 Open Access
Open Access
REVIEW
LncRNAs unraveling their sponge role in glioblastoma and potential therapeutic applications
School of Pharmacy, Binzhou Medical University, Yantai, 264003, China
* Corresponding Author: YUNQI MA. Email:
(This article belongs to the Special Issue: MicroRNA as Biomarkers for Disease Diagnosis and Progression)
BIOCELL 2024, 48(3), 387-401. https://doi.org/10.32604/biocell.2024.048791
Received 18 December 2023; Accepted 02 February 2024; Issue published 15 March 2024
Abstract
Glioblastoma multiforme (GBM), the most common and aggressive primary brain tumor in adults, is the most malignant and still has no cure. However, the novel role of long non-coding RNAs (lncRNAs) in the pathogenesis of glioblastoma is attracting extensive attention. LncRNAs are transcribed RNA molecules over 200 nucleotides long that do not encode proteins. Unlike small non-coding RNAs, such as microRNAs (miRNAs), lncRNAs have more complex secondary and tertiary structures that enable them to interact with DNA, RNA, and proteins and perform multiple regulatory functions. LncRNAs act as molecular sponges, absorbing and sequestering other biomolecules, particularly miRNAs, thereby preventing these molecules from performing their normal functions. LncRNAs influence glioblastoma through gene expression regulation, molecular sponge capacity, epigenetic modulation, and signaling pathway interactions. In glioblastoma, a large number of lncRNAs have been found to be abnormally expressed, affecting tumor growth, invasion and resistance to treatment. Due to its regulatory role and disease-specific expression patterns, lncRNA has become a potential biomarker for glioblastoma and a promising new therapeutic target. This paper discusses the spongy role of lncRNAs in glioblastoma and its potential therapeutic applications, which will lay a foundation for our understanding of glioblastoma biology and the development of new diagnostic and therapeutic strategies in the future.Graphic Abstract
Keywords
Abbreviation
| GBM | Glioblastoma multiforme |
| LncRNAs | Long non-coding RNAs |
| miRNAs | MicroRNAs |
| CNS | Central nervous system |
| WHO | The World Health Organization |
| EGFR | Epidermal growth factor receptor |
| PTEN | Phosphatase and tensin homolog |
| IDH1 | Isocitrate dehydrogenase 1 |
| CeRNAs | Competing endogenous RNAs |
| CSCs | Cancer stem cells |
| DNMT1 | DNA cytosine-5-methyltransferase 1 |
| CEBPA | Enhancer binding protein α |
| CRNDE | Colorectal neoplasia differentially expressed |
| PIWIL4 | Piwi-like RNA-mediated gene silencing 4 |
| MEN | Multiple endocrine neoplasia |
| HOTAIR | HOX transcript antisense RNA |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 |
| VM | Vasculogenic mimicry |
| TME | The tumor microenvironment |
| NF-κB | Nuclear factor Kb |
| CAFs | Cancer-associated fibroblasts |
| ECM | The extracellular matrix |
| Tregs | Regulatory T cells |
| MDSCs | Myeloid-derived suppressor cells |
| TAMs | Tumor-associated macrophages |
| TUG1 | Taurine upregulated gene 1 |
| BET | Bromodomain and extra-terminal |
| RNAi | RNA interference |
| siRNAs | Small interfering RNAs |
| shRNAs | Short hairpin RNAs |
| ASOs | Antisense oligonucleotides |
| DNMTs | DNA methyltransferases |
| TETs | Ten-eleven translocation proteins |
| SAM | S-adenosylmethionine |
| SAH | S-adenosylhomocysteine |
| PI3K | The phosphatidylinositol 3-kinasemammalian target of rapamycin |
| mTOR | Mammalian target of rapamycin |
| Ras | Rat sarcoma virus |
| ERK | Extracellular signal-regulated kinase |
| hnRNPK | Heterogeneous nuclear ribonucleoprotein K |
| PRC2 | Polycomb repressive complex 2 |
| RNP | RNA-protein |
| PTENP1 | Phosphatase and tensin homolog pseudogene 1 |
| HOTAIR | HOX transcript antisense RNA |
| XIST | X inactive specific transcript |
| MALAT1 | Metastasis associated in lung adenocarcinoma transcript 1 |
| BCSC | Breast cancer stem cell |
| TRG | T-cell receptor gamma |
| AS1 | Antisense RNA 1 |
| PSMB8 | Proteasome (prosome, macropain) subunit, beta type, 8 |
| HOXA-AS2 | Homeobox (HOX) A cluster antisense RNA 2 |
| HOXA-AS3 | HOXA Cluster Antisense RNA 3 |
| OXCT1 | Oxoacid CoA transferase 1 |
| Tmem 161b | Transmembrane protein 161b |
| SNHG11 | Small nucleolar RNA Host Gene 11 |
| GAPLINC | Gastric adenocarcinoma predictive long intergenic non-coding RNA |
| MEG3 | Maternally expressed gene 3 |
| PVT1 | Plasmacytoma variant translocation 1 |
| UCA1 | Urothelial carcinoma antigen 1 (UCA1) |
| TPT1-AS1 | Tumor Protein Translationally Controlled 1 (TPT1) Antisense RNA 1 |
| GLS | Generalized least squares (GLS) |
| TUG1 | Taurine upregulated 1 |
| EZH2 | Enhancer of zeste homolog 2 |
| PNCK | Pregnancy up-regulated non-ubiquitous calmodulin kinase |
| IκBα | Inhibitor of nuclear factor kappa B α |
| PD-L1 | Programmed death-ligand 1 |
| CTLA-4 | Cytotoxic T-lymphocyte-associated protein 4 |
| MIR155HG | MIR155 host gene |
| PCED1B-AS1 | PCED1B antisense RNA 1 |
| SNORD76 | C/D box snoRNA U76 |
General introduction to glioblastoma
Glioblastoma, also known as glioblastoma multiforme (GBM), is an incurable primary brain tumor that is highly prevalent and aggressive in adults [1,2]. It originates from glial cells or their precursors in the central nervous system (CNS) and is characterized by its rapid growth, diffusive infiltration into the surrounding brain tissues, and resistance to traditional therapies.
The World Health Organization (WHO) classifies glioblastoma as a Grade IV astrocytoma, the most malignant form on a scale ranging from I to IV [3]. Histologically, glioblastoma exhibits various cellular morphologies (hence the term “multiforme”) and features such as necrosis and microvascular proliferation. Despite this heterogeneity [4,5], all glioblastomas share a common inclination for invasiveness, resistance to cell death, and robust blood vessel formation, collectively contributing to their notorious clinical behavior. The ability to invade surrounding tissue is a crucial factor in the malignant progression of the tumor [6].
Clinically, glioblastoma typically presents symptoms that indicate increased pressure within the skull, focal neurological deficits, or seizures, which are often determined by the tumor’s size and location. Due to its infiltrative nature, complete surgical removal is typically not feasible, resulting in a poor prognosis. Even with current standard treatments involving maximal safe surgical resection, radiotherapy, and chemotherapy with temozolomide [7], the median survival rate for glioblastoma patients is approximately 15 months [8], highlighting the urgent need for more effective therapies.
From a molecular perspective, glioblastoma is characterized by several well-documented genetic alterations. These include amplification and mutation of the epidermal growth factor receptor (EGFR), loss or mutation of the tumor suppressor genes phosphatase and tensin homolog (PTEN) and TP53, and mutation of isocitrate dehydrogenase 1 (IDH1), among others [9]. These genetic changes disrupt normal cell signaling pathways, resulting in uncontrolled cell growth, resistance to cell death, formation of new blood vessels, and invasion. Additionally, the role of non-coding RNAs, particularly long non-coding RNAs (lncRNAs), in glioblastoma pathogenesis is gaining attention and providing new insights into the molecular complexity underlying this devastating disease.
Brief on long non-coding RNAs (lncRNAs)
Recent advances in whole-genome and transcriptome sequencing technologies have revolutionized our understanding of the human genome. We now know that a significant portion of the genome is transcribed, but only a fraction of those transcripts actually encode proteins. One prominent group of transcribed RNA molecules is the long non-coding RNAs (lncRNAs), which are characterized by their length of more than 200 nucleotides and lack of protein-coding potential [10].
LncRNAs are an intriguing aspect of the genome’s “dark matter” and exhibit distinct characteristics that differentiate them from other types of non-coding RNAs. Unlike small non-coding RNAs like microRNAs (miRNAs), lncRNAs possess more intricate secondary and tertiary structures, enabling them to interact with DNA, RNA, and proteins and perform diverse regulatory functions.
The functional versatility of lncRNAs is vast, encompassing roles in gene expression regulation at both the transcriptional and post-transcriptional levels. They can modulate protein activity [11], alter protein localization, and act as signals, decoys, guides, or scaffolds, influencing crucial biological processes like cell growth, differentiation, and apoptosis [12–14]. Additionally, lncRNAs can exert their regulatory influence through epigenetic mechanisms, such as DNA methylation and histone modification.
Of particular interest is the ability of lncRNAs to function as “molecular sponges,” sequestering other biomolecules, notably miRNAs, and thereby interfering with their normal functions. These lncRNAs are often termed competing endogenous RNAs (ceRNAs).
Furthermore, the dysregulation of lncRNAs has been implicated in various diseases, including cancers [15]. In glioblastoma, for example, a multitude of lncRNAs have been found to be abnormally expressed, impacting tumor growth, invasion, and responsiveness to treatments.
Importance of lncRNA in cancer biology
The essential roles played by lncRNAs in cancer biology have garnered significant attention in recent years due to their contributions to the regulation of key cellular processes that are frequently perturbed in malignancies.
LncRNAs play a crucial role in fine-tuning gene expression. They exert their influence through various mechanisms at both the transcriptional and post-transcriptional levels, influencing chromatin modification, transcriptional activation or repression, and mRNA stability. For instance, lncRNAs can guide chromatin-modifying enzymes to specific genomic sites, leading to alterations in gene expression patterns. These regulatory activities can impact multiple genes and signaling pathways associated with cancer development, including cell proliferation, apoptosis, and metastasis [16].
The function of lncRNAs as “sponges” for miRNAs, also known as competing endogenous RNA (ceRNA) activity [17,18], has been identified as a novel mechanism of gene regulation. By sequestering and deactivating miRNAs, lncRNAs can prevent the inhibition of their target mRNAs by these miRNAs, indirectly modulating gene expression. This intricate network of lncRNA-miRNA-mRNA can impact various cancer characteristics, including tumor growth, invasiveness, and resistance to therapy [19].
LncRNAs also play a pivotal role in shaping the epigenetic landscape of cells, affecting patterns of DNA methylation and histone modification. DNA methylation, which is dynamically regulated during development and meticulously maintained in somatic cells, is one of the most significant epigenetic mechanisms for gene expression regulation. Aberrant DNA methylation patterns are strongly linked to human diseases, including cancer. A growing body of evidence supports the notion that lncRNAs mediate DNA methylation in both normal and pathological conditions. Firstly, lncRNAs can attract or repel DNA modifiers such as DNA methyltransferases and ten-eleven translocation proteins (DNMTs/TETs) to specific gene targets. Secondly, lncRNAs can regulate DNMT activity by controlling the levels of the DNMT cofactor S-adenosylmethionine and S-adenosylhomocysteine (SAM/SAH). Lastly, lncRNAs can influence the expression of DNMTs/TETs themselves at multiple levels [20]. These alterations can result in the silencing of tumor suppressor genes or the activation of oncogenes, contributing to cancer initiation and progression [21].
Interaction with signaling pathways
LncRNAs can disrupt the normal functioning of signaling pathways, often leading to the abnormal activation of pathways that facilitate cell survival, proliferation, and migration. For instance, lncRNAs have been implicated in the regulation of crucial cancer-related pathways, such as the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR), Wnt/β-catenin, and rat sarcoma virus (Ras) and extracellular signal-regulated kinase (ERK) pathways.
Implication in cancer stem cells
Emerging evidence has suggested a role for lncRNAs in the maintenance and differentiation of cancer stem cells (CSCs), a subset of cells within tumors that are associated with therapy resistance and disease recurrence. By influencing the self-renewal and pluripotency of CSCs, lncRNAs contribute to tumor heterogeneity and progression [22].
In the case of glioblastoma, numerous lncRNAs have been found to be dysregulated, exerting diverse functions in tumor development and progression. Understanding the mechanisms through which these lncRNAs impact glioblastoma pathogenesis is vital for the development of novel diagnostic and therapeutic strategies.
Despite advancements in surgical techniques, radiotherapy, and chemotherapy, glioblastoma, the most aggressive primary brain tumor, still has a poor prognosis. Recent studies have revealed the involvement of non-coding RNAs, specifically long non-coding RNAs (lncRNAs), in glioblastoma pathogenesis, shedding light on its complex molecular and cellular heterogeneity.
One interesting mechanism by which lncRNAs function is by acting as molecular ‘sponges,’ interacting with and sequestering other biomolecules such as microRNAs (miRNAs). This interaction significantly affects various aspects of glioblastoma biology, including cell proliferation, apoptosis, migration, angiogenesis, and immune evasion.
Despite the growing research in this field, a comprehensive understanding of the sponge function of lncRNAs in glioblastoma remains unclear. Hence, the goal of this review is fourfold:
(1) To provide a comprehensive overview of how lncRNAs act as molecular sponges in glioblastoma, explaining their roles and mechanisms.
(2) To emphasize the impact of this sponge activity on the different characteristics of glioblastoma, from tumorigenesis to resistance against therapy.
(3) To discuss the potential of lncRNAs as diagnostic and prognostic biomarkers for glioblastoma.
(4) To explore the challenges and therapeutic potential of targeting lncRNA-miRNA interactions in glioblastoma.
Through this review, we aim to highlight the significance of the lncRNA-mediated sponge effect in glioblastoma, stimulating further research in this promising field. Additionally, we hope to facilitate discussions on how this knowledge can be utilized to enhance the diagnosis and treatment of glioblastoma.
lncRNA Biogenesis and Function
The process of long non-coding RNA (lncRNA) biogenesis closely resembles that of mRNA, but with notable differences that underscore their unique regulatory functions. LncRNA genes are transcribed by RNA polymerase II (Pol II), and like mRNAs, their primary transcripts undergo 5′ capping, splicing, and polyadenylation [23,24].
The transcription of lncRNAs can occur in different genomic regions, including intergenic regions (known as long intergenic non-coding RNAs or lincRNAs), antisense strands of protein-coding genes, and intronic regions. Importantly, the transcription of lncRNAs is often controlled by mechanisms and factors that also regulate protein-coding genes, such as promoter and enhancer elements, transcription factors, and epigenetic modifications. For instance, an lncRNA transcribed from the enhancer binding protein α (CCAAT/CEBPA) locus activates CEBPA mRNA transcription by inhibiting DNA cytosine-5-methyltransferase 1 (DNMT1) [25–27]. In addition, p53-regulated lncRNAs (lincRNA-p21, PANDA, lincRNA-PINT) suppress target gene transcription by interacting with transcription factors heterogeneous nuclear ribonucleoprotein K (hnRNPK) or polycomb repressive complex 2 (PRC2) [28–30].
Splicing, polyadenylation, and stability
In contrast to mRNA molecules, lncRNAs generally undergo less efficient alternative splicing and have a lower number of exons. These characteristics, along with their lower expression levels and limited cellular distribution, pose challenges for their detection and analysis. Polyadenylation, a common feature of lncRNAs, can influence their stability and cellular localization. However, some lncRNAs lack polyadenylation and instead rely on the presence of triple-helical structures at their 3′ ends to maintain stability.
The cellular localization of lncRNAs varies widely. While some transcripts are predominantly found in the nucleus, others are primarily located in the cytoplasm [31–33]. This diverse localization reflects the different functions of lncRNAs: nuclear lncRNAs can regulate gene expression at the transcriptional level or modulate chromatin structure [34–36], while cytoplasmic lncRNAs [37,38] can impact mRNA stability and translation [39–41] or serve as miRNA sponges.
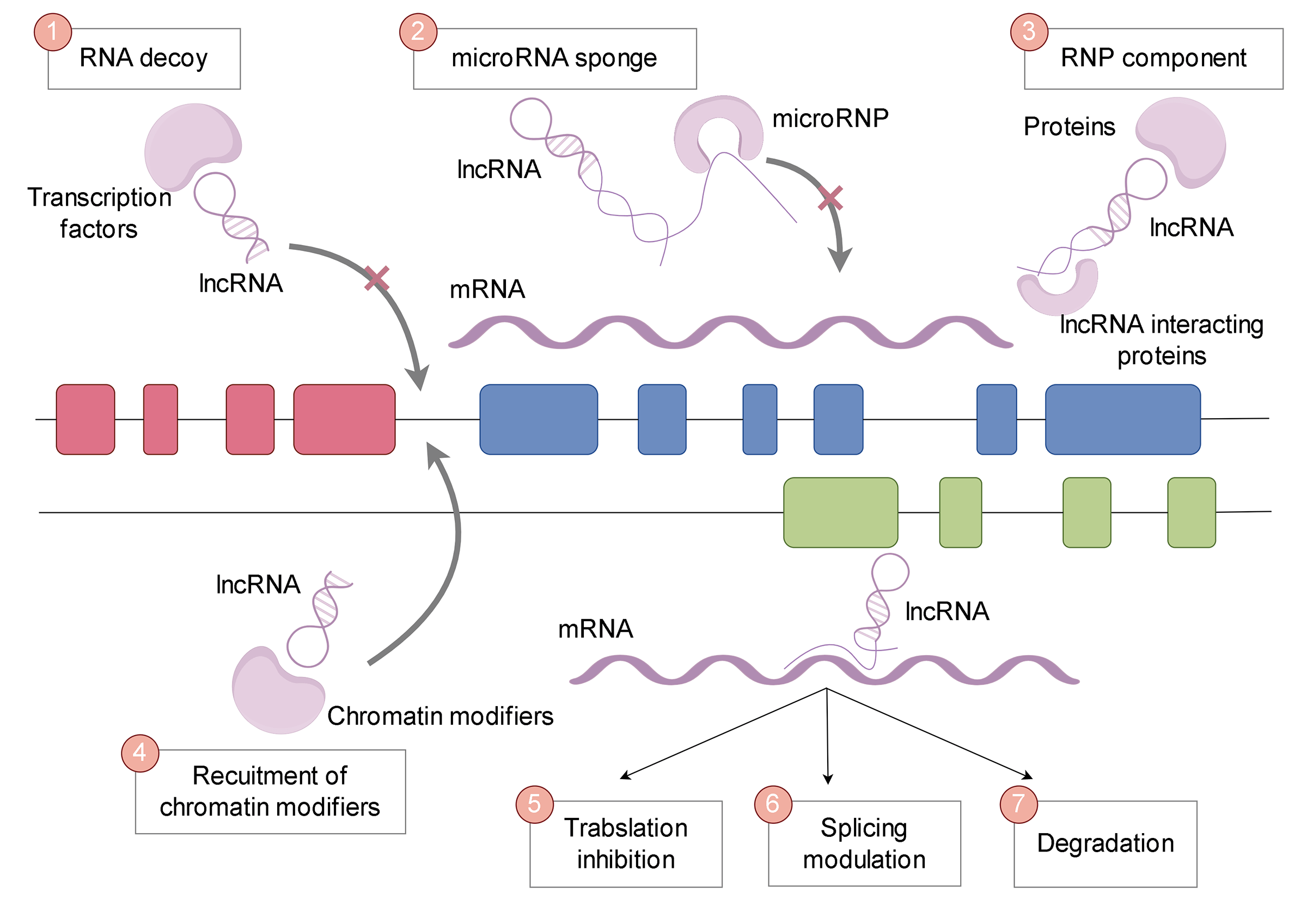
Long non-coding RNAs (lncRNAs) exhibit a multitude of functional capabilities that contribute to their crucial roles in cellular processes and disease states (Fig. 1). These functions can be broadly categorized into the following groups:
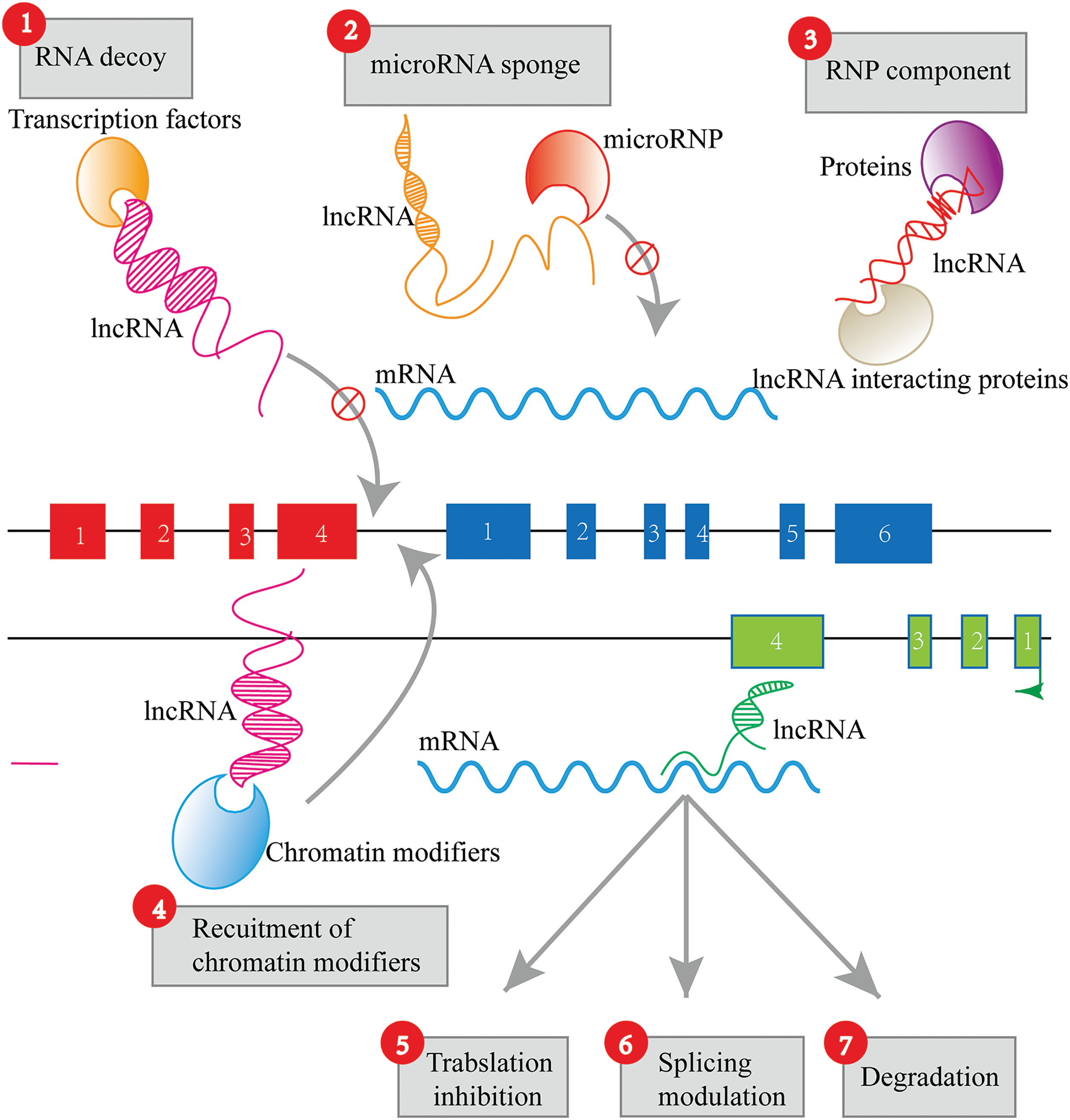
Figure 1: Functions of lncRNAs. LncRNA functions include RNA decoy, microRNA sponge, RNA-protein (RNP) components, recruitment of chromatin modifiers, translation inhibition, splicing regulation, and degradation.
LncRNAs can act as “sponges” or “decoys” by sequestering proteins or miRNAs, preventing them from binding to their intended targets (Fig. 2). This competitive interaction of lncRNAs, known as ceRNA activity, can lead to significant alterations in cellular gene expression profiles and functions. A well-studied example is the lncRNA PTENP1, which acts as a decoy for miRNAs that target the tumor suppressor gene PTEN, thereby influencing cancer progression [42]. Phosphatase and tensin homolog pseudogene 1 (PTENP1) has been demonstrated to be an active transcript that competes with miRNAs, enhancing PTEN protein expression. It exerts a growth-suppressive effect by impeding the binding of miRNAs to the 3′ untranslated region (UTR) of PTEN, thus protecting it from degradation [42].
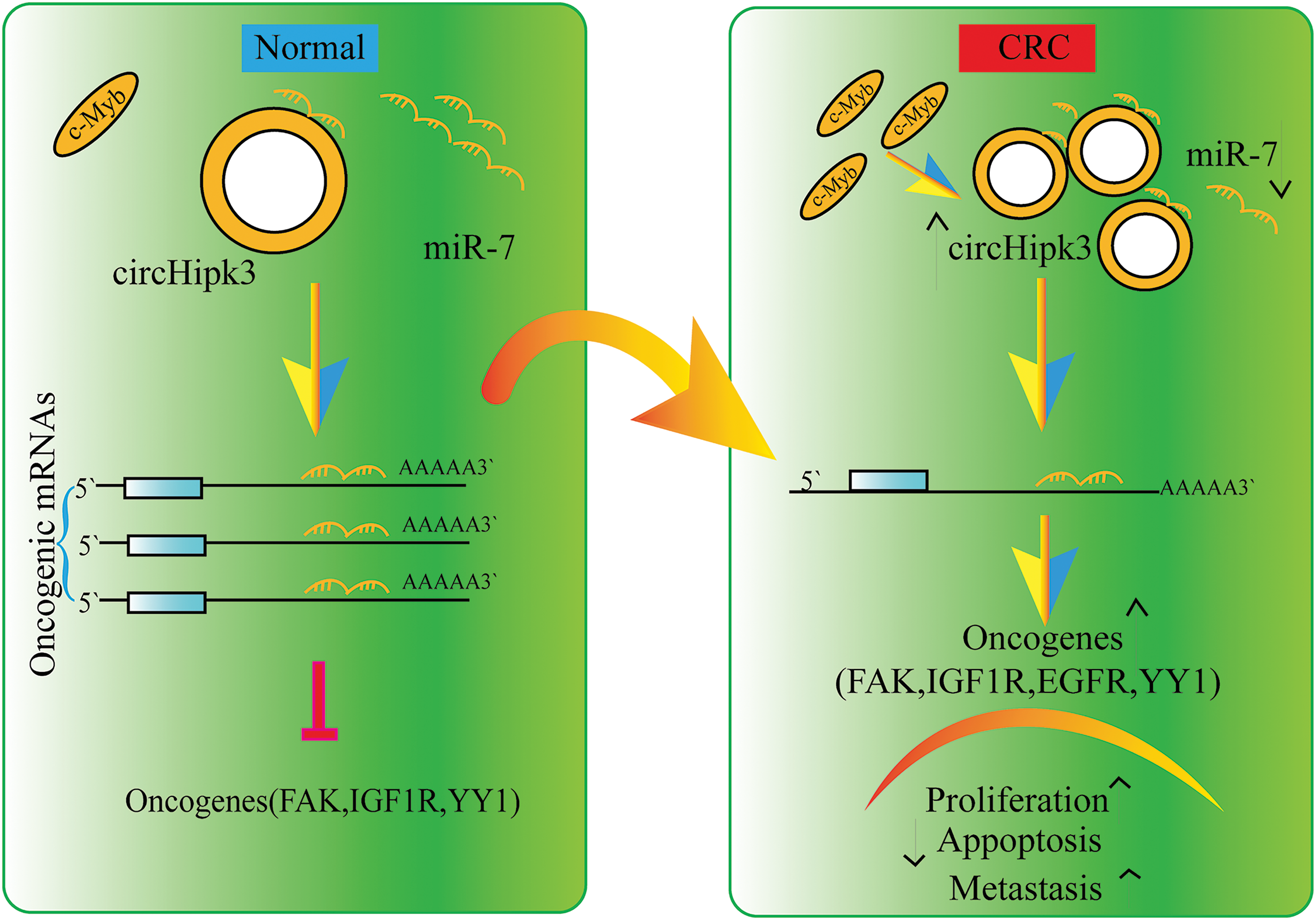
Figure 2: Sponging action of lncRNAs.
Fig. 2 presents the mechanism by which lncRNA functions as a sponge molecule, using circHipk3 and its interaction with miR-7 as a representative example. The figure illustrates how circHipk3 sponges miR-7, competitively inhibiting its binding to oncogenic mRNAs. This inhibition prevents miR-7 from exerting its tumor-suppressive effects, thereby leading to the up-regulation of various oncogenes, such as focal adhesion kinase (FAK), insulin-like growth factor 1 receptor (IGF1R), epidermal growth factor receptor (EGFR), and Yin Yang 1 (YY1). The diagram details the resulting processes of tumor cell proliferation, metastasis, and the promotion of apoptosis inhibition, which are facilitated by the disrupted regulation of these oncogenes. This depiction effectively demonstrates the critical role of lncRNA-miRNA interactions in cancer biology and elucidates the underlying mechanism contributing to tumor progression and malignancy.
LncRNAs have the capability to act as molecular platforms, allowing multiple proteins to come together and form functional complexes. They can also act as navigators, guiding protein complexes to specific targets. These abilities give lncRNAs the power to finely regulate a wide range of biological processes.
Recruitment of chromatin modifiers
LncRNAs have the ability to directly interact with chromatin, aiding in the recruitment of enzymes that modify chromatin to specific genomic locations. Through this interaction, they can induce changes in the chemical modifications of histones, influencing the structure of chromatin and subsequently affecting the expression of genes [43,44]. Well-known examples of lncRNAs that act through this mechanism include HOX transcript antisense RNA (HOTAIR) and X inactive specific transcript (XIST).
Post-transcriptional regulation (translation inhibition, splicing regulation, and degradation)
In the cytoplasm, lncRNAs play a crucial role in post-transcriptional gene regulation. They can impact the stability of mRNA, thereby influencing mRNA splicing, translation efficiency, or degradation. For instance, the lncRNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) modulates alternative splicing by interacting with splicing factors.
In the context of glioblastoma, the diverse functions of lncRNAs provide ample opportunities for understanding disease mechanisms and identifying potential therapeutic targets. Significantly, their role as molecular absorbents and regulators of miRNA activity adds a new layer of intricacy to our understanding of glioblastoma pathogenesis.
A noteworthy aspect of the function of lncRNA that has garnered substantial attention in recent years is its capacity to act as a molecular absorbent. In this role, lncRNAs sequester and gradually release other regulatory RNA or protein molecules from their usual targets, thereby modifying their activity and the resulting cellular processes.
The absorbent function of lncRNAs is particularly apparent in their interactions with microRNAs (miRNAs). MiRNAs are small, non-coding RNAs that typically act as inhibitors at the post-transcriptional level by binding to complementary sequences on target mRNAs. By harboring these miRNA binding sites, lncRNAs can competitively hinder the interactions between miRNAs and their target mRNAs, ultimately increasing the expression of these target genes (Fig. 3). This ceRNA or ‘absorbent’ hypothesis has expanded our knowledge of the regulatory network of gene expression, emphasizing the intricate functional role of lncRNAs in cellular processes.
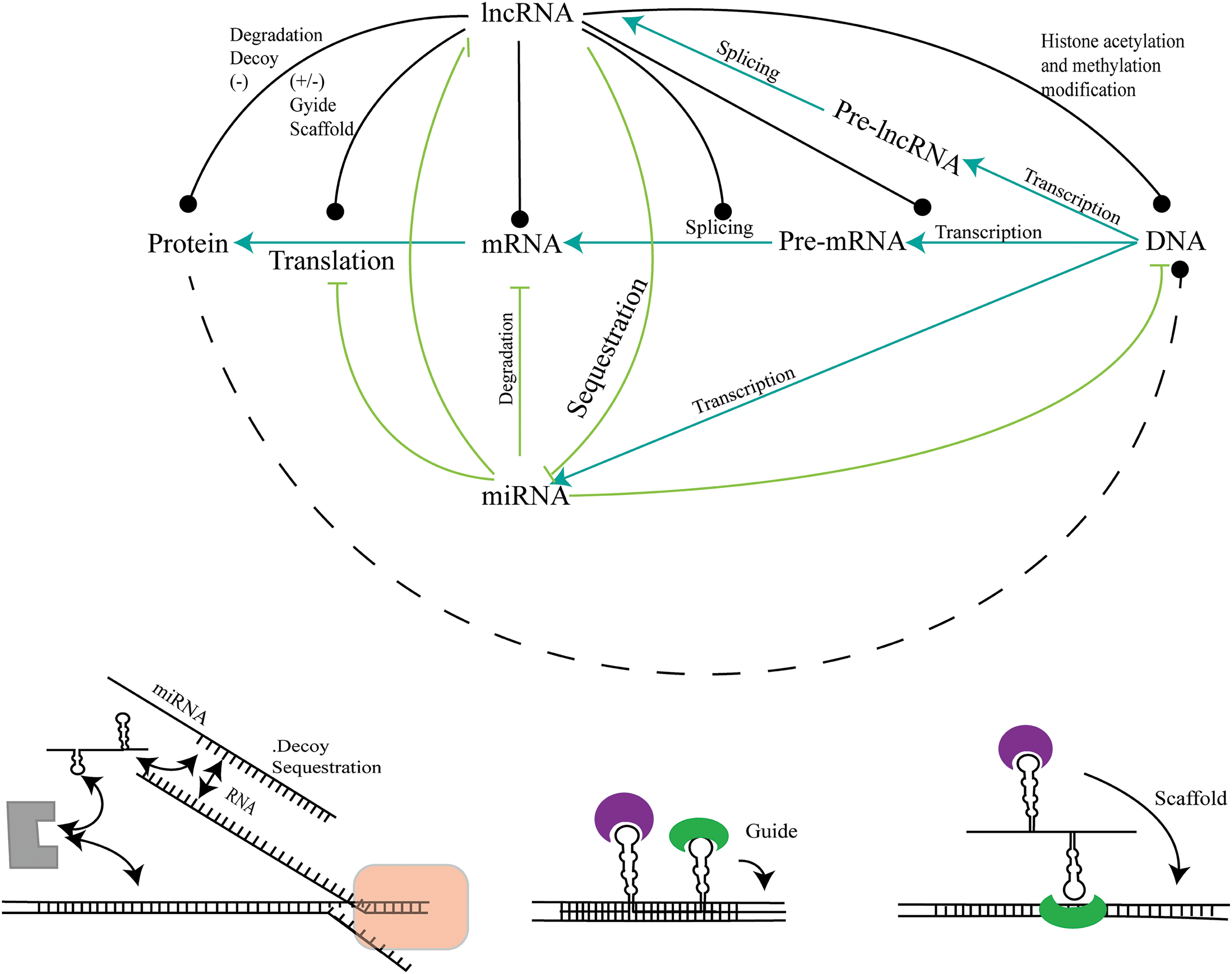
Figure 3: The interaction of lncRNAs and miRNA. The regulatory network formed by the interaction of lncRNAs and miRNA. LncRNAs can regulate proteins, mRNAs, pre-mRNAs, and DNA by interacting with miRNAs, and play roles such as transcriptional regulation, modification, molecular decoy or sponge, scaffolding, and guide.
One well-documented illustration of a sponge lncRNA is H19 in humans. H19, an imprinted oncofetal gene, plays a central role in carcinogenesis. H19 acts as a molecular sponge for let-7 miRNA, a well-known tumor suppressor miRNA, thereby exerting a negative regulatory effect on its activity and amplifying the expression of let-7 target genes, such as the core pluripotency factor LIN28. The enrichment of LIN28 in breast cancer stem cell (BCSC) populations and breast patient samples further supports this finding [45]. Another notable example is the lncRNA PTENP1, which possesses identical miRNA response elements as its associated protein-coding gene PTEN. Through sequestration of PTEN-targeting miRNAs, PTENP1 indirectly enhances PTEN expression, consequently influencing cancer progression.
In the context of glioblastoma, numerous lncRNAs have been identified as molecular sponges, exerting pivotal roles in tumorigenesis, progression, and treatment response. These lncRNAs modulate miRNA activity, thereby impacting various oncogenic or tumor suppressive pathways that regulate cell proliferation, apoptosis, migration, invasion, angiogenesis, and immune response. For example, the lncRNA HOTAIR has been demonstrated to function as a sponge for miR-126 and miR-128 in glioblastoma, facilitating cell proliferation and stemness.
The ability of lncRNAs to act as molecular sponges adds a new layer of complexity to the RNA-mediated regulatory network in glioblastoma. Deciphering this intricate network not only enhances our comprehension of glioblastoma biology but also paves the way for the development of innovative diagnostic tools and therapeutic strategies.
Mechanisms of LncRNA Acting as Sponges in Glioblastoma
Interactions between lncRNA and microRNAs in glioblastoma
Interactions between long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have recently emerged as a crucial regulatory network in glioblastoma. Through these interactions, lncRNAs can act as molecular ‘sponges’, sequestering miRNAs and modulating their activity. This sponging effect significantly impacts glioblastoma pathogenesis, progression, and response to therapy.
LncRNA-microRNA interactions: examples in glioblastoma
Several lncRNAs have been identified as key players in glioblastoma due to their interaction with miRNAs, as depicted in Table 1.
We have identified potential binding sites for miR-126-5p in HOTAIR, a suppressor in glioma [61]. In glioblastoma, HOTAIR acts as a sponge for miR-126 and miR-128, preventing these miRNAs from suppressing their target genes. By competing with miR-126-5p, HOTAIR can modulate generalized least squares (GLS), affecting glioma cell glutamine metabolism, angiogenesis, invasion, and temozolomide resistance [62]. MiR-128 is one of the most commonly found tumor-suppressor miRNAs in brain tissues [63,64]. Consequently, this interaction promotes cell proliferation and the stem-like phenotype of glioblastoma cells.
Colorectal neoplasia differentially expressed (CRNDE) is an lncRNA initially discovered in colorectal cancer, found on chromosome 16, and exerts oncogenic functions in multiple cancers [65]. Another glioblastoma-associated lncRNA, CRNDE, functions as a sponge for miR-384. At the molecular level, the knockdown of CRNDE or overexpression of miR-384 leads to reduced levels of piwi-like RNA-mediated gene silencing 4 (PIWIL4) protein. Moreover, miR-384 targets PIWIL4, which plays an oncogenic role in glioma. This interaction enhances the expression of the miR-384 target gene, PIWIL4, subsequently promoting glioblastoma cell proliferation, migration, and invasion.
NEAT1 is a highly regulated lncRNA in recent pangenomic datasets [66], with tight control over its transcription. It originates from the familial tumor syndrome multiple endocrine neoplasia (MEN) type 1 locus on chromosome 11 and is unique for lacking introns [67]. The alteration in NEAT1 expression has been observed in various human malignancies, including leukemia and ovarian carcinoma, and is known to be crucial for tumorigenesis [68,69]. NEAT1 acts by sequestering miR-449b-5p, resulting in the elevation of c-Met expression, a well-known oncogene in glioblastoma. Sequentially, NEAT1 targets miR-449b, which directly targets c-Met [70]. This interaction has been linked to the stimulation of cell proliferation, migration, and invasion.
Implications for glioblastoma biology and therapy
The involvement of lncRNAs and miRNAs in glioblastoma has widespread consequences on various aspects of tumor biology. By modulating miRNA activity, lncRNAs can influence essential processes such as cell cycle regulation, apoptosis, angiogenesis, cell migration and invasion, stemness, and immune responses [71–73]. As a result, these interactions shape tumor growth, invasiveness, heterogeneity, and the tumor’s response to therapy.
Furthermore, the network of lncRNA-miRNA interactions presents tremendous opportunities for the development of innovative approaches for glioblastoma diagnosis and treatment. LncRNAs and the associated miRNAs have the potential to serve as valuable biomarkers for the detection, prognosis, and prediction of treatment response in glioblastoma. Additionally, targeting these detrimental lncRNA-miRNA interactions using antisense oligonucleotides or small molecule inhibitors can offer novel therapeutic strategies.
In conclusion, the exploration of lncRNA-miRNA interactions in glioblastoma provides captivating insights into the intricate molecular network of this aggressive brain tumor, paving the way for advancements in our understanding and management of this disease.
Specific lncRNA-miRNA networks implicated in glioblastoma
Gaining insights into the mechanisms of tumorigenesis and therapeutic resistance in glioblastoma, the intricate network of interactions between long non-coding RNA (lncRNA) and microRNA (miRNA) reveals novel perspectives [74]. Key regulators of glioblastoma biology have been identified among various lncRNAs, as they interact with specific miRNAs. In this section, we delve into notable lncRNA-miRNA axes implicated in glioblastoma.
HOTAIR (HOX transcript antisense RNA) is a 2.2 kb lncRNA originally identified for its role in orchestrating HOX gene expression [75]. However, it has since been implicated in glioblastoma and other malignancies, exerting a significant role in miRNA regulation. Moreover, HOTAIR contributes to cancer pathogenesis and progression by modulating inflammation and immune signaling [76].
Functioning as a molecular sponge, HOTAIR sequesters miR-126 and miR-128, preventing their interaction with target genes. Consequently, this interaction facilitates cell proliferation and stemness in glioblastoma by upregulating the expression of target genes involved in cell cycle progression and stem cell maintenance [77]. Furthermore, the HOTAIR-miR-126/128 network has been associated with adverse patient prognosis, underscoring its potential as a therapeutic target and prognostic marker.
The interaction between the lncRNA CRNDE and miR-384 modulates the expression of the oncogene PIWIL4 in glioblastoma. By acting as a sponge for miR-384, CRNDE enhances the expression of PIWIL4, thereby promoting cell proliferation, migration, and invasion in glioblastoma [78].
Nuclear paraspeckle assembly transcript 1 (NEAT1) is an lncRNA that localizes to paraspeckles, which are nuclear bodies. These paraspeckles participate in the regulation of gene expression by facilitating mRNA editing and relocation from the nucleus to the cytoplasm [79–81]. Although NEAT1 has multifaceted functions, its role as a miRNA sponge has attracted considerable attention in the context of glioblastoma.
NEAT1, an lncRNA commonly found in glioblastomas, has the ability to trap miR-449b-5p, which ultimately results in higher levels of c-Met, the gene targeted by miR-449b-5p [70]. This interaction plays a pivotal role in enhancing cell proliferation, migration, and invasion, thereby increasing the aggressiveness of glioblastomas. The NEAT1-miR-449b-5p network, similar to HOTAIR, has been linked to unfavorable clinical outcomes, underscoring its significance in both the biology and treatment of glioblastomas.
The lncRNA taurine upregulated 1 (TUG1) has been discovered to interact with miR-144 within glioblastoma. By acting as a sponge for miR-144, TUG1 enhances the expression of EZH2, a gene involved in histone methylation and gene suppression [82]. This interaction stimulates cell proliferation, migration, and invasion, highlighting the oncogenic role of TUG1 in glioblastoma.
The analysis of these networks comprising of long non-coding RNAs and microRNAs provides a comprehensive comprehension of the underlying molecular mechanisms in glioblastoma pathogenesis and progression. Remarkably, these networks also present promising targets for therapeutic intervention. For instance, the use of antisense oligonucleotides or small molecule inhibitors to disrupt these detrimental interactions between long non-coding RNAs and microRNAs could establish novel therapeutic strategies for tackling glioblastoma.
Mechanisms of how these lncRNA-miRNA networks influence glioblastoma progression
The interactions between lncRNAs and miRNAs play a crucial role in the progression of glioblastoma by affecting various cellular processes, such as cell proliferation, migration, invasion, angiogenesis, apoptosis, and treatment resistance. This section focuses on the mechanisms of how specific lncRNA-miRNA networks impact these processes in glioblastoma.
Several lncRNA-miRNA networks promote cell proliferation in glioblastoma. For example, the HOTAIR-miR-126/128 and NEAT1-miR-449b-5p networks increase the expression of genes promoting cell cycle progression, enhancing cell proliferation. Similarly, the CRNDE-miR-384 and TUG1-miR-144 networks bolster cell proliferation by upregulating oncogenes such as PIWIL4 and enhancer of zeste homolog 2 (EZH2), respectively.
Influence on migration and invasion
Numerous long non-coding RNAs (lncRNAs) regulate cell migration and invasion, which are critical processes in glioblastoma invasiveness and recurrence. For instance, HOTAIR, CRNDE, and TUG1 sponge miR-126/128, miR-384, and miR-144, respectively, to enhance the expression of target genes involved in cell migration and invasion, thereby promoting the invasive phenotype of glioblastoma cells.
The impact of lncRNA-miRNA networks on angiogenesis, a process crucial for supplying oxygen and nutrients to the growing tumor, is noteworthy. Tumor cells can form microvascular channels, vasculogenic mimicry (VM), to increase nutrient and blood supply in different types of tumors, including GBM. The overexpression of lncRNA HULC significantly enhanced the tubular formation and invasion of human GBM cells and promoted the growth of GBM tumors in mice, while the silencing of lncRNA HULC had the opposite effects. Furthermore, the overexpression of lncRNA HULC increased the EMT process and MMP expression, while its silencing decreased them in GBM [83]. However, the specific lncRNAs involved in glioblastoma angiogenesis via miRNA sponging have yet to be fully elucidated.
Long non-coding RNAs (lncRNAs) can modulate apoptosis, a process of programmed cell death that is often dysregulated in cancer. Some lncRNAs interact with specific miRNAs to alter the expression of apoptosis-related genes, thereby influencing glioblastoma cell survival. For instance, PITPNA-AS1 sponges miR-223-3p to promote EGFR expression, thus activating the PI3K/AKT signaling pathway to accelerate proliferation and inhibit apoptosis of GBM cells [84]. The specifics of such interactions in glioblastoma require further investigation.
Influence on treatment resistance
Recent studies suggest that certain long non-coding RNAs (lncRNAs) may play a role in the resistance of glioblastoma cells to current therapies, including temozolomide, radiation, and targeted therapy. This resistance is often due to the ability of lncRNAs to modulate DNA repair, drug efflux, cell survival pathways, and stemness by interacting with miRNAs.
In conclusion, lncRNA-miRNA networks can influence glioblastoma progression through multiple mechanisms. Understanding these mechanisms enhances our knowledge of glioblastoma biology and highlights the potential of these networks as targets for therapeutic intervention.
Impact on Tumor Microenvironment and Immune Response
Role of lncRNA-miRNA networks in shaping the tumor microenvironment
The tumor microenvironment (TME) is a complex network of neoplastic, immune, stromal, and endothelial cells, along with extracellular matrix and secreted factors. It plays a crucial role in glioblastoma pathogenesis and therapy resistance. Long non-coding RNAs (lncRNAs), particularly through their interactions with microRNAs (miRNAs), can significantly shape the TME, impacting tumor progression, immune evasion, and therapeutic outcomes. There is increasing evidence to support this claim [85]. For instance, CamK-A is significantly expressed in various human cancers and plays a role in remodeling the cancer microenvironment by activating Ca2+-triggered signaling pathways. Mechanistically, CamK-A activates Ca2+/calmodulin-dependent pregnancy up-regulated non-ubiquitous calmodulin kinase (PNCK), which subsequently phosphorylates inhibitor of nuclear factor kappa B α (IκBα) and triggers the activation of calcium-dependent nuclear factor κB (NF-κB). This regulation results in the remodeling of the tumor microenvironment, which includes macrophage recruitment, angiogenesis, and tumor progression [86].
Certain long non-coding RNA (lncRNA)-microRNA (miRNA) networks have been implicated in modulating the immune component of the TME [87]. For instance, some lncRNAs can influence the expression of immune checkpoint molecules, cytokine and chemokine secretion, and the recruitment and function of immune cells, such as T cells, macrophages, and myeloid-derived suppressor cells, by sponging immune-related miRNAs. However, the specifics of these interactions in glioblastoma remain to be fully elucidated.
Interactions between lncRNAs and miRNAs can also impact the function of stromal cells in the tumor microenvironment (TME), including cancer-associated fibroblasts (CAFs) and endothelial cells. For instance, specific lncRNAs can regulate the activation and function of CAFs or modulate angiogenesis by altering endothelial cell behavior through their interaction with miRNAs.
Impacting extracellular matrix remodeling
The remodeling of the extracellular matrix (ECM) [88] is a crucial aspect of the tumor microenvironment (TME) that contributes to tumor progression and therapy resistance. Certain long non-coding RNAs (lncRNAs) can regulate the expression of matrix metalloproteinases [89], collagen [90], and other ECM components by interacting with miRNAs, thereby influencing ECM remodeling.
Contributing to therapy resistance
Recent studies indicate that long non-coding RNA (lncRNA) and microRNA (miRNA) networks may contribute to therapy resistance in glioblastoma by influencing the TME [91]. For example, specific lncRNAs can promote the formation of a pro-tumorigenic and immunosuppressive TME [92], thereby facilitating glioblastoma cell resistance to radiation, chemotherapy, and immunotherapy [93].
In conclusion, the lncRNA-miRNA networks have a significant impact on the TME [94], contributing to glioblastoma pathogenesis and therapy resistance. Further exploration of these interactions will provide valuable insights into glioblastoma biology and highlight new avenues for therapeutic intervention.
Influence on immune evasion in glioblastoma
Glioblastoma, like other cancers, exhibits immune evasion [95]. This is achieved by creating an immunosuppressive tumor microenvironment (TME), which allows glioblastoma cells to evade immune surveillance and avoid destruction by the immune system. Recent studies suggest that lncRNAs play a crucial role in this process by interacting with miRNAs.
Regulation of immune checkpoints and immune cell recruitment
Certain lncRNAs can affect the expression of immune checkpoint molecules, such as programmed death-ligand 1 (PD-L1) [96], cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), MIR155 host gene (MIR155HG), TRG-AS1 and PCED1B antisense RNA 1 (PCED1B-AS1) [97], by interacting with specific microRNAs (miRNAs). In glioblastoma, the lncRNA-miRNA network may upregulate immune checkpoints, inhibiting T cell function and promoting immune evasion. The specific lncRNAs and their interacting miRNAs involved in this regulation have yet to be fully elucidated.
The lncRNA-miRNA networks have the ability to regulate the recruitment and function of different immune cells in the TME, which contributes to the immunosuppressive environment of glioblastoma. For instance, specific lncRNAs can enhance the recruitment of regulatory T cells (Tregs) and myeloid-derived suppressor cells (MDSCs) by sponging certain miRNAs, both of which can suppress antitumor immune responses. Similarly, lncRNA-miRNA networks can influence the polarization of tumor-associated macrophages (TAMs) [98], promoting the M2 phenotype [99] that favors tumor progression and immune evasion.
Influence on cytokine and chemokine secretion
Additionally, cytokines and chemokines secreted by glioblastoma and stromal cells can shape the TME to favor immune evasion. lncRNA-miRNA networks can regulate the expression of these molecules, altering the immune landscape of the TME. For example, some long non-coding RNAs (lncRNAs) can promote the secretion of immunosuppressive cytokines, such as TGF-β [100] and IL-10, by interacting with miRNAs. This can contribute to an immunosuppressive TME.
Impact on antigen presentation and T cell activation
Recent studies indicate that lncRNA-miRNA networks can impact antigen presentation and T cell activation [101], which are essential steps in antitumor immune responses. Certain lncRNAs can modify the expression of MHC class I molecules or co-stimulatory and co-inhibitory molecules [102] by interacting with specific miRNAs, thereby influencing T cell activation and function.
In summary, lncRNA-miRNA networks can significantly influence immune evasion in glioblastoma [103] through multiple mechanisms. Unraveling these interactions offers valuable insights into glioblastoma immunobiology and novel therapeutic targets for enhancing the efficacy of immunotherapies [104].
Potential Therapeutic Implications
LncRNA as diagnostic or prognostic biomarkers
Biomarkers are biological measures that provide critical information about the biological state or condition of an individual. In the context of glioblastoma, several potential diagnostic, prognostic, and predictive biomarkers have been investigated [105]. Biomarkers can assist in the diagnosis, prognosis, and monitoring of therapeutic responses. Long non-coding RNAs (lncRNAs), due to their regulatory roles and disease-specific expression patterns, have emerged as potential biomarkers in glioblastoma.
Several long non-coding RNAs (lncRNAs) have been identified as potential diagnostic biomarkers for glioblastoma due to their differential expression in glioblastoma tissues compared to normal brain tissues. For instance, lncRNA HOTAIR has been found to be upregulated in glioblastoma tissues and can distinguish glioblastoma patients from healthy individuals with significant accuracy. Similarly, other lncRNAs such as C/D box snoRNA U76 (SNORD76) [106], NEAT1 and MALAT1, have shown potential as diagnostic biomarkers.
In addition, lncRNAs can be detected in bodily fluids, such as blood and cerebrospinal fluid, which allows for non-invasive or minimally invasive diagnostic procedures. Their stability in these fluids further enhances their potential as diagnostic biomarkers.
Prognostic and predictive biomarkers
LncRNAs have potential as prognostic biomarkers, providing insights into the likely course or outcome of the disease. Certain lncRNAs expression levels have been correlated with survival rates in glioblastoma patients. For example, higher levels of AGAP2-AS1 [107], HOTAIR, and CRNDE have been associated with poorer prognosis, signifying shorter overall survival. Therefore, these lncRNAs could potentially stratify patients into different risk groups and guide treatment decisions.
Additionally, some lncRNAs may serve as predictive biomarkers, indicating the likely response or resistance to specific therapies. For instance, taurine upregulated gene 1 (TUG1) has a significant regulatory effect on the malignant biological behavior of glioma cells [108]. LncRNA TUG1 has been associated with temozolomide resistance in glioblastoma, indicating its potential as a predictive biomarker for this commonly used chemotherapeutic agent.
In conclusion, lncRNAs have disease-specific expression patterns and regulatory roles, making them promising diagnostic, prognostic, and predictive biomarkers in glioblastoma. However, further research is necessary to validate these findings and explore their clinical utility.
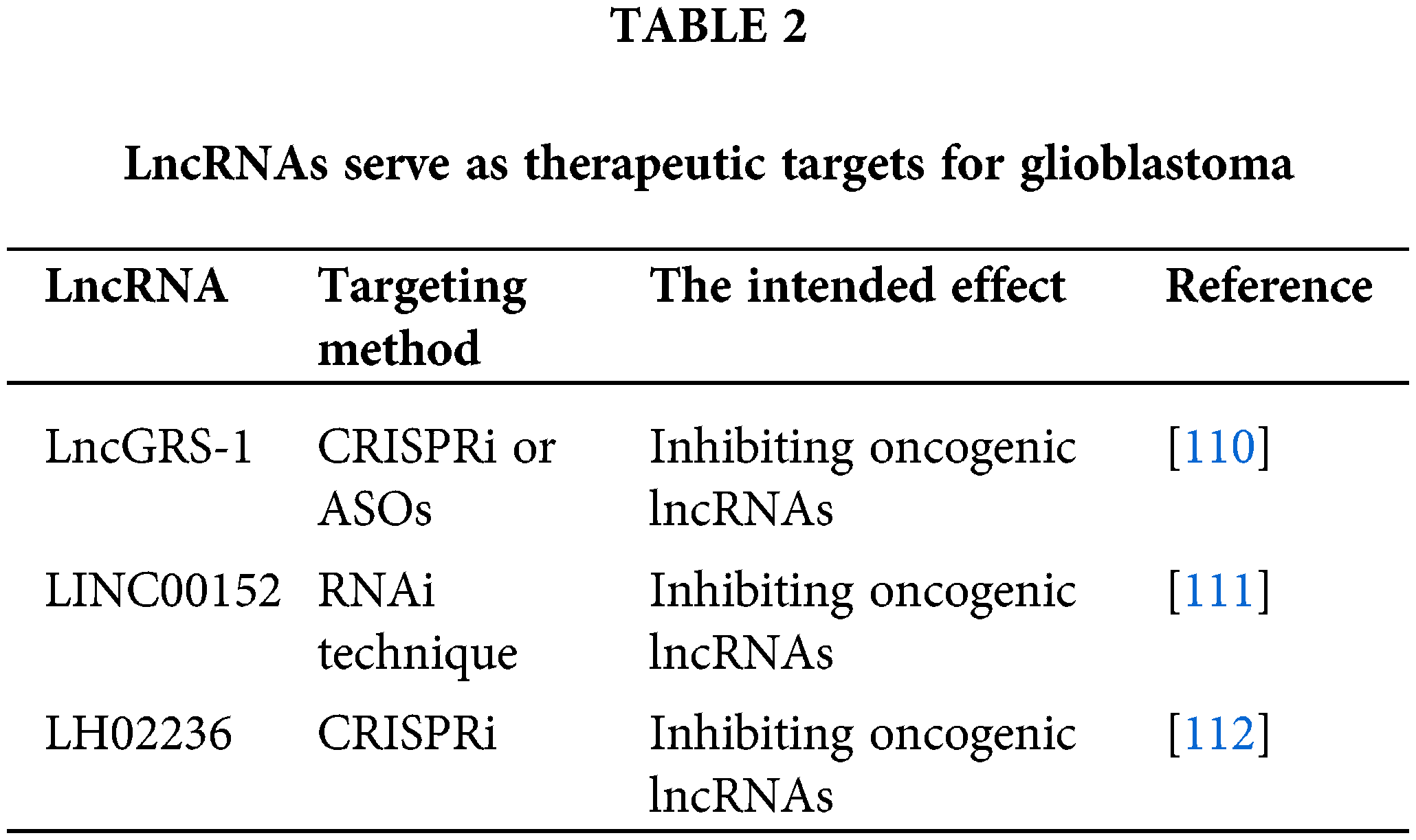
Long non-coding RNAs (lncRNAs) play an integral role in glioblastoma pathogenesis and have potential as biomarkers and therapeutic targets. Various strategies have been proposed to target lncRNAs, and preclinical studies have shown promising results, as shown in Table 2. For instance, bromodomain and extra-terminal (BET) domain proteins have been found to reduce the level of lncRNA HOTAIR [109].
One approach to targeting lncRNAs is to inhibit their expression or function. This can be achieved using RNA interference (RNAi) techniques such as small interfering RNAs (siRNAs) or short hairpin RNAs (shRNAs), antisense oligonucleotides (ASOs), or CRISPR/Cas-based techniques. For example, siRNAs targeting HOTAIR have been shown to inhibit glioblastoma cell proliferation and invasion in vitro and slow tumor growth in vivo.
Disruption of lncRNA-miRNA interactions
Since many long non-coding RNAs (lncRNAs) act as molecular sponges for microRNAs (miRNAs), disrupting these interactions is a potential therapeutic strategy. This can be accomplished by introducing miRNA mimics that compete with the lncRNA for miRNA binding or by using molecules that interfere with the lncRNA-miRNA interaction. However, a comprehensive understanding of the specific lncRNA-miRNA network involved is necessary for this approach.
Targeting lncRNA-associated proteins
Some long non-coding RNAs (lncRNAs) exert their functions by interacting with proteins. Therefore, targeting these lncRNA-associated proteins represents another potential therapeutic strategy. Small molecules, peptides, or antibodies could be designed to disrupt these lncRNA-protein interactions, thereby inhibiting the function of the lncRNA.
Utilizing lncRNAs for gene therapy
Finally, lncRNAs could be used for gene therapy. For example, tumor-suppressive lncRNAs could be delivered to glioblastoma cells using viral vectors or nanoparticle-based delivery systems to suppress tumor growth.
In conclusion, lncRNAs are promising therapeutic targets in glioblastoma, and various strategies have been proposed to target these molecules. However, several challenges persist, such as delivering lncRNA-targeting agents across the blood-brain barrier and the possibility of off-target effects. Therefore, additional research is necessary to refine these strategies and evaluate their safety and effectiveness in preclinical and clinical settings.
This review comprehensively discusses the role of long non-coding RNAs (lncRNAs) in glioblastoma, focusing on their ‘sponge’ function. LncRNAs regulate the activity of microRNAs (miRNAs) to modulate diverse cellular processes.
The discussion begins with a general introduction to glioblastoma and lncRNAs, detailing their relevance in cancer biology. We examined the biogenesis and functions of lncRNAs, highlighting their role as molecular sponges in modulating miRNA activity.
We also explored the interactions between lncRNAs and miRNAs in glioblastoma, focusing on specific lncRNA-miRNA networks implicated in this brain cancer. Detailed explorations of the mechanisms by which lncRNA-miRNA networks influence glioblastoma progression were provided, highlighting the roles of several prominent lncRNAs, such as HOTAIR and NEAT1, in glioblastoma pathogenesis.
The text also examines how lncRNA-miRNA networks shape the tumor microenvironment to promote glioblastoma progression and enable immune evasion. We considered the potential of lncRNAs as diagnostic or prognostic biomarkers in glioblastoma due to their disease-specific expression patterns.
Additionally, we explored the possibility of targeting lncRNAs for therapy. Strategies such as lncRNA inhibition, disruption of lncRNA-miRNA interactions, and lncRNA-based gene therapy were discussed. Despite the challenges, such as functional annotation, technical limitations, and therapeutic delivery, there is optimism about the future of lncRNA research in glioblastoma.
By utilizing integrated approaches, single-cell technologies, and focused efforts on clinical validation, substantial progress is anticipated in this field, bringing us closer to realizing the full potential of lncRNAs in glioblastoma diagnosis, prognosis, and therapy.
Current gaps in understanding and future research directions
As explored in this review, long non-coding RNAs (lncRNAs) play a crucial role in the pathogenesis of glioblastoma by acting as molecular sponges to regulate microRNA activity. However, there are still significant gaps in our understanding that require further research.
Function of many lncRNAs remains unknown
Although the number of identified lncRNAs is increasing, their functional annotation remains a major challenge. To address these issues, further research is needed. The function of many lncRNAs in glioblastoma is still unclear due to their low expression level and high tissue specificity, which pose technical challenges for their detection and quantification. Additionally, distinguishing the functions of lncRNAs from those of their associated small RNAs or protein partners is challenging. An integrated approach may be the key to the future of lncRNA research in glioblastoma. This approach should combine genomic, transcriptomic, and epigenomic data to comprehensively understand the roles of lncRNAs in glioblastoma biology. Future research should focus on uncovering these functions.
Detailed mechanisms of lncRNA-miRNA interactions
Although it is evident that lncRNAs interact with miRNAs in glioblastoma, the precise mechanisms of these interactions remain unclear. Future research should focus on clarifying these mechanisms, which may reveal new therapeutic targets.
Role of lncRNAs in the tumor microenvironment
Our knowledge of the function of lncRNAs in shaping the tumor microenvironment and promoting immune evasion is still limited. Single-cell RNA sequencing technologies could provide insights into the specific roles of lncRNAs in glioblastoma, helping to clarify their functions in different cellular components of the tumor microenvironment. Future research should further investigate this area, possibly utilizing single-cell sequencing technologies to comprehend the cell-type specific functions of lncRNAs.
Clinical utility of lncRNAs as biomarkers or therapeutic targets
The development and optimization of lncRNA-based therapies, including strategies to inhibit oncogenic lncRNAs or replace tumor-suppressive lncRNAs, represent exciting avenues for future research. Combining lncRNA-based therapies with existing treatments may also enhance therapeutic outcomes. Although preclinical studies have shown that lncRNAs can be used as diagnostic, prognostic or predictive biomarkers and therapeutic targets for glioblastoma. In particular, lncRNA DSCAM-AS1 and GATA3-AS1 can serve as examples of this because of their high subtype-specific expression profiles in intracavitary B-like breast cancer [113]. However, its clinical application has not been fully verified. Large-scale, multicenter clinical studies are needed to confirm these findings and assess the safety and efficacy of lncRNA-based therapies.
Optimization of lncRNA-based therapies
Although several strategies for targeting lncRNAs have been proposed, many challenges persist. These include delivering lncRNA-targeting agents across the blood-brain barrier and avoiding off-target effects. Future research should focus on optimizing these strategies.
In conclusion, while our understanding of lncRNAs in glioblastoma has advanced significantly, there are still significant gaps that need to be addressed. Future research that addresses these gaps could significantly enhance our understanding of glioblastoma biology and lead to the development of new diagnostic and therapeutic strategies for this devastating disease.
Acknowledgement: None.
Funding Statement: The study is funded by Binzhou Medical University Research Fund Project (Grant Number BY2021KYQD02).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yunqi Ma; draft manuscript preparation: Caijuan Liu, Xinghao Li, Yuxuan Wu, Jinhui Yang, Menghan Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Wen PY, Weller M, Lee EQ, Alexander BM, Barnholtz-Sloan JS, Barthel FP, et al. Glioblastoma in adults: a society for neuro-oncology (SNO) and European society of neuro-oncology (EANO) consensus review on current management and future directions. Neuro Oncol. 2020;22(8):1073–113. [Google Scholar] [PubMed]
2. Weller M, van den Bent M, Preusser M, Le Rhun E, Tonn JC, Minniti G, et al. EANO guidelines on the diagnosis and treatment of diffuse gliomas of adulthood. Nat Rev Clin Oncol. 2021;18(3):170–86. [Google Scholar] [PubMed]
3. Komori T. Updating the grading criteria for adult diffuse gliomas: beyond the WHO2016CNS classification. Brain Tumor Pathol. 2020;37(1):1–4. [Google Scholar] [PubMed]
4. Jiang Y, Uhrbom L. On the origin of glioma. Ups J Med Sci. 2012;117(2):113–21. [Google Scholar] [PubMed]
5. Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16(7):896–913. [Google Scholar] [PubMed]
6. Roizen MF. Hallmarks of cancer: the next generation. Year Book of Anesthesiology and Pain Management. 2012;2012:646–74. [Google Scholar]
7. Clavreul A, Lemee JM, Soulard G, Rousseau A, Menei P. A simple preoperative blood count to stratify prognosis in isocitrate dehydrogenase-wildtype glioblastoma patients treated with radiotherapy plus concomitant and adjuvant temozolomide. Cancers. 2021;13(22):5778. [Google Scholar] [PubMed]
8. Mega A, Hartmark Nilsen M, Leiss LW, Tobin NP, Miletic H, Sleire L, et al. Astrocytes enhance glioblastoma growth. Glia. 2020;68(2):316–27. [Google Scholar] [PubMed]
9. Kim HJ, Park JW, Lee JH. Genetic architectures and cell-of-origin in glioblastoma. Front Oncol. 2020;10:615400. [Google Scholar] [PubMed]
10. Mattick JS, Amaral PP, Carninci P, Carpenter S, Chang HY, Chen LL, et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat Rev Mol Cell Biol. 2023;24(6):430–47. [Google Scholar] [PubMed]
11. Xu GE, Zhao X, Li G, Gokulnath P, Wang L, Xiao J. The landscape of epigenetic regulation and therapeutic application of N6-methyladenosine modifications in non-coding RNAs. Genes & Dis. 2023;S2352-3042(23)00326-4. [Google Scholar]
12. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. [Google Scholar] [PubMed]
13. Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020;21(3):1027. [Google Scholar] [PubMed]
14. Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89:283–308. [Google Scholar] [PubMed]
15. Bhattacharjee R, Prabhakar N, Kumar L, Bhattacharjee A, Kar S, Malik S, et al. Crosstalk between long noncoding RNA and microRNA in cancer. Cell Oncol. 2023;46(4):885–908. [Google Scholar]
16. Zhang H, Wang Y, Liu X, Li Y. Progress of long noncoding RNAs in anti-tumor resistance. Pathol Res Pract. 2020;216(11):153215. [Google Scholar] [PubMed]
17. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–8. [Google Scholar] [PubMed]
18. Yang N, Liu K, Yang M, Gao X. ceRNAs in cancer: mechanism and functions in a comprehensive regulatory network. J Oncol. 2021;2021:4279039. [Google Scholar] [PubMed]
19. Bai N, Peng E, Qiu X, Lyu N, Zhang Z, Tao Y, et al. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. J Exp Clin Cancer Res. 2018;37(1):172. [Google Scholar] [PubMed]
20. Huang W, Li H, Yu Q, Xiao W, Wang DO. LncRNA-mediated DNA methylation: an emerging mechanism in cancer and beyond. J Exp Clin Cancer Res. 2022;41(1):100. [Google Scholar] [PubMed]
21. Kazimierczyk M, Wrzesinski J. Long non-coding RNA epigenetics. Int J Mol Sci. 2021;22(11):6166. [Google Scholar] [PubMed]
22. Ciafre SA, Russo M, Michienzi A, Galardi S. Long noncoding RNAs and cancer stem cells: dangerous liaisons managing cancer. Int J Mol Sci. 2023;24(3):1828. [Google Scholar] [PubMed]
23. Mushtaq A, Mir US, Hunt CR, Pandita S, Tantray WW, Bhat A, et al. Role of histone methylation in maintenance of genome integrity. Genes. 2021;12(7):1000. [Google Scholar] [PubMed]
24. Gholami A, Farhadi K, Sayyadipour F, Soleimani M, Saba F. Long noncoding RNAs (lncRNAs) in human lymphomas. Genes Dis. 2022;9(4):900–14. [Google Scholar] [PubMed]
25. di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–6. [Google Scholar] [PubMed]
26. Bharti H, Han S, Chang HW, Reinberg D. Polycomb repressive complex 2 accessory factors: rheostats for cell fate decision? Curr Opin Genet Dev. 2023;84(9):102137. [Google Scholar] [PubMed]
27. Yu Y, Li X, Jiao R, Lu Y, Jiang X, Li X. H3K27me3-H3K4me1 transition at bivalent promoters instructs lineage specification in development. Cell Biosci. 2023;13(1):66. [Google Scholar] [PubMed]
28. Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–9. [Google Scholar] [PubMed]
29. Marin-Bejar O, Marchese FP, Athie A, Sanchez Y, Gonzalez J, Segura V, et al. Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol. 2013;14(9):R104. [Google Scholar] [PubMed]
30. Tesfaye E, Martinez-Terroba E, Bendor J, Winkler L, Olivero C, Chen K, et al. The p53 transcriptional response across tumor types reveals core and senescence-specific signatures modulated by long noncoding RNAs. Proc Natl Acad Sci USA. 2021;118(31):2025539118. [Google Scholar]
31. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. [Google Scholar] [PubMed]
32. Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genom. 2007;8(1):39. [Google Scholar]
33. Colognori D, Sunwoo H, Wang D, Wang CY, Lee JT. Xist repeats A and B account for two distinct phases of X inactivation establishment. Dev Cell. 2020;54(1):21–32. [Google Scholar] [PubMed]
34. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–93. [Google Scholar] [PubMed]
35. Jiang B, Yuan Y, Yi T, Dang W. The roles of antisense long noncoding RNAs in tumorigenesis and development through Cis-regulation of neighbouring genes. Biomolecules. 2023;13(4):684. [Google Scholar] [PubMed]
36. Wollny D, Vernot B, Wang J, Hondele M, Safrastyan A, Aron F, et al. Characterization of RNA content in individual phase-separated coacervate microdroplets. Nat Commun. 2022;13(1):2626. [Google Scholar] [PubMed]
37. Carlevaro-Fita J, Rahim A, Guigo R, Vardy LA, Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22(6):867–82. [Google Scholar] [PubMed]
38. Gasparotto E, Burattin FV, di Gioia V, Panepuccia M, Ranzani V, Marasca F, et al. Transposable elements co-option in genome evolution and gene regulation. Int J Mol Sci. 2023;24(3):2610. [Google Scholar] [PubMed]
39. Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. [Google Scholar] [PubMed]
40. Schein A, Zucchelli S, Kauppinen S, Gustincich S, Carninci P. Identification of antisense long noncoding RNAs that function as SINEUPs in human cells. Sci Rep. 2016;6(1):33605. [Google Scholar] [PubMed]
41. Espinoza S, Bon C, Valentini P, Pierattini B, Matey AT, Damiani D, et al. SINEUPs: a novel toolbox for RNA therapeutics. Essays Biochem. 2021;65(4):775–89. [Google Scholar] [PubMed]
42. Ghafouri-Fard S, Khoshbakht T, Hussen BM, Taheri M, Akbari Dilmaghani N. A review on the role of PTENP1 in human disorders with an especial focus on tumor suppressor role of this lncRNA. Cancer Cell Int. 2022;22(1):207. [Google Scholar] [PubMed]
43. Sun Y, Zhang H. A unified mode of epigenetic gene silencing: RNA meets polycomb group proteins. RNA Biol. 2005;2(1):8–10. [Google Scholar] [PubMed]
44. Ma S, Zhang B, LaFave L, Chiang Z, Hu Y, Ding J, et al. Chromatin potential identified by shared single cell profiling of RNA and chromatin. bioRxiv-Genomics. 2020;145:156943. [Google Scholar]
45. Peng F, Li TT, Wang KL, Xiao GQ, Wang JH, Zhao HD, et al. H19/let-7/LIN28 reciprocal negative regulatory circuit promotes breast cancer stem cell maintenance. Cell Death Dis. 2017;8(1):e2569. [Google Scholar] [PubMed]
46. Guo S, King P, Liang E, Guo AA, Liu M. LncRNA HOTAIR sponges miR-301a-3p to promote glioblastoma proliferation and invasion through upregulating FOSL1. Cell Signal. 2022;94:110306. [Google Scholar] [PubMed]
47. Xie H, Shi S, Chen Q, Chen Z. LncRNA TRG-AS1 promotes glioblastoma cell proliferation by competitively binding with miR-877-5p to regulate SUZ12 expression. Pathol Res Pract. 2019;215(8):152476. [Google Scholar] [PubMed]
48. Hu T, Wang F, Han G. LncRNA PSMB8-AS1 acts as ceRNA of miR-22-3p to regulate DDIT4 expression in glioblastoma. Neurosci Lett. 2020;728:134896. [Google Scholar] [PubMed]
49. Shou J, Gao H, Cheng S, Wang B, Guan H. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int. 2021;21(1):39. [Google Scholar] [PubMed]
50. Chen W, Li Q, Zhang G, Wang H, Zhu Z, Chen L. LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. J Cell Mol Med. 2020;24(20):11755–67. [Google Scholar] [PubMed]
51. Zhong C, Yu Q, Peng Y, Zhou S, Liu Z, Deng Y, et al. Novel LncRNA OXCT1-AS1 indicates poor prognosis and contributes to tumorigenesis by regulating miR-195/CDC25A axis in glioblastoma. J Exp Clin Cancer Res. 2021;40(1):123. [Google Scholar] [PubMed]
52. Chen Q, Wang W, Wu Z, Chen S, Chen X, Zhuang S, et al. Over-expression of lncRNA TMEM161B-AS1 promotes the malignant biological behavior of glioma cells and the resistance to temozolomide via up-regulating the expression of multiple ferroptosis-related genes by sponging hsa-miR-27a-3p. Cell Death Discov. 2021;7(1):1390. [Google Scholar]
53. Geng YB, Xu C, Wang Y, Zhang LW. Long non-coding RNA SNHG11 promotes cell proliferation, invasion and migration in glioma by targeting miR-154-5p. Eur Rev Med Pharmacol Sci. 2020;24(9):4901–8. [Google Scholar] [PubMed]
54. Chen HH, Zong J, Wang SJ. LncRNA GAPLINC promotes the growth and metastasis of glioblastoma by sponging miR-331-3p. Eur Rev Med Pharmacol Sci. 23(1):262–70. [Google Scholar]
55. Ren S, Xu Y. AC016405.3, a novel long noncoding RNA, acts as a tumor suppressor through modulation of TET2 by microRNA-19a-5p sponging in glioblastoma. Cancer Sci. 2019;110(5):1621–32. [Google Scholar] [PubMed]
56. Yao Y, Ma J, Xue Y, Wang P, Li Z, Liu J, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359(1):75–86. doi:10.1016/j.canlet.2014.12.051. [Google Scholar] [PubMed] [CrossRef]
57. Momtazmanesh S, Rezaei N. Long non-coding RNAs in diagnosis, treatment, prognosis, and progression of glioma: a state-of-the-art review. Front Oncol. 2021;11:712786. [Google Scholar] [PubMed]
58. Lin H, Zuo D, He J, Ji T, Wang J, Jiang T. Long noncoding RNA WEE2-AS1 plays an oncogenic role in glioblastoma by functioning as a molecular sponge for microRNA-520f-3p. Oncol Res. 2021;28(6):591–603. [Google Scholar] [PubMed]
59. Gao X, Cao Y, Li J, Wang C, He H. LncRNA TPT1-AS1 sponges miR-23a-5p in glioblastoma to promote cancer cell proliferation. Cancer Biother Radiopharm. 2021;36(7):549–55. [Google Scholar] [PubMed]
60. Sun J, Xu L, Zhang Y, Li H, Feng J, Lu X, et al. Long non-coding RNA DPP10-AS1 represses the proliferation and invasiveness of glioblastoma by regulating miR-24-3p/CHD5 signaling pathway. BIOCELL. 2023;47(12):2721–33. [Google Scholar]
61. Li Y, Li Y, Ge P, Ma C. MiR-126 regulates the ERK pathway via targeting KRAS to inhibit the glioma cell proliferation and invasion. Mol Neurobiol. 2017;54(1):137–45. [Google Scholar] [PubMed]
62. Liu L, Cui S, Wan T, Li X, Tian W, Zhang R, et al. Long non-coding RNA HOTAIR acts as a competing endogenous RNA to promote glioma progression by sponging miR-126-5p. J Cell Physiol. 2018;233(9):6822–31. doi:10.1002/jcp.26432. [Google Scholar] [PubMed] [CrossRef]
63. Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6(9):e24248. [Google Scholar] [PubMed]
64. Memari E, Maghsoudi A, Yazdian F, Yousefi M, Mohammadi M. Synthesis of PHB-co-PEI nanoparticles as gene carriers for miR-128-encoding plasmid delivery to U87 glioblastoma cells. Colloids Surf A Physicochem Eng Asp. 2020;599:124898. [Google Scholar]
65. Taheri M, Badrlou E, Hussen BM, Kashi AH, Ghafouri-Fard S, Baniahmad A. Importance of long non-coding RNAs in the pathogenesis, diagnosis, and treatment of prostate cancer. Front Oncol. 2023;13:1123101. [Google Scholar] [PubMed]
66. Choudhry H, Schodel J, Oikonomopoulos S, Camps C, Grampp S, Harris AL, et al. Extensive regulation of the non-coding transcriptome by hypoxia: role of HIF in releasing paused RNApol2. EMBO Rep. 2014;15(1):70–6. [Google Scholar] [PubMed]
67. Faggiano A, Fazzalari B, Mikovic N, Russo F, Zamponi V, Mazzilli R, et al. Clinical factors predicting multiple endocrine neoplasia type 1 and type 4 in patients with neuroendocrine tumors. Genes. 2023;14(9):1782. [Google Scholar] [PubMed]
68. Kim YS, Do JH, Bae S, Bae DH, Ahn WS. Identification of differentially expressed genes using an annealing control primer system in stage III serous ovarian carcinoma. BMC Cancer. 2010;10:576. [Google Scholar] [PubMed]
69. Farzaneh M, Abouali Gale Dari M, Anbiyaiee A, Najafi S, Dayer D, Mousavi Salehi A, et al. Emerging roles of the long non-coding RNA NEAT1 in gynecologic cancers. J Cell Commun Signal. 2023;17(3):531–47. [Google Scholar] [PubMed]
70. Zhen L, Liu YH, Diao HY, Ma J, Yao YL. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumour Biol. 2016;37(1):673–83. [Google Scholar] [PubMed]
71. Mercer TM, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–9. [Google Scholar] [PubMed]
72. Li W, Wang YY, Xiao L, Ding J, Wang L, Wang F, et al. Mysterious long noncoding RNAs and their relationships to human disease. Front Mol Biosci. 2022;9:950408. [Google Scholar] [PubMed]
73. Li Y, Shan G, Teng ZQ, Wingo TS. Editorial: non-coding RNAs and human diseases. Front Genet. 2020;11:523. [Google Scholar] [PubMed]
74. Ebrahimpour A, Sarfi M, Rezatabar S, Tehrani SS. Novel insights into the interaction between long non-coding RNAs and microRNAs in glioma. Mol Cell Biochem. 2021;476(6):2317–35. [Google Scholar] [PubMed]
75. He S, Liu S, Zhu H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol Biol. 2011;11:102. [Google Scholar] [PubMed]
76. Yuan C, Ning Y, Pan Y. Emerging roles of HOTAIR in human cancer. J Cell Biochem. 2020;121(5–6):3235–47. [Google Scholar] [PubMed]
77. Angelopoulou E, Paudel YN, Piperi C. Critical role of HOX transcript antisense intergenic RNA (HOTAIR) in gliomas. J Mol Med. 2020;98(11):1525–46. [Google Scholar] [PubMed]
78. Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24(7):1199–215. [Google Scholar] [PubMed]
79. Murthy UM, Rangarajan PN. Identification of protein interaction regions of VINC/NEAT1/Men epsilon RNA. FEBS Lett. 2010;584(8):1531–5. [Google Scholar] [PubMed]
80. Shen W, Liang XH, Crooke ST. Phosphorothioate oligonucleotides can displace NEAT1 RNA and form nuclear paraspeckle-like structures. Nucleic Acids Res. 2014;42(13):8648–62. [Google Scholar] [PubMed]
81. Yu X, Li Z, Zheng H, Chan MT, Wu WK. NEAT1: a novel cancer-related long non-coding RNA. Cell Prolif. 2017;50(2):12329. [Google Scholar]
82. Cao J, Han X, Qi X, Jin X, Li X. TUG1 promotes osteosarcoma tumorigenesis by upregulating EZH2 expression via miR-144-3p. Int J Oncol. 2017;51(4):1115–23. [Google Scholar] [PubMed]
83. Yin T, Wu J, Hu Y, Zhang M, He J. Long non-coding RNA HULC stimulates the epithelial-mesenchymal transition process and vasculogenic mimicry in human glioblastoma. Cancer Med. 2021;10(15):5270–82. [Google Scholar] [PubMed]
84. Geng S, Tu S, Fu W, Wang J, Bai Z. LncRNA PITPNA-AS1 stimulates cell proliferation and suppresses cell apoptosis in glioblastoma via targeting miR-223-3p/EGFR axis and activating PI3K/AKT signaling pathway. Cell Cycle. 2021;20(19):1988–98. [Google Scholar] [PubMed]
85. di Martino MT, Riillo C, Scionti F, Grillone K, Polera N, Caracciolo D, et al. miRNAs and lncRNAs as novel therapeutic targets to improve cancer immunotherapy. Cancers. 2021;13(7):1587. [Google Scholar] [PubMed]
86. Sang LJ, Ju HQ, Liu GP, Tian T, Ma GL, Lu YX, et al. LncRNA CamK–A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol Cell. 2018;72(1):71–83. [Google Scholar] [PubMed]
87. Martinez-Barriocanal A, Arango D, Dopeso H. PVT1 long non-coding RNA in gastrointestinal cancer. Front Oncol. 2020;10:38. [Google Scholar] [PubMed]
88. Karamanos NK, Theocharis AD, Piperigkou Z, Manou D, Passi A, Skandalis SS, et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021;288(24):6850–912. [Google Scholar] [PubMed]
89. Niland S, Riscanevo AX, Eble JA. Matrix metalloproteinases shape the tumor microenvironment in cancer progression. Int J Mol Sci. 2021;23(1):146. [Google Scholar] [PubMed]
90. Chen D, Chen G, Jiang W, Fu M, Liu W, Sui J, et al. Association of the collagen signature in the tumor microenvironment with lymph node metastasis in early gastric cancer. JAMA Surg. 2019;154(3):e185249. [Google Scholar] [PubMed]
91. Ou A, Yung WKA, Majd N. Molecular mechanisms of treatment resistance in glioblastoma. Int J Mol Sci. 2020;22(1):351. [Google Scholar] [PubMed]
92. Li Z, Meng X, Wu P, Zha C, Han B, Li L, et al. Glioblastoma cell-derived lncRNA-containing exosomes induce microglia to produce complement C5, promoting chemotherapy resistance. Cancer Immunol Res. 2021;9(12):1383–99. [Google Scholar] [PubMed]
93. Mattei V, Santilli F, Martellucci S, Delle Monache S, Fabrizi J, Colapietro A, et al. The importance of tumor stem cells in glioblastoma resistance to therapy. Int J Mol Sci. 2021;22(8):3863. [Google Scholar] [PubMed]
94. Zhang Q, Ren H, Ge L, Zhang W, Song F, Huang P. A review on the role of long non-coding RNA and microRNA network in clear cell renal cell carcinoma and its tumor microenvironment. Cancer Cell Int. 2023;23(1):16. [Google Scholar] [PubMed]
95. Du L, Lee JH, Jiang H, Wang C, Wang S, Zheng Z, et al. Beta-Catenin induces transcriptional expression of PD-L1 to promote glioblastoma immune evasion. J Exp Med. 2020;217(11):20191115. [Google Scholar]
96. Wang L, Yang G, Liu G, Pan Y. Identification of lncRNA signature of tumor-infiltrating T lymphocytes with potential implications for prognosis and chemotherapy of head and neck squamous cell carcinoma. Front Pharmacol. 2021;12:795205. [Google Scholar] [PubMed]
97. Gao Y, Wang X, Dong L, Qu C, Lu Q, Wang P, et al. Identifying immune checkpoint-related lncRNA biomarkers for immunotherapy response and prognosis in cancers. Sci Data. 2023;10(1):663. [Google Scholar] [PubMed]
98. Xin L, Zhou LQ, Liu C, Zeng F, Yuan YW, Zhou Q, et al. Transfer of lncRNA CRNDE in TAM-derived exosomes is linked with cisplatin resistance in gastric cancer. EMBO Rep. 2021;22(12):e52124. [Google Scholar] [PubMed]
99. Scheurlen KM, Snook DL, Gardner SA, Eichenberger MR, Galandiuk S. Macrophage differentiation and polarization into an M2-like phenotype using a human monocyte-like THP-1 leukemia cell line. J Vis Exp. 2021;174:62652. [Google Scholar]
100. Lai XN, Li J, Tang LB, Chen WT, Zhang L, Xiong LX. MiRNAs and lncRNAs: dual roles in TGF-β signaling-regulated metastasis in lung cancer. Int J Mol Sci. 2020;21(4):1193. [Google Scholar] [PubMed]
101. Yao F, Zhan Y, Pu Z, Lu Y, Chen J, Deng J, et al. LncRNAs target ferroptosis-related genes and impair activation of CD4+ T cell in gastric cancer. Front Cell Dev Biol. 2021;9:797339. [Google Scholar] [PubMed]
102. Li G, Kryczek I, Nam J, Li X, Li S, Li J, et al. LIMIT is an immunogenic lncRNA in cancer immunity and immunotherapy. Nat Cell Biol. 2021;23(5):526–37. [Google Scholar] [PubMed]
103. Daubon T, Hemadou A, Romero Garmendia I, Saleh M. Glioblastoma immune landscape and the potential of new immunotherapies. Front Immunol. 2020;11:585616. [Google Scholar] [PubMed]
104. Muir M, Gopakumar S, Traylor J, Lee S, Rao G. Glioblastoma multiforme: novel therapeutic targets. Expert Opin Ther Targets. 2020;24(7):605–14. [Google Scholar] [PubMed]
105. Szopa W, Burley TA, Kramer-Marek G, Kaspera W. Diagnostic and therapeutic biomarkers in glioblastoma: current status and future perspectives. Biomed Res Int. 2017;2017:8013575. [Google Scholar] [PubMed]
106. Latowska J, Grabowska A, Zarebska Z, Kuczynski K, Kuczynska B, Rolle K. Non-coding RNAs in brain tumors, the contribution of lncRNAs, circRNAs, and snoRNAs to cancer development-their diagnostic and therapeutic potential. Int J Mol Sci. 2020;21(19):7001. [Google Scholar] [PubMed]
107. Tian Y, Zheng Y, Dong X. AGAP2-AS1 serves as an oncogenic lncRNA and prognostic biomarker in glioblastoma multiforme. J Cell Biochem. 2019;120(6):9056–62. [Google Scholar] [PubMed]
108. Cao Y, Chai W, Wang Y, Tang D, Shao D, Song H, et al. lncRNA TUG1 inhibits the cancer stem cell‐like properties of temozolomide‐resistant glioma cells by interacting with EZH2. Mol Med Rep. 2021;24(1):533. [Google Scholar] [PubMed]
109. Pastori C, Kapranov P, Penas C, Peschansky V, Volmar CH, Sarkaria JN, et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc Natl Acad Sci U S A. 2015;112(27):8326–31. [Google Scholar] [PubMed]
110. Liu SJ, Malatesta M, Lien BV, Saha P, Thombare SS, Hong SJ, et al. CRISPRi-based radiation modifier screen identifies long non-coding RNA therapeutic targets in glioma. Genome Biol. 2020;21(1):83. [Google Scholar] [PubMed]
111. Yadav G, Kulshreshtha R. Metastasis associated long noncoding RNAs in glioblastoma: biomarkers and therapeutic targets. J Cell Physiol. 2022;237(1):401–20. [Google Scholar] [PubMed]
112. Kathleen Tsung JH, Liu K, Loh E, Attenello F. CRISPR functional screen identifies a novel long noncoding RNA modulating glioblastoma invasion. Neuro Oncol. 2022;24(7):vii29. [Google Scholar]
113. Arriaga-Canon C, Contreras-Espinosa L, Aguilar-Villanueva S, Bargallo-Rocha E, Garcia-Gordillo JA, Cabrera-Galeana P, et al. The clinical utility of lncRNAs and their application as molecular biomarkers in breast cancer. Int J Mol Sci. 2023;24(8):7426. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF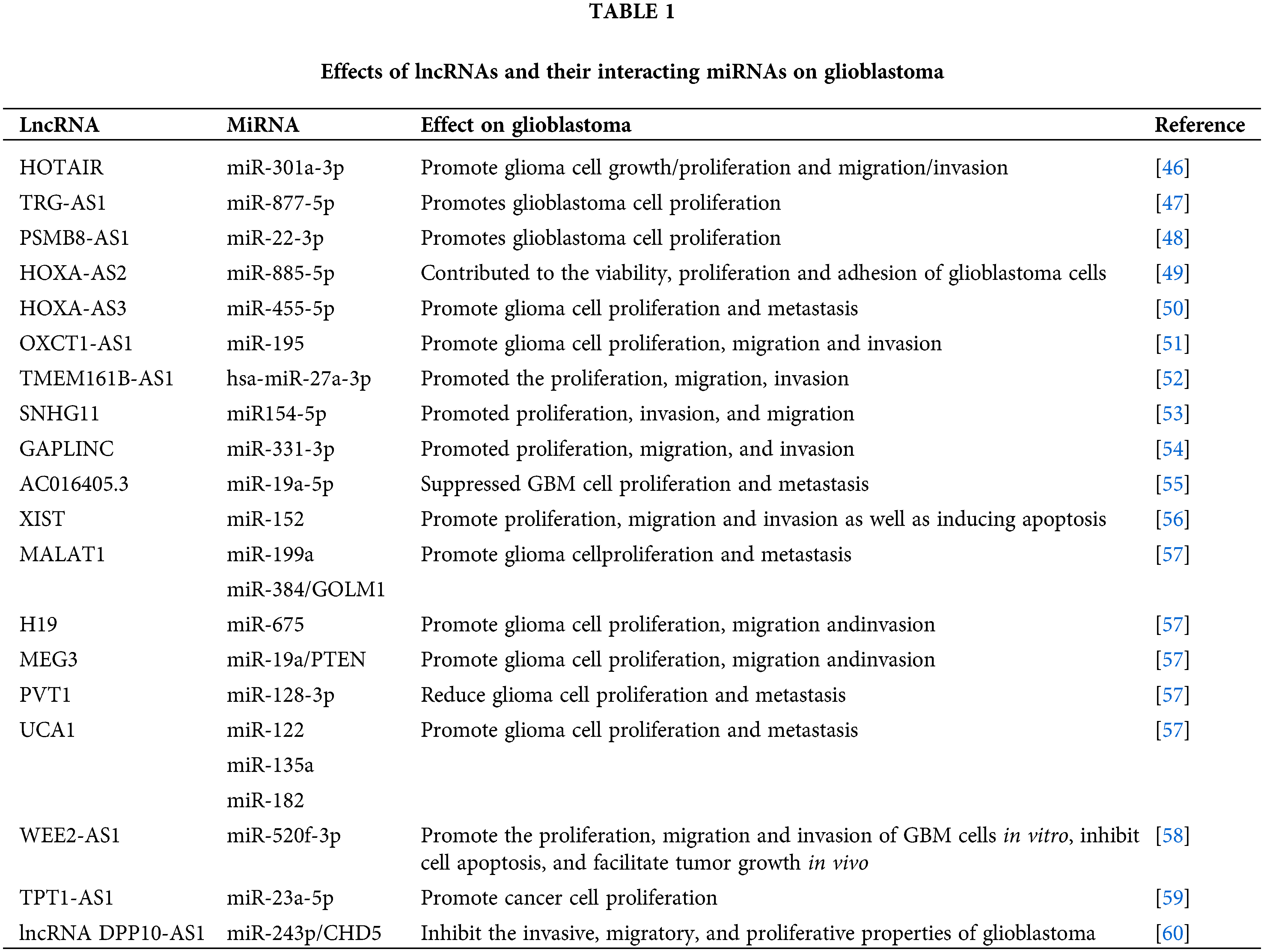
 Downloads
Downloads
 Citation Tools
Citation Tools