 Open Access
Open Access
REVIEW
MicroRNAs modulation in lung cancer: exploring dual mechanisms and clinical prospects
1 Department of Biotechnology, Kohsar University Murree, Murree, Pakistan
2 Macau University of Science and Technology State Key Laboratory of Quality Research in Chinese Medicines, Taipa, Macau
3 Department of Bioinformatics and Biosciences, Capital University of Science and Technology, Islamabad, Pakistan
4 Department of Microbiology, Kohsar University Murree, Murree, Pakistan
* Corresponding Author: SHAHID HUSSAIN. Email:
(This article belongs to the Special Issue: Advances in Biomarker Research: Unveiling the Pathways to Precision Medicine)
BIOCELL 2024, 48(3), 403-413. https://doi.org/10.32604/biocell.2024.044801
Received 09 August 2023; Accepted 22 January 2024; Issue published 15 March 2024
Abstract
The global incidence of lung cancer is marked by a considerably elevated mortality rate. MicroRNAs (miRNAs) exert pivotal influence in the intricate orchestration of gene regulation, and their dysregulation can precipitate dire consequences, notably cancer. Within this context, miRNAs encapsulated in exosomes manifest a diversified impact on the landscape of lung cancer, wherein their actions may either foster angiogenesis, cell proliferation, and metastasis, or counteract these processes. This comprehensive review article discerns potential targets for the prospective development of therapeutic agents tailored for lung cancer. Tumor-suppressive miRNAs, such as miR-204, miR-192, miR-30a, miR-34a, miR-34b, miR-203, and miR-212, exhibit heightened expression and demonstrate the capacity to inhibit cellular proliferation and invasiveness. Conversely, the deleterious effects of tumor-promoting miRNAs like miR-21, miR-106a, miR-155, miR-205, and miR-210 can be attenuated through the application of their respective inhibitors. Distinct miRNAs selectively target various oncogenes, including NUAK Family Kinase 1 (NUAK1), Snail Family Transcriptional Repressor 1 (Snai1), Astrocyte elevated gene-1 (AEG-1), Vimentin, Proliferation and apoptosis adaptor protein 15 (PEA-15/PED), Hypoxia-inducible factor 1-alpha (HIF1), as well as tumor suppressor genes such as phosphatase and tensin homolog (PTEN), Suppressor of cytokine signaling 1 (SOCS1), Tumor protein P53 binding protein 1 (TP53BP1), and PH Domain and Leucine Rich Repeat Protein Phosphatase 2 (PHLP22). This investigative approach proves invaluable in elucidating the specific miRNAs implicated in the deregulation of crucial genes pivotal to the pathogenesis of cancer.Keywords
The mortality rate of lung cancer is 1.3 million worldwide with equal prevalence in men and women. Based on pathology, there are two categories of lung cancer; non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC) with their prevalence rate of 80.4% and 16.8%, respectively [1,2]. While many factors contribute to lung cancer development like viral infection, smoking tobacco, and radiation, the mechanism involved is still elusive [3]. Individuals who have a higher risk of developing lung cancer are believed to derive the most advantage from undergoing lung cancer screening. There exist various methods to identify these high-risk individuals [4]. MiRNAs are 22nt-long gene products present in diverse organisms, serving a crucial role in regulating mRNA coding and degradation. They achieve this by partially pairing with specific sites on mRNA molecules. Importantly, this binding primarily occurs in the untranslated region of mRNA [5,6].
Long precursor RNAs are expressed within the nucleus, serving as precursors for miRNAs. Within the nucleus, these precursors undergo processing and are then transported to the cytoplasm via an exportin-5-dependent mechanism [7]. In cytoplasm, an enzyme DICER cleaves miRNA [8], resulting in a miRNA of about 17–24 nt long. Association of miRNAs and RNA induced silencing complex cause’s interference of RNA [9]. It has been proposed that miRNAs take part in various biological events like stress resistance, cell division, apoptosis, and fat metabolism by regulating the expression of genes [10]. Normal miRNAs play a pivotal role in various cellular activities by regulating multiple mRNAs with a single miRNA. However, if there is dysregulation in miRNA function, it can lead to severe repercussions. In cancer, the loss of tumor-suppressive miRNAs leads to the expression of oncogenes targeted by these miRNAs. There is an increase in the expression of oncogenic miRNAs (oncomirs) which, in turn, inhibit genes involved in suppressing tumors. Exosomal miRNAs play a crucial role in modifying the lung cancer environment, potentially facilitating angiogenesis, invasion, metastasis, progression, and resistance to therapy [11]. This breakthrough paved the way for extensive research aimed at comprehending how oncomirs target genes and at restoring miRNAs responsible for suppressing tumors [12]. MiRNAs play a crucial role in lung cancer development by regulating oncogenes and tumor suppressor genes at the post-transcriptional stage. Dysregulation of various types of miRNAs is linked to lung cancer, and their biological significance has been clarified through various assays [13]. The objective of this review was to analyze role of various exosomes derived miRNAs in lung cancer and to identify potential candidate genes which may be used in lung cancer therapy.
The process of miRNA formation is highly conserved throughout evolution and involves a series of steps orchestrated by two RNase III enzymes, Drosha and Dicer as shown in Fig. 1 [14]. Initially, RNA polymerase II (RNA Pol II) transcribes the primary miRNA transcript (pri-miRNA), which is then processed by Drosha within the nucleus, shaping it into a ∼60–100 nt hairpin structure known as precursor-miRNA (pre-miRNA) [15]. Once cleaved by Drosha, the pre-miRNA is transported out of the nucleus through interactions involving Exportin-5 and Ran-GTP. Subsequently, Dicer catalyzes further processing of the pre-miRNA [16], leading to a cleavage event generating a ∼22 nt double-stranded (ds) RNA product containing both the mature miRNA guide strand and the passenger strand. The mature miRNA then facilitates the assembly of the RNA-induced silencing complex (RISC), a large protein complex, onto specific regions within the 3′-untranslated region (3′-UTR) of target genes [17]. MiRNA targets are selected through a process involving imperfect matching between the miRNA and a recognition site on the 3′-UTR of the target mRNA. This imperfect pairing allows a single miRNA to potentially target tens to hundreds of mRNAs [18]. When the miRNA associates with the RISC, it leads to the suppression of the target gene by either encouraging mRNA degradation or inhibiting translation [9]. By repressing these targets, miRNAs trigger significant alterations in gene expression patterns that are known to influence various aspects of biology, such as developmental timing, differentiation, proliferation, cell death, and metabolism [19].
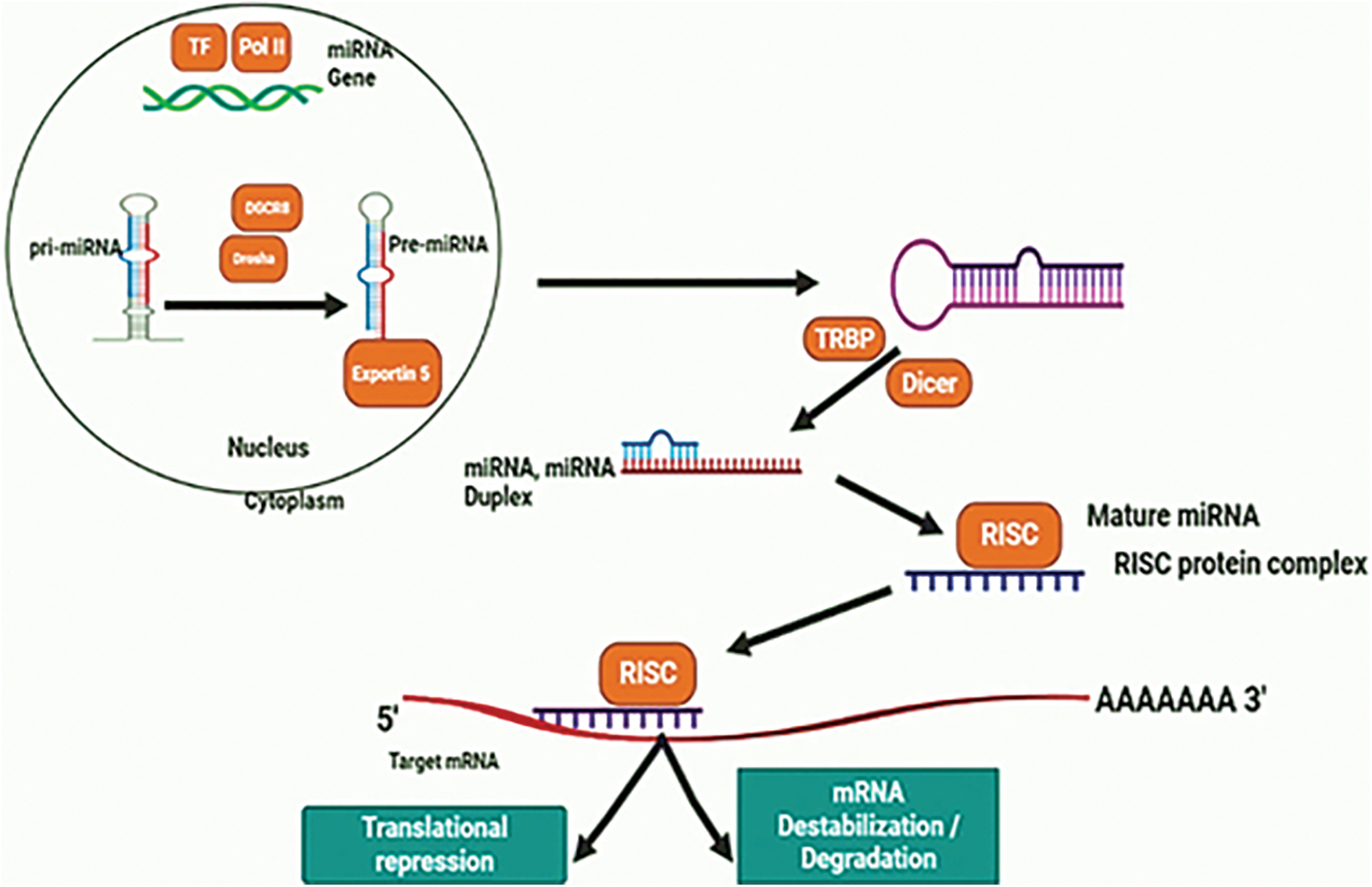
Figure 1: Biogenesis mechanism of miRNA.
Exosomes Derived miRNAs and Lung Cancer Progression
Exosomes originating from cancerous cells have been associated with tumor growth through intercellular communication pathways [20–22]. Exosomes play a crucial role in intercellular communication systems, particularly in disorders like cancer, as well as in maintaining normal cellular balance or homeostasis [23]. The study conducted by Rabinowits et al. established a correlation between tumor-derived exosomes and exosome-packaged miRNAs in the development of lung cancer. This research involved 27 lung cancer patients and 9 control subjects, ranging from 21 to 80 years old. Analysis of circulating exosomes revealed the presence of small RNAs and miRNAs. The average concentration of exosomes in patients was 2.85 mg/mL, while in controls, it was 0.77 mg/mL. Moreover, the average miRNA concentration in patients was 158.6 ng/mL, compared to 68.1 ng/mL in controls. Differences in the ranks of exosomes and miRNAs among patients suggest their association with lung cancer. Further exploration indicated that the miRNA signatures found in exosomes were similar to those found in tumor tissues, indicating their potential as biomarkers for screening tests [24].
Zhang and colleagues established a link between miR-21 and the PTEN gene. They found that miR-21, when overexpressed, is present in numerous cancers and targets tumor suppressor genes, thereby promoting cancer progression. High levels of miR-21 were observed in tumor tissues, and an inverse correlation was identified between miR-21 and the PTEN protein. Their investigation demonstrated that miR-21 initially reduces PTEN expression after transcription in transfected cells, leading to increased cell growth. Moreover, they discovered that suppressing miR-21 expression in NSCLC cell lines led to a decrease in PTEN protein levels. When the NSCLC cell line was transfected with a miR-21 inhibitor, it exhibited reduced growth and invasive characteristics. These findings suggest that targeting miR-21 with an inhibitor could be a potential therapeutic strategy for treating NSCLC [25]. Besides PTEN, other targets of miR-21 are Programmed Cell Death 4 (PDCD4) [26], reversion-inducing cysteine-rich protein with kazal motifs (RECK) [27], Nuclear Factor I B (NFIB) [28], Tropomyosin 1 (TPM1) [29] and maspin [30]. Most of these targeted genes are tumor suppressors.
This molecule was found to target PTEN expression, resulting in the downregulation of PTEN due to heightened miR-106a expression. In NSCLC, the relationship between miR-106a and PTEN expression showed an inverse correlation. To further confirm the impact of miR-106a, researchers employed a miR-106a inhibitor. When this inhibitor was used to transfect NSCLC cells, there was a noticeable decrease in cell proliferation, invasion, and migration. This demonstrates the potential therapeutic value of targeting miR-106a as a strategy for combating NSCLC [31].
By down regulation of the SOCS1 gene, miR-155 displays its oncogenic properties causing cell proliferation [32] and its anti-apoptotic effect is expressed through blocking caspase 3-action [33]. It also suppresses pro-apoptotic genes like TP53BP1 [34]. It modifies the SMAD family member 4 (Smad4)/Tumor growth factor beta (TGF-β) pathways. Moreover, miR-155 targets Ras Homolog Family Member A (RhoA), which contributes significantly to the interference of cells [35].
The miR-205 molecule shows elevated levels in various tumor types and directly suppresses the expression of PTEN and PHLPP2 genes. Its overexpression is triggered by Nuclear factor-kappa B (NF-κB), leading to the transactivation of the miR-205 gene. Moreover, miR-205 stimulates the Protein kinase B (PKB, also known as AKT mammalian target of rapamycin (mTOR) and AKT/Forkhead Box O3 (FOXO3a) signaling pathways. In NSCLC, miR-205 promotes both angiogenesis and cell proliferation in laboratory settings (in vivo and in vitro). However, the adverse effects of miR-205 were mitigated by using RNA interference to silence its expression. Additionally, the malignant impact of miR-205 could be counteracted by employing AKT inhibitors. Furthermore, the influence of miR-205 could also be diminished by restoring the expression of PTEN/PHLPP2, administering rapamycin treatment, or upregulating FOXO3a [36].
miR-210 is involved in targeting HIF1 activity, leading to the upregulation of HIF1 expression. In the later stages of NSCLC, miR-210 exhibits increased expression. It contributes to the loss of mitochondrial membrane potential. Cells displaying heightened levels of miR-210 showed an increase in transcripts associated with apoptosis and mitochondrial dysfunction. Among its targets, miR-210 affects SDHD, a transcript responsible for encoding the subunit D of the succinate dehydrogenase complex (SDH) within the electron transport chain. When SDHD is knocked down, it results in mitochondrial alterations. Therefore, the activity of miR-210 influences cell survival, metabolism, and HIF1 activity due to its impact on mitochondrial function and SDHD expression [37].
MiRNA-221/222 is a non-coding microRNA widely distributed in eukaryotes and profoundly involved in the post-transcriptional regulation of gene expression [38]. miR-221/222 plays important regulatory roles in cancer development and progression, both in promoting and suppressing cancer [39]. Molecular epidemiology studies have shown that hundreds of genes are involved in lung carcinogenesis. Researchers in this study showed the restraining actions of miR-221 and miR-222 in lung cancer cells. Suppression of lung cancer growth and development by miR-221 or miR-222 through S-phase cell cycle arrest, with apoptosis partially attributed to DNA double-strand breaks (DSBs). Their results showed that miR-221 or miR-222 could increase sensitivity to S-phase targeting drugs such as gemcitabine and cisplatin, but had no effect on M-phase targeting drugs such as paclitaxel [40]. Two tumor suppressor miRNAs, miR-221 and miR-222, enhanced the tumorigenic phenotype of H460 lung cancer cells associated with resistance to invasive TRAIL-induced apoptosis through suppression of PTEN and Metalloproteinase inhibitor 3 (TIMP3). miR-221 and miR-130a control airway branching and pulmonary micro vascular development by targeting the developing vasculature. It was found that augmented ranks of miR-221 or lessened ranks of miR130a in lung cultures lead to disruption of the vascular network associated with restricted airway branching [41]. miR-221/222 plays an important role in tyrosine kinase inhibitor (TKI) resistance in NSCLC. Epidermal growth factor receptor (EGFR) and MET Proto-Oncogene, Receptor Tyrosine Kinase (MET) regulate miR-221/222 expression by targeting peptide-activating factor 1 (APAF1), regulating gefitinib-induced apoptosis and NSCLC tumorigenesis [42]. Exosomal miR-222-3p stimulated the growth, migration, invasion, and susceptibility to gemcitabine in NSCLC by focusing on SOCS3 [43]. Over expression of miR-221/222 in invasive NSCLC cells induced TRAIL resistance and promoted cell migration by targeting PTEN and TIMP3 [44]. In addition to serving as a significant epigenetic mechanism in the control of CSC self-renewal and maintenance, the miR-221/222-Reck-Notch1 axis offers possible targets in this regard [45].
Ten isoforms of the let-7 family, the earliest known human miRNA, function as tumor suppressors by interacting with a variety of mRNAs encoding oncogenes, including RAS [46]. Let-7c is a member of the let-7 family and works by destroying germinal-center kinase-like kinase (GLK, also known as MAP4K3) and Integrin beta-3 (ITGβ3) to prevent NSCLC cells from migrating and invading. Low let-7 expression was associated with poor prognosis, metastases, venous assault, and advancement of TNM (Tumor, Node, Metastasis) stage in patients with NSCLC [47]. Let-7g efficiently induces cell cycle arrest and cell death in mouse lung cancer cells expressing K-RasG12D by targeting [35] the Kirsten rat sarcoma viral oncogene homolog (KRAS) oncogene [48]. Reduced expression of let-7 in tumor cells will hasten the progression of cancer because disruptions to regulatory networks, such as the Ras GTPase, occur. A down regulated in let-7a has been found in NSCLC and binds to the 3′ UTR of DICER [49]. Previous studies have shown targets of exosomes miRNAs and thus effecting the targeted genes and involve in regulation of various cellular activities as shown in Table 1.
Exosomes Derived miRNAs and Tumor Suppression
Several miRNAs have been found to have a tumor suppression effects in NSCLS.
MiR-204 is downregulated in NSCLC [59]. It was noted that migration and proliferation of cells are inhibited upon overexpression of miR-204, with simultaneous promotion of apoptosis [60]. Excessive expression of NUAK1 is observed in NSCLC cells and is associated with increased metastasis and invasiveness in human NSCLC. NUAK1 overexpression triggers the phosphorylation of p70 ribosomal S6 kinase 1 (p70S6K1) and eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1), alongside influencing mTOR phosphorylation. Furthermore, reduced levels of miR-204 correlate with enhanced progression, migration, and invasiveness in NSCLC. The clinical relevance lies in the downregulation of miR-204 and the concurrent upregulation of NUAK1 in NSCLC. While miR-204 acts as a suppressor, limiting tumor growth and impeding tumor cell invasion by decreasing NUAK1 expression, the presence of NUAK1 suggests its potential as a therapeutic target for NSCLC treatments [61].
In the context of NSCLC, the activation of miR-192 led to the suppression of Retinoblastoma 1 (RB1). Interestingly, nicotine therapy in NSCLC cell lines resulted in a reduction of miR-192 levels. This reduction in miR-192 directly affected RB1, as miR-192 serves as a direct target for RB1 regulation. Upon exposure to nicotine, RB1 expression increased in NSCLC cells. The decrease in miR-192 expression following nicotine treatment was followed by enhanced cell proliferation and a transition from an epithelial to a mesenchymal state (EMT) in these cells. Restoring miR-192 expression countered this effect by reducing RB1 levels, thereby neutralizing the impact of nicotine on NSCLC cell growth and the epithelial-to-mesenchymal transition [62].
NSCLC has the potential to metastasize, spreading to other regions of the body through either the bloodstream or the lymphatic system. This metastatic spread can involve distant organs like the bones, liver, brain, or other areas of the lungs. The blood vessels and lymphatic channels provide pathways for these cancerous cells to travel and establish secondary tumors in other parts of the body [63]. miR-30a demonstrated its potential as a tumor suppressor. By targeting Snai1 [64], miR-30a hindered the epithelial-to-mesenchymal transition (EMT) in NSCLC cell lines, effectively inhibiting this transition process. Moreover, in various in vitro experiments, the increased expression of miR-30a was shown to inhibit cell migration, invasion, and detached spread in multiple cell lines associated with NSCLC. These findings demonstrates the role of miR-30a as a tumor suppressor in NSCLC, emphasizing its ability to impede EMT and the aggressive behavior of cancer cells such as migration, invasion, and spread [65]. Liu observed that miR-30a targets not only Snai1 but also AEG1 (astrocyte elevated gene-1) and vimentin. Interestingly, an increase in the levels of Snai1, AEG1, and vimentin coincided with a decrease in miR-30a expression. AEG1, upon binding to miR-30a, weakens the inhibitory effect of miR-30a on vimentin and Snai1. This intricate interaction suggests that AEG1 plays a role in disrupting the suppressive impact of miR-30a on vimentin and Snai1, contributing to the regulation of these targets involved in processes such as epithelial-to-mesenchymal transition (EMT) in various cancer contexts, including NSCLC [66]. Hence, miR-30a is an efficient biomarker for NSCLC patient’s diagnosis and prognosis, and due to its tumor suppressor activity, it possesses the capacity to be a therapeutic target. for NSCLC [67].
MiR-34 is an analytical factor for patients of NSCLC. The miR-34a and miR-34b suffer down regulation in tumor cells. Patients with low expression of miR-34a exhibited a high frequency of p53 mutations. miR-34a levels alternated because of methylation of the miRNA 34a gene’s promoter region [68]. In response to genotoxic stress, p53 directly induces the transcription of miR-34, to have the cell cycle arrested or apoptosis [69]. A large number of proteins’ expression is regulated by p53 via miR-34. Over expression of miR-34a leads to a decline in proliferation and activation of apoptosis in tumor cells, hence suggesting the role of miR-34a as a tumor suppressor [70]. Furthermore, In NSCLC patients who underwent curative surgery, low levels of miR-34a were correlated with a higher risk of recurrence [70].
MiR-203 has been designated as tumor suppressive miRNA. Lung tumor cells express miR-203 at a downregulated level as compared with healthy surrounding tissues. PKCα (protein kinase C alpha) is a direct target of miR-203. The mRNA levels of PKCα were remarkably high in lung cancer tissues. miR-203 exhibited an inverse correlation with PKCα in lung cancer. By silencing PKCα via miR-203, cell apoptosis was modulated. Moreover, cell invasion and cell proliferation were inhibited by miR-203, as an outcome of silencing PKCα. Therapeutic approaches to enhance miR-203 or silence PKCα are likely to benefit lung cancer patients [71].
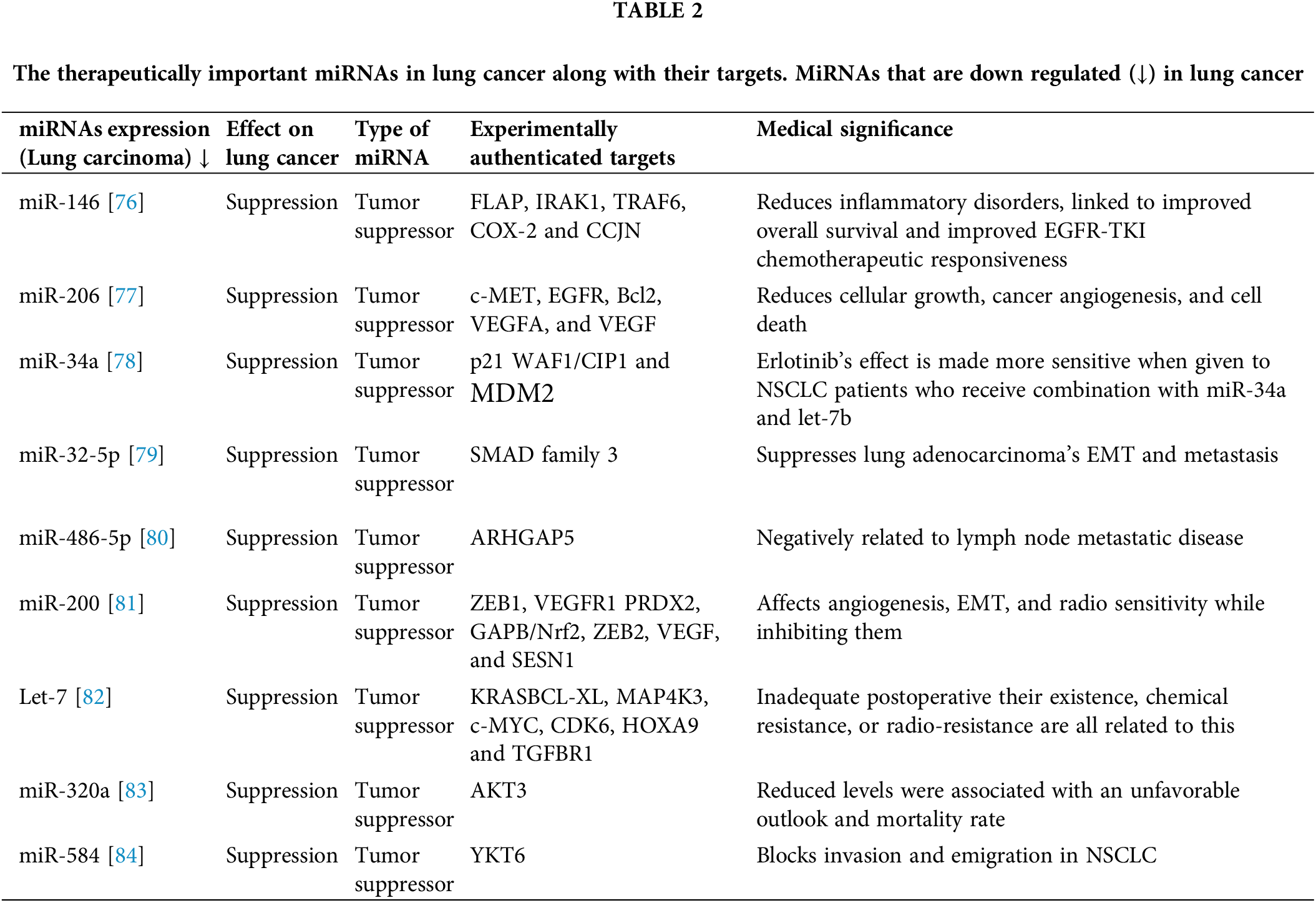
MiR-212 negates the expression of Phosphoprotein Enriched in Astrocytes 15 (PEA-15) also known as PED. Expression of miR-212 increases cell death in NSCLC, by inducing the ligand that induces apoptosis in relation to TRAIL. Alternatively, inhibition of miR-212 leads to a surge of PED protein. Hence, miR-212 is a tumor suppressor and is a candidate therapeutic agent for treating NSCLC [72]. PED is an anti-apoptotic molecule that targets metabolic activities and cell growth [73]. PED overexpression is associated with many different kinds of tumors including lung cancer [74]. PEDs can block intrinsic and extrinsic apoptotic pathways [75]. From the previous studies, some of the miRNAs as shown in Table 2 are down regulated in lung cancer and are considered to be therapeutically important.
Exosomal miRNA as Predictive Biomarkers for Lung Cancer
Identifying predictive biomarkers for the recurrence of NSCLC after curative resection is vital for effective cancer treatment. Functional miRNAs found in plasma and serum exosomes show promise as stable, non-invasive indicators to assess the risk of cancer recurrence in patients [85]. It has been established that Exosomal miR-29a-3p and miR-150-5p are dosage exposure biomarkers that may be utilized as predictive biomarkers of toxicity or response [86]. Serum Exosomal miR-146a-5p may be employed as a biomarker to track treatment resistance and enhance Cisplatin efficacy predictions in NSCLC [86]. Exosomal miR-425-3p shows potential as a biomarker for evaluating the clinical response of NSCLC patients to platinum-based chemotherapy [86]. According to multiple studies, there are different subtypes of lung cancer, and different miRNA expression signatures can distinguish between lung tumors with better and worse prognoses. MiRNAs circulate in bodily fluids, they may be useful biomarkers for both patient prognosis and the early identification of lung cancer [87]. Various bodily fluids can include potential biomarkers for early lung cancer, but blood is the most abundant and accessible source [88]. Among the potential candidates for such biomarkers include metabolites, free nucleic acids, and serum proteins [89]. Circulating tumor cells and circulating free DNA are two additional potential lung cancer biomarkers [90]. A novel potential set of biomarkers for lung cancer involves blood metabolites and lipids. Various cells release exosomes, and in cancer patients, particularly those with lung cancer, exosome levels in the blood are notably higher compared to healthy individuals. Recent discoveries suggest that cancer patients may have up to 109 vesicles per milliliter of blood. Serum or plasma-derived exosomes present an intriguing avenue for exploring lung cancer biomarkers, discussed extensively in several publications. Quantitative PCR and next-generation sequencing (NGS) represent the primary analytical techniques frequently employed to detect miRNAs in these investigations. The search for a lung cancer biomarker has led to numerous studies on miRNAs, a class of short endogenous non-coding RNAs spanning 18–24 nucleotides that regulate target genes. Presently, over 2500 mature miRNAs have been identified in humans [91]. miRNA profiles from bodily fluids or tissue can also be used to identify the best treatments for treating NSCLC and as predictive biomarkers for how sensitive NSCLC tumors are to specific therapy. For instance, via controlling EGFR expression, miR-143-3p, which play a role in the migration, encroachment and proliferation of NSCLC cells [88].
MiRNAs as Therapeutic in Lungs Cancer
MiRNAs are vital participants in the control of genes and, consequently, the development of tumors. MiRNAs open up brand-new avenues for lung cancer research in terms of biomarker development, detection, and treatment, an illustration is shown in Fig. 2. MiRNAs control as much as 30% of the genes that code for proteins in humans, causing them one among the more common groupings. of regulatory genes in humans due to their capacity to target numerous genes within a passageway [92]. When lung cancer samples from NSCLC adenocarcinoma patients were evaluated using miRNA microarray data, high expression of miR-155 was a highly unfavorable prognostic variable. They verified miR-17, miR-21, and miR-155 in three additional cohorts of individuals with NSCLC adenocarcinoma. Based on the microarrays results they ultimately proposed that a lower prognosis is linked to increased levels of miR-21 [93]. The expression levels of several miRNAs in a cohort of 357 stage I NSCLC patients discovered the predictive usefulness of two miRNA signatures. It was demonstrated that miR-21 was elevated in fresh frozen NSCLC tissues compared to comparable nearby noncancerous tissues and that this over expression corresponded with the operating system. Mature miR-21 prognostic usefulness in NSCLC was initially demonstrated in 2008 [94]. Two meta-analyses that combined the information from several researchers concluded that miR-21 over expression was a distinct prognostic factor for NSCLC [76]. Four miRNAs (miR-155, -21, -34, and let-7) appeared more frequently in a meta-analysis study that looked at 141 prognostic miRNAs. In particular, miR-21 and miR-155 were discovered to represent separate prognostic indicators connected to low mortality and lung cancer resurgence [95].
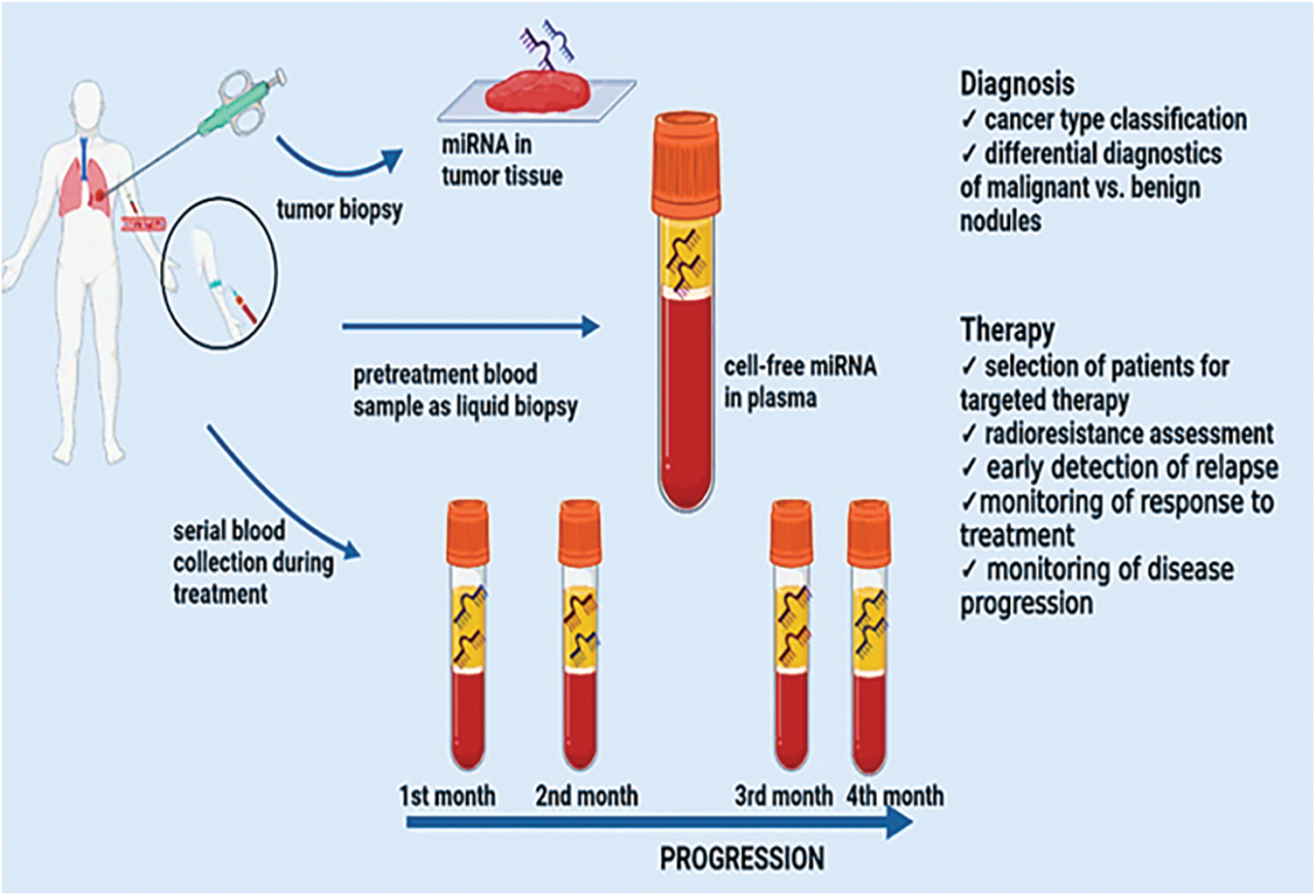
Figure 2: miRNAs as biomarkers and therapeutic targets in lung tumors.
Since then, several researches have demonstrated that high levels of miR-21 in tissue and plasma are associated with a worse prognosis in NSCLC patients and may serve as a separate prognostic marker [96]. Lung cancer patients can be distinguished from non-smokers with 69.4% sensitivity and 78.3% specificity by having higher levels of the circulating miR-21, miR-155, and miR-145, and this pattern is linked to poor survival. miR-21 has been discovered to be considerably higher in the serum of NSCLC patients with poor survival, suggesting that serum miR-21 may be a possible diagnostic biomarker for the diagnosis of NSCLC [97]. Higher miR-21 expression is associated with shorter survival time and disease recurrence [98]. Sixty healthy controls, forty-two patients with non-cancerous lung illness, and 126 patients with early-stage NSCLC provided plasma samples. Five of these plasma miRNAs miR-20a, miR-145, miR-21, miR-223, and miR-221 have the potential to be useful biomarkers for early NSCLC screening [99]. As miRNAs are highly stable in biological samples, there is a chance that they will be used as prognostic, accurate, and diagnostic biomarkers for different kinds of malignancies [100]. The use of biomarkers that are not invasive in screening programs and for keeping track of suspicious nodules that were found by computerized tomography. Numerous types of research have focused on finding miRNAs with sensitivity and specificity ranging from 60% to 100% that can distinguish between NSCLC patients and healthy donors or those with non-neoplastic disorders [101]. Serum miR-21 has been found to be significantly greater in NSCLC patients who have a lower prognosis, indicating that serum miR-21 may be a potential predictive biomarker for NSCLC identification. Recurrence of the disease and a shorter period of survival are linked to higher level of expression of miR-21 in individuals with postoperatively lung cancer, according to research. Blood levels of miR-21 significantly dropped following successful procedures [102]. Sixty healthy controls, forty patients with non-cancerous lung illness, and 126 patients with early-stage NSCLC provided plasma samples. Five of these plasma miRNAs—miR-20a, miR-145, miR-21, miR-223, and miR-221—have the potential to be useful biomarkers for early NSCLC screening [103]. A general overview for the development of miRNA-based drug for lung cancer is shown in Fig. 3.
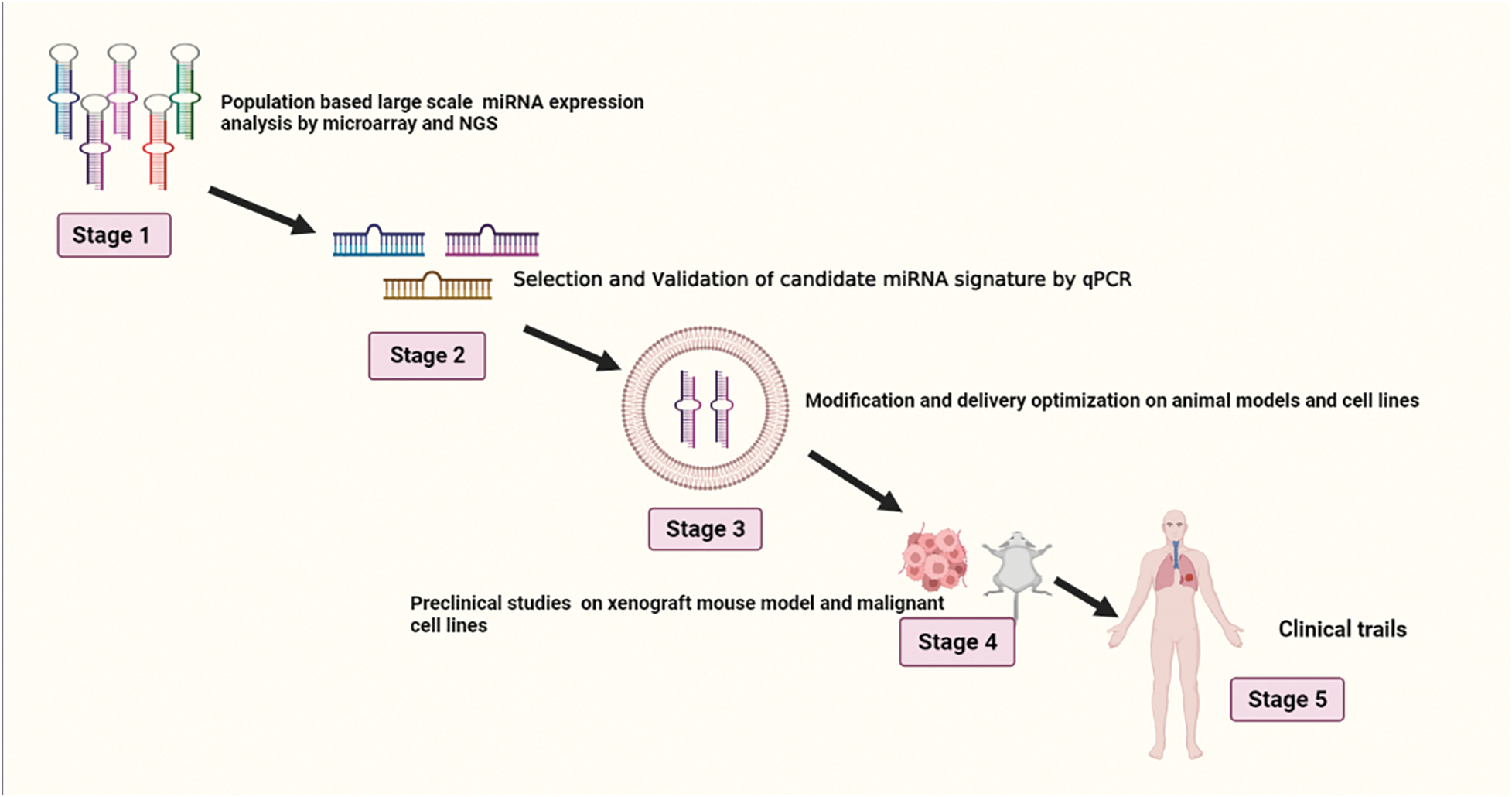
Figure 3: Illustrates stages of development in miRNA-based drug design for lung cancer.
Current Limitations and Future Perspectives
Although miRNA has a lot of potential for therapy, there are some drawbacks, including the need to maintain stability and circulation time, endosomal escape, poor delivery specificity, and off-target effects. These issues must be resolved if miRNA-based therapy is to become more effective [104]. The ability of miRNA expression to serve as a lung cancer predictive and diagnostic tool is greatly influenced by numerous pre- and analytical factors. Some of these are intrinsic to miRNAs; they include nucleotide additions or deletions, Single Nucleotide Polymorphisms (SNPs), isomers, and the GC (guanine and cytosine) content of miRNAs. These modifications have an impact on miRNA recovery and sequence, which in turn has a major impact on miRNA profiling [105]. To finalize a viable miRNA, a potential delivery agent, a highly precise targeting ligand, and an endosomolytic drug for its preclinical and in vitro assessment, substantial optimization is needed. Understanding the many miRNA delivery methods, their benefits, and their drawbacks is therefore crucial.
Lung cancer stands as a formidable contributor to global mortality rates. Its onset often involves the dysregulation of vital genetic mechanisms, marked by the downregulation of tumor suppressor genes or the overexpression of oncogenes. MiRNAs, intricate regulators of gene expression, play a pivotal role in this intricate landscape. Exosomes derived from lung cancer tumors harbor a diverse repertoire of miRNAs, exhibiting both oncogenic and tumor-suppressive characteristics. Tumor-suppressive miRNAs, if augmented within the context of lung cancer, hold promise in curtailing cell proliferation and impeding metastasis. Conversely, the strategic application of inhibitors targeting oncomirs emerges as a potential avenue for developing effective cancer therapies. Progressing towards a cure for lung cancer involves multifaceted approaches. One such approach entails the augmentation of tumor suppressor genes like PTEN, SOCS1, and PHLP22, aiming to reinstate their functionality. Alternatively, the downregulation of oncogenes such as NUAK1, Snai1, AEG1, Vimentin, and PEA-15/PED represents another viable strategy to impede cancer progression.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design by Shahid Hussain and Habib Bukhari. Muhammad Adnan Shereen and Aiman Fatima contributed in contents collection. Shaukat Iqbal Malik and Sundas Ijaz helped in Figures and Tables. Xing-Xing Fan contributed in manuscript preparation. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Padinharayil H, Varghese J, John MC, Rajanikant GK, Wilson CM, Al-Yozbaki M, et al. Non-small cell lung carcinoma (NSCLCimplications on molecular pathology and advances in early diagnostics and therapeutics. Genes Dis. 2023;10(3):960–89. [Google Scholar] [PubMed]
2. Hussain S, Malik SI. Proliferative effects of lung cancer cells derived exosomes on T cells. Braz Arch Biol Technol. 2022;65:e22210234. doi:10.1590/1678-4324-2022210234. [Google Scholar] [CrossRef]
3. Saab S, Zalzale H, Rahal Z, Khalifeh Y, Sinjab A, Kadara H. Insights into lung cancer immune-based biology, prevention, and treatment. Front Immunol. 2020;11:159. [Google Scholar] [PubMed]
4. Adams SJ, Stone E, Baldwin DR, Vliegenthart R, Lee P, Fintelmann FJ. Lung cancer screening. Lancet. 2023;401(10374):390–408. [Google Scholar] [PubMed]
5. Hill M, Tran N. miRNA interplay: mechanisms and consequences in cancer. Dis Model Mech. 2021;14(4):dmm047662. [Google Scholar] [PubMed]
6. Hussain S, Malik S. Effects of tumor-derived exosomes on T cells markers expression. Braz J Biol. 2022;84:e250556. doi:10.1590/1519-6984.250556. [Google Scholar] [PubMed] [CrossRef]
7. de las Mercedes Carro M, Grimson A, Cohen PE. Small RNAs and their protein partners in animal meiosis. Curr Top in Dev Bio; 2023;151:245–79. [Google Scholar]
8. Vergani-Junior CA, Tonon-da-Silva G, Inan MD, Mori MA. DICER: structure, function, and regulation. Biophys Rev. 2021;13(6):1081–90. [Google Scholar] [PubMed]
9. Iwakawa H, Tomari Y. Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol Cell. 2022;82(1):30–43. [Google Scholar] [PubMed]
10. Zhang X, Wang L, Li H, Zhang L, Zheng X, Cheng W. Crosstalk between noncoding RNAs and ferroptosis: new dawn for overcoming cancer progression. Cell Death Dis. 2020;11(7):580. [Google Scholar] [PubMed]
11. Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, et al. Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441–51. [Google Scholar] [PubMed]
12. Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020;8:409. [Google Scholar] [PubMed]
13. Tang S, Li S, Liu T, He Y, Hu H, Zhu Y, et al. MicroRNAs: emerging oncogenic and tumor-suppressive regulators, biomarkers and therapeutic targets in lung cancer. Cancer Lett. 2021;502:71–83. [Google Scholar] [PubMed]
14. Tomasello L, Distefano R, Nigita G, Croce CM. The microRNA family gets wider: the IsomiRs classification and role. Front Cell Dev Biol. 2021;9:668648. [Google Scholar] [PubMed]
15. Ergin K, Çetinkaya R. Regulation of microRNAs. miRNomics. 2022;1–32. [Google Scholar]
16. de Rooij LA, Mastebroek DJ, Ten Voorde N, van der Wall E, van Diest PJ, Moelans CB. The microRNA Lifecycle in Health and Cancer. Canc. 2022;14(23):5748. [Google Scholar]
17. Wang P, Zhou Y, Richards AM. Effective tools for RNA-derived therapeutics: siRNA interference or miRNA mimicry. Therano. 2021;11(18):8771–96. [Google Scholar]
18. Yang B, McJunkin K. CRISPR screening strategies for microRNA target identification. FEBS J. 2020;287(14):2914–22. [Google Scholar] [PubMed]
19. Hussen BM, Hidayat HJ, Salihi A, Sabir DK, Taheri M, Ghafouri-Fard S. MicroRNA: a signature for cancer progression. Biomed Pharmacother. 2021;138(24):111528. [Google Scholar] [PubMed]
20. Nicolini A, Ferrari P, Biava PM. Exosomes and cell communication: from tumour-derived exosomes and their role in tumour progression to the use of exosomal cargo for cancer treatment. Cancers. 2021;13(4):822. [Google Scholar] [PubMed]
21. Hussain S, Fatima A, Fan XX, Malik SI. The biological importance of cells secreted exosomes. Pakistan J Pharm Sci. 2021;34(6):2273–9. [Google Scholar]
22. Hussain S, Bokhari SEZ, Fan XX, Iqbal S. The role of exosomes derived miRNA in cancer. J of Pak Med Assoc. 2021;71(7):1856–61. [Google Scholar]
23. Gurunathan S, Kang MH, Kim JH. A comprehensive review on factors influences biogenesis, functions, therapeutic and clinical implications of exosomes. Int J Nanomed. 2021;16:1281–312. [Google Scholar]
24. Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10(1):42–6. [Google Scholar] [PubMed]
25. Zhang JG, Wang JJ, Zhao F, Liu Q, Jiang K, Yang GH. MicroRNA-21 (miR-21) represses tumor suppressor PTEN and promotes growth and invasion in non-small cell lung cancer (NSCLC). Clin Chim Acta. 2010;411(11–12):846–52. [Google Scholar] [PubMed]
26. Chen Y, Knösel T, Kristiansen G, Pietas A, Garber ME, Matsuhashi S, et al. Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol. 2003;200(5):640–6. [Google Scholar] [PubMed]
27. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28(17):5369–80. [Google Scholar] [PubMed]
28. Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378(3):492–504. [Google Scholar] [PubMed]
29. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282(19):14328–36. [Google Scholar] [PubMed]
30. Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18(3):350–9. [Google Scholar] [PubMed]
31. Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu FQ, et al. miR-106a promotes growth and metastasis of non-small cell lung cancer by targeting PTEN. Int J Clin Exp Pathol. 2015;8(4):3827–34. [Google Scholar] [PubMed]
32. Yuan Y, Ye J, Zhang X, Liu Z. LncRNA CASC2 regulate cell proliferation and invasion by targeting miR-155/SOCS1 axis in hepatocellular carcinoma. J Oncol. 2023;2023:8457112. doi:10.1155/2023/8457112. [Google Scholar] [PubMed] [CrossRef]
33. Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67(22):10782–8. [Google Scholar] [PubMed]
34. Sirajudeen F, Malhab LJ, Bustanji Y, Shahwan M, Alzoubi KH, Semreen MH, et al. Exploring the potential of rosemary derived compounds (rosmarinic and carnosic acids) as cancer therapeutics: current knowledge and future perspectives. Biomol Thera. 2024;32(1):38–55. [Google Scholar]
35. Kong W, Yang H, He L, Zhao JJ, Coppola D, Dalton WS, et al. MicroRNA-155 is regulated by the transforming growth factor β/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA. Mol Cell Biol. 2008;28(22):6773–84. [Google Scholar] [PubMed]
36. Cai J, Fang L, Huang Y, Li R, Yuan J, Yang Y, et al. miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73(17):5402–15. [Google Scholar] [PubMed]
37. Puissegur M, Mazure N, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, et al. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18(3):465–78. [Google Scholar] [PubMed]
38. Song Q, An Q, Niu B, Lu X, Zhang N, Cao X. Role of miR-221/222 in Tumor development and the underlying mechanism. J Oncol. 2019;2019:7252013. doi:10.1155/2019/7252013. [Google Scholar] [PubMed] [CrossRef]
39. Wu X, Huang J, Yang Z, Zhu Y, Zhang Y, Wang J, et al. MicroRNA-221-3p is related to survival and promotes tumour progression in pancreatic cancer: a comprehensive study on functions and clinicopathological value. Can Cell Int. 2020;20(1):1–25. [Google Scholar]
40. Yamashita R, Sato M, Kakumu T, Hase T, Yogo N, Maruyama E, et al. Growth inhibitory effects of miR-221 and miR-222 in non-small cell lung cancer cells. Cancer Med. 2015;4(4):551–64. [Google Scholar] [PubMed]
41. Mujahid S, Nielsen HC, Volpe MV. MiR-221 and miR-130a regulate lung airway and vascular development. PLoS One. 2013;8(2):e55911. [Google Scholar] [PubMed]
42. Garofalo M, Romano G, Di Leva G, Nuovo G, Jeon YJ, Ngankeu A, et al. Retracted article: EGFR and MET receptor tyrosine kinase-altered microRNA expression induces tumorigenesis and gefitinib resistance in lung cancers. Nature Med. 2012;18(1):74–82. [Google Scholar]
43. Campos A, Sharma S, Obermair A, Salomon C. Extracellular vesicle-associated miRNAs and chemoresistance: a systematic review. Can. 2021;13(18):4608. [Google Scholar]
44. Jiang F, Huang Q, Nian J, Yang S. miR-221/222/PUMA axis promotes oral squamous cell carcinoma apoptosis. Open J Stom. 2023;13(10):367–83. [Google Scholar]
45. Wang Y, Dai W, Chu X, Yang B, Zhao M, Sun Y. Metformin inhibits lung cancer cells proliferation through repressing microRNA-222. Biotechnol Lett. 2013;35:2013–9. [Google Scholar] [PubMed]
46. Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. [Google Scholar] [PubMed]
47. Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342(1):43–51. [Google Scholar] [PubMed]
48. Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci. 2008;105(10):3903–8. [Google Scholar] [PubMed]
49. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–22. [Google Scholar] [PubMed]
50. Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci. 2012;109(31):E2110–6. [Google Scholar] [PubMed]
51. Hsu Y, Hung J, Chang W, Lin Y, Pan Y, Tsai P, et al. Hypoxic lung cancer-secreted exosomal miR-23a increased angiogenesis and vascular permeability by targeting prolyl hydroxylase and tight junction protein ZO-1. Oncogene. 2017;36(34):4929–42. [Google Scholar] [PubMed]
52. Hsu YL, Hung JY, Chang WA, Jian SF, Lin YS, Pan YC, et al. Hypoxic lung-cancer-derived extracellular vesicle microRNA-103a increases the oncogenic effects of macrophages by targeting PTEN. Mol Ther. 2018;26(2):568–81. [Google Scholar] [PubMed]
53. Cui H, Seubert B, Stahl E, Dietz H, Reuning U, Moreno-Leon L, et al. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2015;34(28):3640–50. [Google Scholar] [PubMed]
54. Rana S, Malinowska K, Zöller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15(3):281–95, IN14–31. [Google Scholar] [PubMed]
55. Aylon Y, Bublik D, Moskovits N, Toperoff G, Azaiza D, Biagoni F, et al. Reactivation of epigenetically silenced miR-512 and miR-373 sensitizes lung cancer cells to cisplatin and restricts tumor growth. Cell Death Differ. 2015;22(8):1328–40. [Google Scholar] [PubMed]
56. Berchem G, Noman MZ, Bosseler M, Paggetti J, Baconnais S, Le Cam E, et al. Hypoxic tumor-derived microvesicles negatively regulate NK cell function by a mechanism involving TGF-β and miR23a transfer. Oncoimmunol. 2016;5(4):e1062968. [Google Scholar]
57. Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31(17):3513–23. [Google Scholar] [PubMed]
58. Liu Y, Luo F, Wang B, Li H, Xu Y, Liu X, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–35. [Google Scholar] [PubMed]
59. Liang CY, Li ZY, Gan TQ, Fang YY, Gan BL, Chen WJ, et al. Downregulation of hsa-microRNA-204-5p and identification of its potential regulatory network in non-small cell lung cancer: RT-qPCR, bioinformatic-and meta-analyses. Respir Res. 2020;21(1):1–27. [Google Scholar]
60. Yang F, Bian Z, Xu P, Sun S, Huang Z. MicroRNA-204-5p: a pivotal tumor suppressor. Cancer Med. 2023;12(3):3185–200. [Google Scholar] [PubMed]
61. Imam JS, Plyler JR, Bansal H, Prajapati S, Bansal S, Rebeles J, et al. Genomic loss of tumor suppressor miRNA-204 promotes cancer cell migration and invasion by activating AKT/mTOR/Rac1 signaling and actin reorganization. PLoS One. 2012;7(12):e52397. [Google Scholar] [PubMed]
62. Du X, Qi F, Lu S, Li Y, Han W. Nicotine upregulates FGFR3 and RB1 expression and promotes non-small cell lung cancer cell proliferation and epithelial-to-mesenchymal transition via downregulation of miR-99b and miR-192. Biomed Pharmacother. 2018;101(1):656–62. [Google Scholar] [PubMed]
63. Popper H. Primary tumor and metastasis—sectioning the different steps of the metastatic cascade. Transl Lung Cancer Res. 2020;9(5):2277–300. [Google Scholar] [PubMed]
64. Huang F, Li Y, Guan L, Hu Y, Zeng M. MiR-30a inhibits silica dust-induced epithelial-mesenchymal transition by targeting Snail. Toxicol In Vitro. 2023;92:105657. [Google Scholar] [PubMed]
65. Zhang T, Zhou Y, You B, You Y, Yan Y, Zhang J, et al. miR-30a-5p inhibits epithelial-to-mesenchymal transition by targeting CDK6 in nasal polyps. Am J Rhinol Allergy. 2021;35(2):152–63. [Google Scholar] [PubMed]
66. Liu K, Guo L, Guo Y, Zhou B, Li T, Yang H, et al. AEG-1 3′-untranslated region functions as a ceRNA in inducing epithelial-mesenchymal transition of human non-small cell lung cancer by regulating miR-30a activity. Eur J Cell Biol. 2015;94(1):22–31. [Google Scholar] [PubMed]
67. Wani JA, Majid S, Imtiyaz Z, Rehman MU, Alsaffar RM, Shah NN, et al. MiRNAs in lung cancer: diagnostic, prognostic, and therapeutic potential. Diagnostics. 2022;12(7):1610. [Google Scholar] [PubMed]
68. Garinet S, Didelot A, Denize T, Perrier A, Beinse G, Leclere JB, et al. Clinical assessment of the miR-34, miR-200, ZEB1 and SNAIL EMT regulation hub underlines the differential prognostic value of EMT miRs to drive mesenchymal transition and prognosis in resected NSCLC. Br J Cancer. 2021;125(11):1544–51. [Google Scholar] [PubMed]
69. Cui D, Qu R, Liu D, Xiong X, Liang T, Zhao Y. The cross talk between p53 and mTOR pathways in response to physiological and genotoxic stresses. Front Cell Dev Biol. 2021;9:775507. [Google Scholar] [PubMed]
70. Naghizadeh S, Mohammadi A, Duijf PH, Baradaran B, Safarzadeh E, Cho WCS, et al. The role of miR-34 in cancer drug resistance. J Cell Physiol. 2020;235(10):6424–40. [Google Scholar] [PubMed]
71. Wang C, Wang X, Liang H, Wang T, Yan X, Cao M, et al. miR-203 inhibits cell proliferation and migration of lung cancer cells by targeting PKCα. PLoS One. 2013;8(9):e73985. [Google Scholar] [PubMed]
72. Incoronato M, Garofalo M, Urso L, Romano G, Quintavalle C, Zanca C, et al. MiR-212 increases tumor necrosis factor-related apoptosis-inducing ligand sensitivity in non-small cell lung cancer by targeting the antiapoptotic protein PED. Cancer Res. 2010;70(9):3638–46. [Google Scholar] [PubMed]
73. Condorelli G, Vigliotta G, Cafieri A, Trencia A, Andalò P, Oriente F, et al. PED/PEA-15: an anti-apoptotic molecule that regulates FAS/TNFR1-induced apoptosis. Oncogene. 1999;18(31):4409–15. [Google Scholar] [PubMed]
74. Park J, Tacam MJ, Chauhan G, Cohen EN, Gagliardi M, Iles LR, et al. Nonphosphorylatable PEA15 mutant inhibits epithelial-mesenchymal transition in triple-negative breast cancer partly through the regulation of IL-8 expression. Br Can Res Treat. 2021;189(2):333–45. [Google Scholar]
75. Wu Y, Li X, Chen M, Liu Z, Zhang X, Zheng S, et al. Phosphorylation of PED/PEA-15 at Ser116 and phosphorylation of p27 at Thr187 indicates a poor prognosis in hepatocellular carcinoma. Onco Lett. 2021;21(3):177. [Google Scholar] [PubMed]
76. Cornett AL, Lutz CS. Regulation of COX-2 expression by miR-146a in lung cancer cells. RNA. 2014;20(9):1419–30. [Google Scholar] [PubMed]
77. Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y, et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6(28):25533–74. [Google Scholar] [PubMed]
78. Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70(14):5923–30. [Google Scholar] [PubMed]
79. Zhang JX, Yang W, Wu JZ, Zhou C, Liu S, Shi HB, et al. MicroRNA-32-5p inhibits epithelial-mesenchymal transition and metastasis in lung adenocarcinoma by targeting SMAD family 3. J Cancer. 2021;12(8):2258–67. [Google Scholar] [PubMed]
80. Weidle UH, Birzele F, Nopora A. MicroRNAs as potential targets for therapeutic intervention with metastasis of non-small cell lung cancer. Cancer Genom Proteom. 2019;16(2):99–119. [Google Scholar]
81. Lee JS, Ahn YH, Won HS, Sun DS, Kim YH, Ko YH. Prognostic role of the microRNA-200 family in various carcinomas: a systematic review and meta-analysis. BioMed Res Int. 2017;2017(3):1–11. [Google Scholar]
82. Wang X, Cao L, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review). Oncol Lett. 2012;3(5):955–60. [Google Scholar] [PubMed]
83. Khandelwal A, Sharma U, Barwal TS, Seam RK, Gupta M, Rana MK, et al. Circulating miR-320a acts as a tumor suppressor and prognostic factor in non-small cell lung cancer. Front Oncol. 2021;11:645475. [Google Scholar] [PubMed]
84. Lee SB, Park YS, Sung JS, Lee JW, Kim B, Kim YH. Tumor suppressor miR-584-5p inhibits migration and invasion in smoking related non-small cell lung cancer cells by targeting YKT6. Cancers. 2021;13(5):1159. [Google Scholar] [PubMed]
85. Yang Z, Fu S, Li Y, Liang Y, Hao M, Guo R, et al. Exosome as non-invasive prognostic and diagnostic biomarker and nanovesicle for targeted therapy of non-small cell lung carcinoma. Chem Eng J. 2023;480:148160. doi:10.1016/j.cej.2023.148160. [Google Scholar] [CrossRef]
86. Durendez-Saez E, Torres-Martinez S, Calabuig-Farinas S, Meri-Abad M, Ferrero-Gimeno M, Camps C. Exosomal microRNAs in non-small cell lung cancer. Trans Cancer Res. 2021;10(6):3128–39. [Google Scholar]
87. Dama E, Melocchi V, Mazzarelli F, Colangelo T, Cuttano R, Di Candia L, et al. Non-coding RNAs as prognostic biomarkers: a miRNA signature specific for aggressive early-stage lung adenocarcinomas. Non-Coding RNA. 2020;6(4):48. [Google Scholar] [PubMed]
88. Gayosso-Gómez LV, Ortiz-Quintero B. Circulating microRNAs in blood and other body fluids as biomarkers for diagnosis, prognosis, and therapy response in lung cancer. Diagnostics. 2021;11(3):421. [Google Scholar]
89. Cueto AFV, Álvarez L, García M, Álvarez-Barrios A, Artime E, Cueto LFV, et al. Candidate glaucoma biomarkers: from proteins to metabolites, and the pitfalls to clinical applications. Biol. 2021;10(8):763. [Google Scholar]
90. Peng M, Huang Q, Yin W, Tan S, Chen C, Liu W, et al. Circulating tumor DNA as a prognostic biomarker in localized non-small cell lung cancer. Front Oncol. 2020;10:561598. [Google Scholar] [PubMed]
91. You L, Fan Y, Liu X, Shao S, Guo L, Noreldeen HA, et al. Liquid chromatography—mass spectrometry—based tissue metabolic profiling reveals major metabolic pathway alterations and potential biomarkers of lung cancer. J Proteome Res. 2020;19(9):3750–60. [Google Scholar] [PubMed]
92. Li M, Marin-Muller C, Bharadwaj U, Chow KH, Yao Q, Chen C. MicroRNAs: control and loss of control in human physiology and disease. Wor J of surg. 2009;33:667–84. [Google Scholar]
93. Raczkowska J, Bielska A, Krętowski A, Niemira M. Extracellular circulating miRNAs as potential non-invasive biomarkers in non-small cell lung cancer patients. Front Oncol. 2023;13:1209299. [Google Scholar] [PubMed]
94. Markou A, Tsaroucha EG, Kaklamanis L, Fotinou M, Georgoulias V, Lianidou ES. Prognostic value of mature microRNA-21 and microRNA-205 overexpression in non-small cell lung cancer by quantitative real-time RT-PCR. Clin Chem. 2008;54(10):1696–704. [Google Scholar] [PubMed]
95. Zhu W, Xu B. MicroRNA-21 identified as predictor of cancer outcome: a meta-analysis. PLoS One. 2014;9(8):e103373. [Google Scholar] [PubMed]
96. Wang W, Li X, Liu C, Zhang X, Wu Y, Diao M, et al. MicroRNA-21 as a diagnostic and prognostic biomarker of lung cancer: a systematic review and meta-analysis. Biosci Rep. 2022;42(5):BSR20211653. [Google Scholar] [PubMed]
97. Tang D, Shen Y, Wang M, Yang R, Wang Z, Sui A, et al. Identification of plasma microRNAs as novel noninvasive biomarkers for early detection of lung cancer. Eur J Cancer Prev. 2013;22(6):540–8. [Google Scholar] [PubMed]
98. Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, et al. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol. 2012;29(2):618–26. [Google Scholar] [PubMed]
99. Geng Q, Fan T, Zhang B, Wang W, Xu Y, Hu H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir Res. 2014;15(1):1–9. [Google Scholar]
100. Condrat CE, Thompson DC, Barbu MG, Bugnar OL, Boboc A, Cretoiu D, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2):276. [Google Scholar] [PubMed]
101. Xu J, Wu C, Che X, Wang L, Yu D, Zhang T, et al. Circulating microRNAs, miR-21, miR-122, and miR-223, in patients with hepatocellular carcinoma or chronic hepatitis. Mol Carcinog. 2011;50(2):136–42. [Google Scholar] [PubMed]
102. Lee JH, Voortman J, Dingemans AMC, Voeller DM, Pham T, Wang Y, et al. MicroRNA expression and clinical outcome of small cell lung cancer. PLoS One. 2011;6(6):e21300. [Google Scholar] [PubMed]
103. Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lun Can. 2016;7:19–27. doi:10.2147/LCTT.S60341. [Google Scholar] [PubMed] [CrossRef]
104. Segal M, Slack FJ. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15(9):987–91. [Google Scholar] [PubMed]
105. Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools