 Open Access
Open Access
ARTICLE
Bioinformatics analysis and experimental validation of cystathionine-gamma-lyase as a potential prognosis biomarker in hepatocellular carcinoma
1 Department of Gastroenterology and Hepatology, Beijing You’an Hospital Affiliated with Capital Medical University, Beijing, 100069, China
2 Beijing Institute of Hepatology, Beijing You’an Hospital Affiliated with Capital Medical University, Beijing, 100069, China
* Corresponding Authors: SHANSHAN WANG. Email: ; HUIGUO DING. Email:
BIOCELL 2024, 48(3), 463-471. https://doi.org/10.32604/biocell.2024.048244
Received 01 December 2023; Accepted 04 January 2024; Issue published 15 March 2024
Abstract
Background: Hepatocellular carcinoma (HCC) is a common malignant tumor with poor prognosis and high mortality worldwide. Although cystathionine-gamma-lyase (CSE) plays an important role in the development of multiple tumors, the clinical implication and potential mechanisms of CSE in HCC development remain elusive. Methods: In our study, the CSE expression in HCC was analyzed in Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) datasets and further confirmed by RT-qPCR and immunohistochemistry assays in HCC samples. Furthermore, the associations between CSE expression and HCC malignancy as well as survival were analyzed in GSE14520 and validated in HCC patients. Finally, the biological functions of CSE in HCC cells was assessed by CCK-8, flow cytometry and Western blotting. Results: Lower transcriptional and proteomic CSE expressions were found in HCC tissues in contrast to adjacent normal tissues. Decreased CSE mRNA expression was significantly associated with advanced clinicopathological features and poor outcomes in HCC patients from public database and our cohort. Following univariate and multivariate analyses of GSE14520 data showed that CSE expression was an independent prognostic indicator for the overall survival (OS) and recurrence-free survival (RFS) of HCC patients. In vitro experiments further explained that CSE might trigger HCC cell apoptosis by H2S. Conclusion: In summary, the present study identified the relationship between CSE expression and HCC malignancy as well as OS and RFS, indicating that CSE might be a potential prognostic biomarker and a novel therapeutic target for HCC.Keywords
Abbreviations
| AFP | Alpha-fetoprotein |
| BCLC | Barcelona Clinic Liver Cancer |
| CSE | Cystathionine-gamma-lyase |
| EGFR | Epidermal growth factor receptor |
| GEO | Gene Expression Omnibus |
| HBsAg | Hepatitis B surface antigen |
| HCC | Hepatocellular carcinoma |
| MAPK | Mitogen-activated protein kinase |
| OS | Overall survival |
| RFS | Recurrence-free survival |
| TCGA | The Cancer Genome Atlas |
Hepatocellular carcinoma (HCC) is among the most prevalent malignancies and is frequently detected late in clinical practice [1,2]. According to the World Health Organization, HCC is the sixth most common cancer and the third leading cause of cancer-related death worldwide, with a global incidence and mortality rate of around 906,000 and 830,000, respectively [3]. China’s HCC incidence and mortality rates are also growing, with approximately 410,000 new cases and 391,000 deaths in 2020 [3]. The effective treatment for HCC includes surgery, liver transplantation, radiotherapy, and immunological and targeted therapy. A revolution in the treatment of advanced HCC has been made possible by various targeted drugs that extend patients’ survival. However, the overall life expectancy of HCC patients with sorafenib or regorafenib therapy remains poor [4–6]. Identifying potential biomarkers involved in the molecular pathogenesis of HCC is crucial for early diagnosis, prevention, and treatment.
Cystathionine-gamma-lyase (CSE, also known as CTH), encoded by the gene cth, is an important enzyme involved in the transsulfuration pathway, cysteine catabolism and H2S generation. CSE deficiency can lead to oxidative stress, aberrant stress responses, vascular deficits and hyperhomocysteinemia, which is vital in the pathogenesis of neurodegenerative, cardiovascular, pulmonary and cancerous diseases [7–10]. Differential expression and dysfunction of CSE have been reported in various cancers [11–14]. Knowledge about CSE was highly expressed in some tumors, such as ovarian cancer [11], thyroid cancer [12], melanoma [13] and breast adenocarcinoma [14], and CSE promoted proliferation, invasion, and migration of tumor cells and reduced tumor cell apoptosis. CSE expression was downregulated in tumorous tissues in renal cancer [15,16], prostate cancer and urothelial carcinoma [17] as demonstrated by several studies. CSE may serve as a tumor suppressor by promoting apoptosis. Recently, it has been reported that CSE overexpression was observed in HepG2 and PLC/PRF/5 cells, and CSE/H2S could contribute to a resistance of the induction of hepatoma cells apoptosis by epidermal growth factor receptor (EGFR) signal pathway [18]. However, the clinical implications of CSE expression in HCC have not been investigated.
Therefore, the main goal of this work was to analyze the CSE expression and its association with survival in HCC samples from a public database and validate it in our cohort. We further investigated the role of the CSE in HCC cell proliferation and apoptosis to elucidate possible mechanisms in HCC development.
Our study identified the relationship between CSE expression and HCC malignancy as well as OS and RFS, which provides novel evidence for a importance role of CSE in HCC progression and its potential as a prognostic biomarker.
We acquired gene expression data and clinical information of HCC from Gene Expression Omnibus (GEO) (GSE14520, GSE45114 and GSE60502; http://www.ncbi.nlm.nih.gov/geo) and The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov). For clinicopathological characteristics and survival analysis in GSE14520, patients were divided into low (CSE gene expression ≤4.837, N = 124) and high gene expression groups (CSE gene expression >4.837, N = 123) according to the median CSE mRNA expression level in tumorous tissues. Besides, the metastasis risk is determined based on the expression of Gene signature, which is composed of the expression profiles of 161 genes in HCC specimens [19].
Tumorous and adjacent non-tumorous liver tissues in this study were obtained from 45 HCC patients undergoing hepatectomy at Beijing You’an Hospital, affiliated with Capital Medical University from July 2020 to January 2022. HCC diagnosis was pathologically confirmed. The study protocol was approved by the Ethics Committee of Beijing You’an Hospital Affiliated to Capital Medical University (Approval No. LL-2020- 009-K). All enrolled patients provided written informed consent. Clinical samples obtained from patients were used to examine the expression of CSE mRNA and protein. Detailed specimen clinical information and their associations with CSE mRNA expression are summarized in Table 1.
Human normal heptical cell line HL-7702 and two HCC cell lines, HLE and Hep3B cells, were obtained from the Beijing Institute of Hepatology. Dulbecco’s modified Eagle’s medium (DMEM, 11995065, Gibco, Paisley, UK) supplemented with 10% fetal bovine serum (FBS, 10099141C, Gibco, Paisley, UK) and Penicillin-Streptomycin (15140122, Gibco, Paisley, UK) was used to culture the cell lines at 37°C in a 5% CO2 incubator. For experiments, cells plated on 6-well plates were treated with D,L-propargylglycine (PGG) (10 mM; P7888, Sigma, USA) or NaHS (10−3 M; 161527, Sigma, USA).
Quantitative real-time reverse transcription PCR (RT-qPCR)
RT-qPCR were used to evaluate CSE and GAPDH expression. Isolation of total RNA was performed using the TRIzol reagent (15596026, Thermo Fisher Scientific, Carlsbad, CA, USA) and subsequently reverse transcription was carried out with High-Capacity cDNA Reverse Transcription Kits (4368814, Thermo Fisher Scientific, Waltham, MA, USA). RT-qPCR was conducted using the TB Green (RR420A, Takara, Japan) on an ABIV7 machine (GX-XVI R2, Applied Biosystems Life Tech, USA). The 2−ΔΔCt method was used to quantitatively analyze relative mRNA expression levels. Primers of homo sapien used for RT-qPCR were as follows: GAPDH forward were 5′-TGAAGGTCGGAGTCAACGGA-3′ and reverse were 5′-CCTGGAAGATGGTGATGGGAT-3′; CSE forward were 5′-AAGACGCCTCCTCACAAGGT-3′ and reverse were 5′-ATATTCAAAACCCGAGTGCTGG-3′.
Briefly, cells were lysed in ice-cold RIPA lysis buffer (89901, Thermo Fisher Scientific, Waltham, MA, USA) with protease inhibitors, then 40 μg of cell protein were separated using 12% SDS-PAGE and then transferred onto a PVDF membrane. Then membranes were incubated with primary antibodies against GAPDH (#2118, CST, Beverly, MA, USA) or CSE (12217-1-AP, Proteintech, Chicago, USA) overnight at 4°C (dilution ratio 1:1000). Membranes were subsequently incubated with secondary antibodies (#7074, CST, Beverly, MA, USA) for 1 h at room temperature after blocking. By incubating with an enhanced chemiluminescence system (T4580, Thermo Scientific, Waltham, MA, USA), a target protein signal was visualized.
IHC analyses were performed using an HRP kit (DS-0003, Zhongshan Golden Bridge Biotechnology, Beijing, China). Sections of tumor tissue embedded in paraffin were incubated with CSE antibody (1:200; 12217-1-AP, proteintech, Chicago, USA). The mean optical density values of IHC were analyzed using Image-Pro Plus 6.0 software.
The H2S release amount was measured using a Hydrogen sulfide Assay Kit (JEB-11780, Jin Yibai Biological Technology, Nanjing, China) following the manufacturer’s guidelines.
HCC cells proliferation was analyzed using Cell count KIT-8 assay (CCK8; M4839, AbMole, USA) according to the instruction. The percentage of cell viability was calculated as [(A450 sample-background)/(A450 control-backgroud)] × 100%.
An Annexin V-FITC/PI Apoptosis Detection Kit (40302ES20, Yeasen, Shanghai, China) was used to evaluate cell apoptosis. Briefly, cells were collected and incubated with 5 µL Annexin V-FITC and 10 µL PI for 15 min after treatment, detected within one hour by flow cytometry.
Statistical analyses were performed using the SPSS software (version 23.0). Statistical significance was determined using the Student’s t-test, ANOVA-test, χ2 test or Fisher’s exact test. Survival analyses were performed using the Kaplan–Meier method and the log rank test. Cox proportional hazard model was used to find independent predictors for HCC patients. Statistical difference was considered significant at p ≤ 0.05.
Decreased expression of the CSE gene in HCC tissues in multiple cohorts
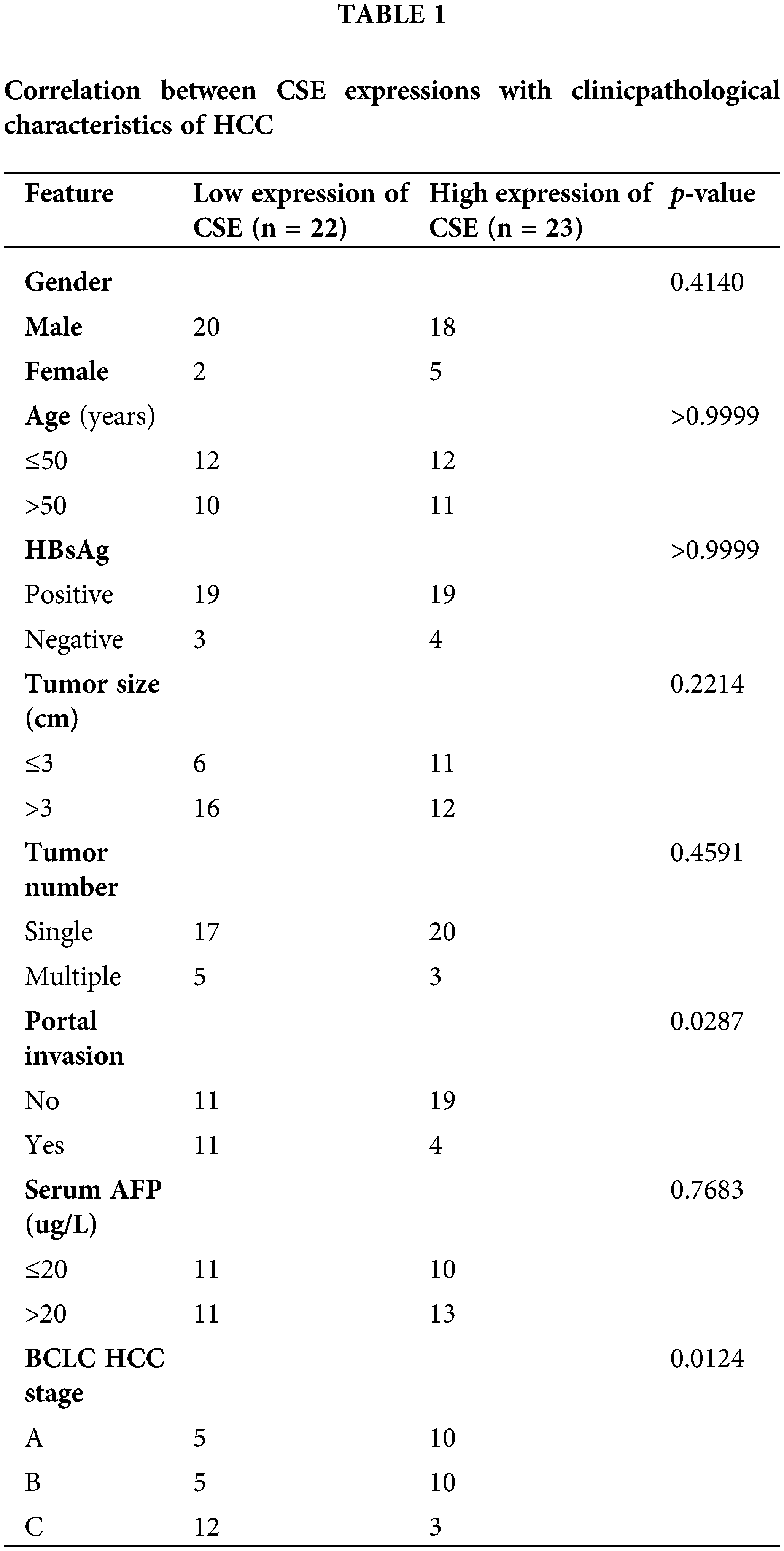
We initially evaluated CSE mRNA expression levels based on RNA-sequence data in multiple HCC studies from GEO and TCGA databases. Analysis of GSE14520, GSE45114, GSE60502 and TCGA databases revealed that CSE mRNA expression was significantly reduced in tumorous tissues relative to adjacent non-tumorous tissues (p all < 0.0001), as shown in Fig. 1. These results suggested that CSE transcriptional levels decreased in HCC tissues compared to normal tissues.
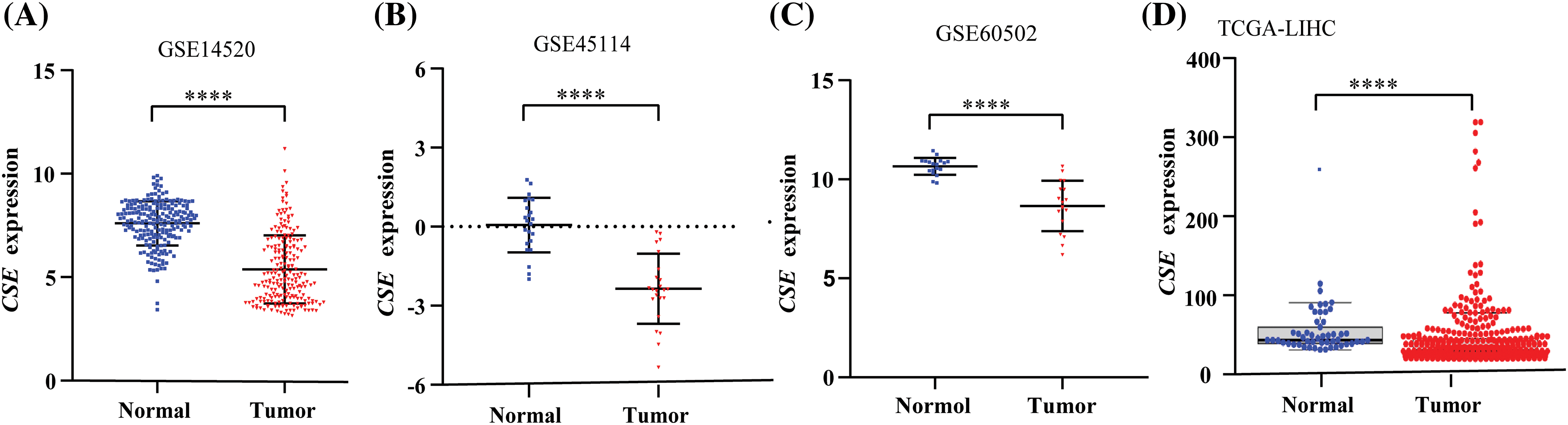
Figure 1: CSE expression in HCC tumor tissues and adjacent normal tissues in multiple cohorts. (A–C) CSE mRNA expression was lowly expressed in cancerous tissues compared with adjacent non-cancerous tissues in GSE14520 (N = 214; ****p < 0.0001), GSE45114 (N = 23; **** p < 0.0001) and GSE60502 (N = 18; ****p < 0.0001). (D) Transcriptional level of CSE expression was observed significantly decreased in HCC tissues relative to adjacent normal tissues in TCGA cohort (****p < 0.0001). Statistical significance with a p-value less than 0.05 was determined by the Student’s t-test.
CSE expression was associated with malignant clinicopathological features and poor clinical outcomes in the GEO cohort
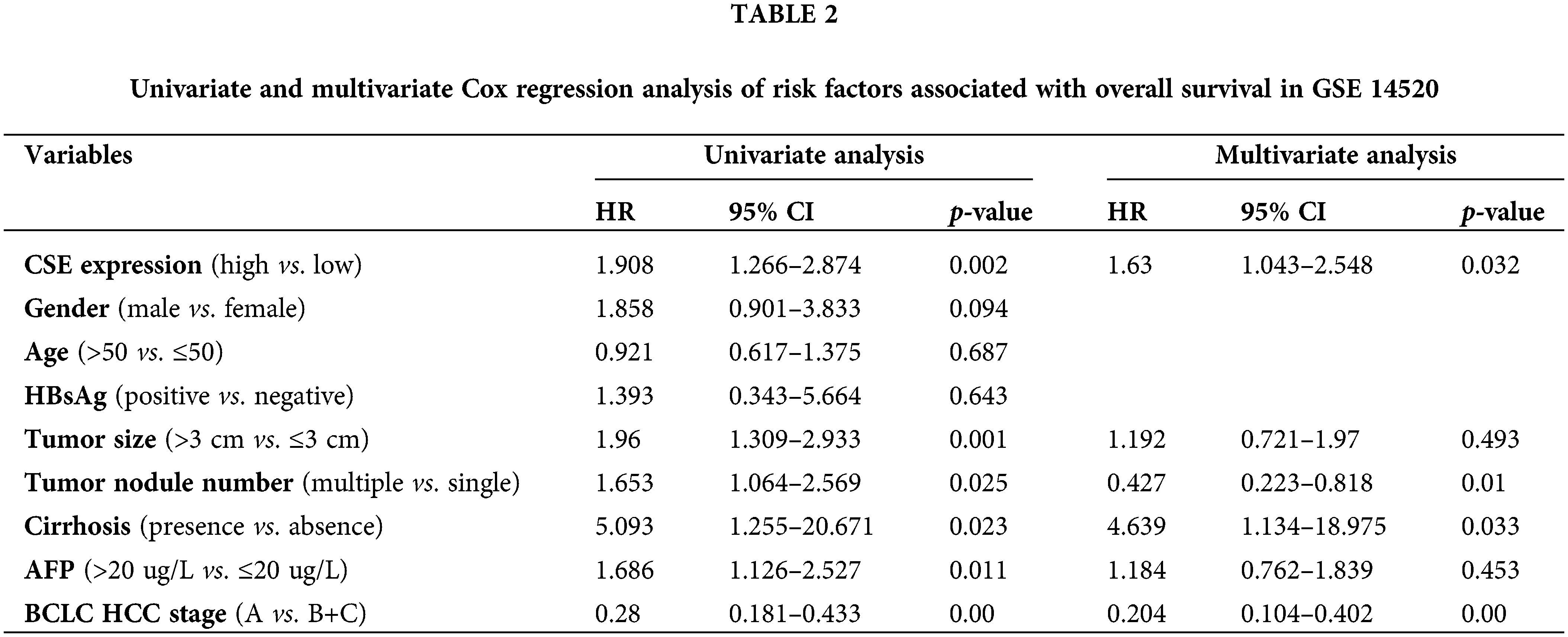
We next examined the association of CSE expression with clinicopathological indexes in GSE14520. As shown in Fig. 2, the clinicopathological association analysis in the 247 HCCs showed that the low CSE expression was closely correlated with advanced clinicopathological features, including high Barcelona Clinic Liver Cancer (BCLC) stage (Fig. 2A; p < 0.01), high alpha-fetoprotein (AFP) levels (Fig. 2B; p < 0.0001), greater tumor multinodular (Fig. 2C; p < 0.05) and even higher metastasis risk (Fig. 2D; p < 0.0001). Furthermore, we assessed the correlations between CSE expression and the survival outcomes in GSE14520. The Kaplan-Meier analysis found that HCC patients with low CSE expression showed reductions in overall survival (OS; Log-rank, 9.77; p = 0.002; Fig. 2E) and recurrence-free survival (RFS; Log-rank, 4.042; p = 0.044; Fig. 2F). Furthermore, the Cox proportional hazard model evaluated prognostic factors for HCC patient survival in GSE14520. As indicated in Tables 2 and 3, Univariate Cox regression analysis found that tumor size and BCLC HCC stage were significant both in OS and RFS. Low CSE expression correlated with poor OS (HR = 1.908; p = 0.002; Table 2) and poor RFS (HR = 1.411; p = 0.046; Table 3). In multivariate Cox regression analysis, traditional prognostic factors such as tumor nodule number, cirrhosis and BCLC HCC stage were relevant to OS (Table 2). Moreover, gender and BCLC HCC stage were closely associated with RFS (Table 3). Low CSE expression was significantly related to poor OS in HCC patients (HR = 1.63; p = 0.032; Table 2). These findings indicated that low CSE expression could be an independent prognostic predictor for HCC patients.
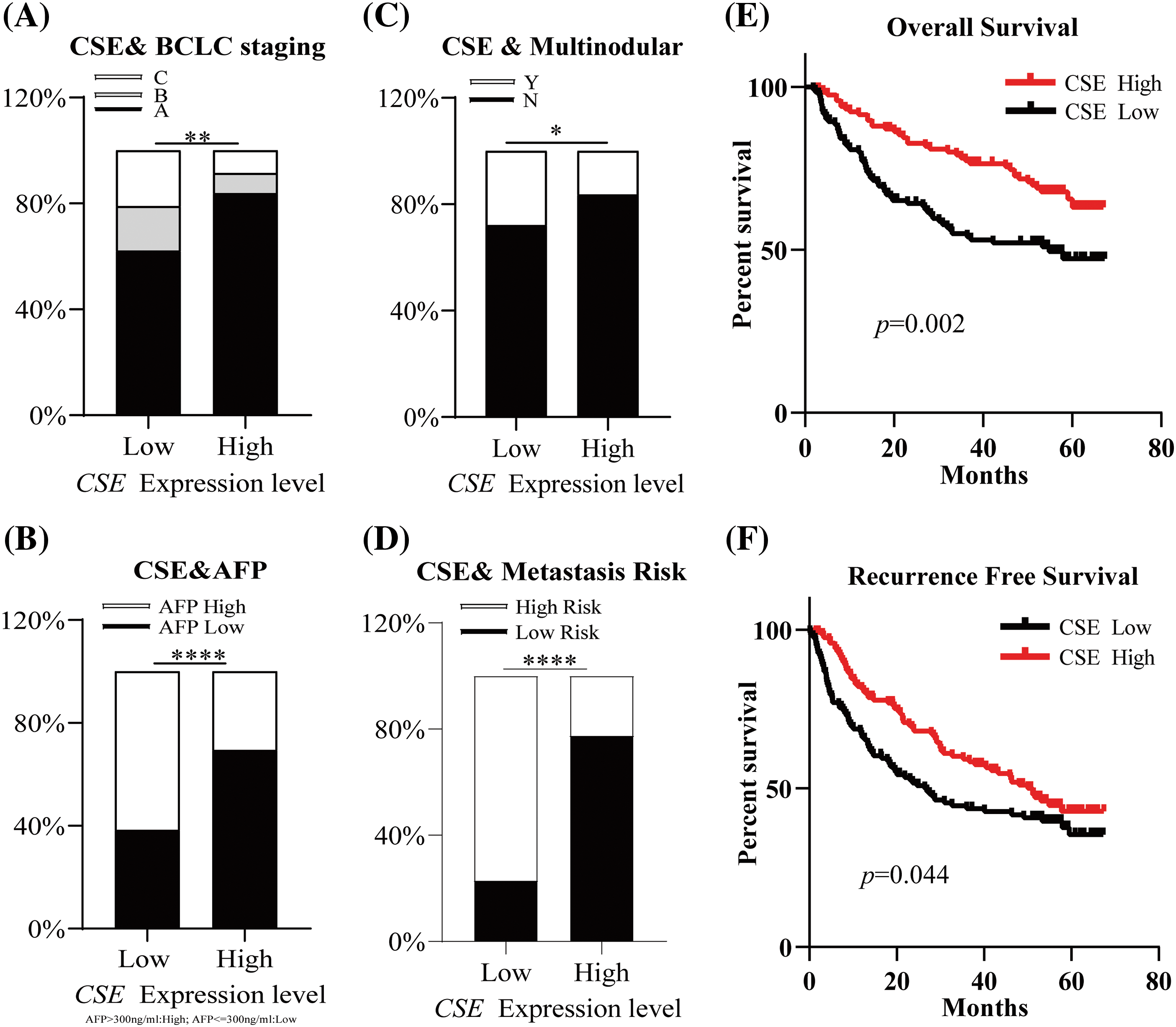
Figure 2: CSE mRNA expressions significantly was correlated with advanced tumor features and poor clinical outcomes in HCC patients from GSE14520. (A–D) Transcriptional expression of CSE was significantly correlated with Barcelona Clinic Liver Cancer (BCLC) stage (p < 0.01), alpha-fetoprotein (AFP) levels (p < 0.0001), tumor multinodular (p < 0.05) and metastasis risk (p < 0.0001). Statistical significance with a p-value less than 0.05 was determined by χ2 test or Fisher’s exact test. (E–F) Survival analyses by Kaplan–Meier method and the log rank test indicated that patients with low CSE mRNA expression showed poor overall survival (OS; p = 0.002) and recurrence-free survival (RFS; p = 0.044) in 247 HCC patients (All *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
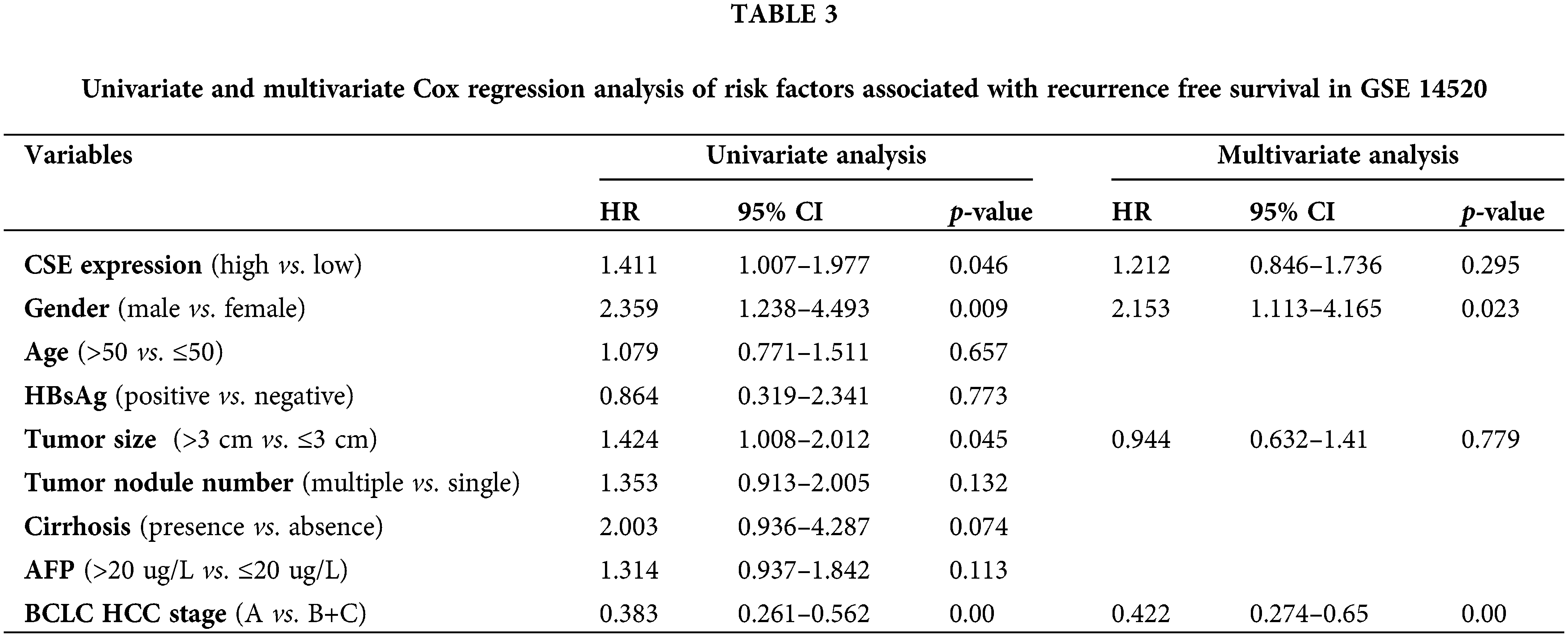
CSE suppressed proliferation and promoted apoptosis in HCC cells
To explore the CSE biological function in HCC progression, we compared its expression in HCC cell lines to that of an immortalized human normal hepatic cell line. Western blotting results showed that CSE protein expression was decreased in HCC cell lines, including HLE and Hep3B cells, compared to human hepatic immortalized cell line HL-7702 (p < 0.05; Fig. 3A).
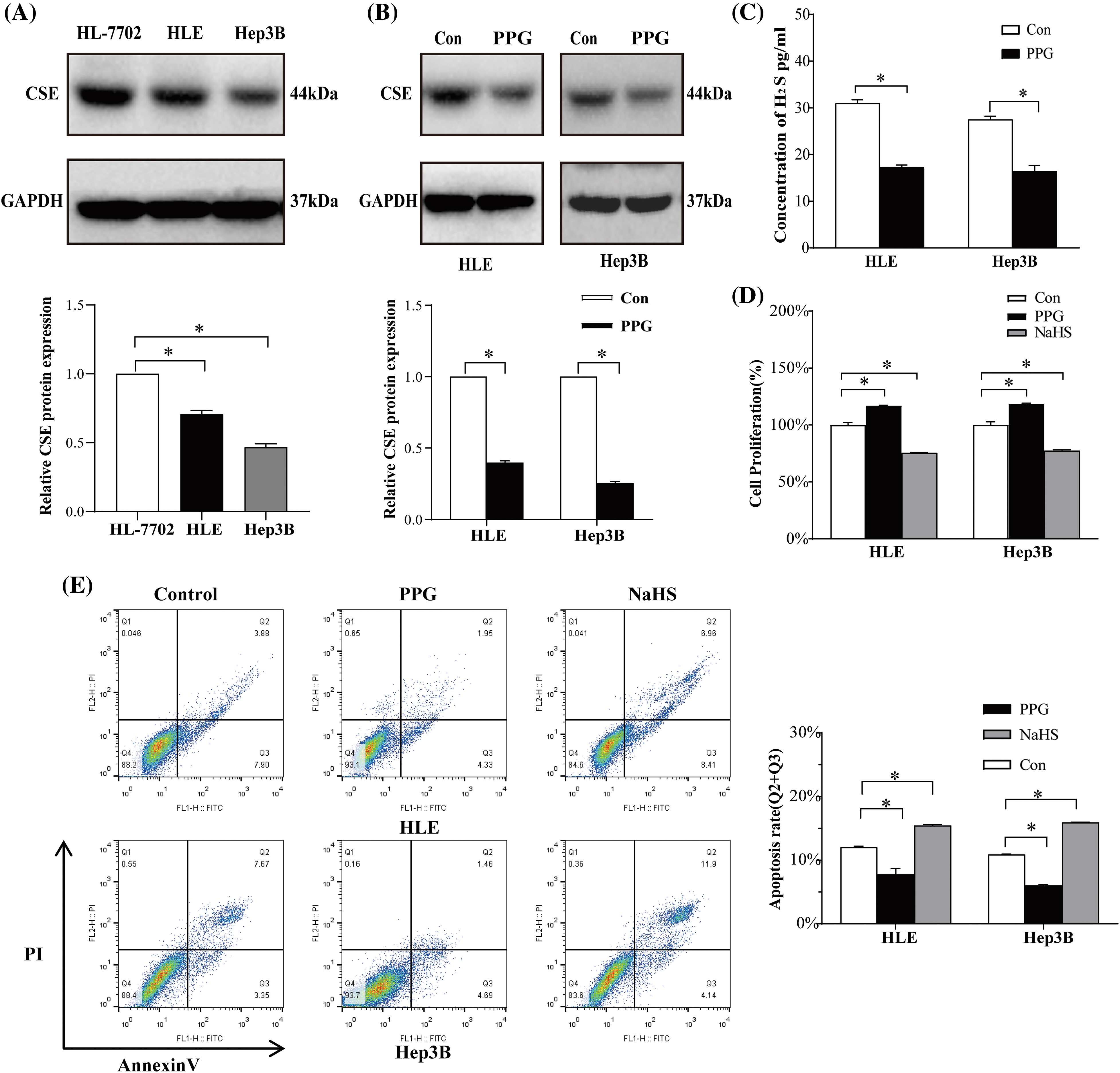
Figure 3: CSE promote proliferation and suppress HCC cells apoptosis. (A) Western blotting analysis of CSE and its column plot for HLE and Hep3B cells using ANOVA-test. (B) CSE analysis by Western blotting and its column plot for the HLE and Hep3B cells treated with PPG using Students’ t-test. (C) ELISA analysis of H2S in HLE and Hep3B cells treated with PPG using Students’ t-test. (D) CCK-8 analysis of proliferation of HLE and Hep3B cells treated with PPG or NaHS using ANOVA-test. (E) Flow cytometric analysis of apoptosis of HLE and Hep3B cells treated with PPG or NaHS using ANOVA-test. The results from three independent experiments were expressed as mean ± SD (n = 3), *p < 0.05.
To further investigate the CSE effect in HCC cell proliferation and apoptosis, we next blocked CSE expression with PPG, a specific CSE blocker. We also found that PPG notably reduced CSE expression and H2S in two HCC cell lines by Western blotting (p < 0.05; Fig. 3B) and ELISA (p < 0.05; Fig. 3C). CCK-8 results displayed that the cell proliferation vitality was induced When CSE was blocked but was greatly reduced in HLE and Hep3B cells treated by NaHS, as an exogenous H2S donor (p < 0.05; Fig. 3D). Flow cytometric study revealed that PPG lowered apoptotic rates in HLE and Hep3B cells relative to the control group, but NaHS significantly enhanced apoptosis (p < 0.05; Fig. 3E). On the other side, the effect of CSE was also confirmed through transfection of the cse gene into HCC cells, which indicated that CSE overexpression prevented proliferation and promoted apoptosis in HLE and Hep3B cells by H2S production (Suppl. Fig. S1). These data suggest that CSE may play an important role in HCC cells proliferation and apoptosis.
Validation of CSE expression in association with poor outcome in our cohort
To confirm the CSE transcriptional level differences in HCC, RT-qPCR was performed on 45 paired tumor samples from our cohort. As shown in Fig. 4A, CSE mRNA expression was identified to be downregulated in tumorous tissues relative to non-tumorous tissues (p < 0.05). To evaluate CSE protein expression in HCC tumor samples, we performed IHC staining and found that CSE protein expression was much lower in tumor tissues than in adjacent normal tissues (p < 0.05; Fig. 4B). We further verified the association between CSE mRNA expression, tumor characteristics, and survival outcomes in 45 HCC patients. According to the median value of CSE mRNA expression in tumorous tissues, we divided 45 patients into a group with high CSE expression (CSE mRNA expression ≥0.99, N = 23) and a group with low CSE expression (CSE mRNA expression <0.99, N = 22). As shown in Table 1, the low CSE expression group exhibited greater portal invasion and higher BCLC stage than the high CSE expression group (p all < 0.05). Additionally, we noted that the CSE low-expression group exhibited poor OS (Log-rank, 3.882; p = 0.048; Fig. 4C) and RFS (Log-rank, 3.226; p = 0.073; Fig. 4D). According to these findings, low CSE expression significantly predicted advanced clinicopathological features and poor clinical outcomes in HCC patients.
Figure 4: CSE expression and it is association with survival outcomes in our cohort. (A) CSE mRNA expression difference in 45 paired tumor and normal tissues by RT-qPCR assay using Students’ t-test. (B) Immunohistochemistry staining of CSE in carcinoma tissue and carcinoma adjacent tissue using Students’ t-test (Scale bars: 40 μm). (C–D) Relationship between CSE mRNA expression level and OS (p = 0.048) and RFS (p = 0.073) in 45 HCC patients using Kaplan–Meier method and the log rank test. *p < 0.05.
Increasing evidence suggests that CSE functions in liver pathology and liver disorders, influencing multiple biological processes, including cysteine catabolism, hepatic lipid metabolism, oxidative stress, mitochondrial bioenergetics, nonalcoholic fatty liver disease, cirrhosis, and liver cancer [20–23]. CSE is highly expressed in the healthy liver [24,25] and, indeed, is diminished in a mouse model for hepatic ischemia-reperfusion injury, nonalcoholic fatty liver disease, hepatitis, and fibrosis [26–29]. Defects in CSE have been shown to promote pro-inflammatory cytokines in the liver and exacerbate acute hepatitis and liver fibrosis by reducing H2S release from cysteine in the liver [28]. Another study revealed that CSE expression was decreased in both an in vitro cell model and an in vivo animal model of nonalcoholic fatty liver disease, and inhibition of CSE activity accelerated intracellular lipid accumulation. It worsened lipogenesis, inflammation, and fibrosis [29]. However, the clinical implications of CSE expression in HCC have not been reported. The current study revealed that CSE was weakly expressed in tumorous tissues compared to adjacent non-tumorous tissues both in databases and HCC patients and was also decreased in HCC cell lines, HLE and Hep3B cells. Our studies from both in vivo and in vitro indicated that CSE was significantly reduced in HCC and could be used as a potential biomarker for HCC.
The prognostic value of CSE expression in thyroid cancer, pancreatic adenocarcinoma, and prostate cancer has been studied [12,30,31]. However, its prognostic value in other cancers, including HCC, remains unknown. It has been reported that CSE overexpression was correlated with the TNM stage in prostate cancer, and patients with high CSE expression had a shorter OS than those with low expression [30]. In our study, CSE expression in GSE14520 was notably associated with the tumor’s malignancy, such as BCLC stage, AFP levels, tumor multinodular and metastasis risk. Furthermore, reduced CSE expression was significantly correlated with shorter OS and RSF time by univariate and multivariate analysis of GSE14520. Our cohort confirmed that decreased CSE expression was significantly associated with advanced clinicopathological characteristics and poor clinical survival, suggesting that low CSE levels may be an independent biomarker for clinical survival in HCC patients.
As the essential enzyme for H2S generation, CSE exerted both pro-apoptotic and anti-apoptotic effects on tumor growth, according to an increasing number of studies [15,18,32]. For example, CSE was shown to be overexpressed in hepatoma HepG2 and PLC/PRF/5 cells. Reduced expression of CSE/H2S could suppress the excessive growth of HCC cells by stimulating mitochondrial apoptosis and suppressing the EGFR signal pathway [18]. It has also been reported that suppression of CSE/H2S in Nasopharyngeal carcinoma (NPC) cells restrained tumor growth through promoting apoptosis and inhibiting proliferation and angiogenesis via ROS-mediated mitogen-activated protein kinase (MAPK) and PI3K/AKT/mTOR pathways [32]. However, another study showed that in clear cell renal cell carcinoma, inhibition of H2S-producing enzymes, mainly CSE might contribute to the suppression of apoptosis by endogenous H2S [15]. Recently, a study found that CSE/H2S was impaired in urothelial carcinoma and overexpression of CSE/H2S inhibited cell proliferation and promoted apoptosis [17]. Consistent with the apoptosis-inducting effect in tumor of CSE/H2S, the results of our study also indicated that CSE might contribute to enhancing apoptosis and suppressing proliferation via H2S in HCC cells. These findings suggested that the CSE deregulation and its significant correlation with malignant phenotype and poor prognosis of HCC might be due to its crucial role in tumor cell apoptosis.
In conclusion, our findings revealed that CSE expression was decreased in HCC. Additionally, we found that decreased CSE expression was associated with poor clinicopathological features and clinical survival in HCC patients. Further, our study suggests a novel hypothesis that CSE may impact the progress of HCC through its effects on HCC apoptosis by H2S, illuminating that CSE/H2S could act as a potential target for HCC treatment. However, this study has some limitations, and further clinical or experimental investigation will be needed to explore the underlying mechanism.
Acknowledgement: None.
Funding Statement: This study was supported by Beijing Municipal Science & Technology Commission to Huiguo Ding (Z221100007422002) and Beijing Hospitals Authority Youth Programme to Shanshan Wang (QML20211701).
Author Contributions: Huiguo Ding designed and supervised the study; Yanan Ma and Shanshan Wang conducted experiments or interpreted the data; Yanan Ma wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: The study protocol (Approval No. LL-2020-009-K) was approved by the Ethics Committee of Beijing You’an Hospital Affiliated to Capital Medical University. The informed consent was obtained from all enrolled patients.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Cinnamon E, Pikarsky E. Are we ready for targeted therapy combinations in HCC? Gut. 2020;69(4):613–4. doi:10.1136/gutjnl-2019-319780. [Google Scholar] [PubMed] [CrossRef]
2. Global Burden of Disease 2019 Cancer Collaboration, Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019: a systematic analysis for the global burden of disease study 2019. JAMA Oncol. 2022;8(3):420–44. doi:10.1001/jamaoncol.2021.6987. [Google Scholar] [PubMed] [CrossRef]
3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi:10.3322/caac.21660. [Google Scholar] [PubMed] [CrossRef]
4. Kelley RK, Rimassa L, Cheng AL, Kaseb A, Qin S, Zhu AX, et al. Cabozantinib plus atezolizumab versus sorafenib for advanced hepatocellular carcinoma (COSMIC-312a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2022;23(8):995–1008. doi:10.1016/S1470-2045(22)00326-6. [Google Scholar] [PubMed] [CrossRef]
5. Ji F, Nguyen MH. Cabozantinib plus atezolizumab in advanced hepatocellular carcinoma and the role of adjuvant antiviral therapy. Lancet Oncol. 2022;23(8):962–3. doi:10.1016/S1470-2045(22)00383-7. [Google Scholar] [PubMed] [CrossRef]
6. Cammarota A, Zanuso V, D’Alessio A, Pressiani T, Personeni N, Rimassa L. Cabozantinib plus atezolizumab for the treatment of advanced hepatocellular carcinoma: shedding light on the preclinical rationale and clinical trials. Expert Opin Investig Drugs. 2022;31(4):401–13. doi:10.1080/13543784.2022.2032641. [Google Scholar] [PubMed] [CrossRef]
7. Paul BD. Cysteine metabolism and hydrogen sulfide signaling in Huntington’s disease. Free Radic Biol Med. 2022;186:93–8. doi:10.1016/j.freeradbiomed.2022.05.005. [Google Scholar] [PubMed] [CrossRef]
8. Bełtowski J, Kowalczyk-Bołtuć J. Hydrogen sulfide in the experimental models of arterial hypertension. Biochem Pharmacol. 2023;208:115381. doi:10.1016/j.bcp.2022.115381. [Google Scholar] [PubMed] [CrossRef]
9. Chen CJ, Cheng MC, Hsu CN, Tain YL. Sulfur-containing amino acids, hydrogen sulfide, and sulfur compounds on kidney health and disease. Metabolites. 2023;13(6):688. doi:10.3390/metabo13060688. [Google Scholar] [PubMed] [CrossRef]
10. Munteanu C, Turnea MA, Rotariu M. Hydrogen sulfide: an emerging regulator of oxidative stress and cellular homeostasis-a comprehensive one-year review. Antioxid. 2023;12(9):1737. doi:10.3390/antiox12091737. [Google Scholar] [PubMed] [CrossRef]
11. Honda K, Hishiki T, Yamamoto S, Yamamoto T, Miura N, Kubo A, et al. On-tissue polysulfifide visualization by surface-enhanced Raman spectroscopy benefits patients with ovarian cancer to predict post-operative chemosensitivity. Redox Biol. 2021;41:101926. doi:10.1016/j.redox.2021.101926. [Google Scholar] [PubMed] [CrossRef]
12. Xu S, Pan J, Cheng X, Zheng J, Wang X, Guan H, et al. Diallyl trisulfide, a H2S donor, inhibits cell growth of human papillary thyroid carcinoma KTC-1 cells through a positive feedback loop between H2S and cystathionine-gamma-lyase. Phytother Res. 2020;34(5):1154–65. doi:10.1002/ptr.6586. [Google Scholar] [PubMed] [CrossRef]
13. Xu M, Zhang L, Song S, Pan L, Muhammad Arslan I, Chen Y, et al. Hydrogen sulfide: recent progress and perspectives for the treatment of dermatological diseases. J Adv Res. 2020;27(2):11–7. doi:10.1016/j.jare.2020.02.003. [Google Scholar] [PubMed] [CrossRef]
14. Liu Y, Wang L, Zhang X, Deng Y, Pan L, Li H, et al. A novel cystathionine γ-lyaseinhibitor, I194496, inhibits the growth and metastasis of human TNBC via downregulating multiple signaling pathways. Sci Rep. 2021;11(1):8963. doi:10.1038/s41598-021-88355-9. [Google Scholar] [PubMed] [CrossRef]
15. BrezaJr J, Soltysova A, Hudecova S, Penesova A, Szadvari I, Babula P, et al. Endogenous H2S producing enzymes are involved in apoptosis induction in clear cell renal cell carcinoma. BMC Cancer. 2018;18(1):591. doi:10.1186/s12885-018-4508-1. [Google Scholar] [PubMed] [CrossRef]
16. Roubenne L, Marthan R, Le Grand B, Guibert C. Hydrogen sulfide metabolism and pulmonary hypertension. Cells. 2021;10(6):1477. doi:10.3390/cells10061477. [Google Scholar] [PubMed] [CrossRef]
17. Panza E, Bello I, Smimmo M, Brancaleone V, Mitidieri E, Bucci M, et al. Endogenous and exogenous hydrogen sulfide modulates urothelial bladder carcinoma development in human cell lines. Biomed Pharmacother. 2022;151:113137. doi:10.1016/j.biopha.2022.113137. [Google Scholar] [PubMed] [CrossRef]
18. Pan Y, Ye S, Yuan D, Zhang J, Bai Y, Shao C. Hydrogen sulfide (H2S)/cystathionine γ-lyase (CSE) pathway contributes to the proliferation of hepatoma cells. Mutat Res. 2014;763–764:10–8. doi:10.1016/j.mrfmmm.2014.03.002. [Google Scholar] [PubMed] [CrossRef]
19. Roessler S, Jia HL, Budhu A, Forgues M, Ye QH, Lee JS, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70(24):10202–12. doi:10.1158/0008-5472.CAN-10-2607. [Google Scholar] [PubMed] [CrossRef]
20. Kimura H. Signalling by hydrogen sulfide and polysulfides via protein S-sulfuration. Br J Pharmacol. 2020;177(4):720–33. doi:10.1111/bph.14579. [Google Scholar] [PubMed] [CrossRef]
21. Yang H, Tan M, Gao Z, Wang S, Lyu L, Ding H. Role of hydrogen sulfide and hypoxia in hepatic angiogenesis of portal hypertension. J Clin Transl Hepatol. 2023;11(3):675–81. doi:10.14218/JCTH.2022.00217. [Google Scholar] [PubMed] [CrossRef]
22. Yang N, Liu Y, Li T, Tuo Q. Role of hydrogen sulfide in chronic diseases. DNA Cell Biol. 2020;39(2):187–96. doi:10.1089/dna.2019.5067. [Google Scholar] [PubMed] [CrossRef]
23. Szlęzak D, Bronowicka-Adamska P, Hutsch T, Ufnal M, Wróbel M. Hypertension and aging affect liver sulfur metabolism in rats. Cells. 2021;10(5):1238. doi:10.3390/cells10051238. [Google Scholar] [PubMed] [CrossRef]
24. Sun HJ, Wu ZY, Nie XW, Wang XY, Bian JS. Implications of hydrogen sulfide in liver pathophysiology: mechanistic insights and therapeutic potential. J Adv Res. 2020;27:127–35. doi:10.1016/j.jare.2020.05.010. [Google Scholar] [PubMed] [CrossRef]
25. Lu X, Ding Y, Liu H, Sun M, Chen C, Yang Y, et al. The role of hydrogen sulfide regulation of autophagy in liver disorders. Int J Mol Sci. 2022;23(7):4035. doi:10.3390/ijms23074035. [Google Scholar] [PubMed] [CrossRef]
26. Chen L, Lin B, Yang J, Zhong L, Xiong X, Wang X. Hydrogen sulfide alleviates ischemia induced liver injury by repressing the SPHK1/S1P pathway. Ann Transl Med. 2023;11(2):73. doi:10.21037/atm-22-6460. [Google Scholar] [PubMed] [CrossRef]
27. Lee JH, Im SS. Function of gaseous hydrogen sulfide in liver fibrosis. BMB Rep. 2022;55(10):481–7. doi:10.5483/BMBRep.2022.55.10.124. [Google Scholar] [PubMed] [CrossRef]
28. Xu W, Cui C, Cui C, Chen Z, Zhang H, Cui Q, et al. Hepatocellular cystathionine γ lyase/hydrogen sulfide attenuates nonalcoholic fatty liver disease by activating farnesoid X receptor. Hepatol. 2022;76(6):1794–810. doi:10.1002/hep.32577. [Google Scholar] [PubMed] [CrossRef]
29. Ali A, Zhang Y, Fu M, Pei Y, Wu L, Wang R, et al. Cystathionine gamma-lyase/H2S system suppresses hepatic acetyl-CoA accumulation and nonalcoholic fatty liver disease in mice. Life Sci. 2020;252:117661. doi:10.1016/j.lfs.2020.117661. [Google Scholar] [PubMed] [CrossRef]
30. Wang X, Li Y, Li Z, Lin S, Wang H, Sun J, et al. Mitochondrial calciumuniporter drives metastasis and confers a targetable cystine dependency in pancreatic cancer. Cancer Res. 2022;82(12):2254–68. doi:10.1158/0008-5472.CAN-21-3230. [Google Scholar] [PubMed] [CrossRef]
31. Lu WC, Saha A, Yan W, Garrison K, Lamb C, Pandey R, et al. Enzyme-mediated depletion of serum l-Met abrogates prostate cancer growth via multiple mechanisms without evidence of systemic toxicity. Proc Natl Acad Sci USA. 2020;117(23):13000–11. doi:10.1073/pnas.1917362117. [Google Scholar] [PubMed] [CrossRef]
32. Zhang Q, Gao Y, Zhang Y, Jing M, Wang D, Wang Y, et al. Cystathionine γ-lyase mediates cell proliferation, migration, and invasion of nasopharyngeal carcinoma. Oncogene. 2022;41(49):5238–52. doi:10.1038/s41388-022-02512-6. [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
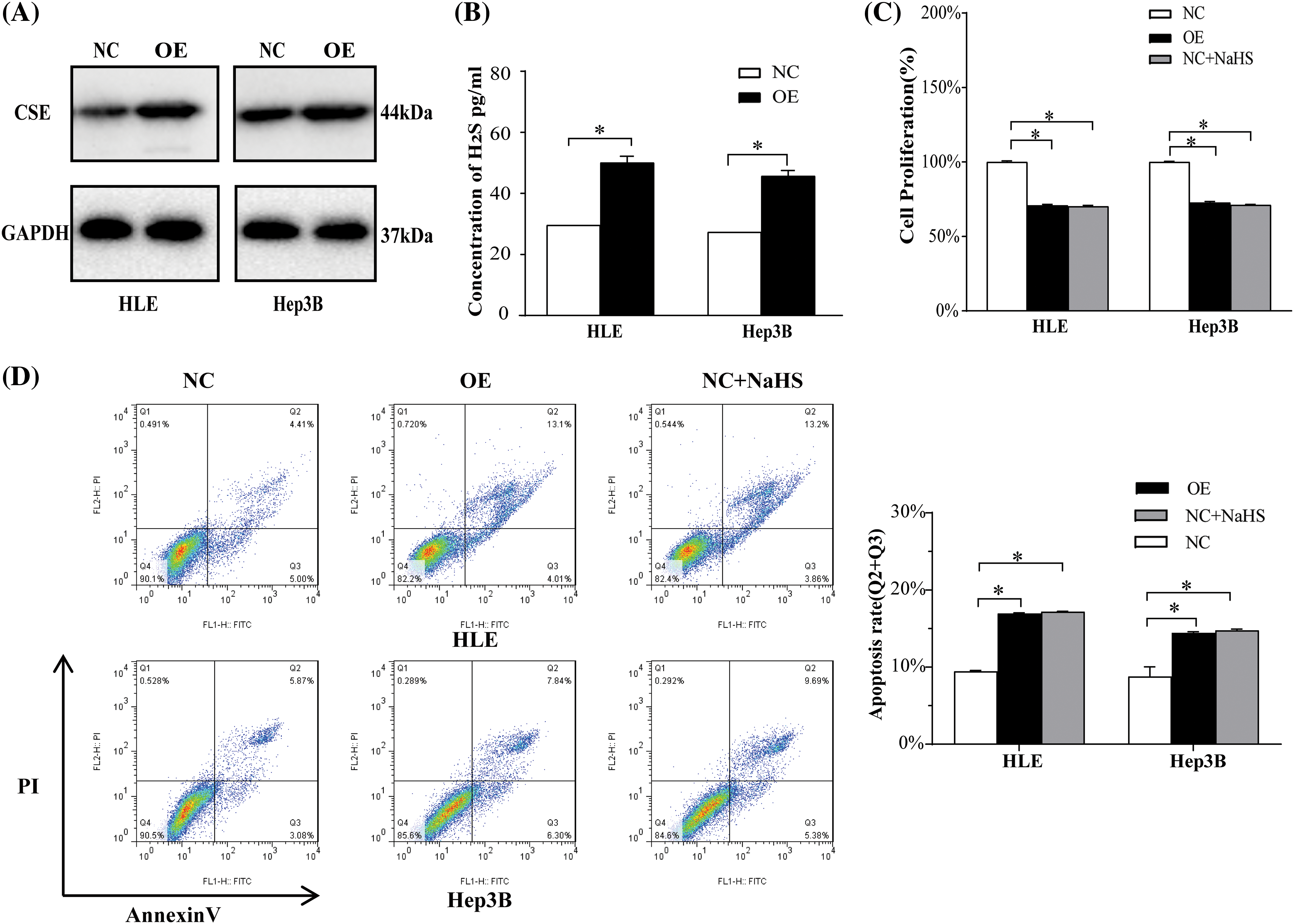
Figure S1: The effects of CSE overexpression in HCC cell proliferation and apoptosis. (A) Western blotting analysis of CSE protein expression in HLE and Hep3B cells transfected with CSE-expressing plasmid using Students’ t-test. (B) ELISA analysis of H2S in HLE and Hep3B cells transfected with CSE-expressing plasmid using Students’ t-test. (C) CCK-8 analysis of proliferation of HLE and Hep3B cells transfected with CSE-expressing plasmid or NaHS using ANOVA-test. (D) Flow cytometric analysis of apoptosis of HLE and Hep3B cells transfected with CSE-expressing plasmid or NaHS using ANOVA-test. The results from three independent experiments were expressed as mean ± SD (n = 3), *p < 0.05.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools