 Open Access
Open Access
REVIEW
Revolutionizing stem cell research: unbiased insights through single-cell sequencing
Institute of Stomatology, First Medical Center, Chinese PLA General Hospital, Beijing, 100853, China
* Corresponding Authors: YI JIANG. Email: ; QUAN SHI. Email:
# These authors contributed equally to this work
BIOCELL 2024, 48(11), 1531-1542. https://doi.org/10.32604/biocell.2024.054278
Received 23 May 2024; Accepted 12 July 2024; Issue published 07 November 2024
Abstract
Stem cells have shown great application potential in wound repair, tissue regeneration, and disease treatment. Therefore, a full understanding of stem cells and their related regulatory mechanisms in disease treatment is conducive to improving the therapeutic effect of stem cells. However, thus far, there are still many unsolved mysteries in the field of stem cells due to technical limitations, which hinder the in-depth exploration of stem cells and their wide clinical application. Single-cell sequencing (SCS) has provided very powerful and unbiased insights into cell gene expression profiles at the single-cell level, bringing exciting results to the stem cell field. At present, SCS has been widely applied in the field of stem cells, covering various aspects, including lineage tracing the development of stem cells, identifying new stem cell types, exploring cellular heterogeneity, and identifying internal functional subpopulations. In this paper, we focus on the latest research progress and discuss the application of SCS technology in stem cells.Keywords
List of Abbreviations
| SCS | Single-cell sequencing |
| ESCs | Embryonic stem cells |
| ASCs | Adult stem cells |
| MSCs | Mesenchymal stem/stromal cells |
| DE | Definitive endoderm |
| HSCs | Hematopoietic stem cells |
| HECs | Hemogenic endothelial cells |
| scRNA-seq | Single-cell RNA sequencing |
| AGM | Aorta–gonad–mesonephros |
| CSs | Carnegie stages |
| BM | Bone marrow |
| SSCs | Skeleton stem cells |
| eSSPCs | Embryonic skeletal stem/progenitor cells |
| ISCT | The International Society of Cell Therapy |
| ADSCs | Adipose mesenchymal stem cells |
| BMSCs | Bone marrow stem cells |
| DMSCs | Dermis derived MSCs |
| hucMSCs | Human umbilical cord derived MSCs |
| ECM | Extracellular matrix |
| WJ-MSCs | Wharton’s jelly mesenchymal stem cells |
| hDPSCs | Human dental pulp stem cells |
| hPDLSCs | Human periodontal ligament stem cells |
| TFs | Transcription factors |
| esMSCs | ESC-derived mesenchymal stem cells |
| SASP | Senescence-associated secretory phenotype |
| VSCs | Vascular stem cells |
| SMCs | Smooth muscle cells |
| VSPCs | Vascular stem/progenitor cells |
| EMCN | Endomucin |
| PROCR | Protein C receptor |
| RUNX1T1 | RUNX1 partner transcriptional co-repressor 1 |
| PDPN | Podoplanin |
| PI3K | Phosphoinositide3-kinase |
| Akt | RAC-alpha serine/threonine-protein kinase |
| GSK3β | Glycogen synthase kinase-3β |
| Spi1 | Spleen focus forming virus proviral integration oncogene |
| Gata1 | GATA binding protein 1 |
| Egr3 | Early growth response 3 |
| Bmi1 | B-cell-specific Moloney murine leukemia virus insertion region 1 |
| Gfi1b | Growth factor independence 1b |
| PRUNE2 | Prune homolog 2 |
| DIO2 | Iodothyronine deiodinase 2 |
| CPA4 | Carboxypeptidase A4 |
| PRKAA2 | Protein kinase AMP-activated catalytic subunit alpha 2 |
| DMD | Dystrophin |
| DDAH1 | Dimethylarginine dimethylaminohydrolase 1 |
| YAP | Yes-associated protein |
| VEGF | Vascular endothelial growth factor |
| LRRC75A | Leucine-rich repeat-containing 75A |
| PDGF | Platelet derived growth factor |
| CXCL12 | C-X-C motif chemokine ligand 12 |
| GAS6 | Growth arrest-specific protein 6 |
| PRDM1 | PR domain zinc finger protein 1 |
Cells are the basic units of the structure and function of organisms. Each cell has a unique phenotype and function, including morphology, genome, epigenome, and other aspects. Therefore, research at the level of single cells will bring new perspectives and different results. In general, traditional high-throughput sequencing can only be carried out at the bulk-population level, which means the average of the gene expression of cell populations or only represents the life activity information of the dominant cells in number [1,2]. However, it cannot accurately reflect information about the heterogeneity of cells in the sample and may lead to ignoring the differences in gene expression regulation between cells [3].
Over the past decade, genomic analyses of single cells have become possible based on the development of sequencing technology, cell isolation, and whole genome amplification technology. Single-cell sequencing (SCS) has emerged as a revolutionary and powerful new set of technology that sequences the genome at the single-cell level [4–6]. In brief, SCS is promising biotechnology to sequence the genome, transcriptome, and epigenome at the single-cell level with high throughput and use bioinformatics analysis to mine the biological significance of genes at the single-cell level to reveal the gene structure and gene expression status of a single cell. The results of SCS can reflect the heterogeneity between cells and trace the lineage path of cells or the evolutionary relationships of the cells [7,8]. At present, the research achievements of SCS technology involve various research fields and various cell types, such as the tumor microenvironment [9,10], immunotherapy [11,12], embryonic development [13], disease occurrence and development mechanisms [14,15], and stem cell biology [16,17], which not only bring new and gratifying results to related fields but also bring novel inspiration, insights and solutions to solve problems in biomedicine.
Stem cells are multipotent cells with self-renewal ability, which can be divided into embryonic stem cells (ESCs) and adult stem cells (ASCs) [18]. As undifferentiated and immature cells, stem cells have the potential to repair and even regenerate various tissues and organs. Therefore, since the discovery of stem cells, their related research has been widely studied, and stem cells are considered to have broad application prospects in regenerative medicine [19,20], developmental biology [21], pharmacology [22], and other fields [19,23]. However, it is undeniable that thus far, the related research on stem cells still has limitations, and it has not reached expectations in some fields, which hinders researchers’ further research on stem cells and the wide clinical application that can be achieved [18,24,25]. For example, the mechanism and regulation of the origin, development, and differentiation of stem cells are still not fully understood; the lack of in-depth understanding of the functional heterogeneity of stem cells has seriously hampered the effective and reproducible clinical applications of stem cells. Therefore, it is urgent to find relevant solutions to improve the understanding of stem cells and help realize the clinical application of stem cells.
SCS technology has been successfully and widely used in related research in stem cells, which has laid a good technical foundation and provided the possibility to solve the above problems (Fig. 1). Using SCS technology has successfully helped us in-depth study the origin and development of stem cells [26,27], confirm the heterogeneity of stem cells under different states [16,28], discover the functional subpopulations of stem cells and find key regulatory genes [29,30]. The relevant research results provide a basis for further promoting the application of stem cells and enable researchers to recognize the microscopic appearance of stem cells from a new perspective [17,31,32]. More importantly, researchers have created online resources using single-cell sequencing data, which can help scientists identify genetic networks and functions in many kinds of individual stem cells. In this paper, we discuss the application of SCS technology in stem cells and highlight challenges and limitations in the application of SCS in stem cell-related studies. In contrast to other articles that focus on the technology itself, we place more emphasis on new conclusions and insights into the field of stem cells based on SCS.
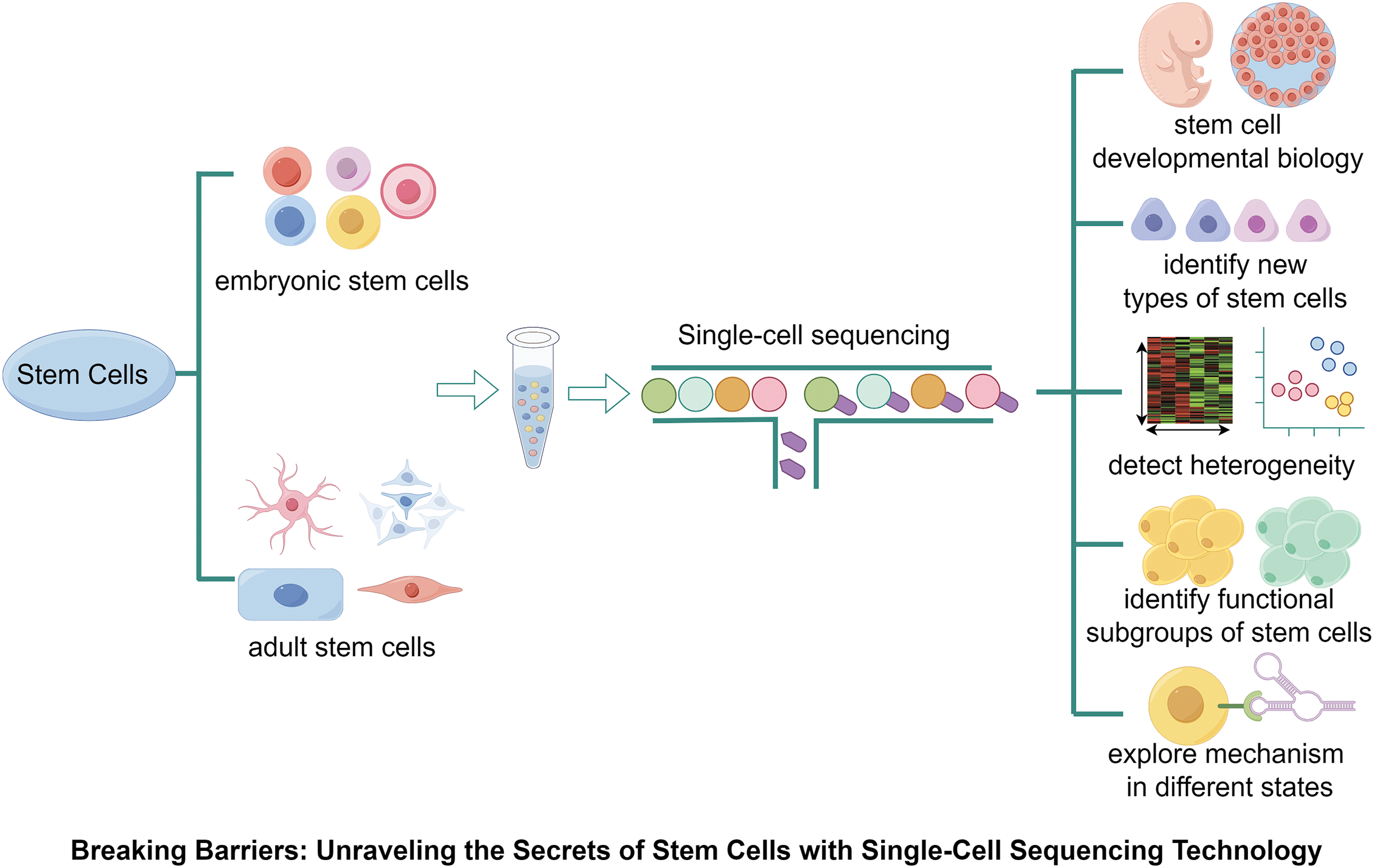
Figure 1: Application of single-cell sequencing technology in stem cell biology. SCS has been applied in a variety of stem cells, which has successfully helped us in-depth study the origin and development of stem cells, confirm the heterogeneity of stem cells under different states, discover the functional subpopulations/heterogeneity of stem cells, and find key regulatory genes.
Biological Applications of the SCS in Stem Cells
Application of SCS in stem cell developmental biology
At present, multiple types of stem cells have been discovered and studied, but due to technological limitations, scarcity of tissue samples, or insufficient sample cell size, the development and origin of some types of stem cells have not been fully studied, which remains an unsolved mystery in this field [33]. The emergence of lineage-tracing approaches in SCS technology has compensated for technological limitations and further improved research on the development and origin of stem cells [34].
In general, it is believed that mesenchymal stem/stromal cells (MSCs) come from a mesodermal origin, and the same applies to MSCs isolated from endodermal organs [35]. However, this concept has been challenged. Based on the SCS and related experiments, it has been proven that human pluripotent stem cell-derived endoderm progenitors could differentiate into MSCs, which emerged from definitive endoderm (DE) during in vitro culture [36]. DE-MSCs express classic MSC markers, including CD44, CD73, CD105, and CD271, have inflammatory regulatory effects in an ulcerative colitis mouse model [36]. Therefore, with the help of the SCS, these results indicated that definitive endodermal cells can be a source of MSCs for disease treatment.
Hematopoietic stem cells (HSCs) have the self-renew ability and undergo multilineage differentiation in the human body, playing a critical role in maintaining the function of the blood system and in the treatment of hematology and immune system diseases [37,38]. However, due to technical limitations and material rarity, the current understanding of the human early embryonic hematopoietic system and HSC development is still insufficient, which hinders the formation of truly functional HSCs induced in vitro for scientific research or disease treatment and has become the focus and difficulty in the field of hematopoietic development research [39–41]. To precisely understand the cellular and molecular programs and interactions that underlie the generation of the first HSCs from hemogenic endothelial cells (HECs), Zeng et al. based on a 10× genomics platform, performed single-cell RNA sequencing (scRNA-seq) on cells in the aorto-gonado-mesonephric (AGM) region of different Carnegie stages (CS). The first genome-level gene expression landscape covering the transition of endothelial cells to HSCs during human embryogenesis was constructed [42]. They found that CD44 could be used as a cell surface marker to enrich HECs from AGM ECs, and the group of CD44+ cells present in the embryos with the time window of HSC production showed clear arterial characteristics. The researchers accurately analyzed the multiple stages of HEC-targeted HSC fate transformation and revealed a set of genes specifically upregulated in the HEC stage, including endomucin (EMCN), protein C receptor (PROCR), and RUNX1 partner transcriptional co-repressor 1 (RUNX1T1), which tend towards transient upregulated expression upon the moment hemogenic fate choice of arterial ECs. The related results reveal that the process of arterialization is a necessary pathway for HECs to obtain functional HSCs and arterial-featured HECs as the source of the first batch of HSCs in the AGM region.
Zeng et al.’s study focused on the early development of HSCs, while other studies used a single-cell transcriptomics technique to track the development and migration of HSCs from early pregnancy to birth and to clarify the relationship between HSCs and HECs [43]. The molecular characteristics of HSCs in the AGM region of CS14-CS15 embryos were RUNX1+HOXA9+MLLT3+MECOM+HLF+SPINK2+. With the development of embryos, HSCs are enriched from the AGM, placenta, yolk sac, umbilical cord, and yolk sac vascular region to the fetal liver at CS17 [43]. This means that HSCs grow in multiple tissues outside the embryo before entering the circulatory system or colonizing the liver. In addition, by comparing AGM (CS13-17) SCS data with embryo and yolk sac (CS10-11) data, it was found that HECs that could transform into HSCs were derived from metabolically quiescent IL33+ALDH1A1+ arterial endothelium. In addition, a single-cell transcriptomic landscape of human fetal bone marrow (BM) and spleen was constructed, which indicated that the first functional HSCs are present in human fetal BM at 12 weeks post-conception but not in the spleen [44]. These data fully establish a detailed molecular map of human HSC development and help us track HSCs as they emerge from hematopoietic endothelial tissue and migrate through different sites during development. Undeniably, SCS will not only help to understand the ins and outs of HSCs but also lay a foundation for further understanding the molecular mechanism of related blood diseases. Moreover, SCS makes it possible to successfully generate fully functional HSCs in vitro [34].
SCS helps identify new types of stem cells
Stem cells have been found in many tissues or organs and have been extracted for study both in vivo and in vitro. With the deepening of research and the development of experimental technology, new types of stem cells are still being discovered, identified, and isolated, especially with the application of single-cell sequencing technology, which provides us with a powerful tool. Stem cells in the thymus [45], bone [46], and other tissues have been found based on SCS technology [45–47] (Table 1).
It has long been thought that the thymus does not contain “true” epithelial stem cells but only progenitor cells produced during fetal development. However, a recent study detected the presence of true epithelial stem cells with atypical signatures in the human thymus for the first time [45]. Thymic epithelial stem cells exhibit unique transcription spectrum and phenotype characteristics, including pleiotropic multilineage efficacy, which can generate a variety of cell types not previously considered to have a common origin, such as epithelial cells, myoid cells, and neuroendocrine cells. In addition, they found niches of stem cells in two locations in the thymus: the subcapsular and perivascular spaces around the medulla [45]. In the analysis of salivary gland single cells in embryonic tissue, a new type of salivary gland striated duct-derived stem cell with the ability to regenerate acinar groups was discovered, which obtained the expression of many genes associated with stem cell-like identity, including proliferation, self-renewal and DNA repair [47].
In addition to glandular tissues, stem cells in the skeleton have also been discovered based on SCS. In 2015, scientists found evidence of the existence of mouse skeleton stem cells (mSSCs) through lineage tracking and cloning analysis [48,49]. Recently, with the help of scRNA-seq, human skeletal stem cells (hSSCs) have been identified and analyzed for the first time [46]. The signature proteins of hSSCs are Podoplanin (PDPN), CD146, CD73 and CD164. Purified PDPN+ CD146− CD73+ CD164+ hSSCs have the ability to self-renew in vitro, and multilineage ossicles can be differentiated after transplantation under the mouse renal capsule [46]. As a result, hSSCs lineally transition into early bone, cartilage, and stroma progenitors, which then give rise to osteoprogenitors and chondroprogenitors prior to eventually forming bone, cartilage, and stroma. Notably, hSSCs do not differentiate into adipocytes. Further quantitative analysis showed that there were differences in gene expression during the differentiation of fetal and adult hSSCs, which could partly explain the different proportions of cartilage and bone in ossicles formed from different hSSCs [46]. Another study mapped a comprehensive human embryonic skeletogenesis cell and identified a group of embryonic skeletal stem/progenitor cells (eSSPCs) located in the embryonic perichondrium by scRNA-seq transcriptomic and functional analyses [50]. The eSPPC-associated markers PDGFRAlow/− PDPN+ CADM1+ were also identified [50]. These studies helped by SCS are of great significance for further understanding human bone development and damage repair mechanisms, as well as developing novel stem cell-based therapies to promote bone regeneration.
SCS can effectively detect cellular heterogeneity in inter/intra stem cells
Stem cells have cellular heterogeneity, which is reflected not only between stem cells from different tissues or periods but also between single cells within the same kind of stem cell, including the cell cycle, cell state (apoptosis, senescence, etc.), and gene expression [29,52–54]. Deep exploration of the heterogeneity of stem cells is important for achieving stem cell-based disease treatment and tissue regeneration. However, there is still a lack of a full understanding of the heterogeneity of stem cells at present. For example, multiple MSC-based clinical trials have been registered aimed at developing MSCs-based disease treatment therapy, but only limited clinical conversion applications have been achieved, which may partly be attributed to cellular heterogeneity [55,56]. Fortunately, the research in this field has attracted attention and has been extensively studied using SCS technology.
The minimal criteria of MSCs had been published by the International Society of Cell Therapy (ISCT) in 2006, despite it is widely used and accepted, it cannot reflect the heterogeneity of MSCs from different tissues [57]. Although cell surface marker expression and multidirectional differentiation potential indicate that MSCs are both homogenous populations, using scRNA-seq, we can clearly identify extreme variability in gene expression levels between individual cells within a certain MSC type. Researchers have even explored novel potential markers for MSC purification to demonstrate cellular heterogeneity and related biological roles [16]. A study with more than 130,000 single-MSC transcriptomes from 4 kinds of human tissue-derived stem cells, including adipose mesenchymal stem cells (ADSCs), bone marrow stem cells (BMSCs), dermis-derived MSCs (DMSCs) and umbilical cord-derived MSCs (hucMSCs) was constructed and analyzed at the single-cell level, confirmed that MSCs from different tissues have prominent transcriptomic heterogeneity [58]. Genes related to lineage differentiation, immune regulation, and senescence are heterogeneously expressed in different tissue-derived MSCs. In addition, these results indicated that the extracellular matrix (ECM) highly contributes to heterogeneity between different MSCs. MSCs from the dermis or umbilical cord have stronger antiaging properties, and a specific subpopulation of hucMSCs has advantages in immunomodulation properties, which is similar to other studies comparing gene expression models between human Wharton’s jelly MSCs (WJ-MSCs) and BMSCs using scRNA-seq [59]. Human dental pulp stem cells (hDPSCs) and human periodontal ligament stem cells (hPDLSCs) are both derived from teeth isolated from dental pulp and the periodontal ligament, respectively [60]. However, analysis at the single-cell level revealed that hDPSCs mainly exhibited osteogenic and neurogenic populations, while hPDLSCs, mainly comprised osteogenic and myofibroblastic populations [61]. These results can be used as valuable resources and compelling evidence for the development of the “most suitable/effective stem cell”-based therapy.
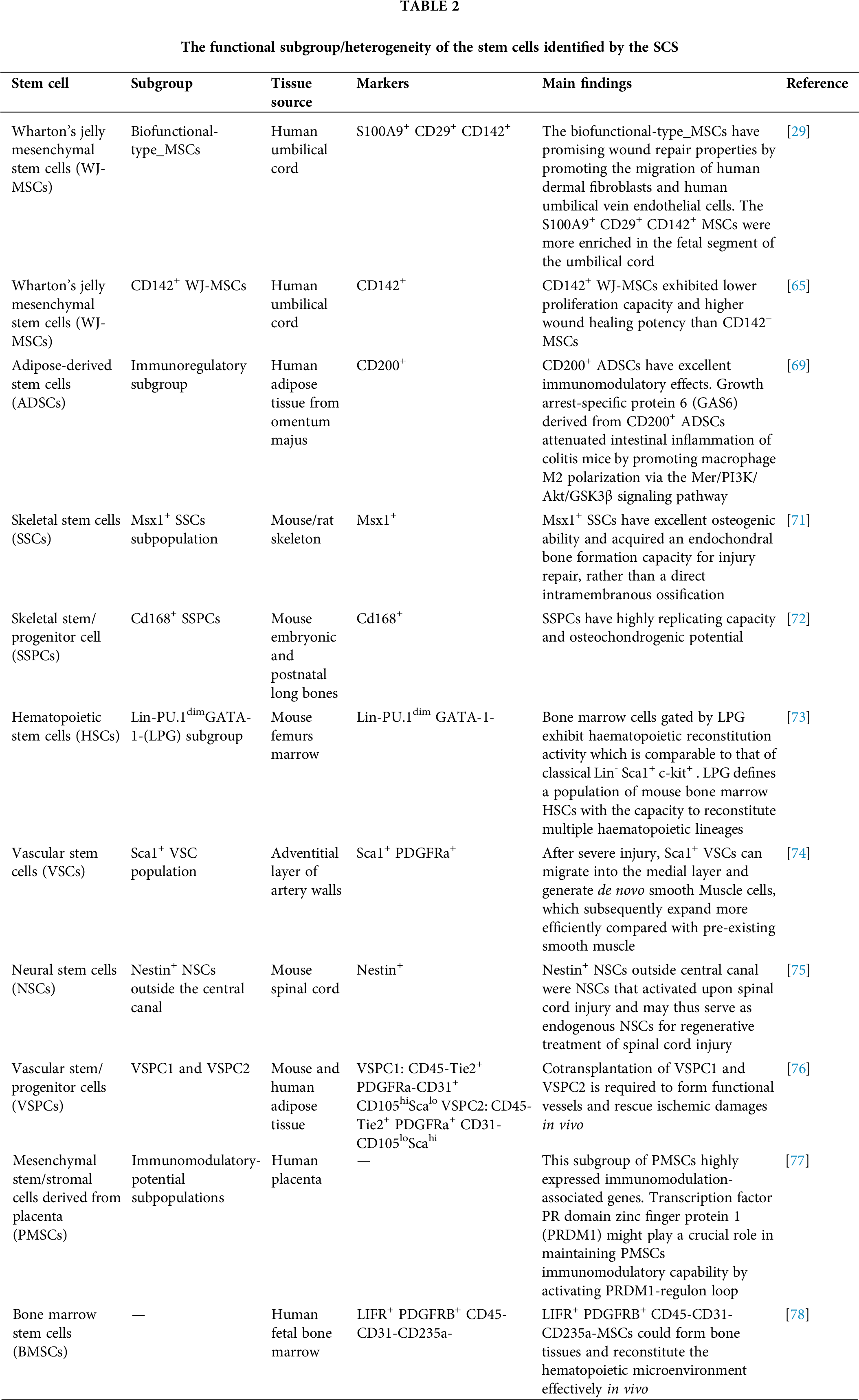
In addition to exploring the heterogeneity between stem cells of different tissue origins at a deeper level, SCS has also been widely applied to analyze the heterogeneity of a kind of stem cells, which depends on the ability of SCS technology to examine gene expression profiles at the level of individual cells in an unbiased manner (Table 2). To date, multiple stem cells, such as ADSCs [62], hucMSCs [63], cartilage stem cells [28], and BMSCs [16], have been detected to be heterogeneous by SCS. This heterogeneity is reflected in multiple aspects; for example, the proliferation and aging status of their internal cells are not completely uniform; when exerting biological functions and disease treatment effects, not all cells but a certain subgroup of stem cells play a crucial role. To avoid the impact of in vitro culture on cell characteristics, Zhang et al. investigated freshly isolated, uncultured hucMSCs by scRNA-seq [64]. Although the isolated MSCs meet the stem cell surface labeling criteria defined by ISCT, the scRNA-seq data reveal that the cells can be separated into different groups based on differentially expressed genes. One group was enriched in immune regulation, muscle cell proliferation and differentiation, stemness, and oxidative stress, while the main functions of the other group were extracellular matrix production, osteoblast and chondrocyte differentiation, and bone and cartilage growth. Another study also found that there are several distinct subpopulations of hucMSCs that exhibit diverse functional characteristics related to proliferation, development, and inflammatory responses [65]. In addition, by sorting the cells with the expression of CD142, it was shown that despite CD142− hucMSCs only accounting for approximately 20% of the total cells, this subgroup presented higher proliferation capacity and “wound healing” potential. These results of single-cell sequencing compensate for the current one-sided definition of stem cells by ISCT at the molecular and functional level; on the other hand, they also help us to identify heterogeneity more completely.
SCS helps identify functional subgroups of stem cells
As not fully differentiated cells, stem cells have multidifferentiation abilities and can play a critical role in tissue repair or disease treatment. However, we have also mentioned above that a large amount of evidence indicates that stem cells, especially MSCs, have intrinsic heterogeneity in phenotype and function [66], which divides stem cells into subgroups with different functions. This is also one of the biological bases for MSCs to have multidirectional differentiation ability, immune regulation ability, and other biological characteristics. These functional subsets contribute to improving the therapeutic efficiency and stability of stem cells [67]. Using single-cell sequencing to identify the functional subsets of stem cells and their surface molecular markers, gene expression characteristics, and other information will help overcome the limitations of heterogeneity and promote stem cell-based disease therapy to a new level [68] (Table 2).
After scRNA-seq analysis and gene ontology analysis of ADSCs, five subgroups of ADSCs were identified, including multipotential differentiation subgroup, self-regulatory subgroup, self-renewal subgroup, immunoregulatory subgroup, and metabolism- and hematopoiesis-associated subgroup [69]. The “immunoregulatory subgroup” was marked by the expression of CD200 (CD200+ ADSCs) mainly enriched in genes related to immune regulation, such as negative regulation of the T-cell receptor signaling pathway and tumor necrosis factor-mediated signaling pathway [69]. More importantly, in vitro and in vivo studies revealed that CD200+ ADSCs attenuated intestinal inflammation in colitis mice by promoting macrophage M2 polarization via the Mer/phosphoinositide3-kinase (PI3K)/RAC-alpha serine/threonine-protein kinase (Akt)/glycogen synthase kinase-3β (GSK3β) signaling pathway, which offers a new treatment for inflammatory bowel diseases [69]. Similarly, the heterogeneity of WJ-MSCs also hinders their clinical translation [70]. Recently, a study mapped the first comprehensive large-scale spatial and single-cell transcriptomic atlas to decipher the heterogeneity of WJ-MSCs, and a superior functional subgroup screening scheme for stem cells with high wound healing ability was established [29]. Single-cell transcriptomics analysis identifies 4 WJ-MSCs subpopulations, including the proliferative subgroup, niche-supporting subgroup, metabolism-related subgroup, and biofunctional-type subgroup. Importantly, the biofunctional-type subgroup, with S100A9, CD29, and CD142 as marker genes, has been proven to have superior wound repair ability after functional verification in vivo and in vitro. Finally, it was found that the S100A9+CD29+CD142+ subgroup was enriched in the fetal end of the human umbilical cord, which indicates that biological functional stem cells from the fetal end may be an ideal MSCs source for wound treatment. This study defined the functional subgroups of WJ-MSCs and provided an outstanding resource and solution for further development of MSCs-based cell therapy. Moreover, the functional subgroup of SSCs and markers have also been studied, such as the CD168+ skeletal stem/progenitor cell population and Msx1+ skeletal stem cells, both of which have excellent osteogenic or chondrogenic ability [71,72].
SCS can describe the relevant functions of stem cell subpopulations by identifying cell markers and related molecules, as well as by detecting and identifying relevant transcription factors (TFs). The scRNA-seq analyze results from HSCs with Lin−Sca1+c-kit+ population have two HSC clusters, which can be distinguished by spleen focus forming virus proviral integration oncogene (Spi1) and GATA binding protein 1 (Gata1) expression level [73]. One cluster highly expresses the myeloid and lymphoid lineage-specific TF genes early growth response 3 (Egr3) and Gata3, indicating the multipotency of this cluster; while the other one is characterized by upregulated expression of the erythroid lineage-specific TF genes, such as B-cell-specific Moloney murine leukemia virus insertion region 1 (Bmi1), growth factor independence 1b (Gfi1b) and Gata1, which indicates the erythroid-biased differentiation potential [73].
In short, these studies based on the SCS exploring the subgroup of stem cells define the functional subpopulations inside stem cells from a new perspective. At same time, it also reflects the consistency of gene expression and function for the stem cells and their subgroup. These results also suggest that in future research and clinical practice, it may not be the choice of which stem cells are more suitable for the treatment of a certain disease but the choice of which functional subgroup of stem cells is more appropriate (Fig. 2).
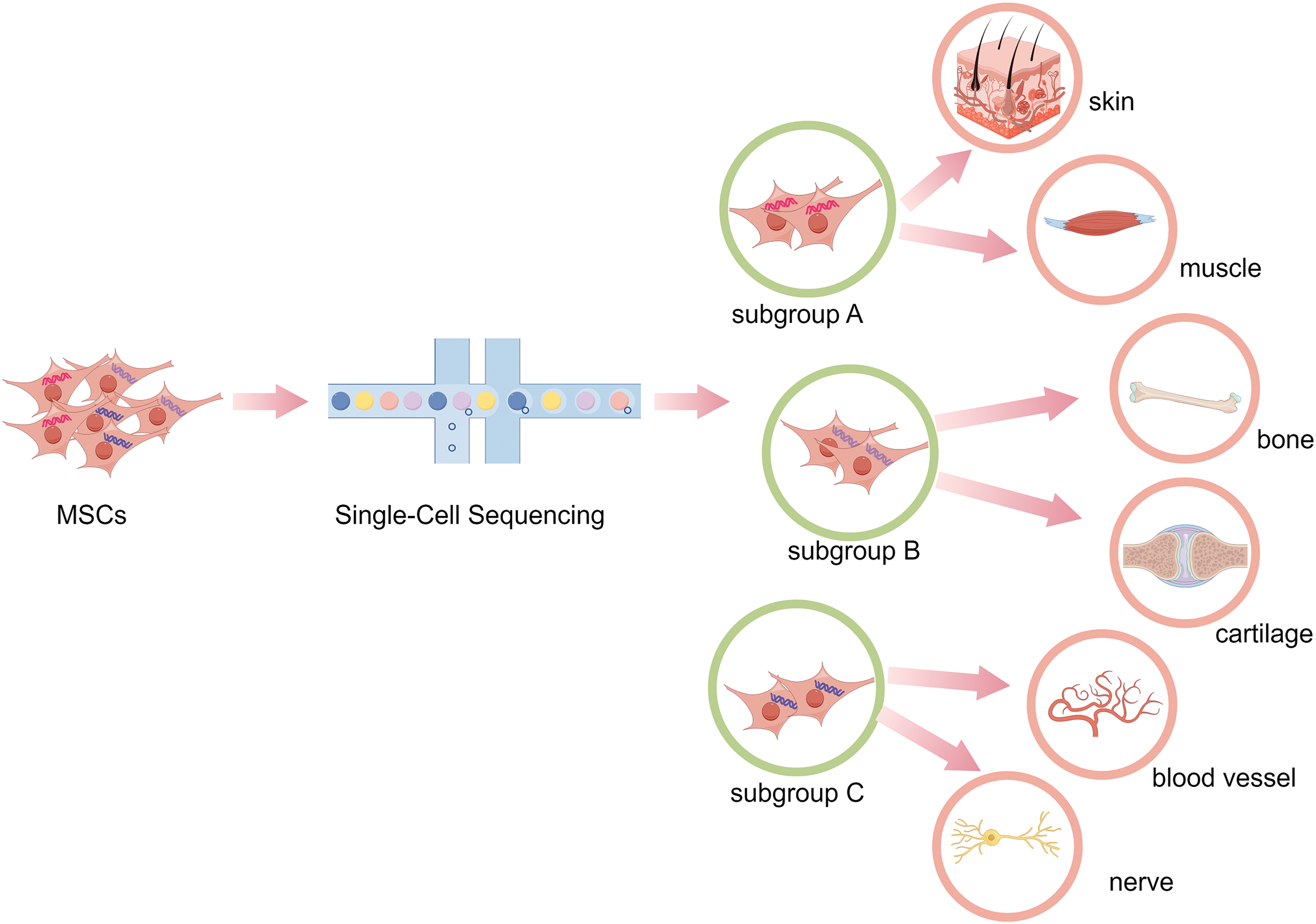
Figure 2: Using single-cell sequencing to identify the heterogeneity and functional subgroups of MSCs. Through single-cell sequencing, the functional subgroups of MSCs are analyzed, so as to select suitable functional MSCs subgroup to treat a certain disease or tissue regeneration to exert the desired effect.
SCS explores the function and mechanism of stem cells in different states or microenvironments
The biological function of stem cells can change with multigeneration culture, microenvironment changes and disease state, but the specific mechanism and significance are still unclear, which hinders the regulation of the cell state and plays a role in disease treatment [79–82]. Routine sequencing is performed at the bulk-population level and may obscure critical and tiny information. With the help of SCS, we can more comprehensively analyze the molecular mechanisms by which stem cells play a role in physiological and pathological processes and the expression of related genes. More importantly, it will provide help for the future to achieve precise regulation of stem cells to play an ideal role. At present, the research on the function of stem cells based on SCS has covered many aspects, such as aging [83], angiogenesis [84], osteogenesis [85], and drug sensitivity [86].
In general, to achieve the needed cell numbers for clinical application, stem cells typically need to be expanded in vitro for a long time, which will cause the stem cells to enter a senescent phenotype [87,88]. A study based on the SCS first analyzed the process of replicative senescence of healthy proliferative ESC-derived mesenchymal stem cells (esMSCs) in vitro [89]. In the process of expansion, esMSCs successively transform into cells characterized by the expression of metabolic and oxidative stress genes, enter a presenescent state characterized by the expression of endoplasmic reticulum stress and p53 regulation of age-related genes, and finally transform into cells undergoing aging [89]. Subsequently, this population splits into either deep senescent cells with strong senescence-associated secretory phenotype (SASP) secretions or into cells with SASP and oncogene signatures. The results of this single-cell sequencing analysis revealed the different aging subsets in the process of stem cell replication senescence and provided a time-sequential cell subpopulation transition process. Another study established a “TF-miRNA‒target” regulatory network to describe the replicative senescence of MSCs cultured in vitro [83]. Seven gene signatures, including Prune homolog 2 (PRUNE2), iodothyronine deiodinase 2 (DIO2), carboxypeptidase A4 (CPA4), protein kinase AMP-activated catalytic subunit alpha 2 (PRKAA2), dystrophin (DMD), dimethylarginine dimethylaminohydrolase 1 (DDAH1) and GATA6 have been considered possible candidate markers for estimating MSCs in vitro senescence. In addition, pathways related to oxygen and ECM might play pivotal roles in regulating the aging and anti-aging characteristics of MSCs. These results of single-cell sequencing analysis revealed the different aging subsets in the process of stem cell replication senescence, provided a time-sequential cell subpopulation transition process, and described relevant markers and regulatory networks, providing new insights as well as evidence to further identify and intervene in the process of replication senescence of stem cells. However, there are still many unsolved mysteries about stem cell senescence, such as senescent drift of the stem cells [90,91]. In future research, it is expected to obtain answers to these problems with the help of SCS technology, by which to guide the accurate and effective application of stem cells.
Angiogenesis and vessel repair are accompanied by a variety of physiological and pathological processes, especially during tissue/organ repair and regeneration [92–94]. Moreover, peripheral vascular disease and ischemia remain clinical problems worldwide [95]. Although many studies have been devoted to the role of stem cells in this process, the mechanisms involved are not fully understood. Rapid regeneration and repair of smooth muscle after vascular injury is crucial for blood vessels. By means of SCS, it was found that resident vascular stem cells (VSCs) characterized by Sca1+PDGFRa+ in the outer cortex of arteries, which can generate new smooth muscle cells (SMCs) after the injury response, and Yes-associated protein (YAP) is needed during the vascular repair process [74]. Using single-cell transcriptome analysis, a cell subpopulation associated with vascular endothelial growth factor (VEGF) secretion in human BMSCs under ischemia was identified, which also highlighted the critical role of leucine-rich repeat-containing 75A (LRRC75A) in inducing VEGF secretion under ischemia [96]. In another study, two vascular stem/progenitor cells (VSPCs) were identified from ADSCs, including VSPC1 which forms stunted vessels, and VSPC2, which could form both vessels and fat [76]. Only cotransplantation of VSPC1 and VSPC2 can effectively form functional vessels and improve perfusion in a mouse hindlimb ischemia model [76]. scRNA-seq was used to characterize distinct vessel-forming populations and their interactions, and a complex signaling regulation of VSPC1 to VSPC2 populations, including Platelet derived growth factor (PDGF), C-X-C motif chemokine ligand 12 (CXCL12) and other signals were revealed. Moreover, PDGF signals may act as unidirectional stromal cues from the VSPC1 subfraction to guide VSPC2 form vessels. These results acquired through the SCS are encouraging and promote the field of stem cell therapy.
As an emerging technology, SCS can achieve single-cell level research on stem cell types, cell heterogeneity and the role of different biological processes and has achieved meaningful results. Nevertheless, we think that there are still some limitations in this field: 1. Currently, the number of applications of SCS technology in the field of stem cells is still limited, so the accuracy and effectiveness of current results still require time and research to verify. 2. The state of stem cells is influenced by multiple factors [97], including tissue source and culture environment. In different studies, these factors will inevitably affect the accuracy and scalability of SCS results, especially for stem cells cultured in vitro. 3. Although multiple studies have been conducted on stem cells using SCS, it is well known that stem cells function through the combined action of the microenvironment, surrounding cells, and other factors [82,98]. However, research on this aspect is relatively limited and weak at present, which hinders our in-depth understanding of the function and mechanism of stem cells, which needs to be explored further.
Through the literature review mentioned above, we can see that SCS technology has been widely used in the research of stem cells, covering various aspects, such as stem cell genesis, development, identification, and application. By summarizing the current relevant literature, it has been shown that SCS technology can effectively track the development of stem cells and draw developmental maps, identify new stem cell types, explore internal heterogeneity of stem cells at individual cell level, identify the internal functional subpopulations of stem cells, enhance their biological function and disease treatment efficiency, and analyze the biological roles and molecular mechanisms of stem cells in different states or physiological processes. Compared to conventional population-level sequencing techniques such as transcriptomics, RNA-seq, and microarrays, SCS has elevated our understanding and application of stem cells to a new level, providing a strong theoretical basis for stem cell-based disease treatment and regenerative medicine. In this article, we briefly summarize the current application of single-cell sequencing technology in the field of stem cells. Considering the relevant progress and limitations of SCS technology in stem cells, for next step, it is necessary to further combine the current SCS research on stem cells with multiomics analysis, such as spatial transcriptomics and DNA methylation omics, to obtain more comprehensive and accurate results. In addition, more attention should be given to the changes and mechanisms of stem cells in various microenvironments while coordinating with simulated in vivo experiments, animal model experiments, and other validation results to promote the transformation and application of relevant results in the clinic.
Acknowledgement: None.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Quan Shi and Yi Jiang; draft manuscript preparation: Hao Wu and Na Huo; review and editing: Hao Wu, Na Huo, Situo Wang and Ziwei Liu; visualization: Situo Wang and Ziwei Liu; supervision: Quan Shi and Yi Jiang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y. Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med. 2022;12(3):e694. [Google Scholar] [PubMed]
2. Li X, Wang CY. From bulk, single-cell to spatial RNA sequencing. Int J Oral Sci. 2021;13(1):36. [Google Scholar] [PubMed]
3. Hedlund E, Deng Q. Single-cell RNA sequencing: technical advancements and biological applications. Mol Aspects Med. 2018;59:36–46. [Google Scholar] [PubMed]
4. Evrony GD, Hinch AG, Luo C. Applications of single-cell DNA sequencing. Annu Rev Genomics Hum Genet. 2021;22:171–97. [Google Scholar] [PubMed]
5. Chang X, Zheng Y, Xu K. Single-Cell RNA sequencing: technological progress and biomedical application in cancer research. Mol Biotechnol. 2024;66(7):1497–591. [Google Scholar] [PubMed]
6. Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6(5):377–82. doi:10.1038/nmeth.1315. [Google Scholar] [PubMed] [CrossRef]
7. Li M, Yan T, Wang M, Cai Y, Wei Y. Advances in single-cell sequencing technology and its applications in triple-negative breast cancer. Breast Cancer: Target. 2022;14:465–74. doi:10.2147/BCTT.S388534. [Google Scholar] [PubMed] [CrossRef]
8. Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018;14(8):479–92. doi:10.1038/s41581-018-0021-7. [Google Scholar] [PubMed] [CrossRef]
9. Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J, et al. Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol. 2021;14(1):91. doi:10.1186/s13045-021-01105-2. [Google Scholar] [PubMed] [CrossRef]
10. Cai Z, Chen L, Chen S, Fang R, Chen X, Lei W. Single-cell RNA sequencing reveals pro-invasive cancer-associated fibroblasts in hypopharyngeal squamous cell carcinoma. Cell Commun Signal. 2023;21(1):292. doi:10.1186/s12964-023-01312-z. [Google Scholar] [PubMed] [CrossRef]
11. Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018;18(1):35–45. [Google Scholar] [PubMed]
12. Christodoulou MI, Zaravinos A. Single-cell analysis in immuno-oncology. Int J Mol Sci. 2023;24(9):8422. [Google Scholar] [PubMed]
13. Shangguan Y, Li C, Lin H, Ou M, Tang D, Dai Y, et al. Application of single-cell RNA sequencing in embryonic development. Genomics. 2020;112(6):4547–51. [Google Scholar] [PubMed]
14. Li Q, Wang M, Zhang S, Jin M, Chen R, Luo Y, et al. Single-cell RNA sequencing in atherosclerosis: mechanism and precision medicine. Front Pharmacol. 2022;13:977490. [Google Scholar] [PubMed]
15. Voigt AP, Mullin NK, Stone EM, Tucker BA, Scheetz TE, Mullins RF. Single-cell RNA sequencing in vision research: insights into human retinal health and disease. Prog Retin Eye Res. 2021;83:100934. [Google Scholar] [PubMed]
16. Wang Z, Li X, Yang J, Gong Y, Zhang H, Qiu X, et al. Single-cell RNA sequencing deconvolutes the in vivo heterogeneity of human bone marrow-derived mesenchymal stem cells. Int J Biol Sci. 2021;17(15):4192–206. [Google Scholar] [PubMed]
17. Wen L, Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016;17:71. [Google Scholar] [PubMed]
18. Zakrzewski W, Dobrzyński M, Szymonowicz M, Rybak Z. Stem cells: past, present, and future. Stem Cell Res Ther. 2019;10(1):68. [Google Scholar] [PubMed]
19. Chen X, Liu B, Li C, Wang Y, Geng S, Du X, et al. Stem cell-based therapy for COVID-19. Int Immunopharmacol. 2023;124:110890. [Google Scholar] [PubMed]
20. Goradel NH, Hour FG, Negahdari B, Malekshahi ZV, Hashemzehi M, Masoudifar A, et al. Stem cell therapy: a new therapeutic option for cardiovascular diseases. J Cell Biochem. 2018;119(1):95–104. [Google Scholar] [PubMed]
21. Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19(10):1129–55. [Google Scholar] [PubMed]
22. Ebert AD, Svendsen CN. Human stem cells and drug screening: opportunities and challenges. Nat Rev Drug Discov. 2010;9(5):367–72. [Google Scholar] [PubMed]
23. Hoang DM, Pham PT, Bach TQ, Ngo ATL, Nguyen QT, Phan TTK, et al. Stem cell-based therapy for human diseases. Signal Transduct Target Ther. 2022;7(1):272. [Google Scholar] [PubMed]
24. Levy O, Kuai R, Siren EMJ, Bhere D, Milton Y, Nissar N, et al. Shattering barriers toward clinically meaningful MSC therapies. Sci Adv. 2020;6(30):eaba6884. [Google Scholar] [PubMed]
25. Galipeau J, Sensébé L. Mesenchymal stromal cells: clinical challenges and therapeutic opportunities. Cell Stem Cell. 2018;22(6):824–33. [Google Scholar] [PubMed]
26. Nishimura K, Yang S, Lee KW, Ásgrímsdóttir ES, Nikouei K, Paslawski W, et al. Single-cell transcriptomics reveals correct developmental dynamics and high-quality midbrain cell types by improved hESC differentiation. Stem Cell Reports. 2023;18(1):337–53. doi:10.1016/j.stemcr.2022.10.016. [Google Scholar] [PubMed] [CrossRef]
27. Yan L, Yang M, Guo H, Yang L, Wu J, Li R, et al. Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol. 2013;20(9):1131–9. doi:10.1038/nsmb.2660. [Google Scholar] [PubMed] [CrossRef]
28. Qi L, Wang J, Chen X, Ding Y, Ling B, Wang W, et al. Single-cell transcriptomics reveals variable trajectories of CSPCs in the progression of osteoarthritis. Heliyon. 2022;8(11):e11148. doi:10.1016/j.heliyon.2022.e11148. [Google Scholar] [PubMed] [CrossRef]
29. Chen P, Tang S, Li M, Wang D, Chen C, Qiu Y, et al. Single-cell and spatial transcriptomics decodes Wharton’s jelly-derived mesenchymal stem cells heterogeneity and a subpopulation with wound repair signatures. Adv Sci. 2023;10(4):e2204786. doi:10.1002/advs.202204786. [Google Scholar] [PubMed] [CrossRef]
30. Qu R, He K, Fan T, Yang Y, Mai L, Lian Z, et al. Single-cell transcriptomic sequencing analyses of cell heterogeneity during osteogenesis of human adipose-derived mesenchymal stem cells. Stem Cells. 2021;39(11):1478–88. doi:10.1002/stem.3442. [Google Scholar] [PubMed] [CrossRef]
31. Matsushita Y, Ono W, Ono N. Synergy of single-cell sequencing analyses and in vivo lineage-tracing approaches: a new opportunity for stem cell biology. BIOCELL. 2022;46(5):1157–62. doi:10.32604/biocell.2022.018960. [Google Scholar] [CrossRef]
32. Chen T, Li J, Jia Y, Wang J, Sang R, Zhang Y, et al. Single-cell sequencing in the field of stem cells. Curr Genomics. 2020;21(8):576–84. [Google Scholar] [PubMed]
33. Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, Medvinsky A. Human haematopoietic stem cell development: from the embryo to the dish. Development. 2017;144(13):2323–37. [Google Scholar] [PubMed]
34. Zhang P, Li X, Pan C, Zheng X, Hu B, Xie R, et al. Single-cell RNA sequencing to track novel perspectives in HSC heterogeneity. Stem Cell Res Ther. 2022;13(1):39. [Google Scholar] [PubMed]
35. Introna M, Golay J. Tolerance to bone marrow transplantation: do mesenchymal stromal cells still have a future for acute or chronic GvHD? Front Immunol. 2020;11:609063. [Google Scholar] [PubMed]
36. Zhang Y, Yi Y, Xiao X, Hu L, Xu J, Zheng D, et al. Definitive endodermal cells supply an in vitro source of mesenchymal stem/stromal cells. Commun Biol. 2023;6(1):476. [Google Scholar] [PubMed]
37. Wilkinson AC, Igarashi KJ, Nakauchi H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat Rev Genet. 2020;21(9):541–54. [Google Scholar] [PubMed]
38. Skulimowska I, Sosniak J, Gonka M, Szade A, Jozkowicz A, Szade K. The biology of hematopoietic stem cells and its clinical implications. Febs J. 2022;289(24):7740–59. [Google Scholar] [PubMed]
39. Xiang Y, Sugimura R. Single-cell approaches to deconvolute the development of HSCs. Cells. 2021;10(11):2876. [Google Scholar] [PubMed]
40. Weijts B, Yvernogeau L, Robin C. Recent advances in developmental hematopoiesis: diving deeper with new technologies. Front Immunol. 2021;12:790379. [Google Scholar] [PubMed]
41. Shi G, Zhang P, Zhang X, Li J, Zheng X, Yan J, et al. The spatiotemporal heterogeneity of the biophysical microenvironment during hematopoietic stem cell development: from embryo to adult. Stem Cell Res Ther. 2023;14(1):251. doi:10.1186/s13287-023-03464-8. [Google Scholar] [PubMed] [CrossRef]
42. Zeng Y, He J, Bai Z, Li Z, Gong Y, Liu C, et al. Tracing the first hematopoietic stem cell generation in human embryo by single-cell RNA sequencing. Cell Res. 2019;29(11):881–94. doi:10.1038/s41422-019-0228-6. [Google Scholar] [PubMed] [CrossRef]
43. Calvanese V, Capellera-Garcia S, Ma F, Fares I, Liebscher S, Ng ES, et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature. 2022;604(7906):534–40. doi:10.1038/s41586-022-04571-x. [Google Scholar] [PubMed] [CrossRef]
44. Zheng Z, He H, Tang XT, Zhang H, Gou F, Yang H, et al. Uncovering the emergence of HSCs in the human fetal bone marrow by single-cell RNA-seq analysis. Cell Stem Cell. 2022;29(11):1562–79.e7. doi:10.1016/j.stem.2022.10.005. [Google Scholar] [PubMed] [CrossRef]
45. Ragazzini R, Boeing S, Zanieri L, Green M, D’Agostino G, Bartolovic K, et al. Defining the identity and the niches of epithelial stem cells with highly pleiotropic multilineage potency in the human thymus. Dev Cell. 2023;58(22):2428–46.e9. doi:10.1016/j.devcel.2023.08.017. [Google Scholar] [PubMed] [CrossRef]
46. Chan CKF, Gulati GS, Sinha R, Tompkins JV, Lopez M, Carter AC, et al. Identification of the human skeletal stem cell. Cell. 2018;175(1):43–56.e21. doi:10.1016/j.cell.2018.07.029. [Google Scholar] [PubMed] [CrossRef]
47. Ehnes DD, Alghadeer A, Hanson-Drury S, Zhao YT, Tilmes G, Mathieu J, et al. Sci-seq of human fetal salivary tissue introduces human transcriptional paradigms and a novel cell population. Front Dent Med. 2022;3:887057. doi:10.3389/fdmed.2022.887057. [Google Scholar] [PubMed] [CrossRef]
48. Chan CK, Seo EY, Chen JY, Lo D, McArdle A, Sinha R, et al. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160(1–2):285–98. doi:10.1016/j.cell.2014.12.002. [Google Scholar] [PubMed] [CrossRef]
49. Worthley DL, Churchill M, Compton JT, Tailor Y, Rao M, Si Y, et al. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160(1–2):269–84. doi:10.1016/j.cell.2014.11.042. [Google Scholar] [PubMed] [CrossRef]
50. He J, Yan J, Wang J, Zhao L, Xin Q, Zeng Y, et al. Dissecting human embryonic skeletal stem cell ontogeny by single-cell transcriptomic and functional analyses. Cell Res. 2021;31(7):742–57. doi:10.1038/s41422-021-00467-z. [Google Scholar] [PubMed] [CrossRef]
51. Kameishi S, Umemoto T, Matsuzaki Y, Fujita M, Okano T, Kato T, et al. Characterization of rabbit limbal epithelial side population cells using RNA sequencing and single-cell qRT-PCR. Biochem Biophys Res Commun. 2016;473(3):704–9. [Google Scholar] [PubMed]
52. Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22(5):627–38. [Google Scholar] [PubMed]
53. Hayashi Y, Ohnuma K, Furue MK. Pluripotent stem cell heterogeneity. Adv Exp Med Biol. 2019;1123:71–94. [Google Scholar] [PubMed]
54. Tyurin-Kuzmin PA, Hayashi Y, Kulebyakin K. Editorial: functional heterogeneity of stem cells. Front Cell Dev Biol. 2023;11:1179911. [Google Scholar] [PubMed]
55. Olmedo-Moreno L, Aguilera Y, Baliña-Sánchez C, Martín-Montalvo A, Capilla-González V. Heterogeneity of in vitro expanded mesenchymal stromal cells and strategies to improve their therapeutic actions. Pharmaceutics. 2022;14(5):1112. [Google Scholar] [PubMed]
56. Galderisi U, Peluso G, Di Bernardo G. Clinical trials based on mesenchymal stromal cells are exponentially increasing: where are We in recent years? Stem Cell Rev Rep. 2022;18(1):23–36. [Google Scholar] [PubMed]
57. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8(4):315–7. [Google Scholar] [PubMed]
58. Wang Z, Chai C, Wang R, Feng Y, Huang L, Zhang Y, et al. Single-cell transcriptome atlas of human mesenchymal stem cells exploring cellular heterogeneity. Clin Transl Med. 2021;11(12):e650. [Google Scholar] [PubMed]
59. Barrett AN, Fong CY, Subramanian A, Liu W, Feng Y, Choolani M, et al. Human Wharton’s jelly mesenchymal stem cells show unique gene expression compared with bone marrow mesenchymal stem cells using single-cell RNA-sequencing. Stem Cells Dev. 2019;28(3):196–211. [Google Scholar] [PubMed]
60. Nel S, Durandt C, Murdoch C, Pepper MS. Determinants of dental pulp stem cell heterogeneity. J Endod. 2022;48(10):1232–40. [Google Scholar] [PubMed]
61. Lee S, Chen D, Park M, Kim S, Choi YJ, Moon SJ, et al. Single-Cell RNA sequencing analysis of human dental pulp stem cell and human periodontal ligament stem cell. J Endod. 2022;48(2):240–8. doi:10.1016/j.joen.2021.11.005. [Google Scholar] [PubMed] [CrossRef]
62. Zhou W, Lin J, Zhao K, Jin K, He Q, Hu Y, et al. Single-cell profiles and clinically useful properties of human mesenchymal stem cells of adipose and bone marrow origin. Am J Sports Med. 2019;47(7):1722–33. doi:10.1177/0363546519848678. [Google Scholar] [PubMed] [CrossRef]
63. Wang Q, Li J, Wang S, Deng Q, Wang K, Dai X, et al. Single-cell transcriptome profiling reveals molecular heterogeneity in human umbilical cord tissue and culture-expanded mesenchymal stem cells. Febs J. 2021;288(18):5311–30. doi:10.1111/febs.15834. [Google Scholar] [PubMed] [CrossRef]
64. Zhang S, Wang JY, Li B, Yin F, Liu H. Single-cell transcriptome analysis of uncultured human umbilical cord mesenchymal stem cells. Stem Cell Res Ther. 2021;12(1):25. doi:10.1186/s13287-020-02055-1. [Google Scholar] [PubMed] [CrossRef]
65. Sun C, Wang L, Wang H, Huang T, Yao W, Li J, et al. Single-cell RNA-seq highlights heterogeneity in human primary Wharton’s jelly mesenchymal stem/stromal cells cultured in vitro. Stem Cell Res Ther. 2020;11(1):149. doi:10.1186/s13287-020-01660-4. [Google Scholar] [PubMed] [CrossRef]
66. McLeod CM, Mauck RL. On the origin and impact of mesenchymal stem cell heterogeneity: new insights and emerging tools for single cell analysis. Eur Cell Mater. 2017;34:217–31. [Google Scholar] [PubMed]
67. Gao Q, Wang L, Wang S, Huang B, Jing Y, Su J. Bone marrow mesenchymal stromal cells: identification, classification, and differentiation. Front Cell Dev Biol. 2021;9:787118. [Google Scholar] [PubMed]
68. Li Z, Zhang C, Weiner LP, Zhang Y, Zhong JF. Molecular characterization of heterogeneous mesenchymal stem cells with single-cell transcriptomes. Biotechnol Adv. 2013;31(2):312–7. [Google Scholar] [PubMed]
69. Liang ZX, Liu HS, Xiong L, Zeng ZW, Zheng XB, Kang L, et al. GAS6 from CD200+ adipose-derived stem cells mitigates colonic inflammation in a macrophage-dependent manner. J Crohns Colitis. 2023;17(2):289–301. [Google Scholar] [PubMed]
70. Leng Z, Li L, Zhou X, Dong G, Chen S, Shang G, et al. Novel insights into the stemness and immune privilege of mesenchymal stem cells from human Wharton jelly by single-cell RNA sequencing. Med Sci Monit. 2022;28:e934660. [Google Scholar] [PubMed]
71. Zhang X, Jiang W, Xie C, Wu X, Ren Q, Wang F, et al. Msx1+ stem cells recruited by bioactive tissue engineering graft for bone regeneration. Nat Commun. 2022;13(1):5211. [Google Scholar] [PubMed]
72. Hao RC, Li ZL, Wang FY, Tang J, Li PL, Yin BF, et al. Single-cell transcriptomic analysis identifies a highly replicating Cd168+ skeletal stem/progenitor cell population in mouse long bones. J Genet Genomics. 2023;50(9):702–12. [Google Scholar] [PubMed]
73. Xu H, Tan S, Zhao Y, Zhang L, Cao W, Li X, et al. Lin-PU.1dimGATA-1− defines haematopoietic stem cells with long-term multilineage reconstitution activity. Cell Prolif. 2023;56(11):e13490. [Google Scholar] [PubMed]
74. Tang J, Wang H, Huang X, Li F, Zhu H, Li Y, et al. Arterial Sca1+ vascular stem cells generate de novo smooth muscle for artery repair and regeneration. Cell Stem Cell. 2020;26(1):81–96.e4. [Google Scholar] [PubMed]
75. Shu M, Xue X, Nie H, Wu X, Sun M, Qiao L, et al. Single-cell RNA sequencing reveals Nestin+ active neural stem cells outside the central canal after spinal cord injury. Sci China Life Sci. 2022;65(2):295–308. [Google Scholar] [PubMed]
76. Zhao L, Lee AS, Sasagawa K, Sokol J, Wang Y, Ransom RC, et al. A combination of distinct vascular stem/progenitor cells for neovascularization and ischemic rescue. Arterioscler Thromb Vasc Biol. 2023;43(7):1262–77. [Google Scholar] [PubMed]
77. Li J, Wang Q, An Y, Chen X, Xing Y, Deng Q, et al. Integrative single-cell RNA-Seq and ATAC-Seq analysis of mesenchymal stem/stromal cells derived from human placenta. Front Cell Dev Biol. 2022;10:836887. [Google Scholar] [PubMed]
78. Zhang P, Dong J, Fan X, Yong J, Yang M, Liu Y, et al. Characterization of mesenchymal stem cells in human fetal bone marrow by single-cell transcriptomic and functional analysis. Signal Transduct Target Ther. 2023;8(1):126. [Google Scholar] [PubMed]
79. Li B, Li J, Li B, Ouchi T, Li L, Li Y, et al. A single-cell transcriptomic atlas characterizes age-related changes of murine cranial stem cell niches. Aging Cell. 2023;22(11):e13980. [Google Scholar] [PubMed]
80. Fraile M, Eiro N, Costa LA, Martín A, Vizoso FJ. Aging and mesenchymal stem cells: basic concepts, challenges and strategies. Biology. 2022;11(11):1678. [Google Scholar] [PubMed]
81. Afkhami H, Mahmoudvand G, Fakouri A, Shadab A, Mahjoor M, Komeili Movahhed T. New insights in application of mesenchymal stem cells therapy in tumor microenvironment: pros and cons. Front Cell Dev Biol. 2023;11:1255697. [Google Scholar] [PubMed]
82. Xue Z, Liao Y, Li Y. Effects of microenvironment and biological behavior on the paracrine function of stem cells. Genes Dis. 2024;11(1):135–47. [Google Scholar] [PubMed]
83. Yang Y, Zhang W, Wang X, Yang J, Cui Y, Song H, et al. A passage-dependent network for estimating the in vitro senescence of mesenchymal stromal/stem cells using microarray, bulk and single cell RNA sequencing. Front Cell Dev Biol. 2023;11:998666. [Google Scholar] [PubMed]
84. Wakabayashi T, Naito H. Cellular heterogeneity and stem cells of vascular endothelial cells in blood vessel formation and homeostasis: insights from single-cell RNA sequencing. Front Cell Dev Biol. 2023;11:1146399. [Google Scholar] [PubMed]
85. Liu Y, Chen Y, Li XH, Cao C, Zhang HX, Zhou C, et al. Dissection of cellular communication between human primary osteoblasts and bone marrow mesenchymal stem cells in osteoarthritis at single-cell resolution. Int J Stem Cells. 2023;16(3):342–55. [Google Scholar] [PubMed]
86. Shen K, Chen B, Gao W. Integrated single-cell RNA sequencing analysis reveals a mesenchymal stem cell-associated signature for estimating prognosis and drug sensitivity in gastric cancer. J Cancer Res Clin Oncol. 2023;149(13):11829–47. [Google Scholar] [PubMed]
87. Weng Z, Wang Y, Ouchi T, Liu H, Qiao X, Wu C, et al. Mesenchymal stem/stromal cell senescence: hallmarks, mechanisms, and combating strategies. Stem Cells Transl Med. 2022;11(4):356–71. [Google Scholar] [PubMed]
88. Neri S, Borzì RM. Molecular mechanisms contributing to mesenchymal stromal cell aging. Biomolecules. 2020;10(2):340. [Google Scholar] [PubMed]
89. Taherian Fard A, Leeson HC, Aguado J, Pietrogrande G, Power D, Gómez-Inclán C, et al. Deconstructing heterogeneity of replicative senescence in human mesenchymal stem cells at single cell resolution. Geroscience. 2024;46(1):999–1015. [Google Scholar] [PubMed]
90. Smirnova A, Yatsenko E, Baranovskii D, Klabukov I. Mesenchymal stem cell-derived extracellular vesicles in skin wound healing: the risk of senescent drift induction in secretome-based therapeutics. Mil Med Res. 2023;10(1):60. [Google Scholar] [PubMed]
91. Siraj Y, Galderisi U, Alessio N. Senescence induces fundamental changes in the secretome of mesenchymal stromal cells (MSCsimplications for the therapeutic use of MSCs and their derivates. Front Bioeng Biotechnol. 2023;11:1148761. doi:10.3389/fbioe.2023.1148761. [Google Scholar] [PubMed] [CrossRef]
92. Mohamad Yusoff F, Higashi Y. Mesenchymal stem/stromal cells for therapeutic angiogenesis. Cells. 2023;12(17):2162. doi:10.3390/cells12172162. [Google Scholar] [PubMed] [CrossRef]
93. Hosseini M, Shafiee A. Vascularization of cutaneous wounds by stem cells. Prog Mol Biol Transl Sci. 2023;199(3):327–50. doi:10.1016/bs.pmbts.2023.03.002. [Google Scholar] [PubMed] [CrossRef]
94. Morley RL, Sharma A, Horsch AD, Hinchliffe RJ. Peripheral artery disease. BMJ. 2018;360:j5842. doi:10.1136/bmj.j5842. [Google Scholar] [PubMed] [CrossRef]
95. Jeyaraman M, Nagarajan S, Maffulli N, R PP, Jeyaraman N, N A. Stem cell therapy in critical limb ischemia. Cureus. 2023;15(7):e41772. doi:10.7759/cureus.41772. [Google Scholar] [PubMed] [CrossRef]
96. Miura T, Kouno T, Takano M, Kuroda T, Yamamoto Y, Kusakawa S, et al. Single-cell RNA-Seq reveals LRRC75A-expressing cell population involved in VEGF secretion of multipotent mesenchymal stromal/stem cells under ischemia. Stem Cells Transl Med. 2023;12(6):379–90. [Google Scholar] [PubMed]
97. Zhou Y, Tsai TL, Li WJ. Strategies to retain properties of bone marrow-derived mesenchymal stem cells ex vivo. Ann N Y Acad Sci. 2017;1409(1):3–17. [Google Scholar] [PubMed]
98. Kusuma GD, Carthew J, Lim R, Frith JE. Effect of the microenvironment on mesenchymal stem cell paracrine signaling: oopportunities to engineer the therapeutic effect. Stem Cells Dev. 2017;26(9):617–31. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF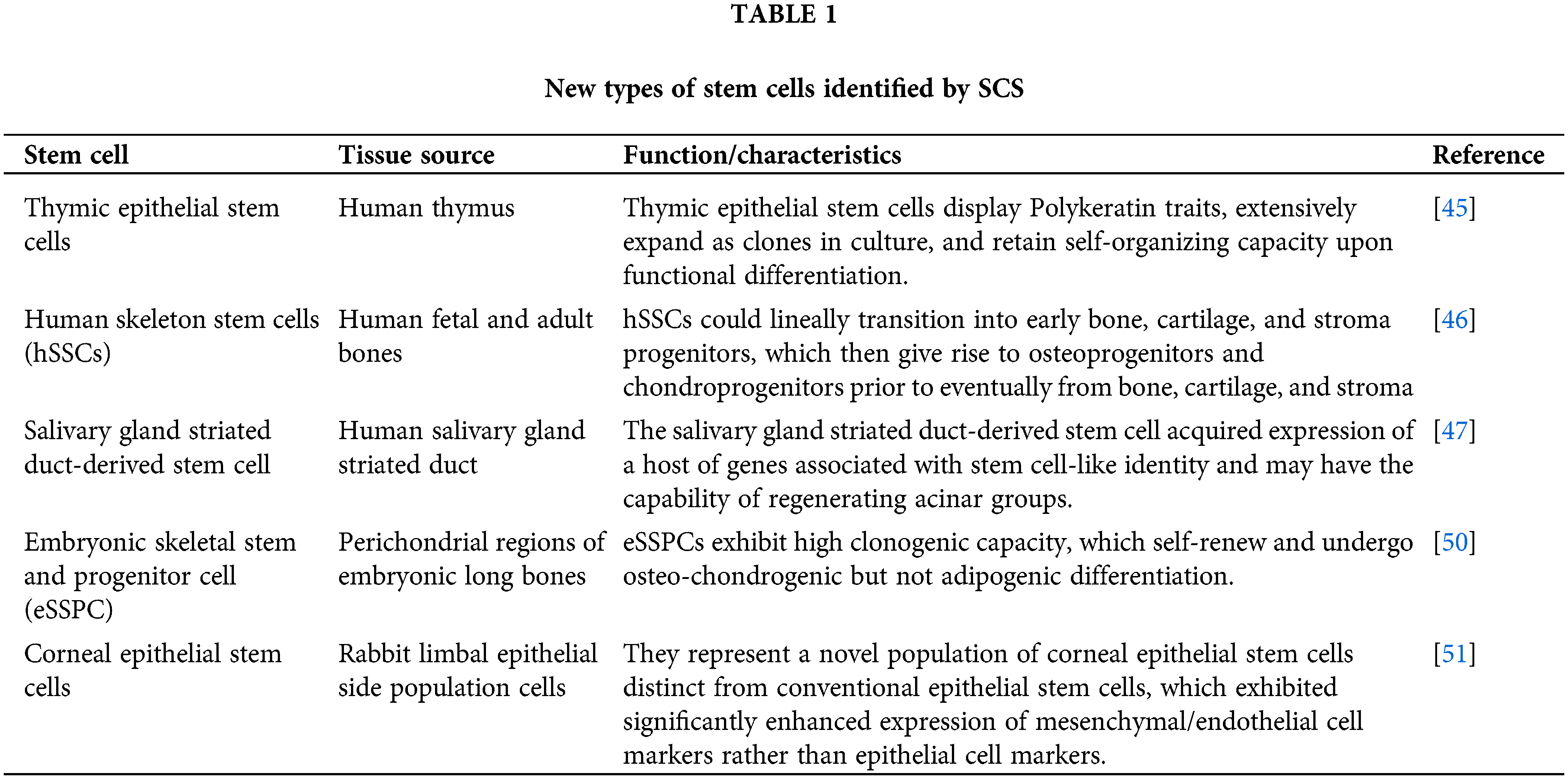
 Downloads
Downloads
 Citation Tools
Citation Tools