 Open Access
Open Access
REVIEW
Unraveling the RAGE axis in pulmonary disorders: Mechanisms and therapeutical potential
1 Key Laboratory of Drug-Targeting and Drug Delivery System of the Education Ministry and Sichuan Province, Sichuan Engineering Laboratory for Plant-Sourced Drug and Sichuan Research Center for Drug Precision Industrial Technology, West China School of Pharmacy, Sichuan University, Chengdu, 610041, China
2 National Clinical Research Center for Respiratory Disease, State Key Laboratory of Respiratory Disease, Guangzhou Institute of Respiratory Health, The First Affiliated Hospital of Guangzhou Medical University, Guangzhou, 510120, China
* Corresponding Authors: TAO JIA. Email: ; ZIFENG YANG. Email:
BIOCELL 2024, 48(12), 1721-1734. https://doi.org/10.32604/biocell.2024.055753
Received 06 July 2024; Accepted 12 September 2024; Issue published 30 December 2024
Abstract
The Receptor for Advanced Glycation End Products (RAGE) is a multiligand receptor of the immunoglobulin superfamily, notably highly expressed in the lungs. Its interaction with a variety of ligands, including advanced glycation end products (AGEs), S100 proteins, and high mobility group box 1 (HMGB1), activates multiple signaling pathways that are pivotal in the pathogenesis of numerous pulmonary diseases and comorbidities. However, comprehensive reviews on the role of ligands-RAGE signaling in specific lung diseases are rare. This review aims to elucidate the mechanisms by which RAGE-mediated signaling pathways either provide protective or pathogenic effects in pulmonary diseases, focusing on its key regulatory roles in asthma, chronic obstructive pulmonary disease (COPD), acute respiratory distress syndrome (ARDS), pulmonary fibrosis (PF), Lung Cancer and COVID-19 Pneumonia, as well as to discuss its therapeutic potential in the specific context of lung diseases.Keywords
Abbreviation
| RAGE | Receptor for Advanced Glycation End Products |
| AGEs | Advanced glycation end products |
| COPD | Chronic obstructive pulmonary disease |
| ARDS | Acute respiratory distress syndrome |
| PF | Pulmonary fibrosis |
| HMGB1 | High mobility group box 1 |
| esRAGE | Endogenous secretory RAGE |
| sRAGE | Soluble RAGE |
| CML | Nε-Carboxymethyl-Lysine |
| CEL | Nε-(carboxyethyl)lysine |
| MG-H1 | Methylglyoxal-derived hydroimidazolone-1 |
| ADAM10 | A disintegrin and metalloproteinase 10 |
| MMP9 | Matrix metalloprotein 9 |
| MMPs | Matrix metalloproteinases |
| AGE-R | AGE-receptor complex |
| ICAM-1 | Intercellular cell adhesion molecule-1 |
| NF-κB | Recombinant Nuclear Factor Kappa B |
| JNK | c-Jun N-terminal kinase |
| Rac1 | Ras-related C3 botulinum toxin substrate 1 |
| BMM model | Bone Marrow Macrophage model |
| TLR4 | Toll-like receptor 4 |
| ERK | Extracellular regulated protein kinases |
| MAPK | Mitogen-activated protein kinase |
| Jak | Janus Kinase |
| ROS | Reactive oxygen species |
| Cdc42 | Cell Division Cycle Protein 42 |
| DIAPH1 | Diaphanous-related formin 1 |
| TIRAP | Toll/interleukin-1 receptor domain-containing adapter protein |
| P38 | p38 mitogen-activated protein kinase |
| P13K | Phosphatidylinositol 3-kinase |
| CRP | C-reactive protein |
| IL-6 | Interleukin 6 |
| Stat3 | Signal transducer and activator of transcription3 |
| MCP-1 | Monocyte Chemotactic Protein-1 |
| VACM-1 | Vascular Cell Adhesion Molecule-1 |
| PAMPs | Pathogen-associated molecular patterns |
| DAMPs | Damage-associated molecular patterns |
| TSLP | Thymic stromal lymphopoietin |
| FEV1 | Forced Expiratory Volume in 1 second |
| NSCLC | Non-small cell lung cancer |
| NOX-4 | NADPH oxidase 4 |
| AKT/PKB | Protein Kinase B |
| KRAS | Kirsten rat sarcoma viral oncogene |
| FGFR | Fibroblast growth factor receptor |
| LSCC | Lung squamous cell carcinoma |
| TGF-β1 | Transforming growth factor-β1 |
| PDGF | Platelet-derived growth factor |
| AQP-5 | Aquaporin-5 |
The Receptor for Advanced Glycation End Products (RAGE) is a multiligand cell surface receptor belonging to the immunoglobulin superfamily. The discovery of RAGE was initially made by Neeper with his colleagues in 1992 during their investigation into the atherosclerotic process, where they demonstrated its ligand-binding function [1]. RAGE is a multiligand cell surface protein belonging to the immunoglobulin superfamily, which plays a pivotal role in inflammatory, and both innate and adaptive immune responses. RAGE is capable of binding a wide range of ligands, including advanced glycosylation end products (AGEs), S100 proteins, inflammatory mediators, and DNA and RNA fragments. The role of this process is crucial in various physiological activities, including immune response, tissue homeostasis, and recovery and regeneration after damage in organisms [2], and it is also linked to many diseases associated with aging and inflammation, such as diabetes, tumors, and bone-related diseases [3,4]. RAGE serves as a promising biomarker for diagnosing, and evaluating prognosis and therapeutic responses. In the last decade, many RAGE inhibitors have been developed, and some of them have entered the evaluation phase in the clinic [5]. RAGE inhibitors show high therapeutical potentiality, including anti-inflammatory, and anti-tumor effects, modulation of neurotoxic substances, and immunomodulatory actions [2].
The aberrant ligands-RAGE signaling has been associated with many diseases, in particular, vasculopathy due to diabetes, neurodegenerative diseases, and tumorigenesis [6,7]. Recent studies emphasized that RAGE is expressed at the highest level in lung tissues and plays an important role in the formation and progression of various lung diseases, including chronic obstructive pulmonary disease, pulmonary fibrosis, acute lung injury, and lung cancer [8]. However, systematic examination of the role of RAGE receptors and their mediated signaling in lung disease onset, development, and disease progression has rarely been reported. This paper focuses on deciphering RAGE-mediated signaling in lung-related diseases and defining therapeutical potential. First, we highlight the progress in studying RAGE receptor expression and function in both lung physiological and pathological contexts. Second, we discuss the mechanisms of the RAGE signaling pathway in different lung diseases, particularly its key regulatory roles in the inflammatory response, fibrosis, and tumor formation with progression. Finally, we discuss the current status and prospects of research on RAGE receptors as potential therapeutic targets for lung diseases. An in-depth understanding of the role of RAGE receptors and their signaling pathways in lung diseases will aid in developing new diagnostic and therapeutic strategies.
RAGE structure, function and distribution in lungs
The RAGE structure consists of a V-type structural domain located outside the N-terminal cell, two C-type structural domains, a transmembrane structural domain, and a C-terminal cytoplasmic tail. These structural domains interact with numerous proteins related to signaling from outside to the cellular nucleus, likely transcriptional regulation. The N-terminal V-type structural domain is regarded as the ligand attachment point, while the C-terminal cytoplasmic structural domain plays an irreplaceable role in signaling [9].
Two types of extracellular secretory RAGE have been identified, which are endogenous secretory RAGE (esRAGE) and soluble RAGE (sRAGE) [10]. esRAGE is RAGE mRNA generated by indicated RNA splicing regulation, while sRAGE is a product produced by releasing extracellular tissues after protein cleavage of full-length RAGE by the disintegrin and metalloproteinase 10 (ADAM10) and matrix metalloprotein 9 (MMP9) [11]. Current studies have emphasized that secreted RAGE has multiple biological functions, such as signaling, transcriptional regulation, translation initiation, and immunosuppressive effects. Although sRAGE do not possess the properties of transmembrane regions and cytoplasmic structural domains, they retain the ability to bind to ligands and thus can function as decoy receptors that function as competitive inhibitors, thereby preventing ligand binding to RAGE or other receptors [12]. In addition, the substance has been reported to have the ability to regulate directly apoptosis and related gene expression. Collectively, sRAGE does not solely act as a RAGE decoy receptor; it also initiates signaling and triggers an inflammatory response [13].
RAGE is highly expressed in a variety of tissues during embryonic development, and its levels decrease significantly in most tissues upon reaching adulthood, except notably in the lungs [14]. Immunohistochemical studies reveal that RAGE expression remains significantly elevated in lung tissues compared to other organs [15,16]. This elevated expression is particularly pronounced in types I and II alveolar epithelial cells, alveolar macrophages, bronchial epithelial cells, and endothelial cells, particularly accentuated in type I alveolar epithelial cells [17]. Several studies highlight RAGE’s pivotal role in regulating the proliferation of type I alveolar epithelial cells, which is essential for maintaining alveolar function and stability. Additionally, RAGE mediates the adhesion dynamics between these cells and basement membranes [18]. Consequently, RAGE is recognized as a critical factor influencing the initiation and progression of various lung-related diseases, such as lung cancer and pulmonary fibrosis.
RAGE is a multi-ligand receptor
RAGE can bind to various ligands, including advanced glycosylation end products (AGEs), S100 proteins [19], inflammatory mediators, Amyloid β, and specific DNA or RNA fragments (Table 1) [10,20–22].
AGEs are a collective term for a group of compounds generated from amino acid residues of proteins and lipids through non-enzymatic glycosylation and autoxidation [23]. AGEs are resistant to degradation by proteasomes and lysosomes, leading to their penetration and accumulation in tissues and triggering a series of inflammatory response pathways. The precise mechanism of receptor interactions in regulating this pathway has not been fully clarified; however, studies suggest that AGEs are highly involved in the regulation of various inflammatory cells for the expression of cytokines, as well as the regulation of immune function. The interaction between AGEs and RAGE has been shown to induce the formation of matrix metalloproteinases (MMPs) and activate multiple signaling pathways, thereby promoting rapid inflammatory responses and tumor progression [24]. Besides RAGE, other receptors, such as the AGE-receptor complex (AGE-R), play crucial roles in the clearance of AGEs, contributing significantly to cellular homeostasis and functional integrity across different tissues and organs [25].
High mobility group box-1 (HMGB1) is a chromatin-binding protein that plays a vital role in nucleosome stability, DNA replication, transcription, and repair. HMGB1 can translocate from the nucleus to the extracellular space, where it mediates various biological functions impacting numerous signaling pathways involved in tumor formation and progression. HMGB1 can bind to RAGE, activating pathways such as intercellular cell adhesion molecule-1 (ICAM-1), Nuclear Factor Kappa B (NF-κB), c-Jun N-terminal kinase (JNK), and ras-related C3 botulinum toxin substrate 1 (Rac1), which enhance tumor growth, infiltration, and metastasis by promoting cell adhesion and tumor vascularization [26]. Additionally, HMGB1 can activate extracellular matrix degradation mechanisms like matrix metalloproteinase MMP-2, accelerating tumor metastasis. HMGB1 also serves as an immunomodulator, inducing immune responses against different antigens or antibodies [27]. In the Bone Marrow Macrophage (BMM) model, researchers typically employ macrophages isolated from murine or other experimental animal bone marrow to investigate immune responses, inflammatory mechanisms, and cell signaling pathways [28]. Within this framework, Toll-like receptor4 (TLR4) is central to initial signaling events (e.g., extracellular regulated protein kinases (ERK) and NF-κB activation) of HMGB1, while RAGE regulates subsequent response events. HMGB1 is expressed at high levels in various tumor tissues and has significant roles in both malignant and non-malignant tumors, such as lung cancer. Furthermore, HMGB1 can induce polarized co-distribution of RAGE and β2 integrins in neutrophils, playing a key role in neutrophil recruitment associated with inflammation [29].
The S100 family consists of small Ca2+-binding proteins with over 20 different members, each exhibiting specific distribution and function across various tissues. The binding of RAGE and S100 protein was initially demonstrated through in vitro cellular immunoprecipitation assays [30]. While numerous studies have explored the regulation and signaling mechanisms of these proteins, systematic studies on interactions between different isoforms and RAGE are very limited. S100A8/A9 binds to the V and C1 domains of RAGE, protecting the VC1 region from proteolytic cleavage and continuously activating downstream pathways such as mitogen-activated protein kinase (MAPK), NF-κB, and Janus Kinase (JAKs). This activation triggers inflammatory responses that promote tumor growth, invasiveness, and metastasis [31]. Additionally, S100A8/A9 exert immunomodulatory effects by inducing apoptosis or inhibiting the cell cycle. It stimulates the release of soluble RAGE (sRAGE) and dampens RAGE’s canonical signaling in tumor dissemination [32].
RAGE-initiated signaling pathway
Upon binding to its ligands, the RAGE receptor activates multiple signaling pathways critical for cell proliferation, apoptosis, inflammatory responses, and metabolic regulation. RAGE activation of the NF-κB signaling pathway leads to increased expression of inflammatory factors. NF-κB activation promotes cell proliferation and inhibits apoptosis, contributing to tumor growth [35]. Additionally, RAGE regulates cell proliferation and differentiation via the MAPK signaling pathway, which includes JNK and p38 signaling nodes (Fig. 1). Activation of these pathways can lead to cell growth, survival, and inflammatory responses [36]; RAGE can activate the Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway, which plays an important role in cell proliferation, differentiation, and immune responses (Fig. 1). The activation of the JAK/STAT pathway is closely related to tumor cell growth and metastasis [37]. Furthermore, RAGE-mediated signaling can increase the generation of intracellular reactive oxygen species (ROS), leading to oxidative stress (Fig. 1). Oxidative stress is implicated not only in cell damage and apoptosis but also in the development of chronic diseases and cancer [38].
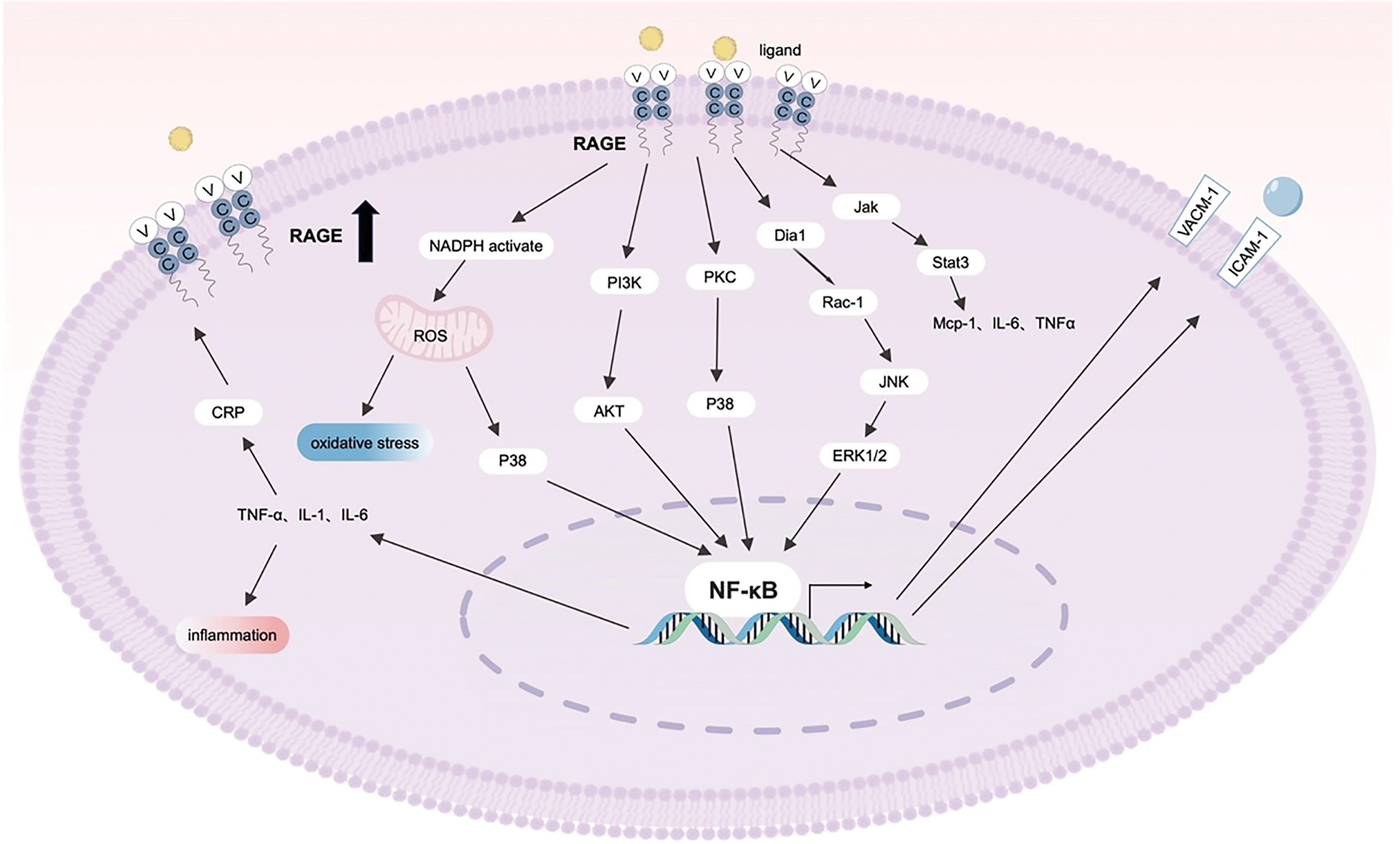
Figure 1: Diagram of the RAGE-mediated signaling pathway activated by the RAGE receptor upon binding to its various ligands, including the MAPK-NF-κB and JAK/STAT pathways, as well as pathways associated with oxidative stress. The RAGE receptor triggers a series of intracellular signaling cascades by binding to ligands such as AGEs and S100 proteins, ultimately regulating gene transcriptional activity. Abbreviations: Jak: Janus Kinase, PI3K: Phosphatidylinositol 3-Kinase, AKT: Serine/Threonine Protein Kinase B, NADPH: Nicotinamide Adenine Dinucleotide Phosphate, ROS: Reactive Oxygen Species, CRP: C-Reactive Protein, p38: p38 mitogen-activated protein kinase, PKC: Protein Kinase C, Rac-1: Ras-related C3 botulinum toxin substrate 1, JNK: c-Jun N-terminal Kinase, ERK1/2: Extracellular Regulated Protein Kinases, IL-6: Interleukin 6, Stat3: Signal transducer and activator of transcription3, MCP-1: Monocyte Chemotactic Protein-1, TNF-α: Tumor Necrosis Factor-alpha, NF-κB: Recombinant Nuclear Factor Kappa B, VACM-1: Vascular Cell Adhesion Molecule-1, ICAM-1: Intercellular Cell Adhesion Molecule-1. The images were generated using Biorender.
RAGE binds to proteins associated with cellular stress and damage, including AGEs, HMGB1, the S100 family, and Aβ [12]. Elevated extracellular levels of RAGE ligands predominantly activate RAGE expression through NF-κB activation. This interaction establishes a positive feedback mechanism, wherein increased RAGE activation further enhances NF-κB activity, resulting in sustained upregulation of inflammatory responses and RAGE expression [39]. This loop could perpetuate chronic inflammation and significantly contribute to the progression of various diseases, including cancer, diabetes, and neurodegenerative disorders.
In addition to binding to AGEs, RAGE interacts with a variety of other ligands, such as the S100/calreticulin family, HMGB1, β-folded fibrils, lysophosphatidic acid, β-amyloid, transthyretin, serum amyloid A, and β2-integrin. These interactions activate multiple mechanisms that can also accelerate the downstream activation of NF-κB and initiate similar positive feedback mechanisms leading to sustained RAGE expression, which further enhances NF-κB activation, creating a continuous inflammatory response [14]. Moreover, RAGE signaling activation contributes to the formation of ROS, pro-inflammatory cytokine release, pro-thrombotic factor production, and the expression of cell adhesion molecules. It has been demonstrated that VC1 domains efficiently bind to multiple negatively charged RAGE ligands, extending from low molecular weight AGEs, such as CML and CEL, to macromolecular structures like β-amyloid peptides and specific proteins such as the S100/calmodulin family and HMGB1 [40]. VC1 conjugates also can activate Rac1/Cdc42 of the Rho GTPase family, p38-mitogen-activated protein kinases (p38-MAPK) (Fig. 1). The intracellular domains of RAGE bind to key signaling molecules, which play integral roles in signal transduction. These include Diaphanous-related formin 1 (DIAPH1), Toll/interleukin-1 receptor domain-containing adapter protein (TIRAP), and extracellular signal-regulated kinase 1/2 (ERK1/2) [41,42].
RAGE and Lung-Related Diseases
Asthma, also known as bronchial asthma, is a chronic inflammatory disease of the airways characterized by diverse etiologies and clinical symptoms, triggered by a complex interplay of genetic and environmental factors [43]. Asthma involves various cells and components, with chronic airway inflammation being a central feature. This inflammation leads to structural and physiological alterations in lung tissue and function, impacting prognosis and quality of life [44]. In the pathological mechanism of asthma, pattern recognition receptors (PRRs) such as RAGE are reported to play a key role [45].
RAGE not only recognizes pathogen-associated molecular patterns (PAMPs) but also damage-associated molecular patterns (DAMPs) such as HMGB1 [46]. PAMPs are molecular structures typically produced by microorganisms such as viruses and bacteria, which trigger inflammatory responses by activating the immune system [47]. In asthma patients, exposure to certain environmental pathogens can trigger PAMPs, thereby activating the immune response in the airways, leading to airway inflammation and overreaction. Meanwhile, DAMPs are endogenous molecules released by damaged or dying cells, such as HMGB1, which can also exacerbate airway inflammation and pathological changes by activating the immune system [48]. By recognizing these diverse ligands, RAGE can stimulate the release of various cytokines, including thymic stromal lymphopoietin (TSLP), IL-1β, IL-33, and IL-25. This stimulation, in turn, triggers and regulates the activation and differentiation of dendritic cells (DCs), T-lymphocytes, and B-lymphocytes, further perpetuating the inflammatory response in asthmatic airways [49].
Peng et al. reported that abnormalities in the expression of epithelial barrier proteins, catenin, and E-cadherin, in toluene diisocyanate (TDI)-induced Asthma may be modulated via the RAGE-ERK axis [50], highlighting aberrant RAGE-mediated β-catenin signaling as a crucial factor in airway inflammation. Moreover, the RAGE’s ligand, HMGB1, is likely to play a significant role in that process. HMGB1, an inflammation-associated cytokine mediator, stimulates neutrophil aggregation and the release of pro-inflammatory factors. These factors are primarily produced by immune cells such as macrophages and monocytes [51]. HMGB1 can also modulate dendritic cell function by binding to receptors on their surface. This interaction enhances the antigen-presenting capability of dendritic cells, thereby facilitating T cell activation. Activated T cells, particularly Th17 cells, subsequently secrete inflammatory mediators such as IL-17, which further exacerbates the inflammatory response in asthma [52].
Clinical studies have shown that the development of asthma airway inflammation is associated with HMGB1 and RAGE. HMGB1 can serve as an important indicator for assessing treatment efficacy relative to asthma severity. Increased HMGB1 expression in the sputum of asthma patients correlates with disease severity and lung inflammatory cell counts (Fig. 2) [53]. Another study highlights the crucial role of S100A8/A9 in airway remodeling and inflammation in asthmatics [54]. Experiments using house dust mites to induce airway inflammation in rats showed that RAGE knockout rats did not exhibit asthma-like changes compared to wild-type rats [55]. Similarly, by using ovalbumin to induce airway sensitization in mice, Akirav et al. observed that RAGE accelerates T cell growth and differentiation by activating T cells [56].
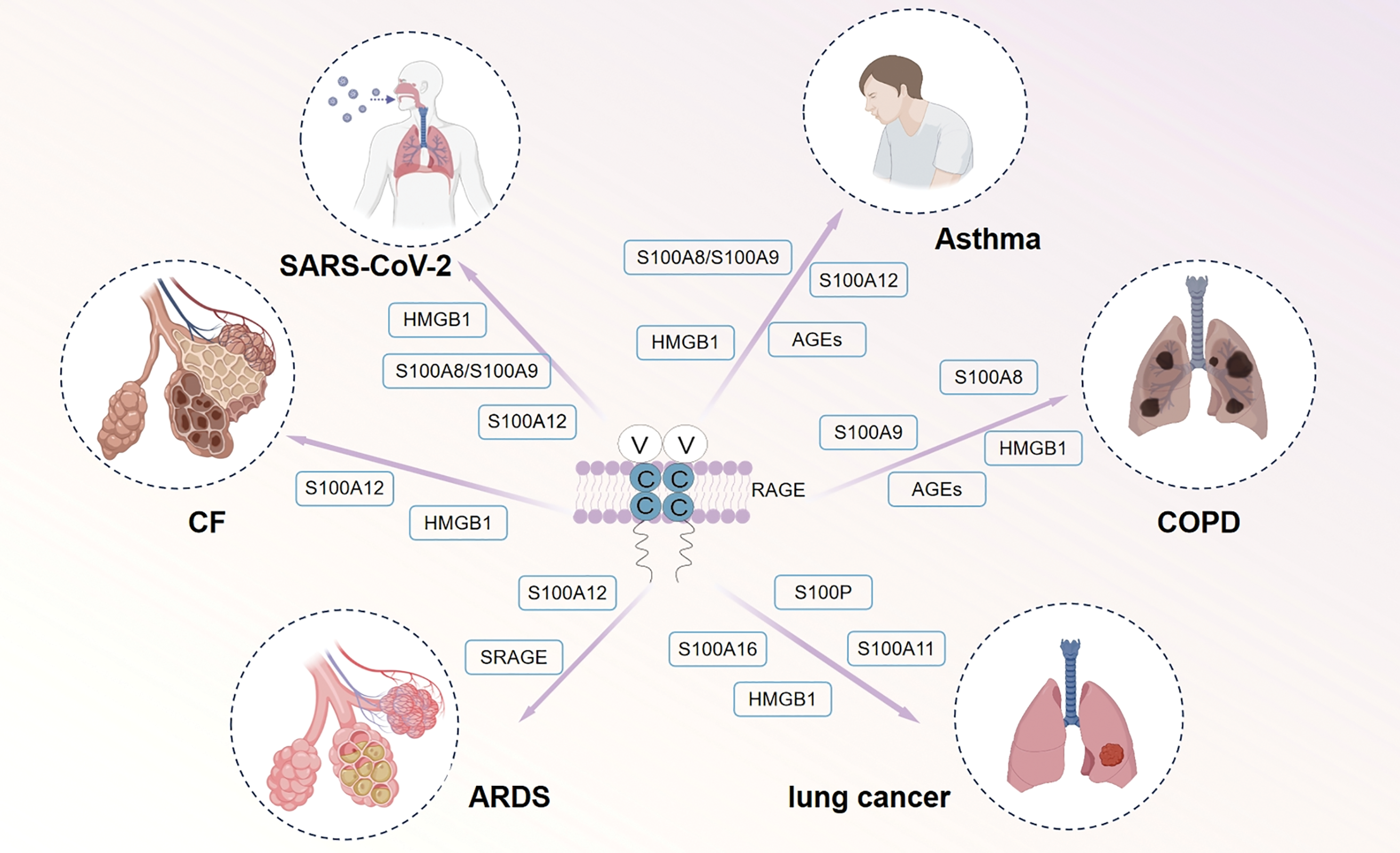
Figure 2: Involvement of multiple RAGE ligands in various human lung-related diseases, including Asthma, COPD, Lung Cancer, ARDS and PF. The coexistence of S100 protein, AGEs, and HMGB1 with RAGE in patients diagnosed with asthma, COPD, ARDS, PF and SARS-CoV-2 can elicit an inflammatory response, thereby potentially leading to alterations in lung tissue structure and physiological function. Elevated levels of sRAGE in the plasma of individuals at high risk for ARDS serve as a promising predictive and diagnostic biomarker for this condition. Moreover, the release of S100 protein and HMGB1 from cancer patients can induce cellular activation and contribute to tumor invasion and migration. In addition, S100A12 levels are significantly elevated in severe COVID-19 patients and can serve as a marker for identifying severe cases. HMGB1 is considered a prospective biomarker for the severity of COVID-19 and a potential therapeutic target. Abbreviations: PF: Pulmonary Fibrosis, COPD: Chronic Obstructive Pulmonary Disease, ARDS: Acute Respiratory Distress Syndrome, HMGB1: High-Mobility Group Box 1, AGEs: Advanced Glycation End Products, sRAGE: soluble Receptor for Advanced Glycation End Products. The images were generated using Biorender.
Current asthma treatments primarily rely on medications that control symptoms; however, these are often limited in efficacy or associated with significant side effects in some patients. Considering the significant role of the HMGB1/RAGE axis in asthma-related inflammation, targeting RAGE and its downstream pathways presents a promising therapeutic strategy. However, current research is mainly based on animal models and in vitro experiments, with limited evidence from (large-scale) clinical trials. Thus, while preliminary findings are promising, further studies or validation through clinical trials is necessary. Moreover, a comprehensive understanding of RAGE and its downstream pathways across different stages of inflammation and asthma phenotypes can enhance clinical treatment outcomes. This knowledge would assist clinicians in selecting more precise treatment strategies, facilitating personalized medicine, improving patient outcomes, reducing disease recurrence, and enhancing quality of life. Additionally, in-depth research into RAGE’s downstream signaling pathways may uncover new potential drug targets, providing a foundation for pharmaceutical companies to develop novel therapeutics. This approach not only expands the therapeutic scope of existing medications but also increases the specificity and efficacy of treatments. Future studies should focus on designing and conducting large-scale, randomized controlled trials to confirm the efficacy and safety of RAGE antagonists and HMGB1 inhibitors in asthma patients. Additionally, delving into the specific mechanisms of RAGE and its downstream pathways in asthma inflammation and clarifying their roles in different Asthma inflammatory stages is crucial.
Chronic obstructive pulmonary disease (COPD)
Chronic obstructive pulmonary disease (COPD) is a widespread, preventable, and treatable condition primarily characterized by airway inflammation and structural remodeling [57]. With the increasing understanding of COPD, more attention is being paid to its various complications, such as respiratory failure, heart failure, pulmonary edema, hypoxemia, and cardiac arrhythmias. Despite extensive research, the exact pathogenesis of COPD remains elusive. Core mechanisms include chronic inflammation, imbalances between proteases and anti-proteases, oxidative stress, and increased aberrant apoptosis [58]. RAGE and its associated ligands play significant roles in the pathogenesis of COPD. And genetic factors also influence RAGE function, with polymorphisms in the gene which encoding RAGE being linked to decreased lung function and higher COPD risk [59]. A clinical study also confirm that the G82S polymorphism of RAGE gene increases the risk of COPD among Chinese smokers [60].
The expression of RAGE and its ligand HMGB1 was reported to be significantly enhanced in both sputum and pathological samples from COPD patients (Fig. 2) [61]. Elevated levels of AGEs were also detected in lung tissues and small airways of COPD patients (Fig. 2) [62]. Additionally, it is evidenced that an increasing trend in the number of AGEs in the skin of COPD patients compared to healthy smoking and non-smoking controls [63]. Meanwhile, the expression of sRAGE has been found to be decreased in patients with COPD according to numerous scientific studies. sRAGE functions as a decoy receptor that sequesters circulating AGEs, thereby inhibiting their binding to cell surface receptors and subsequently reducing the inflammatory response mediated by RAGE signaling [8,64,65]. Research has demonstrated that reduced levels of sRAGE are associated with the severity of COPD and, to some extent, with lung function. In a study by Gopal et al., serum levels of sRAGE and Forced Expiratory Volume in 1 s (FEV1) were tested in 88 patients with COPD. The results showed a significant association between total plasma sRAGE levels and FEV1, suggesting that sRAGE can be an important indicator for assessing disease status and severity in COPD patients [66]. Overall, the involvement of RAGE signaling in COPD underscores its significance in disease mechanisms and potential as a target for therapeutic interventions.
Lung cancer remains one of the most lethal diseases globally, with its rising incidence posing significant threats to human health. High-risk factors include cigarette smoking, environmental smoke, genetic factors, air pollution, and occupational exposures such as asbestos and radon [67]. RAGE is implicated in the pathogenesis and progression of non-small cell lung cancer (NSCLC) [68]. Unlike other solid tumors like hepatocellular carcinoma, RAGE expression was reported to be significantly reduced in NSCLC [69]. However, it may play a crucial role in chronic inflammation and oxidative stress within the tumor microenvironment, contributing to cancer progression.
NADPH oxidase 4 (NOX-4), a member of the NOX family, is involved in the formation of endogenous ROS and the inflammatory response. NOX-4 proteins may act as downstream effectors of RAGE in chronic inflammation, affecting the tumor microenvironment. The interaction between RAGE and NOX-4 mediates chronic inflammation, which can increase cancer risk [70]. RAGE also engages in ligand-receptor binding with S100 family proteins, initiating a cascade of biochemical reactions. When extracellular S100P binds to RAGE on the cell surface, it triggers downstream signaling cascades that influence the cell growth cycle and activity (Fig. 2). This interaction is crucial, particularly in mediating drug resistance and invasive metastasis of tumors [71].
Studies on lung adenocarcinoma have demonstrated that S100A11 expression is significantly increased in tissues with the Kirsten rat sarcoma viral oncogene (KRAS) gene variant, promoting cell proliferation. And both S100A11 and S100A16 are associated with the prognosis of lung adenocarcinoma, with S100A16 being prominent in high-stage lung cancer (Fig. 2) [72]. Recent study suggests that the extracellular segment of the S100 receptor (esFc) acts as a significant trigger for lung cancer metastasis upon stimulation by S100A8/A9. Therefore, disrupting this interaction may help in preventing cancer metastasis [73]. Moreover, Choi et al. [74] identified a serum protein level threshold of 0.058 ng/mL for S100B when examining lung cancer associated with brain metastases. This threshold exhibited a sensitivity of 89%, specificity of 43%, and overall accuracy of 51% in diagnosing patients with brain metastases. When combined with autoantibody levels of S100B, the overall diagnostic accuracy improved to 62.5%. Interestingly, S100B can bind to RAGE, promoting cell growth by activating the fibroblast growth factor receptor (FGFR1) in a bFGF-dependent manner [75], while aberrant bFGF-FGFR1 is reported to be highly involved in the tumor initiation progression and drug resistance in lung squamous cell carcinoma (LSCC) [76]. Further exploration of RAGE signaling in the specific context of LSCC might provide new targets to conquer this specific type of LSCC, which is still absent of targeted therapy [77].
It was demonstrated that a significant reduction in RAGE expression levels among non-small cell lung cancer patients [78]. Kobayashi et al. investigated RAGE expression in 182 non-small cell lung cancer tissues, revealing either absent or low RAGE expression in 147 samples, which correlated positively with patient survival [79]. Consistently, increasing RAGE expression notably inhibited lung cancer cell growth and tumorigenesis [80]. Lung cancer development involves substantial microenvironmental changes that impact cancer cell differentiation and metastasis. Mechanically, reduced RAGE expression in lung cancer disrupts cellular and extracellular matrix interactions, enhancing cell proliferation, migration, and metastasis [81]. Notably, Oczypok et al. observed highest RAGE expression in type I alveolar epithelial cells compared to lower levels in bronchial epithelial cells of lung cancer tissues. This differential expression may enhance cancer cell proliferative capacity rather than suppress apoptosis, potentially evading immune recognition and fostering immune tolerance [22]. Chronic inflammation and oxidative stress mediated by RAGE-associated factors like NF-κB further exacerbate tumorigenesis by impairing normal cell interactions and apoptosis [82].
Unexpectedly, despite decreased RAGE expression in lung cancer, the levels of its ligands such as AGEs tend to increase due to negative feedback regulation and the high metabolic activity of tumor tissues, promoting tumor progression [69]. Conversely, sRAGE levels decrease in lung cancer tissues compared to normal and benign lesions, showing a negative correlation with lymph node metastasis degree, thereby holding promise for lung cancer detection and prognostic evaluation [83].
Collectively, RAGE signaling plays a complex role in lung cancer, involving chronic inflammation, oxidative stress, and interactions with various proteins and ligands. Despite reduced RAGE expression in lung cancer, there is an increase in its ligands, suggesting a potential feedback mechanism. Furthermore, sRAGE shows promise as a biomarker for lung cancer, offering avenues for future therapeutic and diagnostic strategies. Deep elucidating the role of RAGE-mediated signaling in different subtypes of lung cancer may provide new directions for precision therapies.
Acute respiratory distress syndrome (ARDS)
Acute respiratory distress syndrome (ARDS) is a prevalent inflammatory lung injury characterized by increased pulmonary edema and respiratory failure triggered by hypoxemia [84]. Clinically, ARDS manifests as persistent or recurrent wheezing, cough, varying degrees of chest tightness, and shortness of breath. Severe dyspnea, progressive cyanosis, and wet rhonchi in the lungs are key distinguishing symptoms. Death by asphyxia can result from thickening of the alveolar walls due to the deposition of large amounts of extracellular exudate in the alveolar cavities, leading to impaired ventilation [85]. Alveolar fluid is transported to alveolar epithelial cells primarily by sodium ions via sodium channels, followed by the NA+-K+ ATPase within the basement membrane that pumps these sodium ions to the interstitium, creating a localized osmotic gradient [86]. Aquaporin-5 (AQP-5) channels in the alveolar epithelium facilitate water transport, effectively removing fluid from the alveoli. These pathways are crucial for maintaining pulmonary circulation homeostasis. The active transport of ions and fluids by the alveolar epithelium is vital for keeping the alveoli dry and ensuring efficient gas exchange [87]. Besides, anions, cations, and other small molecules in lung tissue can enter the alveolar lumen through cell membranes. When lung damage occurs, the ion transport pathways are disrupted, which reduces the efficiency of alveolar fluid clearance. This leads to the accumulation of water and sodium, ultimately resulting in pulmonary edema.
RAGE is highly expressed in the lungs, particularly on the basement membrane of type I alveolar epithelial cells, and is also present in type II alveolar epithelial cells [10]. Additionally, polymorphisms in the sRAGE gene (AGER) play a crucial role in predicting the development of ARDS. Specifically, the AGER SNP rs2070600 (Ser/Ser) is associated with an increased risk of ARDS and elevated plasma sRAGE levels (Fig. 2). In high-risk intensive care patient populations, elevated plasma sRAGE levels can help identify individuals more susceptible to ARDS and predict their prognosis [88]. Mechanical degeneration of epithelial and endothelial cells can trigger a significant release of sRAGE in the short term when lung tissue is compromised. These cells can modulate their oxygen requirements through secretion or direct action on other organelles, leading to a range of pathophysiologic changes, including airway obstruction, alveolar damage, and inflammatory responses. During treatment, serum levels of sRAGE gradually decrease, primarily due to reduced inflammation, and research indicates that sRAGE can be rapidly cleared by the kidneys [89]. Elevated sRAGE levels correlate with ARDS severity, serving as a biomarker. Future treatments may focus on modulating sRAGE-mediated signaling, improving early detection, prognosis, and personalized therapies for treating ARDS.
Englert et al. reported significantly lower RAGE protein levels in alveolar lavage fluid and tissue homogenates from PF patients compared to healthy individuals [90]. Experimental models of pulmonary fibrosis in mice, induced by bleomycin and asbestos, also showed decreased RAGE expression, suggesting its possible protective role in the context of complex inflammatory response leading to pulmonary fibrosis due to excessive extracellular matrix deposition around blood vessels [91]. Further studies revealed that RAGE knockout mice naturally exhibit altered lung fibrosis compared to wild-type mice. Both lung injury and sham-operated groups showed multiple pro-fibrotic cytokines compared to normal mice, suggesting involvement in lung tissue remodeling. This indicates RAGE’s protective role in pulmonary fibrosis development [21]. He et al. assessed pro-fibrotic cytokines like transforming growth factor-beta1 (TGF-β1) and platelet-derived growth factor (PDGF) in RAGE knockout and wild-type mice, finding no increasing trend in RAGE knockout mice [92]. Ramsgaard et al. found similar asbestos-induced pulmonary fibrosis effects in both RAGE knockout and wild-type mice [21]. However, Morbini used immunohistochemistry to examine RAGE expression in smokers’ fibrosis and healthy tissue samples, finding increased RAGE in alveolar cells, bronchial epithelial cells, and endothelial cells [93].
The role of RAGE in pulmonary fibrosis is debated, with animal experiments yielding varied conclusions. Nevertheless, most studies show a gradual decrease in RAGE expression in both patients and animal models of pulmonary fibrosis. Whether this is due to a decrease in the RAGE gene itself or damage to type I alveolar epithelial cells remains unclear, necessitating further investigation.
SARS-CoV-2 was first reported in 2019 and led to the global COVID-19 pandemic. Patients infected with this virus may be asymptomatic or present with moderate to severe clinical symptoms, primarily manifesting as pulmonary infections such as pneumonia, respiratory failure, and death [94].
Research indicates that RAGE can act as a functional receptor for SARS-CoV-2, binding to the S1 receptor-binding domain of the virus. The interaction between RAGE and the Spike protein facilitates the infection of monocytes, and the RAGE pathway is crucial for SARS-CoV-2 infection [95]. Basile’s research reveals that SARS-CoV-2 infection causes a significant upregulation of the PI3K-AKT-mTOR pathway, primarily evidenced by extensive modification of protein phosphorylation sites [96]. Studies reveal that downstream MAPK signaling molecules in COVID-19 patients, such as ERK1/2 and p38, show significantly elevated phosphorylation levels, indicating that the MAPK signaling pathway is significantly activated [97].
Numerous studies have shown that many ligands of RAGE play a significant role in the disease progression of COVID-19 patients. Elevated levels of S100A8, S100A9, and HMGB1 are observed in COVID-19 patients. Holms discovered that S100A8/A9 can bind to RAGE, activating downstream pathways and leading to the release of IL-6 [98]. The secreted IL-6 can further activate STAT3, which stimulates the expression of S100A8/A9 in monocytes, creating a positive feedback loop that amplifies inflammation. S100A12, another ligand of RAGE, is primarily activated in severe patients and shows increased expression in lung tissues. Its exact role remains unclear, but research indicates that compared to healthy controls, COVID-19 patients have higher levels of S100A12, and its levels correlate positively with disease severity [99]. Mester proposed that S100A12 could serve as a biomarker for severe cases and suggested that serological measurement of S100A12 might improve the selection of antibiotics and antiviral drugs for COVID-19 patients [100]. HMGB1 plays a role in COVID-19 patients like its role in other diseases. It can form complexes with DNA and RNA, which then bind to mRNA and Toll-like receptors, initiating pro-inflammatory responses. This interaction increases the output of pro-inflammatory factors, creating a harmful feedback loop [101]. It has been reported that HMGB1 may serve as a prognostic marker for the severity of COVID-19 and as a potential therapeutic target [102].
AGE mediates the interaction between inflammatory proteins and connective tissue, making them more susceptible to damage from immune system imbalance. Inflammation is currently understood as a complex process induced by various types of cytokines, including interleukins, tumor necrosis factors, and other inflammatory mediators, as well as regulatory substances like the complement system. RAGE is expressed in multiple inflammation-related cells. When AGE is activated, it triggers the activation of several downstream signaling pathways, which can ultimately lead to tissue damage due to inflammatory responses. Published studies suggest that the AGE-RAGE signaling pathway may be associated with the severity of COVID-19 [103–105]. Furthermore, some researchers propose that this signaling pathway could serve as a novel anti-inflammatory drug target for clinical application. Multiple studies have found that the AGE-RAGE signaling pathway may be associated with the therapeutic effects of various traditional Chinese herbs on COVID-19. It has been confirmed that some traditional Chinese medicines, such as Astragalus injection, Honeysuckle injection, and Baicalin injection, have antiviral replication-inhibiting and immune-modulating effects [106]. These findings suggest a close connection between the AGE-RAGE signaling pathway and the pathogenic mechanisms of SARS-CoV-2.
RAGE Inhibitor and Therapeutical Potential in Lung Related Diseases
RNA interference (RNAi) based therapy has been explored to reduce RAGE expression [107]. In addition, methods to antagonize ligands-RAGE signaling include using sRAGE decoy molecules, as well as small molecule inhibitors such as FPS-ZM1, which selectively and competitively binds to RAGE to prevent signaling ligands’ interaction [108,109]. Other approaches include the use of RAGE antibodies and low molecular weight heparin to inhibit RAGE activity.
While the current development of RAGE inhibitors has primarily focused on diseases such as Alzheimer’s disease and cancer [109–112], the potential of RAGE inhibitors in treating specific lung-related diseases has received less attention. Therefore, we herein summarized specific clinical studies focusing on RAGE inhibiting molecules for treating lung or its associated diseases (Table 2). These clinical trials have shown promising results, which suggest that targeting RAGE could be a promising therapeutic strategy for various lung-related diseases.
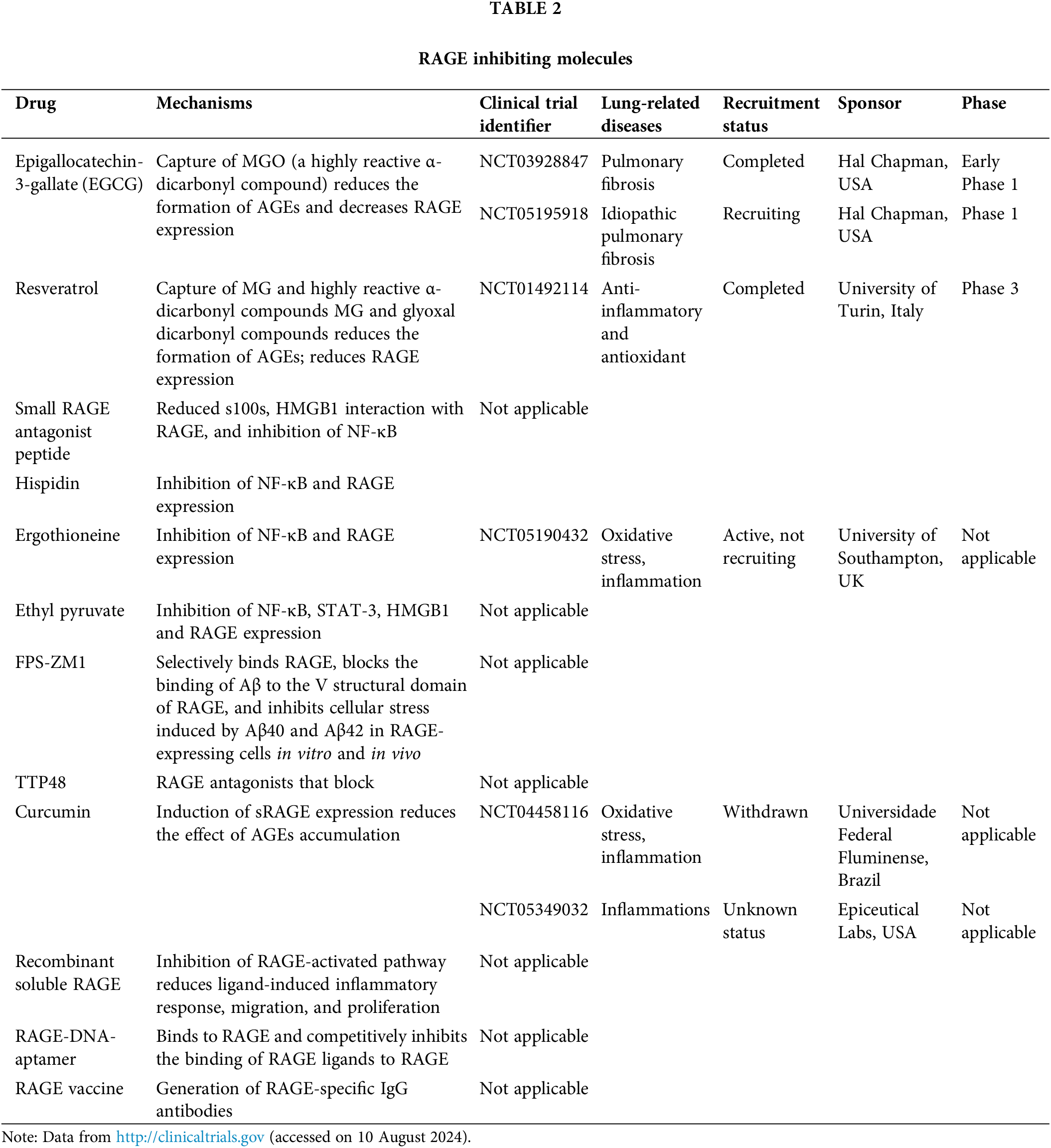
RAGE exhibits notable expression in pulmonary tissues under physiological conditions, distinguishing it from other organ systems. Its regulatory influence in lung-related disorders such as lung cancer, asthma, and COPD is increasingly acknowledged. The modulation of ligands-RAGE signaling in these diseases underscores its significant role, with outcomes varying markedly depending on the specific context of lung pathology.
Inflammatory responses are prominently initiated by RAGE activation, highlighting its pivotal role in conditions like asthma and COPD. Activation of this receptor commonly amplifies inflammatory reactions, a shared characteristic observed in diseases including diabetes, neurodegenerative disorders, and cardiovascular diseases. Through the NF-κB signaling pathway, RAGE enhances the expression of multiple inflammatory mediators, perpetuating chronic inflammation crucial to disease progression. Additionally, RAGE activation escalates intracellular ROS, triggering oxidative stress that plays a pivotal role in cellular damage and apoptosis across diverse cell types such as endothelial cells, macrophages, and neuronal cells.
In lung diseases like lung cancer and pulmonary fibrosis, RAGE’s presence may confer a protective role by exerting anti-inflammatory or immunosuppressive effects through modulation of immune responses and interaction with genes involved in airway inflammation. Further investigations are necessary to elucidate its precise mechanisms. We proposed future research efforts should prioritize:
1. Elucidating the specific mechanisms of ligands-RAGE signaling in either promoting or mitigating lung-related diseases to identify novel therapeutic targets.
2. Assessing the potential of RAGE and its various ligands as biomarkers for specific lung diseases and complex pulmonary comorbidities, considering their varied expression levels and critical roles across numerous lung diseases.
3. Developing potent and selective inhibitors targeting RAGE’s specific ligands, which may offer enhanced safety profiles compared to broad RAGE inhibitors, represents a promising avenue for clinical translation in therapeutic intervention.
While the initial objective of this review is to explore recent discoveries of AGE-RAGE signaling axis in the specific context of lung diseases from both basic research and clinical advancements, thus, to provide valuable references for future studies, it is important to note that certain limitations remain, including:
First, the scope of this review primarily centers on established mechanisms of RAGE interaction and signaling, potentially overlooking emerging insights into less studied ligands and novel regulatory mechanisms. Expanding the exploration of these newly discovered pathways could provide a more holistic understanding of the complexities of RAGE signaling. Furthermore, while this review predominantly discusses RAGE’s role in lung-related diseases, it’s important to acknowledge that the complexity of lung diseases extends beyond the conditions discussed here, such as asthma, COPD, pulmonary fibrosis, ARDS, and lung cancer. Other respiratory conditions, including bronchitis and pneumonia, are influenced by a multitude of factors. Notably, various viruses and bacteria may induce different lung injuries and are potentially linked to varying degrees of dysregulation in the AGE-RAGE signaling pathway, even though related mechanistic studies and clinical investigations are still lacking.
The existing literature lacks sufficient evidence supporting the feasibility and efficacy of clinical applications related to this topic. This oversight may lead to an oversimplified understanding of the underlying mechanisms governing RAGE’s functionality. We believe that although existing research has highlighted the significance of RAGE in lung diseases, further exploration is warranted to elucidate its specific mechanisms. Exploring the mechanisms of the AGE-RAGE pathway in the initiation and progression of specific lung diseases, particularly through clinical research and trials-driven basic studies, holds great significance for developing more effective targeted therapeutic strategies of specific lung diseases.
In conclusion, the multifaceted involvement of RAGE in lung diseases underscores its potential as both a therapeutic target and biomarker, necessitating further exploration to fully harness its clinical potential.
Acknowledgement: None.
Funding Statement: This research was funded by Science and Technology Department of Sichuan Province 2023NSFSC0130 and 2023NSFSC1992, Startup Foundation for Advanced Talents of Sichuan University (YJ2021124), the Fundamental Research Funds for the Central Universities, and sponsored by Open Project of State Key Laboratory of Respiratory Disease (SKLRD-OP-202501).
Author Contributions: Study conception and design: Tao Jia, Shuochen Pang; draft manuscript preparation: Shuochen Pang; review and editing: Tao Jia, Shuochen Pang, Zifeng Yang; visualization: Shuochen Pang; supervision: Tao Jia, Zifeng Yang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Dong H, Zhang Y, Huang Y, Deng H. Pathophysiology of RAGE in inflammatory diseases. Front Immunol. 2022;13:931473. doi:10.3389/fimmu.2022.931473. [Google Scholar] [CrossRef]
2. Hudson BI, Lippman ME. Targeting RAGE signaling in inflammatory disease. Annu Rev Med. 2018;69(1):349–64. doi:10.1146/annurev-med-041316-085215. [Google Scholar] [CrossRef]
3. Chen L, Duan Z, Tinker L, Sangi-Haghpeykar H, Strickler H, Ho GYF, et al. A prospective study of soluble receptor for advanced glycation end-products and colorectal cancer risk in postmenopausal women. Cancer Epidemiol. 2016;42(Suppl. 3):115–23. doi:10.1016/j.canep.2016.04.004. [Google Scholar] [CrossRef]
4. Wu X-Q, Zhang D-D, Wang Y-N, Tan Y-Q, Yu X-Y, Zhao Y-Y. AGE/RAGE in diabetic kidney disease and ageing kidney. Free Radical Biol Med. 2021;171(6):260–71. doi:10.1016/j.freeradbiomed.2021.05.025. [Google Scholar] [CrossRef]
5. Shen CY, Lu CH, Wu CH, Li KJ, Kuo YM, Hsieh SC, et al. The development of maillard reaction, and advanced glycation end product (AGE)-receptor for AGE (RAGE) signaling inhibitors as novel therapeutic strategies for patients with AGE-related diseases. Molecules. 2020;25(23):5591. doi:10.3390/molecules25235591. [Google Scholar] [CrossRef]
6. Kim OY, Song J. The importance of BDNF and RAGE in diabetes-induced dementia. Pharmacol Res. 2020;160:105083. doi:10.1016/j.phrs.2020.105083. [Google Scholar] [CrossRef]
7. Koerich S, Parreira GM, de Almeida DL, Vieira RP, de Oliveira ACP. Receptors for advanced glycation end products (RAGEpromising targets aiming at the treatment of neurodegenerative conditions. Curr Neuropharmacol. 2023;21(2):219. doi:10.2174/1570159X20666220922153903. [Google Scholar] [CrossRef]
8. Sharma A, Kaur S, Sarkar M, Sarin B, Changotra H. The AGE-RAGE axis and RAGE genetics in chronic obstructive pulmonary disease. Clin Rev Allergy Immunol. 2021;60:244–58. doi:10.1007/s12016-020-08815-4. [Google Scholar] [CrossRef]
9. Plotkin LI, Essex AL, Davis HM. RAGE signaling in skeletal biology. Curr Osteoporos Rep. 2019;17(1):16–25. doi:10.1007/s11914-019-00499-w. [Google Scholar] [CrossRef]
10. Salehi M, Amiri S, Ilghari D, Hasham LFA, Piri H. The remarkable roles of the receptor for advanced glycation end products (RAGE) and its soluble isoforms in COVID-19: the importance of RAGE pathway in the lung injuries. Indian J Clin Biochem. 2023;38(2):159–71. doi:10.1007/s12291-022-01081-5. [Google Scholar] [CrossRef]
11. Braley A, Kwak T, Jules J, Harja E, Landgraf R, Hudson BI. Regulation of receptor for advanced glycation end products (RAGE) ectodomain shedding and its role in cell function. J Biol Chem. 2016;291(23):12057–73. doi:10.1074/jbc.M115.702399. [Google Scholar] [CrossRef]
12. Chuah YK, Basir R, Talib H, Tie TH, Nordin N. Receptor for advanced glycation end products and its involvement in inflammatory diseases. Int J Inflam. 2013;2013:403460. [Google Scholar] [PubMed]
13. Zhou M, Zhang Y, Shi L, Li L, Zhang D, Gong Z, et al. Activation and modulation of the AGEs-RAGE axis: implications for inflammatory pathologies and therapeutic interventions—a review. Pharmacol Res. 2024;206:107282. doi:10.1016/j.phrs.2024.107282. [Google Scholar] [CrossRef]
14. Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, et al. Understanding RAGE, the receptor for advanced glycation end products. J Mol Med. 2005;83(11):876–86. doi:10.1007/s00109-005-0688-7. [Google Scholar] [CrossRef]
15. Rojas A, Gonzalez I, Morales MA. SARS-CoV-2-mediated inflammatory response in lungs: should we look at RAGE? Inflamm Res. 2020;69:641–3. doi:10.1007/s00011-020-01353-x. [Google Scholar] [CrossRef]
16. Reynaert NL, Vanfleteren LE, Perkins TN. The AGE-RAGE axis and the pathophysiology of multimorbidity in COPD. J Clin Med. 2023;12(10):3366. doi:10.3390/jcm12103366. [Google Scholar] [CrossRef]
17. Uchida T, Shirasawa M, Ware LB, Kojima K, Hata Y, Makita K, et al. Receptor for advanced glycation end-products is a marker of type I cell injury in acute lung injury. Am J Respir Crit Care Med. 2006;173(9):1008–15. doi:10.1164/rccm.200509-1477OC. [Google Scholar] [CrossRef]
18. Wu S, Mao L, Li Y, Yin Y, Yuan W, Chen Y, et al. RAGE may act as a tumour suppressor to regulate lung cancer development. Gene. 2018;651(30):86–93. doi:10.1016/j.gene.2018.02.009. [Google Scholar] [CrossRef]
19. Zhan X, Wu R, Kong X-H, You Y, He K, Sun X-Y, et al. Elevated neutrophil extracellular traps by HBV-mediated S100A9-TLR4/RAGE-ROS cascade facilitate the growth and metastasis of hepatocellular carcinoma. Cancer Commun. 2023;43(2):225–45. doi:10.1002/cac2.12388. [Google Scholar] [CrossRef]
20. Onyeagucha BC, Mercado-Pimentel ME, Hutchison J, Flemington EK, Nelson MA. S100P/RAGE signaling regulates microRNA-155 expression via AP-1 activation in colon cancer. Exp Cell Res. 2013;319(13):2081–90. doi:10.1016/j.yexcr.2013.05.009. [Google Scholar] [CrossRef]
21. Ramsgaard L, Englert JM, Tobolewski J, Tomai L, Fattman CL, Leme AS, et al. The role of the receptor for advanced glycation end-products in a murine model of silicosis. PLoS One. 2010;5(3):e9604. doi:10.1371/journal.pone.0009604. [Google Scholar] [CrossRef]
22. Oczypok EA, Perkins TN, Oury TD. All the “RAGE” in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr Respir Rev. 2017;23:40–9. [Google Scholar] [PubMed]
23. Pal R, Bhadada SK. AGEs accumulation with vascular complications, glycemic control and metabolic syndrome: a narrative review. Bone. 2023;176:116884. doi:10.1016/j.bone.2023.116884. [Google Scholar] [CrossRef]
24. Ishiguro H, Nakaigawa N, Miyoshi Y, Fujinami K, Kubota Y, Uemura H. Receptor for advanced glycation end products (RAGE) and its ligand, amphoterin are overexpressed and associated with prostate cancer development. Prostate. 2005;64(1):92–100. doi:10.1002/pros.v64:1. [Google Scholar] [CrossRef]
25. Ott C, Jacobs K, Haucke E, Navarrete Santos A, Grune T, Simm A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014;2:411–29. doi:10.1016/j.redox.2013.12.016. [Google Scholar] [CrossRef]
26. Taguchi A, Blood DC, del Toro G, Canet A, Lee DC, Qu W, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature. 2000;405(6784):354–60. doi:10.1038/35012626. [Google Scholar] [CrossRef]
27. Fiuza C, Bustin M, Talwar S, Tropea M, Gerstenberger E, Shelhamer JH, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood. 2003;101(7):2652–60. doi:10.1182/blood-2002-05-1300. [Google Scholar] [CrossRef]
28. Alam MI, Mae M, Farhana F, Oohira M, Yamashita Y, Ozaki Y, et al. NLRP3 inflammasome negatively regulates RANKL-induced osteoclastogenesis of mouse bone marrow macrophages but positively regulates it in the presence of lipopolysaccharides. Int J Mol Sci. 2022;23(11):6096. doi:10.3390/ijms23116096. [Google Scholar] [CrossRef]
29. Orlova VV, Choi EY, Xie C, Chavakis E, Bierhaus A, Ihanus E, et al. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 2007;26(4):1129–39. doi:10.1038/sj.emboj.7601552. [Google Scholar] [CrossRef]
30. Zhu L, Ito T, Nakahara T, Nagae K, Fuyuno Y, Nakao M, et al. Upregulation of S100P, receptor for advanced glycation end products and ezrin in malignant melanoma. J Dermatol. 2013;40(12):973–9. doi:10.1111/1346-8138.12323. [Google Scholar] [CrossRef]
31. Robinson MJ, Tessier P, Poulsom R, Hogg N. The S100 family heterodimer, MRP-8/14, binds with high affinity to heparin and heparan sulfate glycosaminoglycans on endothelial cells. J Biol Chem. 2002;277(5):3658–65. doi:10.1074/jbc.M102950200. [Google Scholar] [CrossRef]
32. Ghavami S, Rashedi I, Dattilo BM, Eshraghi M, Chazin WJ, Hashemi M, et al. S100A8/A9 at low concentration promotes tumor cell growth via RAGE ligation and MAP kinase-dependent pathway. J Leukoc Biol. 2008;83(6):1484–92. doi:10.1189/jlb.0607397. [Google Scholar] [PubMed] [CrossRef]
33. Taguchi K, Fukami K. RAGE signaling regulates the progression of diabetic complications. Front Pharmacol. 2023;14:1128872. doi:10.3389/fphar.2023.1128872. [Google Scholar] [CrossRef]
34. Arakawa S, Suzuki R, Kurosaka D, Ikeda R, Saito M. Mass spectrometric quantitation of AGEs and enzymatic crosslinks in human cancellous bone. Sci Rep. 2020;10(1):18774. doi:10.1038/s41598-020-75923-8. [Google Scholar] [CrossRef]
35. Yang X, Zeng J, Xie K, Su S, Guo Y, Zhang H, et al. Advanced glycation end product-modified low-density lipoprotein promotes pro-osteogenic reprogramming via RAGE/NF-κB pathway and exaggerates aortic valve calcification in hamsters. Mol Med. 2024;30(1):76. doi:10.1186/s10020-024-00833-8. [Google Scholar] [CrossRef]
36. Moulahoum H, Ghorbanizamani F, Khiari Z, Toumi M, Benazzoug Y, Tok K, et al. Artemisia alleviates AGE-induced liver complications via MAPK and RAGE signaling pathways modulation: a combinatorial study. Mol Cell Biochem. 2022;477(10):2345–57. doi:10.1007/s11010-022-04437-w. [Google Scholar] [CrossRef]
37. Qureshy Z, Johnson DE, Grandis JR. Targeting the JAK/STAT pathway in solid tumors. J Cancer Metastasis Treat. 2020;6. doi:10.20517/2394-4722.2020.58. [Google Scholar] [CrossRef]
38. Sukjamnong S, Chen H, Saad S, Santiyanont R. Fimbristylis ovata and Artemisia vulgaris extracts inhibited AGE-mediated RAGE expression, ROS generation, and inflammation in THP-1 cells. Toxicol Res. 2022;38(3):331–43. doi:10.1007/s43188-021-00114-0. [Google Scholar] [CrossRef]
39. Ahmad S, Khan H, Siddiqui Z, Khan MY, Rehman S, Shahab U, et al. AGEs, RAGEs and s-RAGE; friend or foe for cancer. Semin Cancer Biol. 2018;49(4):44–55. doi:10.1016/j.semcancer.2017.07.001. [Google Scholar] [CrossRef]
40. Xue J, Ray R, Singer D, Böhme D, Burz DS, Rai V, et al. The receptor for advanced glycation end products (RAGE) specifically recognizes methylglyoxal-derived AGEs. Biochemistry. 2014;53(20):3327–35. doi:10.1021/bi500046t. [Google Scholar] [CrossRef]
41. Hudson BI, Kalea AZ, del Mar Arriero M, Harja E, Boulanger E, D’Agati V, et al. Interaction of the RAGE cytoplasmic domain with diaphanous-1 is required for ligand-stimulated cellular migration through activation of Rac1 and Cdc42. J Biol Chem. 2008;283(49):34457–68. doi:10.1074/jbc.M801465200. [Google Scholar] [CrossRef]
42. Manigrasso MB, Pan J, Rai V, Zhang J, Reverdatto S, Quadri N, et al. Small molecule inhibition of ligand-stimulated RAGE-DIAPH1 signal transduction. Sci Rep. 2016;6(1):22450. doi:10.1038/srep22450. [Google Scholar] [CrossRef]
43. Nguyen V, Zhang Q, Pan F, Jin Q, Sun M, Tangthianchaichana J, et al. Zi-Su-Zi decoction improves airway hyperresponsiveness in cough-variant asthma rat model through PI3K/AKT1/mTOR, JAK2/STAT3 and HIF-1α/NF-κB signaling pathways. J Ethnopharmacol. 2023;314:116637. doi:10.1016/j.jep.2023.116637. [Google Scholar] [CrossRef]
44. Gu XF, Chen XM, Chen HJ, Xu TT, Qiu ZW, Sun DD, et al. The role of S100A8/RAGE and Caveolin-1 and the effect of roxithromycin on their expression in a rat model of neutrophilic asthma. Chin J Tuberc Resp. 2019;42(11):845–51 (In Chinese). [Google Scholar]
45. Brandt EB, Lewkowich IP. RAGE-induced asthma: a role for the receptor for advanced glycation end-products in promoting allergic airway disease. J Allergy Clin Immunol. 2019;144(3):651–3. doi:10.1016/j.jaci.2019.06.012. [Google Scholar] [CrossRef]
46. Singh H, Agrawal DK. Therapeutic potential of targeting the HMGB1/RAGE axis in inflammatory diseases. Molecules. 2022;27(21):7311. doi:10.3390/molecules27217311. [Google Scholar] [CrossRef]
47. Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu Rev Pathol. 2020;15(1):493–518. doi:10.1146/annurev-pathmechdis-012419-032847. [Google Scholar] [CrossRef]
48. Kim YH, Lim JO, Kim JS, Kim BY, Pyun BJ, Lee SJ, et al. Protease allergen-induced HMGB1 contributes to NLRC4 inflammasome-mediated inflammation in experimental asthma. Allergy. 2023;78(5):1387–92. doi:10.1111/all.15668. [Google Scholar] [CrossRef]
49. Dou L, Wang W, Wang J, Zhang X, Hu X, Zheng W, et al. miR-3934 regulates the apoptosis and secretion of inflammatory cytokines of basophils via targeting RAGE in asthma. Allergy, Asthma Clin Immunol. 2022;18(1):66. doi:10.1186/s13223-022-00704-z. [Google Scholar] [CrossRef]
50. Peng X, Huang M, Zhao W, Lan Z, Wang X, Yuan Y, et al. RAGE mediates airway inflammation via the HDAC1 pathway in a toluene diisocyanate-induced murine asthma model. BMC Pulm Med. 2022;22(1):61. doi:10.1186/s12890-022-01832-3. [Google Scholar] [CrossRef]
51. Qian B, Huang H, Cheng M, Qin T, Chen T, Zhao J. Mechanism of HMGB1-RAGE in Kawasaki disease with coronary artery injury. Eur J Med Res. 2020;25(1):8. doi:10.1186/s40001-020-00406-5. [Google Scholar] [CrossRef]
52. Sun J, Jiang Y, Li L, Li R, Ling F, Du X, et al. HMGB1/RAGE signaling regulates Th17/IL-17 and its role in bronchial epithelial-mesenchymal transformation. Curr Mol Med. 2023;24:1401–12. [Google Scholar]
53. Watanabe T, Asai K, Fujimoto H, Tanaka H, Kanazawa H, Hirata K. Increased levels of HMGB-1 and endogenous secretory RAGE in induced sputum from asthmatic patients. Respir Med. 2011;105(4):519–25. doi:10.1016/j.rmed.2010.10.016. [Google Scholar] [CrossRef]
54. Halayko AJ, Ghavami S. S100A8/A9: a mediator of severe asthma pathogenesis and morbidity? Can J Physiol Pharmacol. 2009;87(10):743–55. doi:10.1139/Y09-054. [Google Scholar] [CrossRef]
55. Milutinovic PS, Alcorn JF, Englert JM, Crum LT, Oury TD. The receptor for advanced glycation end products is a central mediator of asthma pathogenesis. Am J Pathol. 2012;181(4):1215–25. doi:10.1016/j.ajpath.2012.06.031. [Google Scholar] [CrossRef]
56. Akirav EM, Henegariu O, Preston-Hurlburt P, Schmidt AM, Clynes R, Herold KC. The receptor for advanced glycation end products (RAGE) affects T cell differentiation in OVA induced asthma. PLoS One. 2014;9(4):e95678. doi:10.1371/journal.pone.0095678. [Google Scholar] [CrossRef]
57. Celli B, Fabbri L, Criner G, Martinez FJ, Mannino D, Vogelmeier C, et al. Definition and nomenclature of chronic obstructive pulmonary disease: time for its revision. Am J Respir Crit Care Med. 2022;206(11):1317–25. doi:10.1164/rccm.202204-0671PP. [Google Scholar] [CrossRef]
58. Zhao X, Zhang Q, Zheng R. The interplay between oxidative stress and autophagy in chronic obstructive pulmonary disease. Front Physiol. 2022;13:1004275. doi:10.3389/fphys.2022.1004275. [Google Scholar] [CrossRef]
59. Castaldi PJ, Cho MH, San José Estépar R, McDonald ML, Laird N, Beaty TH, et al. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190(4):399–409. doi:10.1164/rccm.201403-0569OC. [Google Scholar] [CrossRef]
60. Li Y, Yang C, Ma G, Gu X, Chen M, Chen Y, et al. Association of polymorphisms of the receptor for advanced glycation end products gene with COPD in the Chinese population. DNA Cell Biol. 2014;33(4):251–8. doi:10.1089/dna.2013.2303. [Google Scholar] [CrossRef]
61. Lin L, Li J, Song Q, Cheng W, Chen P. The role of HMGB1/RAGE/TLR4 signaling pathways in cigarette smoke-induced inflammation in chronic obstructive pulmonary disease. Immun Inflammation Dis. 2022;10(11):e711. doi:10.1002/iid3.711. [Google Scholar] [CrossRef]
62. Marin-Oto M, Sanz-Rubio D, Santamaría-Martos F, Benitez I, Simon AL, Forner M, et al. Soluble RAGE in COPD, with or without coexisting obstructive sleep apnoea. Respir Res. 2022;23(1):163. doi:10.1186/s12931-022-02092-9. [Google Scholar] [CrossRef]
63. Chen L, Sun X, Zhong X. Role of RAGE and its ligand HMGB1 in the development of COPD. Postgrad Med. 2022;134(8):763–75. doi:10.1080/00325481.2022.2124087. [Google Scholar] [CrossRef]
64. Röhl A, Baek SH, Kachroo P, Morrow JD, Tantisira K, Silverman EK, et al. Protein interaction networks provide insight into fetal origins of chronic obstructive pulmonary disease. Respir Res. 2022;23(1):69. doi:10.1186/s12931-022-01963-5. [Google Scholar] [CrossRef]
65. Yonchuk JG, Silverman EK, Bowler RP, Agustí A, Lomas DA, Miller BE, et al. Circulating soluble receptor for advanced glycation end products (sRAGE) as a biomarker of emphysema and the RAGE axis in the lung. Am J Respir Crit Care Med. 2015;192(7):785–92. doi:10.1164/rccm.201501-0137PP. [Google Scholar] [CrossRef]
66. Gopal P, Reynaert NL, Scheijen JL, Schalkwijk CG, Franssen FM, Wouters EF, et al. Association of plasma sRAGE, but not esRAGE with lung function impairment in COPD. Respir Res. 2014;15(1):24. doi:10.1186/1465-9921-15-24. [Google Scholar] [CrossRef]
67. Chen P, Liu Y, Wen Y, Zhou C. Non-small cell lung cancer in China. Cancer Commun. 2022;42(10):937–70. doi:10.1002/cac2.v42.10. [Google Scholar] [CrossRef]
68. Yu YX, Pan WC, Cheng YF. Silencing of advanced glycosylation and glycosylation and product-specific receptor (RAGE) inhibits the metastasis and growth of non-small cell lung cancer. Am J Transl Res. 2017;9(6):2760–74. [Google Scholar] [PubMed]
69. Faruqui T, Khan MS, Akhter Y, Khan S, Rafi Z, Saeed M, et al. RAGE inhibitors for targeted therapy of cancer: a comprehensive review. Int J Mol Sci. 2023;24(1):266. [Google Scholar]
70. Liao YF, Yin S, Chen ZQ, Li F, Zhao B. High glucose promotes tumor cell proliferation and migration in lung adenocarcinoma via the RAGE‐NOXs pathway. Mol Med Rep. 2018;17(6):8536–41. [Google Scholar] [PubMed]
71. Gibadulinova A, Tothova V, Pastorek J, Pastorekova S. Transcriptional regulation and functional implication of S100P in cancer. Amino Acids. 2011;41(4):885–92. doi:10.1007/s00726-010-0495-5. [Google Scholar] [CrossRef]
72. Saito K, Kobayashi M, Nagashio R, Ryuge S, Katono K, Nakashima H, et al. S100A16 is a prognostic marker for lung adenocarcinomas. Asian Pac J Cancer Prev. 2015;16(16):7039–44. doi:10.7314/APJCP.2015.16.16.7039. [Google Scholar] [PubMed] [CrossRef]
73. Kinoshita R, Sato H, Yamauchi A, Takahashi Y, Inoue Y, Sumardika IW, et al. exSSSRs (extracellular S100 soil sensor receptors)-Fc fusion proteins work as prominent decoys to S100A8/A9-induced lung tropic cancer metastasis. Int J Cancer. 2018;144(12):3138–45. [Google Scholar] [PubMed]
74. Choi H, Puvenna V, Brennan C, Mahmoud S, Wang XF, Phillips M, et al. S100B and S100B autoantibody as biomarkers for early detection of brain metastases in lung cancer. Transl Lung Cancer Res. 2016;5(4):413–9. doi:10.21037/tlcr. [Google Scholar] [CrossRef]
75. Riuzzi F, Sorci G, Donato R. S100B protein regulates myoblast proliferation and differentiation by activating FGFR1 in a bFGF-dependent manner. J Cell Sci. 2011;124(14):2389–400. doi:10.1242/jcs.084491. [Google Scholar] [CrossRef]
76. Li C, Kuang K, Du J, Eymin B, Jia T. Far beyond anti-angiogenesis: benefits for anti-basicFGF therapy in cancer. Biochim et Biophys Acta—Mol Cell Res. 2022;1869(7):119253. doi:10.1016/j.bbamcr.2022.119253. [Google Scholar] [CrossRef]
77. Pan Y, Han H, Labbe KE, Zhang H, Wong KK. Recent advances in preclinical models for lung squamous cell carcinoma. Oncogene. 2021;40(16):2817–29. doi:10.1038/s41388-021-01723-7. [Google Scholar] [CrossRef]
78. Stav D, Bar I, Sandbank J. Usefulness of CDK5RAP3, CCNB2, and RAGE genes for the diagnosis of lung adenocarcinoma. Int J Biol Markers. 2007;22(2):108–13. doi:10.1177/172460080702200204. [Google Scholar] [CrossRef]
79. Kobayashi S, Kubo H, Suzuki T, Ishizawa K, Yamada M, He M, et al. Endogenous secretory receptor for advanced glycation end products in non-small cell lung carcinoma. Am J Respir Crit Care Med. 2007;175(2):184–9. doi:10.1164/rccm.200602-212OC. [Google Scholar] [CrossRef]
80. Kalea AZ, See F, Harja E, Arriero M, Schmidt AM, Hudson BI. Alternatively spliced RAGEv1 inhibits tumorigenesis through suppression of JNK signaling. Cancer Res. 2010;70(13):5628–38. doi:10.1158/0008-5472.CAN-10-0595. [Google Scholar] [CrossRef]
81. Mukherjee TK, Mukhopadhyay S, Hoidal JR. Implication of receptor for advanced glycation end product (RAGE) in pulmonary health and pathophysiology. Respir Physiol Neurobiol. 2008;162(3):210–5. doi:10.1016/j.resp.2008.07.001. [Google Scholar] [CrossRef]
82. Rojas A, Figueroa H, Morales E. Fueling inflammation at tumor microenvironment: the role of multiligand/RAGE axis. Carcinogenesis. 2010;31(3):334–41. doi:10.1093/carcin/bgp322. [Google Scholar] [PubMed] [CrossRef]
83. Jing R, Cui M, Wang J, Wang H. Receptor for advanced glycation end products (RAGE) soluble form (sRAGEa new biomarker for lung cancer. Neoplasma. 2010;57(1):55. doi:10.4149/neo_2010_01_055. [Google Scholar] [PubMed] [CrossRef]
84. Bos LD, Ware LB. Acute respiratory distress syndrome: causes, pathophysiology, and phenotypes. The Lancet. 2022;400(10358):1145–56. doi:10.1016/S0140-6736(22)01485-4. [Google Scholar] [CrossRef]
85. Meyer NJ, Gattinoni L, Calfee CS. Acute respiratory distress syndrome. The Lancet. 2021;398(10300):622–37. doi:10.1016/S0140-6736(21)00439-6. [Google Scholar] [CrossRef]
86. Han J, Li H, Bhandari S, Cao F, Wang XY, Tian C, et al. Maresin conjugates in tissue regeneration 1 improves alveolar fluid clearance by up-regulating alveolar ENaC, Na, K-ATPase in lipopolysaccharide-induced acute lung injury. J Cell Mol Med. 2020;24(8):4736–47. doi:10.1111/jcmm.15146. [Google Scholar] [CrossRef]
87. Blondonnet R, Audard J, Belville C, Clairefond G, Lutz J, Bouvier D, et al. RAGE inhibition reduces acute lung injury in mice. Sci Rep. 2017;7(1):7208. doi:10.1038/s41598-017-07638-2. [Google Scholar] [CrossRef]
88. Jabaudon M, Berthelin P, Pranal T, Roszyk L, Godet T, Faure J-S, et al. Receptor for advanced glycation end-products and ARDS prediction: a multicentre observational study. Sci Rep. 2018;8(1):2603. doi:10.1038/s41598-018-20994-x. [Google Scholar] [CrossRef]
89. Yang Q, Yan CS, Wang YF, Li XY, Zhang WM, Xiao Y, et al. Serial changes in biomarkers in pulmonary and extrapulmonary acute respiratory distress syndrome. Zhonghua Yi Xue Za Zhi. 2019;99(36):2816–9 (In Chinese). [Google Scholar] [PubMed]
90. Englert JM, Hanford LE, Kaminski N, Tobolewski JM, Tan RJ, Fattman CL, et al. A role for the receptor for advanced glycation end products in idiopathic pulmonary fibrosis. Am J Pathol. 2008;172(3):583–91. doi:10.2353/ajpath.2008.070569. [Google Scholar] [PubMed] [CrossRef]
91. Queisser MA, Kouri FM, Königshoff M, Wygrecka M, Schubert U, Eickelberg O, et al. Loss of RAGE in pulmonary fibrosis: molecular relations to functional changes in pulmonary cell types. Am J Respir Cell Mol Biol. 2008;39(3):337–45. doi:10.1165/rcmb.2007-0244OC. [Google Scholar] [CrossRef]
92. He M, Kubo H, Ishizawa K, Hegab AE, Yamamoto Y, Yamamoto H, et al. The role of the receptor for advanced glycation end-products in lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2007;293(6):L1427–36. doi:10.1152/ajplung.00075.2007. [Google Scholar] [CrossRef]
93. Morbini P, Villa C, Campo I, Zorzetto M, Inghilleri S, Luisetti M. The receptor for advanced glycation end products and its ligands: a new inflammatory pathway in lung disease? Mod Pathol. 2006;19(11):1437–45. doi:10.1038/modpathol.3800661. [Google Scholar] [CrossRef]
94. Borges do Nascimento IJ, Cacic N, Abdulazeem HM, von Groote TC, Jayarajah U, Weerasekara I, et al. Novel coronavirus infection (COVID-19) in humans: a scoping review and meta-analysis. J Clin Med. 2020;9(4):941. doi:10.3390/jcm9040941. [Google Scholar] [CrossRef]
95. Angioni R, Bonfanti M, Caporale N, Sánchez-Rodríguez R, Munari F, Savino A, et al. RAGE engagement by SARS-CoV-2 enables monocyte infection and underlies COVID-19 severity. Cell Rep Med. 2023;4(11):101266. doi:10.1016/j.xcrm.2023.101266. [Google Scholar] [CrossRef]
96. Basile MS, Cavalli E, McCubrey J, Hernández-Bello J, Muñoz-Valle JF, Fagone P, et al. The PI3K/Akt/mTOR pathway: a potential pharmacological target in COVID-19. Drug Discov Today. 2022;27(3):848–56. doi:10.1016/j.drudis.2021.11.002. [Google Scholar] [CrossRef]
97. Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C, et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11):1317–29. doi:10.1182/blood.2020007214. [Google Scholar] [CrossRef]
98. Holms RD. Long COVID (PASC) is maintained by a self-sustaining pro-inflammatory TLR4/RAGE-Loop of S100A8/A9 > TLR4/RAGE signalling, inducing chronic expression of IL-1b, IL-6 and TNFa: anti-inflammatory ezrin peptides as potential therapy. Immuno. 2022;2(3):512–33. doi:10.3390/immuno2030033. [Google Scholar] [CrossRef]
99. Murphy SL, Halvorsen B, Holter JC, Huse C, Tveita A, Trøseid M, et al. Circulating markers of extracellular matrix remodelling in severe COVID-19 patients. J Intern Med. 2023;294(6):784–97. doi:10.1111/joim.v294.6. [Google Scholar] [CrossRef]
100. Mester P, Keller D, Kunst C, Räth U, Rusch S, Schmid S, et al. High serum S100A12 as a diagnostic and prognostic biomarker for severity, multidrug-resistant bacteria superinfection and herpes simplex virus reactivation in COVID-19. Viruses. 2024;16(7):1084. doi:10.3390/v16071084. [Google Scholar] [CrossRef]
101. Chen R, Huang Y, Quan J, Liu J, Tang D. HMGB1 as a potential biomarker and therapeutic target for severe COVID-19. Heliyon. 2020;6(12):e05672. doi:10.1016/j.heliyon.2020.e05672. [Google Scholar] [CrossRef]
102. Wulandari S, Hartono WT. The role of HMGB1 in COVID-19-induced cytokine storm and its potential therapeutic targets: a review. Immunology. 2023;169(2):117–31. doi:10.1111/imm.v169.2. [Google Scholar] [CrossRef]
103. Ahirwar AK, Kaim K, Ahirwar P, Kumawat R. The second wave of COVID-19 results in outbreak of mucormycosis: diabetes and immunological perspective. Horm Mol Biol Clin Investig. 2022;43(3):353–5. doi:10.1515/hmbci-2021-0072. [Google Scholar] [CrossRef]
104. Wolszczak-Biedrzycka B, Dorf J, Matowicka-Karna J, Wojewódzka-Żeleźniakowicz M, Żukowski P, Zalewska A, et al. Significance of nitrosative stress and glycoxidation products in the diagnosis of COVID-19. Sci Rep. 2024;14(1):9198. doi:10.1038/s41598-024-59876-w. [Google Scholar] [CrossRef]
105. Pedreañez A, Mosquera-Sulbaran JA, Tene D. Role of the receptor for advanced glycation end products in the severity of SARS-CoV-2 infection in diabetic patients. Diabetol Int. 2024. doi:10.1007/s13340-024-00746-1. [Google Scholar] [CrossRef]
106. Cao LH, Jia XY, He HJ, Wang ZZ, Zhao YY, Yang X, et al. Using a system pharmacology method to search for the potential targets and pathways of Yinqiaosan against COVID-19. J Healthc Eng. 2022;2022:9248674. doi:10.1155/2022/9248674. [Google Scholar] [CrossRef]
107. Hong J, Ku SH, Lee MS, Jeong JH, Mok H, Choi D, et al. Cardiac RNAi therapy using RAGE siRNA/deoxycholic acid-modified polyethylenimine complexes for myocardial infarction. Biomaterials. 2014;35(26):7562–73. doi:10.1016/j.biomaterials.2014.05.025. [Google Scholar] [CrossRef]
108. Lutterloh EC, Opal SM. Antibodies against RAGE in sepsis and inflammation: implications for therapy. Expert Opin Pharmacother. 2007;8(9):1193–6. doi:10.1517/14656566.8.9.1193. [Google Scholar] [CrossRef]
109. Nishad R, Meshram P, Singh AK, Reddy GB, Pasupulati AK. Activation of Notch1 signaling in podocytes by glucose-derived AGEs contributes to proteinuria. BMJ Open Diabetes Res Care. 2020;8(1):e001203. doi:10.1136/bmjdrc-2020-001203. [Google Scholar] [CrossRef]
110. Zhang IY, Zhou H, Liu H, Zhang L, Gao H, Liu S, et al. Local and systemic immune dysregulation alters glioma growth in hyperglycemic mice. Clin Cancer Res. 2020;26(11):2740–53. doi:10.1158/1078-0432.CCR-19-2520. [Google Scholar] [CrossRef]
111. Zhang C, Wang L, Xu Y, Huang Y, Huang J, Zhu J, et al. Discovery of novel dual RAGE/SERT inhibitors for the potential treatment of the comorbidity of Alzheimer’s disease and depression. Eur J Med Chem. 2022;236(289):114347. doi:10.1016/j.ejmech.2022.114347. [Google Scholar] [CrossRef]
112. Cai C, Dai X, Zhu Y, Lian M, Xiao F, Dong F, et al. A specific RAGE-binding peptide biopanning from phage display random peptide library that ameliorates symptoms in amyloid β peptide-mediated neuronal disorder. Appl Microbiol Biotechnol. 2016;100(2):825–35. doi:10.1007/s00253-015-7001-7. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools