 Open Access
Open Access
ARTICLE
Overexpression of RUNX1 mitigates dexamethasone-induced impairment of osteogenic differentiation and oxidative stress injury in bone marrow mesenchymal stem cells by promoting alpha-2 macroglobulin transcription
1 Department of Bone Joint and Sports Medicine, Fuzhou Hospital of Traditional Chinese Medicine Affiliated to Fujian University of Traditional Chinese Medicine, Fuzhou, 350001, China
2 Nursing Department, The First Affiliated Hospital of Fujian Medical University, Fuzhou, 350004, China
3 Nursing Department, National Regional Medical Center, Binhai Campus of the First Affiliated Hospital of Fujian Medical University, Fuzhou, 350004, China
4 Department of Orthopedic Surgery, The First Affiliated Hospital of Fujian Medical University, Fuzhou, 350004, China
5 Department of Sports Medicine, National Regional Medical Center, Binhai Campus of the First Affiliated Hospital of Fujian Medical University, Fuzhou, 350004, China
* Corresponding Author: SHANHONG FANG. Email:
BIOCELL 2024, 48(2), 205-216. https://doi.org/10.32604/biocell.2023.045109
Received 17 August 2023; Accepted 24 October 2023; Issue published 23 February 2024
Abstract
Introduction: Dexamethasone (Dex) caused impaired osteoblast differentiation and oxidative stress (OS) in bone marrow mesenchymal stem cells (BMSCs). This work sought to elucidate the precise molecular pathway through which Dex influences osteogenic differentiation (OD) and OS in BMSCs. Methods: The expression of Runt-related transcription factor 1 (RUNX1) and alpha-2 macroglobulin (A2M) was assessed in Dex-treated BMSCs using qRT-PCR and Western Blot. Following the functional rescue experiments, cell proliferation was determined by MTT assay, reactive oxygen species (ROS) expression by DCFH-DA fluorescent probe, lactate dehydrogenase (LDH), superoxide dismutase (SOD), catalase (CAT), and glutathione peroxidase (Gpx) expression by kits, OD by alkaline phosphatase (ALP) staining and activity quantification, and the expression of OD-related proteins RUNX2, collagen type 1 alpha 1 (COL1A1), and osteocalcin (OCN) by qRT-PCR and Western Blot. The binding of RUNX1 to A2M was initially analyzed through Jaspar website and subsequently verified by dual-luciferase reporter and ChIP assays. Results: Dex-treated BMSCs had low RUNX1 and A2M expression. Dex treatment apparently elevated ROS and LDH levels, diminished cell proliferation rate and SOD, CAT, and Gpx expression, lightened intensity of ALP staining, and declined calcified nodules, ALP activity, and RUNX2, COL1A1, and OCN expression in BMSCs, which was counterweighed by RUNX1 or A2M overexpression. RUNX1 positively targeted A2M. A2M knockdown effectively nullified the ameliorative effects of RUNX1 overexpression on impaired OD and OS injury in Dex-induced BMSCs. Conclusions: Overexpression of RUNX1 attenuated Dex-induced impaired OD and OS injury in BMSCs by promoting A2M transcription.Keywords
Abbreviations
| Dex | Dexamethasone |
| OS | Oxidative stress |
| BMSCs | Bone marrow mesenchymal stem cells |
| OD | Osteogenic differentiation |
| RUNX1 | Runt-related transcription factor 1 |
| A2M | Alpha-2 macroglobulin |
| ROS | Reactive oxygen species |
| LDH | Lactate dehydrogenase |
| SOD | Superoxide dismutase |
| CAT | Catalase |
| Gpx | Glutathione peroxidase |
| ALP | Alkaline phosphatase |
| COL1A1 | Collagen type 1 alpha 1 |
| OCN | Osteocalcin |
| PBS | Phosphate-buffered saline |
| ARS | Alizarin red staining |
| MTT | 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| ChIP | Chromatin immunoprecipitation |
| lgG | Immunoglobulin G |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
Dexamethasone (Dex), a glucocorticoid agonist introduced to clinics in 1961, has been extensively applied in the treatment of diverse disorders. These include joint disorders, cancers, eye disorders, allergies, the coronavirus disease 2019, and autoimmune disorders [1,2]. Glucocorticoid drugs including Dex have directly adverse effects on osteocytes, reduce the rate of new bone formation and lead to a heightened danger of osteoporosis and fracture in users [3]. Dex is known to contribute to a loss in bone mass and an increase in bone marrow adiposity. Furthermore, it has the potential to skew the differentiation of bone marrow mesenchymal stem cells (BMSCs) towards the adipocyte lineage rather than osteoblasts [4]. Moreover, many evidences indicated that Dex induced oxidative stress (OS) [5–7], while OS can inhibit OD of BMSCs and served as a primary mechanism in the pathogenesis of impaired bone formation [8]. Notably, Dex at a certain concentration can inhibit OD [9,10]. Hence, the concrete mechanism by which Dex influences OD and OS in BMSCs deserves to be queried.
Runt-related transcription factor 1 (RUNX1) was unveiled to protect lymphoma cells against apoptosis induced by Dex [11]. RUNX1 serves as a vital transcription factor known for its significant regulatory roles in diverse cellular processes, including proliferation, differentiation, DNA damage response, and tissue growth [12]. During the last decades, investigations on the functions of RUNX1 were primarily concentrated in acute leukemia and cancers, like hepatocellular carcinoma, pancreatic cancer, and colorectal cancer [13–16]. It was disclosed that RUNX1 facilitated the osteogenic ability and suppresses adipogenesis in BMSCs via the Wnt/β-catenin pathway [17]. On contrary, loss of RUNX1 suppressed OD of osteogenic induction medium-induced MC3T3-E1 cells [18]. Besides, RUNX1 participated in the repression of microRNA (miR)-18-5p on hypoxia-induced myocardial apoptosis and OS [19].
Furthermore, as a heterodimeric transcription factor, RUNX1 can bind to the core elements of numerous promoters and enhancers [20]. Predictions based on databases revealed that RUNX1 bound to alpha-2 macroglobulin (A2M) promoter and modulated A2M transcription. A2M, a multifunctional protein, is primarily renowned as an inhibitor of broad-spectrum proteases and can shift proteolysis to small substrates and facilitate the binding of damaged extracellular proteins, growth factors, and cytokines and cell migration [21]. A2M reduced the number of osteoclasts in the mandibular bone marrow and apoptosis of bone marrow cells and protects BMSCs from apoptosis and oxidative damage in advanced osteoradionecrosis [22]. Reference [23] pointed out that the cooperation between A2M and clozapine protected neural tissue against injury caused by OS.
In summary, a hypothesis was formulated suggesting that RUNX1 influenced OD and OS in BMSCs after Dex treatment by regulating A2M transcription. This work intended to elucidate the concrete actions of RUNX1/A2M on OD and OS in Dex-treated BMSCs for the purpose of providing a precious clue for the treatment of osteoporosis.
BMSCs (CP-H166, Procell, Wuhan, China) were incubated at 37°C in Dulbecco’s modified eagle medium (Gibco, Grand Island, NY, USA) encompassing fetal bovine serum (10%) and penicillin/streptomycin (1%) under 5% CO2 [24].
Flow cytometry was utilized to assess the expression of surface antigens on BMSCs. Specifically, the third-generation BMSCs growing to approximately 80% were trypsin digested for 1 min and subsequently centrifuged for 5 min at 1000 g to eliminate the supernatant. Following resuspension in phosphate-buffered saline (PBS), the cells were counted and adjusted to l × l07 cells/mL. The cells were incubated on ice for 15 min with 100 μL cell suspension and antibodies against CD44, CD73, CD29, CD34, and CD45 in the dark. Following washing with l mL PBS, the cells were centrifuged. Following washing with l mL PBS, the cells were centrifuged for 5 min at 1000 g to remove the supernatant. Subsequently, the cells were treated with 200 μL PBS, and the expression of cell surface antigens was examined by flow cytometry (Beckman Coulter, Chaska, MN, USA) [25].
In addition, the cells were respectively identified for lipogenic, osteogenic, and chondrogenic differentiation ability using alizarin red staining (ARS) solution (GP1055, Servicebio, Wuhan, China), oil red O staining solution (ab150678, Abcam, Cambridge, UK), and alcian blue staining solution (BP-DL241, SenBeiJia Biological Technology, Nanjing, China). Next, only cells from the third generation were selected for subsequent experimentations.
Induction of osteogenesis, adipogenesis, and chondrogenesis and Dex treatment of BMSCs
BMSCs were osteogenically induced referring to the preceding literature [26]. Specifically, the third-generation BMSCs were seeded in 24-well culture plates (5 × 104 cells/well). The stock solution was aspirated and discarded after the BMSCs adhered to the wall. The osteogenic induction was then performed with complete α-minimum essential medium encompassing β-glycerophosphate (10 mM, Merck KGaA, Darmstadt, Germany) and ascorbic acid (20 μM, Merck KGaA) instead of the original medium. The old fluid was aspirated, discarded, and refilled with new fluid every three days. OD ability was determined via ARS, alkaline phosphatase (ALP) activity assay, and protein expression of osteogenic marker genes. The induction of adipogenesis was conducted utilizing adipogenic induction medium (PD-016, Procell, China). The induction medium and maintenance medium are cultured alternately, and then the maintenance medium is continuously cultured until the lipid droplets become large enough. Oil red O staining was used to detect adipocytes [25]. As for chondrogenic induction, BMSCs (1 × 106) were seeded onto 12-well plates, and then induced using the StemPro™ chondrogenic differentiation kit (A1007101, Thermo Fisher Scientific, Wilmington, DE, USA) for 0, 7, and 14 days. The medium was refreshed every 4 days. Finally, BMSCs were stained using alcian blue [27].
The osteogenically differentiated BMSCs were treated with reference to the preceding literature [26]. Briefly, the osteogenic medium of BMSCs was enriched with 5 μm Dex, and then the cells were cultivated for 7 days before subsequent experimentations. Dex was not included in the control group.
Vectors for Overexpression (oe) (oe-RUNX1, oe-A2M, and oe-negative control [NC]) and low expression vectors (short hairpin RNA [sh]-A2M and sh-NC) were attained from Hanbio (Shanghai, China). The transfections were performed as per the specifications, with subsequent experimentations performed at 48 h of transfection.
ALP staining was performed following the instructions of the BCIP/NBT ALP Color Development Kit (C3206, Beyotime, Shanghai, China). To elaborate, the cells underwent a series of steps: they were rinsed 3–5 times with PBS buffer, subsequently fixed at room temperature for 30 min in a 4% paraformaldehyde solution and washed another 3 times with PBS buffer. Next, the cells were covered with appropriate volume of BCIP/NBT staining working solution to cover all cells and incubated at room temperature for 30 min in the dark. Following incubation, the working solution was discarded. The cells were washed 1–2 times with distilled water to terminate the color development reaction, followed by photography and observation under the microscope (Olympus).
ALP activity assay was performed following the instructions of the ALP assay kit (P0321S, Beyotime). The cells were washed 3 times with PBS buffer, lysed for 30 min with a 1% Triton X-100 solution on ice, and centrifuged for 5 min at 4°C and 7500 rpm. The supernatant was collected and thoroughly mixed and incubated for 15 min with working solution, 50 μL buffer, and 50 L substrate solution at 37°C. After terminating the reaction with 150 L chromogenic agent, the absorbance at 405 nm was measured [28].
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay
The cells were seeded in 96-well plates in three replicates and treated with MTT solution (10 μL/well, M6494, Thermo Fisher Scientific, Wilmington, DE, USA) for 2 h. After termination of the reaction with dimethyl sulfoxide (Sigma-Aldrich, St Louis, MO, USA), the optical density value at 490 nm was determined using a microplate reader (Model 680, Bio-Rad, Hercules, CA, USA) [29,30].
Detection of reactive oxygen species (ROS)
ROS content in cells was assessed as per the previous literature [31]. Briefly, the ROS content was evaluated following the manuals of the ROS fluorescent probe (DCFH-DA molecular probe, Sigma-Aldrich). The cell supernatant was incubated for 30 min with 10 μmol/LDCFH-DA probe (2 mL) at 37°C in darkness. After oxidation, DCFH-DA was converted into DCFH with high fluorescence, which exhibited a green fluorescence in the cytoplasm. A fluorescent microscope (Olympus) was employed to acquire fluorescent images.
Lactate dehydrogenase (LDH) detection
The LDH assay kit (A020-2-2, Jiancheng, Nanjing, China) was employed for the determination of LDH, a marker of injury. Briefly, serum samples were added to a 96-well plate and operated following the protocols of the kit, and optical density values were evaluated at 450 nm on the microplate reader [32].
Detection of superoxide dismutase (SOD) and catalase (CAT)
The levels of OS factors CAT and SOD were evaluated using the SOD (A001-3-2, Jiancheng) and CAT (A007-1-1, Jiancheng) assay kits. Briefly, the supernatant was attained via cell centrifugation and executed following the protocols of the kits [26].
Glutathione peroxidase (Gpx) detection
Gpx concentration in cells was calculated following the manuals of the Gpx assay kit (A005-1-2, Jiancheng). Specifically, 1 × 108 cells were seeded onto the plates, followed by PBS washing, added with 3 volf 5% sulfosalicylic acid solution, and centrifuged for 10 min at 10,000 g. The supernatant underwent 5-min incubation with 150 μL Gpx mixture at room temperature and 20-min incubation with 50 μL diluted NADPH solution before examining the absorbance at 412 nm using the microplate reader (Model 680, Bio-Rad).
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total cellular RNA was isolated using the RNeasy mini kit (Qiagen, Valencia, CA, USA) with the concentration and purity determined using the Nano Drop micro-nucleic acid assay and reverse transcribed into cDNA using the RT kit (RR047A, Takara, Tokyo, Japan). qRT-PCR was conducted using the SYBR® Premix Ex TaqTM II and a real-time fluorescent quantitative PCR instrument (ABI 7500, Applied Biosystems, Foster City, CA, USA) with three replicates for each quantitative PCR reaction. Data were performed using the 2−ΔΔCt method with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal reference. Primer sequences are provided in Table 1.
Cells were harvested by trypsinization, lysed by enhanced Radio-Immunoprecipitation assay lysis solution with protease inhibitors. The protein concentration was determined by the bicinchoninic acid protein quantification kit (Boster, Wuhan, China). The proteins received separation by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transfer to polyvinylidene fluoride membranes, which were closed for 2 h by 5% bovine serum albumin at room temperature. The membranes received probing at 4°C with diluted primary antibodies (1:1000, Abcam Inc., Cambridge, UK) against RUNX1 (ab272456), A2M (ab109422), RUNX2 (ab236639), osteocalcin (OCN, ab93876), collagen type 1 alpha 1 (COL1A1, ab138492), and GAPDH (ab8245) before washing. The membranes were then probed for at room temperature with horseradish peroxidase-labeled secondary antibodies (ab6728 and ab6721, 1:2000, Abcam) before being washed for 10 min with Tris-buffered saline with Tween 20 for 3 times. Next, the membranes were developed with an electrogenerated chemiluminescence solution (P0018FS, Beyotime) before examination under a chemiluminescence imaging system (Bio-rad) and analysis by Quantity One v4.6.2 software. The relative protein content was calculated as the grayscale value of the corresponding protein bands/the grayscale value of the GAPDH protein bands.
Dual-luciferase reporter assay
The binding sites of RUNX1 to A2M were predicted by the Jaspar website (http://jaspar.genereg.net/) before designing and synthesizing the wild (wt) and mutant (mut) sequences of the binding sites. The wt-A2M and mut-A2M sequences received insertion into the luciferase reporter gene vectors (pGL3-Basic) before co-transfection with oe-RUNX1 or oe-NC into HEK 293T cells. The cell fluorescence activity was measured using the dual-luciferase reporter kit (Promega, Madison, WI, USA) and a luminometer (Turner Biosystems, Sunnyvale, CA, USA).
Chromatin immunoprecipitation (ChIP)
Cells received 10-min formaldehyde fixation to generate DNA-protein crosslinks. With a setting of 10 s sonication and 10 s interval for 15 cycles, a sonicator was employed to break the cells and interrupt the chromatin into fragments. The fragments received 10-min centrifugation at 12000 g and 4°C to attain the supernatant. The supernatant was divided into two tubes and respectively incubated with immunoglobulin G (lgG) antibodies (ab172730, Abcam) and the target protein-specific antibodies against RUNX1 (ab272456, Abcam) overnight at 4°C for sufficient binding. The DNA-protein complexes were precipitated with Protein Agarose/Sepharose and centrifuged for 5 min at 12,000 g and the supernatant were discarded. The non-specific complexes were washed, de-crosslinked overnight at 65°C, and purified by phenol/chloroform extraction to retrieve DNA fragments. qPCR was performed with A2M-specific primers to test the binding of RUNX1 to A2M.
The data were statistically analyzed with the GraphPad Prism7 software and presented as mean ± standard deviation. Comparisons between two groups were conducted using t-test and comparisons among multiple groups were conducted using one-way analysis of variance with Tukey’s multiple comparison test. p < 0.05 was considered statistically significant.
Dex led to impaired OD and OS injury in BMSCs with low RUNX1 expression
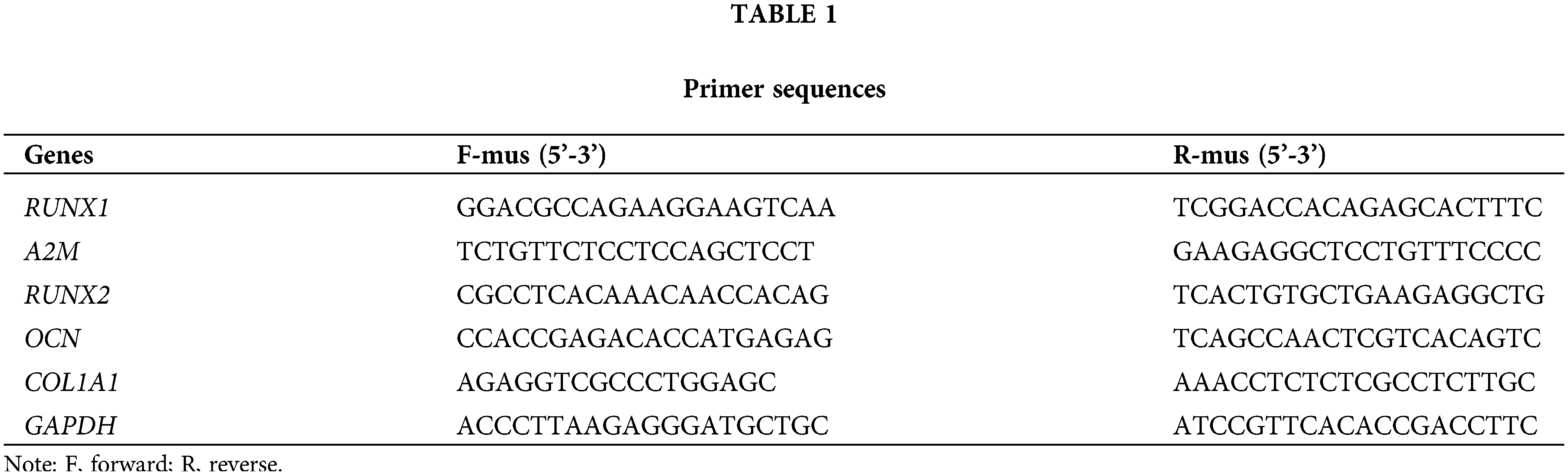
Firstly, the purchased BMSCs were identified, which displayed a classical long shuttle or shuttle shape of BMSCs (Fig. 1A). The differentiation results demonstrated that BMSCs were positive for ARS, oil red O staining, and alcian blue staining when respectively cultured in osteogenic, lipogenic, and chondrogenic induction media (Fig. 1B). Flow cytometry results revealed that a positive expression of positive cell surface markers (CD29, CD73, and CD44) and a negative expression of negative cell surface markers (CD34 and CD45) in BMSCs (Fig. 1C). The aforementioned findings suggested that the purchased BMSCs were of high purity and homogeneity and were available for subsequent experiments.
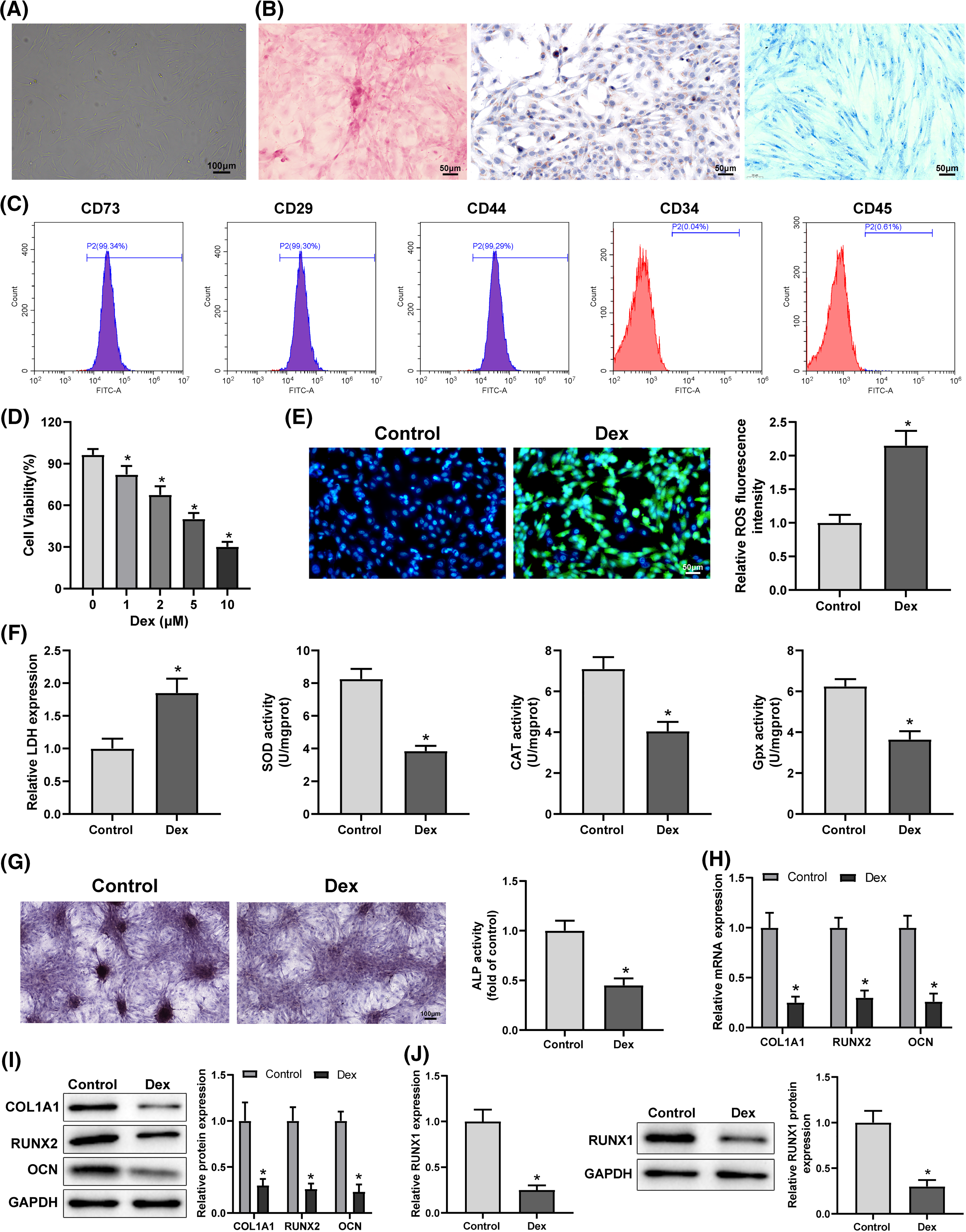
Figure 1: Dex caused low RUNX1 expression, impaired OD, and OS injury in BMSCs. Note: (A) morphological observation of BMSCs; (B) identification of OD (alizarin red staining), lipogenic differentiation (oil red O staining), and chondrogenic differentiation (alcian blue staining) of BMSCs; (C) flow cytometry to examine expression of BMSC markers CD29, CD73, and CD44 and non-BMSC markers CD45 and CD34. Then, BMSCs were treated with 5 μM Dex. (D) MTT assay to evaluate BMSC proliferation; (E) DCFH-DA molecular probe to detect ROS expression in BMSCs; (F) kits to examine the expression of OS indicators LDH, SOD, CAT, and Gpx in BMSCs; (G) ALP staining and ALP activity assay to assess OD of BMSCs; (H and I) qRT-PCR and Western Blot to measure the expression of OD-related proteins RUNX2, COL1A1, and OCN in BMSCs; (J) qRT-PCR and Western Blot to measure RUNX1 expression in BMSCs. Data were shown as mean ± standard deviation, N = 3. Comparisons between two groups were conducted using t-tests. *p < 0.05 compared with the control group. BMSCs, bone marrow mesenchymal stem cells; Dex, dexamethasone; RUNX1, Runt-related transcription factor 1; ROS, reactive oxygen species; LDH, lactate dehydrogenase; SOD, superoxide dismutase; CAT, catalase; Gpx, glutathione peroxidase; ALP, alkaline phosphatase; COL1A1, collagen type 1 alpha 1; OCN, osteocalcin.
Subsequently, the impacts of Dex on OD and OS in BMSCs were examined. MTT assay results exhibited that the increased concentrations of Dex substantially restrained BMSC proliferation ability, and the inhibition of cell viability was close to 50% after 5 μM Dex treatment (Fig. 1D, *p < 0.05), and 5 μM Dex was chosen to treat BMSCs in the follow-up experiments. The detection results of DCFH-DA fluorescent probes revealed that Dex treatment dramatically elevated ROS level in BMSCs (Fig. 1E, *p < 0.05). The detection results of the kits manifested that Dex treatment apparently increased LDH expression but decreased SOD, CAT, and Gpx expression in BMSCs (Fig. 1F, *p < 0.05). The results of ALP staining and activity assay to determine OD showed that Dex treatment resulted in a markedly lighter intensity of ALP staining and a remarkable reduction in calcified nodules and ALP activity (Fig. 1G, *p < 0.05). The results of Western Blot and qRT-PCR for OD-related protein expression presented that Dex treatment pronouncedly diminished RUNX2, COL1A1, and OCN expression (Figs. 1H and 1I, *p < 0.05). In addition, Western Blot and qRT-PCR results highlighted that Dex treatment dramatically reduced RUNX1 expression in BMSCs (Fig. 1J, *p < 0.05). These results indicated that Dex treatment resulted in impaired OD and OS injury in BMSCs with low RUNX1 expression.
Overexpression of RUNX1 suppressed Dex-induced impaired OD and OS injury in BMSCs
Dex-treated BMSCs were transfected with oe-RUNX1 or oe-NC to clarify the action of RUNX1 in Dex-induced impairment of OD and OS injury. The transfection efficiency was tested, which exhibited that oe-RUNX1 transfection considerably upregulated RUNX1 expression in Dex-treated BMSCs (Fig. 2A, *p < 0.05). Assays of OS in BMSCs demonstrated that oe-RUNX1 transfection notably lowered ROS and LDH levels, and upregulated cell proliferation rate and SOD, CAT, and Gpx levels in Dex-treated BMSCs (Figs. 2B–2D, *p < 0.05). Assays of OD of BMSCs manifested that oe-RUNX1 transfection resulted in a prominently darker intensity of ALP staining and a conspicuous elevation in calcified nodules, ALP activity, and RUNX2, COL1A1, and OCN levels in Dex-treated BMSCs (Figs. 2E and 2F, *p < 0.05). These findings indicated that overexpression of RUNX1 repressed Dex-induced impaired OD and OS injury in BMSCs.
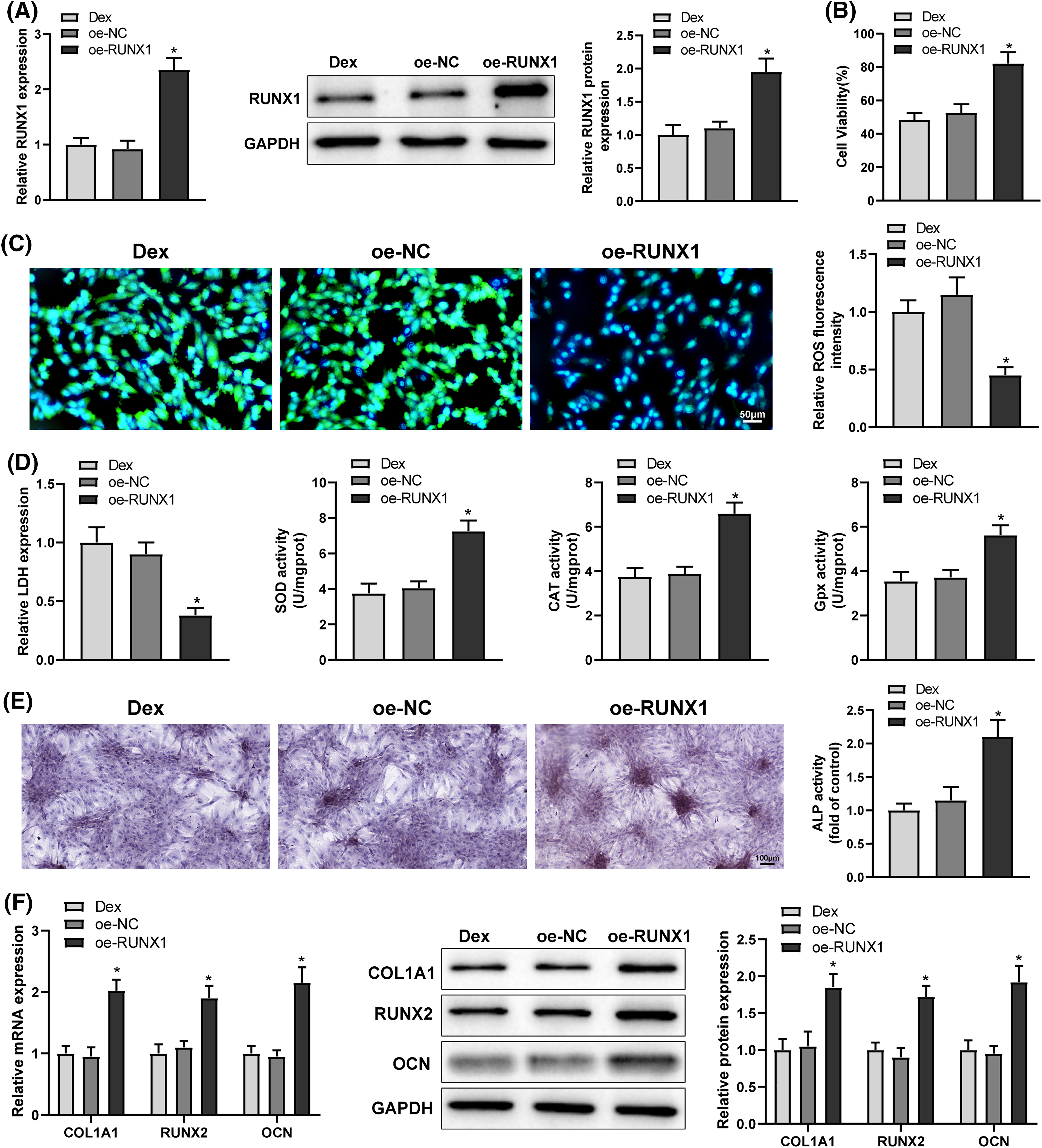
Figure 2: Dex-induced impairment of OD and OS injury in BMSCs was inhibited by RUNX1 overexpression. Note: Dex-treated BMSCs were transfected with oe-RUNX1. (A) qRT-PCR and Western Blot to detect RUNX1 expression in Dex-treated BMSCs; (B) MTT assay to measure BMSC proliferation; (C) DCFH-DA molecular probe to evaluate ROS expression in Dex-treated BMSCs; (D) kits to examine LDH, SOD, CAT, and Gpx expression in Dex-treated BMSCs; (E) ALP staining and ALP activity assay to determine OD of Dex-treated BMSCs; (F) qRT-PCR and Western Blot to detect RUNX2, COL1A1, and OCN expression in Dex-treated BMSCs. Data were shown as mean ± standard deviation, N = 3. Comparisons among multiple groups were conducted using one-way analysis of variance with Tukey’s multiple comparison test. *p < 0.05 compared with the oe-NC group. BMSCs, bone marrow mesenchymal stem cells; Dex, dexamethasone; RUNX1, Runt-related transcription factor 1; ROS, reactive oxygen species; LDH, lactate dehydrogenase; SOD, superoxide dismutase; CAT, catalase; Gpx, glutathione peroxidase; ALP, alkaline phosphatase; COL1A1, collagen type 1 alpha 1; OCN, osteocalcin.
RUNX1 bound to A2M promoter and promoted A2M transcription
Analyses of the hTFtarget and Jaspar websites identified that the transcription factor RUNX1 bound to A2M promoter and modulated A2M transcription (Fig. 3A). Western Blot and qRT-PCR results exhibited that A2M protein and mRNA expression was signally reduced in Dex-treated BMSCs (Fig. 3B, *p < 0.05). Therefore, we hypothesized that RUNX1 might engage in Dex-induced impaired OD and OS injury in BMSCs by modulating A2M transcription. Dual-luciferase reporter assays presented that oe-RUNX1 dramatically enhanced the luciferase activity of wt-A2M relative to oe-NC, whereas the luciferase activity of mut-A2M was not noticeably altered by oe-RUNX1 or oe-NC (Fig. 3C, *p < 0.05). ChIP assays displayed a pronounced increase in the A2M gene promoter bound by RUNX1 vs. lgG (Fig. 3D, *p < 0.05). Afterwards, Dex-treated BMSCs were transfected with oe-RUNX1 alone or combined with sh-A2M. Western Blot and qRT-PCR results manifested that oe-RUNX1 transfection strikingly upregulated A2M expression (*p < 0.05), while further sh-A2M transfection abolished the promotion of A2M expression by oe-RUNX1 transfection (Figs. 3E and 3F, #p < 0.05). These results indicated that RUNX1 bound to the A2M promoter and boosted A2M transcription.
Figure 3: RUNX1 bound to A2M and promoted A2M transcription. Note: (A) Jaspar website to predict the binding of RUNX1 to A2M; (B) qRT-PCR and Western Blot to measure A2M expression in Dex-treated BMSCs; (C) dual-luciferase reporter assay to verify the binding of RUNX1 to A2M; (D) ChIP assay to verify the binding of RUNX1 to A2M; (E and F) qRT-PCR and Western Blot to measure A2M expression in Dex-treated BMSCs after transfection of oe-RUNX1 alone or combined with sh-A2M. Data were shown as mean ± standard deviation, N = 3. Comparisons between two groups were conducted using t-test and comparisons among multiple groups were conducted using one-way analysis of variance with Tukey’s multiple comparison test. *p < 0.05 compared with the control group; #p < 0.05 compared with the oe-NC or IgG group; &p < 0.05 compared with the oe-RUNX1 + sh-NC group. BMSCs, bone marrow mesenchymal stem cells; Dex, dexamethasone; RUNX1, Runt-related transcription factor 1; A2M, alpha-2 macroglobulin.
Overexpression of A2M2 inhibited Dex-induced impaired OD and OS injury in BMSCs
To identify the function of A2M in Dex-induced impaired OD and OS injury, Dex-treated BMSCs were transfected with oe-A2M. The transfection efficiency was examined, and the results exhibited that oe-A2M transfection significantly boosted A2M expression in BMSCs (Fig. 4A, *p < 0.05). The results of OS experiments displayed that oe-A2M transfection considerably diminished ROS and LDH levels and augmented cell proliferation rate and SOD, CAT, and Gpx levels in Dex-treated BMSCs (Figs. 4B–4D, *p < 0.05). The results of OD experiments revealed that oe-A2M transfection resulted in a markedly darker intensity of ALP staining and a noticeable enhancement in calcified nodules, ALP activity, and RUNX2, COL1A1, and OCN expression in Dex-treated BMSCs (Figs. 4E and 4F, *p < 0.05). These findings suggested that overexpression of A2M attenuated Dex-induced impaired OD and OS injury in BMSCs.
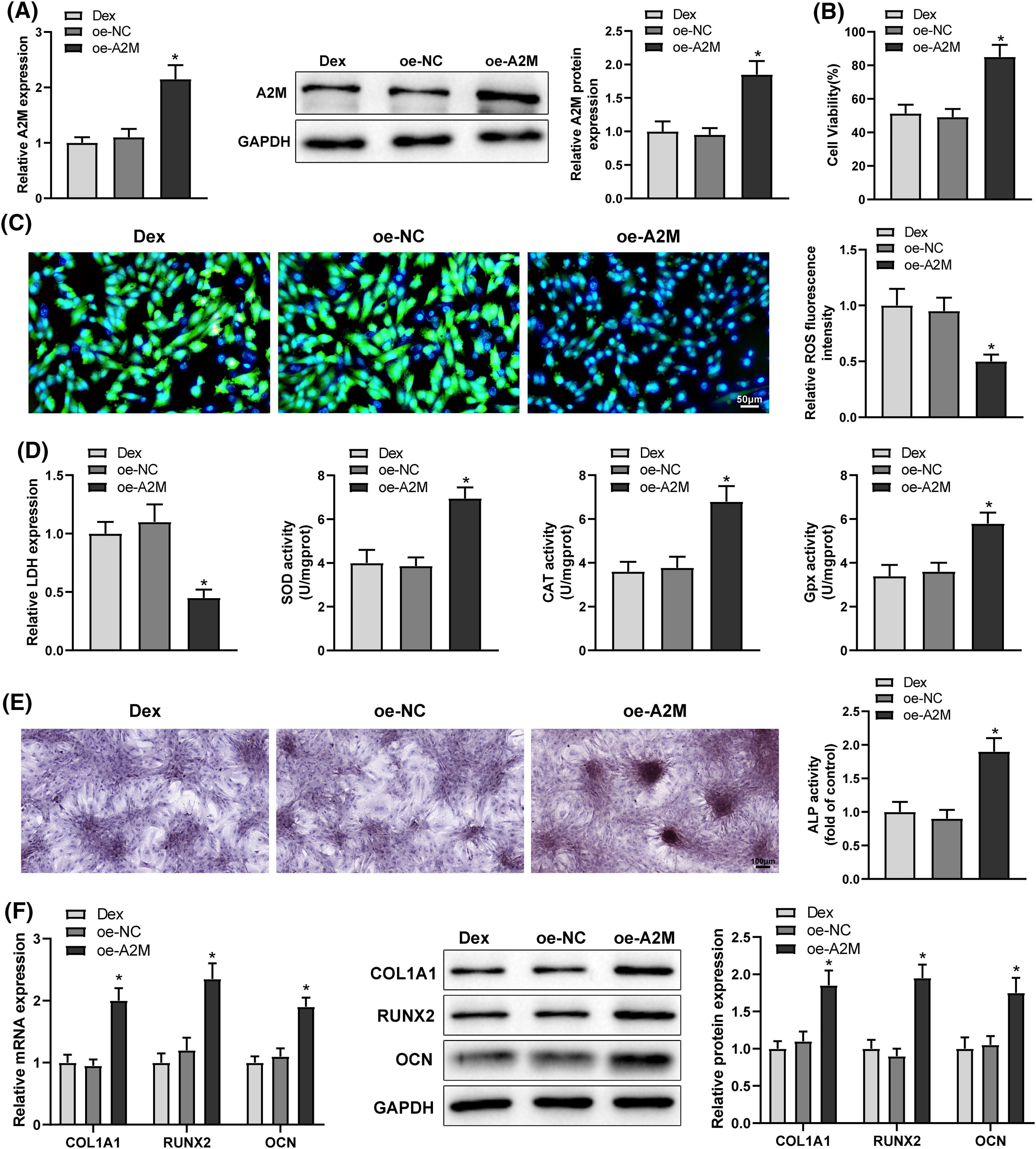
Figure 4: Overexpression of A2M alleviated Dex-induced impairment of OD and OS injury in BMSCs. Note: oe-A2M was transfected into Dex-treated BMSCs. (A) qRT-PCR and Western Blot to determine A2M expression; (B) MTT assay to assess cell proliferation; (C) DCFH-DA molecular probe to detect ROS expression; (D) kits to detect LDH, SOD, CAT, and Gpx expression; (E) ALP staining and ALP activity assay to measure OD; (F) qRT-PCR and Western Blot to determine RUNX2, COL1A1, and OCN expression in Dex-treated BMSCs. Data were shown as mean ± standard deviation, N = 3. Comparisons among multiple groups were conducted using one-way analysis of variance with Tukey’s multiple comparison test. *p < 0.05 compared with the oe-NC group. BMSCs, bone marrow mesenchymal stem cells; Dex, dexamethasone; RUNX1, Runt-related transcription factor 1; A2M, alpha-2 macroglobulin; ROS, reactive oxygen species; LDH, lactate dehydrogenase; SOD, superoxide dismutase; CAT, catalase; Gpx, glutathione peroxidase; ALP, alkaline phosphatase; COL1A1, collagen type 1 alpha 1; OCN, osteocalcin.
Overexpression of RUNX1 attenuated Dex-induced impaired OD and OS injury in BMSCs by promoting A2M transcription
To ascertain whether RUNX1 affected OD and OS in Dex-treated BMSCs by boosting A2M transcription, Dex-treated BMSCs were transfected with oe-RUNX1 alone or in combination with sh-A2M. Corresponding experimental results highlighted that sh-A2M transfection reversed the mitigating effect of oe-RUNX1 transfection on impaired OD and OS injury in Dex-induced BMSCs (Figs. 5A–5E, #p < 0.05). Taken together, RUNX1 overexpression attenuated Dex-induced impaired OD and OS injury in BMSCs by increasing A2M transcription.
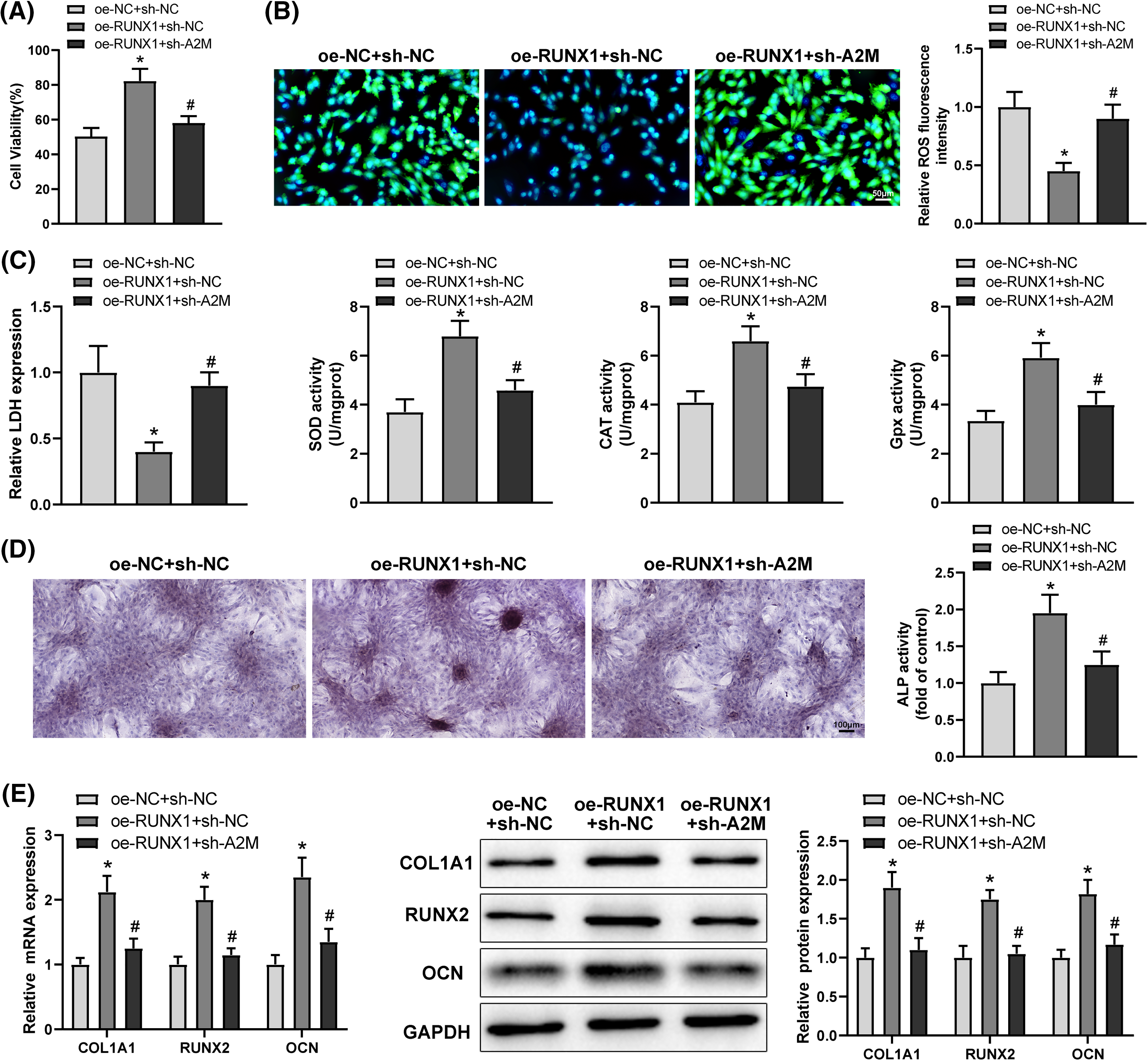
Figure 5: Overexpression of RUNX1 alleviated Dex-induced impairment of OD and OS injury in BMSCs via promotion of A2M transcription. Note: Dex-treated BMSCs were transfected with oe-RUNX1 alone or combined with sh-A2M. (A) MTT assay to determine cell proliferation; (B) DCFH-DA molecular probe to examine ROS expression; (C) kits to evaluate LDH, SOD, CAT, and Gpx expression; (D) ALP staining and ALP activity assay to assess OD; (E) qRT-PCR and Western Blot to examine RUNX2, COL1A1, and OCN expression in Dex-treated BMSCs. Data were shown as mean ± standard deviation, N = 3. Comparisons among multiple groups were conducted using one-way analysis of variance with Tukey’s multiple comparison test. *p < 0.05 compared with the oe-NC + sh-NC group; #p < 0.05 compared with the oe-RUNX1 + sh-NC group. BMSCs, bone marrow mesenchymal stem cells; Dex, dexamethasone; RUNX1, Runt-related transcription factor 1; A2M, alpha-2 macroglobulin; ROS, reactive oxygen species; LDH, lactate dehydrogenase; SOD, superoxide dismutase; CAT, catalase; Gpx, glutathione peroxidase; ALP, alkaline phosphatase; COL1A1, collagen type 1 alpha 1; OCN, osteocalcin.
One serious side effect of long-term Dex use is the elevated danger of osteoporosis or even osteonecrosis of the femoral head, which can result in substantial suffering to the patients [33]. High levels of OS after exposure to glucocorticoid generate excessive ROS that impairs DNA and cell membranes, lead impaired osteoblast differentiation, and even enhance apoptosis of BMSCs [26]. However, the particular molecular pathways via which Dex impacts OD and OS in BMSCs are unknown and warrant further investigation. This work disclosed that overexpression of RUNX1 attenuated Dex-induced impaired OD and OS injury in BMSCs by promoting A2M transcription.
Dex treatment of BMSCs resulted in a reduced intensity of ALP staining, decreased calcified nodules, ALP activity, and RUNX2, COL1A1, and OCN expression, enhanced ROS and LDH levels, and decreased cell proliferation rate and SOD, CAT, and Gpx expression in BMSCs. Since ALP, OCN, and COL1A1 are bone-specific matrix proteins related to bone formation, and RUNX2 is an essential transcriptional modulator of osteoblast differentiation [34]. OS is classified as a disturbance of the pro/antioxidant balance and excessively produces high ROS and reactive nitrogen species that are detrimental to cells [35]. LDH, SOD, CAT and Gpx are anti-oxidative and oxidative metabolites and enzymes [36], which are indicators of OS. This work indicated that Dex treatment hindered osteogenic development and caused OS injury in BMSCs, which is compatible with the findings of [26].
RUNX1 was documented to enhance glucocorticoid resistance in leukemia by enhancing long noncoding RNA (lncRNA) HOTAIRM1 expression [37]. RUNX1 is a member of the RUNX proteins (otherwise called master regulators of embryonic development), which are a family of transcription factors that exert critical functions in diverse biological processes, such as lineage determination and cell differentiation, apoptosis, and proliferation [38]. Our data revealed that RUNX1 expression was low in Dex-treated BMSCs. RUNX1 restrains adipogenesis and promoted osteoblast lineage commitment, osteoblast differentiation and proliferation, and bone formation by mediating several signaling pathways that participate in bone formation [39]. Overexpression of lncRNA maternally expressed gene 3 inactivates the miR-129-5p/RUNX1 axis to foster differentiation of BMSCs to chondrocytes [40]. Mesenchymal stem cells can differentiate into chondrocytes more easily when RUNX1 is over’ expressed [41]. Similarly, this work elucidated that overexpression of RUNX1 prevented Dex from impairing BMSCs’ ability to differentiate into osteoblasts. However, there has not been any prior discussion of RUNX1’s effect on OS in BMSCs. This work novelly disclosed that overexpression of RUNX1 inhibited Dex-induced OS injury in BMSCs.
Transcription factors are the proteins that bind to gene-specific sequences to modulate gene expression and transcription and perform a vital function in modulating cell biological activities [42]. In this work, analyses of the hTFtarget and Jaspar websites, as well as the dual-luciferase reporter and ChIP assays identified that the transcription factor RUNX1 bound to A2M promoter and enhanced A2M transcription. This work revealed that Dex-treated BMSCs had low A2M expression. A former paper illustrated that A2M plays a crucial function in osteogenesis by increasing the immediate availability of osteogenic growth peptide, which non-covalently complexes with thermosensitive and high molecular weight osteogenic growth peptide binding proteins, thereby stimulating bone formation [43]. Deer antler extract may inhibit the signaling network generated by Apof, A2M, Serpina3n, and their interacting regulators to orchestrate bone remodeling and formation, upsetting osteoblasts’ homeostasis [44]. As an enhancer of osteoblast differentiation, adrenocorticotropic hormone upregulates A2M to facilitate OD of BMSCs [45]. Likewise, this work uncovered that overexpression of A2M enhanced the OD of Dex-treated BMSCs. Moreover, A2M upregulates antioxidant enzyme (SOD2 and heme oxygenase-1) activities and resulted in a decrease in ROS and 8-hydroxy-2′-deoxyguanosine levels, thereby protecting BMSCs against OS injury [46]. A2M enhances the proliferative rate and OD and diminishes autophagy and apoptosis in BMSCs after irradiation through an antioxidant pathway [47]. Notably, this work found that overexpression of A2M attenuated Dex-induced OS injury in BMSCs. Besides, knockdown of A2M abolished the mitigating effect of RUNX1 overexpression on impaired OD and OS injury in Dex-induced BMSCs.
This research showed for the first time that RUNX1 bound to A2M and controlled its transcription. This work also unveiled that overexpression of RUNX1 repressed Dex-induced impairment of OD and OS injury in BMSCs by promoting A2M transcription. Since reduction in osteoblast proliferation and differentiation is a feature of osteoporosis and OS acts in osteoporosis pathogenesis [29], this work is expected to shed a novel light on the management of Dex-induced osteoporosis. However, the current study had the problem of not being validated in vivo. More comprehensive investigations about the effects of RUNX1/A2M on Dex-induced BMSCs in the animal setting and the modulation of other target genes are required in the future research.
Acknowledgement: Not applicable.
Funding Statement: Thanks for the grants from the Natural Science Foundation of Fujian Province (No. 2023J011558), the Innovation of Science and Technology of Fujian Province (No. 2021Y9098) and Fujian Provincial Finance Project (No. BPB-2022FSH).
Author Contributions: HQJ conceived the ideas. HQJ and ZHX designed the experiments. HQJ and ZHX performed the experiments. HQJ analyzed the data. HQJ provided critical materials. HQJ and ZHX wrote the manuscript. FSH supervised the study. All the authors have read and approved the final version for publication.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare there is no conflict of interests.
References
1. Andreakos E, Papadaki M, Serhan CN. Dexamethasone, pro-resolving lipid mediators and resolution of inflammation in COVID-19. Allerg. 2021;76:626–8. doi:10.1111/all.14595. [Google Scholar] [PubMed] [CrossRef]
2. Madamsetty VS, Mohammadinejad R, Uzieliene I, Nabavi N, Dehshahri A, García-Couce J, et al. Dexamethasone: insights into pharmacological aspects, therapeutic mechanisms, and delivery systems. ACS Biomater Sci Eng. 2022;8(5):1763–90. doi:10.1021/acsbiomaterials.2c00026. [Google Scholar] [PubMed] [CrossRef]
3. He HP, Gu S. The PPAR-γ/SFRP5/Wnt/β-catenin signal axis regulates the dexamethasone-induced osteoporosis. Cytokine. 2021;143(46):155488. doi:10.1016/j.cyto.2021.155488. [Google Scholar] [PubMed] [CrossRef]
4. Li J, Zhang N, Huang X, Xu J, Fernandes JC, Dai K, et al. Dexamethasone shifts bone marrow stromal cells from osteoblasts to adipocytes by C/EBPalpha promoter methylation. Cell Death Dis. 2013;4(10):e832. doi:10.1038/cddis.2013.348. [Google Scholar] [PubMed] [CrossRef]
5. Eid Y, Kirrella AA, Tolba A, El-Deeb M, Sayed S, El-Sawy HB, et al. Dietary pomegranate by-product alleviated the oxidative stress induced by dexamethasone in laying hens in the pre-peak period. Animals. 2021;11(4):1022. doi:10.3390/ani11041022. [Google Scholar] [PubMed] [CrossRef]
6. Ulla A, Uchida T, Miki Y, Sugiura K, Higashitani A, Kobayashi T, et al. Morin attenuates dexamethasone-mediated oxidative stress and atrophy in mouse C2C12 skeletal myotubes. Arch Biochem Biophys. 2021;704(4):108873. doi:10.1016/j.abb.2021.108873. [Google Scholar] [PubMed] [CrossRef]
7. Zhu Z, Wang X, Wang Z, Zhao Z, Zhou P, Gao X. Neobavaisoflavone protects osteoblasts from dexamethasone- induced oxidative stress by upregulating the CRNDE-mediated Nrf2/HO-1 signaling pathway. Drug Develop Res. 2021;82(7):1044–54. doi:10.1002/ddr.21811. [Google Scholar] [PubMed] [CrossRef]
8. Yang Y, Sun Y, Mao WW, Zhang H, Ni B, Jiang L. Oxidative stress induces downregulation of TP53INP2 and suppresses osteogenic differentiation of BMSCs during osteoporosis through the autophagy degradation pathway. Free Radic Bio Med. 2021;166(3):226–37. doi:10.1016/j.freeradbiomed.2021.02.025. [Google Scholar] [PubMed] [CrossRef]
9. Song M, Zhao D, Wei S, Liu C, Liu Y, Wang B, et al. The effect of electromagnetic fields on the proliferation and the osteogenic or adipogenic differentiation of mesenchymal stem cells modulated by dexamethasone. Bioelectromagnet. 2014;35(7):479–90. doi:10.1002/bem.21867. [Google Scholar] [PubMed] [CrossRef]
10. Tang J, Qing MF, Li M, Gao Z. Dexamethasone inhibits BMP7-induced osteogenic differentiation in rat dental follicle cells via the PI3K/AKT/GSK-3β/β-catenin pathway. Int J Med Sci. 2020;17(17):2663–72. doi:10.7150/ijms.44231. [Google Scholar] [PubMed] [CrossRef]
11. Kilbey A, Terry A, Wotton S, Borland G, Zhang Q, Mackay N, et al. Runx1 orchestrates sphingolipid metabolism and glucocorticoid resistance in lymphomagenesis. J Cell Biochem. 2017;118(6):1432–41. doi:10.1002/jcb.25802. [Google Scholar] [PubMed] [CrossRef]
12. Zhang D, Liang C, Li P, Yang L, Hao Z, Kong L, et al. Runt-related transcription factor 1 (Runx1) aggravates pathological cardiac hypertrophy by promoting p53 expression. J Cell Mol Med. 2021;25(16):7867–77. doi:10.1111/jcmm.16704. [Google Scholar] [PubMed] [CrossRef]
13. Li Q, Lai Q, He C, Fang Y, Yan Q, Zhang Y, et al. RUNX1 promotes tumour metastasis by activating the Wnt/β-catenin signalling pathway and EMT in colorectal cancer. J Exp Clin Cancer Res. 2019;38(1):334. doi:10.1186/s13046-019-1330-9. [Google Scholar] [PubMed] [CrossRef]
14. Liu S, Xie F, Gan L, Peng T, Xu X, Guo S, et al. Integration of transcriptome and cistrome analysis identifies RUNX1-target genes involved in pancreatic cancer proliferation. Genomics. 2020a;112(6):5343–55. doi:10.1016/j.ygeno.2020.11.010. [Google Scholar] [PubMed] [CrossRef]
15. Liu C, Xu D, Xue B, Liu B, Li J, Huang J. Upregulation of RUNX1 suppresses proliferation and migration through repressing VEGFA expression in hepatocellular carcinoma. Pathol Oncol Res. 2020b;26(2):1301–11. doi:10.1007/s12253-019-00694-1. [Google Scholar] [PubMed] [CrossRef]
16. Tang X, Sun L, Wang G, Chen B, Luo F. RUNX1: a regulator of NF-κB signaling in pulmonary diseases. Curr Protein Peptide Sci. 2018;19:172–8. doi:10.2174/1389203718666171009111835. [Google Scholar] [PubMed] [CrossRef]
17. Luo Y, Zhang Y, Miao G, Zhang Y, Liu Y, Huang Y. Runx1 regulates osteogenic differentiation of BMSCs by inhibiting adipogenesis through Wnt/β-catenin pathway. Arch Oral Biol. 2019;97:176–84. doi:10.1016/j.archoralbio.2018.10.028. [Google Scholar] [PubMed] [CrossRef]
18. Liu J, Chang X, Dong D. MicroRNA-181a-5p curbs osteogenic differentiation and bone formation partially through impairing runx1-dependent inhibition of AIF-1 transcription. Endocrinol Metab. 2023;38(1):156–73. doi:10.3803/EnM.2022.1516. [Google Scholar] [PubMed] [CrossRef]
19. Li P, Jia XY. MicroRNA-18-5p inhibits the oxidative stress and apoptosis of myocardium induced by hypoxia by targeting RUNX1. Eur Rev Med Pharmacol Sci. 2022;26:432–9. doi:10.26355/eurrev_202201_27867. [Google Scholar] [PubMed] [CrossRef]
20. Hong M, He J, Li D, Chu Y, Pu J, Tong Q, et al. Runt-related transcription factor 1 promotes apoptosis and inhibits neuroblastoma progression in vitro and in vivo. J Exp Clin Canc Res. 2020;39(1):52. doi:10.1186/s13046-020-01558-2. [Google Scholar] [PubMed] [CrossRef]
21. Vandooren J, Itoh Y. Alpha-2-macroglobulin in inflammation, immunity and infections. Front Immunol. 2021;12:803244. doi:10.3389/fimmu.2021.803244. [Google Scholar] [PubMed] [CrossRef]
22. Li J, Yin P, Chen X, Kong X, Zhong W, Ge Y, et al. Effect of α2- macroglobulin in the early stage of jaw osteoradionecrosis. Int J Oncol. 2020;57(1):213–22. doi:10.3892/ijo.2020.5051. [Google Scholar] [PubMed] [CrossRef]
23. Sunderic M, Vasovic T, Milcic M, Miljevic C, Nedic O, Nikolic MR, et al. Antipsychotic clozapine binding to alpha-2-macroglobulin protects interacting partners against oxidation and preserves the anti-proteinase activity of the protein. Int J Biol Macromol. 2021;183(2019):502–12. doi:10.1016/j.ijbiomac.2021.04.155. [Google Scholar] [PubMed] [CrossRef]
24. Chen J, Cui Z, Wang Y, Lyu L, Feng C, Feng D, et al. Cyclic polypeptide D7 protects bone marrow mesenchymal cells and promotes chondrogenesis during osteonecrosis of the femoral head via growth differentiation factor 15-mediated redox signaling. Oxid Med Cell Longev. 2022;2022(65):3182368. doi:10.1155/2022/3182368. [Google Scholar] [PubMed] [CrossRef]
25. Duan P, Wang H, Yi X, Zhang H, Chen H, Pan Z. C/EBPα regulates the fate of bone marrow mesenchymal stem cells and steroid-induced avascular necrosis of the femoral head by targeting the PPARγ signalling pathway. Stem Cell Res Ther. 2022;13(1):342. doi:10.1186/s13287-022-03027-3. [Google Scholar] [PubMed] [CrossRef]
26. Chen L, Wang BZ, Xie J, Zhang RY, Jin C, Chen WK, et al. Therapeutic effect of SIRT3 on glucocorticoid-induced osteonecrosis of the femoral head via intracellular oxidative suppression. Free Radic Biol Med. 2021;176(1):228–40. doi:10.1016/j.freeradbiomed.2021.07.016. [Google Scholar] [PubMed] [CrossRef]
27. He JY, Cheng M, Ye JL, Peng CH, Chen J, Luo B, et al. YY1-induced lncRNA XIST inhibits cartilage differentiation of BMSCs by binding with TAF15 to stabilizing FUT1 expression. Regen Ther. 2022;20(6):41–50. doi:10.1016/j.reth.2022.02.002. [Google Scholar] [PubMed] [CrossRef]
28. Sun Y, Zhu Y, Liu X, Chai Y, Xu J. Morroniside attenuates high glucose-induced BMSC dysfunction by regulating the Glo1/AGE/RAGE axis. Cell Proliferat. 2020;53(8):e12866. doi:10.1111/cpr.12866. [Google Scholar] [PubMed] [CrossRef]
29. Li Y, Zhang Y, Zhang X, Lu W, Liu X, Hu M, et al. Aucubin exerts anti-osteoporotic effects by promoting osteoblast differentiation. Aging. 2020;12(3):2226–45. doi:10.18632/aging.102742. [Google Scholar] [PubMed] [CrossRef]
30. Lv J, Hao YN, Wang XP, Lu WH, Xie LY, Niu D. Bone marrow mesenchymal stem cell-derived exosomal miR-30e-5p ameliorates high-glucose induced renal proximal tubular cell pyroptosis by inhibiting ELAVL1. Renal Fail. 2023;45(1):2177082. doi:10.1080/0886022X.2023.2177082. [Google Scholar] [PubMed] [CrossRef]
31. Leng Y, Wu Y, Lei S, Zhou B, Qiu Z, Wang K, et al. Inhibition of HDAC6 activity alleviates myocardial ischemia/reperfusion injury in diabetic rats: potential role of peroxiredoxin 1 acetylation and redox regulation. Oxid Med Cell Longev. 2018;2018:9494052. doi:10.1155/2018/9494052. [Google Scholar] [PubMed] [CrossRef]
32. Xiao R, Wang Q, Peng J, Yu Z, Zhang J, Xia Y. BMSC-derived exosomal Egr2 ameliorates ischemic stroke by directly upregulating SIRT6 to suppress notch signaling. Mol Neurobiol. 2023;60:1–17. doi:10.1007/s12035-022-03037-5. [Google Scholar] [PubMed] [CrossRef]
33. Hu H, Li Z, Lu M, Yun X, Li W, Liu C, et al. Osteoactivin inhibits dexamethasone-induced osteoporosis through up-regulating integrin β1 and activate ERK pathway. Biomed Pharmacother. 2018;105:66–72. doi:10.1016/j.biopha.2018.05.051. [Google Scholar] [PubMed] [CrossRef]
34. Hu H, Zhao C, Zhang P, Liu Y, Jiang Y, Wu E, et al. miR-26b modulates OA induced BMSC osteogenesis through regulating GSK3β/β-catenin pathway. Exp Mol Pathol. 2019;107(13):158–64. doi:10.1016/j.yexmp.2019.02.003. [Google Scholar] [PubMed] [CrossRef]
35. Daenen K, Andries A, Mekahli D, Van Schepdael A, Jouret F, Bammens B. Oxidative stress in chronic kidney disease. Pediatr Nephrol. 2019;34(6):975–91. doi:10.1007/s00467-018-4005-4. [Google Scholar] [PubMed] [CrossRef]
36. Huang B, Qian C, Ding C, Meng Q, Zou Q, Li H. Fetal liver mesenchymal stem cells restore ovarian function in premature ovarian insufficiency by targeting MT1. Stem Cell Res Ther. 2019;10(1):362. doi:10.1186/s13287-019-1490-8. [Google Scholar] [PubMed] [CrossRef]
37. Liang L, Gu W, Li M, Gao R, Zhang X, Guo C, et al. The long noncoding RNA HOTAIRM1 controlled by AML1 enhances glucocorticoid resistance by activating RHOA/ROCK1 pathway through suppressing ARHGAP18. Cell Death Dis. 2021;12(7):702. doi:10.1038/s41419-021-03982-4. [Google Scholar] [PubMed] [CrossRef]
38. Lin TC. RUNX1 and cancer. Biochim Biophys Acta (BBA)—Rev Cancer. 2022;1877:188715. doi:10.1016/j.bbcan.2022.188715. [Google Scholar] [PubMed] [CrossRef]
39. Tang CY, Wu M, Zhao D, Edwards D, McVicar A, Luo Y, et al. Runx1 is a central regulator of osteogenesis for bone homeostasis by orchestrating BMP and WNT signaling pathways. PLoS Genet. 2021;17(1):e1009233. doi:10.1371/journal.pgen.1009233. [Google Scholar] [PubMed] [CrossRef]
40. Zhu J, Fu Q, Shao J, Peng J, Qian Q, Zhou Y, et al. Over-expression of MEG3 promotes differentiation of bone marrow mesenchymal stem cells into chondrocytes by regulating miR-129-5p/RUNX1 axis. Cell Cycle. 2021;20(1):96–111. doi:10.1080/15384101.2020.1863043. [Google Scholar] [PubMed] [CrossRef]
41. Wang Y, Belflower RM, Dong YF, Schwarz EM, O’Keefe RJ, Drissi H. Runx1/AML1/Cbfa2 mediates onset of mesenchymal cell differentiation toward chondrogenesis. J Bone Miner Res. 2005;20:1624–36. doi:10.1359/JBMR.050516. [Google Scholar] [PubMed] [CrossRef]
42. Lv M, Liu AJ, Li QW, Su P. Progress on the origin, function and evolutionary mechanism of RHR transcription factor family. Yi Chuan. 2021;43:215–25. doi:10.16288/j.yczz.20-332 (In Chinese). [Google Scholar] [PubMed] [CrossRef]
43. Bari E, Roato I, Perale G, Rossi F, Genova T, Mussano F, et al. Biohybrid bovine bone matrix for controlled release of mesenchymal stem/stromal cell lyosecretome: a device for bone regeneration. Int J Mol Sci. 2021;22(8):4064. doi:10.3390/ijms22084064. [Google Scholar] [PubMed] [CrossRef]
44. Yao B, Gao H, Liu J, Zhang M, Leng X, Zhao D. Identification of potential therapeutic targets of deer antler extract on bone regulation based on serum proteomic analysis. Mol Biol Rep. 2019;46:4861–72. doi:10.1007/s11033-019-04934-0. [Google Scholar] [PubMed] [CrossRef]
45. Sadeghi F, Vahednia E, Naderi Meshkin H, Kerachian MA. The effect of adrenocorticotropic hormone on alpha-2-macroglobulin in osteoblasts derived from human mesenchymal stem cells. J Cell Mol Med. 2020;24(8):4784–90. doi:10.1111/jcmm.15152. [Google Scholar] [PubMed] [CrossRef]
46. Li ZH, Hu H, Zhang XY, Liu GD, Ran B, Zhang PG, et al. MiR-291a-3p regulates the BMSCs differentiation via targeting DKK1 in dexamethasone-induced osteoporosis. The Kaohsiung J Med Sci. 2020;36:35–43. doi:10.1002/kjm2.12134. [Google Scholar] [PubMed] [CrossRef]
47. Liu Y, Cao W, Kong X, Li J, Chen X, Ge Y, et al. Protective effects of α-2-macroglobulin on human bone marrow mesenchymal stem cells in radiation injury. Mol Med Rep. 2018;18:4219–28. doi:10.3892/mmr.2018.9449. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools