 Open Access
Open Access
ARTICLE
Epigenetic regulation of ABCG2 promoter methylation in adolescents with hyperuricemia
1 Department of Nephrology and Rheumatology, Yantian District People’s Hospital, Shenzhen, 518000, China
2 Department of Urology, the Fifth Affiliated Hospital of Zhengzhou University, Zhengzhou, 450000, China
3 Department of Biology, Chemistry, Pharmacy, Free University of Berlin, Berlin, 14195, Germany
* Corresponding Authors: CHEN LI. Email: ; ZHIXIAN PAN. Email:
BIOCELL 2024, 48(12), 1805-1813. https://doi.org/10.32604/biocell.2024.056431
Received 23 July 2024; Accepted 02 October 2024; Issue published 30 December 2024
Abstract
Background: Hyperuricemia is a metabolic disorder which is characterized by increased serum uric acid levels, which can contribute to serious health issues such as gout, cardiovascular disease, and kidney damage. Epigenetic modifications, for example, DNA methylation, exert a crucial function in gene regulation and have been implicated in various metabolic disorders. The ATP-Binding Cassette Subfamily G Member 2 (ABCG2) gene is involved in uric acid excretion, and its expression can be influenced by methylation of its promoter region. Methods: This study involved the design of three guide RNA (gRNA) sequences targeting specific CpG sites within the ABCG2 promoter region. Using the Clustered Regularly Interspaced Short Palindromic Repeats/dead Cas9-Ten-Eleven Translocation 1 (CRISPR/dCas9-TET1) system, these gRNAs were employed to guide targeted demethylation of the ABCG2 promoter in cell models. A non-targeting gRNA served as a negative control. The methylation status of the ABCG2 promoter and its effect on gene expression were assessed using bisulfite sequencing and qRT-PCR. Results: Among the gRNAs tested, gRNA2 and gRNA3 effectively guided the dCas9-TET1 complex to the ABCG2 promoter, resulting in significant demethylation. gRNA2 showed the most pronounced effect, leading to a substantial increase in ABCG2 expression. Clinical data analysis revealed that adolescents with hyperuricemia had higher uric acid levels compared to healthy controls, and a higher proportion of the hyperuricemia group reported a high-protein diet, suggesting a link between diet and ABCG2 methylation. Conclusion: The findings demonstrate that targeted demethylation of the ABCG2 promoter can significantly upregulate its expression, which may help modulate uric acid levels. These results indicate that dietary factors, such as a high-protein diet, could influence ABCG2 methylation and thus impact hyperuricemia. Advanced research is necessary to explore the therapeutic potential of aiming at epigenetic modifications for the treatment of hyperuricemia.Keywords
Abbreviation
| ABCG2 | ATP-Binding Cassette Subfamily G Member 2 |
| CRISPR/dCas9-TET1 | Clustered Regularly Interspaced Short Palindromic Repeats/dead Cas9-Ten-Eleven Translocation 1 |
| gRNA | guide RNA |
| RRBS | Reduced Representation Bisulfite Sequencing |
| ALT | Alanine Aminotransferase |
| AST | Aspartate Aminotransferase |
| Cr | Creatinine |
| FPG | Fasting Plasma Glucose |
| TC | Total Cholesterol |
| TG | Triglycerides |
| DMEM | Dulbecco’s Modified Eagle Medium |
| FBS | Fetal Bovine Serum |
| ELISA | Enzyme-Linked Immunosorbent Assay |
| SD | Standard Deviation |
| SPSS | Statistical Package for the Social Sciences |
| E2F1 | E2F Transcription Factor 1 |
| DNMT3A | DNA Methyltransferase 3 Alpha |
| ACTB | Actin Beta |
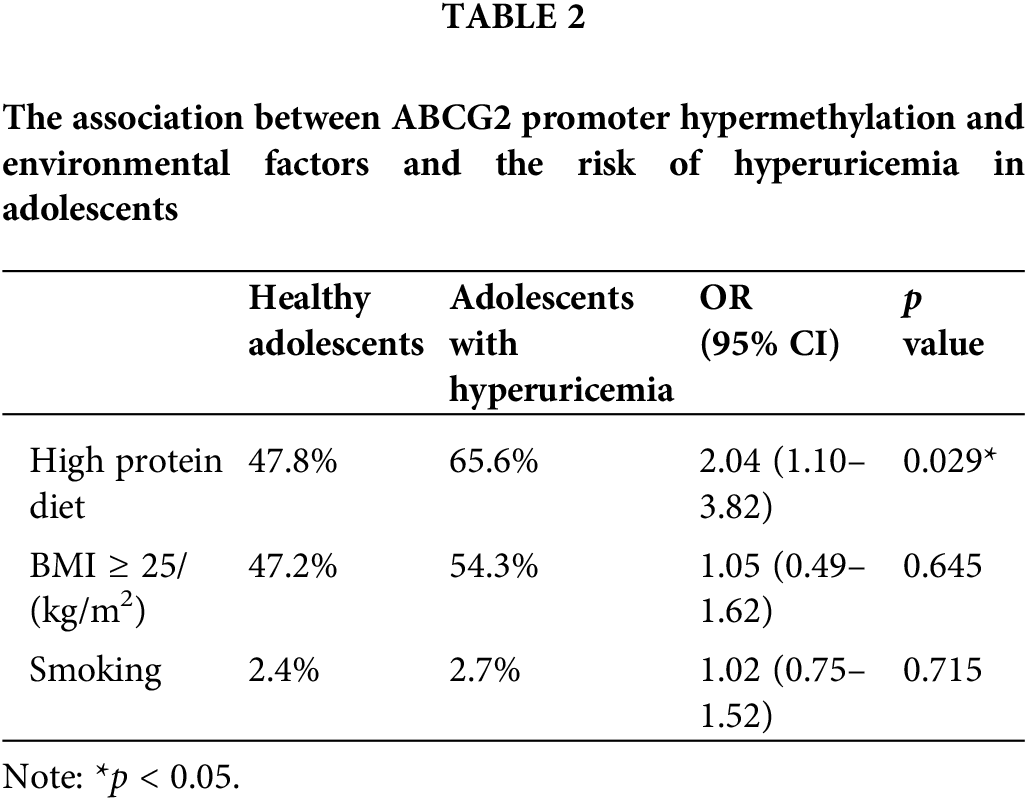
Hyperuricemia is a metabolic disorder characterized by increased levels of uric acid in the blood, resulting from abnormalities in purine metabolism [1]. This condition is increasingly being recognized as a significant health concern due to its association with severe complications such as cardiovascular, cerebrovascular, and renal diseases [2]. These comorbidities substantially contribute to morbidity and mortality, highlighting the urgency of addressing hyperuricemia’s underlying mechanisms and potential interventions [3]. Epidemiological studies have shown that the prevalence of hyperuricemia in mainland China is approximately 13.3%. Alarmingly, this condition is becoming more prevalent among adolescents aged 10 to 18 years [4,5]. This rising trend in adolescent hyperuricemia warrants comprehensive research to understand its pathogenesis and relationship with environmental and lifestyle elements such as smoking, alcohol consumption, high-protein diets, and obesity, which have all been linked to the development of hyperuricemia [6].
The ATP-binding cassette subfamily G member 2 (ABCG2) gene has emerged as a critical player in uric acid transport. Identified through multiple genome-wide association studies (GWAS) as a gene significantly associated with uric acid levels, ABCG2 is primarily expressed on the apical membrane of proximal tubule cells in the kidneys, facilitating uric acid excretion [7]. Dysfunction or reduced expression of ABCG2 has been linked to the decrease of uric acid clearance, leading to high serum uric acid levels and the subsequent development of hyperuricemia and gout [8]. Therefore, understanding the regulation of ABCG2 expression and function is vital for developing therapeutic strategies for hyperuricemia.
A crucial role in regulating gene expression is played by DNA methylation without altering the fundamental DNA sequence [9]. This modification consists of increasing a methyl group to the cytosine base within CpG dinucleotides, which often results in transcriptional repression [10]. Methylation predominantly occurs in CpG islands, which are regions rich in CG dinucleotides located near gene promoters. Approximately 60% of human genes have CpG islands in their promoters, and methylation in these regions can significantly influence gene expression [11]. In the context of disease, aberrant DNA methylation patterns have been identified as crucial factors in the pathogenic mechanism of various conditions, including cancers, diabetes, and hypertension [12]. Given the critical role of DNA methylation in gene regulation, we selected specific CpG sites within the ABCG2 promoter region for analysis. These sites were chosen because they are located near known transcription factor binding sites, and earlier studies have indicated that the methylation status of these sites is closely associated with the regulation of ABCG2 gene expression [13]. According to these specific sites, we aim to uncover how changes in methylation affect ABCG2 expression and function, particularly in the pathophysiology of hyperuricemia. Recently, advancements in CRISPR/Cas9 technology have made precise epigenetic modifications possible, offering a novel approach to studying disease mechanisms and developing therapeutic strategies [14].
This study aims to explore the methylation levels of the ABCG2 promoter in adolescents with hyperuricemia and to illustrate the relationship between ABCG2 promoter methylation and uric acid levels. We hypothesize that hypermethylation of the ABCG2 promoter in adolescents with hyperuricemia leads to reduced expression of the ABCG2 transporter, resulting in impaired excretion of uric acid and an elevated level of serum uric acid. This research will utilize reduced representation bisulfite sequencing (RRBS) to assess methylation at specific CpG sites within the ABCG2 promoter and correlate these methylation patterns with clinical data. To test this hypothesis, we will collect whole blood samples from adolescents with hyperuricemia. The CRISPR/dCas9 system, specifically the dead Cas9 (dCas9) variant fused with TET1, a ten-eleven translocation methylcytosine dioxygenase, was used to target and modify methylation at specific sites within the ABCG2 promoter. This approach will enable us to assess the functional consequences of altered methylation on ABCG2 expression and uric acid levels in cell models of hyperuricemia. The expression levels of ABCG2 following methylation modification will be analyzed using quantitative real-time PCR (RT-qPCR), and uric acid levels will be measured using specific assay kits. Bioinformatics tools will predict potential interactions involving ABCG2, thereby exploring the broader impact of ABCG2 methylation on cellular function.
Via investigating the epigenetic regulation of ABCG2 in adolescents with hyperuricemia, this study aims to provide new insights into the mechanisms driving this condition and to determine potential biomarkers for early diagnosis and targets for therapeutic intervention. Our findings could pave the way for novel strategies to prevent and treat hyperuricemia in the pediatric population, ultimately improving health outcomes and quality of life for affected individuals.
Sample collection and DNA extraction
We obtained whole blood samples from two groups: 35 healthy adolescents and 35 adolescents diagnosed with hyperuricemia. The subjects were selected depending on strict inclusion and exclusion criteria to ensure homogeneity within the groups. All participants were aged between 10 to 18 years, consistent with the World Health Organization’s definition of adolescence. The research was performed in line with ethical guidelines and approved by the Ethics Committee of Yantian District People’s Hospital with approval number 2023-135. All participants or their guardians gave their informed consent. Genomic DNA was extracted from whole blood samples using the QIAamp DNA Blood Mini Kit (Qiagen, 51106, Hilden, Germany) according to the manufacturer’s instructions. The concentration and purity of the extracted DNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, ND-2000, Waltham, MA, USA). We stored DNA samples at −80°C until further analysis.
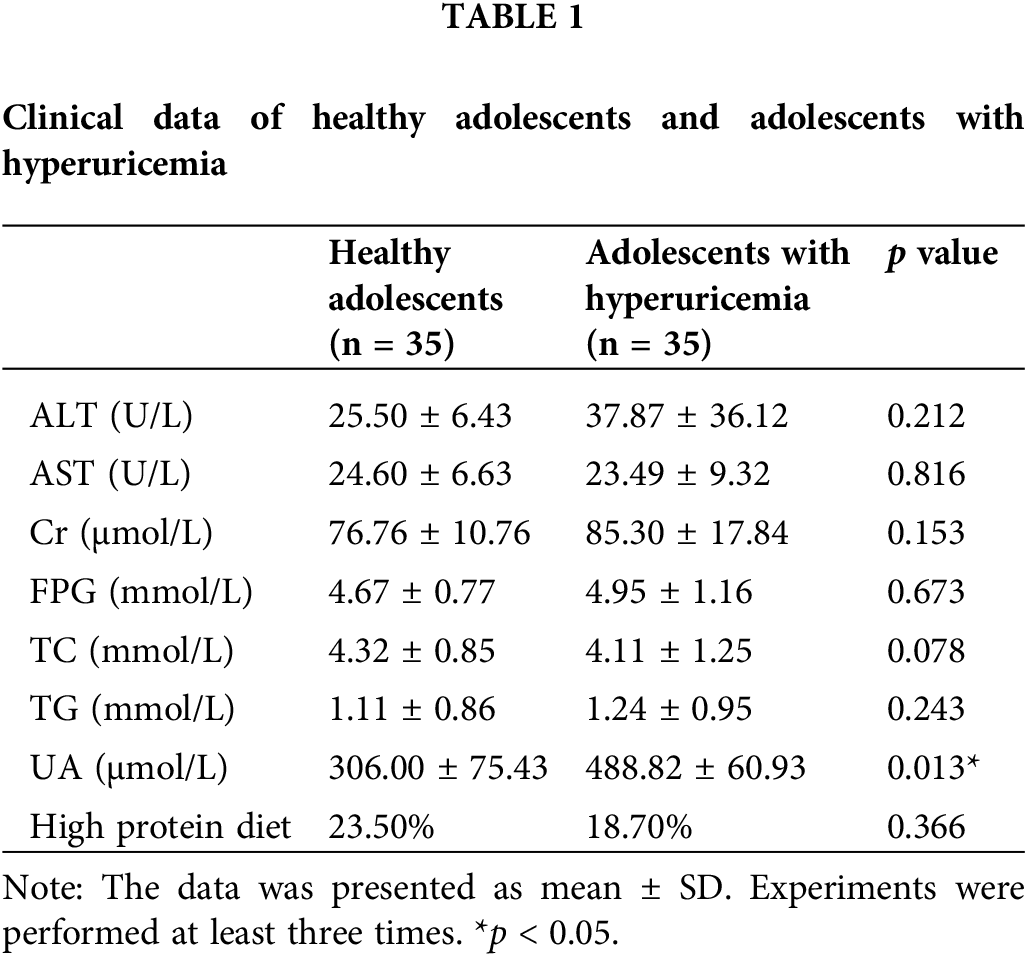
Clinical data including serum uric acid levels, alanine aminotransferase (ALT), aspartate aminotransferase (AST), creatinine (Cr), fasting plasma glucose (FPG), total cholesterol (TC), triglycerides (TG), and dietary habits were collected. High-protein diet was defined as consuming dairy products on a daily basis, more than two servings of beans or eggs per week, and a daily intake of meat, fish, or poultry. Statistical analysis of these data was performed to contrast the differences between the adolescents with hyperuricemia group and the healthy control group.
Reduced representation bisulfite sequencing (RRBS)
Reduced Representation Bisulfite Sequencing (RRBS) was utilized to analyze methylation patterns at CpG-rich regions, particularly targeting CpG islands within the ABCG2 promoter. This technique enables high-resolution mapping of DNA methylation at single-nucleotide levels. Genomic DNA was digested with the MspI restriction enzyme (New England Biolabs, R0106S, Ipswich, MA, USA), which recognizes the CCGG sequence and cleaves at sites rich in CpG dinucleotides. Digested DNA fragments were then end-repaired, adenylated at the 3′ ends, and ligated to methylated sequencing adapters. Bisulfite treatment was employed by using the EZ DNA Methylation-Gold Kit (Zymo Research, D5006, Irvine, CA, USA) to convert unmethylated cytosines to uracils while leaving 5-methylcytosines unaffected. The treated DNA was then expanded using PCR to enrich for CpG-containing regions. The bisulfite-converted DNA libraries were quantified using a Qubit 4 Fluorometer (Thermo Scientific, Q33226, Waltham, MA, USA) and assessed for quality using an Agilent 2100 Bioanalyzer (Agilent Technologies, G2939BA, Santa Clara, CA, USA). Methylation levels at individual CpG sites within the ABCG2 promoter were quantified and compared between the adolescents with hyperuricemia and healthy control groups. Statistical analysis was utilized to identify differentially methylated regions. The correlation between ABCG2 promoter methylation and clinical/environmental factors was also analyzed.
CRISPR/dCas9 system for DNA methylation editing
The CRISPR/dCas9 system was utilized to target and modify the methylation status of the ABCG2 promoter. We employed a dead Cas9 (dCas9) variant fused with the TET1 enzyme to achieve targeted demethylation [15]. Guide RNA (gRNA) sequences targeting the methylated sites within the ABCG2 promoter were designed using the CRISPR-ERA web tool [16]. Three gRNA sequences (gRNA1: TGCAGTGGCACGGTCTTGGG, gRNA2: CCCAAAGTGCTGGGATTACA, gRNA3: CCAGGCTGGTCTTGAACTCC) were synthesized and tested for their efficiency in guiding the dCas9-TET1 complex to the target sites. The dCas9-TET1 fusion construct was cloned into a plasmid vector under the regulation of a Cytomegalovirus (CMV) promoter for high expression level (Addgene plasmid #84472). The gRNAs were promoted by the U6 promoter.
AML12 (mouse liver cells) and HK-2 (human kidney cells) cells were cultured in DMEM/F12 (Gibco, 11320033, Grand Island, NY, USA) and DMEM medium (Gibco, 11995065, Grand Island, NY, USA), respectively, added with 10% fetal bovine serum (FBS, Gibco, 10099141, Grand Island, NY, USA), 1% penicillin-streptomycin (Thermo Fisher Scientific, 15140122, Waltham, MA, USA), and sustained at 37°C in a 5% CO2 incubator. Mycoplasma contamination was routinely monitored using a MycoAlert Mycoplasma Detection Kit (Lonza, LT07-318, Rockland, ME, USA) to ensure that the cells remained free of contamination throughout the experiments. Cells were co-transfected with dCas9-TET1 and gRNA plasmids using Lipofectamine 2000 (Thermo Scientific, 11668019, Waltham, MA, USA) according to the manufacturer’s instructions. Cells were obtained after transfection and genomic DNA was derived. The efficiency of methylation editing was assessed by bisulfite PCR.
AML12 and HK-2 cells were dealt with 1 mM adenosine (Sigma-Aldrich, A4036, St. Louis, MO, USA) and 1 mM xanthine (Sigma-Aldrich, X7375, St. Louis, MO, USA) for 24 h [17]. Control cells were treated with vehicle only. The treatment induced intracellular uric acid accumulation, mimicking hyperuricemia. Intracellular uric acid levels were evaluated by a Uric Acid Assay Kit (Abcam, ab65344, Cambridge, UK). Cell lysates were prepared, and uric acid concentrations were determined colorimetrically following the protocol of the manufacturer. Absorbance was measured at 570 nm using a Synergy H1 Microplate Reader (BioTek, Winooski, VT, USA).
Total RNA was extracted from treated cells using the TRIzol reagent (Invitrogen, 15596026, Carlsbad, CA, USA). RNA concentration and purity were measured via a NanoDrop 2000 spectrophotometer (Thermo Scientific, ND-2000, Waltham, MA, USA). cDNA was manufactured from 1 µg of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad, 1708891, Hercules, CA, USA). RT-qPCR was carried out using the PowerUp SYBR Green PCR Master Mix (Applied Biosystems, A25742, Foster City, CA, USA) on a StepOnePlus Real-Time PCR System (Applied Biosystems, 4376600, Foster City, CA, USA). Primers for ABCG2 and housekeeping genes (GAPDH and β-actin) were designed using Primer3 software (version 4.1.0). The sequences for ABCG2 were: forward primer 5′-TTTCCAAGCATCCAGGTGTATC-3′ and reverse primer 5′-GGATTGCTTCACTGGAACAGC-3′. For GAPDH, the sequences were: forward primer 5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse primer 5′-GAAGATGGTGATGGGATTTC-3′. For β-actin, the sequences were: forward primer 5′- AGAGCTACGAGCTGCCTGAC-3′ and reverse primer 5′-AGC ACTGTGTTGGCGTACAG-3′. These primers, all of which are derived from the human species, were used to quantify and detect the expression of the ABCG2 gene and the housekeeping genes GAPDH and β-actin. Relative gene expression levels were calculated using the 2−ΔΔCt method.
Uric acid levels in cell culture supernatants were measured using the Uric Acid Assay Kit (Abcam, ab65344, Cambridge, UK). The assay involved enzymatic conversion of uric acid to allantoin and hydrogen peroxide, with the latter reacting with a probe to produce a colorimetric signal. Absorbance was read at 570 nm. Uric acid concentrations were normalized to cell number, and comparisons were made between treated and control groups.
Bioinformatics and protein interaction analysis
Bioinformatics tools such as STRING (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string-db.org) (accessed on 01 October 2024) [18] and BioGRID (Biological General Repository for Interaction Datasets, https://thebiogrid.org) (accessed on 01 October 2024) [19] were used to predict potential protein interactions with ABCG2. Genes and proteins with high interaction scores were selected for further study. ELISA kits (R&D Systems, DNMT3A: DY5219, ACTB: DY2100, ABCG2: DY995, E2F1: DY4124, Minneapolis, MN, USA) were used to levels of the protein levels of DNMT3A, ACTB, ABCG2, and E2F1 in HK-2 cell lysates. We lysed the cells and measured the protein concentrations depending on the protocols of the manufacturer. We read absorbance at 450 nm and normalized protein levels to total protein content.
All experimental data from three independent experiments were presented as mean ± standard deviation (SD). All statistical data were analyzed via SPSS 22.0 software (SPSS Inc., Chicago, IL, USA). To analyze data we used an independent samples t-test. A two-tailed value of p < 0.05 was regarded as statistically meaningful.
A total of 35 healthy adolescents and 35 adolescents with hyperuricemia were recruited for this study. Analysis of clinical data between the two groups revealed significant differences in uric acid levels, with the hyperuricemia group displaying notably higher levels by comparison with the healthy control group (Table 1, p < 0.05). Other parameters such as ALT, AST, Cr, FPG, TC, and TG showed no significant disparities between the groups. Notably, the proportion of participants adhering to a high-protein diet was significantly higher in the hyperuricemia group, suggesting a potential link between high-protein diets and the methylation of the ABCG2 promoter (Table 2, p < 0.05).
Methylation analysis of ABCG2 promoter
The reduced representation bisulfite sequencing (RRBS) method was utilized to evaluate the methylation status of the ABCG2 promoter in both healthy adolescents and adolescents with hyperuricemia. The analysis identified four methylation sites on the ABCG2 promoter. Among these, the methylation level of Site 1 was conspicuously elevated in the hyperuricemia group by comparison with the healthy group (p < 0.05), while no meaningful differences were observed for Sites 2, 3, and 4 (Fig. 1A–D). The overall methylation level of the ABCG2 promoter was also conspicuously higher in the hyperuricemia group (p < 0.05), indicating a potential role of hypermethylation in the pathogenesis of adolescent hyperuricemia (Fig. 1E).
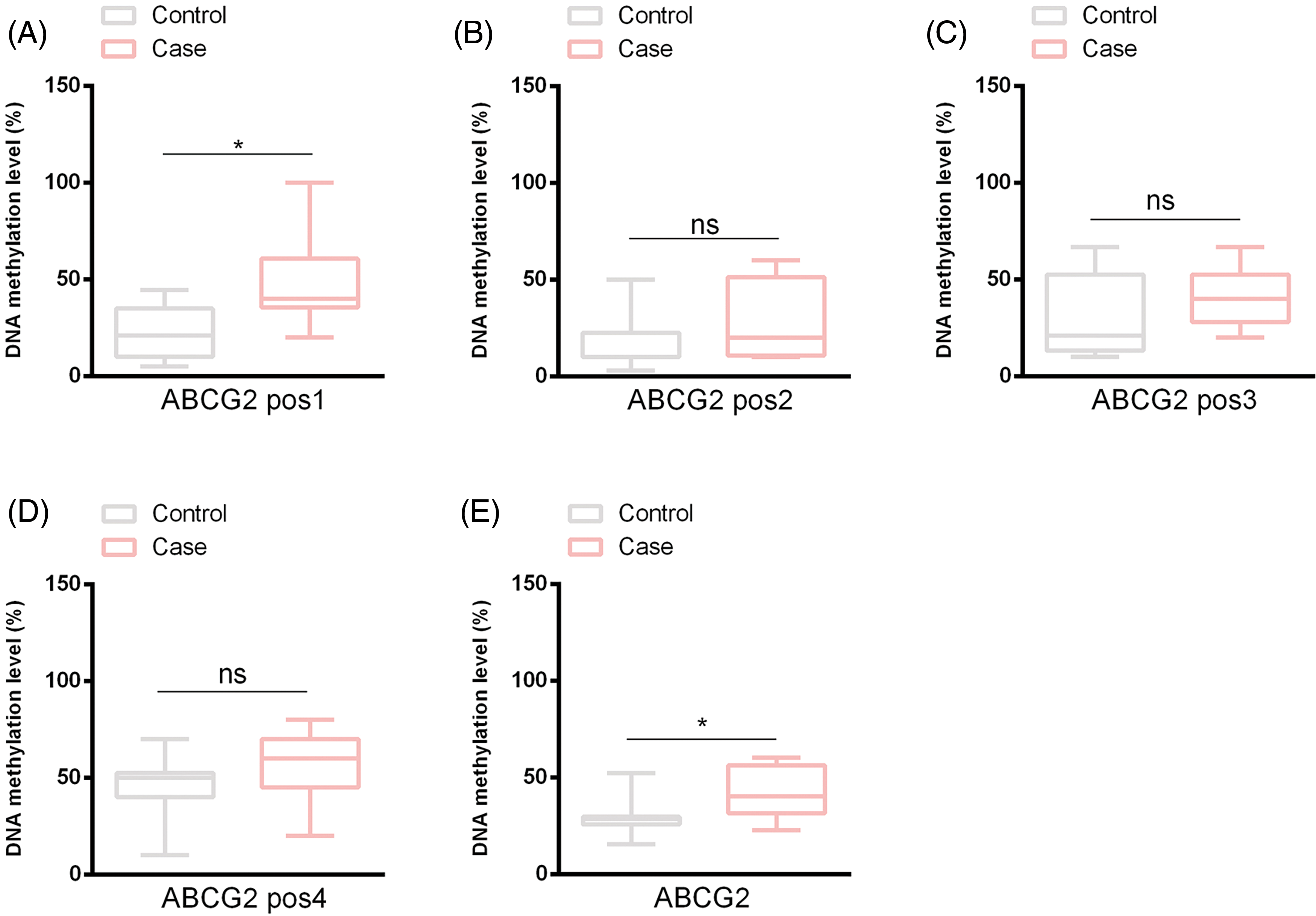
Figure 1: ABCG2 promoter methylation levels were measured in control and hyperuricemia groups. This diagram illustrates the DNA methylation levels at four specific positions (pos1, pos2, pos3, and pos4) within the ATP-Binding Cassette Subfamily G Member 2 (ABCG2) promoter, as well as the overall methylation levels, in both control and hyperuricemia groups. (A) shows the methylation at Position 1, (B) shows the methylation at Position 2, (C) shows the methylation at Position 3, (D) shows the methylation at Position 4, and (E) shows the overall methylation levels. ns indicates non-significant differences between the compared groups. All data are indicated as mean ± Standard Deviation (SD) (*p < 0.05).
CRISPR/dCas9-TET1 mediated demethylation
To explore the functional effects of ABCG2 promoter methylation, we designed three gRNA sequences targeting methylation site 1 of the ABCG2 promoter. A non-targeting gRNA was employed as a negative control for ensuring that the observed effects were specific to the targeting of the ABCG2 promoter (Fig. 2). Among these, gRNA2 and gRNA3 effectively guided the dCas9-TET1 complex to the ABCG2 promoter, resulting in significant demethylation (Fig. 3A). gRNA2 showed the most pronounced effect and was selected for further studies. In the AML12 cell model, demethylation of the ABCG2 promoter via the gRNA2/dCas9-TET1 system led to a remarkable increase in ABCG2 expression and a concomitant decrease in uric acid levels (p < 0.05) (Fig. 3B,C). Similar outcomes were witnessed in the HK-2 cell model, where demethylation also resulted in elevated ABCG2 expression and reduced uric acid levels (p < 0.05) (Fig. 3D,E).
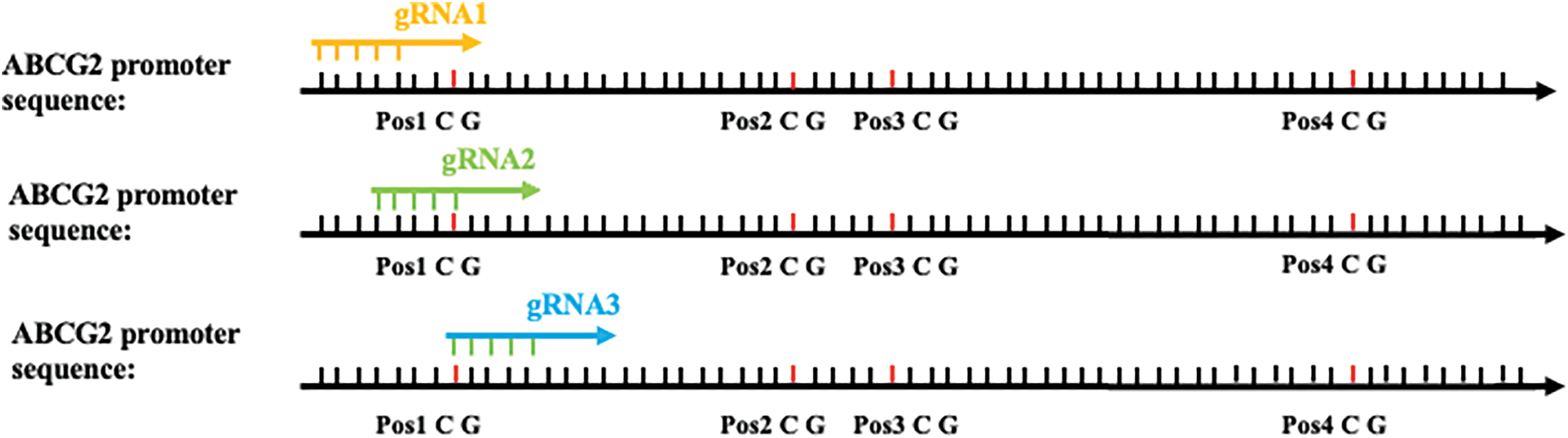
Figure 2: The promoter sequence of the ABCG2 gene and the specific sites targeted in this study. The highlighted areas indicate CpG sites, and arrows mark the binding sites for gRNA1, gRNA2, and gRNA3. These sites were selected for their proximity to known transcription factor binding sites and their potential role in regulating ABCG2 expression through methylation.
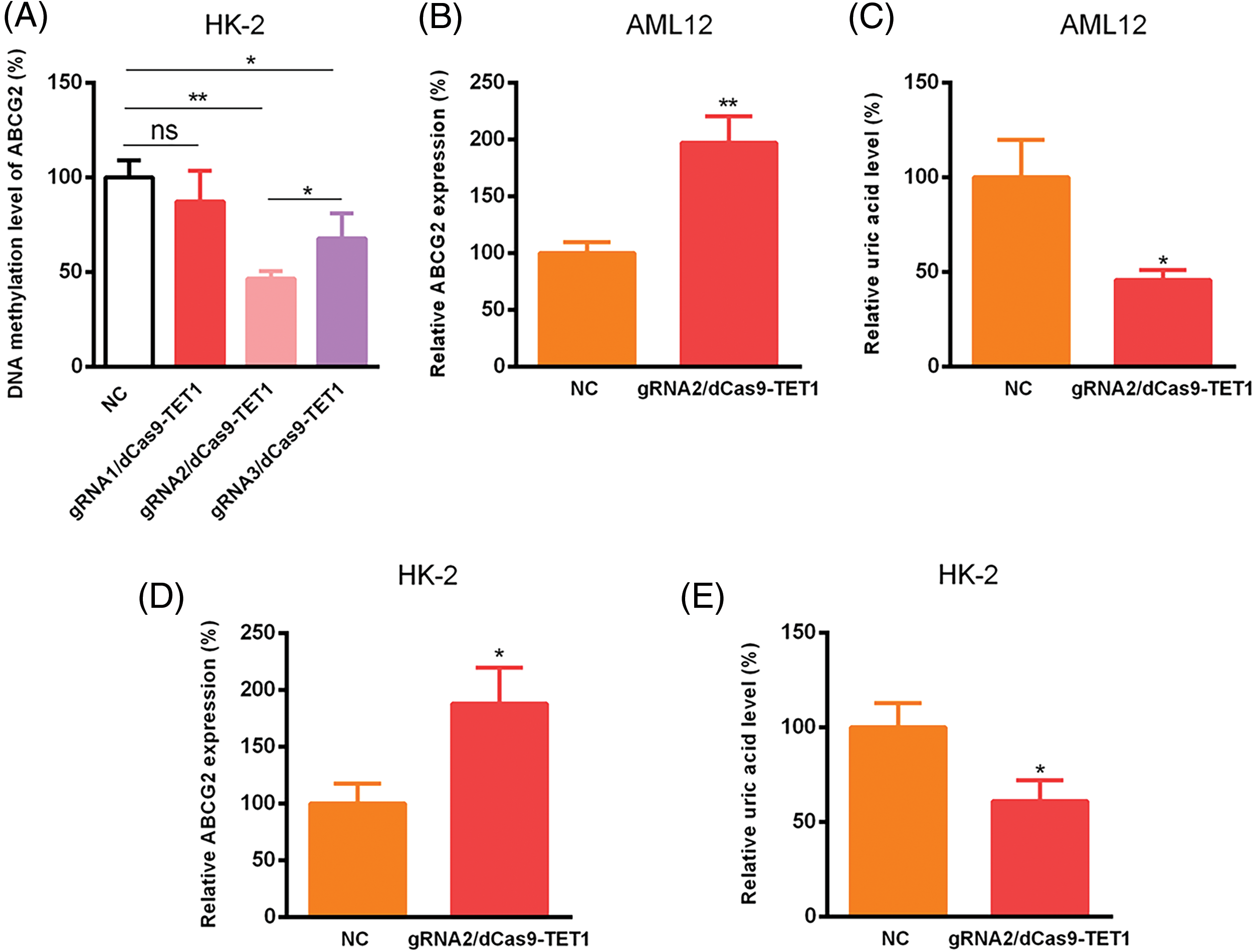
Figure 3: Relative ABCG2 and uric acid levels expression was detected utilizing CRISPR/dCas9-TET1. (A) DNA methylation of ABCG2 was tested using CRISPR/dCas9-TET1, with a non-targeting gRNA (nc) used as a negative control. Relative ABCG2 expression was presented in AML12 (B) and HK-2 cells (D). Relative uric acid levels were shown in AML12 (C) and HK-2 cells (E). ns indicates non-significant differences between the compared groups. All data are expressed as mean ± SD (*p < 0.05, **p < 0.01).
Bioinformatics analysis using STRING and BioGRID databases predicted several genes and proteins interacting with ABCG2. Based on the scoring and relevant literature, we selected DNMT3A, ACTB, and E2F1 for further investigation. ELISA assays demonstrated that in the high-uric acid HK-2 cell model, ABCG2 expression was significantly reduced while DNMT3A, ACTB, and E2F1 levels were markedly increased (p < 0.05). Following the demethylation of the ABCG2 promoter, expression levels of ABCG2, DNMT3A, ACTB, and E2F1 all significantly increased (p < 0.05) (Fig. 4A,B). Previous studies have identified DNMT3A as a crucial factor in gene promoter methylation and E2F1 as an activating transcription factor promotes the expression of DNMT3A via binding with the promoter of DNMT3A expression [20]. Lots of studies demonstrated that DNMT3A promotes hypermethylation in various diseases [21]. Our results suggest that E2F1 may enhance DNMT3A expression by binding to its promoter, thereby contributing to the hypermethylation of the ABCG2 promoter in adolescents with hyperuricemia.
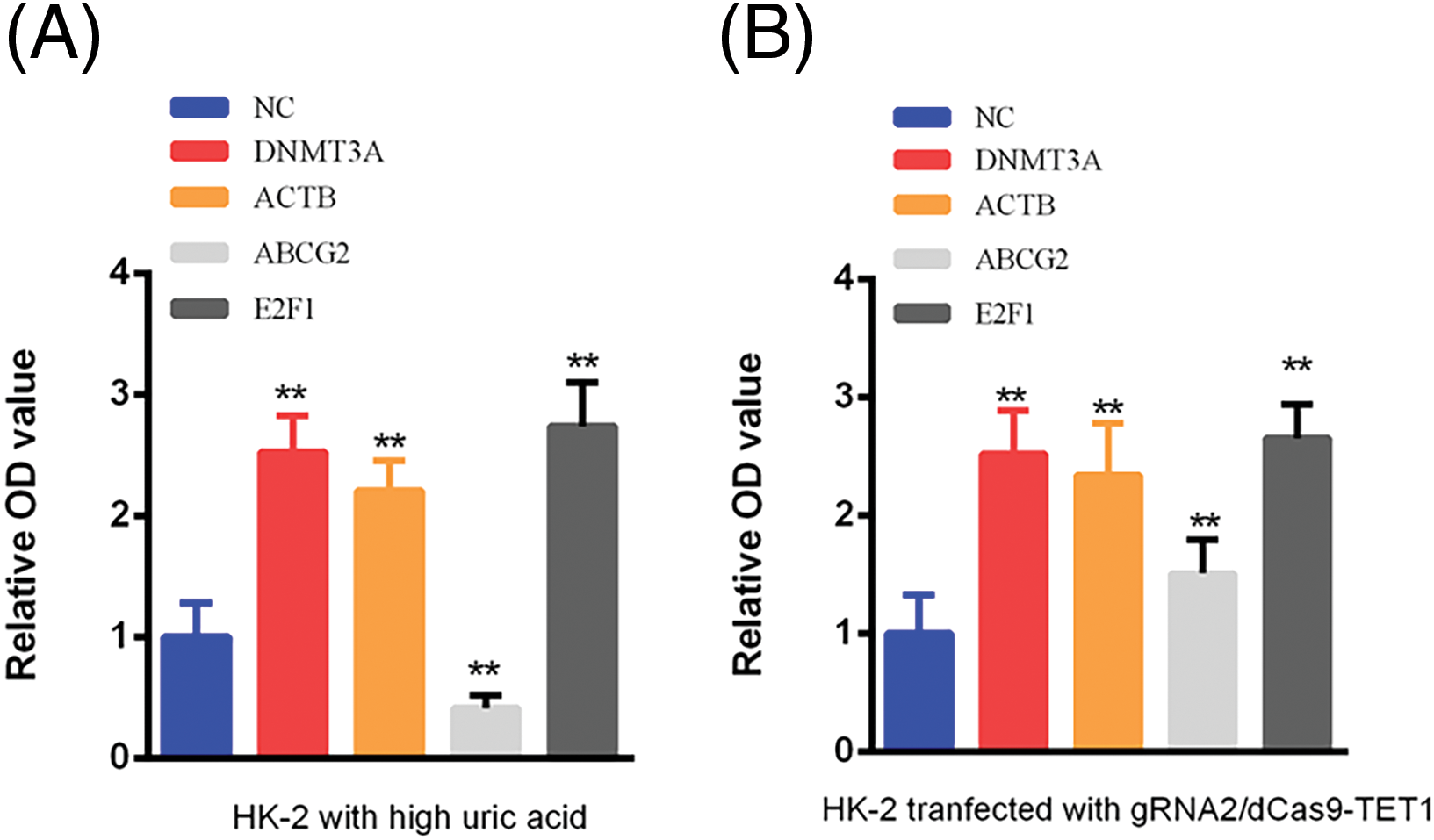
Figure 4: Expression of DNMT3A, ACTB, ABCG2, and E2F1 in Control and Hyperuricemia Groups Measured by ELISA. (A) The expression of DNMT3A, ACTB, ABCG2 and E2F1 was detected in HK-2 with hyperuricemia. (B) The expression of DNMT3A, ACTB, ABCG2 and E2F1 was tested in HK-2 transfected with gRNA2/dCas9-TET1. NC represents the negative control group, which refers to samples that did not receive specific treatment, serving as a baseline reference. All data are shown as mean ± SD (**p < 0.01).
The ABCG2 transporter, situated on the apical membrane of the human proximal tubule [22], appears to be essential for urate excretion in this region. Research has demonstrated that single nucleotide polymorphisms (SNPs) like p.Q141K are connected to conditions such as hyperuricemia and/or gout [23]. This particular mutation leads to a reduction in transport efficiency [24]. Although there has been substantial research on hyperuricemia in adults, studies focusing on this condition in young people are still lacking [25]. Therefore, our aim was to explore the prevalence of hyperuricemia among youth in China and to identify the related risk factors. Unlike SNP, DNA methylation plays a essential role in the regulation of gene expression without altering the underlying DNA sequence [26]. The studies about the relation between DNA methylation and hyperuricemia are still few [27]. The present study explores the relationship between the methylation levels of the ABCG2 promoter and uric acid levels in adolescents with hyperuricemia. Our results illuminate the potential epigenetic mechanisms underlying the regulation of uric acid transport and the development of hyperuricemia in this population. Our analysis of clinical data revealed that adolescents with hyperuricemia exhibited significantly higher uric acid levels compared to healthy controls, while other clinical parameters, such as ALT, AST, Cr, FPG, TC, and TG, showed no statistical differences. Notably, the proportion of a high-protein diet was significantly higher in the hyperuricemia group, suggesting that dietary habits might influence ABCG2 methylation and subsequent uric acid levels by increasing purine intake and uric acid production. This aligns with previous studies highlighting the role of environmental factors, such as diet, in modulating gene expression through epigenetic modifications [28]. The RRBS method identified four methylation sites within the ABCG2 promoter. Among these, Site 1 showed significantly higher methylation levels in the hyperuricemia group compared to controls, while Sites 2, 3, and 4 did not exhibit notable differences. The overall methylation level of the ABCG2 promoter was significantly elevated in adolescents with hyperuricemia. This increased methylation likely contributes to the suppression of ABCG2 expression, impairing uric acid excretion and leading to elevated blood uric acid levels. Our results are consistent with previous findings that DNA methylation often inhibits gene expression, thereby affecting.
The use of the CRISPR/dCas9-TET1 system to target and demethylate the ABCG2 promoter demonstrated that gRNA2 was the most effective in reducing methylation at Site 1. This reduction in methylation corresponded with a significant increase in ABCG2 expression and a concomitant decrease in uric acid levels in both AML12 and HK-2 cell models. This indicates that the methylation status of the ABCG2 promoter directly affects its expression and function. These findings highlight the specific biological significance of the chosen CRISPR-Cas9 target sites within the ABCG2 promoter. By targeting these key CpG sites (gRNA1: TGCAGTGGCACGGTCTTGGG, gRNA2: CCCAAAGTGCTGGGATTACA, gRNA3: CCAGGCTGGTCTTGAACTCC), we were able to effectively modulate the expression of the ABCG2 gene. The methylation status at these sites likely influences the binding of transcription factors, thereby altering gene expression levels. This aligns with findings from other studies utilizing CRISPR-Cas9 technology for targeted epigenetic modifications, demonstrating the potential of this technology for precise gene regulation and correction of aberrant epigenetic marks [14,28]. Moreover, such targeted demethylation strategies may offer novel therapeutic avenues for managing hyperuricemia by restoring the function of key uric acid transporters.
Our study demonstrates that increased methylation of the ABCG2 promoter is associated with reduced gene expression and elevated uric acid levels in adolescents with hyperuricemia. This finding underscores the critical role of DNA methylation in regulating ABCG2 expression. However, the epigenetic regulation of ABCG2 is not limited to DNA methylation but also involves other mechanisms such as histone modifications and non-coding RNAs. Studies have shown that histone acetylation and methylation can alter chromatin structure, influencing the accessibility of transcription factors to the ABCG2 promoter and thus modulating gene transcription [29]. Additionally, microRNAs and long non-coding RNAs play significant roles in the regulation of ABCG2 expression by binding to mRNA, affecting its stability and translation [30,31]. These multi-layered epigenetic regulatory mechanisms collectively control ABCG2 gene expression, highlighting the complexity of its regulation. This complex regulatory network suggests that future therapeutic approaches for hyperuricemia may need to combine multiple epigenetic interventions to effectively restore normal ABCG2 function and manage uric acid levels.
Further analysis using ELISA revealed that in hyperuricemic HK-2 cells, the expression levels of DNMT3A, ACTB, and E2F1 were significantly increased, while ABCG2 expression was markedly reduced. Following the inhibition of ABCG2 promoter methylation, the expression of DNMT3A, ACTB, and E2F1 significantly increased, alongside an increase in ABCG2 expression. Studies have illustrated that DNMT3A regulates the expression of specific genes by methylating their promoter regions, thereby affecting cell differentiation and development [32]. Besides, the activating transcription factor E2F1 promoted DNMT3A expression through combining with its promoter [33]. These results suggest a regulatory feedback loop where DNMT3A-mediated methylation of the ABCG2 promoter is influenced by transcription factors such as E2F1. The elevated expression of DNMT3A and E2F1 in hyperuricemic cells implies their involvement in the epigenetic regulation of ABCG2.
Our study draws attention to the significance of epigenetic modifications in the regulation of genes involved in uric acid metabolism. The correlation between high-protein diets and increased ABCG2 methylation suggests that lifestyle and dietary interventions could play a crucial role in managing hyperuricemia. Further research is warranted to explore the broader epigenetic landscape influencing uric acid levels and to identify additional environmental factors that may impact these epigenetic changes. The successful utilization of the CRISPR/dCas9-TET1 system highlights the potential for epigenome editing as a therapeutic strategy. Future studies should aim to refine these techniques and assess their long-term efficacy and safety in vivo. Additionally, exploring the interactions between DNMT3A, E2F1, and other transcription factors may provide deeper insights into the complex regulatory networks governing gene expression in hyperuricemia. While our study provides valuable insights, it has several limitations. The sample size was relatively small, and larger cohorts are needed to validate our findings. Additionally, the study focused on specific methylation sites within the ABCG2 promoter, and a more comprehensive analysis of the entire promoter region and other regulatory elements could reveal additional epigenetic modifications. Finally, while in vitro models provide controlled environments for studying gene expression, in vivo studies are necessary to confirm the physiological relevance of our findings.
In conclusion, our study demonstrates that increased methylation of the ABCG2 promoter is associated with reduced gene expression and elevated uric acid levels in adolescents with hyperuricemia. These findings emphasize the critical role of epigenetic regulation in uric acid metabolism and suggest potential therapeutic targets for managing hyperuricemia. Advanced study is needed to broaden our horizons of the epigenetic mechanisms involved and to develop effective strategies for early diagnosis and treatment of hyperuricemia in adolescents.
Acknowledgement: None.
Funding Statement: This research was supported by the Science and Technology Plan Program of Yantian District of Shenzhen (YTWS20200208). Project funding department is Shenzhen Yantian District Science and Technology Innovation Bureau. The authors thank all the donors whose names were not included in the author list, but who participated in this program.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zhixian Pan; data collection: Xueting Huang; analysis and interpretation of results: Chaojie Xu and Chen Li; draft manuscript preparation: Xueting Huang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: This study involves human data and was conducted in accordance with ethical guidelines. The study protocol was approved by the Ethics Committee of Yantian District People’s Hospital, with approval number 2023-135. Informed consent was obtained from all participants or their guardians.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Pai HL, Lin DP, Chang HH. Current updates for hyperuricemia and gout in age-related macular degeneration. FASEB J. 2024;38(10):e23676. doi:10.1096/fsb2.v38.10. [Google Scholar] [CrossRef]
2. Brikman S, Serfaty L, Abuhasira R, Schlesinger N, Bieber A, Rappoport N. A machine learning-based prediction model for gout in hyperuricemics: a nationwide cohort study. Rheumatology. 2024;63(9):2411–7. doi:10.1093/rheumatology/keae273. [Google Scholar] [CrossRef]
3. Skare TL, Freire de Carvalho J. Gout, hyperuricemia, and ear function: a systematic review. Isr Med Assoc J. 2024;26(5):318–23. [Google Scholar]
4. Liu R, Han C, Wu D, Xia X, Gu J, Guan H, et al. Prevalence of hyperuricemia and gout in mainland China from 2000 to 2014: a systematic review and meta-analysis. Biomed Res Int. 2015;2015:762820. [Google Scholar]
5. Zhang M, Zhu X, Wu J, Huang Z, Zhao Z, Zhang X, et al. Prevalence of hyperuricemia among Chinese adults: findings from two nationally representative cross-sectional surveys in 2015–16 and 2018–19. Front Immunol. 2021;12:791983. [Google Scholar]
6. MacFarlane LA, Kim SC. Gout: a review of nonmodifiable and modifiable risk factors. Rheum Dis Clin North Am. 2014;40(4):581–604. doi:10.1016/j.rdc.2014.07.002. [Google Scholar] [CrossRef]
7. Liu J, Yang W, Li Y, Wei Z, Dan X. ABCG2 rs2231142 variant in hyperuricemia is modified by SLC2A9 and SLC22A12 polymorphisms and cardiovascular risk factors in an elderly community-dwelling population. BMC Med Genet. 2020;21(1):54. doi:10.1186/s12881-020-0987-4. [Google Scholar] [CrossRef]
8. Ristic B, Sikder MOF, Bhutia YD, Ganapathy V. Pharmacologic inducers of the uric acid exporter ABCG2 as potential drugs for treatment of gouty arthritis. Asian J Pharm Sci. 2020;15(2):173–80. doi:10.1016/j.ajps.2019.10.002. [Google Scholar] [CrossRef]
9. Bogdanović O, Lister R. DNA methylation and the preservation of cell identity. Curr Opin Genet Dev. 2017;46:9–14. doi:10.1016/j.gde.2017.06.007. [Google Scholar] [CrossRef]
10. Tang L, Ye H, Hong Q, Wang L, Wang Q, Wang H, et al. Elevated CpG island methylation of GCK gene predicts the risk of type 2 diabetes in Chinese males. Gene. 2014;547(2):329–33. doi:10.1016/j.gene.2014.06.062. [Google Scholar] [CrossRef]
11. Zhu H, Zhu H, Tian M, Wang D, He J, Xu T. DNA methylation and hydroxymethylation in cervical cancer: diagnosis, prognosis and treatment. Front Genet. 2020;11:347. doi:10.3389/fgene.2020.00347. [Google Scholar] [CrossRef]
12. Shen J, Su J, Chu X, Yu X, Gao X. DNMT3A-mediated DNA methylation and transcription inhibition of FZD5 suppresses lung carcinogenesis. Heliyon. 2024;10(9):e29733. doi:10.1016/j.heliyon.2024.e29733. [Google Scholar] [CrossRef]
13. To KK, Zhan Z, Bates SE. Aberrant promoter methylation of the ABCG2 gene in renal carcinoma. Mol Cell Biol. 2006;26(22):8572–85. doi:10.1128/MCB.00650-06. [Google Scholar] [CrossRef]
14. Goell JH, Hilton IB. CRISPR/Cas-based epigenome editing: advances, applications, and clinical utility. Trends Biotechnol. 2021;39(7):678–91. doi:10.1016/j.tibtech.2020.10.012. [Google Scholar] [CrossRef]
15. Zhang W, Wang H, Chen S, Fan X, Liu Y, Shi S, et al. Reactivation of methylation-silenced PAX1 inhibits cervical cancer proliferation and migration via the WNT/TIMELESS pathway. Mol Carcinog. 2024;63(7):1349–61. doi:10.1002/mc.v63.7. [Google Scholar] [CrossRef]
16. Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. CRISPR-ERA: a comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015;31(22):3676–8. doi:10.1093/bioinformatics/btv423. [Google Scholar] [CrossRef]
17. Gao T, Xu J, Xiao Y, Li J, Hu W, Su X, et al. Therapeutic effects and mechanisms of N-(9,10-anthraquinone-2-ylcarbonyl) xanthine oxidase inhibitors on hyperuricemia. Front Pharmacol. 2022;13:950699. doi:10.3389/fphar.2022.950699. [Google Scholar] [CrossRef]
18. Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638–46. doi:10.1093/nar/gkac1000. [Google Scholar] [CrossRef]
19. Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30(1):187–200. doi:10.1002/pro.v30.1. [Google Scholar] [CrossRef]
20. Al-Kharashi LA, Bakheet T, AlHarbi WA, Al-Moghrabi N, Aboussekhra A. Eugenol modulates genomic methylation and inactivates breast cancer-associated fibroblasts through E2F1-dependent downregulation of DNMT1/DNMT3A. Mol Carcinog. 2021;60(11):784–95. doi:10.1002/mc.v60.11. [Google Scholar] [CrossRef]
21. Bhargavan B, Chhunchha B, Kubo E, Singh DP. DNA methylation as an epigenetic mechanism in the regulation of LEDGF expression and biological response in aging and oxidative stress. Cell Death Discov. 2024;10(1):296. doi:10.1038/s41420-024-02076-2. [Google Scholar] [CrossRef]
22. Dehghan A, Köttgen A, Yang Q, Hwang SJ, Kao WL, Rivadeneira F, et al. Association of three genetic loci with uric acid concentration and risk of gout: a genome-wide association study. Lancet. 2008;372(9654):1953–61. doi:10.1016/S0140-6736(08)61343-4. [Google Scholar] [CrossRef]
23. Yamagishi K, Tanigawa T, Kitamura A, Köttgen A, Folsom AR, Iso H. The rs2231142 variant of the ABCG2 gene is associated with uric acid levels and gout among Japanese people. Rheumatology. 2010;49(8):1461–5. doi:10.1093/rheumatology/keq096. [Google Scholar] [CrossRef]
24. Woodward OM, Köttgen A, Coresh J, Boerwinkle E, Guggino WB, Köttgen M. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A. 2009;106(25):10338–42. doi:10.1073/pnas.0901249106. [Google Scholar] [CrossRef]
25. Vourdoumpa A, Paltoglou G, Mertzanian A, Sertedaki A, Sakou II, Karanasios S, et al. Challenges in the management of patients with HNF1B MODY and multisystem manifestations: the cases of two adolescent boys. Hormones. 2024;23(3):439–45. doi:10.1007/s42000-024-00580-9. [Google Scholar] [CrossRef]
26. Jeddi F, Faghfuri E, Mehranfar S, Soozangar N. The common bisulfite-conversion-based techniques to analyze DNA methylation in human cancers. Cancer Cell Int. 2024;24(1):240. doi:10.1186/s12935-024-03405-2. [Google Scholar] [CrossRef]
27. Li L, Zhang Y, Zeng C. Update on the epidemiology, genetics, and therapeutic options of hyperuricemia. Am J Transl Res. 2020;12(7):3167–81. [Google Scholar]
28. McCutcheon SR, Rohm D, Iglesias N, Gersbach CA. Epigenome editing technologies for discovery and medicine. Nat Biotechnol. 2024;42(8):1199–217. doi:10.1038/s41587-024-02320-1. [Google Scholar] [CrossRef]
29. Neganova ME, Klochkov SG, Aleksandrova YR, Aliev G. Histone modifications in epigenetic regulation of cancer: perspectives and achieved progress. Semin Cancer Biol. 2022;83:452–71. doi:10.1016/j.semcancer.2020.07.015. [Google Scholar] [CrossRef]
30. Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75(6):1374–9. doi:10.1124/mol.108.054163. [Google Scholar] [CrossRef]
31. Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017;8(38):64638–50. doi:10.18632/oncotarget.v8i38. [Google Scholar] [CrossRef]
32. Cuautle DG, Donna S, Cieri MB, Villarreal A, Ramos AJ. Pathological remodeling of reactive astrocytes: involvement of DNA methylation and downregulation of homeostatic genes. J Neurochem. 2024;168(9):2935–55. [Google Scholar]
33. Song H, Zhang Y, Liu J, Liu W, Luo B. Activation of DNA methyltransferase 3a by Epstein-Barr nuclear antigen 1 in gastric carcinoma. Dig Liver Dis. 2022;54(7):973–83. doi:10.1016/j.dld.2021.06.004. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools