 Open Access
Open Access
REVIEW
An overview of autophagy in the differentiation of dental stem cells
State Key Laboratory of Oral & Maxillofacial Reconstruction and Regeneration, National Clinical Research Center for Oral Diseases, Shaanxi Clinical Research Center for Oral Diseases, Department of Pediatric Dentistry, School of Stomatology, The Fourth Military Medical University, Xi’an, 710032, China
* Corresponding Authors: LULU WANG. Email: ; LI-AN WU. Email:
(This article belongs to the Special Issue: Perspectives on Stem Cells and Regenerative Medicine)
BIOCELL 2024, 48(1), 47-64. https://doi.org/10.32604/biocell.2023.045591
Received 01 September 2023; Accepted 02 November 2023; Issue published 30 January 2024
Abstract
Dental stem cells (DSCs) have attracted significant interest as autologous stem cells since they are easily accessible and give a minimal immune response. These properties and their ability to both maintain self-renewal and undergo multi-lineage differentiation establish them as key players in regenerative medicine. While many regulatory factors determine the differentiation trajectory of DSCs, prior research has predominantly been based on genetic, epigenetic, and molecular aspects. Recent evidence suggests that DSC differentiation can also be influenced by autophagy, a highly conserved cellular process responsible for maintaining cellular and tissue homeostasis under various stress conditions. This comprehensive review endeavors to elucidate the intricate regulatory mechanism and relationship between autophagy and DSC differentiation. To achieve this goal, we dissect the intricacies of autophagy and its mechanisms. Subsequently, we elucidate its pivotal roles in impacting DSC differentiation, including osteo/odontogenic, neurogenic, and angiogenic trajectories. Furthermore, we reveal the regulatory factors that govern autophagy in DSC lineage commitment, including scaffold materials, pharmaceutical cues, and the extrinsic milieu. The implications of this review are far-reaching, underpinning the potential to wield autophagy as a regulatory tool to expedite DSC-directed differentiation and thereby promote the application of DSCs within the realm of regenerative medicine.Keywords
The extensively investigated mesenchymal stem cells (MSCs) were collected from several adult tissues, such as bone marrow, adipose tissue, dental tissue, and birth-derived tissues [1]. However, MSCs obtained from bone marrow showed certain shortcomings, including pain, morbidity, and limited cell number [2]. MSCs from adipose tissue isolated via lipoaspirates or lipectomy faced the disadvantages of techniques and the limitation of donor age [3]. Currently, there is growing evidence that dental stem cells (DSCs) have several distinct advantages that make them an ideal candidate for regenerative medicine. Specifically, these cells are accessible and reliable, conventionally isolated from discarded teeth or periodontal tissue, obviating invasive procedures. More importantly, DSCs exhibit multi-lineage differentiation, low immunogenicity, circumventing ethical concerns, and heightening the viability of autologous transplants.
It is now understood that DSCs can be readily obtained from tissues originating from the oral region [4]. To date, numerous different types of human DSCs have been identified and named according to their origin, including dental pulp stem cells (DPSCs) [5], stem cells from human exfoliated deciduous teeth (SHED) [6], stem cells from apical papilla (SCAPs) [7], periodontal ligament stem cells (PDLSCs) [8], dental follicle cells (DFCs) [9], and gingival mesenchymal stem cells (GMSCs) [10]. The main characteristics of DSCs are shown in Table 1. However, DPSCs exhibit a higher proliferation potential, superior neuroprotective property and neurogenetic ability as compared with MSCs from bone marrow, adipose tissue, and umbilical cord [11].
DSCs belong to the MSC category, mirroring the characteristics of MSCs, including self-renewal ability and multi-lineage differentiation potential [12,13]. It has been demonstrated that DSCs possess the abilities of dentinogenesis, cementogenesis, osteogenesis, adipogenesis, chondrogenesis, myogenesis, angiogenesis, neurogenesis, and hepatogenesis when provided with the appropriate inductive signals [14]. In addition, DSCs exert potent immunomodulatory functions [14,15]. Their easy availability and advantages position them as promising entities in various biomedical fields, including regenerative dentistry, osseous reconstruction, and neural tissue regeneration [16]. Therefore, a comprehensive exploration of the regulatory mechanisms underlying DSC differentiation into specific cellular and tissue types is imperative.
The differentiation process of DSCs is significantly influenced by a range of regulatory factors, with previous studies primarily focusing on genetic, epigenetic, and molecular aspects [17–19]. Recently, the role of autophagy in DSC differentiation has emerged as a research hotspot. Autophagy typically orchestrates cellular homeostasis by degrading redundant or detrimental cytoplasmic constituents, particularly during stress conditions. Dysregulated autophagy has been linked to various clinical diseases, including cancer, cardiovascular diseases, inflammation, metabolic syndromes, neurodegeneration, and musculoskeletal disorders [20]. In dental medicine, autophagy participates in the physiology and pathophysiology of teeth, including tooth development, pulp aging, periapical lesions, and stress adaptation [21,22]. It also reportedly participates in suppressing inflammation within the dentin-pulp complex, dental pulp, and periapical tissues [22,23]. Additionally, autophagy is intricately connected to stem cell developmental processes encompassing self-renewal, lineage commitment, and cell aging [24,25].
Mammalian autophagic pathways can be classified into three types based on morphological and mechanistic features: macroautophagy [75], microautophagy [76], and chaperone-medicated autophagy (CMA) [77]. Among these, macroautophagy is the predominant system and has been extensively investigated. Macroautophagy involves membrane elongation, resulting in the formation of the autophagosome that has a double-membraned structure [78]. Microautophagy involves membrane internalization, generating intraluminal vesicles [79,80]. The above two pathways undergo selective or non-selective processes that transport cytoplasmic components into lysosomes to be degraded. However, CMA could directly and selectively transport cytosolic materials into lysosomes without membrane deformation [77,81].
Recent research has corroborated that autophagy activation is involved during cell differentiation [82,83]. Autophagy inhibition could turn young MSCs into a relatively aged state that exhibits degenerative changes [84]. In an in vivo study, autophagy activation could restore bone loss in aged mice, suggesting autophagy might be a future target for clinical treatment of age-related bone loss [84]. Thus, it is highly conceivable that autophagy plays a pivotal role during stem cell differentiation compared to their undifferentiated state. However, excessive autophagic activity may lead to cell function dysregulation and cell death [85–88], accentuating the challenge of appropriately harnessing autophagy in directing stem cell differentiation. Recent studies have substantiated autophagy modulation as pivotal for DSC lineage commitment and potentially pivotal in maxillofacial tissue regeneration, yet no comprehensive review has been reported.
This review aims to document recent advances in understanding the intricate regulatory mechanisms and relationships between autophagy and DSC differentiation. We conducted a comprehensive search for relevant articles based on up-to-date databases, including PubMed, Google Scholar, and Web of Science. The keywords used in our search included “DSCs”, “DPSCs”, “SHED”, “SCAPs”, “DFCs”, “GMSCs”, “DPLSCs”, “autophagy”, “mitophagy”, “cell differentiation”, and related terms. We applied no publication date limits and incorporated articles that published in English. We screened the titles, abstracts, and full texts of the identified articles to select articles related to the topic of autophagy in DSC differentiation. Studies with insufficient or poor-quality data and duplicates were excluded. Our final selection included articles that contributed valuable insights into the role of autophagy in the differentiation of dental stem cells. This review delineates the autophagy mechanism, emphasizes its function and regulation in DSC differentiation, and provides insights into the regulatory mechanisms that can be harnessed as targets in DSC-driven regenerative medicine. Furthermore, this review outlines a potential approach to the precise regulation of autophagy in stem cell differentiation, serving as a guide for DSC-based research and promoting their application in tissue reconstruction and regeneration.
The Process and Mechanism of Autophagy
Macroautophagy, henceforth referred to as autophagy, represents a self-degradation and recycling mechanism, serving as a “housekeeping” function by eliminating dysfunctional organelles, pathogenic proteins, and intracellular pathogens [89,90]. It is a dynamic process that can be briefly subdivided into five phases: initiation, nucleation, phagophore expansion, sealing and cargo sequestration, and autolysosome formation (see Fig. 1) [75,91].
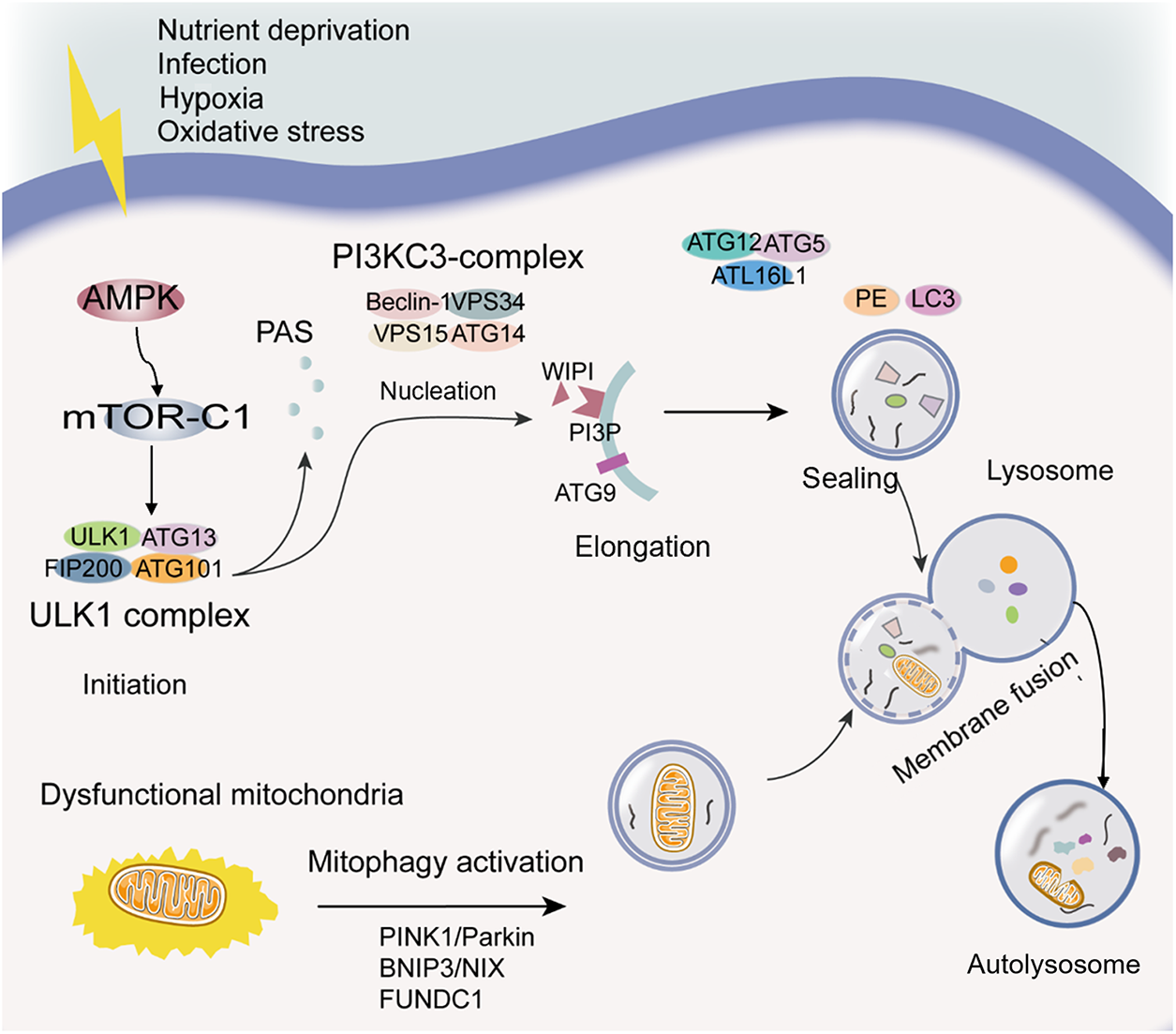
Figure 1: Diagram of the autophagy process. Initiation: The initiation of the autophagic process can be induced by various stress conditions, which result in AMPK activation, mTOR-C1 inactivation, and ULK1 complex phosphorylation. Nucleation: PI3KC3-complex phosphorylation and PI3P generation contribute to phagophore nucleation. Elongation: WIPI binding to PI3P and the transmembrane protein ATG9-containing vesicles contribute to the elongation of the phagophore membrane. Sealing: Two ubiquitin-like protein conjugation systems (ATG12 conjugates with ATG5 and ATL16L1, LC3 conjugates with PE) are required for elongation and closure of the phagophore membrane, and its cargo sequestration can be nonselective or selective. The selective autophagy is related to LC3 and LC3 homologs. Autolysosome formation: The autophagosome outer membrane fuses with the lysosomal membrane to form a single-membrane autophagic body termed the autolysosome. Mitophagy: Mitophagy, targeted dysfunctional mitochondria, is a particular type of autophagy. PINK1/Parkin, BNIP3/NIX, and FUNDC1 signaling pathways are mainly mitophagy regulatory pathways. Abbreviations: AMPK, adenosine monophosphate-activated protein kinase; ATG, autophagy-related proteins; BNIP3/NIX, BCL2 interacting protein 3/NIP3-like protein X signaling; FUNDC1, FUN14 domain containing 1; LC3, microtubule-associated protein 1 light chain 3; mTOR-C1, the mechanistic target of rapamycin complex 1; PAS, phagophore assembly site; PE, phosphatidylethanolamine; PI3KC3, the class III PI3K; PI3P, phosphatidylinositol-3-phosphate; PINK1/Parkin, the phosphatase and tensin homolog-induced putative kinase 1/E3 ubiquitin ligases Parkin signaling; ULK1, Unc-51-like kinase 1; WIPI, WD-repeat domain phosphoinositide-interacting.
The process of autophagosome biogenesis is intricate and is typically initiated by a wide range of cellular events, including oxidative stress, infection, nutrient deficiency, and hypoxia, among others [92]. These events lead to the recruitment of approximately 16–20 core autophagy-related (ATG) proteins to the phagophore assembly site (PAS), acting at specific stages of autophagosome initiation or formation [93,94]. Current evidence suggests that adenosine monophosphate-activated protein kinase (AMPK) activation is triggered in response to energy insufficiency-induced stress, leading to the inhibitory phosphorylation of the mechanistic target of rapamycin complex 1 (mTOR-C1). AMPK-induced autophagy activation can also circumvent mTOR and directly phosphorylate key proteins in initiation, such as Unc-51-like kinase 1 (ULK1) and ATG13. Notably, the ULK1 complex, comprising ULK1, ATG13, FIP200, and ATG101, holds pivotal importance in autophagy initiation, prompting phagophore nucleation through the phosphorylation of components within the class III PI3K (PI3KC3) complex I (PI3KC3-C1 consists of Beclin 1, VPS34, VPS15, and ATG14) [92,93]. Beclin 1, a critical component in the PI3KC3 complex, plays a vital role in initiating the autophagic process [95]. Besides, it has been reported that PI3KC3-C1 leads to phosphatidylinositol-3-phosphate (PI3P) generation to act in autophagosome nucleation [93].
The phagophore is a specific structure that arises from the membrane generated during nucleation. It has been established that the involvement of WD-repeat domain phosphoinositide-interacting (WIPI) proteins and related entities occurs during the early phases of membrane elongation via PI3P binding [91]. Furthermore, the transmembrane protein ATG9-containing vesicles contribute to the elongation of the phagophore membrane by delivering various cellular membranes such as the plasma membrane, mitochondrial membrane, recycling endosomes, and the Golgi complex membrane [92]. Over time, the phagophore membrane elongates, enclosing a portion of the cytosol, leading to membrane expansion and sealing into a characteristic double-membraned spherical vesicle known as the autophagosome [92]. Studies have indicated that two ubiquitin-like protein conjugation systems are integral to phagophore membrane elongation and closure: ATG12 conjugates with ATG5 and ATL16L1, and members of the ubiquitin-like protein LC3 (microtubule-associated protein 1 light chain 3) subfamily, part of the ATG8 protein family, conjugate with membrane-resident phosphatidylethanolamine (PE) [89,93].
This autophagosome entraps substances, excess or potentially hazardous cytoplasmic entities, and intracellular pathogens for subsequent degradation as autophagic cargo [93]. While autophagy has been considered primarily as a non-selective degradation pathway, it plays a critical role in the digestion of specific cargoes like proteins and organelles. LC3 and its homologs are pivotal in sequestering specifically labeled autophagic substrates into autophagosomes through cargo receptors containing the LC3-interacting region during selective autophagy. Conversely, nutrient scarcity initiates bulk autophagy, which is relatively nonselective, engulfing a range of cytoplasmic substrates [75]. The closure of the autophagosomal membrane marks the completion of autophagosome formation.
Subsequently, the outer membrane of the autophagosome fuses with the lysosomal membrane, resulting in the formation of a single-membraned autophagic body termed the autolysosome. Within the lysosomal lumen, acidic hydrolases facilitate the degradation of the autolysosome, releasing salvaged nutrients back into the cytoplasm for cellular reuse [92].
According to the selectivity towards degradation substrates, autophagy can be divided into mitophagy, reticulophagy, ribophagy, pexophagy, etc. [96]. Recently, mitophagy, an autophagic process that targets mitochondria, has attracted significant interest as a specialized form of autophagy. It maintains mitochondrial quality and homeostasis by selectively eliminating dysfunctional or surplus mitochondria [97]. Mitochondrial dysfunction can be assessed in several ways, such as: mitochondrial membrane potentia (ΔΨm) measured using fluorescent probes like JC-1 or TMRM, the elevated ROS levels detected by DHE (dihydroethidium) or MitoSOX, abnormal mitochondrial morphology and mass assessed by electron microscopy and staining with fluorescent dyes like MitoTracker, and functional assay including those measuring ATP production, oxygen consumption, and mitochondrial respiratory chain activity, which can provide insight into mitochondrial function and dysfunction [98,99]. In addition, laser-induced autofluorescence could be used to detect mitochondrial dysfunction in vitro [100].
Mitochondrial dysfunction plays a crucial role in the autophagy cascade. Membrane depolarization of mitochondria is a trigger of autophagy [101]. The cellular mitochondria of ROS production and oxidative modification may be primary targets of autophagy [102]. Mitophagy is initiated when mitochondrion becomes damaged or impaired. Mitophagy, a specialized form of autophagy, is the selective targeting and elimination of damaged mitochondria. Hence, mitochondria contribute to prosurvival functions of autophagy. Currently, regulatory pathways of mitophagy primarily revolve around the phosphatase and tensin homolog-induced putative kinase 1 (PINK1)/E3 ubiquitin ligases Parkin signaling, BCL2 interacting protein 3 (BNIP3)/NIP3-like protein X (NIX) signaling, and FUN14 domain containing 1 (FUNDC1) signaling [25,103].
Briefly, autophagy/mitophagy is a self-digestive process that is conserved in eukaryotic cells and responsible for maintaining cellular homeostasis through degrading of harmful components in cells. Previous research has found pharmacological inhibition of autophagy via chloroquine (late-stage autophagy inhibitor, already in clinical use) may be a promising strategy for alleviating pathologic bone loss induced by glucocorticoids and by ovariectomy in murine models [104], suggesting late autophagy regulation may play a protective role in a hostile environment.
Effects of Autophagy on DSC Differentiation
Stem cell-based tissue engineering holds promise for more robust tissue repair and regeneration. A mounting body of evidence underscores the correlation between autophagy and DSC differentiation (see Table 2), especially osteo/odontogenic differentiation, neuronal differentiation, and vasculogenic differentiation.
Osteo/odontogenic differentiation
In recent years, significant emphasis has been placed on the osteogenic and odontogenic differentiation abilities of DSCs. Given the shared components and properties of bone and dentin, we attempt in this section to describe the potential impact of autophagy on osteogenic/odontogenic differentiation and mineralization of DSCs, offering a tantalizing prospect for tissue regeneration.
Recent in vitro investigations have predominantly focused on autophagy’s ultrastructure and the expression levels of autophagy-related genes in stem cells cultured in osteo/odontogenic induction conditions. In this regard, human dental pulp cells (DPCs) have become a research hotspot. During odontoblastic differentiation, the expression of dentin matrix protein-1 (DMP-1) and dentin sialoprotein (DSP) is steadily increased, paralleled by autophagosome and autophagolysosome formation. Furthermore, there is a notable shift from cytosolic LC3-I to the autophagosome marker LC3-II, albeit with diminished levels of the selective autophagy cargo receptor p62 [105]. A similar trend was observed in the osteoblast differentiation of DPSCs. Core ATG proteins (ATG3 and ATG7) are upregulated, while the autophagy-inhibiting factor mTOR is significantly downregulated [106]. These findings collectively indicate autophagy activation during the osteo/odontogenic differentiation process.
The prevailing consensus suggests that autophagy activation could potentially potentiate osteoblast and odontoblast mineralization, whereas its inhibition might yield opposite effects [107–111]. It has been corroborated that autophagy facilitates type I collagen synthesis, thereby modulating the osteoblastic differentiation of periodontal ligament (PDL) cells [112]. Moreover, Nollet et al. [113] reported that autophagy bodies of living cells can be used as carriers to secrete apatite crystals into the extracellular matrix. Additionally, autophagy interfaces with signaling pathways to amplify cell differentiation and mineralization. In this respect, research has shown that autophagy fosters odontoblastic differentiation via the phosphatidylinositol 3-kinase (PI3K)/Akt signaling while simultaneously attenuating stemness in DPCs under odontogenic induction conditions [105]. AMPK activation has been reported to promote osteogenic differentiation of DPSCs through different approaches: early mTOR inhibition-mediated autophagy and later activation of the Akt/mTOR signaling axis [114] suggesting that the differentiation ability of DSCs is closely associated with the regulatory effects of autophagy.
The emphasis should be placed on the distinct autophagic shifts occurring at different osteo/odontogenic differentiation stages. Activation or inhibition of autophagy barely affected early differentiation markers. During extended cultures with an osteogenic differentiation medium, autophagy markers experience significant downregulation. Under these circumstances, autophagy activation significantly mitigates biomineralization ability of DFCs, while autophagy inhibition yields a divergent effect [115]. However, another report proves that initiating autophagy could enhance early osteogenic activity via AMPK/mTOR signaling pathway [116]. This temporal regulation and stem cell type might account for the discrepancies between the inhibition and stimulation effects of autophagy in response to mineralization induction. Essentially, by accelerating the hard tissue formation potential of DSCs, the precise modulation of autophagy may emerge as a crucial factor in promoting dentinogenesis and osteogenesis.
Although most studies have focused on the abilities of DSCs to differentiate into odontoblasts or osteoblasts, it should be borne in mind that DSCs also possess a powerful neurogenic differentiation capacity [11,60]. Originating from the neural crest, DSCs express distinct neural markers such as nestin, glial fibrillary acidic protein (GFAP), β-tubulin III, synaptophysin, and S100 protein [7,117]. Furthermore, it has been reported that human DPSCs exhibit higher neurotrophin levels and more extensive innervation than human bone marrow-derived MSCs [118], positioning DSCs as a promising candidate for neural tissue regeneration and repair [119].
Upon 7 days of DPSC cultivation in neural differentiation-specific media, differentiated cells adopt a neural-like pyramidal morphology characterized by short dendrite projections and elongated axon-like extensions, accompanied by a significant elevation in neural-specific markers [120]. Prateeksha et al. [120] revealed significant upregulation of autophagy and mitophagy-associated markers during neural differentiation (ND) of DPSCs. Enhancing autophagy and mitophagy levels notably enhanced ND, underscoring the pivotal role of autophagy and mitophagy in directing DPSCs toward neural commitment. Another research has demonstrated that the autophagy inducer, 5-Azacytidine, enhances neuronal differentiation of MSCs [121], there remains a paucity of studies investigating the role of autophagy in stem cell neuronal differentiation, warranting further exploration of the underlying mechanisms.
It is now understood that mitochondria, which are the targets of autophagy, play a pivotal role in providing energy during cell differentiation. Previous studies have observed heightened mitochondrial membrane potential, increased mitochondrial DNA, and elongated mitochondria during the neuronal differentiation of SHED. Inhibition of mitochondrial activity has been shown to impede neuronal differentiation, emphasizing the significance of mitochondrial function [122]. Additionally, increased mitochondrial respiration and spare respiratory capacity have been observed in neuron-like differentiated cells [120]. Upon depletion of mitochondrial fission factor (MFF) in SHED differentiating into dopaminergic neurons, the marker protein of mitochondria biogenesis was downregulated, and neurons showed impaired neurite outgrowth and excessive reactive oxygen species (ROS) [123]. It has been confirmed that autophagy is an important regulator in fighting against excessive ROS-induced impairment [124,125]. These findings suggest the crucial function of mitochondria in the neuronal differentiation of stem cells.
Therefore, it is unsurprising that mitochondrial dysfunction has been implicated in various human diseases, including neurodevelopmental and neurodegenerative disorders. Impaired neurite development, linked to mitochondrial dysfunction, has been observed in dopaminergic neurons differentiated from stem cells obtained from children with neurodegenerative diseases like Rett syndrome [126], autism spectrum disorder (ASD) [127], attention deficit hyperactivity disorder (ADHD) [128], and Down syndrome [129], as well as DPSCs obtained from a patient with metabotropic dysplasia and multiple neuropsychiatric symptoms [130], suggesting mitochondria might be a potential therapeutic target of neuro-related diseases. Nevertheless, further investigations are warranted to fully elucidate the role and mechanism of mitophagy in neuro-related disorders.
First of all, we should make clear the difference between vasculogenesis and angiogenesis, which are two subsequent processes of the development of blood vessels system. Angiogenesis refers to the establishment of new blood vessels from the preexisting vasculatures, occurs throughout life, and consists of four stages. Vasculogenesis is the formation and development of the vascular system, including the formation of blood vessels from endothelial cells during embryogenesis (see Fig. 2) [131,132].
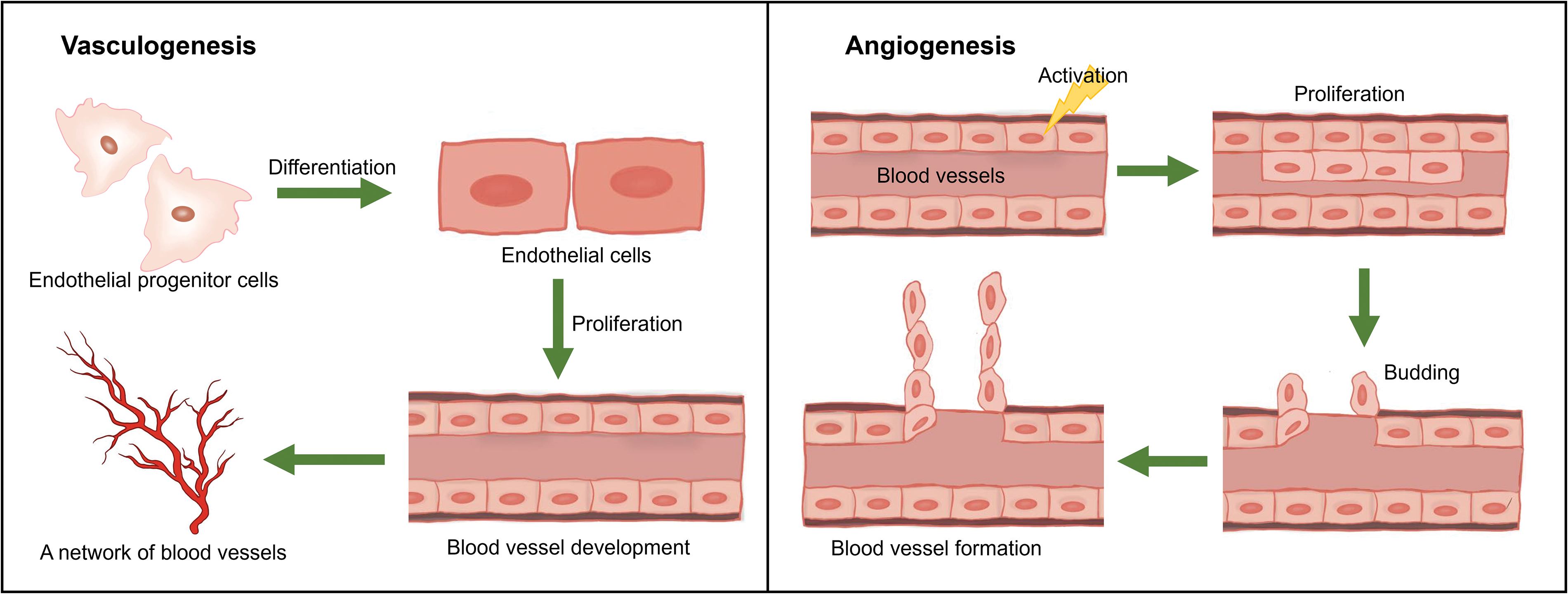
Figure 2: Hypothetical figure showing the processes of vasculogenesis and angiogenesis.
The induction of angiogenesis remains a pivotal challenge in tissue engineering and regeneration. DSCs have demonstrated the ability to differentiate into vasculogenic endothelial cells capable of generating functional blood vessels [133]. DSCs have been increasingly recognized for their therapeutic potential in dental pulp and periodontal tissue engineering.
In primary DPSCs and SHED, a specific sub-population characterized by high vascular endothelial growth factor receptor 1 (VEGFR1) expression has been identified, enabling these cells to respond to vasculogenic stimuli [134]. However, they lack VEGFR2, CD31, and VE-cadherin expression [135,136]. When subjected to endothelial differentiation medium supplemented with VEGF, DPSCs progressively express endothelial cell-related markers (e.g., VEGFR2, VE-cadherin, CD31) [137]. Upon transplantation into murine hosts, DPSCs differentiate into functional CD31-positive mature blood vessels [135,136], underscoring the angiogenic potential of DPSCs for tissue regeneration. Despite these promising findings, the precise mechanisms governing vascular differentiation induction remain largely unclear.
Emerging evidence has linked autophagy to the process of angiogenesis. For instance, colocalization of endothelial marker CD31 and autophagic marker LC3 has been observed in regenerated pulp-like tissue, suggesting the potential involvement of autophagy in pulp revascularization [138]. This hypothesis is corroborated by findings indicating that autophagy regulates endothelial cell processing and maturation [139]. In another study, the angiogenic effect of erythropoietin on PDLSCs was linked to autophagy activation under an inflammatory microenvironment modulated through the Akt/Erk1/2/BAD signaling pathway [140]. This novel approach presents a promising strategy for angiogenic tissue engineering.
It has been reported that PDLSCs can contribute to vasculogenesis not only through direct differentiation into endotheliocytes but also by secreting pro-angiogenic factors. In an inflammatory milieu, increased autophagy levels in PDLSCs correlated with heightened secretion of pro-angiogenic cytokines, including angiopoietin and basic fibroblast growth factor (bFGF), ultimately leading to enhanced tube formation [141]. Autophagy’s role in bolstering the paracrine function of PDLSCs increases their pro-angiogenic potential, offering a pathway to expedite blood circulation establishment within DSC-based tissue-engineered constructs. In summary, an autophagy-centered approach to angiogenic differentiation holds considerable therapeutic potential for dental pulp and periodontal tissue regeneration.
Currently, published data are conflicting about the potential of DSC adipogenic differentiation in vitro. DSCs could differentiate into adipocytes under suitable conditions, which was confirmed by oil red O staining of the accumulated lipid droplets [142]. However, a recent study disclosed that DPSCs cultured in the adipogenic induction medium had a limited capacity to differentiate into adipocytes, which is supported by their gene expression profile [143]. There is no doubt that DSCs still possess adipogenic differentiation potential. Adipocytes are fat-storing cells with limited clinical application, but they could be used for further molecular research. However, no literature related to autophagy in adipogenic differentiation of DSCs has been retrieved. The impact of autophagy on the adipogenic differentiation ability is worth exploring.
Several studies have investigated the chondrogenic potential of DSCs. After 21 days of chondrogenic induction, toluidine blue staining could be used to assess glycosaminoglycans production, and alcian blue and picrosirius red staining could evaluate collagen deposition. Gene expression analysis has confirmed the presence of chondrogenic markers, including Sox9, Comp, and Acan, during chondrogenic differentiation of DPSCs [144]. It has been reported that the chondrogenic differentiation potential of SCAPs enhanced by distal-less homeobox 5 (DLX5) and homeobox C8 (HOXC8) [145]. Fas cell surface death receptor ligand (FasL) stimulation is implicated in supporting the chondrogenic differentiation of DPSCs [146]. Chondrogenic differentiation is integral to cartilage repair and regeneration. It is a pity that how autophagy regulates chondrocyte differentiation remains still unclear.
DSCs hold significant potential for myogenic differentiation [147,148]. Furthermore, DPSCs can differentiate into insulin-producing cells [149] and hepatic-like cells [150]. The effects of autophagy on the multi-lineage differentiation ability of DSCs are worth exploring, and reports on the function and regulation of autophagy in the DSC differentiation into the above tissue-like cells are still lacking.
Furthermore, it is worth noting the concept of hormesis, which has gained prominence in the study of stem cell biology. Hormesis is a biphasic dose/concentration response. It displays a low-dose/concentration stimulation and a high-dose/concentration inhibition [151–153]. The hormetic response is typically demonstrated during preclinical investigations, markedly enhancing therapeutic applications through selecting optimal hormetic doses in clinical trials. Recently, hormesis and its potential relevance to dental stem cells have extensively been studied, especially in cell proliferation and differentiation [154–156]. Additional researches are still needed to investigate the implications of hormesis on dental stem cell behavior and its potential applications in regenerative dentistry.
Regulation of Autophagy in DSC Differentiation
The differentiation of DSCs is influenced by various factors involving autophagy pathways. In the field of tissue engineering, scaffold materials play a pivotal role in impacting DSC differentiation. Additionally, drug stimuli function as signaling agents, orchestrating DSC differentiation. The external environment surrounding DSCs must also be considered. This comprehensive array of factors impacting autophagy-mediated DSC differentiation is elaborated upon in the subsequent sections (see Fig. 3).
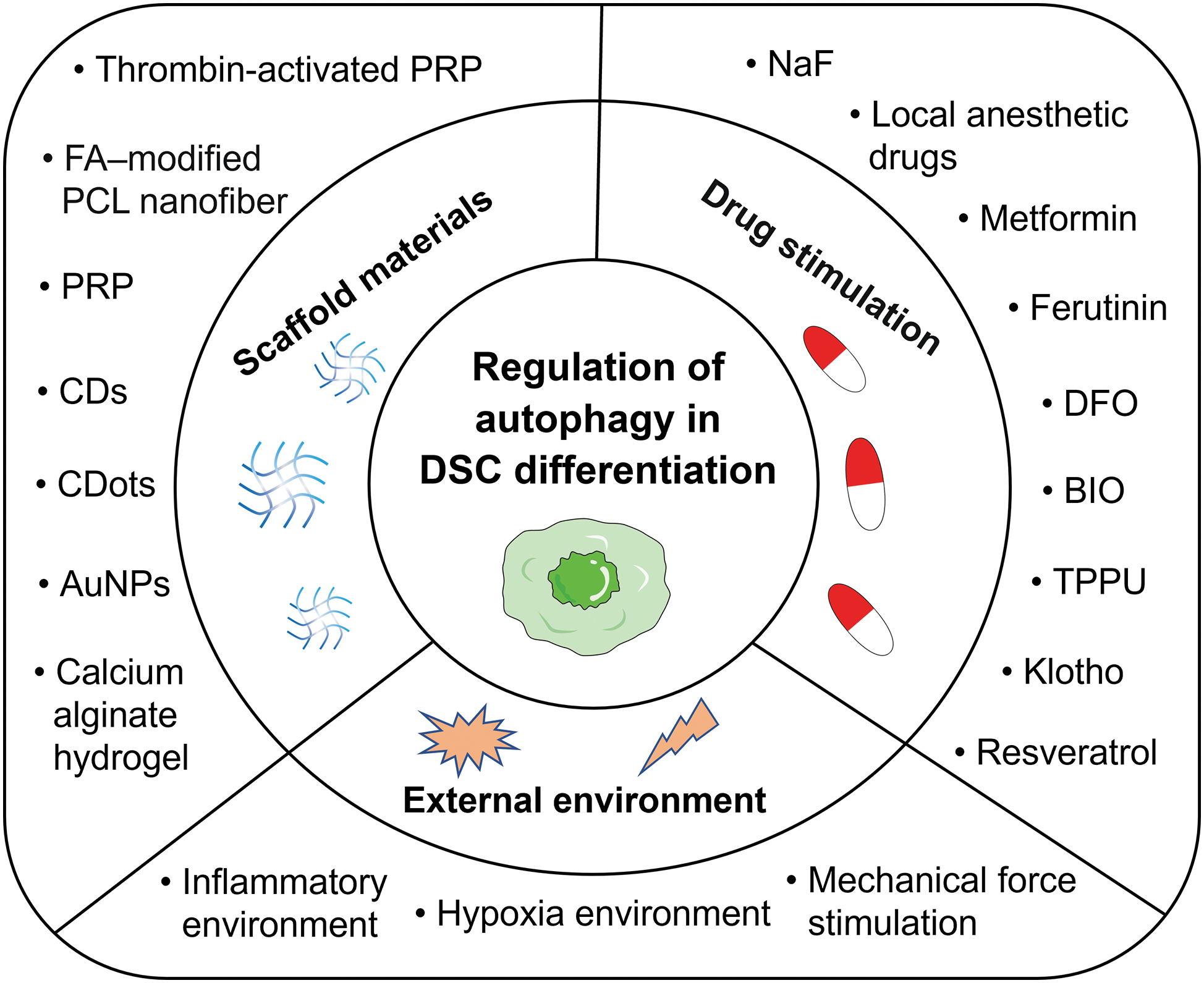
Figure 3: An overview of the major factors that regulate autophagy in the differentiation of DSCs. These factors can be mainly divided into three categories: scaffold materials, drug stimulation, and external environment. Abbreviations: AuNPs, gold nanoparticles; BIO, 6-bromoindirubin-3’-oxime; CDs, carbon nanodots; CDots, carbon dots; DFO, deferoxamine; DSCs, dental stem cells; FA-modified PCL nanofiber, fluorapatite-modified polycaprolactone nanofiber; NaF, sodium fluoride; PRP, platelet-rich plasma; TPPU, a soluble epoxide hydrolase inhibitor.
An in-depth exploration of natural and synthetic biomaterials suitable for scaffold construction in oral tissue regeneration has been outlined in prior reviews [16,157]. Experiments indicated that the nanopore structure and the pore size were found to be important for regulating the expression of autophagy pathway components (LC3A/B, Beclin-1, Atg3, Atg7, and P62) and the osteogenic pathways [158]. The nano-textured surface of biomaterials induced a stronger autophagic response, resulting in stronger osteogenesis [159]. In addition, numerous studies have corroborated the potential of specific materials to facilitate DSC differentiation through autophagy, although the precise underlying mechanisms remain partly obscured.
Tissue-derived scaffold: Platelet-rich plasma (PRP) has been effectively utilized as a tissue scaffold during clinical practice in recent years to regenerate pulp-like tissue and stimulate root development during regenerative endodontic treatment for immature teeth [160]. Notably, PRP exhibits potential for bone regeneration, with PRP-induced autophagy playing a crucial role in driving substantial osteogenic differentiation and mineralization in DPCs [109]. Furthermore, thrombin-activated PRP has been shown to augment autophagy-mediated osteogenic differentiation and viability of PDLSCs [110]. These results suggest that autophagy might be a mechanism underlying PRP-mediated osteogenic differentiation and mineralization of DSCs.
Nanomaterials scaffold: Recently, bioactive nanomaterials have been widely proposed to regulate DSC differentiation in tissue regeneration. Carbon nanodots (CDs) have been recognized for enhancing DSC differentiation by activating autophagy, exemplified by metformin-based CDs promoting efficient odontogenic differentiation of DPSCs [161]. Carbon dots (CDots) have been found to enhance osteo/odontogenic differentiation of DPSCs through reasonable autophagy activation via the PI3K/Akt/mTOR signaling pathway, contributing to the effective regeneration of dentin-pulp complex and blood vessels in vivo [162]. Graphene oxide quantum dots (GOQDs) bolster DPSC mineralization by modulating autophagy induced by ROS [163]. Moreover, fluorapatite (FA)-modified polycaprolactone (PCL) nanofibers induce DPSC differentiation and mineralization, potentially influenced by insulin, hedgehog, and Wnt signal pathways interconnected with and/or mediated by cell autophagy [164]. Gold nanoparticles (AuNPs) have been implicated in affecting PDL cells osteogenic differentiation via autophagy [165]. In vivo experiments have also shown that treatment with PDLSC sheets and 45 nm AuNPs increased bone structure and collagen formation, mediated by autophagy pathway activation [111]. AuNPs treatment has also been observed to rescue PDLSC osteogenic potential under inflammatory conditions by restoring the inflammation-compromised autophagy-lysosome system [166]. Hence, the development of autophagy-regulating materials holds promise for promoting tissue repair and regeneration through DSC differentiation.
Hydrogel scaffold: The combination of biomaterials with signaling factors and DSCs in a hydrogel scaffold holds significant promise for tissue regeneration. Each component yields distinct effects; when combined, they may work synergistically to enhance treatment outcomes. For instance, calcium alginate hydrogel, DPSCs, and fibroblast growth factor 21 (FGF21) have shown remarkable effectiveness in promoting recovery from spinal cord injuries. This occurs through multiple mechanism, where DPSCs differentiate into neurons, and FGF21 inhibits autophagy via the AMPK-mTOR-LC3 pathway, particularly during the chronic recovery phase. Additionally, the calcium ions within this hydrogel facilitate self-adhesion through cadherin, while alginic acid helps prevent calcium overload injuries by absorbing excess calcium ions [167]. Indeed, biomaterials loaded with signaling molecules represent a potent strategy for balancing autophagy and DSC differentiation in tissue injury repair.
Autophagy’s vital role in DSC differentiation provides the basis for exploring autophagy-regulating agents beneficial for dental pulp and periodontal tissue reconstruction. Rapamycin is a commonly used autophagy enhancer, while 3-methyladenine (3-MA) and the antimalarial drug chloroquine serve as autophagy inhibitors [91]. Additionally, numerous drugs have been shown to impact cell differentiation by modulating autophagy pathways.
Chemical drugs: Sodium fluoride (NaF), a key component of fluoride toothpaste and mouthwash, has been reported to participate in tooth formation and mineralization processes. Low concentrations of NaF modulate autophagy in SCAPs, promoting osteo/odontogenic differentiation and suggesting NaF-induced autophagy as a regulator of SCAP differentiation [174].
Interestingly, local anesthetic drugs frequently used in dental treatment, like Ubistesin, Ubistesin Forte, Septanest, Scandonest, and Xylocaine, have potential impacts on dentin repair and regeneration by inducing autophagy in DPCs [169].
Metformin, a widely used hypoglycemic drug, enhances the biocompatibility of polydopamine-templated hydroxyapatite (tHA). Combined with tHA, metformin increases the viability and osteogenic ability of PDLSCs through the AMPK/mTOR signaling pathway, regulating autophagy [176].
Deferoxamine (DFO) is an iron chelator clinically used in treating iron overload. It has been reported that during differentiation and mineralization of DPSCs, DFO-induced autophagy could significantly contribute to the repair ability of DPSCs through activation of the hypoxia-inducible factor-1α (HIF-1α)/BNIP3 pathway in a ROS-dependent manner [171].
Natural drugs: Ferutinin, a natural non-steroidal phytoestrogen, catalyzes osteoblast differentiation of DPSCs via KLF2-mediated autophagy/mitophagy, suggesting its potential for regenerative bone therapy [170].
Resveratrol, a naturally occurring polyphenol, has been reported to facilitate the osteogenic differentiation process of GMSCs cultured in the induction differentiation medium through the AMPK-Beclin-1 pro-autophagic pathway. This effect appeared to be intricately linked to resveratrol’s role as both an autophagy inducer and a promoter of MSC osteogenic differentiation [184].
Molecule drugs: 6-bromoindirubin-3’-oxime (BIO), a promising small molecule candidate with the ability to influence Wnt signaling, has been reported to effectively promote the odonto/osteogenic differentiation of DPSCs while simultaneously reducing their adipogenic differentiation potential. Analysis of RNA sequencing data from DPSCs treated with BIO revealed differential expression of genes associated with autophagy-modulated pathways [172].
Klotho, an anti-aging protein, has been found to protect PDLSC viability in harsh environments. Moreover, it facilitated bone regeneration following transplantation in vivo using a rat model of cranial bone defect by maintaining a low level of autophagic activity in PDLSCs. In contrast, abnormally elevated autophagy activity was observed in locally transplanted PDLSCs that did not undergo Klotho pretreatment [182].
Despite their great potential for autophagy regulation, these drugs have not been developed for this purpose and lacked specificity. Thus, it is important to design and develop clinically targeted autophagy modulators based on the unique autophagic features associated with each specific disease.
The critical importance of an optimal environment for differentiated transplanted cells is well recognized, with harsh microenvironments compromising their stemness and impeding their therapeutic efficacy in tissue regeneration engineering. Satisfactory therapeutic outcomes in patients with conditions such as pulpitis, periodontitis, and orthodontic treatment remain elusive, potentially limited by the inflammatory milieu and insufficient oxygen supply within the cell transplant site. Consequently, a comprehensive exploration of the role and regulatory factors of autophagy in the intricate and variable transplant setting becomes imperative.
Inflammatory environment: Research substantiates that an inflammatory microenvironment triggers mitochondrial damage, activating mitophagy via the PINK1-Parkin pathway to degrade impaired mitochondria [185]. The role of autophagy in DSC differentiation is multifaceted, as evidenced by reports that autophagy prompted by lipopolysaccharide (LPS) stimulation contributes to the attenuation of SCAP osteo/odontogenic differentiation capacity [175]. Dang et al. [173] revealed that LPS-induced inflammatory conditions trigger excessive autophagy in DPSCs, suppressing osteogenic differentiation, an effect reversed by the soluble epoxide hydrolase inhibitor TPPU. Intriguingly, low concentrations of tumor necrosis factor-α (TNF-α) fostered osteogenic differentiation of PDLSCs through autophagy activation via AKT/mTOR signaling inhibition [186]. Hence, it is highly conceivable that DSC differentiation is intricately connected to the activation level of autophagy driven by inflammation. Nonetheless, contrasting studies have shown that autophagy enhances odontoblast differentiation by suppressing NF-κB activation within an inflammatory milieu [177]. Additionally, the anti-inflammatory factor interleukin-37 (IL-37) augmented osteo/odontogenic differentiation of DPSCs via activated autophagy [187]. These discrepancies most likely stem from variations in the cell lines used and the level of inflammatory stimulation. Taken together, these findings unveil fresh perspectives into the mechanisms safeguarding the dentin-pulp complex during inflammatory intrusion, offering potential therapeutic implications for caries, pulpitis, and dental pulp regeneration within inflammatory contexts.
Mechanical force environment: During the course of mechanical force-induced tooth movement, the periodontal tissues undergo the development of a sterile inflammatory microenvironment. This environment is marked by heightened levels of inflammatory cytokines and chemokines, as well as increased activity of immune cells associated with inflammation [188,189]. PDLSC autophagy induced by mechanical compressive force has been reported to facilitate M1-like macrophage polarization by inhibiting the AKT signaling pathway in periodontal tissues, thereby fostering inflammatory bone remodeling and promoting orthodontic tooth movement [190]. However, observations by Chen et al. [191] suggested that orthodontic force-triggered autophagy primarily occurs on the pressured side of PDL tissues, reducing the speed of orthodontic tooth movement by suppressing osteoclastogenesis. This discrepancy likely arises from the multifaceted roles of autophagy under different compressive force levels. Zheng et al. [178] observed heightened autophagy levels due to mechanical tension force, effectively promoting the osteogenic differentiation of PDLSCs. Recent research has unveiled the ability of the combination of mechanical stimuli and biochemical signals to influence cellular fate through modulation of the autophagy pathway. Notably, the induction of PDLSC autophagy under orthodontic compressive force was found to be orchestrated by long non-coding RNA (lncRNA) FER1L4 via the AKT/FOXO3 pathway [179]. These findings highlight autophagy modulation as a prospective strategy to bolster periodontal tissue reconstruction while also offering a promising avenue to expedite orthodontic tooth movement.
Hypoxia environment: Hypoxia, often triggered by inflammation stemming from bacterial infection or trauma, is a common occurrence in injured tissues. Certain researchers have uncovered the protective role of autophagy under hypoxic conditions. A previous report highlighted FUNDC1’s role as a receptor that mediated mitophagy in response to hypoxia [103]. Diminishing FUNDC1 expression undermined hypoxia-induced proliferation, migration, and odontoblastic differentiation of DPCs, suggesting that FUNDC1-mediated autophagy potentially governs the biological behavior of DPCs [168]. Hypoxia stimulates lactate production, a glycolysis end product. Lactate has been reported to inhibit autophagy and subsequently hinder the osteogenic differentiation of PDLSCs via the MCT1-mTOR signaling pathway [192]. A20, also recognized as TNF-α-inducible protein 3 (TNFAIP3), functions as a potent anti-inflammatory enzyme. In a hypoxic microenvironment, A20 was found to repress osteoclast differentiation of PDLSCs by downregulating autophagy [180]. Importantly, circCDK8 was found to exhibit an inhibitory effect of osteogenic differentiation via autophagy induction [181]. These findings indicate the close relationship between osteogenic/osteoclast differentiation and autophagy levels, highlighting autophagy’s potential as a novel therapeutic target for bone loss in periodontitis.
High glucose environment: In addition, the osteogenic ability of PDLSCs is impaired by high glucose environments [193]. Elevation of autophagy partly reversed the detrimental impact of high glucose conditions on PDLSCs, highlighting the protective role of autophagy in preserving cellular function. In a diabetic rat periodontal trauma model, the periodontium tissue exhibited partial recovery in the autophagy-enhanced cell injection group, offering stronger evidence for the regulatory role of autophagy in vivo [194]. This finding sheds light on the mechanistic understanding of tissue regeneration in periodontitis associated with diabetes, potentially offering a fresh perspective for treating and researching periodontitis in diabetic patients.
LncRNAs are an abundant class of RNAs that do not encode proteins but play an important regulatory role in gene expression. A recent study uncovered the regulatory role of lncRNA insulin-like growth factor binding protein 7-antisense 1 (IGFBP7-AS1) in odontogenic differentiation of SHED through autophagy, indicating its potential as a gene target in the regeneration of dental hard tissue and the dental-pulp complex [195]. Although the gene expression levels regulated by lncRNAs can be broadly categorized into epigenetic, transcriptional, and post-transcriptional regulation, their specific role in autophagy regulation remains unclear. Additionally, cellular communication network factor 1 (CCN1), a pivotal matricellular protein, facilitated osteogenesis in PDLSCs through autophagy and the MAPK/ERK pathway [196]. CCN1 exhibits promising regulatory potential in tissue regeneration, playing multifaceted roles in cell differentiation, adhesion, and migration and serving as a key participant in bone development, osteoblast differentiation, and the remodeling of the extracellular matrix.
DSCs hold immense potential for stem cell-based medical therapies, necessitating an in-depth comprehension of their differentiation mechanisms. Based on the available literature, a viable avenue exists for enhancing DSC regenerative potential via autophagy modulation. Serving as a self-degrading and recycling system, autophagy actively participates in DSC differentiation, including osteo/odontogenic, neurogenic, and angiogenic differentiation, influenced by factors considered in tissue regeneration, such as scaffold materials, drug stimulation, and the external environment. A comprehensive synthesis of current knowledge concerning autophagy’s role and regulation in DSC differentiation could significantly advance research into DSC-based regeneration. However, despite escalating interest and investigation into autophagy’s contribution to DSC differentiation, there is still a considerable gap in our overall comprehension of autophagy’s influence and the underlying regulatory mechanisms regulating its implications within this context.
The differentiation ability of DSCs is significantly affected by the transplantation environment mediated by the autophagy pathway. Furthermore, most autophagy modulators have limitations such as poor target specificity, low stability, low concentration, and limited drug action duration. Addressing these challenges could be achieved by a drug delivery system that leverages the controlled drug release properties of scaffold materials through intelligent response to the microenvironment. This system, which incorporates autophagy modulators and DSCs for in-situ implantation, offers an innovative and effective approach for repair and reconstruction [183]. This method introduces a novel perspective, demonstrating that autophagy modulators can be safely and efficiently applied in harsh environments through scaffold-based delivery. This innovative approach paves a fresh path toward precise and rigorous control of autophagy during stem cell differentiation. Nevertheless, given the complexity and significance of autophagy in DSC differentiation for tooth and bone tissue regeneration, further research is needed to explore strategies for the precise and rigorous regulation of autophagy.
Acknowledgement: We sincerely thank for the help of team members of Dr. Wu.
Funding Statement: This research was funded by grants from the National Natural Science Foundation of China (Nos. 81771095, 82071235), Key R&D Program of Shaanxi Province (2017SF-103, 2021KWZ-26, and 2023-JC-ZD-56) and State Key Laboratory of Military Stomatology (2020ZA01).
Author Contributions: The authors confirm contribution to the paper as follows: the conception of the study: LLW, LAW; wrote the manuscript: XTZ; revised the manuscript: TJJ, XWL, CFL. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Berebichez-Fridman R, Montero-Olvera PR. Sources and clinical applications of mesenchymal stem cells: state-of-the-art review. Sultan Qaboos Univ Med J. 2018;18(3):e264–e77. [Google Scholar] [PubMed]
2. Huang GT, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88(9):792–806. [Google Scholar] [PubMed]
3. Choudhery MS, Badowski M, Muise A, Pierce J, Harris DT. Donor age negatively impacts adipose tissue-derived mesenchymal stem cell expansion and differentiation. J Transl Med. 2014;12(1):8. [Google Scholar] [PubMed]
4. Chen J, Zheng X, Rao N, Huang Y, Liu J, Li Y. Key markers and epigenetic modifications of dental-derived mesenchymal stromal cells. Stem Cells Int. 2021;2021(2):5521715. [Google Scholar] [PubMed]
5. Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97(25):13625–30. [Google Scholar] [PubMed]
6. Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci USA. 2003;100(10):5807–12. [Google Scholar] [PubMed]
7. Sonoyama W, Liu Y, Yamaza T, Tuan RS, Wang S, Shi S. Characterization of the apical papilla and its residing stem cells from human immature permanent teeth: a pilot study. J Endod. 2008;34(2):166–71. [Google Scholar] [PubMed]
8. Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364(9429):149–55. [Google Scholar] [PubMed]
9. Morsczeck C, Gotz W, Schierholz J, Zeilhofer F, Kuhn U, Mohl C. Isolation of precursor cells (PCs) from human dental follicle of wisdom teeth. Matrix Biol. 2005;24(2):155–65. [Google Scholar] [PubMed]
10. Zhang Q, Shi S, Liu Y, Uyanne J, Shi Y, Shi S. Mesenchymal stem cells derived from human gingiva are capable of immunomodulatory functions and ameliorate inflammation-related tissue destruction in experimental colitis. J Immunol. 2009;183(12):7787–98. [Google Scholar] [PubMed]
11. Lan X, Sun Z, Chu C, Boltze J, Li S. Dental pulp stem cells: an attractive alternative for cell therapy in ischemic stroke. Front Neurol. 2019;10:824. [Google Scholar] [PubMed]
12. Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D. Minimal criteria for defining multipotent mesenchymal stromal cells. The international society for cellular therapy position statement. Cytotherapy. 2006;8(4):315–7. [Google Scholar] [PubMed]
13. Martellucci S, Manganelli V, Santacroce C, Santilli F, Piccoli L, Sorice M. Role of prion protein-EGFR multimolecular complex during neuronal differentiation of human dental pulp-derived stem cells. Prion. 2018;12(2):117–26. [Google Scholar] [PubMed]
14. Li B, Ouchi T, Cao Y, Zhao Z, Men Y. Dental-derived mesenchymal stem cells: state of the art. Front Cell Dev Biol. 2021;9:654559. [Google Scholar] [PubMed]
15. Li P, Ou Q, Shi S, Shao C. Immunomodulatory properties of mesenchymal stem cells/dental stem cells and their therapeutic applications. Cell Mol Immunol. 2023;2023(6):1–12. [Google Scholar]
16. Granz CL, Gorji A. Dental stem cells: the role of biomaterials and scaffolds in developing novel therapeutic strategies. World J Stem Cells. 2020;12(9):897–921. [Google Scholar] [PubMed]
17. Liu G, Ma S, Zhou Y, Lu Y, Jin L, Wang Z. Signaling pathways in dental stem cells during their maintenance and differentiation. In: Şahin F, Doğan A, Demirci S, editors. Dental Stem Cells. Cham: Springer International Publishing; 2016. p. 69–92. [Google Scholar]
18. Hussain A, Tebyaniyan H, Khayatan D. The role of epigenetic in dental and oral regenerative medicine by different types of dental stem cells: a comprehensive overview. Stem Cells Int. 2022;2022(7):5304860. [Google Scholar] [PubMed]
19. Zhang H, Fu H, Fang H, Deng Q, Huang H, Hou D. Epigenetic regulation of methylation in determining the fate of dental mesenchymal stem cells. Stem Cells Int. 2022;2022:5015856. [Google Scholar] [PubMed]
20. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P. Autophagy in major human diseases. EMBO J. 2021;40(19):e108863. [Google Scholar] [PubMed]
21. Huang HY, Wang WC, Lin PY, Huang CP, Chen CY, Chen YK. The roles of autophagy and hypoxia in human inflammatory periapical lesions. Int Endod J. 2018;51(S2):e125–45. [Google Scholar] [PubMed]
22. Yang S, Fan W, Li Y, Liu Q, He H, Huang F. Autophagy in tooth: physiology, disease and therapeutic implication. Cell Biochem Funct. 2021;39(6):702–12. [Google Scholar] [PubMed]
23. Zhang L, Chen Z. Autophagy in the dentin-pulp complex against inflammation. Oral Dis. 2018;24(1–2):11–3. [Google Scholar] [PubMed]
24. Chen X, He Y, Lu F. Autophagy in stem cell biology: a perspective on stem cell self-renewal and differentiation. Stem Cells Int. 2018;2018(9):9131397. [Google Scholar] [PubMed]
25. Lin Q, Chen J, Gu L, Dan X, Zhang C, Yang Y. New insights into mitophagy and stem cells. Stem Cell Res Ther. 2021;12(1):452. [Google Scholar] [PubMed]
26. Morsczeck C, Reichert TE. Dental stem cells in tooth regeneration and repair in the future. Expert Opin Biol Ther. 2018;18(2):187–96. [Google Scholar] [PubMed]
27. Yasui T, Mabuchi Y, Morikawa S, Onizawa K, Akazawa C, Nakagawa T. Isolation of dental pulp stem cells with high osteogenic potential. Inflamm Regen. 2017;37(1):8. [Google Scholar] [PubMed]
28. Anitua E, Troya M, Zalduendo M. Progress in the use of dental pulp stem cells in regenerative medicine. Cytotherapy. 2018;20(4):479–98. [Google Scholar] [PubMed]
29. Xia K, Chen Z, Chen J, Xu H, Xu Y, Yang T. RGD- and VEGF-mimetic peptide epitope-functionalized self-assembling peptide hydrogels promote dentin-pulp complex regeneration. Int J Nanomedicine. 2020;15:6631–47. [Google Scholar] [PubMed]
30. Nakashima M, Iohara K, Murakami M, Nakamura H, Sato Y, Ariji Y. Pulp regeneration by transplantation of dental pulp stem cells in pulpitis: a pilot clinical study. Stem Cell Res Ther. 2017;8(1):61. [Google Scholar] [PubMed]
31. Li Y, Zhao S, Nan X, Wei H, Shi J, Li A. Repair of human periodontal bone defects by autologous grafting stem cells derived from inflammatory dental pulp tissues. Stem Cell Res Ther. 2016;7(1):141. [Google Scholar] [PubMed]
32. Syed-Picard FN, Du Y, Lathrop KL, Mann MM, Funderburgh ML, Funderburgh JL. Dental pulp stem cells: a new cellular resource for corneal stromal regeneration. Stem Cells Transl Med. 2015;4(3):276–85. [Google Scholar] [PubMed]
33. Nicola FC, Rodrigues LP, Crestani T, Quintiliano K, Sanches EF, Willborn S. Human dental pulp stem cells transplantation combined with treadmill training in rats after traumatic spinal cord injury. Braz J Med Biol Res. 2016;49(9):e5319. [Google Scholar] [PubMed]
34. Yang C, Li X, Sun L, Guo W, Tian W. Potential of human dental stem cells in repairing the complete transection of rat spinal cord. J Neural Eng. 2017;14(2):026005. [Google Scholar] [PubMed]
35. Chamieh F, Collignon AM, Coyac BR, Lesieur J, Ribes S, Sadoine J. Accelerated craniofacial bone regeneration through dense collagen gel scaffolds seeded with dental pulp stem cells. Sci Rep. 2016;6(1):38814. [Google Scholar] [PubMed]
36. Giuliani A, Manescu A, Langer M, Rustichelli F, Desiderio V, Paino F. Three years after transplants in human mandibles, histological and in-line holotomography revealed that stem cells regenerated a compact rather than a spongy bone: biological and clinical implications. Stem Cells Transl Med. 2013;2(4):316–24. [Google Scholar] [PubMed]
37. Nagpal A, Kremer KL, Hamilton-Bruce MA, Kaidonis X, Milton AG, Levi C. TOOTH (The open study of dental pulp stem cell therapy in humansstudy protocol for evaluating safety and feasibility of autologous human adult dental pulp stem cell therapy in patients with chronic disability after stroke. Int J Stroke. 2016;11(5):575–85. [Google Scholar] [PubMed]
38. Gandia C, Armiñan A, García-Verdugo JM, Lledó E, Ruiz A, Miñana MD. Human dental pulp stem cells improve left ventricular function, induce angiogenesis, and reduce infarct size in rats with acute myocardial infarction. Stem Cells. 2008;26(3):638–45. [Google Scholar] [PubMed]
39. Gnanasegaran N, Govindasamy V, Simon C, Gan QF, Vincent-Chong VK, Mani V. Effect of dental pulp stem cells in MPTP-induced old-aged mice model. Eur J Clin Invest. 2017;47(6):403–14. [Google Scholar] [PubMed]
40. Bar JK, Lis-Nawara A, Grelewski PG. Dental pulp stem cell-derived secretome and its regenerative potential. Int J Mol Sci. 2021;22(21):12018. [Google Scholar] [PubMed]
41. Zhang N, Chen B, Wang W, Chen C, Kang J, Deng SQ. Isolation, characterization and multi-lineage differentiation of stem cells from human exfoliated deciduous teeth. Mol Med Rep. 2016;14(1):95–102. [Google Scholar] [PubMed]
42. Ishkitiev N, Yaegaki K, Imai T, Tanaka T, Nakahara T, Ishikawa H. High-purity hepatic lineage differentiated from dental pulp stem cells in serum-free medium. J Endod. 2012;38(4):475–80. [Google Scholar] [PubMed]
43. Okada M, Ishkitiev N, Yaegaki K, Imai T, Tanaka T, Fukuda M. Hydrogen sulphide increases hepatic differentiation of human tooth pulp stem cells compared with human bone marrow stem cells. Int Endod J. 2014;47(12):1142–50. [Google Scholar] [PubMed]
44. Yang X, Ma Y, Guo W, Yang B, Tian W. Stem cells from human exfoliated deciduous teeth as an alternative cell source in bio-root regeneration. Theranostics. 2019;9(9):2694–711. [Google Scholar] [PubMed]
45. Xuan K, Li B, Guo H, Sun W, Kou X, He X. Deciduous autologous tooth stem cells regenerate dental pulp after implantation into injured teeth. Sci Transl Med. 2018;10(455):eaaf3227. [Google Scholar] [PubMed]
46. Gomes JA, Geraldes Monteiro B, Melo GB, Smith RL, Cavenaghi Pereira da Silva M, Lizier NF. Corneal reconstruction with tissue-engineered cell sheets composed of human immature dental pulp stem cells. Invest Ophthalmol Vis Sci. 2010;51(3):1408–14. [Google Scholar] [PubMed]
47. Nicola FDC, Marques MR, Odorcyk F, Arcego DM, Petenuzzo L, Aristimunha D. Neuroprotector effect of stem cells from human exfoliated deciduous teeth transplanted after traumatic spinal cord injury involves inhibition of early neuronal apoptosis. Brain Res. 2017;1663(8):95–105. [Google Scholar] [PubMed]
48. Zheng Y, Liu Y, Zhang CM, Zhang HY, Li WH, Shi S. Stem cells from deciduous tooth repair mandibular defect in swine. J Dent Res. 2009;88(3):249–54. [Google Scholar] [PubMed]
49. Hirata M, Ishigami M, Matsushita Y, Ito T, Hattori H, Hibi H. Multifaceted therapeutic benefits of factors derived from dental pulp stem cells for mouse liver fibrosis. Stem Cells Transl Med. 2016;5(10):1416–24. [Google Scholar] [PubMed]
50. Wang J, Wang X, Sun Z, Wang X, Yang H, Shi S. Stem cells from human-exfoliated deciduous teeth can differentiate into dopaminergic neuron-like cells. Stem Cells Dev. 2010;19(9):1375–83. [Google Scholar] [PubMed]
51. Cantore S, Ballini A, de Vito D, Martelli FS, Georgakopoulos I, Almasri M. Characterization of human apical papilla-derived stem cells. J Biol Regul Homeost Agents. 2017;31(4):901–10. [Google Scholar] [PubMed]
52. Li G, Han N, Zhang X, Yang H, Cao Y, Wang S. Local injection of allogeneic stem cells from apical papilla enhanced periodontal tissue regeneration in minipig model of periodontitis. Biomed Res Int. 2018;2018(3):3960798. [Google Scholar] [PubMed]
53. Hilkens P, Bronckaers A, Ratajczak J, Gervois P, Wolfs E, Lambrichts I. The angiogenic potential of DPSCs and SCAPs in an in vivo model of dental pulp regeneration. Stem Cells Int. 2017;2017(2):2582080. [Google Scholar] [PubMed]
54. Kim BC, Jun SM, Kim SY, Kwon YD, Choe SC, Kim EC. Engineering three dimensional micro nerve tissue using postnatal stem cells from human dental apical papilla. Biotechnol Bioeng. 2017;114(4):903–14. [Google Scholar] [PubMed]
55. Tanaka Y, Sonoda S, Yamaza H, Murata S, Nishida K, Hama S. Suppression of AKT-mTOR signal pathway enhances osteogenic/dentinogenic capacity of stem cells from apical papilla. Stem Cell Res Ther. 2018;9(1):334. [Google Scholar] [PubMed]
56. Tomokiyo A, Wada N, Maeda H. Periodontal ligament stem cells: regenerative potency in periodontium. Stem Cells Dev. 2019;28(15):974–85. [Google Scholar] [PubMed]
57. Queiroz A, Albuquerque-Souza E, Gasparoni LM, de França BN, Pelissari C, Trierveiler M. Therapeutic potential of periodontal ligament stem cells. World J Stem Cells. 2021;13(6):605–18. [Google Scholar] [PubMed]
58. Feng F, Akiyama K, Liu Y, Yamaza T, Wang TM, Chen JH. Utility of PDL progenitors for in vivo tissue regeneration: a report of 3 cases. Oral Dis. 2010;16(1):20–8. [Google Scholar] [PubMed]
59. Ha SH, Choung PH. MSM promotes human periodontal ligament stem cells differentiation to osteoblast and bone regeneration. Biochem Biophys Res Commun. 2020;528(1):160–7. [Google Scholar] [PubMed]
60. Mohebichamkhorami F, Fattahi R, Niknam Z, Aliashrafi M, Khakpour Naeimi S, Gilanchi S. Periodontal ligament stem cells as a promising therapeutic target for neural damage. Stem Cell Res Ther. 2022;13(1):273. [Google Scholar] [PubMed]
61. Zhao Q, Li G, Wang T, Jin Y, Lu W, Ji J. Human periodontal ligament stem cells transplanted with nanohydroxyapatite/chitosan/gelatin 3D porous scaffolds promote jaw bone regeneration in swine. Stem Cells Dev. 2021;30(10):548–59. [Google Scholar] [PubMed]
62. Karamzadeh R, Baghaban Eslaminejad M, Sharifi-Zarchi A. Comparative in vitro evaluation of human dental pulp and follicle stem cell commitment. Cell J. 2017;18(4):609–18. [Google Scholar] [PubMed]
63. Bi R, Lyu P, Song Y, Li P, Song D, Cui C. Function of dental follicle progenitor/stem cells and their potential in regenerative medicine: from mechanisms to applications. Biomolecules. 2021;11(7):997. [Google Scholar] [PubMed]
64. Li X, Yang C, Li L, Xiong J, Xie L, Yang B. A therapeutic strategy for spinal cord defect: human dental follicle cells combined with aligned PCL/PLGA electrospun material. Biomed Res Int. 2015;2015:197183. [Google Scholar] [PubMed]
65. Chen Z, Gan L, Chen X, Zheng J, Shi S, Wu L. LncRNA HOTAIRM1 promotes dental follicle stem cell-mediated bone regeneration by regulating HIF-1α/KDM6/EZH2/H3K27me3 axis. J Cell Physiol. 2023;238(7):1542–57. [Google Scholar] [PubMed]
66. Guo W, Chen L, Gong K, Ding B, Duan Y, Jin Y. Heterogeneous dental follicle cells and the regeneration of complex periodontal tissues. Tissue Eng Part A. 2012;18(5–6):459–70. [Google Scholar] [PubMed]
67. Luo X, Yang B, Sheng L, Chen J, Li H, Xie L. CAD based design sensitivity analysis and shape optimization of scaffolds for bio-root regeneration in swine. Biomaterials. 2015;57(3):59–72. [Google Scholar] [PubMed]
68. Du L, Yang P, Ge S. Isolation and characterization of human gingiva-derived mesenchymal stem cells using limiting dilution method. J Dent Sci. 2016;11(3):304–14. [Google Scholar] [PubMed]
69. Fawzy El-Sayed KM, Paris S, Becker ST, Neuschl M, De Buhr W, Sälzer S. Periodontal regeneration employing gingival margin-derived stem/progenitor cells: an animal study. J Clin Periodontol. 2012;39(9):861–70. [Google Scholar] [PubMed]
70. Rao SR, Subbarayan R, Dinesh MG, Arumugam G, Raja ST. Differentiation of human gingival mesenchymal stem cells into neuronal lineages in 3D bioconjugated injectable protein hydrogel construct for the management of neuronal disorder. Exp Mol Med. 2016;48(2):e209. [Google Scholar] [PubMed]
71. Fawzy El-Sayed KM, Mekhemar MK, Beck-Broichsitter BE, Bähr T, Hegab M, Receveur J. Periodontal regeneration employing gingival margin-derived stem/progenitor cells in conjunction with IL-1ra-hydrogel synthetic extracellular matrix. J Clin Periodontol. 2015;42(5):448–57. [Google Scholar] [PubMed]
72. Wang F, Yu M, Yan X, Wen Y, Zeng Q, Yue W. Gingiva-derived mesenchymal stem cell-mediated therapeutic approach for bone tissue regeneration. Stem Cells Dev. 2011;20(12):2093–102. [Google Scholar] [PubMed]
73. Xu QC, Wang ZG, Ji QX, Yu XB, Xu XY, Yuan CQ. Systemically transplanted human gingiva-derived mesenchymal stem cells contributing to bone tissue regeneration. Int J Clin Exp Pathol. 2014;7(8):4922–9. [Google Scholar] [PubMed]
74. Zhang Q, Nguyen PD, Shi S, Burrell JC, Cullen DK, Le AD. 3D bio-printed scaffold-free nerve constructs with human gingiva-derived mesenchymal stem cells promote rat facial nerve regeneration. Sci Rep. 2018;8(1):6634. [Google Scholar] [PubMed]
75. Galluzzi L, Green DR. Autophagy-independent functions of the autophagy machinery. Cell. 2019;177(7):1682–99. [Google Scholar] [PubMed]
76. Oku M, Sakai Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. Bioessays. 2018;40(6):e1800008. [Google Scholar] [PubMed]
77. Kaushik S, Cuervo AM. The coming of age of chaperone-mediated autophagy. Nat Rev Mol Cell Biol. 2018;19(6):365–81. [Google Scholar] [PubMed]
78. Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020;21(8):439–58. [Google Scholar] [PubMed]
79. Schuck S. Microautophagy—distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci. 2020;133(17):e31652. [Google Scholar]
80. Wang L, Klionsky DJ, Shen HM. The emerging mechanisms and functions of microautophagy. Nat Rev Mol Cell Biol. 2023;24(3):186–203. [Google Scholar] [PubMed]
81. Yamamoto H, Matsui T. Molecular mechanisms of macroautophagy, microautophagy, and chaperone-mediated autophagy. J Nippon Med Sch. 2023;1–26. [Google Scholar]
82. Sotthibundhu A, Promjuntuek W, Liu M, Shen S, Noisa P. Roles of autophagy in controlling stem cell identity: a perspective of self-renewal and differentiation. Cell Tissue Res. 2018;374(2):205–16. [Google Scholar] [PubMed]
83. Zhou J, He H, Zhang JJ, Liu X, Yao W, Li C. ATG7-mediated autophagy facilitates embryonic stem cell exit from naive pluripotency and marks commitment to differentiation. Autophagy. 2022;18(12):2946–68. [Google Scholar] [PubMed]
84. Ma Y, Qi M, An Y, Zhang L, Yang R, Doro DH. Autophagy controls mesenchymal stem cell properties and senescence during bone aging. Aging Cell. 2018;17(1):ii29. [Google Scholar]
85. D’Arcy MS. Cell death: a review of the major forms of apoptosis, necrosis and autophagy. Cell Biol Int. 2019;43(6):582–92. [Google Scholar] [PubMed]
86. Yang M, Wei H. Anesthetic neurotoxicity: apoptosis and autophagic cell death mediated by calcium dysregulation. Neurotoxicol Teratol. 2017;60(7):59–62. [Google Scholar] [PubMed]
87. Nah J, Zhai P, Huang CY, Fernández AF, Mareedu S, Levine B. Upregulation of Rubicon promotes autosis during myocardial ischemia/reperfusion injury. J Clin Invest. 2020;130(6):2978–91. [Google Scholar] [PubMed]
88. Xu X, Wang J, Xia Y, Yin Y, Zhu T, Chen F. Autophagy, a double-edged sword for oral tissue regeneration. J Adv Res. 2023;1–19. [Google Scholar]
89. Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24(1):24–41. [Google Scholar] [PubMed]
90. Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188(1):53–67. [Google Scholar] [PubMed]
91. Galluzzi L, Bravo-San Pedro JM, Levine B, Green DR, Kroemer G. Pharmacological modulation of autophagy: therapeutic potential and persisting obstacles. Nat Rev Drug Discov. 2017;16(7):487–511. [Google Scholar] [PubMed]
92. Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19(6):349–64. [Google Scholar] [PubMed]
93. Levine B, Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. [Google Scholar] [PubMed]
94. Matoba K, Noda NN. Structural catalog of core Atg proteins opens new era of autophagy research. J Biochem. 2021;169(5):517–25. [Google Scholar] [PubMed]
95. Wang J. Beclin 1 bridges autophagy, apoptosis and differentiation. Autophagy. 2008;4(7):947–8. [Google Scholar] [PubMed]
96. Okamoto K. Organellophagy: eliminating cellular building blocks via selective autophagy. J Cell Biol. 2014;205(4):435–45. [Google Scholar] [PubMed]
97. Onishi M, Yamano K, Sato M, Matsuda N, Okamoto K. Molecular mechanisms and physiological functions of mitophagy. EMBO J. 2021;40(3):e104705. [Google Scholar] [PubMed]
98. Wiemerslage L, Lee D. Quantification of mitochondrial morphology in neurites of dopaminergic neurons using multiple parameters. J Neurosci Methods. 2016;262(3):56–65. [Google Scholar] [PubMed]
99. Kuznetsov AV, Javadov S, Margreiter R, Hagenbuchner J, Ausserlechner MJ. Analysis of mitochondrial function, structure, and intracellular organization in situ in cardiomyocytes and skeletal muscles. Int J Mol Sci. 2022;23(4):2252. [Google Scholar] [PubMed]
100. Raghushaker CR, Chandra S, Chakrabarty S, Kabekkodu SP, Satyamoorthy K, Mahato KK. Detection of mitochondrial dysfunction in vitro by laser-induced autofluorescence. J Biophotonics. 2019;12(11):e201900056. [Google Scholar] [PubMed]
101. Lyamzaev KG, Tokarchuk AV, Panteleeva AA, Mulkidjanian AY, Skulachev VP, Chernyak BV. Induction of autophagy by depolarization of mitochondria. Autophagy. 2018;14(5):921–4. [Google Scholar] [PubMed]
102. Minibayeva F, Dmitrieva S, Ponomareva A, Ryabovol V. Oxidative stress-induced autophagy in plants: the role of mitochondria. Plant Physiol Biochem. 2012;59:11–9. [Google Scholar] [PubMed]
103. Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–22. [Google Scholar] [PubMed]
104. Lin NY, Chen CW, Kagwiria R, Liang R, Beyer C, Distler A. Inactivation of autophagy ameliorates glucocorticoid-induced and ovariectomy-induced bone loss. Ann Rheum Dis. 2016;75(6):1203–10. [Google Scholar] [PubMed]
105. Park SY, Cho HS, Chung KH, Lee BN, Kim SH, Kim WJ. Inactivation of PI3K/Akt promotes the odontoblastic differentiation and suppresses the stemness with autophagic flux in dental pulp cells. J Dent Sci. 2022;17(1):145–54. [Google Scholar] [PubMed]
106. Maity J, Deb M, Greene C, Das H. KLF2 regulates dental pulp-derived stem cell differentiation through the induction of mitophagy and altering mitochondrial metabolism. Redox Biol. 2020;36(5411):101622. [Google Scholar] [PubMed]
107. Ji F, Zhu L, Pan J, Shen Z, Tao J. hsa_circ_0026827 Promotes osteoblast differentiation of human dental pulp stem cells through the Beclin1 and RUNX1 signaling pathways by sponging miR-188-3p. Front Cell Dev Biol. 2020;8:1626. [Google Scholar]
108. Cho HS, Park SY, Kim SM, Kim WJ, Jung JY. Autophagy-related protein MAP1LC3C plays a crucial role in odontogenic differentiation of human dental pulp cells. Tissue Eng Regen Med. 2021;18(2):265–77. [Google Scholar] [PubMed]
109. Xu H, Xu F, Zhao J, Zhou C, Liu J. Platelet-rich plasma induces autophagy and promotes regeneration in human dental pulp cells. Front Bioeng Biotechnol. 2021;9:659742. [Google Scholar] [PubMed]
110. Xu Y, Wang X, Liu W, Lu W. Thrombin-activated platelet-rich plasma enhances osteogenic differentiation of human periodontal ligament stem cells by activating SIRT1-mediated autophagy. Eur J Med Res. 2021;26(1):105. [Google Scholar] [PubMed]
111. Zhang Y, Wang P, Wang Y, Li J, Qiao D, Chen R. Gold nanoparticles promote the bone regeneration of periodontal ligament stem cell sheets through activation of autophagy. Int J Nanomedicine. 2021;16:61–73. [Google Scholar] [PubMed]
112. Nakamura T, Yamashita M, Ikegami K, Suzuki M, Yanagita M, Kitagaki J. Autophagy facilitates type I collagen synthesis in periodontal ligament cells. Sci Rep. 2021;11(1):1291. [Google Scholar] [PubMed]
113. Nollet M, Santucci-Darmanin S, Breuil V, Al-Sahlanee R, Cros C, Topi M. Autophagy in osteoblasts is involved in mineralization and bone homeostasis. Autophagy. 2014;10(11):1965–77. [Google Scholar] [PubMed]
114. Pantovic A, Krstic A, Janjetovic K, Kocic J, Harhaji-Trajkovic L, Bugarski D. Coordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cells. Bone. 2013;52(1):524–31. [Google Scholar] [PubMed]
115. Pieles O, Hartmann M, Morsczeck C. AMP-activated protein kinase and the down-stream activated process of autophagy regulate the osteogenic differentiation of human dental follicle cells. Arch Oral Biol. 2021;122:104951. [Google Scholar] [PubMed]
116. Wang G, Luo J, Qiao Y, Zhang D, Liu Y, Zhang W. AMPK/mTOR pathway is involved in autophagy induced by magnesium-incorporated TiO2 surface to promote BMSC osteogenic differentiation. J Funct Biomater. 2022;13(4):221. [Google Scholar] [PubMed]
117. Martens W, Wolfs E, Struys T, Politis C, Bronckaers A, Lambrichts I. Expression pattern of basal markers in human dental pulp stem cells and tissue. Cells Tissues Organs. 2012;196(6):490–500. [Google Scholar] [PubMed]
118. Pagella P, Miran S, Neto E, Martin I, Lamghari M, Mitsiadis TA. Human dental pulp stem cells exhibit enhanced properties in comparison to human bone marrow stem cells on neurites outgrowth. FASEB J. 2020;34(4):5499–511. [Google Scholar] [PubMed]
119. Wang D, Wang Y, Tian W, Pan J. Advances of tooth-derived stem cells in neural diseases treatments and nerve tissue regeneration. Cell Prolif. 2019;52(3):e12572. [Google Scholar] [PubMed]
120. Prateeksha P, Naidu P, Das M, Barthels D, Das H. KLF2 regulates neural differentiation of dental pulp-derived stem cells by modulating autophagy and mitophagy. Stem Cell Rev Rep. 2023;19(8):2886–900. [Google Scholar] [PubMed]
121. Sotthibundhu A, Muangchan P, Phonchai R, Promjantuek W, Chaicharoenaudomrung N, Kunhorm P. Autophagy promoted neural differentiation of human placenta-derived mesenchymal stem cells. In Vivo. 2021;35(5):2609–20. [Google Scholar] [PubMed]
122. Kato H, Thi Mai Pham T, Yamaza H, Masuda K, Hirofuji Y, Han X. Mitochondria regulate the differentiation of stem cells from human exfoliated deciduous teeth. Cell Struct Funct. 2017;42(2):105–16. [Google Scholar] [PubMed]
123. Sun X, Dong S, Kato H, Kong J, Ito Y, Hirofuji Y. Mitochondrial calcium-triggered oxidative stress and developmental defects in dopaminergic neurons differentiated from deciduous teeth-derived dental pulp stem cells with MFF insufficiency. Antioxidants. 2022;11(7):1361. [Google Scholar] [PubMed]
124. Kongara S, Karantza V. The interplay between autophagy and ROS in tumorigenesis. Front Oncol. 2012;2:171. [Google Scholar] [PubMed]
125. Li L, Tan J, Miao Y, Lei P, Zhang Q. ROS and autophagy: interactions and molecular regulatory mechanisms. Cell Mol Neurobiol. 2015;35(5):615–21. [Google Scholar] [PubMed]
126. Hirofuji S, Hirofuji Y, Kato H, Masuda K, Yamaza H, Sato H. Mitochondrial dysfunction in dopaminergic neurons differentiated from exfoliated deciduous tooth-derived pulp stem cells of a child with Rett syndrome. Biochem Biophys Res Commun. 2018;498(4):898–904. [Google Scholar] [PubMed]
127. Nguyen HTN, Kato H, Masuda K, Yamaza H, Hirofuji Y, Sato H. Impaired neurite development associated with mitochondrial dysfunction in dopaminergic neurons differentiated from exfoliated deciduous tooth-derived pulp stem cells of children with autism spectrum disorder. Biochem Biophys Rep. 2018;16:24–31. [Google Scholar] [PubMed]
128. Nguyen Nguyen HT, Kato H, Sato H, Yamaza H, Sakai Y, Ohga S. Positive effect of exogenous brain-derived neurotrophic factor on impaired neurite development and mitochondrial function in dopaminergic neurons derived from dental pulp stem cells from children with attention deficit hyperactivity disorder. Biochem Biophys Res Commun. 2019;513(4):1048–54. [Google Scholar] [PubMed]
129. Sun X, Kato H, Sato H, Han X, Hirofuji Y, Kato TA. Dopamine-related oxidative stress and mitochondrial dysfunction in dopaminergic neurons differentiated from deciduous teeth-derived stem cells of children with down syndrome. FASEB Bioadv. 2022;4(7):454–67. [Google Scholar] [PubMed]
130. Sun X, Kato H, Sato H, Torio M, Han X, Zhang Y. Impaired neurite development and mitochondrial dysfunction associated with calcium accumulation in dopaminergic neurons differentiated from the dental pulp stem cells of a patient with metatropic dysplasia. Biochem Biophys Rep. 2021;26:100968. [Google Scholar] [PubMed]
131. Dudley AC, Griffioen AW. Pathological angiogenesis: mechanisms and therapeutic strategies. Angiogenesis. 2023;26(3):313–47. [Google Scholar] [PubMed]
132. Payne S, Neal A, de Val S. Transcription factors regulating vasculogenesis and angiogenesis. Dev Dyn. 2023;1–31. [Google Scholar]
133. Xu JG, Gong T, Wang YY, Zou T, Heng BC, Yang YQ. Inhibition of TGF-β signaling in SHED enhances endothelial differentiation. J Dent Res. 2018;97(2):218–25. [Google Scholar] [PubMed]
134. Bergamo MT, Zhang Z, Oliveira TM, Nör JE. VEGFR1 primes a unique cohort of dental pulp stem cells for vasculogenic differentiation. Eur Cell Mater. 2021;41:332–44. [Google Scholar] [PubMed]
135. Zhang Z, Nör F, Oh M, Cucco C, Shi S, Nör JE. Wnt/β-catenin signaling determines the vasculogenic fate of postnatal mesenchymal stem cells. Stem Cells. 2016;34(6):1576–87. [Google Scholar] [PubMed]
136. Zhang Z, Oh M, Sasaki JI, Nör JE. Inverse and reciprocal regulation of p53/p21 and Bmi-1 modulates vasculogenic differentiation of dental pulp stem cells. Cell Death Dis. 2021;12(7):644. [Google Scholar] [PubMed]
137. Sasaki JI, Zhang Z, Oh M, Pobocik AM, Imazato S, Shi S. VE-cadherin and anastomosis of blood vessels formed by dental stem cells. J Dent Res. 2020;99(4):437–45. [Google Scholar]
138. Yang JW, Zhang YF, Wan CY, Sun ZY, Nie S, Jian SJ. Autophagy in SDF-1α-mediated DPSC migration and pulp regeneration. Biomaterials. 2015;44(3):11–23. [Google Scholar] [PubMed]
139. Torisu T, Torisu K, Lee IH, Liu J, Malide D, Combs CA. Autophagy regulates endothelial cell processing, maturation and secretion of von Willebrand factor. Nat Med. 2013;19(10):1281–7. [Google Scholar] [PubMed]
140. Huang D, Lei J, Li X, Jiang Z, Luo M, Xiao Y. Erythropoietin activates autophagy to regulate apoptosis and angiogenesis of periodontal ligament stem cells via the Akt/ERK1/2/BAD signaling pathway under inflammatory microenvironment. Stem Cells Int. 2022;2022(4):9806887. [Google Scholar] [PubMed]
141. Wei W, An Y, An Y, Fei D, Wang Q. Activation of autophagy in periodontal ligament mesenchymal stem cells promotes angiogenesis in periodontitis. J Periodontol. 2018;89(6):718–27. [Google Scholar] [PubMed]
142. Luke AM, Patnaik R, Kuriadom S, Abu-Fanas S, Mathew S, Shetty KP. Human dental pulp stem cells differentiation to neural cells, osteocytes and adipocytes—an in vitro study. Heliyon. 2020;6(1):e03054. [Google Scholar] [PubMed]
143. Fracaro L, Senegaglia AC, Herai RH, Leitolis A, Boldrini-Leite LM, Rebelatto CLK. The expression profile of dental pulp-derived stromal cells supports their limited capacity to differentiate into adipogenic cells. Int J Mol Sci. 2020;21(8):2753. [Google Scholar] [PubMed]
144. Longoni A, Utomo L, van Hooijdonk IE, Bittermann GK, Vetter VC, Kruijt Spanjer EC. The chondrogenic differentiation potential of dental pulp stem cells. Eur Cell Mater. 2020;39:121–35. [Google Scholar] [PubMed]
145. Yang H, Cao Y, Zhang J, Liang Y, Su X, Zhang C. DLX5 and HOXC8 enhance the chondrogenic differentiation potential of stem cells from apical papilla via LINC01013. Stem Cell Res Ther. 2020;11(1):271. [Google Scholar] [PubMed]
146. Pisciotta A, Bertani G, Bertoni L, di Tinco R, de Biasi S, Vallarola A. Modulation of cell death and promotion of chondrogenic differentiation by Fas/FasL in human dental pulp stem cells (hDPSCs). Front Cell Dev Biol. 2020;8:279. [Google Scholar] [PubMed]
147. Xie Y, Shen G. MicroRNA-139-5p elevates skeletal myogenic differentiation of human adult dental pulp stem cells through Wnt/β-catenin signaling pathway. Exp Ther Med. 2018;16(4):2835–42. [Google Scholar] [PubMed]
148. Jiang W, Wang D, Alraies A, Liu Q, Zhu B, Sloan AJ. Wnt-GSK3 β/β-catenin regulates the differentiation of dental pulp stem cells into bladder smooth muscle cells. Stem Cells Int. 2019;2019:1–13. [Google Scholar]
149. Yagi Mendoza H, Yokoyama T, Tanaka T, Ii H, Yaegaki K. Regeneration of insulin-producing islets from dental pulp stem cells using a 3D culture system. Regen Med. 2018;13(6):673–87. [Google Scholar] [PubMed]
150. Chen YK, Huang AH, Chan AW, Lin LM. Human dental pulp stem cells derived from cryopreserved dental pulp tissues of vital extracted teeth with disease demonstrate hepatic-like differentiation. J Tissue Eng Regen Med. 2016;10(6):475–85. [Google Scholar] [PubMed]
151. Calabrese EJ, Baldwin LA. Defining hormesis. Hum Exp Toxicol. 2002;21(2):91–7. [Google Scholar] [PubMed]
152. Mattson MP. Hormesis defined. Ageing Res Rev. 2008;7(1):1–7. [Google Scholar] [PubMed]
153. Calabrese EJ, Mattson MP. Hormesis provides a generalized quantitative estimate of biological plasticity. J Cell Commun Signal. 2011;5(1):25–38. [Google Scholar] [PubMed]
154. Calabrese EJ. Human periodontal ligament stem cells and hormesis: enhancing cell renewal and cell differentiation. Pharmacol Res. 2021;173:105914. [Google Scholar] [PubMed]
155. Calabrese EJ. Hormesis and dental apical papilla stem cells. Chem Biol Interact. 2022;357(2):109887. [Google Scholar] [PubMed]
156. Calabrese EJ, Agathokleous E, Dhawan G, Kapoor R, Calabrese V. Human dental pulp stem cells and hormesis. Ageing Res Rev. 2022;73(35):101540. [Google Scholar] [PubMed]
157. Matichescu A, Ardelean LC, Rusu LC, Craciun D, Bratu EA, Babucea M. Advanced biomaterials and techniques for oral tissue engineering and regeneration—a review. Materials. 2020;13(22):5303. [Google Scholar] [PubMed]
158. Chen Z, Ni S, Han S, Crawford R, Lu S, Wei F. Nanoporous microstructures mediate osteogenesis by modulating the osteo-immune response of macrophages. Nanoscale. 2017;9(2):706–18. [Google Scholar] [PubMed]
159. Li L, Yang S, Xu L, Li Y, Fu Y, Zhang H. Nanotopography on titanium promotes osteogenesis via autophagy-mediated signaling between YAP and β-catenin. Acta Biomater. 2019;96(2):674–85. [Google Scholar] [PubMed]
160. Ulusoy AT, Turedi I, Cimen M, Cehreli ZC. Evaluation of blood clot, platelet-rich plasma, platelet-rich fibrin, and platelet pellet as scaffolds in regenerative endodontic treatment: a prospective randomized trial. J Endod. 2019;45(5):560–6. [Google Scholar] [PubMed]
161. Lu J, Li R, Ni S, Xie Y, Liu X, Zhang K. Metformin carbon nanodots promote odontoblastic differentiation of dental pulp stem cells by pathway of autophagy. Front Bioeng Biotechnol. 2022;10:1002291. [Google Scholar] [PubMed]
162. Liu L, Li X, Bu W, Jin N, Meng Y, Wang Y. Carbon dots enhance extracellular matrix secretion for dentin-pulp complex regeneration through PI3K/Akt/mTOR pathway-mediated activation of autophagy. Mater Today Bio. 2022;16:100344. [Google Scholar] [PubMed]
163. Li X, Liu H, Yu Y, Ma L, Miao L. Graphene oxide quantum dots-induced mineralization via the reactive oxygen species-dependent autophagy pathway in dental pulp stem cells. J Biomed Nanotechnol. 2020;16(6):965–74. [Google Scholar] [PubMed]
164. Guo T, Cao G, Li Y, Zhang Z, Nor JE, Clarkson BH. Signals in stem cell differentiation on fluorapatite-modified scaffolds. J Dent Res. 2018;97(12):1331–8. [Google Scholar] [PubMed]
165. Zhang Y, Kong N, Zhang Y, Yang W, Yan F. Size-dependent effects of gold nanoparticles on osteogenic differentiation of human periodontal ligament progenitor cells. Theranostics. 2017;7(5):1214–24. [Google Scholar] [PubMed]
166. Yin Y, Tian BM, Li X, Yu YC, Deng DK, Sun LJ. Gold nanoparticles targeting the autophagy-lysosome system to combat the inflammation-compromised osteogenic potential of periodontal ligament stem cells: from mechanism to therapy. Biomaterials. 2022;288:121743. [Google Scholar] [PubMed]
167. Zhu S, Ying Y, Wu Q, Ni Z, Wang Z. Alginate self-adhesive hydrogel combined with dental pulp stem cells and FGF21 repairs hemisection spinal cord injury via apoptosis and autophagy mechanisms. Chem Eng J. 2021;2021(3):130827. [Google Scholar]
168. Liu Y, Chen L, Gong Q, Jiang H, Huang Y. Hypoxia‐induced mitophagy regulates proliferation, migration and odontoblastic differentiation of human dental pulp cells through FUN14 domain‐containing 1. Int J Mol Med. 2022;49(5):72. [Google Scholar] [PubMed]
169. Zhuang H, Hu D, Singer D, Walker JV, Nisr RB, Tieu K. Local anesthetics induce autophagy in young permanent tooth pulp cells. Cell Death Discov. 2015;1(1):15024. [Google Scholar] [PubMed]
170. Maity J, Barthels D, Sarkar J, Prateeksha P, Deb M, Rolph D. Ferutinin induces osteoblast differentiation of DPSCs via induction of KLF2 and autophagy/mitophagy. Cell Death Dis. 2022;13(5):452. [Google Scholar] [PubMed]
171. Wang X, Wu TT, Jiang L, Rong D, Zhu YQ. Deferoxamine-induced migration and odontoblast differentiation via ROS-dependent autophagy in dental pulp stem cells. Cell Physiol Biochem. 2017;43(6):2535–47. [Google Scholar] [PubMed]
172. Kornsuthisopon C, Rochanavibhata S, Nowwarote N, Tompkins KA, Sukarawan W, Osathanon T. 6-bromoindirubin-3’-oxime regulates colony formation, apoptosis, and odonto/osteogenic differentiation in human dental pulp stem cells. Int J Mol Sci. 2022;23(15):8676. [Google Scholar] [PubMed]
173. Dang H, Chen W, Chen L, Huo X, Wang F. TPPU inhibits inflammation-induced excessive autophagy to restore the osteogenic differentiation potential of stem cells and improves alveolar ridge preservation. Sci Rep. 2023;13(1):1574. [Google Scholar] [PubMed]
174. Pan Y, Li Z, Wang Y, Yan M, Wu J, Beharee RG. Sodium fluoride regulates the osteo/odontogenic differentiation of stem cells from apical papilla by modulating autophagy. J Cell Physiol. 2019;234(9):16114–24. [Google Scholar] [PubMed]
175. Lei S, Liu XM, Liu Y, Bi J, Zhu S, Chen X. Lipopolysaccharide downregulates the osteo-/odontogenic differentiation of stem cells from apical papilla by inducing autophagy. J Endod. 2020;46(4):502–8. [Google Scholar] [PubMed]
176. Yang Z, Gao X, Zhou M, Kuang Y, Xiang M, Li J. Effect of metformin on human periodontal ligament stem cells cultured with polydopamine-templated hydroxyapatite. Eur J Oral Sci. 2019;127(3):210–21. [Google Scholar] [PubMed]
177. Pei F, Wang HS, Chen Z, Zhang L. Autophagy regulates odontoblast differentiation by suppressing NF-κB activation in an inflammatory environment. Cell Death Dis. 2016;7(3):e2122. [Google Scholar] [PubMed]
178. Zheng J, Xu B, Yang K. Autophagy regulates osteogenic differentiation of human periodontal ligament stem cells induced by orthodontic tension. Stem Cells Int. 2022;2022(1):2983862. [Google Scholar] [PubMed]
179. Huang Y, Liu H, Guo R, Han Y, Yang Y, Zhao Y. Long non-coding RNA FER1L4 mediates the autophagy of periodontal ligament stem cells under orthodontic compressive force via AKT/FOXO3 pathway. Front Cell Dev Biol. 2021;9:631181. [Google Scholar] [PubMed]
180. Yan K, Wu C, Ye Y, Li L, Wang X, He W. A20 inhibits osteoclastogenesis via TRAF6-dependent autophagy in human periodontal ligament cells under hypoxia. Cell Prolif. 2020;53(3):e12778. [Google Scholar] [PubMed]
181. Zheng J, Zhu X, He Y, Hou S, Liu T, Zhi K. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann N Y Acad Sci. 2021;1485(1):56–70. [Google Scholar] [PubMed]
182. Niu Q, Chen H, Ou Q, Yang S, Peng Y, Xie Y. Klotho enhances bone regenerative function of hPDLSCs via modulating immunoregulatory function and cell autophagy. J Orthop Surg Res. 2023;18(1):400. [Google Scholar] [PubMed]
183. Li XC, Zhao Y, Peng HR, Gu DA, Liu C, Ren SS. Robust intervention for oxidative stress-induced injury in periodontitis via controllably released nanoparticles that regulate the ROS-PINK1-Parkin pathway. Front Bioeng Biotechnol. 2022;10:8882. [Google Scholar]
184. Vidoni C, Ferraresi A, Secomandi E, Vallino L, Gardin C, Zavan B. Autophagy drives osteogenic differentiation of human gingival mesenchymal stem cells. Cell Commun Signal. 2019;17(1):98. [Google Scholar] [PubMed]
185. Yang F, Li Y, Duan H, Wang H, Pei F, Chen Z. Activation of mitophagy in inflamed odontoblasts. Oral Dis. 2019;25(6):1581–8. [Google Scholar] [PubMed]
186. Qi Y, Luan Y, Wang L. Low concentrations of tumor necrosis factor-alpha promote human periodontal ligament stem cells osteogenic differentiation by activation of autophagy via inhibition of AKT/mTOR pathway. Mol Biol Rep. 2023;50(4):3329–39. [Google Scholar]
187. Li N, Yan M, Chen Y, Wang Y, Yu J. Extracellular IL-37 promotes osteogenic and odontogenic differentiation of human dental pulp stem cells via autophagy. Exp Cell Res. 2021;407(1):112780. [Google Scholar] [PubMed]
188. Garlet TP, Coelho U, Silva JS, Garlet GP. Cytokine expression pattern in compression and tension sides of the periodontal ligament during orthodontic tooth movement in humans. Eur J Oral Sci. 2007;115(5):355–62. [Google Scholar] [PubMed]
189. Yan Y, Liu F, Kou X, Liu D, Yang R, Wang X. T Cells are required for orthodontic tooth movement. J Dent Res. 2015;94(10):1463–70. [Google Scholar] [PubMed]
190. Jiang N, He D, Ma Y, Su J, Wu X, Cui S. Force-induced autophagy in periodontal ligament stem cells modulates M1 macrophage polarization via AKT signaling. Front Cell Dev Biol. 2021;9:666631. [Google Scholar] [PubMed]
191. Chen L, Mo S, Hua Y. Compressive force-induced autophagy in periodontal ligament cells downregulates osteoclastogenesis during tooth movement. J Periodontol. 2019;90(10):1170–81. [Google Scholar] [PubMed]
192. Luo Y, Gou H, Chen X, Li L, Wang X, Xu Y. Lactate inhibits osteogenic differentiation of human periodontal ligament stem cells via autophagy through the MCT1-mTOR signaling pathway. Bone. 2022;162(17038):116444. [Google Scholar] [PubMed]
193. Tan J, Zhou Y, Luo J, Wu X, Liu H, Wang W. High glucose inhibits the osteogenic differentiation of periodontal ligament stem cells in periodontitis by activating endoplasmic reticulum stress. Ann Transl Med. 2022;10(4):204. [Google Scholar] [PubMed]
194. Zhang K, Liu F, Jin D, Guo T, Hou R, Zhang J. Autophagy preserves the osteogenic ability of periodontal ligament stem cells under high glucose conditions in rats. Arch Oral Biol. 2019;101(5):172–9. [Google Scholar] [PubMed]
195. Wang D, Zhu N, Xie F, Qin M, Wang Y. Long non-coding RNA IGFBP7-AS1 promotes odontogenic differentiation of stem cells from human exfoliated deciduous teeth through autophagy: an in vitro study. Arch Oral Biol. 2022;141(15):105492. [Google Scholar] [PubMed]
196. Li Z, Wu Z, Xi X, Zhao F, Liu H, Liu D. Cellular communication network factor 1 interlinks autophagy and ERK signaling to promote osteogenesis of periodontal ligament stem cells. J Periodontal Res. 2022;57(6):1169–82. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF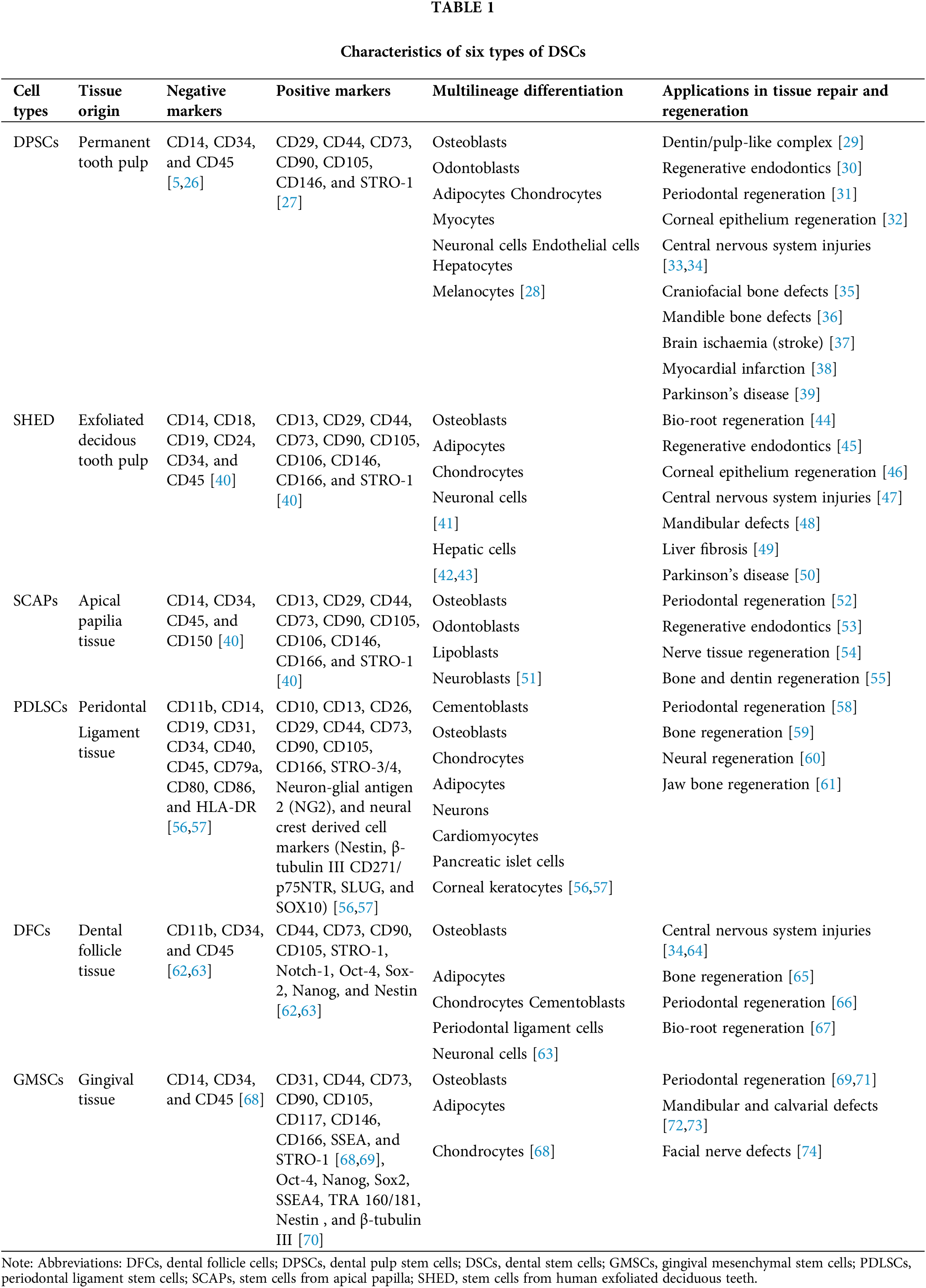
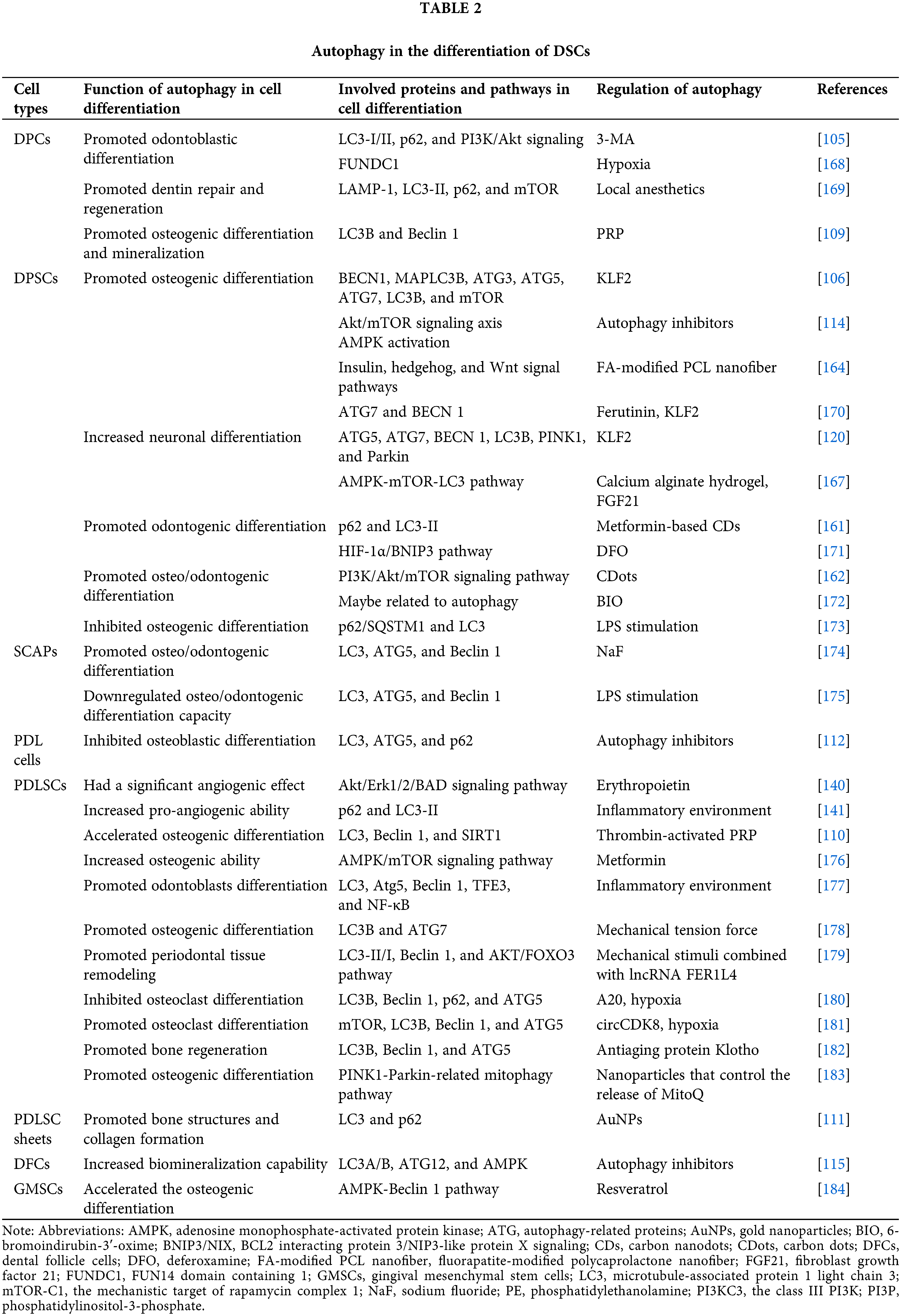
 Downloads
Downloads
 Citation Tools
Citation Tools