 Open Access
Open Access
REVIEW
Mitochondrial-epigenetic crosstalk as an integrative standpoint into gut microbiome dysbiosis and related diseases
1 Department of Structural and Functional Biology, UNESP, São Paulo State University, Institute of Biosciences, Botucatu, São Paulo, 18618-689, Brazil
2 Departamento de Tecnología Médica, Universidad Maimónides, Ciudad Autónoma de Buenos Aires, Buenos Aires, C1405, Argentina
3 Área de Farmacología, Departamento de Patología, Facultad de Ciencias Médicas, Universidad Nacional de Cuyo, Mendoza, 5500, Argentina
4 Instituto de Medicina y Biología Experimental de Cuyo (IMBECU), Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET), Mendoza, 5500, Argentina
* Corresponding Author: WALTER MANUCHA. Email:
(This article belongs to the Special Issue: Gut Microbiota in Human Health: Exploring the Complex Interplay)
BIOCELL 2024, 48(10), 1429-1442. https://doi.org/10.32604/biocell.2024.053478
Received 01 May 2024; Accepted 12 August 2024; Issue published 02 October 2024
Abstract
The interplay between mitochondria, epigenetics, and the microbiota is intricately linked to both health and disease. Within our cells, a complex molecular dance occurs, where these components intertwine in a mesmerizing ballet that plays a decisive role in our health. Mitochondria, beyond being energy powerhouses, modulate nuclear gene expression through messengers like reactive oxidative stress (ROS) and calcium. Epigenetics, acting as the molecular conductor, regulates the expression of both nuclear and mitochondrial genes through modifications like DNA methylation. The intestinal microbiota itself produces short-chain fatty acids (SCFAs) that influence mitochondrial activity. SCFA-induced epigenetic modifications, like histone acetylation, impact mitochondrial function which may lead to disease. Mitochondrial dysfunction generates retrograde signals that alter nuclear gene expression, as evidenced by increased histone H3 lysine 27 acetylation (H3K27ac) in genes essential for neuronal differentiation and mitochondrial reprogramming. Alterations in the mitochondrial-nuclear-microbiota axis are associated with diseases including diabetes, neurodegeneration, and cancer. Modulating the intestinal microbiota with probiotics or prebiotics can restore balance while intervening in mitochondrial pathways, which can be a therapeutic strategy. Additionally, using epigenetic agents like histone deacetylase (HDAC) inhibitors can reprogram gene expression and improve mitochondrial function. Finally, the present review aims to explore the central interplay between mitochondria, epigenetics modifications, and microbiota in a complex and dynamic molecular context that plays a fundamental role in human health. Specifically, it will examine the impact of microbiome components and metabolites generated from normobiosis and dysbiosis on mitochondria and epigenetic modifications across different diseases and metabolic conditions. This integrated understanding of the molecular players and their interactions provides a deeper perspective on how to promote health and potentially combat disease.Keywords
Abbreviations
| 5mC | 5-methylcytosine |
| CoA | acetyl-CoA |
| AFAP1 | Actin Filament-Associated Protein 1 |
| AT | Adipose tissue |
| AREG | Amphiregulin |
| ALX4 | Aristaless-Like Homeobox 4 |
| GABA | γ-aminobutyric acid |
| ASD | Autism spectrum disorder |
| BMI | Body mass index |
| CNS | Central nervous system |
| TCA | Citric acid cycle |
| CoQ10 | Coenzyme Q10 |
| CpG | Cytosine-guanine dinucleotide |
| EZH2 | Enhancer of Zeste Homolog 2 |
| GF | Germ-free |
| GSK3β | Glycogen Synthase Kinase-3 Beta |
| GPCRs | G-protein-coupled receptors |
| GALT | Gut-associated lymphoid tissue |
| HAT | Histone acetyltransferase |
| HDAC | Histone deacetylases |
| HDM | Histone demethylase |
| H3K27ac | Histone H3 Lysine 27 acetylation |
| IPA | Indole-3-propionic acid |
| IFN-γ | Interferon Gamma |
| LSD-1 | Lysine-specific demethylase-1 |
| mtDNA | Mammalian mitochondrial DNA |
| MPC | Mitochondrial pyruvate carrier |
| Tfam | Mitochondrial transcription factor A |
| NPY | Neuropeptide Y |
| Nrf2 | Nuclear factor-erythroid 2-related factor 2 |
| ncRNA | Non-coding RNA |
| NF-κB | Nuclear factor-κB |
| OXPHOS | Oxidative phosphorylation system |
| PD | Parkinson’s disease |
| PENK | Proenkephalin |
| ROS | Reactive oxidative stress |
| SAM | S-adenosyl methionine |
| SEPT9 | Septin 9 |
| SCFAs | Short-chain fatty acids |
| SFRP1,2,3 | Secreted Frizzled Related Protein 1, 2, and 3 |
| NaBt | Sodium butyrate |
| TCA | Tricarboxylic acid cycle |
| T2D | Type 2 diabetes |
| VN | Vagus nerve |
| WIF1 | WNT Inhibitory Factor 1 |
The human body harbors a vast and intricate microbial ecosystem encompassing trillions of microorganisms. This diverse community, predominantly composed of bacteria but also including archaea, viruses, and eukaryotes, flourishes within the gastrointestinal tract, reaching its peak density in the intestine. It is crucial to distinguish between “microbiota” and “microbiome”; the former refers to the assemblage of microbes themselves, while the latter encompasses their collective genetic makeup [1]. The gut microbiota plays a pivotal role in shaping the host’s health status. Its composition, characterized by various species’ abundance, dominance, and viability, exhibits dynamism and is susceptible to dietary choices, lifestyle factors, and environmental influences [2]. These microbial populations’ richness and diversity are essential for maintaining overall well-being.
Recent advancements have unveiled the gut microbiome’s centrality in a multitude of physiological processes, underscoring its significance in human health. It contributes significantly to food digestion, nutrient absorption and utilization, and the biosynthesis of vital molecules such as vitamins (K and B complex), proteins, and bile acids. Notably, the gut microbiome is responsible for the generation of short-chain fatty acids (SCFAs), which exert regulatory effects on host physiology [2]. Furthermore, this microbial community facilitates the breakdown of dietary polysaccharides that would otherwise remain undigested. The gut microbiota is also the main antigenic stimulus responsible for the development, activation, and function of gut-associated lymphoid tissue (GALT) [3] which modulates innate and mucosal immunity, thereby influencing gut barrier function, gut motility, and the suppression of pathogenic microbial expansion [2].
Intriguingly, recent studies suggest that the gut microbiome can extend its influence beyond the gut, regulating host gene expression, circadian rhythms, drug efficacy and toxicity, and even aspects of the central nervous system (CNS) [4]. As reviewed by Dicks [5], gut bacteria synthesize neurotransmitters such as dopamine, histamine, serotonin, norepinephrine, and γ-aminobutyric acid (GABA), to communicate with the CNS through afferent vagus nerve (VN) fibers. In response, via VN efferent fibers, the brain sends signals back to enteroendocrine and enterochromaffin cells in the intestinal epithelium, and to mucosal immune system cells to improve the integrity of the gut wall and inhibit the release of pro-inflammatory cytokine to reduce peripheral inflammation, keeping the gut microbiome in a balanced state.
This interplay between the gut microbiota, and the host fosters cooperation and functional stability within the gut ecosystem, aptly termed the holobiont [6]. This concept underscores the intimate and dynamic relationship between the host and its associated microbiota, highlighting its indispensable role in host physiology and health [6]. The holobiont framework challenges the notion of the gut microbiota as a mere collection of independent microbes, instead portraying it as a complex system characterized by coevolution and reciprocal interactions between the host and its microbial inhabitants. For instance, the gut microbiota can influence the expression of host genes, while host genetics can shape the composition and function of the microbiota. The microbiota potentially affects host behavior, social interactions, and even the evolution of the host species [7].
There is a remarkable interindividual variability in the human gut microbiota. Each individual has a diversity of bacteria to perform the same biological function, this functional redundancy is exhibited by robust microbiotas [8]. The establishment of this diversity begins early in life and changes by various factors such as gestational age, mode of delivery, infant feeding practices, weaning, and exposure to environmental factors (e.g., antibiotics). While the gut microbiota composition exhibits a degree of stability in adulthood, factors like body mass index (BMI), lifestyle choices, exercise frequency, ethnicity dietary, and cultural habits [9] (Fig. 1).
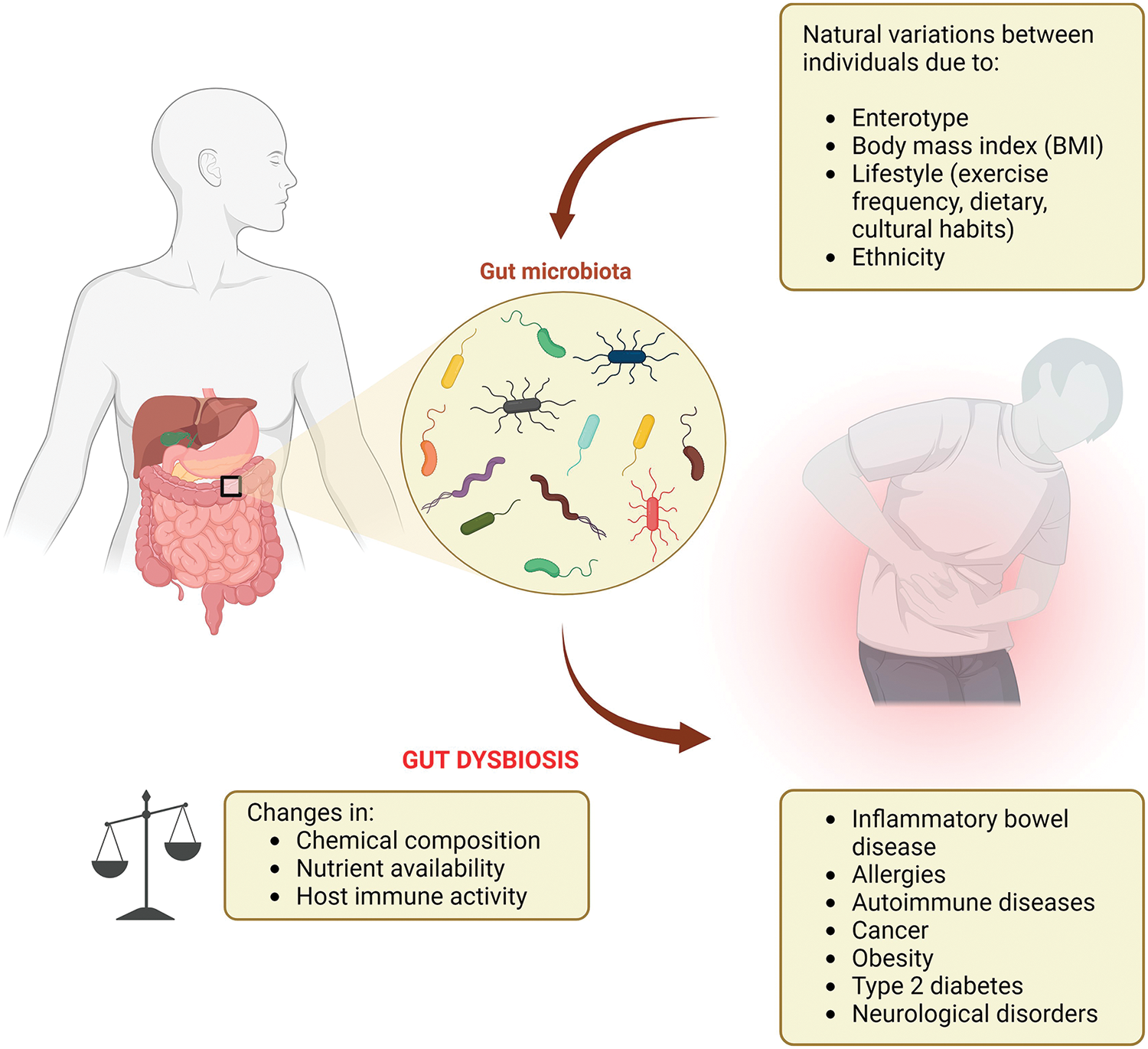
Figure 1: Gut microbiota, dysbiosis, and the association with human diseases.
Particularly regarding ethnicity, it can be seen as a manifestation of multiple lifestyle factors, which have a low effect singly, but collectively are significant to result in a unique profile of gut microbiota that distinguishes ethnic groups [10]. The microbiota can also be changed by a host’s social environment as microbes can be transmitted through different social contact [11]. Consequently, there is no single “optimal” gut microbiota composition, as it is unique to everyone, but can be considered stable if, after a disturbance, has resistance to change its composition, resilience to restore the initial composition, and functional redundancy to recover the initial function despite compositional changes [8].
The human gastrointestinal tract presents a spectrum of physiological variations along its length, encompassing the small intestine and colon. These variations are characterized by gradients in chemical composition nutrient availability, and compartmentalized host immune activity, all known to influence bacterial community composition. Microbial cell populations are most abundant within the gastrointestinal tract, with lower abundances observed in the oral cavity and stomach, potentially attributed to the harsher acidic environment in these regions [12,13]. The number of microorganisms progressively increases along the small intestine, reaching a peak in the colon, where the low oxygen tension (O2) favors the establishment of a dense anaerobic microbial community [12,13].
Disruption or alteration in the composition and function of the gut microbiota, known as gut dysbiosis, can have significant consequences for both intestinal and extra-intestinal health [14] (Fig. 1). The potential implications of gut dysbiosis are vast and alarming, spanning a broad spectrum of diseases, including inflammatory bowel disease, irritable bowel syndrome, allergies, autoimmune diseases, obesity, type 2 diabetes (T2D), and even neurological disorders (Fig. 1). These understandings may offer insights into potential therapeutic strategies for modulating the mitochondrial-nuclear-microbiota axis to promote health while avoiding these afflictive diseases.
While the precise mechanisms underlying the link between gut dysbiosis and disease remain under investigation, several hypotheses have been proposed. One theory suggests that an altered microbiota can trigger an inappropriate immune response, leading to chronic inflammation and tissue damage [15–17]; this disruption is common in inflammatory bowel diseases, autoimmune diseases, and even cancer. Another hypothesis posits that dysbiosis can disrupt nutrient metabolism and energy storage, potentially contributing to the development of obesity and T2D [18,19]. Furthermore, dysbiosis has been associated with alterations in the gut-brain axis, which may play a role in developing neurological disorders [20,21]. Gut microbiota, mitochondria, and neurodegeneration have been recently explored concerning Alzheimer’s disease, amyotrophic lateral sclerosis, Huntington’s disease [22,23], and age-related cognitive decline [24,25]. This review will explore the impact of microbiome components and microbiome-derived metabolites on mitochondrial metabolism and epigenetic regulation across various pathological contexts.
Altered Gut Microbiome and Epigenetics: Crosslink with Potential Diseases
Emerging evidence underscores a fascinating bidirectional communication between the gut microbiome and the epigenome. Epigenetics encompasses a range of stimuli that induce alterations in gene expression patterns without modifying the underlying DNA sequence itself. These alterations can be transient or even persist across generations. Fundamental epigenetic mechanisms include DNA methylation, histone modifications, non-coding RNA (ncRNA) regulation, and chromosomal remodeling [22,26].
DNA methylation involves the enzymatic addition of a methyl group to a cytosine residue within the DNA sequence, mediated by DNA methyltransferases and requiring S-adenosylmethionine as a cofactor. This modification primarily targets cytosines within a cytosine-guanine dinucleotide context (CpG). The conversion transforms cytosine to 5-methylcytosine (5mC), and CpG-rich regions, known as CpG islands, are scattered unevenly throughout the genome. DNA methylation can repress gene expression by hindering the binding of transcription factors to the DNA. DNA methylation patterns are generated during development and often exhibit tissue specificity [24]. Disruptions in these patterns have been linked to a variety of diseases, including cancer, neurological disorders, and cardiovascular disease [27].
Histone modification refers to alterations in the proteins that package DNA. Post-translational modifications of histones, such as acetylation, methylation, sumoylation, phosphorylation, etc., can influence chromatin structure and potentially impact gene transcription [28,29]. These modifications can alter chromatin accessibility and thereby regulate gene expression. Similar to DNA methylation, histone modifications occur along with development and can be influenced by environmental factors. Notably, dysregulation of histone modifications has been implicated in various diseases, including cancer, neurological disorders, and inflammaging-associated hypertension [30–32]. Histone methylation significantly influences mitochondrial function through crosstalk between the nucleus and mitochondria. This modification involves the addition of methyl groups to histone proteins, primarily on lysine and arginine residues, altering chromatin structure and gene expression. Dysbiosis-associated changes in histone methylation can lead to functional implications such as altering mitochondrial biogenesis, cellular energy homeostasis, and cellular health, thus contributing to the pathogenesis of various diseases.
The ncRNA regulation involves the modulation of gene expression by RNAs lacking protein-coding capacity. These ncRNAs function as inhibitors of gene expression by binding to messenger RNA, thereby preventing their translation into proteins. Conversely, ncRNAs can enhance gene expression by interacting with DNA and influencing chromatin structure [33]. Perturbations in ncRNA regulation have been associated with various diseases, including cancer and cardiovascular disease [34,35].
More importantly, both dysregulation of the gut microbiome and epigenetic modifications have been implicated in various diseases, including obesity [36], inflammatory bowel disease [37], and cancer [38]. Microbial metabolites mediate epigenetic changes that contribute to metabolic disorders by affecting intestinal permeability, immune and inflammatory responses, and insulin resistance; these metabolites can also lead to dysbiosis in the microbiota by directly interacting with G-protein-coupled receptors (GPCRs) [39]. Thus, diet, the gut microbiota, and epigenetics changes may be of significant value when considering the development of metabolic diseases like others (Fig. 2). The potential of deciphering the mechanisms underlying this communication is immense, as it may pave the way for the development of innovative strategies for preventing and treating these conditions.
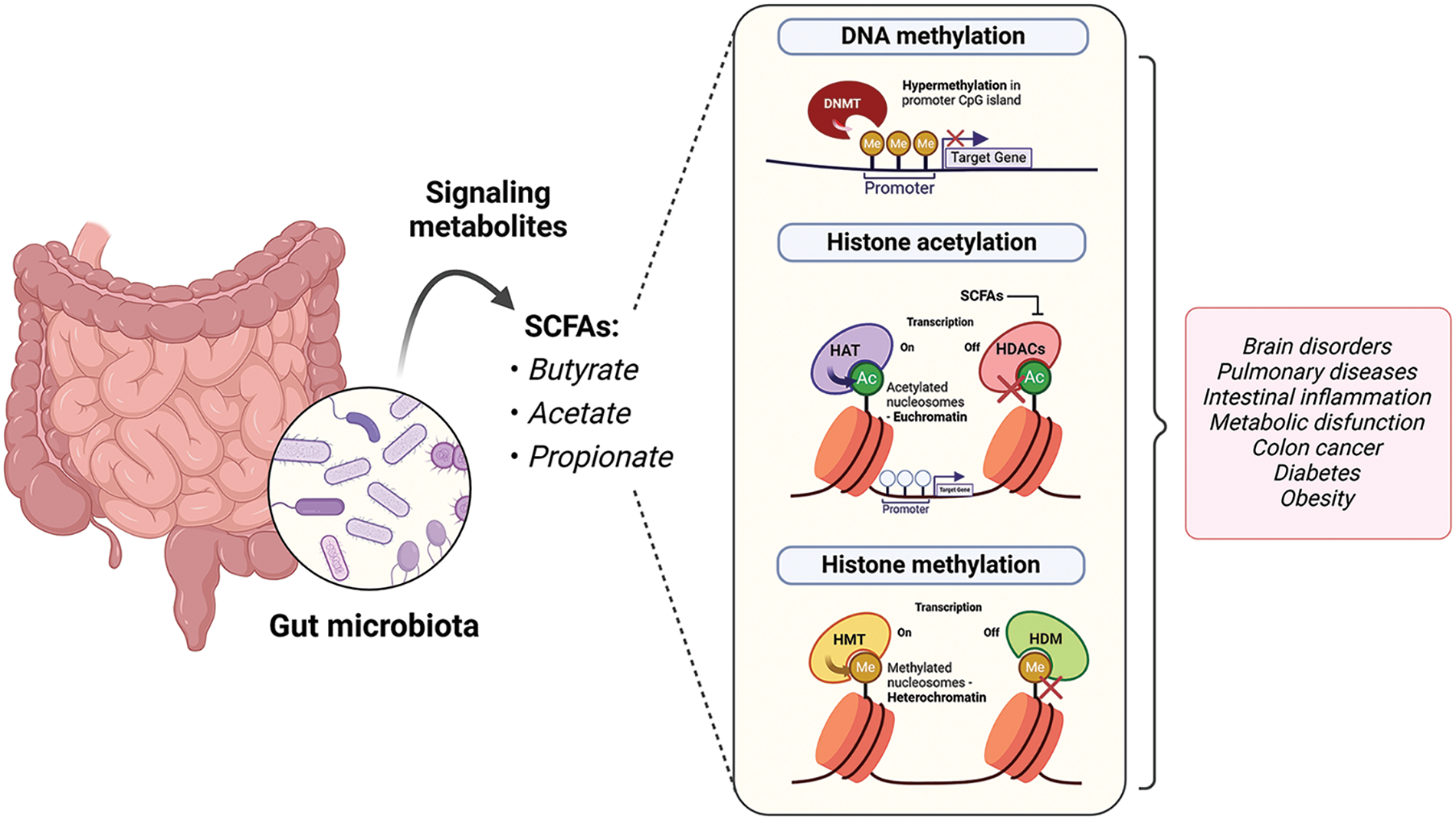
Figure 2: Intestinal microbiota and SCFAs can affect the epigenome through different ways of action by interacting with epigenetic substrates or chromatin-modifying enzymes. These regulations have been documented in multiple diseases. Acetyl (Ac); DNA methyltransferase (DNMT); histone acetyltransferase (HAT); histone deacetylases (HDACs); histone demethylase (HDM); histone methyltransferase (HMT); methyl (Me); short-chain fatty acids (SCFAs).
A randomized controlled trial involving 69 male and female health volunteers showed that anthocyanin-rich functional ingredients could improve cognitive function, working memory, and eye dryness via modulation of the gut microbiome and histone acetylation [40]. Considering cognitive function, a recent study revealed a potential involvement of Alzheimer’s disease (AD)-associated dysbiosis with the development of cognitive deficits [41]. Using AD transgenic mice or administrating feces of AD transgenic mice to wild-type mice, the researchers identified gut microbiota dysbiosis, Tau phosphorylation, and cognitive impairment in the wild-type mice; all these manifestations were reversed following gavage with Lactobacillus and Bifidobacterium. The epigenetic mechanisms underlying these changes have been attributed to the butyric acid-mediated acetylation of Glycogen Synthase Kinase-3 beta (GSK3β) at lysine 15 which regulated its phosphorylation finally impacting Tau phosphorylation. More recently, Behrens et al. [42] showed an improvement in depressive-like behavior of rats after supplementation with sodium propionate (50 mM). These effects were particularly associated with the modulation of gut bacterial communities and attenuation of the epigenetic dysregulation via histone 3 epigenetic regulation in the brain of stressed animals.
Considering fecal microbiota transplant (FMT), a larger gut microbiota shift was obtained with allogenic FMT, indicating that the introduction of new species can modulate the plasma metabolome and epigenome. For instance, Prevotella amplicon sequence variants correlated with methylation of the Actin Filament-Associated Protein 1 (AFAP1) gene, impacting mitochondrial function, insulin sensitivity, and peripheral insulin resistance, suggesting FMT as promising to ameliorate insulin sensitivity in metabolic syndrome [43].
Recent studies indicate that microbe-derived metabolites can significantly impact host metabolism by serving as epigenetic regulators. Butyrate, a metabolite produced by intestinal bacteria, functions as a histone deacetylase inhibitor and has been documented to enhance memory and learning in animal models. Notably, butyrate and lipopolysaccharide (LPS) generated from transplanted gut microbiota of children with type 1 diabetes (T1D) have protective and deleterious effects, respectively, on pancreatic islet structure and function in a streptozotocin-induced T1D mice model [44]. While LPS enhanced inflammatory response, butyrate activated insulin1 and 2 gene expressions. Diabetic dysbiosis is a novel key factor related to diabetic complications. By administering butyrate to non-obese T2D MKR mice, Noureldein et al. [45] evaluated alterations in colon shortening and inflammation; they observed that dysbiosis associated with T2D is accompanied by reduced bacteroid fragilis and butyrate-forming bacteria, and the inadequate secretion of butyrate alleviated HDAC3 inhibition and altered colon permeability. By contrast, treatment with butyrate restored the normal levels of inflammatory markers and reduced ROS production which may attenuate diabetic complications. Interestingly, colon tissue of diabetic mice showed overexpression of inflammatory cytokines (interleukin (IL)-1β) and NADPH oxidase (NOX)4 which was associated with dysbiosis and lower levels of butyrate-forming bacteria [46]. A cohort study (95 adults) showed that gut microbiome remains stable in individuals during the early stages of diabetic chronic kidney disease with a predominance of phyla Firmicutes and Bacteroidetes [47].
In colon cancer cells, butyrate is more available than glucose and serves as an HDAC inhibitor, thus preventing cell proliferation and inducing apoptosis [48]. Previous reports found that colorectal cancer (CRC)-associated microbiota can enhance the number of hypermethylated genes in murine colonic mucosa compared to health control microbiota; Secreted Frizzled Related Protein 1, 2, and 3 (SFRP1,2,3), Proenkephalin (PENK), Aristaless-Like Homeobox 4 (ALX4), WNT Inhibitory Factor 1 (WIF1), Septin 9 (SEPT9), and Neuropeptide Y (NPY) gene promoters were hypermethylated in CRC in contrast to normal tissue or components of fecal donors and blood methylation levels of Wif1, PENK, and NPY were closely related to CRC dysbiosis [49]. New findings revealed that the CRC microbiome programs DNA methylation of host cells mainly by interfering with methyl donor metabolism [50]. Researchers also showed associations between methylation profiles of CRC-related genes and specific bacterial taxa, emphasizing their role in physiological homeostasis and in contributing to CRC development. In human colonic biopsies, ulcerative colitis and Crohn’s disease have been extensively associated with DNA methylation involving microbiota composition, disease classification, and inflammatory status [51].
The gut-lung axis is another target of investigation in lung disorders. To investigate if gut microbiota contributes to tuberculosis and host immunity, Yang et al. [52] studied pulmonary inflammation in a mouse model of mycobacterium tuberculosis infection. They found a downregulation of gut bacteria-regulated long-non-coding RNA (lncRNA), termed lncRNA-CGB, during dysbiosis while a direct regulator of lncRNA-CGB was Bacteroides fragilis. Importantly, lncRNA-CGB interacted with Enhancer of Zeste Homolog 2 (EZH2) to negatively regulate H3K27 tri-methylation (H3K27Me3) epigenetic programming resulting in Interferon Gamma (IFN-γ) expression and immune protection against tuberculosis.
An interesting study by Cuomo et al. [53] showed that methylation signatures of human host fecal DNA (HFD) are correlated with gut dysbiosis and inflammation in children with autism spectrum disorder (ASD). Particularly, most methylated genes in HFD were associated with inflammatory response and immune cells; this methylation-based algorithm could also be used to explore other brain disorders and intestinal inflammatory diseases. New evidence indicating that paternal microbiome perturbations affect offspring’s parameters (e.g., low birth weight, growth restriction, and premature mortality) is linked to the gut-germline axis where dysbiosis is mostly associated with altered testicular metabolite profiles, impaired leptin signaling, and remapped small RNA payloads in sperm [54]. Using obese wild-type C57BL/6NCrl mice and transgenic mice with an altered sperm epigenome, their descendants were characterized for metabolic profile and the histone 3 lysine 4 trimethylated (H3K4me3) showed a pivotal role in the paternal transmission of metabolic dysfunction [55]; although this study focused on evaluating the metabolic alterations driven by a high-fat diet, it is important to note that the microbiota could also be affected.
In the context of a high-fat diet, the development of obesity has been prevented by changes in microbiota-derived signals and intestinal epithelial HDAC3 [56]. The regulation of HDAC3 by the microbiota occurs via the integration of multiple molecular signals. Compared to control microbiota, the obesity-related microbiota transfer induced increased enrichment of histone H3 lysine 27 acetylation (H3K27Ac) at metabolic genes in colonic intestinal epithelial cells [57]. Under microbial colonization, histone modifications occur in different immune cells through different regulatory mechanisms (for details refer to the paper by Woo et al. [58]).
Gut Microbiota Dysbiosis and the Role of Mitochondria
The human gut microbiome, teeming with trillions of microorganisms, produces a vast array of small metabolites that significantly impact mitochondrial homeostasis, cellular ROS levels, and ultimately, intestinal, and metabolic health [59] (Fig. 3). Mitochondria are essential organelles responsible for ATP generation through the mitochondrial oxidative phosphorylation system (OXPHOS) during substrate oxidation. In addition to energy production, these organelles play a multifaceted role in metabolism, participating in sugar, fat, and protein breakdown (catabolism) while also contributing to the synthesis of essential biomolecules (anabolism) such as glucose, fatty acids, and amino acids. Additionally, mitochondria regulate ionic balance, influence cell death pathways, and contribute to ROS generation [60].
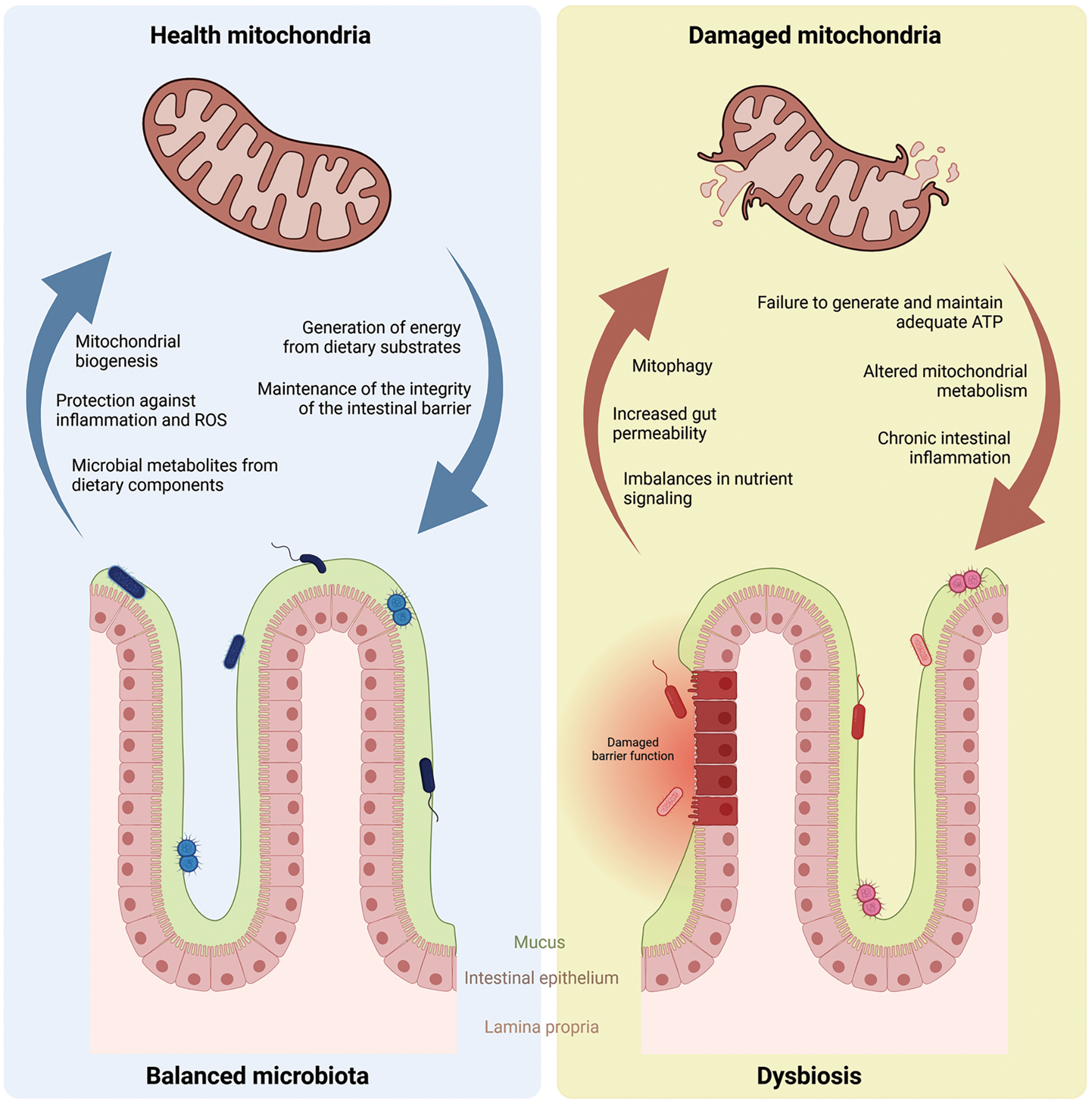
Figure 3: Mitochondrial-microbiota integration under balanced and unbalanced (dysbiosis) conditions.
The molecular crosstalk between the gut microbiota and mitochondria is a topic of intense research, with evidence suggesting a bidirectional communication channel (Fig. 3). The intestinal microbiota can influence mitochondrial function and biogenesis through the production of specific metabolites, such as butyrate that prevented autophagy in colonocytes [61,62]. Conversely, mitochondrial function can, in turn, modulate the composition and activity of the gut microbiota [63].
Diet plays a central role in this fascinating interplay. Dietary choices have a profound impact on the composition and function of the gut microbiota, and this influence is implicated in the pathogenesis of various metabolic disorders, including obesity, T2D, and metabolic syndrome [64,65]. Furthermore, mitochondria, being the cornerstone of cellular energy metabolism, are highly responsive to dietary components [66,67].
Metabolic disorders are often characterized by dysregulated energy metabolism and chronic low-grade inflammation (Fig. 3). Oxidative stress and inflammation are key players in the development of these conditions, with significant consequences for mitochondrial function [68,69]. Given their critical role in energy generation, mitochondria are particularly susceptible to inflammation-induced oxidative stress. They can sense inflammatory signals and are among the first organelles affected by systemic inflammation [70].
Mitochondrial dysfunction arises when these organelles fail to generate and maintain adequate ATP levels due to imbalances in nutrient signaling, energy production, and oxidative respiration (Fig. 3). Consequently, mitochondria play a significant role in the development of obesity-related metabolic disorders. Their core function is cellular energy metabolism, achieved through the oxidation of carbohydrates, lipids, and proteins [71]. More recently, a review by Colangeli et al. [72] showed the mechanisms whereby gut microbiota and microbial metabolites are capable of shaping host metabolism, adipogenesis, and white adipose tissue inflammation, and, particularly, in obesity, this dysregulation can affect adipose tissue (AT) metabolism via direct and indirect effects on AT mitochondria.
The capacity of diet to shape the composition and diversity of the gut microbiota is well-known. Several dietary components have been identified as beneficial for promoting a healthy and diverse gut microbiome. Dietary fiber, abundant in whole grains, fruits, and vegetables, functions as a prebiotic, selectively nourishing and promoting the growth of beneficial bacteria [73]. Additionally, fermented foods like yogurt and kefir, rich in probiotics, introduce live beneficial microorganisms directly into the gut [74,75]. Polyphenols, found in various plant-based foods, can promote the growth of beneficial bacteria while inhibiting the growth of harmful species [76,77]. Moreover, omega-3 fatty acids, commonly found in fatty fish and certain nuts and seeds, have been associated with a more favorable gut microbiota profile [78].
Certain dietary components can also positively influence mitochondrial function. Antioxidant-rich foods, including colorful fruits and vegetables, can counteract oxidative stress and protect mitochondria from damage. Coenzyme Q10 (CoQ10), present in meat, poultry, fish, nuts, and seeds, is involved in mitochondrial energy production and can enhance mitochondrial function. Other natural compounds, particularly polyphenols, possess antioxidant properties that enable them to inhibit the activation of nuclear factor-κB (NF-κB) and the expression of target genes associated with inflammation [79]. In addition to inhibiting inflammation, oxidative stress, and metabolic dysfunction linked to gut dysbiosis, polyphenols improve blood-brain barrier (BBB) permeability, thus protecting against the onset of brain diseases; they can activate nuclear factor-erythroid 2-related factor 2 (Nrf2) and sirtuin-1 pathways to inhibit ROS generation [79]. Furthermore, foods rich in essential nutrients such as B vitamins and magnesium provide cofactors necessary for mitochondrial metabolism and ATP production [80]. Thus, the modulation of mitochondrial function represents one of the mechanisms by which bioactive compounds exert health benefits.
The gut microbiota has been implicated in the pathogenesis of metabolic disorders. Dysbiosis-associated changes in microbial metabolites and gut barrier function can influence dietary energy extraction, adipose tissue metabolism, and host inflammatory responses (Fig. 3). These interactions can disrupt energy homeostasis, promote fat storage (adiposity), and contribute to obesity development [81]. Dysbiotic microbiota profiles are often associated with metabolic endotoxemia, chronic low-grade inflammation, and insulin resistance [14,18,81]. Gut microbiota dysbiosis and mitochondrial dysfunction are also associated with chronic intestinal inflammation and colorectal cancer. Studies have suggested that gut microbiota signal to the mitochondria of epithelial cells and immune cells thus altering mitochondrial metabolism, immune cell activation, inflammasome assembly, and epithelial barrier function [82].
Diets high in saturated fats and deficient in fiber are linked to an imbalanced gut microbiota characterized by a decline in beneficial bacteria and a surge in potentially harmful microbes [83]. These shifts in the microbial community composition can significantly impact the host’s metabolism and immune function [84]. For example, dysbiosis induced by a high-fat diet in mice promoted changes in the production of microbial metabolite that compromise mitochondrial function in the colonic epithelium, resulting in mitochondrial dysfunction and associated metabolic disorders such as inflammatory bowel disease [85].
The relationship between diet, the gut microbiota, and mitochondria is highly complex and plays a crucial role in a multitude of aspects of human health and disease (Fig. 3). Favorable dietary components that nurture a diverse and balanced gut microbiota can indirectly influence mitochondrial function by producing microbial metabolites, such as SCFAs which serve as valuable energy substrates. Conversely, optimal mitochondrial function contributes to gut health by maintaining the integrity of the intestinal barrier and regulating immune responses [86].
Modulating dietary habits to promote a beneficial gut microbiota composition while simultaneously enhancing mitochondrial function holds promise for preventing and managing various metabolic disorders. Strategies to modulate the gut microbiota through dietary interventions, including prebiotic and probiotic supplementation, increased dietary fiber intake, and personalized nutrition plans, are emerging as potential allies to mitigate these disorders. Additionally, therapeutic approaches targeting the gut microbiota, such as fecal microbiota transplantation and using microbially derived metabolites, are also possibilities for disease management.
Mitochondrial-Epigenetic Interactions are Closely Linked to Dysfunctional Microbiota and Associated Diseases
Mammalian mitochondrial DNA (mtDNA) encodes limited components for protein synthesis. Most mitochondrial proteins are encoded by nuclear DNA and transported into the mitochondria [87]. This necessitates a tightly coordinated communication between the nucleus and mitochondria, termed “mitonuclear communication” [88,89]. The nucleus governs mitochondrial biogenesis and activity by regulating the expression of mitochondrial genes (anterograde signaling). Conversely, mitochondria signal back to the nucleus (retrograde response) through energy cues, ROS production, and calcium signaling, thereby influencing cellular function and metabolic reprogramming. This bidirectional communication ensures cellular homeostasis and adaptation to various challenges.
Within this scenario, the nuclear epigenome, which controls gene expression patterns without altering the DNA sequence, can modulate mitochondrial function. Conversely, mitochondria can initiate epigenetic modifications in the nucleus [90,91]. Epigenetic modifications on nuclear DNA play a crucial role in the expression of genes encoding mitochondrial proteins. For instance, lysine-specific demethylase-1 (LSD-1), an enzyme that removes methyl groups from histones, i.e., a histone demethylase (HDM), is essential for expressing a third of nuclear-encoded mitochondrial genes in the liver [92]. This highlights the significance of histone methylation and HDMs in regulating mitochondrial biogenesis and function.
Microglia, the immune cells residing in the CNS, are essential to maintaining neuronal networks. They do this by eliminating apoptotic cells and dysfunctional synapses during healthy brain function. However, they can also be activated by disease, injury, or infection, assuming an immune role [93,94]. What is truly interesting is the influence of the gut microbiota on microglial maturation and function especially through the production of SCFAs, a byproduct of bacterial fermentation. Among these SCFAs, acetate has emerged as a key player, promoting microglial maturation, and regulating metabolic homeostasis [95]. This underscores the significant role of gut microbiota in microglial function and, by extension, brain health.
The absence of gut microbiota throughout the lifespan of germ-free (GF) mice leads to profound epigenetic changes in microglia. These changes involve alterations in histone 3 lysine 9 acetylation (H3K9ac) and H3K4me3, which in turn regulate microglial proliferation, morphology, activation, and even metabolic functions, including mitochondrial density [95]. The implications of these changes are significant, as GF mice displayed microglia with dysfunctional mitochondria, likely due to reduced brain acetate levels. Notably, supplementation with acetate restored these mitochondrial abnormalities, suggesting a crucial connection between gut microbiota, epigenetics changes, microglial function, and neuronal protection. Acetate, in the form of acetyl-CoA, can fuel the citric acid cycle (TCA) and ATP production and potentially contribute to nuclear histone acetylation, providing a possible mechanism for this functional link. These findings reinforce the delicate balance in our body’s systems and the potential consequences of any disruption.
As previously mentioned, SCFAs, generated by gut bacteria during fermentation, can influence histone modifications by inhibiting histone deacetylases (HDACs) (Fig. 4). This finding is particularly intriguing in the context of recent research linking diet to ASD [96]. Studies using PC12 cells, a model system for studying neuronal processes, have shown that propionic and butyric acids, two common SCFAs, can induce broad changes in gene expression profiles. These alterations include neurotransmitter systems, neuronal adhesion molecules, inflammation, oxidative stress, lipid metabolism, and mitochondrial function–all aspects relevant to ASD pathophysiology [96].
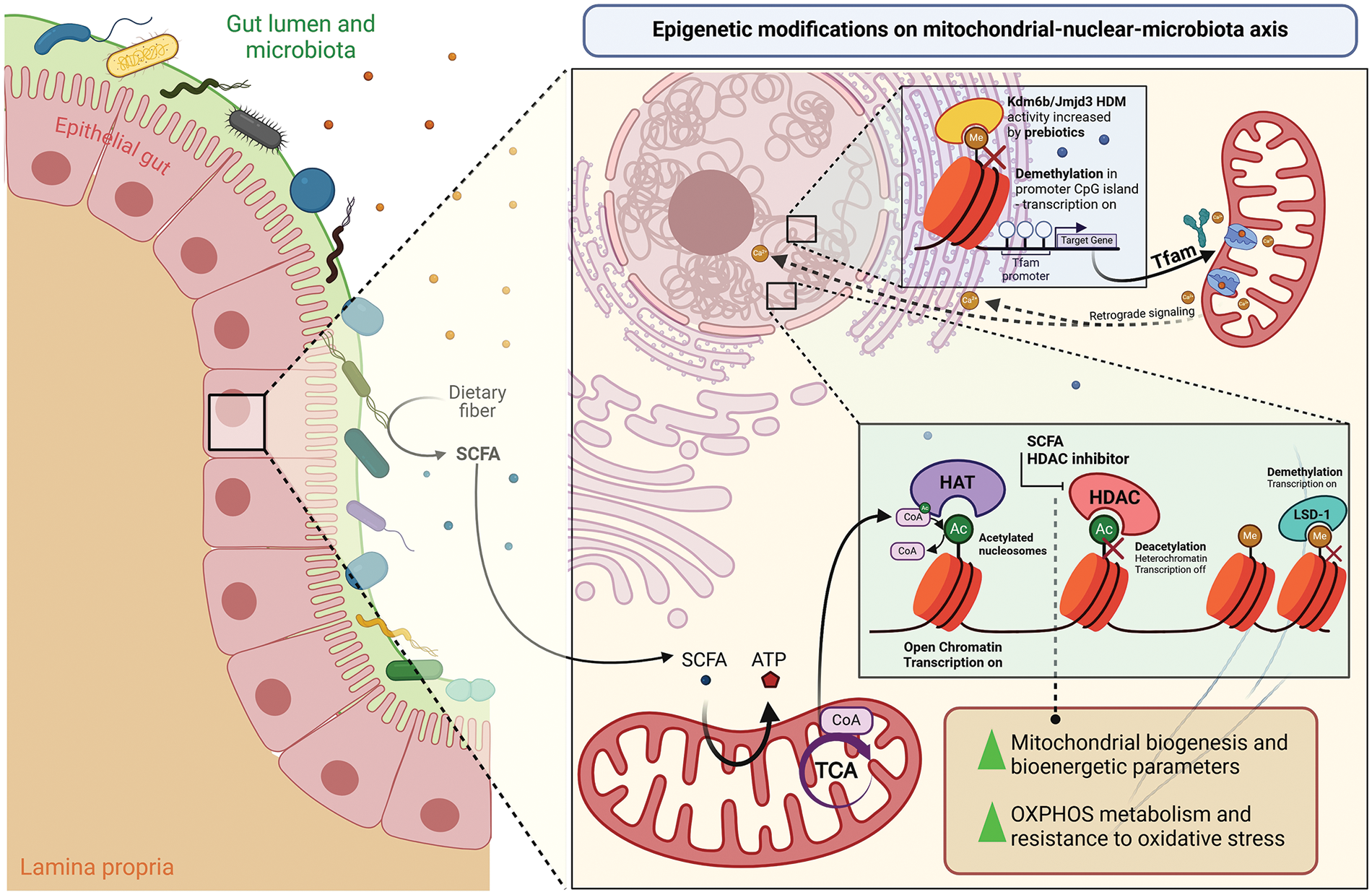
Figure 4: The diet promotes epigenetic changes in the mitochondrial-nuclear-microbiota axis. The intestinal microbiota produces short-chain fatty acids (SCFA) through the fermentation of dietary fibers. SCFAs such as acetate, in the form of acetyl-CoA (CoA), in turn, can be used in the mitochondrial machinery to generate ATP in the tricarboxylic acid cycle (TCA) or, together with histone acetyltransferase (HAT), to promote epigenetic acetylation of nucleosomes, opening chromatin to activate nuclear gene transcription. SCFAs like sodium butyrate also act to inhibit histone deacetylases (HDAC), preventing deacetylation to maintain specific acetylation of histone H3 lysine 27 (H3K27ac) to increase mitochondrial biogenesis, bioenergetic potential, and also improving the metabolism of the oxidative phosphorylation system (OXPHOS) and the resistance to reactive oxidative stress (ROS). Other epigenetic modifications on nuclear DNA act on methyl groups to prevent transcription, maintaining a condensed form of chromatin. Through demethylation, methyl groups are removed from histones to activate the transcription of essential genes encoding mitochondrial proteins. In this way, lysine-specific demethylase-1 (LSD-1) is a crucial enzyme that plays a role in liver nuclear cells, while lysine (K)-specific demethylase 6B/Jumonji domain-containing protein-3 (Kdm6b/Jmjd3) also has histone demethylase (HDM) activity in osteoblasts that allows exposure of the region promoter of mitochondrial transcription factor A (Tfam) to the expression of its target gene through the influence of prebiotics intake. Tfam, in turn, alters the Ca2+ flux in the mitochondria membrane, which acts in retrograde signaling to regulate the Ca2+ flux in the endoplasmic reticulum and nucleus according to the metabolic state.
Accumulating evidence has demonstrated the role of the gut microbiome on amygdala development in infancy, suggesting differential regulation in the frontal cortex and other brain regions [97]. These processes include the regulation of microRNA-451, 14-3-3, cytochrome P450 (CYP)1B1, and the melatonergic pathways. Decreased monoamine oxidase activity, increased levels of miR-451 and CYP1B1, along with reduced 14-3-3 activity, inhibit the synthesis of N-acetyl serotonin and melatonin. This contributes to the hyperserotonemia often seen in ASD, affecting mitochondrial function and the content of released exosomes. Considering Parkinson’s disease (PD), the nuclear and mitochondrial genome, epigenome, and the host gut microbiome have been examined in the onset and progression of PD since they revealed potential biomarkers to predict early stages of the disease and interventions with personalized treatment [98]. For instance, non-coding RNAs are involved with PD-associated regulatory mechanisms and metagenomic analyses may be an important approach for early diagnosis.
Further studies delve deeper into the mechanisms by which SCFAs might influence brain health. Research on embryonic hippocampal progenitor cells and PC12-NeuroD6 cells (expressing high levels of NeuroD6, a protein regulating neural progenitor proliferation) has shown that sodium butyrate (NaBt), an HDAC inhibitor, triggers a specific H3K27ac in genes critical for neuronal differentiation and mitochondrial reprogramming [99]. These epigenetic changes promote mitochondrial biogenesis, enhance OXPHOS metabolism, and improve mitochondrial bioenergetic parameters (Fig. 4). Moreover, they confer resistance to oxidative stress, a crucial factor during neuronal differentiation. These findings suggest that epigenetic modulation of the mitochondrial pool before neurotrophic signaling can enhance mitochondrial biogenesis, thereby improving the efficiency of neural progenitor cells to differentiate into neurons [99].
Notably, recent research suggests a potential link between epigenetic modifications and major depressive disorder [100]. Yu et al. [100] utilized affinity enrichment and high-resolution liquid chromatography-tandem mass spectrometry to identify acetylated proteins in the hippocampus of gut microbiota-absent mice. A total of 986 lysine acetylation sites in 543 proteins were identified. These proteins were involved in various biological functions, primarily located in mitochondria, and most of the altered pathways showed disturbances in OXPHOS and the tricarboxylic acid cycle. These findings suggest that lysine acetylation alterations may contribute to mitochondrial dysfunction, potentially linking gut microbiota to brain function and behavior.
Since the communication between the nucleus and mitochondria is bidirectional, mitochondria respond to nuclear signals and can also signal back to the nucleus, influencing gene expression through epigenetic modifications. They regulate the nuclear supply of acetyl-CoA and S-adenosyl methionine (SAM), essential for DNA and histone modifications. Mitochondrial damage can trigger retrograde signaling, triggering specific epigenetic changes, and altering gene expression [101]. Various mediators, including ROS, fatty acids, and TCA cycle metabolites, participate in this retrograde signaling pathway. These mitochondrial-derived metabolites can induce epigenetic changes in the nucleus, thereby enhancing or repressing gene expression. This may involve differential regulation of cellular metabolism, tissue repair, and disease progression, ultimately contributing to cellular homeostasis [102].
Studies on liver injury further exemplify the concept of mitochondrial dysfunction leading to epigenetic changes via retrograde signaling. In a murine model, a loss of mitochondrial transcription factor A, which promotes mitochondrial dysfunction, triggered the secretion of the growth factor amphiregulin (AREG) from hepatocytes. Mitochondrial dysfunction-responsive enhancers of the AREG gene are activated, leading to chromatin remodeling and epigenetic reprogramming. This suggests that, in response to stress, retrograde signaling can induce transcriptional and epigenetic changes to promote tissue repair through the secretion of growth factors from dysfunctional mitochondria [101].
Behera et al. [103] investigated how probiotics affect mitochondrial biogenesis and bone health through histone methylation in obese mice fed a high-fat diet. Treatment with probiotics or indole-3-propionic acid (IPA) significantly increased trabecular bone volume and bone mechanical strength while preventing gut inflammation in obese mice. From a mechanistic standpoint, probiotics treatment upregulated mitochondrial transcription factor A (Tfam) expression in osteoblasts by enhancing Kdm6b/Jmjd3 histone demethylase activity, thereby reducing H3K27me3 epigenetic methylation at the Tfam promoter (Fig. 4). Moreover, obese mice overexpressing Tfam did not exhibit obesity-induced reductions in glucose uptake, mitochondrial biogenesis, or mineralization in osteoblasts when fed a high-fat diet.
Recent research shows how the gut microbiome can influence immune T-cell function through a previously unknown pathway involving mitochondrial metabolism and epigenetics changes [104]. Studies show that inhibiting the mitochondrial pyruvate carrier (MPC) in CD8+ T cells, a type of immune cell, promotes their differentiation into memory T cells. Memory T cells are crucial for long-term immunity. Interestingly, this differentiation is driven by a “mitochondrial retrograde pathway” involving a metabolic-epigenetic axis.
The inhibition of MPC in CD8+ T cells does not significantly alter their immediate effector function or energy status. However, it does trigger a metabolic shift, favoring acetyl-CoA production through glutamine and fatty acid oxidation. This increase in acetyl-CoA is then linked to the acetylation of a specific site (lysine residue 27) on histone H3. Histone acetylation is an epigenetic modification that influences gene expression. In this case, it promotes chromatin accessibility on genes associated with memory T cell differentiation, allowing these genes to be more readily expressed. This epigenetic reprogramming, triggered by the metabolic shift caused by MPC inhibition, ultimately enhances the T cells’ capacity to fight tumors in adoptive cell transfer therapy.
A healthy gut microbiome contributes to normal mitochondrial function in immune cells by providing essential metabolites for cellular energy production and signaling pathways. However, a dysbiotic microbiome, characterized by an imbalance in bacterial communities, can disrupt mitochondrial function [105]. This disruption can manifest in several ways, including increased reliance on aerobic glycolysis (a less efficient way of producing energy), reduced OXPHOS, and altered fatty acid oxidation. Additionally, a dysbiotic microbiome can lead to changes in mitochondrial membrane permeability and resistance to cell death in immune cells. This could be impactful since butyrate, a SCFA produced by gut bacteria, has been shown to affect mitochondrial respiration and cell death, improving intestinal inflammation [106].
Given the potential of mitochondrial-derived metabolic signals to induce epigenetic modifications in response to stress or damage, interventions aimed at a dysbiotic gut microbiome could be a game-changing strategy. By fostering a healthy gut microbiota, we might be able to avert epigenetic changes triggered by altered mitochondrial retrograde signaling. This could potentially lead to enhanced immune function and overall health, underscoring the practical implications of this data collection for clinical settings.
An imbalance in gut microbial communities (e.g., dysbiosis) has been linked to various metabolic disorders like T2D, obesity, and metabolic syndrome. However, the exact mechanisms connecting these entities remain under investigation. In this context, it has proposed a two-way communication between the gut microbiota and mitochondria, playing significant metabolic roles. The gut microbiome can influence mitochondrial function through metabolite production, while optimal mitochondrial function supports gut health. The interplay between the gut microbiome and epigenetics changes might also contribute to the development of metabolic diseases.
Epigenetic modifications, which can be influenced by environmental factors including diet and the microbiome, regulate gene expression without altering the DNA sequence itself (Fig. 4). Understanding this complex regulatory network between gut microbiota, mitochondria, and epigenetics modifications has the potential to reveal novel therapeutic approaches for preventing and managing metabolic diseases. Since mitochondrial signals can trigger epigenetic changes, modifying a dysbiotic gut microbiome could be valuable in restoring homeostasis and attenuating the risk of various metabolic and inflammatory disorders. Some food compounds can protect the organism from age-related diseases by acting on the epigenetic level and slowing down aging–in doing this the physiology of the microbiome is also modified [107]. Dietary interventions that promote a healthy gut community, such as prebiotics, probiotics, increased fiber intake, and personalized nutrition, hold promise for mitigating metabolic disorders and improving patients’ quality of life (Fig. 4). Also, polyphenols may interact with the gut microbiota where they undergo extensive metabolism to produce bioactive molecules [108]; these molecules, in turn, act on transcription factors that are crucial for mitochondrial biogenesis, antioxidant systems, DNA repair, and glucose and lipid homeostasis.
Understanding the crosstalk between the microbiome, mitochondria, and epigenetic regulation is essential for unraveling the complex mechanisms underlying human health and disease. While existing literature has established the link between gut dysbiosis and metabolic disorders, the exact mechanisms underlying these connections remain incompletely understood. The relationship between gut microbiota and metabolic disorders is well-documented; however, there is a significant gap in comprehensive studies that integrate mitochondrial function and epigenetic modifications into this framework. In vivo and in vitro analyses might lead to the development of novel therapeutic strategies targeting these interactions to prevent or treat a wide range of disorders.
Acknowledgement: The authors thank Silvia G. Barbuzza, M.Ed. (Coordinator English Department, FFyL-UNCUyo) for revising and editing for grammatical, syntax, and stylistic errors, following the APA Style Guide.
Funding Statement: This work was supported by grants from the Research and Technology Council of Cuyo University (SECyT), Mendoza, Argentina, and from ANPCyT FONCyT, both of which were awarded to Walter Manucha, Grant No. PICT 2000 Serie A 4000.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Vinícius Augusto Simão, Luiz Gustavo de Almeida Chuffa, León Ferder, Felipe Inserra, Walter Manucha; data collection: Vinícius Augusto Simão, Luiz Gustavo de Almeida Chuffa, León Ferder, Felipe Inserra, Walter Manucha; analysis and interpretation of results: Vinícius Augusto Simão, Luiz Gustavo de Almeida Chuffa, León Ferder, Felipe Inserra, Walter Manucha; draft manuscript preparation: Vinícius Augusto Simão, Luiz Gustavo de Almeida Chuffa, Walter Manucha. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Schmidt TSB, Raes J, Bork P. The human gut microbiome: from association to modulation. Cell. 2018;172(6):1198–215. doi:10.1016/j.cell.2018.02.044. [Google Scholar] [PubMed] [CrossRef]
2. Adak A, Khan MR. An insight into gut microbiota and its functionalities. Cell Mol Life Sci. 2019;76(3):473–93. doi:10.1007/s00018-018-2943-4. [Google Scholar] [PubMed] [CrossRef]
3. Gaboriau-Routhiau M-CMV. Influence of resident intestinal microflora on the development and functions of the gut-associated lymphoid tissue. Microb Ecol Health Dis. 2001;13(2):65–86. doi:10.1080/08910600120558. [Google Scholar] [CrossRef]
4. Liu J, Tan Y, Cheng H, Zhang D, Feng W, Peng C. Functions of gut microbiota metabolites, current status and future perspectives. Aging Dis. 2022;13(4):1106–26. doi:10.14336/AD.2022.0104. [Google Scholar] [PubMed] [CrossRef]
5. Dicks LMT. Gut bacteria and neurotransmitters. Microorganisms. 2022;10(9):1838. doi:10.3390/microorganisms10091838. [Google Scholar] [PubMed] [CrossRef]
6. Schneider T. The holobiont self: understanding immunity in context. Hist Philos Life Sci. 2021;43(3):99. doi:10.1007/s40656-021-00454-y. [Google Scholar] [PubMed] [CrossRef]
7. Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab. 2017;26(1):110–30. doi:10.1016/j.cmet.2017.05.008. [Google Scholar] [PubMed] [CrossRef]
8. Moya A, Ferrer M. Functional redundancy-induced stability of gut microbiota subjected to disturbance. Trends Microbiol. 2016;24(5):402–13. doi:10.1016/j.tim.2016.02.002. [Google Scholar] [PubMed] [CrossRef]
9. Rinninella E, Raoul P, Cintoni M, Franceschi F, Miggiano GAD, Gasbarrini A, et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi:10.3390/microorganisms7010014. [Google Scholar] [PubMed] [CrossRef]
10. Dwiyanto J, Hussain MH, Reidpath D, Ong KS, Qasim A, Lee SWH, et al. Ethnicity influences the gut microbiota of individuals sharing a geographical location: a cross-sectional study from a middle-income country. Sci Rep. 2021;11(1):2618. doi:10.1038/s41598-021-82311-3. [Google Scholar] [PubMed] [CrossRef]
11. Raulo A, Allen BE, Troitsky T, Husby A, Firth JA, Coulson T, et al. Social networks strongly predict the gut microbiota of wild mice. ISME J. 2021;15(9):2601–13. doi:10.1038/s41396-021-00949-3. [Google Scholar] [PubMed] [CrossRef]
12. Dieterich W, Schink M, Zopf Y. Microbiota in the gastrointestinal tract. Med Sci. 2018;6(4):116. doi:10.3390/medsci6040116. [Google Scholar] [PubMed] [CrossRef]
13. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nat Rev Microbiol. 2015;14(1):20–32. doi:10.1038/nrmicro3552. [Google Scholar] [PubMed] [CrossRef]
14. Di Tommaso N, Gasbarrini A, Ponziani FR. Intestinal barrier in human health and disease. Int J Environ Res Public Health. 2021;18(23):12836. doi:10.3390/ijerph182312836. [Google Scholar] [PubMed] [CrossRef]
15. Scheithauer TPM, Rampanelli E, Nieuwdorp M, Vallance BA, Verchere CB, van Raalte DH, et al. Gut microbiota as a trigger for metabolic inflammation in obesity and type 2 diabetes. Front Immunol. 2020;11:571731. doi:10.3389/fimmu.2020.571731. [Google Scholar] [PubMed] [CrossRef]
16. Martel J, Chang SH, Ko YF, Hwang TL, Young JD, Ojcius DM. Gut barrier disruption and chronic disease. Trends Endocrinol Metabol. 2022;33(4):247–65. doi:10.1016/j.tem.2022.01.002. [Google Scholar] [PubMed] [CrossRef]
17. Mou Y, Du Y, Zhou L, Yue J, Hu X, Liu Y, et al. Gut microbiota interact with the brain through systemic chronic inflammation: implications on neuroinflammation, neurodegeneration, and aging. Front Immunol. 2022;13:796288. doi:10.3389/fimmu.2022.796288. [Google Scholar] [PubMed] [CrossRef]
18. Amabebe E, Robert FO, Agbalalah T, Orubu ESF. Microbial dysbiosis-induced obesity: role of gut microbiota in homoeostasis of energy metabolism. Br J Nutr. 2020;123(10):1127–37. doi:10.1017/S0007114520000380. [Google Scholar] [PubMed] [CrossRef]
19. Huda MN, Kim M, Bennett BJ. Modulating the microbiota as a therapeutic intervention for type 2 diabetes. Front Endocrinol. 2021;12:632335. doi:10.3389/fendo.2021.632335. [Google Scholar] [PubMed] [CrossRef]
20. Cryan JF, O. KJ, Cowan M, Sandhu CS, S.Bastiaanssen KV, Boehme TF, et al. The microbiota-gut-brain axis. Physiol Rev. 2019;99:1877–2013. doi:10.1152/physrev.00018.2018.-The. [Google Scholar] [CrossRef]
21. Margolis KG, Cryan JF, Mayer EA. The microbiota-gut-brain axis: from motility to mood. Gastroenterol. 2021;160(5):1486–501. doi:10.1053/j.gastro.2020.10.066. [Google Scholar] [PubMed] [CrossRef]
22. El-Sayed A, Aleya L, Kamel M. Microbiota and epigenetics: promising therapeutic approaches? Environ Sci Pollut Res. 2021;28:49343–61. doi:10.1007/s11356-021-15623-6/Published. [Google Scholar] [CrossRef]
23. Borbolis F, Mytilinaiou E, Palikaras K. The crosstalk between microbiome and mitochondrial homeostasis in neurodegeneration. Cells. 2023;12(3):429. doi:10.3390/cells12030429. [Google Scholar] [PubMed] [CrossRef]
24. Meng H, Meng H, Cao Y, Qin J, Song X, Zhang Q, et al. DNA methylation, its mediators and genome integrity. Int J Biol Sci. 2015;11(5):604–17. doi:10.7150/ijbs.11218. [Google Scholar] [PubMed] [CrossRef]
25. Prajapati SK, Shah R, Alford N, Mishra SP, Jain S, Hansen B, et al. The triple alliance: microbiome, mitochondria, and metabolites in the context of age-related cognitive decline and Alzheimer’s disease. J Gerontol-Ser A Biol Sci Med Sci. 2023;78(12):2187–202. doi:10.1093/gerona/glad226. [Google Scholar] [PubMed] [CrossRef]
26. Martín Giménez VM, Chuffa LGA, Simão VA, Reiter RJ, Manucha W. Protective actions of vitamin D, anandamide and melatonin during vascular inflammation: epigenetic mechanisms involved. Life Sci. 2022;288(1):120191. doi:10.1016/j.lfs.2021.120191. [Google Scholar] [PubMed] [CrossRef]
27. Zhang L, Chang C, Lu Q. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253(5):3–55. doi:10.1007/978-981-15-3449-2_1. [Google Scholar] [PubMed] [CrossRef]
28. Agarwal P, Morriseau TS, Kereliuk SM, Doucette CA, Wicklow BA, Dolinsky VW. Maternal obesity, diabetes during pregnancy and epigenetic mechanisms that influence the developmental origins of cardiometabolic disease in the offspring. Crit Rev Clin Lab Sci. 2018;55(2):71–101. doi:10.1080/10408363.2017.1422109. [Google Scholar] [PubMed] [CrossRef]
29. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(3):245–54. doi:10.1038/ng1089. [Google Scholar] [PubMed] [CrossRef]
30. Smith B, Carregari V. Histone modifications in neurological disorders. Adv Exp Med Biol. 2022;1382:95–107. doi:10.1007/978-3-031-05460-0_. [Google Scholar] [CrossRef]
31. Simão VA, Ferder L, Manucha W, Chuffa LGA. Epigenetic mechanisms involved in inflammaging-associated hypertension. Curr Hypertens Rep. 2022;24(11):547–62. doi:10.1007/s11906-022-01214-4. [Google Scholar] [PubMed] [CrossRef]
32. Xu X, Peng Q, Jiang X, Tan S, Yang Y, Yang W, et al. Metabolic reprogramming and epigenetic modifications in cancer: from the impacts and mechanisms to the treatment potential. Exp Mol Med. 2023;55(7):1357–70. doi:10.1038/s12276-023-01020-1. [Google Scholar] [PubMed] [CrossRef]
33. Yamada L, Chong S. Epigenetic studies in developmental origins of health and disease: pitfalls and key considerations for study design and interpretation. J Dev Orig Health Dis. 2017;8(1):30–43. doi:10.1017/S2040174416000507. [Google Scholar] [PubMed] [CrossRef]
34. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2017;18(1):5–18. doi:10.1038/nrc.2017.99. [Google Scholar] [PubMed] [CrossRef]
35. Poller W, Dimmeler S, Heymans S, Zeller T, Haas J, Karakas M, et al. Non-coding RNAs in cardiovascular diseases: diagnostic and therapeutic perspectives. Eur Heart J. 2018;39(29):2704–16. doi:10.1093/eurheartj/ehx165. [Google Scholar] [PubMed] [CrossRef]
36. Ling C, Rönn T. Epigenetics in human obesity and type 2 diabetes. Cell Metab. 2019;29(5):1028–44. doi:10.1016/j.cmet.2019.03.009. [Google Scholar] [PubMed] [CrossRef]
37. Annese V. Genetics and epigenetics of IBD. Pharmacol Res. 2020;159:104892. doi:10.1016/j.phrs.2020.104892. [Google Scholar] [PubMed] [CrossRef]
38. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi:10.1016/j.cell.2012.06.013. [Google Scholar] [PubMed] [CrossRef]
39. Li D, Li Y, Yang S, Lu J, Jin X, Wu M. Diet-gut microbiota-epigenetics in metabolic diseases: from mechanisms to therapeutics. Biomed Pharmacother. 2022;153:113290. doi:10.1016/j.biopha.2022.113290. [Google Scholar] [PubMed] [CrossRef]
40. Wattanathorn J, Tong-Un T, Thukham-Mee W, Paholpak P, Rangseekhajee P. A randomized, double-blind, placebo-controlled study of an anthocyanin-rich functional ingredient on cognitive function and eye dryness in late adulthood volunteers: roles of epigenetic and gut microbiome modulations. Nutrients. 2023;15(16):3499. doi:10.3390/nu15163499. [Google Scholar] [PubMed] [CrossRef]
41. Zhang Y, Shen Y, Liufu N, Liu L, Li W, Shi Z, et al. Transmission of Alzheimer’s disease-associated microbiota dysbiosis and its impact on cognitive function: evidence from mouse models and human patients. Res Sq. 2023;28(10):4421–37. doi:10.21203/rs.3.rs-2790988/v1. [Google Scholar] [PubMed] [CrossRef]
42. Behrens LMP, Gasparotto J, Rampelotto PH, Escalona MAR, da Silva L dos S, Carazza-Kessler FG, et al. Sodium propionate oral supplementation ameliorates depressive-like behavior through gut microbiome and histone 3 epigenetic regulation. J Nutrit Biochem. 2024;130(1):109660. doi:10.1016/j.jnutbio.2024.109660. [Google Scholar] [PubMed] [CrossRef]
43. van der Vossen EWJ, Bastos D, Stols-Gonçalves D, de Goffau MC, Davids M, Pereira JPB, et al. Effects of fecal microbiota transplant on DNA methylation in subjects with metabolic syndrome. Gut Microbes. 2021;13(1):1993513. doi:10.1080/19490976.2021.1993513. [Google Scholar] [PubMed] [CrossRef]
44. Yuan X, Wang R, Han B, Sun CJ, Chen R, Wei H, et al. Functional and metabolic alterations of gut microbiota in children with new-onset type 1 diabetes. Nat Commun. 2022;13(1):6356. doi:10.1038/s41467-022-33656-4. [Google Scholar] [PubMed] [CrossRef]
45. Noureldein MH, Bitar S, Youssef N, Azar S, Eid AA. Butyrate modulates diabetes-linked gut dysbiosis: epigenetic and mechanistic modifications. J Mol Endocrinol. 2020;64(1):29–42. doi:10.1530/JME-19-0132. [Google Scholar] [PubMed] [CrossRef]
46. Noureldein M, Nawfal R, Bitar S, Maxwell SS, Khurana I, Kassouf HK, et al. Intestinal microbiota regulates diabetes and cancer progression by IL-1β and NOX4 dependent signaling cascades. Cell Mol Life Sci. 2022;79(9):502. doi:10.1007/s00018-022-04485-x. [Google Scholar] [PubMed] [CrossRef]
47. Lecamwasam A, Nelson TM, Rivera L, Ekinci EI, Saffery R, Dwyer KM. Gut microbiome composition remains stable in individuals with diabetes-related early to late stage chronic kidney disease. Biomed. 2020;9(1):1–16. doi:10.3390/biomedicines. [Google Scholar] [CrossRef]
48. Haque S, Raina R, Afroze N, Hussain A, Alsulimani A, Singh V, et al. Microbial dysbiosis and epigenetics modulation in cancer development–a chemopreventive approach. Semin Cancer Biol. 2022;86(7464):666–81. doi:10.1016/j.semcancer.2021.06.024. [Google Scholar] [PubMed] [CrossRef]
49. Sobhani I, Bergsten E, Couffin S, Amiot A, Nebbad B, Barau C, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci U S A. 2019;116(48):24285–95. doi:10.1073/pnas.1912129116. [Google Scholar] [PubMed] [CrossRef]
50. Liu Z, Zhang Q, Zhang H, Yi Z, Ma H, Wang X, et al. Colorectal cancer microbiome programs DNA methylation of host cells by affecting methyl donor metabolism. Genome Med. 2024;16(1):77. doi:10.1186/s13073-024-01344-1. [Google Scholar] [PubMed] [CrossRef]
51. Ryan FJ, Ahern AM, Fitzgerald RS, Laserna-Mendieta EJ, Power EM, Clooney AG, et al. Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat Commun. 2020;11(1):1512. doi:10.1038/s41467-020-15342-5. [Google Scholar] [PubMed] [CrossRef]
52. Yang F, Yang Y, Chen L, Zhang Z, Liu L, Zhang C, et al. The gut microbiota mediates protective immunity against tuberculosis via modulation of lncRNA. Gut Microbes. 2022;14(1):2029997. doi:10.1080/19490976.2022.2029997. [Google Scholar] [PubMed] [CrossRef]
53. Cuomo M, Coretti L, Costabile D, Della Monica R, De Riso G, Buonaiuto M, et al. Host fecal DNA specific methylation signatures mark gut dysbiosis and inflammation in children affected by autism spectrum disorder. Sci Rep. 2023;13(1):18197. doi:10.1038/s41598-023-45132-0. [Google Scholar] [PubMed] [CrossRef]
54. Argaw-Denboba A, Schmidt TSB, Di Giacomo M, Ranjan B, Devendran S, Mastrorilli E, et al. Paternal microbiome perturbations impact offspring fitness. Nature. 2024;629(8012):652–9. doi:10.1038/s41586-024-07336-w. [Google Scholar] [PubMed] [CrossRef]
55. Pepin AS, Lafleur C, Lambrot R, Dumeaux V, Kimmins S. Sperm histone H3 lysine 4 tri-methylation serves as a metabolic sensor of paternal obesity and is associated with the inheritance of metabolic dysfunction. Mol Metab. 2022;59(5):101463. doi:10.1016/j.molmet.2022.101463. [Google Scholar] [PubMed] [CrossRef]
56. Kuang Z, Wang Y, Li Y, Ye C, Ruhn KA, Behrendt CL, et al. The intestinal microbiota programs diurnal rhythms in host metabolism through histone deacetylase 3. Sciences. 2019;365(6460):1428–34. doi:10.1126/science.aaw3134. [Google Scholar] [PubMed] [CrossRef]
57. Qin Y, Roberts JD, Grimm SA, Lih FB, Deterding LJ, Li R, et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 2018;19(1):1–14. doi:10.1186/s13059-018-1389-1. [Google Scholar] [PubMed] [CrossRef]
58. Woo V, Alenghat T. Epigenetic regulation by gut microbiota. Gut Microbes. 2022;14(1):2022407. doi:10.1080/19490976.2021.2022407. [Google Scholar] [PubMed] [CrossRef]
59. Ballard JWO, Towarnicki SG. Mitochondria, the gut microbiome and ROS. Cell Signal. 2020;75:109737. doi:10.1016/j.cellsig.2020.109737. [Google Scholar] [PubMed] [CrossRef]
60. Brand MD, Orr AL, Perevoshchikova IV, Quinlan CL. The role of mitochondrial function and cellular bioenergetics in ageing and disease. Br J Dermatol. 2013;169(s2):1–8. doi:10.1111/bjd.12208. [Google Scholar] [PubMed] [CrossRef]
61. Franco-Obregón A, Gilbert JA. The microbiome-mitochondrion connection: common ancestries, common mechanisms, common goals. MSystems. 2017;2(3):e00018–17. doi:10.1128/msystems.00018-17. [Google Scholar] [PubMed] [CrossRef]
62. Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–26. doi:10.1016/j.cmet.2011.02.018. [Google Scholar] [PubMed] [CrossRef]
63. Clark A, Mach N. The crosstalk between the gut microbiota and mitochondria during exercise. Front Physiol. 2017;8:319. doi:10.3389/fphys.2017.00319. [Google Scholar] [PubMed] [CrossRef]
64. Muñoz-Garach A, Diaz-Perdigones C, Tinahones FJ. Gut microbiota and type 2 diabetes mellitus. Endocrinol Nutric. 2016;63(10):560–8. doi:10.1016/j.endoen.2016.07.004. [Google Scholar] [CrossRef]
65. Cuevas-Sierra A, Ramos-Lopez O, Riezu-Boj JI, Milagro FI, Martinez JA. Diet, gut microbiota, and obesity: links with host genetics and epigenetics and potential applications. Adv Nutrit. 2019;10(3):S17–30. doi:10.1093/advances/nmy078. [Google Scholar] [PubMed] [CrossRef]
66. Greenhill C. Diet composition affects mitochondria trafficking in adipose tissue. Nat Rev Endocrinol. 2022;18(11):655. doi:10.1038/s41574-022-00755-7. [Google Scholar] [PubMed] [CrossRef]
67. Kyriazis ID, Vassi E, Alvanou M, Angelakis C, Skaperda Z, Tekos F, et al. The impact of diet upon mitochondrial physiology. Int J Mol Med. 2022;50(5):1–26. doi:10.3892/ijmm.2022.5191. [Google Scholar] [PubMed] [CrossRef]
68. Montgomery MK, Turner N. Mitochondrial dysfunction and insulin resistance: an update. Endocr Connect. 2015;4(1):R1–15. doi:10.1530/EC-14-0092. [Google Scholar] [PubMed] [CrossRef]
69. Fernández-Sánchez A, Madrigal-Santillán E, Bautista M, Esquivel-Soto J, Morales-González Á., Esquivel-Chirino C, et al. Inflammation, oxidative stress, and obesity. Int J Mol Sci. 2011;12(5):3117–32. doi:10.3390/ijms12053117. [Google Scholar] [PubMed] [CrossRef]
70. Gilkerson R. A disturbance in the force: cellular stress sensing by the mitochondrial network. Antioxidants. 2018;7(10):126. doi:10.3390/antiox7100126. [Google Scholar] [PubMed] [CrossRef]
71. Bhatti JS, Bhatti GK, Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders—a step towards mitochondria based therapeutic strategies. Biochim Biophys Acta Mol Basis Dis. 2017;1863(5):1066–77. doi:10.1016/j.bbadis.2016.11.010. [Google Scholar] [PubMed] [CrossRef]
72. Colangeli L, Escobar Marcillo DI, Simonelli V, Iorio E, Rinaldi T, Sbraccia P, et al. The crosstalk between gut microbiota and white adipose tissue mitochondria in obesity. Nutrients. 2023;15(7):1723. doi:10.3390/nu15071723. [Google Scholar] [PubMed] [CrossRef]
73. Makki K, Deehan EC, Walter J, Bäckhed F. The impact of dietary fiber on gut microbiota in host health and disease. Cell Host Microbe. 2018;23(6):705–15. doi:10.1016/j.chom.2018.05.012. [Google Scholar] [PubMed] [CrossRef]
74. Dimidi E, Cox SR, Rossi M, Whelan K. Fermented foods: definitions and characteristics, impact on the gut microbiota and effects on gastrointestinal health and disease. Nutrients. 2019;11(8):1806. doi:10.3390/nu11081806. [Google Scholar] [PubMed] [CrossRef]
75. Wieërs G, Belkhir L, Enaud R, Leclercq S, Philippart de Foy JM, Dequenne I, et al. How probiotics affect the microbiota. Front Cell Infect Microbiol. 2020;9:454. doi:10.3389/fcimb.2019.00454. [Google Scholar] [PubMed] [CrossRef]
76. Cardona F, Andrés-Lacueva C, Tulipani S, Tinahones FJ, Queipo-Ortuño MI. Benefits of polyphenols on gut microbiota and implications in human health. J Nutrit Biochem. 2013;24(8):1415–22. doi:10.1016/j.jnutbio.2013.05.001. [Google Scholar] [PubMed] [CrossRef]
77. Wan MLY, Co VA, El-Nezami H. Dietary polyphenol impact on gut health and microbiota. Crit Rev Food Sci Nutr. 2021;61(4):690–711. doi:10.1080/10408398.2020.1744512. [Google Scholar] [PubMed] [CrossRef]
78. Costantini L, Molinari R, Farinon B, Merendino N. Impact of omega-3 fatty acids on the gut microbiota. Int J Mol Sci. 2017;18(12):2645. doi:10.3390/ijms18122645. [Google Scholar] [PubMed] [CrossRef]
79. Scuto M, Rampulla F, Reali GM, Spanò SM, Trovato Salinaro A, Calabrese V. Hormetic nutrition and redox regulation in gut-brain axis disorders. Antioxidants. 2024;13(4):484. doi:10.3390/antiox13040484. [Google Scholar] [PubMed] [CrossRef]
80. Du J, Zhu M, Bao H, Li B, Dong Y, Xiao C, et al. The role of nutrients in protecting mitochondrial function and neurotransmitter signaling: implications for the treatment of depression, PTSD, and suicidal behaviors. Crit Rev Food Sci Nutr. 2016;56(15):2560–78. doi:10.1080/10408398.2013.876960. [Google Scholar] [PubMed] [CrossRef]
81. Moszak M, Szulińska M, Bogdański P. You are what you eat—the relationship between diet, microbiota, and metabolic disorders—a review. Nutrients. 2020;12(4):1096. doi:10.3390/nu12041096. [Google Scholar] [PubMed] [CrossRef]
82. Jackson DN, Theiss AL. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes. 2020;11(3):285–304. doi:10.1080/19490976.2019.1592421. [Google Scholar] [PubMed] [CrossRef]
83. Malesza IJ, Malesza M, Walkowiak J, Mussin N, Walkowiak D, Aringazina R, et al. High-fat, western-style diet, systemic inflammation, and gut microbiota: a narrative review. Cells. 2021;10(11):3164. doi:10.3390/cells10113164. [Google Scholar] [PubMed] [CrossRef]
84. Weiss GA, Hennet T. Mechanisms and consequences of intestinal dysbiosis. Cell Mol Life Sci. 2017;74(16):2959–77. doi:10.1007/s00018-017-2509-x. [Google Scholar] [PubMed] [CrossRef]
85. Lee JY, Cevallos SA, Byndloss MX, Tiffany CR, Olsan EE, Butler BP, et al. High-fat diet and antibiotics cooperatively impair mitochondrial bioenergetics to trigger dysbiosis that exacerbates pre-inflammatory bowel disease. Cell Host Microbe. 2020;28(2):273–84. doi:10.1016/j.chom.2020.06.001. [Google Scholar] [PubMed] [CrossRef]
86. Vezza T, Abad-Jiménez Z, Marti-Cabrera M, Rocha M, Víctor VM. Microbiota-mitochondria inter-talk: a potential therapeutic strategy in obesity and type 2 diabetes. Antioxidants. 2020;9(9):1–21. doi:10.3390/antiox9090848. [Google Scholar] [PubMed] [CrossRef]
87. Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271(50):31763–6. doi:10.1074/jbc.271.50.31763. [Google Scholar] [PubMed] [CrossRef]
88. Todorov IN, Todorov GI. Multifactorial nature of high frequency of mitochondrial DNA mutations in somatic mammalian cells. Biochemistry. 2009;74(9):962–70. doi:10.1134/S000629790909003X. [Google Scholar] [PubMed] [CrossRef]
89. Couvillion MT, Soto IC, Shipkovenska G, Churchman LS. Synchronized mitochondrial and cytosolic translation programs. Nature. 2016;533(7604):499–503. doi:10.1038/nature18015. [Google Scholar] [PubMed] [CrossRef]
90. Quirós PM, Mottis A, Auwerx J. Mitonuclear communication in homeostasis and stress. Nat Rev Mol Cell Biol. 2016;17(4):213–26. doi:10.1038/nrm.2016.23. [Google Scholar] [PubMed] [CrossRef]
91. Matilainen O, Quirós PM, Auwerx J. Mitochondria and epigenetics–crosstalk in homeostasis and stress. Trends Cell Biol. 2017;27(6):453–63. doi:10.1016/j.tcb.2017.02.004. [Google Scholar] [PubMed] [CrossRef]
92. Cao Y, Tang L, Du K, Paraiso K, Sun Q, Liu Z, et al. Anterograde regulation of mitochondrial genes and FGF21 signaling by hepatic LSD1. JCI Insight. 2021;6(17):410. doi:10.1172/jci.insight.147692. [Google Scholar] [PubMed] [CrossRef]
93. Woodburn SC, Bollinger JL, Wohleb ES. The semantics of microglia activation: neuroinflammation, homeostasis, and stress. J Neuroinflammation. 2021;18(1):1–16. doi:10.1186/s12974-021-02309-6. [Google Scholar] [PubMed] [CrossRef]
94. Borst K, Dumas AA, Prinz M. Microglia: Immune and non-immune functions. Immunity. 2021;54(10):2194–208. doi:10.1016/j.immuni.2021.09.014. [Google Scholar] [PubMed] [CrossRef]
95. Erny D, Dokalis N, Mezö C, Castoldi A, Mossad O, Staszewski O, et al. Microbiota-derived acetate enables the metabolic fitness of the brain innate immune system during health and disease. Cell Metab. 2021;33(11):2260–76. doi:10.1016/j.cmet.2021.10.010. [Google Scholar] [PubMed] [CrossRef]
96. Nankova BB, Agarwal R, MacFabe DF, La Gamma EF. Enteric bacterial metabolites propionic and butyric acid modulate gene expression, including CREB-dependent catecholaminergic neurotransmission, in PC12 cells-Possible relevance to autism spectrum disorders. PLoS One. 2014;9(8):e103740. doi:10.1371/journal.pone.0103740. [Google Scholar] [PubMed] [CrossRef]
97. Seo M, Anderson G. Gut-amygdala interactions in autism spectrum disorders: developmental roles via regulating mitochondria, exosomes, immunity and microRNAs. Curr Pharm Des. 2019;25(41):4344–56. doi:10.2174/1381612825666191105102545. [Google Scholar] [PubMed] [CrossRef]
98. Cabral GF, Schaan AP, Cavalcante GC, Sena-Dos-santos C, de Souza TP, Souza Port’S NM, et al. Nuclear and mitochondrial genome, epigenome and gut microbiome: emerging molecular biomarkers for Parkinson’s disease. Int J Mol Sci. 2021;22(18):9839. doi:10.3390/ijms22189839. [Google Scholar] [PubMed] [CrossRef]
99. Uittenbogaard M, Brantner CA, Chiaramello A. Epigenetic modifiers promote mitochondrial biogenesis and oxidative metabolism leading to enhanced differentiation of neuroprogenitor cells. Cell Death Dis. 2018;9(3):360. doi:10.1038/s41419-018-0396-1. [Google Scholar] [PubMed] [CrossRef]
100. Yu Y, Wang H, Rao X, Liu L, Zheng P, Li W, et al. Proteomic profiling of lysine acetylation indicates mitochondrial dysfunction in the hippocampus of gut microbiota-absent mice. Front Mol Neurosci. 2021;14:594332. doi:10.3389/fnmol.2021.594332. [Google Scholar] [PubMed] [CrossRef]
101. Hino Y, Nagaoka K, Oki S, Etoh K, Hino S, Nakao M. Mitochondrial stress induces AREG expression and epigenomic remodeling through c-JUN and YAP-mediated enhancer activation. Nucleic Acids Res. 2022;50(17):9765–79. doi:10.1093/nar/gkac735. [Google Scholar] [PubMed] [CrossRef]
102. Kim DK, Jeong H, Bae J, Cha MY, Kang M, Shin D, et al. Aβ-induced mitochondrial dysfunction in neural progenitors controls KDM5A to influence neuronal differentiation. Exp Mol Med. 2022;54(9):1461–71. doi:10.1038/s12276-022-00841-w. [Google Scholar] [PubMed] [CrossRef]
103. Behera J, Ison J, Voor MJ, Tyagi N. Probiotics stimulate bone formation in obese mice via histone methylations. Theranostics. 2021;11(17):8605–23. doi:10.7150/thno.63749. [Google Scholar] [PubMed] [CrossRef]
104. Wenes M, Jaccard A, Wyss T, Maldonado-Pérez N, Teoh ST, Lepez A, et al. The mitochondrial pyruvate carrier regulates memory T cell differentiation and antitumor function. Cell Metab. 2022;34(5):731–46. doi:10.1016/j.cmet.2022.03.013. [Google Scholar] [PubMed] [CrossRef]
105. Weber-Stiehl S, Järke L, Castrillón-Betancur JC, Gilbert F, Sommer F. Mitochondrial function and microbial metabolites as central regulators of intestinal immune responses and cancer. Front Microbiol. 2022;13:919424. doi:10.3389/fmicb.2022.919424. [Google Scholar] [PubMed] [CrossRef]
106. Hinrichsen F, Hamm J, Westermann M, Schröder L, Shima K, Mishra N, et al. Microbial regulation of hexokinase 2 links mitochondrial metabolism and cell death in colitis. Cell Metab. 2021;33(12):2355–66. doi:10.1016/j.cmet.2021.11.004. [Google Scholar] [PubMed] [CrossRef]
107. Gadecka A, Bielak-Zmijewska A. Slowing down ageing: the role of nutrients and microbiota in modulation of the epigenome. Nutrients. 2019;11(6):1251. doi:10.3390/nu11061251. [Google Scholar] [PubMed] [CrossRef]
108. Sorrenti V, Fortinguerra S, Caudullo G, Buriani A. Deciphering the role of polyphenols in sports performance: from nutritional genomics to the gut microbiota toward phytonutritional epigenomics. Nutrients. 2020;12(5):1265. doi:10.3390/nu12051265. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools