 Open Access
Open Access
REVIEW
Advances in micropillar arrays in cellular biomechanics detection and tissue engineering
Institute of Biomedical Engineering, West China School of Basic Medical Sciences & Forensic Medicine, Sichuan University, Chengdu, 610041, China
* Corresponding Author: YE ZENG. Email:
BIOCELL 2024, 48(11), 1521-1529. https://doi.org/10.32604/biocell.2024.055410
Received 09 April 2024; Accepted 29 July 2024; Issue published 07 November 2024
Abstract
Cellular biomechanical features contributed to the occurrence and development of various physiological and pathological phenomena. Micropillar arrays have emerged as an important tool for both the assessment and manipulation of cellular biomechanical characteristics. This comprehensive review provides an in-depth understanding of the fabrication methodologies of micropillar arrays and their applications in deciphering and fine-tuning cellular biomechanical properties and the innovative experimental platforms including organ-on-a-chip and organoids-on-a-chip. This review provides novel insights into the potential of micropillar technology, poised to update the landscape of stem cell research and tissue engineering.Keywords
Cellular biomechanics, a fascinating field of study, exerts influences on a myriad of cellular behaviors and is involved in the occurrence and development of human diseases [1,2]. From the control of cell migration [3,4] to the orchestration of differentiation biological processes [5] and even shaping intracellular responses to external stimuli [6,7], the regulatory role of cellular biomechanics in cell function is indisputable. In tissue engineering, the ability to finely tune and modulate these behaviors represents a potential goldmine for regenerative therapies. While the classical approach has long relied on chemical cues to direct stem cell fate [8], recent research has thrown the spotlight on the role of mechanical cues, such as micropatterns, in shaping and guiding cellular behavior.
Mechanical cues, stemming from micropatterned surfaces, trigger a cascade of events within cells. They guide cell contact, induce reorganization of the cell’s internal cytoskeleton, and even the deformation of the nucleus including orientation, morphology, Lamin A/C levels, and chromatin condensation degree [9,10]. What makes this even more intriguing is the diverse cellular responses observed depending on the specific geometric configuration of the micropatterns.
Micropillar arrays, consisting of precisely engineered cylindrical or pillar-like structures on substrates, present a controlled platform for exploring cellular responses to mechanical cues [11]. When mesenchymal stem cells (MSCs) were grown on triangular prism micropatterns, the well-structured F-actin fibers span the entire cell, efficiently transmitting compressive forces to the nucleus and resulting in the flattening of the nucleus and a stretching of nuclear pores, enhancing the nuclear import of YES-associated protein (YAP). Conversely, the story takes a different turn when cells grow on cylinder micropatterns, which exert a comparatively less compressive force on.
The nucleus, leading to a reduced translocation of YAP into the nucleus and a corresponding decrease in the secretion of angiogenic growth factors. The enhancement of the paracrine activity of MSCs facilitated tissue regeneration [9]. Micropillar arrays have become an exciting avenue for investigating and understanding how cells respond to mechanical cues, arrays represent a cutting-edge tool in the modulation of cellular behavior through biomechanical stimuli, opening up exciting new horizons in the field of tissue engineering and regenerative medicine (Fig. 1). Utilizing multilayer microfluidic chips integrated with polydimethylsiloxane (PDMS) micropillar arrays has been harnessed for the cultivation of specific brain region organoids derived from human induced pluripotent stem cells (hiPSCs) [12].
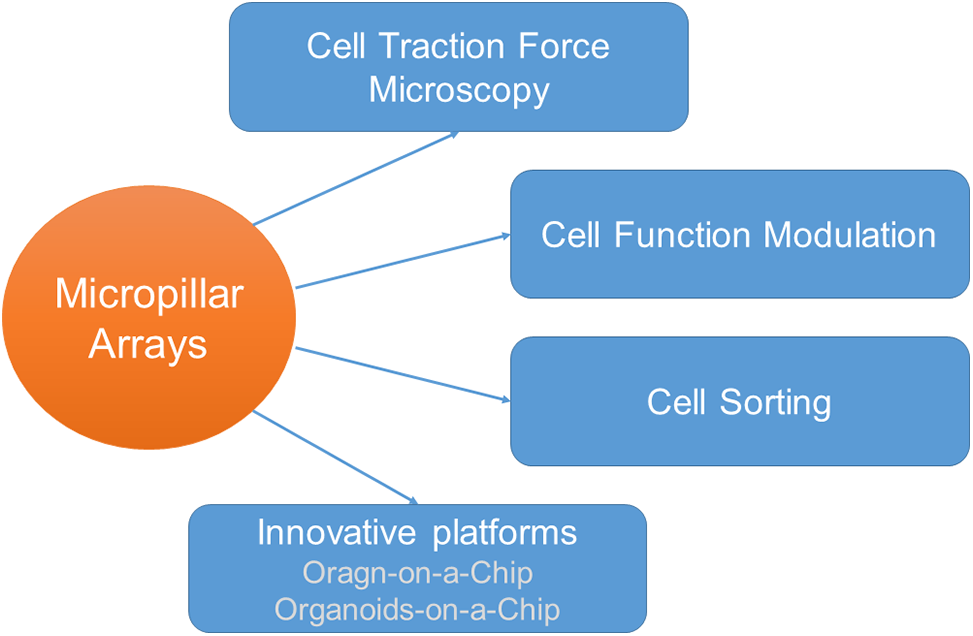
Figure 1: Application of micropillar arrays in cellular biomechanics detection and tissue engineering.
This review aimed to introduce the applications, fabrication techniques, and underlying mechanisms of micropillar arrays in cellular biomechanics detection and tissue engineering. We analyzed their utility in quantifying cell traction forces, assessing cellular mechanical properties, and guiding cell separation and stem cell differentiation. Additionally, we reviewed the innovative platforms enabled by micropillar arrays, including organ-on-a-chip and organoids-on-a-chip systems.
Fabrication of Micropillar Arrays
Soft lithography and micromolding
Soft lithography [13,14] and micromolding [15,16] are two well-established techniques for fabricating micropillar arrays. Micromolding involves casting a polymer material into a mold to create micropillar structures, offering high reproducibility and scalability for large-scale production [17].
Soft lithography, which emerged in the late 1990s, utilizes elastomeric materials like PDMS to create micro-holes as molds for micropillar arrays, which provide high resolution and precise control of micropillar dimensions [18].
Soft lithography replaces traditional lithography with elastic molds, mainly PDMS, for producing microstructures or micromolds. This technology offers flexibility and advantages such as the ability to create complex multi-layer or three-dimensional structures, even on irregular surfaces. Currently, soft lithography is applied in biology, particularly for controlling molecular composition on surfaces for cell fate control [19,20].
3D printing and nanofabrication
3D printing enables layer-by-layer deposition of materials to create micropillar arrays with complex architectures [21–23], offering versatility in terms of materials used and allowing for the incorporation of gradient properties within the micropillars.
Nanofabrication techniques, such as electron beam lithography and nanoimprint lithography, provide sub-micron resolution, enabling the creation of nanoscale micropillar arrays [24,25]. These techniques are particularly valuable for investigating cellular responses at the nanoscale.
Microfluidic devices can generate monodisperse emulsions containing precursor materials that solidify into micropillars upon exposure to Ultraviolet (UV) light or thermal curing [26–29]. This controlled assembly and scalability approach allows for the precise control of micropillar size, spacing, and composition. Furthermore, microfluidic platforms can facilitate the integration of biochemical cues or multiple cell types to construct complex micropillar arrays [30,31]. The fabrication techniques mentioned above are illustrated in Fig. 2.
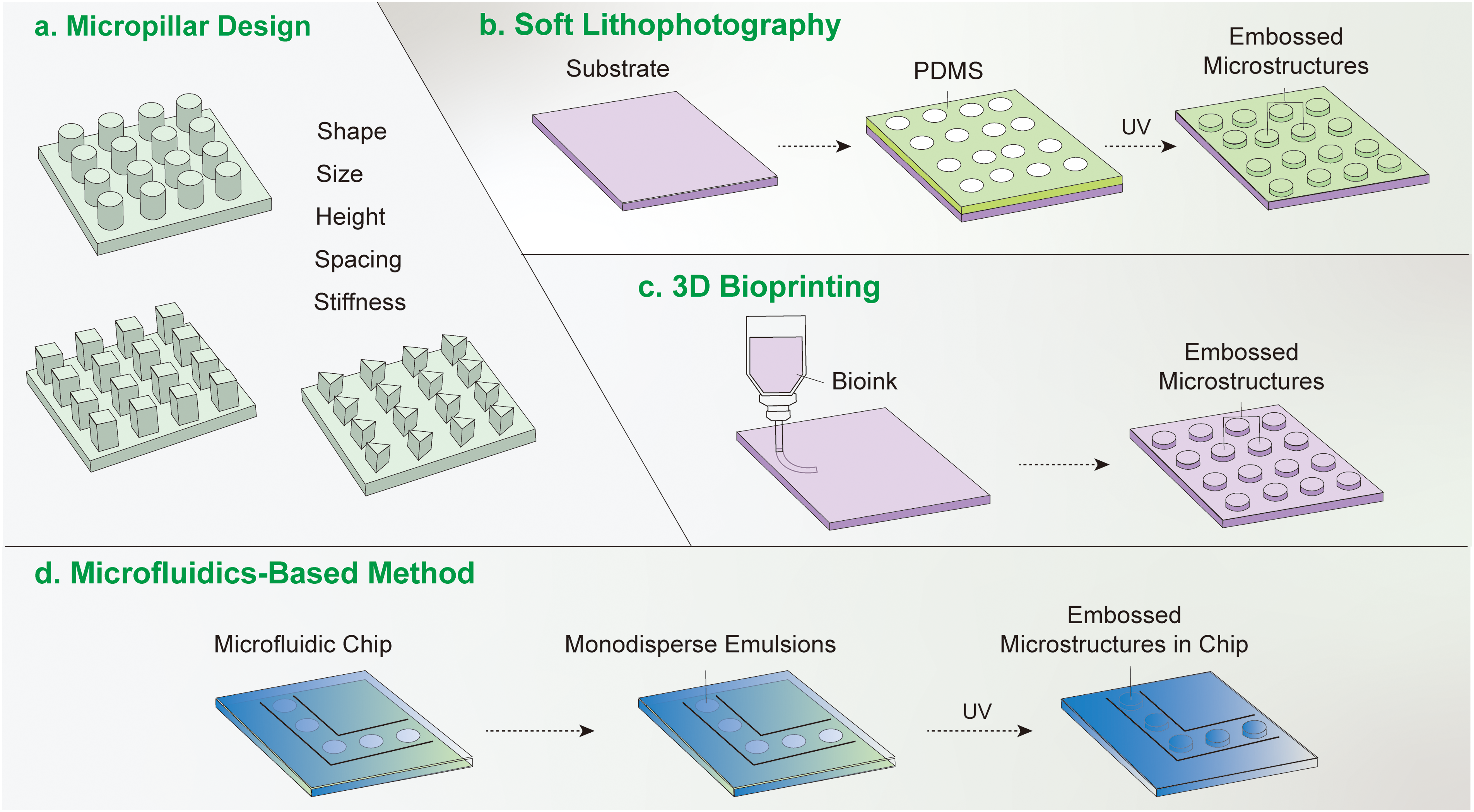
Figure 2: Methods of micropillar fabrication. (a) Considerations for micropillar preparation. Various parameters must be considered when preparing micropillars, encompassing their shape, size, height, spacing, and mechanical stiffness. (b) Soft lithography method. In the soft lithography approach, a substrate is coated with a photoresist. Ultraviolet (UV) light is then directed through a predefined irradiation mask, leading to the formation of an ordered micropillar array on the substrate. (c) 3D bioprinting technique. The 3D bioprinting method involves extruding material from a designated container onto a substrate, ultimately resulting in the creation of a micropillar array on the substrate. (d) Microfluidic-based method. The microfluidic-based approach follows a sequence of steps, commencing with the coating of a silicon plate with SU-8 photoresist. Subsequent UV irradiation creates a template, which is then employed as a mold for applying a polydimethylsiloxane (PDMS) precursor. The template is subsequently removed through thermoplastic molding, yielding the desired micropillar array.
A comprehensive analysis of how micropillar array design parameters affect cellular behavior and outcomes in tissue engineering has been reviewed [9,10]. For instance, the formation of adhesion patches is significantly affected by the dimensions of nanopillars. When the side length of square nanopillars on a substrate was less than 333 nm, both the expression of integrins and the formation of adhesion patches were dramatically reduced [32]. Shorter micropillars exhibit higher basal stiffness while increasing the height of micropillars reduces their basal stiffness. These variations in stiffness affect the area of adhesion plaques formed by MSCs, with smaller adhesion areas correlating with an increased propensity for adipogenic differentiation. MSCs on nanopillars with spacing less than 140 nm had faster skeleton formation and greater ability to differentiate towards osteogenic lineages. The spacing and height of the micropillars affected the cell migration speed [32]. When the spacing of the micropillars was 3 μm, the cells hardly moved. When the spacing increased from 3 to 5 μm, the cells became elongated, with a migration speed of 0.18 μm/min. When the height decreased from 20 to 10 μm, the cell migration speed was decreased from 0.23 to 0.84 μm/min [33].
Cell Traction Force Microscopy Using Micropillar Arrays
Cell traction force microscopy (TFM) relies on measuring the deformation of the substrate caused by cell-generated forces and subsequently inferring the traction forces based on the mechanical properties of the substrate and its deformation [34–36]. This method enables the quantification of forces exerted by cells on their surrounding substrate, which is particularly valuable for studying cell-matrix interactions, mechanotransduction, and cellular responses to physical cues. There are two TFM approaches, including fluorescent magnetic bead-based TFM and micropillar-based TFM.
Fluorescent magnetic bead-based TFM
Cells grown on the substrate in which fluorescent magnetic beads are embedded. The interaction of the cells with the beads causes deformation of the substrate in response to the forces exerted by the cells. The deformation of the substrate is tracked by imaging the displacement of the beads, allowing the cell-generated forces to be calculated. This method offers high spatial resolution and is suitable for studying single-cell forces and their distribution [37].
PDMS micropillar arrays provide another platform for cell traction force measurement (Fig. 3). The pillars within the array deform in response to traction force, and the extent of pillar deformation is correlated with the magnitude of forces. Micropillar arrays can be fabricated with micron or nanometer scale sizes, which leverages the flexibility of PDMS to accurately capture the traction force over a large area [38,39].
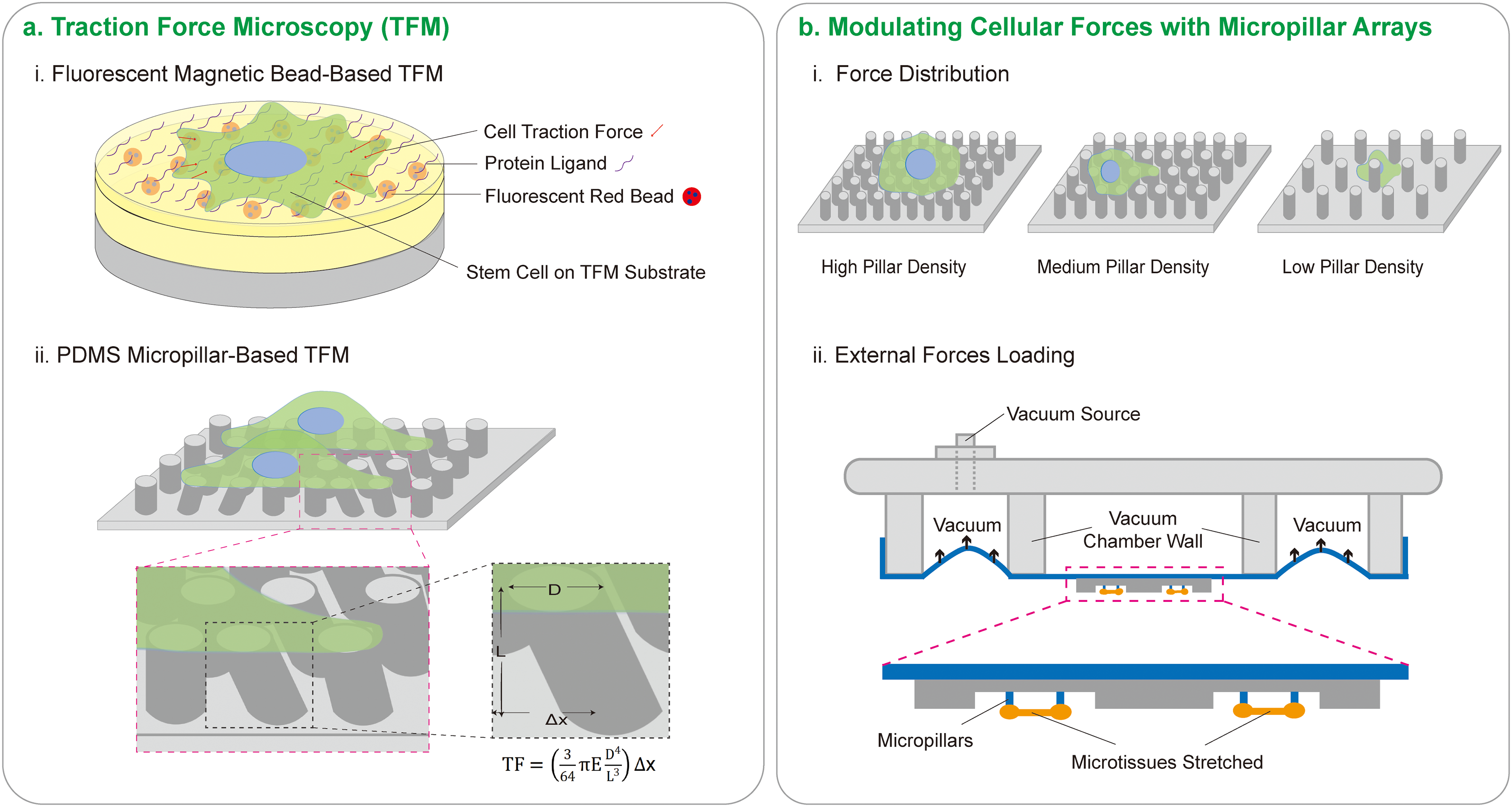
Figure 3: Cellular traction force detection and regulation. (a) Traction force microscopy (TFM) employs two primary methods: (i) Fluorescent magnetic bead-based TFM. Cells are cultured on a substrate with a known elastic modulus and are pre-labeled with fluorescent tracer particles. As cells contract, they exert traction forces that deform the substrate. These mechanical forces adhere to the principles of elastic mechanics. Utilizing a laser confocal microscope to monitor the displacement of fluorescent tracer particles, the degree of elastic deformation in the substrate can be quantified. Ultimately, employing an inverse algorithm, data pertaining to the traction forces exerted by the cells can be extracted. (ii) PDMS Micropillar-based TFM. Cell contraction causes deformation in PDMS micropillars. The extent of deformation in these micropillars can be observed and measured using an optical microscope. An appropriate equation can then be applied to calculate the force exerted by the cell on the micropillars. (b) Modulating cell forces with micropillar arrays. (i) Impact of micropillar array density on force distribution. At high densities, cells form focal adhesions predominantly on the micropillar surfaces. At medium densities, focal adhesions are observed on both the upper surfaces and sides of the micropillars. Conversely, at low densities, cells tend to occupy the inter-pillar spaces. (ii) External forces loading. Cells can be stretched by creating negative pressure that stretches the micropillar arrays on the membrane. Alteration in cells can be monitored using an inverted microscope, facilitating the observation of cellular responses to the mechanical application.
However, the arrangement and composition of micropillar arrays modulate the cell traction force. Cells exhibit distinct force distributions on regions of different pillar densities. Specifically, cells tend to exert lower forces on regions of higher pillar density and higher forces on regions of lower density, with cells showing a preference to migrate towards regions of increasing pillar density [40,41]. Magnetic micropillars containing magnetic particles can alter pillar spacing and induce changes in cell attachment area and migration [10].
In other works, micropillar was thought to have a limited effect on cells. Within the heart-on-a-chip platform, photolithographically patterned Au microelectrodes are combined with printed 3D conductive polymer micropillars (200 µm in size). The low Young’s modulus micropillars effectively prevent any deformation of the tissue during compression. Tissue itself, slightly displaces the pillars during the contraction [42]. The micropillar arrays were also used in external forces loading such as stretch [10].
Although the deformation of the micropillar arrays provides a method for cell traction force quantification, the micropillars may also influence the generation of cellular forces, and this complex interplay among micropillar, extracellular matrix (ECM) and cell and how to accurately quantify the cell traction forces remains to be investigated.
Cell Properties and Micropillar Arrays
Cellular biomechanical properties
Micropillar arrays offer the means to assess cell spreading and cell membrane fluidity [43]. Also, micropillar arrays influence the viscoelastic properties of cells and their mechanical behavior. Primary human MSCs (hMSCs) were observed to be softer compared to human fibroblasts on smooth surfaces, while MSCs on micropillar arrays tended to exhibit linear growth, transitioning from a viscoelastic fluid property to a viscoelastic solid, indicating an increase in cell stiffness [44].
Micropillar arrays offer a 3D microenvironment that supports cell growth. Micropillar arrays have shown the potential to elongate and align cells. MSCs are adept at sensing matrix stiffness, pillar length, and the concavity/convexity of pillar surfaces, leading to specific cellular responses. PDMS micropillar arrays with varying stiffness and geometry can be tailored to induce elongation and alignment in hMSCs and cardiomyocytes. These anisotropic micropillars can accelerate elongation rates compared to traditional 2D microenvironments [45]. Coating PDMS surfaces with proteins, such as collagen or fibronectin, modulate the cell spreading area and cellular sensitivity via control of gene expression levels [46,47].
Focal adhesions (FAs), transmembrane protein complexes on the cell surface, detect the micropillars’ topographical and mechanical properties, relaying these signals to the cell’s interior. The cytoskeleton, tethered to both the intracellular FAs and the nucleus, acts as a conduit for mechanical signals, inducing alterations in nuclear and chromatin structure, and regulating ECM-related gene expression and protein secretion [48,49]. This feedback mechanism between the cell and the ECM sustains mechanical homeostasis and governs cell fates [50,51].
Moreover, the broader microenvironment shapes stem cell responses to micropillar arrays. The interplay between soluble factors, neighboring cells, and extracellular matrix composition all contribute to the signaling networks involved in determining stem cell fate.
Cell sorting using micropillar arrays
Micropillar arrays are combined with fluid dynamics to separate cells using deterministic laminar flow and asymmetrically distributed arrays of micropillars, where cells smaller than a critical size flow through the micropillar gaps while larger cells move laterally and undergo a lateral offset [52]. When nickel micropillars are integrated into microfluidic devices, programmatically controlled pneumatic microvalves manipulate fluid flow, achieving separation [53,54].
By adjusting the size and shape of the micropillars, the cell capture rate can be improved. For example, changing the orientation of the triangular micropillar arrays creates a smoother hydrodynamic gradient and prolongs the contact time between the cells and the immune-modified micropillars, facilitating the capture of circulating tumor cells (CTCs) from blood [54,55]. The antibody-modified micropillars had been developed to enrich and recognize circulating nucleated red blood cells from maternal blood [56], which is expected to become a next-generation non-invasive prenatal diagnostic technology.
Innovative Platforms: Organ-on-a-Chip and Organoids-on-a-Chip
The advancement of organ-on-a-chip systems benefits from a deeper understanding of cellular heterogeneity in complex and dynamic in vivo microenvironments [57]. The Chip-on-a-Transwell platform was used to investigate the microphysiological systems in vitro [58].
Organ-on-a-chip technology employs microfluidic devices to replicate the tissue-tissue interfaces, spatiotemporal chemical gradients, and dynamic mechanical microenvironments in vivo, enabling specific physiological studies and the development of dedicated in vitro disease models (Fig. 4). As documented, organ-on-a-chip systems have been utilized to study endothelial differentiation of hiPSCs, incorporating quadrilateral micropillars to provide shear protection and enhance the physiological relevance [59,60]. Organ-on-a-chip systems can mimic the structure and function of alveolar-capillary interfaces and simulate conditions such as pulmonary oedema [61], providing alternatives to traditional cell culture models and animal testing, as well as applications in drug development and toxicology. Moreover, microfluidic chips with PDMS micropillar arrays have been used to develop in vitro blood-brain barrier (BBB) models incorporating the human brain microvascular endothelial cells (hBMECs), human pericytes (hPCs), and human astrocytes (hACs) [62], offering a tool to study drug permeability and neurological disorders.
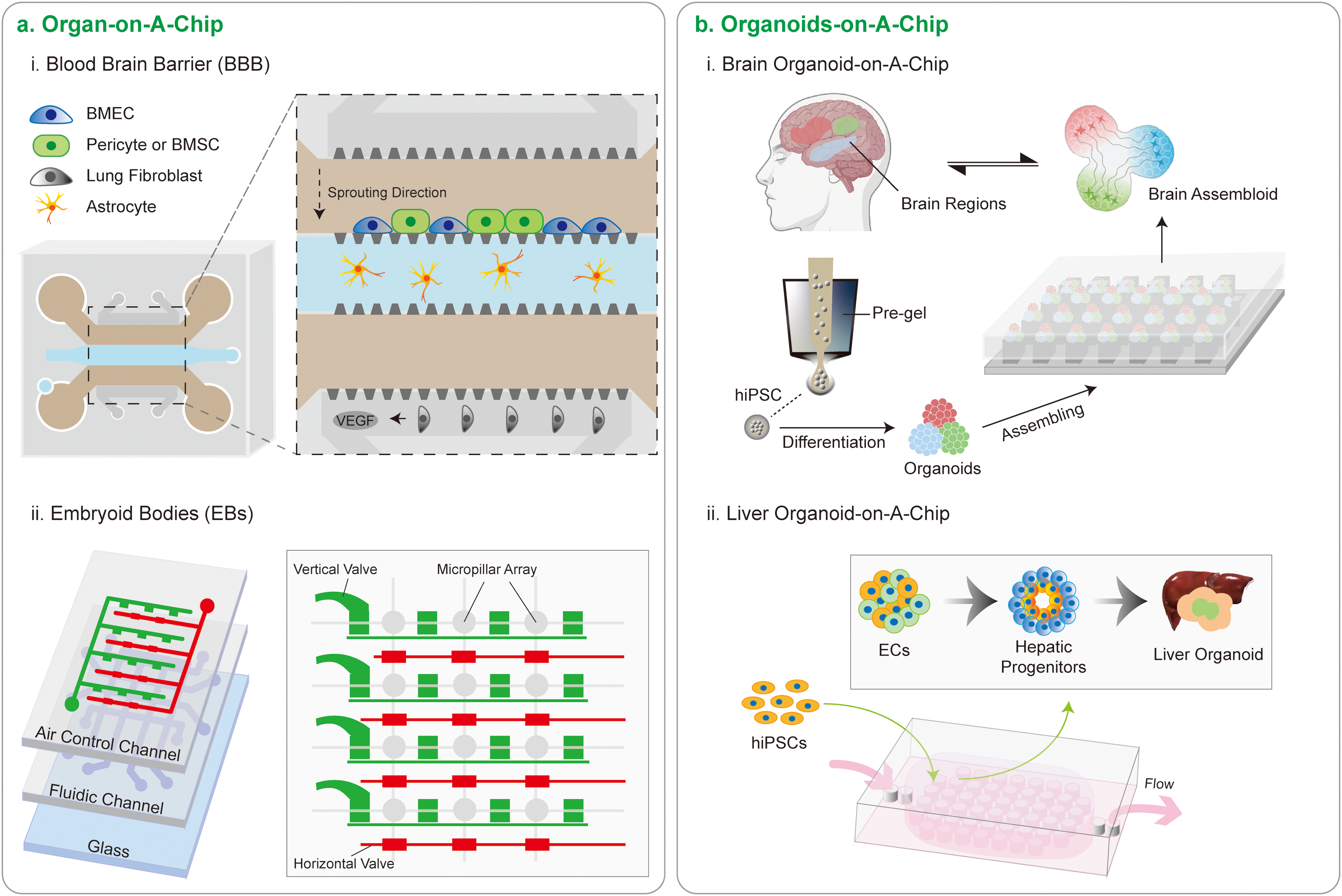
Figure 4: Micropillars advancing cutting-edge chip design. (a) Two organ-on-a-chips. (i) Directed sprouting of brain microvascular endothelial cells (BMECs). Organ-on-a-chip technology, exemplified by a vertically integrated microfluidic chip, mimics physiological organ behavior. In this context, the chip enables the directed sprouting of BMECs by localized delivery of vascular endothelial growth factor (VEGF) from lung fibroblasts. Notably, bone marrow mesenchymal stem cells (BMSCs) function as perivascular pericytes, displaying superior vessel-constrictive capabilities compared to human pericytes, thereby aiding the formation of a robust blood-brain barrier. Additionally, astrocytes play a pivotal role in maturing the capillary network. (ii) Regulating embryonic stem (ES) Cell fate. An integrated microfluidic culture device, featuring a 4 × 4 microfluidic arrangements with air control channels and fluidic channels, incorporates micropillar arrays (300 μm in diameter). These micropillars, spaced 12 μm apart, facilitate ES cell docking. This microfluidic culture device, equipped with shear-protected micropillar arrays, offers substantial potential for uniform cell docking and forming of embryoid bodies (EBs). (b) Two organoids-on-a-chips. (i) Brain organoids-on-a-chip. Human induced pluripotent stem cells (hiPSCs), encapsulated in hydrogels, undergo controlled differentiation to form organoids representing distinct brain regions. These organoids can be precisely assembled using micropillar arrays, enabling the creation of fused organoids that closely mimic the complexity of the brain, providing a robust platform for studying intricate brain structures and functions within a controlled in vitro environment. (ii) Liver organoids-on-a-chip. The organoids-on-a-chip facilitates the formation of human liver organoids through hiPSC-based EBs cultured on perfusable micropillar arrays. This approach enables in situ hepatic differentiation, long-term 3D culture, and the generation of liver-like organs.
Manufacturing limitations are a major obstacle to current advances in organ-on-a-chip. While laser-induced methods offer higher precision engineering capabilities, costs remain prohibitive. New methods such as 3D printing and injection molding show substantial potential for organ-on-a-chip manufacturing [57].
As an emerging field, organoid microarrays aim to facilitate the manipulation and control of organoids, enabling a comprehensive reflection of the complex internal environment in vivo. Organoid microarrays are characterized by their high throughput and high sensitivity microstructures, capable of integrating a series of experimental processes such as organoid sorting, cultivation, observation, stimulus induction, detection, and analysis. For instance, a micropillar concentrator (for capturing volatile carbonyl metabolites with high efficiency) was integrated with a lung organ chip for the identification of cellular carbonyl metabolites [63]. HiPSC-derived multi-organoids-on-chip system (co-cultured liver and heart organoids on chip) can reflect the drug metabolism and responses at multi-organ levels [64]. These advanced platforms are applied across various biological fields, including the construction of developmental or disease models, drug discovery and development, immune response therapy, and microbial infections. This integration enhances the predictability of clinical treatment plans and significantly improves experimental efficiency.
The synergy between micropillar arrays and microfluidic organoid microarrays presents a powerful tool for both clinical and basic research (Fig. 4). Using multilayer microfluidic chips with PDMS micropillar arrays, brain-like organoids derived from hiPSCs were constructed and represent multiregional brain regions such as the cortex, hippocampus, and thalamus [12]. Multiregional brain organoids-on-a-chip offer a unique opportunity to investigate the interactions between different brain regions and their contribution to neurological disorders [12].
The integration of micropillar arrays with microfluidic chips allows precise control of the spatial arrangement and mechanical properties of the organoids and mimics mechanical cues such as compression and shear stress. By harnessing the power of micropillar arrays, researchers can create more accurate and physiologically relevant models of organs, paving the way for new discoveries in life.
Micropillar arrays integrated with stem cell research hold great promise for breakthroughs in regenerative medicine. The interaction between stem cells and micropillars initiates complex signaling cascades that intricately shape cellular responses. Simultaneously, the stem cell microenvironment plays a pivotal role in governing their responses to micropillars. While our current understanding of how micropillars influence stem cell behavior is substantial, unveiling the mechanotransduction pathways and dissecting cellular responses to multidimensional cues, and the nuanced interplay among soluble factors, extracellular matrix composition, and neighboring cells continue to intrigue researchers deeply.
Unlocking the molecular machinery responsible for translating mechanical cues into specific cellular behaviors remains a hard pursuit. Scrutinizing mechanosensitive ion channels, cytoskeletal elements, and mechanotransduction molecules can shed light on the genetic and epigenetic changes triggered by micropillar arrays. Employing advanced techniques like single-cell sequencing and chromatin accessibility profiling holds promise for comprehensively understanding these alterations. Progress hinges on deciphering mechanotransduction mechanisms, probing complex cell-cell interactions, understanding microenvironmental influences, and dissecting the interplay of multidimensional cues.
Exploring how microenvironmental factors intersect with micropillar array effects could unveil complex signaling networks orchestrating cellular behaviors. The synergy between physical cues from micropillars and bioactive molecules presents an enticing frontier. Functionalizing micropillar surfaces with such molecules could lead to synergistic effects, finely tuning stem cell responses.
Advancing micropillar technology stands to benefit greatly from integrating cutting-edge methodologies. Harnessing microfluidics for precise biochemical delivery, live-cell imaging for real-time observation, and dynamic modulation of mechanical forces for time-resolved studies open new vistas. High-throughput screening platforms that streamline the discovery of materials and treatments bridge the gap between research and practice. Investigating co-culture scenarios, such as combining mesenchymal stem cells (MSCs) with immune or endothelial cells in the presence of micropillar arrays, offers a promising avenue for insights into determinants of stem cell fate, immune modulation, and angiogenesis. Moreover, the integration of micropillar arrays with 3D bioprinting and organ-on-a-chip models presents novel avenues for research and application.
Despite great advancements in micropillar arrays, there remain other challenges. Future research should focus on refining in vitro models to better match in vivo physiological conditions, integrating more complex cell types, and recreating the cell-cell interactions within microenvironments. The development of novel materials that minimize drug and nutrient uptake while maintaining flexibility and biocompatibility is also crucial. The biocompatibility of materials used in micropillar arrays is critical for their application. PDMS, commonly used for its optical clarity and flexibility, has limitations such as non-specific absorption of small molecules [65]. To address these issues, alternative materials such as polyurethanes and cyclic olefin polymers could be explored.
Furthermore, ethical considerations, particularly in stem cell research, need to be addressed to ensure the responsible development and application of micropillar technologies. This includes adherence to ethical guidelines and maintaining transparency in research practices, including consent, privacy, and the long-term impact of regenerative therapies on patients.
Due to the reproducibility and precision of organoid microarrays, they are poised to revolutionize medical research, potentially replacing animal and 2D cellular experiments. These chips could become essential tools for preclinical research, offering more functional, integrated, automated, and personalized solutions. The future development of organoid chips is likely to focus on creating human organoid biomimetic chips composed of multiple organoids connected by pipelines [64]. Achieving this will require extensive research on the population ratios, differentiation conditions, and spatial distributions among various cell types, as well as the determination of amplification factors to simulate the complex physiological environment in the body.
In summary, micropillar arrays are a useful platform for cellular biomechanics and tissue engineering research. The development of this platform will unlock novel possibilities for understanding and manipulating cellular behavior, driving innovative strategies in regenerative medicine, disease modeling, and personalized therapies.
Acknowledgement: None.
Funding Statement: Supported by the National Natural Science Foundation of China (No. 12272246) and the Key Research and Development Projects in Sichuan Province (No. 2023YFS0075).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Ye Zeng; draft manuscript preparation: Ye Zeng, Xueling He; review and editing: Ye Zeng, Xueling He, Linlu Jin, Yixue Qin, Jian Zhong, Zhi Ouyang; visualization: Ye Zeng, Jian Zhong, Zhi Ouyang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Jiang Z-L. Mechanobiology research in China. Mechanobiol Med. 2023;1(1):100002. doi:10.1016/j.mbm.2023.100002. [Google Scholar] [CrossRef]
2. Zhong J, Ouyang Z, Shen J, Zeng Y. Improvement of hemodynamics in mesenteric microcirculation in septic shock rats by anisodamine and anisodine. Mechanobiol Med. 2023;1(1):100006. doi:10.1016/j.mbm.2023.100006. [Google Scholar] [CrossRef]
3. Isomursu A, Park KY, Hou J, Cheng B, Mathieu M, Shamsan GA, et al. Directed cell migration towards softer environments. Nat Mater. 2022;21(9):1081–90. doi:10.1038/s41563-022-01294-2. [Google Scholar] [PubMed] [CrossRef]
4. Ren H, Chen J, Huang K, Qi Y-X. Platelet-derived microvesicles drive vascular smooth muscle cell migration via forming podosomes and promoting matrix metalloproteinase-9 activity. Mechanobiol Med. 2023;1(1):100003. doi:10.1016/j.mbm.2023.100003. [Google Scholar] [CrossRef]
5. Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, et al. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nat Mater. 2014;13(10):979–87. doi:10.1038/nmat4051. [Google Scholar] [PubMed] [CrossRef]
6. Gjorevski N, Nikolaev M, Brown TE, Mitrofanova O, Brandenberg N, DelRio FW, et al. Tissue geometry drives deterministic organoid patterning. Science. 2022;375(6576):eaaw9021. doi:10.1126/science.aaw9021. [Google Scholar] [PubMed] [CrossRef]
7. Shou J, Huo Y. Changes of calcium cycling in HFrEF and HFpEF. Mechanobiol Med. 2023;1(1):100001. doi:10.1016/j.mbm.2023.100001. [Google Scholar] [CrossRef]
8. Xie W, Wei X, Kang H, Jiang H, Chu Z, Lin Y, et al. Static and dynamic: evolving biomaterial mechanical properties to control cellular mechanotransduction. Adv Sci. 2023;10(9):e2204594. doi:10.1002/advs.202204594. [Google Scholar] [PubMed] [CrossRef]
9. Li Y, Zhong Z, Xu C, Wu X, Li J, Tao W, et al. 3D micropattern force triggers YAP nuclear entry by transport across nuclear pores and modulates stem cells paracrine. Natl Sci Rev. 2023;10(8):nwad165. doi:10.1093/nsr/nwad165. [Google Scholar] [PubMed] [CrossRef]
10. Long Y, Sun Y, Jin L, Qin Y, Zeng Y. Micropillars in biomechanics: role in guiding mesenchymal stem cells differentiation and bone regeneration. Adv Mater Interfaces. 2024;11(2):2300703. doi:10.1002/admi.202300703. [Google Scholar] [CrossRef]
11. Xie N, Xiao C, Shu Q, Cheng B, Wang Z, Xue R, et al. Cell response to mechanical microenvironment cues via Rho signaling: from mechanobiology to mechanomedicine. Acta Biomater. 2023;159:1–20. doi:10.1016/j.actbio.2023.01.039. [Google Scholar] [PubMed] [CrossRef]
12. Zhu Y, Zhang X, Sun L, Wang Y, Zhao Y. Engineering human brain assembloids by microfluidics. Adv Mater. 2023;35(14):e2210083. doi:10.1002/adma.202210083. [Google Scholar] [PubMed] [CrossRef]
13. Handrea-Dragan IM, Botiz I, Tatar AS, Boca S. Patterning at the micro/nano-scale: polymeric scaffolds for medical diagnostic and cell-surface interaction applications. Colloids Surf B Biointerfaces. 2022;218:112730. doi:10.1016/j.colsurfb.2022.112730. [Google Scholar] [PubMed] [CrossRef]
14. Shen Y, Wang C, Liu Z, Zhang X, Su R, Wang Y, et al. Multicomponent structural color membrane based on soft lithography array for high-sensitive Raman detection. J Colloid Interface Sci. 2023;652:518–28. doi:10.1016/j.jcis.2023.08.066. [Google Scholar] [PubMed] [CrossRef]
15. Kim H, Bae C, Kook YM, Koh WG, Lee K, Park MH. Mesenchymal stem cell 3D encapsulation technologies for biomimetic microenvironment in tissue regeneration. Stem Cell Res Ther. 2019;10(1):51. doi:10.1186/s13287-018-1130-8. [Google Scholar] [PubMed] [CrossRef]
16. Jang W, Song EL, Mun SJ, Bong KW. Efficient isolation of encoded microparticles in a degassed micromold for highly sensitive and multiplex immunoassay with signal amplification. Biosens Bioelectron. 2024;261(10):116465. doi:10.1016/j.bios.2024.116465. [Google Scholar] [PubMed] [CrossRef]
17. Tien J, Dance YW. Microfluidic biomaterials. Adv Healthc Mater. 2021;10(4):e2001028. doi:10.1002/adhm.202001028. [Google Scholar] [PubMed] [CrossRef]
18. Cutarelli A, Ghio S, Zasso J, Speccher A, Scarduelli G, Roccuzzo M, et al. Vertically-aligned functionalized silicon micropillars for 3D culture of human pluripotent stem cell-derived cortical progenitors. Cells. 2019;9(1):88. doi:10.3390/cells9010088. [Google Scholar] [PubMed] [CrossRef]
19. Whitesides GM, Ostuni E, Takayama S, Jiang X, Ingber DE. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng. 2001;3(1):335–73. doi:10.1146/annurev.bioeng.3.1.335. [Google Scholar] [PubMed] [CrossRef]
20. Stasiowski E, O’Laughlin R, Holness S, Csicsery N, Hasty J, Hao N. A microfluidic platform for screening gene expression dynamics across yeast strain libraries. Bio Protoc. 2023;13(22):e4883. doi:10.21769/BioProtoc.4883. [Google Scholar] [PubMed] [CrossRef]
21. Sanchez-Rubio A, Jayawarna V, Maxwell E, Dalby MJ, Salmeron-Sanchez M. Keeping it organized: multicompartment constructs to mimic tissue heterogeneity. Adv Healthc Mater. 2023;12(17):e2202110. doi:10.1002/adhm.v12.17. [Google Scholar] [CrossRef]
22. Cortelli G, Grob L, Patruno L, Cramer T, Mayer D, Fraboni B, et al. Determination of stiffness and the elastic modulus of 3D-printed micropillars with atomic force microscopy-force spectroscopy. ACS Appl Mater Interfaces. 2023;15(5):7602–9. doi:10.1021/acsami.2c21921. [Google Scholar] [PubMed] [CrossRef]
23. Chliara MA, Elezoglou S, Zergioti I. Bioprinting on organ-on-chip: development and applications. Biosensors. 2022;12(12):1135. doi:10.3390/bios12121135. [Google Scholar] [PubMed] [CrossRef]
24. Otsuka H. Nanofabrication technologies to control cell and tissue function in three-dimension. Gels. 2023;9(3):203. doi:10.3390/gels9030203. [Google Scholar] [PubMed] [CrossRef]
25. Ermis M, Antmen E, Hasirci V. Micro and Nanofabrication methods to control cell-substrate interactions and cell behavior: a review from the tissue engineering perspective. Bioact Mater. 2018;3(3):355–69. [Google Scholar] [PubMed]
26. Bi W, Cai S, Lei T, Wang L. Implementation of blood-brain barrier on microfluidic chip: recent advance and future prospects. Ageing Res Rev. 2023;87:101921. doi:10.1016/j.arr.2023.101921. [Google Scholar] [PubMed] [CrossRef]
27. Lee S, Chung M, Jeon NL. BBB-on-a-chip: modeling functional human blood-brain barrier by mimicking 3D brain angiogenesis using microfluidic chip. Methods Mol Biol. 2022;2492:251–63. doi:10.1007/978-1-0716-2289-6. [Google Scholar] [CrossRef]
28. Banik S, Uchil A, Kalsang T, Chakrabarty S, Ali MA, Srisungsitthisunti P, et al. The revolution of PDMS microfluidics in cellular biology. Crit Rev Biotechnol. 2023;43(3):465–83. doi:10.1080/07388551.2022.2034733. [Google Scholar] [PubMed] [CrossRef]
29. Zhu Z, Lv Z, Wang L, Tan H, Xu Y, Li S, et al. A pump-free paper/PDMS hybrid microfluidic chip for bacteria enrichment and fast detection. Talanta. 2024;275:126155. doi:10.1016/j.talanta.2024.126155. [Google Scholar] [PubMed] [CrossRef]
30. Lan Y, Zhou Y, Wu M, Jia C, Zhao J. Microfluidic based single cell or droplet manipulation: methods and applications. Talanta. 2023;265:124776. doi:10.1016/j.talanta.2023.124776. [Google Scholar] [PubMed] [CrossRef]
31. Wang Y, Wang Y, Wang X, Sun W, Yang F, Yao X, et al. Label-free active single-cell encapsulation enabled by microvalve-based on-demand droplet generation and real-time image processing. Talanta. 2024;276(4):126299. doi:10.1016/j.talanta.2024.126299. [Google Scholar] [PubMed] [CrossRef]
32. Fu J, Wang YK, Yang MT, Desai RA, Yu X, Liu Z, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7(9):733–6. doi:10.1038/nmeth.1487. [Google Scholar] [PubMed] [CrossRef]
33. Hui J, Pang SW. Cell migration on microposts with surface coating and confinement. Biosci Rep. 2019; 19;39(2):BSR20181596. doi:10.1042/BSR20181596. [Google Scholar] [PubMed] [CrossRef]
34. Joshi R, Han SB, Cho WK, Kim DH. The role of cellular traction forces in deciphering nuclear mechanics. Biomater Res. 2022;26(1):43. doi:10.1186/s40824-022-00289-z. [Google Scholar] [PubMed] [CrossRef]
35. Rajendran AK, Sankar D, Amirthalingam S, Kim HD, Rangasamy J, Hwang NS. Trends in mechanobiology guided tissue engineering and tools to study cell-substrate interactions: a brief review. Biomater Res. 2023;27(1):55. doi:10.1186/s40824-023-00393-8. [Google Scholar] [PubMed] [CrossRef]
36. Quan Y, Huang Z, Wang Y, Liu Y, Ding S, Zhao Q, et al. Coupling of static ultramicromagnetic field with elastic micropillar-structured substrate for cell response. Mater Today Bio. 2023;23:100831. doi:10.1016/j.mtbio.2023.100831. [Google Scholar] [PubMed] [CrossRef]
37. Legant WR, Choi CK, Miller JS, Shao L, Gao L, Betzig E, et al. Multidimensional traction force microscopy reveals out-of-plane rotational moments about focal adhesions. Proc Natl Acad Sci U S A. 2013;110(3):881–6. doi:10.1073/pnas.1207997110. [Google Scholar] [PubMed] [CrossRef]
38. Tan JL, Tien J, Pirone DM, Gray DS, Bhadriraju K, Chen CS. Cells lying on a bed of microneedles: an approach to isolate mechanical force. Proc Natl Acad Sci U S A. 2003;100(4):1484–9. doi:10.1073/pnas.0235407100. [Google Scholar] [PubMed] [CrossRef]
39. Ghassemi S, Meacci G, Liu S, Gondarenko AA, Mathur A, Roca-Cusachs P, et al. Cells test substrate rigidity by local contractions on submicrometer pillars. Proc Natl Acad Sci U S A. 2012;109(14):5328–33. doi:10.1073/pnas.1119886109. [Google Scholar] [PubMed] [CrossRef]
40. Bucaro MA, Vasquez Y, Hatton BD, Aizenberg J. Fine-tuning the degree of stem cell polarization and alignment on ordered arrays of high-aspect-ratio nanopillars. ACS Nano. 2012;6(7):6222–30. doi:10.1021/nn301654e. [Google Scholar] [PubMed] [CrossRef]
41. Wang Z, Geng Y. Unidirectional cell crawling model guided by extracellular cues. J Biomech Eng. 2015;137(3):10. doi:10.1115/1.4029301. [Google Scholar] [PubMed] [CrossRef]
42. Wu Q, Zhang P, O’Leary G, Zhao Y, Xu Y, Rafatian N, et al. Flexible 3D printed microwires and 3D microelectrodes for heart-on-a-chip engineering. Biofabrication. 2023;15(3):035023. doi:10.1088/1758-5090/acd8f4. [Google Scholar] [PubMed] [CrossRef]
43. Shin TH, Lee DY, Ketebo AA, Lee S, Manavalan B, Basith S, et al. Silica-coated magnetic nanoparticles decrease human bone marrow-derived mesenchymal stem cell migratory activity by reducing membrane fluidity and impairing focal adhesion. Nanomaterials. 2019;9(10):1475. doi:10.3390/nano9101475. [Google Scholar] [PubMed] [CrossRef]
44. Spagnol ST, Lin WC, Booth EA, Ladoux B, Lazarus HM, Dahl KN. Early passage dependence of mesenchymal stem cell mechanics influences cellular invasion and migration. Ann Biomed Eng. 2016;44(7):2123–31. doi:10.1007/s10439-015-1508-z. [Google Scholar] [PubMed] [CrossRef]
45. Alapan Y, Younesi M, Akkus O, Gurkan UA. Anisotropically Stiff 3D micropillar niche induces extraordinary cell alignment and elongation. Adv Healthc Mater. 2016;5(15):1884–92. doi:10.1002/adhm.201600096. [Google Scholar] [PubMed] [CrossRef]
46. Saez A, Anon E, Ghibaudo M, du Roure O, Di Meglio JM, Hersen P, et al. Traction forces exerted by epithelial cell sheets. J Phys Condens Matter. 2010;22(19):194119. doi:10.1088/0953-8984/22/19/194119. [Google Scholar] [PubMed] [CrossRef]
47. Kuddannaya S, Chuah YJ, Lee MH, Menon NV, Kang Y, Zhang Y. Surface chemical modification of poly(dimethylsiloxane) for the enhanced adhesion and proliferation of mesenchymal stem cells. ACS Appl Mater Interfaces. 2013;5(19):9777–84. doi:10.1021/am402903e. [Google Scholar] [PubMed] [CrossRef]
48. Keung AJ, Kumar S, Schaffer DV. Presentation counts: microenvironmental regulation of stem cells by biophysical and material cues. Annu Rev Cell Dev Biol. 2010;26:533–56. doi:10.1146/annurev-cellbio-100109-104042. [Google Scholar] [PubMed] [CrossRef]
49. Ferrai C, Schulte C. Mechanotransduction in stem cells. Eur J Cell Biol. 2024;103(2):151417. doi:10.1016/j.ejcb.2024.151417. [Google Scholar] [PubMed] [CrossRef]
50. Mishra J, Chakraborty S, Niharika, Roy A, Manna S, Baral T, et al. Mechanotransduction and epigenetic modulations of chromatin: role of mechanical signals in gene regulation. J Cell Biochem. 2024;125(3):e30531. doi:10.1002/jcb.v125.3. [Google Scholar] [CrossRef]
51. Vivo M, Rosti V, Cervone S, Lanzuolo C. Chromatin plasticity in mechanotransduction. Curr Opin Cell Biol. 2024;88:102376. doi:10.1016/j.ceb.2024.102376. [Google Scholar] [PubMed] [CrossRef]
52. Seder I, Kim DM, Hwang SH, Sung H, Kim DE, Kim SJ. Microfluidic chip with movable layers for the manipulation of biochemicals. Lab Chip. 2018;18(13):1867–74. doi:10.1039/C8LC00382C. [Google Scholar] [PubMed] [CrossRef]
53. Liu YJ, Guo SS, Zhang ZL, Huang WH, Baigl D, Xie M, et al. A micropillar-integrated smart microfluidic device for specific capture and sorting of cells. Electrophoresis. 2007;28(24):4713–22. doi:10.1002/elps.v28:24. [Google Scholar] [CrossRef]
54. Descamps L, Le Roy D, Deman AL. Microfluidic-based technologies for CTC isolation: a review of 10 years of intense efforts towards liquid biopsy. Int J Mol Sci. 2022;23(4):1981. doi:10.3390/ijms23041981. [Google Scholar] [PubMed] [CrossRef]
55. Loutherback K, D’Silva J, Liu L, Wu A, Austin RH, Sturm JC. Deterministic separation of cancer cells from blood at 10 mL/min. AIP Adv. 2012;2(4):42107. doi:10.1063/1.4758131. [Google Scholar] [PubMed] [CrossRef]
56. Zhang H, Yang Y, Li X, Shi Y, Hu B, An Y, et al. Frequency-enhanced transferrin receptor antibody-labelled microfluidic chip (FETAL-Chip) enables efficient enrichment of circulating nucleated red blood cells for non-invasive prenatal diagnosis. Lab Chip. 2018;18(18):2749–56. doi:10.1039/C8LC00650D. [Google Scholar] [PubMed] [CrossRef]
57. Wang H, Ning X, Zhao F, Zhao H, Li D. Human organoids-on-chips for biomedical research and applications. Theranostics. 2024;14(2):788–818. doi:10.7150/thno.90492. [Google Scholar] [PubMed] [CrossRef]
58. Cheng JW, Sip CG, Lindstedt PR, Boitano R, Bluestein BM, Gamble LJ, et al. “Chip-on-a-transwell” devices for user-friendly control of the microenvironment of cultured cells. ACS Appl Bio Mater. 2019;2(11):4998–5011. doi:10.1021/acsabm.9b00672. [Google Scholar] [PubMed] [CrossRef]
59. Lee JM, Kim JE, Borana J, Chung BH, Chung BG. Dual-micropillar-based microfluidic platform for single embryonic stem cell-derived neuronal differentiation. Electrophoresis. 2013;34(13):1931–838. doi:10.1002/elps.201200578. [Google Scholar] [PubMed] [CrossRef]
60. Lee JM, Kim JE, Kang E, Lee SH, Chung BG. An integrated microfluidic culture device to regulate endothelial cell differentiation from embryonic stem cells. Electrophoresis. 2011;32(22):3133–7. doi:10.1002/elps.201100161. [Google Scholar] [PubMed] [CrossRef]
61. Farhang Doost N, Srivastava SK. A comprehensive review of organ-on-a-chip technology and its applications. Biosensors. 2024;14(5):225. doi:10.3390/bios14050225. [Google Scholar] [PubMed] [CrossRef]
62. Kim S, Lee S, Lim J, Choi H, Kang H, Jeon NL, et al. Human bone marrow-derived mesenchymal stem cells play a role as a vascular pericyte in the reconstruction of human BBB on the angiogenesis microfluidic chip. Biomaterials. 2021;279:121210. doi:10.1016/j.biomaterials.2021.121210. [Google Scholar] [PubMed] [CrossRef]
63. Liu J, Qin M, Shi Y, Jiang R, Wang Z, Zhang L, et al. Volatile carbonyl metabolites analysis of nanoparticle exposed lung cells in an organ-on-a-chip system. Talanta. 2024;274(3):126066. doi:10.1016/j.talanta.2024.126066. [Google Scholar] [PubMed] [CrossRef]
64. Yin F, Zhang X, Wang L, Wang Y, Zhu Y, Li Z, et al. HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab Chip. 2021;21(3):571–81. doi:10.1039/D0LC00921K. [Google Scholar] [PubMed] [CrossRef]
65. Pattanayak P, Singh SK, Gulati M, Vishwas S, Kapoor B, Chellappan DK, et al. Microfluidic chips: recent advances, critical strategies in design, applications and future perspectives. Microfluid Nanofluidics. 2021;25(12):99. doi:10.1007/s10404-021-02502-2. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools