 Open Access
Open Access
REVIEW
Autophagy and circadian rhythms: interactions and clinical implications
1 State Key Laboratory of Oral & Maxillofacial Reconstruction and Regeneration, National Clinical Research Center for Oral Diseases, Shaanxi Clinical Research Center for Oral Diseases, Department of Pediatric Dentistry, School of Stomatology, The Fourth Military Medical University, Xi’an, 710032, China
2 Department of Stomatology, The 969th Hospital, Joint Logistics Support Force of the Chinese People’s Liberation Army, Hohhot, 010000, China
3 Department of Stomatology, General Hospital of Tibetan Military Command, Lhasa, 850000, China
4 Key Laboratory of Shaanxi Province for Craniofacial Precision Medicine Research, Clinical Research Center of Shaanxi Province for Dental and Maxillofacial Diseases, Department of Pediatric Dentistry, College of Stomatology, Xi’an Jiaotong University, Xi’an, 710000, China
5 Department of Neurobiology and Institute of Neurosciences, School of Basic Medicine, Fourth Military Medical University, Xi’an, 710032, China
* Corresponding Authors: YUJIANG CHEN. Email: ; LULU WANG. Email:
# Co-first authors
BIOCELL 2024, 48(1), 33-45. https://doi.org/10.32604/biocell.2023.031638
Received 07 August 2023; Accepted 07 November 2023; Issue published 30 January 2024
Abstract
Autophagy is a widespread biological process that controls cellular growth, survival, development, and death. Circadian rhythm is a recurring reaction of living organisms and behaviors to variations in surrounding brightness and obscurity. Most of the fundamental physiological processes in mammals, such as the sleep-wake pattern and the rhythm of nutrition and energy metabolism, are governed by circadian rhythms. Research has indicated that autophagy exhibits a specific circadian pattern in both normal and abnormal conditions. Autophagy can modulate circadian rhythms by breaking down proteins that regulate the circadian clock. The potential regulatory connection between the two has been a popular subject of clinical and fundamental research. Understanding the interaction between circadian rhythm and autophagy could potentially lead to the development of novel approaches for disease treatment in the future. The present analysis presented a summary of the molecular processes implicated in the interplay between autophagy and circadian rhythm, as well as the pathological importance of the disrupted regulatory association between these two phenomena.Keywords
Autophagy is a ubiquitous biological process that controls cellular growth, survival, development, and death. Cellular homeostasis and survival heavily rely on autophagy, which is intricately linked to numerous conditions including liver disease, neurodegenerative disorders, myopathy, heart disease, cancer, and immune system dysregulation [1,2]. Autophagy is commonly triggered by different stressors, such as endoplasmic reticulum stress (ERS), growth factor withdrawal, nutrient deprivation, mitochondrial dysfunction, and inflammatory responses [3–5].
An individual’s circadian rhythm refers to a recurring alteration in conduct, bodily functions, and internal conditions, enduring for about 24 h [6,7]. These rhythms exist at different levels within the biological realm, including broad aspects like the pattern of sleep and wakefulness, as well as minute occurrences like the periodic variations in biomolecular quantities [8–10]. In mammals, the daily fluctuations are controlled by the oscillations of the hypothalamic clock in the suprachiasmatic nucleus (SCN) and peripheral clocks. These clocks synchronize with the light and feeding schedules, respectively [11,12].
It has been implied that autophagy is regulated by circadian rhythms. Recently, there has been a growing interest in examining the daily variations in physiological processes, with a particular focus on the circadian rhythm of important regulatory molecules like mTOR and ATG in the autophagic process [13,14]. The central gene BMAL1/CLOCK of the circadian clock mechanism demonstrates the capacity to trigger gene transcription linked to the control of autophagy and the circadian cycle [13]. Nevertheless, the risk of diseases can also be heightened due to autophagy malfunction caused by pathogenic factors [15].
Likewise, autophagy controls circadian rhythms. Autophagy plays a crucial part in the pineal gland and the production of melatonin during the circadian rhythm, with both functions being regulated by autophagy [16]. Recent studies have shown that autophagy plays a crucial role in controlling the essential components of the circadian clock [17]. In the case of mice that lacked ATG7, it was observed that hepatic CRY1 levels increased while blood glucose levels decreased, suggesting that autophagic degradation of CRY1 may play a role in regulating blood glucose levels. CRY1 has been suggested to interact with LC3 through its LIR motif, leading to degradation through autophagy [18].
In summary, circadian rhythm and autophagy coregulate each other at the cellular level. This bidirectional interaction between circadian rhythm and autophagy has significant implications in disease treatments. Understanding this relationship provides opportunities to develop new therapeutic strategies by combining circadian and autophagy modulators for better treatment of cancers, metabolic disorders and neurodegenerative diseases [19–21].
Autophagy and Circadian Rhythm
Autophagy performs several important cellular functions including recycling nutrients, removing damaged organelles, fighting intracellular pathogens, and clearing protein aggregates [22]. During autophagy, portions of the cytoplasm and organelles are sequestered into double-membrane vesicles called autophagosomes. The formation of autophagosomes is a highly regulated multi-step process that involves initiation, nucleation, elongation and closure of the isolation membrane [22].
There are three different types of autophagy: macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA). In autophagy, lysosomes degrade abnormal organelles and proteins. The procedure includes the use of double-layered sacs, referred to as autophagosomes or autophagic vacuoles, that specifically engulf and remove denatured proteins or damaged organelles. The complex procedure involves multiple phases, such as commencement, vesicle formation, vesicle enlargement, and finalization [23–25]. On the one hand, during macroautophagy, after autophagosomes are formed, lysosomes are involved in the degradation of phagocytic organelles, cytoplasmic proteins, and other substances, resulting in the production of autolysosomes [5]. On the other hand, microautophagy entails the transfer of substances into lysosomes and their subsequent breakdown by the direct protrusion, invagination, or separation of lysosomes. The interaction between chaperone proteins and cytosolic target substrates in CMA is facilitated by HSC70, a 70 kDa heat shock cognate protein. HSC70 contains pentapeptide motifs similar to Lys-Phe-Glu-Arg-Gln (KFERQ). Afterwards, HSC70 aids in the transfer of goods into the lysosomal lumen by relying on the receptor LAMP2A, which is associated with the lysosome membrane [1]. Ultimately, the target cargo is degraded by hydrolytic enzymes in all three forms of autophagy (Fig. 1).
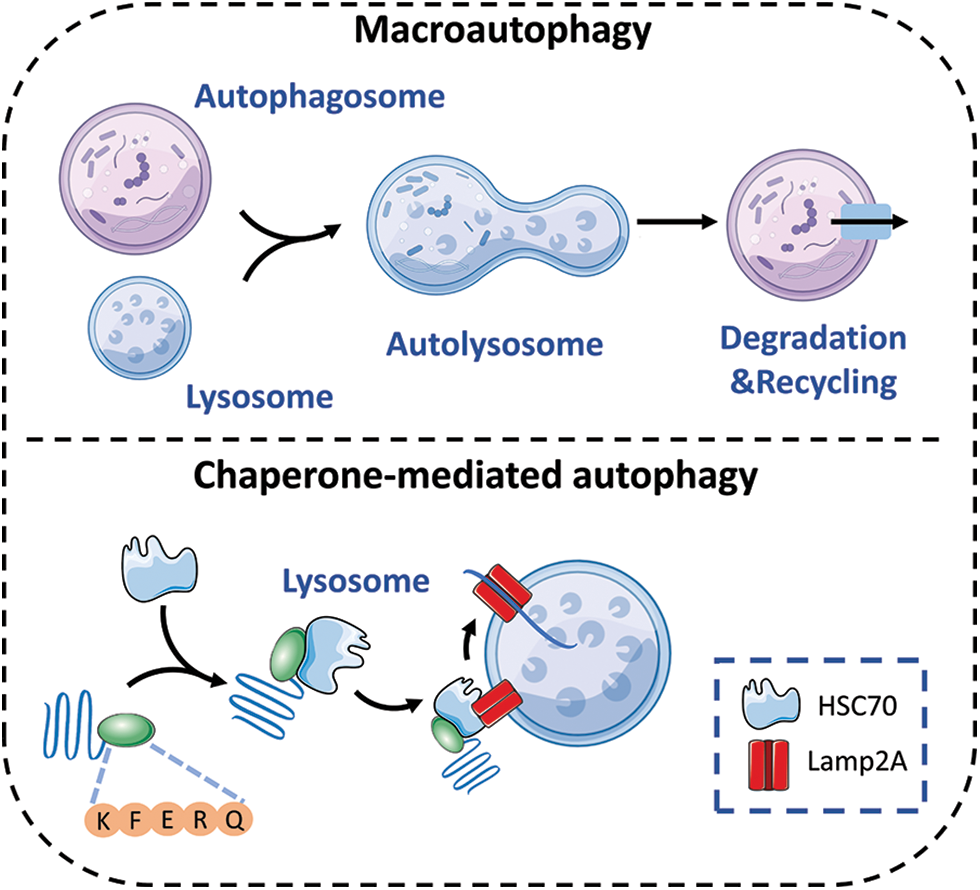
Figure 1: The autophagy processes that are impacted by circadian rhythms consist of macroautophagy and chaperone-mediated autophagy.
The specific regulation of autophagy involves multiple genes. Autophagy has been extensively studied, with the recognition of at least 30 autophagy-associated genes (ATGs) in yeast and the detection of various ATG counterparts in mammals, which are known to regulate autophagy [26–28]. At the beginning of autophagy, the activation of the ATG1 complex takes place. The ATG1 complex in yeast consists of the ATG17-ATG31-ATG29 subcomplex, ATG1, and ATG13. The UNC-51-like kinase (ULK) complex, which is present in mammals, is composed of ULK1 or ULK2 (mammalian ATG1 homologs), ATG13 (a potential counterpart of yeast ATG17), RB1-induced frizzled protein 1 (RB1CC1), and ATG101 [28,29]. In the course of vesicle formation, it is crucial to stimulate the PI3K complex, consisting of Vps34, Beclin-1, and ATG14, for nucleation of vesicles. The main function of this intricate structure is to enlist ATG proteins to the assembly locations of autophagosomes (PAS). During the process of vesicle expansion and finalization, the sequestration membrane experiences elongation with the participation of two UBI binding systems, namely the ATG12-ATG5-ATG16 complex and phosphatidylethanolamine (PE)-microtubule-associated protein 1 light chain 3 (LC3, also known as ATG8). In the final stages of autophagy, mature autophagosomes combine with lysosomes, leading to the creation of autolysosomes. Within these autolysosomes, the enclosed cargo is broken down with the help of hydrolytic enzymes. Afterwards, the final outcome is released into the cytosol [30,31].
Mechanisms of the circadian rhythm
Many physiological and pathological processes exhibit circadian rhythms, and these rhythms are regulated by an autonomous oscillatory rhythm known as the circadian clock. The circadian clock controls fundamental functions like organism behavior and activity, typically following a circadian rhythm of around 24 h [32,33]. The SCN, located in the front part of the hypothalamus, acts as the central point of the mammalian circadian rhythm, consisting of approximately 15,000 nerve cells [32]. On the other hand, the peripheral circadian clock is located in peripheral organs and tissues, and has the ability to independently control specific functions to some extent [34,35]. Although the suprachiasmatic nucleus (SCN) clock plays a crucial role in coordinating overall circadian rhythms, tissue-specific cell-autonomous molecular clocks can regulate rhythmic processes within peripheral tissues [32,36].
Circadian rhythms consist of a collection of core circadian clock genes and clock control genes (CCGs), which contribute to both positive and negative feedback [7,37]. The primary clock genes consist of CLOCK, BMAL1 (also referred to as ARNTL), Cryptochrome (CRY1/2), Period (PER1/2), REV-ERB, and ROR. Likewise, the circadian clock genes (CCGs) contain Dbp (D-box binding protein), Hlf (hepatic leukemia factor), Tef (thyrotroph embryonic factor), and Dec1 (Stra13 or Sharp2) [34,35]. At the molecular level, the CLOCK and BMAL1 proteins interact to create a clock-activated complex, which aids in the transcription of CCG. Furthermore, these genes are responsible for producing clock suppressor proteins, specifically PERs, CRYs, and REV-ERBs proteins. The presence of these inhibitory proteins enables the reduction of CLOCK/BMAL1-dependent transcription. The circadian expression of around 10%–15% of mammalian genes is achieved through the combined effect of core clock genes, local transcription factors, and chromatin-remodeling enzymes, resulting in cell- or tissue-specific gene rhythmic expressions [38]. The dynamic equilibrium between clock activators and clock inhibitory complexes leads to the circadian expression of approximately 10%–15% of mammalian genes [39,40].
Herein, the specific mechanism is described. To begin with, it is crucial to mention that BMAL1 and CLOCK are essential transcription factors that come together to create a heterodimer. Subsequently, this heterodimer initiates the activation of PER and CRY genes’ transcription. After being translated in the cytoplasm, the PER and CRY proteins move to the nucleus and combine with each other to form dimers. The purpose of this process is to suppress and hinder the function of the BMAL1/CLOCK heterodimer [41]. As a result, these heterodimers move to the nucleus and suppress the transcriptional activity of BMAL1/CLOCK heterodimers on target genes located downstream. Ultimately, this process establishes a negative feedback regulatory loop [42,43]. The proteasome in the cytoplasm breaks down the PER protein, but the PER/CRY heterodimers can move to the nucleus and stop PER degradation, thus hindering the BMAL1/CLOCK complex’s function [44]. The decrease in levels of PER and CRY heterodimeric proteins reduces the inhibitory impact on the transcriptional activity of CLOCK/BMAL1, resulting in the commencement of a fresh transcription-translation cycle that governs the organism’s circadian rhythm. Furthermore, the interaction between BMAL1 and CLOCK heterodimers is involved in the regulation of the transcription of REV-ERBα/β and RORα/β genes, which are classified as orphan nuclear receptors. The REV-ERB protein, when translated, competes with the ROR protein for the ROR response element (RRE) found in the binding site of the BMAL1 promoter region. This competition in binding results in either the activation or inhibition of BMAL1 transcription, depending on the specific protein it attaches to. The ROR protein stimulates the transcription of BMAL1, whereas REV-ERB suppresses the transcription of BMAL1, creating an additional regulatory loop for the circadian rhythm. As shown in Fig. 2 [13,34,35,45].
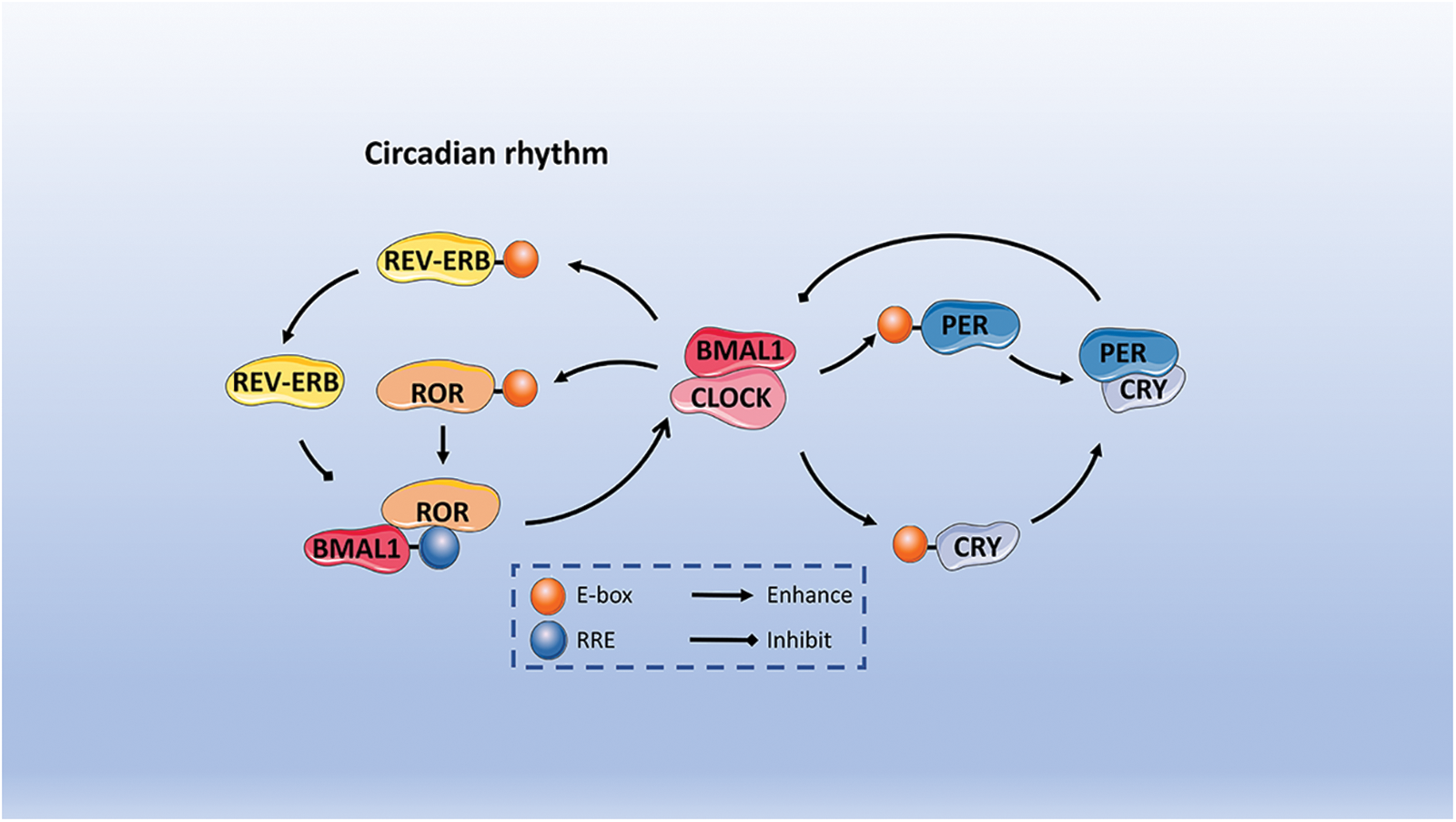
Figure 2: The description of circadian rhythms includes both positive and negative feedback. Initially, BMAL1 and CLOCK serve as fundamental transcription factors that create heterodimers which bind to E-boxes in order to initiate the transcription process of PER and CRY genes. Following translation in the cytoplasm, PER and CRY proteins join together to suppress and hinder the function of the BMAL1/CLOCK heterodimer. REV-ERB and ROR protein compete for the ROR response element. The ROR protein triggers the transcription of BMAL1 by attaching to the ROR response element (RRE) found within BMAL1. Conversely, REV-ERB hinders the transcription of BMAL1, thereby establishing an additional regulatory loop for the circadian rhythm.
Circadian Regulation of Autophagy
Regulation of macroautophagy by circadian rhythm
Circadian rhythms have been shown to regulate macroautophagy, which is the predominant form of autophagy according to numerous studies [46–49]. The regulation of autophagy is often studied using genes that are regulated by the circadian rhythm. The summary of the characteristics and role of these rhythmic genes can be found in the subsequent section.
REV-ERBα is a key factor linking autophagy to the circadian clock. It has been reported that obvious circadian rhythmicity is exhibited in autophagy. For instance, the circadian activation of TFEB and TFE3, pivotal regulators of autophagy and lysosomal biogenesis, is closely linked to REV-ERBα [50,51]. Pariollaud et al. found that the inhibition of rhythmic diurnal variation occurred upon knocking down REV-ERBα, resulting in a significant increase in inflammatory cytokines in the mouse lung. This observation suggests that circadian clock molecules play a role in mediating REV-ERBα, thereby promoting an inflammatory response and exacerbating the progression of chronic obstructive pulmonary disease (COPD) [52]. The impairment of autophagy in emphysema is mediated by perinuclear accumulation of TFEB and promotes inflammatory-oxidative stress responses [53]. TFEB and TFE3 have the ability to engage with REV-ERBα and contribute to the rhythmic manifestation of autophagy, consequently governing lung homeostasis [54]. The circadian clock genes can regulate the suppressor of autophagy, mTOR, through two mechanisms: firstly, the core clock protein PER2 can impede mTORC1 activity to induce autophagy [55], and secondly, clock-related transcription factors can positively modulate mTORC1 to hinder autophagy and phosphorylation of the clock protein BMAL1 [56]. The binding of TFE3 and REV-ERB to the promoter regions of genes related to circadian, autophagic, and lysosomal biogenesis (such as LAMP1, ATG3/5 and CTSL) was observed. The presence of Alpha, TFEB, and TFE3 plays a crucial role in regulating autophagy, lysosome biogenesis, and cellular homeostasis, particularly in relation to circadian rhythms. Additionally, it can also regulate clock genes involved in autophagy and the expression of REV-ERBα [50].
Researchers have conducted studies aiming to control autophagy and address diseases through the targeting of REV-ERB. The utilization of the REV-ERB-specific agonist SR9009 has been found to elicit REV-ERB-dependent anti-small cell lung cancer effects by inhibiting autophagy both in vivo and in vitro. The regulation of autophagy by REV-ERBα plays a significant role in this process. The primary autophagy gene, ATG5, is a direct downstream target of REV-ERBα, and the interaction between REV-ERBα and ATG5 may impede autophagic activity, leading to the cytotoxic effects of SR9009 in SCLC cells [57]. Similar conclusions have been confirmed in multiple studies that reported its effectiveness against cancer and liver fibrosis via pharmacological modulation of circadian mechanisms. Additionally, previous research has demonstrated that SR9011 plays a crucial role in initiating apoptotic responses in malignant cells. These REV-ERB agonists possess selective anticancer properties that effectively impede cancer cell growth and enhance survival rates in experimental animals, all while minimizing detrimental effects on normal tissues [9,45,58–60]. Moreover, the circadian clock gene REV-ERBs have the potential to alleviate follicular dysplasia by inhibiting granulosa cell apoptosis and promoting proliferation through the regulation of mitochondrial biogenesis and autophagy in polycystic ovary syndrome [9,61]. However, some researchers have also found that the effect of REV-ERBs is not completely positive. For example, REV-ERBα upregulation exacerbated steatosis in alcoholic fatty liver (AFL) by regulating autophagy and promoting the progression of AFL [62]. The expression of REV-ERBα is heightened by stressors associated with diabetes, leading to the suppression of autophagy and the viability of β-cells. Conversely, the administration of REV-ERBα agonists impedes the flow of autophagy, viability, and insulin secretion, indicating the significant involvement of REV-ERBα activation in the modulation of β-cells in the face of diabetic stressors [63].
The protein CLOCK/BMAL1 serves as a fundamental regulator of the circadian clock. Recent research has demonstrated that autophagy, a cellular process, is initiated by reactive oxygen species (ROS) in conditions of low oxygen levels (hypoxia). Furthermore, the overexpression of CLOCK in human umbilical vein endothelial cells (HUVECs) during hypoxia results in an augmentation of autophagosomes. Conversely, the suppression of CLOCK expression counteracts these effects. Additionally, pre-treatment with 3-methyladenine (3-MA) only mitigates autophagy without influencing ROS generation [64]. C/EBPβ, a transcription factor regulated by the circadian rhythm, stimulates the expression of autophagic genes including Ulk1, Gabarapl1, LC3B, and Bnip3, as well as protein degradation [54]. In zebrafish, the CLOCK-BMAL1 heterodimers bind to E-boxes to modulate the transcription of C/EBPβ, which subsequently governs the transcription of autophagy-related genes in an indirect manner [65]. In atherosclerosis, the downregulation of CLOCK and circadian dysregulation promoted vascular smooth muscle cell apoptosis by attenuating defects in autophagy [66]. In investigations concerning myocardial ischemia/reperfusion injury, it has been observed that the regulation of autophagy by the HDAC3/SIRT1 pathway through BMAL1 also exhibits a protective impact [67]. A study conducted on diabetic mice demonstrated that melatonin, in a dose-dependent manner, augmented the expression of proteins associated with the SIRT1-BMAL1 pathway and enhanced autophagy, thereby mitigating cerebral ischemia-reperfusion (I/R) injury [68]. The study revealed that the suppression of circadian gene CLOCK expression resulted in the reversal of cisplatin resistance in SKOV3/DDP cell lines of ovarian cancer. This effect was attributed to the modulation of protein expression of resistance genes associated with autophagy. Consequently, the CLOCK gene emerges as a promising candidate for targeted gene therapy in cases of drug-resistant ovarian cancer [69].
The clock genes CRY and PER exhibit negative regulation and are implicated in the regulation of autophagy. Through the PI3K/AKT/mTOR pathway, the upregulation of the clock gene PER2 impedes the progression of oral squamous cell carcinoma (OSCC) by inducing autophagy. Consequently, PER2 holds promise as a valuable therapeutic marker for OSCC [70]. In OSCC tissues, the expression of the clock gene PER1 was found to be significantly reduced. Furthermore, the silencing of PER1 in SCC15 OSCC cells was observed to have a significant inhibitory effect on autophagy, while simultaneously activating the AKT/mTOR pathway, thereby promoting the progression of OSCC [71]. Additionally, it was discovered that PER2 functions as a scaffold protein, facilitating the binding of tuberous sclerosis complex 1 (Tsc1), Raptor, and mTOR. This interaction leads to the specific inhibition of mTORC1 complex activity and the activation of autophagy. Consequently, a novel role for PER2 in the regulation of the mTORC1 pathway has been unveiled [72]. The expression of PER2 decreased with aging, which might be due to decreasing autophagy [73]. Neuronal PAS domain protein 2 (NPAS2) is a fundamental gene involved in the regulation of circadian clock rhythm. Research has demonstrated that NPAS2 plays a beneficial role in mitigating myocardial ischemia/reperfusion (I/R) injury in rats by modulating the CX3CL1 pathway and autophagy regulation. NPAS2 can interact with CRY2 to promote CX3CL1 signaling pathway transcriptional activity. Additionally, NPAS2 exerts its influence on the downstream AKT/mTOR pathway to suppress autophagy, thereby leading to an improvement in cardiac I/R injury in rats [74].
Mitophagy is regulated by circadian rhythms
Cellular function is significantly reliant on the oxidative metabolism and quality control of mitochondria, rendering mitophagy an attractive subject of study in the field of autophagy research. The disruption of mitophagy adversely affects clock gene activity, resulting in an excessive buildup of mitochondrial reactive oxygen species, structural impairment of mitochondria, and subsequent cardiac dysfunction. The restoration of autophagy/mitophagy has the potential to repair autophagy defects and mitochondrial damage in cardiomyocytes lacking the CLOCK gene. The cytoprotective effect of CLOCK is nullified when autophagy is inhibited through ATG7 knockdown. CLOCK has the ability to regulate mitophagy in cardiomyocytes, thereby modulating adaptive stress responses crucial for cell survival. Compensatory mechanisms involving CLOCK may prove beneficial in mitigating cardiac injury in individuals experiencing circadian rhythm disturbances [75].
Chaperone-mediated autophagy (CMA) is regulated by circadian rhythms
Researchers found that circadian rhythms and CMAs are interdependent. The regulation of circadian rhythms is governed by the degradation of core clock proteins by chaperone-mediated autophagy (CMA) in both central and peripheral systems. Disruption of the circadian cycle occurs when CMAs are lost in vivo. Conversely, the circadian clock modulates CMA activity in a tissue-specific manner and controls protein transcription at distinct circadian phases [76]. The circadian rhythms govern the oscillatory fluctuations in protein abundance via the clock mechanism to maintain physiological functions in accordance with the light-dark cycle. Moreover, the timely degradation of these proteins by various proteolytic systems is crucial for ensuring the strength and adaptability of circadian fluctuations. Researchers found that the circadian activity of CMA in mouse tissue cells was regulated by BMAL1 at the transcriptional level. Specifically, BMAL1 regulated the diurnal variation of CMA activity by inhibiting LAMP2A, and the degree to which CMA rhythmicity was regulated by BMAL1 in the liver, renal tubules, glomeruli, and heart tissues was inconsistent. Circadian rhythm disturbances can lead to the loss of CMA rhythmicity, resulting in marked changes in cellular protein expressions [77].
Regulation of Circadian Rhythm by Autophagy
Regulation of circadian rhythm by macroautophagy
In contrast to the extensive research on the circadian regulation of autophagy, the evidence supporting the regulation of circadian rhythms by autophagy is relatively limited. However, it is increasingly acknowledged that circadian clocks and their associated clock genes play crucial roles in various physiological and pathological processes. Therefore, it has been postulated by researchers that the autophagy system governing circadian clock could serve as a crucial focal point for addressing these diseases [13,78]. Lysosomal targets, namely BMAL1, CLOCK, REV-ERBα, and CRY1, have the potential to function as circadian proteins, while macrophage autophagy exerts an influence on the circadian clock through the selective degradation of these associated proteins.
Here we start with these genes that receive autophagy regulation and briefly describe the process how autophagy regulates biological rhythms. It has been reported that CRY1, which acts as a gluconeogenic inhibitor, undergoes periodic autophagic degradation during starvation. This is dependent on gluconeogenesis, similar to blood glucose maintenance. The consumption of a diet high in fat has been observed to expedite the process of autophagic degradation of CRY1, ultimately resulting in hyperglycemia associated with obesity. CRY1 possesses multiple LC3-interacting region (LIR) motifs, which aid in the interaction between cargo proteins and the autophagosome marker LC3. This interaction plays a crucial role in the regulation of circadian glycemic control through the degradation of CRY1. Consequently, targeting the LIR motifs presents a promising avenue for the management of hyperglycemia [18]. It has also been suggested that the dynamic balance and reciprocal interaction between TFEB/TFE3 and REV-ERBα may be responsible for the regulation and oscillations of autophagy and metabolic genes in the diurnal cycle [51]. Studies have also verified the regulation of autophagy on the circadian clock by other means. For example, researchers have found that bromide inhibited clock gene expression and autophagy processes in rat cardiomyocytes, while rapamycin (an autophagy inducer) significantly restored this inhibition [79]. The overexpression of PER1/2 has been shown to impede the formation of cancer cell colonies, suggesting that targeting PER2 could be a potential therapeutic approach for the treatment of malignant tumors. Research findings have demonstrated that suppressing autophagy in the HCT 116 human colon cancer cell line leads to a notable elevation in PER2 protein expression and a substantial reduction in cancer cell clones [80].
Regulation of circadian rhythms by CMA
CMA can rhythmically degrade clock proteins and remodel circadian rhythms through selective autophagy. Most clock proteins such as BMAL1, CLOCK, REV-ERBα, CRY1, and RORα carry a KFERQ-like pentapeptide motif for lysosomal targeting of CMA, with BMAL1 and CLOCK being the most compatible. There are targeting motifs similar to KFERQ that preferentially bind to CMA active lysosomes, HSC70 and LAMP2A and can be directly translocated into free lysosomes. Studies have displayed that CMA inhibition can lead to circadian rhythm disorders in mice. For example, Lamp2A KO mice have abnormal circadian gene transcription cycles. CMA disorder causes BMAL1 degradation obstruction and is sufficient to change the periodic cycle characteristics of clock protein. Experiments have also observed the phenotype of disordered circadian rhythm in aging mice and suggested that age-dependent CMA decline may be one of the causes of circadian rhythm disorders in elderly organisms [77]. The regulatory relationship between ferroptosis and autophagy, termed “clockophagy”, is elucidated as the selective degradation of the core clock protein BMAL1 by autophagy. SQSTM1 acts as a receptor protein to guide REV-ERB autophagic degradation and is essential for ferroptosis [81].
Co-regulation of autophagy with circadian rhythm
Autophagy and circadian rhythm generally present a relationship of mutual regulation, but some studies have also found that the intervention of diseased patients or animal models led to the dual regulation of autophagy and circadian rhythm, in which the specific mechanism remains to be explored.
For example, the expression of the core clock genes PER2 and REV-ERBα in the adipose tissues of overweight subjects increased with weight loss, and the clock gene and LC3 expression levels are closely related to the degree of weight loss [82]. Acrylamide (ACR) is a persistent neurotoxin within the brain that hampers the magnitude of oscillations in clock genes, namely BMAL1, CRY2, and REV-ERBβ, or induces alterations in the timing of clock genes, specifically CLOCK, PER2, and REV-ERBα. Consequently, this phenomenon augments the nocturnal accumulation of autophagosomes, instigating detrimental consequences such as nerve cell damage, apoptosis, and ultimately culminating in cognitive decline [9]. In the pathogenesis of Parkinson’s disease, gene mutations in mitochondrial ligases MUL1 and PARKIN caused circadian and circadian clock disturbances and prolonged periods of locomotor rhythms, increased total activity, and shortened sleep duration at night. The expression levels of clock genes PER and CLOCK are changed in mutants, and ATG5 expression decreases in neurons in the brain [83]. Based on the idea that REV-ERBβ is upregulated in various tumor cells, the investigators identified ARN5187 through compound screening. ARN5187 has a dual inhibitory effect against REV-ERB-mediated transcriptional regulation and autophagy, and this was reported to be effective in killing cancer cells [84].
Regulatory Mechanisms of Autophagy and Circadian Rhythm in Diseases and Clinical Implications
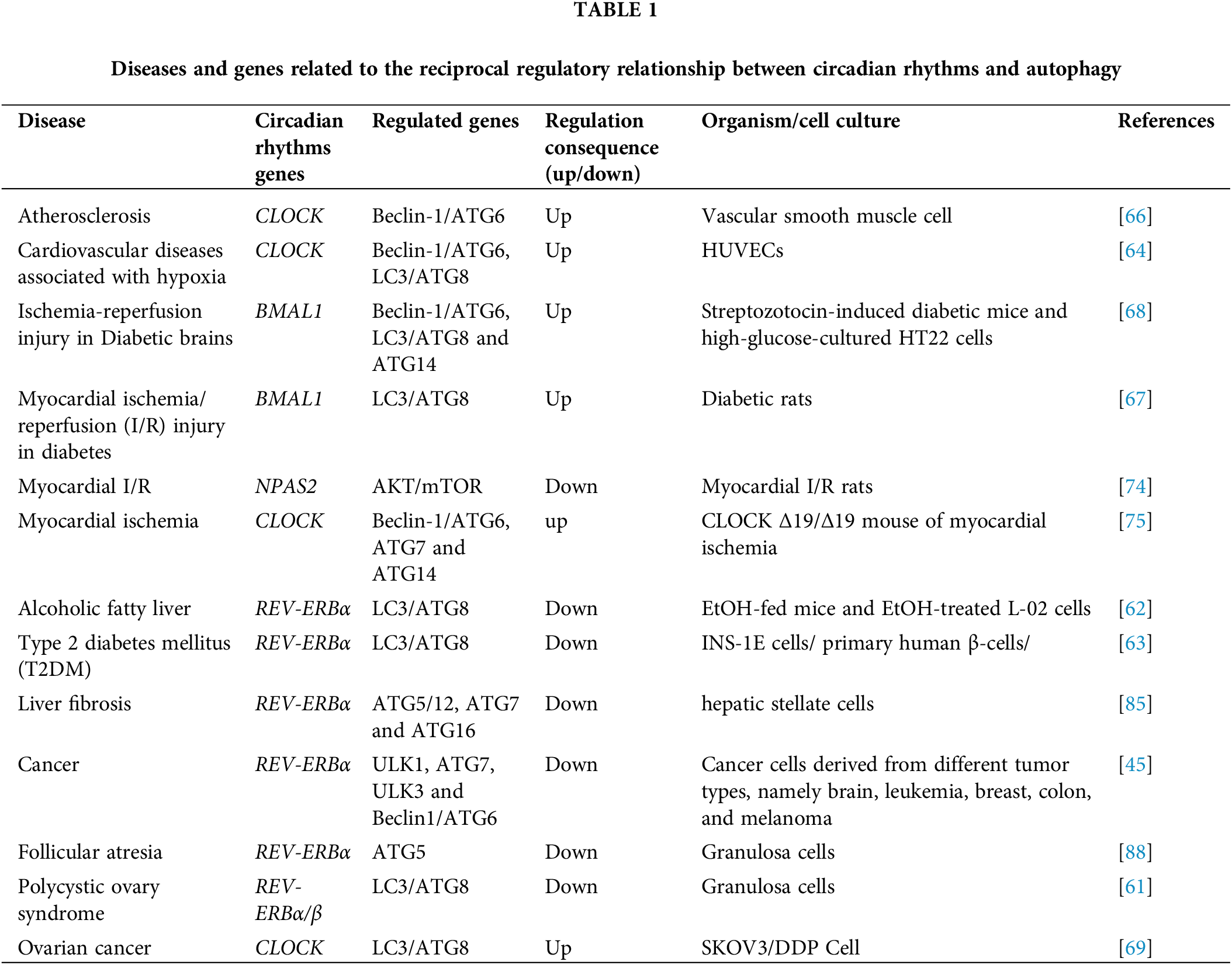
Circadian rhythms and autophagy are essential physiological processes with a bidirectional regulatory relationship. Imbalances in this regulation contribute to many diseases. Understanding this relationship is crucial for uncovering disease mechanisms and potential therapies. As shown in Fig. 3 and Table 1, we will briefly describe the significance of this regulatory process in these diseases.
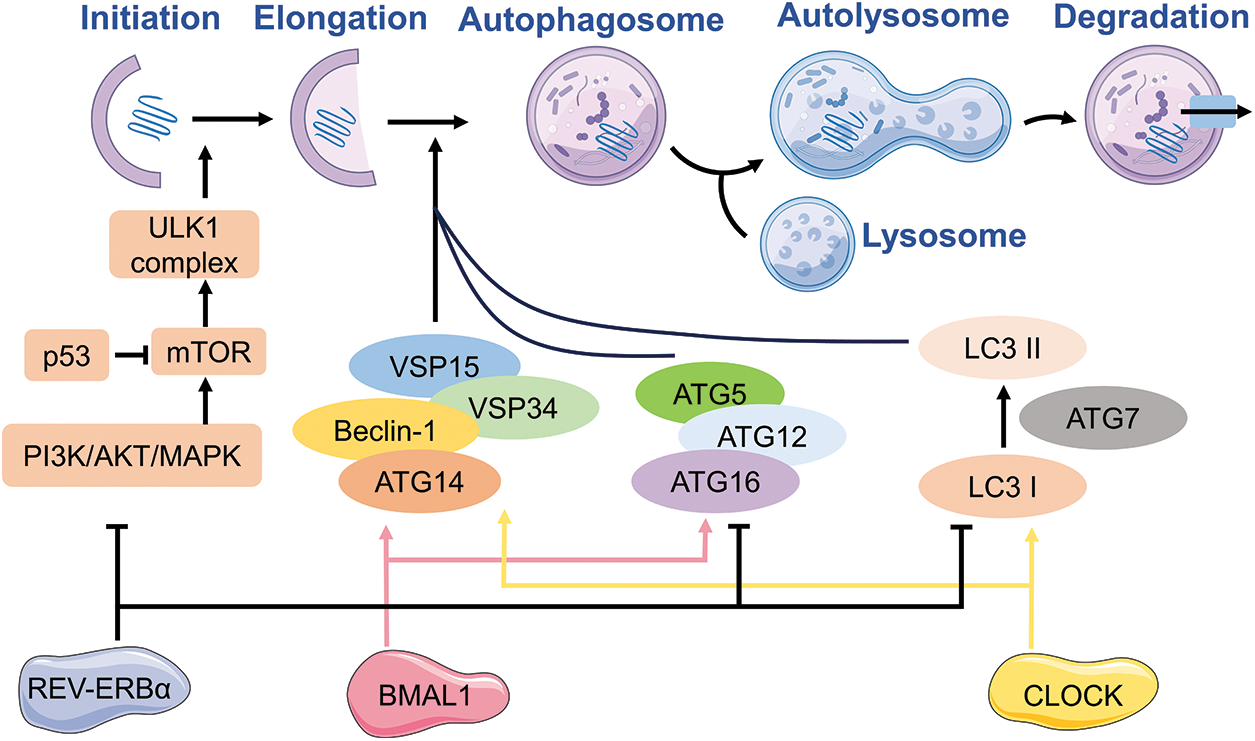
Figure 3: The autophagy processes that are impacted primarily consist of macroautophagy and chaperone-mediated autophagy. Table 1 will provide a detailed discussion on the genes implicated in the interplay of autophagy and circadian rhythm.
Cardiovascular and cerebrovascular diseases
Circadian rhythms and autophagy have raised concerns in cardiovascular disease over the last decade. The expression of rhythmic genes fluctuates with the diurnal variation of the vascular physiological state, suggesting that vascular rhythm plays a key role. The circadian clock directly regulates endothelial cell synthesis, secretion, and phenotypic switching of vascular smooth muscle cells, thereby regulating vasodilation and contraction [48]. Downregulation of CLOCK and circadian rhythm disturbance in vascular smooth muscle cells can promote apoptosis and vascular calcification by attenuating autophagy when atherosclerosis occurs [66]. In myocardial ischemic diseases, CLOCK regulates cardiomyocyte survival and adaptive responses by affecting mitophagy. Thus, the appropriate upregulation of CLOCK expression under ischemia-hypoxia conditions may help to reduce myocardial injury [64,75]. In addition, in diabetes-induced brain I/R injury, melatonin can modulate the SIRT1-BMAL1 pathway to improve cell survival, induce antioxidant and anti-apoptotic effects, and enhance autophagy to improve prognosis [68]. Histone deacetylase 3 (HDAC3) and silent information modulator 1 (SIRT1) are enzymes involved in the deacetylation of histones, which in turn regulate crucial metabolic pathways and exert significant influence in the context of diabetes and myocardial ischemia/reperfusion (I/R) injury. Targeting the HDAC3/SIRT1 pathway-mediated regulation of autophagy via BMAL1 effectively ameliorates myocardial I/R injury in diabetic rats [67]. NPAS2 is one of the core genes controlling circadian clock rhythm. The myocardial I/R injury rat model displayed a decreased expression of NPAS2 and increased reperfusion time. In contrast, the overexpression of NPAS2 could regulate the downstream AKT/mTOR pathway to inhibit autophagy, thereby improving myocardial injury in rats [74].
Increasing evidence suggests that circadian clock disruption during chronic lung disease is crucial in increasing oxidative stress, inflammatory responses, metabolic imbalance, mucus secretion, dysregulation of autophagy, and altering lung function [58]. The dysregulation of oscillations and expression of circadian clock genes such as BMAL1, PER1, and PER2 have been associated with lung inflammatory diseases, and targeting the association of circadian rhythms and autophagy is a promising therapeutic strategy [78]. In addition, TFEB and TFE3 act as critical regulators of autophagy, lysosome biogenesis, and cellular circadian rhythms, and they regulate clock genes involved in autophagy and REV-ERBα expression [50]. The TFE3 and REV-ERBα binding sites exist in the promoter regions of genes involved in circadian rhythm, autophagy, and lysosomal biogenesis (e.g., LAMP1, ATG3/5, and CTSL). Chromatin co-immunoprecipitation analysis revealed that there were several ChIP-seq peaks of TFEB and TFE3 in the REV-ERBα promoter region [50].
Digestive and endocrine system disorders
The circadian rhythm system is related to various physiological processes (e.g., body metabolism and hormone secretion) and diseases (e.g., fatty liver, obesity, metabolic syndrome) [41,47,49]. Hepatic gluconeogenesis is one of the important modes of blood glucose regulation. CRY1 can play the role of gluconeogenesis inhibitors, and autophagy can selectively degrade CRY1 to regulate circadian rhythm and blood glucose. Therefore, the LC3 interaction region (LIR) motif in CRY1 and its protein sequence may be potential targets for controlling blood glucose in the future [18]. The failure of pancreatic β-cells in individuals with type 2 diabetes mellitus (T2DM) is linked to the compromised regulation of autophagy, a process that governs the development, function, and survival of β-cells by eliminating misfolded proteins and damaged organelles. Previous studies have established a connection between REV-ERBα and disrupted autophagy, as well as the survival of β-cells under diabetic circumstances. Researchers have discovered that the reduction of REV-ERBα expression offers partial protection against inflammation and glucotoxicity-induced β-cell failure [63].
Alcoholic fatty liver (AFL) is a hepatic disorder resulting from the excessive intake of alcohol, manifesting as hepatic steatosis. Recent investigations have revealed that REV-ERBα can intensify hepatic steatosis in alcoholic liver disease and facilitate the advancement of AFL through the regulation of autophagy. These findings propose an innovative mechanism for the regulation of steatosis in AFL and indicate that targeting REV-ERBα could hold promise as a therapeutic approach for AFL [62]. In studies related to liver fibrosis, the REV-ERB agonist SR9009 was found to be effective in regulating autophagy and attenuating liver fibrosis [85].
The disruption of the circadian clock has been linked to a range of human diseases, including cancer [21,86]. The observed abnormal expression of clock genes in numerous tumors is a consequence of overall changes in the cancer cell transcriptome and could potentially enhance cancer cell survival. Several studies have suggested that manipulating clock-related proteins through pharmacological means could serve as a promising approach in combating cancer [59,60]. REV-ERB is an important regulatory target in various cancers and inflammatory diseases [87], and researchers proposed the REV-ERB-specific agonist SR9009 as a first-line/second-line treatment for small cell lung cancer. The specific mechanism of SR9009 is to activate REV-ERBα, which in turn regulates the core autophagy gene ATG5, affects autophagy, and produces cytotoxicity in SCLC cells [57]. Similar studies (including SR9009 and SR9011) have also been validated in various benign and malignant tumors such as glioblastoma, leukemia, breast cancer, colon cancer, and melanoma [45]. In addition to this, certain researchers have explored the concurrent dual suppression of REV-ERBβ and autophagy as a novel pharmacological strategy to elicit cytotoxicity in malignant cells [84]. The suppression of the circadian gene CLOCK has been observed to counteract cisplatin resistance in SKOV3/DDP cell lines of ovarian cancer by modulating the expression of autophagy-associated resistance genes. Consequently, CLOCK genes hold promise as a prospective therapeutic target for drug-resistant ovarian cancer [69].
Polycystic ovary syndrome (PCOS) is an endocrine disorder characterized by a multifaceted pathophysiology and frequently leading to infertility due to anovulation in women. Researchers have observed disturbances in circadian rhythms in individuals with PCOS. In PCOS mouse models, an intervention utilizing the REV-ERBα/β agonist SR9009 effectively improved follicular dysplasia by stimulating the generation of new mitochondria and suppressing autophagy. These findings highlight the potential of SR9009 as a therapeutic agent for managing PCOS [61]. The rhythmic expression of CLOCK genes was observed in rodent ovarian tissue and granulosa cells. The study revealed that core clock genes (BMAL1, PER2, and REV-ERB) and autophagy-related gene (ATG5) displayed rhythmic expression patterns within a 24-hour period in mouse ovaries and primary granulosa cells. The clock genes affect granulosa cell function by regulating granulosa cell autophagy through REV-ERB-mediated ATG5 inhibition [88].
Autophagy exhibits rhythmic activation in a clock-dependent fashion within the physiological state of the organism, thereby exerting an influence on circadian rhythms through the degradation of circadian proteins [41]. Recently, many studies have demonstrated that disturbances in the regulatory relationship between autophagy and circadian rhythms can lead to disease development [89–91]. At present, the mechanism of the regulatory relationship between the two is not fully clarified, and its impact on the disease may involve all aspects, such as inflammation, oxidative stress, aging, and dysfunctional synthesis and secretion. This review discussed the interaction and mechanism between autophagy and circadian rhythm, introduced the molecules affecting autophagy and circadian rhythm, and summarized the related diseases, targets, and drugs.
Maintaining a certain autophagy rhythm enables regular clearance of damaged material from cells. Autophagy assumes a crucial function in the growth, development, and physiological functioning of the body [92,93]. Perturbation of this intricate mechanism results in the accumulation of aberrant cellular constituents, thereby instigating pathological manifestations within the cells and the organism as a whole [94–96]. Rhythmic dysregulation of autophagy has been implicated in the cellular accumulation of misfolded proteins, resulting in alterations in membrane permeability, increased production of reactive oxygen species (ROS), and the degradation of mitochondria and DNA. These processes ultimately contribute to the development of metabolic disorders, neurodegenerative conditions, and the aging process [97,98]. The circadian regulation of autophagy is believed to play a crucial role in preventing the buildup of abnormal cellular components, which are known to be key factors in the pathogenesis of specific diseases.
Despite the limited number of studies investigating the regulation of circadian rhythm through autophagy, it is noteworthy that this process forms a reciprocal relationship, commonly referred to as a “feedback loop,” with the regulation of autophagy by the aforementioned rhythmic genes. Moreover, this intricate interplay assumes a pivotal role in numerous physiological and pathological processes [13,78]. For example, lysosomes in macrophages selectively degrade circadian proteins such as BMAL1, CLOCK, REV-ERBα, and CRY1 [18]. The dynamic balance and reciprocal interaction between the key transcription factors (TFEB/TFE3 and REV-ERBα) regulating autophagy may be responsible for the regulation and oscillations of autophagy and metabolic genes in the diurnal cycle [51]. CMA can remove clock proteins and remodel circadian rhythms by selective autophagy. Most clock proteins such as BMAL1, CLOCK, REV-ERBα, CRY1, and RORα carry pentapeptide motifs for lysosomal targeting of CMA, and CMA obstruction can lead to circadian rhythm disturbances in mice and alter the cycling properties of circadian rhythms [77]. The discovery of “clockophagy” has highlighted the importance of autophagy for circadian rhythm regulation [81].
In conclusion, the feedback loop formed by the close coupling of autophagic protein degradation and the biological clock plays an important role in the processes of growth and development, maturation and aging, and physiological and pathological processes. This interaction can reshape intracellular protein composition and normal cell function and maintain metabolic homeostasis. In addition, it can also affect the occurrence and development of diseases by regulating cellular inflammation, oxidative stress, cell degeneration, and other processes. The prevalence of circadian rhythm disruptions is on the rise in contemporary society. The incongruity between external stimuli and the internal circadian clock results in the perturbation of the biological timekeeping system.
The advent of modern electronic devices, such as smartphones and computer screens, exposes individuals to nocturnal light exposure, thereby inducing desynchronization of their intrinsic circadian rhythms. The emission of short-wavelength light by these electronic devices constitutes a significant contributing factor, which has been associated with the development of metabolic syndrome and the manifestation of sleepiness. Furthermore, sleep disruption and engagement in shift work are considerably consequential occurrences that can engender an augmented susceptibility to cardiovascular disease [99,100].
In future scientific research, the following several directions may be the focus of study. Firstly, further elucidate the molecular mechanisms of interaction between circadian rhythm and autophagy. Currently it is still not fully clear how circadian clock genes and autophagy genes regulate each other at the molecular level. More research is needed to map out the detailed signaling pathways and feedback loops. Secondly, explore how disruption of circadian-autophagy interaction contributes to disease development. Studies can investigate whether and how conditions like metabolic disorders and neurodegeneration are linked to deregulation of this interaction. This can provide new insights into disease pathogenesis. Thirdly, develop circadian-autophagy dual targeting therapeutic strategies. Based on understanding the interaction mechanism, researchers can design treatments that modulate both circadian rhythm and autophagy for better treatment of diseases found to involve disrupted interaction between the two processes.
Conducting a thorough investigation into the correlation between autophagy and circadian rhythms will facilitate an enhanced comprehension of various ailments, such as cancer, neurodegeneration, aging, and metabolic disorders. Although numerous investigations have been conducted in cellular and animal models, limited advancements have been achieved in clinical trials. Future work should focus on elucidating the complex regulatory relationship in diseases, especially the impact of specific external factors (e.g., inflammatory microenvironment) and mechanical signals (e.g., cell environment) on the related mechanisms. Gaining insights into the underlying mechanism governing the interplay between autophagy and circadian rhythm holds the potential to yield novel therapeutic approaches for the management of diseases in clinical application.
Acknowledgement: We thank all members of the research team for helpful discussion. We are grateful to funding from the National Natural Science Foundation of China and Department of Science and Technology of Shaanxi Province, China.
Funding Statement: This current study was funded by the National Natural Science Foundation of China (Code No. 82100954/81800924); Natural Science Foundation of Inner Mongolia Autonomous Region (Code No. 2023QN08026); Key Research and Development Projects of Shaanxi Province (Code No. 2022KW-12); The Basic and Natural Science Research Program of Shaanxi Province (Code No. 2022JQ-915); Key Research and Development Program of Tibet Autonomous Region (XZ202001ZY0059G); New Technology and New Business Project of the Third Affiliated Hospital of Air Force Military Medical University (Code No. LX2021-416).
Author Contributions: The authors confirm contribution to the paper as follows: Tiankai Di, Zhifei Zhou and Fen Liu jointly drafted the manuscript. Yujiang Chen and Lulu Wang contributed to the in-depth discussion and revised the manuscript. All authors approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15(7):713–20. doi:https://doi.org/10.1038/ncb2788. [Google Scholar] [PubMed] [CrossRef]
2. Ge Y, Huang M, Yao YM. Autophagy and proinflammatory cytokines: interactions and clinical implications. Cytokine Growth Factor Rev. 2018;43:38–46. doi:https://doi.org/10.1016/j.cytogfr.2018.07.001. [Google Scholar] [PubMed] [CrossRef]
3. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi:https://doi.org/10.1038/nature06639. [Google Scholar] [PubMed] [CrossRef]
4. Codogno P, Mehrpour M, Proikas-Cezanne T. Canonical and non-canonical autophagy: variations on a common theme of self-eating? Nat Rev Mol Cell Biol. 2011;13(1):7–12. doi:https://doi.org/10.1038/nrm3249. [Google Scholar] [PubMed] [CrossRef]
5. Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13(5):495–504. doi:https://doi.org/10.1016/j.cmet.2011.04.004. [Google Scholar] [PubMed] [CrossRef]
6. Greco CM, Sassone-Corsi P. Personalized medicine and circadian rhythms: opportunities for modern society. J Exp Med. 2020;217(6):e20200702. doi:https://doi.org/10.1084/jem.20200702. [Google Scholar] [PubMed] [CrossRef]
7. Kim YH, Lazar MA. Transcriptional control of circadian rhythms and metabolism: a matter of time and space. Endocr Rev. 2020;41(5):707–32. doi:https://doi.org/10.1210/endrev/bnaa014. [Google Scholar] [PubMed] [CrossRef]
8. Ryzhikov M, Eubanks A, Haspel JA. Measuring diurnal rhythms in autophagic and proteasomal flux. J Vis Exp. 2019;2019(151):e60133. doi:https://doi.org/10.3791/60133. [Google Scholar] [PubMed] [CrossRef]
9. Tan X, Ye J, Liu W, Zhao B, Shi X, Zhang C, et al. Acrylamide aggravates cognitive deficits at night period via the gut-brain axis by reprogramming the brain circadian clock. Arch Toxicol. 2019;93(2):467–86. doi:https://doi.org/10.1007/s00204-018-2340-7. [Google Scholar] [PubMed] [CrossRef]
10. van Dyck L, Casaer MP. Intermittent or continuous feeding: any difference during the first week? Curr Opin Crit Care. 2019;25(4):356–62. doi:https://doi.org/10.1097/MCC.0000000000000617. [Google Scholar] [PubMed] [CrossRef]
11. Dreyer AP, Martin MM, Fulgham CV, Jabr DA, Bai L, Beshel J, et al. A circadian output center controlling feeding: fasting rhythms in Drosophila. PLoS Genet. 2019;15(11):e1008478. doi:https://doi.org/10.1371/journal.pgen.1008478. [Google Scholar] [PubMed] [CrossRef]
12. Paul S, Hanna L, Harding C, Hayter EA, Walmsley L, Bechtold DA, et al. Output from VIP cells of the mammalian central clock regulates daily physiological rhythms. Nat Commun. 2020;11(1):1453. doi:https://doi.org/10.1038/s41467-020-15277-x. [Google Scholar] [PubMed] [CrossRef]
13. Maiese K. Moving to the rhythm with clock (circadian) genes, autophagy, mTOR, and SIRT1 in degenerative disease and cancer. Curr Neurovasc Res. 2017;14(3):299–304. doi:https://doi.org/10.2174/1567202614666170718092010. [Google Scholar] [PubMed] [CrossRef]
14. Kim H, Williams D, Qiu Y, Song Z, Yang Z, Kimler V, et al. Regulation of hepatic autophagy by stress-sensing transcription factor CREBH. FASEB J. 2019;33(7):7896–914. doi:https://doi.org/10.1096/fj.201802528R. [Google Scholar] [PubMed] [CrossRef]
15. Rijo-Ferreira F, Takahashi JS. Genomics of circadian rhythms in health and disease. Genome Med. 2019;11(1):82. doi:https://doi.org/10.1186/s13073-019-0704-0. [Google Scholar] [PubMed] [CrossRef]
16. Ge W, Yan ZH, Wang L, Tan SJ, Liu J, Reiter RJ, et al. A hypothetical role for autophagy during the day/night rhythm-regulated melatonin synthesis in the rat pineal gland. J Pineal Res. 2021;71(1):e12742. doi:https://doi.org/10.1111/jpi.12742. [Google Scholar] [PubMed] [CrossRef]
17. Jeong K, He B, Nohara K, Park N, Shin Y, Kim S, et al. Dual attenuation of proteasomal and autophagic BMAL1 degradation in ClockΔ19/+ mice contributes to improved glucose homeostasis. Sci Rep. 2015;5:12801. doi:https://doi.org/10.1038/srep12801. [Google Scholar] [PubMed] [CrossRef]
18. Toledo M, Batista-Gonzalez A, Merheb E, Aoun ML, Tarabra E, Feng D, et al. Autophagy regulates the liver clock and glucose metabolism by degrading CRY1. Cell Metab. 2018;28(2):268–81.E4. doi:https://doi.org/10.1016/j.cmet.2018.05.023. [Google Scholar] [PubMed] [CrossRef]
19. Lasorsa F, Rutigliano M, Milella M, Ferro M, Pandolfo SD, Crocetto F, et al. Cellular and molecular players in the tumor microenvironment of renal cell carcinoma. J Clin Med. 2023;12(12):3888. doi:https://doi.org/10.3390/jcm12123888. [Google Scholar] [PubMed] [CrossRef]
20. Lasorsa F, di Meo NA, Rutigliano M, Milella M, Ferro M, Pandolfo SD, et al. Immune checkpoint inhibitors in renal cell carcinoma: molecular basis and rationale for their use in clinical practice. Biomedicines. 2023;11(4):1071. doi:https://doi.org/10.3390/biomedicines11041071. [Google Scholar] [PubMed] [CrossRef]
21. Loizzo D, Pandolfo SD, Rogers D, Cerrato C, di Meo NA, Autorino R, et al. Novel insights into autophagy and prostate cancer: a comprehensive review. Int J Mol Sci. 2022;23(7):3826. doi:https://doi.org/10.3390/ijms23073826. [Google Scholar] [PubMed] [CrossRef]
22. Klionsky DJ, Abdel-Aziz AK, Abdelfatah S, Abdellatif M, Abdoli A, Abel S, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy. 2021;17(1):1–382. doi:https://doi.org/10.1080/15548627.2020.1797280. [Google Scholar] [PubMed] [CrossRef]
23. Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–41. doi:https://doi.org/10.1016/j.cell.2011.10.026. [Google Scholar] [PubMed] [CrossRef]
24. Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330(6009):1344–8. doi:https://doi.org/10.1126/science.1193497. [Google Scholar] [PubMed] [CrossRef]
25. Füllgrabe J, Klionsky DJ, Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15(1):65–74. doi:https://doi.org/10.1038/nrm3716. [Google Scholar] [PubMed] [CrossRef]
26. Ohsumi Y. Historical landmarks of autophagy research. Cell Res. 2014;24(1):9–23. doi:https://doi.org/10.1038/cr.2013.169. [Google Scholar] [PubMed] [CrossRef]
27. Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53(3):471–83. doi:https://doi.org/10.1016/j.molcel.2013.12.011. [Google Scholar] [PubMed] [CrossRef]
28. Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9(3):424–5. doi:https://doi.org/10.4161/auto.22931. [Google Scholar] [PubMed] [CrossRef]
29. Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15(7):741–50. doi:https://doi.org/10.1038/ncb2757. [Google Scholar] [PubMed] [CrossRef]
30. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171(4):603–14. doi:https://doi.org/10.1083/jcb.200507002. [Google Scholar] [PubMed] [CrossRef]
31. Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9(10):1142–51. doi:https://doi.org/10.1038/ncb1634. [Google Scholar] [PubMed] [CrossRef]
32. Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–49. doi:https://doi.org/10.1146/annurev-physiol-021909-135821. [Google Scholar] [PubMed] [CrossRef]
33. Schibler U, Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111(7):919–22. doi:https://doi.org/10.1016/S0092-8674(02)01225-4. [Google Scholar] [PubMed] [CrossRef]
34. Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Nat Acad Sci USA. 2014;111(45):16219–24. doi:https://doi.org/10.1073/pnas.1408886111. [Google Scholar] [PubMed] [CrossRef]
35. Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi:https://doi.org/10.1016/B978-0-12-387690-4.00006-4. [Google Scholar] [PubMed] [CrossRef]
36. Saini C, Brown SA, Dibner C. Human peripheral clocks: applications for studying circadian phenotypes in physiology and pathophysiology. Front Neurol. 2015;6:95. doi:https://doi.org/10.3389/fneur.2015.00095. [Google Scholar] [PubMed] [CrossRef]
37. Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(suppl_2):R271–7. doi:https://doi.org/10.1093/hmg/ddl207. [Google Scholar] [PubMed] [CrossRef]
38. Grimaldi B, Nakahata Y, Kaluzova M, Masubuchi S, Sassone-Corsi P. Chromatin remodeling, metabolism and circadian clocks: the interplay of CLOCK and SIRT1. Int J Biochem Cell Biol. 2009;41(1):81–6. doi:https://doi.org/10.1016/j.biocel.2008.08.035. [Google Scholar] [PubMed] [CrossRef]
39. Richards J, Gumz ML. Mechanism of the circadian clock in physiology. Am J Physiol Regul Integr Comp Physiol. 2013;304(12):R1053–64. doi:https://doi.org/10.1152/ajpregu.00066.2013. [Google Scholar] [PubMed] [CrossRef]
40. Hardin PE, Panda S. Circadian timekeeping and output mechanisms in animals. Curr Opin Neurobiol. 2013;23(5):724–31. doi:https://doi.org/10.1016/j.conb.2013.02.018. [Google Scholar] [PubMed] [CrossRef]
41. Wang X, Xu Z, Cai Y, Zeng S, Peng B, Ren X, et al. Rheostatic balance of circadian rhythm and autophagy in metabolism and disease. Front Cell Dev Biol. 2020;8:616434. doi:https://doi.org/10.3389/fcell.2020.616434. [Google Scholar] [PubMed] [CrossRef]
42. Jahanban-Esfahlan R, Mehrzadi S, Reiter RJ, Seidi K, Majidinia M, Baghi HB, et al. Melatonin in regulation of inflammatory pathways in rheumatoid arthritis and osteoarthritis: involvement of circadian clock genes. Br J Pharmacol. 2018;175(16):3230–8. doi:https://doi.org/10.1111/bph.13898. [Google Scholar] [PubMed] [CrossRef]
43. Gabryelska A, Sochal M, Turkiewicz S, Białasiewicz P. Relationship between HIF-1 and circadian clock proteins in obstructive sleep apnea patients-preliminary study. J Clin Med. 2020;9(5):1599. doi:https://doi.org/10.3390/jcm9051599. [Google Scholar] [PubMed] [CrossRef]
44. Rabinovich-Nikitin I, Lieberman B, Martino TA, Kirshenbaum LA. Circadian-regulated cell death in cardiovascular diseases. Circulation. 2019;139(7):965–80. doi:https://doi.org/10.1161/CIRCULATIONAHA.118.036550. [Google Scholar] [PubMed] [CrossRef]
45. Sulli G, Rommel A, Wang X, Kolar MJ, Puca F, Saghatelian A, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature. 2018;553(7688):351–5. doi:https://doi.org/10.1038/nature25170. [Google Scholar] [PubMed] [CrossRef]
46. Wu P, Cheng J, Chen L, Xiang J, Pan Y, Zhang Y, et al. Nr1d1 affects autophagy in the skeletal muscles of juvenile Nile tilapia by regulating the rhythmic expression of autophagy-related genes. Fish Physiol Biochem. 2020;46(3):891–907. doi:https://doi.org/10.1007/s10695-019-00757-9. [Google Scholar] [PubMed] [CrossRef]
47. Li S, Lin JD. Transcriptional control of circadian metabolic rhythms in the liver. Diabetes Obes Metab. 2015;17 Suppl 1(01):33–8. doi:https://doi.org/10.1111/dom.12520. [Google Scholar] [PubMed] [CrossRef]
48. Huang H, Li Z, Ruan Y, Feng W, Chen J, Li X, et al. Circadian rhythm disorder: a potential inducer of vascular calcification? J Physiol Biochem. 2020;76(4):513–24. doi:https://doi.org/10.1007/s13105-020-00767-9. [Google Scholar] [PubMed] [CrossRef]
49. Tarquini R, Mazzoccoli G. Clock genes, metabolism, and cardiovascular risk. Heart Fail Clin. 2017;13(4):645–55. doi:https://doi.org/10.1016/j.hfc.2017.05.001. [Google Scholar] [PubMed] [CrossRef]
50. Pastore N, Vainshtein A, Herz NJ, Huynh T, Brunetti L, Klisch TJ, et al. Nutrient-sensitive transcription factors TFEB and TFE3 couple autophagy and metabolism to the peripheral clock. EMBO J. 2019;38(12):e101347. doi:https://doi.org/10.15252/embj.2018101347. [Google Scholar] [PubMed] [CrossRef]
51. Pastore N, Ballabio A. Keeping the autophagy tempo. Autophagy. 2019;15(10):1854–6. doi:https://doi.org/10.1080/15548627.2019.1645545. [Google Scholar] [PubMed] [CrossRef]
52. Pariollaud M, Gibbs JE, Hopwood TW, Brown S, Begley N, Vonslow R, et al. Circadian clock component REV-ERBα controls homeostatic regulation of pulmonary inflammation. J Clin Invest. 2018;128(6):2281–96. doi:https://doi.org/10.1172/JCI93910. [Google Scholar] [PubMed] [CrossRef]
53. Bodas M, Patel N, Silverberg D, Walworth K, Vij N. Master autophagy regulator transcription factor EB regulates cigarette smoke-induced autophagy impairment and chronic obstructive pulmonary disease-emphysema pathogenesis. Antioxid Redox Signal. 2017;27(3):150–67. doi:https://doi.org/10.1089/ars.2016.6842. [Google Scholar] [PubMed] [CrossRef]
54. Ma D, Panda S, Lin JD. Temporal orchestration of circadian autophagy rhythm by C/EBPβ. EMBO J. 2011;30(22):4642–51. doi:https://doi.org/10.1038/emboj.2011.322. [Google Scholar] [PubMed] [CrossRef]
55. Kijak E, Pyza E. TOR signaling pathway and autophagy are involved in the regulation of circadian rhythms in behavior and plasticity of L2 interneurons in the brain of Drosophila melanogaster. PLoS One. 2017;12(2):e0171848. doi:https://doi.org/10.1371/journal.pone.0171848. [Google Scholar] [PubMed] [CrossRef]
56. Dadon-Freiberg M, Chapnik N, Froy O. REV-ERBα alters circadian rhythms by modulating mTOR signaling. Mol Cell Endocrinol. 2021;521:111108. doi:https://doi.org/10.1016/j.mce.2020.111108. [Google Scholar] [PubMed] [CrossRef]
57. Shen W, Zhang W, Ye W, Wang H, Zhang Q, Shen J, et al. SR9009 induces a REV-ERB dependent anti-small-cell lung cancer effect through inhibition of autophagy. Theranostics. 2020;10(10):4466–80. doi:https://doi.org/10.7150/thno.42478. [Google Scholar] [PubMed] [CrossRef]
58. Giri A, Wang Q, Rahman I, Sundar IK. Circadian molecular clock disruption in chronic pulmonary diseases. Trends Mol Med. 2022;28(6):513–27. doi:https://doi.org/10.1016/j.molmed.2022.04.002. [Google Scholar] [PubMed] [CrossRef]
59. REV-ERB agonists block autophagy in cancer cells. Cancer Discov. 2018;8(3):261. doi:https://doi.org/10.1158/2159-8290.Cd-rw2018-011. [Google Scholar] [PubMed] [CrossRef]
60. Ercolani L, Ferrari A, De Mei C, Parodi C, Wade M, Grimaldi B. Circadian clock: time for novel anticancer strategies? Pharmacol Res. 2015;100:288–95. doi:https://doi.org/10.1016/j.phrs.2015.08.008. [Google Scholar] [PubMed] [CrossRef]
61. Sun L, Tian H, Xue S, Ye H, Xue X, Wang R, et al. Circadian clock genes REV-ERBs inhibits granulosa cells apoptosis by regulating mitochondrial biogenesis and autophagy in polycystic ovary syndrome. Front Cell Dev Biol. 2021;9:658112. doi:https://doi.org/10.3389/fcell.2021.658112. [Google Scholar] [PubMed] [CrossRef]
62. Liu Q, Xu L, Wu M, Zhou Y, Yang J, Huang C, et al. Rev-erbα exacerbates hepatic steatosis in alcoholic liver diseases through regulating autophagy. Cell Biosci. 2021;11(1):129. doi:https://doi.org/10.1186/s13578-021-00622-4. [Google Scholar] [PubMed] [CrossRef]
63. Brown MR, Laouteouet D, Delobel M, Villard O, Broca C, Bertrand G, et al. The nuclear receptor REV-ERBα is implicated in the alteration of β-cell autophagy and survival under diabetogenic conditions. Cell Death Dis. 2022;13(4):353. doi:https://doi.org/10.1038/s41419-022-04767-z. [Google Scholar] [PubMed] [CrossRef]
64. Tang X, Lin C, Guo D, Qian R, Li X, Shi Z, et al. Clock promotes endothelial damage by inducing autophagy through reactive oxygen species. Oxid Med Cell Longev. 2016;2016:9591482. doi:https://doi.org/10.1155/2016/9591482. [Google Scholar] [PubMed] [CrossRef]
65. Huang G, Zhang F, Ye Q, Wang H. The circadian clock regulates autophagy directly through the nuclear hormone receptor Nr1d1/Rev-erbα and indirectly via Cebpb/(C/ebpβ) in zebrafish. Autophagy. 2016;12(8):1292–309. doi:https://doi.org/10.1080/15548627.2016.1183843. [Google Scholar] [PubMed] [CrossRef]
66. Guo Z, Yu B, Li X, Yang X, Wang C, Fan L. Circadian misalignment promotes vascular smooth muscle cell apoptosis via defective autophagy. J Mol Histol. 2021;52(4):799–808. doi:https://doi.org/10.1007/s10735-021-10000-6. [Google Scholar] [PubMed] [CrossRef]
67. Qiu Z, Ming H, Zhang Y, Yu Y, Lei S, Xia ZY. The protective role of bmal1-regulated autophagy mediated by HDAC3/SIRT1 pathway in myocardial ischemia/reperfusion injury of diabetic rats. Cardiovasc Drugs Ther. 2022;36(2):229–43. doi:https://doi.org/10.1007/s10557-021-07159-1. [Google Scholar] [PubMed] [CrossRef]
68. Liu L, Cao Q, Gao W, Li BY, Zeng C, Xia Z, et al. Melatonin ameliorates cerebral ischemia-reperfusion injury in diabetic mice by enhancing autophagy via the SIRT1-BMAL1 pathway. FASEB J. 2021;35(12):e22040. doi:https://doi.org/10.1096/fj.202002718RR. [Google Scholar] [PubMed] [CrossRef]
69. Sun Y, Jin L, Sui YX, Han LL, Liu JH. Circadian gene CLOCK affects drug-resistant gene expression and cell proliferation in ovarian cancer SKOV3/DDP cell lines through autophagy. Cancer Biother Radiopharm. 2017;32(4):139–46. doi:https://doi.org/10.1089/cbr.2016.2153. [Google Scholar] [PubMed] [CrossRef]
70. Liu H, Gong X, Yang K. Overexpression of the clock gene Per2 suppresses oral squamous cell carcinoma progression by activating autophagy via the PI3K/AKT/mTOR pathway. J Cancer. 2020;11(12):3655–66. doi:https://doi.org/10.7150/jca.42771. [Google Scholar] [PubMed] [CrossRef]
71. Yang G, Yang Y, Tang H, Yang K. Loss of the clock gene Per1 promotes oral squamous cell carcinoma progression via the AKT/mTOR pathway. Cancer Sci. 2020;111(5):1542–54. doi:https://doi.org/10.1111/cas.14362. [Google Scholar] [PubMed] [CrossRef]
72. Wu R, Dang F, Li P, Wang P, Xu Q, Liu Z, et al. The circadian protein period2 suppresses mTORC1 activity via recruiting Tsc1 to mTORC1 complex. Cell Metab. 2019;29(3):653–67.E6. doi:https://doi.org/10.1016/j.cmet.2018.11.006. [Google Scholar] [PubMed] [CrossRef]
73. Kalfalah F, Janke L, Schiavi A, Tigges J, Ix A, Ventura N, et al. Crosstalk of clock gene expression and autophagy in aging. Aging. 2016;8(9):1876–95. doi:https://doi.org/10.18632/aging.101018. [Google Scholar] [PubMed] [CrossRef]
74. Huang J, Qing W, Pan Y. NPAS2 ameliorates myocardial ischaemia/reperfusion injury in rats via CX3CL1 pathways and regulating autophagy. Aging. 2021;13(16):20569–84. doi:https://doi.org/10.18632/aging.203445. [Google Scholar] [PubMed] [CrossRef]
75. Rabinovich-Nikitin I, Rasouli M, Reitz CJ, Posen I, Margulets V, Dhingra R, et al. Mitochondrial autophagy and cell survival is regulated by the circadian Clock gene in cardiac myocytes during ischemic stress. Autophagy. 2021;17(11):3794–812. doi:https://doi.org/10.1080/15548627.2021.1938913. [Google Scholar] [PubMed] [CrossRef]
76. Kaushik S, Juste YR, Cuervo AM. Circadian remodeling of the proteome by chaperone-mediated autophagy. Autophagy. 2022;18(5):1205–7. doi:https://doi.org/10.1080/15548627.2022.2038503. [Google Scholar] [PubMed] [CrossRef]
77. Juste YR, Kaushik S, Bourdenx M, Aflakpui R, Bandyopadhyay S, Garcia F, et al. Reciprocal regulation of chaperone-mediated autophagy and the circadian clock. Nat Cell Biol. 2021;23(12):1255–70. doi:https://doi.org/10.1038/s41556-021-00800-z. [Google Scholar] [PubMed] [CrossRef]
78. Hu Y, He T, Zhu J, Wang X, Tong J, Li Z, et al. The link between circadian clock genes and autophagy in chronic obstructive pulmonary disease. Med Inflamm. 2021;2021:2689600. doi:https://doi.org/10.1155/2021/2689600. [Google Scholar] [PubMed] [CrossRef]
79. Jiang Y, Gu Y, Xu H, Tian X, Zhang X, Xu X, et al. Bromide impairs the circadian clock and glycolytic homeostasis via disruption of autophagy in rat H9C2 cardiomyocytes. BMC Mol Cell Biol. 2020;21(1):44. doi:https://doi.org/10.1186/s12860-020-00289-8. [Google Scholar] [PubMed] [CrossRef]
80. Hua H, Wang Y, Wan C, Liu Y, Zhu B, Wang X, et al. Inhibition of tumorigenesis by intratumoral delivery of the circadian gene mPer2 in C57BL/6 mice. Cancer Gene Ther. 2007;14(9):815–8. doi:https://doi.org/10.1038/sj.cgt.7701061. [Google Scholar] [PubMed] [CrossRef]
81. Yang M, Chen P, Liu J, Zhu S, Kroemer G, Klionsky DJ, et al. Clockophagy is a novel selective autophagy process favoring ferroptosis. Sci Adv. 2019;5(7):eaaw2238. doi:https://doi.org/10.1126/sciadv.aaw2238. [Google Scholar] [PubMed] [CrossRef]
82. Pivovarova O, Gögebakan Ö., Sucher S, Groth J, Murahovschi V, Kessler K, et al. Regulation of the clock gene expression in human adipose tissue by weight loss. Int J Obes. 2016;40(6):899–906. doi:https://doi.org/10.1038/ijo.2016.34. [Google Scholar] [PubMed] [CrossRef]
83. Doktór B, Damulewicz M, Pyza E. Effects of MUL1 and PARKIN on the circadian clock, brain and behaviour in Drosophila Parkinson’s disease models. BMC Neurosci. 2019;20(1):24. doi:https://doi.org/10.1186/s12868-019-0506-8. [Google Scholar] [PubMed] [CrossRef]
84. De Mei C, Ercolani L, Parodi C, Veronesi M, Lo Vecchio C, Bottegoni G, et al. Dual inhibition of REV-ERBβ and autophagy as a novel pharmacological approach to induce cytotoxicity in cancer cells. Oncogene. 2015;34(20):2597–608. doi:https://doi.org/10.1038/onc.2014.203. [Google Scholar] [PubMed] [CrossRef]
85. Thomes PG, Brandon-Warner E, Li T, DonohueJr. TM, Schrum LW. Reverb agonist and TGF-β similarly affect autophagy but differentially regulate hepatic stellate cell fibrogenic phenotype. Int J Biochem Cell Biol. 2016;137–47. doi:https://doi.org/10.1016/j.biocel.2016.11.007. [Google Scholar] [PubMed] [CrossRef]
86. Aveta A, Cilio S, Contieri R, Spena G, Napolitano L, Manfredi C, et al. Urinary MicroRNAs as biomarkers of urological cancers: a systematic review. Int J Mol Sci. 2023;24(13):10846. doi:https://doi.org/10.3390/ijms241310846. [Google Scholar] [PubMed] [CrossRef]
87. Ryzhikov M, Ehlers A, Steinberg D, Xie W, Oberlander E, Brown S, et al. Diurnal rhythms spatially and temporally organize autophagy. Cell Rep. 2019;26(7):1880–92.E6. doi:https://doi.org/10.1016/j.celrep.2019.01.072. [Google Scholar] [PubMed] [CrossRef]
88. Zhang J, Zhao L, Li Y, Dong H, Zhang H, Zhang Y, et al. Circadian clock regulates granulosa cell autophagy through NR1D1-mediated inhibition of ATG5. Am J Physiol Cell Physiol. 2022;322(2):C231–45. doi:https://doi.org/10.1152/ajpcell.00267.2021. [Google Scholar] [PubMed] [CrossRef]
89. Rožman P. The potential of non-myeloablative heterochronous autologous hematopoietic stem cell transplantation for extending a healthy life span. Gerosci. 2018;40(3):221–42. doi:https://doi.org/10.1007/s11357-018-0027-x. [Google Scholar] [PubMed] [CrossRef]
90. Sarker MR, Franks SF. Efficacy of curcumin for age-associated cognitive decline: a narrative review of preclinical and clinical studies. Gerosci. 2018;40(2):73–95. doi:https://doi.org/10.1007/s11357-018-0017-z. [Google Scholar] [PubMed] [CrossRef]
91. Dong Y, Digman MA, Brewer GJ. Age- and AD-related redox state of NADH in subcellular compartments by fluorescence lifetime imaging microscopy. Gerosci. 2019;41(1):51–67. doi:https://doi.org/10.1007/s11357-019-00052-8. [Google Scholar] [PubMed] [CrossRef]
92. Tan P, Ye Y, Mao J, He L. Autophagy and immune-related diseases. Adv Exp Med Biol. 2019;1209:167–79. doi:https://doi.org/10.1007/978-981-15-0606-2. [Google Scholar] [CrossRef]
93. Keller CW, Münz C, Lünemann JD. Autophagy pathways in CNS myeloid cell immune functions. Trends Neurosci. 2020;43(12):1024–33. doi:https://doi.org/10.1016/j.tins.2020.09.003. [Google Scholar] [PubMed] [CrossRef]
94. Pacheco Y, Lim CX, Weichhart T, Valeyre D, Bentaher A, Calender A. Sarcoidosis and the mTOR, Rac1, and autophagy triad. Trends Immunol. 2020;41(4):286–99. doi:https://doi.org/10.1016/j.it.2020.01.007. [Google Scholar] [PubMed] [CrossRef]
95. Li F, Zhang C, Tang Z, Zhang L, Dai Z, Lyu S, et al. A plant RNA virus activates selective autophagy in a UPR-dependent manner to promote virus infection. New Phytol. 2020;228(2):622–39. doi:https://doi.org/10.1111/nph.16716. [Google Scholar] [PubMed] [CrossRef]
96. Wu X, Nagy LE. MLKL contributes to Western diet-induced liver injury through inhibiting autophagy. Autophagy. 2020;16(7):1351–2. doi:https://doi.org/10.1080/15548627.2020.1760624. [Google Scholar] [PubMed] [CrossRef]
97. Abdrakhmanov A, Gogvadze V, Zhivotovsky B. To eat or to die: deciphering selective forms of autophagy. Trends Biochem Sci. 2020;45(4):347–64. doi:https://doi.org/10.1016/j.tibs.2019.11.006. [Google Scholar] [PubMed] [CrossRef]
98. Gu Y, Han J, Jiang C, Zhang Y. Biomarkers, oxidative stress and autophagy in skin aging. Ageing Res Rev. 2020;59:101036. doi:https://doi.org/10.1016/j.arr.2020.101036. [Google Scholar] [PubMed] [CrossRef]
99. Crnko S, Du Pré BC, Sluijter JPG, Van Laake LW. Circadian rhythms and the molecular clock in cardiovascular biology and disease. Nat Rev Cardiol. 2019;16(7):437–47. doi:https://doi.org/10.1038/s41569-019-0167-4. [Google Scholar] [PubMed] [CrossRef]
100. Strohmaier S, Devore EE, Zhang Y, Schernhammer ES. A review of data of findings on night shift work and the development of DM and CVD events: a synthesis of the proposed molecular mechanisms. Curr Diab Rep. 2018;18(12):132. doi:https://doi.org/10.1007/s11892-018-1102-5. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools