 Open Access
Open Access
REVIEW
Extracellular vesicles as brain tumor biomarkers
1 Institute of Fundamental Medicine and Biology, Kazan Federal University, Kazan, 420008, Russia
2 Department of Genetics and Biotechnology, St. Petersburg State University, St. Petersburg, 199034, Russia
3 Division of Medical and Biological Sciences, Tatarstan Academy of Sciences, Kazan, 420111, Russia
* Corresponding Author: ALBERT RIZVANOV. Email:
BIOCELL 2024, 48(12), 1667-1681. https://doi.org/10.32604/biocell.2024.058490
Received 13 September 2024; Accepted 28 November 2024; Issue published 30 December 2024
Abstract
Aggressive malignant brain tumors have a poor prognosis, and early detection can significantly improve treatment effectiveness and increase patient survival rates. Various methods are available for diagnosing brain tumors, with biopsy being one of the primary options. However, a biopsy is an invasive procedure that carries a risk of brain damage, highlighting the need for safer alternatives. One promising non-invasive method is liquid biopsy, which involves extracting extracellular vesicles (EVs) from different biological fluids. Most cell types can produce and release extracellular vesicles. EVs isolated from bodily fluids, along with the molecules they carry—such as proteins, nucleic acids, and lipids—can be used to diagnose brain tumors. This approach has the potential to replace labor-intensive and expensive diagnostic methods that can adversely affect patient health. This review discusses recent advancements in the use of EVs as biomarkers for diagnosing brain tumors.Keywords
Gliomas are primary brain tumors, derived from glial stem cells or precursors. Gliomas are typically classified into four grades according to the World Health Organization (WHO) classification system, with grades I and II being benign, and grades III and IV malignant [1,2]. The main types of gliomas include pilocytic astrocytoma (Grade I), anaplastic astrocytomas (Grade II), oligodendrogliomas (Grade III), and glioblastomas (GBM) (Grade IV), the latter being the most aggressive and lethal subtype of glioma with a poor prognosis [3,4]. Gliomas are the most common type of malignant neoplasm of the central nervous system in adults, accounting for about 80% of all primary brain tumors. Gliomas are a heterogeneous group of tumors originating from glial cells and characterized by diffuse growth and infiltration of brain tissue, which significantly complicates their therapy [5].
Currently, detecting tumors at their early developmental stages remains a challenge [6]. One such approach is biopsy, a diagnostic test involving the removal of a small amount of tissue from an organ for microscopic examination. Tissue biopsy for brain tumors typically carries a much higher risk of complications compared to most other tumors. For instance, hemorrhage or brain swelling may damage healthy brain tissue, leading to disability or death. Liquid biopsy offers a non-invasive diagnostic method that enables early detection of tumors [7]. In body fluids such as blood, cerebrospinal fluid (CSF), and urine, extracellular nucleic acids (e.g., circulating free DNA or RNA (cfDNA/cfRNA)), extracellular vesicles (EVs), and circulating tumor cells (CTCs) can be detected. Liquid biopsy is currently used in observational and interventional studies involving patients with GBM to monitor tumor progression or response to treatment [8].
One common type of liquid biopsy is the clinical application of cfDNA analysis. Circulating tumor DNA (ctDNA) is a component of extracellular DNA released by malignant tumors into the bloodstream and other body fluids. The levels of cfDNA are typically low, especially in patients with localized disease, necessitating highly sensitive detection and quantification methods [9]. A major challenge in this area of research is that tumor-derived cfDNA can constitute as little as 0.01% of total cfDNA, making detection particularly difficult in the early stages of cancer. Digital polymerase chain reaction (PCR) and next-generation sequencing (NGS) are the most popular methods for cfDNA analysis, although they have some limitations. Digital PCR can only detect known variants and analyze a limited number of variants in a single reaction. Meanwhile, NGS remains a relatively expensive and time-consuming method and also requires bioinformatics expertise for data analysis and interpretation [10].
CTCs are tumor cells that have detached from the primary tumor, are extravasated into the bloodstream, and circulate within it. Detecting CTCs among blood cells is a challenging task [11]. For instance, in the blood of patients with GBM, only one or a few cells may be found in a 10-mL blood sample, and they exhibit low stability in circulation. These factors make CTCs far from representing the molecular structure of the entire tumor in a real clinical scenario, where blood sampling is limited to small volumes [12].
Another form of liquid biopsy is the isolation and analysis of EVs. EVs are membrane-bound particles that are secreted by all types of cells [13].
Based on various characteristics, such as size, biogenesis, cellular origin, morphology, and content, EVs are divided into four main classes: small exosomes of endosomal origin (50–150 nm), medium-sized microvesicles from the plasma membrane (100–1000 nm), large oncosomes (1000–10,000 nm), and apoptotic bodies (500–4000 nm) (Fig. 1) [14–16]. It is important to note that according to the literature, it is customary to divide vesicles by their sizes, but there is no clear division. Exosomes and microvesicles can have similar sizes.
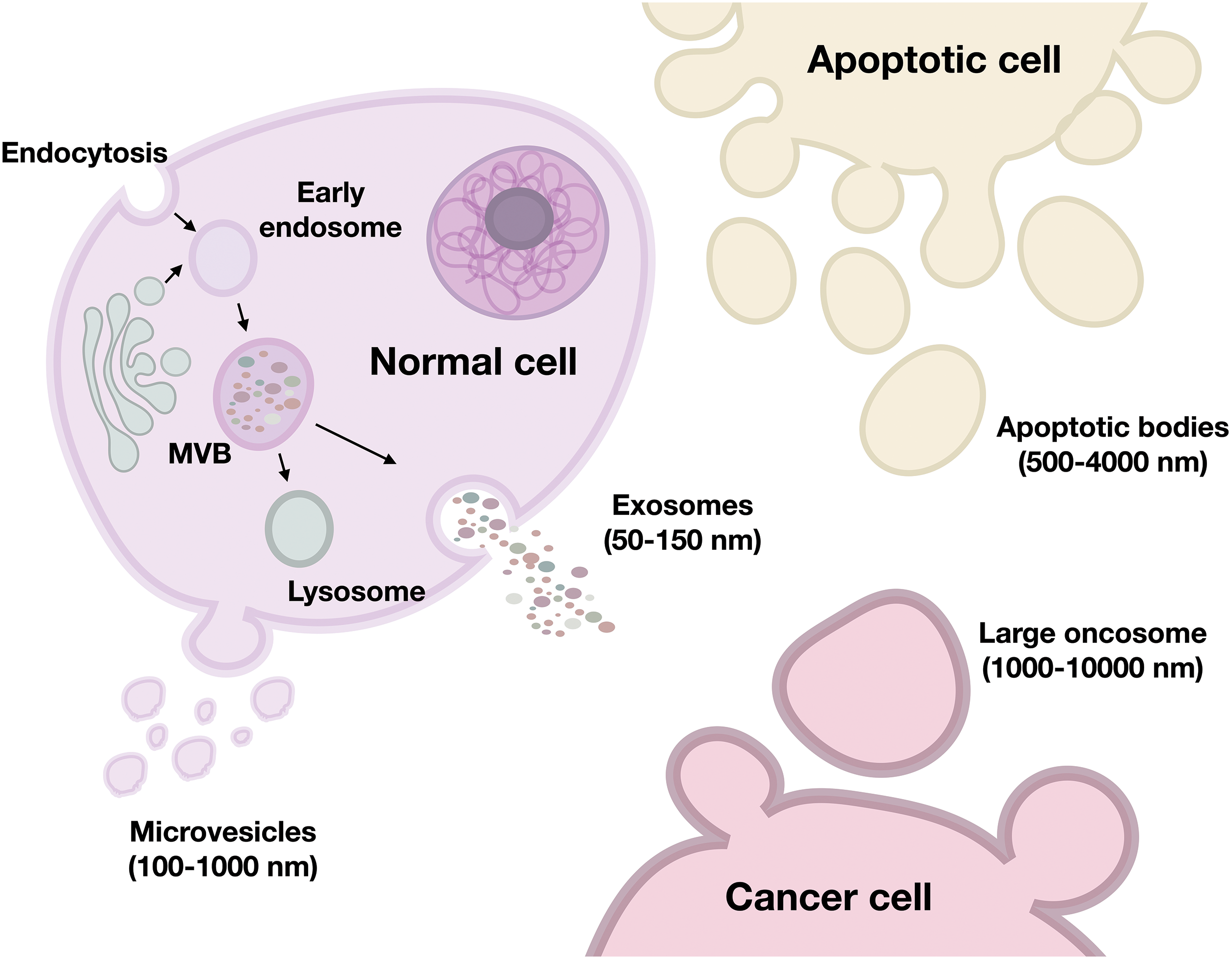
Figure 1: Types of EVs secreted by cells. MVB—multivesicular body. The figure was created using Keynote.
EVs offer advantages over ctDNA and CTCs as tools for liquid biopsy. EVs have a bilayer membrane structure that provides stability, and they also effectively preserve biological information from source cells [6]. Furthermore, EVs reflect the real-time status of the disease, providing insights into the biological and functional nature of the tumor. They can be used for accurate diagnosis and monitoring of the disease status, enabling a personalized approach to therapy (Fig. 2) [5].
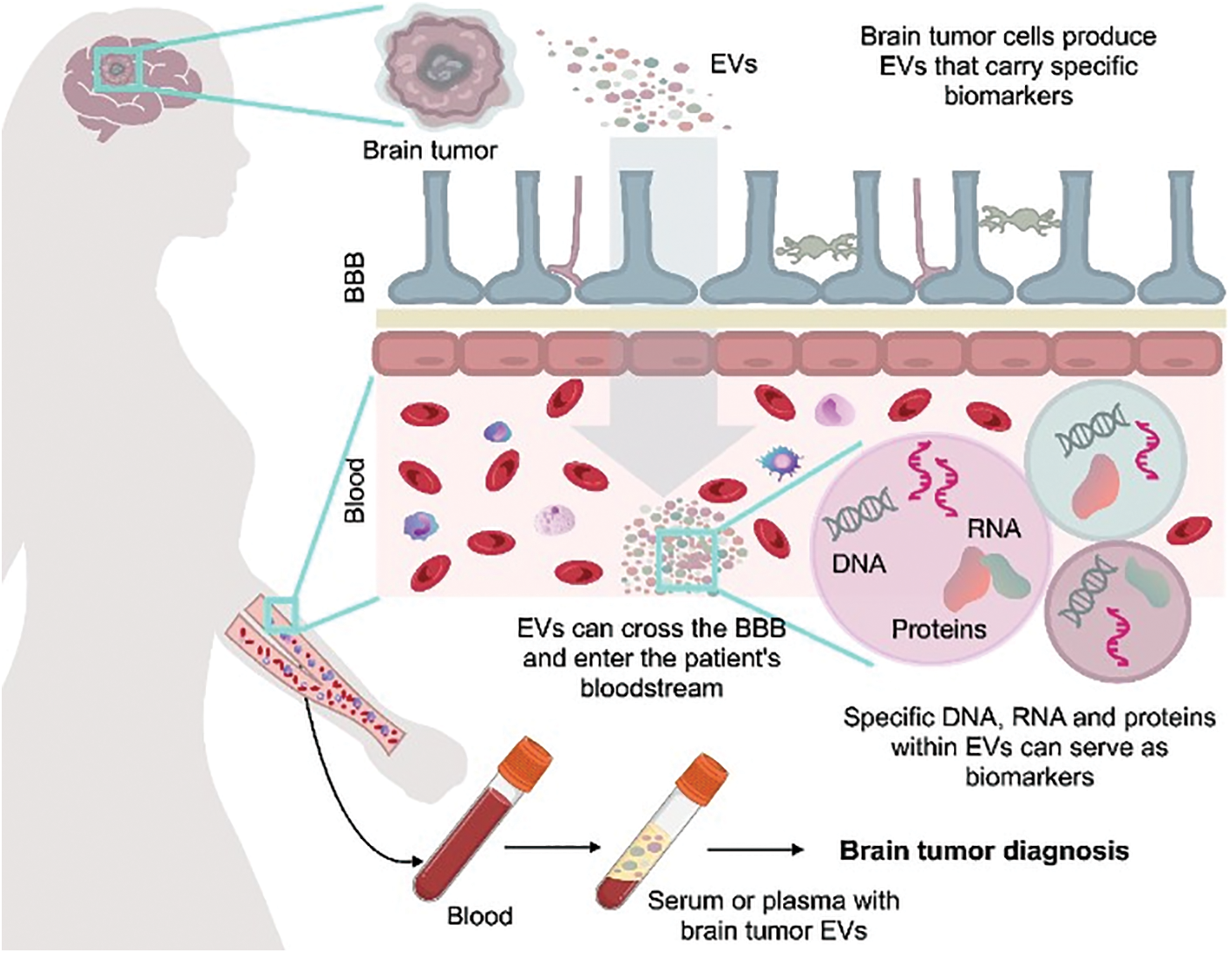
Figure 2: Possibility of using vesicles for diagnostics of brain cancer. Potential for using blood-derived vesicles that carry biomarkers to diagnose brain cancer. BBB—blood–brain barrier. The figure was created using Keynote.
It can be hypothesized that using EVs, isolated through liquid biopsy, primarily from human blood, could replace labor-intensive and expensive cancer detection methods, thereby reducing the impact on patient health [7,17].
EVs can be detected using various diagnostic methods, including isolation from different human secretions such as saliva, blood plasma, breast milk, ejaculate, CSF, and urine [18–21]. It is assumed that EVs play a role in cell-to-cell communication, including in pathological processes associated with various types of cancer [22–26]. EVs can cross the blood-brain barrier (BBB), and since vesicles from different tissues may have specific components in their composition, there is evidence that they can be detected due to this. It has been found that vesicles produced by cancer cells contain specific RNA and protein sequences that allow for the identification of their oncogenic origin [26–30]. In studies of EVs and plasma particles from cancer patients in the first and fourth stages of cancer progression, it was observed that cancer could be detected at early stages using tumor-associated extracellular vesicle and particle (EVP) proteins [7]. These properties of EVs potentially allow their use as biomarkers for brain tumors.
According to the literature, the production of both exosomes and microvesicles increases in damaged or malignant cells. Aggressive and highly differentiated tumor cells generate a greater number of EVs compared to their normal cellular equivalents [31]. However, there is an opposite case associated with a decrease in exosome production from colorectal cancer cells due to hyperoxia. Hypoxia, on the other hand, promotes the release of exosomes [32]. EVs derived from tumor cells contain distinct pro-tumorigenic components that regulate several aspects of cancer development. For instance, they carry pro-tumorigenic proteins such as growth factors (mutant epidermal growth factor receptor (EGFR)) and oncogenes (e.g., Kirsten rat sarcoma (KRAS)), as well as RNA that targets the transcriptional machinery of recipient cells, promoting oncogenesis [33]. Substantial evidence indicates that EVs significantly influence the formation and metastasis of GBM. EVs derived from GBM play a crucial role in oncogenesis, tumor microenvironment alterations, angiogenesis, immune response, invasion, subtype specification, and chemoresistance by transferring oncogenic proteins and nucleic acids [1].
In this review, we aim to present the current state of knowledge on the potential applications of EV-based biomarkers. We focus on brain tumor biomarkers, analyze potential vesicular tumor biomarkers, describe methods for vesicle isolation, and discuss techniques for detecting extracellular vesicle biomarkers.
The potential use of EVs as biomarkers for brain tumors was first explored in 2008 [26]. Since then, extensive research has investigated the relationship between vesicles and brain cancer. (Table 1). To date, a fairly large number of glioma biomarkers are already known (Fig. 3).
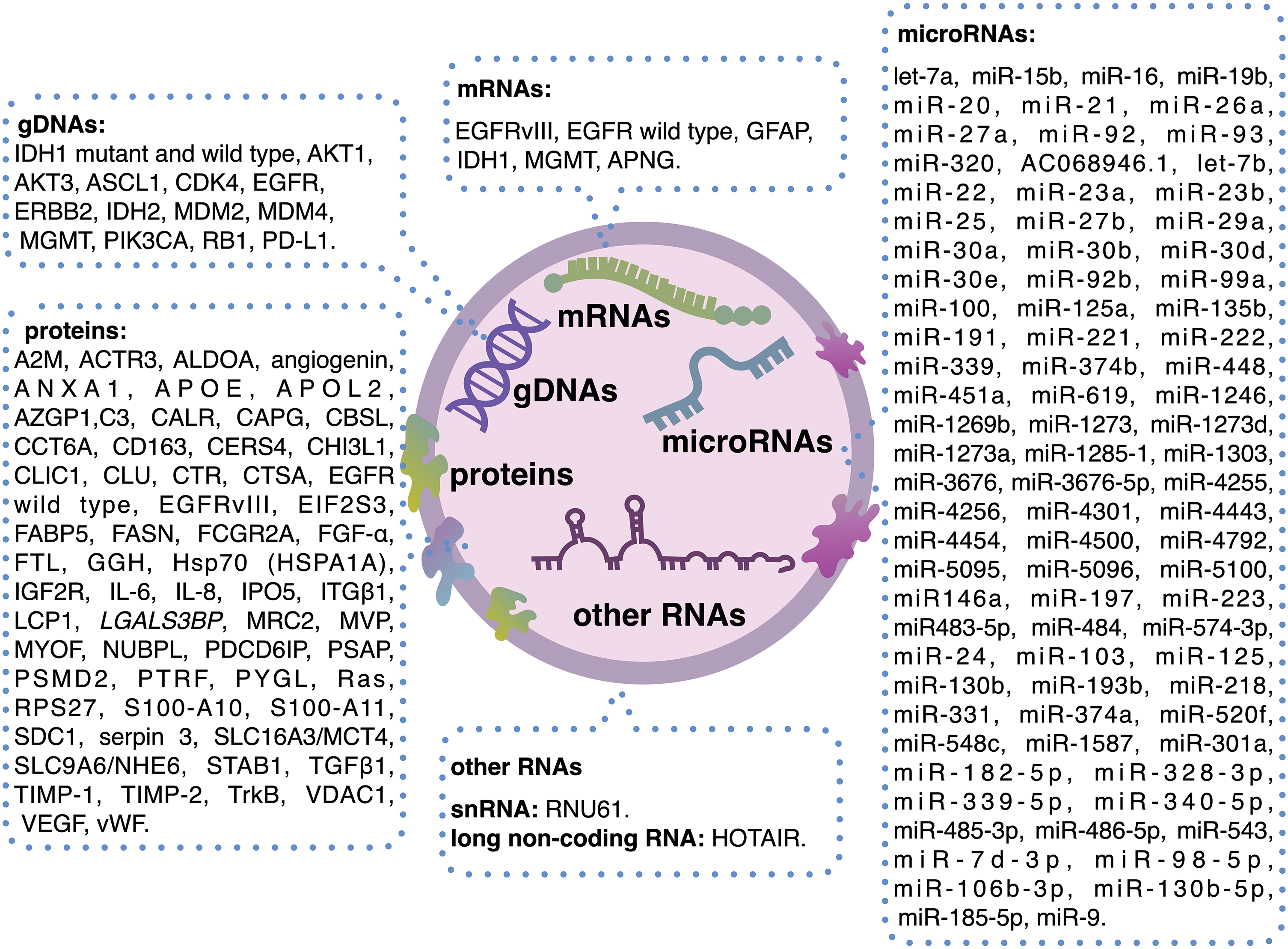
Figure 3: Brain tumor biomarkers containing EVs. The figure was created using Keynote.
The initial study, conducted in 2008 by Al-Nedawi et al., discovered that aggressive human brain tumors (gliomas) often express a truncated and oncogenic form of the EGFR, known as EGFRvIII (EGFR variant III). This receptor can be transported via microvesicles released into the bloodstream, thereby, the presence of such vesicles can indicate the existence of a malignant brain tumor. However, this study did not examine the role of microvesicles as markers, focusing instead on horizontal transfer, colony formation ability, and increased cell proliferation induced by EGFRvIII [26]. Later, in 2018, research on EGFRvIII and EGFR as markers revealed that the mRNA of these proteins is present in very small quantities within vesicles, requiring RNA amplification via PCR for detection. It is important to note that these molecules were not found in all patients, and correlated with the progression of GBM. In one-third of patients with grade IV GBM, this marker was detected, whereas less than 10% of samples from patients with grade III showed this marker. Additionally, in some cases, gene expression was found in biopsy tissue samples but not in vesicles isolated from blood [58]. Another study on EGFRvIII, conducted by Choi et al., investigated the impact of non-vesicular EGFRvIII on the protein composition of EVs. It was found that EGFRvIII expression was associated with the production of vesicles with low expression of CD81 and syntenin-1 (SDCBP), a complete absence of CD82, and higher expression of CD9, compared to vesicles from cells that did not express EGFRvIII. Moreover, there were significant changes in the proteome of vesicles produced by EGFRvIII-positive cells, suggesting a greater potential for using these vesicles as biomarkers [27], but some of these markers and the increase or decrease of their expression might be hardly enough to define the cells of origin in vivo.
A study by Hallal et al. in 2019 investigated the proteome of vesicles isolated from neurosurgical aspirates of GBM and grades II and III gliomas. They identified 298 distinct proteins that differentiate high-grade glioblastoma from low-grade glioma. Among these, 133 proteins were more prevalent in high-grade GBM patients, while 165 were more prevalent in low-grade glioma patients. Additionally, 19 specific marker proteins were identified, 15 of which were unique to GBM S100 calcium-binding protein (S100)-A10, S100A11, plastin-2 (LCP1), chitinase-3-like protein 1 (CHI3L1), chloride intracellular channel protein 1 (CLIC1), eukaryotic translation initiation factor 2 subunit 3 (EIF2S3), scavenger receptor cysteine-rich type 1 protein M130 (CD163), mannose receptor C type 2, apolipoprotein (APO)-L2, stabilising 1 (STAB1), 40S ribosomal protein S27 (RPC27), glycogen phosphorylase, liver form (PYGL), low-affinity immunoglobulin gamma Fc region receptor II-a (FCGR2A), macrophage-capping protein (CAPG), monocarboxylate transporter 4 (SLC16A3/MCT4), and 4 were specific to low-grade glioma ceramide synthase 4 (CERS4), cystathionine beta-synthase-like protein (CBSL), iron-sulfur protein NUBPL, also known as nucleotide-binding protein-like, sodium/hydrogen exchanger 6 (SLC9A6/NHE6). These findings suggest that vesicles could be used to determine the type and severity of brain tumors [28]. In a follow-up study in 2020, Hallal and her team discovered several vesicular protein markers specific to grade III and higher GBMs, astrocytomas, and oligodendroglioma. For instance, the cell division cycle 40 (CDC40) protein was found in samples from patients with grade III astrocytomas, while the tyrosylprotein sulfotransferase 2 (TPST2) protein was identified in patients with grade III oligodendrogliomas. However, other proteins, including axin interactor, dorsalization associated (AIDA), Rho guanine nucleotide exchange factor 10 (ARHGEF10), BCL2 interacting protein 3 like (BNIP3L), FYN binding protein 1 (FYB1), lysine methyltransferase 2D (KMT2D), microtubule associated protein 7 (MAP7), microtubule-associated serine/threonine kinase family member 4 (MAST4), phosphodiesterase 8A (PDE8A), RNA polymerase II subunit D (POLR2D), renin binding protein (RENBP), and solute carrier family 25 member 17 (SLC25A17) were found in patients with grade IV GBM. These findings complicate the use of these proteins as standalone markers for GBM, and their potential use as indicators of disease severity requires further investigation [29]. Similarly, Vineesh Indira Chandran et al. identified the protein syndecan-1 in vesicles, which is specific to patients with astrocytoma. Since syndecan-1 levels are significantly higher in patients with grade IV multiform GBM compared to those with grade II GBM, this marker could potentially differentiate disease severity. However, the study did not mention whether this marker is present in vesicles from patients with grade II GBM, leaving its value as a standalone marker in question [60].
In another 2019 proteomic study, researchers found that vesicles isolated from the plasma of patients with GBM and other brain tumors had elevated levels of proteins ferritin light chain (FTL), von Willebrand factor (vWF), alpha-2-glycoprotein 1, zinc-binding (AZGP1), serpin 3, complement component 3 (C3) and APOE, which may play a role in the pathophysiology of GBM. The study revealed that GBM releases all types of vesicles, in which immunoblotting identified the presence of the CD9 antigen. While the vesicles’ size showed no difference between control and patient groups, their concentration in the blood was significantly higher in GBM patients. Since the vesicles from healthy individuals and patients appear identical, additional protein isolation methods are required for analysis, which again complicates the use of vesicles as tumor markers [30].
A recent proteomic study of EVs derived from blood plasma, conducted in 2021 by Rana et al., identified potential markers for glioma. Protein groups of 34, 21, and 14 were discovered, with differential prevalence exceeding 1.3-fold across various glioma grades. Additionally, the transcription factors Yin Yang 1 (YY1) and signal transducer and activator of transcription (STAT)-1 were identified as potential regulators of various proteins in the early stages of glioma. Elevated levels of galectin-3 were also observed in samples from patients with grades I, II, and III gliomas compared to the control group. The lectin galactoside-binding soluble 3-binding protein (LGALS3BP) was similarly present in increased amounts in these patients’ samples [66].
It should be noted that microRNA in vesicles can be biomarkers. The article by Jafari et al. describes a fairly large number of exosomal microRNAs (ExomiR) that can play a role in tumor diagnosis [67]. For example, miR-1246, obtained from CSF, can be used as a possible diagnostic biomarker for gliomas [68].
An intriguing discovery was made in a 2015 study by Rui Shi et al. The microRNA molecule miR-21 was identified in exosomes derived from CSF. Their findings showed a correlation between the presence of gliomas and elevated miRNA levels. miR-21 expression from CSF exosomes was significantly higher in both high-grade and low-grade glioma groups, compared to the control group. These results suggest that miR-21 could serve as a biomarker for brain tumors [45].
Another marker was found among patients with variant disease severity in 2023 by Dobra et al. They examined 222 patients with various brain lesions, including meningioma, GBM, and brain metastases from non-small cell lung cancer. The study revealed higher levels of matrix metalloproteinase-9 (MMP-9) in EVs isolated from the blood serum of patients with malignant tumors. A correlation was observed between the degree of malignancy and tumor aggressiveness, and MMP-9 levels. Furthermore, MMP-9 levels decreased following surgical resection, therapy, or in cases of recurrence. High MMP-9 levels were also associated with lower survival rates in patients. Since levels of MMP-9 do not depend on gender or age, but directly correlate with the degree of tumor malignancy, this marker has significant prognostic and diagnostic potential [69].
Also of considerable interest is a 2019 study by Cumba Garcia et al. In which, exosomes isolated from blood were subjected to sequential density gradient ultracentrifugation (DGU). Although no specific markers were identified, it was found that with one-stage DGU, exosomes from patients with wild-type glioma were significantly smaller and more abundant in the blood, compared to those from healthy individuals. Additionally, exosomes from patients with grade IV GBM contained lower levels of cytokines interferon (IFN)-γ, interleukin (IL)-10, and IL-3 compared to healthy individuals. This research showed that the diagnostic potential of vesicles lies not only in the transport of specific markers but also in the differential composition of various non-tumor-specific biomolecules [70].
In conclusion, various studies on EV biomarkers indicate a substantial number of potential markers that can be derived from biological fluids and used as biomarkers for brain tumors. However, there still remains the challenge of vesicle identification and the search for specific biomarkers, highlighting the need for further research.
Isolation of Extracellular Vesicles
In order to utilize EVs as biomarkers for oncological diseases, it is essential to be able to isolate them. EVs from tumor cells have been shown to cross the BBB and enter the bloodstream [71], which highlights the importance of isolating vesicles from serum and plasma [72]. Based on the literature, circulating EVs can cross the BBB bidirectionally, from the bloodstream to the brain and vice versa, where they can be more easily detected [71]. For instance, Garcia-Romero et al. demonstrated that all three types of EVs secreted by human glioma cells can cross an intact BBB and be detected in peripheral blood, providing a minimally invasive method for their detection [50]. Transcytosis has been implicated as a key active mechanism for the passage of BBB-crossing molecules. However, despite extensive research, direct visualization of transcytosis across the blood-brain barrier in mammals, including humans, remains elusive. Evidence for transcytosis has been obtained in zebrafish studies, but their neurovascular unit differs from the more complex mammalian structure, raising concerns about the full applicability of these findings. Therefore, a comprehensive approach, incorporating both in vitro and in vivo studies, is still necessary to gain a more complete understanding of the transcytosis process across the BBB [73].
To obtain serum and plasma samples, vacutainers with different anticoagulants can be used. Mari Palviainen et al. investigated the extent to which anticoagulants affect the concentration, cellular origin, and protein composition of EVs. For serum collection, anticoagulants such as acid-citrate-dextrose (ACD), citrate, or ethylenediaminetetraacetic acid (EDTA), as well as samples without anticoagulants, were used in the study. The research showed that serum samples indeed contain significantly higher concentrations of platelet-derived EVs compared to plasma samples and that serum-derived EVs bind more annexin V. The number of platelet-derived EVs was also higher when using citrate anticoagulant, compared to ACD and EDTA. This suggests that citrate inhibits platelet activation less effectively than ACD and EDTA, leading to the formation of platelet EVs in blood samples during and after collection. This is crucial, as the uncontrolled release of platelet-derived EVs affects biomarker detection; for example, it has been shown that platelet-derived EVs distort miRNA populations in patient and control samples [74]. Moreover, there is evidence that it is better to use plasma to determine biomarkers for EVs instead of serum since the latter represents a higher concentration of platelet-derived EVs [74]. Therefore, to reduce confounding factors potentially arising from platelet-released EVs, plasma is the preferred source of EVs for biomarker detection, whereas serum is used to study cell-free microRNAs [75].
The presence and amount of plasma exosomes correlate with tumor mass [76], as well as with cancer progression and overall patient survival [77]. Interestingly, plasma analysis can be performed at different time points (before treatment, one month post-surgery, and after six months following therapy), allowing monitoring of EVs’ quantities at various stages of treatment [78]. This repeated plasma sampling approach is also effective for tracking treatment progress in brain cancer or monitoring disease progression [77].
EVs can be obtained through centrifugation/ultracentrifugation. For example, in a study by Erika Bandini et al., exosome isolation from blood plasma involved a series of centrifugation steps with progressively increasing speeds to separate larger vesicles and particles. Another crucial step is filtration using centrifugal filter devices [78]. The pore size of filters may vary, the supernatant can be filtered through a 0.45 μm or 0.22 μm filter. The final purification step for exosomes involves ultracentrifugation. After removing the supernatant, the resulting pellet is examined [79].
Washing the samples is also an important step. Following ultracentrifugation, the sample is washed with a large volume of phosphate-buffered saline [77].
Additional purification of EVs can be achieved using size-exclusion chromatography (SEC). However, this method is primarily applied to verify the purity of the isolated exosome preparation [78,80]. In SEC, samples are filtered through a porous stationary phase. Sample components with smaller hydrodynamic radii pass through the pores more quickly and are thus eluted faster, whereas components with larger radii do not enter the pores and freely exit the column [81]. During the development of this method, starch, and water were initially used to form the pores in the column, but over time, various other compounds have been utilized, such as polymer dextran (Sephadex), agarose (Sepharose), and polyacrylamide (Sephacryl or Bio-Gel) [82]. Various columns can be used in SEC. For instance, Brennan et al. used IZON qEV columns in their study, which enabled the separation of EVs larger than 70 nm from smaller particles and proteins that took longer to pass through the column [83].
Another method for EV isolation is magnetic immunoprecipitation. In the case of immunomagnetic capture, a biotinylated antibody against the target antigen is attached to the surface of streptavidin-coated magnetic beads. The antibody-coated beads are then incubated with the sample from which exosomes need to be isolated. The main advantage of this method over enzyme-linked immunosorbent assay (ELISA) is that the beads provide a larger surface area for capturing exosomes, resulting in higher isolation efficiency [84].
Various techniques are employed for the analysis of EV size, morphology, and quantification. Among such techniques are nanoparticle tracking analysis (NTA), tunable resistive pulse sensing (TRPS), and transmission electron microscopy [85]. NTA aids in detecting the content and size of EVs [79], while their morphology can be examined using transmission electron microscopy [79]. Western blotting is used to confirm the purification of EVs [79], by analyzing the expression of exosomal markers [78].
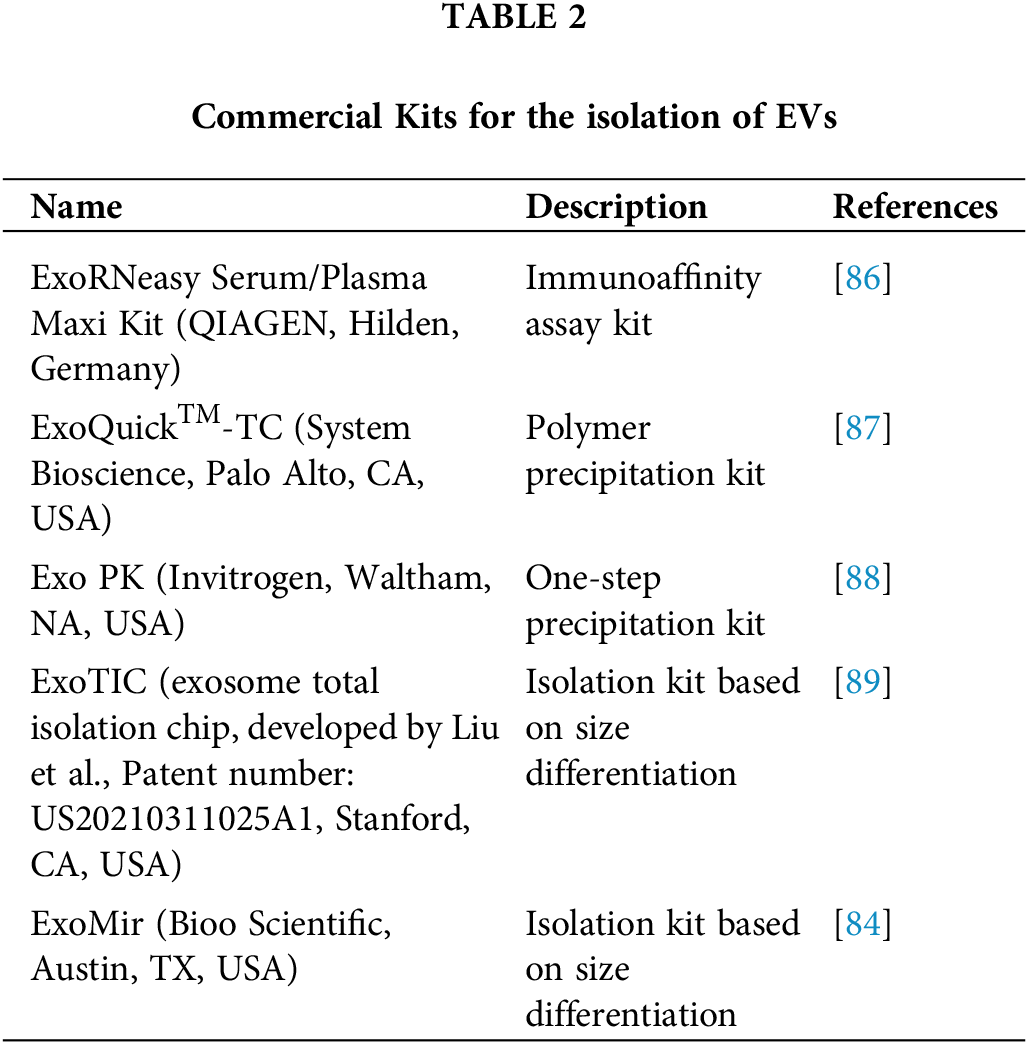
Specialized kits for the isolation of EVs have also been developed (Table 2).
Therefore, reliable methods for EV isolation are currently available, enabling their use as biomarkers for tumor diseases.
Currently, brain tumor detection relies on the symptomatic presentation of the tumor, magnetic resonance imaging (MRI), and invasive tissue biopsy, which can lead to a delayed diagnosis. Therefore, there is an urgent need to develop diagnostic tools that enable timely and non-invasive assessment of brain tumors [90]. Analysis of EVs, derived from patient blood, could serve as a potential solution to this problem. ELISA is commonly used for identifying these vesicular biomarkers (Fig. 4).
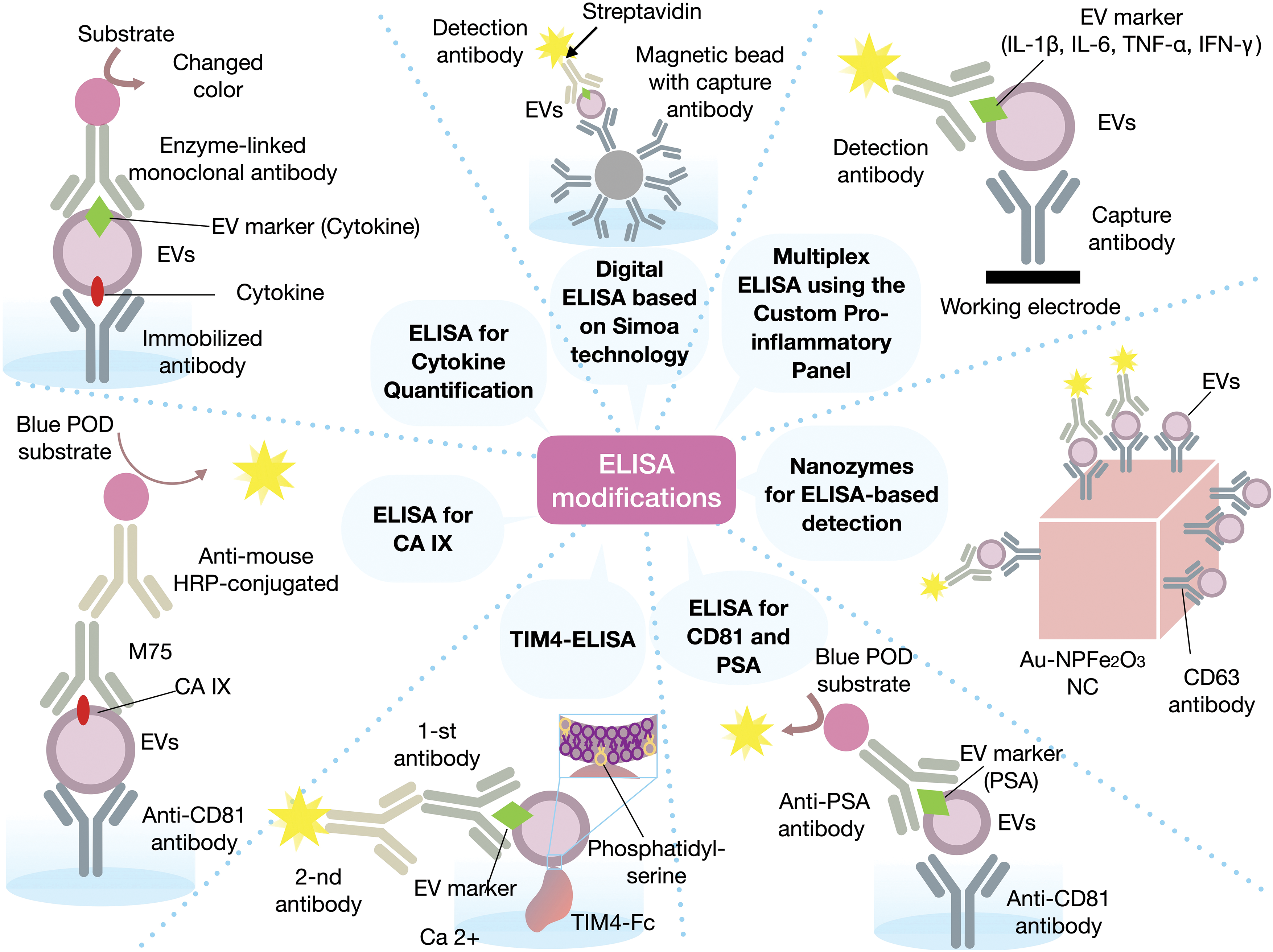
Figure 4: Various modifications used in the ELISA method. CA IX—carbonic anhydrase 9, CD63—cluster of differentiation 63, CD81—cluster of differentiation 81, HRP—horseradish peroxidase, IFN-γ—interferon gamma, IL-1β—interleukin 1 beta, IL-6—interleukin 6, PSA—prostate-specific antigen, Simoa—single molecule array, TNF-α—tumor necrosis factor alpha, TIM-4—T-cell membrane protein 4. The figure was created using Keynote.
ELISA is a method utilized for the quantification of peptides and proteins [91]. All techniques employing enzymes to detect antigen-antibody reactions are generally referred to as enzyme immunoassays (EIA/ELISA) [91].
The immunocapture-based ELISA for EVs/exosomes was first described in 2009. Logozzi et al. developed a sandwich ELISA to capture and quantify exosomes in plasma, based on the expression of housekeeping proteins (CD63 and Rab-5b) and a tumor-associated marker (caveolin-1). This method enabled the sensitive detection and quantification of EVs isolated from human tumor cell culture supernatants and plasma from SCID mice xenografted with human melanoma. It was found that plasma exosomes expressing CD63 or caveolin-1 were significantly elevated in melanoma patients, compared to healthy donors [76]. This study demonstrated that immunocapture-based ELISA allows for the simultaneous detection and quantification of both exosome-specific antigens and tumor antigens on EVs.
The immunocapture-based ELISA method involves coating a polypropylene surface of a 96-well plate with specific antibodies against an exosomal antigen. Subsequently, exosomes are incubated in the wells to facilitate capture. The plate is then washed, and a detection antibody either directly or indirectly conjugated with an enzyme, is added. Upon addition of the appropriate substrate, a signal is generated that can be read using a microplate spectrophotometer. Furthermore, the detection antibody can be targeted either at an exosomal antigen, or specific disease markers, making this method applicable for disease diagnosis and monitoring [92].
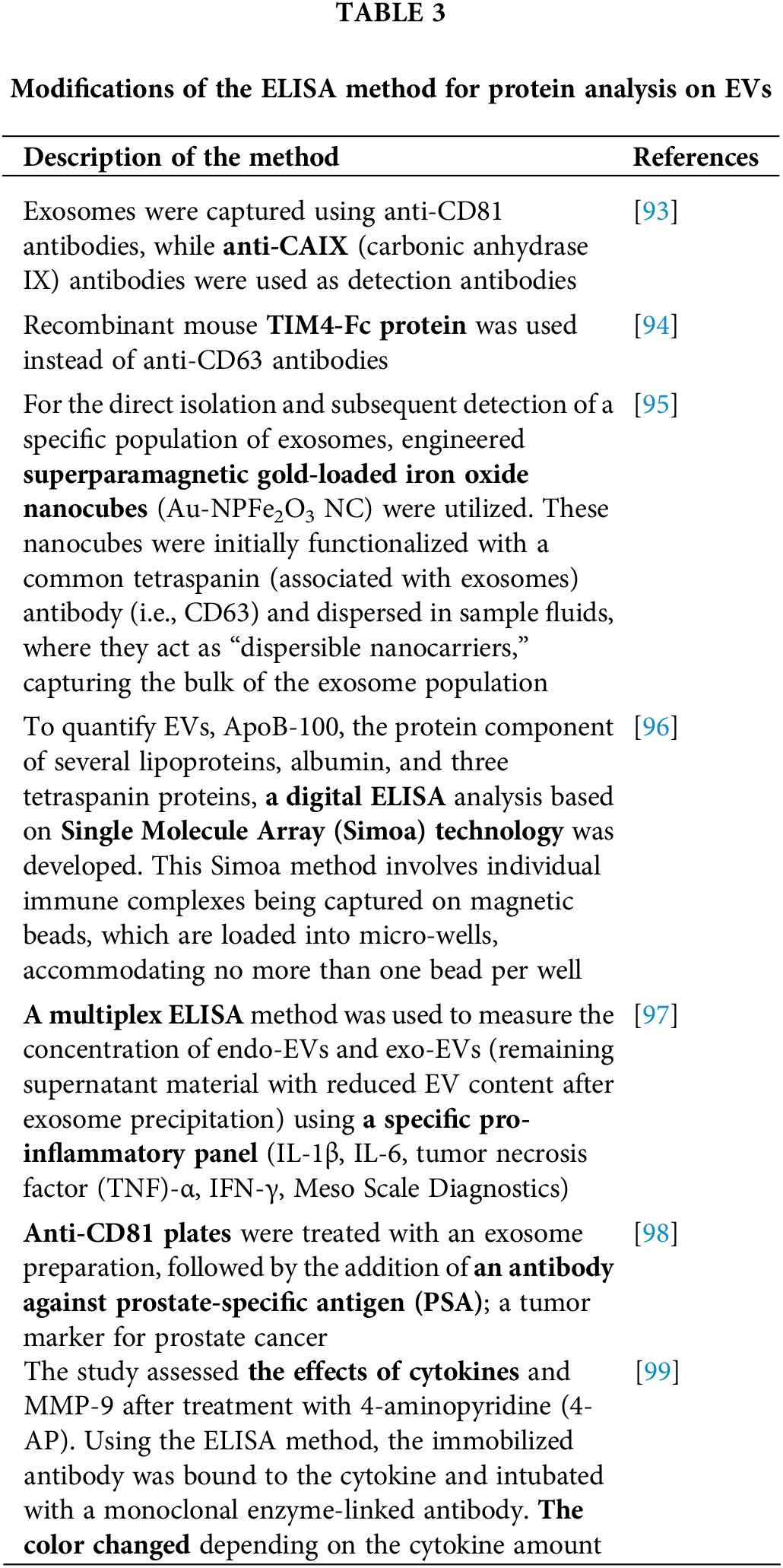
Following the adaptation of the ELISA method for protein analysis on EVs, this technique gained widespread popularity and became broadly used for detecting tumor biomarkers (Table 3).
ELISA is not the only method for detecting biomarkers on EVs. For instance, there is a rapid lateral magnetic flux immunoassay test called the lateral flow immunoassay (LFIA). LFIA utilizes antibodies immobilized on a specialized nitrocellulose membrane, which react with the analyte and migrate via capillary effect. “Sandwich complexes” of the target protein and free-capture antibodies also form and accumulate in two regions to form the test and control lines. In a melanoma study, the capture targets were tetraspanins CD9, CD63, and CD81; common biomarkers of exosomes [100]. This method can vary depending on experimental conditions, such as the type of disease being investigated. It is crucial to select the appropriate antibodies for EV-associated antigens relevant to the specific cancer type. LFIA has been employed for diagnosing melanoma [100,101], colorectal cancer [85], ovarian cancer [102], prostate cancer [103,104], and cervical cancer [105]. LFIA could also be effective for diagnosing and monitoring brain tumors with the proper EV isolation methods and antibody selection.
The addition of silver [103,105] and gold nanoparticles [100,104] improves the accuracy and efficacy of LFIA [100].
Furthermore, there is the option of using the time-resolved fluorescent microsphere (TRFM) method. These microspheres are conjugated with an antibody against CA199, a serum biomarker useful in cancer diagnostics and progression monitoring. Researchers have detected signals using ultraviolet light, testing the method on serum from healthy individuals and liver cancer patients, noting a 60% efficiency in differentiation, with potential for method improvement in the future [106].
ELISA and LFIA can be used together to compare results or confirm the effectiveness of each method for specific studies [100].
Unlike other immunoassays, LFIA is a rapid test that can be conducted in an extremely short time, down to several minutes. A primary limitation is that LFIA examines larger vesicles rather than exosomes, and may lack precision [100], which is less productive for glioma research where accuracy is crucial due to limited non-invasive diagnostic methods. Nonetheless, LFIA can provide preliminary information about the patient’s condition and an initial cancer assessment.
A recent study involving RNA sequencing of serum EVs from a large cohort of patients with isocitrate dehydrogenase (IDH)-wildtype GBM and healthy individuals aimed to discover new tumor biomarkers. The research showed that serum EV biomarker panels could predict/diagnose tumor tissue status in terms of IDH1 mutation, methylguanine methyltransferase (MGMT) promoter methylation, telomerase reverse transcriptase (TERT) promoter mutation, and p53 mutation, with high sensitivity and specificity for GBM. RNA sequencing of serum EVs from preoperative glioblastoma patients vs. controls identified 569 differentially expressed genes (differential gene expression (DEG), false discovery rate (FDR) <0.05). Based on these DEGs, serum EV biomarker panels were developed for IDH1 wild-type glioblastoma (96% sensitivity/80% specificity), MGMT promoter methylation (91% sensitivity/73% specificity), p53 gene mutation (100% sensitivity/89% specificity) and TERT promoter mutation (89% sensitivity/100% specificity). This study showed that a panel of tumor biomarkers based on EVs derived from the serum of patients with GBM can be used clinically at the genetic level to diagnose GBM, monitor tumor burden, and assess tumor progression and response to therapy [107].
Jalali et al. developed a multiplex fluidic device with integrated nanocavity microchips (MoSERS microchip), enabling molecular profiling of distinct EVs in small fluid volumes (<10 µL), via surface-enhanced Raman spectroscopy (SERS). This device can provide information on the surface properties and composition of EVs for early detection and monitoring of cellular transformation during tumor progression and therapy. The MoSERS microchip was applied to identify and distinguish signals from distinct EVs derived from benign glial cells (NHA), glioma cells (U373, U87), and glioma stem cells (GSC83, GSC1005, and GSC1123). These cell lines were either wild-type or modified to express relevant molecular changes, such as EGFRvIII and phosphatase and tensin homolog (PTEN), which are naturally lost during GBM progression, and MGMT, a marker and effector of GBM resistance to temozolomide. After that, to maximize the method’s ability to distinguish SERS spectra with minor differences, and achieve more precise and sensitive multidimensional EV identification, SERS of individual EVs was combined with deep learning methods based on computational neural network (CNN). The CNN was then applied to recognize EVs derived from blood samples of GBM patients. The CNN approach allowed for determining the probability that each EV carried specific oncogenic markers, including EGFR amplification, EGFRvIII mutation, and MGMT expression. The study found that combining CNN with MoSERS improved diagnostic accuracy (87%) in detecting GBM mutations in the blood samples of 12 patients, compared to clinical pathological tests. Accordingly, MoSERS demonstrates the potential for molecular stratification of cancer patients using circulating EVs [108].
Two patents include panels for the application of EVs as biomarkers for brain tumors. The first patent (RU2712223C2) relates to laboratory diagnostics and oncology, focusing on the detection of tumors in patients in vitro. It has been found that the protein transmembrane 9 superfamily member 4 (TM9SF4) is overexpressed in tumor-derived EVs. Results from a sandwich ELISA, where 40, 20, 10, and 5 μg of exosomes, purified via ultracentrifugation from conditioned medium of the GBM cell line (U87), are captured by an antibody against TM9SF4 and detected by an antibody against CD9, demonstrate that these biomarkers are expressed on the exosomal membrane. This indicates that the sandwich ELISA can be utilized to detect exosomes from malignant peripheral nerve sheath tumors (MPNST) or other solid tumors such as GBM. Therefore, the analysis of the TM9SF4 protein can be used to determine the presence of tumors or the state of tumor transformation in patients in vitro.
Another patent (US20150293101) was granted for an invention related to the diagnosis and prognosis of cancer, as well as for monitoring tumor progression by detecting the activity of oncogenic proteins and mediators in microvesicles. More specifically, the oncogenic or EV-associated proteins can be EGFR and human epidermal growth factor receptor (HER)-2, or HER-2 and HER-3, or HER-2 and EGFR2, or EGFRvIII and HER-2. For cancer diagnosis and prognosis, the amount of oncogenic EV-associated protein was evaluated before and after treatment. As for monitoring tumor progression, the level of phosphorylation of the oncogenic EV-associated protein was also assessed before and after treatment, where an increase in phosphorylation levels indicates tumor progression and treatment inefficacy. Accordingly, the quantity of oncogenic EV-associated proteins and their phosphorylation levels can serve as reliable tumor biomarkers.
Currently, numerous methods are available for analyzing the composition of EVs, enabling their potential application as biomarkers for the diagnosis and prognosis of brain tumors.
In recent years, there has been significant interest in using EVs as biomarkers for brain tumors. They can be isolated from various biological fluids, allowing for their use in liquid biopsies. Unlike traditional biopsies, liquid biopsy is considered a safe and non-invasive method. Tumor-derived EVs hold great potential as biomarkers, with current protocols developed for isolation and various methods for analyzing their contents. However, challenges persist in identifying vesicles and searching for specific biomarkers, requiring further research. While most studies concentrate on identifying specific biomarkers, there is a hypothesis that vesicles from tumor cells may differ in the composition of non-tumor-specific biomarkers. Despite vesicles having great potential as biomarkers, they may be hard to identify in patients with low grade GBM. Also there is a concern that vesicles from tumor and normal cells can be indistinguishable, and on the other hand they can be associated with cancer and other diseases both. Therefore, extensive research is needed to fully utilize EVs as tumor biomarkers, a task that needs significant time and effort.
Acknowledgement: None.
Funding Statement: This paper has been supported by the Kazan Federal University Strategic Academic Leadership Program (PRIORITY-2030).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zarema Gilazieva; preparation of manuscript and tables: Daniil Moldavskii, Ekaterina Luzina, Aisylu Kadyrova; preparation of the drawing: Alisa Shaimardanova; editing of the manuscript Shaza Issa, Daniil Moldavskii, Valeriya Solovyeva; supervision: Albert Rizvanov. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Chen Y, Jin Y, Wu N. Role of tumor-derived extracellular vesicles in glioblastoma. Cells. 2021;10(3):512. doi:10.3390/cells10030512. [Google Scholar] [PubMed] [CrossRef]
2. Lin Y, Wu Z. Hypoxia-inducible factor 1α (HIF-1α)-activated Gli1 induces invasion and EMT by H3K4 methylation in glioma cells. Oncologie. 2023;25(1):71–9. doi:10.1515/oncologie-2023-0004. [Google Scholar] [CrossRef]
3. Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–20. doi:10.1007/s00401-016-1545-1. [Google Scholar] [PubMed] [CrossRef]
4. Biserova K, Jakovlevs A, Uljanovs R, Strumfa I. Cancer stem cells: significance in origin, pathogenesis and treatment of glioblastoma. Cells. 2021;10(3):621. doi:10.3390/cells10030621. [Google Scholar] [PubMed] [CrossRef]
5. Indira Chandran V, Gopala S, Venkat EH, Kjolby M, Nejsum P. Extracellular vesicles in glioblastoma: a challenge and an opportunity. npj Precis Oncol. 2024;8(1):103. doi:10.1038/s41698-024-00600-2. [Google Scholar] [PubMed] [CrossRef]
6. Connal S, Cameron JM, Sala A, Brennan PM, Palmer DS, Palmer JD, et al. Liquid biopsies: the future of cancer early detection. J Transl Med. 2023;21(1):118. doi:10.1186/s12967-023-03960-8. [Google Scholar] [PubMed] [CrossRef]
7. Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, et al. Extracellular vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044–1061.e18. doi:10.1016/j.cell.2020.07.009. [Google Scholar] [PubMed] [CrossRef]
8. Eibl RH, Schneemann M. Liquid biopsy and glioblastoma. Explor Target Anti-Tumor Ther. 2023;4(1):28–41. doi:10.37349/etat.2023.00121. [Google Scholar] [PubMed] [CrossRef]
9. Chin RI, Chen K, Usmani A, Chua C, Harris PK, Binkley MS, et al. Detection of solid tumor molecular residual disease (MRD) using circulating tumor DNA (ctDNA). Mol Diagn Ther. 2019;23(3):311–31. doi:10.1007/s40291-019-00390-5. [Google Scholar] [PubMed] [CrossRef]
10. Urabe F, Kosaka N, Ito K, Kimura T, Egawa S, Ochiya T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am J Physiol Cell Physiol. 2020;318(1):C29–39. doi:10.1152/ajpcell.00280.2019. [Google Scholar] [PubMed] [CrossRef]
11. Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, et al. Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther. 2021;6(1):404. doi:10.1038/s41392-021-00817-8. [Google Scholar] [PubMed] [CrossRef]
12. Westphal M, Lamszus K. Circulating biomarkers for gliomas. Nat Rev Neurol. 2015;11(10):556–66. doi:10.1038/nrneurol.2015.171. [Google Scholar] [PubMed] [CrossRef]
13. Welsh JA, Goberdhan DCI, O’Driscoll L, Buzas EI, Blenkiron C, Bussolati B, et al. Minimal information for studies of extracellular vesicles (MISEV2023from basic to advanced approaches. J Extracell Vesicles. 2024;13(2):e12404. Erratum in: J Extracell Vesicles. 2024; 13(5e12451. doi:10.1002/jev2.v13.2. [Google Scholar] [CrossRef]
14. Menck K, Sivaloganathan S, Bleckmann A, Binder C. Microvesicles in cancer: small size, large potential. Int J Mol Sci. 2020;21(15):5373. doi:10.3390/ijms21155373. [Google Scholar] [PubMed] [CrossRef]
15. Gilazieva Z, Chulpanova D, Ponomarev A, Filin I, Garanina E, Rizvanov A, et al. Comparative analysis of natural and cytochalasin B-induced membrane vesicles from tumor cells and mesenchymal stem cells. Curr Issues Mol Biol. 2022;44(11):5363–78. doi:10.3390/cimb44110363. [Google Scholar] [PubMed] [CrossRef]
16. Gilazieva Z, Ponomarev A, Rizvanov A, Solovyeva V. The dual role of mesenchymal stromal cells and their extracellular vesicles in carcinogenesis. Biology. 2022;11(6):813. doi:10.3390/biology11060813. [Google Scholar] [PubMed] [CrossRef]
17. Choi WWY, Sánchez C, Li JJ, Dinarvand M, Adomat H, Ghaffari M, et al. Extracellular vesicles from biological fluids as potential markers in castration resistant prostate cancer. J Cancer Res Clin Oncol. 2023;149(8):4701–17. doi:10.1007/s00432-022-04391-6. [Google Scholar] [PubMed] [CrossRef]
18. Akers JC, Ramakrishnan V, Yang I, Hua W, Mao Y, Carter BS, et al. Optimizing preservation of extracellular vesicular miRNAs derived from clinical cerebrospinal fluid. Cancer Biomark Sect Dis Markers. 2016;17(2):125–32. doi:10.3233/CBM-160609. [Google Scholar] [PubMed] [CrossRef]
19. Liu Z, Cauvi DM, Bernardino EMA, Lara B, Lizardo RE, Hawisher D, et al. Isolation and characterization of human urine extracellular vesicles. Cell Stress Chaperones. 2018;23(5):943–53. doi:10.1007/s12192-018-0902-5. [Google Scholar] [PubMed] [CrossRef]
20. Höög JL, Lötvall J. Diversity of extracellular vesicles in human ejaculates revealed by cryo-electron microscopy. J Extracell Vesicles. 2015;4(1):28680. doi:10.3402/jev.v4.28680. [Google Scholar] [PubMed] [CrossRef]
21. Iwai K, Yamamoto S, Yoshida M, Shiba K. Isolation of extracellular vesicles in saliva using density gradient ultracentrifugation. Methods Mol Biol. 2017;1660:343–50. doi:10.1007/978-1-4939-7253-1_27. [Google Scholar] [CrossRef]
22. Chulpanova DS, Rizvanov AA, Solovyeva VV. The role of cancer stem cells and their extracellular vesicles in the modulation of the antitumor immunity. Int J Mol Sci. 2023;24(1):395. doi:10.3390/ijms24010395. [Google Scholar] [PubMed] [CrossRef]
23. Yang Q, Liu J, Wu B, Wang X, Jiang Y, Zhu D. Role of extracellular vesicles in osteosarcoma. Int J Med Sci. 2022;19(8):1216–26. doi:10.7150/ijms.74137. [Google Scholar] [PubMed] [CrossRef]
24. Wang D, Ming X, Xu J, Xiao Y. Circ_0009910 shuttled by exosomes regulates proliferation, cell cycle and apoptosis of acute myeloid leukemia cells by regulating miR-5195-3p/GRB10 axis. Hematol Oncol. 2021 Aug;39(3):390–400. doi:10.1002/hon.2874. [Google Scholar] [PubMed] [CrossRef]
25. Simionescu N, Zonda R, Petrovici AR, Georgescu A. The multifaceted role of extracellular vesicles in glioblastoma: microRNA nanocarriers for disease progression and gene therapy. Pharmaceutics. 2021;13(7):988. doi:10.3390/pharmaceutics13070988. [Google Scholar] [PubMed] [CrossRef]
26. Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–24. doi:10.1038/ncb1725. [Google Scholar] [PubMed] [CrossRef]
27. Choi D, Montermini L, Kim DK, Meehan B, Roth FP, Rak J. The impact of oncogenic EGFRvIII on the proteome of extracellular vesicles released from glioblastoma cells. Mol Cell Proteomics. 2018;17(10):1948–64. doi:10.1074/mcp.RA118.000644. [Google Scholar] [PubMed] [CrossRef]
28. Hallal S, Russell BP, Wei H, Lee MYT, Toon CW, Sy J, et al. Extracellular vesicles from neurosurgical aspirates identifies chaperonin containing TCP1 subunit 6A as a potential glioblastoma biomarker with prognostic significance. Proteomics. 2019;19(1–2):e1800157. [Google Scholar] [PubMed]
29. Hallal S, Azimi A, Wei H, Ho N, Lee MYT, Sim HW, et al. A comprehensive proteomic SWATH-MS workflow for profiling blood extracellular vesicles: a new avenue for glioma tumour surveillance. Int J Mol Sci. 2020;21(13):4754. doi:10.3390/ijms21134754. [Google Scholar] [PubMed] [CrossRef]
30. Osti D, Del Bene M, Rappa G, Santos M, Matafora V, Richichi C, et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25(1):266–76. doi:10.1158/1078-0432.CCR-18-1941. [Google Scholar] [PubMed] [CrossRef]
31. Bamankar S, Londhe VY. The rise of extracellular vesicles as new age biomarkers in cancer diagnosis: promises and pitfalls. Technol Cancer Res Treat. 2023;22:15330338221149266. doi:10.1177/15330338221149266. [Google Scholar] [PubMed] [CrossRef]
32. To KKW, Cho WCS. Exosome secretion from hypoxic cancer cells reshapes the tumor microenvironment and mediates drug resistance. Cancer Drug Resist. 2022;5(3):577–94. doi:10.20517/cdr. [Google Scholar] [CrossRef]
33. Stavrou A, Ortiz A. Extracellular vesicles: a novel tool in nanomedicine and cancer treatment. Cancers. 2022;14(18):4450. doi:10.3390/cancers14184450. [Google Scholar] [PubMed] [CrossRef]
34. Hallal S, Ebrahimkhani S, Shivalingam B, Graeber MB, Kaufman KL, Buckland ME. The emerging clinical potential of circulating extracellular vesicles for non-invasive glioma diagnosis and disease monitoring. Brain Tumor Pathol. 2019;36(2):29–39. [Google Scholar] [PubMed]
35. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. doi:10.1038/ncb1800. [Google Scholar] [PubMed] [CrossRef]
36. Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106(10):3794–9. doi:10.1073/pnas.0804543106. [Google Scholar] [PubMed] [CrossRef]
37. Graner MW, Alzate O, Dechkovskaia AM, Keene JD, Sampson JH, Mitchell DA, et al. Proteomic and immunologic analyses of brain tumor exosomes. FASEB J Off Publ Fed Am Soc Exp Biol. 2009;23(5):1541–57. doi:10.1096/fj.08-122184. [Google Scholar] [PubMed] [CrossRef]
38. Noerholm M, Balaj L, Limperg T, Salehi A, Zhu LD, Hochberg FH, et al. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer. 2012;12(1):22. doi:10.1186/1471-2407-12-22. [Google Scholar] [PubMed] [CrossRef]
39. Li CCY, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10(8):1333–44. doi:10.4161/rna.25281. [Google Scholar] [PubMed] [CrossRef]
40. Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. BEAMing and Droplet digital PCR analysis of Mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2(7):e109. doi:10.1038/mtna.2013.28. [Google Scholar] [PubMed] [CrossRef]
41. Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSFa platform for glioblastoma biomarker development. PLoS One. 2013;8(10):e78115. doi:10.1371/journal.pone.0078115. [Google Scholar] [PubMed] [CrossRef]
42. Manterola L, Guruceaga E, Gállego Pérez-Larraya J, González-Huarriz M, Jauregui P, Tejada S, et al. A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro-Oncol. 2014;16(4):520–7. doi:10.1093/neuonc/not218. [Google Scholar] [PubMed] [CrossRef]
43. Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun. 2015;6(1):6999. doi:10.1038/ncomms7999. [Google Scholar] [PubMed] [CrossRef]
44. Akers JC, Ramakrishnan V, Kim R, Phillips S, Kaimal V, Mao Y, et al. miRNA contents of cerebrospinal fluid extracellular vesicles in glioblastoma patients. J Neurooncol. 2015;123(2):205–16. doi:10.1007/s11060-015-1784-3. [Google Scholar] [PubMed] [CrossRef]
45. Shi R, Wang PY, Li XY, Chen JX, Li Y, Zhang XZ, et al. Exosomal levels of miRNA-21 from cerebrospinal fluids associated with poor prognosis and tumor recurrence of glioma patients. Oncotarget. 2015;6(29):26971–81. doi:10.18632/oncotarget.4699. [Google Scholar] [PubMed] [CrossRef]
46. Pinet S, Bessette B, Vedrenne N, Lacroix A, Richard L, Jauberteau MO, et al. TrkB-containing exosomes promote the transfer of glioblastoma aggressiveness to YKL-40-inactivated glioblastoma cells. Oncotarget. 2016;7(31):50349–64. doi:10.18632/oncotarget.10387. [Google Scholar] [PubMed] [CrossRef]
47. Mallawaaratchy DM, Hallal S, Russell B, Ly L, Ebrahimkhani S, Wei H, et al. Comprehensive proteome profiling of glioblastoma-derived extracellular vesicles identifies markers for more aggressive disease. J Neurooncol. 2017;131(2):233–44. doi:10.1007/s11060-016-2298-3. [Google Scholar] [PubMed] [CrossRef]
48. Luhtala N, Aslanian A, Yates JR, Hunter T. Secreted glioblastoma nanovesicles contain intracellular signaling proteins and active ras incorporated in a farnesylation-dependent manner. J Biol Chem. 2017;292(2):611–28. doi:10.1074/jbc.M116.747618. [Google Scholar] [PubMed] [CrossRef]
49. Yang JK, Yang JP, Tong J, Jing SY, Fan B, Wang F, et al. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J Neurooncol. 2017;131(2):255–65. doi:10.1007/s11060-016-2308-5. [Google Scholar] [PubMed] [CrossRef]
50. García-Romero N, Carrión-Navarro J, Esteban-Rubio S, Lázaro-Ibáñez E, Peris-Celda M, Alonso MM, et al. DNA sequences within glioma-derived extracellular vesicles can cross the intact blood-brain barrier and be detected in peripheral blood of patients. Oncotarget. 2017;8(1):1416–28. doi:10.18632/oncotarget.13635. [Google Scholar] [PubMed] [CrossRef]
51. Akers JC, Hua W, Li H, Ramakrishnan V, Yang Z, Quan K, et al. A cerebrospinal fluid microRNA signature as biomarker for glioblastoma. Oncotarget. 2017;8(40):68769–79. doi:10.18632/oncotarget.18332. [Google Scholar] [PubMed] [CrossRef]
52. Figueroa JM, Skog J, Akers J, Li H, Komotar R, Jensen R, et al. Detection of wild-type EGFR amplification and EGFRvIII mutation in CSF-derived extracellular vesicles of glioblastoma patients. Neuro-Oncol. 2017;19(11):1494–502. doi:10.1093/neuonc/nox085. [Google Scholar] [PubMed] [CrossRef]
53. Figueroa J, Phillips LM, Shahar T, Hossain A, Gumin J, Kim H, et al. Exosomes from glioma-associated mesenchymal stem cells increase the tumorigenicity of glioma stem-like cells via transfer of miR-1587. Cancer Res. 2017;77(21):5808–19. doi:10.1158/0008-5472.CAN-16-2524. [Google Scholar] [PubMed] [CrossRef]
54. Lan F, Qing Q, Pan Q, Hu M, Yu H, Yue X. Serum exosomal miR-301a as a potential diagnostic and prognostic biomarker for human glioma. Cell Oncol Dordr. 2018;41(1):25–33. doi:10.1007/s13402-017-0355-3. [Google Scholar] [PubMed] [CrossRef]
55. Ricklefs FL, Alayo Q, Krenzlin H, Mahmoud AB, Speranza MC, Nakashima H, et al. Immune evasion mediated by PD-L1 on glioblastoma-derived extracellular vesicles. Sci Adv. 2018;4(3):eaar2766. doi:10.1126/sciadv.aar2766. [Google Scholar] [PubMed] [CrossRef]
56. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17(1):74. doi:10.1186/s12943-018-0822-0. [Google Scholar] [PubMed] [CrossRef]
57. Huang K, Fang C, Yi K, Liu X, Qi H, Tan Y, et al. The role of PTRF/Cavin1 as a biomarker in both glioma and serum exosomes. Theranostics. 2018;8(6):1540–57. doi:10.7150/thno.22952. [Google Scholar] [PubMed] [CrossRef]
58. Manda SV, Kataria Y, Tatireddy BR, Ramakrishnan B, Ratnam BG, Lath R, et al. Exosomes as a biomarker platform for detecting epidermal growth factor receptor-positive high-grade gliomas. J Neurosurg. 2018;128(4):1091–101. doi:10.3171/2016.11.JNS161187. [Google Scholar] [PubMed] [CrossRef]
59. Ebrahimkhani S, Vafaee F, Hallal S, Wei H, Lee MYT, Young PE, et al. Deep sequencing of circulating exosomal microRNA allows non-invasive glioblastoma diagnosis. npj Precis Oncol. 2018;2(1):28. doi:10.1038/s41698-018-0071-0. [Google Scholar] [PubMed] [CrossRef]
60. Indira Chandran V, Welinder C, Månsson AS, Offer S, Freyhult E, Pernemalm M, et al. Ultrasensitive immunoprofiling of plasma extracellular vesicles identifies syndecan-1 as a potential tool for minimally invasive diagnosis of glioma. Clin Cancer Res. 2019;25(10):3115–27. doi:10.1158/1078-0432.CCR-18-2946. [Google Scholar] [PubMed] [CrossRef]
61. Ricklefs FL, Maire CL, Matschke J, Dührsen L, Sauvigny T, Holz M, et al. FASN is a biomarker enriched in malignant glioma-derived extracellular vesicles. Int J Mol Sci. 2020;21(6):1931. doi:10.3390/ijms21061931. [Google Scholar] [PubMed] [CrossRef]
62. Geng L, Xu J, Zhu Y, Hu X, Liu Y, Yang K, et al. Targeting miR-9 in glioma stem cell-derived extracellular vesicles: a novel diagnostic and therapeutic biomarker. Transl Oncol. 2022;22:101451. doi:10.1016/j.tranon.2022.101451. [Google Scholar] [PubMed] [CrossRef]
63. Dufrusine B, Capone E, Ponziani S, Lattanzio R, Lanuti P, Giansanti F, et al. Extracellular LGALS3BP: a potential disease marker and actionable target for antibody-drug conjugate therapy in glioblastoma. Mol Oncol. 2023;17(8):1460–73. doi:10.1002/mol2.v17.8. [Google Scholar] [CrossRef]
64. Alberti G, Sánchez-López CM, Marcilla A, Barone R, Caruso Bavisotto C, Graziano F, et al. Hsp70 and calcitonin receptor protein in extracellular vesicles from glioblastoma multiforme: biomarkers with putative roles in carcinogenesis and potential for differentiating tumor types. Int J Mol Sci. 2024;25(6):3415. doi:10.3390/ijms25063415. [Google Scholar] [PubMed] [CrossRef]
65. Hallal SM, Tűzesi Á, Sida LA, Xian E, Madani D, Muralidharan K, et al. Glioblastoma biomarkers in urinary extracellular vesicles reveal the potential for a liquid gold biopsy. Br J Cancer. 2024;130(5):836–51. doi:10.1038/s41416-023-02548-9. [Google Scholar] [PubMed] [CrossRef]
66. Rana R, Chauhan K, Gautam P, Kulkarni M, Banarjee R, Chugh P, et al. Plasma-derived extracellular vesicles reveal galectin-3 binding protein as potential biomarker for early detection of glioma. Front Oncol. 2021;11:778754. doi:10.3389/fonc.2021.778754. [Google Scholar] [PubMed] [CrossRef]
67. Jafari A, Karimabadi K, Rahimi A, Rostaminasab G, Khazaei M, Rezakhani L, et al. The emerging role of exosomal miRNAs as biomarkers for early cancer detection: a comprehensive literature review. Technol Cancer Res Treat. 2023;22:15330338231205999. doi:10.1177/15330338231205999. [Google Scholar] [PubMed] [CrossRef]
68. Qian M, Wang S, Guo X, Wang J, Zhang Z, Qiu W, et al. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene. 2020;39(2):428–42. doi:10.1038/s41388-019-0996-y. [Google Scholar] [PubMed] [CrossRef]
69. Dobra G, Gyukity-Sebestyén E, Bukva M, Harmati M, Nagy V, Szabó Z, et al. MMP-9 as prognostic marker for brain tumours: a comparative study on serum-derived small extracellular vesicles. Cancers. 2023;15(3):712. doi:10.3390/cancers15030712. [Google Scholar] [PubMed] [CrossRef]
70. Cumba Garcia LM, Peterson TE, Cepeda MA, Johnson AJ, Parney IF. Isolation and analysis of plasma-derived exosomes in patients with glioma. Front Oncol. 2019;9:651. doi:10.3389/fonc.2019.00651. [Google Scholar] [PubMed] [CrossRef]
71. Saint-Pol J, Gosselet F, Duban-Deweer S, Pottiez G, Karamanos Y. Targeting and crossing the blood-brain barrier with extracellular vesicles. Cells. 2020;9(4):851. doi:10.3390/cells9040851. [Google Scholar] [PubMed] [CrossRef]
72. Hawari FI, Rouhani FN, Cui X, Yu ZX, Buckley C, Kaler M, et al. Release of full-length 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc Natl Acad Sci U S A. 2004;101(5):1297–302. doi:10.1073/pnas.0307981100. [Google Scholar] [PubMed] [CrossRef]
73. Ramos-Zaldívar HM, Polakovicova I, Salas-Huenuleo E, Corvalán AH, Kogan MJ, Yefi CP, et al. Extracellular vesicles through the blood-brain barrier: a review. Fluids Barriers CNS. 2022;19(1):60. doi:10.1186/s12987-022-00359-3. [Google Scholar] [PubMed] [CrossRef]
74. Palviainen M, Saraswat M, Varga Z, Kitka D, Neuvonen M, Puhka M, et al. Extracellular vesicles from human plasma and serum are carriers of extravesicular cargo-Implications for biomarker discovery. PLoS One. 2020;15(8):e0236439. doi:10.1371/journal.pone.0236439. [Google Scholar] [PubMed] [CrossRef]
75. Zhang X, Takeuchi T, Takeda A, Mochizuki H, Nagai Y. Comparison of serum and plasma as a source of blood extracellular vesicles: increased levels of platelet-derived particles in serum extracellular vesicle fractions alter content profiles from plasma extracellular vesicle fractions. PLoS One. 2022;17(6):e0270634. doi:10.1371/journal.pone.0270634. [Google Scholar] [PubMed] [CrossRef]
76. Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4(4):e5219. doi:10.1371/journal.pone.0005219. [Google Scholar] [PubMed] [CrossRef]
77. Rodríguez Zorrilla S, Pérez-Sayans M, Fais S, Logozzi M, Gallas Torreira M, García García A. A pilot clinical study on the prognostic relevance of plasmatic exosomes levels in oral squamous cell carcinoma patients. Cancers. 2019;11(3):429. doi:10.3390/cancers11030429. [Google Scholar] [PubMed] [CrossRef]
78. Bandini E, Rossi T, Scarpi E, Gallerani G, Vannini I, Salvi S, et al. Early detection and investigation of extracellular vesicles biomarkers in breast cancer. Front Mol Biosci. 2021;8:732900. doi:10.3389/fmolb.2021.732900. [Google Scholar] [PubMed] [CrossRef]
79. Hu H, Ling B, Shi Y, Wu H, Zhu B, Meng Y, et al. Plasma exosome-derived SENP1 may be a potential prognostic predictor for melanoma. Front Oncol. 2021;11:685009. doi:10.3389/fonc.2021.685009. [Google Scholar] [PubMed] [CrossRef]
80. Campos-Silva C, Cáceres-Martell Y, Sánchez-Herrero E, Sandúa A, Beneitez-Martínez A, González Á, et al. A simple immunoassay for extracellular vesicle liquid biopsy in microliters of non-processed plasma. J Nanobiotechnol. 2022;20(1):72. doi:10.1186/s12951-022-01256-5. [Google Scholar] [PubMed] [CrossRef]
81. De Sousa KP, Rossi I, Abdullahi M, Ramirez MI, Stratton D, Inal JM. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2023;15(1):e1835. doi:10.1002/wnan.1835. [Google Scholar] [PubMed] [CrossRef]
82. Akbar A, Malekian F, Baghban N, Kodam SP, Ullah M. Methodologies to isolate and purify clinical grade extracellular vesicles for medical applications. Cells. 2022;11(2):186. doi:10.3390/cells11020186. [Google Scholar] [PubMed] [CrossRef]
83. Brennan K, Martin K, FitzGerald SP, O’Sullivan J, Wu Y, Blanco A, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep. 2020;10(1):1039. doi:10.1038/s41598-020-57497-7. [Google Scholar] [PubMed] [CrossRef]
84. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019;8(7):727. doi:10.3390/cells8070727. [Google Scholar] [PubMed] [CrossRef]
85. Moyano A, Serrano-Pertierra E, Duque JM, Ramos V, Teruel-Barandiarán E, Fernández-Sánchez MT, et al. Magnetic lateral flow immunoassay for small extracellular vesicles quantification: application to colorectal cancer biomarker detection. Sensors. 2021;21(11):3756. doi:10.3390/s21113756. [Google Scholar] [PubMed] [CrossRef]
86. Enderle D, Spiel A, Coticchia CM, Berghoff E, Mueller R, Schlumpberger M, et al. Characterization of RNA from exosomes and other extracellular vesicles isolated by a novel spin column-based method. PLoS One. 2015;10(8):e0136133. doi:10.1371/journal.pone.0136133. [Google Scholar] [PubMed] [CrossRef]
87. Balaj L, Atai NA, Chen W, Mu D, Tannous BA, Breakefield XO, et al. Heparin affinity purification of extracellular vesicles. Sci Rep. 2015;5(1):10266. doi:10.1038/srep10266. [Google Scholar] [PubMed] [CrossRef]
88. Paolini L, Zendrini A, Di Noto G, Busatto S, Lottini E, Radeghieri A, et al. Residual matrix from different separation techniques impacts exosome biological activity. Sci Rep. 2016;6(1):23550. doi:10.1038/srep23550. [Google Scholar] [PubMed] [CrossRef]
89. Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, et al. The exosome total isolation chip. ACS Nano. 2017;11(11):10712–23. doi:10.1021/acsnano.7b04878. [Google Scholar] [PubMed] [CrossRef]
90. Cilibrasi C, Simon T, Vintu M, Tolias C, Samuels M, Mazarakis NK, et al. Definition of an inflammatory biomarker signature in plasma-derived extracellular vesicles of glioblastoma patients. Biomedicines. 2022;10(1):125. doi:10.3390/biomedicines10010125. [Google Scholar] [PubMed] [CrossRef]
91. Aydin S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides. 2015;72:4–15. doi:10.1016/j.peptides.2015.04.012. [Google Scholar] [PubMed] [CrossRef]
92. Logozzi M, Di Raimo R, Mizzoni D, Fais S. Immunocapture-based ELISA to characterize and quantify exosomes in both cell culture supernatants and body fluids. Methods Enzymol. 2020;645:155–80. doi:10.1016/bs.mie.2020.06.011. [Google Scholar] [PubMed] [CrossRef]
93. Logozzi M, Mizzoni D, Capasso C, Del Prete S, Di Raimo R, Falchi M, et al. Plasmatic exosomes from prostate cancer patients show increased carbonic anhydrase IX expression and activity and low pH. J Enzyme Inhib Med Chem. 2020;35(1):280–8. doi:10.1080/14756366.2019.1697249. [Google Scholar] [PubMed] [CrossRef]
94. Ma Y, Yoshida T, Matoba K, Kida K, Shintani R, Piao Y, et al. Identification of small compounds regulating the secretion of extracellular vesicles via a TIM4-affinity ELISA. Sci Rep. 2021;11(1):13471. doi:10.1038/s41598-021-92860-2. [Google Scholar] [PubMed] [CrossRef]
95. Boriachek K, Masud MK, Palma C, Phan HP, Yamauchi Y, Hossain MSA, et al. Avoiding pre-isolation step in exosome analysis: direct isolation and sensitive detection of exosomes using gold-loaded nanoporous ferric oxide nanozymes. Anal Chem. 2019;91(6):3827–34. doi:10.1021/acs.analchem.8b03619. [Google Scholar] [PubMed] [CrossRef]
96. Ter-Ovanesyan D, Gilboa T, Budnik B, Nikitina A, Whiteman S, Lazarovits R, et al. Improved isolation of extracellular vesicles by removal of both free proteins and lipoproteins. eLife. 2023;12:e86394. doi:10.7554/eLife.86394. [Google Scholar] [PubMed] [CrossRef]
97. Zhang X, Hsueh MF, Huebner JL, Kraus VB. TNF-α carried by plasma extracellular vesicles predicts knee osteoarthritis progression. Front Immunol. 2021;12:758386. doi:10.3389/fimmu.2021.758386. [Google Scholar] [PubMed] [CrossRef]
98. Logozzi M, Angelini DF, Giuliani A, Mizzoni D, Di Raimo R, Maggi M, et al. Increased plasmatic levels of PSA-expressing exosomes distinguish prostate cancer patients from benign prostatic hyperplasia: a prospective study. Cancers. 2019;11(10):1449. doi:10.3390/cancers11101449. [Google Scholar] [PubMed] [CrossRef]
99. Saadat F, Zareighane Z, Safavifar F, Jalali SZ, Berahmeh A, Khorramizadeh MR. The repression of matrix metalloproteinases and cytokine secretion in glioblastoma by targeting K+ channel. Basic Clin Neurosci. 2021;12(6):737–44. doi:10.32598/bcn. [Google Scholar] [CrossRef]
100. Oliveira-Rodríguez M, López-Cobo S, Reyburn HT, Costa-García A, López-Martín S, Yáñez-Mó M, et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J Extracell Vesicles. 2016;5:31803. doi:10.3402/jev.v5.31803. [Google Scholar] [PubMed] [CrossRef]
101. López-Cobo S, Campos-Silva C, Moyano A, Oliveira-Rodríguez M, Paschen A, Yáñez-ó M, et al. Immunoassays for scarce tumour-antigens in exosomes: detection of the human NKG2D-Ligand, MICA, in tetraspanin-containing nanovesicles from melanoma. J Nanobiotechnology. 2018;16(1):47. doi:10.1186/s12951-018-0372-z. [Google Scholar] [PubMed] [CrossRef]
102. Bayoumy S, Hyytiä H, Leivo J, Talha SM, Huhtinen K, Poutanen M, et al. Glycovariant-based lateral flow immunoassay to detect ovarian cancer-associated serum CA125. Commun Biol. 2020;3(1):460. doi:10.1038/s42003-020-01191-x. [Google Scholar] [PubMed] [CrossRef]
103. Kim HM, Kim J, Bock S, An J, Choi YS, Pham XH, et al. Silver-assembled silica nanoparticles in lateral flow immunoassay for visual inspection of prostate-specific antigen. Sensors. 2021;21(12):4099. doi:10.3390/s21124099. [Google Scholar] [PubMed] [CrossRef]
104. Wu JC, Chuang YH, Wei YC, Hsieh CC, Pong YH, Su YR, et al. Validating METCAM/MUC18 as a novel biomarker to predict the malignant potential of prostate cancer at an early stage by using a modified gold nanoparticles-based lateral flow immunoassay. Diagn Basel Switz. 2021;11(3):443. doi:10.3390/diagnostics11030443. [Google Scholar] [PubMed] [CrossRef]
105. Xia J, Liu Y, Ran M, Lu W, Bi L, Wang Q, et al. The simultaneous detection of the squamous cell carcinoma antigen and cancer antigen 125 in the cervical cancer serum using nano-Ag polydopamine nanospheres in an SERS-based lateral flow immunoassay. RSC Adv. 2020;10(49):29156–70. doi:10.1039/D0RA05207H. [Google Scholar] [PubMed] [CrossRef]
106. Jiao X, Peng T, Liang Z, Hu Y, Meng B, Zhao Y, et al. Lateral flow immunoassay based on time-resolved fluorescence microspheres for rapid and quantitative screening CA199 in human serum. Int J Mol Sci. 2022;23(17):9991. doi:10.3390/ijms23179991. [Google Scholar] [PubMed] [CrossRef]
107. Mut M, Adiguzel Z, Cakir-Aktas C, Hanalioğlu Ş, Gungor-Topcu G, Kiyga E, et al. Extracellular-vesicle-based cancer panels diagnose glioblastomas with high sensitivity and specificity. Cancers. 2023;15(15):3782. doi:10.3390/cancers15153782. [Google Scholar] [PubMed] [CrossRef]
108. Jalali M, Del Real Mata C, Montermini L, Jeanne O, Hosseini I, Gu I, et al. MoS2-plasmonic nanocavities for Raman spectra of single extracellular vesicles reveal molecular progression in glioblastoma. ACS Nano. 2023;17(13):12052–71. doi:10.1021/acsnano.2c09222. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF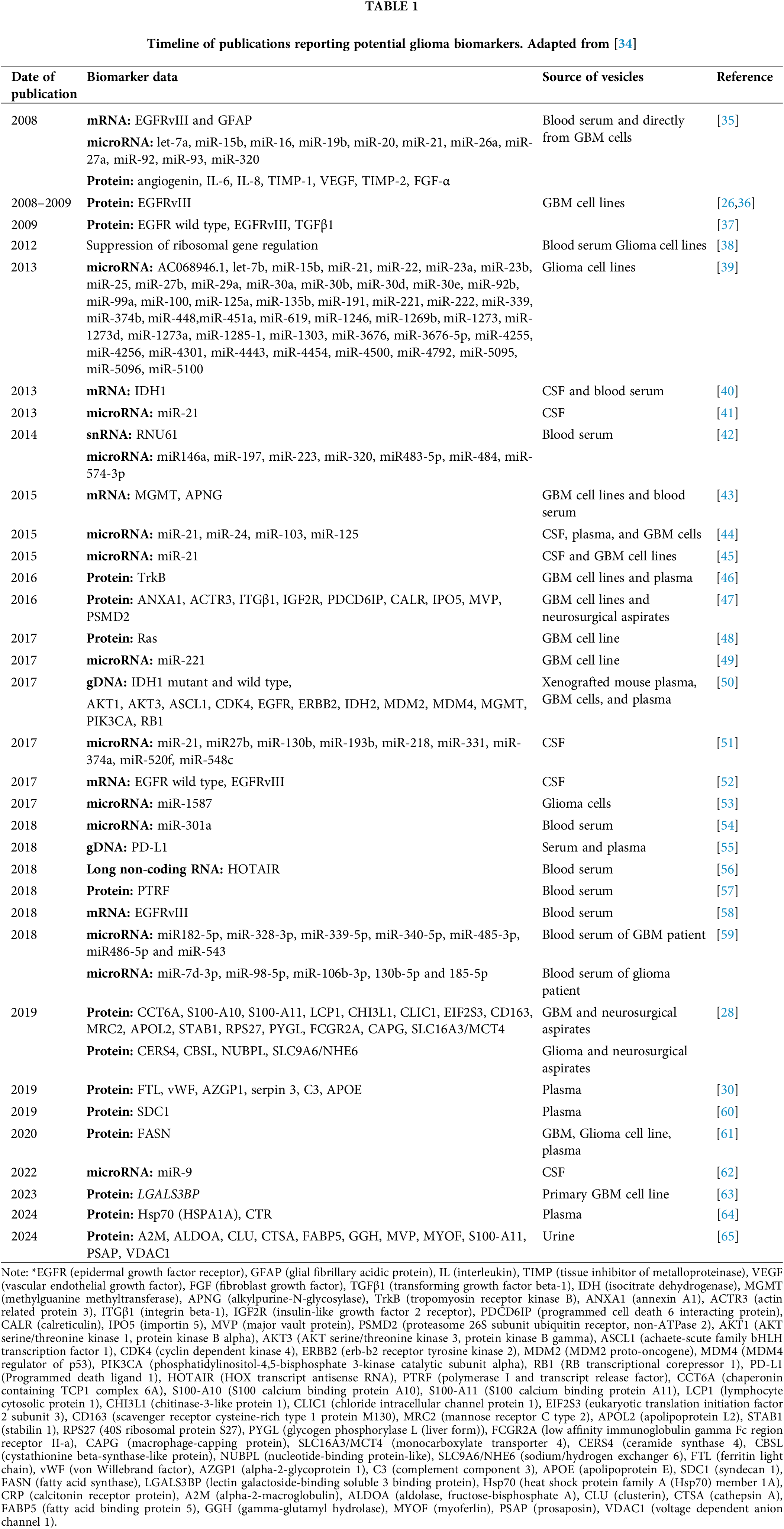
 Downloads
Downloads
 Citation Tools
Citation Tools