 Open Access
Open Access
REVIEW
The roles and mechanisms of miRNA in HBV-HCC carcinogenesis: Why no therapeutic agents after 30 years?
1 Hepatitis Diversity Research Unit, School of Internal Medicine, University of the Witwatersrand, Johannesburg, 2050, South Africa
2 School of Laboratory Medicine and Molecular Sciences, University of KwaZulu-Natal, Durban, 4041, South Africa
3 Africa HepatoPancreatoBiliary Cancer Consortium (AHPBCC), Mayo Clinic, Jacksonville, MN 55902, USA
4 Centre for Clinical Research (UQCCR), Faculty of Medicine, University of Queensland, Brisbane, NSW2580, Australia
5 Basic Research Laboratory, Centre for Cancer Research, National Cancer Institute, Leidos Biomedical Research, Inc., Frederick Nat. Lab. for Cancer Research, Frederick, MD 240, USA
6 Department of Hepatobiliary and Pancreatic Surgery, The First Affiliated Hospital of Soochow University, Suzhou, 215100, China
7 Department of General Surgery, Wujin Hospital Affiliated with Jiangsu University, Zhenjiang, 212013, China
* Corresponding Authors: KURT SARTORIUS. Email: ; YUNJIE LU. Email:
(This article belongs to the Special Issue: Non-coding RNAs (ncRNAs) in Human Diseases)
BIOCELL 2024, 48(11), 1543-1567. https://doi.org/10.32604/biocell.2024.055505
Received 28 June 2024; Accepted 11 September 2024; Issue published 07 November 2024
Abstract
Hepatitis B-associated hepatocellular carcinoma (HBV-HCC) remains an intractable high-mortality solid tumor cancer that accounted for 42% of global HCC cases in 2019. Despite some developments in systemic therapy, only a small subset of late-stage HCC patients responds positively to recently developed therapeutic innovations. MicroRNAs (miRNAs) act as an ancillary epigenetic system that can regulate genome expression in all cancer pathways including HCC. The molecular mechanisms of miRNA regulation in cancer pathogenesis offered researchers a new approach that was widely hoped would translate into miRNA-based drugs and diagnostics. Thirty years on, miRNA-based diagnostic and therapeutic agents for HCC remain a work-in-progress (WIP) and no current miRNA HCC clinical trial has progressed to Phase 4. The question remains why this is the case after 30 years and what is the way forward. The major findings and contribution of this paper are that it illustrates the complexity of the HBV-miRNA interactome in HBV-HCC in all cellular processes, as well as the ancillary role of miRNA in the epigenetic and immune systems. This is combined with a review of the outcomes and problems of clinical trials, to explain why miRNA therapeutics and diagnostics have not progressed to approved drugs or serum-based diagnostic tests. The way forward suggests a radical rethink might be so that involves the incorporation of AI, bioinformatics, and nanotechnology to solve the problem.Keywords
Schematic: The roles and mechanisms of miRNA in HBV-HCC carcinogenesis: Why no therapeutic agents after 30 years?
• Introduction to miRNA biology and dysregulation in HBV-HCC
• MiRNA regulation in the pre-malignant HBV-HCC environment
• MiRNA regulation of cellular processes in HBV-HCC
• MiRNA as an ancillary epigenetic system
• MiRNA interaction with the immune system
• MiRNA therapeutic developments and problems
• MiRNA diagnostic biomarker developments and problems
• The way forward
• Conclusion and limitations
In 2019, hepatitis B-associated hepatocarcinoma (HBV-HCC) still accounted for 42% of global incidence despite the global ASR per 100,000 declining from 3.557 to 2.478 between 1990 and 2019 [1,2]. Moreover, in the same period, the incidence of HBV-HCC rose significantly in Australasia, Central Asia, Eastern Europe, Asia Pacific, and North America [1]. Surgical resection remains the most effective intervention for early-stage disease while new developments deploying a combination of immunotherapy and targeted therapy (atezolizumab plus bevacizumab) have yielded modest results [3,4]. Despite these developments in systemic therapy, only a small subset of late-stage HCC patients respond positively to these innovations [5]. Recent evidence also suggests postoperative patients (resection) with HBV-HCC have a worse prognosis than non HBV-HCC cases [6]. MicroRNAs (miRNAs), first identified in the 1990s, act as an ancillary epigenetic system that is capable of fine-tuning mRNA translation in all solid tumor cancer pathways including HCC [7]. In the non-coding RNA (ncRNA) HBV-HCC interactome, multiple miRNAs regulate host and viral gene expression in HBV-HCC pathogenesis from early HBV infection in the pre-malignant environment (PME) to the onset of HCC [8]. The miRNA regulatory interactome in HBV-HCC, which regulates host and viral genomic expression, includes its interaction with the mainstream epigenetic machinery, the innate and adaptive immune systems, and another ncRNA like long non-coding RNA (lncRNA) and circular RNA (circRNA) [9–11]. The molecular role of miRNA in cancer pathogenesis offered researchers a new approach based on regulating oncogenic mRNA expression. It was, thus, widely hoped that miRNA-based therapeutics and diagnostics would translate into novel systemic drugs and liquid biopsy based diagnostic biomarkers to detect early-stage HCC [12,13]. Thirty years on, and thousands of research studies later, miRNA-based therapeutics and diagnostics for HBV-HCC remain a work-in-progress (WIP) and a recent systematic review indicates no miRNA HCC clinical trial has progressed to Stage 4 [14]. The question remains why this is the case after 30 years and what is the way forward?
To answer this question, this paper first reviews the roles and molecular mechanisms of miRNA in HBV-HCC pathogenesis to illustrate the complexity of the miRNA interactome. The paper then provides an explanation that contributes to understanding why miRNA-based therapeutics and diagnostics have not translated into approved drugs and diagnostic tests. More specifically, the paper first introduces the regulatory mechanism of miRNAs and how their expression is dysregulated in HBV-HCC pathogenesis. The regulatory role of miRNAs is then outlined in the pre-malignant micro-environment (PME) of chronic hepatitis B infection (CHB), inflammation, and the onset of fibrosis. Next, we review the multiplicity of the Hepatitis B x protein (HBx) dysregulated miRNA, and their molecular mechanisms, in the HCC tumor microenvironment (TME) including cell proliferation, apoptosis, angiogenesis, invasion-migration, and metastasis in their respective HBV-HCC signaling pathways. The complexity of the miRNA interactome is further illustrated by the regulatory role of miRNAs as an ancillary epigenetic system before examining miRNA interaction with immune response in HBV-HCC pathogenesis. The paper then assesses the status quo of miRNA-based therapeutic and diagnostic developments before outlining the problems that explain why this field is still in the work-in-progress phase. A way forward is then discussed and a conclusion is developed highlighting the contributions and limitations of this paper.
The Function and Dysregulation of miRNA
Dynamic combinations of miRNAs act as a homeostatic ancillary epigenetic system that fine-tune mRNA translation in all cellular processes [15–17] and a recent estimate indicated that 2300 classified human miRNAs are under the transcriptional control of ~1000 genes [18]. In order to exert a regulatory role, miRNAs form Argonaut (AG) based RNA-induced silencing complexes (RISCs) that bind to specific mRNA through complementarity sequences that result in either the inhibition of mRNA translation and/or de-adenylation followed by mRNA decay [19]. The degree of silencing and/or degradation of miRNA-mRNA translation is a function of the degree of complementarity of their sequence specific bases in the 5′UTR region with the corresponding target mRNA 3′UTR region [20]. It has been demonstrated that intracellular miRNA silencing often occurs at the surface of the endoplasmic reticulum (ER) because this is the primary location of mRNA protein synthesis and RISC loading. Mature miRNAs in the intracellular space can be packaged and transported in a variety of protein and membranous bodies to regulate mRNA expression in the cytosol [16]. Mature miRNAs can also be exported to the extracellular space (serum) in a range of vesicles including exosomes, micro-vesicles, apoptotic bodies, high-density lipoprotein (HDL) complexes, and other AG complexes [21]. Solid evidence has emerged that these extracellular miRNAs contain paracrine mRNA silencing capability [22], however, the extent of their regulatory ability remains in question [23,24].
The molecular mechanisms of miRNA dysregulation in HBV-HCC pathogenesis include genomic alterations, mainstream epigenetic expression, defects in the miRNA biogenesis machinery, immune response, co-expression with host genes, the presence of carcinogens, viral infection [25–27] and other competing ncRNA like lncRNA and circRNA [28]. HBV expression of viral proteins like HBx, for instance, dysregulates multiple miRNAs in every stage of pathogenesis by inducing host genomic and epigenetic changes from early infection in the PME to the onset of HBV-HCC [8]. Dysregulation can also occur at any stage in the miRNA transcription machinery from the early pri-miRNA stage to the production and packaging of mature miRNAs because of various processing defects [29]. In HBV-HCC pathogenesis, dysregulated miRNAs can potentially regulate both pro and anti-oncogenic expression [30].
The Regulatory Role of miRNA in the Pre-Malignant Micro-Environment (PME)
An understanding of the role of miRNA regulation in every stage of HBV-HCC pathogenesis, from the PME to the tumor-microenvironment (TME), is especially important because chronic hepatitis B infection (CHB) triggers inflammation, injury, and tissue replacement that collectively dysregulates multiple miRNAs [8,31]. For example, one study indicated >80 dysregulated miRNAs in the PME as a result of CHB-induced inflammation including important miRNAs like miR-195a-5p/-1974/-203a/-21/-22/-210/-34a/-451/-548d-5p/-654/-711/-760/-767-3p [8]. Interestingly, many of these miRNAs are also dysregulated in HBV-HCC serum [32]. MiRNA regulation in the PME is both complex and (often) contradictory and three miRNAs, namely, miR-155/-122/-let-7 illustrate some of these regulatory complexities in early stage CHB and inflammation. In one feedback loop, the HBx protein can upregulate miR-155 expression to subdue HBV replication by repressing CCAAT-enhancer-binding protein (C/EBP) induced enhancement of HBV core promoters [33,34]. Pro-inflammatory cytokines in the PME can also upregulate miR-155 expression which can repress tumor suppressors like PTEN and C/EBPβ to promote pro-oncogenic progress in the PME [33]. HBV infection can downregulate miR-122 which, in turn, can also repress HBV replication by modulating cell cycle controls like cyclin G1 and by binding to HBV mRNA and HBx [35–37]. This important liver miRNA also acts as an anti-inflammatory agent by repressing inflammatory cytokines like Interleukin 6 (IL-6) and Tumor necrosis factor-alpha (TNF-α) [38], however, these inflammatory cytokines can induce C-MYC inhibition of miR-122 expression in CHB [39]. In early-stage HBV infection, all the let-7 family members have been reported as significantly downregulated by the HBx protein resulting in the reduced modulation of multiple pro-inflammatory cytokines, interleukins, activated Kupffer cells, and liver-derived macrophages [40,41] that promote a wide range of cellular responses in injured liver tissue [42,43].
A hallmark of HBV-HCC is the presence of fibrosis/cirrhosis that often precedes the onset of HBV-HCC for many years. In the inflammation–fibrosis axis, fibrogenesis is orchestrated by a complex network of common cytokine-mediated signaling pathways that regulate the activation of hepatic stellate cells (HSCs) and downstream extracellular matrix (ECM) proteins (see Fig. 1). These cytokines include Transforming growth factor-beta (TGF-β), platelet-derived growth factor (PDGF), TNF-α, interferons (IFNα/β), and interleukins (IL-1/6/17) [42,43]. One study showed that >75 dysregulated miRNA could potentially regulate liver fibrogenesis in the PME illustrating a range of miRNA that directly target the TGF-β pathway in fibrogenesis including miR-17-5p/-122-5p/-181b/-21/-23a-5p/-26b/-27a-b/-30/-34a/-148a/-150/-151b/-152/-214-3p/-221-5p/-222/-29a-b/-33a/-942/-192/-200 [8]. Upregulated miRNA can either repress or attenuate fibrogenesis. Upregulated miR-17-5p/-21/33a promote fibrogenesis by repressing the SMAD 7 inhibitor [44–47] and miR-29a-b/-214-5p attenuate fibrogenesis by repressing growth factors and TGF-β signaling [48,49]. Important downregulated miRNA like miR-29a/-29b/-34a/-122 mostly fail to repress fibrogenesis because they have a reduced ability to repress collagens, growth factors (GFs), and SMAD 4 [40,44,50].
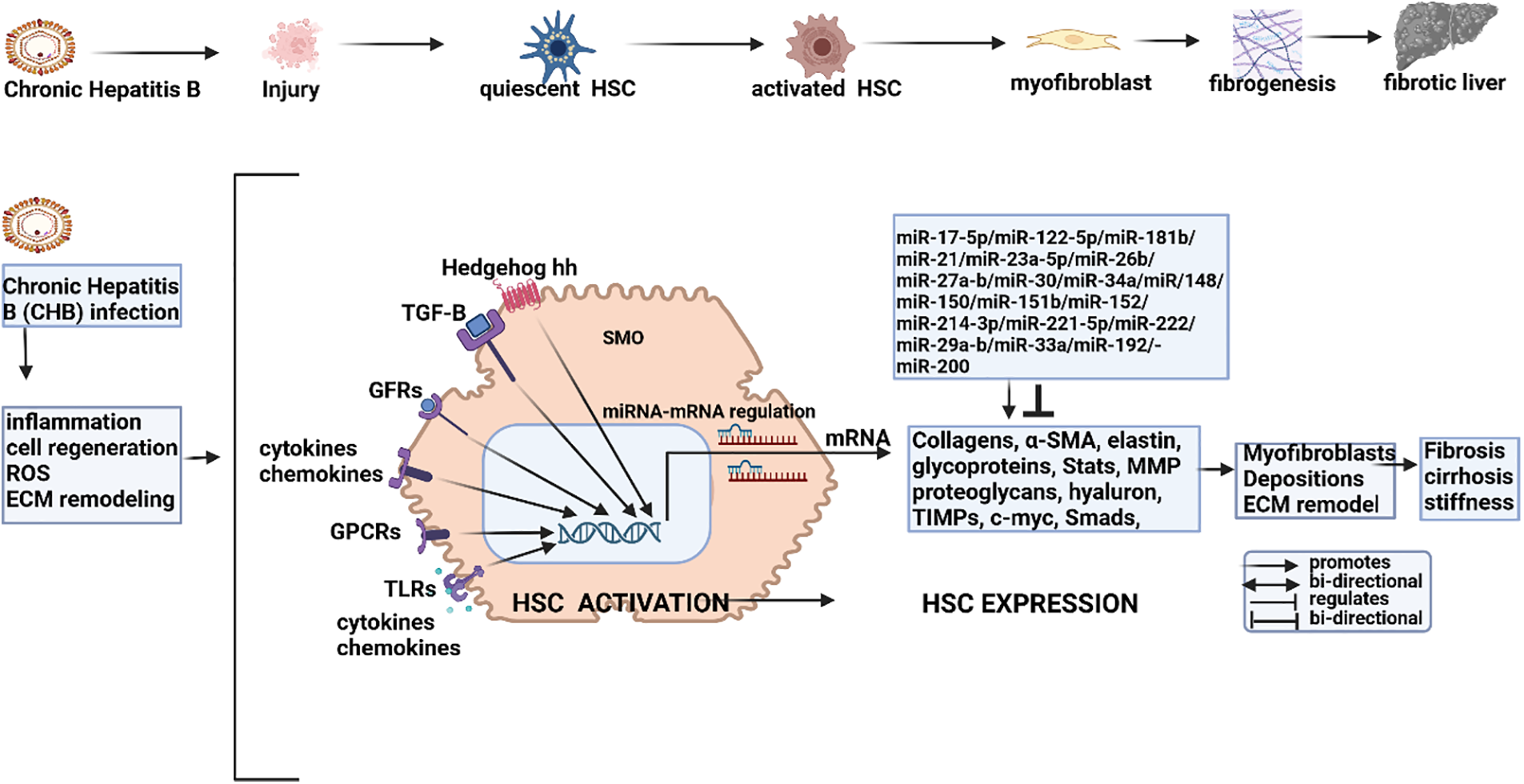
Figure 1: miRNA modulation in HBV-induced fibrogenesis. Chronic Hepatitis B infection (CHB) induces inflammation and injury to activate quiescent hepatic stellate cells (HSCs) to promote fibrogenic materials like myofibroblasts and fibrosis. CHB also promotes cell regeneration, oxidative stress (ROS), and ECM remodeling. Collectively, these conditions trigger the activation of HSCs by promoting the expression of growth factors (GFs), protein-coupled receptors (GPCRs), cytokines and chemokines, signaling pathways (e.g., TGF-B and Hedgehog) and toll-like receptors (TLRs). Activated HSCs promote the transcription of a wide range of fibrogenic mRNA including collagens, α-SMA, glycoproteins, metalloproteinases (MMPs), proteoglycans, tissue inhibitors of metalloproteinases (TIMPs) and onco-proteins like β-catenin and C-MYC. These mRNA, in turn, are modulated by ncRNA (miRNA/lncRNA/circ-RNA) that, in turn, are modulated by the mainstream epigenetic machinery including DNA methyl transferases (DNMTs), histone de-acetyltransferases (HDACs), histone acetyltransferases (HATs), histone methyl transferases (HMTs) and histone de-methylation transferases (HDMTs). The transcription of fibrotic depositions and ECM remodeling is thus modulated by a combination of ncRNA and the mainstream epigenic machinery to determine fibrogenesis, ECM remodeling, and eventually advanced fibrosis/cirrhosis (Figures done in Biorender).
The three important miRNAs, namely, miR-155/-122/let-7a, not only modulate viral expression and inflammation but also play a regulatory role in liver fibrosis. Upregulated miR-155 can invoke pro-inflammatory induced liver fibrogenesis [51] via the Signal transducer and activator of the transcription 3 (STAT3) pathway [52] and promote liver fibrosis by repressing suppressor of cytokine signaling 1 (SOCS1) [53]. miR-122 can promote fibrosis by repressing Insulin-like growth factor 1 receptor (IGF1R), Cyclin G1 (CCNG1) and Prolyl 4-hydroxylase subunit alpha-1(P4HA1) to induce hepatic stellate cell (HSC) activation and the expression of collagen [54] and let-7 family members can regulate TGF-β expression that activates HSCs and the expression of multiple fibrogenic materials [55]. Illustrating the complexity of miRNA regulation in this microenvironment, HBx can suppress p53-led transcription of the miR-192/-200 cluster that then reduces its regulation of Zinc Finger E-Box Binding Homeobox 1/2 (ZEB1/2) resulting in a reduction in E-cadherin in the WNT/β-catenin pathway that is often an early feature of fibrosis/epithelial-mesenchymal transition (EMT) in the PME [56–58].
Molecular Mechanisms of miRNA Regulation in HBV-HCC Pathways
This section illustrates how miRNAs regulate cell proliferation, apoptosis, angiogenesis, migration-invasion-metastasis, and cell cycle in the main HBV-HCC pathways and networks (see Figs. 2 and 3, Table 1). Collectively, miRNAs regulate oncogenic or tumor suppressor expression in multiple pathways and networks in HBV-HCC pathogenesis including the TP53, PI3K/MAPK, JAK/STAT, WNT/ β-catenin, TGF-B/SMAD, SAPK/JNK and NF-κB signaling pathways [59]. In HBV-HCC pathogenesis, moreover, chronic hepatitis B infection dysregulates both host gene expression, as well as miRNAs to (stealthily) balance viral replication with immune evasion [9]. In many cases, miRNAs recorded as dysregulated in the PME remain dysregulated in HBV-HCC pathogenesis, however, their mRNA targets and direction of dysregulation are often different [32].
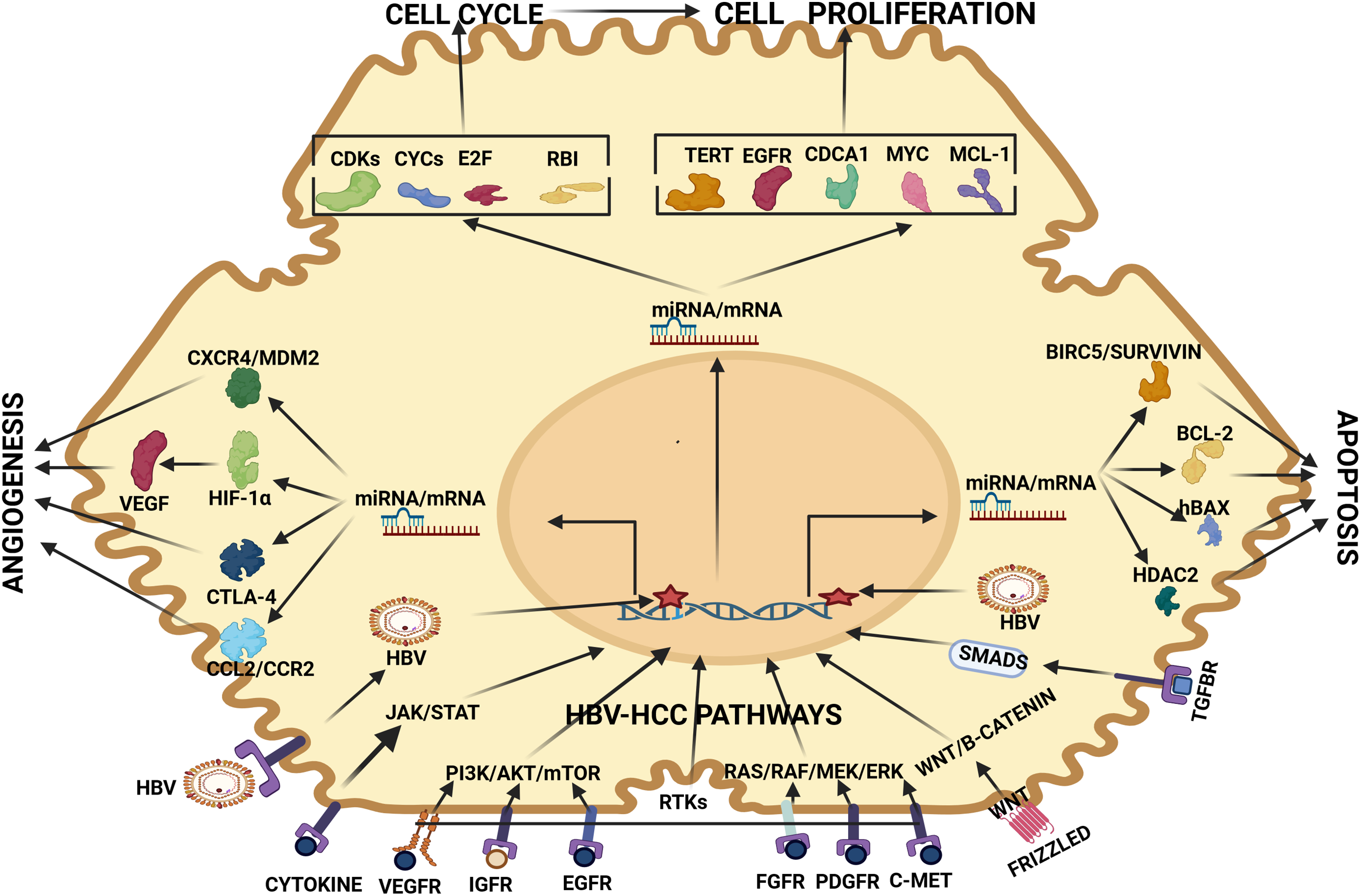
Figure 2: miRNA regulation of HBV-HCC pathways. HBV-HCC: The HBV virion enters the host cell and integrates into the host DNA causing genomic changes. In tandem, the TME activates HBV-HCC pathways like JAK/STAT, PI3K/AKT, RAS/RAF, WNT/β-catenin and TGF-SMAD via multiple cell receptors to activate miRNA/mRNA expression to influence cell cycle, proliferation, apoptosis, and angiogenesis. HBV-HCC Receptors include VEGFR, IGFR, EGFR, FGFR, PDGFR, C-MET, HGFR, WNT that promote transcription of mRNA/miRNA to influence Cell Cycle mRNA: miRNA regulates cell cycle mRNA including CYCs, CDKs, E2F and RB1, Cell proliferation mRNA: miRNA regulate mRNA like TERT, EGFR, CDCA1, C-MYC, MCL-1, and ARL4C, Apoptosis mRNA: miRNA regulate mRNA like BIRC5/Survivin, BCL-2, hBAX, HDAC2 to influence apoptosis and Angiogenesis mRNA: miRNA regulates CXCR4/MDM2, CTLA-4, HIF-1α and VEGF to influence angiogenesis. Abbreviations: VEGFR: Vascular endothelial growth factor receptor; IGFR: Insulin-like growth factor receptor; EGFR: Epidermal growth factor receptor; FGFR: Fibroblast growth factor receptor; PDGFR: Platelet-derived growth factor receptor; C-MET: Receptor tyrosine kinase; TGF-BR: Transforming growth factor-beta receptor; PI3K: Phosphatidylinositol 3-kinases; AKT: Protein kinase B; mTOR: mammalian target of rapamycin; RAS: Rat sarcoma virus; RAF: Rapidly Accelerated Fibrosarcoma; MEK: Mitogen-activated protein kinase; ERK: Extracellular signal-regulated kinase; WNT: Wingless and Int-1 SMADs: Suppressor of mother against decapentaplegic; HBV: Hepatitis B virus; HDAC2: Histone de-acetylase 2; hBAX: Bcl-2 Associated X-protein; BCL-2: B-cell lymphoma 2; BIRC-5: Baculoviral IAP Repeat Containing 5; MCL-1: Myeloid cell leukemia-1; MYC: Myelocytomatosis oncogene; CDCA-1: Cell division associated 1 TERT: Telomerase reverse transcriptase; CDKs: Cyclin Dependent Kinases; CYCs: Cyclins; E2F: Eukaryote 2 transcription factor; RB1: Retinoblastoma protein 1; CXCR4: Chemokine receptor type 4; MDM2: Mouse double minute 2 homolog; HIF-1α: Hypoxia inducible factor 1α; CTLA-4: Cytotoxic T lymphocyte antigen-4; CCL2: Chemokine (C-C motif) ligand 2; CCR2: CC chemokine receptor 2 (Figures done in Biorender).
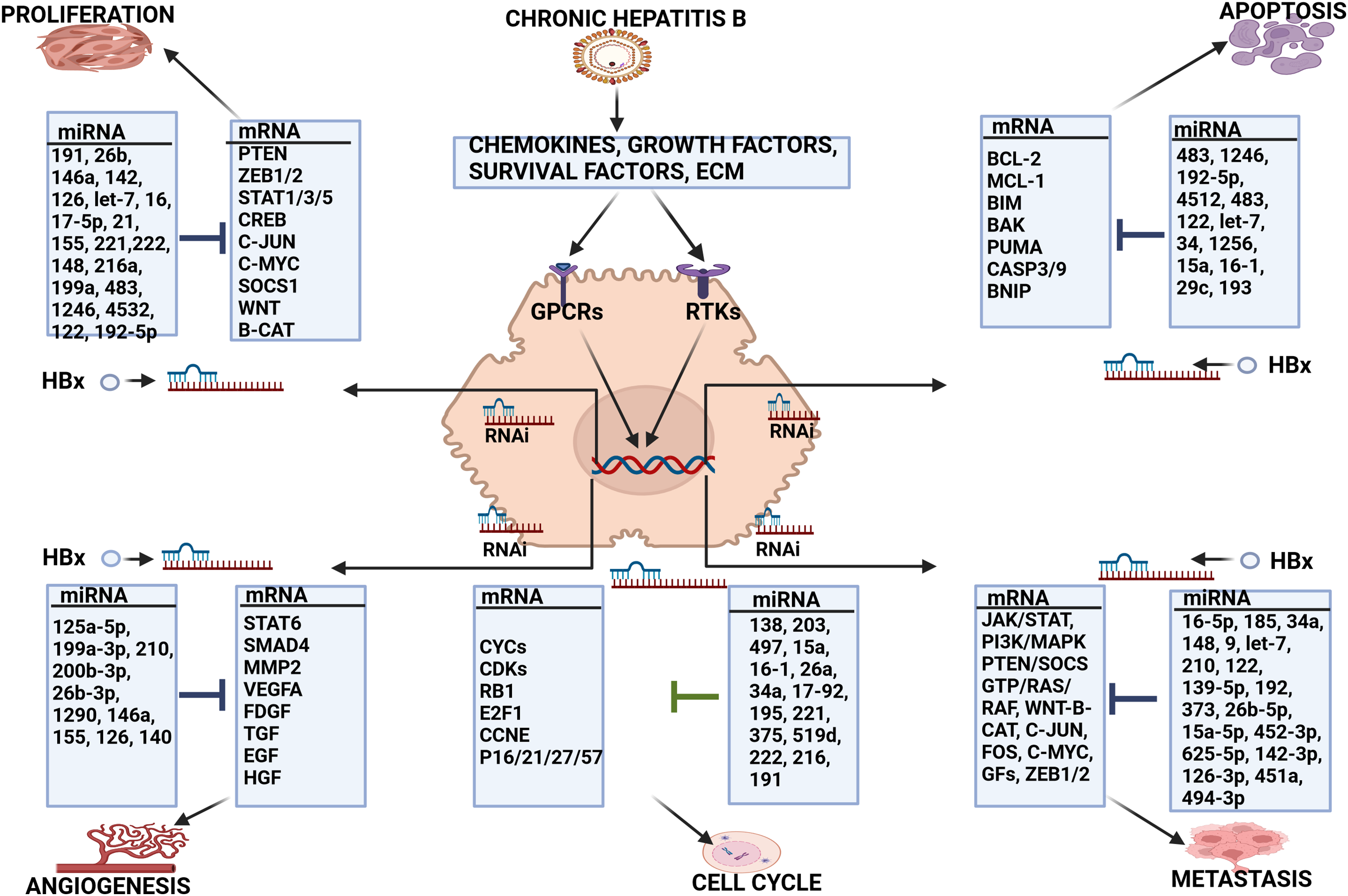
Figure 3: miRNA regulation of HBV-HCC pathogenesis. CHB infection promotes the expression of a wide range of chemokines, growth and survival factors and changes in the extra-cellular matrix (ECM) and the HBx protein can dysregulate miRNA expression to influence a wide range of cellular processes. Cell Proliferation: HBx dysregulated miR-191/-26b/-146a/-142/-126/let-7/-16/-17-5p/-21/-155/-221/-222/-148/-216a/-199a/-483/-1246/-4532/-122/-192-5p regulate PTEN/ ZEB1-2/STAT1,3,5/CREB/C-JUN/C-MYC/SOCS1/WNT/β-catenin expression to influence proliferation. Apoptosis: HBx dysregulated miR-483/-1246/-192-5p/-4512/-122/let-7/-34/-1256/-15a/-16-1/-29c/-193 collectively regulate BCL-2/MCL-1/BIM/BAK/PUMA/CASP3/CASP9/BNIP to influence apoptosis. Angiogenesis: HBx dysregulated miR-125a-5p/-199a-3p/-210/-200b-3p/-26b-3p/1290/-146a/-155/-126/-140 can collectively modulate STAT/SMAD/MMP2/VEGF/FDGF/TGF/EGF/HGF to influence angiogenesis. Cell Cycle: HBx dysregulated miR-138/-203/-497/-15a/-16-1/-26a/-34a/-17-92/-195/-221/-375/-519d/-222/-216/-191modulate CYCs/CDKs/RB1/E2F1/CCNE/P16/21/27/57. Metastasis: HBx dysregulated miR-16-5p/-185/-34a/-148/-9/-let-7/-210/-122/-139-5p/-192/-373/-26b-5p/-15a-5p/-452-3p/-625-5p/-142-3p/-126-3p/-451a/-494-3 that regulate JAK/STAT/PI3K/MAPK/PTEN/SOCS1/GTP/RAS/RAF/WNT/β-catenin/C-JUN/-C-FOS/C-MYC/GFs/ZEB1-2 to influence invasion and metastasis. Abbreviations: Extra cellular matrix (ECM), Receptor tyrosine kinase (RTK), G protein coupled receptors (GPCR) (Figures done in Biorender).
Cell cycle and miRNA regulation
A key regulator of cell cycle processes is cyclin dependent kinase (CDK) activity and CDKs are activated by specific cyclins (CYCs) that accumulate during different stages of the cell cycle [60]. CDK activity activates cell cycle-regulated transcription to initiate cell cycle entry and its progression through the pre-replicative G1 phase and the post-replicative G2 phase [61]. Commitment to replication initiation and S phase entry is closely linked to the activation of the E2F transcriptional network [62] which, in turn. is regulated by the key tumor suppressor RB1 in HCC [63]. Other transcription repressors in HCC including p16/21/27/57 are closely linked to the p53 protein [64,65]. CHB infection can fundamentally dysregulate cell cycle regulators and the HBx protein, for example, can promote the cell cycle by downregulating TGF-β to promote G1/S to G2/M [66], as well as attenuating cell cycle progression by upregulating p21/27 induced downregulation of CDK activity, and by disrupting E2F1 inhibition by repressing RB1 [67].
Although multiple miRNAs have been linked directly to the regulation of cell cycle control, a bioinformatics approach suggests the five most important miRNA/mRNA hubs in HBV-HCC include miR-195-5p/Cyclin-dependent kinase 1 (CDK1), miR-5589-3p/Cyclin B1 (CCNB1), let-7c-3p/Cyclin-dependent kinase regulatory subunit 2 (CKS2), miR-195-5p/Cyclin E1 (CCNE1) and miR-30c-2-3p/CCNE1 [68]. Other examples in HBV-HCC (see Table 1) include miR-138/-203/-497/-15a/-16-1/-26a/-34a that can all directly regulate cyclins and cyclin-dependent kinase activity including CYCD/CYCE/CDK2-4-6 [69,70]. In addition, miR-17-92/-195/-221/-375 can directly regulate cell cycle tumor suppressors RB1/G1 [71,72], miR-519d/-216/-17-92/-221/-222 can target transcription repressors including p16/21/26/57 [73,74]. Other miRNA regulating cell cycle controls include miR-191/-26b/-146a/-142/-126/let-7/-16-5p that can directly target a wide range of cell cycle machinery including both activators and repressors like CDK1NA/CDK2/CCNE/RB1/E2F1 [32]. Further illustrating the complexity of molecular mechanisms involving miRNA regulation of cell cycle, various studies show that the HBx can dysregulate multiple miRNA regulating cell cycle controls including downregulating miR-138/-15a/16-1/-17-92/-375/-216/-222/-let-7 expression and upregulating miR-203/-221/-146a [8] (see Fig. 3, Table 1).
Cell proliferation and miRNA regulation
Cell proliferation is regulated by multiple miRNAs that are dysregulated by the HBx protein in various HBV-HCC pathways including the PI3K/MAPK, JAK/STAT, WNT/β-catenin, SAPK/JNK/TGF-B/SMAD and TP53 suppressor network (see Table 1). Cell proliferation in HBV-HCC pathogenesis is invariably increased by upregulated onco-protein expression, downregulated tumor suppressor genes, the dysregulation of cell cycle controls, and a range of HBV proteins [138,139]. Simultaneously, multiple miRNAs also become dysregulated often failing to regulate onco-protein mRNA or alternatively by repressing tumor suppressor expression [8]. Onco-expression and proliferation in HCC pathways are typically precipitated by a wide range of growth factors (GFs) including TGF-B/FGF/EGF/HGF/IGF, as well as multiple onco-proteins including JAKs/SMADs/C-MYC/C-JUN/C-FOS/RAS/MTOR/KRAS and CTNN1B amongst many more [140]. Other tumor suppressor targets include p21/p27/p16/E-CAD/AXIN1-2/APC/SOCS1-3/KLF6 and PTEN [141]. The HBx protein can both stimulate and repress proliferation by targeting growth factors like CTGF [142], as well as by dysregulating C-MYC, TGF-α, FAS, and RAS/RAF pathway expression [8,67]. Examples of HBx dysregulated miRNA regulation in cell proliferation include let-7/miR-191/-26b/-146a/-142/-126/-16 that target RAS/RAF/ERK/C-JUN/C-FO S/C-MYC in the PI3K/MAPK pathway [32,84] and miR-637/-483/-1246/-192-5p/-4532/-122/-let-7 that target JAK1/STAT3/5 in the JAK/STAT signaling pathway [32,87,88]. In the WNT/β-catenin pathway, cell proliferation can be regulated by miR-122/-148/-30/-320a/-145/-214 that target onco-protein expression of B-CATENIN/C-MYC/C-JUN [90,91,96,97]. Examples of tumor suppressor regulation include miR-155/221 which targets SOCS1/3 in the JAK/STAT pathway [85,86,143], and miR-17-5p/-21/-155/-221/-222/-148 which can target the tumor suppressor PTEN expression in the PI3K/MAPK pathway [81,82]. Other examples of miRNA repression of tumor suppressors include miR-135/-106b/-155/-200/-34 which represses the tumor suppressor APC [80] and miR-21 regulates Programmed cell death protein 4 (PDCD4) in the WNT/β-catenin pathway [80,89]. Further illustrating the complex HBx-miRNA-HCC interactome, many cell proliferation miRNA are also dysregulated by the HBx protein including miR-192-5p/-122/-let-7/-146a/-148/-30c/-145/-155/-221/-222/-17-5p/-21 [8] (see Fig. 3, Table 1).
Apoptosis and miRNA regulation
HBV-HCC pathogenesis typically involves the upregulation of anti-apoptotic proteins including NF-κB/BCL-2/BCL-XL and MCL-1, however, the TGF-β pathway can be stimulated at the cirrhosis stage to promote apoptosis by activating SMAD3 mediated BCL-2 downregulation [138]. Typically HBx dysregulated miRNA like miR-122/-let-7/-34/-29c/-125b/-15a/-16-1 target multiple anti-apoptotic targets like BCL2/BCL-XL/MCL1/CASP9/CASP3 [75,76] but all of these miRNA are reported as frequently downregulated by the HBx protein [8]. Illustrating some of the complexities in HCC pathogenesis, pro-apoptotic proteins like BAX/BCL-XS can be both downregulated [144] and upregulated while simultaneously being regulated by dysregulated miRNA [145]. In the TP53 network, miR-483/-145/-122/-519d/-221 can regulate apoptosis by regulating targets like PUMA/MDM2/P21, as well as their downstream targets like B-cell lymphoma 2 (BCL2)/B-cell lymphoma-extra large (BCL-XL)/Myeloid cell leukemia-1 (MCL1)/CASP9/CASP3. Simultaneously, miR-221/-519d can regulate PTEN expression in the PI3K/MAPK pathways to influence apoptosis [146]. In this microenvironment, the HBx protein can also promote both anti and pro-apoptotic expression. The HBx protein, for example, can promote MCL-1/BCL-2 expression to repress pro-apoptotic BAX activation of CASP9/3 [147], as well as induce the pro-apoptotic expression of TRAIL-R2 (DR5) [148] (see Fig. 3, Table 1).
HBV-HCC is sometimes characterized as a highly angiogenic cancer that is characterized by hypoxia and the overexpression of VEGF [149] and the HBx protein is capable of promoting pro-angiogenic expression by upregulating IL-6/COX2 [150]. VEGF expression is regulated by oncogenic gene mutations, hormones, cytokines, and various signaling molecules like nitric oxide and MAPKs [67]. Moreover, VEGF may be released by stromal cells and from the ECM via MMP-9-mediated proteolysis [151,152]. Multiple pro-angiogenic growth factors like PDGF/FGF/TGF-α and TGF-β/HGF/EGF/VEGF are triggered in HCC pathogenesis, as well as other pro-angiogenic factors including Angiopoietin-2 (ANG2)/IL-4/IL-6 and IL-8 [153]. Simultaneously, both anti-angiogenic signaling-inducing angiostatins, endostatins, and thrombostatins can be orchestrated in the NOTCH pathway while pro-angiogenic signaling can be induced in the PI3K/AKT and WNT-B-CAT pathways [154–156]. Multiple HBx dysregulated miRNAs including miR-125a-5p/-199a-3p/-210/-200b-3p/-26b-3p/-1290/-146a/-155/-126/-140 modulate angiogenic growth factors and their receptors like VEGF/HGF/PDGF/EGF (see Table 1). In addition, in HBV-HCC pathogenesis, many of these miRNA are simultaneously dysregulated by the HBx protein including mIR-125a-5p/-199a-3p/-200b-3p/-146a/-155 [8] suggesting an increased or reduced ability to influence pro or anti-angiogenic mRNA expression (see Fig. 3, Table 1).
Migration/Invasion/metastasis and miRNA
It is difficult to isolate consistent hub miRNA-mRNA patterns in advanced HBV-HCC because of the elevated level of miRNA dysregulation and aberrant expression in multiple cancer pathways including PI3K/MAPK, JAK/STAT, WNT/B-CATENIN, SAPK/JNK, TGF-B/SMAD, NOTCH, Hedgehog (HH) and TP53 networks [157]. Understandably, because of the multiplicity of miRNA/mRNA dysregulation, no meta-studies illustrate a comprehensive list of miRNAs specifically related to the advanced stages of HBV-HCC pathogenesis. However, multiple studies have identified HBx dysregulated miRNA and its targets in the migration/invasion and metastasis phases of HBV-HCC (see Fig. 3, Table 1). The accumulation of β-catenin has been widely associated with multiple cancers including HCC [157] and the dysregulation of miRNA expression targeting key mRNA in the WNT/B-CATENIN pathway appears to be a common feature in advanced stages of HBV-HCC oncogenesis [158]. Various studies identify important miRNA like miR-148a-b/-122/-139-5p/-192/-373 that directly target SNAIL/β-catenin/ZEB 1-2/E-CADHEREN and LRP6 (see Table 1). Interestingly, the ubiquitous role of the HBx protein is also illustrated in this pathway. HBx, for instance, can activate the WNT/β-catenin pathway via modulation of WNT1 to activate SRC kinase to promote β-catenin [159]. HBx can also bind to APC from interacting with it and obstruct its regulation of the APC/AXIN/GSK3b complex to promote β-catenin expression [160].
In HBV-HCC related migration and invasion, HBx dysregulated miRNAs like downregulated miR-16-5p have a reduced ability to regulate IGF1R mRNA which plays an important role in this aspect of HCC pathogenesis [109]. Invasion and migration can also be increased by upregulated miRNA like miR-34a-5p which can attenuate the expression of transcription factor YY1 to mediate MYCT1 [112]. Interestingly, hypoxia-induced migration and EMT can also be influenced by miR-210-5p/-3p [118]. On the other hand, repressed miR-26b-5p expression can fail to modulate EMT, migration, and invasion as a result of an inability to repress SMAD1 [125]. Other examples of studies illustrating miRNA regulation of migration and invasion in HBV-HCC pathogenesis include HBx dysregulated miR-452-5p [128,129], miR-625-3p [130,131], miR-15a-5p [126,127].
Multiple miRNAs modulate HBV-HCC induced metastasis and the following examples are merely illustrations rather than purporting to provide a comprehensive list (see Table 1). Downregulated miR-142-3p in HBV-HCC, for instance, fails to repress HMGβ1 resulting in the reduced attenuation of metastasis [132]. Similarly, downregulated miR-126-3p also fails to repress LRP6 and PIK3R2 expression to promote metastasis and angiogenesis [134]. Similarly, repressed expression of miR-451a fails to regulate YWHAZ and ADAM10 thus promoting metastasis and EMT [135,136] while upregulated miR-494-3p augments enhance HCC metastasis by targeting BMAL1 [137]. Additional example of miRNAs influencing metastasis in HCC pathogenesis include miR-96-5p [161–164], miR-1246 [95,165–168] and miR-210-3p [101,118,169,170].
miRNA as an Ancillary Epigenetic System
MicroRNAs can be classified as an ancillary epigenetic system because they are able to regulate gene expression at a post-transcriptional level, as well as because they are directly connected to the mainstream epigenetic machinery via upstream and downstream regulatory loops [171,172]. In this regard, miRNAs regulate gene expression, rather than silence it, by attaching themselves to complementary 5′mRNA to repress a small percentage of mRNA translation [20]. It differs, therefore, from the mainstream epigenetic machinery in terms of the degree of silencing, as well as the molecular mechanism employed [173]. In all cancers, the mainstream epigenetic machinery is either activated or de-activated by multiple clinical, environmental or genomic changes that interact with miRNA expression [174]. In HBV-HCC pathogenesis, the HBx protein can manipulate the expression of DNMTs, HMTs, HDMTs, HATs and HDACs [175]. The important role of the HBx protein in HBV-HCC pathogenesis is illustrated by its ability to upregulate DNMTs like DNMT1, DNMT3A1, and DNMT3A2, as well as selectively promote regional hypermethylation of specific tumor suppressor genes [176]. Similarly, the HBx can promote the silencing of mRNA expression by inducing histone deacetylases (HDACs) which are made up of zinc-dependent HDACs (HDAC1-11) and non-zinc-dependent HDACs or sirtuins (Sirt1-7) [177]. Conversely, HBx can upregulate mRNA expression by promoting HATs which include four families including GNATs (Gcn5, PCAF, Hat1, Elp3 and Hpa2), p300/CBP (p300 and CBP), MYST (Esa1, MOF, Sas2, Sas3, MORF, Tip60 and Hbo1) and Rtt10 [178]. Similar to HATs and HDACs, the HBx protein can promote histone methyl transferases (HMTs) and histone dimethyl transferases (HDMs) that are responsible for the cycling of methyl groups in histone tails [179]. In particular, the HBx protein can interfere with the Polycomb Repressive Complex 2 (PRC2) to induce H3K27me3-mediated gene expression silencing by targeting PRC2 proteins like embroyonic ectoderm development (EED), enhancer of Zeste 1/2 (EZH1/2) and suppressor of Zeste 12 (SUZ12) [180].
In many cases, HBV infection promotes epigenetic/miRNA feedback loops that include upstream epigenetic manipulation of miRNA expression to target downstream epigenetic targets [171,181] (see Table 2). HBx-dysregulated miRNAs, therefore, can be regarded as ancillary epigenetic regulators in HBV-HCC pathogenesis that interact with the mainstream epigenetic machinery. Examples of the interaction of the mainstream epigenetic machinery with miRNA regulation in HBV-HCC includes the dysregulation of miR-17-92 family members by the HDAC SAHA who can regulate downstream DNMT activity [182] while miR-221 can be significantly increased by hypermethylation of HDAC 6 expression [183,184]. Two important epi-miRNAs, miR-29a/b, can be upregulated by HBx induced HATs to repress downstream DNMT1/3A to reduce PTEN silencing and exert an anti-oncogenic influence [185–189]. Other examples of HBV induced upstream epigenetic regulation of miRNA expression includes the HBx recruitment of the epigenetic protein EZH2 to downregulate miR-139-5p [190]. Illustrating the complexity of these interactions, HBx dysregulated miRNA can repress multiple mRNA targets that include proto-oncogenes, tumor suppressors and epigenetic proteins (see Table 2).
DNA methylation in HBV-HCC pathogenesis, for instance, can downregulate miR-1/-122/-124/-132/-/148/-200/-205 (see Fig. 2). Conversely, HATs or HDAC inhibitors can upregulate miR-224/-29/-155/-17-92, and HMTs can downregulate let-7c/miR-101/-125b/-139-5p (see Fig. 2). In many cases, these same miRNAs regulate downstream epigenetic targets. HBx induced HMT, for instance, can attenuate let-7c expression that fails to regulate downstream epigenetic proteins like EED/EZH1-2/-SUZ12 that promote HMT [190–192]. HBx induced DNMT1 can downregulate miR-124 which reduces its repression of the PRC2 protein, EZH2 which promotes HMT [190,198,199]. Conversely, HBV infection can promote hypermethylation of tumour suppressor ZHX2 that upregulates miR-155 that increases its modulation of downstream PRC2 epigenetic proteins [34] (see Fig. 4).
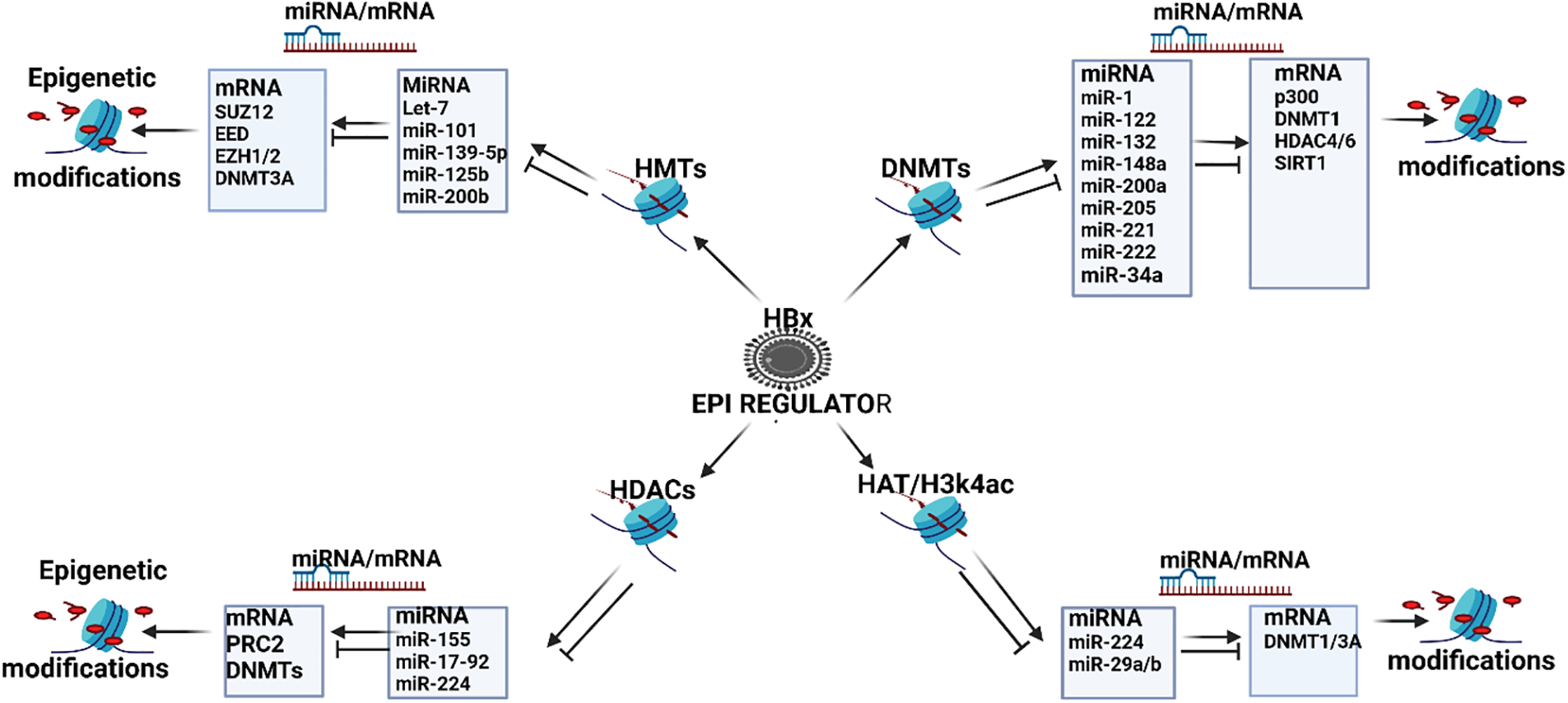
Figure 4: HBx promoted epigenetic regulation of miRNAs in HBV-HCC. The HBx protein can modulate the mainstream epigenetic machinery including Histone methyl transferases (HMTs) that can dysregulate miRNA like let-7/-miR-101/-125b/-139-5p/-200b that, in turn, modulate downstream epigenetic proteins like EED, EZH1/2 and SUZ12 to influence a range of downstream HMTs, as well as DNMT3a to activate/silence gene expression in all the HBV-HCC pathways. DNA Methyl Transferases (DNMTs): The HBx protein can regulate DNMTs that dysregulate miRNA like miR-1/-122/-132/-148a/-200a/-2 05/-221/-222/-34a that regulate downstream epigenetic proteins including the HAT E1A-binding protein (p300), DNMT1, HDAC4/6, and SIRT1. Histone deacetylases (HDACs): The HBx protein can also influence a range of HDACs that can dysregulate miRNA like miR-155/-17-92/-224 that modulate downstream PRC2 proteins, as well as DNMT expression. Histone acetylases (HATs): The HBx protein can influence HATs that can dysregulate miRNA like miR-224/-29a/-29b that regulate downstream DNMT1/3A silencing (Figures done in Biorender).
miRNA Interaction with Immune Response in HBV-HCC Pathogenesis
The immune system is influenced by an interactome of genes whose expression is modulated by extracellular signaling, the mainstream epigenetic machinery, multiple transcription/splicing factors, translational protein modifiers, and miRNAs [9]. MiRNA regulation of both the innate and adaptive immune system in HBV-HCC has been well documented. In the innate immune system, for instance, granulopoiesis can be modulated by miR-155/-21/-223/-21/-196b/130 [221] while miRNA like miR-181a/-150 and Let-7 can regulate natural killer (NK) cell expression [221]. Macrophage output, for example, can be modulated by miR-155/-146a/-124/-125b/-21/-9 and let-7e [221] while dendritic cell (DC) update and differentiation can be modulated by miR-155 [222] and miR-21/-34a, respectively [223]. In the adaptive immune system, miR-17-92/-181a can have a regulatory role in T-cell development [224,225] while B-cell development can be modulated by miR-181/-150/-212/-132/-17-92/-34a/-21/-148/-125b/146a/155 [221]. To provide a more detailed explanation of miRNA immune system regulation than the simplistic examples above, four key HBx dysregulated miRNAs, namely, miR-21/-181a/17-92/miR-155 illustrate the respective molecular mechanisms (see Fig. 5).
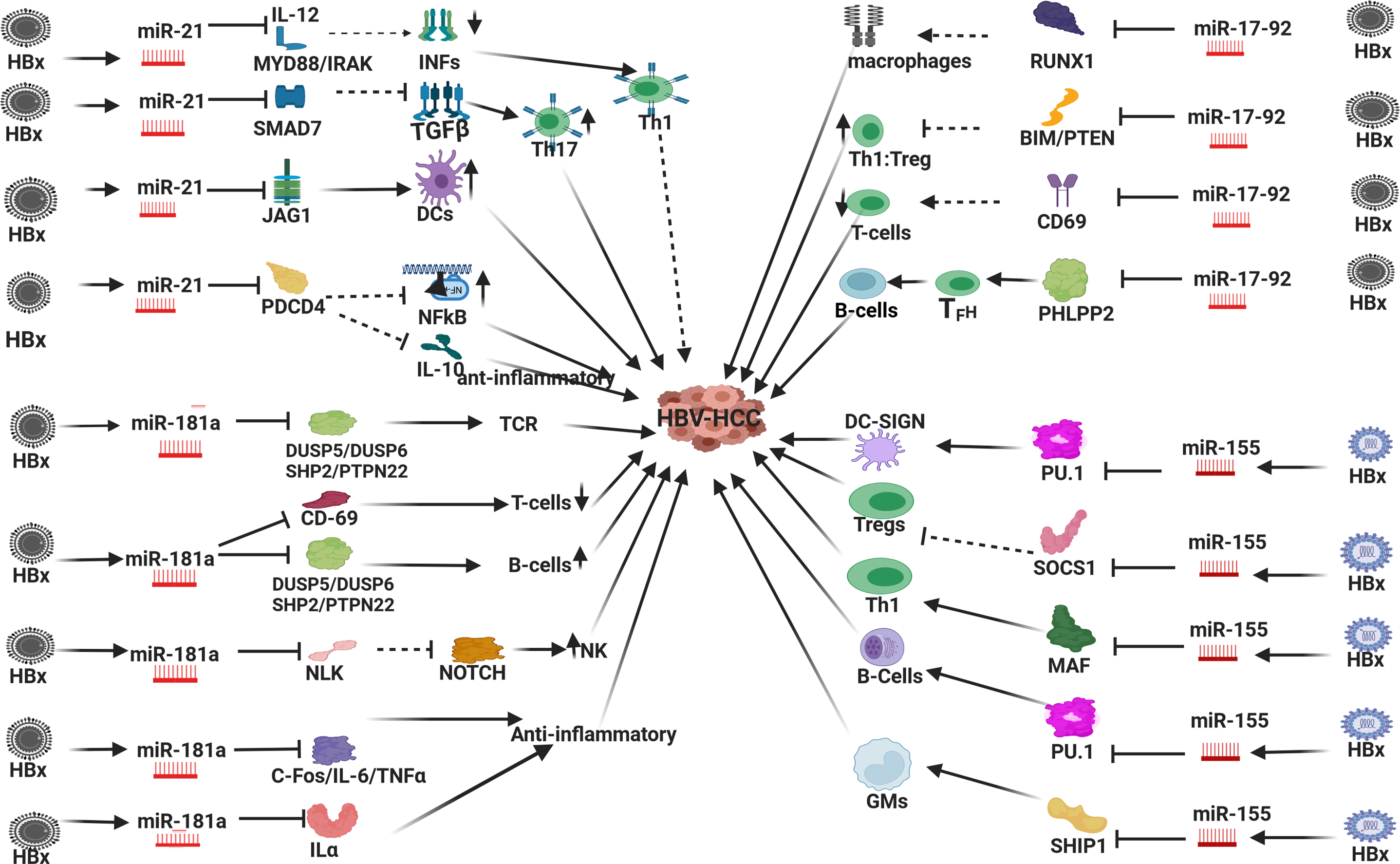
Figure 5: HBx dysregulated miRNA and immune response in HBV-HCC. In HBV-HCC pathogenesis the HBx virion dysregulates miR-21/-181a/-17-92 family/-155 which influences immune expression in both the innate and adaptive immune systems: HBx dysregulated miR-21: can reduce Th1:Th2 ratio by targeting MYD88/IL-12 induction of INFs to promote Th1; it can also promote Th17 expression by suppressing SMAD7 which is a negative regulator of TGFβ; miR-21 can also increase DC output by repressing JAG1; in T-cells, miR-21 can also influence macrophage output by repressing PDCD4 to reduce its repression of pro-inflammatory NF-κB ledsignalingg; miR-21 repression of PDCD4 can also upregulate of the anti-inflammatory IL-10. HBx dysregulated miR-181a: miR-181a can repress DUSP5/6/SHP2/PTPN22 to promote T-cell receptor expression and B-cell output; miR-181a can repress CD-69 to attenuate T-cell production; miR-181a can repress NLK which fails to modulate NOTCH induced NKs; miR-181a can repress C-FOS/IL-6/TNFα/Interleukin 1α (ILα) to promote an anti-inflammatory response influencing monocyte and DC expression. HBx dysregulated miR-17-92: miR-17-92 can repress RUNX1 to attenuate macrophage expression; miR-17-92 can repress PTEN/BIM to influence Th1:Treg ratio; miR-17-92 can repress CD-69 to attenuate T-cell production; miR-17-92 can repress PHLPP2 to promote TFH promotion of B-cell output. HBx upregulated miR-155: miR-155 can repress PU.1 to promote DCs; miR-155 can repress SOCS1 that fails to repress Treg expression; miR-155 can repress MAF mRNA to promote Th1 differentiation; miR-155 can repress PU.1 to promote B-cell expression; miR-155 can repress phosphoinositide phosphatase 1 (SHIP1) to promote granulocyte-macrophage (GM) population (Figures done in Biorender).
HBx induced miR-21 in HBV-HCC can influence immuno-expression in a number of ways: In the innate immune system HBx upregulated miR-21 can exert both pro and anti-inflammatory influences due to its modulation of the transcription inhibitor PDCD4 which influences macrophage activity. First, miR-21 repression of transcription inhibitor PDCD4 can promote pro-inflammatory NF-κB led signaling, and conversely, upregulated miR-21 repression of PDCD4 augments the promotion of anti-inflammatory IL-10 expression [226,227]. HBx upregulated miR-21 can also regulate monocyte-derived dendritic cell (MDDC) differentiation by attenuating JAG1 and WNT1 [223]. In the adaptive immune system, HBx upregulated miR-21 can promote Th17 differentiation by representing SMAD-7, an inhibitor of TGF-β signaling [228]. This miRNA also suppresses SMAD7 to promote TGF-β led promotion of Th17 differentiation [228], as well as represses IL-12, which can induce Th1 responses to attenuate IFNγ production resulting in a diminuation of the Th1:Th2 ratio in T-cell production [229]. This HBx upregulated miRNA can also attenuate IFN by repressing MYD88/IRAK to influence HBV replication [230]. Finally, miR-21 can promote NFκB activation and TNF-α and IFNγ production in activated T-cells to promote inflammation in the HBV TME [231]. These examples illustrate that miR-21 expression can exert multiple pro- and anti-inflammatory responses [227] in both the innate and adaptive immune systems.
In the innate immune system, HBx upregulated miR-181 can regulate inflammatory responses by repressing IL-1a to exert an anti-inflammatory response influencing the expression of monocytes and macrophages [232]. This miRNA can also repress the inflammatory response in dendritic cells (DCs) cells by targeting FOS [233], as well as modulate an anti-inflammatory response by targeting IL-6 and TNFα [234,235]. HBx upregulated miR-181a can also promote natural killer (NK) cell output by upregulating NOTCH signaling as a result of repressing NLK [236]. In the adaptive immune system, miR-181 can stimulate T-cell receptors (TCRs) by repressing DUSP5/DUSP6/SHP2/PTPN22. Conversely, this upregulated miRNA can also repress T-cell expression by repressing CD69 [225]. The overexpression of miR-181 can also skew haematopoiesis towards the development of B-cells at the expense of T-cells by repressing DUSP5/6/SHP2 and PTPN22 [237].
In a further example, the HBx upregulated miR-17-92 family members also play a prominent role in modulating immune response in HBV-HCC pathogenesis. This miRNA can reduce monocyte production by repressing RUNX1 which then fails to promote Granulocyte colony stimulating factor receptor (G-CSFR) expression. Conversely, RUNX1 can also repress miR-17-92 expression and promote monocyte differentiation [238]. CSFR stimulation is linked to increased macrophage activity, inflammation, tissue remodeling, and HCC [239,240]. In the adaptive immune system, the upregulated expression of miR-17-92 can repress the tumour suppressor PTEN to promote Th1 response vs. Treg generation [241]. In addition, miR-17-92 family members can also target CD69 resulting in reduced T-cell output [242]. Upregulated miR-17-92 can also suppress BIM to promote B-cell development [243] and miR-17-92 family members can modulate the migration of CD4+ T cells into B cell follicles by repressing PHLPP2 to promote T-follicular helper (TFH) cell differentiation [244].
The miR-155 family is a crucially important multifunctional regulator of immune responses including inflammation, hematopoietic lineage differentiation, and the onset of carcinogenesis [245]. This important miRNA is expressed by macrophages, dendritic cells (DCs), B cells, T cells, and progenitor/stem cell populations. In the presence of healthy cases, the expression of miR-155 in immune cells is low until their activation by antigens, Toll-like receptor (TLR) ligands, and inflammatory cytokines, which rapidly promote their expression [246,247]. In summary, this important miRNA plays a major role in regulating cytokine production, inflammation, and myeloid and lymphoid differentiation and has a unique ability to modulate the transcriptome of activated myeloid and lymphoid cells [248].
The Status quo of miRNA Therapeutics and Diagnostics: Problems and the Way Forward
In this section, we review the status quo of clinical trials for miRNA deployed as therapeutic and diagnostic agents before outlining some of the problems confronting this field of research and making suggestions for the way forward (see Table 3). In the previous sections of this paper, it is clear after 30 years of miRNA research in HCC pathogenesis, that we have a detailed understanding of the molecular mechanisms of miRNA as a regulatory agent. This detailed understanding includes the molecular mechanisms of miRNA biogenesis, function, and dysregulation, as well as their regulatory targets and role in HBV-HCC pathogenesis from the PME to the TME. This understanding shows precisely how miRNA regulates mRNA in cell proliferation, apoptosis, angiogenesis, migration, invasion, and metastasis. There is also a detailed understanding of their role as an ancillary epigenetic agent, as well as their interactive regulatory role in the innate and adaptive immune systems. The question remains, if we have this detailed understanding of miRNA regulation in HCC pathogenesis, why after 30 years have we not been able to develop miRNA drugs or biomarkers outside of a laboratory setting? To date, no approved miRNA-based drug or commercially adapted biomarker has been developed. An evaluation of miRNA clinical trials for HCC suggests a much higher proportion were established to find diagnostic biomarkers than to develop miRNA drugs. The following HCC miRNA clinical trials highlight some of the problems (see Table 3).
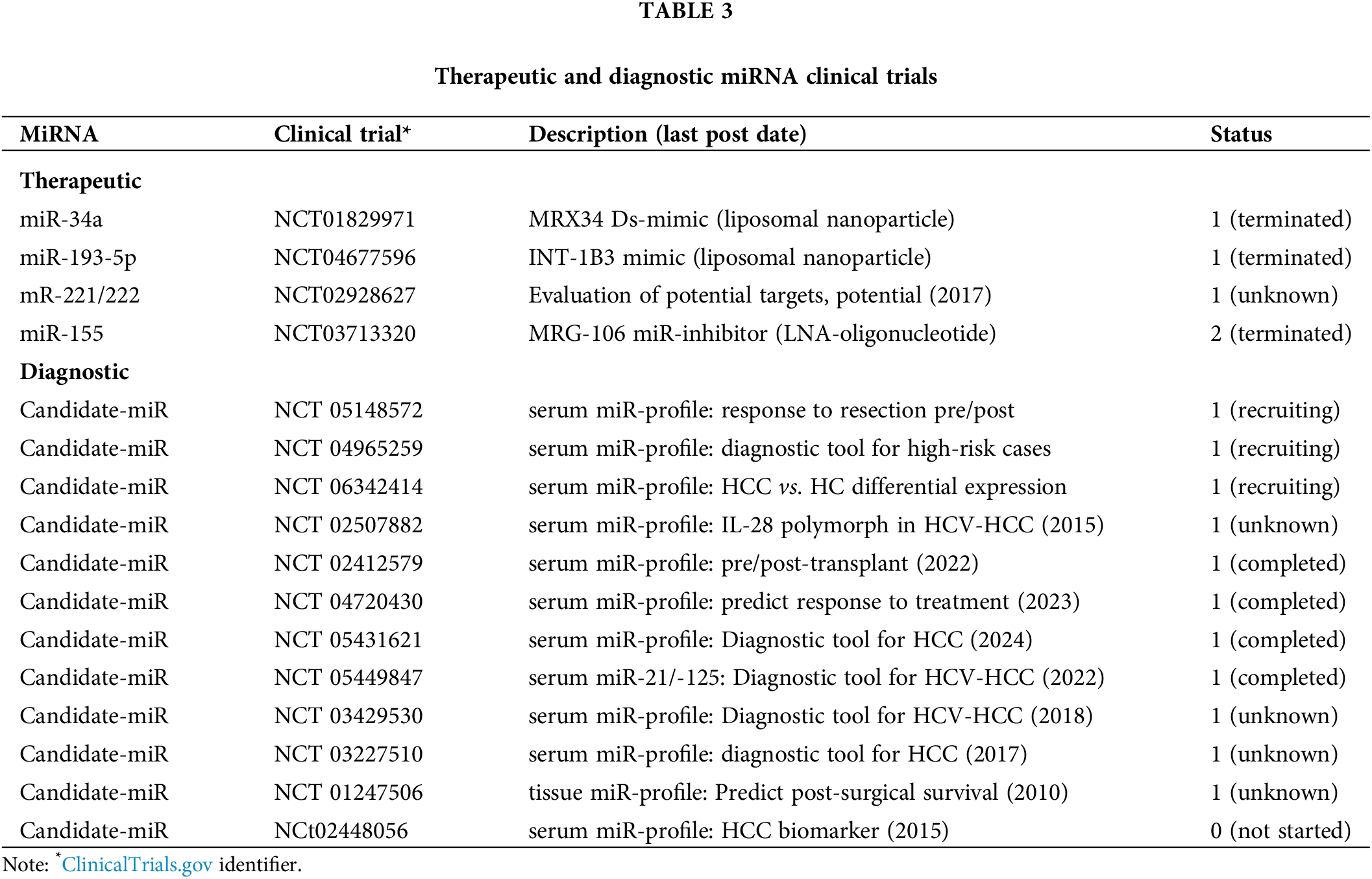
HCC clinical trials testing miRNA as therapeutic agents
This section examines the results of clinical trials for miR-34a/-193-5p/-221/-222/-155. A Phase 1 clinical trial using a miR-34a mimic (NCT01829971) developed a therapeutic agent (MRX34) that used a synthetic double-stranded (ds) miR-34a mimic that was designed to be encapsulated in a liposomal nanoparticle in order to translate this drug to be used in a clinical application. This miRNA has been extensively tested as a therapeutic agent for HCC in multiple experiments involving animal and cell-line studies [249,250]. The importance of this miRNA in HCC pathogenesis is illustrated in this study where it is cited as playing a regulatory role in the PME (fibrogenesis), cell cycle, proliferation, apoptosis, metastasis, as well as interacting with the mainstream epigenetic machinery (see Tables 1 and 2). The Phase 1 clinical trial of MRX34 was the first-in-human clinical trial of miRNA therapy that was deployed for a range of cancers including primary liver cancer. The clinical trial was abandoned as a result of toxic immune mediated events and not as a result of delivery issues [251]. Although the tumor suppressor miR-34a mimic in this trial successfully reduced expression in a range of onco-targets, it elicited an unintended adverse immune response [252]. From a translational perspective, this drug is designed upregulate the expression of targets like MAP4K4 and CCL22 mRNA in HBV-HCC to repress tumor growth in a microenvironment where the HBx protein represses miR-34a [250,253]. A second (more promising) ongoing clinical trial for HCC in Phase 1, developed a drug INT-1B3 that packaged a miR-193-5p miRNA mimic in a lipid nanoparticle (NCT046775996). To date, this drug has demonstrated that it can regulate cell proliferation and from a translational perspective is designed to target CDK2 controls and be delivered directly to tumors in vivo [254] but the ClinicalTrial.gov website now lists this trial as terminated. Other trials involving miRNA specifically for HCC appear inconclusive. One trial deploying miR-221/-222 intends to measure circulating miRNA expression in healthy controls vs. HCC cases to evaluate their therapeutic potential (NCT02928627) but the last comment was posted on 25/10/2017, no publications have been recorded and the status is unknown. An interesting clinical trial (NCT 03713320), designed for solid tumors in general (not specific to HCC), involved the drug MRG-106 which successfully deployed a miR-155 inhibitor using an LNA-modified oligonucleotide. This trial progressed to Phase 2 when the trial was abandoned due to business related reasons and appears to have been successfully tolerated by patients [255]. Interestingly, it has been argued that deploying miRNA inhibitors results in fewer unintended consequences than miRNA mimics because the targeted mRNA can be made more specific [252]. Although this clinical trial was not specifically targeted for HCC, this oncogenic miRNA plays an important role in HCC pathogenesis (see Tables 1, 2), especially regarding its regulatory role in both the innate and adaptive immune system (see Fig. 5). In summation, therefore, no miRNA clinical trial has progressed to stage 4 and therefore miRNA therapeutics remain in the work-in-progress phase (https://classic.clinicaltrials.gov/ct2/results) (accessed on 10 September 2024).
In general, a list of challenges in developing successful miRNA drugs include degradation by in vivo nucleases, poor cell membrane penetration or getting trapped in the endosome, poor binding affinity for complementary sequences, poor delivery to desired target tissues, off-target and unwanted toxicities and activation of unintended immune responses [256]. To a large extent, delivery problems are less of a problem and, despite a detailed understanding of how miRNA regulate single gene targets, a research gap exists with respect to unintended consequences of off-target regulation and resultant toxic effects [252]. Simultaneously, a central problem of developing systemic therapy for HCC, especially in advanced stages, is that a multiplicity of driver genes, genes, cancer pathways, and miRNA become simultaneously dysregulated [32,257,258]. Collectively, this illustrates that single miRNA mimics could be simplistic tools given that their regulatory role can be categorized as an ancillary epigenetic tool that finetunes mRNA expression [171]. Simultaneously, one miRNA can have >100 mRNA targets making the problems of unintended consequences difficult to avoid [252]. In addition, miRNA regulation in HBV-HCC pathogenesis involves a dynamic orchestra of miRNAs, rather than any single miRNA, that is simultaneously regulated by viral proteins, the mainstream epigenetic machinery, other ncRNA like lncRNA and circRNA, and the innate and adaptive immune systems [28,259]. A further research gap that needs to be better understood is the reasons for the degree of heterogeneity between miRNA expression in tumor tissue vs. serum. For instance, one HCC study demonstrated that miR-100-5p/-99a-5p/-455-3p/-10a-5p/-30b-5p/204/5p/let-7a-3p were upregulated in plasma and downregulated in tumor tissue suggesting that miRNA can be selectively released from the TME into serum [260]. The differential miRNA expression between tissue and serum is illustrated in multiple cancers including breast and colorectal cancers [261,262] and this needs to be better understood when selecting candidate miRNA for therapeutic or diagnostic purposes. Simultaneously, in HBV-HCC pathogenesis, it is possible that the interests of the hepatitis B virus contradict other aspects of pathogenesis in both the PME and the TME [8]. A research gap indicates that the role of the HBV virion and its resultant expression need to be extended to a more detailed understanding of how this virus interacts with miRNA in HBV-HCC pathogenesis from the PME to the onset of the tumor environment. In conclusion, the multiplicity of interactions, targets, and mechanisms, illustrated by this paper, highlights a central problem confronting miRNA therapeutics, namely, unintended mRNA regulation because a single miRNA can regulate multiple mRNA targets [263]. These problems appear to have influenced current miRNA clinical trials to focus on the potential of miRNA as diagnostic agents rather than their therapeutic potential. In this regard, siRNA therapeutics appear to have progressed further than miRNA and typically target mRNA in a more efficient way than miRNA [264]. Although both ncRNA silence mRNA, one miRNA can regulate multiple different mRNA targets to varying degrees depending on the complementarity of their binding RNA sequences whereas siRNA is designed to bind to a single mRNA target with perfect complementarity [264,265].
HCC diagnostic clinical trials
Currently registered miRNA HCC clinical trials indicate 12 out of 13 trials are for diagnostic purposes and only one for a therapeutic outcome (https://clinicaltrials.gov/search) (accessed on 10 September 2024). These clinical trials mostly focus on the detection of early stage HCC, evaluating the implications of therapeutic interventions like resections/transplants or as a prognostic tool [252]. Of the 12 registered clinical trials, three are in the recruiting phase and nine have been completed or their status is unknown. In cases that are still recruiting, NCT 05148572 will develop circulating miRNA profiles pre vs. post resection patients to evaluate the efficacy of this therapy. NCT 04965259 will determine serum miRNA expression profiles for high-risk vs. healthy patients to develop an early-stage biomarker and NCT06342414 will also attempt to develop a liquid biopsy based on suitable circulating miRNA candidates as an early-stage biomarker. The outcomes of the other nine clinical trials appear to have produced indeterminate results with no publications generated. The following Phase 1 clinical trials and the last date an update was posted (in brackets) are all listed as completed with no publications including NCT 02412579 (2022), NCT 04720430 (2023), NCT 05431621 (2024) and NCT 05449847 (2022). NCT 02412579 investigated serum miRNA profile to test response to liver transplant, NCT04720430 investigated serum miRNA profile to determine response to pre-transplant bridging treatment, NCT 05431621 attempted to develop serum based biomarker for early stage gastric cancers (including HCC), and NCT 05449847 tested miR-21/-125 as biomarker for early stage HCV-HCC. Multiple clinical trials indicate unknown status including with date of last posted comment in brackets NCT 02507882 (2015), NCT 03429530 (2018), NCT 03227510 (2017), and NCT 01247506 (2010). NCT 02507882 was designed to test serum miRNA profiles of IL-28B polymorphism in HCV-HCC, NCT 03429530 was designed as a serum based tool to diagnose HCV-HCC, NCT 03227510 was designed as a serum based miRNA biomarker for HCC and NCT 01247506 involved tissue based miRNA profiling to determine HCC progression. None of these trials developed any publications.
In summary, no commercially adopted miRNA based diagnostic tools have been adopted. In a meta study involving HBV-HCC cases, miR-125b was listed as the most consistently identified miRNA with high levels of specificity and accuracy [266] while let-7 and miR-122 family members are consistently listed as having high Area under Curves (AUCs), specificity and sensitivity in China (AUC = 0.905) [267], South Africa (AUC = 0.9420) [32] and India [268]. Interestingly, let-7a which is listed as an important regulator of HBV-HCC (see Table 1) is currently being tested in a clinical trial to diagnose non-Hodgkin’s lymphoma and acute leukemia (ClinicalTrials.gov identifier: NCT05477667) [252]. Similarly to miRNA clinical trials for therapeutic outcomes, no HCC miRNA clinical trial has progressed to stage 4 concerning its use as a commercially used biomarker (https://classic.clinicaltrials.gov/ct2/results) and this field of research also remains in the work-in-progress phase.
Despite multiple very similar studies, there is limited overlap between these concerning candidate miRNA or miRNA panels. In 32 breast cancer studies, for example, 143 dysregulated miRNAs were identified as potential biomarkers. Of these, 100 miRNAs were only identified in a single study, and in 10 studies only miR-21/-155 was reported as upregulated in three and two studies respectively even though seven of the ten studies used the same method of normalization while other studies showed these two miRNA as downregulated [269]. Furthermore, many candidate miRNA with high levels of specificity are deregulated in multiple cancers. For example, a promising candidate like miR-141 is dysregulated in prostate, breast, colon, HCC, and lupus and, incidentally, has been reported as upregulated during pregnancy [24]. This led researchers to question whether common factors in carcinogenesis like inflammation or injured tissue are the primary triggers of differential miRNA expression in these studies leading to the question are miRNAs potential biomarkers of neoplasms or general disease [270]. A central research gap is evident, namely, in identifying biomarker miRNA that is specific to HBV-HCC and only to HBV-HCC.
Another central problem of overlap between miRNA-based diagnostics is the dynamic nature of miRNA expression that is constantly fluctuating and highly sensitive to any change in the tumor microenvironment, as well as the differential nature of the patient profile including circadian rhymes, diet, and exercise [271]. The differential patient profile including sex, smoking status, and patient specific physiological conditions (diet, active exercise, comorbidities) that intimately affect miRNA expression profiles with potential errors that can be compounded by the modality of sample preparation, RNA extraction, and differences in outcomes from purchased kits, as well as choice of statistical methods used to normalize miRNA expression [271]. Other examples causing limited overlap between similar studies include using miRNA reads vs. using unique molecular index (UMI) counts that are different [272]. A further issue is the differential expression of miRNA of circulating miRNA compared to tissue-based miRNA expression that is not fully understood [261]. A research gap, therefore, will be to stabilize miRNA expression across overlap studies to control for multiple confounding variables.
Although miRNA research is yet to be translated into tangible therapeutic and diagnostic outcomes, it remains an important opportunity to better detect and manage HCC. Future research is vital to ensure the development of dynamic miRNA panels tailored for patient specific characteristics. To do this, it is not inconceivable that future research might need to incorporate AI based approaches to develop algorithms for predicting miRNA targets that can work alongside current miRNA bioinformatics software [273,274]. Given the lack of control with respect to the unintended consequences of miRNA mimics, tailored delivery sites for miRNA based drugs could reduce systemic unintended consequences [252,254]. Alternatively, tailored packages of upregulated miRNAs can be acutely targeted by miRNA inhibitors or possibly technology to shield selected target tumor suppressor mRNA from overexpressed onco-miRNAs [275]. A new approach is needed for the development of miRNA-based therapeutics given the current depressing status quo illustrated by a lack of promising clinical trials. Refining the targeted ability of miRNA is underlined by siRNA based therapeutics that appear to have progressed to two approved drugs and a number of stage 111 clinical trials including one for breast cancer [263]. Similarly, future research on miRNA as diagnostic agents will need to be able to accommodate the heterogenous nature of patient characteristics, especially in the PME and early-stage HCC pathogenesis. Early detection is vital to provide the most promising prognosis for HCC and miRNA diagnostics in this stage have not been developed beyond Alpha-fetoprotein (AFP) and ultrasound testing which has many limitations remains widely used [276]. The molecular mechanisms resulting in the production and release of miRNA into plasma/serum by tumor cells need to be better understood to determine whether cancer cells selectively release miRNA into serum [277].
HBV-HCC incidence still accounts for a large proportion of global HCC incidence and its idiosyncratic features demand differential therapeutic innovations due to the role of the HBV virion. CHB infection influences HBV-HCC pathogenesis from early-stage infection, inflammation, and fibrosis in the PME to the onset of HBV-HCC. CHB infection simultaneously dysregulates multiple cellular processes, mRNA and miRNA expression to influence cell proliferation, angiogenesis, apoptosis, invasion, and metastasis in a wide range of cancer pathways.
This paper makes the following contributions. First, it summarizes key miRNA aspects of its molecular mechanisms in HBV-HCC pathogenesis including its role in cellular processes, its role as an ancillary epigenetic system, and its interaction with the immune system, importantly it covers an important gap that illustrates miRNA regulation in the PME as a result of CHB infection. This offers an opportunity in HBV-HCC pathogenesis to potentially deploy miRNA in a microenvironment where genomic instability is less pronounced and/or specific aspects of pathogenesis like inflammation or fibrosis can be targeted. Second, the review illustrates the complexity of the HBV-miRNA interactome to attempt to answer the question as to why miRNA therapeutics and diagnostics remain a work-in-progress. Importantly, this is supported by evidence of a lack of successful clinical trials. The paper, therefore, promotes the idea of a rethink as to how this problem can be solved because it becomes clear that the multiplicity of dysregulated gene expression in HBV-HCC is unlikely to be regulated by the deployment of individual miRNA.
We acknowledge there are many limitations in this paper. First, the paper only really provides simplistic snapshots of the complexity of the miRNA interactome, and bioinformatics diagrams of the interaction of a single miRNA can involve 100s of gene targets that are not captured in this paper. Second, this paper does not unpack its interaction with a wide range of other ncRNA like lncRNA and circRNA. In the miRNA interactome with epigenetic machinery, for instance, multiple ncRNA regulate each other, as well as downstream epigenetic targets. Adding to this complexity, upstream epigenetic machinery can modulate downstream ncRNA and all of these players can be regulated by a range of HBV proteins (HBx) that are simultaneously attempting to balance the virus’s replication and evading host immune response.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this review article.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Kurt Sartorius; draft manuscript preparation: Kurt Sartorius; review and editing: Benn Sartorius, Cherie Winkler, Anil Chuturgoon, Anna Kramvis, Ping An,Weigang Zhang; visualization: Kurt Sartorius, Anna Kramvis, Ping An, Weigang Zhang; supervision: Yunjie Lu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Veracruz N, Gish RG, Cheung R, Chitnis AS, Wong RJ. Global incidence and mortality of hepatitis B and hepatitis C acute infections, cirrhosis and hepatocellular carcinoma from 2010 to 2019. J Viral Hepat. 2022;29(5):352–65. doi:10.1111/jvh.v29.5. [Google Scholar] [CrossRef]
2. Kim D. Changing etiology and epidemiology of hepatocellular carcinoma: Asia and worldwide. J Liver Cancer. 2024;24(1):62–70. [Google Scholar] [PubMed]
3. Girardi DM, Sousa LP, Miranda TA, Haum FN, Pereira GC, Pereira AA. Systemic therapy for advanced hepatocellular carcinoma: current stand and perspectives. Cancers. 2023;15(6):1680. doi:10.3390/cancers15061680. [Google Scholar] [PubMed] [CrossRef]
4. Mandlik DS, Mandlik SK, Choudhary HB. Immunotherapy for hepatocellular carcinoma: current status and future perspectives. World J Gastroenterol. 2023;29(6):1054. doi:10.3748/wjg.v29.i6.1054. [Google Scholar] [PubMed] [CrossRef]
5. Yang C, Zhang H, Zhang L, Zhu AX, Bernards R, Qin W, et al. Evolving therapeutic landscape of advanced hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2023;20(4):203–22. doi:10.1038/s41575-022-00704-9. [Google Scholar] [CrossRef]
6. Zhao Y, Jia Y, Qi S, Wu C, Wu J, Zhang R, et al. Comparison of postoperative prognosis among HBV-Related, HCV-Related, and Non-HBV Non-HCV hepatocellular carcinomas: a systematic review and meta-analysis. Hepat Mon. 2022;22(1):e121820. [Google Scholar]
7. Kanwal N, Al Samarrai OR, Al-Zaidi HMH, Mirzaei AR, Heidari MJ. Comprehensive analysis of microRNA (miRNA) in cancer cells. Cell Mol Biomed Rep. 2023;3(2):89–97. doi:10.55705/cmbr.2022.364591.1070. [Google Scholar] [CrossRef]
8. Sartorius K, Makarova J, Sartorius B, An P, Winkler C, Chuturgoon A, et al. The regulatory role of microRNA in hepatitis-B virus-associated hepatocellular carcinoma (HBV-HCC) pathogenesis. Cells. 2019;8(12):1504. doi:10.3390/cells8121504. [Google Scholar] [PubMed] [CrossRef]
9. Sartorius K, Swadling L, An P, Makarova J, Winkler C, Chuturgoon A, et al. The multiple roles of hepatitis B virus X protein (HBx) dysregulated microRNA in hepatitis B virus-associated hepatocellular carcinoma (HBV-HCC) and immune pathways. Viruses. 2020;12(7):746. doi:10.3390/v12070746. [Google Scholar] [PubMed] [CrossRef]
10. Xiong D-D, Dang Y-W, Lin P, Wen D-Y, He R-Q, Luo D-Z, et al. A circRNA-miRNA–mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16:1–21. [Google Scholar]
11. Zhao X, Bai Z, Li C, Sheng C, Li H. Identification of a novel eight-lncRNA prognostic signature for HBV-HCC and analysis of their functions based on coexpression and ceRNA networks. Biomed Res Int. 2020;2020(1):8765461. [Google Scholar] [PubMed]
12. Kp A, Kaliaperumal K, Sekar D. microRNAs and their therapeutic strategy in phase I and phase II clinical trials. Epigenomics. 2024;16(4):259–271. doi:10.2217/epi-2023-0363. [Google Scholar] [PubMed] [CrossRef]
13. Saini V, Dawar R, Suneja S, Gangopadhyay S, Kaur C. Can microRNA become next-generation tools in molecular diagnostics and therapeutics? A systematic review. Egypt J Med Hum Genet. 2021;22:1–9. [Google Scholar]
14. Ito M, Miyata Y, Okada M. Current clinical trials with non-coding RNA-based therapeutics in malignant diseases: a systematic review. Transl Oncol. 2023;31:101634. doi:10.1016/j.tranon.2023.101634. [Google Scholar] [PubMed] [CrossRef]
15. Vidigal JA, Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends cell biol. 2015;25(3):137–47. doi:10.1016/j.tcb.2014.11.004. [Google Scholar] [PubMed] [CrossRef]
16. Turchinovich A, Samatov T, Tonevitsky A, Burwinkel B. Circulating miRNAs: cell-cell communication function? The origin, function and diagnostic potential of extracellular microRNA in human body fluids. Front Genet. 2014;5:30. [Google Scholar] [PubMed]
17. Turchinovich A, Tonevitsky AG, Burwinkel B. Extracellular miRNA: a collision of two paradigms. Trends Biochem Sci. 2016;41(10):883–92. doi:10.1016/j.tibs.2016.08.004. [Google Scholar] [PubMed] [CrossRef]
18. Alles J, Fehlmann T, Fischer U, Backes C, Galata V, Minet M, et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019;47(7):3353–64. doi:10.1093/nar/gkz097. [Google Scholar] [PubMed] [CrossRef]
19. Frédérick PM, Simard MJ. Regulation and different functions of the animal microRNA-induced silencing complex. WIREs RNA. 2022;13(4):e1701. doi:10.1002/wrna.v13.4. [Google Scholar] [CrossRef]
20. Naeli P, Winter T, Hackett AP, Alboushi L, Jafarnejad SM. The intricate balance between microRNA-induced mRNA decay and translational repression. FEBS J. 2023;290(10):2508–24. doi:10.1111/febs.v290.10. [Google Scholar] [CrossRef]
21. Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010;38(20):7248–59. doi:10.1093/nar/gkq601. [Google Scholar] [PubMed] [CrossRef]
22. Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA, et al. Intracellular and extracellular microRNA: an update on localization and biological role. Prog Histochem Cytochem. 2016;51(3–4):33–49. [Google Scholar]
23. Nassar W, El-Ansary M, Fayyad T, Aziz MA. Extracellular micro-RNAs in health and disease: basic science, biogenesis and release. Am J Mol Biol. 2016;6(1):1–11. doi:10.4236/ajmb.2016.61001. [Google Scholar] [CrossRef]
24. Witwer KW. Circulating microRNA biomarker studies: pitfalls and potential solutions. Clin Chem. 2015;61(1):56–63. doi:10.1373/clinchem.2014.221341. [Google Scholar] [PubMed] [CrossRef]
25. Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18(3):215–22. doi:10.1097/PPO.0b013e318250c001. [Google Scholar] [PubMed] [CrossRef]
26. Gulyaeva LF, Kushlinskiy NE. Regulatory mechanisms of microRNA expression. J Transl Med. 2016;14(1):143. doi:10.1186/s12967-016-0893-x. [Google Scholar] [PubMed] [CrossRef]
27. Xu J, An P, Winkler CA, Yu Y. Dysregulated microRNAs in hepatitis B virus-related hepatocellular carcinoma: potential as biomarkers and therapeutic targets. Front Oncol. 2020;10:1271. doi:10.3389/fonc.2020.01271. [Google Scholar] [PubMed] [CrossRef]
28. Han T-S, Hur K, Cho H-S, Ban HS. Epigenetic associations between lncRNA/circRNA and miRNA in hepatocellular carcinoma. Cancers. 2020;12(9):2622. doi:10.3390/cancers12092622. [Google Scholar] [PubMed] [CrossRef]
29. Starega-Roslan J, Koscianska E, Kozlowski P, Krzyzosiak WJ. The role of the precursor structure in the biogenesis of microRNA. Cell Mol Life Sci. 2011;68(17):2859–71. doi:10.1007/s00018-011-0726-2. [Google Scholar] [PubMed] [CrossRef]
30. Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Makarova J. The biological and diagnostic role of miRNA’s in hepatocellular carcinoma. Front Biosci (Landmark Ed). 2018;23(9):1701–20. doi:10.2741/4668. [Google Scholar] [PubMed] [CrossRef]
31. Seki E, Schwabe RF. Hepatic inflammation and fibrosis: functional links and key pathways. Hepatology. 2015;61(3):1066–79. doi:10.1002/hep.27332. [Google Scholar] [PubMed] [CrossRef]
32. Sartorius K, Sartorius B, Winkler C, Chuturgoon A, Shen T-W, Zhao Y, et al. Serum microRNA profiles and pathways in hepatitis B-Associated hepatocellular carcinoma: a South African study. Int J Mol Sci. 2024;25(2):975. doi:10.3390/ijms25020975. [Google Scholar] [PubMed] [CrossRef]
33. Wang B, Majumder S, Nuovo G, Kutay H, Volinia S, Patel T, et al. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice. Hepatology. 2009;50(4):1152–61. doi:10.1002/hep.23100. [Google Scholar] [PubMed] [CrossRef]
34. Song X, Tan S, Wu Z, Xu L, Wang Z, Xu Y, et al. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int J Cancer. 2018;143(12):3120–30. doi:10.1002/ijc.v143.12. [Google Scholar] [CrossRef]
35. Arataki K, Hayes CN, Akamatsu S, Akiyama R, Abe H, Tsuge M, et al. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J Med Virol. 2013;85(5):789–98. doi:10.1002/jmv.v85.5. [Google Scholar] [CrossRef]
36. Nakamura M, Kanda T, Jiang X, Haga Y, Takahashi K, Wu S, et al. Serum microRNA-122 and Wisteria floribunda agglutinin-positive Mac-2 binding protein are useful tools for liquid biopsy of the patients with hepatitis B virus and advanced liver fibrosis. PLoS One. 2017;12(5):e0177302. doi:10.1371/journal.pone.0177302. [Google Scholar] [PubMed] [CrossRef]
37. Bandiera S, Pfeffer S, Baumert TF, Zeisel MB. miR-122-a key factor and therapeutic target in liver disease. J Hepatol. 2015;62(2):448–57. doi:10.1016/j.jhep.2014.10.004. [Google Scholar] [PubMed] [CrossRef]
38. Hsu S-H, Wang B, Kota J, Yu J, Costinean S, Kutay H, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–83. doi:10.1172/JCI63539. [Google Scholar] [PubMed] [CrossRef]
39. Li C, Deng M, Hu J, Li X, Chen L, Ju Y, et al. Chronic inflammation contributes to the development of hepatocellular carcinoma by decreasing miR-122 levels. Oncotarget. 2016;7(13):17021–34. doi:10.18632/oncotarget.7740. [Google Scholar] [PubMed] [CrossRef]
40. Li G, Cai G, Li D, Yin W. MicroRNAs and liver disease: viral hepatitis, liver fibrosis and hepatocellular carcinoma. Postgrad Med J. 2014;90(1060):106–12. doi:10.1136/postgradmedj-2013-131883. [Google Scholar] [PubMed] [CrossRef]
41. Jiang X, Kanda T, Wu S, Nakamura M, Miyamura T, Nakamoto S, et al. Regulation of microRNA by hepatitis B virus infection and their possible association with control of innate immunity. World J Gastroenterol. 2014;20(23):7197. doi:10.3748/wjg.v20.i23.7197. [Google Scholar] [PubMed] [CrossRef]
42. Elsharkawy AM, Mann DA. Nuclear factor-κB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46(2):590–7. doi:10.1002/(ISSN)1527-3350. [Google Scholar] [CrossRef]
43. Zhou W-C, Zhang Q-B, Qiao L. Pathogenesis of liver cirrhosis. World J Gastroenterol. 2014;20(23):7312. doi:10.3748/wjg.v20.i23.7312. [Google Scholar] [PubMed] [CrossRef]
44. Noetel A, Kwiecinski M, Elfimova N, Huang J, Odenthal M. microRNA are central players in anti-and profibrotic gene regulation during liver fibrosis. Front Physiol. 2012;3(49):1–6. doi:10.3389/fphys.2012.00049. [Google Scholar] [CrossRef]
45. Zhang Z, Zha Y, Hu W, Huang Z, Gao Z, Zang Y, et al. The autoregulatory feedback loop of microRNA-21/programmed cell death protein 4/activation protein-1 (miR-21/PDCD4/AP-1) as a driving force for hepatic fibrosis development. J Biol Chem. 2013;288(52):37082–93. doi:10.1074/jbc.M113.517953. [Google Scholar] [PubMed] [CrossRef]
46. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi:10.1053/j.gastro.2007.05.022. [Google Scholar] [PubMed] [CrossRef]
47. Huang C-F, Sun C-C, Zhao F, Zhang Y-D, Li D-J. miR-33a levels in hepatic and serum after chronic HBV-induced fibrosis. J Gastroenterol. 2015;50(4):480–90. doi:10.1007/s00535-014-0986-3. [Google Scholar] [PubMed] [CrossRef]
48. Chen L, Charrier A, Zhou Y, Chen R, Yu B, Agarwal K, et al. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology. 2014;59(3):1118–29. doi:10.1002/hep.26768. [Google Scholar] [PubMed] [CrossRef]
49. Izawa T, Horiuchi T, Atarashi M, Kuwamura M, Yamate J. Anti-fibrotic role of miR-214 in thioacetamide-induced liver cirrhosis in rats. Toxicol Pathol. 2015;43(6):844–51. doi:10.1177/0192623315573587. [Google Scholar] [PubMed] [CrossRef]
50. Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, et al. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53(1):209–18. doi:10.1002/hep.23922. [Google Scholar] [PubMed] [CrossRef]
51. Bala S, Csak T, Saha B, Zatsiorsky J, Kodys K, Catalano D, et al. The pro-inflammatory effects of miR-155 promote liver fibrosis and alcohol-induced steatohepatitis. J Hepatol. 2016;64(6):1378–87. doi:10.1016/j.jhep.2016.01.035. [Google Scholar] [CrossRef]
52. Bala S, Zhuang Y, Nagesh PT, Catalano D, Zivny A, Wang Y, et al. Therapeutic inhibition of miR-155 attenuates liver fibrosis via STAT3 signaling. Mol Ther-Nucleic Acids. 2023;33:413–27. doi:10.1016/j.omtn.2023.07.012. [Google Scholar] [PubMed] [CrossRef]
53. Tan L, Jiang W, Lu A, Cai H, Kong L, Editors. miR-155 aggravates liver ischemia/reperfusion injury by suppressing SOCS1 in mice. Transpl P. 2018;50(10):3831–9. doi:10.1016/j.transproceed.2018.08.060. [Google Scholar] [PubMed] [CrossRef]
54. Lou G, Yang Y, Liu F, Ye B, Chen Z, Zheng M, et al. MiR-122 modification enhances the therapeutic efficacy of adipose tissue-derived mesenchymal stem cells against liver fibrosis. J Cell Mol Med. 2017;21(11):2963–73. doi:10.1111/jcmm.13208. [Google Scholar] [PubMed] [CrossRef]
55. Song J, Lv H, Liu B, Hao M, Taylor HS, Zhang X, et al. Let-7 suppresses liver fibrosis by inhibiting hepatocyte apoptosis and TGF-β production. Mol Metab. 2023;78(3):101828. doi:10.1016/j.molmet.2023.101828. [Google Scholar] [PubMed] [CrossRef]
56. Schulz WA. Molecular biology of human cancers: an advanced student’s textbook. New York, USA: Springer; 2005. [Google Scholar]
57. Murakami Y, Toyoda H, Tanaka M, Kuroda M, Harada Y, Matsuda F, et al. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS One. 2011;6(1):e16081. doi:10.1371/journal.pone.0016081. [Google Scholar] [PubMed] [CrossRef]
58. Xie K-L, Zhang Y-G, Liu J, Zeng Y, Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4(12):1176–1192. doi:10.7150/thno.8715. [Google Scholar] [PubMed] [CrossRef]
59. Wang Y, Deng B. Hepatocellular carcinoma: molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev. 2023;42(3):629–52. doi:10.1007/s10555-023-10084-4. [Google Scholar] [PubMed] [CrossRef]
60. Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079–93. doi:10.1242/dev.091744. [Google Scholar] [PubMed] [CrossRef]
61. Matthews HK, Bertoli C, de Bruin RA. Cell cycle control in cancer. Nat Rev Mol Cell Biol. 2022;23(1):74–88. doi:10.1038/s41580-021-00404-3. [Google Scholar] [PubMed] [CrossRef]
62. Ye W-Y, Lu H-P, Li J-D, Chen G, He R-Q, Wu H-Y, et al. Clinical implication of E2F transcription factor 1 in hepatocellular carcinoma tissues. Cancer Biother Radiopharm. 2023;38(10):684–707. [Google Scholar] [PubMed]
63. Chand V, Liao X, Guzman G, Benevolenskaya E, Raychaudhuri P. Hepatocellular carcinoma evades RB1-induced senescence by activating the FOXM1-FOXO1 axis. Oncogene. 2022;41(30):3778–90. doi:10.1038/s41388-022-02394-8. [Google Scholar] [PubMed] [CrossRef]
64. Shamloo B, Usluer S. p21 in cancer research. Cancers. 2019;11(8):1178. doi:10.3390/cancers11081178. [Google Scholar] [PubMed] [CrossRef]
65. Guo H, Lv Y, Tian T, Hu TH, Wang WJ, Sui X, et al. Downregulation of p57 accelerates the growth and invasion of hepatocellular carcinoma. Carcinogenesis. 2011;32(12):1897–904. doi:10.1093/carcin/bgr220. [Google Scholar] [PubMed] [CrossRef]
66. Xia Y, Cheng X, Li Y, Valdez K, Chen W, Liang TJ. Hepatitis B virus deregulates the cell cycle to promote viral replication and a premalignant phenotype. J Virol. 2018;92(19). doi:10.1128/JVI.00722-18. [Google Scholar] [PubMed] [CrossRef]
67. Sivasudhan E, Blake N, Lu Z, Meng J, Rong R. Hepatitis B viral protein HBx and the molecular mechanisms modulating the hallmarks of hepatocellular carcinoma: a comprehensive review. Cells. 2022;11(4):741. doi:10.3390/cells11040741. [Google Scholar] [PubMed] [CrossRef]
68. Huang D-P, Zeng Y-H, Yuan W-Q, Huang X-F, Chen S-Q, Wang M-Y, et al. Bioinformatics analyses of potential miRNA-mRNA regulatory axis in HBV-related hepatocellular carcinoma. Int J Med Sci. 2021;18(2):335–46. doi:10.7150/ijms.50126. [Google Scholar] [PubMed] [CrossRef]
69. Wang W, Zhao L-J, Tan Y-X, Ren H, Qi Z-T. MiR-138 induces cell cycle arrest by targeting cyclin D3 in hepatocellular carcinoma. Carcinogenesis. 2012;33(5):1113–20. doi:10.1093/carcin/bgs113. [Google Scholar] [PubMed] [CrossRef]
70. Furuta M, Kozaki K-I, Tanimoto K, Tanaka S, Arii S, Shimamura T, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8(3):e60155. doi:10.1371/journal.pone.0060155. [Google Scholar] [PubMed] [CrossRef]
71. Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP, Zhuang SM. MicroRNA-195 suppresses tumorigenicity and regulates G1/S transition of human hepatocellular carcinoma cells. Hepatology. 2009;50(1):113–21. doi:10.1002/hep.22919. [Google Scholar] [PubMed] [CrossRef]
72. Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20(12):1603–14. doi:10.1038/cdd.2013.125. [Google Scholar] [PubMed] [CrossRef]
73. Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci. 2010;107(1):264–9. doi:10.1073/pnas.0907904107. [Google Scholar] [PubMed] [CrossRef]
74. Yang Y-F, Wang F, Xiao J-J, Song Y, Zhao Y-Y, Cao Y, et al. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int J Clin Exp Med. 2014;7(4):893. [Google Scholar]
75. Mizuguchi Y, Takizawa T, Yoshida H, Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: a systematic review. Hepatol Res. 2015;46(5):391–406. [Google Scholar] [PubMed]
76. Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu X-D. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288(25):18484–93. doi:10.1074/jbc.M113.458158. [Google Scholar] [PubMed] [CrossRef]
77. Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, et al. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51(3):836–45. [Google Scholar] [PubMed]
78. Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52(5):698–704. doi:10.1016/j.jhep.2009.12.024. [Google Scholar] [PubMed] [CrossRef]
79. Pepe F, Visone R, Veronese A. The glucose-regulated miR-483-3p influences key signaling pathways in cancer. Cancers. 2018;10(6):181. doi:10.3390/cancers10060181. [Google Scholar] [PubMed] [CrossRef]
80. Callegari E, Gramantieri L, Domenicali M, D’abundo L, Sabbioni S, Negrini M. MicroRNAs in liver cancer: a model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22(1):46–57. doi:10.1038/cdd.2014.136. [Google Scholar] [PubMed] [CrossRef]
81. Fu X, Wen H, Jing L, Yang Y, Wang W, Liang X, et al. Micro RNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI 3K/Akt pathway. Cancer Sci. 2017;108(4):620–31. doi:10.1111/cas.13177. [Google Scholar] [PubMed] [CrossRef]
82. Wong QW, Ching AK, Chan AW, Choy K-W, To K-F, Lai PB, et al. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin Cancer Res. 2010;16(3):867–75. doi:10.1158/1078-0432.CCR-09-1840. [Google Scholar] [PubMed] [CrossRef]
83. Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, et al. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70(12):5184–93. doi:10.1158/0008-5472.CAN-10-0145. [Google Scholar] [PubMed] [CrossRef]
84. Akula SM, Abrams SL, Steelman LS, Emma MR, Augello G, Cusimano A, et al. RAS/RAF/MEK/ERK, PI3K/PTEN/AKT/mTORC1 and TP53 pathways and regulatory miRs as therapeutic targets in hepatocellular carcinoma. Expert Opin Ther Targets. 2019;23(11):915–29. doi:10.1080/14728222.2019.1685501. [Google Scholar] [PubMed] [CrossRef]
85. Chen Z, Ma T, Huang C, Hu T, Li J. The pivotal role of microRNA-155 in the control of cancer. J Cell Physiol. 2014;229(5):545–50. doi:10.1002/jcp.24492. [Google Scholar] [PubMed] [CrossRef]
86. Gui Y, Yeganeh M, Donates Y, Tobelaim W, Chababi W, Mayhue M, et al. Regulation of MET receptor tyrosine kinase signaling by suppressor of cytokine signaling 1 in hepatocellular carcinoma. Oncogene. 2015;34(46):5718–28. doi:10.1038/onc.2015.20. [Google Scholar] [CrossRef]
87. Wang Y, Lu Y, Toh ST, Sung W-K, Tan P, Chow P, et al. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53(1):57–66. doi:10.1016/j.jhep.2009.12.043. [Google Scholar] [PubMed] [CrossRef]
88. Lokau J, Schoeder V, Haybaeck J, Garbers C. Jak-stat signaling induced by interleukin-6 family cytokines in hepatocellular carcinoma. Cancers. 2019;11(11):1704. doi:10.3390/cancers11111704. [Google Scholar] [CrossRef]
89. Pu J, Xu Z, Nian J, Fang Q, Yang M, Huang Y, et al. M2 macrophage-derived extracellular vesicles facilitate CD8+ T cell exhaustion in hepatocellular carcinoma via the miR-21-5p/YOD1/YAP/β-catenin pathway. Cell Death Discov. 2021;7(1):182. doi:10.1038/s41420-021-00556-3. [Google Scholar] [PubMed] [CrossRef]
90. Xu J, Zhu X, Wu L, Yang R, Yang Z, Wang Q, et al. MicroRNA-122 suppresses cell proliferation and induces cell apoptosis in hepatocellular carcinoma by directly targeting Wntβ-catenin pathway. Liver Int. 2012;32(5):752–60. doi:10.1111/j.1478-3231.2011.02750.x. [Google Scholar] [PubMed] [CrossRef]
91. Zhang J, Zeng C, Xu L, Gong J, Fang J, Zhuang S. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2014;33(31):4069–76. doi:10.1038/onc.2013.369. [Google Scholar] [PubMed] [CrossRef]
92. Zhang J-G, Shi Y, Hong D-F, Song M, Huang D, Wang C-Y, et al. MiR-148b suppresses cell proliferation and invasion in hepatocellular carcinoma by targeting WNT1/β-catenin pathway. Sci Rep. 2015;5(1):8087. doi:10.1038/srep08087. [Google Scholar] [PubMed] [CrossRef]
93. Zhang J, Zhang H, Liu J, Tu X, Zang Y, Zhu J, et al. miR-30 inhibits TGF-β1-induced epithelial-to-mesenchymal transition in hepatocyte by targeting Snail1. Biochem Biophys Res Commun. 2012;417(3):1100–5. doi:10.1016/j.bbrc.2011.12.121. [Google Scholar] [PubMed] [CrossRef]
94. Shen G, Jia H, Tai Q, Li Y, Chen D. miR-106b downregulates adenomatous polyposis coli and promotes cell proliferation in human hepatocellular carcinoma. Carcinogenesis. 2012;34(1):211–9. [Google Scholar] [PubMed]
95. Huang J-L, Fu Y-P, Gan W, Liu G, Zhou P-Y, Zhou C, et al. Hepatic stellate cells promote the progression of hepatocellular carcinoma through microRNA-1246-RORα-Wnt/β-Catenin axis. Cancer Lett. 2020;476:140–51. doi:10.1016/j.canlet.2020.02.012. [Google Scholar] [CrossRef]
96. Xia H, Ooi LLP, Hui KM. MiR-214 targets β-catenin pathway to suppress invasion, stem-like traits and recurrence of human hepatocellular carcinoma. PLoS One. 2012;7(9):e44206. doi:10.1371/journal.pone.0044206. [Google Scholar] [PubMed] [CrossRef]
97. Wu S, Chen S, Lin N, Yang J. Long non-coding RNA SUMO1P3 promotes hepatocellular carcinoma progression through activating Wnt/β-catenin signalling pathway by targeting miR-320a. J Cell Mol Med. 2020;24(5):3108–16. doi:10.1111/jcmm.v24.5. [Google Scholar] [CrossRef]
98. Hill L, Browne G, Tulchinsky E. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer. 2013;132(4):745–54. doi:10.1002/ijc.v132.4. [Google Scholar] [CrossRef]
99. Liu J, Tao C. Overexpression of miRNA-125a-5p inhibits the growth and angiogenesis of hepatocellular carcinoma by regulating the expression of VEGF-A. Biotechnol Biotechnol Equip. 2019;33(1):1116–25. doi:10.1080/13102818.2019.1640073. [Google Scholar] [CrossRef]
100. Ghosh A, Dasgupta D, Ghosh A, Roychoudhury S, Kumar D, Gorain M, et al. MiRNA199a-3p suppresses tumor growth, migration, invasion and angiogenesis in hepatocellular carcinoma by targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 2017;8(3):e2706. doi:10.1038/cddis.2017.123. [Google Scholar] [CrossRef]
101. Lin X-J, Fang J-H, Yang X-J, Zhang C, Yuan Y, Zheng L, et al. Hepatocellular carcinoma cell-secreted exosomal microRNA-210 promotes angiogenesis in vitro and in vivo. Mol Ther-Nucleic Acids. 2018;11:243–52. doi:10.1016/j.omtn.2018.02.014. [Google Scholar] [PubMed] [CrossRef]
102. Moh-Moh-Aung A, Fujisawa M, Ito S, Katayama H, Ohara T, Ota Y, et al. Decreased miR-200b-3p in cancer cells leads to angiogenesis in HCC by enhancing endothelial ERG expression. Sci Rep. 2020;10(1):10418. doi:10.1038/s41598-020-67425-4. [Google Scholar] [PubMed] [CrossRef]
103. Wang Y, Sun B, Sun H, Zhao X, Wang X, Zhao N, et al. Regulation of proliferation, angiogenesis and apoptosis in hepatocellular carcinoma by miR-26b-5p. Tumor Biol. 2016;37(8):10965–79. doi:10.1007/s13277-016-4964-7. [Google Scholar] [PubMed] [CrossRef]
104. Wang Q, Wang G, Niu L, Zhao S, Li J, Zhang Z, et al. Exosomal miR-1290 promotes angiogenesis of hepatocellular carcinoma via targeting SMEK1. J Oncol. 2021;2021:1–13. doi:10.1155/2021/6617700. [Google Scholar] [PubMed] [CrossRef]
105. Zhu K, Pan Q, Zhang X, Kong L-Q, Fan J, Dai Z, et al. MiR-146a enhances angiogenic activity of endothelial cells in hepatocellular carcinoma by promoting PDGFRA expression. Carcinogenesis. 2013;34(9):2071–9. doi:10.1093/carcin/bgt160. [Google Scholar] [CrossRef]
106. Matsuura Y, Wada H, Eguchi H, Gotoh K, Kobayashi S, Kinoshita M, et al. Exosomal miR-155 derived from hepatocellular carcinoma cells under hypoxia promotes angiogenesis in endothelial cells. Dig Dis Sci. 2019;64(3):792–802. doi:10.1007/s10620-018-5380-1. [Google Scholar] [PubMed] [CrossRef]
107. Hu M-H, Ma C-Y, Wang X-M, Ye C-D, Zhang G-X, Chen L, et al. MicroRNA-126 inhibits tumor proliferation and angiogenesis of hepatocellular carcinoma by down-regulating EGFL7 expression. Oncotarget. 2016;7(41):66922–34. doi:10.18632/oncotarget.11877. [Google Scholar] [PubMed] [CrossRef]
108. Hou Z-H, Xu X-W, Fu X-Y, Zhou L-D, Liu S-P, Tan D-M. Long non-coding RNA MALAT1 promotes angiogenesis and immunosuppressive properties of HCC cells by sponging miR-140. Am J Physiol-Cell Physiol. 2020;318(3):C649–63. doi:10.1152/ajpcell.00510.2018. [Google Scholar] [PubMed] [CrossRef]
109. Cheng B, Ding F, Huang C, Xiao H, Fei F, Li J. Role of miR-16-5p in the proliferation and metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(1):137–45. [Google Scholar] [PubMed]
110. Li B, Zhou J, Luo Y, Tao K, Zhang L, Zhao Y, et al. Suppressing circ_0008494 inhibits HSCs activation by regulating the miR-185-3p/Col1a1 axis. Front Pharmacol. 2022;13:1050093. doi:10.3389/fphar.2022.1050093. [Google Scholar] [PubMed] [CrossRef]
111. Zhi Q, Zhu J, Guo X, He S, Xue X, Zhou J, et al. Metastasis-related miR-185 is a potential prognostic biomarker for hepatocellular carcinoma in early stage. Biomed Pharmacother. 2013;67(5):393–8. doi:10.1016/j.biopha.2013.03.022. [Google Scholar] [PubMed] [CrossRef]
112. Xu X-P, Peng X-Q, Yin X-M, Liu Y, Shi Z-Y. miR-34a-5p suppresses the invasion and metastasis of liver cancer by targeting the transcription factor YY1 to mediate MYCT1 upregulation. Acta Histochem. 2020;122(6):151576. doi:10.1016/j.acthis.2020.151576. [Google Scholar] [PubMed] [CrossRef]
113. Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275(1):44–53. doi:10.1016/j.canlet.2008.09.035. [Google Scholar] [PubMed] [CrossRef]
114. Ding G, Peng Z, Shang J, Kang Y, Ning H, Mao C. LincRNA-p21 inhibits invasion and metastasis of hepatocellular carcinoma through miR-9/E-cadherin cascade signaling pathway molecular mechanism. Onco Targets Ther. 2017;3241–7. doi:10.2147/OTT. [Google Scholar] [CrossRef]
115. Sun Z, Han Q, Zhou N, Wang S, Lu S, Bai C, et al. MicroRNA-9 enhances migration and invasion through KLF17 in hepatocellular carcinoma. Mol Oncol. 2013;7(5):884–94. doi:10.1016/j.molonc.2013.04.007. [Google Scholar] [CrossRef]
116. Ji J, Zhao L, Budhu A, Forgues M, Jia H-L, Qin L-X, et al. Let-7g targets collagen type I α2 and inhibits cell migration in hepatocellular carcinoma. J Hepatol. 2010;52(5):690–7. doi:10.1016/j.jhep.2009.12.025. [Google Scholar] [PubMed] [CrossRef]
117. Chen K-J, Hou Y, Wang K, Li J, Xia Y, Yang X-Y, et al. Reexpression of let-7g microRNA inhibits the proliferation and migration via K-Ras/HMGA2/snail axis in hepatocellular carcinoma. Biomed Res Int. 2014;2014(1):742417. [Google Scholar]
118. Li X, Wu H, Wu M, Feng Y, Wu S, Shen X, et al. Hypoxia-related miR-210-5p and miR-210-3p regulate hypoxia-induced migration and epithelial-mesenchymal transition in hepatoma cells. Int J Clin Exp Med. 2019;12:5096–104. [Google Scholar]
119. Liang H-W, Wang N, Wang Y, Wang F, Fu Z, Yan X, et al. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J Hepatol. 2016;64(2):278–91. doi:10.1016/j.jhep.2015.09.013. [Google Scholar] [PubMed] [CrossRef]
120. Wong CCL, Wong CM, Tung EKK, Au SLK, Lee JMF, Poon RTP, et al. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140(1):322–31. doi:10.1053/j.gastro.2010.10.006. [Google Scholar] [CrossRef]
121. Qiu G, Lin Y, Zhang H, Wu D. miR-139-5p inhibits epithelial-mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Commun. 2015;463(3):315–21. doi:10.1016/j.bbrc.2015.05.062. [Google Scholar] [CrossRef]
122. Georges SA, Biery MC, Kim S-Y, Schelter JM, Guo J, Chang AN, et al. Coordinated regulation of cell cycle transcripts by p53-inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68(24):10105–12. doi:10.1158/0008-5472.CAN-08-1846. [Google Scholar] [PubMed] [CrossRef]
123. Lian J, Jing Y, Dong Q, Huan L, Chen D, Bao C, et al. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7(3):2672. doi:10.18632/oncotarget.6603. [Google Scholar] [PubMed] [CrossRef]
124. Arzumanyan A, Friedman T, Kotei E, Ng IO, Lian Z, Feitelson MA. Epigenetic repression of E-cadherin expression by hepatitis B virus x antigen in liver cancer. Oncogene. 2012;31(5):563–72. doi:10.1038/onc.2011.255. [Google Scholar] [CrossRef]
125. Wang Y, Sun B, Zhao X, Zhao N, Sun R, Zhu D, et al. Twist1-related miR-26b-5p suppresses epithelial-mesenchymal transition, migration and invasion by targeting SMAD1 in hepatocellular carcinoma. Oncotarget. 2016;7(17):24383–401. doi:10.18632/oncotarget.8328. [Google Scholar] [PubMed] [CrossRef]
126. He Y, Huang H, Jin L, Zhang F, Zeng M, Wei L, et al. CircZNF609 enhances hepatocellular carcinoma cell proliferation, metastasis, and stemness by activating the Hedgehog pathway through the regulation of miR-15a-5p/15b-5p and GLI2 expressions. Cell Death Dis. 2020;11(5):358. doi:10.1038/s41419-020-2441-0. [Google Scholar] [PubMed] [CrossRef]
127. Li Y, Li D, Yang Y, Wang J. miR-15a-5p regulates liver cancer cell migration, apoptosis and cell cycle progression by targeting transcription factor E2F3. Crit Rev Eukaryot Gene Expr. 2022;32(6):1–10. doi:10.1615/CritRevEukaryotGeneExpr.2022042503. [Google Scholar] [PubMed] [CrossRef]
128. Zheng J, Cheng D, Wu D, Wang L, Qu F, Wu X, et al. MiR-452-5p mediates the proliferation, migration and invasion of hepatocellular carcinoma cells via targeting COLEC10. Pers Med. 2021;18(2):97–106. doi:10.2217/pme-2020-0027. [Google Scholar] [PubMed] [CrossRef]
129. Zhu L, Yang N, Chen J, Zeng T, Yan S, Liu Y, et al. LINC00052 upregulates EPB41L3 to inhibit migration and invasion of hepatocellular carcinoma by binding miR-452-5p. Oncotarget. 2017;8(38):63724–37. doi:10.18632/oncotarget.18892. [Google Scholar] [PubMed] [CrossRef]
130. Wang J, Zhu Y, Ai X, Wan H, Jia W, Chu J, et al. Long noncoding RNA 02027 inhibits proliferation, migration and invasion of hepatocellular carcinoma via miR-625-3p/PDLIM5 pathway. J Gene Med. 2023;25(6):e3485. doi:10.1002/jgm.3485; [Google Scholar] [CrossRef]
131. Chen S. LINC00852 regulates cell proliferation, invasion, migration and apoptosis in hepatocellular carcinoma Via the miR-625/E2F1 Axis. Cell Mol Bioeng. 2022;15(2):207–17. doi:10.1007/s12195-021-00714-8. [Google Scholar] [PubMed] [CrossRef]
132. Fu Y, Sun L-Q, Huang Y, Quan J, Hu X, Tang D, et al. miR-142-3p inhibits the metastasis of hepatocellular carcinoma cells by regulating HMGB1 gene expression. Curr Mol Med. 2018;18(3):135–41. doi:10.2174/1566524018666180907161124. [Google Scholar] [PubMed] [CrossRef]
133. He C, Liu Z, Jin L, Zhang F, Peng X, Xiao Y, et al. lncRNA TUG1-mediated miR-142-3p downregulation contributes to metastasis and the epithelial-to-mesenchymal transition of hepatocellular carcinoma by targeting ZEB1. Cell Physiol Biochem. 2018;48(5):1928–41. doi:10.1159/000492517. [Google Scholar] [PubMed] [CrossRef]
134. Du C, Lv Z, Cao L, Ding C, Gyabaah O-AK, Xie H, et al. MiR-126-3p suppresses tumor metastasis and angiogenesis of hepatocellular carcinoma by targeting LRP6 and PIK3R2. J Transl Med. 2014;12(1):1–11. doi:10.1186/s12967-014-0259-1. [Google Scholar] [PubMed] [CrossRef]
135. Wei G-Y, Hu M, Zhao L, Guo W-S. MiR-451a suppresses cell proliferation, metastasis and EMT via targeting YWHAZ in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23(12):5158–67. [Google Scholar] [PubMed]
136. Xu Y, Lai Y, Cao L, Li Y, Chen G, Chen L, et al. Human umbilical cord mesenchymal stem cells-derived exosomal microRNA-451a represses epithelial-mesenchymal transition of hepatocellular carcinoma cells by inhibiting ADAM10. RNA Biol. 2021;18(10):1408–23. doi:10.1080/15476286.2020.1851540. [Google Scholar] [PubMed] [CrossRef]
137. Yang Y, Yang T, Zhao Z, Zhang H, Yuan P, Wang G, et al. Down-regulation of BMAL1 by miR-494-3p promotes hepatocellular carcinoma growth and metastasis by increasing GPAM-mediated Lipid biosynthesis. Int J Biol Sci. 2022;18(16):6129–44. doi:10.7150/ijbs.74951. [Google Scholar] [PubMed] [CrossRef]
138. Gungor MZ, Uysal M, Senturk S. The bright and the dark side of TGF-β signaling in hepatocellular carcinoma: mechanisms, dysregulation, and therapeutic implications. Cancers. 2022;14(4):940. doi:10.3390/cancers14040940. [Google Scholar] [PubMed] [CrossRef]
139. Medhat A, Arzumanyan A, Feitelson MA. Hepatitis B x antigen (HBx) is an important therapeutic target in the pathogenesis of hepatocellular carcinoma. Oncotarget. 2021;12(24):2421–33. doi:10.18632/oncotarget.28077. [Google Scholar] [CrossRef]
140. Shoraka S, Hosseinian SM, Hasibi A, Ghaemi A, Mohebbi SR. The role of hepatitis B virus genome variations in HBV-related HCC: effects on host signaling pathways. Front Microbiol. 2023;14:1213145. doi:10.3389/fmicb.2023.1213145. [Google Scholar] [PubMed] [CrossRef]
141. Martin J, Dufour J-F. Tumor suppressor and hepatocellular carcinoma. World J Gastroenterol. 2008;14(11):1720. doi:10.3748/wjg.14.1720. [Google Scholar] [PubMed] [CrossRef]
142. Guo G-H, Tan D-M, Zhu PA, Liu F. Hepatitis B virus X protein promotes proliferation and upregulates TGF-beta1 and CTGF in human hepatic stellate cell line, LX-2. Hepatobiliary pancreat Dis Int: HBPD INT. 2009;8(1):59–64. [Google Scholar]
143. Khan MGM, Ghosh A, Variya B, Santharam MA, Ihsan AU, Ramanathan S, et al. Prognostic significance of SOCS1 and SOCS3 tumor suppressors and oncogenic signaling pathway genes in hepatocellular carcinoma. BMC Cancer. 2020;20(1):1–18. doi:10.1186/s12885-020-07285-3. [Google Scholar] [PubMed] [CrossRef]
144. Singh AK, Kumar R, Pandey AK. Hepatocellular carcinoma: causes, mechanism of progression and biomarkers. Curr Chem Genom Transl Med. 2018;12(1):9–26. doi:10.2174/2213988501812010009. [Google Scholar] [PubMed] [CrossRef]
145. Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, Masaki T. MicroRNAs in the pathogenesis of hepatocellular carcinoma: a review. Cancers. 2021;13(3):514. doi:10.3390/cancers13030514. [Google Scholar] [PubMed] [CrossRef]
146. Pollutri D, Gramantieri L, Bolondi L, Fornari F. TP53/MicroRNA interplay in hepatocellular carcinoma. Int J Mol Sci. 2016;17(12):2029. doi:10.3390/ijms17122029. [Google Scholar] [PubMed] [CrossRef]
147. Shen L, Zhang X, Hu D, Feng T, Li H, Lu Y, et al. Hepatitis B virus X (HBx) play an anti-apoptosis role in hepatic progenitor cells by activating Wnt/β-catenin pathway. Mol Cell Biochem. 2013;383(1–2):213–22. doi:10.1007/s11010-013-1769-5. [Google Scholar] [PubMed] [CrossRef]
148. Kong F, You H, Zhao J, Liu W, Hu L, Luo W, et al. The enhanced expression of death receptor 5 (DR5) mediated by HBV X protein through NF-kappaB pathway is associated with cell apoptosis induced by (TNF-α related apoptosis inducing ligand) TRAIL in hepatoma cells. Virol J. 2015;12(1):1–9. doi:10.1186/s12985-015-0416-z. [Google Scholar] [PubMed] [CrossRef]
149. Zhu AX, Duda DG, Sahani DV, Jain RK. HCC and angiogenesis: possible targets and future directions. Nat Rev Clin Oncol. 2011;8(5):292–301. doi:10.1038/nrclinonc.2011.30. [Google Scholar] [PubMed] [CrossRef]
150. Zhang J, Chen WN. Inhibition of HBV-induced angiogenesis by ibuprofen: role of HBx. Interv Med Appl Sci. 2012;4(1):21–31. [Google Scholar]
151. Fukumura D, Xavier R, Sugiura T, Chen Y, Park E-C, Lu N, et al. Tumor induction of VEGF promoter activity in stromal cells. Cell. 1998;94(6):715–25. doi:10.1016/S0092-8674(00)81731-6. [Google Scholar] [CrossRef]
152. Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2(10):737–44. doi:10.1038/35036374. [Google Scholar] [PubMed] [CrossRef]
153. Semela D, Dufour J-F. Angiogenesis and hepatocellular carcinoma. J Hepatol. 2004;41(5):864–80. doi:10.1016/j.jhep.2004.09.006. [Google Scholar] [PubMed] [CrossRef]
154. Li W, Tan D, Zhang Z, Liang JJ, Brown RE. Activation of Akt-mTOR-p70S6K pathway in angiogenesis in hepatocellular carcinoma. Oncol Rep. 2008;20(4):713–9. [Google Scholar]
155. Qu B, Liu BR, Du YJ, Chen J, Cheng YQ, Xu W, et al. Wnt/β‐catenin signaling pathway may regulate the expression of angiogenic growth factors in hepatocellular carcinoma. Oncol Lett. 2014;7(4):1175–8. doi:10.3892/ol.2014.1828. [Google Scholar] [PubMed] [CrossRef]
156. Morse MA, Sun W, Kim R, He AR, Abada PB, Mynderse M, et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res. 2019;25(3):912–20. doi:10.1158/1078-0432.CCR-18-1254. [Google Scholar] [PubMed] [CrossRef]
157. Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1):S84–101. doi:10.1016/j.jhep.2016.02.021. [Google Scholar] [PubMed] [CrossRef]
158. Rana MA, Ijaz B, Daud M, Tariq S, Nadeem T, Husnain T. Interplay of Wnt β-catenin pathway and miRNAs in HBV pathogenesis leading to HCC. Clin Res Hepatol Gastroenterol. 2019;43(4):373–86. doi:10.1016/j.clinre.2018.09.012. [Google Scholar] [CrossRef]
159. Cha MY, Kim CM, Park YM, Ryu WS. Hepatitis B virus X protein is essential for the activation of Wnt/β-catenin signaling in hepatoma cells. Hepatology. 2004;39(6):1683–93. doi:10.1002/(ISSN)1527-3350. [Google Scholar] [CrossRef]
160. Hsieh A, Kim H-S, Lim S-O, Yu D-Y, Jung G. Hepatitis B viral X protein interacts with tumor suppressor adenomatous polyposis coli to activate Wnt/β-catenin signaling. Cancer Lett. 2011;300(2):162–72. doi:10.1016/j.canlet.2010.09.018. [Google Scholar] [PubMed] [CrossRef]
161. Iwai N, Yasui K, Tomie A, Gen Y, Terasaki K, Kitaichi T, et al. Oncogenic miR-96-5p inhibits apoptosis by targeting the caspase-9 gene in hepatocellular carcinoma. Int J Oncol. 2018;53(1):237–45. doi:10.3892/ijo.2018.4369. [Google Scholar] [CrossRef]
162. Yuan F, Tang Y, Cao M, Ren Y, Li Y, Yang G, et al. Identification of the hsa_circ_0039466/miR-96-5p/FOXO1 regulatory network in hepatocellular carcinoma by whole-transcriptome analysis. Ann Transl Med. 2022;10(14):769. doi:10.21037/atm-22-3147. [Google Scholar] [PubMed] [CrossRef]
163. Assal RA, El Tayebi HM, Hosny KA, Esmat G, Abdelaziz AI. A pleiotropic effect of the single clustered hepatic metastamiRs miR-96-5p and miR-182-5p on insulin-like growth factor II, insulin-like growth factor-1 receptor and insulin-like growth factor-binding protein-3 in hepatocellular carcinoma. Mol Med Rep. 2015;12(1):645–50. doi:10.3892/mmr.2015.3382. [Google Scholar] [PubMed] [CrossRef]
164. Matsui T, Hamada-Tsutsumi S, Naito Y, Nojima M, Iio E, Tamori A, et al. Identification of microRNA-96-5p as a postoperative, prognostic microRNA predictor in nonviral hepatocellular carcinoma. Hepatol Res. 2022;52(1):93–104. doi:10.1111/hepr.13674. [Google Scholar] [PubMed] [CrossRef]
165. Moshiri F, Salvi A, Gramantieri L, Sangiovanni A, Guerriero P, De Petro G, et al. Circulating miR-106b-3p, miR-101-3p and miR-1246 as diagnostic biomarkers of hepatocellular carcinoma. Oncotarget. 2018;9(20):15350–64. doi:10.18632/oncotarget.24601. [Google Scholar] [CrossRef]
166. Chai S, Ng KY, Tong M, Lau EY, Lee TK, Chan KW, et al. Octamer 4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in liver cancer stem cells. Hepatology. 2016;64(6):2062–76. doi:10.1002/hep.28821. [Google Scholar] [PubMed] [CrossRef]
167. Chuma M, Toyoda H, Matsuzaki J, Saito Y, Kumada T, Tada T, et al. Circulating microRNA-1246 as a possible biomarker for early tumor recurrence of hepatocellular carcinoma. Hepatol Res. 2019;49(7):810–22. doi:10.1111/hepr.13338. [Google Scholar] [PubMed] [CrossRef]
168. Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai C, et al. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC cancer. 2014;14(1):1–11. doi:10.1186/1471-2407-14-616. [Google Scholar] [PubMed] [CrossRef]
169. Morishita A, Fujita K, Iwama H, Chiyo T, Fujihara S, Oura K, et al. Role of microRNA-210-3p in hepatitis B virus-related hepatocellular carcinoma. Am J Physiol-Gastrointest Liver Physiol. 2020;318(3):G401–G9. doi:10.1152/ajpgi.00269.2019. [Google Scholar] [PubMed] [CrossRef]
170. You R, Yang Y, Yin G, Jiang H, Lu Y, Gui L, et al. CPEB2 suppresses hepatocellular carcinoma epithelial-mesenchymal transition and metastasis through regulating the HIF-1α/miR-210-3p/CPEB2 axis. Pharmaceutics. 2023;15(7):1887. doi:10.3390/pharmaceutics15071887. [Google Scholar] [PubMed] [CrossRef]
171. Sartorius K, An P, Winkler C, Chuturgoon A, Li X, Makarova J, et al. The epigenetic modulation of cancer and immune pathways in hepatitis B virus-associated hepatocellular carcinoma: the influence of HBx and miRNA dysregulation. Front Immunol. 2021;12:661204. doi:10.3389/fimmu.2021.661204. [Google Scholar] [PubMed] [CrossRef]
172. Mangiavacchi A, Morelli G, Orlando V. Behind the scenes: how RNA orchestrates the epigenetic regulation of gene expression. Front Cell Dev Biol. 2023;11:1123975. doi:10.3389/fcell.2023.1123975. [Google Scholar] [PubMed] [CrossRef]
173. Yao Q, Chen Y, Zhou X. The roles of microRNAs in epigenetic regulation. Curr Opin Chem Biol. 2019;51:11–7. doi:10.1016/j.cbpa.2019.01.024. [Google Scholar] [PubMed] [CrossRef]
174. Herceg Z, Vaissière T. Epigenetic mechanisms and cancer: an interface between the environment and the genome. Epigenetics. 2011;6(7):804–19. doi:10.4161/epi.6.7.16262. [Google Scholar] [PubMed] [CrossRef]
175. Park IY, Sohn BH, Yu E, Suh DJ, Chung YH, Lee JH, et al. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132(4):1476–94. doi:10.1053/j.gastro.2007.01.034. [Google Scholar] [PubMed] [CrossRef]
176. Zhang D, Guo S, Schrodi SJ. Mechanisms of DNA methylation in virus-host interaction in hepatitis b infection: pathogenesis and oncogenetic properties. Int J Mol Sci. 2021;22(18):9858. doi:10.3390/ijms22189858. [Google Scholar] [PubMed] [CrossRef]
177. Shon JK, Shon BH, Park IY, Lee SU, Fa L, Chang KY, et al. Hepatitis B virus-X protein recruits histone deacetylase 1 to repress insulin-like growth factor binding protein 3 transcription. Virus Res. 2009;139(1):14–21. doi:10.1016/j.virusres.2008.09.006. [Google Scholar] [PubMed] [CrossRef]
178. Zeisel MB, Guerrieri F, Levrero M. Host epigenetic alterations and hepatitis B virus-associated hepatocellular carcinoma. J Clin Med. 2021;10(8):1715. doi:10.3390/jcm10081715. [Google Scholar] [PubMed] [CrossRef]
179. Wang D-Y, An S-H, Liu L, Bai S-S, Wu K-X, Zhu R, et al. Hepatitis B virus X protein influences enrichment profiles of H3K9me3 on promoter regions in human hepatoma cell lines. Oncotarget. 2016;7(51):84883–92. doi:10.18632/oncotarget.12751. [Google Scholar] [PubMed] [CrossRef]
180. Zhang H, Xing Z, Mani SKK, Bancel B, Durantel D, Zoulim F, et al. RNA helicase DEAD box protein 5 regulates Polycomb repressive complex 2/Hox transcript antisense intergenic RNA function in hepatitis B virus infection and hepatocarcinogenesis. Hepatology. 2016;64(4):1033–48. doi:10.1002/hep.28698. [Google Scholar] [PubMed] [CrossRef]
181. Dandri M, Editor. Epigenetic modulation in chronic hepatitis B virus infection. In: Seminars in immunopathology. Berlin Heidelberg, Germany: Springer; 2020. [Google Scholar]
182. Yang H, Lan P, Hou Z, Guan Y, Zhang J, Xu W, et al. Histone deacetylase inhibitor SAHA epigenetically regulates miR-17-92 cluster and MCM7 to upregulate MICA expression in hepatoma. Br J Cancer. 2015;112(1):112–21. doi:10.1038/bjc.2014.547. [Google Scholar] [PubMed] [CrossRef]
183. Bae HJ, Jung KH, Eun JW, Shen Q, Kim HS, Park SJ, et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol. 2015;63(2):408–19. doi:10.1016/j.jhep.2015.03.019. [Google Scholar] [PubMed] [CrossRef]
184. Fornari F, Milazzo M, Galassi M, Callegari E, Veronese A, Miyaaki H, et al. p53/mdm2 feedback loop sustains miR-221 expression and dictates the response to anticancer treatments in hepatocellular carcinoma. Mol Cancer Res. 2014;12(2):203–16. doi:10.1158/1541-7786.MCR-13-0312-T. [Google Scholar] [PubMed] [CrossRef]
185. Kwon JJ, Factora TD, Dey S, Kota J. A systematic review of miR-29 in cancer. Mol Ther-Oncolytics. 2019;12(Suppl 1):173–94. doi:10.1016/j.omto.2018.12.011. [Google Scholar] [PubMed] [CrossRef]
186. Parpart S, Roessler S, Dong F, Rao V, Takai A, Ji J, et al. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology. 2014;60(3):872–83. doi:10.1002/hep.27200. [Google Scholar] [PubMed] [CrossRef]
187. Sarkar N, Chakravarty R. Hepatitis B virus infection, microRNAs and liver disease. Int J Mol Sci. 2015;16(8):17746–62. doi:10.3390/ijms160817746. [Google Scholar] [CrossRef]
188. Morales S, Monzo M, Navarro A. Epigenetic regulation mechanisms of microRNA expression. Biomol Concepts. 2017;8(5–6):203–12. doi:10.1515/bmc-2017-0024. [Google Scholar] [PubMed] [CrossRef]
189. Wong CM, Wei L, Law CT, Ho DWH, Tsang FHC, Au SLK, et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology. 2016;63(2):474–87. doi:10.1002/hep.28304. [Google Scholar] [PubMed] [CrossRef]
190. Au SLK, Wong CCL, Lee JMF, Fan DNY, Tsang FH, Ng IOL, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56(2):622–31. doi:10.1002/hep.25679. [Google Scholar] [PubMed] [CrossRef]
191. Zhu X, Wu L, Xu J, Yang R, Wu F. Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res. 2011;39(6):2323–9. doi:10.1177/147323001103900631. [Google Scholar] [PubMed] [CrossRef]
192. Ma L, Chua M-S, Andrisani O, So S. Epigenetics in hepatocellular carcinoma: an update and future therapy perspectives. World J Gastroenterol. 2014;20(2):333. doi:10.3748/wjg.v20.i2.333. [Google Scholar] [PubMed] [CrossRef]
193. Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation mediated silencing of microRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68(13):5049–58. doi:10.1158/0008-5472.CAN-07-6655. [Google Scholar] [PubMed] [CrossRef]
194. Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, et al. Modulation of hepatitis B virus replication and hepatocyte differentiation by microRNA-1. Hepatology. 2011;53(5):1476–85. doi:10.1002/hep.24195. [Google Scholar] [PubMed] [CrossRef]
195. Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, et al. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25(2):439–46. doi:10.1016/j.cellsig.2012.10.013. [Google Scholar] [PubMed] [CrossRef]
196. Song K, Han C, Zhang J, Lu D, Dash S, Feitelson M, et al. Epigenetic regulation of microRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology. 2013;58(5):1681–92. doi:10.1002/hep.26514. [Google Scholar] [CrossRef]
197. Jung CJ, Iyengar S, Blahnik KR, Ajuha TP, Jiang JX, Farnham PJ, et al. Epigenetic modulation of miR-122 facilitates human embryonic stem cell self-renewal and hepatocellular carcinoma proliferation. PLoS One. 2011;6(11):e27740. doi:10.1371/journal.pone.0027740. [Google Scholar] [PubMed] [CrossRef]
198. Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, et al. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133(3):660–70. doi:10.1002/ijc.28068. [Google Scholar] [PubMed] [CrossRef]
199. Furuta M, Kozaki K-I, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31(5):766–76. doi:10.1093/carcin/bgp250. [Google Scholar] [PubMed] [CrossRef]
200. Coppola N, de Stefano G, Panella M, Onorato L, Iodice V, Minichini C, et al. Lowered expression of microRNA-125a-5p in human hepatocellular carcinoma and up-regulation of its oncogenic targets sirtuin-7, matrix metalloproteinase-11, and c-Raf. Oncotarget. 2017;8(15):25289–99. doi:10.18632/oncotarget.15809. [Google Scholar] [CrossRef]
201. Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57(3):1055–67. doi:10.1002/hep.26101. [Google Scholar] [CrossRef]
202. Ngo-Yin Fan D, Ho-Ching Tsang F, Hoi-Kam Tam A, Leung-Kuen Au S, Chak-Lui Wong C, Wei L, et al. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57(2):637–47. doi:10.1002/hep.26083. [Google Scholar] [PubMed] [CrossRef]
203. White CA, Pone EJ, Lam T, Tat C, Hayama KL, Li G, et al. Histone deacetylase inhibitors upregulate B cell microRNAs that silence AID and Blimp-1 expression for epigenetic modulation of antibody and autoantibody responses. J Immunol. 2014;193(12):5933–50. doi:10.4049/jimmunol.1401702. [Google Scholar] [PubMed] [CrossRef]
204. Wang Z, Wu Z, Huang P. The function of miRNAs in hepatocarcinogenesis induced by hepatitis B virus X protein. Oncol Rep. 2017;38(2):652–64. doi:10.3892/or.2017.5716. [Google Scholar] [PubMed] [CrossRef]
205. Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, et al. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell signal. 2013;25(5):1037–43. doi:10.1016/j.cellsig.2013.01.019. [Google Scholar] [PubMed] [CrossRef]
206. Long X-R, He Y, Huang C, Li J. MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in hepatocellular carcinogenesis. Int J Oncol. 2014;44(6):1915–22. doi:10.3892/ijo.2014.2373. [Google Scholar] [PubMed] [CrossRef]
207. Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51(3):881–90. doi:10.1002/hep.23381. [Google Scholar] [PubMed] [CrossRef]
208. Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52(1):60–70. doi:10.1002/hep.23660. [Google Scholar] [PubMed] [CrossRef]
209. Ji Y, Fioravanti J, Zhu W, Wang H, Wu T, Hu J, et al. miR-155 harnesses Phf19 to potentiate cancer immunotherapy through epigenetic reprogramming of CD8+ T cell fate. Nat Commun. 2019;10(1):1–12. doi:10.1038/s41467-019-09882-8. [Google Scholar] [PubMed] [CrossRef]
210. Moon IY, Choi JH, Chung JW, Jang ES, Jeong SH, Kim JW. MicroRNA‐20 induces methylation of hepatitis B virus covalently closed circular DNA in human hepatoma cells. Mol Med Rep. 2019;20(3):2285–93. doi:10.3892/mmr.2019.10435. [Google Scholar] [PubMed] [CrossRef]
211. Yuan JH, Yang F, Chen BF, Lu Z, Huo XS, Zhou WP, et al. The histone deacetylase 4/SP1/microrna-200a regulatory network contributes to aberrant histone acetylation in hepatocellular carcinoma. Hepatology. 2011;54(6):2025–35. doi:10.1002/hep.24606. [Google Scholar] [PubMed] [CrossRef]
212. Zhang L, Yang F, Yuan J-H, Yuan S-X, Zhou W-P, Huo X-S, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2012;34(3):577–86. doi:10.1093/carcin/bgs381. [Google Scholar] [PubMed] [CrossRef]
213. Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, et al. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15(11):IN24–IN6. doi:10.1593/neo.131362. [Google Scholar] [PubMed] [CrossRef]
214. Wang Y, Toh HC, Chow P, Chung AY, Meyers DJ, Cole PA, et al. MicroRNA-224 is up-regulated in hepatocellular carcinoma through epigenetic mechanisms. FASEB J. 2012;26(7):3032–41. doi:10.1096/fj.11-201855. [Google Scholar] [PubMed] [CrossRef]
215. Wang Y, Lee CG. Role of miR-224 in hepatocellular carcinoma: a tool for possible therapeutic intervention? Epigenomics. 2011;3(2):235–43. doi:10.2217/epi.11.5. [Google Scholar] [PubMed] [CrossRef]
216. Zhuang C, Wang P, Huang D, Xu L, Wang X, Wang L, et al. A double-negative feedback loop between EZH2 and miR-26a regulates tumor cell growth in hepatocellular carcinoma. Int J Oncol. 2016;48(3):1195–204. doi:10.3892/ijo.2016.3336. [Google Scholar] [CrossRef]
217. Zhang X, Zhang X, Wang T, Wang L, Tan Z, Wei W, et al. MicroRNA-26a is a key regulon that inhibits progression and metastasis of c-Myc/EZH2 double high advanced hepatocellular carcinoma. Cancer Lett. 2018;426(6):98–108. doi:10.1016/j.canlet.2018.04.005. [Google Scholar] [PubMed] [CrossRef]
218. Ma D-N, Chai Z-T, Zhu X-D, Zhang N, Zhan D-H, Ye B-G, et al. MicroRNA-26a suppresses epithelial-mesenchymal transition in human hepatocellular carcinoma by repressing enhancer of zeste homolog 2. J Hematol Oncol. 2016;9(1):1–10. doi:10.1186/s13045-015-0229-y. [Google Scholar] [PubMed] [CrossRef]
219. Kim HS, Shen Q, Nam SW. Histone deacetylases and their regulatory microRNAs in hepatocarcinogenesis. J Korean Med Sci. 2015;30(10):1375–80. doi:10.3346/jkms.2015.30.10.1375. [Google Scholar] [PubMed] [CrossRef]
220. Chamani F, Sadeghizadeh M, Masoumi M, Babashah S. Evaluation of miR-34 family and DNA methyltransferases 1, 3A, 3B gene expression levels in hepatocellular carcinoma following treatment with dendrosomal nanocurcumin. Asian Pac J Cancer Prev. 2016;17(S3):219–24. doi:10.7314/APJCP.2016.17.S3.219. [Google Scholar] [PubMed] [CrossRef]
221. Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279–94. doi:10.1038/nri.2016.40. [Google Scholar] [PubMed] [CrossRef]
222. Ceppi M, Pereira PM, Dunand-Sauthier I, Barras E, Reith W, Santos MA, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc Natl Acad Sci. 2009;106(8):2735–40. doi:10.1073/pnas.0811073106. [Google Scholar] [PubMed] [CrossRef]
223. Hashimi ST, Fulcher JA, Chang MH, Gov L, Wang S, Lee B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood. 2009;114(2):404–14. doi:10.1182/blood-2008-09-179150. [Google Scholar] [PubMed] [CrossRef]
224. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–14. doi:10.1038/ni1575. [Google Scholar] [PubMed] [CrossRef]
225. Li Q-J, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129(1):147–61. doi:10.1016/j.cell.2007.03.008. [Google Scholar] [PubMed] [CrossRef]
226. Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’leary JJ, Ruan Q, et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11(2):141. doi:10.1038/ni.1828. [Google Scholar] [PubMed] [CrossRef]
227. Sheedy FJ. Turning 21: induction of miR-21 as a key switch in the inflammatory response. Front Immunol. 2015;6(15):19. doi:10.3389/fimmu.2015.00019. [Google Scholar] [PubMed] [CrossRef]
228. Murugaiyan G, da Cunha AP, Ajay AK, Joller N, Garo LP, Kumaradevan S, et al. MicroRNA-21 promotes Th17 differentiation and mediates experimental autoimmune encephalomyelitis. J Clin Investig. 2015;125(3):1069–80. doi:10.1172/JCI74347. [Google Scholar] [PubMed] [CrossRef]
229. Lu TX, Hartner J, Lim E-J, Fabry V, Mingler MK, Cole ET, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-γ pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187(6):3362–73. doi:10.4049/jimmunol.1101235. [Google Scholar] [PubMed] [CrossRef]
230. Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, et al. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9(4):e1003248. doi:10.1371/journal.ppat.1003248. [Google Scholar] [PubMed] [CrossRef]
231. Ando Y, Yang G-X, Kenny TP, Kawata K, Zhang W, Huang W, et al. Overexpression of microRNA-21 is associated with elevated pro-inflammatory cytokines in dominant-negative TGF-β receptor type II mouse. J Autoimmun. 2013;41:111–9. doi:10.1016/j.jaut.2012.12.013. [Google Scholar] [PubMed] [CrossRef]
232. Xie W, Li M, Xu N, Lv Q, Huang N, He J, et al. MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One. 2013;8(3):e58639. doi:10.1371/journal.pone.0058639. [Google Scholar] [PubMed] [CrossRef]
233. Wu C, Gong Y, Yuan J, Zhang W, Zhao G, Li H, et al. microRNA-181a represses ox-LDL-stimulated inflammatory response in dendritic cell by targeting c-Fos. J Lipid Res. 2012;53(11):2355–63. doi:10.1194/jlr.M028878. [Google Scholar] [PubMed] [CrossRef]
234. Prieto J. Inflammation, HCC and sex: IL-6 in the centre of the triangle. J hepatol. 2008;48(2):380–1. doi:10.1016/j.jhep.2007.11.007. [Google Scholar] [PubMed] [CrossRef]
235. Xie W, Li Z, Li M, Xu N, Zhang Y. miR-181a and inflammation: miRNA homeostasis response to inflammatory stimuli in vivo. Biochem Biophys Res Commun. 2013;430(2):647–52. doi:10.1016/j.bbrc.2012.11.097. [Google Scholar] [PubMed] [CrossRef]
236. Cichocki F, Felices M, McCullar V, Presnell SR, Al-Attar A, Lutz CT, et al. Cutting edge: microRNA-181 promotes human NK cell development by regulating Notch signaling. J Immunol. 2011;187(12):6171–5. doi:10.4049/jimmunol.1100835. [Google Scholar] [PubMed] [CrossRef]
237. Chen C-Z, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–6. doi:10.1126/science.1091903. [Google Scholar] [PubMed] [CrossRef]
238. Fontana L, Pelosi E, Greco P, Racanicchi S, Testa U, Liuzzi F, et al. MicroRNAs 17-5p-20a–106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9(7):775–87. doi:10.1038/ncb1613. [Google Scholar] [PubMed] [CrossRef]
239. Dougherty G, Duncan MB, Rohlman CE, Rehman A, Thakur P. The role of CSF1 in hepatocellular carcinoma-recruited macrophages. Fed Am Societies Exp Biol. 2013;27(S1):794–19. [Google Scholar]
240. Sasaki M, Tsuneyama K, Ishikawa A, Nakanuma Y. Intrahepatic cholangiocarcinoma in cirrhosis presents granulocyte and granulocyte-macrophage colony-stimulating factor. Hum Pathol. 2003;34(12):1337–44. doi:10.1016/j.humpath.2003.07.012. [Google Scholar] [PubMed] [CrossRef]
241. Jiang S, Li C, Olive V, Lykken E, Feng F, Sevilla J, et al. Molecular dissection of the miR-17-92 cluster’s critical dual roles in promoting Th1 responses and preventing inducible Treg differentiation. Blood. 2011;118(20):5487–97. doi:10.1182/blood-2011-05-355644. [Google Scholar] [PubMed] [CrossRef]
242. Blevins R, Bruno L, Carroll T, Elliott J, Marcais A, Loh C, et al. microRNAs regulate cell-to-cell variability of endogenous target gene expression in developing mouse thymocytes. PLoS Genet. 2015;11(2):e1005020. doi:10.1371/journal.pgen.1005020. [Google Scholar] [PubMed] [CrossRef]
243. Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–74. doi:10.1016/j.cell.2008.02.020. [Google Scholar] [PubMed] [CrossRef]
244. Kang SG, Liu W-H, Lu P, Jin HY, Lim HW, Shepherd J, et al. MicroRNAs of the miR-17∼92 family are critical regulators of TFH differentiation. Nat Immunol. 2013;14(8):849–57. doi:10.1038/ni.2648. [Google Scholar] [PubMed] [CrossRef]
245. Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim et Biophys Acta (BBA)-Mol Basis Dis. 2009;1792(6):497–505. doi:10.1016/j.bbadis.2009.02.013. [Google Scholar] [CrossRef]
246. O’connell RM, Rao DS, Chaudhuri AA, Baltimore D. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–22. doi:10.1038/nri2708. [Google Scholar] [PubMed] [CrossRef]
247. O’Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci. 2007;104(5):1604–9. doi:10.1073/pnas.0610731104. [Google Scholar] [PubMed] [CrossRef]
248. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–11. doi:10.1126/science.1139253; [Google Scholar] [CrossRef]
249. Yang F, Li Q-J, Gong Z-B, Zhou L, You N, Wang S, et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13(1):77–86. doi:10.7785/tcrt.2012.500364. [Google Scholar] [PubMed] [CrossRef]
250. Ou Q, Wang G, Li B, Li W-F. Decreased miR-34a promotes growth by regulating MAP4K4 in hepatitis B virus related hepatocellular carcinoma. Int J Clin Exp Med. 2017;10(2):2523–31. [Google Scholar]
251. Peltier HJ, Kelnar K, Bader AG. Effects of MRX34, a liposomal miR-34 mimic, on target gene expression in human white blood cells (hWBCsqRT-PCR results from a first-in-human trial of microRNA cancer therapy. Ann Oncol. 2016;27:vi531. doi:10.1093/annonc/mdw392.16. [Google Scholar] [CrossRef]
252. Kim T, Croce CM. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp Mol Med. 2023;55(7):1314–21. doi:10.1038/s12276-023-01050-9. [Google Scholar] [PubMed] [CrossRef]
253. Yang P, Li Q-J, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22(3):291–303. doi:10.1016/j.ccr.2012.07.023. [Google Scholar] [PubMed] [CrossRef]
254. Telford BJ, Yahyanejad S, de Gunst T, den Boer HC, Vos RM, Stegink M, et al. Multi-modal effects of 1B3, a novel synthetic miR-193a-3p mimic, support strong potential for therapeutic intervention in oncology. Oncotarget. 2021;12(5):422–39. doi:10.18632/oncotarget.27894. [Google Scholar] [PubMed] [CrossRef]
255. Querfeld C, Foss FM, Pinter-Brown LC, Porcu P, William BM, Pacheco T, et al. Phase 1 study of the safety and efficacy of MRG-106, a synthetic inhibitor of microRNA-155, in CTCL patients. Blood. 2017;130(Suppl_1):820. doi:10.1182/blood.V130.Suppl_1.820.820. [Google Scholar] [CrossRef]
256. Segal M, Slack FJ. Challenges identifying efficacious miRNA therapeutics for cancer. Expert Opin Drug Discov. 2020;15(9):987–91. doi:10.1080/17460441.2020.1765770. [Google Scholar] [PubMed] [CrossRef]
257. Porta-Pardo E, Valencia A, Godzik A. Understanding oncogenicity of cancer driver genes and mutations in the cancer genomics era. FEBS Lett. 2020;594(24):4233–46. doi:10.1002/1873-3468.13781. [Google Scholar] [PubMed] [CrossRef]
258. Nault JC, Martin Y, Caruso S, Hirsch TZ, Bayard Q, Calderaro J, et al. Clinical impact of genomic diversity from early to advanced hepatocellular carcinoma. Hepatology. 2020;71(1):164–82. doi:10.1002/hep.30811. [Google Scholar] [PubMed] [CrossRef]
259. Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–65. doi:10.1016/j.cell.2013.03.043. [Google Scholar] [PubMed] [CrossRef]
260. Bai X, Liu Z, Shao X, Wang D, Dong E, Wang Y, et al. The heterogeneity of plasma miRNA profiles in hepatocellular carcinoma patients and the exploration of diagnostic circulating miRNAs for hepatocellular carcinoma. PLoS One. 2019;14(2):e0211581. doi:10.1371/journal.pone.0211581. [Google Scholar] [PubMed] [CrossRef]
261. Zhu J, Zheng Z, Wang J, Sun J, Wang P, Cheng X, et al. Different miRNA expression profiles between human breast cancer tumors and serum. Front Genet. 2014;5:149. doi:10.3389/fgene.2014.00149. [Google Scholar] [PubMed] [CrossRef]
262. Gmerek L, Martyniak K, Horbacka K, Krokowicz P, Scierski W, Golusinski P, et al. MicroRNA regulation in colorectal cancer tissue and serum. PLoS One. 2019;14(8):e0222013. doi:10.1371/journal.pone.0222013. [Google Scholar] [PubMed] [CrossRef]
263. Zhang S, Cheng Z, Wang Y, Han T. The risks of miRNA therapeutics: in a drug target perspective. Drug Des Devel Ther. 2021;15:721–33. doi:10.2147/DDDT.S288859. [Google Scholar] [PubMed] [CrossRef]
264. Hu B, Zhong L, Weng Y, Peng L, Huang Y, Zhao Y, et al. Therapeutic siRNA: state of the art. Signal Transduct Target Ther. 2020;5(1):101. doi:10.1038/s41392-020-0207-x. [Google Scholar] [PubMed] [CrossRef]
265. Lam JK, Chow MY, Zhang Y, Leung SW. siRNA versus miRNA as therapeutics for gene silencing. Mol Ther-Nucleic Acids. 2015;4:e252. doi:10.1038/mtna.2015.23. [Google Scholar] [PubMed] [CrossRef]
266. Jin X, Cai C, Qiu Y. Diagnostic value of circulating microRNAs in hepatitis B virus-related hepatocellular carcinoma: a systematic review and meta-analysis. J Cancer. 2019;10(20):4754–64. doi:10.7150/jca.32833. [Google Scholar] [PubMed] [CrossRef]
267. Rui T, Zhang X, Guo J, Xiang A, Tang N, Liu J, et al. Serum-exosome-derived miRNAs serve as promising biomarkers for HCC diagnosis. Cancers. 2022;15(1):205. doi:10.3390/cancers15010205. [Google Scholar] [PubMed] [CrossRef]
268. Yousuf T, Dar SB, Bangri SA, Choh NA, Rasool Z, Shah A, et al. Diagnostic implication of a circulating serum-based three-microRNA signature in hepatocellular carcinoma. Front Genet. 2022;13:929787. doi:10.3389/fgene.2022.929787. [Google Scholar] [PubMed] [CrossRef]
269. Leidner RS, Li L, Thompson CL. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 2013;8(3):e57841. doi:10.1371/journal.pone.0057841. [Google Scholar] [PubMed] [CrossRef]
270. Egidi MG, Cochetti G, Serva MR, Guelfi G, Zampini D, Mechelli L, et al. Circulating microRNAs and kallikreins before and after radical prostatectomy: are they really prostate cancer markers? Biomed Res Int. 2013;2013(1):241780–11. doi:10.1155/2013/241780. [Google Scholar] [PubMed] [CrossRef]
271. Precazzini F, Detassis S, Imperatori AS, Denti MA, Campomenosi P. Measurements methods for the development of MicroRNA-based tests for cancer diagnosis. Int J Mol Sci. 2021;22(3):1176. doi:10.3390/ijms22031176. [Google Scholar] [PubMed] [CrossRef]
272. Saunders K, Bert AG, Dredge BK, Toubia J, Gregory PA, Pillman KA, et al. Insufficiently complex unique-molecular identifiers (UMIs) distort small RNA sequencing. Sci Rep. 2020;10(1):14593. doi:10.1038/s41598-020-71323-0. [Google Scholar] [PubMed] [CrossRef]
273. Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Brief bioinform. 2019;20(5):1836–52. doi:10.1093/bib/bby054. [Google Scholar] [PubMed] [CrossRef]
274. Lu H, Zhang J, Cao Y, Wu S, Wei Y, Yin R. Advances in applications of artificial intelligence algorithms for cancer-related miRNA research. Zhejiang da xue xue bao. Yi xue ban = J Zhejiang Univ Med Sci. 2024;53(2):231–43 (In Chinese). [Google Scholar]
275. Wang Z. The principles of MiRNA-masking antisense oligonucleotides technology. MicroRNA Cancer: Methods Protocols. 2011;676:43–9. doi:10.1007/978-1-60761-863-8. [Google Scholar] [CrossRef]
276. Johnson P, Zhou Q, Dao DY, Lo YD. Circulating biomarkers in the diagnosis and management of hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2022;19(10):670–81. doi:10.1038/s41575-022-00620-y. [Google Scholar] [PubMed] [CrossRef]
277. Pigati L, Yaddanapudi SC, Iyengar R, Kim D-J, Hearn SA, Danforth D, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515. doi:10.1371/journal.pone.0013515. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF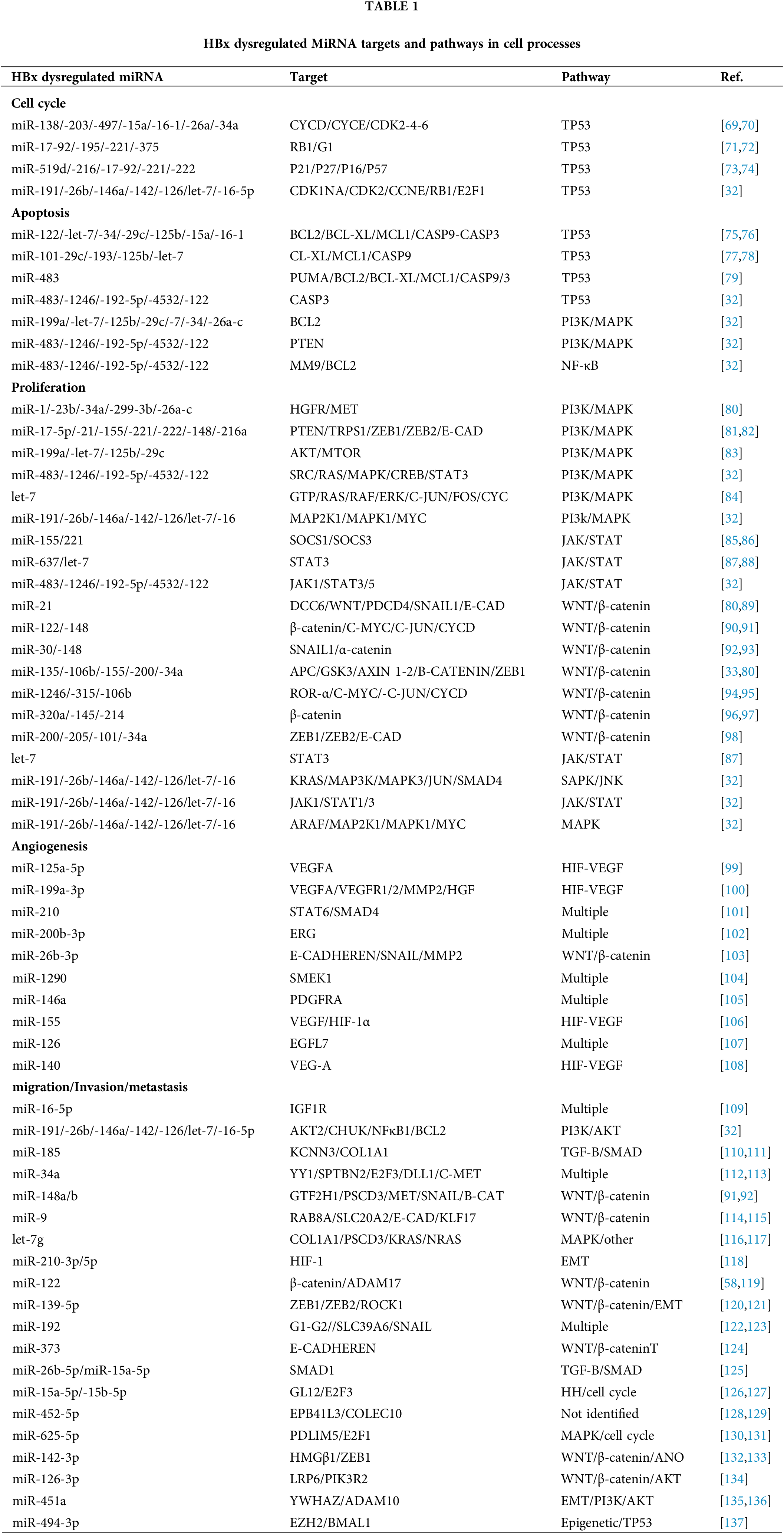
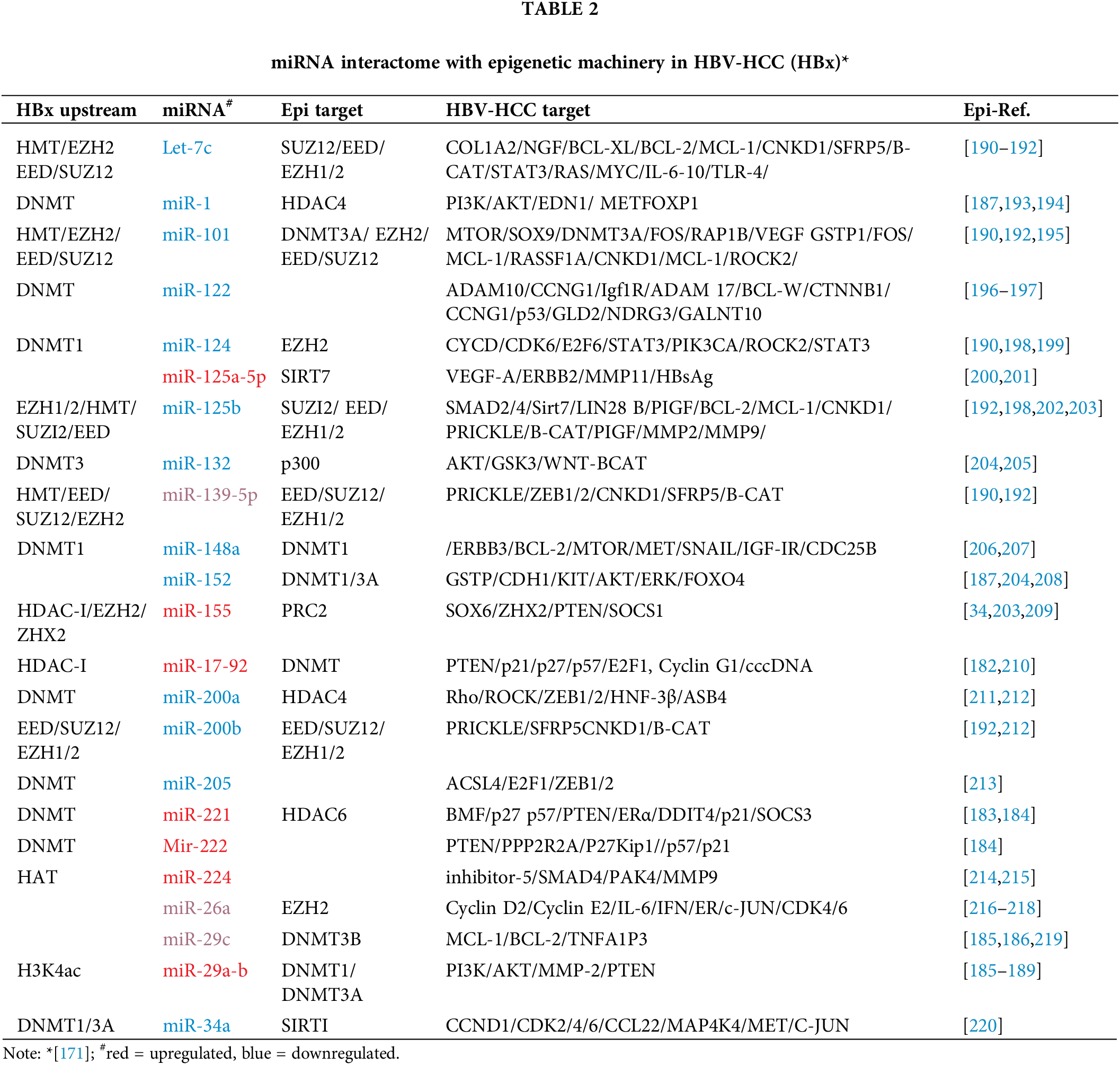
 Downloads
Downloads
 Citation Tools
Citation Tools