 Open Access
Open Access
ARTICLE
LncRNA LOC103694972 promotes fibrosis of NRK-49F cells by regulating STAT3-dependent Smad/CTGF pathway via targeting miR-29c-3p
1 Department of Vascular and Tumor Intervention, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
2 Department of Intensive Care Unit, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
3 Department of Nursing Research, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
4 Department of Intensivie Care Unit, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
5 Department of Endocrinology, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
6 Department of Preventive Treatment Center, The First Hospital of Hunan University of Chinese Medicine, Changsha, 410007, China
* Corresponding Author: XINYU CHEN. Email:
BIOCELL 2024, 48(3), 501-511. https://doi.org/10.32604/biocell.2023.030854
Received 27 April 2023; Accepted 18 October 2023; Issue published 15 March 2024
Abstract
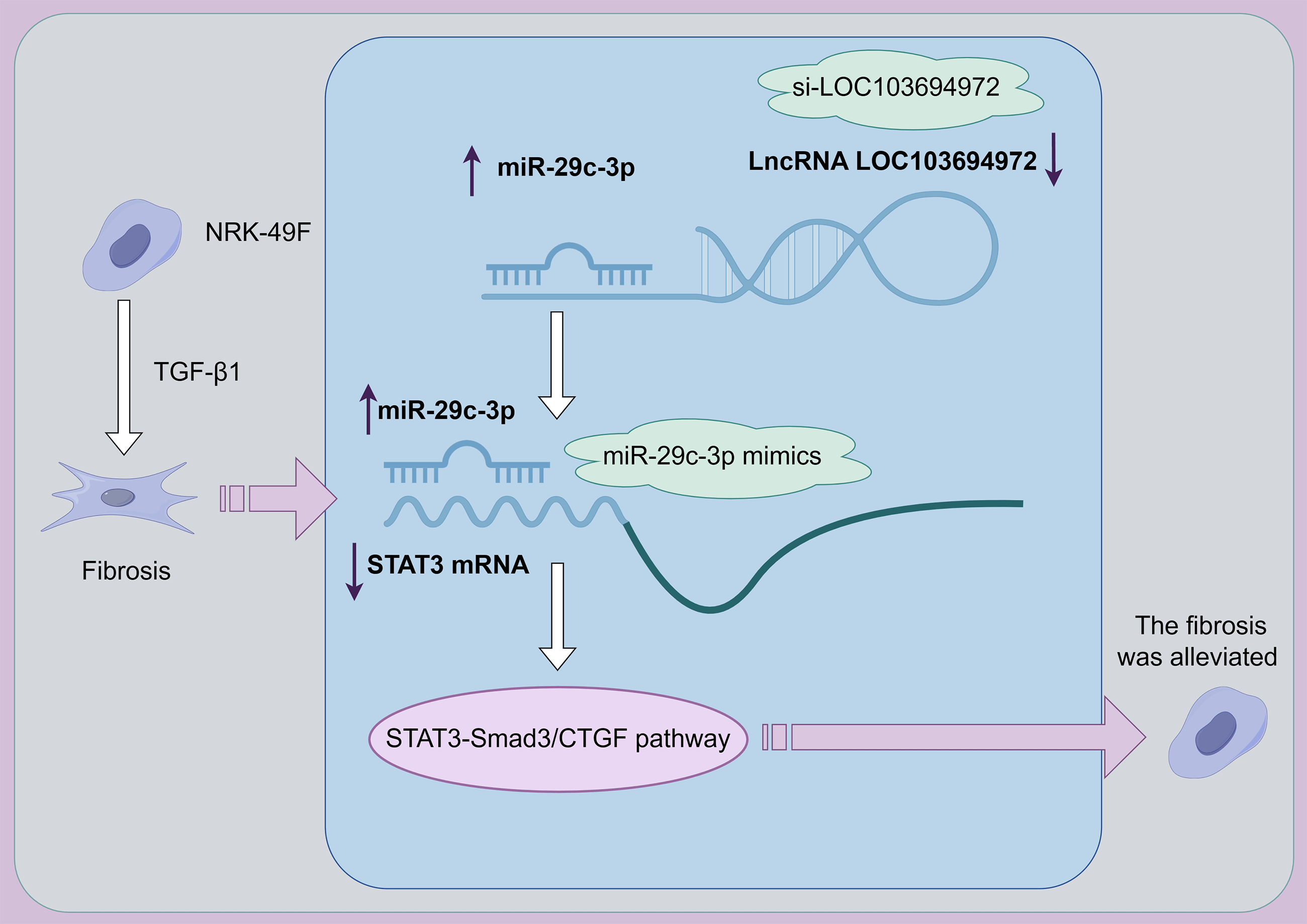
Background: Renal fibrosis is an important process in the development of chronic kidney disease. Understanding the pathogenesis and finding effective treatments for renal fibrosis is crucial. This study aims to investigate whether a newly discovered long non-coding RNA (lncRNA) called LOC103694972 could be a potential target for treating fibrosis of NRK-49F cells. Methods:: LncRNA Chip was used to identify differentially expressed lncRNAs between TGF-β1-induced NRK-49F cells and normal cells. The dual-luciferase assay confirmed the binding between miR-29c-3p and signal transducer and activator of transcription (STAT3), as well as between miR-29c-3p and lncRNA LOC103694972. Si-LOC103694972 and miR-29c-3p mimic were then transfected into TGF-β1-induced NRK-49F cells. Results: The study found that LOC103694972 was highly expressed in TGF-β1-induced NRK-49F cells. These cells exhibited increased cell length and activity compared to the control group. The expression levels of Collagen I, α-Smooth muscle actin (α-SMA), and tissue inhibitor of metalloproteinase (TIMP-1) were increased, while matrix Metalloproteinase 2 (MMP2) and matrix Metalloproteinase 9 (MMP9) expression was decreased. However, transfection with si-LOC103694972 and miR-29c-3p mimics restored cell morphology and reduced cell viability. This led to a decrease in the levels of Collagen I, α-SMA, and TIMP-1, as well as an increase in MMP2 and MMP9 expression. Additionally, TGF-β1-induced NRK-49F cells transfected with miR-29c-3p mimics activated the STAT3-Smad3/CTGF pathway. Conclusion: Based on these findings, lncRNA LOC103694972 shows promise as a target for treating renal fibrosis. It negatively regulates miR-29c-3p and activates the STAT3-Smad3/CTGF pathway.Graphic Abstract
Keywords
Chronic kidney disease (CKD) causes an increasing social burden, with high prevalence and high mortality [1]. In the early stages of CKD, patients may not experience any symptoms, but as the disease progresses, complications such as renal insufficiency may arise [2,3]. Presently, the only available clinical treatments for CKD are conservative measures and alternative therapies [2]. Hence, it is crucial to urgently explore the pathogenesis and treatment of CKD. Renal fibrosis, a prevalent progressive process in CKD and end-stage renal disease, involves myofibroblasts as the primary effector cells, and the presence of agglomerated lesions in the renal interstitium represents a significant pathological characteristic of fibrosis [4]. α-Smooth muscle actin (α-SMA) and Collagen I are markers of myofibroblasts. Renal fibrosis develops through the accumulation of scar tissue in the parenchyma [3,5], which correlates with the process of intrinsic cell fibrosis and stiffening in the kidney [6]. Previous studies have identified various mechanisms linked to renal fibrosis, yet no definitive strategy has been established to effectively delay its progression [3,5].
Non-coding RNA (ncRNA) refers to RNA that does not code proteins, including miRNAs, lncRNAs and circ-RNAs. These types of RNA do not require translation into proteins to carry out their biological functions [7]. LncRNA, a subtype of ncRNA, is characterized by its length, which is typically greater than 200 nucleotides. Advancements in chip technology have led to the discovery of numerous lncRNAs that are implicated in renal fibrosis [3,8]. Currently, several lncRNAs have been confirmed to have significant associations with renal fibrosis. Overexpression of LncRNA lnc-ISG20 has been shown to decrease miR-486-5p levels, thereby promoting renal fibrosis [9]. Overexpression of lncRNA GAS5 has been found to suppress renal fibrosis through the regulation of Smad3/miRNA-142-5p aixs [10]. In our study, we have identified multiple lncRNAs, including lncRNA LOC103694972, and investigated its mechanism of action in NRK-49F cell fibrosis through lncRNA chip analysis.
Transforming growth factor-β (TGF-β) is widely recognized as a crucial regulator of renal fibrosis [11]. In various kidney diseases, TGF-β expression in the kidney is increased and positively correlated with the degree of fibrosis [12]. miR-29c, a key target in fibrotic diseases, is down-regulated in conditions such as renal fibrosis, liver fibrosis, and pulmonary fibrosis, and is considered to possess anti-fibrotic effects [13]. As a pleiotropic transcription factor, signal transducer and activator of transcription (STAT3) induces several fibrotic gene expressions, including TGF-β [14]. Studies have revealed that miR-29c-3p can suppress the growth of cardiac fibroblasts by directly targeting STAT3 expression [15]. Connective tissue growth factor (CTGF) is a well-known gene that promotes fibrosis, and its expression is up-regulated in renal fibrosis [16]. In fibroblasts, TGF-β1-induced up-regulation of CTGF depends on Smad3, while STAT3 is involved in the activation of CTGF [17]. The deposition of extracellular matrix (ECM) proteins played a central role in fibrosis [17]. TGF-β1 can activate Smad3, leading to ECM synthesis [18]. Furthermore, TGF-β1 inhibits the degradation of extracellular matrix (ECM) by suppressing metalloproteinases (MMPs) and inducing tissue inhibitors of metalloproteinases (TIMPs).
This study aimed to investigate the role and mechanism of lncRNA LOC103694972 in regulating fibrosis in NRK-49F cells. Specifically, it explored the interactions between miR-29c-3p, lncRNA LOC103694972, and STAT3. This study aims to elucidate the effectiveness of lncRNA LOC103694972 targeted therapy for cell fibrosis.
NRK-49F cells (Shanghai Tongpai Biotechnology Co., Ltd., Shanghai, China) were cultured in DMEM containing 15% FBS and 1% penicillin-streptomycin. The cells were incubated at 37°C in a 5% CO2 environment with saturated humidity. Initially, the NRK-49F cells were divided into two groups: the Control group and the TGF-β1 group. The cells in the Control group were cultured normally, while the TGF-β1 group cells were treated with 10 ng/mL TGF-β1 for 48 h. Subsequently, the cells were divided into four groups, the Control group, the TGF-β1 group, the TGF-β1+si-NC group, and the TGF-β1+si-LOC103694972 group. The Control group and TGF-β1 group were treated as before. Cells in the si-NC+TGF-β1 group and the si-LOC103694972+TGF-β1 group were transfected with si-NC plasmid and si-LOC103694972 plasmid, respectively, for 48 h, followed by treatment with 10 ng/mL TGF-β1 for an additional 48 h. The sequences of the si-NC is 5′-UUCUCCGAACGUGUCACGUTT-3′. And the sequences of the si-LOC103694972 is 5′-CACTTAACGTCACGCTAGT-3′. The plasmids were purchased from Honorgene. After the predetermined time, the cell morphology was observed under a microscope. Subsequently, the cells and cell culture supernatant were collected for further testing.
After collecting the cells from each group, they were immediately frozen in liquid nitrogen and subsequently stored at a temperature of −80°C. The quality of RNA and its concentration were assessed using the NanoDrop ND-1000 system. Additionally, the samples were labeled using the array start flash RNA labeling kit. The chip and RNA were then subjected to hybridization using the Agilent SureHyb system. The Agilent DNA Microarray Scanner was employed to wash and scan the chip, and the signal values were collected using the Agilent Feature Extraction software.
Cell counting kit 8 (CCK8) assay
The cells were seeded into 96-well plates and subjected to the designated treatments as per the grouping mentioned above. Upon reaching the predefined time point, the culture medium was aspirated and discarded using a pipette gun. Subsequently, each well was supplemented with 10 μL of a cck8 working solution (NU679, Tongren Chemical Research Institute, kumamoto, Japan) and 90 μL of serum-free DMEM medium. The plates were then placed in a multifunctional microplate reader, and the absorbance at 450 nm was measured after a 4-h incubation period.
Quantitative real-time PCR (RT-qPCR)
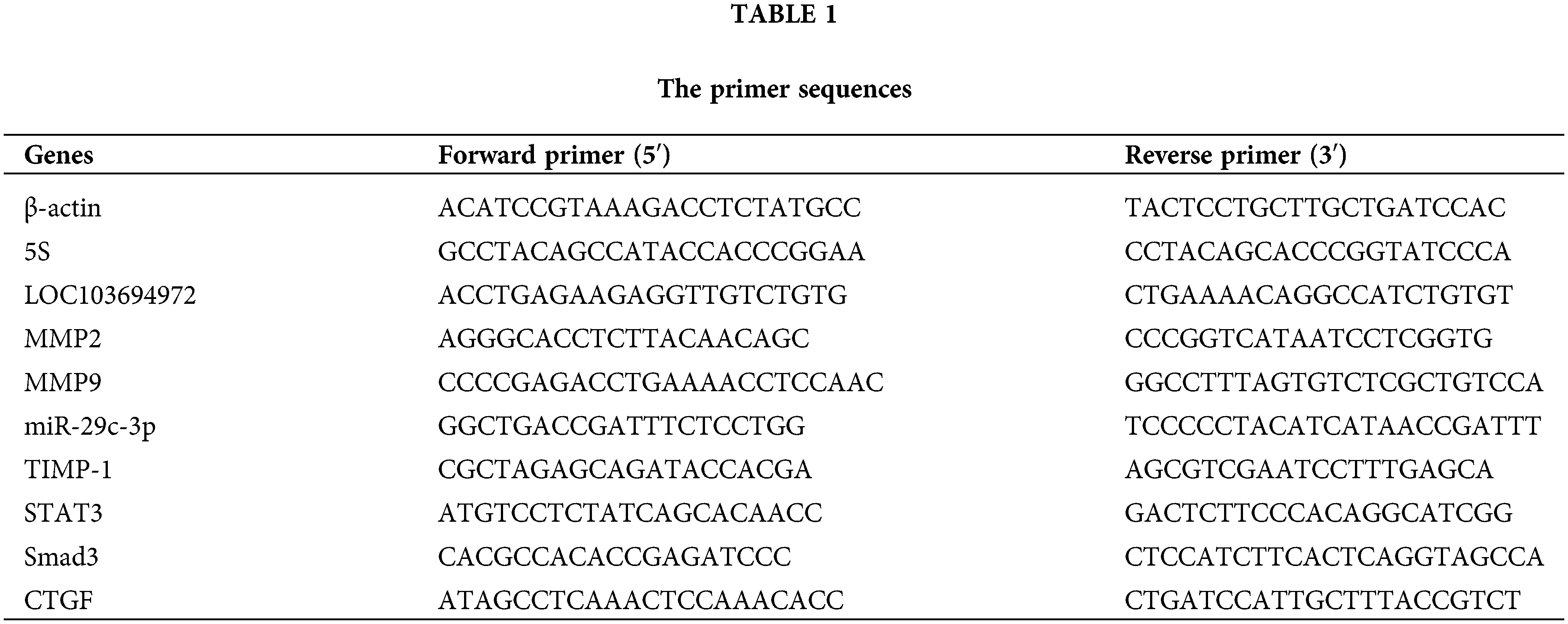
Trizol (15596026, Thermofisher, USA) was employed to extract total RNA from cells. miRNA was reverse-transcribed into cDNA using the miRNA cDNA Synthesis kit (CW2141, CWBIO, Beijing, China), while mRNA was reverse-transcribed into cDNA using the HiFscript cDNA Synthesis kit (CW2569, CWBIO, Beijing, China). All primers used were synthesized by Sangon Biotech and their sequences are provided in Table 1. The samples were mixed with UltraSYBR Mixture (CW2601, CWBIO, Beijing, China) to create a mixture, and the relative expression of RNAs was analyzed on the QuantStudio1 Real-Time PCR System (Thermo Fisher, Massachusetts, USA). The relative RNA levels were calculated using the 2−ΔΔCt method. β-actin served as an internal reference for mRNA and lncRNA, while 5S was used as an internal reference for miRNA.
The cells were lysed with RIPA lysis buffer after collection. BCA protein quantification kits were used to measure the protein content in the cells. The proteins were then separated using SDS-PAGE electrophoresis and transferred to a PVDF membrane. The PVDF membrane was incubated overnight at 4°C with primary antibodies MMP9 (ab76003, Abcam, Cambridge, UK), MMP2 (ab76003, Abcam, Cambridge, UK), TIMP-1 (ab211926, Abcam, Cambridge, UK), CollagenI (ab270993, Abcam, Cambridge, UK), α-SMA (ab21027, Abcam, Cambridge, UK), STAT3 (ab109085, Abcam, Cambridge, UK), Smad3 (ab40854, Abcam, Cambridge, UK), p-STAT3 (ab267373, Abcam, Cambridge, UK), p-Smad3 (ab52903, Abcam, Cambridge, UK), and β-actin (66009-1-Ig, Proteintech, Chicago, USA) as the internal control. Subsequently, the membranes were incubated with appropriate secondary antibodies HRP goat Anti-Rabbit IgG, HRP goat anti-mouse IgG, and HRP goat anti-Goat IgG at 37°C for 60 min. Lastly, the membranes were incubated with ECL chemiluminescence solution (Abiowell, Changsha, China) for 1 min, and then images were captured using a chemiluminescence imaging system instrument.
miRanda (http://www.bioinformatics.com.cn/local_miranda_miRNA_target_prediction_120) and TargetScan v.80 (https://www.targetscan.org/vert_80/) were explored to predict the targeted binding site. We employed the dual-luciferase reporter plasmids psiCHECK-2-LOC972 and psiCHECK-2-LOC972-Mut for LncRNA, as well as the dual-luciferase reporter plasmids pHG-MirTarget-STAT3-3U and pHG-MirTarget-STAT3-MUT-3U for STAT3. Additionally, we transfected the miR-29c-3p overexpression plasmid miR-29c-3p mimics and the corresponding empty plasmid miR-NC into the cells. These plasmids were obtained from (HonorGene, Changsha, China).
The dual-luciferase detection kit was used according to the provided protocols to lyse the different cell groups with PLB cell lysate. Subsequently, 20 μL of the resulting cell lysates were mixed with 100 μL of LARII solution, and the activity of firefly luciferase was measured using a dual-luciferase detection instrument. Then, 100 μL of Stop&Glo detection solution was added to the mixed solution and subjected to the double luciferase detection instrument to measure the luciferase activity of the sea kidney.
Enzyme-linked immunosorbent (Elisa) assay
The cell culture supernatant from each cell group was collected. The concentrations of MMP2 (CSB-E07411r, Cusabio, Wuhan, China), MMP9 (CSB-E08008r, Cusabio, Wuhan, China), and TIMP-1 (CSB-E08005r, Cusabio, Wuhan, China) in the cell supernatant were determined using ELISA Kits. The absorbance values of each group were measured using a multifunctional enzyme labeling analyzer (Huisong, Shenzhen, China).
Immunofluorescence (IF) staining
The cells were evenly seeded on the slides and grew adherently after 24 h. After fixation with 4% paraformaldehyde, the cells underwent three washes with PBS. Next, the permeabilization of cells was achieved with 0.3% Triton for 30 min, followed by blocking with 5% BSA for 60 min at 37°C. The cells were then incubated overnight at 4°C with primary antibodies Collagen I (14695-1-AP, Proteintech, Chicago, USA) and α-SMA (BM0002, BOSTER, Wuhan, China). Following this, the cells were exposed to secondary antibodies CoraLite488-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (SA00013-2, proteintech, Chicago, USA) and CoraLite488-conjugated Affinipure Goat Anti-Mouse IgG (H+L) (SA00013-1, proteintech, Chicago, USA) for 90 min at 37°C. Nuclei were stained with DAPI (AWI0331a, Abiowell, Changsha, China) for 5 min. After mounting the slides with buffered glycerol, they were observed and photographed using a microscope.
In cell experiments, data was analyzed using GraphPad Prism 9. The Shapiro-Wilk test was employed to determine conformity of the data to a normal distribution. The t-test was used to compare data between two groups, while one-way ANOVA was used to compare differences between more than two groups. Statistical significance was defined as p < 0.05. In miRNA-seq, the original data was standardized and analyzed using GeneSpring GX v12.1 software. Differentially expressed lncRNAs and mRNAs between the two samples were identified using a Fold change ≥2.0 criteria.
TGF-β1 was employed to induce NRK-49F cells for the purpose of cell morphology modification. Following induction, cell morphology was observed using a microscope (Fig. 1A). The elongated cell morphology observed after TGF-β1 induction indicated cellular fibrosis. Fig. 1B demonstrated that Collagen I and α-SMA expressions were upregulated in the TGF-β1 group compared to the Control group, suggesting the transformation of fibroblasts into myofibroblasts induced by TGF-β1. Additionally, the results depicted in Fig. 1C displayed a decrease in fibrosis-related cytokines matrix metalloproteinase 2 (MMP2) and matrix Metalloproteinase 9 (MMP9) levels, along with an increase in tissue inhibitor of metalloproteinase (TIMP-1) content after TGF-β1 induction. These findings further elucidated the cellular fibrosis process.
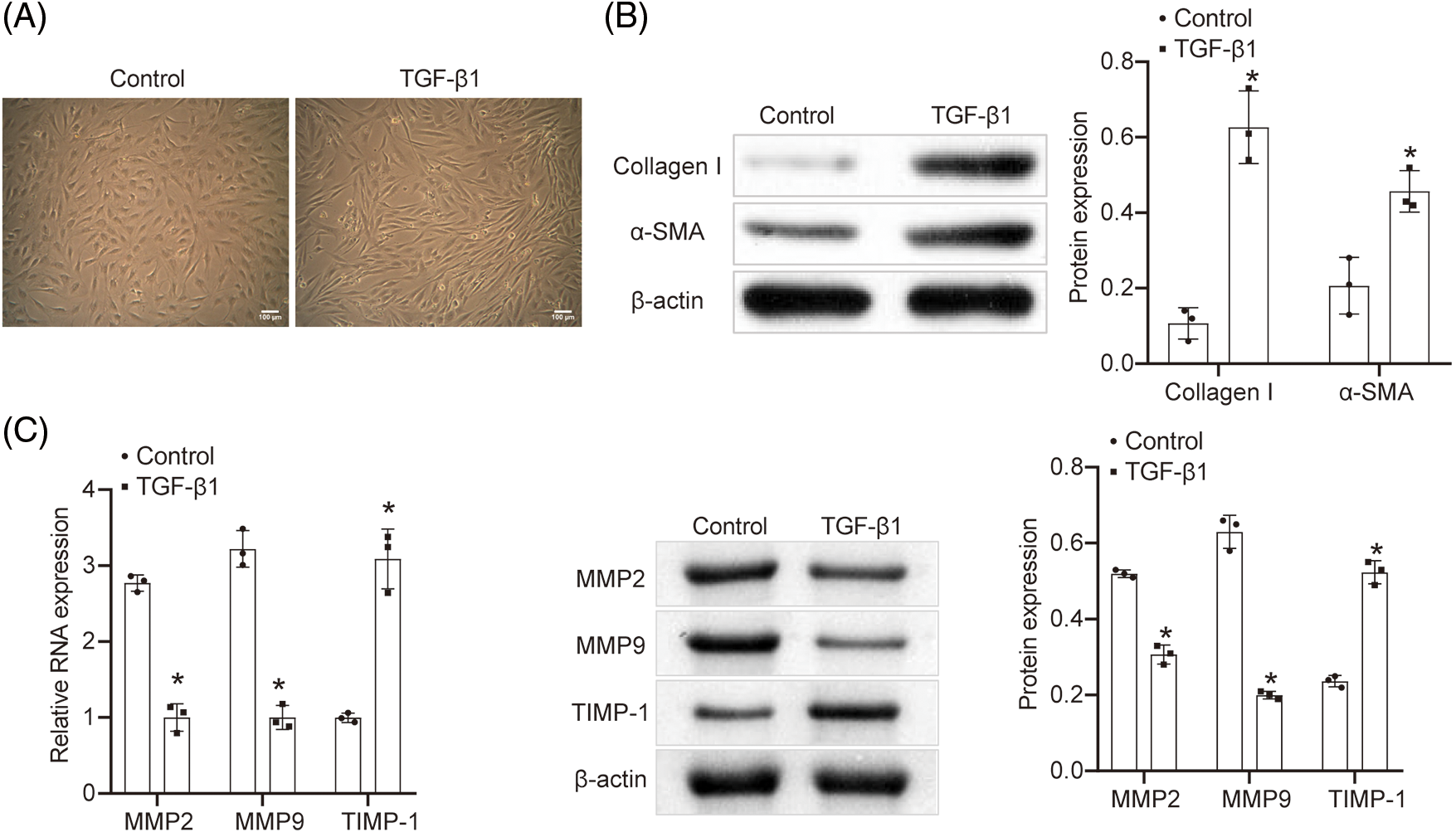
Figure 1: Construction of fibrosis model in vitro. (A) Cell morphology was observed under the microscope (scale bar = 100 μm). n = 3. (B) WB was utilized to detect the content of Collagen I and α-SMA. n = 3. (C) qRT-PCR and WB were explored to determine the content of MMP2, MMP9, and TIMP-1. n = 3. *p < 0.05 vs. the Control group.
Construction of fibrosis model in vitro
The regulatory relationship between lncRNA LOC103694972 and miR-29c-3p
To identify differentially expressed long non-coding RNAs (lncRNAs) in fibrotic cells, we isolated the cell mRNA samples from the Control group and the TGF-β1 group, and performed lncRNA chip analysis for detection. Fig. 2A displays a total of 135 differentially expressed lncRNAs (Fold change ≥ 2.0). Additionally, Fig. 2B exhibits 14 up-regulated lncRNAs in the TGF-β1 group with Fold change ≥ 3.0. Among these, the lncRNA LOC103694972 showed significant differential expression and was chosen as the target gene for subsequent experimental verification. We used miRanda and TargetScan v.80 to predict the binding sites of lncRNA LOC103694972 and miR-29c-3p (Fig. 2C). The results from dual-luciferase experiments confirmed the binding activity of miR-29c-3p mimics with LOC103694972 WT (Fig. 2D), indicating a targeting relationship between miR-29c-3p and lncRNA LOC103694972. Subsequently, we analyzed the expressions of lncRNA LOC103694972 and miR-29c-3p (Fig. 2E). Compared to the Control group, lncRNA LOC103694972 expression was up-regulated in the TGF-β1 group, while miR-29c-3p expression was down-regulated. Correlation analysis demonstrated a negative correlation between the expressions of lncRNA LOC103694972 and miR-29c-3p (Fig. 2F). Overall, our findings suggest that lncRNA LOC103694972 negatively regulates miR-29c-3p.
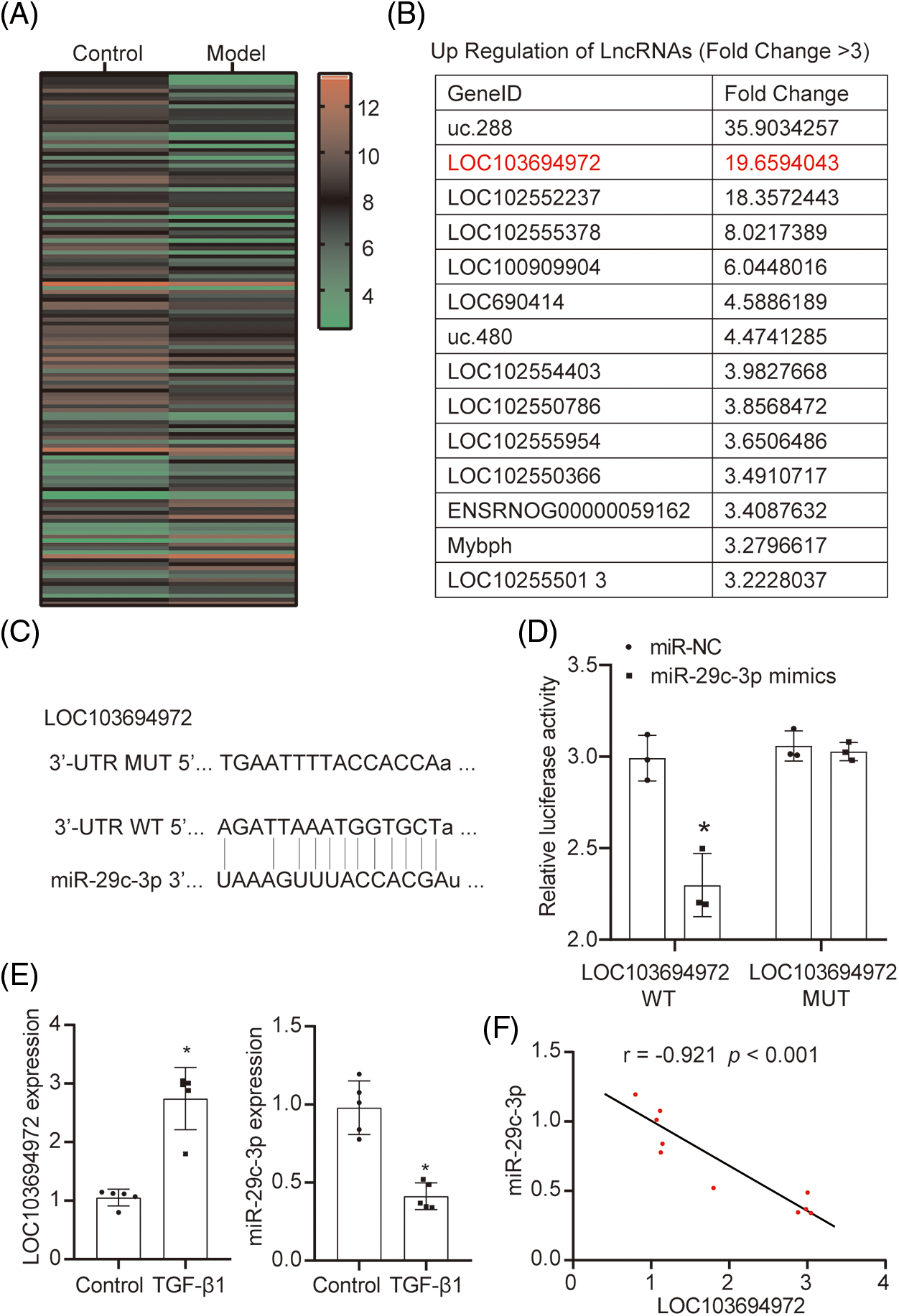
Figure 2: The regulatory relationship between lncRNA LOC103694972 and miR-29c-3p. (A and B) Differentially expressed lncRNAs was screened out by lncRNA chip. n = 1. (C) Prediction of the targeted binding site between lncRNA LOC103694972 and miR-29c-3p by miRanda and TargetScan v.80. (D) Dual-luciferase assay between lncRNA LOC103694972 and miR-29c-3p. n = 3. (E) qRT-PCR to detected lncRNA LOC103694972 and miR-29c-3p expressions. n = 5. (F) Correlation analysis between lncRNA LOC103694972 and miR-29c-3p expression. n = 10. *p < 0.05 vs. the Control group.
The effect of lncRNA LOC103694972 on NRK-49F fibrosis
In order to investigate the impact of lncRNA LOC103694972 on fibrotic NRK-49F cells, we conducted a silencing experiment on cells induced by TGF-β1 (Fig. 3A). Cell morphology was observed, and it was found that the length of cells in the TGF-β1+si-LOC103694972 group showed similarities to that of the Control cells. The expression levels of lncRNA LOC103694972 and miR-29c-3p were measured in each group (Fig. 3B). In the TGF-β1+si-LOC103694972 group, there was a decrease in lncRNA LOC103694972 expression, while miR-29c-3p expression increased compared to the negative control. This indicates successful knockdown of lncRNA LOC103694972, and also suggests a negative correlation between the expression of lncRNA LOC103694972 and miR-29c-3p. The cell activity in the TGF-β1+si-LOC103694972 group was higher than in the TGF-β1+si-NC group (Fig. 3C). In the TGF-β1+si-LOC103694972 group, the levels of the myofibroblast markers Collagen I and α-SMA, as well as the matrix metalloproteinase inhibitor TIMP-1, were lower compared to the TGF-β1+si-NC group. The levels of matrix metalloproteinase MMP2 and MMP9 increased in the TGF-β1+si-LOC103694972 group compared to the TGF-β1+si-NC group. This suggests that the extent of fibrosis in TGF-β1-induced NRK-49F cells was reduced (Figs. 3D–3F).
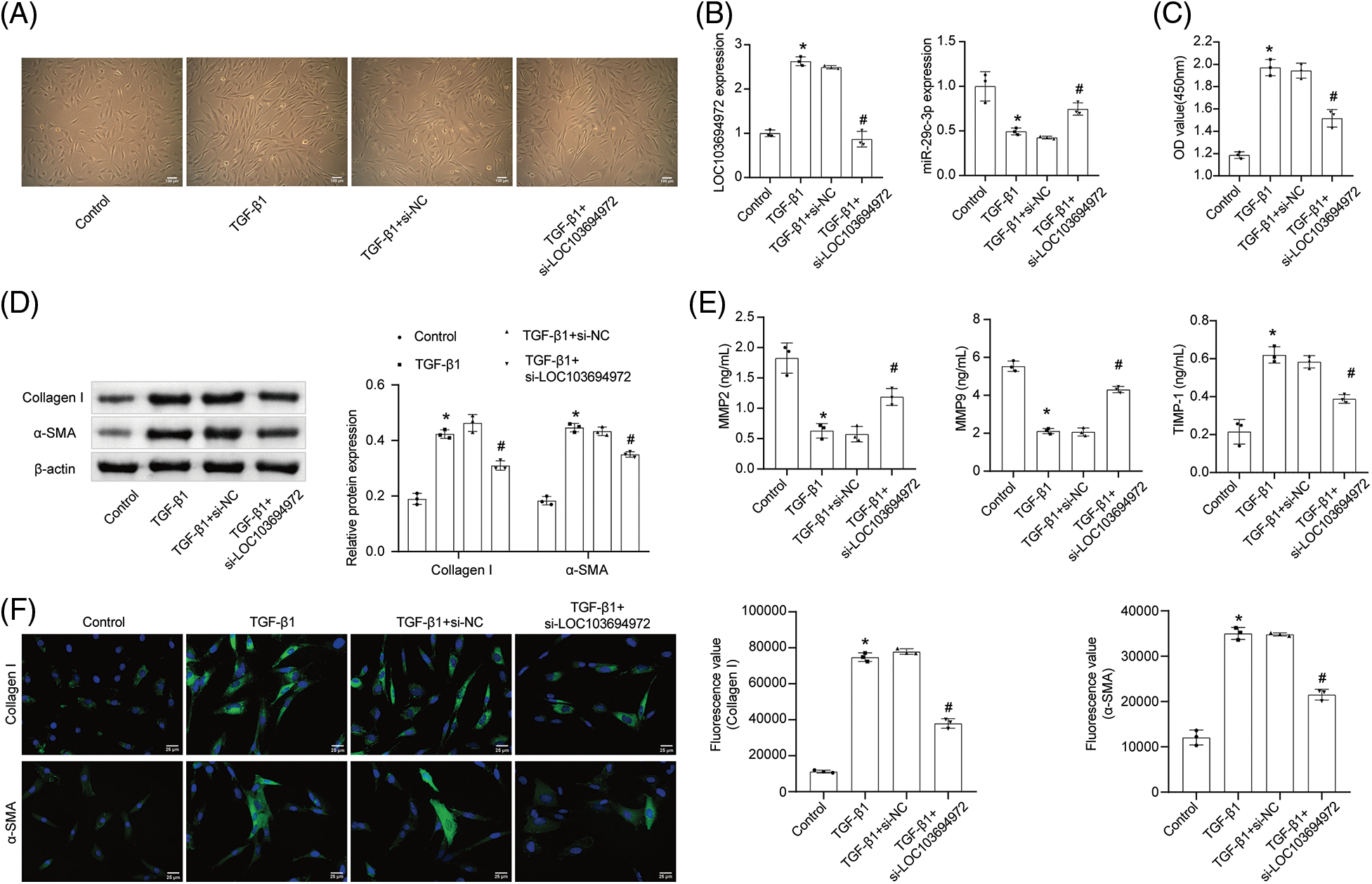
Figure 3: The effect of lncRNA LOC103694972 on NRK-49F fibrosis. (A) Cell morphology was observed under the microscope (scale bar = 100 μm). n = 3. (B) qRT-PCR analysis of lncRNA LOC103694972 and miR-29c-3p expression. n = 3. (C) Cell proliferation was examined by CCK8 assay. n = 3. (D) WB assay of Collagen I and α-SMA expression. n = 3. (E) qRT-PCR analysis of MMP2, MMP9, and TIMP-1 expression. n = 3. (F) IF was used to detected the expression of Collagen I and α-SMA in cells (scale bar = 25 μm). n = 3. *p < 0.05 vs. the Control group. #p < 0.05 vs. the TGF-β1+si-NC group.
Targeting relationship between miR-29c-3p and STAT3
We utilized TargetScan v.80 to predict the specific binding site between miR-29c-3p and STAT3 (Fig. 4A). Subsequently, a dual-luciferase assay was performed to determine the existence of a targeting relationship between miR-29c-3p and STAT3. Comparing the relative luciferase activity, we observed that the miR-29c-3p mimics+STAT3 WT group displayed lower levels compared to the miR-NC+STAT3 WT group. Notably, there was no significant difference observed in the relative luciferase activity between the miR-NC+STAT3 MUT group and miR-29c-3p mimics+STAT3 MUT group (Fig. 4B).
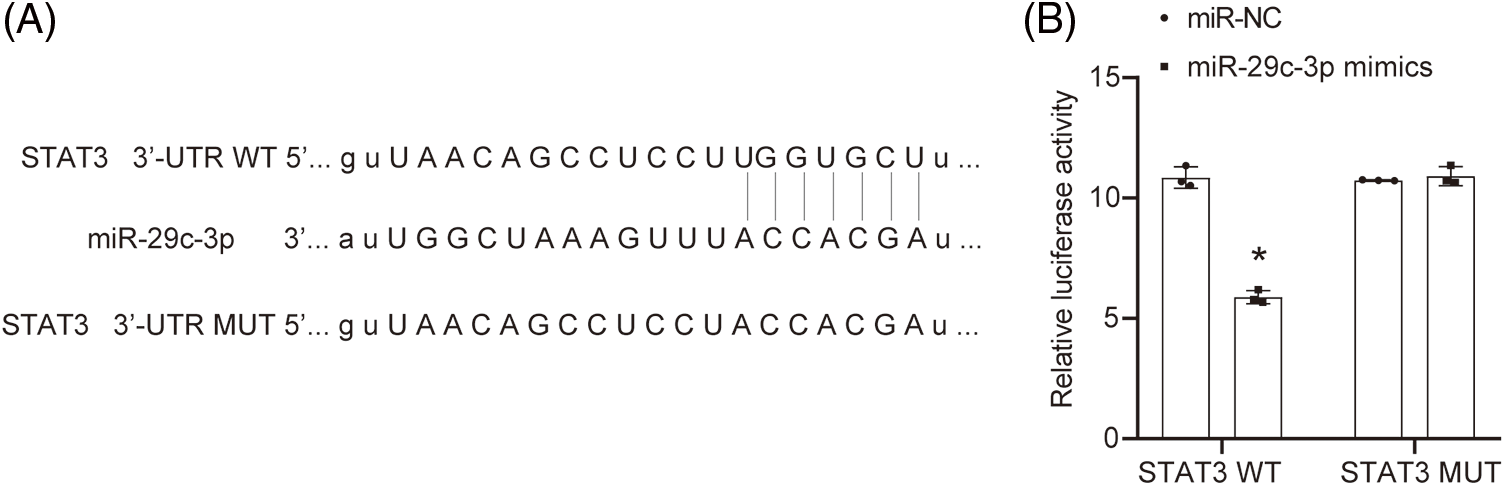
Figure 4: Targeting relationship between miR-29c-3p and STAT3. (A) The targeted binding site between miR-29c-3p and STAT3 by TargetScan v.80. (B) The binding between miR-29c-3p and STAT3 was demonstrated by dual-luciferase assay. n = 3. *p < 0.05 vs. the miR-NC group.
The effect of miR-29c-3p on fibrosis of NRK-49F
To demonstrate the role of miR-29c-3p in fibrotic cells, we conducted an overexpression experiment on TGF-β1-induced NRK-49F cells. Initially, we observed the cell morphology of all groups using a microscope (Fig. 5A). It was observed that the cells exhibited elongated and slender characteristics compared to their previous state. However, after treatment with miR-29c-3p mimics, the cell length returned to its original state. We then performed qRT-PCR to confirm the successful transfection (Fig. 5B). Additionally, we used miR-NC transfected with miR-29c-3p mimics to validate the reliability of the transfection. Furthermore, we conducted a CCK8 assay to assess cell proliferation (Fig. 5C). It was observed that the proliferation ability of NRK-49F cells increased after treatment with TGF-β1, whereas the introduction of miR-29c-3p mimics inhibited their proliferation. Following transfection with miR-29c-3p mimics, the levels of Collagen I, α-SMA, and TIMP-1 in NRK-49F cells decreased, while the levels of MMP2 and MMP9 increased (Figs. 5D, 5E). Consequently, cell fibrosis was alleviated.
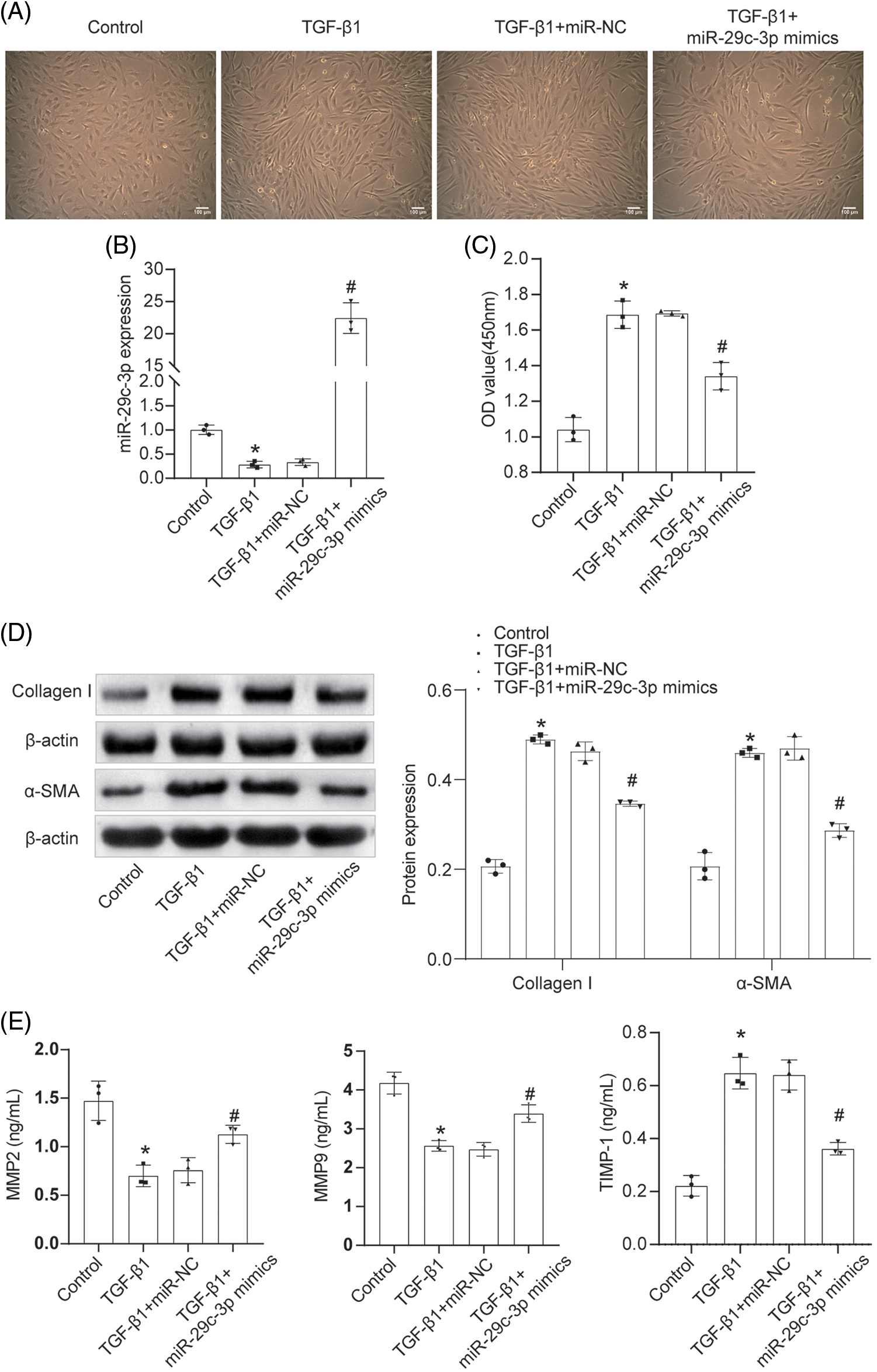
Figure 5: The effect of miR-29c-3p on fibrosis of NRK-49F. (A) Cell morphology was observed under the microscope (scale bar = 100 μm). n = 3. (B) The expression of miR-29c-3p in each group was assayed by qRT-PCR. n = 3. (C) The proliferation of cells was examined by CCK8 assay. n = 3. (D) WB was used to detect Collagen I and α-SMA expressions in each group. n = 3. (E) qRT-PCR assay of MMP2, MMP9, and TIMP-1 expression. n = 3. *p < 0.05 vs. the Control group. #p < 0.05 vs. the TGF-β1+miR-NC group.
The effect of miR-29c-3p on STAT3-Smad3/CTGF pathway
After inducing TGF-β1 on NRK-49F cells, we proceeded with the miR-29c-3p overexpression treatment. Subsequently, we assessed the mRNA expression of STAT3, Smad3, and CTGF in each cell group (Figs. 6A, 6B). Additionally, we analyzed the protein levels of p-STAT3 and p-Smad3 (Fig. 6B). Interestingly, compared to the Control group, the levels of STAT3, Smad3, and CTGF increased in the TGF-β1 group. Importantly, the TGF-β1+miR-29c-3p mimics group exhibited reduced levels of STAT3, Smad3, and CTGF. Furthermore, we evaluated the protein levels of STAT3, p-STAT3, Smad3, p-Smad3, and CTGF. Notably, TGF-β1 upregulated the expression of p-STAT3/STAT3, p-Smad3/Smad3, and CTGF. Conversely, treatment with miR-29c-3p mimics resulted in decreased p-STAT3/STAT3, p-Smad3/Smad3, and CTGF expression. Collectively, our findings suggest that TGF-β1 activated the STAT3-Smad3/CTGF signaling pathway, while miR-29c-3p mimics act as an effective target fibrosis in TGF-β1-induced NRK-49F cells by modulating the STAT3-Smad3/CTGF pathway.
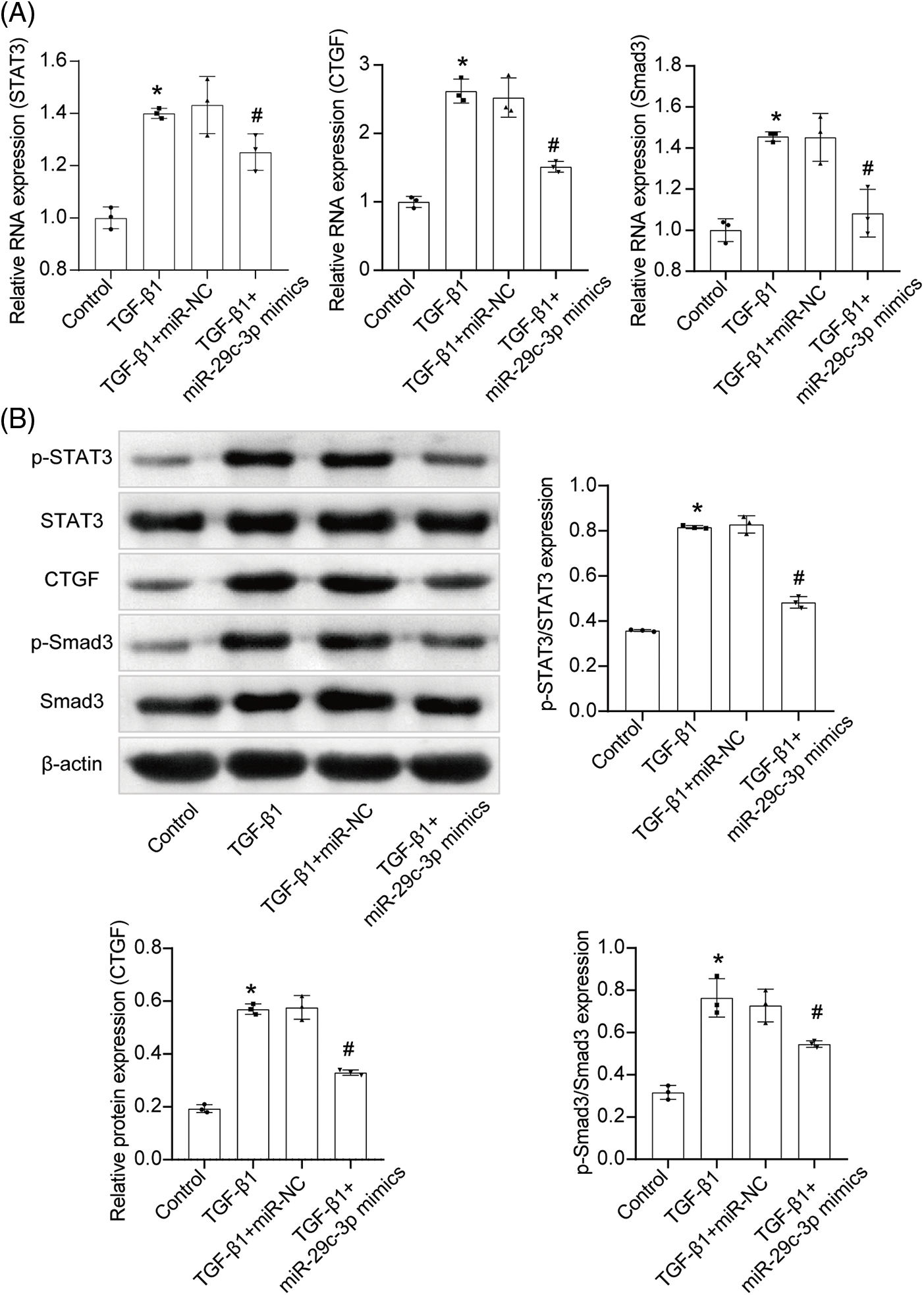
Figure 6: The effect of miR-29c-3p on STAT3-Smad3/CTGF pathway. (A) qRT-PCR assay of STAT3, CTGF and Smad3 expressions. n = 3. (B) WB assay of p-STAT3, STAT3, CTGF, p-Smad3 and Smad3 expressions. n = 3. *p < 0.05 vs. the Control group. #p < 0.05 vs. the TGF-β1+miR-NC group.
Renal fibrosis, an intermediate process in most kidney diseases, has garnered significant attention in terms of treatment. The fibrosis model induced by TGF-β1 in NRK-49F cells is a commonly used in vitro model for fibrosis [19–21]. In this study, we constructed an in vitro fibrosis model and observed significant elongation of the cell morphology. Collagen I and α-SMA were employed as markers to assess the transdifferentiation of fibroblasts into myofibroblasts [22,23]. Our study successfully up-regulated Collagen I and α-SMA, indicating the deposition of interstitial collagen and increased myofibroblasts. The down-regulation of matrix metalloproteinases MMP2 and MMP9 contributed to the accumulation of extracellular matrix (ECM), while the matrix metalloproteinase inhibitor TIMP-1 inhibited the activity of MMP2 [24,25]. Upon TGF-β1 interference, NRK-49F cells exhibited down-regulation of MMP2 and MMP9, as well as up-regulation of TIMP-1. These results collectively demonstrated the effective intervention of TGF-β1.
In recent years, the role of lncRNAs as epigenetic regulatory factors has gained prominence [26]. Numerous studies have highlighted the ability of lncRNAs to regulate diseases through the ceRNA network [27]. LncRNAs, such as NR_038323, MALAT1, and GAS5, have been identified as effective factors in the treatment of renal fibrosis [27–29]. In our study, we utilized lncRNA chip analysis to identify a highly expressed lncRNA, LOC103694972, in TGF-β1-induced NRK-49F cells. To verify the impact of lncRNA LOC103694972 on fibrosis in NRK-49F cells, we knocked down this lncRNA and observed the restoration of cell morphology. Furthermore, compared to the TGF-β1 group, the TGF-β1+si-LOC103694972 group exhibited down-regulation of Collagen I, α-SMA, and TGF-β1, along with up-regulation of MMP2 and MMP9. These findings indicated a mitigation of cell fibrosis.
Moreover, we found that miR-29c-3p was located downstream of lncRNA LOC103694972, and the two could be targeted to bind. The expression of the two was negatively correlated. miR-29c-3p was down-regulated in NRK-49F cells induced by TGF-β1. Gholaminejad et al. [30] used bioinformatics analysis to study the miRNA expression profile of renal fibrosis and found a series of marker miRNAs of renal fibrosis, including miR-29c-3p. Later, Sun et al. [31] verified that the down-regulation of miR-29c-3p could induce renal fibrosis in vivo and vitro. This was consistent with our research results. When miR-29c-3p was overexpressed, the fibrosis of NRK-49F cells induced by TGF-β1 was relieved.
STAT3 has been implicated in the regulation of fibrosis-related genes [32], and studies have shown that miR-29c-3p can negatively regulate STAT3 [15,33]. To confirm this relationship, we conducted dual-luciferase verification, which confirmed that STAT3 is indeed the target gene of miR-29c-3p. Our research demonstrated that TGF-β1 triggered the activation of STAT3, leading to increased expression of p-STAT3 [33]. However, upon overexpression of miR-29c-3p, the expression of p-STAT3 was down-regulated. Smad3 plays a crucial role in TGF-β1-induced fibrosis, and the activation of the Smad3 signaling pathway is a common response in various cells to TGF-β1 [34,35]. In our study, TGF-β1 treatment of NRK-49F cells resulted in the phosphorylation and activation of Smad3. However, the overexpression of miR-29c-3p inhibited the expression of p-Smad3. CTGF serves as a downstream target gene of Smad3 [36,37], and Lei et al. discovered that MKL1 could directly bind to the CTGF promoter by interacting with Smad3 to activate CTGF transcription [38]. Our research established that TGF-β1 mediated the activation of CTGF, but this activation was suppressed by overexpression of miR-29c-3p. In summary, the findings suggest that overexpression of miR-29c-3p could alleviate TGF-β1-induced fibrosis in NRK-49F cells by inhibiting the STAT3-Smad3/CTGF pathway.
The left–right determination factor (Lefty) plays a certain role in the TGF-β1-mediated transformation of fibroblasts into myofibroblasts. TGF-β1 is a transforming growth factor that has been shown to promote the transformation of fibroblasts into myofibroblasts. Zhang et al. [19] studied the effect of exogenous Lefty-1 on TGF-β1-induced transformation of NRK-49F cells and found that Lefty-1 can inhibit the transformation of fibroblasts into myofibroblasts. Specifically, Lefty can inhibit the expression of TGF-β1 receptors or block the activation of TGF-β1 receptors to inhibit the transmission of TGF-β1 signals and reduce the transformation of fibroblasts into myofibroblasts. Therefore, Lefty plays a negative regulatory role in the TGF-β1-mediated transformation of fibroblasts into myofibroblasts and plays an important role in balancing and regulating this process.
It is worth noting that this study investigated the activating role of TGF-β1 on the STAT3-Smad3/CTGF pathway in fibrosis of NRK-49F cells. However, some other studies have shown that TGF-β1 can also transmit signals through non-Smad pathways. Understanding these signal transductions can provide a more comprehensive understanding of the mechanism of renal fibrosis. Non-Smad pathways include the MAPK pathway and the PI3K/AKT pathway, among others. Studies by Wang et al. [39] and Zhu et al. [40] have shown that TGF-β1 can mediate the MAPK pathway in renal interstitial fibrosis and mediate the epithelial-mesenchymal transition (EMT). In addition, TGF-β1 can also affect the balance of cell survival and apoptosis through the PI3K/AKT pathway, affecting the development of renal tubulointerstitial fibrosis [41,42]. In summary, the occurrence of renal fibrosis is influenced by complex cross-talk between multiple pathways. Further research on these cross-talk mechanisms and regulations will contribute to a deeper understanding of the pathogenesis of renal fibrosis and provide a theoretical basis for the development of new treatment strategies.
Although our study has provided a new perspective on the treatment of renal fibrosis, it is still unknown whether lncRNA LOC103694972 can be used as a clinical treatment target. Therefore, our future research aims to explore the clinical significance of lncRNA LOC103694972 in renal fibrosis in more detail.
During our study, we discovered a novel lncRNA called LOC103694972. We found that silencing LOC103694972 could enhance the expression of miR-29c-3p. Furthermore, increasing the expression of miR-29c-3p significantly inhibited the activity of STAT3. Through the STAT3-Smad3/CTGF pathway, miR-29c-3p demonstrated the ability to alleviate fibrosis in NRK-49F cells. These findings provide a fresh perspective for understanding the underlying mechanisms of renal fibrosis.
Acknowledgement: The authors would like to acknowledge the FiGDRAW software.
Funding Statement: This work was supported by the Hunan Provincial Education Department General Project Research Fund (No. 20C1412), the Hunan Graduate Scientific Research Innovation Project (No. CX2018B474), and the National Famous Elderly Chinese Medicine Experts Xinyu Chen Inheritance Workshop Construction Project (No. [2022] 75).
Author Contributions: Designed the study, supervised the data collection: Yan Li and Huzhi Cai; Analyzed the data, interpreted the data: Xiaoling Peng, Youhui Liu and Qingyang Chen; Prepared the manuscript for publication and reviewed the draft of the manuscript: Xiangdong Lin and Xinyu Chen. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Matsushita K, Ballew SH, Wang AY, Kalyesubula R, Schaeffner E, Agarwal R. Epidemiology and risk of cardiovascular disease in populations with chronic kidney disease. Nat Rev Nephrol. 2022;18:696–707. doi:10.1038/s41581-022-00616-6. [Google Scholar] [PubMed] [CrossRef]
2. Ammirati AL. Chronic kidney disease. Rev Assoc Med Bras. 2020;66:S3–9. doi:10.1590/1806-9282.66.S1.3. [Google Scholar] [PubMed] [CrossRef]
3. Evans M, Lewis RD, Morgan AR, Whyte MB, Hanif W, Bain SC, et al. A narrative review of chronic kidney disease in clinical practice: current challenges and future perspectives. Adv Ther. 2022;39:33–43. doi:10.1007/s12325-021-01927-z. [Google Scholar] [PubMed] [CrossRef]
4. Wang WJ, Chen XM, Cai GY. Cellular senescence and the senescence-associated secretory phenotype: potential therapeutic targets for renal fibrosis. Exp Gerontol. 2021;151:111403. doi:10.1016/j.exger.2021.111403. [Google Scholar] [PubMed] [CrossRef]
5. Humphreys BD. Mechanisms of renal fibrosis. Annu Rev Physiol. 2018;80:309–26. doi:10.1146/annurev-physiol-022516-034227. [Google Scholar] [PubMed] [CrossRef]
6. Yan H, Xu J, Xu Z, Yang B, Luo P, He Q. Defining therapeutic targets for renal fibrosis: exploiting the biology of pathogenesis. Biomed Pharmacother. 2021;143:112115. doi:10.1016/j.biopha.2021.112115. [Google Scholar] [PubMed] [CrossRef]
7. Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncrna biology. Adv Exp Med Biol. 2017;1008:283–323. doi:10.1007/978-981-10-5203-3. [Google Scholar] [CrossRef]
8. Guo Y, Li G, Gao L, Cheng X, Wang L, Qin Y, et al. Exaggerated renal fibrosis in lncRNA Gas5-deficient mice after unilateral ureteric obstruction. Life Sci. 2021;264(1):118656. doi:10.1016/j.lfs.2020.118656. [Google Scholar] [PubMed] [CrossRef]
9. Duan YR, Chen BP, Chen F, Yang SX, Zhu CY, Ma YL, et al. LncRNA lnc-ISG20 promotes renal fibrosis in diabetic nephropathy by inducing AKT phosphorylation through miR-486-5p/NFAT5. J Cell Mol Med. 2021;25:4922–37. doi:10.1111/jcmm.16280. [Google Scholar] [PubMed] [CrossRef]
10. Zhang YY, Tan RZ, Yu Y, Niu YY, Yu C. LncRNA gas5 protects against TGF-β-induced renal fibrosis via the Smad3/miRNA-142-5p axis. Am J Physiol–Renal Physiol. 2021b;321:F517–26. doi:10.1152/ajprenal.00085.2021. [Google Scholar] [PubMed] [CrossRef]
11. Gifford CC, Tang J, Costello A, Khakoo NS, Nguyen TQ, Goldschmeding R, et al. Negative regulators of TGF-β1 signaling in renal fibrosis; pathological mechanisms and novel therapeutic opportunities. Clin Sci. 2021;135:275–303. doi:10.1042/CS20201213. [Google Scholar] [PubMed] [CrossRef]
12. Gu YY, Liu XS, Huang XR, Yu XQ, Lan HY. TGF-β in renal fibrosis: triumphs and challenges. Future Med Chem. 2020;12(9):853–66. doi:10.4155/fmc-2020-0005. [Google Scholar] [PubMed] [CrossRef]
13. He Y, Huang C, Lin X, Li J. MicroRNA-29 family, a crucial therapeutic target for fibrosis diseases. Biochimie. 2013;95(7):1355–9. doi:10.1016/j.biochi.2013.03.010. [Google Scholar] [PubMed] [CrossRef]
14. Li Y, Guo F, Huang R, Ma L, Fu P. Natural flavonoid pectolinarigenin alleviated kidney fibrosis via inhibiting the activation of TGFβ/SMAD3 and JAK2/STAT3 signaling. Int Immunopharmacol. 2021;91:107279. doi:10.1016/j.intimp.2020.107279. [Google Scholar] [PubMed] [CrossRef]
15. Ye S, Yang C, Wang C, Jin A, Zhang Q, Zhang S. MiR-29c-3p inhibits proliferation and migration in rat primary cardiac fibroblasts via interacting with STAT3. Panminerva Med. 2023;65(2):199–204. doi:10.23736/S0031-0808.20.03975-0. [Google Scholar] [PubMed] [CrossRef]
16. Xu D, Chen PP, Zheng PQ, Yin F, Cheng Q, Zhou ZL, et al. KLF4 initiates sustained YAP activation to promote renal fibrosis in mice after ischemia-reperfusion kidney injury. Acta Pharmacol Sin. 2021;42(3):436–50. doi:10.1038/s41401-020-0463-x. [Google Scholar] [PubMed] [CrossRef]
17. Zhang SM, Wei CY, Wang Q, Wang L, Lu L, Qi FZ. M2- polarized macrophages mediate wound healing by regulating connective tissue growth factor via AKT, ERK1/2, and STAT3 signaling pathways. Mol Biol Rep. 2021;48(9):6443–56. doi:10.1007/s11033-021-06646-w. [Google Scholar] [PubMed] [CrossRef]
18. Ma TT, Meng XM. TGF-β/Smad and renal fibrosis. Adv Exp Med Biol. 2019;1165:347–64. doi:10.1007/978-981-13-8871-2. [Google Scholar] [CrossRef]
19. Zhang L, Zhang J, Xu C, Zhou X, Wang W, Zheng R, et al. Lefty-1 alleviates TGF-β1-induced fibroblast-myofibroblast transdifferentiation in NRK-49F cells. Drug Design, Dev Ther. 2015;9:4669–78. doi:10.2147/dddt.S86770. [Google Scholar] [PubMed] [CrossRef]
20. Wang JL, Chen CW, Tsai MR, Liu SF, Hung TJ, Yu Ju H, et al. Antifibrotic role of PGC-1α-siRNA against TGF-β1-induced renal interstitial fibrosis. Exp Cell Res. 2018;370(1):160–7. doi:10.1016/j.yexcr.2018.06.016. [Google Scholar] [PubMed] [CrossRef]
21. Li N, Wang Z, Gao F, Lei Y, Li Z. Melatonin ameliorates renal fibroblast-myofibroblast transdifferentiation and renal fibrosis through miR-21-5p regulation. J Cell Mol Med. 2020;24(10):5615–28. doi:10.1111/jcmm.15221. [Google Scholar] [PubMed] [CrossRef]
22. Xia ZE, Xi JL, Shi L. 3,3’-diindolylmethane ameliorates renal fibrosis through the inhibition of renal fibroblast activation in vivo and in vitro. Renal Fail. 2018;40(1):447–54. doi:10.1080/0886022X.2018.1490322. [Google Scholar] [PubMed] [CrossRef]
23. Wei Q, Su J, Dong G, Zhang M, Huo Y, Dong Z. Glycolysis inhibitors suppress renal interstitial fibrosis via divergent effects on fibroblasts and tubular cells. Am J Physiol–Renal Physiol. 2019;316(6):F1162–72. doi:10.1152/ajprenal.00422.2018. [Google Scholar] [PubMed] [CrossRef]
24. Zhang G, Kang Y, Zhou C, Cui R, Jia M, Hu S, et al. Amelioratory effects of testosterone propionate on age-related renal fibrosis via suppression of TGF-β1/Smad signaling and activation of Nrf2-ARE signaling. Sci Rep. 2018;8(1):10726. doi:10.1038/s41598-018-29023-3. [Google Scholar] [PubMed] [CrossRef]
25. Gao Y, Yuan D, Gai L, Wu X, Shi Y, He Y, et al. Saponins from panax japonicus ameliorate age-related renal fibrosis by inhibition of inflammation mediated by NF-κB and TGF-β1/Smad signaling and suppression of oxidative stress via activation of Nrf2-ARE signaling. J Ginseng Res. 2021;45(3):408–19. doi:10.1016/j.jgr.2020.08.005. [Google Scholar] [PubMed] [CrossRef]
26. Jia H, Ma T, Hao C. Identification of candidate lncRNA biomarkers for renal fibrosis: a systematic review. Life Sci. 2020;262:118566. doi:10.1016/j.lfs.2020. [Google Scholar] [CrossRef]
27. Liu P, Zhang B, Chen Z, He Y, Du Y, Liu Y, et al. M6A-induced lncRNA MALAT1 aggravates renal fibrogenesis in obstructive nephropathy through the miR-145/FAK pathway. Aging. 2020;12:5280–99. doi:10.18632/aging.102950. [Google Scholar] [PubMed] [CrossRef]
28. Ge Y, Wang J, Wu D, Zhou Y, Qiu S, Chen J, et al. LncRNA NR_038323 suppresses renal fibrosis in diabetic nephropathy by targeting the miR-324-3p/DUSP1 axis. Mol Ther Nucleic Acids. 2019;17:741–53. doi:10.1016/j.omtn.2019.07.007. [Google Scholar] [PubMed] [CrossRef]
29. Wang W, Jia YJ, Yang YL, Xue M, Zheng ZJ, Wang L, et al. LncRNA GAS5 exacerbates renal tubular epithelial fibrosis by acting as a competing endogenous RNA of miR-96-5p. Biomed Pharmacother. 2020a;121:109411. doi:10.1016/j.biopha.2019.109411. [Google Scholar] [PubMed] [CrossRef]
30. Gholaminejad A, Abdul Tehrani H, Gholami Fesharaki M. Identification of candidate microRNA biomarkers in renal fibrosis: a meta-analysis of profiling studies. Biomark. 2018;23:713–24. doi:10.1080/1354750X.2018.1488275. [Google Scholar] [PubMed] [CrossRef]
31. Sun CM, Zhang WY, Wang SY, Qian G, Pei DL, Zhang GM. Fer exacerbates renal fibrosis and can be targeted by miR-29c-3p. Open Med. 2021;16(1):1378–85. doi:10.1515/med-2021-0319. [Google Scholar] [PubMed] [CrossRef]
32. Chen W, Yuan H, Cao W, Wang T, Chen W, Yu H, et al. Blocking interleukin-6 trans-signaling protects against renal fibrosis by suppressing STAT3 activation. Theranostics. 2019;9(14):3980–91. doi:10.7150/thno.32352. [Google Scholar] [PubMed] [CrossRef]
33. Wu Y, Xu J, Xu J, Cheng J, Jiao D, Zhou C, et al. Lower serum levels of miR-29c-3p and miR-19b-3p as biomarkers for Alzheimer’s disease. Tohoku J Exp Med. 2017;242(2):129–36. doi:10.1620/tjem.242.129. [Google Scholar] [PubMed] [CrossRef]
34. Ying H, Fang M, Hang QQ, Chen Y, Qian X, Chen M. Pirfenidone modulates macrophage polarization and ameliorates radiation-induced lung fibrosis by inhibiting the TGF-β1/Smad3 pathway. J Cell Mol Med. 2021;25(18):8662–75. doi:10.1111/jcmm.16821. [Google Scholar] [PubMed] [CrossRef]
35. Wu W, Wang X, Yu X, Lan HY. Smad3 signatures in renal inflammation and fibrosis. Int J Biol Sci. 2022;18(7):2795–806. doi:10.7150/ijbs.71595. [Google Scholar] [PubMed] [CrossRef]
36. Liang JN, Zou X, Fang XH, Xu JD, Xiao Z, Zhu JN, et al. The Smad3-miR-29b/miR-29c axis mediates the protective effect of macrophage migration inhibitory factor against cardiac fibrosis. Biochim Biophys Acta (BBA)–Mol Basis Dis. 2019;1865(9):2441–50. doi:10.1016/j.bbadis.2019.06.004. [Google Scholar] [PubMed] [CrossRef]
37. Zheng H, Ji J, Zhao T, Wang E, Zhang A. Exosome-encapsulated miR-26a attenuates aldosterone-induced tubulointerstitial fibrosis by inhibiting the CTGF/Smad3 signaling pathway. Int J Mol Med. 2023;51(2):11. doi:10.3892/ijmm.2022.5214. [Google Scholar] [PubMed] [CrossRef]
38. Mao L, Liu L, Zhang T, Wu X, Zhang T, Xu Y. MKL1 mediates TGF-β-induced CTGF transcription to promote renal fibrosis. J Cell Physiol. 2020;235(5):4790–803. doi:10.1002/jcp.29356. [Google Scholar] [PubMed] [CrossRef]
39. Wang S, Zhou Y, Zhang Y, He X, Zhao X, Zhao H, et al. Roscovitine attenuates renal interstitial fibrosis in diabetic mice through the TGF-β1/p38 MAPK pathway. Biomed Pharmacother. 2019;115:108895. doi:10.1016/j.biopha.2019.108895. [Google Scholar] [PubMed] [CrossRef]
40. Zhu JH, Wang L, Ma ZX, Duan JA, Tao JH. Rehmannia glutinosa libosch and cornus officinalis sieb herb couple ameliorates renal interstitial fibrosis in CKD rats by inhibiting the TGF-β1/MAPK signaling pathway. J Ethnopharmacol. 2023;318:117039. doi:10.1016/j.jep.2023.117039. [Google Scholar] [PubMed] [CrossRef]
41. Wang Z, Chen Z, Li B, Zhang B, Du Y, Liu Y, et al. Curcumin attenuates renal interstitial fibrosis of obstructive nephropathy by suppressing epithelial-mesenchymal transition through inhibition of the TLR4/NF-кB and PI3K/AKT signalling pathways. Pharm Biol. 2020;58(1):828–37. doi:10.1080/13880209.2020.1809462. [Google Scholar] [PubMed] [CrossRef]
42. Li D, Yu K, Feng F, Zhang Y, Bai F, Zhang Y, et al. Hydroxychloroquine alleviates renal interstitial fibrosis by inhibiting the PI3K/AKT signaling pathway. Biochem Biophys Res Commun. 2022;610:154–61. doi:10.1016/j.bbrc.2022.04.058. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools