 Open Access
Open Access
ARTICLE
Ganoderic acid A ameliorates renal fibrosis by suppressing the expression of NPC1L1
Department of Nephrology and Environmental Occupational Health, West China School of Public Health and West China Fourth Hospital, Sichuan University, Chengdu, 610041, China
* Corresponding Author: LINSHEN XIE. Email:
# These authors contributed equally to this work
BIOCELL 2024, 48(11), 1625-1638. https://doi.org/10.32604/biocell.2024.055029
Received 14 June 2024; Accepted 29 August 2024; Issue published 07 November 2024
Abstract
Objective: The study aimed to explore the protective mechanism of Ganoderic acid A (GAA) in renal fibrosis and to verify that GAA can ameliorate renal fibrosis by regulating the Niemann-pick C1-like 1 (NPC1L1) gene. Methods: Transforming growth factor beta1 (TGF-β1) was used to treat Human Kidney-2 (HK-2) cells to establish a renal fibrosis model. The differentially expressed genes in the control (CTRL) group, TGF-β1 group, and TGF-β1 + GAA group were screened via transcriptome sequencing technology and verified by qPCR and Western blot experiments. The NPC1L1 gene overexpression plasmid was constructed. The expression levels of N-cad, E-cad, and Slug-related proteins in CTRL, TGF-β1, TGF-β1+GAA (25 μg/mL), and TGF-β1+GAA (25 μg/mL) + NPC1L1 Overexpression (OE) groups were detected by qPCR and Western blot analysis. Western blot analysis was used to identify the extracellular matrix-associated proteins Tenascin-C, α-SMA, and fibrosis-related protein Collagen I. Fibrosis marker protein Fibronectin was detected and quantified by immunofluorescence. Results: Transcriptomic sequencing revealed that TGF-β1 stimulation led to 267 differentially regulated genes, with 118 up-regulated and 149 down-regulated, while further modulation of 213 genes, comprising 112 up-regulated and 101 down-regulated genes, was observed in the GAA intervention group. The target gene in these processes was found to be NPC1L1 by investigations using Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). qPCR and Western blot results confirmed that TGF-β1 increased NPC1L1 expression, which was attenuated by GAA. Additionally, TGF-β1 upregulated N-cad and Slug. However, GAA reversed this effect and NPC1L1 overexpression partially rescued the GAA effect. TGF-β1 also decreased E-cad expression, reversed by GAA, and NPC1L1 overexpression antagonized this reversal. Furthermore, TGF-β1 promoted Collagen I, α-SMA, and Tenascin-C expression, and GAA reduced these levels, effects that were reversed by NPC1L1 overexpression. Immunofluorescence results showed that TGF-β1 increased fibronectin expression, which was decreased by GAA, and increased by NPC1L1 overexpression. Conclusion: GAA ameliorates renal fibrosis by antagonizing NPC1L1 gene expression inhibiting epithelial-mesenchymal transition and reducing extracellular matrix formation.Keywords
Chronic kidney disease (CKD) is marked by chronic impairment of renal structure or function, which can be caused by various factors [1]. Its onset is insidious, with a long incubation period, making it challenging to prevent and treat [2]. Consequently, CKD has become a major worldwide public health issue [3]. CKD typically does not show obvious symptoms in the early stages. However, as it progresses to the middle and late stages, patients may experience a series of symptoms related to uremia due to the progressive decline in renal function. At this stage, patients often require expensive renal replacement therapy such as hemodialysis or peritoneal dialysis to stay alive [4]. At present, the global median prevalence of chronic kidney disease is 9.5%, with an interquartile range spanning from 5.9% to 11.7% [5]. End-stage kidney disease (ESKD) represents the terminal, irreversible phase of chronic kidney disease (CKD), during which kidney replacement therapy (KRT) becomes essential for survival. Globally, ESKD presents a significant public health issue and imposes a substantial economic load on healthcare systems.
Renal fibrosis is considered to be the pathway involved in the progression of almost all types of CKD to ESRD [6], characterized by epithelial-to-mesenchymal transition (EMT) and extracellular collagen accumulation [7]. Fibrosis serves as a dynamic biosynthetic reparative mechanism that safeguards tissues from injury [8]. Nevertheless, in cases of a chronic persistent injury, the affected tissue remains deprived of oxygen and blood flow, leading to ongoing inflammation. The onset of inflammation triggers the secretion of a variety of factors that stimulate fibroblast proliferation and amplify the inflammatory process [9]. Additionally, it stimulates the activation of a significant number of immune cells, which further contribute to the deterioration of the renal tissue. Owing to the inherent incapacity to achieve comprehensive tissue repair or regeneration, there occurs a substitution of normal tissue with fibrous tissue, culminating in renal fibrosis and ultimately progressing to renal failure [10]. However, current pharmacotherapeutic interventions aiming to mitigate or reverse renal fibrosis exhibit limited efficacy in curbing or reversing the progressive nature of this pathological process [11,12].
Recent studies have focused on investigating natural substances as potential substitutes for preventing and treating fibrosis [13]. Ganoderma leucocontextum is a newly discovered species within the Ganoderma genus, first identified in Tibet in 2014. It is native to Southwestern China, especially the Tibetan region [14]. The Ganoderma leucocontextum is a distinct variation of Ganoderma leucocontextum found in the Northwest Plateau of China. It is abundant in active substances such as Ganoderma lucidum polypeptides, Ganoderma leucocontextum polysaccharides, triterpenic acids, and alkaloids [15]. Research has demonstrated that the extract derived from Ganoderma leucocontextum shows abundant synergistic properties, including anti-inflammatory, anti-fibrotic, and neuroprotective benefits [16–18]. Ganoderic acid A (GAA), an essential bioactive triterpenoid derived from the Ganoderma leucocontextum, exhibits potent anticancer, antioxidant, anti-inflammatory, hepatoprotective, nephroprotective, and pulmonary protective activities [19]. Studies have shown that the alcohol extract of Ganoderma leucocontextum can significantly enhance the therapy of silicosis by improving pulmonary inflammation and inhibiting the fibrosis process [20]. In terms of anti-liver fibrosis, Ganoderma leucocontextum polysaccharides have been shown to have a wide range of anti-liver fibrotic effects, and the underlying mechanism may be that liver fibrosis is affected by the TGF-β/Smad signaling pathway [21]. Hence, the therapeutic impact of natural substances in reducing fibrosis is well acknowledged. However, the mechanism by which GAA plays a role in influencing renal fibrosis has not been clearly elucidated.
Niemann-Pick C1-like 1 (NPC1L1), a transmembrane entity, is essential for the absorption of cholesterol within the intestinal tract [22]. NPC1L1 gene is highly expressed and functionally localized in the apical membrane of intestinal brush epithelial cells in the proximal jejunum of the small intestine. This localization is crucial for its role in facilitating the absorption of cholesterol from the diet [23]. In addition, the NPC1L1 gene is also expressed on the tubule membrane of human hepatocytes and contributes to sterol reabsorption and enterohepatic circulation [24]. In conclusion, the NPC1L1 gene plays an important role in cholesterol absorption and metabolic pathways. Kidney disease is closely related to the disruption of human cholesterol homeostasis, which is determined by the energy metabolism pathway of renal tubular epithelial cells, which determines their energy supply mainly from fatty acid oxidation [25]. Therefore, metabolic pathways play a key role in the pathogenesis of kidney disease.
Metabolic disturbances, such as disruptions in glucose and lipid metabolism, oxidative stress, and malfunctioning of mitochondria, play a role in the progression of kidney diseases by initiating damage to the tubules, apoptosis, fibrosis, and the formation of scar tissue. Disruption in metabolic pathways, including glucose and lipid metabolism [26]. Studying the intricate connection between immunological and metabolic pathways in renal illnesses has the potential to provide new therapeutic possibilities for preventing and treating kidney disease.
Combined with our research group’s previous findings on the treatment of renal fibrosis by active ingredients in Ganoderma leucocontextum [20], we hypothesized that Ganoderic acid A may improve renal lipid metabolism by down-regulating NPC1L1 gene, thus alleviating the progression of renal fibrosis.
Our research is designed to find out the protective mechanism of GAA intervention against TGF-β1-induced renal fibrosis.
Human Kidney-2 (HK-2) cells (Abiowell Biotechnology, AW-CNH063, Changsha, China) (STR identification) were purchased from Changsha Abiwell Biotechnology Co., Ltd. (Changsha, China). All utilized cells were tested for contamination, and were free from mycoplasma contamination. Cells were cultured in Minimum Essential Medium (MEM) (Gibco, 41500034, Shanghai, China) containing 10% Fetal Bovine Serum (Superfine), Uruguay (MedChemExpress, HY-T1000, Shanghai, China) and 1% Penicillin-Streptomycin Solution, 100X (Solarbio, P1400, Beijing, China) at 37°C, 5% CO2 incubator (Rundu Biotech, Herocell 240, Shanghai, China). The current study was structured to encompass three distinct experimental groups. The control group comprised HK-2 cells cultured under standard conditions; TGF-β1 (MedChemExpress, HY-P70543) stimulation group, involved the cultivation of HK-2 cells with the addition of 10 ng/mL of TGF-β1 for a 24-h duration; GAA intervention group, was identical to the TGF-β1 stimulation group, except for the concurrent addition of 25 μg/mL of GAA (Topscience, T6S1141, Shanghai, China) to the culture medium for the same 24-h period.
The NPC1L1 overexpression plasmid was synthesized and constructed by Suzhou Hongxun Biotechnology Co., Ltd. (Suzhou, China). Cells were cultured until they reached 80% confluence, at which point the existing culture medium was aspirated. The cells were then enumerated and diluted to a concentration of 5 × 105/mL. 1 mL of this cell suspension was put into each well of a 6-well plate, resulting in a density of 5 × 105/mL. The plate was placed at a temperature of 37°C and a carbon dioxide concentration of 5% for a duration of one night, followed by transfection the subsequent day. To prepare the transfection mixture, 5 μL of plasmid DNA at a concentration of 2 μg/mL was combined with 250 μL of serum-free medium and thoroughly mixed. Separately, 5 μL of Lipofectamine™ 2000 (Invitrogen, 11668500, Shanghai, China) was added to another 250 μL of serum-free medium and gently mixed, followed by a 5-min incubation period. Then the diluted plasmid and Lipofectamine™ 2000 were combined and incubated at room temperature for 20 min to facilitate the formation of the plasmid complex. This complex was added to the culture plate wells, which already contained cells in serum-free medium, bringing the total volume to 2 mL per well. Following an incubation period of 4–6 h, the transfection complexes were aspirated, and the cells were refed with a complete medium. The cells were then incubated for an additional 48 h before proceeding with subsequent experimental analyses.
Following the experimental manipulation, the culture medium was carefully removed. Subsequently, the crawling fragments were subjected to two thorough washes using sterile phosphate-buffered saline (PBS). The treated cells were taken and removed from the old medium and washed twice with PBS. Thereafter, a neutral general-purpose tissue fixative (Servicebio, G1101, Wuhan, China) was introduced and the mixture was allowed to stand at room temperature for a fixation period of 10 min. Subsequently, the fixative was meticulously decanted, and the specimens were subjected to three rinses with PBS to eliminate any remaining fixative. After the slide is slightly dried, use a histochemistry pen to draw circles in the middle of the coverslip at the position where the cells were uniformly dispersed. Next, 100 μL of the permeabilization solution was added within the marked area and incubated at ambient temperature for a period of 20 min. The cells are then rinsed thrice with PBS, with each wash for 5 min. Blocking is performed using 3% BSA (Servicebio, GC305006) for a duration of 30 min at room temperature. Shake off the blocking solution gently, add the prepared primary antibody Fibronectin Monoclonal antibody, 1:300, (Wuhan Sanying, 66042-1-Ig, Wuhan, China) dropwise to the cell well plate, incubate at 4°C overnight, and then wash 3 times, 5 min each. After adding the secondary antibody, Goat Anti-Mouse IgG (Servicebio, GB25301), diluted at a ratio of 1:500, incubate the solution for 50 min at room temperature. Following the primary staining, the nuclei are counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (Servicebio, G1012). Finally, use an antifluorescence quenching retardant (Servicebio, G1401) as the mountant. Acquire images. The expression level of the corresponding protein is expressed as an integral absorbance value.
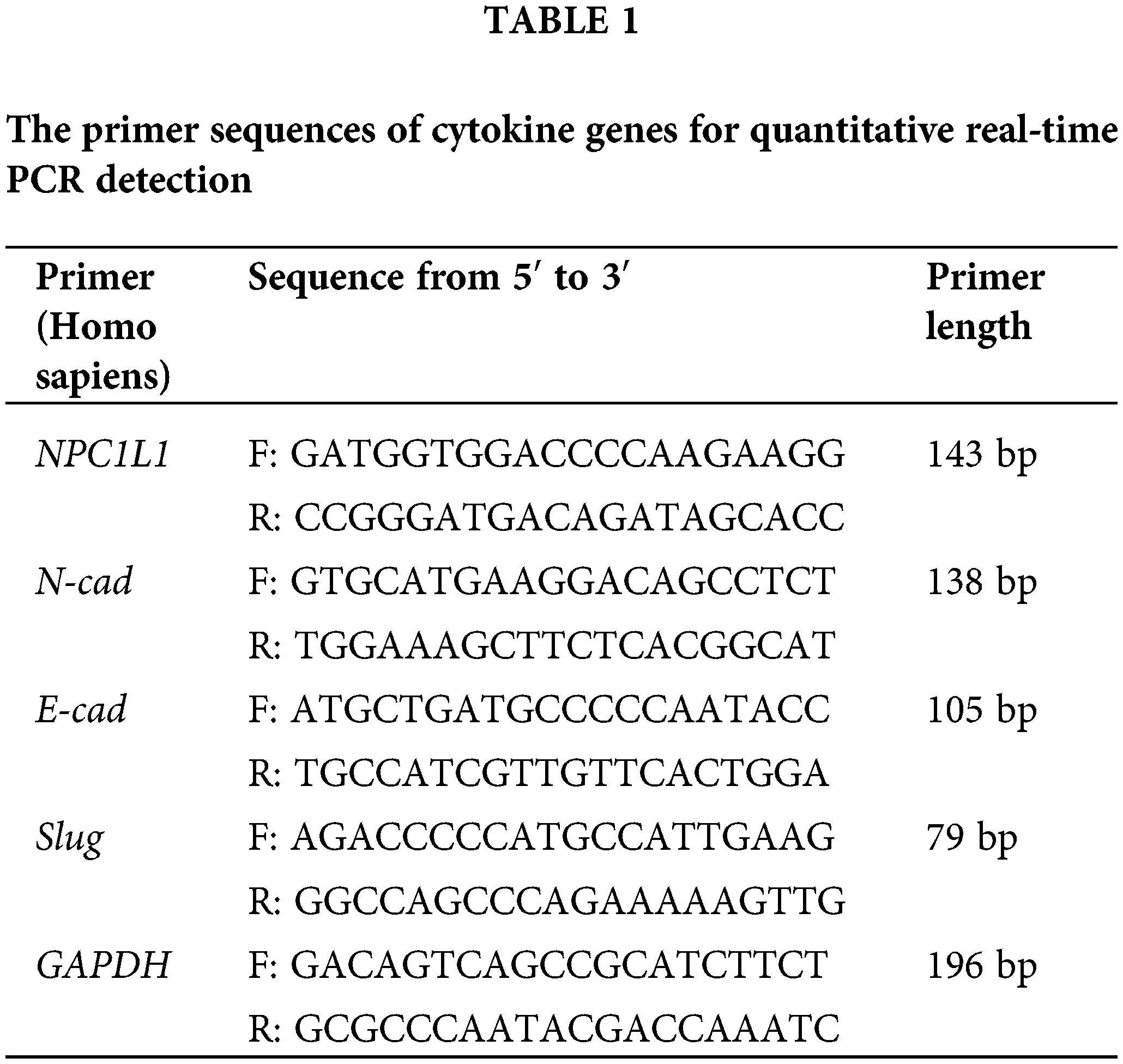
After collecting 1 × 106 cells, 1 mL of Triquick Reagent (Trizol Substitute) (Solarbio, R1100) was introduced to the cell suspension and incubated for 5 min to ensure complete lysis. Subsequently, 200 μL of chloroform was added, and the mixture was thoroughly vortexed before being allowed to stand at room temperature for 2 min. The sample was then centrifuged at 4°C and 12,000 g for 10 min. The supernatant was carefully transferred to a fresh tube, an equal volume of isopropanol was added, and the solution was mixed thoroughly before being placed at −20°C for a 10-min incubation period. RNA pellets were collected and subjected to two washes with 75% ethanol, followed by air drying on a clean bench. The dried pellets were then resuspended in 20–60 μL of DEPC-treated water. cDNA was synthesized via reverse transcription, and quantitative PCR (qPCR) was performed using SYBR (Vazyme Biotech, Q712-02, Nanjing, China) under the specified conditions: The relative expression of the target gene was determined by calculating the ratio of the cycle threshold (CT) values of the target gene to those of the internal reference gene, with each sample analyzed in triplicate. The primer sequences for both the target and reference genes are provided in the accompanying Table 1.
We utilized a protein extraction kit (Solarbio, BC3710) to isolate cellular proteins. Protein concentration was determined using the Pierce™ BCA Protein Assay Reagent A (Thermo Scientific, 23228, Shanghai, China), after which Pierce™ BCA Protein Assay Reagent B (Thermo Scientific, 23224) was added. Subsequently, 5 × SDS sample buffer was introduced to the protein mixture, which was then heated to 100°C for a duration of 5 min. Protein was loaded at 50 μg for SDS-PAGE electrophoresis at 150 V for 30–40 min when the bromophenol blue indicator strip was electrophoresis to the bottom of the gel to end the electrophoresis. The eBlot™ L1 Fast Wet Transfer Kit (Genscript, L00686C, Nanjing, China) and the eBlot L1 Transfer Concentration Kit (Genscript, L00726C) were used for transfer. After the transfer process, the Polyvinylidene fluoride (PVDF) membrane (Millipore, IPVH00010, Darmstadt, Germany) was subjected to a 5-min rinse in TBST (Servicebio, G2150-1L). Thereafter, incubated with a TBST solution containing 5% skimmed milk powder for one hour at ambient temperature, followed by three serial washes using TBST, 5 min each. The membrane was subsequently probed with the following primary antibodies: NPC1L1 (Abclonal, A10049, Wuhan, China), diluted at a ratio of 1:3000; N-cadherin antibody (Wuhan Sanying, 22018-1-AP), diluted at a ratio of 1:5000; E-cadherin antibody (Wuhan Sanying, 20874-1-AP), diluted at a ratio of 1:50,000; Slug antibody (Wuhan Sanying, 12129-1-AP), diluted at a ratio of 1:10,000 and GAPDH (Wuhan Sanying, 60004-1-Ig), diluted at a ratio of 1:300,000. Each of these primary antibodies was allowed to bind to the membrane for 1 h at room temperature. Post-incubation, the membrane was subjected to three additional washes of 5 min each to remove any unbound antibodies.
HRP Conjugated AffiniPure Goat Anti-rabbit IgG (Boster Biological Technology, BA1054, Wuhan, China), diluted at a ratio of 1:20,000, or HRP Conjugated AffiniPure Goat Anti-mouse IgG (Boster Biological Technology, BA1051), diluted at a ratio of 1:20,000, was then incubated at room temperature for 40 min. Subsequently, the membrane was subjected to three 7-min washes. Finally, the ultra-sensitive ECL chemiluminescence substrate kit (Biosharp, BL520B, Hefei, China) was used for chemiluminescence, and the automatic chemiluminescence image analysis system was used for imaging. The optical density of the protein bands was quantified using Image-Pro Plus 6.0 software (Media Cybernetics, Rockville, MD, USA).
The extracted RNA was qualitatively tested, and after the sample was qualified, the eukaryotic mRNA was enriched with magnetic beads with Oligo (dT). The purified mRNA is fragmented and reverse transcribed, and quality control is performed to generate sequencing libraries. Libraries were sequenced on Illumina Novaseq 6000/MGISEQ-T7. The transcriptome sequencing and analysis of this study were commissioned by Shanghai Zhongke New Life Biotechnology Co., Ltd. (China). The raw sequencing data is first evaluated for quality.HISAT2 software was used (http://daehwankimlab.github.io/hisat2/, accessed on 6 February 2020) to sequence align the Clean Reads to the indicated genome.DESeq2 (http://bioconductor.org/packages/release/bioc/html/DESeq2.html, accessed on 26 April 2023) was used for differential expression analysis of genes between groups, and the default screening threshold for differentially expressed genes was: |log2FoldChange| > 1 and p adjusted <0.05. Utilizing the clusterProfiler R package (Bioconductor,4.12.2), we conducted an enrichment analysis of Gene Ontology (GO) functions and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Significance in enrichment was determined at a p-value cutoff of <0.05, indicative of substantial enrichment for GO or KEGG functions. The Search Tool for the Retrieval of Interacting Genes (STRING) database (accessed via https://www.string-db.org/, accessed on 26 July 2023) was employed primarily for mapping protein-protein interaction networks corresponding to the genes of interest. In cases where the species was not represented within the STRING database, we initially aligned the target gene set sequences using blastx (with a threshold of e value <1e-10) against the protein sequences of reference species that are included in the database. Subsequently, the protein interaction data from these reference species were utilized to construct the interaction network.
Statistical analyses were conducted using Prism (Graphpad software Inc., Graphpad 9.0.0, San Diego, CA, USA). The presented data correspond to the average of three separate experimental replicates and are depicted as mean ± standard deviation (SD). To assess the significance of differences among multiple groups, a one-way analysis of variance (ANOVA) was applied, complemented by Tukey’s post-hoc test. p-value < 0.05 was deemed statistically significant.
Data acquisition differentially expressed genes (DEGs) identification
The volcano plot showed the distribution and up and down-regulation of the two groups of differential genes (Fig. 1A,B). Each data point in these plots corresponds to an individual gene. The genes that exhibit no significant difference are represented by gray dots. The genes that are significantly up-regulated are denoted by red dots, while those that are significantly down-regulated are indicated by blue dots. This classification is based on the statistical criteria of p-value < 0.05 and |log2 FC| >1.0. The differential gene cluster map delineates the variability among genes across the two groups (Fig. 1C,D). Cluster analysis outcomes revealed the differential expression profiles of genes within various samples. In this representation, red signifies genes with elevated expression levels, whereas blue denotes genes with reduced expression levels. The genes with altered expression levels under different treatments, along with their log2FoldChanges are listed in Table 2.
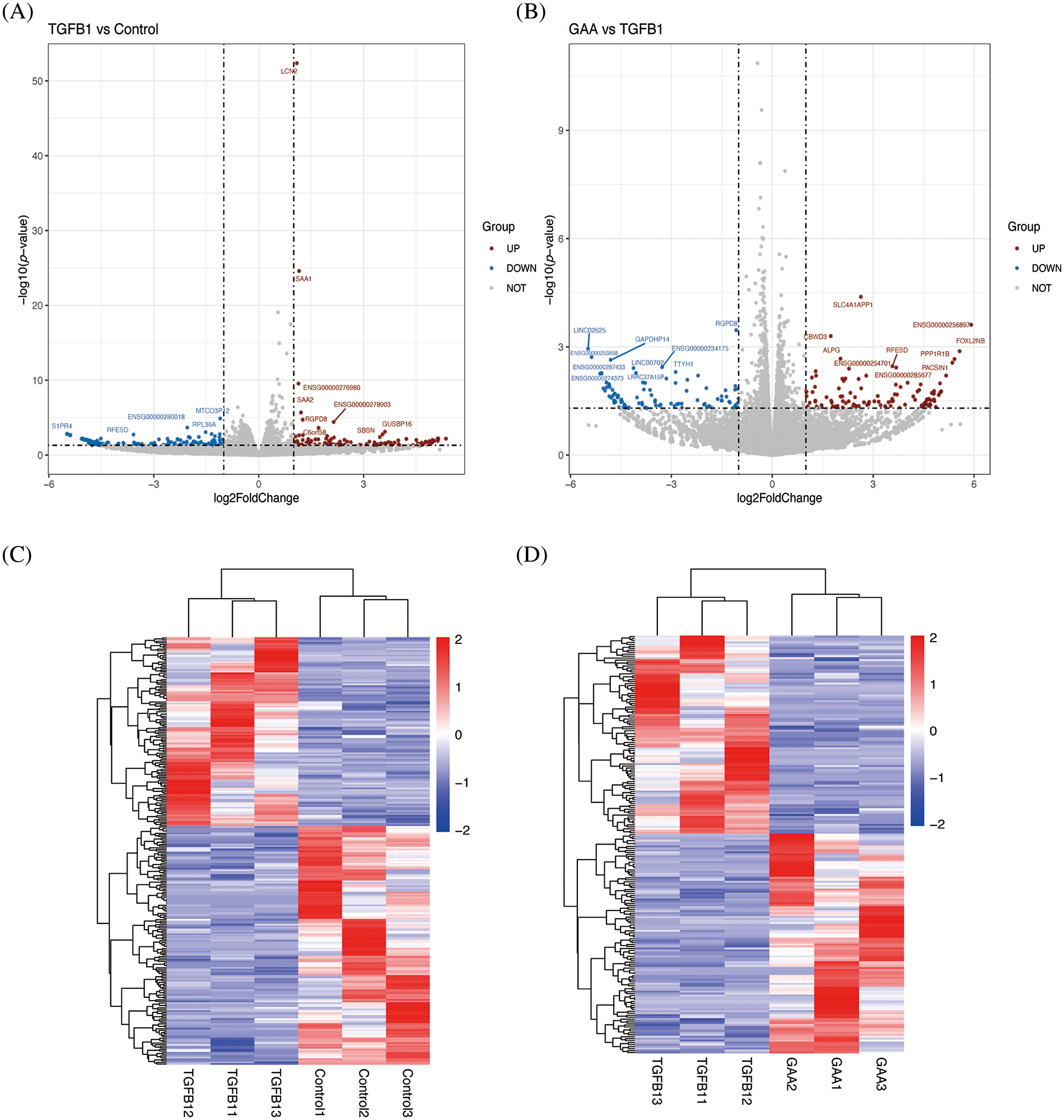
Figure 1: The volcano plots and heat map show DEGs expression between the TGF-β1 group vs. the control group and the TGF-β1 + GAA group vs. the TGF-β1 group. (A) A volcano plot comparing DEGs expression in the TGF-β1 group relative to the control group. (B) The volcano plot of DEGs expression between the TGF-β1 + GAA group and TGF-β1 group. (C) The heat map of DEGs expression between the TGF-β1 group and control group. (D) The heat map of DEGs expression between the TGF-β1 + GAA group and TGF-β1 group. Criteria: p-value < 0.05 and |log2 FC| > 2.0. DEGs: Differentially expressed genes; TGF-β1: Transforming growth factor beta1; GAA: Ganoderic acid A.
GO functional enrichment and KEGG indicated the enrichment of lipid metabolism-related pathways
The functional enrichment analysis of differentially expressed genes is categorized into three classes based on GO functional entries: Biological Process (BP), Cellular Component (CC), and Molecular Function (MF). The top 10 functional entries under each of the three major GO categories, sorted by p value, were selected for representation in the form of a bar chart. This accounts for a total of 30 functional entries. Compared to the control group, the upregulated DEGs in the TGF-β1 group primarily involve molecular functions related to temperature−gated cation channel activity, formimidoyltetrahydrofolate cyclodeaminase activity, glutamate formimidoyltransferase activity, cholesterol 25−hydroxylase activity. Biological Process mainly involve leukocyte chemotaxis, acute-phase response, cell chemotaxis, leukocyte migration, etc. Cell components include eukaryotic 48S preinitiation complex, extracellular exosome, high−density lipoprotein particle, etc. (Fig. 2A). The KEGG enrichment scatter plot provides a vivid illustration of the most significantly enriched pathways among the top 20 entries. The graphical representation uses a color gradient to denote the Rich Factor, where a deeper shade of red signifies a higher value, indicating a stronger enrichment. The magnitude of the scatter points is proportional to the number of DEGs associated with each pathway. The KEGG enrichment results revealed that the upregulated DEGs pathways are significantly enriched in pathways such as Glycosaminoglycan biosynthesis-keratan sulfate, primary bile acid biosynthesis, and histidine metabolism (Fig. 2B).
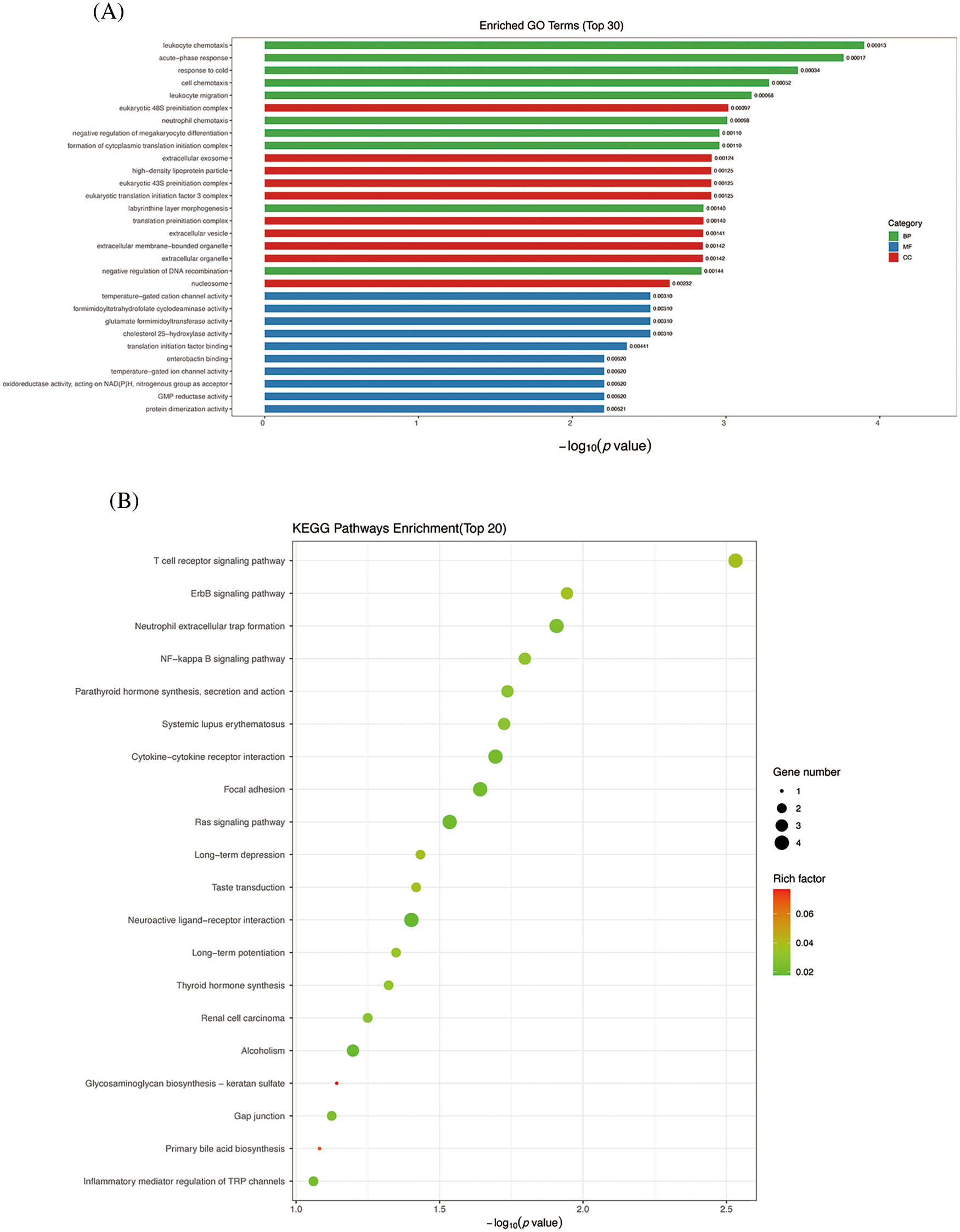
Figure 2: GO enrichment analysis and KEGG analysis of DEGs between TGF-β1 group and control group. (A) GO enrichment analysis. (B) KEGG enrichment analyses. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Similarly, it can be observed that, comparing the TGF-β + GAA intervention group to the TGF-β group, Molecular functions the downregulated DEGs involved are predominantly associated with low-density lipoprotein binding, protein-lipid complex binding, lipoprotein particle binding, and cholesterol 25-hydroxylase. The cellular component primarily involves the glomerular endothelium fenestra (Fig. 3A). KEGG enrichment analyses in Fig. 3B showed that the differential genes are significantly enriched in the primary bile acid biosynthesis pathway (Fig. 3B).
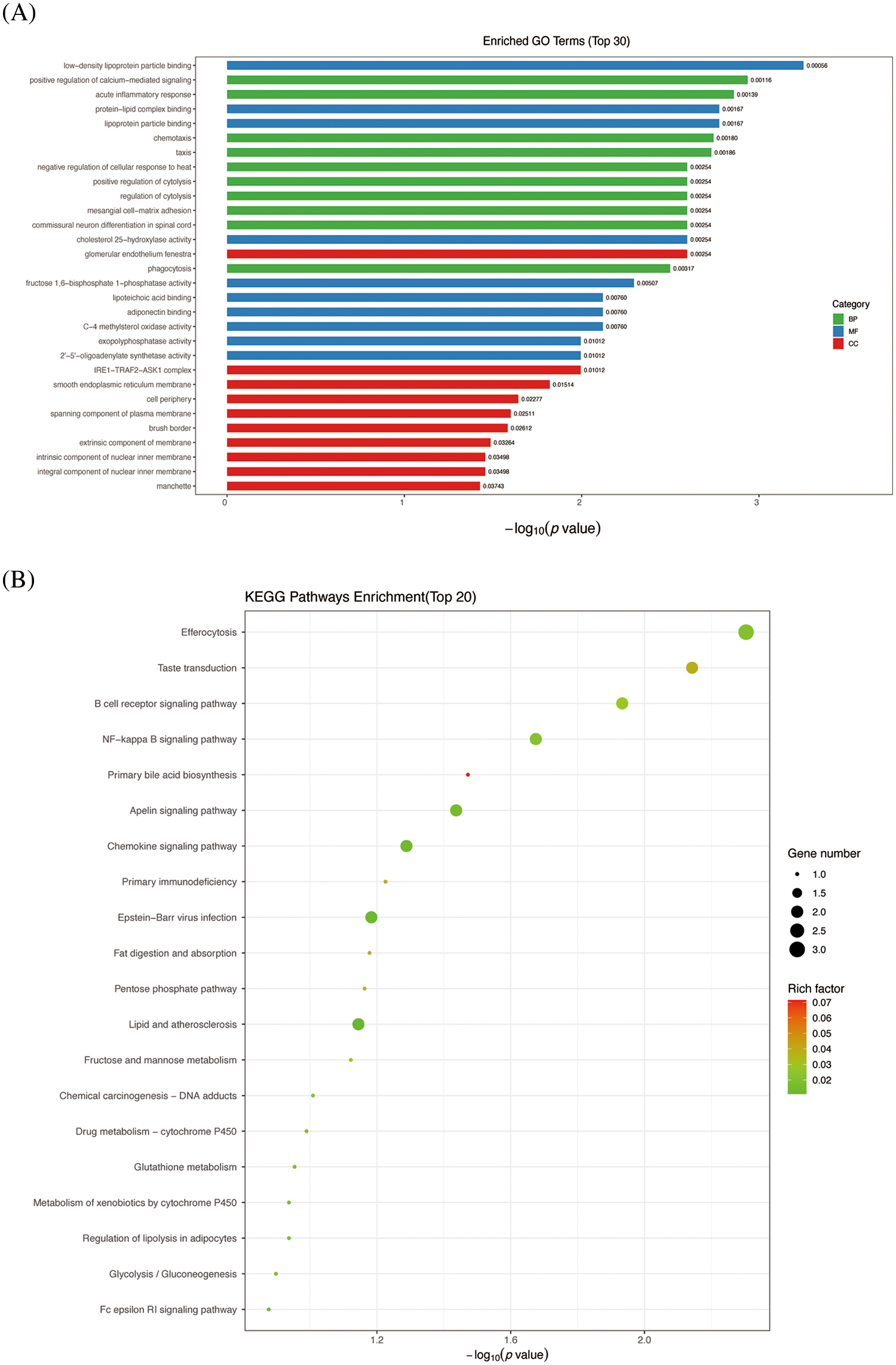
Figure 3: GO enrichment analysis and KEGG analysis of DEGs between TGF-β1 + GAA group and TGF-β1 group. (A) GO enrichment analysis. (B) KEGG enrichment analyses. GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes.
NPC1L1 was screened as differential gene, qPCR and WB proved that GAA can inhibit renal fibrosis by regulating NPC1L1 gene
To explore the inner relationship between GAA intervention and renal fibrosis, heat map was involved to visualize the expression of genes in different samples (Fig. 4A). In contrast to the control group, the TGF-β stimulation group exhibited differential regulation of 267 genes, with 118 genes upregulated and 149 genes downregulated. Compared with the TGF-β intervention group, TGF-β + GAA intervention group showed 213 genes with altered expression levels, comprising 112 upregulated and 101 downregulated genes. Among them, 28 genes were found to be co-regulated, as depicted in Fig. 4B. To identify hub genes among the 28 co-regulated genes, a protein-protein interaction (PPI) network was constructed using the STRING database, as illustrated in Fig. 4C. In conjunction with prior transcriptomic findings, we identified the differentially expressed gene NPC1L1 for initial validation. Expression of NPC1L1 was measured by qPCR in Human Kidney-2 (HK-2) cells treated with TGF-β1 and GAA. The results indicated a marked upregulation of NPC1L1 expression following TGF-β1 treatment, which was subsequently mitigated by GAA (Fig. 4D). Followed by Western blot validation, the expression of NPC1L1 protein was also increased in TGF-β1 group, while inhibited after GAA intervention (Fig. 4E,F).
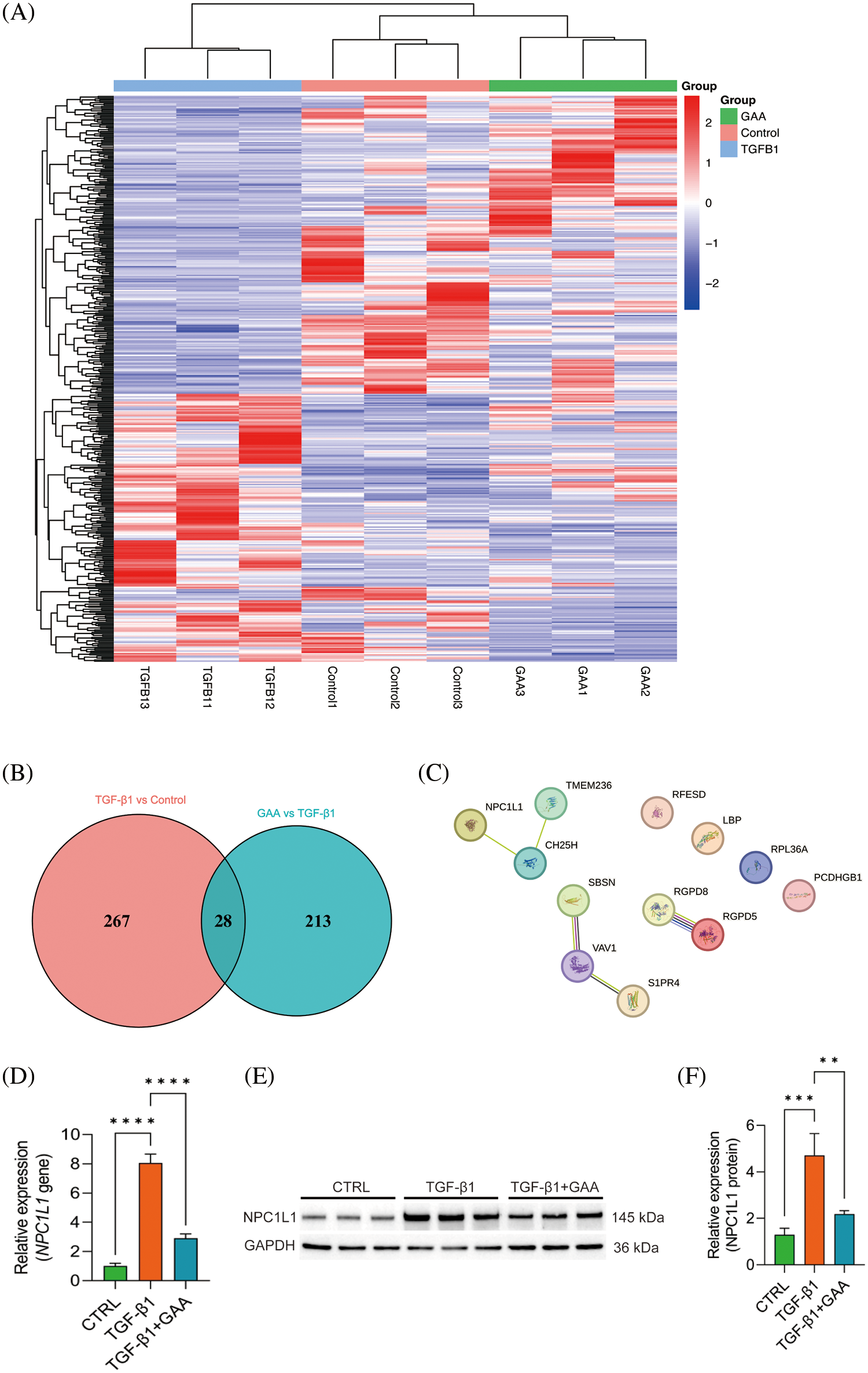
Figure 4: DEGs of GAA targeted in renal fibrosis. (A) Clustering heat map of DEGs in three groups. (B) Venn diagram showing 28 DEGs between TGF-β1 stimulation group and TGF-β1 + GAA group. (C) PPI network related to the hub genes identified. (D) Validation of NPC1L1 expression treated by TGF-β1 and GAA through qPCR assay. (E) Western blot verified TGF-β1 stimulation promoted NPC1L1 expression, and inhibited by GAA. (F) Expression of NPC1L1 protein was measured by Western blot in E. **p < 0.01; ***p < 0.001; ****p < 0.0001.
GAA inhibits EMT by regulating NPC1L1 expression
The results of qPCR and Western blot showed that compared with the control group, TGF-β1 stimulation promoted the expression of N-cadherin and Slug genes while significantly reduced the expression of the E-cadherin gene. Compared with the TGF-β1 group, the expression of the E-cadherin gene was significantly promoted, whereas the expression of N-cadherin and Slug was significantly reduced. After overexpression of NPC1L1, the effects of GAA intervention were significantly reversed (Fig. 5A). Western blot showed that compared to the control group, TGF-β1 stimulation significantly reduced E-cadherin protein expression, while promoted the expression of N-cadherin, α-SMA, and Slug. After GAA intervention, the expression of E-cadherin protein increased, N-cadherin, α-SMA and Slug expression decreased. After overexpression of NPC1L1, the effects of GAA intervention were was reversed again (Fig. 5B,C). The results of Western blot were in line with the qPCR results, which indicates a conclusion that GAA can effectively inhibit EMT by regulating NPC1L1 expression.
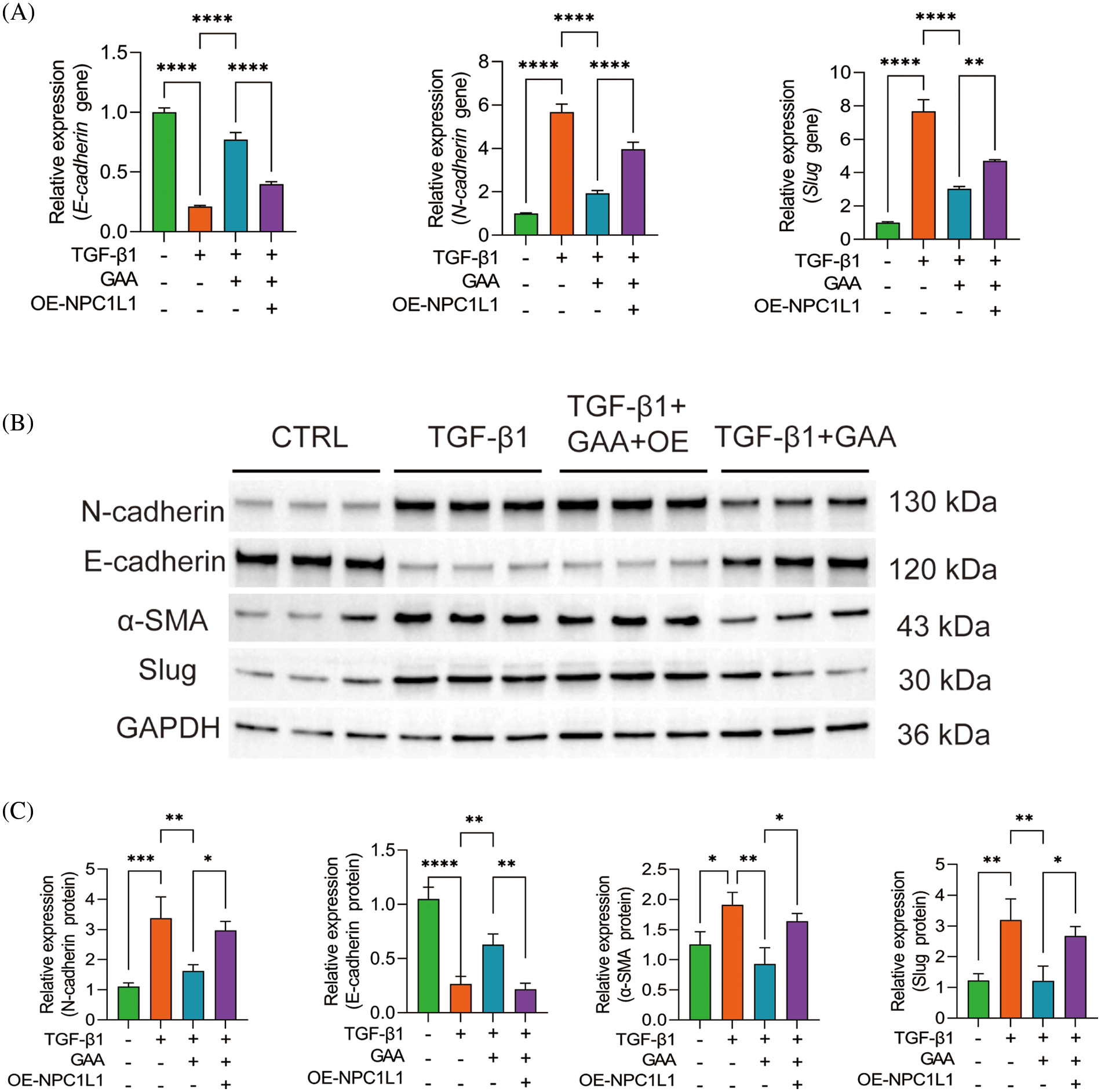
Figure 5: GAA inhibited EMT by regulating NPC1L1 expression. (A) The expression levels of E-cadherin, N-cadherin, Slug genes under different treatments. (B) Western blot analysis of N-cadherin, E-cadherin, α-SMA and Slug proteins under different treatments. (C) The expression levels of N-cadherin, E-cadherin, α-SMA and Slug proteins in different groups. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
GAA affects renal fibrosis by regulating NPC1L1 expression
Western blot analyses revealed that stimulation with TGF-β1 augmented the expression of Collagen I and Tenascin-C proteins, compared to the control group (Fig. 6A,B). Notably, protein expression levels of Collagen I and Tenascin-C were significantly diminished in the TGF-β1 + GAA group. Conversely, following the overexpression of NPC1L1, the TGF-β1 + GAA group exhibited enhanced protein expression of Collagen I and Tenascin-C. We verified the expression levels of Fibronectin in HK-2 cells with CTRL, TGF-β1, TGF-β1 + GAA, TGF-β1 + GAA + OE by immunofluorescence staining (Fig. 6C). The findings demonstrated that GAA intervention led to a reduction in TGF-β1-mediated Fibronectin expression, and Fibronectin expression notably increased after overexpression of NPC1L1 (Fig. 6D).
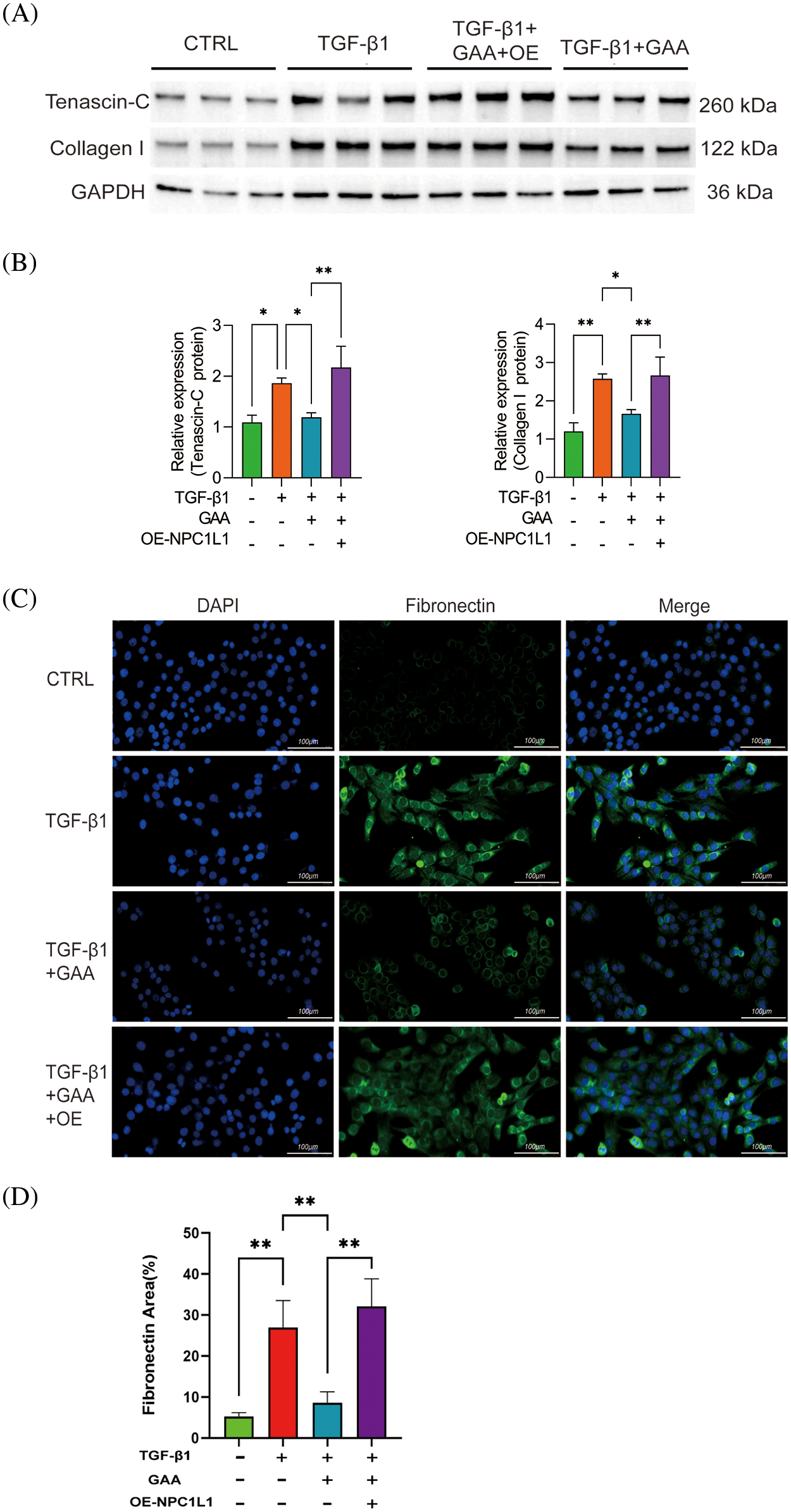
Figure 6: GAA affects renal fibrosis by regulating NPC1L1 expression. (A) Western blot results about expression of Tenascin-C and Collagen I proteins. (B) The expression levels of Tenascin-C and Collagen I proteins under different treatments. (C) Immunofluorescence detection of Fibronectin in Human Kidney-2 (HK-2) cells under varying treatments. (D) Quantitative results of immunofluorescence detection of Fibronectin. *p < 0.05, **p < 0.01. Scale bar = 100 μm.
Renal fibrosis constitutes a pivotal pathway in the progression of CKD pathophysiology [7]. The overexpression of cytokines and vasoactive chemicals is crucial in the development of renal fibrosis. Due to the synergistic impact of these factors, ECM undergoes degradation and becomes imbalanced, leading to a significant buildup of ECM in the kidneys, ultimately ending in renal fibrosis [27]. GAA, a prominent bioactive compound found in Ganoderma leucocontextum, has demonstrated a diverse range of biological effects. Nevertheless, the potential mechanism in which GAA impacts renal fibrosis remains unclear. Therefore, we performed this experiment to clarify it.
During the process of kidney damage, a sequence of intrinsic healing mechanisms were triggered. Several mechanisms contribute to this process, including the increased production of profibrotic proteins, the growth of myofibroblasts, the excessive accumulation of ECM in the renal interstitium, and tubular epithelial-mesenchymal transition (EMT) [28]. TGF-β1 is recognized as a pivotal mediator in the pathogenesis of renal fibrosis [29]. TGF-β1, renowned as the primary fibrogenic cytokine, is instrumental in both the onset and advancement of renal fibrosis through orchestrating a complex interplay of multiple signaling mechanisms [30], which is also a commonly used stimulator in tissue fibrosis models. Therefore, we established a HK-2 model of renal fibrosis by TGF-β1 induction.
EMT in the tubular structure represents a sophisticated mechanism, in which morphological shift from epithelial to mesenchymal cell constitutes a pivotal transformation [31]. When the kidneys sustain damage, the renal tubular epithelial cells undergo a transformation into activated fibroblasts. These fibroblasts then release a significant quantity of collagen and fibronectin, leading to the deposition of a substantial amount of ECM and ultimately ending in fibrosis [32]. E-cadherin (E-cad) and N-cadherin(N-cad) belong to the same cadherin superfamily [33]. E-cadherin serves as a distinctive marker for renal tubular epithelial cells, playing a crucial role in the linkage of the actin cytoskeleton. It is a pivotal adhesion molecule responsible for upholding the polarity and integrity of tight junctions within these epithelial cells of the renal tubules [34]. Studies have shown that the decrease in E-cad expression is a crucial step in the process of EMT, and this decrease results in a reduction in the adhesion between cells [35–37]. During the process of EMT, there was a decrease in the expression of E-cad, while the level of N-cad, which is a marker for interstitial cells, showed a considerable increase [38]. This finding was also corroborated in our investigation.
α-smooth muscle actin (α-SMA) is a characteristic protein expressed by myofibroblasts, which directly reflects the degree of fibrosis in kidney tissue [39]. Myofibroblasts are cells that can contract and express α-SMA. They play a role in normal wound healing and contribute to the formation of scar tissue by producing collagen [40]. The significant accumulation and transformation of myofibroblasts in the renal interstitium is a crucial mechanism for ECM synthesis in renal fibrosis [32]. Slug is a transcription factor that induces EMT and is also the upstream transcription factor of E-cad [41], which has a recognized role in EMT induction. The expression of Slug is conventionally inversely related to the expression of E-cadherin [6]. We increased the correlation between slug protein and E-cad, N-cad, and α-SMA proteins to indicate EMT. This is also confirmed by our findings. Following TGF-β1 stimulation, there was a marked enhancement in the protein levels of N-cadherin, α-SMA, and Slug, with a corresponding decrease in E-cadherin expression when compared to the control group. Treatment with GAA counteracted this phenomenon by boosting E-cadherin levels and concurrently inhibiting the overexpression of N-cadherin, α-SMA, and Slug.
TenascinC is a group of glycoproteins found in the extracellular matrix. They are created in excess during the process of repairing tissue injury and fibrosis [42]. Collagen I and III are the primary components of the ECM and can serve as marker proteins to indicate the formation of the ECM [43]. Fibronectin is a major component of the ECM in renal fibrosis, and its increased synthesis promotes ECM accumulation [44]. The findings of this study revealed that following GAA intervention, the TGF-β1 + GAA group exhibited a marked reduction in the expression levels of Collagen I and Tenascin-C genes and proteins, as compared to the group subjected to TGF-β1 stimulation alone. Simultaneously, the immunofluorescence findings likewise confirmed the related conclusions. The expression of TGF-β1-induced Fibronectin was decreased following GAA intervention, in comparison to the TGF-β1 stimulation group. The impact of GAA intervention in enhancing renal fibrosis has been initially validated.
NPC1L1 is a pivotal protein involved in the intracellular transport of cholesterol, which plays a critical role in the regulation of dietary cholesterol absorption and the modulation of circulating low-density lipoprotein (LDL) cholesterol levels [45,46]. Cholesterol is a fundamental molecule that integral to the architecture of cellular membranes and provides the foundational material for the biosynthesis of bile acids and steroid hormones. Dysregulation of cholesterol metabolism is associated with a plethora of metabolic disorders. NPC1L1 is a critical regulator of cholesterol and lipid metabolism, functioning pivotal in sustaining the equilibrium of cholesterol homeostasis within the organism [47]. Studies have shown that inhibition of NPC1L1 expression can significantly reduce blood cholesterol levels. Research has demonstrated that suppressing the expression of NPC1L1 can effectively decrease levels of cholesterol in the bloodstream. Some studies [48–50] have found that renal lipid metabolism disorders can promote the progression of renal fibrosis, so regulating renal lipid metabolism disorders can be used as the main target for the treatment of renal fibrosis [51].
Alterations in lipid metabolism have been implicated in a variety of metabolic disorders [52–55]. Although the kidneys are not considered metabolic organs, renal tubular cells have a high baseline energy consumption and primarily rely on fatty acid oxidation for energy [52]. The high energy requirement of renal proximal tubular cells, along with their limited glycolytic capabilities, necessitates the utilization of free fatty acid oxidation as the primary energy source for these cells [25]. When renal trauma happens, the capacity for fatty acid oxidation in renal tubular epithelial cells is compromised, concomitant with an elevation in glycolytic activity. This metabolic shift results in an enhanced generation of reactive oxygen species (ROS) and the upregulation of pro-inflammatory and pro-fibrotic mediators, thereby aggravates renal fibrosis [52–54]. NPC1L1 has the ability to regulate the process of transcription genes that are involved in lipid synthesis [56,57]. Furthermore, it can activate the enzymatic activity of fatty acid synthase, which ultimately leads to an augmentation in the levels of free fatty acids. Subsequent to the overexpression of the NPC1L1 gene, an increment in the expression of fibrosis-associated marker proteins was observed, corroborating our prior findings.
While our study offers valuable insights into the anti-fibrotic impacts and the mechanisms by which GAA operates, there are certain limitations. First, using in vitro HK2 cell models is insufficient to properly replicate the intricate etiology of human renal fibrosis. Secondly, in the mechanism of GAA’s regulation of the NPC1L1 gene on renal fibrosis, we hypothesized that the NPC1L1 gene affects renal fibrosis by regulating lipid synthesis and affecting lipid metabolism pathways. However, we did not test for lipid peroxidation and inflammatory responses. In addition, the pathological mechanism of renal fibrosis is complex, primarily involving cell EMT, deposition of ECM, infiltration of inflammatory cells, and other factors. We examined the fundamental mechanisms of GAA in EMT and ECM deposition pathways, but did not explore the involvement of NPC1L1 in the development of other inflammatory pathways.
To summarize, we initially confirmed the anti-renal fibrosis impact of GAA using qPCR and Western blot analysis. We then investigated how GAA regulates the NPC1L1 gene’s influence on the epithelial-mesenchymal transition and extracellular matrix formation in renal tubular cells by producing an overexpressed NPC1L1. This study revealed how GAA affects the protective impact of TGF-β1-induced renal fibrosis, and established a theoretical foundation for using GAA to ameliorate renal fibrosis.
Acknowledgement: The participants who generously dedicated their time and effort to contribute to our study are greatly appreciated. We are also grateful to the laboratory personnel for their invaluable technical support and to the funding agency for providing the requisite resources to facilitate the completion of this research.
Funding Statement: This work was sponsored by Key Research and Development Project of Science and Technology Department of Tibet (No. XZ202201ZY0033G).
Author Contributions: The authors confirm contribution to the paper as follows: draft manuscript preparation: Tianyun Han; data collection: Zhong Li; analysis and interpretation of results: Luoning Zhang; study conception and design: Linshen Xie. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Group KDIGODW. 2022. KDIGO 2022 clinical practice guideline for diabetes management in chronic kidney disease. Kidney Int. 2022;102(5S):S1–127. [Google Scholar]
2. Charles C, Ferris AH. Chronic kidney disease. Prim Care. 2020;47(4):585–95. doi:10.1016/j.pop.2020.08.001. [Google Scholar] [PubMed] [CrossRef]
3. Ammirati AL. Chronic kidney disease. Rev Assoc Med Bras (1992). 2020;66(Suppl 1):s3–9. [Google Scholar]
4. Ruiz-Ortega M, Rayego-Mateos S, Lamas S, Ortiz A, Rodrigues-Diez RR. Targeting the progression of chronic kidney disease. Nat Rev Nephrol. 2020;16(5):269–88. doi:10.1038/s41581-019-0248-y. [Google Scholar] [PubMed] [CrossRef]
5. Bello AK, Okpechi IG, Levin A, Ye F, Damster S, Arruebo S, et al. An update on the global disparities in kidney disease burden and care across world countries and regions. Lancet Glob Health. 2024;12(3):e382–95. doi:10.1016/S2214-109X(23)00570-3. [Google Scholar] [PubMed] [CrossRef]
6. Rayego-Mateos S, Campillo S, Rodrigues-Diez RR, Tejera-Muñoz A, Marquez-Exposito L, Goldschmeding R, et al. Interplay between extracellular matrix components and cellular and molecular mechanisms in kidney fibrosis. Clin Sci. 2021;135(16):1999–2029. [Google Scholar]
7. Vanhove T, Goldschmeding R, Kuypers D. Kidney fibrosis: origins and interventions. Transplantation. 2017;101(4):713–26. [Google Scholar] [PubMed]
8. Geng XQ, Ma A, He JZ, Wang L, Jia YL, Shao GY, et al. Ganoderic acid hinders renal fibrosis via suppressing the TGF-β/Smad and MAPK signaling pathways. Acta Pharmacol Sin. 2020;41(5):670–7. [Google Scholar] [PubMed]
9. Wang X, Gaur M, Mounzih K, Rodriguez HJ, Qiu H, Chen M, et al. Inhibition of galectin-3 post-infarction impedes progressive fibrosis by regulating inflammatory profibrotic cascades. Cardiovasc Res. 2023;119(15):2536–49. [Google Scholar] [PubMed]
10. Yu HX, Lin W, Yang K, Wei LJ, Chen JL, Liu XY, et al. Transcriptome-based network analysis reveals hirudin potentiates anti-renal fibrosis efficacy in UUO rats. Front Pharmacol. 2021;12:741801. [Google Scholar] [PubMed]
11. Evans M, Lewis RD, Morgan AR, Whyte MB, Hanif W, Bain SC, et al. A narrative review of chronic kidney disease in clinical practice: current challenges and future perspectives. Adv Ther. 2022;39(1):33–43. doi:10.1007/s12325-021-01927-z. [Google Scholar] [PubMed] [CrossRef]
12. Webster AC, Nagler EV, Morton RL, Masson P. Chronic kidney disease. Lancet. 2017;389(10075):1238–52. doi:10.1016/S0140-6736(16)32064-5. [Google Scholar] [PubMed] [CrossRef]
13. Chen DQ, Feng YL, Cao G, Zhao YY. Natural products as a source for antifibrosis therapy. Trends Pharmacol Sci. 2018;39(11):937–52. doi:10.1016/j.tips.2018.09.002. [Google Scholar] [PubMed] [CrossRef]
14. Li T-H, Hu H-P, Deng W-Q, Wu S-H, Wang D-M, Tsering T. Ganoderma leucocontextum, a new member of the G. lucidum complex from southwestern China. Mycoscience. 2015;56(1):81–5. doi:10.1016/j.myc.2014.03.005. [Google Scholar] [CrossRef]
15. Gao X, Qi J, Ho C-T, Li B, Mu J, Zhang Y, et al. Structural characterization and immunomodulatory activity of a water-soluble polysaccharide from Ganoderma leucocontextum fruiting bodies. Carbohydr Polym. 2020;249(2):116874. doi:10.1016/j.carbpol.2020.116874. [Google Scholar] [PubMed] [CrossRef]
16. Gao X, Qi J, Ho C-T, Li B, Xie Y, Chen S, et al. Purification, physicochemical properties, and antioxidant activities of two low-molecular-weight polysaccharides from Ganoderma leucocontextum fruiting bodies. Antioxidants. 2021;10(7):1145. doi:10.3390/antiox10071145. [Google Scholar] [PubMed] [CrossRef]
17. Wang K, Bao L, Xiong W, Ma K, Han J, Wang W, et al. Lanostane triterpenes from the tibetan medicinal mushroom ganoderma leucocontextum and their inhibitory effects on HMG-CoA reductase and α-Glucosidase. J Nat Prod. 2015;78(8):1977–89. doi:10.1021/acs.jnatprod.5b00331. [Google Scholar] [PubMed] [CrossRef]
18. Zhao ZZ, Chen HP, Li ZH, Dong ZJ, Bai X, Zhou ZY, et al. Leucocontextins A-R, lanostane-type triterpenoids from Ganoderma leucocontextum. Fitoterapia. 2016;109:91–8. doi:10.1016/j.fitote.2015.12.004. [Google Scholar] [PubMed] [CrossRef]
19. Ma J-Q, Zhang Y-J, Tian Z-K. Anti-oxidant, anti-inflammatory and anti-fibrosis effects of ganoderic acid A on carbon tetrachloride induced nephrotoxicity by regulating the Trx/TrxR and JAK/ROCK pathway. Chem Biol Interact. 2021;344:109529. doi:10.1016/j.cbi.2021.109529. [Google Scholar] [PubMed] [CrossRef]
20. Qian R, Zhang LN, Xu YY, Sun DL, Wang LQ, Chen XX, et al. Network pharmacology reveals the effect and mechanism of Ganoderma leucocontextum ethanol extract on improving inflammatory response in silicosis lungs. Chin Occup Med. 2024;51(1):6–15 (In Chinese). [Google Scholar]
21. Peng H, Zhong L, Cheng L, Chen L, Tong R, Shi J, et al. Ganoderma lucidum: current advancements of characteristic components and experimental progress in anti-liver fibrosis. Front Pharmacol. 2022;13:1094405. [Google Scholar] [PubMed]
22. Altmann SW, Davis Jr HR, Zhu L-J, Yao X, Hoos LM, Tetzloff G, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303(5661):1201–4. [Google Scholar] [PubMed]
23. Huang C-S, Yu X, Fordstrom P, Choi K, Chung BC, Roh S-H, et al. Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition. Sci Adv. 2020;6(25):eabb1989. [Google Scholar] [PubMed]
24. Jia L, Betters JL, Yu L. Niemann-pick C1-like 1 (NPC1L1) protein in intestinal and hepatic cholesterol transport. Annu Rev Physiol. 2011;73:239–59. [Google Scholar] [PubMed]
25. van der Rijt S, Leemans JC, Florquin S, Houtkooper RH, Tammaro A. Immunometabolic rewiring of tubular epithelial cells in kidney disease. Nat Rev Nephrol. 2022;18(9):588–603. [Google Scholar] [PubMed]
26. Zhu Z, Hu J, Chen Z, Feng J, Yang X, Liang W, et al. Transition of acute kidney injury to chronic kidney disease: role of metabolic reprogramming. Metabolism. 2022;131:155194. doi:10.1016/j.metabol.2022.155194. [Google Scholar] [PubMed] [CrossRef]
27. Liao Q, Dong Y, Li B, Qin J, Cao Y, Tu W, et al. Promotion of liver fibrosis by Y-box binding protein 1 via the attenuation of transforming growth factor-beta 3 transcription. Ann Transl Med. 2023;11(6):259. doi:10.21037/atm-23-835. [Google Scholar] [PubMed] [CrossRef]
28. Eddy AA. Overview of the cellular and molecular basis of kidney fibrosis. Kidney Int Suppl. 2014;4(1):2–8. doi:10.1038/kisup.2014.2. [Google Scholar] [PubMed] [CrossRef]
29. Chalkia A, Gakiopoulou H, Theohari I, Foukas PG, Vassilopoulos D, Petras D. Transforming growth factor-β1/Smad signaling in glomerulonephritis and its association with progression to chronic kidney disease. Am J Nephrol. 2021;52(8):653–65. doi:10.1159/000517619. [Google Scholar] [PubMed] [CrossRef]
30. Gu Y-Y, Liu X-S, Huang X-R, Yu X-Q, Lan H-Y. Diverse role of TGF-β in kidney disease. Front Cell Dev Biol. 2020;8:123. doi:10.3389/fcell.2020.00123. [Google Scholar] [PubMed] [CrossRef]
31. Li Y, Luo C, Zeng Y, Zheng Z, Tao D, Liu Q, et al. Renal fibrosis is alleviated through targeted inhibition of IL-11-induced renal tubular epithelial-to-mesenchymal transition. Am J Pathol. 2023;193(12):1936–52. doi:10.1016/j.ajpath.2023.07.005. [Google Scholar] [PubMed] [CrossRef]
32. Li L, Fu H, Liu Y. The fibrogenic niche in kidney fibrosis: components and mechanisms. Nat Rev Nephrol. 2022;18(9):545–57. doi:10.1038/s41581-022-00590-z. [Google Scholar] [PubMed] [CrossRef]
33. Wei J, Wu L, Yang S, Zhang C, Feng L, Wang M, et al. E-cadherin to N-cadherin switching in the TGF-β1 mediated retinal pigment epithelial to mesenchymal transition. Exp Eye Res. 2022;220:109085. doi:10.1016/j.exer.2022.109085. [Google Scholar] [PubMed] [CrossRef]
34. Xie Y, Lan F, Zhao J, Shi W. Hirudin improves renal interstitial fibrosis by reducing renal tubule injury and inflammation in unilateral ureteral obstruction (UUO) mice. Int Immunopharmacol. 2020;81(3):106249. doi:10.1016/j.intimp.2020.106249. [Google Scholar] [PubMed] [CrossRef]
35. Gwon MG, An HJ, Kim JY, Kim WH, Gu H, Kim HJ, et al. Anti-fibrotic effects of synthetic TGF-β1 and Smad oligodeoxynucleotide on kidney fibrosis in vivo and in vitro through inhibition of both epithelial dedifferentiation and endothelial-mesenchymal transitions. FASEB J. 2020;34(1):333–49. doi:10.1096/fj.201901307RR. [Google Scholar] [PubMed] [CrossRef]
36. Uchikado Y, Natsugoe S, Okumura H, Setoyama T, Matsumoto M, Ishigami S, et al. Slug Expression in the E-cadherin preserved tumors is related to prognosis in patients with esophageal squamous cell carcinoma. Clin Cancer Res. 2005;11(3):1174–80. doi:10.1158/1078-0432.1174.11.3. [Google Scholar] [CrossRef]
37. Zhitnyak IY, Rubtsova SN, Litovka NI, Gloushankova NA. Early events in actin cytoskeleton dynamics and E-cadherin-mediated cell-cell adhesion during epithelial-mesenchymal transition. Cells. 2020;9(3):578. doi:10.3390/cells9030578. [Google Scholar] [PubMed] [CrossRef]
38. Teng S, Liu G, Li L, Ou J, Yu Y. CUX1 promotes epithelial-mesenchymal transition (EMT) in renal fibrosis of UUO model by targeting MMP7. Biochem Biophys Res Commun. 2022;608(1):128–34. doi:10.1016/j.bbrc.2022.03.097. [Google Scholar] [PubMed] [CrossRef]
39. Zhao L, Zhao J, Wang X, Chen Z, Peng K, Lu X, et al. Serum response factor induces endothelial-mesenchymal transition in glomerular endothelial cells to aggravate proteinuria in diabetic nephropathy. Physiol Genomics. 2016;48(10):711–8. doi:10.1152/physiolgenomics.00082.2016. [Google Scholar] [PubMed] [CrossRef]
40. Yu W, Song J, Chen S, Nie J, Zhou C, Huang J, et al. Myofibroblast-derived exosomes enhance macrophages to myofibroblasts transition and kidney fibrosis. Ren Fail. 2024;46(1):2334406. doi:10.1080/0886022X.2024.2334406. [Google Scholar] [PubMed] [CrossRef]
41. Wirsik NM, Ehlers J, Mäder L, Ilina EI, Blank AE, Grote A, et al. TGF-β activates pericytes via induction of the epithelial-to-mesenchymal transition protein SLUG in glioblastoma. Neuropathol Appl Neurobiol. 2021;47(6):768–80. doi:10.1111/nan.12714. [Google Scholar] [PubMed] [CrossRef]
42. Xie Q, Zhang M, Mao X, Xu M, Liu S, Shang D, et al. Matrix protein Tenascin-C promotes kidney fibrosis via STAT3 activation in response to tubular injury. Cell Death Dis. 2022;13(12):1044. doi:10.1038/s41419-022-05496-z. [Google Scholar] [PubMed] [CrossRef]
43. Li K, Zhang Y, Zhao W, Wang R, Li Y, Wei L, et al. DPP8/9 inhibition attenuates the TGF-β1-induced excessive deposition of extracellular matrix (ECM) in human mesangial cells via Smad and Akt signaling pathways. Toxicol Lett. 2024;395(12):1–10. doi:10.1016/j.toxlet.2024.03.001. [Google Scholar] [PubMed] [CrossRef]
44. Lok SW, Yiu WH, Li H, Xue R, Zou Y, Li B, et al. The PAR-1 antagonist vorapaxar ameliorates kidney injury and tubulointerstitial fibrosis. Clin Sci. 2020;134(21):2873–91. doi:10.1042/CS20200923. [Google Scholar] [PubMed] [CrossRef]
45. Xu C, Fu F, She Y, Xu C. NPC1L1 plays a novel role in nonalcoholic fatty liver disease. ACS Omega. 2023;8(51):48586–9. doi:10.1021/acsomega.3c07337. [Google Scholar] [PubMed] [CrossRef]
46. Hu M, Yang F, Huang Y, You X, Liu D, Sun S, et al. Structural insights into the mechanism of human NPC1L1-mediated cholesterol uptake. Sci Adv. 2021;7(29):eabg3188. doi:10.1126/sciadv.abg3188. [Google Scholar] [PubMed] [CrossRef]
47. Xiao J, Dong L-W, Liu S, Meng F-H, Xie C, Lu X-Y, et al. Bile acids-mediated intracellular cholesterol transport promotes intestinal cholesterol absorption and NPC1L1 recycling. Nat Commun. 2023;14(1):6469. doi:10.1038/s41467-023-42179-5. [Google Scholar] [PubMed] [CrossRef]
48. Eddy AA, Liu E, McCulloch L. Interstitial inflammation and fibrosis in rats with diet-induced hypercholesterolemia. Kidney Int. 1996;50(4):1139–49. doi:10.1038/ki.1996.421. [Google Scholar] [PubMed] [CrossRef]
49. Zhang Z, Zhang B, Jiang X, Yu Y, Cui Y, Luo H, et al. Hyocholic acid retards renal fibrosis by regulating lipid metabolism and inflammatory response in a sheep model. Int Immunopharmacol. 2023;122(10):110670. doi:10.1016/j.intimp.2023.110670. [Google Scholar] [PubMed] [CrossRef]
50. Lin P-H, Duann P. Dyslipidemia in kidney disorders: perspectives on mitochondria homeostasis and therapeutic opportunities. Front Physiol. 2020;11:1050. doi:10.3389/fphys.2020.01050. [Google Scholar] [PubMed] [CrossRef]
51. Miguel V, Tituaña J, Herrero JI, Herrero L, Serra D, Cuevas P, et al. Renal tubule Cpt1a overexpression protects from kidney fibrosis by restoring mitochondrial homeostasis. J Clin Investig. 2021;131(5):e140695. [Google Scholar] [PubMed]
52. Kang HM, Ahn SH, Choi P, Ko Y-A, Han SH, Chinga F, et al. Defective fatty acid oxidation in renal tubular epithelial cells has a key role in kidney fibrosis development. Nat Med. 2015;21(1):37–46. [Google Scholar] [PubMed]
53. Jiao B, Qu L. The interplay between immune and metabolic pathways in kidney disease. Cells. 2023;12(12):1584. [Google Scholar] [PubMed]
54. Castro BBA, Foresto-Neto O, Saraiva-Camara NO, Sanders-Pinheiro H. Renal lipotoxicity: insights from experimental models. Clin Exp Pharmacol Physiol. 2021;48(12):1579–88. [Google Scholar] [PubMed]
55. Aron-Wisnewsky J, Warmbrunn MV, Nieuwdorp M, Clément K. Metabolism and metabolic disorders and the microbiome: the intestinal microbiota associated with obesity, lipid metabolism, and metabolic health—pathophysiology and therapeutic strategies. Gastroenterology. 2021;160(2):573–99. [Google Scholar] [PubMed]
56. Nihei W, Nagafuku M, Hayamizu H, Odagiri Y, Tamura Y, Kikuchi Y, et al. NPC1L1-dependent intestinal cholesterol absorption requires ganglioside GM3 in membrane microdomains. J Lipid Res. 2018;59(11):2181–7. doi:10.1194/jlr.M089201. [Google Scholar] [PubMed] [CrossRef]
57. Jia L, Ma Y, Haywood J, Jiang L, Xue B, Shi H, et al. NPC1L1 deficiency suppresses ileal fibroblast growth factor 15 expression and increases bile acid pool size in high-fat-diet-fed mice. Cells. 2021;10(12):3468. doi:10.3390/cells10123468. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools