 Open Access
Open Access
ARTICLE
CMTM6 deletion affects chemoresistance and macrophage M2 polarization in colorectal cancer cells
1 Department of Medical Oncology, The Third Medical Center of Chinese PLA General Hospital, Beijing, 100039, China
2 Department of Oncology, People’s Hospital of Ningxia Hui Autonomous Region, Yinchuan, 750000, China
* Corresponding Author: GE YOU. Email:
# Both of the two authors contributed equally and all authors agree to define them as co-first authors
BIOCELL 2024, 48(2), 229-237. https://doi.org/10.32604/biocell.2023.045030
Received 15 August 2023; Accepted 02 November 2023; Issue published 23 February 2024
Abstract
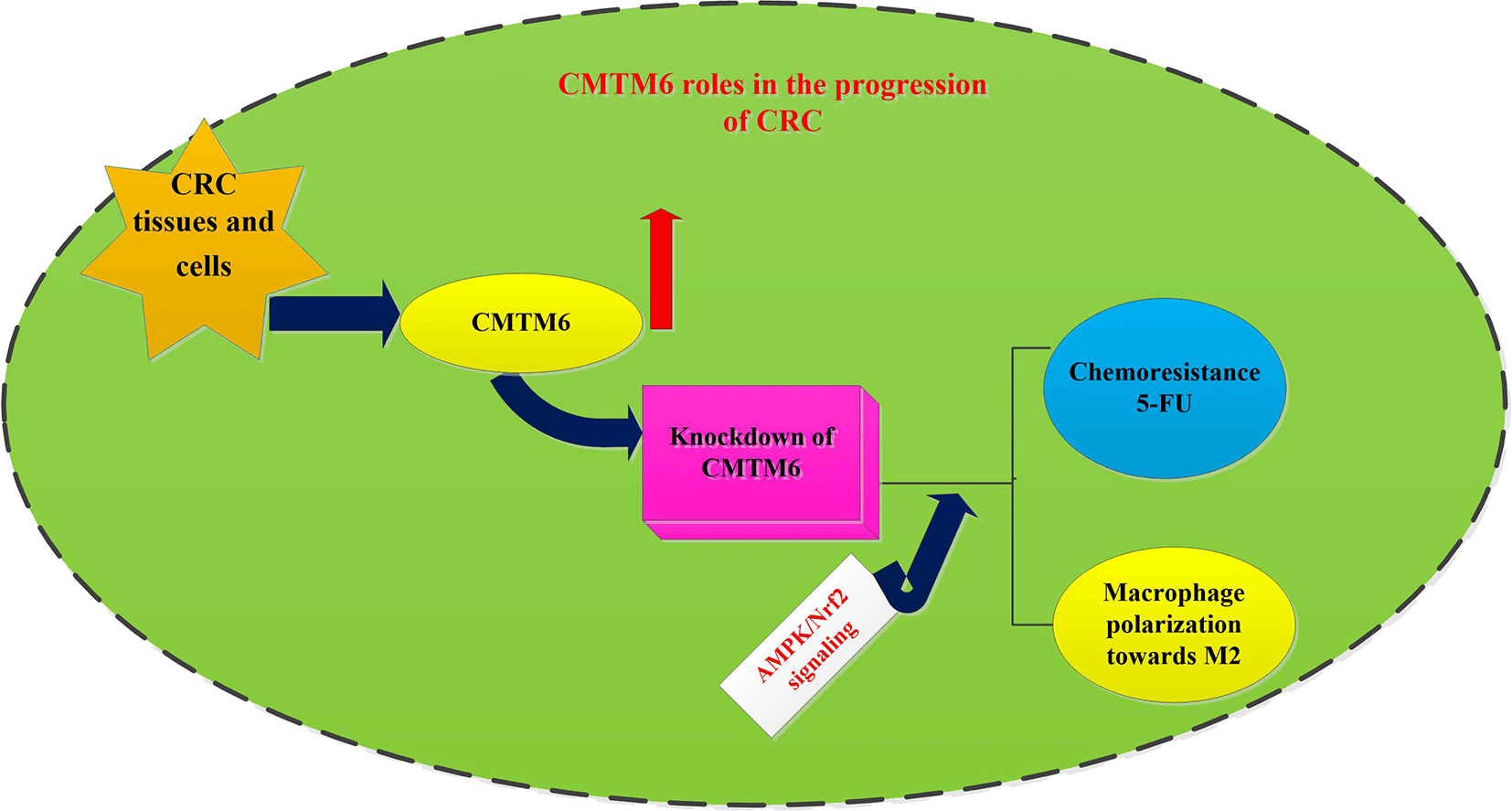
Background: Colorectal cancer (CRC) constitutes the leading cause of death worldwide. Chemoresistance and tumor immune evasion are critical contributors to therapeutic failure in cancer patients. CKLF-like MARVEL transmembrane domain-containing 6 (CMTM6) is aberrantly expressed in various cancers and can regulate tumor immunity. However, its role in chemoresistance and tumor immunity of CRC is not well understood. Methods: Online bioinformatics tools were used to analyze expression and prognosis of CMTM6 in CRC patients. CRC cells were transfected with si-CMTM6. Subsequently, the effects on CRC cell viability and chemoresistance were investigated by CCK-8 assay and flow cytometer. Furthermore, CRC cell-induced macrophage recruitments and polarization were also explored. Results: High expression of CMTM6 was detected in CRC tissues and cells, and its expression was associated with the prognosis of patients with CRC. Notably, CMTM6 knockdown suppressed cancer cell viability. Moreover, CMTM6 loss enhanced cancer cell sensitivity to 5-FU by reducing cell viability and enhancing apoptosis and activity of caspase-3. Importantly, CMTM6 loss inhibited CRC cells-evoked macrophage recruitments and polarization towards M2 phenotype by increasing the ratio of CD206+ cells and expression of M2 macrophage markers (arginase1, IL-10, CD206 and TGF-β). Intriguingly, targeting CMTM6 activated the AMPK singling and suppressed the subsequent activation of the NF-κB pathway. Blockage of the AMPK signaling reversed the suppressive efficacy of CMTM6 loss in 5-FU resistance and cancer cells-evoked macrophage M2-like polarization. Conclusion: CMTM6 may affect the progression of CRC by regulating chemoresistance and macrophage-related immune evasion, implying a promising target to overcome chemoresistance and immune escape in the treatment of CRC.Graphic Abstract
Keywords
Colorectal cancer (CRC) is the third most prevalently diagnosed carcinoma and constitutes an important barrier to life expectancy and a leading cause of death. National Center for Health Statistics (NCHS) reveals that CRC is expected to account for 9% of all new cancer-related cases in men in 2020 [1,2]. Nowadays, there are approximately 900,000 deaths of CRC annually [3]. Although colorectal cancer has been traditionally recognized as a malignancy of older individuals, the incidence of CRC is rising in young people in recent years [4]. For patients with CRC, surgery is the cornerstone for CRC therapy while the subsequent chemotherapy (e.g., 5-fluorouracil, 5-FU) constitutes the major strategy to improve the prognosis of patients with CRC [5]. Nevertheless, the majority of advanced CRC patients will eventually develop acquired chemoresistance, leading to a 5-year survival rate of less than 10% [6,7]. Therefore, illuminating the mechanism lying beneath chemoresistance is urgent for developing a new therapeutic strategy to overcome chemoresistance in CRC.
Except for the traditional strategy of directly regulating the behavior of cancer cells, increasing interests have focused on the key function of tumor microenvironment (TME) in cancer. It is a fact that cancer progression is not only caused by the genetic modification of cancer cells, but also involves the surrounding microenvironment. As one of the most abundant types of cells in TME, tumor-associated macrophages are often derived from circulating monocytes and recruited by cancer cells and polarized to cancer-promoting M2 phenotype from tumor-killing M1 phenotype [8]. Accumulating evidence supports that macrophage M2 polarization will facilitate the malignancy of cancer cells and the progression of cancers, including CRC [8–10]. Notably, tumor-associated macrophages (TAM) are frequently related to the prognosis in a variety of cancers. Intriguingly, emerging evidence has implicated TAM in chemoresistance [11]. For instance, induction of macrophage M2 phenotype facilitates CRC cell resistance to 5-FU [12]. Currently, deciphering the mechanism underlying TAM and chemoresistance is a promising therapeutic approach for CRC.
CKLF-like MARVEL transmembrane domain-containing 6 (CMTM6) belongs to the CMTM6 family and is widely expressed in a variety of cells, including cancer cells. In recent years, CMTM6 is proved to be aberrantly expressed and predicts poor prognosis in various cancers, such as triple-negative breast carcinoma [13], gastric carcinoma [14], and head and neck squamous cell carcinoma [15]. Furthermore, high expression of CMTM6 is associated with cancer recurrence in hepatocellular carcinoma [16]. Abundant vitro and vivo experiments substantiate that CMTM6 is implicated in multiple malignant behaviors of cancer cells, such as cell proliferation, invasion and migration [17,18]. For instance, CMTM6 increases cell migration, invasion and epithelial-mesenchymal transition in hepatocellular cancer [18]. Of interest, CMTM6 can act as a regulator of immune evasion-related PD-L1 and restrain T-cell-mediated anti-tumor immunity [15]. Intriguingly, an emerging study reveals that oral squamous cell carcinoma cell-secreted CMTM6 induces macrophage M2-like polarization [19]. However, up to now, the role of CMTM6 in CRC remains unclear. The present research sought to clarify the roles and lying mechanism of CMTM6 in chemoresistance and macrophage polarization in CRC.
Normal human colorectal epithelial cells (NCM460) and CRC cell lines (HT29, HCT116 and Lovo) were obtained from BeNa Culture Collection (BNCC; Henan, China) and ATCC (Manassas, Virginia, USA), respectively. All cells were maintained in RPMI 1640 medium (Sigma–Aldrich Chemicals, St. Louis, MO, USA) containing 10% fetal bovine serum and streptomycin-penicillin (100 U/ml). For incubation, all cells were housed under 5% CO2 at 37°C.
The down-regulation of CMTM6 and AMPK in CRC cells was conducted using the transfection of siRNAs objecting to CMTM6 and AMPK. The targeted siRNA and scrambled RNA (known as the negative control, si-NC) sequences were prepared by Genomeditech, Co. (Shanghai, China). CRC cells with 2 × 105 cells/well were seeded into 6 well plates. Subsequently, CRC cells were transfected with si-CMTM6-1, si-CMTM6-2, si-AMPK, or the corresponding si-NC using Lipofectamine 3000 (Invitrogen, California, CA, USA). The high inhibition efficiency of si-CMTM6-1 was confirmed and selected for the subsequent experiments. The sequences of siRNA in the current research were as follows: si-CMTM6-1 (5′-CCTCACTGAGCCACTTAAT-3′), si-AMPK (5′-CUGAGUUGCAUAUACUGUA-3′), and si-NC (5′-GACGAGCGGCACGUGCACA-3′). All procedures were conducted as recommended by manufacturer. Forty-eight hours later, qRT-PCR and western blotting were applied to evaluate silencing efficacy.
Macrophage induction, treatment and flow cytometric analysis
CRC cells under si-CMTM6, si-AMPK conditions or not were centrifuged at 2,000 g for 15 min at 4°C. Then, the collected medium was filtrated using a 0.22-mm filter (Millipore, Billerica, MA, USA) to prepare the conditioned medium (CM) from CRC cells. Human THP-1 monocytes were obtained from ATCC and incubated with RPMI medium. To induce M0 macrophage, THP-1 cells were incubated with phorbol 12-myristate 13-acetate (PMA, 100 nM; Sigma) for 24 h. Subsequently, M0 macrophages were incubated with CM from CRC cells. Forty-eight hours later, cells were treated with FITC-conjugated CD206 antibody (as M2 macrophage marker) for 1 h at room temperature. Then, labeled cells were subjected to a flow cytometer with Cell Quest software (Becton Dickinson, San Jose, CA, USA) to evaluate macrophage M2 polarization.
Cells were administrated with the indicated treatments. Then, a TRIzol reagent (Invitrogen) was utilized to prepare the total RNA, followed by RNA quantification using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA). After that, the SuperScript II First-Strand Synthesis System (Invitrogen) was utilized to synthesize first-strand cDNA. To analyze the mRNA expression of CMTM6, CD206, arginase1 (Arg1), IL-10 and TGF-β, a SYBR™ Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) was used to carry out the real-time PCR assay. Specific primer sequences used in the current research were obtained from GenePharma Co., Ltd. (Shanghai, China) and used as follows: CMTM6 (Accession: NM_017801.3; sense, 5′-ACCACAAAGTAGGCCAGATAAG-3′; anti-sense, 5′-ACCGTTGAGCTGTGTGATTTA-3′), CD206 (Accession: NM_002438.4; sense, 5′-GGACGTGGCTGTGGATAAAT-3′; antisense, 5′-ACCCAGAAGACGCATGTAAAG-3′), Arg1 (Accession: NM_001244438.2; sense, 5′-CCCTTTGCTGACATCCCTAAT-3′; antisense, 5′-GGCTGATTCTTCCGTTCTTCT-3′), IL-10 (Accession: NR_168466.1; sense, 5′-GAACCAAGACCCAGACATCAA-3′; antisense, 5′-CCTTCACATAGCCTTGTCCTAC-3′), TGF-β (Accession: M38449.1; sense, 5′-CTGCGGATCTCTGTGTCATT-3′; antisense, 5′-ATGCCGGGCAAAGGAATA-3′), and β-actin (Accession: EF095209.1; sense, 5′-GGCACCACACCTTCTACAAT-3′; antisense, 5′-AACATGATCTGGGTCATCTTCTC-3′). All protocols were conducted as per the instructions of kits. The relative expression levels of these genes were evaluated by the 2–ΔΔCt method and normalized by the internal control of β-actin.
Bioinformatics databases assay
CMTM6 expression was evaluated in CRC tissues and normal specimens by The Cancer Genome Atlas (TCGA) database analysis via online bioinformatics UALCAN database (http://ualcan.path.uab.edu/) and Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database (http://gepia2.cancer-pku.cn/). The correlation between survival and CMTM6 expression was evaluated in patients with CRC by Kaplan–Meier method using an online GEPIA2 database (http://gepia2.cancer-pku.cn/#survival).
All cellular protein samples were extracted using the Radioimmunoprecipitation assay (RIPA) lysis and extraction Buffer (Invitrogen). The prepared protein contents were quantified using a bicinchoninic acid assay (BCA) kit (Thermo Scientific, Waltham, MA, USA). Approximately 30 μg of protein specimens were incubated for 10 min at 95°C and were subsequently loaded onto 10% SDS-PAGE gel. Following the transfer to the PVDF membranes, Tris-buffer saline (TBS) containing 5% no-fat milk was added to the membranes to block the non-specific binding. Approximately 2 h later, membranes were treated overnight with the primary antibodies against human CMTM6 (1:1000; #ab264067), AMPK (1:4000; #ab32508), phospho-AMPK (p-AMPK; 1:6000; #ab92701), p65 NF-κB (1:1000; # ab207297), and p-p65 NF-κB (1:3000; #ab239882) (Abcam, Cambridge, MA, USA) at 4°C. Then, TBST was used to wash the membranes, followed by the treatment with HRP-conjugated secondary antibody for 1 h at room temperature. The binding proteins were then visualized using the ECL reagent and quantified by the ImageJ software.
CRC cells under si-CMTM6, si-AMPK, or 5-FU treatments were collected and treated with 10 μl of CCK-8 reagent (Beyotime, Shanghai, China) at 37°C. Approximately 1 h later, cell viability was determined by measuring the OD values at 450 nm via a microplate reader. All protocols were conducted as recommended by manufacturer.
Cells transfected with siRNAs were exposed to 5-FU (Sigma). Then, cells were treated with Annexin V-FITC (10 μl) and PI (5 μl) avoiding light. Approximately 15 mins later, a flow cytometer (BD Biosciences, San Jose, CA, USA) was utilized to evaluate apoptotic cells. All protocols of apoptotic assay were conducted as per manufacturer’s instructions of Annexin V-FITC Apoptosis Detection Kits (Beyotime).
Evaluation of caspase-3 activity
A commercial Caspase 3 Activity Assay Kit (Beyotime) was used in this study to detect the activity of caspase-3 in CRC cells. Briefly, cells under si-RNA and 5-FU exposure were collected and subsequently lysed with lysis buffer for 15 min. Then, cells were centrifuged at 4°C to collect supernatants. Subsequently, the specimens were treated with 10 μl of specific caspase-3 substrate Ac-DEVD-pNA. Finally, the absorbance at 405 nm was measured to assess the activity of caspase-3.
Cell migration assay was carried out in a 24-well cell culture chamber containing inserts (8 μm pore size) (Corning, Kennebunk, NY, USA). THP-1 monocytes under PMA treatments were induced to M0 macrophage and platted at the bottom chamber. Then, the culture medium containing CM from CRC cells was supplemented into the upper chamber. Forty-eight hours after incubation, the non-migrated cells were gently wiped out. The chamber was then fixed with 4% paraformaldehyde, followed by the staining with 0.1% crystal violet solution. Cell migration was then analyzed by calculating 5 random fields using a light microscope (Nikon, Tokyo, Japan; ×200 magnification).
All results are shown as mean ± standard deviation (SD) and calculated in at least three replicates. The statistical difference was analyzed using the SPSS software (SPSS 19.0, Chicago, IL, USA). The two or multiple comparisons were performed using Student’s t-test and one-way ANOVA with SNK post-hoc test. p < 0.05 was defined as statistical significance.
Analysis of CMTM6 expression and prognosis in CRC tissues
Before investigation of CMTM6 roles in CRC, we first analyzed its expression in CRC tissues and adjacent normal specimens using bioinformatics UALCAN and GEPIA2 database. Intriguingly, UANCAN (Fig. 1A) and GEPIA2 (Fig. 1B) both confirmed the increased expression of CMTM6 in CRC tissues. No obvious difference was observed in CRC tissues with different clinical stages (Fig. 1C). Moreover, patients with low CMTM6 exhibited longer survival times than that in high CMTM6 groups, indicating a potential marker for the prognosis of CRC patients (Fig. 1D). These data indicate the potential roles of CMTM6 in the development of CMTM6.

Figure 1: Detection of CMTM6 expression in tissues and cells in CRC. (A, B) CMTM6 expression in CRC tissues and normal specimens was analyzed by UALCAN (A) and GEPIA2 (B) bioinformatics database. (C, D) CMTM6 expression in tissues with different stages (C) and its correlation with survival (D) were evaluated by Kaplan–Meier method using the online GEPIA2 database. (E, F) The mRNA (E) and protein levels (F) of CMTM6 were determined in NCM460 and CRC cells (Lovo, HCT116 and HT29 cells). n = 4. The statistical difference was evaluated with ANOVA with SNK post-hoc test. *p < 0.05.
High expression of CMTM6 is validated in CRC cells
Compared with the normal colorectal epithelial NCM460 cells, CMTM6 mRNA levels were increased in CRC cells (Lovo, HCT116 and HT29 cells), especially in HCT116 cells (Fig. 1E). Moreover, western blotting assay substantiated the similar up-regulation of CMTM6 protein in CRC cells relative to NCM460 cells (Fig. 1F). The highest expression of CMTM6 was validated in HCT116 cells. Therefore, this cell line was chosen for subsequent experiments.
Inhibition of CMTM6 expression suppresses cancer cell viability and increases cell sensitivity to 5-FU
Transfection with si-CMTM6 into HCT116 cells inhibited transcripts (Fig. 2A) and protein levels of CMTM6 (Fig. 2B). The si-CMTM6-1 exhibited stronger inhibitory efficacy in CMTM6 expression than si-CMTM6-2. Functional assay substantiated that knockdown of CMTM6 dramatically suppressed cell viability relative to the control groups (Fig. 2C). Furthermore, 5-FU treatment inhibited HCT116 cell viability, which was further enhanced when cells were transfected with si-CMTM6-1 (Fig. 2D). Similarly, stimulation with 5-FU induced apoptosis (Fig. 2E) and caspase-3 activity in HCT116 cell (Fig. 2F), and this suppressive efficacy was further elevated after CMTM6 loss. Thus, these findings suggest that CMTM6 loss not only inhibits CRC cell viability but also enhances cell sensitivity to chemoresistance.

Figure 2: Deletion of CMTM6 regulates cell viability and chemoresistance in HCT116 cells. (A) Cells were treated with si-CMTM61, si-CMTM6-2 or si-NC. Approximately 48 h later, the mRNA (A) and protein levels (B) of CMTM6 were determined by the qRT-PCR and western assay. n = 4. (C) Effects of CMTM6 knockdown on cell viability were analyzed. (D–F) Cells transfected with si-CMTM6 were exposed to the indicated concentrations of 5-FU for 24 h. Subsequently, cell viability (D) was determined by CKK-8 assay. Then, cell apoptosis (E) and activity of caspase-3 (F) were assessed in HCT116 cells treated with si-CMTM6 and exposure to 5 μM of 5-FU. n = 3. The statistical difference was assessed by ANOVA with SNK test. *p < 0.05.
Loss of CMTM6 restrains CRC cell ability to recruit and skew macrophage toward the M2 phenotype
Macrophages within tumor microenvironment have been implicated in the development of carcinoma, including CRC. We, therefore, explored the functions of CMTM6 in macrophage recruitment and polarization. As shown in Fig. 3A, THP-1 macrophages exhibited a high positive ratio of CD206+ cells after the cultured with CM from CRC cells, indicating the polarization of macrophage towards M2 phenotype; however, knockdown of CMTM6 reduced CM-induced percentage of CD206+ cells. Moreover, incubation with CM from CRC cells increased the expression of M2 macrophage markers, including CD206 (Fig. 3B), Arg-1 (Fig. 3C), IL-10 (Fig. 3D) and TGF-β (Fig. 3E); however, these increases were reversed following CMTM6 deletion (Figs. 3B–3E), indicating tjat CMTM6 can affect CRC cell ability to skew macrophage towards M2 phenotype. Additionally, si-CMTM6 transfection inhibited CM-induced macrophage migration (Fig. 3F).

Figure 3: CMTM6 knockdown inhibits the ability of CRC cells to recruit and polarize macrophages. (A) THP-1 cells were incubated with 100 nM PMA for 24 h to induce M0 macrophage, and then were treated with CM from si-CMTM6-transfected HCT116 cells, or not. Then, the ratio of CD206+ cells was analyzed by flow cytometry. (B–E) The transcripts of CD206 (B), Arg-1 (C), IL-10 (D) and TGF-β (E) were measured by qRT-PCR. (F) To analyze CRC cell ability to recruit macrophage, macrophages were co-cultured with conditioned medium (CM) from HCT116 cells transfected with si-CMTM6. Then, macrophage migration was evaluated using a Transwell assay. n = 3. The statistical difference was evaluated by ANOVA with SNK test. *p < 0.05 vs. control group. #p < 0.05 vs. CM group.
Downregulation of CMTM6 restores the activation of the AMPK signaling
Mounting evidence has confirmed the implication of AMPK/NF-κB signaling in various malignant processes of cancers, including CRC [20,21]. Here, knockdown of CMTM6 elevated the protein expression of p-AMPK (Figs. 4A and 4B) relative to the control groups, but not in AMPK expression (Figs. 4A and 4C). Moreover, si-CMTM6 transfection significantly diminished protein levels of p-p65 NF-κB (Fig. 4D), but with little change in NF-κB expression (Fig. 4E). These data reveal that CMTM6 suppression may restore the AMPK signaling to inhibit the NF-κB signaling in CRC cells.

Figure 4: CMTM6 downregulation restores the activation of the AMPK signaling. (A) HCT116 cells were treated with si-CMTM6 or si-NC. Approximately 48 h later, the protein levels of p-AMPK, AMPK, p65 NF-κB and p-p65 NF-κB were measured by western. (B–E) The protein bands were quantified by ImageJ software. n = 4. The statistical difference was evaluated by ANOVA with SNK test. *p < 0.05.
Blockage of the AMPK pathway is responsible for CMTM6-mediated chemoresistance and macrophage polarization
To further discern the molecular mechanism underlying CMTM6-mediated chemoresistance and macrophage polarization, we blocked the AMPK signaling by the si-AMPK transfection (Figs. 5A and 5B). Moreover, CMTM6 down-regulation inhibited CRC cell viability, which was offset when blocking the AMPK signaling (Fig. 5C). Especially, loss of CMTM6 antagonized 5-FU-induced inhibition of cell viability (Fig. 5D) and enhanced 5-FU-evoked cell apoptosis (Fig. 5E), which were overturned after si-AMPK transfection. Next, blockage of the AMPK pathway reversed CMTM6 knockdown-mediated inhibition of CM-evoked positive percentage of CD206 (Fig. 5F) and M2 macrophage marker expression (Figs. 5G and 5H) in THP-1 macrophage. Concomitantly, the inhibitory effects of CMTM6 loss on CM-induced macrophage migration were also abrogated after si-AMPK transfection (Fig. 5I).

Figure 5: CMTM6 regulates chemoresistance and macrophage M2-like phenotype by the AMPK signaling. (A, B) The effects of si-AMPK transfection on the activation of the AMPK signaling was determined in HCT116 cells. The statistical difference was performed by Student’s t-test. (C) After transfection with si-CMTM6 and si-AMPK for 48 h, cell viability was then evaluated by CCK-8. (D, E) Cells were treated with si-CMTM6 or si-AMPK under 5-FU (5 μM) conditions. Cell viability and apoptosis were then analyzed. (F–I) Macrophages were co-incubated with CM from HCT116 cells under si-CMTM6 and si-AMPK conditions. Then, the effects on CM-mediated positive percentage of CD206 (F), M2 marker expression of CD206, Arg1 (G), IL-10 and TGF-β (H), and migration (I) of macrophages were determined. n = 3. The statistical difference was assessed by ANOVA with SNK post-hoc test. *p < 0.05.
Despite the enormous improvements over the past few years, CRC remains one of the uppermost causes of cancer-related death and has constituted a great global health threat [3]. In this research, we focused on the regulation of CRC chemoresistance and tumor cell-mediated macrophages by CMTM6. Intriguingly, the present research confirmed high levels of CMTM6 in CRC tissues by online bioinformatics analysis database and found that this high expression was negatively correlated with survival of patients with CRC. Similarly, the increased expression of CMTM6 was also determined in CRC cells relative to the normal colorectal epithelial cells. These findings discover a potential role of CMTM6 in the progression of CRC. Analogously, a recent finding reveals that up-regulation of CMTM6 is associated with shorter survival in patients with gastrointestinal malignancies, including CRC [22]. Intriguingly, the previous findings confirmed that the aberrant expression of CMTM6 is a promising prognostic marker in several carcinomas, such as gastric carcinoma and head and neck squamous cell carcinoma [14,15].
The roles of chemotherapy are expanding and provides a variably effective strategy for unresectable or advanced postoperative patients with cancers. However, the occurrence of chemoresistance constitutes nowadays the uppermost disincentive for chemotherapy failure that accounts for more than 90% of deaths in cancer patients due to relapse and metastasis. More in particular, overcoming chemotherapeutic insensitivity affords a promising intervention to improve the survival of cancer patients [6,7]. 5-FU is a known and indispensable chemotherapeutic drug for CRC and achieves a certain benefit for patients [6]. Herein, one noteworthy observation in the current study was that knockdown of CMTM6 restrained CRC cell viability and elevated cancer cell sensitivity to 5-FU. Of interest, previous findings confirm that CMTM6 not only promotes cancer cell proliferation and invasion, but also enhances cell migration and EMT in oral squamous cell carcinoma and hepatocellular carcinoma [18,23]. Analogously, CMTM6 also drives cisplatin resistance in oral squamous cell carcinoma [24].
Increasing evidence emphasizes the important relationship between therapeutic outcomes and tumor microenvironment (TME) [25]. As the pivotal non-cancer components within TME, macrophages engage in the complex network interactions with tumor cells that will facilitate macrophage polarization from M1-like anti-cancer efficacy to M2-like pro-cancer phenotype. Abundant experiments substantiate that cancer cells can endow macrophage recruitment and M2-like polarization by multiple pathways [26,27]. Notably, M2-like macrophages will in turn promote cancer cell growth, invasion, metastasis, angiogenesis and chemoresistance [28]. In recent years, considerable interest has focused on the prospective strategy for cancer therapy by affecting macrophage polarization towards M2 phenotype [28,29]. Of interest, CMTM6 has been proved to maintain PD-L1 expression, a proverbial immune inhibitor in cancer immune evasion, and can regulate anti-tumor immunity [15,30]. Suppression of CMTM6 enhances CD8+ and CD4+ T-cell infiltration in head and neck squamous cell cancer [15]. Moreover, the expression of CMTM6 is positively associated with PD-L1 and M2 macrophage density in CRC [31]. In parallel with previous studies, the present data uncovered an interesting finding that CMTM6 deletion inhibited CRC cell-evoked macrophage migration and macrophage M2 polarization, indicating the critical roles in cancer immune evasion.
Next, the current data revealed that inhibition of CMTM6 activated the AMPK signaling and blocked subsequent activation of the NF-κB signaling in CRC cells. As an evolutionarily conserved serine/threonine protein kinase, AMPK can act as an energy sensor for energy homeostasis and metabolism regulation. Increasing interest has implicated AMPK in various pathogenic processes of cancer cells, such as cell invasion, chemoresistance and angiogenesis [21,32]. For instance, administration with chitosan oligosaccharide inhibits tumorigenesis of CRC by activating the AMPK and suppressing the NF-κB signaling [33]. Moreover, activating the AMPK pathway blunts gastric cancer cell chemoresistance to 5-FU [20]. Especially, emerging evidence supports the indispensable function of AMPK signaling in tumor immune evasion. For instance, the AMPK-NF-κB signaling is involved in breast carcinoma cell-mediated macrophage polarization towards M2 [34]. The activation of AMPK signaling by astragaloside IV inhibits macrophage M2 polarization, leading to the suppression of lung cancer progression and metastasis [27]. The current data prompted us to hypothesize the involvement of the AMPK signaling in CMTM6-regulated chemoresistance and macrophage polarization. As expected, in this study, blockage of the AMPK overturned the inhibitory effects of CMTM6 down-regulation on 5-FU resistance and macrophage M2 polarization.
Collectively, the present findings corroborated high expression of CMTM6 in cancer tissues and cells in CRC. More importantly, inhibition of CMTM6 restrained CRC cell chemoresistance to 5-FU and cancer cell-induced macrophage recruitments and polarization to M2 by activating the AMPK signaling. Thus, these data highlight that CMTM6 may facilitate the development of CRC by affecting chemoresistance and macrophage polarization, indicating the critical roles of CMTM6 in CRC chemoresistance and immune escape. Therefore, targeting CMTM6 may represent a promising therapeutic strategy for CRC. However, this study only focused on in vitro cellular model and did not analyzed CMTM6 expression in CRC tissues via the immunohistochemistry detection. Therefore, the current results and findings were relatively limited. Does CMTM6 loss exert the ideal anti-tumor efficacy in CRC in animal studies? Whether the AMPK signaling involves in CMTM6-mediated anti-CRC function in vivo. Whether targeting CMTM6 has the ideal potential in the clinical treatment of patients with CRC. These questions will be further elucidated in future studies.
Acknowledgement: None.
Funding Statement: This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: GY; data collection: YX, HYL, GY; analysis and interpretation of results: YX, HYL, GY; draft manuscript preparation: YX. All authors read and approved the final manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590. [Google Scholar] [PubMed] [CrossRef]
2. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. doi:10.3322/caac.21660. [Google Scholar] [PubMed] [CrossRef]
3. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–80. doi:10.1016/S0140-6736(19)32319-0. [Google Scholar] [PubMed] [CrossRef]
4. The Lancet O. Colorectal cancer: A disease of the young? Lancet Oncol. 2017;18(4):413. doi:10.1016/S1470-2045(17)30202-4. [Google Scholar] [PubMed] [CrossRef]
5. Xu D, Liu XF, Yan XL, Wang K, Xing BC. Survival prediction in patients with resectable colorectal liver metastases: Clinical risk scores and tumor response to chemotherapy. Oncol Lett. 2017;14(6):8051–9. doi:10.3892/ol.2017.7191. [Google Scholar] [PubMed] [CrossRef]
6. Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol Ther. 2020;206:107447. doi:10.1016/j.pharmthera.2019.107447. [Google Scholar] [PubMed] [CrossRef]
7. Marin JJ, Sanchez De Medina F, Castano B, Bujanda L, Romero MR, Martinez-Augustin O, et al. Chemoprevention, chemotherapy, and chemoresistance in colorectal cancer. Drug Metab Rev. 2012;44(2):148–72. doi:10.3109/03602532.2011.638303. [Google Scholar] [PubMed] [CrossRef]
8. Mills CD, Lenz LL, Harris RA. A breakthrough: Macrophage-directed cancer immunotherapy. Cancer Res. 2016;76(3):513–6. doi:10.1158/0008-5472.CAN-15-1737. [Google Scholar] [PubMed] [CrossRef]
9. Anderson NR, Minutolo NG, Gill S, Klichinsky M. Macrophage-based approaches for cancer immunotherapy. Cancer Res. 2021;81(5):1201–8. doi:10.1158/0008-5472.CAN-20-2990. [Google Scholar] [PubMed] [CrossRef]
10. Li W, Zhang X, Wu F, Zhou Y, Bao Z, Li H, et al. Gastric cancer-derived mesenchymal stromal cells trigger M2 macrophage polarization that promotes metastasis and EMT in gastric cancer. Cell Death Dis. 2019;10(12):918. doi:10.1038/s41419-019-2131-y. [Google Scholar] [PubMed] [CrossRef]
11. Yin Y, Yao S, Hu Y, Feng Y, Li M, Bian Z, et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin Cancer Res. 2017;23(23):7375–87. doi:10.1158/1078-0432.CCR-17-1283. [Google Scholar] [PubMed] [CrossRef]
12. Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, et al. M2 macrophages confer resistance to 5-fluorouracil in colorectal cancer through the activation of CCL22/PI3K/AKT signaling. Onco Targets Ther. 2019;12:3051–63. doi:10.2147/OTT.S198126. [Google Scholar] [PubMed] [CrossRef]
13. Tian Y, Sun X, Cheng G, Ji E, Yang S, Feng J, et al. The association of CMTM6 expression with prognosis and PD-L1 expression in triple-negative breast cancer. Ann Transl Med. 2021;9(2):131. doi:10.21037/atm-20-7616. [Google Scholar] [PubMed] [CrossRef]
14. Li X, Chen L, Gu C, Sun Q, Li J. CMTM6 significantly relates to PD-L1 and predicts the prognosis of gastric cancer patients. PeerJ. 2020;8:e9536. doi:10.7717/peerj.9536. [Google Scholar] [PubMed] [CrossRef]
15. Chen L, Yang QC, Li YC, Yang LL, Liu JF, Li H, et al. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol Res. 2020;8(2):179–91. doi:10.1158/2326-6066.CIR-19-0394. [Google Scholar] [PubMed] [CrossRef]
16. Muranushi R, Araki K, Yokobori T, Chingunjav B, Hoshino K, Dolgormaa G, et al. High membrane expression of CMTM6 in hepatocellular carcinoma is associated with tumor recurrence. Cancer Sci. 2021;112(8):3314–23. doi:10.1111/cas.15004. [Google Scholar] [PubMed] [CrossRef]
17. Wei L, Wei Q, Yang X, Zhou P. CMTM6 knockdown prevents glioma progression by inactivating the mTOR pathway. Ann Transl Med. 2022;10(4):181. doi:10.21037/atm-21-6894. [Google Scholar] [PubMed] [CrossRef]
18. Huang L, Li X, Liu Y, Liang X, Ye H, Yang C, et al. Curcumin alleviates cerebral ischemia-reperfusion injury by inhibiting NLRP1-dependent neuronal pyroptosis. Curr Neurovasc Res. 2021;18(2):189–96. doi:10.2174/1567202618666210607150140. [Google Scholar] [PubMed] [CrossRef]
19. Pang X, Wang SS, Zhang M, Jiang J, Fan HY, Wu JS, et al. OSCC cell-secreted exosomal CMTM6 induced M2-like macrophages polarization via ERK1/2 signaling pathway. Cancer Immunol Immunother. 2021;70(4):1015–29. doi:10.1007/s00262-020-02741-2. [Google Scholar] [PubMed] [CrossRef]
20. Park JB, Lee JS, Lee MS, Cha EY, Kim S, Sul JY. Corosolic acid reduces 5FU chemoresistance in human gastric cancer cells by activating AMPK. Mol Med Rep. 2018;18(3):2880–8. doi:10.3892/mmr.2018.9244. [Google Scholar] [PubMed] [CrossRef]
21. Tan W, Zhong Z, Carney RP, Men Y, Li J, Pan T, et al. Deciphering the metabolic role of AMPK in cancer multi-drug resistance. Semin Cancer Biol. 2019;56(3):56–71. doi:10.1016/j.semcancer.2018.09.005. [Google Scholar] [PubMed] [CrossRef]
22. Dai M, Lan T, Li X, Xiao B. High expression of CMTM6 is a risk factor for poor prognosis of gastrointestinal tumors: A meta-analysis. Asian J Surg. 2022;46(1):66–72. doi:10.1016/j.asjsur.2022.05.086. [Google Scholar] [PubMed] [CrossRef]
23. Zheng Y, Wang C, Song A, Jiang F, Zhou J, Li G, et al. CMTM6 promotes cell proliferation and invasion in oral squamous cell carcinoma by interacting with NRP1. Am J Cancer Res. 2020;10(6):1691–709. [Google Scholar] [PubMed]
24. Mohapatra P, Shriwas O, Mohanty S, Ghosh A, Smita S, Kaushik SR, et al. CMTM6 drives cisplatin resistance by regulating Wnt signaling through the ENO-1/AKT/GSK3beta axis. JCI Insight. 2021;6(4):e143643. doi:10.1172/jci.insight.143643. [Google Scholar] [PubMed] [CrossRef]
25. Hinshaw DC, Shevde LA. The tumor microenvironment innately modulates cancer progression. Cancer Res. 2019;79(18):4557–66. doi:10.1158/0008-5472.CAN-18-3962. [Google Scholar] [PubMed] [CrossRef]
26. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. 2020;13(1):156. doi:10.1186/s13045-020-00991-2. [Google Scholar] [PubMed] [CrossRef]
27. Xu F, Cui WQ, Wei Y, Cui J, Qiu J, Hu LL, et al. Astragaloside IV inhibits lung cancer progression and metastasis by modulating macrophage polarization through AMPK signaling. J Exp Clin Cancer Res. 2018;37(1):207. doi:10.1186/s13046-018-0878-0. [Google Scholar] [PubMed] [CrossRef]
28. Boutilier AJ, Elsawa SF. Macrophage polarization states in the tumor microenvironment. Int J Mol Sci. 2021;22(13):6995. doi:10.3390/ijms22136995. [Google Scholar] [PubMed] [CrossRef]
29. Mantovani A, Marchesi F, Malesci A, Laghi L, Allavena P. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol. 2017;14(7):399–416. doi:10.1038/nrclinonc.2016.217. [Google Scholar] [PubMed] [CrossRef]
30. Burr ML, Sparbier CE, Chan YC, Williamson JC, Woods K, Beavis PA, et al. CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549(7670):101–5. doi:10.1038/nature23643. [Google Scholar] [PubMed] [CrossRef]
31. Wu X, Lan X, Hu W, Zhang W, Lai X, Xu S, et al. CMTM6 expression in M2 macrophages is a potential predictor of PD-1/PD-L1 inhibitor response in colorectal cancer. Cancer Immunol Immunother. 2021;70(11):3235–48. doi:10.1007/s00262-021-02931-6. [Google Scholar] [PubMed] [CrossRef]
32. Vara-Ciruelos D, Russell FM, Hardie DG. The strange case of AMPK and cancer: Dr. Jekyll or Mr. Hyde? (dagger). Open Biol. 2019;9(7):190099. doi:10.1098/rsob.190099. [Google Scholar] [PubMed] [CrossRef]
33. Mattaveewong T, Wongkrasant P, Chanchai S, Pichyangkura R, Chatsudthipong V, Muanprasat C. Chitosan oligosaccharide suppresses tumor progression in a mouse model of colitis-associated colorectal cancer through AMPK activation and suppression of NF-κB and mTOR signaling. Carbohydr Polym. 2016;145:30–6. doi:10.1016/j.carbpol.2016.02.077. [Google Scholar] [PubMed] [CrossRef]
34. Chiang CF, Chao TT, Su YF, Hsu CC, Chien CY, Chiu KC, et al. Metformin-treated cancer cells modulate macrophage polarization through AMPK-NF-κB signaling. Oncotarget. 2017;8(13):20706–18. doi:10.18632/oncotarget.14982. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools