimic. The transfection of miRNA mimic elevated the cellular levels of miR-30a-5p (Fig. 2A). Upon the transfection of miRNA mimic, the growth rate of both XWLC-05 and NCI-H1650 cells were significantly decreased (Fig. 2B). We also employed an LDH cytotoxicity assay to assess cell death, where an elevation in cell death level was observed following miRNA mimic transfection (Fig. 2C). Moreover, miR-30a-5p mimic effectively suppressed the abilities of XWLC-05 and NCI-H1650 cells to migrate and invade (Figs. 2D and 2E).
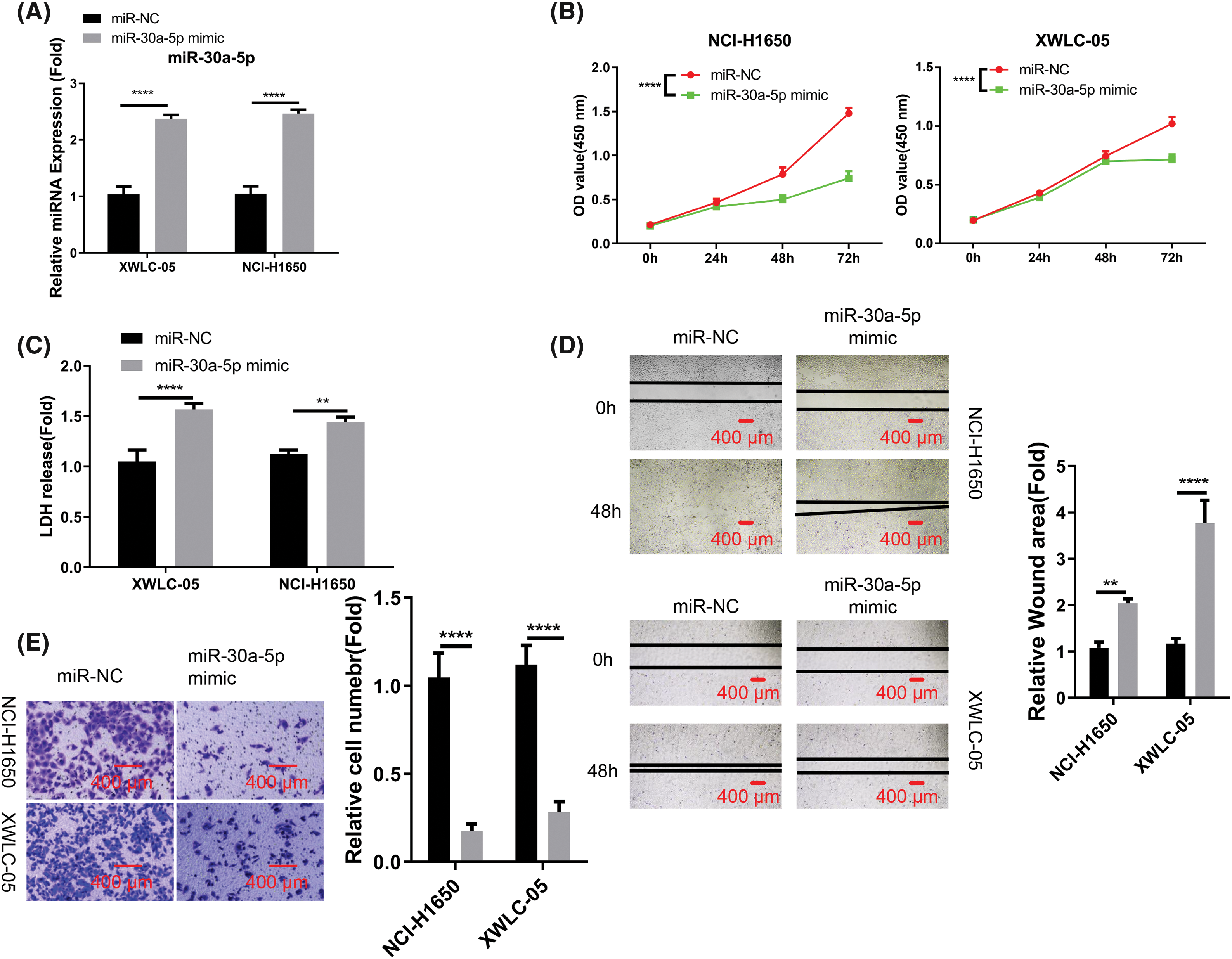
Figure 2: Suppression of aggressive phenotype in lung adenocarcinoma cells by miR-30a-5p mimic transfection. Transfection of miR-NC (negative control for miRNA mimic) or miR-30a-5p mimic was performed on XWLC-05 and NCI-H1650 cells. (A) RT-qPCR analysis of miR-30a-5p in both cells. (B) Assessment of proliferation by cell count kit (CCK)-8 assay. (C) Evaluation of cytotoxicity by lactate dehydrogenase (LDH) assay. (D) Analysis of migration ability by scratch assay. (E) Transwell invasion assay in XWLC-05 and NCI-H1650 cells, and the quantification of the invading cells. n = 3. **p < 0.01; ****p < 0.0001.
miR-30a-5p negatively targets PHTF2 mRNA
In order to find the downstream mRNA regulated by miR-30a-5p, TargetScan online resource was utilized. The analysis revealed potential interaction sequences of miR-30a-5p and the 3′ untranslated region (UTR) of PHTF2 mRNA (Fig. 3A). Following this, luciferase vector harboring the wild-type (WT) or mutated interacting sequences was generated to confirm the interaction between these two molecules. The data of luciferase activity measurement demonstrated that the introduction of miR-30a-5p mimic suppressed WT reporter, while no inhibitory effect was detected on the MUT reporter (Fig. 3B). In the TCGA-LUAD RNA-seq dataset, PHTF2 mRNA levels were increased in lung adenocarcinoma specimens, which is in contrast to the decreased miR-30a-5p expression (Fig. 3C). This finding was also verified by analyzing clinical samples from lung adenocarcinoma patients, where PHTF2 showed an upregulation at the protein and mRNA levels in tumor specimens (Fig. 3D). Moreover, the introduction of miR-30a-5p mimic effectively reduced PHTF2 levels in XWLC-05 and NCI-H1650 cells (Figs. 3E and 3F). Taken together, the results strongly indicate that PHTF2 mRNA is negatively modulated by miR-30a-5p.
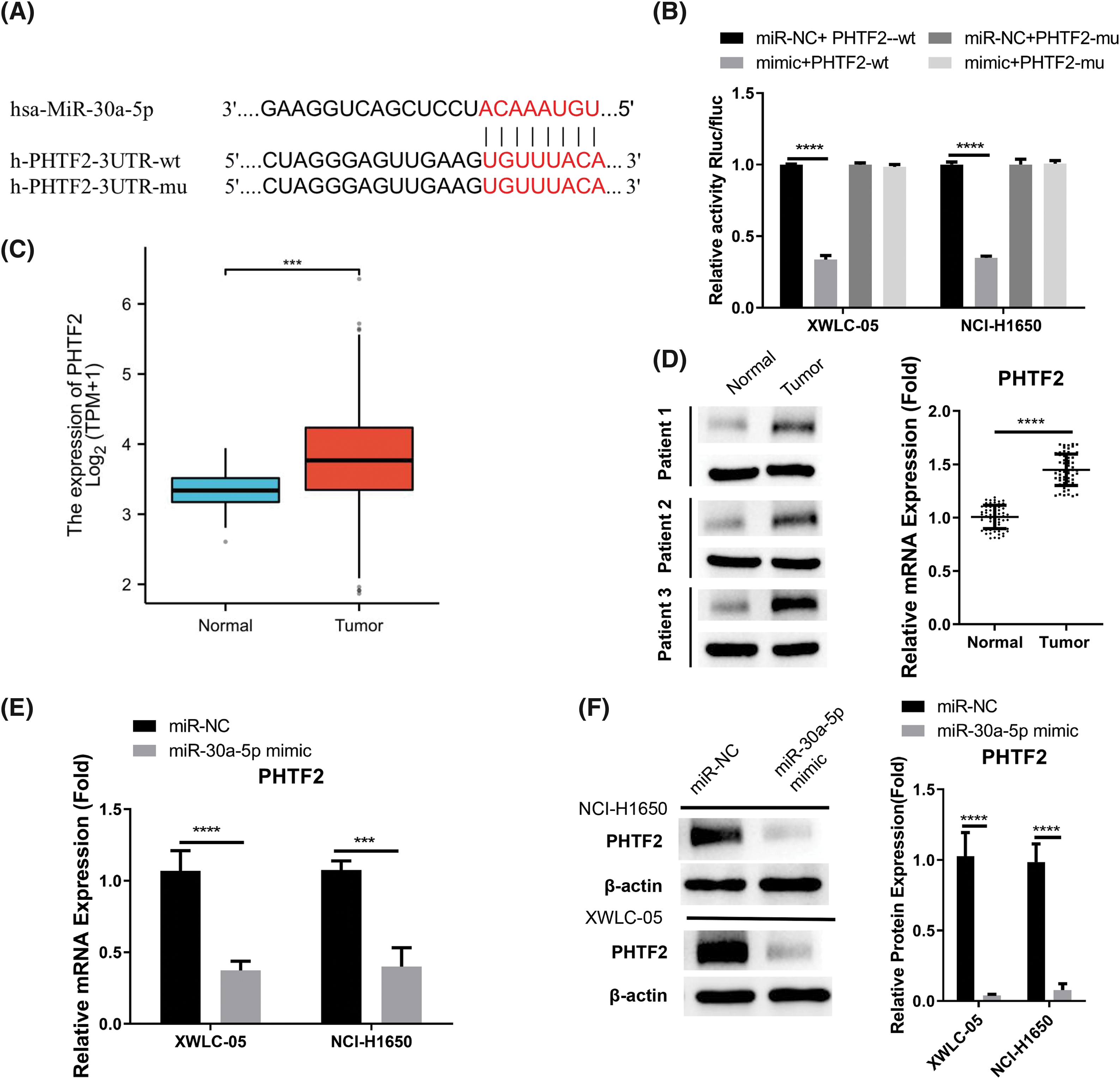
Figure 3: miR-30a-5p negatively targets PHTF2 mRNA. (A) The potential interaction sites of miR-30a-5p and the putative transcription factor 2 of the homeodomain (PHTF2) mRNA 3′ untranslated region (UTR). (B) Luciferase activity measurement was conducted using both wild-type (WT) and mutated (MUT) luciferase reporter. (C) The TCGA-LUAD RNA-seq dataset was utilized to compare the PHTF2 mRNA levels in lung adenocarcinoma and para-cancerous normal specimens. (D) RT-qPCR analysis was performed on 60 pairs of lung adenocarcinoma specimens and the para-cancerous samples to determine the relative mRNA levels of PHTF2. Additionally, Western blot analysis was conducted on 3 pairs of normal and tumor tissues to examine the protein levels of PHTF2. (E and F) The introduction of miR-30a-5p mimic in XWLC-05 and NCI-H1650 cells caused the reduction of PHTF2 expression levels. n = 3. ***p < 0.001; ****p < 0.0001.
PHTF2 overexpression antagonizes the effect of miR-30a-5p mimic
We proceeded to examine whether the role of miR-30a-5p is mediated by PHTF2. For this purpose, we created cells with elevated PHTF2 expression using the lentiviral system (referred to as PHTF2-OE), as well as a control group of cells (OE-NC). We observed that the introduction of miR-30a-5p mimic into OE-NC cells decreased the expression of PHTF2. However, when PHTF2 was overexpressed, PHTF2 expression was largely increased in both XWLC-05 and NCI-H1650 cells (Figs. 4A and 4B). Notably, the inhibitory impact of miR-30a-5p mimic on cell proliferation was largely abrogated in the presence of PHTF2 overexpression (Fig. 4C), and PHTF2 overexpression reduced the extent of cell death after the delivery with miR-30a-5p mimic (Fig. 4D). Meanwhile, PHTF2 overexpression also augmented the migratory and invasive abilities in XWLC-05 and NCI-H1650 cells upon miR-30a-5p mimic delivery (Figs. 4E and 4F). These findings indicate that PHTF2 overexpression counteracts the inhibitory function of miR-30a-5p mimic in lung adenocarcinoma cells.
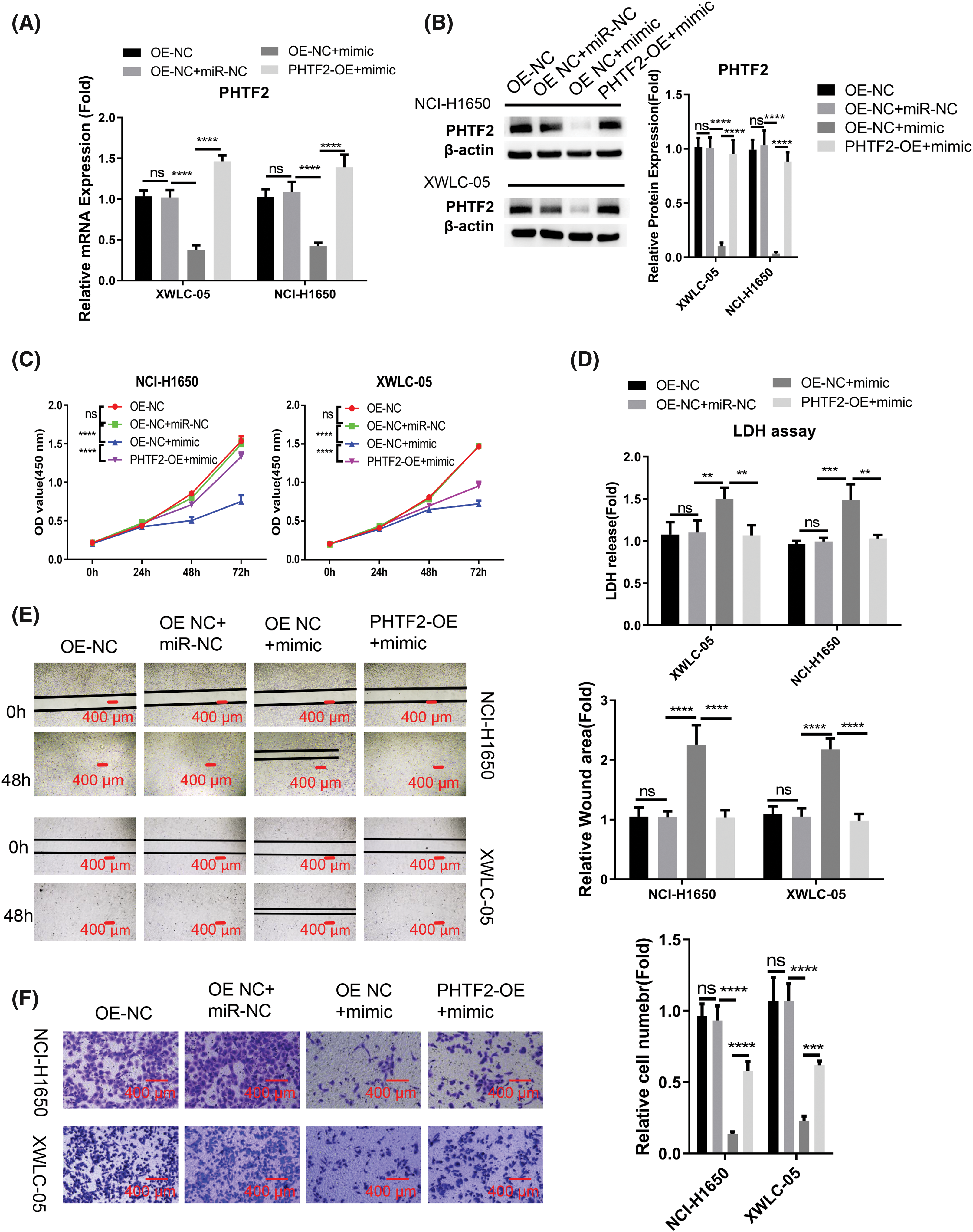
Figure 4: PHTF2 overexpression antagonizes the impact of miR-30a-5p mimic. A lentiviral approach was utilized to generate XWLC-05 and NCI-H1650 cells with stable overexpression of PHTF2 (PHTF2-OE). The control cells were denoted as OE-NC. To assess the interplay of miR-30a-5p and PHTF2, miRNA mimic or the control was introduced into the specified experimental group. (A and B) PHTF2 expression was assessed through RT-qPCR and Western blot analyses in each experimental group. (C) The proliferation assay (CCK-8) was conducted for cell growth analysis. (D) Cytotoxicity (LDH) activity was measured to determine cell death level. (E) The migratory ability of cells was analyzed by the scratch experiment. (F) Transwell invasion experiment was performed to evaluate the invasion potential in each experimental condition. n = 3. **p < 0.01; ***p < 0.001; ****p < 0.0001; ns: no significance.
PHTF2 overexpression antagonizes the anti-tumorigenic effect miR-30a-5p mimic
To verify the interplay of PHTF2 and miR-30a-5p in tumorigenesis, we subcutaneously injected XWLC-05 cells (either OE-NC or PHTF2-OE cells) into nude mice. Additionally, EntransterTM-in vivo-RNA transfection reagent was utilized to deliver either miRNA control (miR-NC) or miR-30a-5p mimic. The administration of miRNA mimic impeded tumor formation in OE-NC cells. However, the overexpression of PHTF2 in PHTF2-OE cells substantially restored tumorigenesis in the nude mice (Fig. 5A). Analysis of Ki-67, a cell proliferation marker, in the tumor tissues indicated that miR-30a-5p administration decreased the proportion of Ki-67-expressing cells. Conversely, PHTF2 overexpression increased the number of Ki-67 positive cells in the presence of miR-30a-5p mimic treatment (Fig. 5B). Moreover, examination of PHTF2 expression levels in the tumor tissues validated our in vitro data (Figs. 5C and 5D). These findings collectively demonstrated that miR-30a-5p serves as a tumor-suppressive factor in lung adenocarcinoma, while PHTF2 acts as a pro-tumorigenic factor.
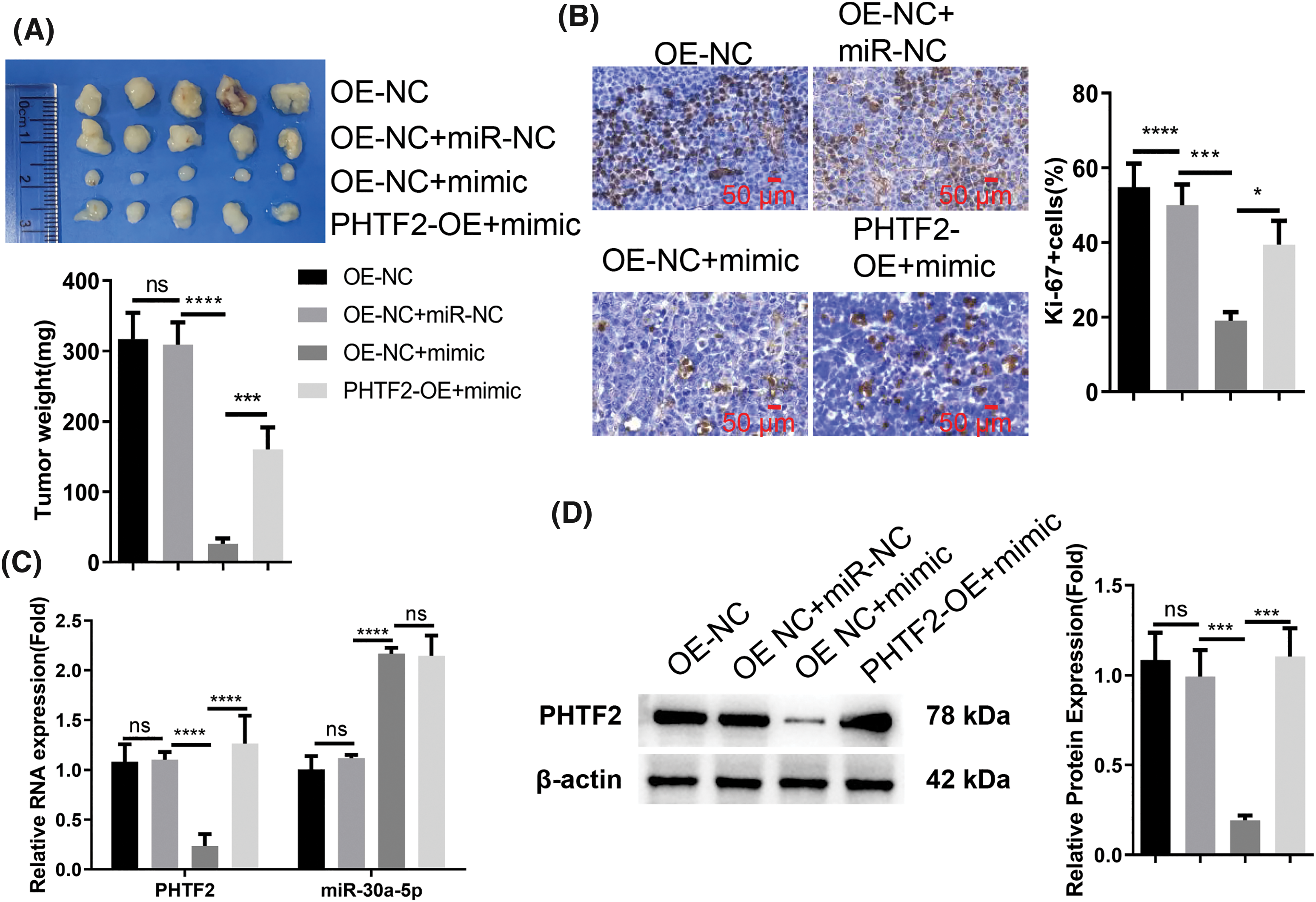
Figure 5: The anti-tumorigenic role of miR-30a-5p mimic on XWLC-05 cells is counteracted by PHTF2 overexpression. For the in vivo tumor formation study, nude mice were inoculated with XWLC-05 cells (either OE-NC or PHTF2-OE cells). EntransterTM-in vivo-RNA transfection reagent was employed for the administration of miRNA control (miR-NC) or miR-30a-5p mimic (n = 6 animals in each group). (A) The xenograft tumor images along with a summary of the tumor mass for each group are presented. (B) The tumor tissues were subject to IHC analysis to assess the proportion of cells expressing the proliferation marker Ki-67. (C) Detection of PHTF2 and miR-30a-5p expression by RT-qPCR in the xenograft tumor samples. (D) Analysis of PHTF2 protein levels by Western blot in the xenograft tumor samples. *p < 0.05; ***p < 0.001; ****p < 0.0001; ns: no significance.
The metastasis of lung adenocarcinoma cells is inhibited by miR-30a-5p, while promoted by PHTF2
Since miR-30a-5p/PHTF2 axis modulates the motility of lung adenocarcinoma cells in vitro, we then established an in vivo model of metastasis by tail injection of XWLC-05 cells carrying the luciferase (OE-NC or PHTF2-OE cells). We also administrated miRNA control (miR-NC) or miR-30a-5p mimic in designated groups. Bioluminescence imaging suggested that when miR-30a-5p mimic was delivered, there were fewer metastases observed in the lung of the mouse injected with OE-NC cells, indicating the suppression of miR-30a-5p on metastasis (Fig. 6A). However, the overexpression of PHTF2 in PHTF2-OE cells promoted metastasis formation in comparison to OE-NC cell injection group after miR-30a-5p mimic treatment (Fig. 6A). We then assessed miR-30a-5p and PHTF2 expressions in the metastases from each experimental group. The data demonstrated that miR-30a-5p mimic administration elevated miR-30a-5p levels but reduced PHTF2 expression in the metastases arising from OE-NC cells. In the metastases formed by PHTF2-OE cells, the expression level of PHTF2 was restored (Figs. 6B and 6C). Therefore, these findings suggest that the metastasis of lung adenocarcinoma could be repressed by miR-30a-5p, while PHTF2 promotes the metastasis.
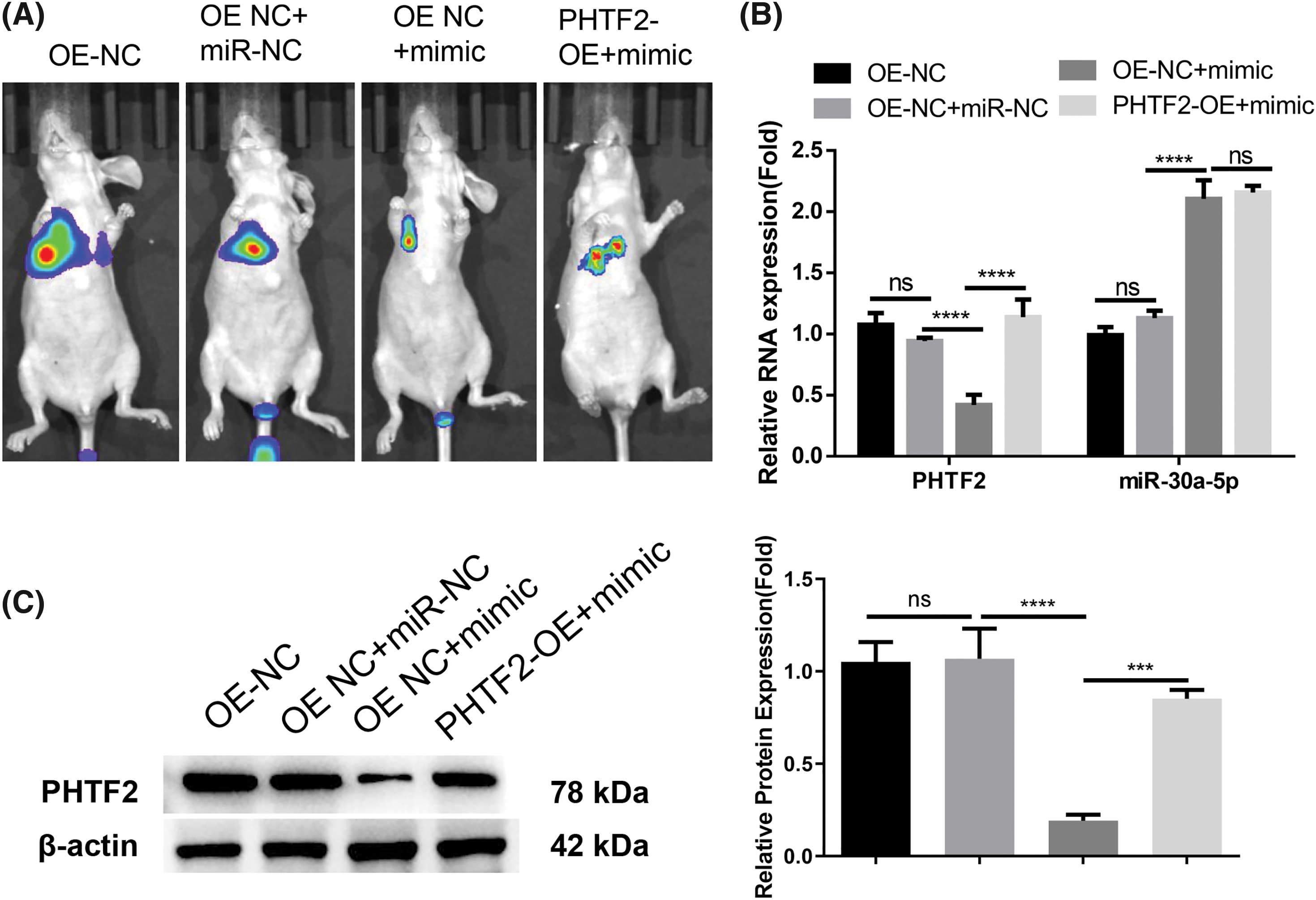
Figure 6: miR-30a-5p inhibits lung adenocarcinoma cell metastasis, while PHTF2 promotes the metastasis. An in vivo model of metastasis was constructed in nude mice using XWLC-05 cells carrying the luciferase (OE-NC or PHTF2-OE cells). Additionally, miRNA control (miR-NC) or miR-30a-5p mimic was administrated to indicated groups (n = 6 in animals each group). (A) The bioluminescence imaging analysis of the lung metastases in each group. (B) Detection of PHTF2 and miR-30a-5p expression by RT-qPCR in the metastatic tissues. (C) Analysis of PHTF2 protein levels by Western blot in the metastatic tissues. ***p < 0.001; ****p < 0.0001; ns: no significance.
In the current investigation, we demonstrated a diminished expression of miR-30a-5p in lung adenocarcinoma specimens. The mimic of miR-30a-5p repressed the aggressive characteristics in lung adenocarcinoma cells, encompassing their cell growth, viability, and motility. Moreover, the introduction of miR-30a-5p mimic hindered tumor formation and metastasis of lung adenocarcinoma cells in nude mice. miR-30a-5p could negatively target PHTF2, which exhibited elevated levels in lung adenocarcinoma. Increasing the expression of PHTF2 counteracted the anti-neoplastic function of miR-30a-5p mimic.
According to our research data, miR-30a-5p is a suppressive factor of tumor growth and is downregulated in lung adenocarcinoma. Our results align with previous studies, which have shown the inhibition of miR-30a-5p on cancer development and metastasis in various malignancies [22–26]. For instance, miR-30a-5p hinders the development and dissemination of breast malignancy by diminishing lactate dehydrogenase A (LDHA) levels and inhibiting aerobic glycolysis [22]. In the case of gallbladder cancer, miR-30a-5p curbs cell proliferation, mobility, and metastasis through suppressing the proto-oncogene E2F Transcription Factor 7 (E2F7) [25]. A correlation has been reported between reduced miR-30a-5p expression and more distant metastases, advanced tumour, node and metastasis stage, and poor tumor differentiation in gastric cancer patients. Furthermore, miR-30a-5p has demonstrated potential as a diagnostic factor in gastric cancer due to its sensitivity and specificity [26]. These findings altogether indicate that miR-30a-5p possesses the ability to modulate various downstream mRNA targets, thereby reducing tumor aggressiveness and restraining metastasis in diverse types of cancers.
Our research further identified miR-30a-5p as a upstream modulator of PHTF2. In lung adenocarcinoma, PHTF2 exhibited an increased expression and its forced expression counteracted the anti-neoplastic effect of miR-30a-5p. The transcription factor family known as PHTF is evolutionarily conserved and it may play a role in mediating stress-induced apoptosis and chemo-sensation across various species [28,29]. There is evidence supporting the overexpression of PHTF1 in acute lymphoblastic leukemia (ALL). Nonetheless, forced PHTF1 expression was found to suppress cell growth and trigger apoptosis in ALL cells [30], indicating the tumor suppressor role of PHTF1 in ALL. However, our data unveiled that PHTF2 overexpression was able to counteract the anti-tumorigenic role of the miR-30a-5p mimic. These observations imply that PHTF2 functions as an oncogenic factor in lung adenocarcinoma. Additionally, a recent report demonstrated that PHTF2 was overexpressed in gastric cancer and its upregulation promotes the biosynthesis of lipids, thereby facilitating the tumorigenesis of gastric cancer cells [27]. Collectively, our findings lead us to conclude that PHTF2 serves as an oncogene in lung adenocarcinoma and its excessive expression can enhance the development and spread of cancer cells.
There are still several unanswered questions regarding the biological roles of miR-30a-5p and PHTF2 in lung adenocarcinoma. It is essential to decipher the mechanism by which miR-30a-5p expression becomes repressed in lung adenocarcinoma. Solving this query may hint on the approaches to target miR-30a-5p expression. Furthermore, future investigation is needed into how PHTF2 regulates the initiation and spread of lung adenocarcinoma. PHTF2 might influence the process of epithelial-mesenchymal transition (EMT) or mediate metabolic reprogramming in lung adenocarcinoma to enhance cancer progression. Importantly, additional collection of clinical samples is necessary to validate whether miR-30a-5p and PHTF2 have the potential to be utilized as predictive biomarkers in lung adenocarcinoma for metastasis.
In conclusion, the decreased level of miR-30a-5p is linked to the upregulation of PHTF2 in patients with lung adenocarcinoma. PHTF2 is a downstream molecule targeted by miR-30a-5p. PHTF2 functions as an oncogenic factor, while miR-30a-5p acts as an anti-tumorigenic miRNA, influencing the tumor formation and metastasis of lung adenocarcinoma. A potential approach to inhibit metastasis in lung adenocarcinoma is through modulating the miR-30a-5p/PHTF2 molecular axis.
Acknowledgement: None.
Funding Statement: This work was supported by the Basic Research Program of Yunnan Province-Joint Project of Kunming Medical University No. 202101AY070001−169.
Author Contributions: Study conception and design: Xicai Wang; data collection: Qingyin Meng, Li Zhuang, Quan Gong; analysis and interpretation of results: Lijuan Zhang, Xianda Huang, Xueqin Li, Shijuan Li, Guoqin Wang; draft manuscript preparation: Lijuan Zhang, Qingyin Meng. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data generated in this study are available upon request to the corresponding author.
Ethics Approval: The use of human samples gained the approval from the Medical Research Ethics Committee of Yunnan Provincial Cancer Hospital (No. KYLX2021-124). All the patients provided the written informed consent. This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Kunming Medical University (No. Kmmu20220583).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Seguin L, Durandy M, Feral CC. Lung adenocarcinoma tumor origin: a guide for personalized medicine. Cancers. 2022;14(7):1759. [Google Scholar] [PubMed]
2. Succony L, Rassl DM, Barker AP, McCaughan FM, Rintoul RC. Adenocarcinoma spectrum lesions of the lung: detection, pathology and treatment strategies. Cancer Treat Rev. 2021;99:102237. [Google Scholar] [PubMed]
3. Chen JW, Dhahbi J. Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci Rep. 2021;11(1):13323. [Google Scholar] [PubMed]
4. Sainz de Aja J, Dost AFM, Kim CF. Alveolar progenitor cells and the origin of lung cancer. J Intern Med. 2021;289(5):629–35. [Google Scholar] [PubMed]
5. He Y, Liu X, Wang H, Wu L, Jiang M, Guo H, et al. Mechanisms of progression and heterogeneity in multiple nodules of lung adenocarcinoma. Small Methods. 2021;5(6):e2100082. [Google Scholar] [PubMed]
6. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24. [Google Scholar] [PubMed]
7. Hua X, Zhao W, Pesatori AC, Consonni D, Caporaso NE, Zhang T, et al. Genetic and epigenetic intratumor heterogeneity impacts prognosis of lung adenocarcinoma. Nat Commun. 2020;11(1):2459. [Google Scholar] [PubMed]
8. Attili I, Del Re M, Guerini-Rocco E, Crucitta S, Pisapia P, Pepe F, et al. The role of molecular heterogeneity targeting resistance mechanisms to lung cancer therapies. Expert Rev Mol Diagn. 2021;21(8):757–66. [Google Scholar] [PubMed]
9. Senosain MF, Massion PP. Intratumor heterogeneity in early lung adenocarcinoma. Front Oncol. 2020;10:349. [Google Scholar] [PubMed]
10. Jiao M, Liu H, Liu X. Transcriptional patterns reveal tumor histologic heterogeneity and immunotherapy response in lung adenocarcinoma. Front Immunol. 2022;13:957751. [Google Scholar] [PubMed]
11. Yu H, Zhang W, Xu XR, Chen S. Drug resistance related genes in lung adenocarcinoma predict patient prognosis and influence the tumor microenvironment. Sci Rep. 2023;13(1):9682. [Google Scholar] [PubMed]
12. Xie S, Wu Z, Qi Y, Wu B, Zhu X. The metastasizing mechanisms of lung cancer: recent advances and therapeutic challenges. Biomed Pharmacother. 2021;138:111450. [Google Scholar] [PubMed]
13. Suzuki HI. Roles of MicroRNAs in disease biology. JMA J. 2023;6(2):104–13. [Google Scholar] [PubMed]
14. Uzuner E, Ulu GT, Gürler SB, Baran Y. The role of MiRNA in cancer: ppathogenesis, diagnosis, and treatment. Methods Mol Biol. 2022;2257:375–422. [Google Scholar] [PubMed]
15. Kobayashi H, Singer RH. Single-molecule imaging of microRNA-mediated gene silencing in cells. Nat Commun. 2022;13(1):1435. [Google Scholar] [PubMed]
16. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci. 2020;21(5):1723. [Google Scholar] [PubMed]
17. Moradi S, Kamal A, Aboulkheyr Es H, Farhadi F, Ebrahimi M, Chitsaz H, et al. Pan-cancer analysis of microRNA expression profiles highlights microRNAs enriched in normal body cells as effective suppressors of multiple tumor types: a study based on TCGA database. PLoS One. 2022;17(4):e0267291. [Google Scholar] [PubMed]
18. Kiran S, Patra A, Verma P, Purkait S, Chhabra G, Guttula PK, et al. Restoration of altered oncogenic and tumor suppressor microRNA expression in breast cancer and colorectal cancer cell using epicatechin. Curr Mol Pharmacol. 2023;16(8):915–26. [Google Scholar] [PubMed]
19. Zhong S, Golpon H, Zardo P, miRNAs in lung cancer Borlak J. A systematic review identifies predictive and prognostic miRNA candidates for precision medicine in lung cancer. Transl Res. 2021;230:164–96. [Google Scholar]
20. Xie Y, Xue C, Guo S, Yang L. MicroRNA-520a suppresses pathogenesis and progression of non-small-cell lung cancer through targeting the RRM2/Wnt Axis. Anal Cell Pathol. 2021;2021:9652420. [Google Scholar]
21. Liu F, Mao Q, Zhu S, Qiu J. MicroRNA-155-5p promotes cell proliferation and invasion in lung squamous cell carcinoma through negative regulation of fibroblast growth factor 9 expression. J Thorac Dis. 2021;13(6):3669–79. [Google Scholar] [PubMed]
22. Li L, Kang L, Zhao W, Feng Y, Liu W, Wang T, et al. miR-30a-5p suppresses breast tumor growth and metastasis through inhibition of LDHA-mediated Warburg effect. Cancer Lett. 2017;400:89–98. [Google Scholar] [PubMed]
23. Outeiro-Pinho G, Barros-Silva D, Aznar E, Sousa AI, Vieira-Coimbra M, Oliveira J, et al. MicroRNA-30a-5pme: a novel diagnostic and prognostic biomarker for clear cell renal cell carcinoma in tissue and urine samples. J Exp Clin Cancer Res. 2020;39(1):98. [Google Scholar] [PubMed]
24. Peng L, Huang X, Qing D, Lu H, Liu X, Chen J, et al. MiR-30a-5p inhibits cell behaviors in esophageal cancer via modulating CBX2. Mutat Res. 2023;826:111818. [Google Scholar] [PubMed]
25. Ye YY, Mei JW, Xiang SS, Li HF, Ma Q, Song XL, et al. MicroRNA-30a-5p inhibits gallbladder cancer cell proliferation, migration and metastasis by targeting E2F7. Cell Death Dis. 2018;9(3):410. [Google Scholar] [PubMed]
26. Soliman SE, Elabd NS, El-Kousy SM, Awad MF. Down regulation of miR-30a-5p and miR-182-5p in gastric cancer: clinical impact and survival analysis. Biochem Biophys Rep. 2021;27:101079. [Google Scholar] [PubMed]
27. Chi Y, Wang H, Wang F, Ding M. PHTF2 regulates lipids metabolism in gastric cancer. Aging. 2020;12(8):6600–10. [Google Scholar] [PubMed]
28. Benton R, Himmel NJ. Structural screens identify candidate human homologs of insect chemoreceptors and cryptic Drosophila gustatory receptor-like proteins. eLife. 2023;12:e85537. [Google Scholar] [PubMed]
29. Oyhenart J, Benichou S, Raich N. Putative homeodomain transcription factor 1 interacts with the feminization factor homolog fem1b in male germ cells. Biol Reprod. 2005;72(4):780–7. [Google Scholar] [PubMed]
30. Huang X, Geng S, Weng J, Lu Z, Zeng L, Li M, et al. Analysis of the expression of PHTF1 and related genes in acute lymphoblastic leukemia. Cancer Cell Int. 2015;15:93. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools