 Open Access
Open Access
ARTICLE
SOX1 promotes osteosarcoma metastasis by modulating TSPAN12 expression
1 Hubei Key Laboratory for Kidney Disease Pathogenesis and Intervention, Hubei Polytechnic University School of Medicine, Huangshi, 435003, China
2 Jiangsu Key Laboratory of Marine Pharmaceutical Compound Screening, College of Pharmacy, Jiangsu Ocean University, Lianyungang, 222005, China
3 Cancer Center and Department of Pharmacology and Toxicology, Medical College of Wisconsin, Milwaukee, WI 53226, USA
* Corresponding Authors: LAN ZHU. Email: ; JING JI. Email:
# These authors contributed equally: Heyi Liu and Wenhao Cheng
(This article belongs to the Special Issue: New Perspectives on Inflammatory Cancer Transformation)
BIOCELL 2024, 48(10), 1465-1473. https://doi.org/10.32604/biocell.2024.052670
Received 10 April 2024; Accepted 12 August 2024; Issue published 02 October 2024
Abstract
Background: Osteosarcoma is the most common primary bone malignancy, with a strong tendency towards local invasion and metastasis. The SRY-Box Transcription Factor 1 (SOX1) gene, a member of the HMG-box family of DNA-binding transcription factors, plays a crucial role in embryogenesis and tumorigenesis. However, its role in osteosarcoma, particularly in relation to metastatic potential, is not well understood. Methods: The GSE14359 dataset containing five samples of conventional osteosarcoma and four samples of lung metastatic osteosarcoma was obtained from the Gene Expression Omnibus (GEO) database and analyzed for differential gene expression using the R language. Gene expression was detected using qPCR and Western blotting. Transcriptional activity was assessed by Luciferase reporter gene assays, and cell metastatic ability was assessed by migration and invasion assays. Results: The study demonstrated that SOX1 binds to a specific response element within the Transmembrane 4 Superfamily Member 12 (TSPAN12) promoter, upregulating TSPAN12 and its associated inflammatory pathways. Silencing TSPAN12 markedly reduces SOX1-mediated osteosarcoma cell invasion and inflammatory response, while TSPAN12 overexpression reverses these effects in SOX1-suppressed cells. Conclusion: In this study, our findings elucidate SOX1’s role in enhancing osteosarcoma metastasis via TSPAN12 upregulation, offering new insights into the molecular mechanisms of osteosarcoma progression.Keywords
Supplementary Material
Supplementary Material FileOsteosarcoma, the most common malignant bone tumor in pediatric and adolescent populations, constitutes approximately 20% of all bone malignancies [1,2]. Originating from stromal cells, these tumors are characterized by the production of osteoid tissue from malignant osteoblasts and exhibit a pronounced propensity for early metastasis [3–5]. The lung represents the most common site for such metastatic spread, with an estimated 20% of patients presenting lung metastases at initial diagnosis [6,7]. Despite considerable advances in osteosarcoma research and therapeutic strategies, the prognosis for affected individuals remains bleak, with no significant improvement in five-year survival rates [8,9]. This is largely attributable to the recurrence and metastasis of the primary tumor following surgical resection [10–12]. Consequently, there is an urgent need to elucidate the molecular mechanisms underlying osteosarcoma metastasis, in order to identify novel biomarkers and therapeutic targets that could potentially improve patient outcomes [13,14].
The transcription factor family sex-determining region Y-box (SRY), encompassed within the high mobility group box (HMG) domain, plays a pivotal role in tumorigenesis and cancer progression [15–17]. The SOX family of transcription factors, critical to cell differentiation and embryogenesis, has been implicated in various cancer pathways, serving both as prognostic indicators and potential therapeutic targets [18,19]. Interestingly, SOX1, a member of this family, exhibits variable expression patterns across different cancers. While high expression levels of SOX1 are associated with poor prognosis in glioblastoma, it appears to exert a suppressive effect on the proliferation and invasion of breast cancer cells when overexpressed [20,21]. Moreover, SOX1 has been shown to regulate the invasion of prostate cancer stem cells by modulating the signal transducer and activator of the transcription 3 (STAT3) signaling pathway, highlighting its complex role in cancer dynamics [22].
Parallel to SOX1, Tetraspanin 12 (TSPAN12), part of the tetraspanin protein family involved in retinal vascular development and tumor progression, has been shown to promote the oncogenic phenotype across a spectrum of tumors [23–26]. It does so by regulating the expression of pro-inflammatory cytokines through the WNT-β-Catenin signaling pathway, underlining its contribution to the malignant behavior of tumor cells [27,28]. The specific research objectives of this study were to investigate the simultaneous overexpression of SOX1 and TSPAN12 in osteosarcoma lung metastasis and to investigate their interactions and potential mechanisms in osteosarcoma metastasis. The study aims to reveal how SOX1 enhances osteosarcoma metastasis through the upregulation of TSPAN12, thus providing a scientific basis for the development of therapeutic interventions targeting this pathway.
For comprehensive molecular characterization, the GSE14359 dataset, encompassing microarray data for five conventional osteosarcoma and four lung metastatic osteosarcoma samples (each with duplicate measurements), was retrieved from the Gene Expression Omnibus (GEO). Differential gene expression analysis employed the limma package in R, setting thresholds at an absolute log fold change (|logFC|) greater than 1.5 and a p-value below 0.05. This approach facilitated the identification of 373 upregulated and 133 downregulated genes in lung metastasis samples compared to primary osteosarcoma specimens. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were conducted to delineate the functional implications of these differentially expressed genes, highlighting a significant association with cell migration, adhesion, and inflammatory signaling pathways.
Osteosarcoma cell lines, namely 143B, U2OS, HOS, MG63, and SAOS-2, alongside the hFOB1.19 normal osteoblast cell line, were procured from the American Type Culture Collection (ATCC, Virginia, USA) and were confirmed to be free of mycoplasma infection. All cell lines were cultured in DMEM/F12 medium (PM150312, Pricella, Wuhan, China) supplemented with 1% penicillin/streptomycin (E607011-0100, Sanon BioEngineering Shanghai, China) and 10% fetal bovine serum (FBS) (FBS-CE500, Nanjing Wobo Biotechnology, Nanjing, China), in a 37°C incubator with a 5% CO2 atmosphere. The cDNAs encoding SOX1 and TSPAN12 were PCR amplified from a HEK 293T cell cDNA library. Full-length cDNAs were constructed by cloning the open reading frames of these genes into the pBABE-3FLAG vector using standard procedures. All constructs were confirmed by sequencing. Targeted knockdown of SOX1 and TSPAN12 was achieved using lentiviral vectors encoding specific short hairpin RNAs (shRNAs), SOX1-shRNA: 5′-GCCAACCAGGACCGGGTCAAA-3′; TSPAN12-shRNA: 5′-CGATCTATTCTTCTGATGCTA-3′ synthesized by Shanghai Zhongze Biotechnology Co., Ltd. (Shanghai, China) and introduced into cells via Lipofectamine 2000 (11568030, Invitrogen, Waltham, MA, USA), following the manufacturer’s guidelines.
Cell viability, clonogenic formation, and cell proliferation
Cell viability was assessed using the Cell Counting Kit-8 (CCK-8) (P24052100142, Targetmol, Shanghai, China). Cells were seeded at a density of 1 × 104 cells per well in a 96-well plate. After 48 h, CCK-8 reagent was added, and absorbance at 450 nm was measured. For clonogenic formation assays, a total of 1 × 105 cells were plated in each well of 6-well plates. After 14 days, colonies were fixed and stained with crystal violet (1:1000 dilution, VS1003, VICMED, Xuzhou, China). Colonies containing more than 50 cells were counted. Cell proliferation was detected using the BrdU assay. Equal amounts of MG63 cells expressing PLKO, shSOX1#1, and shSOX1#2 cells were seeded at a density of 1 × 104 cells per well in 96-well plates. According to the instructions of the BrdU Cell Proliferation Assay Kit (CPK0010, Frdbio, Wuhan, China). Proliferation was quantified by measuring the optical density (OD) at 490 nm for each group using an enzyme marker (BioTek, Synergy Neo2, Winooski, VT, USA).
RNA extraction kit (KR116-02, Tiangen Biochemical Technology Co., Ltd., Beijing, China) was used to extract total RNA from various cells. The reverse transcription kit (FP217-02, Tiangen Biochemical Technology Co., Ltd.) was used to obtain the cDNA and it was stored in a refrigerator at −20°C until use. Reaction procedure: 94°C 10 min; 94°C 30 s, 62°C 30 s, 62°C 30 s, 40 cycles. The reaction system refers to the qPCR kit instructions. β-Actin (Types of PCR primers: human) was used as an internal reference with the following primers: forward 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse 5′-GGCTGTTGTCATACTTCTCATGG-3′; SOX1 (Types of PCR primers: human): forward 5′-CAGTACAGCCCCATCTCCAAC-3′ and reverse 5′-GCGGGCAAGTACATGCTGA-3′; TSPAN12 (Types of PCR primers: human): forward 5′-CCAGAGAAGATTCCGTGAAGTG-3′ and reverse 5′-GTCCCTCATCCAAGCAGAAAC-3′; IL6 (Types of PCR primers: human): forward 5′-ACTCACCTCTTCAGAACGAATTG-3′ and reverse 5′-CCATCTTTGGAAGGTTCAGGTTG-3′; TNF (Types of PCR primers: human): forward 5′-CCTCTCTCTAATCAGCCCTCTG-3′ and reverse 5′-GAGGACCTGGGAGTAGATGAG-3′; CXCL6 (Types of PCR primers: human): forward 5′-AGAGCTGCGTTGCACTTGTT-3′ and reverse 5′-GCAGTTTACCAATCGTTTTGGGG-3′; The 2−ΔΔCt method was used to calculate the relative expression levels of target genes.
Whole-cell extracts were prepared using SDS-containing lysis buffer (abs9117, Abcam, Boston, MA, USA). Equal amounts of protein samples were loaded into SDS-PAGEs and transferred onto polyvinylidene fluoride (PVDF) membranes (IPFL00010, Sigma, St. Louis, MO, USA). After blocking with 5% skim milk in Phosphate-buffered saline Tween (PBST) buffer, the membranes were incubated with the indicated primary antibodies (A0366, ABclonal, Wuhan, China) overnight at 4°C. Following washing with Phosphate-buffered saline Tween (PBST) buffer, the membrane was incubated with the secondary antibody for one hour at 25°C. Immunoreactive bands were visualized using enhanced chemiluminescence reagent (32106, Thermo Scientific, Shanghai, China) with β-Actin serving as a loading control. The membranes were washed with a PBST buffer and incubated with HRP-labeled (1:10000 dilution, A0305, Beyotime, Shanghai, China) secondary antibodies. The primary antibodies used in this study: SOX1 (1:10000 dilution, PA5-23351, Invitrogen), Flag (1:10000 dilution, F1804, Sigma), TSPAN12 (1:5000 dilution, PA5-80195, Invitrogen), β-actin (1:2000 dilution, sc-8432, Santa Cruz, BIOMART).
The promoter region of the TSPAN12 gene and SOX1 gene (2000 bp upstream of the Transcription Start Site) was amplified from the genomic DNA of MG63 cells. For luciferase reporter assays, cells were seeded in 24-well (1 × 105 cells were cultured in each well) plates and transfected with the indicated plasmids using Lipofectamine 2000 (11568030, Invitrogen) for 36 h. Luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega, E1960, Madison, WI, USA). The firefly luciferase luminescence data were normalized by the Renilla luciferase luminescence data.
For migration assay, osteosarcoma cells were inoculated in six-well plates (1 × 105 cells were cultured in each well) with serum-free medium for 24 h. When the cell confluence reached 80%–90%, a 10 μL pipette tip was used to scratch the monolayer to generate gaps. Images were taken at 0 and 36 h for each group. For invasion assay, 20 μL of Matrigel was plated onto the upper chamber of a Transwell insert. Cells (5 × 104) were seeded into the top chamber. The lower chamber was incubated with a culture medium with 10% FBS. 24 h later, the insert was fixed with 4% paraformaldehyde (ZY640017RE, Sigma-Aldrich, Shanghai, China) for 30 min. Non-migrated cells on the top surface of the insert were removed and migrated cells on the lower surface of the insert were stained with 0.05% crystal violet (60506ES60, Sigma-Aldrich, Shanghai, China). Images of cells were taken with a microscope (Nikon T12-N-N, Japan) at 200 × magnification and the relative migration was calculated.
Experimental data, derived from a minimum of three independent experiments, were analyzed using GraphPad Prism 8.0 (GraphPad Software, La Jolla, CA, USA) or R software. Results were expressed as mean ± standard deviation (SD). Comparative analysis between groups was performed using Student’s t-test, with significance levels set at *p < 0.05, **p < 0.01, ***p < 0.001.
Transcriptomic characteristics of osteosarcoma samples with or without lung metastasis
To elucidate the molecular architecture underlying the progression and metastatic dissemination of osteosarcoma, we conducted a comprehensive transcriptomic analysis using the GSE14359 dataset from the Gene Expression Omnibus (GEO). This dataset includes microarray analyses of five conventional osteosarcoma specimens and four osteosarcoma specimens with lung metastases, each replicated for robustness. We applied stringent criteria within the limma package, selecting differentially expressed genes (DEGs) based on an absolute log fold change (|logFC|) greater than 1.5 and a p-value less than 0.05 (Fig. 1A). This analytical approach revealed a significant upregulation of 386 genes and downregulation of 176 genes in the metastatic samples relative to their conventional counterparts (Fig. 1A,B, Tables S1 and S2).
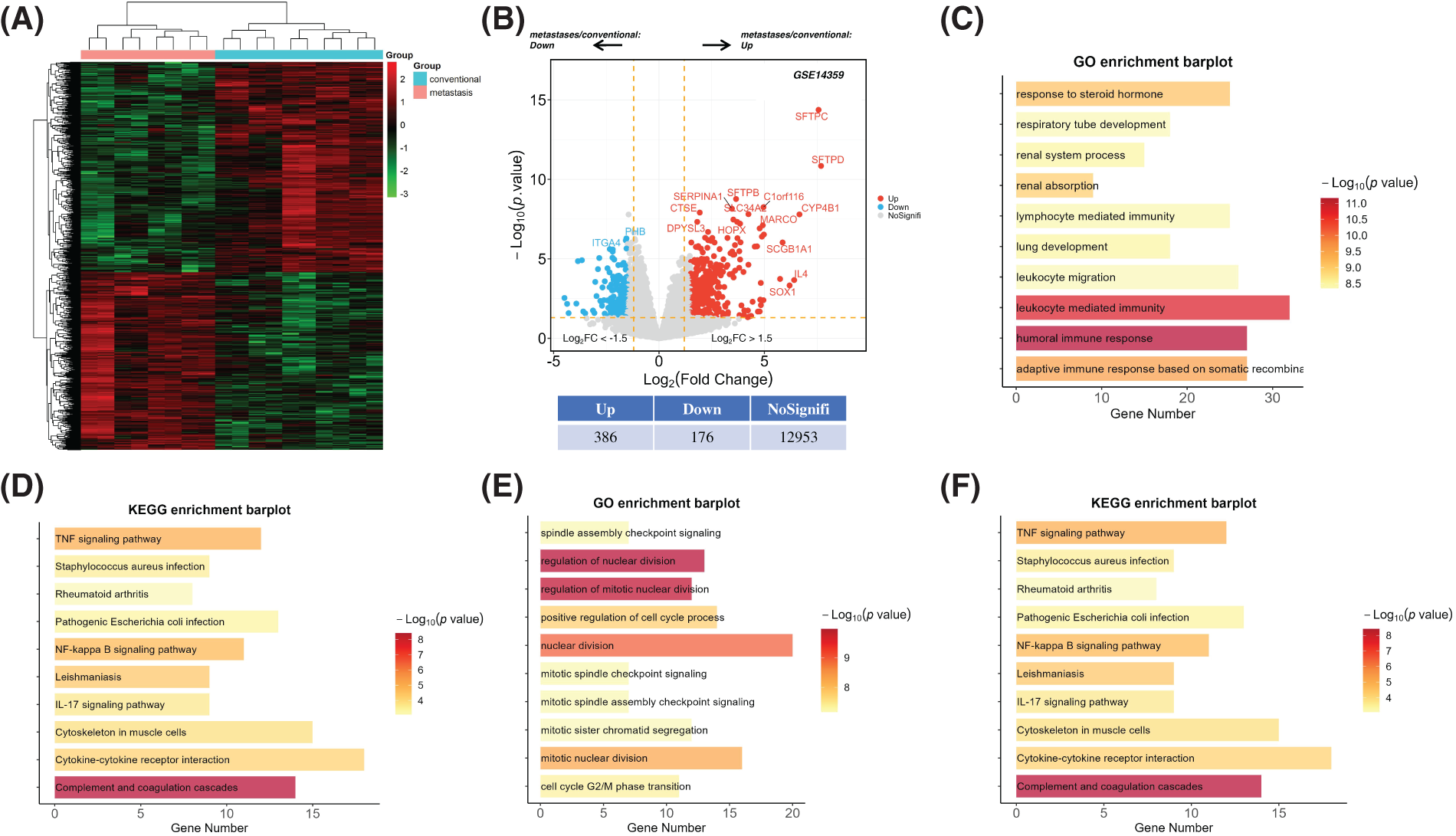
Figure 1: Transcriptomic analysis and functional classification of osteosarcoma samples with and without lung metastases. (A) The heatmap offers a visual comparison of differentially expressed genes (DEGs) between conventional osteosarcoma tissues and their lung metastatic counterparts, utilizing data from the GSE14359 database. This visualization highlights the distinct gene expression patterns characterizing metastatic progression. (B) Volcano plots illustrate the overall shifts in mRNA expression between lung metastases and conventional osteosarcoma samples from the GSE14359 dataset. Upregulated genes are represented by red dots, and downregulated genes by blue dots, with the magnitude of differential expression quantitatively annotated. (C and D) Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses provide a functional enrichment analysis of the 386 genes identified as significantly upregulated. (E and F) Similarly, the functional enrichment of 176 genes found to be downregulated is explored through GO and KEGG analyses, shedding light on the molecular and cellular dynamics potentially impeded in metastatic osteosarcoma.
Subsequent Gene Ontology (GO) analysis illuminated that the augmented gene set predominantly participated in processes integral to cell migration and adhesion (Fig. 1C). Concurrently, pathway interrogation via the Kyoto Encyclopedia of Genes and Genomes (KEGG) elucidated a pronounced enrichment of inflammatory pathways, notably those governed by Tumor Necrosis Factor (TNF) signaling, Nuclear Factor-kappa B (NF-κB), and Interleukin-17 (IL-17) (Fig. 1D). Inversely, the diminished gene set was closely associated with genomic integrity processes, such as chromosome condensation and mitotic checkpoint regulation, a finding mirrored by KEGG pathway analysis highlighting an enrichment in cell cycle and aging pathways (Fig. 1E,F).
Collectively, these insights suggest that dysregulated cell cycle dynamics and exacerbated inflammatory signaling play a pivotal role in facilitating the metastatic journey of osteosarcoma cells to the lungs. This comprehensive transcriptomic landscape delineates the molecular underpinnings of osteosarcoma metastasis and highlights potential molecular targets for therapeutic intervention, offering a refined understanding essential for advancing osteosarcoma research and treatment strategies.
SOX1 is elevated in osteosarcoma lung metastases samples and promotes the metastasis of osteosarcoma cells
Among the upregulated genes identified in osteosarcoma lung metastases, SOX1, a transcription factor from the SOX family, emerges as a focal point of our investigation. This interest is twofold: SOX1 is not only the sole member of the SOX family found to be upregulated in our dataset but also presents a unique opportunity to explore its role in osteosarcoma metastasis (Fig. 2A). Moreover, SOX1 displays a dichotomous role in cancer biology, oscillating between oncogenic and tumor-suppressive functions depending on the cellular context. An in-depth analysis of the Cancer Genome Atlas (TCGA, www.cancer.gov/ccg/research/genome-sequencing/tcga (accessed on 02 April 2024)) database to decode the interplay between SOX1 expression and various signaling pathways revealed a notable association with the HALLMARK_WNT_BETA_CATENIN_SIGNALING pathway across multiple tumor types (Fig. 2B). This suggests that SOX1 may promote tumor progression via this axis.
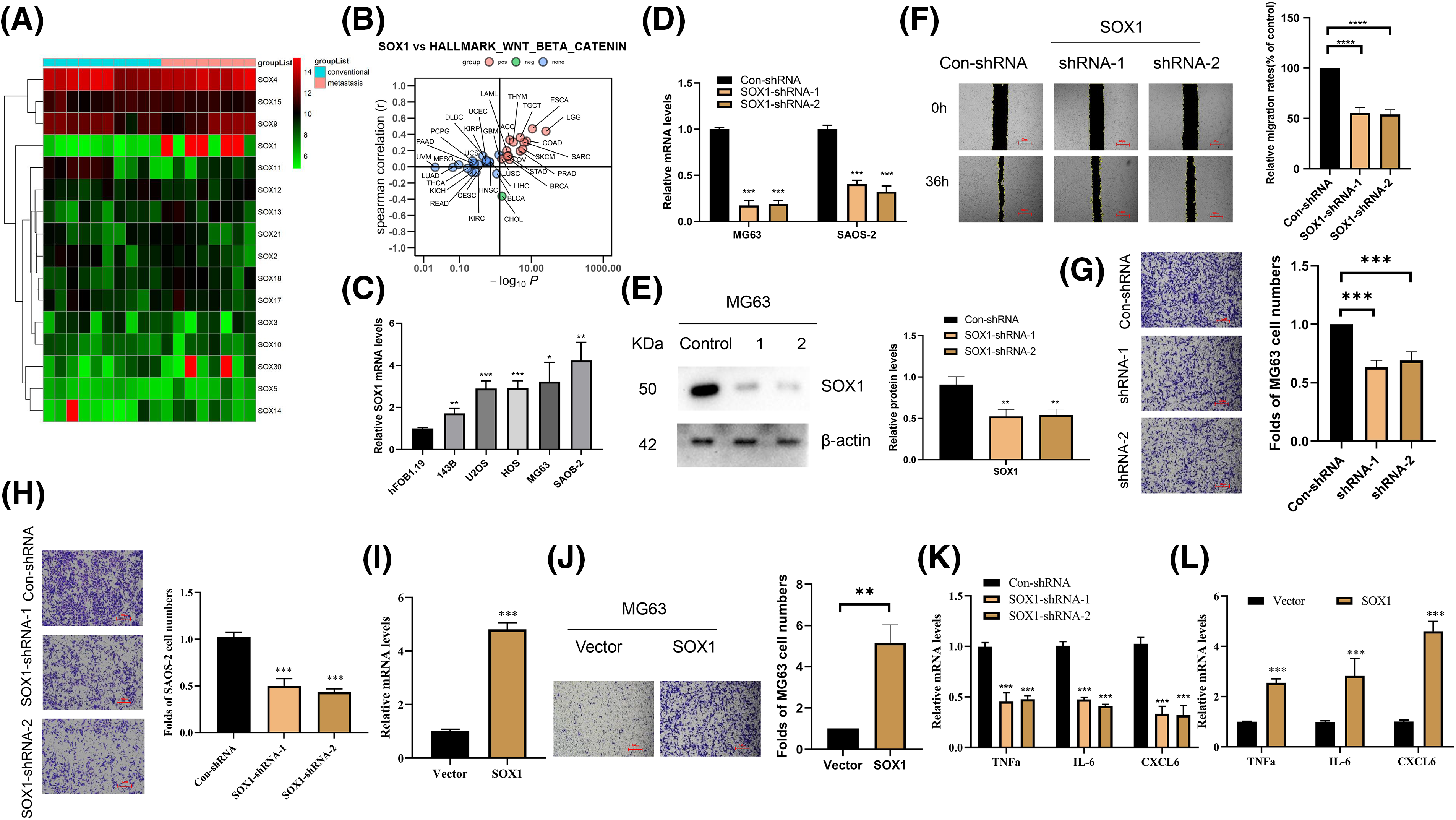
Figure 2: Elevation of SOX1 promotes metastasis of osteosarcoma cells. (A) A heatmap provides a comparative analysis of DEGs within the SOX family, contrasting conventional osteosarcoma tissues against those from lung metastases. This visualization underscores the distinct expression pattern of SOX1 among other family members. (B) A graphical representation of the Spearman correlation analysis shows the association between SOX1 expression and the activation of the Wnt/β-catenin signaling pathway across various tumor types in the TCGA database, with red indicating positive correlation and green denoting negative correlation. (C) Quantitative RT-PCR (RT-qPCR) analysis quantifies SOX1 mRNA levels across osteosarcoma cell lines compared to the hFOB1.19 normal human osteoblast cell line, highlighting SOX1 overexpression in the cancerous cell lines (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 vs. hFOB1.19. (D and E) Evaluation of SOX1 knockdown efficacy in MG63 and SAOS-2 cells via RT-qPCR and Western blot, employing two distinct SOX1-targeting shRNAs (n = 3). **p < 0.01, ***p < 0.001 vs. Con-shRNA. (F) The migratory capacity of SOX1-silenced cells vs. controls is assessed through wound healing assays, illustrating the role of SOX1 in MG63 cell migration (n = 3). ****p < 0.0001 vs. Con-shRNA. (G and H) Transwell assays evaluate the impact of SOX1 silencing on the invasive potential of MG63 and SAOS-2 cells, respectively, revealing reduced invasion in SOX1-silenced cells (n = 3). ***p < 0.001 vs. Con-shRNA. (I) RT-qPCR verifies the overexpression efficiency of SOX1 in MG63 cells (n = 3). (J) Transwell assays further elucidate the pro-invasive effect of SOX1 overexpression on MG63 cells (n = 3). **p < 0.01 vs. Con-shRNA. (K and L) RT-qPCR analysis compares the expression levels of pro-inflammatory cytokines (TNFa, IL-6, and CXCL6) in SOX1-knockdown vs. SOX1-overexpressing osteosarcoma cells, indicating SOX1’s role in modulating inflammatory cytokine expression (n = 3). ***p < 0.001 vs. Con-shRNA.
A comparative analysis highlighted the overexpression of SOX1 in osteosarcoma cell lines compared to the hFOB1.19 normal human osteoblast line (Fig. 2C), suggesting its key role in osteosarcoma pathogenesis. To test this hypothesis, targeted silencing of SOX1 in MG63 and SAOS-2 cells was achieved using two distinct shRNAs. This approach significantly reduced the cells’ migratory and invasive capacities (Fig. 2D–H). In contrast, when SOX1 was overexpressed in MG63 cells, it led to a notable increase in invasiveness (Fig. 2I,J). Furthermore, our CCK-8 assay results indicated that knockdown of SOX1 in MG63 cells did not significantly affect cell proliferation (Fig. S1A). Consistent with these findings, SOX1 knockdown did not result in changes in the number of bromodeoxyuridine (BrdU)-incorporating cells (Fig. S1A). Interestingly, colony formation assays also demonstrated that SOX1 knockdown had minimal impact on cell proliferation (Fig. S1A).
The investigation into SOX1’s role was further extended to understand its mechanistic underpinnings in promoting osteosarcoma metastasis. Bioinformatic analysis revealed an enrichment of inflammatory pathways, particularly those associated with cytokine and cytokine receptor interactions, in metastatic samples. This suggests a mechanism by which SOX1 might enhance osteosarcoma lung metastasis through modulation of the inflammatory milieu. This was empirically supported by the observation that SOX1 silencing led to a significant reduction in the levels of pro-inflammatory cytokines such as TNFa, IL-6, and C-X-C motif chemokine 6(CXCL6) in SAOS-2 cells (Fig. 2K), a trend inversely mirrored by SOX1 overexpression in MG63 cells (Fig. 2L).
Collectively, these findings highlight SOX1 as a key orchestrator of osteosarcoma cell migration and invasion, likely through its regulation of inflammatory signaling pathways. This dual capacity of SOX1 to modulate the cellular transcriptome and influence the inflammatory landscape provides a nuanced understanding of its role in the metastatic progression of osteosarcoma. These insights delineate potential therapeutic avenues aimed at mitigating the spread of this devastating disease.
TSPAN12 is a downstream target gene of SOX1
Given the pivotal role of transcription factors in orchestrating gene expression relevant to cancer metastasis, our investigation focused on SOX1, known for its regulatory capacity within the cellular milieu. Recognizing SOX1’s potential to modulate pro-metastatic genes, we directed our attention towards Tetraspanin 12 (TSPAN12), a gene implicated in enhancing tumor cell migration and invasion. Previous research, including our own, underscores TSPAN12’s elevated expression in metastatic osteosarcoma samples compared to their non-metastatic counterparts (Fig. 3A), along with its involvement in the regulation of inflammatory mediators—TNFa, IL-6, and CXCL6—via the WNT-β-Catenin signaling pathway. This evidence led us to hypothesize TSPAN12 as a functional intermediary through which SOX1 might exert its metastasis-promoting effects in osteosarcoma.
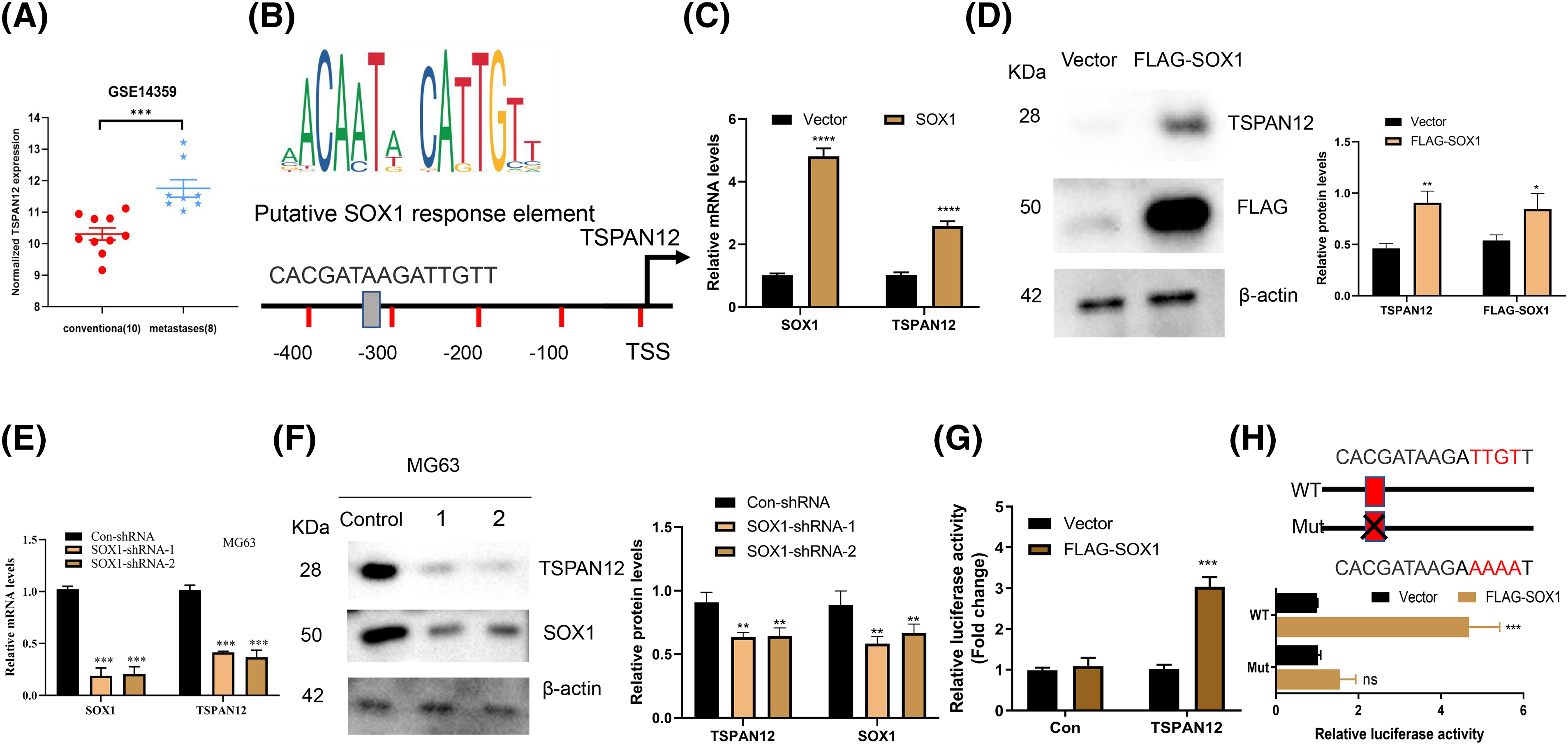
Figure 3: TSPAN12 is a downstream target gene of SOX1 in osteosarcoma. (A) Displays the differential mRNA expression levels of TSPAN12 between conventional osteosarcoma samples and those with lung metastases, as extracted from the GSE14359 dataset, highlighting TSPAN12’s upregulation in metastatic conditions. (B) A schematic diagram presents the TSPAN12 gene promoter region, emphasizing a putative SOX1 binding site proximal to the transcription start site (TSS), suggesting a direct regulatory relationship. (C) Quantitative RT-qPCR analysis measures the mRNA levels of both SOX1 and TSPAN12 in MG63 cells post-transfection with either an empty vector or a SOX1-expressing construct for 36 h, illustrating the effect of SOX1 overexpression on TSPAN12 transcription (n = 3). ****p < 0.0001 vs. vector. (D) Western blot analysis determines TSPAN12 protein levels in MG63 cells following transfection with an empty vector (EV) or a FLAG-tagged SOX1 construct, further substantiating SOX1’s influence on TSPAN12 expression at the protein level (n = 3). *p < 0.05, **p < 0.01 vs. vector. (E and F) Both mRNA and protein expression levels of TSPAN12 are assessed in MG63 cells post-transfection with control or SOX1-targeted shRNAs, demonstrating the regulatory impact of SOX1 knockdown on TSPAN12 expression (n = 3). **p < 0.01, ***p < 0.001 vs. Con-shRNA. (G) Constructs of luciferase reporter vectors containing the TSPAN12 promoter region (ranging from −2000 to 0 bp relative to the TSS) were co-transfected into MG63 cells alongside either an empty vector or a TSPAN12 expression vector to evaluate promoter activity via luciferase assays (n = 3). ***p < 0.001 vs. vector. (H) A comparative luciferase reporter assay involving wild-type (WT) and mutated SOX1 binding site constructs within the TSPAN12 promoter, when co-transfected with a TSPAN12 vector, delineates the specificity of SOX1’s transcriptional activation of TSPAN12, depicted through relative luciferase activities. This sequence of analyses affirms TSPAN12 as a downstream effector of SOX1, highlighting its potential role in osteosarcoma metastasis through transcriptional regulation. nsp > 0.05, ***p < 0.001 vs. vector.
To explore the regulatory relationship between SOX1 and TSPAN12, we commenced with an in-depth analysis of the TSPAN12 promoter region. Utilizing the JASPAR-A database of transcription factor binding profiles (https://jaspar.elixir.no/ (accessed on 02 April 2024)) online database, we identified a potential SOX1 binding motif within this region (Fig. 3B), hinting at TSPAN12’s candidacy as a transcriptional target of SOX1. Subsequent experimental manipulations of SOX1 expression in MG63 cells—both upregulation and suppression—revealed a direct correlation between SOX1 activity and TSPAN12 expression levels (Fig. 3C–F). Overexpression of SOX1 significantly elevated TSPAN12 expression, whereas SOX1 knockdown resulted in the opposite effect, substantiating our hypothesis of TSPAN12’s transcriptional governance by SOX1.
To further validate this regulatory mechanism, we engineered a fluorescent reporter vector incorporating the identified SOX1-responsive element from the TSPAN12 promoter. Functional assays demonstrated that SOX1 activation significantly enhanced the promoter activity of wild-type TSPAN12 (Fig. 3G,H). However, mutation of the SOX1 binding site within the promoter abrogated this effect, conclusively indicating TSPAN12 as a transcriptional target of SOX1 in the context of osteosarcoma (Fig. 3H).
These findings delineate a novel mechanistic pathway through which SOX1 influences osteosarcoma metastasis via the transcriptional regulation of TSPAN12. The identification of this SOX1-TSPAN12 axis provides significant insights into the molecular underpinnings of osteosarcoma dissemination and opens new avenues for targeted therapeutic strategies aimed at disrupting this oncogenic pathway.
SOX1 promoted the invasion of osteosarcoma partially through TSPAN12
Delving further into the molecular interplay between SOX1 and TSPAN12 within the osteosarcoma metastatic cascade, our investigation sought to determine the extent to which TSPAN12 facilitates the SOX1-driven modulation of inflammatory markers and the invasive capability of osteosarcoma cells. Intriguingly, we observed that augmenting TSPAN12 expression could substantially mitigate the reductions in inflammatory factor expression and cellular invasiveness attributed to SOX1 silencing (Fig. 4A–C). Conversely, the silencing of TSPAN12 markedly dampened the upregulation of inflammatory mediators and the enhanced invasive propensity resulting from SOX1 overexpression (Fig. 4D–F).
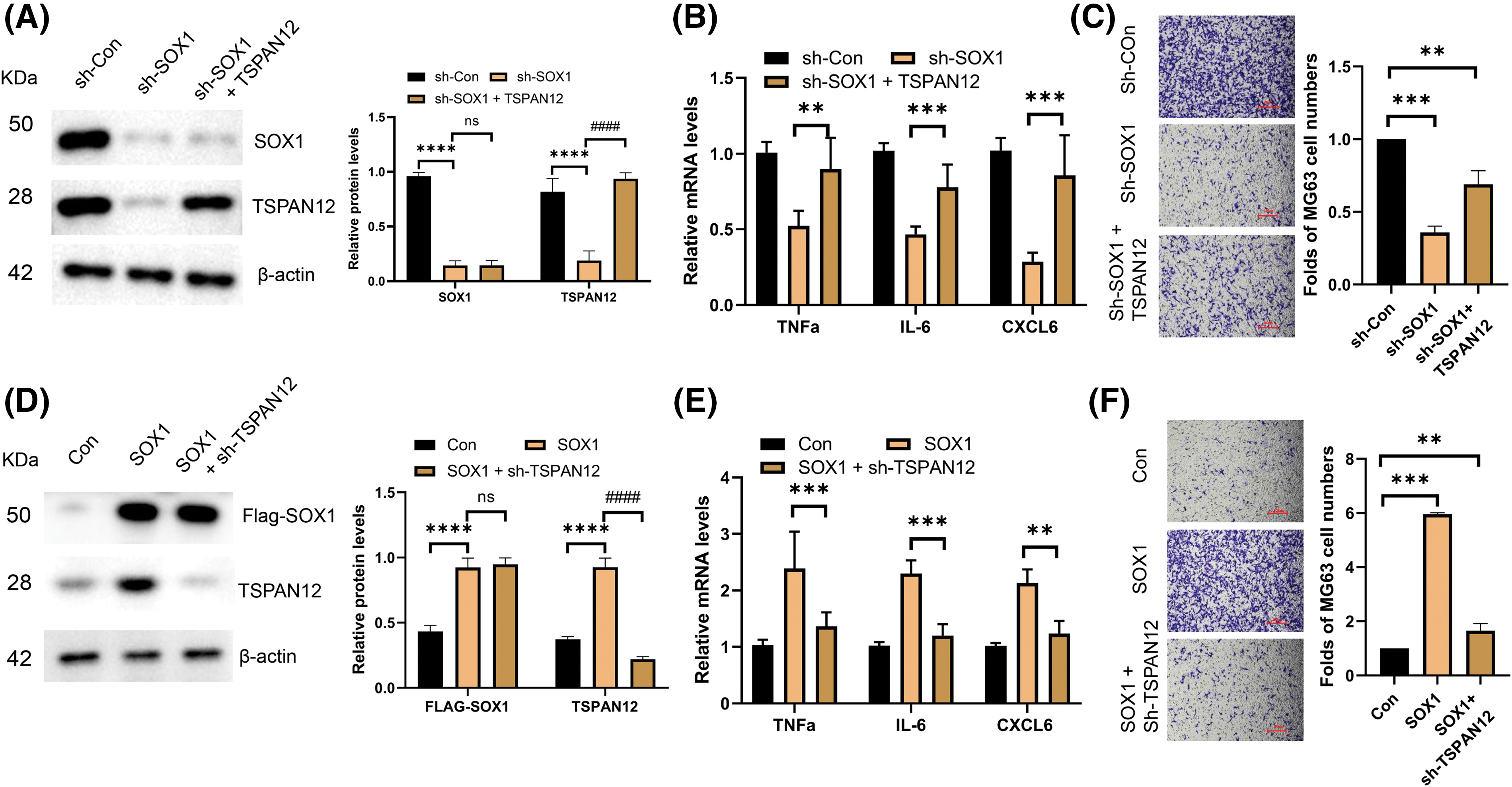
Figure 4: SOX1 promoted the invasion of osteosarcoma partially through TSPAN12. (A) Protein expression analysis by immunoblotting of MG63 cell lysates following transfection with control vectors, SOX1-targeting shRNAs, and a combination of sh-SOX1 and TSPAN12-expressing plasmids (n = 3). ****p < 0.0001 vs. sh-Con, nsp > 0.05, ####p < 0.0001 vs. sh-SOX1. (B) Quantitative RT-qPCR assessment of the inflammatory cytokines TNFa, IL-6, and CXCL6 in MG63 cells illustrates the downstream effects of SOX1 modulation on inflammatory mediator levels (n = 3). **p < 0.01, ***p < 0.001 vs. sh-SOX1. (C) Transwell invasion assay to evaluate the invasive capabilities of MG63 cells post-transfection, correlating SOX1 and TSPAN12 expression levels with cellular invasiveness (n = 3). **p < 0.01, ***p < 0.001 vs. Con-shRNA. (D) Protein expression analysis in MG63 cells transfected with control vectors, SOX1-overexpressing plasmids, and SOX1 plasmids alongside shRNAs targeting TSPAN12 (n = 3). ****p < 0.0001 vs. Con, nsp > 0.05, ####p < 0.0001 vs. SOX1. (E) RT-qPCR analysis in this set quantifies the impact of SOX1 and TSPAN12 expression modulation on the levels of key inflammatory cytokines, providing insight into their regulatory dynamics (n = 3). **p < 0.01, ***p < 0.001 vs. SOX1. (F) Transwell assays to assess the changes in invasive behavior of MG63 cells subjected to the described transfections, highlighting the role of the SOX1-TSPAN12 axis in promoting osteosarcoma invasion (n = 3). **p < 0.01, ***p < 0.001 vs. Con.
These findings illuminate TSPAN12’s crucial role as a downstream effector of SOX1, orchestrating a regulatory axis that significantly influences osteosarcoma’s metastatic behavior. By modulating the expression of key inflammatory factors, this SOX1-TSPAN12 pathway not only underscores the intricate molecular mechanisms driving osteosarcoma progression but also posits TSPAN12 as a pivotal mediator of SOX1’s pro-metastatic function. The elucidation of this pathway offers valuable insights into the complex regulatory networks at play in osteosarcoma metastasis and highlights potential therapeutic targets for curbing the aggressive spread of this malignancy.
In our investigation, we delved into the transcriptomic nuances of osteosarcoma, particularly focusing on the dichotomy between primary tumors and those with lung metastases [29]. This analysis highlighted the transcription factor SOX1, traditionally recognized for its pleiotropic roles across various cellular contexts. Our findings indicate that SOX1 not only serves as a hallmark of lung metastasis in osteosarcoma but also orchestrates a complex regulatory network influencing cell migration and invasion, two critical facets of metastatic dissemination.
The pivotal role of SOX1 in promoting metastatic behavior underscores a paradigm where transcription factors act as arbiters of cellular fate, directing the transition from a localized to a disseminated disease state [30,31]. The elevation of SOX1 in metastatic osteosarcoma samples suggests a pathogenic mechanism where SOX1 could recalibrate the cellular transcriptome towards a metastasis-promoting phenotype. This is particularly intriguing when considering the lack of effect SOX1 silencing had on cellular proliferation, indicating that its primary influence lies in enhancing cellular dynamics conducive to metastasis rather than fostering uncontrolled cell growth.
Further investigation into SOX1’s downstream targets identified TSPAN12 as a gene significantly upregulated in metastatic contexts and directly regulated by SOX1. This discovery was substantiated by the presence of a SOX1 response element within the TSPAN12 promoter region, highlighting a direct transcriptional regulation mechanism. The functional interplay between SOX1 and TSPAN12 extends beyond simple gene activation, encompassing the modulation of inflammatory pathways critical for metastatic progression. The downregulation of key inflammatory cytokines following SOX1 suppression indicates a broader role for SOX1 in shaping the tumor microenvironment, potentially through modulation of immune responses or alteration of cellular adhesion properties, which are essential for metastatic colonization.
The SOX1-TSPAN12 axis elucidates a novel mechanism of metastasis promotion in osteosarcoma, implicating inflammatory regulation as a central theme. This axis not only advances our understanding of the molecular underpinnings of osteosarcoma metastasis but also highlights the nuanced role of transcription factors in cancer progression. Unlike oncogenes that drive uncontrolled proliferation, SOX1’s impact on osteosarcoma appears to be mediated through its ability to prepare the cellular and extracellular landscape for metastatic spread.
This study’s revelations prompt a reevaluation of the potential therapeutic strategies targeting osteosarcoma. By identifying the SOX1-TSPAN12 interaction as a crucial mediator of metastatic behavior, we underscore the potential for interventions aimed at disrupting this axis. The development of therapeutic agents that can inhibit SOX1’s interaction with its target genes or block the downstream effects of TSPAN12 activation offers a promising avenue for limiting the metastatic spread of osteosarcoma. Moreover, the modulation of inflammatory pathways by SOX1 and TSPAN12 suggests that targeting inflammation could serve as an adjunctive strategy in managing osteosarcoma metastasis.
In conclusion, our study contributes to the expanding landscape of cancer biology by delineating the SOX1-TSPAN12 axis and opening new avenues for therapeutic intervention. This work underscores the complexity of metastasis as a biological phenomenon and the intricate molecular choreography that enables it.
This study provides important insights into the role of SOX1 in osteosarcoma metastasis and the mechanism of the SOX1-TSPAN12 axis. Specifically, the study reveals that SOX1, as a transcription factor, may contribute to the metastatic ability of tumors in osteosarcoma cells by regulating the expression of TSPAN12. However, despite these in vitro experimental results demonstrating the underlying molecular mechanisms, the present study still has some limitations, mainly the lack of validation in an in vivo model. In vivo, experiments can more realistically reflect the complex environment in an organism and validate the roles of SOX1 and its downstream pathways under actual physiological conditions. Therefore, future studies need further experiments in animal models to confirm the functions of SOX1 and TSPAN12 in osteosarcoma metastasis, thus providing a more reliable basis for potential therapeutic strategies.
Acknowledgement: None.
Funding Statement: This study was supported by the National Natural Science Foundation of China (82104174), Scientific Research Youth Project of Hubei Polytechnic University: Project No. 19XJK07Q, Hubei Provincial Education Science Planning 2023 Project No. 2023GB085, Jiangsu Province Basic Research Program Natural Science Foundation (Outstanding Youth Fund Project, BK20220063), the Key Program of Basic Science (Natural Science) of Jiangsu Province (22KJA350001), “Huaguo Mountain Talent Plan” of Lianyungang City (Innovative Talents Liu Bin), Qing Lan Project of Jiangsu Universities (Outstanding young backbone teachers, Ji Jing), the Natural Science Foundation of Jiangsu Higher Education Institutions of China (No. 20KJB350008), Priority Academic Program Development of Jiangsu Higher Education Institutions.
Author Contributions: Shaojie Ma, Lan Zhu, Jing Ji were responsible for designing the experiments, providing research ideas, and providing guidance on the direction and methodology of the study. They were also involved in data analysis and interpretation to ensure the scientific validity and accuracy of the study. In addition, significant comments and suggestions for changes were provided during the writing and revision of the manuscript to ensure the quality and completeness of the article. Heyi Liu, and Wenhao Cheng collected, organized and parsed the data from different databases and literatures to support the groundwork of this study. They also utilized real-time PCR to verify the accuracy and reliability of the bioinformatics data to ensure the credibility of the experimental results. Finally, they will also be responsible for writing and proofreading the paper to ensure that the language expression and structure of the paper are in accordance with academic standards. Jingliang He, Luyao Zhang, and Yulu Chen studied the expression regulation mechanism of specific genes through experiments, and investigated its association with cell function and disease relevance. Kadirya Asan, Jiayun Wang, and Qi Gao were responsible for conducting Western Blot experiments and accurately detecting the expression level of target proteins through Western Blot technique. They were also responsible for writing and proofreading the paper to ensure the language expression of the paper was accurate. Seng Wang and Zien Yu were responsible for the cell viability, clone formation and Cell proliferation experiments. The experimental data obtained will also be used and analyzed statistically to verify the credibility of the experimental results. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/biocell.2024.052670.
References
1. Wang J, Zhanghuang C, Tan X, Mi T, Liu J, Jin L, et al. A nomogram for predicting cancer-specific survival of Osteosarcoma and Ewing’s sarcoma in children: a SEER database analysis. Front Public Health. 2022;10:837506. doi:10.3389/fpubh.2022.837506. [Google Scholar] [PubMed] [CrossRef]
2. Ouyang H, Wang Z. Predictive value of the systemic immune-inflammation index for cancer-specific survival of osteosarcoma in children. Front Public Health. 2022;10:879523. doi:10.3389/fpubh.2022.879523. [Google Scholar] [PubMed] [CrossRef]
3. Le Nail LR, Brennan M, Rosset P, Deschaseaux F, Piloquet P, Pichon O, et al. Comparison of tumor- and bone marrow-derived mesenchymal stromal/stem cells from patients with high-grade osteosarcoma. Int J Mol Sci. 2018;19(3):707. [Google Scholar] [PubMed]
4. Mutsaers AJ, Walkley CR. Cells of origin in osteosarcoma: mesenchymal stem cells or osteoblast committed cells? Bone. 2014;62:56–63. doi:10.1016/j.bone.2014.02.003. [Google Scholar] [PubMed] [CrossRef]
5. Ferguson JL, Turner SP. Bone cancer: diagnosis and treatment principles. Am Fam Physician. 2018;98(4):205–13. [Google Scholar] [PubMed]
6. Chiesa AM, Spinnato P, Miceli M, Facchini G. Radiologic assessment of osteosarcoma lung metastases: state of the art and recent advances. Cells. 2021;10(3):553. [Google Scholar] [PubMed]
7. Robl B, Botter SM, Boro A, Meier D, Neri D, Fuchs B. Evaluation of F8-TNF-α in models of early and progressive metastatic osteosarcoma. Transl Oncol. 2017;10(3):419–30. doi:10.1016/j.tranon.2017.02.005. [Google Scholar] [PubMed] [CrossRef]
8. Huang Q, Liang X, Ren T, Huang Y, Zhang H, Yu Y, et al. The role of tumor-associated macrophages in osteosarcoma progression-therapeutic implications. Cell Oncol. 2021;44(3):525–39. doi:10.1007/s13402-021-00598-w. [Google Scholar] [PubMed] [CrossRef]
9. Yu L, Xia K, Gao T, Chen J, Zhang Z, Sun X, et al. The notch pathway promotes osteosarcoma progression through activation of ephrin reverse signaling. Mol Cancer Res. 2019;17(12):2383–94. doi:10.1158/1541-7786.MCR-19-0493. [Google Scholar] [PubMed] [CrossRef]
10. Tan X, Zeng C, Li H, Tan Y, Zhu H. Circ0038632 modulates MiR-186/DNMT3A axis to promote proliferation and metastasis in osteosarcoma. Front Oncol. 2022;12:939994. doi:10.3389/fonc.2022.939994. [Google Scholar] [PubMed] [CrossRef]
11. Geetha R, Iyer S, Keechilat P, Iyer N G, Thankappan KK, Smitha NV. Evaluation of premetastatic changes in lymph nodes (pN0) of oral tongue tumour: a prospective observational study. F1000Res. 2023;12:889. doi:10.12688/f1000research. [Google Scholar] [CrossRef]
12. Tang F, Tie Y, Lan TX, Yang JY, Hong WQ, Chen SY, et al. Surgical treatment of osteosarcoma induced distant pre-metastatic niche in lung to facilitate the colonization of circulating tumor cells. Adv Sci. 2023;10(28):e2207518. doi:10.1002/advs.v10.28. [Google Scholar] [CrossRef]
13. Wang JY, Yang Y, Ma Y, Wang F, Xue A, Zhu J, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. doi:10.1016/j.biopha.2019.109627. [Google Scholar] [PubMed] [CrossRef]
14. Zheng D, Xia K, Yu L, Gong C, Shi Y, Li W, et al. A novel six metastasis-related prognostic gene signature for patients with osteosarcoma. Front Cell Dev Biol. 2021;9:699212. doi:10.3389/fcell.2021.699212. [Google Scholar] [PubMed] [CrossRef]
15. Grimm D, Bauer J, Wise P, Kruger M, Simonsen U, Wehland M, et al. The role of SOX family members in solid tumours and metastasis. Semin Cancer Biol. 2020;67:122–53. doi:10.1016/j.semcancer.2019.03.004. [Google Scholar] [PubMed] [CrossRef]
16. Fu Q, Sun Z, Yang F, Mao T, Gao Y, Wang H. SOX30, a target gene of miR-653-5p, represses the proliferation and invasion of prostate cancer cells through inhibition of Wnt/β-catenin signaling. Cell Mol Biol Lett. 2019;24:71. doi:10.1186/s11658-019-0195-4. [Google Scholar] [PubMed] [CrossRef]
17. Jiang J, Wang Y, Sun M, Luo X, Zhang Z, Wang Y, et al. SOX on tumors, a comfort or a constraint? Cell Death Discov. 2024;10(1):67. doi:10.1038/s41420-024-01834-6. [Google Scholar] [PubMed] [CrossRef]
18. Ye P, Gu R, Zhu H, Chen J, Han F, Nie X. SOX family transcription factors as therapeutic targets in wound healing: a comprehensive review. Int J Biol Macromol. 2023;253:127243. doi: 10.1016/j.ijbiomac.2023.127243. [Google Scholar] [PubMed] [CrossRef]
19. Kumar P, Mistri TK. Transcription factors in SOX family: potent regulators for cancer initiation and development in the human body. Semin Cancer Biol. 2020;67:105–13. doi:10.1016/j.semcancer.2019.06.016. [Google Scholar] [PubMed] [CrossRef]
20. Song L, Liu D, He J, Wang X, Dai Z, Zhao Y, et al. SOX1 inhibits breast cancer cell growth and invasion through suppressing the Wnt/β-catenin signaling pathway. APMIS. 2016;124(7):547–55. doi:10.1111/apm.12543. [Google Scholar] [PubMed] [CrossRef]
21. Fang X, Yoon JG, Li L, Yu W, Shao J, Hua D, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12(1):11. doi:10.1186/1471-2164-12-11. [Google Scholar] [PubMed] [CrossRef]
22. Mathews LA, Hurt EM, Zhang X, Farrar WL. Epigenetic regulation of CpG promoter methylation in invasive prostate cancer cells. Mol Cancer. 2010;9(1):267. doi:10.1186/1476-4598-9-267. [Google Scholar] [PubMed] [CrossRef]
23. Liang G, Meng W, Huang X, Zhu W, Yin C, Wang C, et al. miR-196b-5p-mediated downregulation of TSPAN12 and GATA6 promotes tumor progression in non-small cell lung cancer. Proc Natl Acad Sci U S A. 2020;117(8):4347–57. doi:10.1073/pnas.1917531117. [Google Scholar] [PubMed] [CrossRef]
24. Wang Y, Lai Y, Jiang Z, Li S, Ding X. Five novel dysfunctional variants in the TSPAN12 gene in familial exudative vitreoretinopathy. Exp Eye Res. 2023;234:109574. doi:10.1016/j.exer.2023.109574. [Google Scholar] [PubMed] [CrossRef]
25. Ji G, Liang H, Wang F, Wang N, Fu S, Cui X. TSPAN12 precedes tumor proliferation by cell cycle control in ovarian cancer. Mol Cells. 2019;42(7):557–67. [Google Scholar] [PubMed]
26. Ju Y, Chen T, Ruan L, Zhao Y, Chang Q, Huang X. Mutations in TSPAN12 gene causing familial exudative vitreoretinopathy. Hum Genomics. 2024;18(1):22. doi:10.1186/s40246-024-00589-6. [Google Scholar] [PubMed] [CrossRef]
27. Bruguera ES, Mahoney JP, Weis WI. The co-receptor Tspan12 directly captures norrin to promote ligand-specific beta-catenin signaling. bioRxiv. 2024;63:668. doi:10.1101/2024.02.03.578714. [Google Scholar] [PubMed] [CrossRef]
28. Ding J, Lee SJ, Vlahos L, Yuki K, Rada CC, van Unen V, et al. Therapeutic blood-brain barrier modulation and stroke treatment by a bioengineered FZD4-selective WNT surrogate in mice. Nat Commun. 2023;14(1):2947. doi:10.1038/s41467-023-37689-1. [Google Scholar] [PubMed] [CrossRef]
29. Coleman CN, Mansoura MK, Marinissen MJ, Grover S, Dosanjh M, Brereton HD, et al. Achieving flexible competence: bridging the investment dichotomy between infectious diseases and cancer. BMJ Glob Health. 2020;5(12):e003252. doi:10.1136/bmjgh-2020-003252. [Google Scholar] [PubMed] [CrossRef]
30. Madhu V, Dighe AS, Cui Q, Deal DN. Dual inhibition of activin/nodal/TGF-β and BMP signaling pathways by SB431542 and dorsomorphin induces neuronal differentiation of human adipose derived stem cells. Stem Cells Int. 2016;2016(1):1035374. doi:10.1155/2016/1035374. [Google Scholar] [PubMed] [CrossRef]
31. Chiba S, Lee YM, Zhou W, Freed CR. Noggin enhances dopamine neuron production from human embryonic stem cells and improves behavioral outcome after transplantation into Parkinsonian rats. Stem Cells. 2008;26(11):2810–20. doi:10.1634/stemcells.2008-0085. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools