 Open Access
Open Access
ARTICLE
DNAJA4, DNAJB11 and DNAJC10 induce cell transformation by inhibiting p53 and oncogene-induced senescence
1 Department of Biochemistry and Molecular Biology, Kangwon National University College of Medicine, Chuncheon, 24341, Republic of Korea
2 Department of Biological Sciences, Kangwon National University, Chuncheon, 24341, Republic of Korea
3 Department of Biomedical Sciences, Seoul National University Graduate School, Seoul, 03080, Republic of Korea
4 Genomic Medicine Institute, Medical Research Center, Seoul National University College of Medicine, Seoul, 03080, Republic of Korea
5 Cancer Research Institute, Seoul National University College of Medicine, Seoul, 03080, Republic of Korea
* Corresponding Author: JEONG A. HAN. Email:
BIOCELL 2024, 48(10), 1455-1464. https://doi.org/10.32604/biocell.2024.054538
Received 31 May 2024; Accepted 12 August 2024; Issue published 02 October 2024
Abstract
Objective: Transformation from normal cells to malignant cells is the basis for tumorigenesis. While this cell transformation is known to result from aberrant activation or inactivation of associated genes, these genes have not yet been fully identified. In addition, DNAJs, proteins with a J domain, are known to be molecular co-chaperones, but their cellular functions remain largely unexplored. In this context, we here identified DNAJA4, DNAJB11, and DNAJC10 as pro-transforming genes and elucidated their action mechanisms. Methods: Senescence-associated (SA)-β-galactosidase staining and western blotting were used to analyze cellular senescence and protein expression. Soft agar assay was used to analyze cell transformation. RNA sequencing data was downloaded from the Cancer Genome Atlas (TCGA) database. Results: We observed that overexpression of the three DNAJs inhibited oncogene-induced senescence (OIS) and the p53/p21WAF1/CIP1 pathway under H-RASV12 expression in normal mouse embryonic fibroblasts (MEFs). Additionally, their overexpression inhibited p53-induced senescence. Moreover, overexpression of the three DNAJs induced neoplastic transformation in MEFs under H-RASV12 expression. Furthermore, RNA sequencing analysis showed that the three DNAJs are frequently overexpressed in cancer tissues compared to their matched normal tissues in various human cancers. Conclusion: These results suggest that the three DNAJs are pro-transforming genes whose aberrant overexpression contributes to cell transformation. These results also suggest that the three DNAJs induce cell transformation by inhibiting the senescence function of p53 and OIS. This study may contribute to understanding the molecular basis of cell transformation and the cellular functions of the three DNAJs.Keywords
Cancer cells have common characteristics, including self-mitogenic signaling, the ability to proliferate indefinitely, resistance to density-dependent or contact inhibition, growth in a suspended state (anchorage-independence), and the ability to invade surrounding tissues. The phenomenon in which normal cells acquire these malignant traits is called malignant transformation, and this cellular transformation is the basis for tumorigenesis at the individual level [1,2].
Cellular transformation is known to result from aberrant activation or inactivation of associated genes. For example, aberrant activation of H-RAS [3], K-RAS [3], cyclin E (CCNE) [4], cyclin-dependent kinase 4 (CDK4) [3], or c-MYC [5] by mutations or overexpression, and aberrant inactivation of p53 [3] or Rb [3] by mutations or protein inactivation can induce transformation in human or rodent normal cells. Therefore, identifying transformation-associated genes is essential for understanding the mechanisms of cell transformation and tumorigenesis.
Cellular senescence is a phenomenon in which mitotic cells permanently lose their ability to divide. Senescent cells differ from quiescent cells in that they cannot enter the S phase of the cell cycle even when stimulated by growth factors. In addition, they typically exhibit senescence-associated (SA)-β-galactosidase activity at pH 6.0, and the cells often have a broad and flat shape [6].
During cell transformation, mitogenic oncogenes such as H-RASV12 are activated, allowing normal cells to acquire the characteristic of self-mitogenic signaling [1]. Intriguingly, however, the expression of H-RASV12 alone, a constitutively active form of H-RAS, in human or rodent normal cells generally induces senescence rather than transformation. This phenomenon, in which cellular senescence is induced when some mitogenic oncogenes are activated, is called oncogene-induced senescence (OIS). OIS is considered an important mechanism to prevent cell transformation by permanently arresting the division of dangerous cells in which mitogenic oncogenes are activated [6,7].
In OIS, strong and sustained mitogenic signaling from mitogenic oncogenes leads to replication stress and double-stranded breaks (DSBs) in DNA. This provokes the DNA-damage response (DDR) in cells, including the activation of p53 [8,9].
p53 is a potent tumor suppressor. Mutations in the p53 gene are observed in approximately half of human cancers, and functional inactivation of the p53 protein is frequently observed in the remaining cases. Upon DNA damage, the transcription factor p53 is activated and translocates from the cytosol to the nucleus. In the nucleus, it binds to the promoters of various target genes to induce senescence, apoptosis, or cell cycle arrest. Among these target genes, p21WAF1/CIP1 plays a crucial role in inducing senescence. Therefore, p53 acts as a tumor suppressor by inhibiting the proliferation of cells with damaged DNA and preventing the occurrence of mutations [6,10].
The senescence function of p53 plays a critical role in OIS. The H-RASV12-induced OIS in normal mouse embryonic fibroblasts (MEFs) is accompanied by an increase in p53 expression, where if p53 is inactivated, OIS does not occur and the cells continue to divide [11]. Even in human diploid fibroblasts (HDFs), H-RASV12-induced OIS is accompanied by an increase in p53 expression, where inactivation of p53 often prevents OIS [9].
Therefore, it can be speculated that p53 inactivation under potent mitogenic signals may contribute to cell transformation by evading OIS. Indeed, transformation occurs when p53 is inactivated simultaneously with H-RASV12 expression in MEFs. In the case of HDFs, transformation occurs when both p53 and Rb are inactivated simultaneously with H-RASV12 expression and telomerase activation [3].
Molecular chaperones are proteins that bind to cargo proteins and assist in their transport, folding, or degradation. In most organisms, including humans, heat shock protein 70s (Hsp70s) are major molecular chaperones, and the activity of Hsp70s is regulated by co-chaperones such as DNAJs. DNAJs bind to Hsp70s and stimulate their ATPase activity, thereby stabilizing the interaction between Hsp70s and substrate proteins. DNAJs commonly have a J domain, a specific motif consisting of 70–80 amino acids, which is known to be important for binding to Hsp70s [12]. To date, approximately 50 DNAJs have been identified in the human genome. They are classified into Classes I, II, and III based on the presence or absence of a G/F-rich domain and a zinc finger-like region (ZFLR) in addition to the J domain, and are referred to as DNAJAs, DNAJBs, and DNAJCs, respectively [13,14] (Fig. 1).
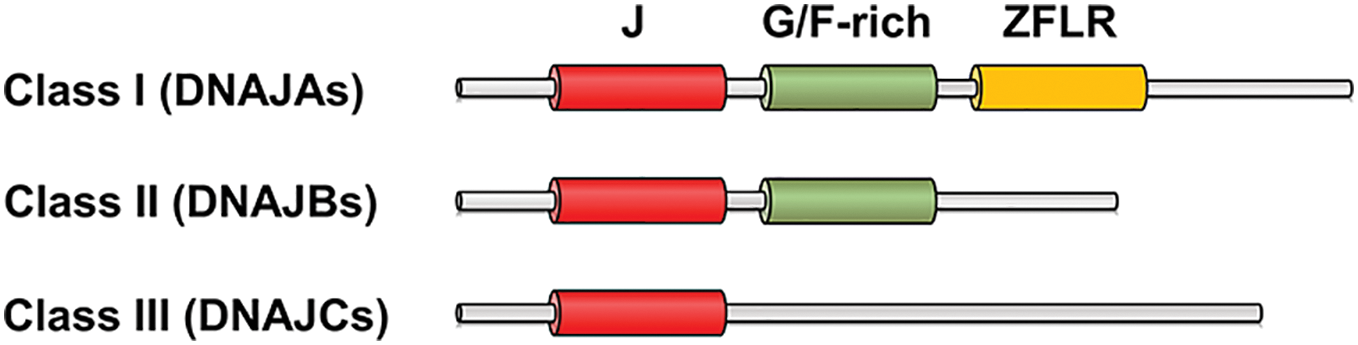
Figure 1: Schematic representation of three classes of DNAJs. J, the J domain; G/F-rich, the G/F-rich domain; ZFLR, the zinc finger-like region.
Among them, DNAJA4 has been reported to assist Hsp70s in preventing the aggregation of luciferase in vitro [15]. DNAJB11 is a luminal protein of the endoplasmic reticulum (ER), where it is known to assist BiP, an ER member of the Hsp70 family, in folding and assembling cargo proteins [16]. DNAJC10, another co-chaperone of BiP, characteristically possesses disulfide reductase activity. DNAJC10 is known to cleave the disulfide bonds of misfolded proteins, thereby accelerating their ER-associated degradation (ERAD) [17,18].
Although identifying transformation-associated genes is essential for understanding the mechanisms of cell transformation and tumorigenesis, these genes have not yet been fully identified. In addition, DNAJs are known to act as co-chaperones for Hsp70s, but their cellular functions remain largely unexplored. In this context, we here investigated the cellular functions of DNAJA4, DNAJB11, and DNAJC10 under strong mitogenic signals using H-RASV12.
The cDNAs of human DNAJA4 and DNAJB11 were obtained from the 21C Frontier Human Gene Bank (KRIBB, Daejeon, Republic of Korea). The cDNA of human DNAJC10 was generously provided by Dr. Billy Tsai from the University of Michigan, USA.
The preparation of mouse embryonic fibroblasts (MEFs) was conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Kangwon National University (Approval No. KW-140521-1). Female ICR mice, 14 days post-coitum (dpc), were obtained from Nara Biotech (Pyungtak, Republic of Korea) and kept in a controlled environment with a stationary temperature of 21 ± 2°C, the humidity of 50 ± 10%, and noise levels below 60 dB. They were fed standard mouse chow (DBL, Rodfeed, Eumseong, Republic of Korea) and had ad libitum access to water (Nara Biotech, Hydrogel, Pyungtak, Republic of Korea). Embryos were extracted from the mice and minced in 100 mm dishes containing Dulbecco’s modified Eagle’s medium (DMEM) (Life Technologies, #12800017, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (AusGeneX, #FBS500-S, Oxenford, Australia). The dishes were then incubated in a 5% CO2 incubator at 37°C. After 2–3 days, the cells were harvested and stored in a liquid nitrogen tank until needed [19]. The cells were cultured in DMEM containing 10% FBS and penicillin-streptomycin (10,000 U/mL) (Life Technologies, #15140-122) in a 5% CO2 incubator at 37°C. There was no mycoplasma contamination in all cells utilized for this study.
MEFs were lysed in a solution containing 150 mM NaCl, 50 mM Tris-HCl, 0.1% sodium dodecyl sulfate (SDS), 0.25% sodium deoxycholate, 1% NP-40, a protease inhibitor cocktail (Sigma-Aldrich, #P2714, Burlington, MA, USA), and 1 mM phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, #P7626). SDS–polyacrylamide gel electrophoresis (PAGE) was carried out to separate proteins. The proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Merck Millipore, IPVH00010, Darmstadt, Germany). The membranes were incubated with 4% skim milk (Seoulmilk, #1985026200513, Yangju, Republic of Korea) in Tris-buffered saline (TBS) for 1 h at room temperature. Then, the membranes were incubated with protein-specific antibodies overnight at 4°C, followed by secondary antibodies for 1 h at room temperature. After washing with TBS, the protein bands were visualized using a detection reagent (AbFrontier, LF-QC0103, Seoul, Republic of Korea) [20]. The protein-specific antibodies used were anti-DNAJA4 (1:500, Abcam, #ab185553, Cambridge, UK), anti-DNAJB11 (1:200, Santa Cruz Biotechnology, #sc-271240, Dallas, TX, USA), anti-DNAJC10 (1:2000, Proteintech, #13101-1-AP, San Diego, CA, USA), anti-p21WAF1/CIP1 (1:50, Santa Cruz Biotechnology, #sc-6246), anti-p53 (1:100, Cell Signaling Technology, #2524, Danvers, MA, USA), anti-Ras (1:200, Calbiochem, #OP40, San Diego, CA, USA), and anti-β-actin (1:4000, Santa Cruz Biotechnology, #sc-47778). The secondary antibodies used were HRP-conjugated goat anti-mouse IgG (1:2000, Invitrogen, #31430, Rockford, IL, USA) and HRP-conjugated goat anti-rabbit IgG (1:1000, Invitrogen, #31460).
cDNAs inserted into the pMSCVpuro vector (Clontech, Mountain View, CA, USA) were transfected into H29D packaging cells [21] using polyethylenimine (PEI) (Polysciences, #23966-2, Warrington, PA, USA). Conditioned media containing retroviruses (viral soups) were collected from days 2 to 4 post-transfection. For viral titration, 293 cells were seeded on 6-well plates at a density of 5 × 103 cells per well. The viral soups were serially diluted and administered to each well with 2 μg/mL polybren (Sigma-Aldrich, #H9268). The cells were incubated with 0.85 μg/mL puromycin (Life Technologies, #A1113803) for 2 days, followed by incubation in a culture medium for an additional 2 days. Then, the total number of foci per well was counted under a light microscope (Olympus, CKX41, Tokyo, Japan), and the plaque-forming units (pfu) per mL of viral soup were calculated. MEFs were infected with 10 multiplicity of infection (MOI) of retroviruses expressing each DNAJ on the 1st day and then infected with 5 MOI of retroviruses expressing H-RASV12 or p53 on the 2nd day in the presence of 2 μg/mL polybren. The MEFs were incubated with 1 μg/mL puromycin for 2 days, followed by incubation in a culture medium for an additional 2 days.
Senescence-associated-β-galactosidase (SA-β-gal) staining
MEFs were fixed in a 3% formaldehyde solution for 5 min at room temperature. The cells were incubated at 37°C for 24 h in a staining solution consisting of 40 mM citric acid/sodium phosphate, pH 6.0, 150 mM sodium chloride, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 2 mM magnesium chloride, and 1 mg/mL X-gal [20]. Then, the cells were photographed under a light microscope (Olympus, CKX41).
MEFs suspended in 0.35% low-melting agarose (Sigma-Aldrich, #A4018) in a culture medium were overlaid on top of 0.5% agarose in 6-well plates at a density of 2.5 × 104 cells per well. After 4 to 5 weeks, foci were photographed under a light microscope (Olympus, CKX41), and the total number of foci per well was counted [22].
Kruskal-Wallis tests were carried out for the entire dataset, followed by Wilcoxon rank-sum tests for specific pairs of groups of interest. The RStudio program (https://www.rstudio.com accessed on 14 July 2019) was used for the statistical analyses. A p-value < 0.05 was considered statistically significant [22].
Overexpression of DNAJA4, DNAJB11, and DNAJC10 prevents oncogene-induced senescence (OIS)
To investigate the cellular functions of DNAJA4, DNAJB11, and DNAJC10 under strong mitogenic signals, we generated retroviruses expressing H-RASV12 (vr-H-RASV12) and the three DNAJs (vr-DNAJs) and infected them into actively proliferating normal mouse embryonic fibroblasts (MEFs).
When MEFs were infected with vr-H-RASV12, it was observed that the cells stopped proliferating and the proportion of SA-β-gal-stained cells increased, showing that H-RASV12 expression in MEFs results in senescence as reported [11]. In contrast, when MEFs were co-infected with vr-H-RASV12 and each vr-DNAJ, it was observed that the cells proliferated normally and the proportion of SA-β-gal-stained cells was restored to the basal level (Fig. 2A–F). These results suggest that overexpression of the three DNAJs prevents OIS under H-RASV12 expression.
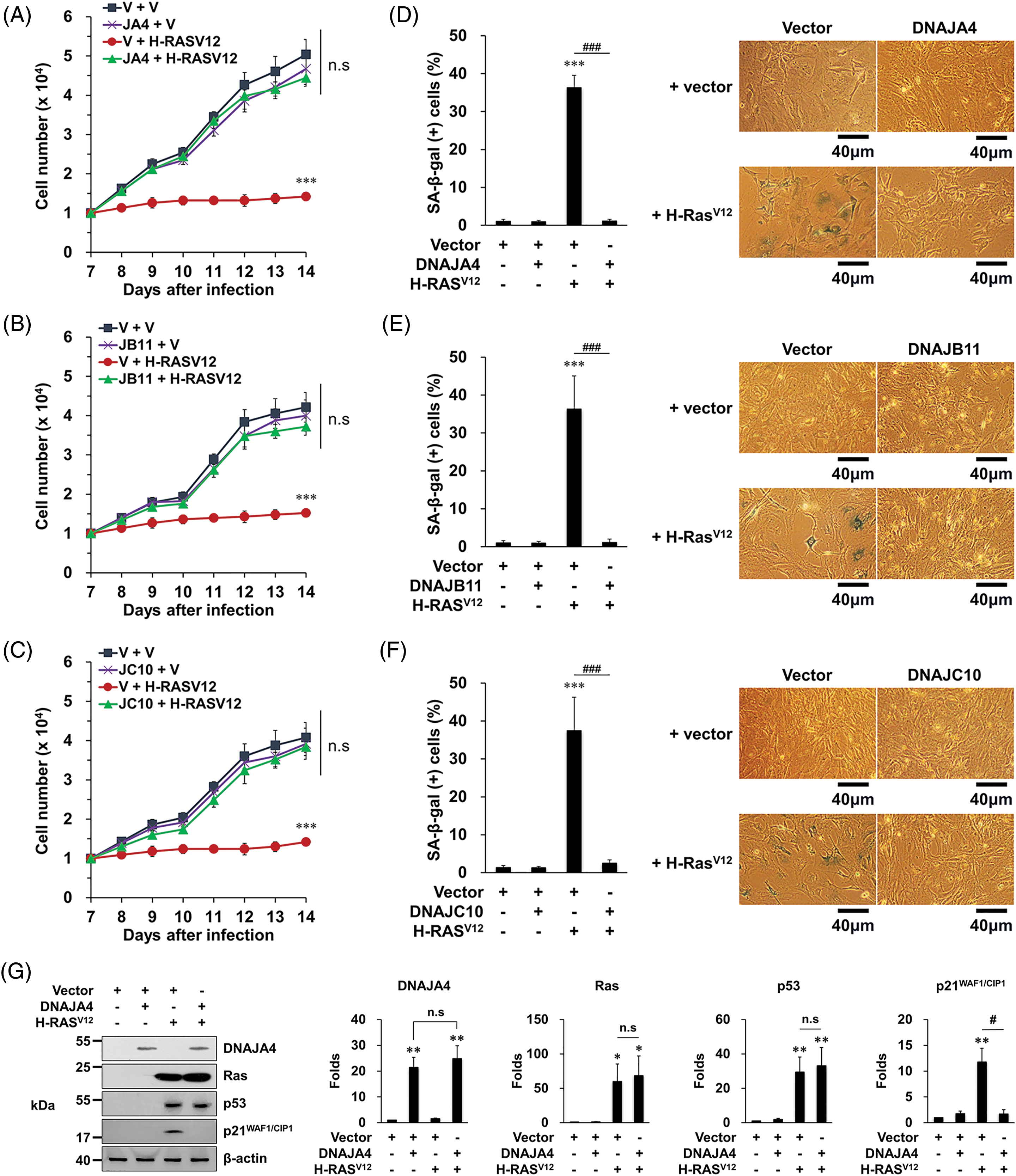
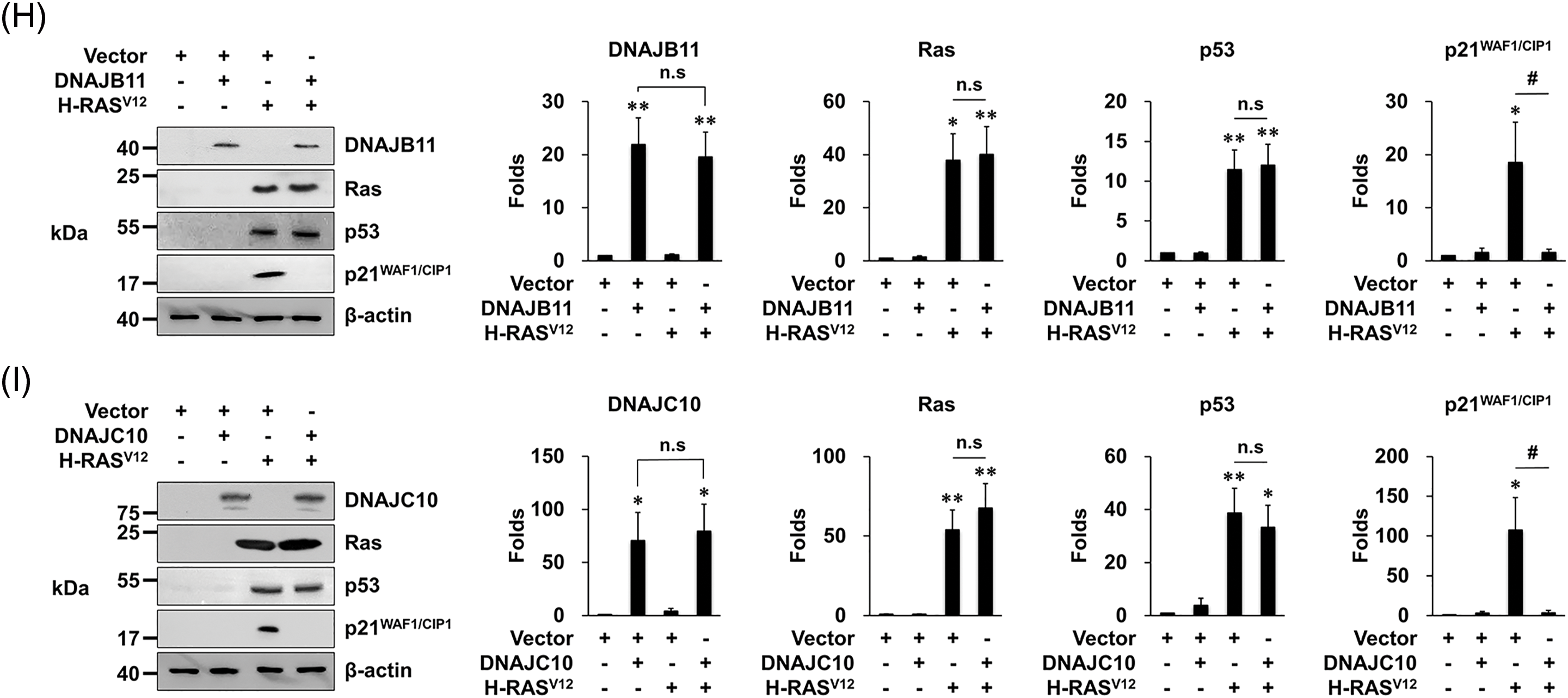
Figure 2: Overexpression of DNAJA4, DNAJB11, and DNAJ10 prevents senescence induced by H-RASV12 in MEFs. (A–C) Cells were transfected with the indicated genes using retroviruses. Then, 1 × 104 cells were seeded on 24-well plates, and the number of cells was counted daily. Data are presented as means ± SD (n = 6; ***p < 0.001 vs. Vector + vector; n.s, not significant at the final day). (D–F) Cells were transfected with the indicated genes using retroviruses. At 14 days after the infection, the cells were incubated with SA-β-gal staining solution, and the percentage of stained cells was calculated. Data are presented as means ± SD (n = 8; ***p < 0.001 vs. Vector; ###p < 0.001, Vector + H-RASV12 vs. each DNAJ + H-RASV12). Scale bars = 40 μm. (G–I) MEFs were transfected with the indicated genes using retroviruses. Western blotting was carried out, and the representative data are shown. β-actin served as a loading control (left panels). The amount of proteins was quantified by densitometry. Data are presented as means ± SE (n = 3; *p < 0.05 and **p < 0.01 vs. Vector; #p < 0.05, Vector + H-RASV12 vs. each DNAJ + H-RASV12; n.s, not significant) (right panels).
Overexpression of DNAJA4, DNAJB11, and DNAJC10 prevents OIS by suppressing the senescence function of p53
It is well known that the p53’s senescence function plays a critical role in OIS [8,9,11]. Since our data showed that overexpression of the three DNAJs prevents OIS, we next investigated whether their overexpression could regulate the p53’s senescence function.
When MEFs were infected with vr-H-RASV12, the expression of p53 as well as p21WAF1/CIP1 increased as reported [11]. In contrast, when MEFs were co-infected with vr-H-RASV12 and each vr-DNAJ, the expression of p21WAF1/CIP1, the senescence-mediating target gene of p53, did not increase although the expression of p53 increased (Fig. 2G–I). These results suggest that overexpression of the three DNAJs prevents OIS by suppressing the p53’s senescence function.
To confirm that the three DNAJs suppress the p53’s senescence function, we examined their role in p53-induced senescence. When MEFs were infected with retroviruses expressing p53 (vr-p53), it was observed that the cells stopped proliferating and the proportion of SA-β-gal-stained cells increased, showing that p53 overexpression induces senescence as reported [23]. In contrast, when MEFs were co-infected with vr-p53 and each vr-DNAJ, it was observed that the cells proliferated normally and the proportion of SA-β-gal-stained cells was restored to the basal level (Fig. 3A–F).
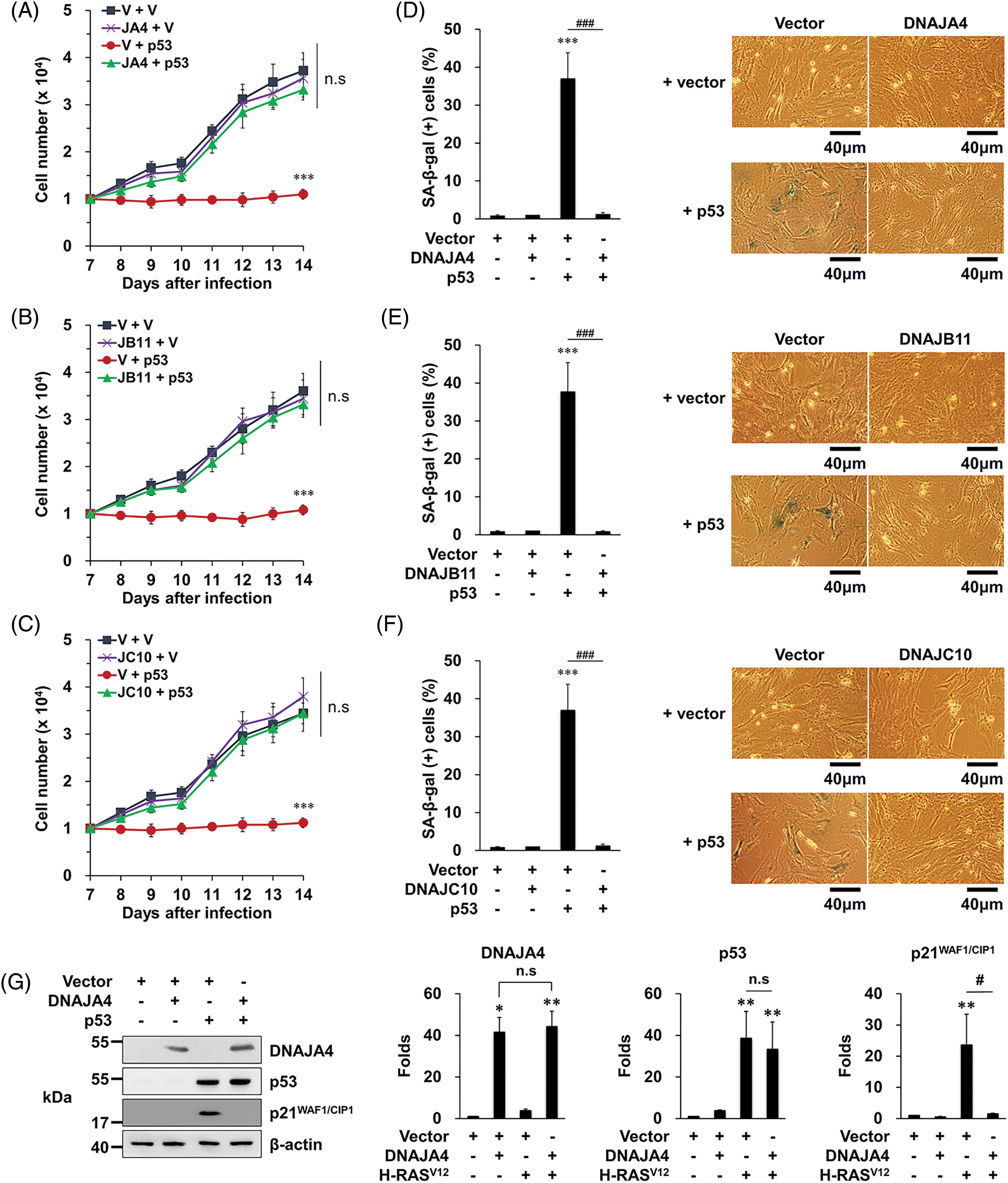
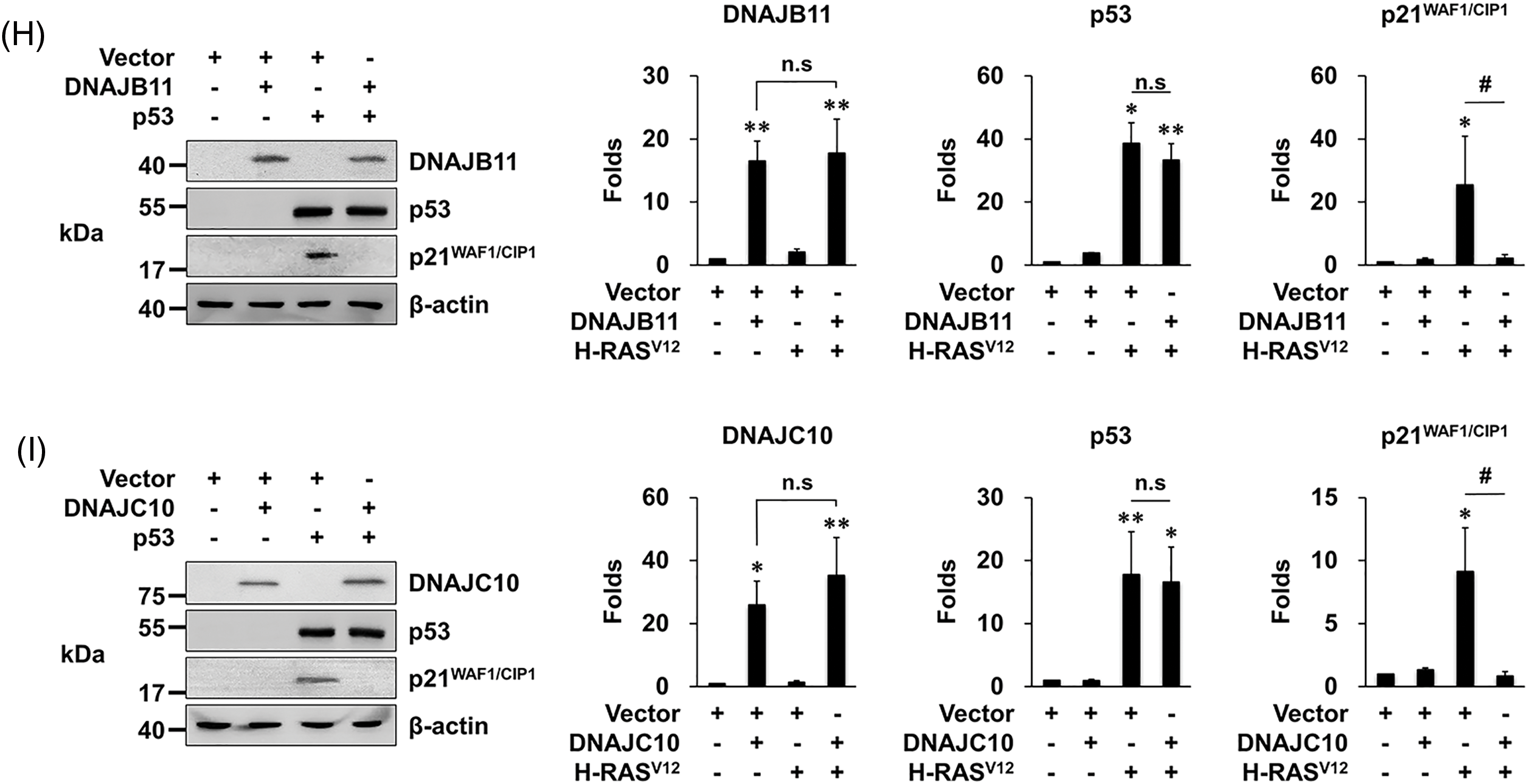
Figure 3: Overexpression of DNAJA4, DNAJB11, and DNAJ10 inhibits p53-induced senescence in MEFs. (A–C) Cells were transfected with the indicated genes using retroviruses. Then, 1 × 104 cells were seeded on 24-well plates, and the number of cells was counted daily. Data are presented as means ± SD (n = 6; ***p < 0.001 vs. Vector + vector; n.s, not significant at the final day). (D–F) Cells were transfected with the indicated genes using retroviruses. At 14 days after the infection, the cells were incubated with SA-β-gal staining solution, and the percentage of stained cells was calculated. Data are presented as means ± SD (n = 6; ***p < 0.001 vs. Vector; ###p < 0.001, Vector + H-RASV12 vs. each DNAJ + H-RASV12). Scale bars = 40 μm. (G–I) MEFs were transfected with the indicated genes using retroviruses. Western blotting was carried out, and the representative data are shown. β-actin served as a loading control (left panels). The amount of proteins was quantified by densitometry. Data are presented as means ± SE (n = 3; *p < 0.05 and **p < 0.01 vs. Vector; #p < 0.05, Vector + H-RASV12 vs. each DNAJ + H-RASV12; n.s, not significant) (right panels).
In addition, when MEFs were infected with vr-p53, the expression of p21WAF1/CIP1 increased as expected, which was almost completely prevented by overexpression of the three DNAJs (Fig. 3G–I). These results demonstrate that the three DNAJs suppress the p53’s senescence function, supporting our conclusion that overexpression of the three DNAJs prevents OIS by suppressing the p53’s senescence function.
Overexpression of DNAJA4, DNAJB11, and DNAJC10 induces neoplastic transformation in MEFs under H-RASV12 expression
It is well known that the escape from OIS contributes to cell transformation [3,4]. Since our data showed that overexpression of the three DNAJs prevents OIS, we next investigated whether their overexpression could regulate cell transformation.
When MEFs were infected with vr-H-RASV12 alone, it was observed that the cells barely formed foci on the soft agar. In contrast, when MEFs were co-infected with vr-H-RASV12 and each vr-DNAJ, it was observed that the cells easily formed foci on the soft agar (Fig. 4). These results suggest that overexpression of the three DNAJs induces neoplastic transformation in normal murine cells by preventing OIS.
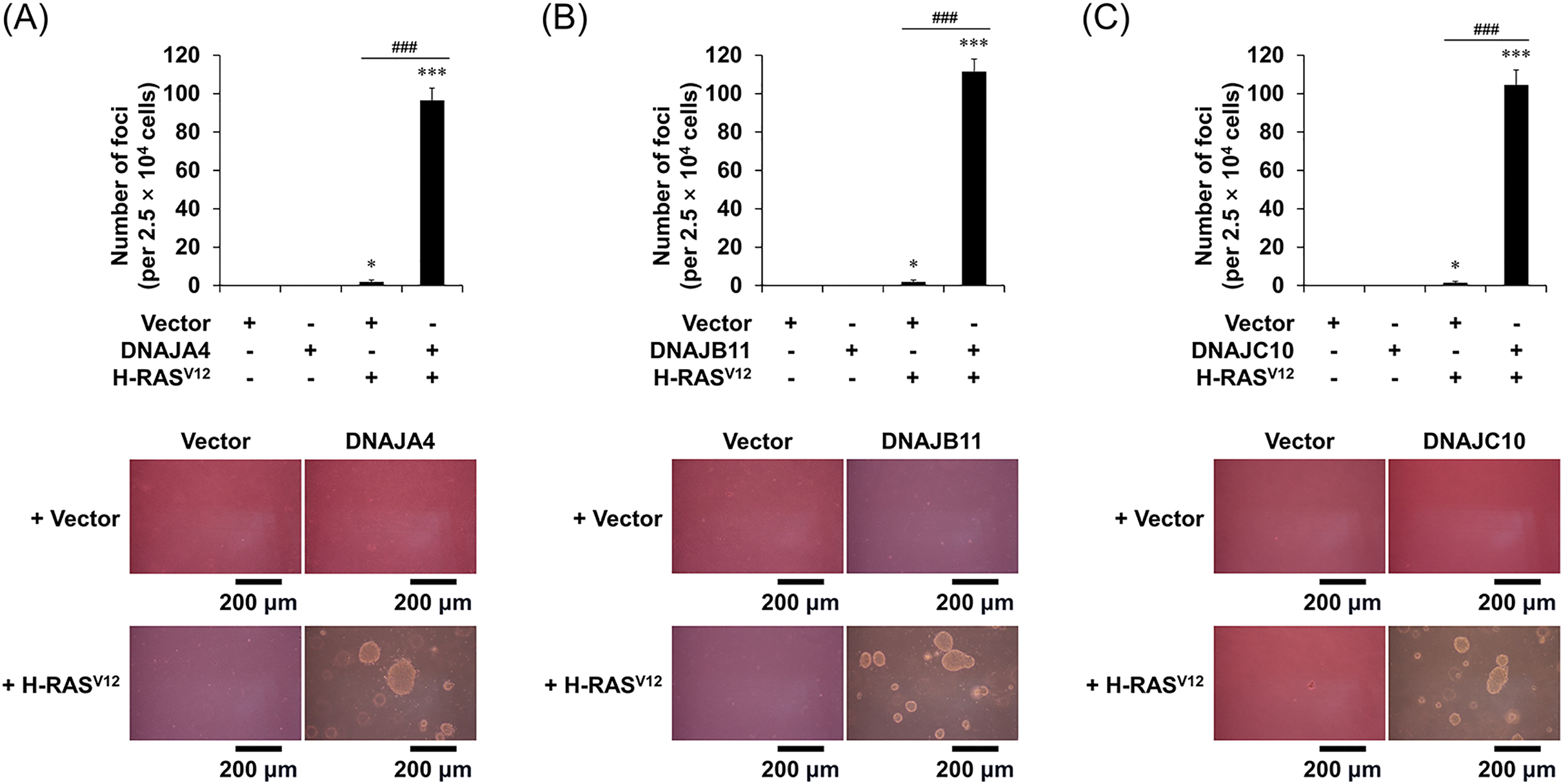
Figure 4: Overexpression of DNAJA4, DNAJB11, and DNAJ10 induces neoplastic transformation under H-RASV12 expression in MEFs. (A–C) Soft agar assays. Cells were transfected with the indicated genes using retroviruses, and 2.5 × 104 cells were overlaid on top of the soft agar. After 4–5 weeks, the total number of foci was counted. Data are presented as means ± SD (n = 6; *p < 0.05, ***p < 0.001 vs. Vector; ###p < 0.001, Vector + H-RASV12 vs. each DNAJ + H-RASV12) (upper panels). Representative data are shown. Scale bars = 200 μm (lower panels).
DNAJA4, DNAJB11, and DNAJC10 are frequently overexpressed in various human cancers
Cellular transformation is the basis for tumorigenesis at the individual level [1,2]. Since our data showed that overexpression of the three DNAJs contributes to cell transformation, we next examined their expression in various human cancers.
For this purpose, we downloaded the RNA sequencing data from the TCGA database [24], and compared the expression of the three DNAJs between cancer tissues and surrounding normal tissues. The results showed that expression of the three DNAJs was frequently increased in a variety of cancer tissues compared to their matched normal tissues with a fold-change ≥ 2.0 (Fig. 5A–C). These results suggest that aberrant overexpression of the three DNAJs may contribute to human tumorigenesis by promoting cell transformation.
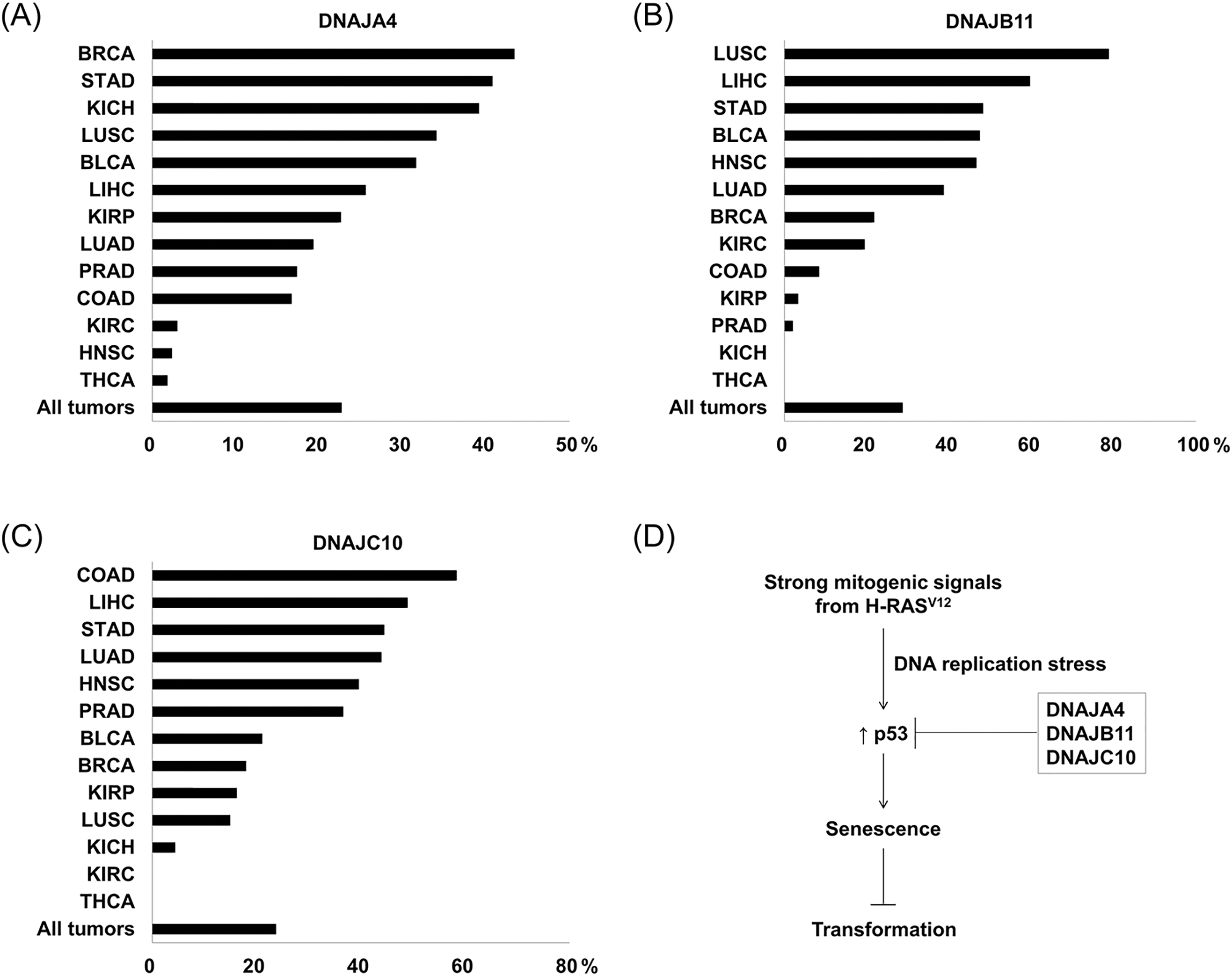
Figure 5: DNAJA4, DNAJB11, and DNAJ10 are frequently overexpressed in various human cancers. (A–C) The percentage of patients with a fold-change ≥ 2.0 in cancer tissues compared to normal tissues. Expression of the three DNAJs was analyzed in cancer tissues and their matched normal tissues when the number of patients was more than 10. BLCA, bladder cancer (n = 19); BRCA, breast cancer (n = 106); COAD, colorectal adenocarcinoma (n = 12); HNSC, head and neck squamous cell carcinoma (n = 43); KICH, kidney chromophobe (n = 23); KIRC, kidney renal clear cell carcinoma (n = 67); KIRP, kidney renal papillary cell carcinoma (n = 31); LIHC, liver hepatocellular carcinoma (n = 47); LUAD, lung adenocarcinoma (n = 57); LUSC, lung squamous cell carcinoma (n = 47); PRAD, prostate adenocarcinoma (n = 52); STAD, stomach adenocarcinoma (n = 27); THCA, thyroid carcinoma (n = 56). (D) A proposed mechanism for the three DNAJs-mediated cell transformation.
Cellular transformation from a normal cell to a malignant cell is the basis for tumorigenesis, and this transformation is known to result from aberrant activation or inactivation of associated genes [1–5]. Therefore, identifying transformation-associated genes and elucidating their action mechanisms are essential for understanding the mechanisms of cell transformation and tumorigenesis. In this context, we here identified DNAJA4, DNAJB11, and DNAJC10 as pro-transforming genes whose aberrant overexpression contributes to cell transformation. In addition, we elucidated their action mechanisms: they suppress the senescence function of p53, thereby preventing oncogene-induced senescence (OIS) and inducing cell transformation (Fig. 5D).
DNAJs, the J domain-containing proteins, are well known to be co-chaperones for Hsp70s [12,14]. However, the cellular functions of DNAJs remain largely unexplored. In this context, we here found novel functions of DNAJA4, DNAJB11, and DNAJC10 under strong mitogenic signals as follows: First, they are pro-transforming factors, as evidenced by the results that they induced neoplastic transformation in normal cells under oncogenic RAS activation (Fig. 4). Second, they are anti-senescence factors. They inhibited H-RASV12-induced senescence as well as p53-induced senescence (Figs. 2 and 3). Third, they are p53 inactivators. They inhibited the p53/p21WAF1/CIP1 pathway as well as p53-induced senescence (Figs. 2 and 3).
To our knowledge, there had been no direct evidence that the three DNAJs regulate cell transformation. Here, we presented both direct and indirect evidence that the three DNAJs contribute to cell transformation. First, they induced neoplastic transformation in normal cells under oncogenic RAS activation. Second, they inhibited OIS, a strong barrier against cell transformation. Third, they inhibited p53, a potent anti-transforming factor.
As for senescence, it has been reported that the proportion of SA-β-gal-stained cells increased in DNAJC10(−/−) MEFs compared to DNAJC10(+/−) MEFs [25]. This result is compatible with our conclusion that DNAJC10 is an anti-senescence factor.
Most pro-transforming genes identified so far are genes that mediate cell proliferation. For example, H-RAS, K-RAS, CCNE, CDK4, or c-MYC are signal transducers, cell cycle regulators, or transcription factors involved in cell division. Meanwhile, some genes are known to have other cellular activities. For example, B-cell leukemia/lymphoma 2 (BCL2) inhibits apoptosis, and mouse double minute 2 homolog (MDM2) or cyclooxygenase-2 (COX-2) inactivates p53 [26–28]. Here, we found that DNAJA4, DNAJB11, and DNAJC10 suppress the p53’ senescence function, thereby preventing OIS and inducing cell transformation. These results suggest that the three DNAJs are pro-transforming genes belonging to the group of p53 inactivators.
At present, it is not clear how DNAJA4, DNAJB11, and DNAJC10 suppress the p53’ senescence function. However, considering the protein-binding properties of DNAJs, it is likely that the three DNAJs might inactivate p53 by binding to it. Additional studies are needed to determine this possibility.
It has been reported that DNAJA4 was overexpressed in acute myeloid leukemia [29], whereas its expression was decreased in nasopharyngeal cancer [30]. DNAJB11 has been reported to be overexpressed in oral cavity, liver, and pancreatic cancer [31–33]. DNAJC10 was overexpressed in liver cancer and glioma [34,35], whereas its expression was reported to be decreased in breast cancer [36]. According to our data, DNAJA4 was overexpressed in more than 10% of patients with breast, stomach, kidney, lung, bladder, liver, prostate, and colon cancer. DNAJB11 was overexpressed in more than 10% of patients with lung, liver, stomach, bladder, head and neck, breast, and kidney cancer. DNAJC10 was overexpressed in more than 10% of patients with colorectal, liver, stomach, head and neck, prostate, bladder, breast, kidney, and lung cancer (Fig. 5A–C). Although all these data show the expression patterns of the three DNAJs in various human cancers, they do not establish a causal relationship between the three DNAJs and human tumorigenesis. Therefore, further studies are required to determine whether the three DNAJs are involved in human tumorigenesis.
Collectively, we here found that DNAJA4, DNAJB11, and DNAJC10 are pro-transforming genes and elucidated their action mechanisms (Fig. 5D). This study may help understand the mechanisms of cell transformation and the cellular functions of the three DNAJs.
Acknowledgement: None.
Funding Statement: This research was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2016R1A2B4012817) and the research grant of Kangwon National University in 2022.
Author Contributions: Study conception and design: Jeong A. Han; data collection: Hyeon Ju Lee, Chang Seop Lee, Si Hoon Kim, Sookyung Kim, Jeong Mi Kim, Seung-Pyo Hong, Hyunjung Lee; analysis and interpretation of results: Sun-Wha Im, Yu-Jin Jung, Jong-Il Kim; draft manuscript preparation: Jeong A. Han. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: This study was conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Kangwon National University (Approval No. KW-140521-1).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi:10.1016/S0092-8674(00)81683-9. [Google Scholar] [PubMed] [CrossRef]
2. Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi:10.1158/2159-8290.CD-21-1059. [Google Scholar] [PubMed] [CrossRef]
3. Akagi T. Oncogenic transformation of human cells: shortcomings of rodent model systems. Trends Mol Med. 2004;10(11):542–8. doi:10.1016/j.molmed.2004.09.001. [Google Scholar] [PubMed] [CrossRef]
4. Karst AM, Jones PM, Vena N, Ligon AH, Liu JF, Hirsch MS, et al. Cyclin E1 deregulation occurs early in secretory cell transformation to promote formation of fallopian tube-derived high-grade serous ovarian cancers. Cancer Res. 2014;74(4):1141–52. doi:10.1158/0008-5472.CAN-13-2247. [Google Scholar] [PubMed] [CrossRef]
5. Doha ZO, Sears RC. Unraveling MYC’s role in orchestrating tumor intrinsic and tumor microenvironment interactions driving tumorigenesis and drug resistance. Pathophysiology. 2023;30(3):400–19. doi:10.3390/pathophysiology30030031. [Google Scholar] [PubMed] [CrossRef]
6. Engler M, Fidan M, Nandi S, Cirstea IC. Senescence in RASopathies, a possible novel contributor to a complex pathophenoype. Mech Ageing Dev. 2021;194:111411. doi:10.1016/j.mad.2020.111411. [Google Scholar] [PubMed] [CrossRef]
7. Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27(20):2801–9. doi:10.1038/sj.onc.1210950. [Google Scholar] [PubMed] [CrossRef]
8. Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–7. doi:10.1038/nature05268. [Google Scholar] [PubMed] [CrossRef]
9. Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–42. doi:10.1038/nature05327. [Google Scholar] [PubMed] [CrossRef]
10. Patil MR, Bihari A. A comprehensive study of p53 protein. J Cell Biochem. 2022;123(12):1891–937. doi:10.1002/jcb.30331. [Google Scholar] [PubMed] [CrossRef]
11. Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. [Google Scholar] [PubMed]
12. Braun JEA. Extracellular chaperone networks and the export of J-domain proteins. J Biol Chem. 2023;299(2):102840. [Google Scholar] [PubMed]
13. Kampinga HH, Craig EA. The HSP70 chaperone machinery: j proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–92. [Google Scholar] [PubMed]
14. Karunanayake C, Page RC. Cytosolic protein quality control machinery: interactions of Hsp70 with a network of co-chaperones and substrates. Exp Biol Med. 2021;246(12):1419–34. [Google Scholar]
15. Hafizur RM, Yano M, Gotoh T, Mori M, Terada K. Modulation of chaperone activities of Hsp70 and Hsp70-2 by a mammalian DnaJ/Hsp40 homolog, DjA4. J Biochem. 2004;135(2):193–200. [Google Scholar] [PubMed]
16. Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16(1):40–50. doi:10.1091/mbc.e04-05-0434. [Google Scholar] [PubMed] [CrossRef]
17. Ushioda R, Hoseki J, Araki K, Jansen G, Thomas DY, Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321(5888):569–72. doi:10.1126/science.1159293. [Google Scholar] [PubMed] [CrossRef]
18. Daverkausen-Fischer L, Pröls F. Dual topology of co-chaperones at the membrane of the endoplasmic reticulum. Cell Death Discov. 2021;7(1):203. doi:10.1038/s41420-021-00594-x. [Google Scholar] [PubMed] [CrossRef]
19. Purazo ML, Ice RJ, Shimpi R, Hoenerhoff M, Pugacheva EN. NEDD9 overexpression causes hyperproliferation of luminal cells and cooperates with HER2 oncogene in tumor initiation: a novel prognostic marker in breast cancer. Cancers. 2023;15(4):1119. doi:10.3390/cancers15041119. [Google Scholar] [PubMed] [CrossRef]
20. Lee HJ, Jung YJ, Lee S, Kim JI, Han JA. DNAJB9 inhibits p53-dependent oncogene-induced senescence and induces cell transformation. Mol Cells. 2020;43(4):397–407. [Google Scholar] [PubMed]
21. Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci U S A. 1996;93(21):11400–6. [Google Scholar] [PubMed]
22. Lee HJ, Kim SR, Jung YJ, Han JA. Cyclooxygenase-2 induces neoplastic transformation by inhibiting p53-dependent oncogene-induced senescence. Sci Rep. 2021;11(1):9853. [Google Scholar] [PubMed]
23. Ferbeyre G, de Stanchina E, Lin AW, Querido E, McCurrach ME, Hannon GJ, et al. Oncogenic ras and p53 cooperate to induce cellular senescence. Mol Cell Biol. 2002;22(10):3497–508. [Google Scholar] [PubMed]
24. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. [Google Scholar] [PubMed]
25. Yamashita R, Fujii S, Ushioda R, Nagata K. Ca2+ imbalance caused by ERdj5 deletion affects mitochondrial fragmentation. Sci Rep. 2021;11(1):20772. [Google Scholar] [PubMed]
26. Rahman N, Khan H, Zia A, Khan A, Fakhri S, Aschner M, et al. Bcl-2 modulation in p53 signaling pathway by flavonoids: a potential strategy towards the treatment of cancer. Int J Mol Sci. 2021;22(21):11315. doi:10.3390/ijms222111315. [Google Scholar] [PubMed] [CrossRef]
27. Timmerman DM, Remmers TL, Hillenius S, Looijenga LHJ. Mechanisms of TP53 pathway inactivation in embryonic and somatic cells-relevance for understanding (germ cell) tumorigenesis. Int J Mol Sci. 2021;22(10):5377. doi:10.3390/ijms22105377. [Google Scholar] [PubMed] [CrossRef]
28. Choi EM, Kim SR, Lee EJ, Han JA. Cyclooxygenase-2 functionally inactivates p53 through a physical interaction with p53. Biochim Biophys Acta. 2009;1793(8):1354–65. doi:10.1016/j.bbamcr.2009.05.006. [Google Scholar] [PubMed] [CrossRef]
29. Zhang H, Xue F, Zhao H, Chen L, Wang T, Wu X. DNA methylation status of DNAJA4 is essential for human erythropoiesis. Epigenomics. 2022;14(20):1249–67. doi:10.2217/epi-2022-0341. [Google Scholar] [PubMed] [CrossRef]
30. Zhang Q, Feng P, Zhu XH, Zhou SQ, Ye ML, Yang XJ, et al. DNAJA4 suppresses epithelial-mesenchymal transition and metastasis in nasopharyngeal carcinoma via PSMD2-mediated MYH9 degradation. Cell Death Dis. 2023;14(10):697. doi:10.1038/s41419-023-06225-w. [Google Scholar] [PubMed] [CrossRef]
31. Kaida A, Iwakuma T. Regulation of p53 and cancer signaling by heat shock protein 40/J-domain protein family members. Int J Mol Sci. 2021;22(24):13527. doi:10.3390/ijms222413527. [Google Scholar] [PubMed] [CrossRef]
32. Pan J, Cao D, Gong J. The endoplasmic reticulum co-chaperone ERdj3/DNAJB11 promotes hepatocellular carcinoma progression through suppressing AATZ degradation. Future Oncol. 2018;14(29):3001–13. doi:10.2217/fon-2018-0401. [Google Scholar] [PubMed] [CrossRef]
33. Liu P, Zu F, Chen H, Yin X, Tan X. Exosomal DNAJB11 promotes the development of pancreatic cancer by modulating the EGFR/MAPK pathway. Cell Mol Biol Lett. 2022;27(1):87. doi:10.1186/s11658-022-00390-0. [Google Scholar] [PubMed] [CrossRef]
34. Cunnea P, Fernandes AP, Capitanio A, Eken S, Spyrou G, Björnstedt M. Increased expression of specific thioredoxin family proteins; a pilot immunohistochemical study on human hepatocellular carcinoma. Int J Immunopathol Pharmacol. 2007;20(1):17–24. doi:10.1177/039463200702000103. [Google Scholar] [PubMed] [CrossRef]
35. Liu F, Tu Z, Liu J, Long X, Xiao B, Fang H, et al. DNAJC10 correlates with tumor immune characteristics and predicts the prognosis of glioma patients. Biosci Rep. 2022;42(1):BSR20212378. doi:10.1042/BSR20212378. [Google Scholar] [PubMed] [CrossRef]
36. Acun T, Senses KM. Downregulation of DNAJC10 (ERDJ5) is associated with poor survival in breast cancer. Breast Cancer. 2020;27(3):483–9. doi:10.1007/s12282-019-01042-6. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools