 Open Access
Open Access
ARTICLE
Analysis of large datasets for identifying molecular targets in intestinal polyps and metabolic disorders
Department of Gastroenterology, The First Affiliated Hospital of Chongqing Medical and Pharmaceutical College, Chongqing, China
* Corresponding Authors: PENG SHI. Email: ; JIE YAO. Email:
(This article belongs to the Special Issue: Advances in Biomarker Research: Unveiling the Pathways to Precision Medicine)
BIOCELL 2024, 48(3), 415-429. https://doi.org/10.32604/biocell.2024.046178
Received 21 September 2023; Accepted 08 January 2024; Issue published 15 March 2024
Abstract
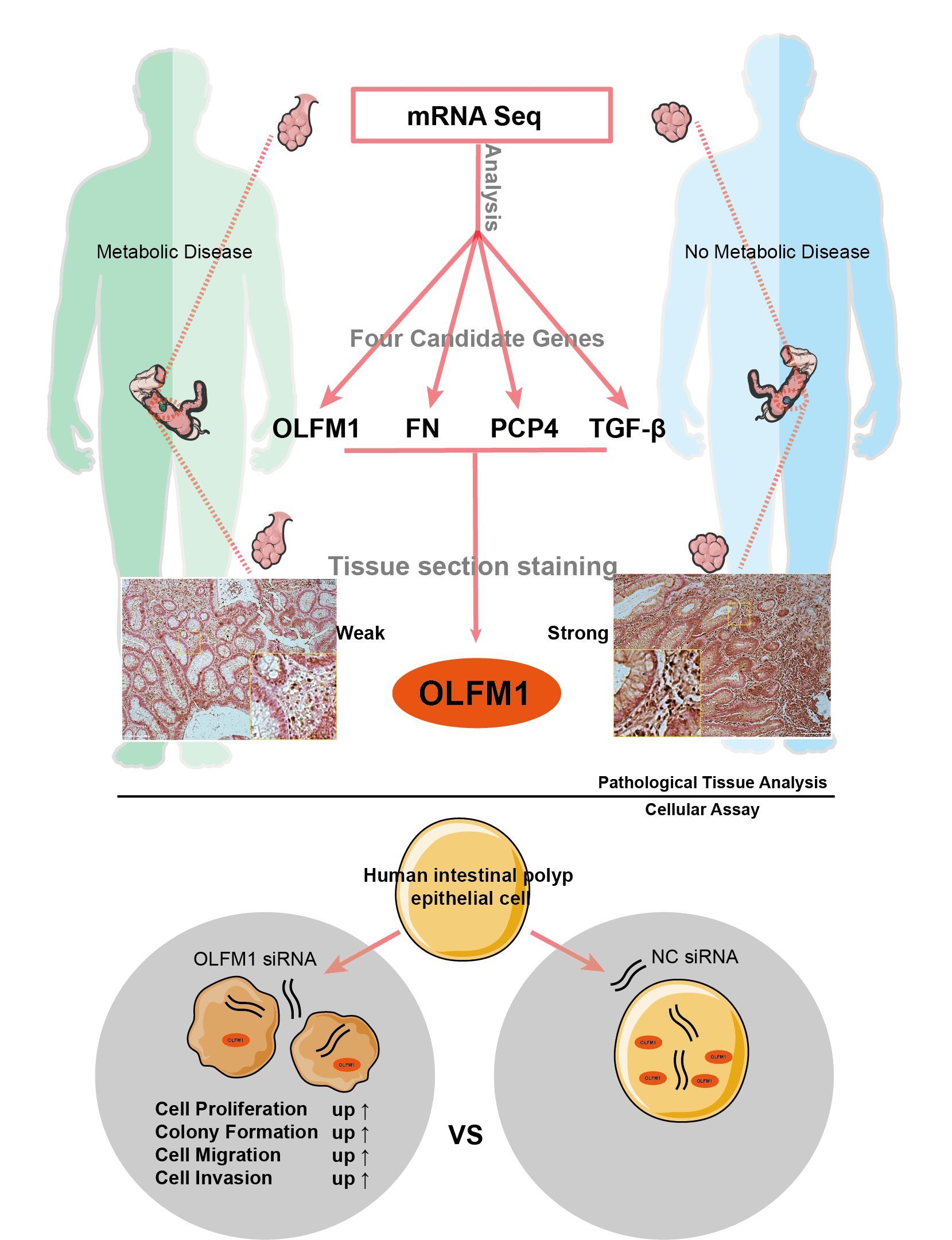
Background: The interrelation between intestinal polyps, metabolic syndrome (MetS), and colorectal cancer (CRC) is a critical area of study. This research focuses on pinpointing potential molecular targets to understand the link between intestinal polyp formation, metabolic irregularities, and CRC progression. Methods: We examined clinical samples from patients with intestinal polyps coexisting with MetS and compared them with samples from patients with standard intestinal polyps. Transcriptome sequencing and public database analysis were employed to identify significant pathways and genes. These targets were then validated through immunohistochemistry (IHC). Following the RNA interference of key target expression, a series of experiments, including the cell counting kit-8 assay, colony formation, wound healing, and Transwell assays, were conducted. Results: Comparative analysis revealed 75 up-regulated and 61 down-regulated differentially expressed genes (DEGs) in the MetS polyp group vs. the control. Kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment suggested these DEGs were primarily associated with cell cycle and mitosis. Integration with comparative toxicogenomics database (CTD) and the cancer genome atlas (TCGA) databases highlighted 44 key CRC-related genes. Protein interaction networks indicated connections of purkinje cell protein 4 (PCP4), olfactomedin 1 (OLFM1), fibronectin 1 (FN1), and transforming growth factor beta 3 (TGF-β3) with the mitogen-activated protein kinase (MAPK) pathway. Tumor correlation studies suggested higher risk associations with FN1, PCP4, and TGF-β3, while OLFM1 was identified as a lower risk gene. Immunohistochemical analysis revealed a decrease in OLFM1 in MetS-associated intestinal polyps. Upon interference with OLFM1 in polyp epithelial cells, there was a significant enhancement in cell proliferation, colony formation, and cell migration and invasion capabilities. Conclusion: Our study highlights a significant decrease in OLFM1 expression in MetS-associated intestinal polyps. And, this reduction in OLFM1 is associated with enhanced cell proliferation, colony formation, and increased cell migration and invasion capabilities. These findings underscore the reduced OLFM1 expression in MetS-associated intestinal polyps may play a crucial role in promoting tumorigenic processes in colorectal pathology. Further research on OLFM1 may provide valuable insights into understanding and targeting MetS-associated intestinal polyps.Graphic Abstract
Keywords
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools