 Open Access
Open Access
ARTICLE
Candidate oncogene placenta specific 8 affects cell growth and cell migration in non-small cell lung cancers
Institute for Medical Biology and Hubei Provincial Key Laboratory for Protection and Application of Special Plants in Wuling Area of China, College of Life Sciences, South-Central Minzu University, Wuhan, 430074, China
* Corresponding Author: LU XUE. Email:
BIOCELL 2024, 48(2), 239-252. https://doi.org/10.32604/biocell.2023.045076
Received 16 August 2023; Accepted 30 November 2023; Issue published 23 February 2024
Abstract
Background: Placenta specific 8 (PLAC8) is a candidate oncogene involved in the development and progression of solid tumors. However, the status of PLAC8 in lung cancer (LC), especially non-small cell lung cancer (NSCLC) is still not lucid. Methods: Tissue microarray analysis (TMA) was performed to detect the expression patterns of PLAC8 in LC tissues and cell lines. Then a series of cellular experiments were performed fto assess cell proliferation, cell cycle profiles, and cell motility to explore the role of PLAC8 in NSCLC-derived cell lines: H1299 and A549. Results: TMA results showed that PLAC8 played complex and even contradictory roles in different LC samples. Detailed cell-based experiments confirmed that PLAC8 could promote cell viability, alter the cell cycle, and accelerate cell mobility by regulating cell cyclins or cadherins, respectively. Conclusions: The study findings indicate that PLAC8 might participate in LC, especially NSCLC, by affecting cellular physiological processes. The outcomes also shed new light on the potential role of PLAC8 as a therapeutic biomarker in NSCLC.Keywords
An earlier version of this article is available online as a preprint on Research Square [1].
Lung cancer (LC) is one of the leading causes of global cancer-related deaths [2–5]. In China, LC has become a severe public health problem and an economic burden because of incorrect diagnosis and high mortality [6,7]. Despite advances in our understanding of the diverse histological types and molecular types of LC [2], there are many challenges. For example, identifying more proto-oncogenes and tumor suppressors involved in lung tumorigenesis is warranted, which necessitates the exploration of novel molecular targets and biomarkers for early diagnosis and treatment of LC.
Placenta specific 8 (PLAC8), also known as Onzin, is a 115-amino acid protein that was first identified from a collection of 15,000 mouse genes with placental and embryonic RNAs [8]. PLAC8 is a small, highly conserved, cysteine-rich protein shown to be involved in the progression of various cancers, including LC, hepatocellular cancer, breast cancer, nasopharyngeal cancer, to name a few [9–13]. For example, down-regulated PLAC8 could promote cell viability, proliferation, and tumor formation via the miR-185-5p/PLAC8/β-catenin axis in hepatocellular carcinoma [10]. In another report, PLAC8 inhibited cell proliferation, cell invasion, and epithelial-mesenchymal transition in oral squamous cell carcinoma [11]. Further, PLAC8 contributed to tamoxifen resistance through the motigen-activated protein kinase (MAPK)/extracellular regulated protein kinases (ERK) pathway in breast cancer [12]. Interestingly, knockout of PLAC8 could increase the sensitivity of nasopharyngeal carcinoma cells to radiotherapy by promoting apoptosis [13]. Taking these findings together, PLAC8 is closely associated with the proliferation, apoptosis, migration, autophagy, chemo-resistance, and radio-sensitivity of several tumor cells. Furthermore, the impact of PLAC8 is heterogeneous and might have opposite effects in different tumors.
Recent studies have also revealed that PLAC8 might play an important role in LC [14,15]. For instance, a report showed that PLAC8 expression was elevated in lung tumors where the alteration of PLAC8 expression could affect tumor growth via the Krüppel-like factors (KLF4)/PLAC8 and Wnt/β-Catenin pathways. Notwithstanding these findings, the research on the involvement of PLAC8 in tumor cell migration and the underlying mechanism are yet to be probed.
Our previous study showed that the knockout of PLAC8 could affect the proliferation and migration of HEK293T [16]. In the current study, we found that PLAC8 expression was decreased in multiple LC cells, while its level in an inflammatory pseudotumor was elevated. Further study revealed that knockout of PLAC8 could inhibit cell growth and reduce cell migration, while overexpression of PLAC8 could promote cell proliferation and enhance cell motility. To summarize, PLAC8 could be an oncogene, which participated in LCs as a complex tumor regulator. Our study also confirmed that certain cell cyclins and epithelial cell adhesion molecules were involved in PLAC8-induced pathological changes in lung tumors. These results might provide novel molecular targets and novel insights for the treatment of LC.
Plasmids, single guide RNAs, and transfection
We established PLAC8 knockout H1299 cell lines employing the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 technology as previously described [16]. Briefly, the CRISPR/Cas9 editing plasmid, PX458 harboring PLAC8-targeted gRNA (5′-CACCGACTCTCTACAGGACCCGATA-3′) that showed editing efficiency in HEK293T cells [16] was transfected into H1299 cells with lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). The monoclonal PLAC8 knockout H1299 cell line was isolated from a single green fluorescent protein (GFP)+ cell and subjected to fluorescence-activated cell sorting (FACS) with an SH800S Cell sorter (Sony, Tokyo, Japan). Once the isolated H1299 cells reached 90%–100% confluence, two independent strains of monoclonal PLAC8 KO cell lines (H2-PLAC8 and H3-PLAC8) were identified and selected for further experiments.
The PLAC8-overexpressing A549 cell lines were constructed as previously described with minor modifications [17]. To summarize, the coding sequence (CDS) of PLAC8 was synthesized by Bio-Transduction Lab, Wuhan, China and was inserted into a hygromycin resistance mammalian expression vector pCMV-3tag-8 (Agilent Technologies, Palo Alto, LA, USA). It was transfected into A549 cells using lipofectamine 3000 (L3000015, Thermo Fisher Scientific, San Jose, CA, USA), while the original vector served as a negative control. The transfected A549 cells were treated with 300 µg/mL hygromycin (1366, BioFroxx, Germany) and incubated all normal A549 cells were all killed to obtain stable PLAC8-overexpressing clones. A stable PLAC8-overexpressing cell line (A549-OE8-2) was finally confirmed by western blotting.
Cell culture, reverse transcription and quantitative real-time polymerase chain reaction (RT-qPCR)
The human lung-derived cell lines (H1299, NCI-H46, 95-D, NCI-H1975, SPC-A-1, SK-MES-1, H125, and A549) were purchased from ScienCell (Shanghai, China). The purchased cell lines and the PLAC8 knockout cell lines (H2-PLAC8 and H3-PLAC8) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) at 37°C in a humid atmosphere containing 5% CO2. The PLAC8 overexpressing cell lines (A549-OE8-1 and A549-OE8-2) were cultured under the same conditions with the addition of 150 µg/mL hygromycin.
Total RNA was extracted from the cells using an RNA extraction kit (Bioteke, Beijing, China) to measure the PLAC8 mRNA levels in human lung-derived cell lines. The template cDNA was then synthesized with a cDNA synthesis kit (Thermo Fisher Scientific, San Jose, CA, USA). The primers used are listed as follows:
GAPDH-F: 5′-TGACTTCAACAGCGACACCCA-3′;
GAPDH-R: 5′-CACCCTGTTGCTGTAGCCAAA-3′;
PLAC8-F: 5′-AATTCAGCAGACACCTCTTCAG-3′;
PLAC8-R: 5′-GCTAAGTTCAGGGACAACATTCA-3′;
The quantitative real-time PCR was performed with SYBR qPCR Mix (Biosharp, Hefei, China) and analysis with a QuantStudio® 5 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) as previously described [18]. The mRNA expression levels of PLAC8 were calculated using the 2−ΔΔCt method and normalized against the expression levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
The immunohistochemical staining was performed as previously described with minor modifications [15]. Briefly, tissue microarrays (TMA) (LC2083) were purchased from Biomax, Derwood, MD, USA, which contained 30 cases of each squamous cell carcinoma and adenocarcinoma, 22 small cell carcinomas, 5 large cell carcinomas, 22 bronchioloalveolar carcinomas, 10 carcinoid, 5 sarcomatoid carcinomas, 30 matched or matched metastatic carcinomas, 10 inflammatory pseudotumors, 20 inflammation tissues, 18 matched or matched adjacent normal tissues and 6 normal tissues. These TMA slides were stained with PLAC8 antibodies (#13885, Cell Signaling Technology, Beverly, MA, USA, 1:1000) using anUltraSensitive™ SP (mouse/rabbit) IHC Kit (KIT-9720, MXB Biotechnologies, Fuzhou, China) according to the manufacturer’s protocol. The stained slides were embedded in neutral balsam (SJ-601, Leica, Heidelberg, Germany), and the bright-field images were photographed under a BX51 microscope (Olympus, Tokyo, Japan).
Cell counting assays were performed to detect the effects of PLAC8 on cell proliferation as previously reported with minor modifications [16,18]. In this assay, the cells were seeded at a density of 2,000 cells/well in 96-well tissue culture plates with 200 µL culture medium. After 4 h incubation with 100 µg of 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma, St Louis, MO, USA) reagent at 37°C, 150 µLdimethyl sulfoxide (DMSO, Sigma, St Louis, MO, USA) was added to each well, and the absorbance at 490 nm was recorded at 0, 24, 48, 96, 120 h, respectively. In the colony formation assay, approximately 200 cells were plated in 6-well plates and cultured for 10 days. The colonies were then fixed with 4% paraformaldehyde and stained with the fast Giemsa Stain kit (40751ES01, Yeasen, Shanghai, China) according to the manufacturer’s protocol. The stained colonies were photographed and counted. We computed the clone formation rate (CFR) using the expression: CFR = clone counts/seeded cell counts × 100%.
The cells were cultured in 60 mm cell culture dishes until they reached 90% confluence. The cells were collected and fixed with 70% ethanol for 2 h on ice to perform the subsequent cell cycle assay. The fixed cells were centrifuged and resuspended with staining buffer (10 mg/mL propidium iodide (PI): 1 mg/ml RNase: phosphate buffered solution (PBS) = 5: 1: 94). To perform the cell apoptosis assay, 1 × 106 cells were collected and stained with an rh Annexin V/FITC Kit (ANT001, AntGene, Wuhan, China) according to the manufacturer’s protocol. The cell cycle distribution and cell apoptosis were analyzed with a BD FACS Calibur Flow Cytometer (BD Biosciences, San Jose, CA, USA) following the manufacturer’s instructions.
Wound healing and invasion assays were performed as previously described with minor modifications [18]. For the wound healing assay, the cells were seeded in 60 mm cell culture dishes and allowed to reach 90% confluence. A wound was then made on the cell layer with a sterile 10 µL pipette tip. The scraped wounds were photographed and analyzed at 0, 12, 24, and 36 h, respectively. The rate of healing was calculated using the expression: rate = (1-wound width/original width) × 100%.
The invasion assay was performed with a 24-well plate Transwell® system with a polycarbonate filter membrane with a pore size of 8.0 μm (Corning Costar, Corning, NY, USA) as previously mentioned [16]. To summarize, 2 × 105 cells resuspended with DMEM containing 1% FBS were seeded on the filter, while the bottom plate was filled with DMEM with 10% FBS and the cells were cultured at 37°C for 48 h. Then, the cells on the top side of the filter were wiped out with a sterile cotton swab, and the migrated cells on the bottom side were fixed with 4% polyoxymethylene and stained with the fast Giemsa Stain kit. Randomly chosen fields (five) were photographed for each well under the microscope, and the number of migrating cells within each field was counted. The cells were lysed in protein lysis buffer (P0013J, Beyotime, Shanghai, China) supplemented with phenylmethanesulfonyl fluoride (PMSF, ST506, Beyotime, Shanghai, China) to perform western blotting. Subsequently, 20 μg of total proteins were separated using 10% SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose (NC) membranes. For PLAC8, we used PLAC8 (E1J2Z) Rabbit mAb (#13885, Cell Signaling Technology, Beverly, MA, USA, 1:1000) while cell cyclins were blotted with indicated antibodies using the Cell Cycle Regulation Antibody Sampler Kit II (#9870, Cell Signaling Technology, Beverly, MA, USA, 1:1000). The bands were then visualized employing the BeyoECL Plus kit (P0018S, Beyotime, Shanghai, China) according to the manufacturer’s protocol. Quantitative analysis was performed with the ImageLab® software (Bio-Rad, Hercules, CA, USA) and β-actin was the internal control.
All statistical analyses were performed using the software origin8.0. All experiments were repeated for at least three times independently. Differences between groups were calculated using a Student’s t-test. Results were expressed as means ± standard deviation (SD) and p < 0.05 was considered to indicate significant differences.
PLAC8 expression is repressed in lung cancers
PLAC8 has been reported to be a candidate oncogene associated with multiple diseases, especially tumors [9]. Previous studies with the oncomine database [19,20] have observed that the expression levels of PLAC8 were significantly decreased in LCs compared with the paired adjacent normal tissues [15]. The expression pattern of PLAC8 was analyzed in human LC tissue microarrays (TMA) to further identify the role of PLAC8 in LC. The PLAC8 staining intensity and the clinicopathological information of the TMAs have been shown in Table 1. We found that the staining signals of PLAC8 in inflammatory pseudotumors, chronic pneumonia, and adjacent normal lung tissue were much stronger than in lung carcinomas.
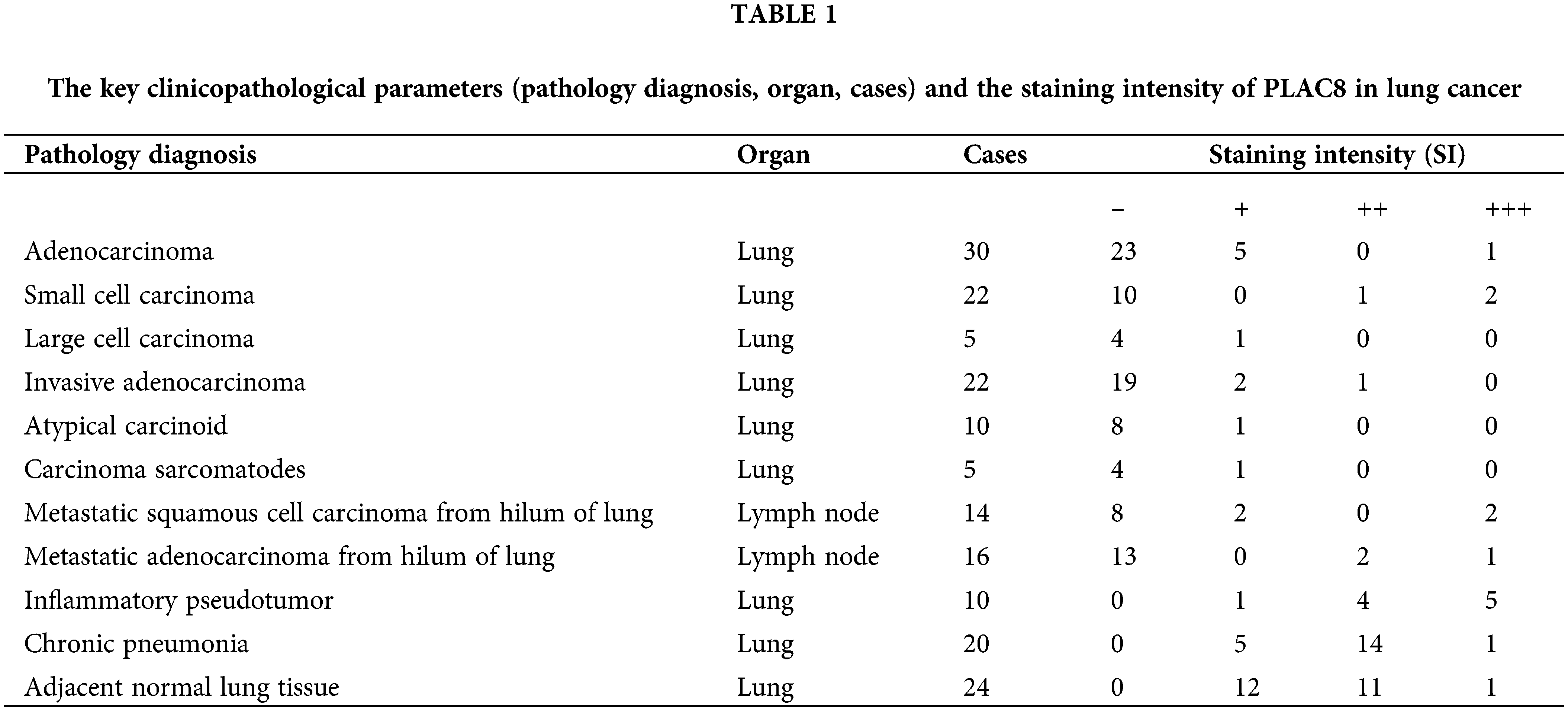
Further statistical analysis showed that the PLAC8 expression in six types of lung tumors (atypical carcinoid, metastatic squamous cell carcinoma, metastatic adenocarcinoma, squamous cell carcinoma, adenocarcinoma, and small cell carcinoma) were much lower compared with adjacent normal lung tissue (Fig. 1A), while the PLAC8 level in inflammatory pseudotumor was higher than that in adjacent normal lung tissue. Four representative images of PLAC8 expression (negative, weak, moderate, and strong) have been shown in Fig. 1B. Taking these results together, the expression of PLAC8 was decreased in LCs and increased in inflammation, which indicated that PLAC8 might play a crucial role in pathological processes.
Figure 1: The protein expression patterns of PLAC8 in lung cancer samples. (A) PLAC8 protein levels were reduced in 5 types of lung cancers (LCs) (atypical carcinoid, metastatic squamous cell carcinoma, metastatic adenocarcinoma, squamous cell carcinoma, and adenocarcinoma) and increased in inflammatory pseudotumors, compared with normal lung tissue. (B) Four representative images have been presented from the immunohistochemical (IHC) analysis of PLAC8 expression (negative, weak, moderate, and strong) in the LC samples.
Knockout of PLAC8 decreased cell proliferation and induced G1 arrest in the H1299 cell line
We next checked the expression levels of PLAC8 in 8 lung-derived cell lines. It was found that PLAC8 was highly expressed in H1299, 95-D, SPC-A-1, and H125 cells, while much lower levels were seen in NCI-H460, NCI-H1975, SK-MES-1, and A549 cells (Fig. 2). These results indicated that PLAC8 might be involved in different signal pathways and influence the pathological processes in different cell lines. To further investigate how PLAC8 played a role in tumorigenesis, H1299 was chosen to construct a PLAC8-knockout cell line, while A549 was chosen to establish a PLAC8-overexpressing cell line.
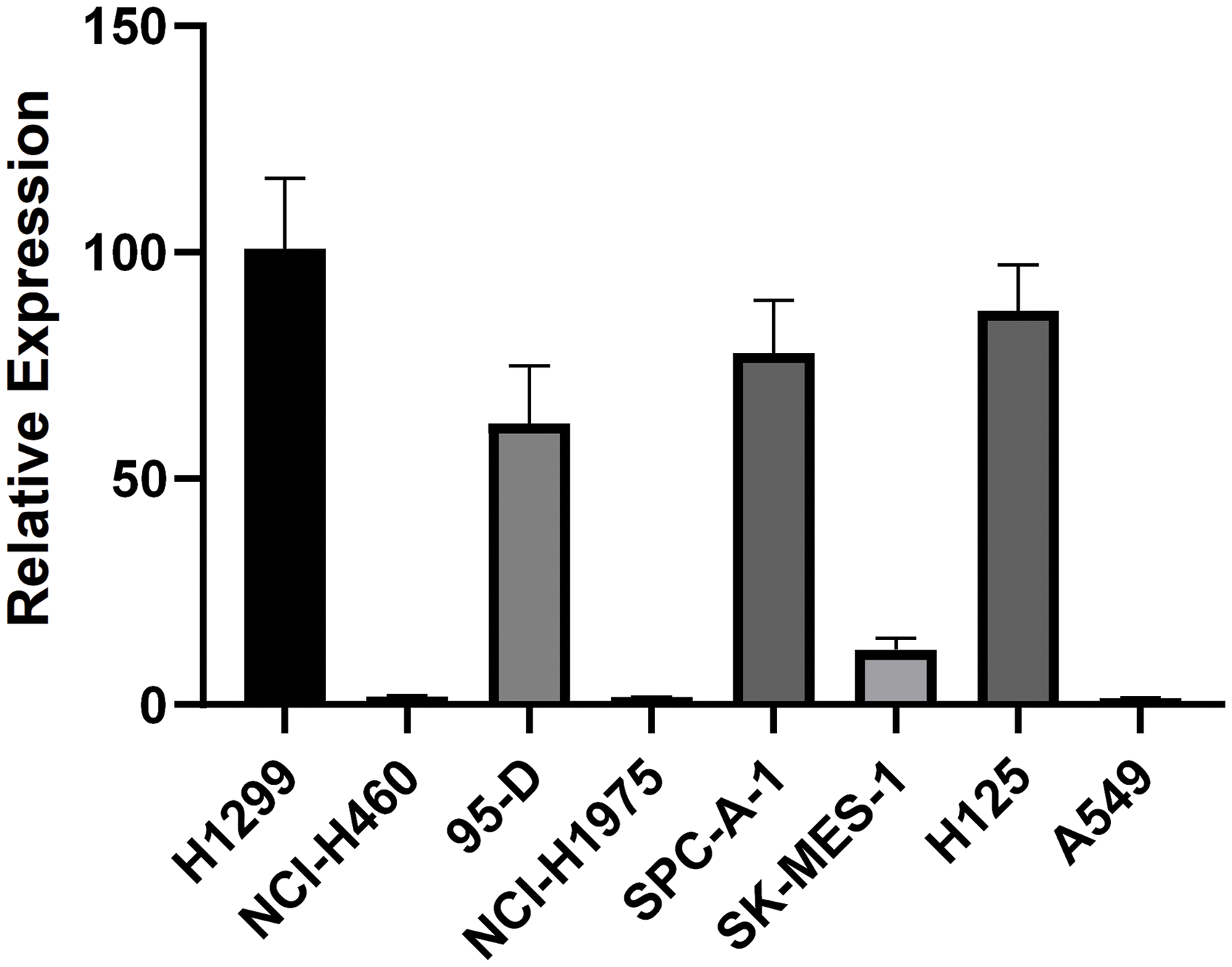
Figure 2: PLAC8 mRNA expression patterns in eight human lung cell lines. The relative mRNA expression of PLAC8 in the human lung cell lines H1299, NCI-H460, 95-D, NCI-H1975, SPC-A-1, SK-MES-1, H125 and A549, respectively. The expression of H1299 was normalized to 1.
Abnormal cell growth is reported to be a typical characteristic of pathological processes. We established two PLAC8 knockout H1299 cell lines. A comparison of mRNA sequences and predicted protein sequences of PLAC8 in H1299 (NM_001130716.2), H2-PLAC8, and H3-PLAC8 cells is shown in Suppl. Fig. S1. The function of PLAC8 in proliferation has been depicted in Fig. 3. As shown in Fig. 3A, the western blotting showed that PLAC8 was highly expressed in H1299, which is consistent with the mRNA expression result. Meanwhile, given that the protein expression of PLAC8 could not be detected in two knockout cell lines (H2-PLAC8 and H3-PLAC8), it indicated that the knockout cell lines were successfully established. Subsequent MTT assays revealed that both H2-PLAC8 and H3-PLAC8 exhibited decreased cell viability (Fig. 3B). Furthermore, colony formation assays showed that knockout of PLAC8 inhibited cell proliferation and growth in H2-PLAC8 and H3-PLAC8 cells (Figs. 3C and 3D). These results indicated that knockout of PLAC8 might play an inhibitory role on the proliferation of H1299 cells.
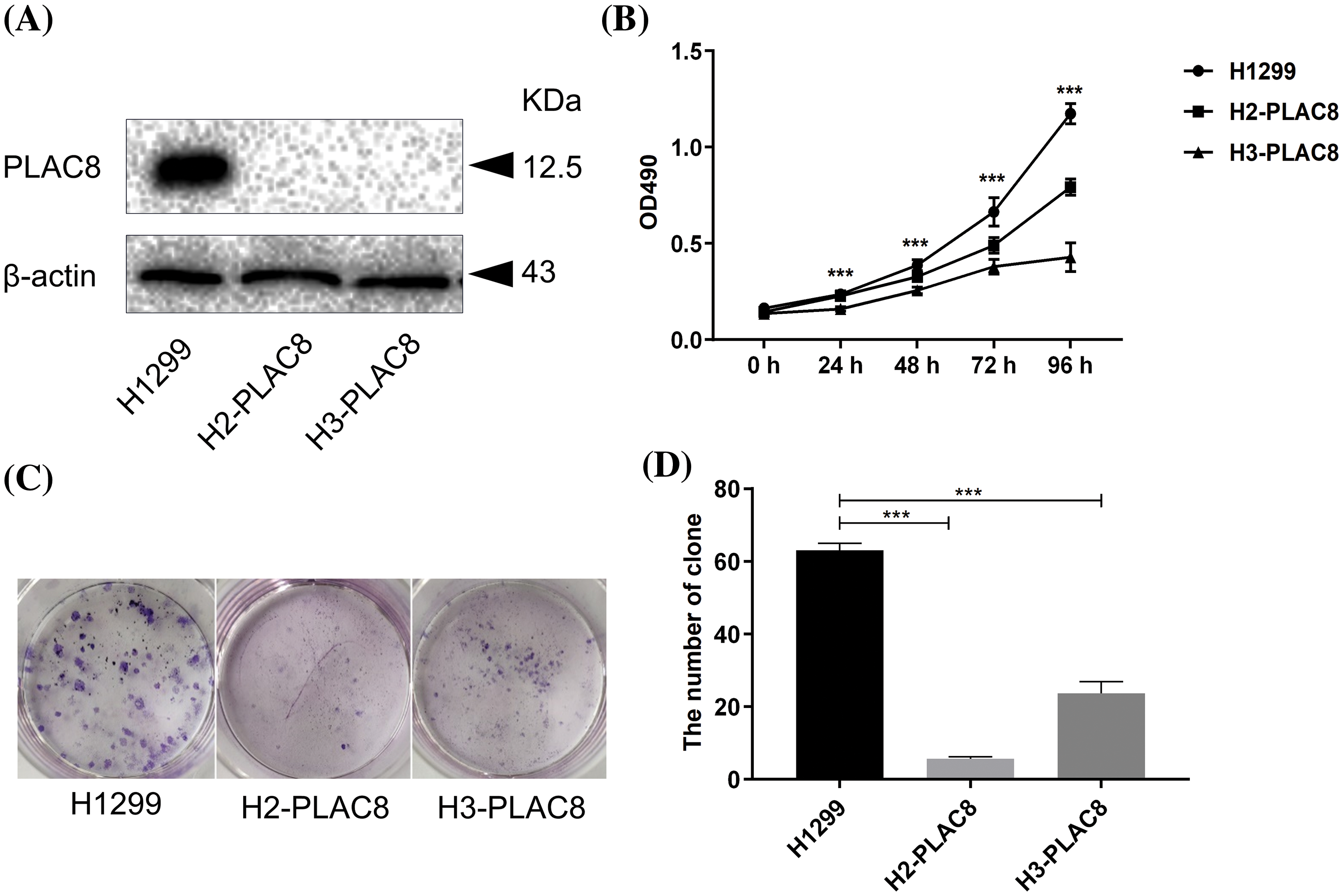
Figure 3: Knockout of PLAC8 inhibits cellular proliferation and colony formation in H1299 cells. (A) Western blotting analysis shows knockout of PLAC8 protein in two knockout cell lines (H2-PLAC8 and H3-PLAC8) compared with the control. (B) Knockout of PLAC8 inhibits cell proliferation, where the value at the starting point (day 1) was set to 1 (***p < 000.1). (C and D) Knockout of PLAC8 inhibits colony formation and the number of colonies has been quantified.
The distribution of the cell cycle phases was then detected and analyzed with flow cytometry. We found that knockout of PLAC8 redistributed the cell cycle in H1299 cells (Fig. 4). The percentage of the cells in the G1 phase was increased while that in the S phase was reduced in H2-PLAC8 and H3-PLAC8 cells (Fig. 4). To further explore the underlying molecular mechanisms, the expression levels of cell cyclins were detected employing western blotting. We found that the expression levels of P27kip1, P21waf/cip1, cyclin-dependent kinase4 (CDK4), and cyclin E2 were increased, while the expression levels of CDK2, cyclin D1, and cyclin D3 were decreased. These results indicated that PLAC8 might affect cell growth by regulating the cell cycle via cell-cyclin-induced G1 arrest.
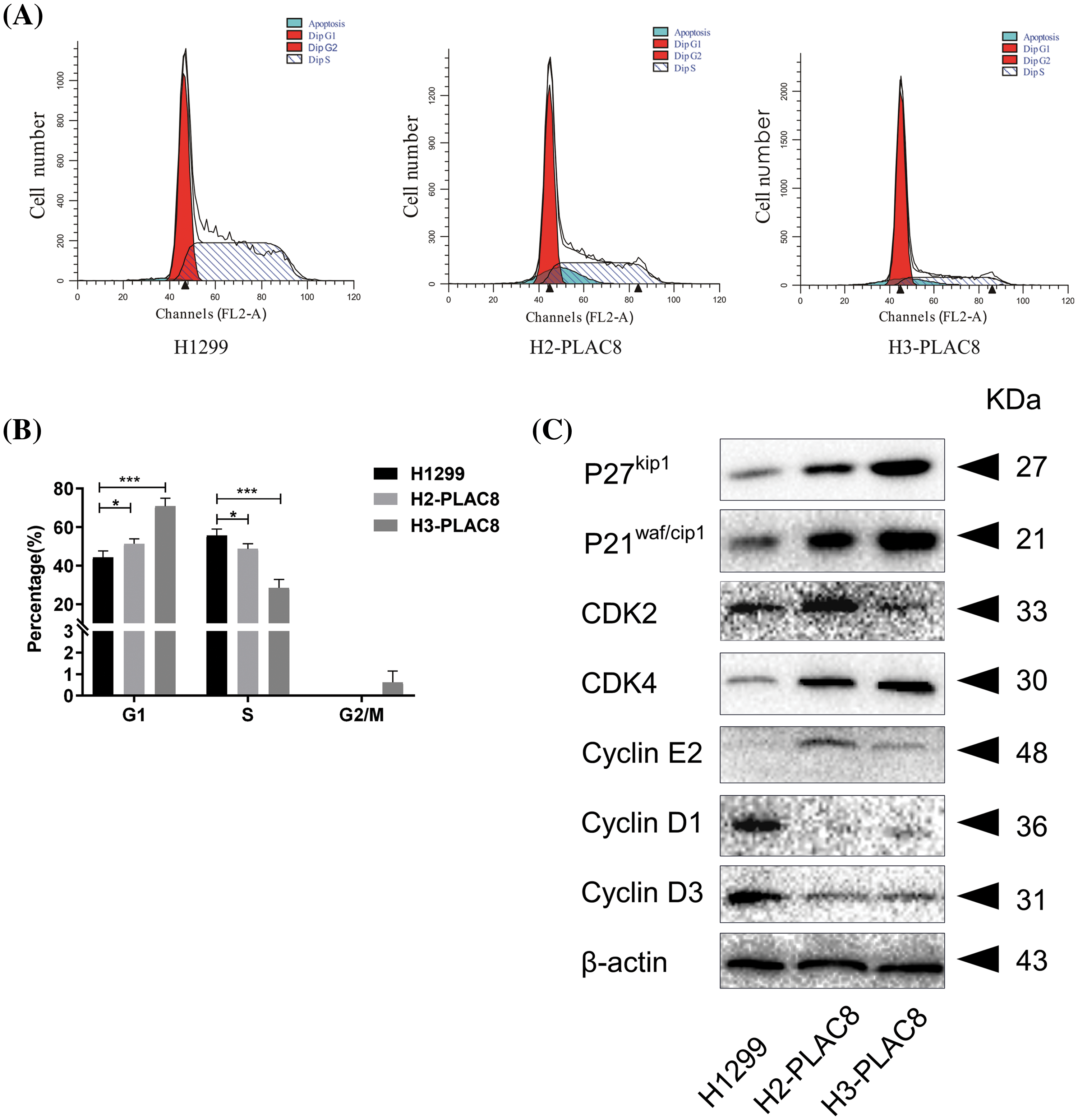
Figure 4: Knockout of PLAC8 induces G1 arrest in H1299 cells. (A and B) Knockout of PLAC8 increases the population of cells in the G1 phase, while reducing the number of cells in the S phase (*p < 0.05, ***p < 000.1). (C) Knockout of PLAC8 alters the expression of cell cycle proteins. Representative blots from three experiments with similar results are shown.
Knockout of PLAC8 suppressed cell motility in the H1299 cell line
The alteration of cell motility is important in metastasis of cancers [15]. We performed a wound-healing assay and invasion assay to determine the effect of PLAC8 on cell motility (Fig. 5). As shown in Figs. 5A and 5B, H2-PLAC8 and H3-PLAC8 cells exhibited significantly less wound closure compared to H1299 cells after 24 h, indicating that the PLAC8 knockout inhibited cell migration. Furthermore, the Transwell® chamber was employed to perform the cell invasion assay as outlined in the materials section. We observed that the number of invading cells in H2-PLAC8 and H3-PLAC8 cells were statistically lower than that in H1299 cells. Moreover, the expression level of E-cadherin, which is a key negative regulator of cell motility, was increased in H2-PLAC8 and H3-PLAC8 cells. These results indicated that PLAC8 might play a positive role in cell motility by regulating E-cadherin.
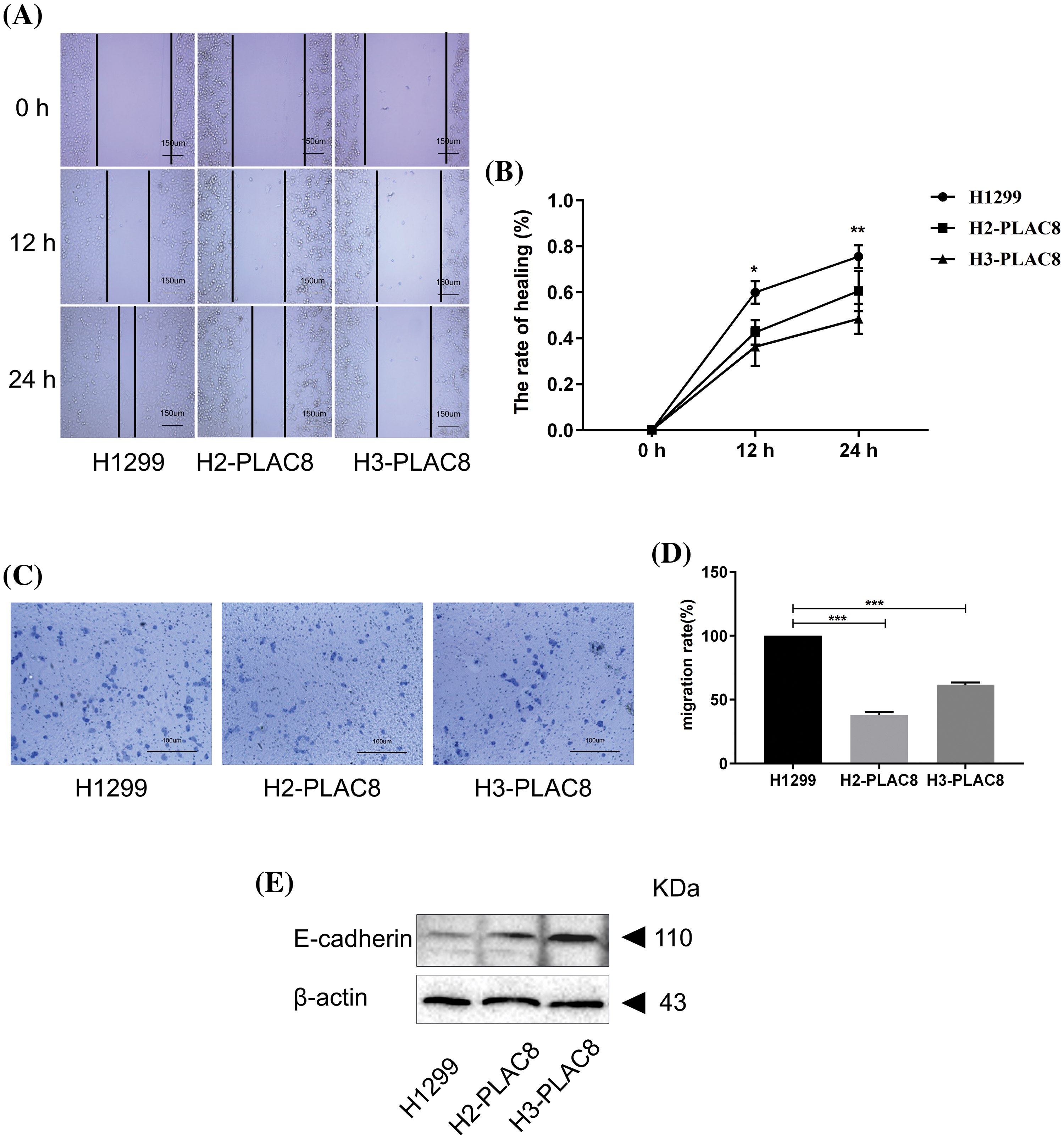
Figure 5: Knockout of PLAC8 inhibits cell migration in H1299 cells. (A and B) Knockout of PLAC8 inhibits cell motility in the wound-healing assay (*p < 0.05, **p < 0.01). (C and D) PLAC8 inhibits cell invasion (***p < 0.001). (E) Knockout of PLAC8 increases the expression of E-cadherin.
PLAC8 overexpresssion promoted cell proliferation and altered cell cycle distribution in the A549 cell line
We then assessed the multifaced roles of PLAC8 in LC in A549 cells by constructing and verifying PLAC8-overpressing cell lines, as shown in Fig. 6A. We successfully constructed a PLAC8-overexpressing cell line (A549-OE8-2), which could stably express PLAC8 protein. The MTT assay and colony formation assay were then also performed in the A549 cell line. We observed that A549-OE8-2 cells exhibited statistically higher cell viability than that of A549 cells (Fig. 6B). Meanwhile, colony formation results also indicated that overexpression of PLAC8 could significantly increase the number of cell colonies in A549 (Figs. 6C and 6D).
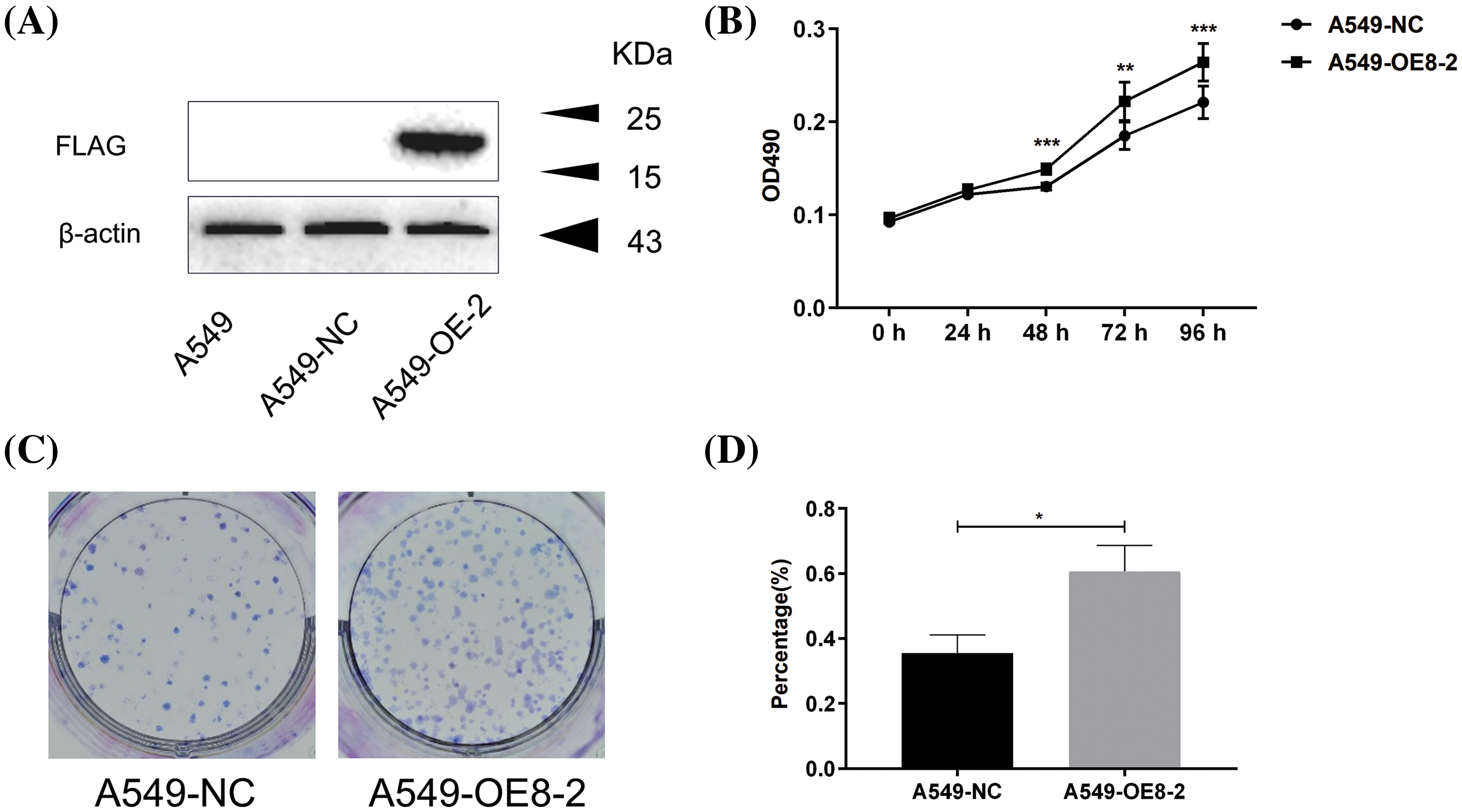
Figure 6: Overexpression of PLAC8 increases cell proliferation in A549 cells. (A) Western blotting shows overexpression of PLAC8 protein in an overexpression cell line (A549-OE8-2) compared with the control. (B) Overexpression of PLAC8 increases cell proliferation (**p < 0.01, ***p < 0.001). (C and D) Knockout of PLAC8 increases the rate of colony formation. The number of colonies has been quantified (*p < 0.05).
Subsequent flow cytometry analysis revealed that PLAC8 overexpression reduced the percentage of cells in the G2/M phase (Figs. 7A and 7B). The western blot showed that up-regulated cell cyclins included cyclin B1, cdc2, P-cdc2, P21waf/cip1, and P-H3, while down-regulated cell cyclins included cyclin E2, CDK2, P27kip1, and P-Wee1 (Fig. 7C). These results demonstrated that PLAC8 might promote proliferation of A549 cells through the altered expression of certain cell cyclins.
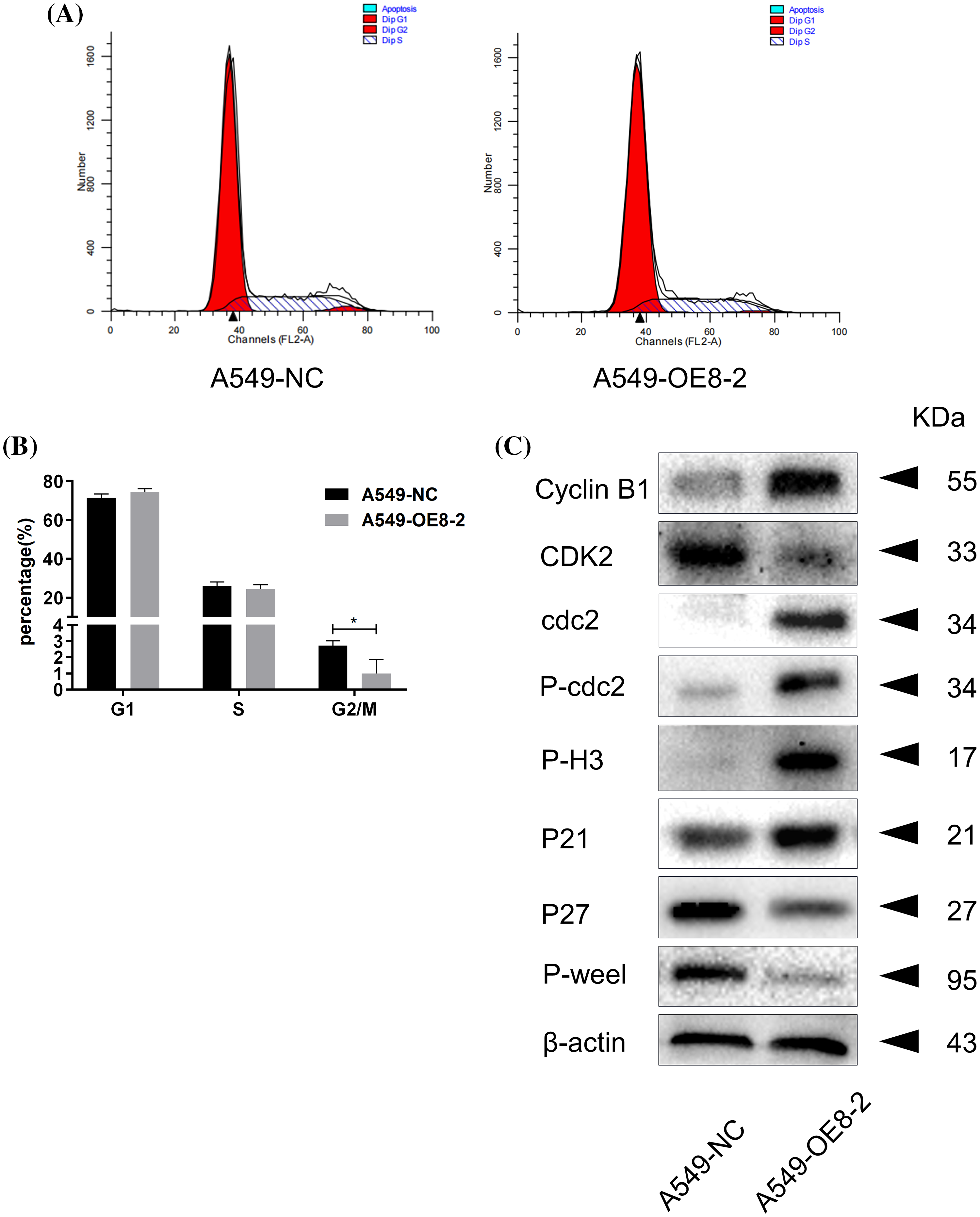
Figure 7: Overexpression of PLAC8 reduces the cell proportion in the G2/M phase in A549 cells. (A and B) Overexpression of PLAC8 reduces the population of cells in the G2/M phase (*p < 0.05). (C) Overexpression of PLAC8 alters the expression of cell cycle proteins. Representative blots from three experiments with similar results are shown.
Overexpressing PLAC8 enhanced cell motility in A549 cell lines
The wound-healing assay results showed that overexpression of PLAC8 could significantly accelerate wound closure in A549 cells compared with the negative control (Figs. 8A and 8B). The overexpression of PLAC8 also increased the number of invading cells in A549-OE8-2 cells, as shown in Figs. 8C and 8D. Moreover, while the expression level of N-cadherin was decreased in A549-OE8-2 cells, that of vimentin was up-regulated (Fig. 8E). Our results indicated that N-cadherin and vimentin might participate in the process of PLAC8-induced cell migration.
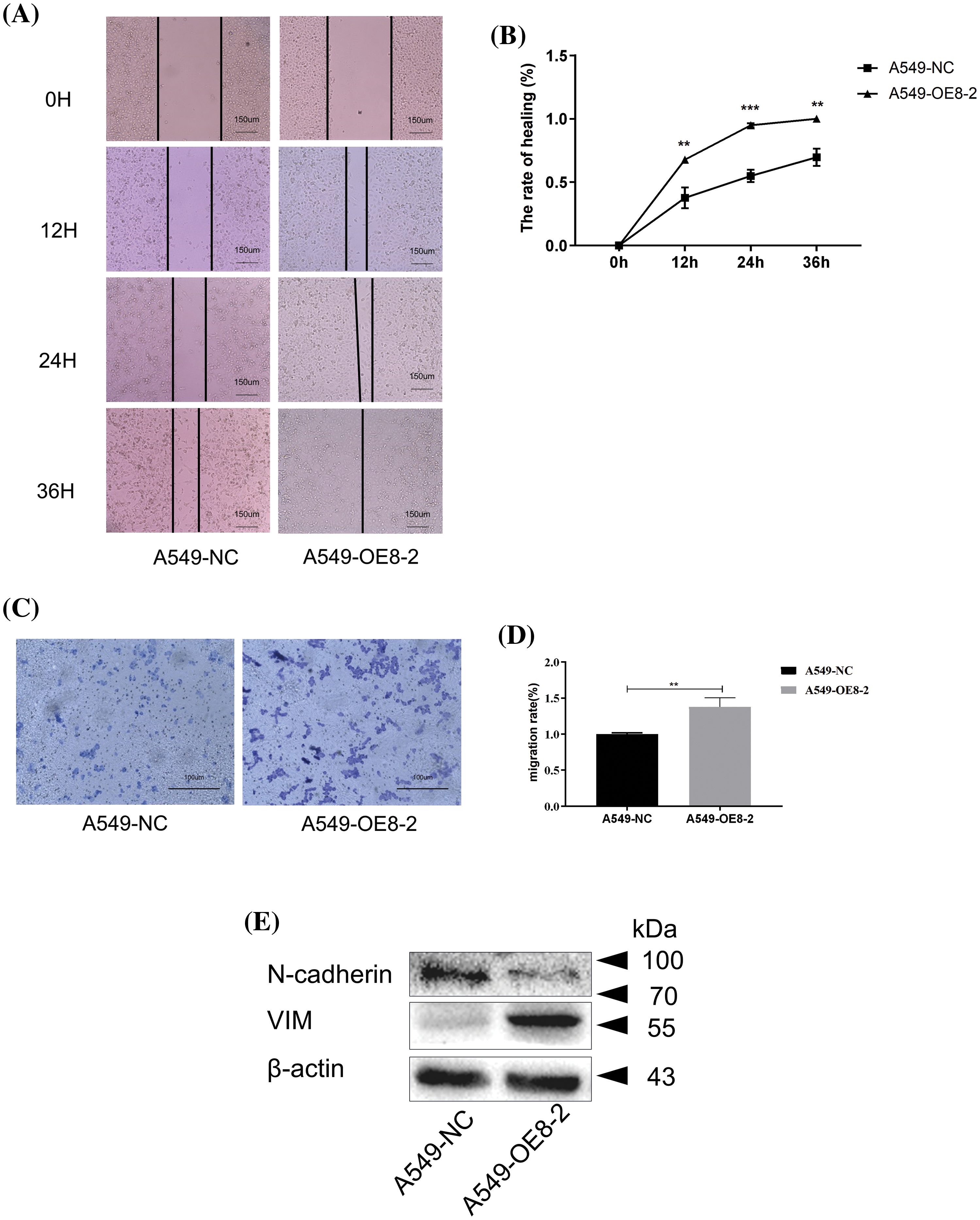
Figure 8: Overexpression of PLAC8 increases cell migration in A549. (A and B) Overexpression of PLAC8 increases cell motility in the wound-healing assay (**p < 0.01, ***p < 0.001). (C and D) Overexpression of PLAC8 increases cell invasion (**p < 0.01). (E) Overexpression of PLAC8 alters the expression of vimentin and N-cadherin.
PLAC8, a candidate oncogene was shown to be involved in several cases of solid tumor formation and metastasis, such as in the liver [10], prostate [21], kidney [22], etc. In the current study, we aimed to explore the role of PLAC8 in LC, given the paucity of reports. Our analysis in the human LC TMA showed that the expression of PLAC8 was decreased in several tumor tissues, while its level was up-regulated in inflammatory pseudotumors. These results indicated that PLAC8 might play complex even opposite roles in the different pulmonary pathological processes. This also provides a new insight into using PLAC8 as a potential biomarker to distinguish between inflammatory pseudotumors and tumors.
To further investigate the role of PLAC8 in LC tumorigenesis and metastasis, the expression levels of PLAC8 in different lung-derived cell lines were detected. We found that PLAC8 was highly expressed in an NSCLC cell line (H1299), a highly metastatic human lung cancer cell line (95-D), and two human lung adenocarcinoma cell lines (SPC-A-1 and H125). However, the expression levels of PLAC8 were much lower in three other NSCLC cell lines (A549, H460, and SK-MES-1), and a human lung adenocarcinoma cell line (H1975) derived from the normal human embryonic lung. It is hard to conclude to explain the complicated mRNA expression patterns of PLAC8 in different LC-derived cell lines and normal lung cell lines. However, this variation is consistent with the above results that PLAC8 plays a complex and even opposite role in different lung-derived cell lines.
The H1299 and A549 used here are both NSCLC cell lines. H1299 was derived from a metastatic lymph node and lacked p53 protein expression, while the A549 line was established from lung cancer tissue where p53 was normally expressed. A previous study indicated that p53 could suppress tumor growth in the case of NSCLC [23,24]. The expression levels of PLAC8 in H1299 were much higher than those in A549. A potential molecular mechanism could be that PLAC8 might participate in NSCLC metastasis as a part of p53 signaling cascades [23]. We chose two NSCLC cell lines, H1299 (p53-null) which highly expresses PLAC8, and A549 (p53-wild-type), s in which the expression level of PLAC8 was much lower.
To investigate the potential tumor suppressor/activator role of PLAC8 in NSCLC, we knocked out PLAC8 protein in H1299 and overexpressed PLAC8 in A549 cells. Our major aim was to assess the proliferative capacity of PLAC8 in LC cells. The cell growth experiments conducted in H1299 and A549 cells indicated that the PLAC8 knockout could inhibit H1299 cell proliferation, while PLAC8 upregulation could promote A549 cell growth. These results were consistent with other studies that studied the role of PLAC8 in LC [14,15]. Our results demonstrated that PLAC8 might be considered as a tumor activator, especially in NSCLC. Cell cycle assays showed that knockout of PLAC8 could accumulate a higher percentage of cells in the G1 phase and reduce the proportion of cells in the S phase. The potential molecular mechanism could be that G1 cyclin-dependent cell cycle inhibitors, including P27kip1 [25,26] and P21waf/cip1 [27,28] were up-regulated and inhibited cell cycle progression at the G1/S interface, resulting in G1 arrest. Furthermore, the expression of regulators necessary for the G1-S transition was also changed. We observed an increase in CDK4 and cyclin E2 levels and a reduction in cyclin D1, cyclin D3, and CDK2 levels in H1299 with PLAC8 knockout. It is well known that the cyclinD-CDK4/6 complex and the cyclin E/CDK2 complex play important roles in the cell cycle transition from the G1 phase to the S phase [29–31]. As shown in Suppl. Fig. S2A, a possible explanation is that the CDK inhibitors P27kip1 and P21waf/cip1 bind and then inactivate the cyclin D/CDK4/6 complex and the cyclin E/CDK2 kinase complex to induce the G1 arrest [32].
Additionally, we also observed alterations of the cell cycle in PLAC8 overexpressing A549 cells. The possible molecular mechanisms have been depicted in Suppl. Fig. S2B. The levels of the G2/M checkpoint regulators, such as cyclin B1 [33], P-cdc2 [34], P21waf/cip1 [35], and P-H3 [36] were up-regulated, while those of P-Wee1 [35], CDK2 [37], and P27kip1 [24,38] were lowered. The potential mechanism for this might be that overexpression of PLAC8 altered the expression of G2/M phase regulators, which lowered the cell proportion in the G2/M phase. Taken together, our results confirmed that the alteration of PLAC8 levels could affect cell growth and cell cycle via imbalanced expression of cell cycle regulators.
Recent studies have revealed that PLAC8 plays a crucial role in the malignant progressions of various cancers [22,39]. Given this background, we further explored the role of PLAC8 in NSCLC metastasis. It was observed that knockout of PLAC8 inhibited H1299 cell mobility along with the up-regulation of E-cadherin levels, which is a transmembrane protein that mediates cell-cell interaction and acts as a tumor suppressor in tumor metastasis [40,41]. On the other hand, PLAC8 overexpression promoted cell migration in A549 cells. Furthermore, the expression level of the cell adhesion molecule N-cadherin [42,43] was down-regulated, while that of the intermediate filament protein vimentin [44,45] was increased. Previous reports have documented that tumor metastasis is often associated with the loss of E-cadherin [46], while N-cadherin and vimentin could promote cell migration [47,48]. Taking these findings and reports together, our investigation hypothesized that overexpression of PLAC8 could affect cell migration and cell invasion via the regulation of cadherins and vimentin levels. Collectively, these results uncovered that PLAC8 could act as a tumor activator in NSCLC. The possible underlying molecular mechanism could be that PLAC8 promotes cell proliferation via the regulation of cell cyclins and accelerates cell migration through the alteration of cadherins and vimentin. Our findings suggest that the targeting of PLAC8 may be a new strategy for LC, especially for NSCLC treatment.
In the current study, we found that PLAC8 could affect cell proliferation and migration in NSCLC-derived cell lines by regulating cell cyclins and cadherins, respectively. Our study also indicated that PLAC8 might play a critical role in the development and progression of LC. Further understanding the novel functions of PLAC8 might open a new chance for detecting and treating LC. However, future animal experiments and preclinical trials are necessary to develop a strategy for the promising usage of PLAC8 in LC therapy.
Acknowledgement: We appreciate all the colleagues at the Institute for Medical Biology for their scientific and technical support.
Funding Statement: This project was supported by the Fund for Key Laboratory Construction of Hubei Province (Grant No. 2018BFC360), the National Natural Science Foundation of China (Grant No. 31101047 to Lu Xue), and “the Fundamental Research Funds for the Central Universities”, South-Central Minzu University (Grant Number: CZQ22013).
Author Contributions: The authors confirm contribution to the paper as follows: LX conceived and designed the experiments; MLZ, JNM, XX, XYG, and HXW performed the experiments; JS and LX analyzed the data and generated the figures; LX wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated and analyzed in this study are available upon reasonable request from the corresponding author.
Ethics Approval: The human tissue microarray (LC2083) analyzed in the current study was a commercial product which was provided by Biomax (Derwood, MD, USA). The detailed information regarding the same can be obtained on the following website: https://www.tissuearray.com/tissue-arrays/tissue-arrays/Regional_Lymph_Nodes_Metastasis/Lung/LC2083.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Zhou ML, Ma JN, Xu X, Gao XY, Wang HX, Shen J, et al. Candidate oncogene placenta specific 8 affect cell growth and cell migration in non-small cell lung cancers. Res Sq. 2023; 1–25. [Google Scholar]
2. Nooreldeen R, Bach H. Current and future development in lung cancer diagnosis. Int J Mol Sci. 2021;22(16):8661. [Google Scholar] [PubMed]
3. Nasim F, Sabath BF, Eapen GA. Lung cancer. Med Clin N Am. 2019;103(3):463–73. [Google Scholar] [PubMed]
4. Bade BC, Dela Cruz CS. Lung cancer 2020: epidemiology, etiology, and prevention. Clin Ches Med. 2020;41(1):1–24. [Google Scholar]
5. Rodriguez-Canales J, Parra-Cuentas E, Wistuba II. Diagnosis and molecular classification of lung cancer. Cancer Res Treat. 2016;170:25–46. [Google Scholar]
6. Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40–6. [Google Scholar] [PubMed]
7. She J, Yang P, Hong Q, Bai C. Lung cancer in China: challenges and interventions. Chest. 2013;143(4):1117–26. [Google Scholar] [PubMed]
8. Galaviz-Hernandez C, Stagg C, de Ridder G, Tanaka TS, Ko MS, Schlessinger D, et al. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene. 2003;309(2):81–9. [Google Scholar] [PubMed]
9. Mao M, Cheng Y, Yang J, Chen Y, Xu L, Zhang X, et al. Multifaced roles of PLAC8 in cancer. Biomark Res. 2021;9(1):73. [Google Scholar] [PubMed]
10. Zou L, Chai J, Gao Y, Guan J, Liu Q, Du JJ. Down-regulated PLAC8 promotes hepatocellular carcinoma cell proliferation by enhancing PI3K/Akt/GSK3β/Wnt/β-catenin signaling. Biomed Pharmacother. 2016;84(2):139–46. [Google Scholar] [PubMed]
11. Wu J, Wang X, Shang A, Vella G, Sun Z, Ji P, et al. PLAC8 inhibits oral squamous cell carcinogenesis and epithelial-mesenchymal transition via the Wnt/β-catenin and PI3K/Akt/GSK3β signaling pathways. Oncol Lett. 2020;20(5):128. [Google Scholar] [PubMed]
12. Mao M, Hu D, Yang J, Chen Y, Zhang X, Shen J, et al. Regulation of tamoxifen sensitivity by the PLAC8/MAPK pathway axis is antagonized by curcumin-induced protein stability change. J Mol Med. 2021;99(6):845–58. [Google Scholar] [PubMed]
13. Shen LJ, Qi CL, Yang R, Huang ML, Zou Y, Jiang Y, et al. PLAC8 gene knockout increases the radio-sensitivity of xenograft tumors in nude mice with nasopharyngeal carcinoma by promoting apoptosis. Am J Transl Res. 2021;13(6):5985–6000. [Google Scholar] [PubMed]
14. Chen W, Wu J, Wang W, Yu L, Xu X. PLAC8 overexpression promotes lung cancer cell growth via Wnt/β-catenin signaling. J Immunol Res. 2022;2022:8854196. [Google Scholar] [PubMed]
15. Jia Y, Ying X, Zhou J, Chen Y, Luo X, Xie S, et al. The novel KLF4/PLAC8 signaling pathway regulates lung cancer growth. Cell Death Dis. 2018;9(6):603. [Google Scholar] [PubMed]
16. Qin XH, Wang HX, Ma L, Shen J, Liu QH, Xue L. Knockout of the placenta specific 8 gene affects the proliferation and migration of human embryonic kidney 293T cell. Cell Biochem Biophys. 2020;78(1):55–64. [Google Scholar] [PubMed]
17. Ha DTT, Glab-Ampai K, Rojsitthisak P, Vajragupta O. Production of human embryonic kidney 293T cells stably expressing C-X-C chemokine receptor type 4 (CXCR4) as a screening tool for anticancer lead compound targeting CXCR4. Life Sci. 2022;303(8):120661. [Google Scholar] [PubMed]
18. Wang HX, Qin XH, Shen J, Liu QH, Shi YB, Xue L. Proteomic analysis reveals that placenta-specific protein 9 inhibits proliferation and stimulates motility of human bronchial epithelial cells. Front Oncol. 2021;11:628480. [Google Scholar] [PubMed]
19. Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. [Google Scholar] [PubMed]
20. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–80. [Google Scholar] [PubMed]
21. Kolluru V, Pal D, Papu John AMS, Ankem MK, Freedman JH, Damodaran C. Induction of Plac8 promotes pro-survival function of autophagy in cadmium-induced prostate carcinogenesis. Cancer Lett. 2017;408:121–9. [Google Scholar] [PubMed]
22. Shi L, Xiao L, Heng B, Mo S, Chen W, Su Z. Overexpression of placenta specific 8 is associated with malignant progression and poor prognosis of clear cell renal cell carcinoma. Int Urol Nephrol. 2017;49(7):1165–76. [Google Scholar] [PubMed]
23. Gupta S, Silveira DA, Mombach JCM. Towards DNA-damage induced autophagy: a Boolean model of p53-induced cell fate mechanisms. DNA Repair. 2020;96:102971. [Google Scholar] [PubMed]
24. Song Z, Yin Y, Hao S, Wei J, Liu B, Huang X, et al. JS‐K induces G2/M phase cell cycle arrest and apoptosis in A549 and H460 cells via the p53/p21WAF1/CIP1 and p27KIP1 pathways. Oncol Rep. 2019;41(6):3475–87. [Google Scholar] [PubMed]
25. Meroni SB, Galardo MN, Rindone G, Gorga A, Riera MF, Cigorraga SB. Molecular mechanisms and signaling pathways involved in sertoli cell proliferation. Front Endocrinol. 2019;10:224. [Google Scholar]
26. Nakayama KI, Hatakeyama S, Nakayama K. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem Bioph Res Co. 2001;282(4):853–60. [Google Scholar]
27. Kuczyk MA, Machtens S, Bokemeyer C, Hradil K, Macheel I, Jetscho V, et al. Prognostic value of p27Kip1 and p21WAF/Cip protein expression in muscle invasive bladder cancer. Oncol Rep. 1999;6(3):687–93. [Google Scholar] [PubMed]
28. Kuczyk MA, Bokemeyer C, Hartmann J, Schubach J, Walter C, Machtens S, et al. Predictive value of altered p27Kip1 and p21WAF/Cip1 protein expression for the clinical prognosis of patients with localized prostate cancer. Oncol Rep. 2001;8(6):1401–7. [Google Scholar] [PubMed]
29. Wang R, Xu K, Gao F, Huang J, Guan X. Clinical considerations of CDK4/6 inhibitors in triple-negative breast cancer. Biochim Biophys Acta Rev Cancer. 2021;1876(2):188590. [Google Scholar] [PubMed]
30. Romero-Pozuelo J, Figlia G, Kaya O, Martin-Villalba A, Teleman AA. Cdk4 and Cdk6 couple the cell-cycle machinery to cell growth via mTORC1. Cell Rep. 2020;31(2):107504. [Google Scholar] [PubMed]
31. Sun L, Huang Y, Wei Q, Tong X, Cai R, Nalepa G, et al. Cyclin E-CDK2 protein phosphorylates plant homeodomain finger protein 8 (PHF8) and regulates its function in the cell cycle. J Biol Chem. 2015;290(7):4075–85. [Google Scholar] [PubMed]
32. Abdel-Wahab N, Weston BS, Roberts T, Mason RM. Connective tissue growth factor and regulation of the mesangial cell cycle: role in cellular hypertrophy. J Am Soc Nephrol. 2002;13(10):2437–45. [Google Scholar] [PubMed]
33. Petrachkova T, Wortinger LA, Bard AJ, Singh J, Warga RM, Kane DA. Lack of Cyclin B1 in zebrafish causes lengthening of G2 and M phases. Dev Biol. 2019;451(2):167–79. [Google Scholar] [PubMed]
34. Stark GR, Taylor WR. Analyzing the G2/M checkpoint. Methods Mol Biol. 2004;280:51–82. [Google Scholar] [PubMed]
35. Pai JT, Hsu MW, Leu YL, Chang KT, Weng MS. Induction of G2/M cell cycle arrest via p38/p21(Waf1/Cip1)-dependent signaling pathway activation by bavachinin in non-small-cell lung cancer cells. Molecules. 2021;26(17):2303-16. [Google Scholar]
36. Xie CL, Zhang D, Lin T, He ZH, Yan QX, Cai Q, et al. Antiproliferative sorbicillinoids from the deep-sea-derived Penicillium allii-sativi. Front Microbiol. 2020;11:636948. [Google Scholar] [PubMed]
37. Chung JH, Bunz F. Cdk2 is required for p53-independent G2/M checkpoint control. PLoS Genet. 2010;6(2):e1000863. [Google Scholar] [PubMed]
38. Fontde Mora J, Uren A, Heidaran M, Santos E. Biological activity of p27kip1 and its amino- and carboxy-terminal domains in G2/M transition of Xenopus oocytes. Oncogene. 1997;15(21):2541–51. [Google Scholar] [PubMed]
39. Huang CC, Shen MH, Chen SK, Yang SH, Liu CY, Guo JW, et al. Gut butyrate-producing organisms correlate to placenta specific 8 protein: importance to colorectal cancer progression. J Adv Res. 2020;22(4):7–20. [Google Scholar] [PubMed]
40. Na TY, Schecterson L, Mendonsa AM, Gumbiner BM. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci USA. 2020;117(11):5931–7. [Google Scholar] [PubMed]
41. Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. J Cell Sci. 2013;126:393–401. [Google Scholar] [PubMed]
42. Cao ZQ, Wang Z, Leng P. Aberrant N-cadherin expression in cancer. Biomed Pharmacother. 2019;118(4):109320. [Google Scholar] [PubMed]
43. Blaschuk OW. N-cadherin antagonists as oncology therapeutics. Philos Trans R Soc Lond B Biol Sci. 2015;370(1661):20140039. [Google Scholar] [PubMed]
44. Satelli A, Li S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell Mol Life Sci. 2011;68(18):3033–46. [Google Scholar] [PubMed]
45. Battaglia RA, Delic S, Herrmann H, Snider NT. Vimentin on the move: new developments in cell migration. F1000Res. 2018;7:1–10. [Google Scholar]
46. Shamir ER, Ewald AJ. Adhesion in mammary development: novel roles for E-cadherin in individual and collective cell migration. Curr Top Dev Biol. 2015;112(12):353–82. [Google Scholar] [PubMed]
47. Choi S, Yu J, Kim W, Park KS. N-cadherin mediates the migration of bone marrow-derived mesenchymal stem cells toward breast tumor cells. Theranostics. 2021;11(14):6786–99. [Google Scholar] [PubMed]
48. Karoii DH, Azizi H, Amirian M. Signaling pathways and protein-protein interaction of vimentin in invasive and migration cells: a review. Cell Reprogram. 2022;24(4):165–74. [Google Scholar] [PubMed]
Supplementary Materials

Figure S1: Comparison of mRNA sequences and predicted protein sequences of PLAC8 in H1299, H2-PLAC8, and H3-PLAC8 cells. (A) Comparsion of PLAC8 mRNA sequences of among H1299, H2-PLAC8, and H3-PLAC8 cell lines. The gRNA sequence has been labeled with a red frame. The stop codons in two mutants have been labeled with blue frames. (B) Comparison of predicted protein sequences of PLAC8 among H1299, H2-PLAC8, and H3-PLAC8 cell lines.
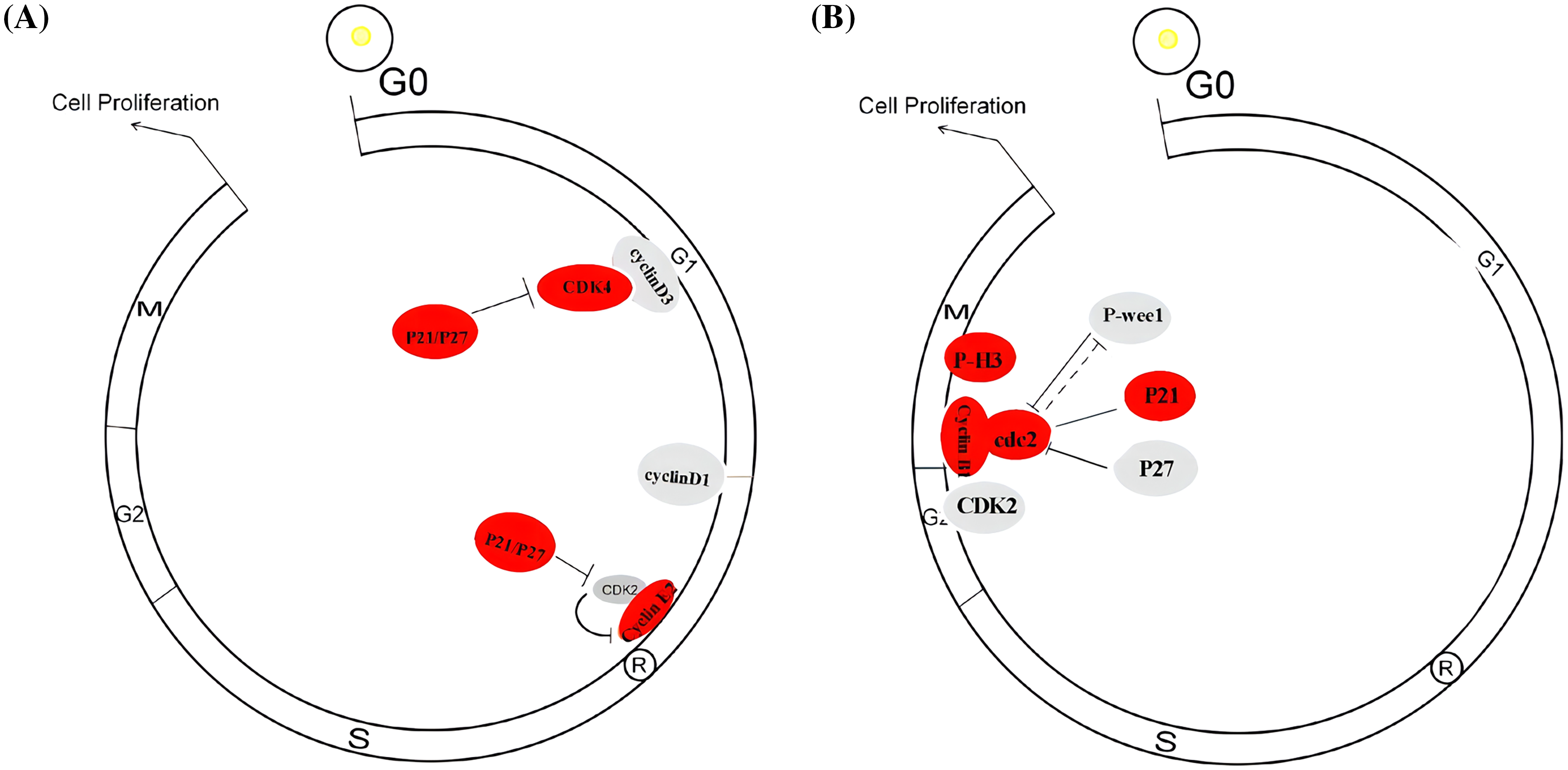
Figure S2: Possible schematic model of how PLAC8 influences cell cycle redistribution. (A) A possible schematic model of how PLAC8 alters the cell cycle in H1299 cells. (B) A possible schematic model of how PLAC8 alters the cell cycle in A549 cells.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools